During heterosexual HIV-1 transmission, a genetic bottleneck occurs in the newly infected individual as the virus passes from the mucosa, leading to systemic infection with a single transmitted HIV-1 clone in the recipient. This bottleneck in the recipient has just been described, and the mechanisms involved in this selection process have not been elucidated.
KEYWORDS: HIV, fitness, transmission
ABSTRACT
Exposure of the genital mucosa to a genetically diverse viral swarm from the donor HIV-1 can result in breakthrough and systemic infection by a single transmitted/founder (TF) virus in the recipient. The highly diverse HIV-1 envelope (Env) in this inoculating viral swarm may have a critical role in transmission and subsequent immune response. Thus, chronic (Envchronic) and acute (Envacute) Env chimeric HIV-1 were tested using multivirus competition assays in human mucosal penile and cervical tissues. Viral competition analysis revealed that Envchronic viruses resided and replicated mainly in the tissue, while Envacute viruses penetrated the human tissue and established infection of CD4+ T cells more efficiently. Analysis of the replication fitness, as tested in peripheral blood mononuclear cells (PBMCs), showed similar replication fitness of Envacute and Envchronic viruses, which did not correlate with transmission fitness in penile tissue. Further, we observed that chimeric Env viruses with higher replication in genital mucosal tissue (chronic Env viruses) had higher binding affinity to C-type lectins. Data presented herein suggest that the inoculating HIV-1 may be sequestered in the genital mucosal tissue (represented by chronic Env HIV-1) but that a single HIV-1 clone (e.g., acute Env HIV-1) can escape this trapped replication for systemic infection.
IMPORTANCE During heterosexual HIV-1 transmission, a genetic bottleneck occurs in the newly infected individual as the virus passes from the mucosa, leading to systemic infection with a single transmitted HIV-1 clone in the recipient. This bottleneck in the recipient has just been described (K. Klein et al., PLoS Pathog 14:e1006754, https://doi.org/10.1371/journal.ppat.1006754), and the mechanisms involved in this selection process have not been elucidated. However, understanding mucosal restriction is of the utmost importance for understanding dynamics of infections and for designing focused vaccines. Using our human penile and cervical mucosal tissue models for mixed HIV infections, we provide evidence that HIV-1 from acute/early infection, compared to that from chronic infection, can more efficiently traverse the mucosal epithelium and be transmitted to T cells, suggesting higher transmission fitness. This study focused on the role of the HIV-1 envelope in transmission and provides strong evidence that HIV transmission may involve breaking the mucosal lectin trap.
INTRODUCTION
In the global spread of human immunodeficiency virus type 1 (HIV-1), the virus is most commonly transmitted during sexual intercourse. For this transmission event, the inoculating virus in vaginal fluids or semen must penetrate physical barriers, such as the mucus and epithelium of the genital mucosal tissue, to establish a propagating infection in susceptible target cells located in the underlying stroma. Regardless of the genital tissue, the risk of HIV-1 transmission per coital act is very low for receptive (0.08%) and insertive (0.04%) penile-vaginal intercourse (1), which is one factor associated with the higher and increasing HIV prevalence in women (2).
Over the years, multiple studies have indicated that a primary HIV-1 infection is initiated by one clone or a small number of transmitted/founder (TF) HIV-1 clones despite the presence of a diverse HIV-1 population in the donor (3, 4). Previous studies have examined the genetic bottlenecks from donor to recipient by describing the HIV in the blood of donors and recipients following transmission. However, there is considerable complexity in these transmission bottlenecks, in that the virus migrates and compartmentalizes in different tissues within the donor. For example, in a female-to-male HIV-1 transmission event, the virus within the female genital tract (FGT) could have been present since the time of acute infection. It is therefore possible that the virus could have evolved in the FGT or likely entered the FGT via the blood. Thus, the HIV-1 that is transmitted from the FGT to the penile tissue in female-to-male transmission may reflect what is transmitted from the vaginal mucosa rather than directly from the blood, in the absence of injury. The virus that infects the penile tissue may have originated from infection of the foreskin, urethra, or the glans tissue and then may pass into the blood, disseminate into different tissues, and migrate into the testes through a barrier similar to the blood-brain barrier. Thus, predicting the efficiency of transmission from donor blood to recipient blood may not relate directly to the actual transmission event, i.e., the virus population moving through various genetic bottlenecks and barriers (5). Different barriers during this transmission process may place the inoculating HIV-1 population under different, possibly distinct selective pressures leading to systemic infection by one predominant HIV-1 clone. We recently provided in vivo evidence that an HIV-1 population and not a single clone infects the vaginal tract during male-to-female transmission and that a possible genetic bottleneck from the vaginal tract to the blood results in systemic infection by a single HIV-1 clone (6). However, it is still not fully understood if this transmission bottleneck is a stochastic event or involves selection for a specific viral function/activity which may be mapped to viral protein structure(s), linear sequence(s), or posttranslational modifications. It is also unclear how genital microbiome, innate immune status, and other host factors could influence this HIV clonal selection during transmission (7–10). For the development of a successful preventive HIV-1 vaccine, it will be critical to obtain in-depth knowledge of the genotypic and phenotypic properties of the transmitted compared to the nontransmitted HIV-1 clones.
When comparing blood-derived HIV-1 from chronic and acute infections, we and others observed no differences in receptor usage, sensitivity to entry inhibitors, and cell tropism (11, 12). Interestingly, the chronic HIV-1, tested as a virus population and not a single clone, appears to have higher replicative fitness and greater host cell entry efficiency than the acute HIV-1, typically a single clone (12, 13). However, many studies have focused on identifying signature sequences of the TF clones, making it distinct in the inoculating HIV-1 swarm from the human donor (14–16). For example, alpha interferon (IFN-α) resistance encoded by the gag gene may be a distinct feature of TF HIV-1, but this has been the subject of debate (9, 11, 17). Typically, TF viruses have been shown to utilize CD4 and CCR5 for cell entry (3). Additionally, fewer potential N-linked glycosylation sites (PNGS) and shorter variable loops have been linked to the TF phenotype (18). One study showed that infectious molecular clones of TF were more infectious and expressed more envelope on the surface than HIV-1 molecular clones derived from chronic disease (17). However, there is still considerable debate about Env properties of TF HIV-1. A reduced glycan shield on TF HIV-1 (9, 19, 20) was thought to improve replicative fitness and/or host cell entry; i.e., it may be a favorable trait for transmission. However, TF clones with fewer N-linked glycan sites had no clear advantage in terms of replicative rate, receptor/coreceptor binding, or entry efficiency over HIV-1 found during chronic disease (11, 12, 21). The factors governing infection by HIV-1 harboring Env with fewer N-linked sites during acute infection remain poorly understood. Clearly, the emergence of HIV-1-specific antibodies following early infection selects for new N-linked sites, especially in the Env hypervariable loops, which corresponds to viral escape from this immune pressure (22). Compared to chronic HIV-1, TF viruses may bind more efficiently to dendritic cells (DCs) for transmission in trans to CD4+ T cells (17, 23), but other studies suggest the opposite (12, 14).
The impact of the Env on the replication efficiency of HIV-1 isolates in primary T cells, macrophages, and DCs has been well described (12, 21), but the role of Env in transmission is poorly understood. Thus, the HIV-1 env genes from individuals during acute/early and chronic disease were analyzed in the context of the same NL4-3 HIV-1 backbone. Acute (Envacute) and chronic (Envchronic) Env chimeric viruses were then tested in multivirus competition assays in human ex vivo penile and cervical tissue to determine if specific Env properties influence the transmission fitness. Differential transmission fitness was compared to replicative fitness in primary CD4+ T cells and C-type lectin binding affinity. C-type lectin receptors are a family of transmembrane and soluble receptors containing carbohydrate recognition domains (24), expressed mostly by myeloid cells. They play critical roles in both innate and adaptive immunity and have been shown to have important protective functioning at mucosal barriers while also facilitating binding and capture of HIV-1 by antigen-presenting cells (APCs) (25). Here, transmission fitness experiments were carried out in penile and cervical tissue following incubation with mannan, a high-mannose polysaccharide, to saturate C-type lectins, found throughout the mucosal tissues by immunofluorescent staining. Based on the studies described herein, increased transmission fitness of HIV-1 may relate to “breaking” the C-type lectin trap in mucosal tissue.
RESULTS
Multivirus infection in human explant tissue as a model for mucosal HIV-1 transmission fitness.
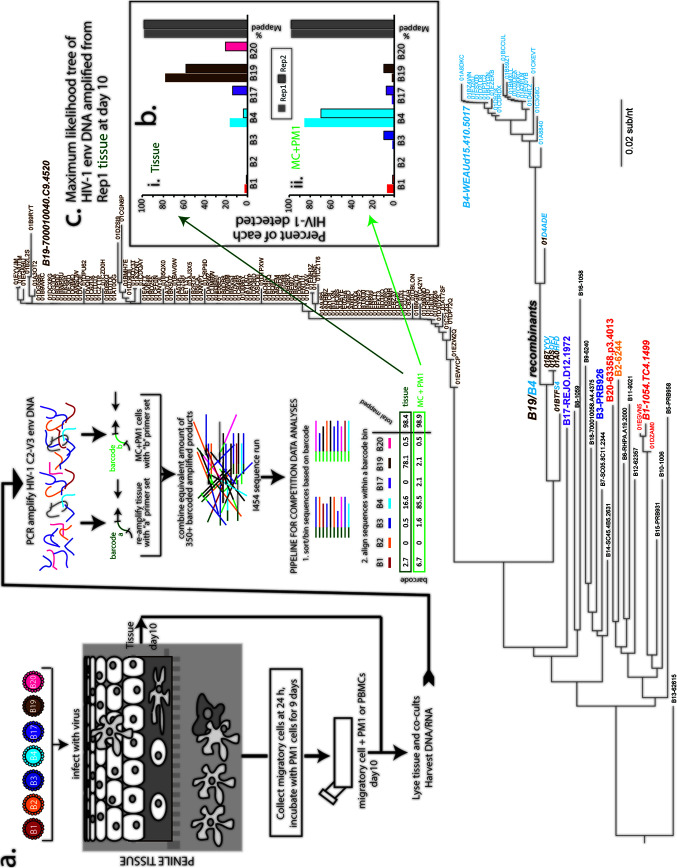
To approximate transmission fitness, we employed an ex vivo tissue model to test the transmissibility of both acute and chronic Env chimeric viruses. Eleven Envacute and three Envchronic viruses that were previously constructed and characterized (12, 26) were used in this study. Phylogenetic analysis and characteristics of all Envacute and Envchronic viruses employed in this study are presented in the phylogenetic tree in Fig. 1c. Titers of all viruses were determined on phytohemagglutinin (PHA)/interleukin 2 (IL-2)-treated peripheral blood mononuclear cells (PBMCs) using the standard TCID50 (50% tissue culture infective dose) method. In the example illustrated in Fig. 1, human penile glans tissue was exposed to a mix of seven Envacute HIV-1 (B1, B2, B3, B4, B17, B19, and B20), each at 150 infectious units (IU), for 3 h and then washed and cultured overnight. At 24 h, migratory cells (MC) were collected and cocultured with PM1 clonal CD4+ T cells. Following 10 days in culture, tissue and MC+PM1 cocultures from the mixed infections were analyzed for relative virus production.
FIG 1.
Schematic of transmission fitness assay and analyses of multivirus infections. (a) Human explant tissue was exposed to a mixture of Envacute and Envchronic viruses for 3 h, washed, and cultured overnight. Cells migrating out of the tissue were collected and cocultured with PM1 CD4+ T cells. After10 days, both tissue and MC+PM1 cocultures were lysed, and DNA was extracted for PCR amplification of the C2-V3 envelope region. After ligation of barcoded oligonucleotides, purification, and quantification, equivalent amounts of products were subject to NGS. Data analysis was performed using SeekDeep, a custom pipeline sorting sequences based on barcodes and aligning sequences within barcode bins. (b) Results of multivirus infections are presented as percent sequence reads of each virus detected in the assay in tissue or MC+PM1 cocultures. Gray bars indicate the percentage of sequences from that barcoded PCR product (representing this multivirus competition) mapped to one of the seven viruses in the competitions. It is important to note that these analyses involve counting HIV isolates based on phylogenetic clustering, where low-frequency mutations do not impact the analyses. This was verified as shown in panel c. The relative amount of each virus in the competition was determined using maximum-likelihood trees of the HIV-1 C2-V3 env DNA amplified from tissue or MC+PM1 cocultures. Env sequence in the maximum likelihood tree of panel c in the bold and larger colored text and the black text represent the C3-V3 gp120 sequences of all the chimeric Env HIV-1 strains that were employed in the various multi-virus competitions used in this study (see below). For the representative multi-virus competition results shown in this figure, the Env chimeric viruses used in this competition are the branch points on the tree labeled with the colored text. The small text labels (text not important) show the individual branches/sequences defined as NGS reads "counted" from the multivirus competitions by SeekDeep. The reference sequences for the viruses in this competition are shown in italic, bold, large text, namely B1-1054.TC4.1499 (red), B2-6244 (yellow), B3-PRB926 (dark blue), B4-WEAUd15.410.5017 (light blue), B17-REJO.D12.1972 (purple), BB19-700010040.C9.4520 (brown), and B20-63358.p3.4013 (orange on the tree but pink for rest of figure). A few recombinant B19/B4 sequences were identified in this competition and were identified as a mixed brown/light blue text label. Of all HIV sequences in the maximum-likelihood tree (c) from the matched-input viruses (B1, B2, B3, B4, B19, and B20), only B1, B4, and B19 replicated in the tissue (see replicate 1 of the tissue in panel b). Less than 1%, or 4 sequences, were recombinants of B4 and B19 with different breakpoints.
As detailed in Materials and Methods, barcoded Env C2-V3 amplicons were sequenced from tissue and MC+PM1 cocultures from each mixed-virus competition. SeekDeep (27) was then used to count sequence reads aligning to each virus added to the competition (Fig. 1a). The relative percentages of the B1, B2, B3, B4, B17, B19, and B20 viruses obtained are shown in Fig. 1b. Additionally, C2-V3 sequences from the same barcoded product were aligned to Envacute, and the maximum-likelihood tree in Fig. 1c shows one representative phylogenetic tree of the C2-V3 sequences amplified from the tissue at 10 days after multivirus infection. As described above, the Env consensus sequence ID labels of viruses used in this study is shown in Fig. 1c (in black text), while the virus ID labels specific for this mixed-virus competition are shown in colored text (Fig. 1c). The maximum-likelihood tree (Fig. 1c) and the SeekDeep analyses (Fig. 1b) were congruent and show that Envacute B19 (nearly 80%) followed by B4 dominated the multivirus competition in tissue, whereas B1 was found at a low percentage. The <1% not mapping to one of the seven HIV-1 isolates were identified as recombinants of B4 and B19 (Fig. 1c).
Despite the high level of B19 replication in the tissue, this virus was not identified in the MC+PM1 cocultures, suggesting that this virus was not carried by the MC out of the tissue. In contrast, the B4 virus was efficiently carried through the tissue to infect and propagate in CD4+ T cells (Fig. 1b). In the subsequent multivirus competitions, we found low levels of stochastic infection by all Envacute and Envchronic HIV-1 of the tissue and transmission to T cells. However, in general, Envacute viruses were more readily transmitted through the penile and cervical tissue to infect T cells, whereas other HIV-1 Env strains (i.e., Envchronic) propagated primarily in the tissue.
To phenotypically characterize the emigrating DCs, tissue blocks were cultured overnight and MC were collected and stained with a multicolor antibody cocktail for flow cytometry. Analysis of the phenotype present within the mDC population derived from penile (n = 5) and cervical (n = 3) tissue revealed that 80% and 28% are CD1c positive, 20% and 12% are CD207 (langerin) positive, and 3.95% and 7.69% are CD209 (DC-SIGN) positive (Fig. 2), which is in accordance with previous findings (28–30).
FIG 2.
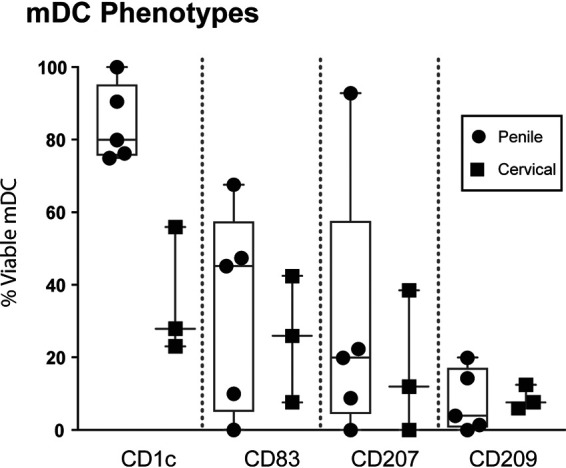
Characterization of the migratory cell phenotype in penile and cervical tissue. The percentages of CD1c-, CD83-, CD207-, and CD209-positive cells of all viable mDCs isolated from penile (n = 5) and cervical (n = 3) tissue were assessed by a multicolor flow panel on an LSRIIFortessa flow cytometer. Box-and-whisker plots show median, minimum/maximum, and 25th/75th percentiles of the percentage of viable cells stratified by CD1c, CD83, CD207, and CD209 expression. Each symbol represents an individual penile or cervical tissue donor.
Higher transmission fitness of acute over chronic Env chimeric viruses in the human penile and cervical tissue transmission model.
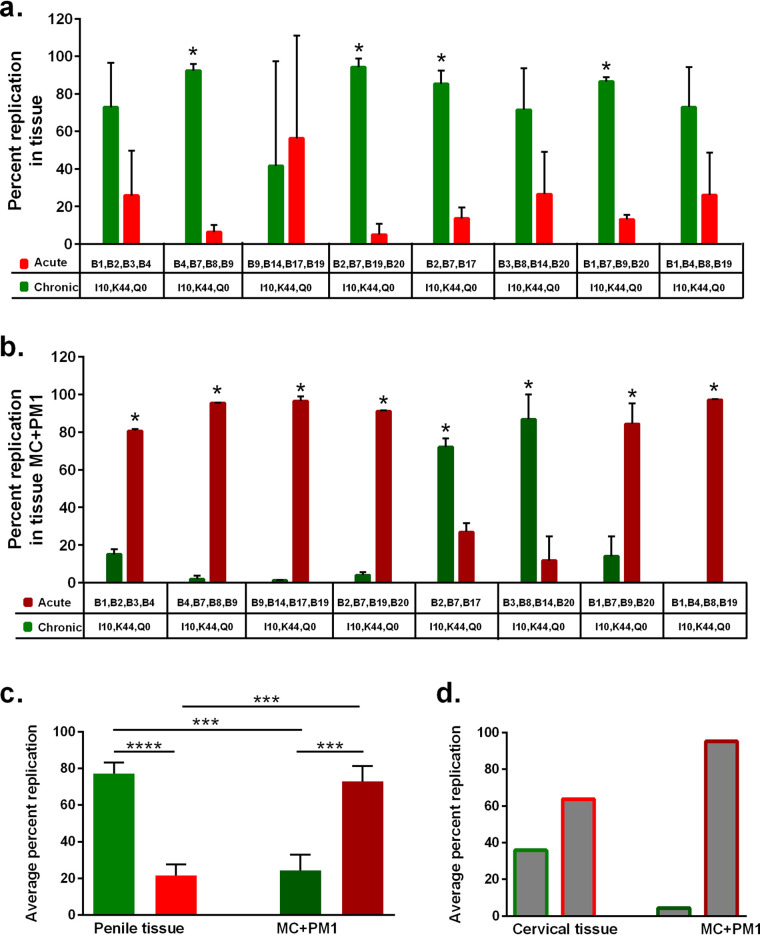
Due to limited availability of cervical tissue, multivirus competitions were predominantly performed in human penile tissue as a model for transmission fitness. Tissue was exposed to a mix of three or four Envacute viruses (B1, B2, B3, B4, B7, B8, B9, B14, B17, B19, and B20) plus three Envchronic viruses (I10, K44, and Q0), each virus at 150 IU. Eight different mixtures of Envacute viruses ensured that all 11 Envacute viruses competed against each other and with the same three Envchronic viruses (Fig. 3). The relative production of each virus in tissue and MC+PM1 multivirus competitions was analyzed as described in Fig. 1. Relative replication of the total Envacute versus Envchronic viruses was compared in tissue and MC+PM1 cocultures (Fig. 3a and b). With the exception of one mixed-virus competition (B9, B14, B17, B19, I10, K44, and Q0), Envchronic outreplicated Envacute in penile tissue (Fig. 3a), to a significant degree in 4 mixed-virus competitions. Even though the chronic virus represents only 43 to 50% of the inoculating virus, chronic virus represented 80% of the virus replicating in the tissue (7 of 8 competitions) (Fig. 3a and c).
FIG 3.
Higher transmission fitness of Envacute versus Envchronic viruses in multivirus competition assays in human tissue. (a) Penile tissues were dissected into 3-mm3 explants and exposed to eight different mixtures of a panel of Envacute viruses and the same three Envchronic viruses. (b) Following overnight culture, migrating cells from the tissue were collected and cocultured with PM1 T cells (MC+PM1). After 10 days in culture, DNA from tissue and MC+PM1 cells was isolated, PCR amplified, and analyzed by NGS. The cumulative percent replication in penile tissue (a) and in MC+PM1 cocultures (b) of Envchronic and Envacute viruses for tissue A and tissue B in each of the eight multivirus panels is shown. (c) The combined average replication of all Envchronic and all Envacute viruses of the eight virus competitions in penile tissues and Envchronic (green bars) and Envacute (red bars) viruses in MC+PM1 cocultures was determined. Each competition was done in triplicates in tissues from two donors. (d) Average replication of Envchronic (bars with green outlines) and Envacute (bars with red outlines) viruses of one competition set in cervical tissue and the corresponding MC+PM1 cocultures were evaluated by NGS. Statistical significance was determined using the Holm-Sidak method (a and b) (*, P < 0.05) and a two-tailed Mann-Whitney test (c and d) (***, P < 0.001; ****, P < 0.0001).
In complete contrast to the preferential Envchronic replication in the tissue, Envacute significantly outreplicated Envchronic HIV in the MC+PM1 cocultures in 6 of the 8 multivirus infections (Fig. 3b and c). For this to occur, Envacute HIV-1 must have been preferentially carried out of the tissue by MC and then transferred to PM1 cells. Combined, Envacute HIV-1 contributed to over 80% of the virus replication in all but two multivirus infections (Fig. 3b and c). In these two multivirus infections (B2, B7, B17, I10, K44, and Q0 and B3, B8, B14, B20, I10, K44, and Q0), Envchronic not only was transmitted to the T cells but also replicated within the tissue (Fig. 3a and b).
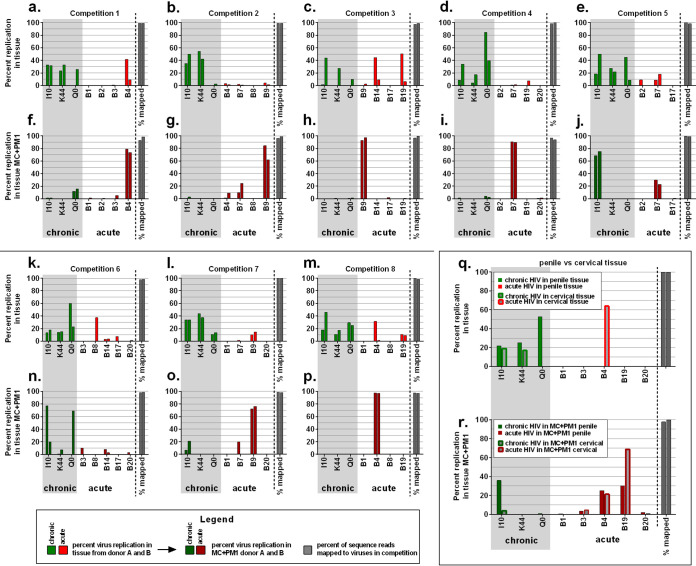
Analyses of individual virus replication in the competitions (Fig. 4), differential transmission fitness of acute HIV-1 was evident in 8 different multivirus infections in penile tissue. Of the 11 Envacute viruses, B4, B7, and B9 consistently showed the highest transmission fitness when combined with the same three Envchronic viruses but different combinations of the other 8 Envacute HIV-1 (Fig. 4f, g, h, i, j, o, and p). When B4, B7, and B9 (Fig. 4b and g) or B7 and B9 (Fig. 4l and o), were in competition together, the acute B9 virus was preferentially transmitted through the tissue by MC to replicate in PM1 cells. In the two competitions where Envchronic was transmitted more efficiently than Envacute (Fig. 4e, j, k, and l), the highly “transmissible” B4 and B9 were not included in the multivirus competitions.
FIG 4.
Multivirus competition assays to assess the transmission fitness across penile and cervical tissue. Penile and cervical explants were exposed to mixtures of three Envchronic viruses (I10, K44, and Q0) and three to five Envacute viruses (B1, B2, B3, B4, B7, B8, B9, B14, B17, B19, and B20) for 3 h. After overnight culture, migrating cells from the tissue were collected and cocultured with CD4+ PM1 T cells (MC+PM1). Following 10 days in culture, both tissue and MC+PM1 cocultures were lysed and PCR amplified, and replication of viruses was analyzed by NGS. (a to p) Percent replication of eight different mixtures of Envchronic and Envacute viruses in tissues from donor A and donor B and Envchronic and Envacute viruses in the corresponding MC+PM1 cocultures. Each graph shows the percentage of sequence reads for donor A and donor B from the NGS analysis mapped to the viruses present in the competitions. (q and r) Side-by-side comparison of Envchronic and Envacute virus replication in penile tissue and Envchronic and Envacute virus replication in cervical tissue (q) and comparison of transmission fitness of Envchronic and Envacute viruses in MC+PM1 derived from penile tissue with transmission fitness of Envchronic and Envacute viruses in MC+PM1 from cervical tissue (r) of one competition set (I10, K44, and Q0 and B1, B3, B4, B19, and B20).
Comparing HIV-1 transmission fitness in an endocervical versus penile glans tissue model.
Penile and endocervical tissues were exposed to I10, K44, QO, B3, B4, B19, and B20 using the experimental procedures described above (Fig. 1). The relative replication in the penile and cervical tissue was similar in that Envacute was again preferentially transmitted through the tissue to infect T cells via MC (Fig. 4q and r). We frequently observed stochastic replication of Envacute in the tissue along with Envchronic in these multivirus competitions. For example, high-level B4 replication was observed in multivirus competitions in the penile tissue (Fig. 4a and m) as well as in the endocervical tissue (Fig. 4q). In contrast, when Envacute viruses with high transmission fitness were present in the multivirus competitions, Envchronic HIV-1 were not transmitted and were almost completely absent in the MC+PM1 cocultures (Fig. 4f, g, h, i, o, p, and r). The patterns of relative HIV-1 replication were similar in experiments using penile and cervical tissue, i.e., Envchronic viruses had sequestered replication in the tissue, while Envacute viruses are preferentially taken up by MC and transferred to T cells (Fig. 3d). Replication of Envacute B4 in the cervical tissue from one of the mixed-virus competitions in one of two donors highlights the stochastic replication of HIV-1 in tissue with some mixed-virus competitions. Nonetheless, Envacute HIV-1 viruses were still preferentially transmitted by the migratory cells from the cervical tissue to free CD4+ T cells (Fig. 3d).
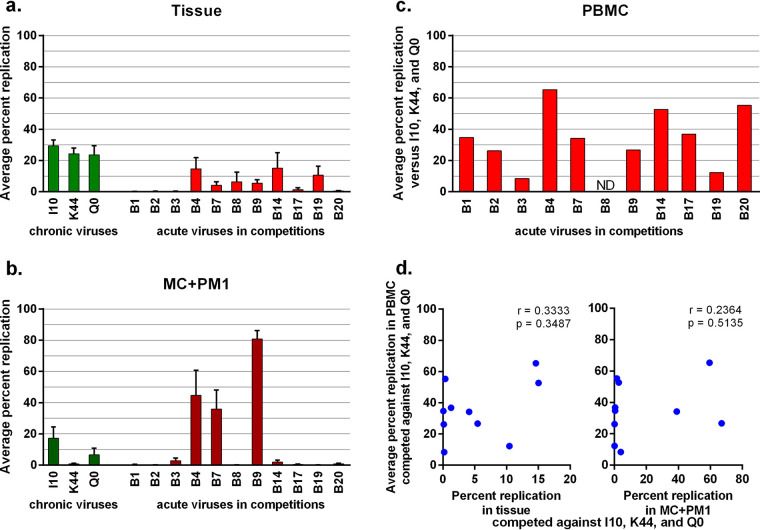
Transmission fitness does not correlate with replicative fitness in peripheral blood mononuclear cells.
The average relative fitness of each virus in the tissue or transmitted by MC to the T cells is shown in Fig. 5a and b (respectively) from the total of all 8 multivirus competitions performed in penile tissues from two donors. Again, B4, B7, and B9 had the highest transmission fitness using this penile explant model. We had previously reported on the replicative fitness of each acute virus in competition against the same three Envchronic viruses (I10, K44, and Q0) in PHA- and IL-2-treated PBMCs (21, 31) (Fig. 5c). Dual-virus competitions in PBMCs showed that all Envacute viruses could replicate with lower or higher fitness than the competing Envchronic viruses (Fig. 5c). Replication of B1, B2, B3, B17, and B20 was low or undetected in both tissue and MC+PM1 cocultures in the penile tissue model (Fig. 5a and b), whereas the same stocks replicated efficiently in PBMCs (Fig. 5c). In Fig. 5d, the average replicative fitness of the Envacute viruses in PBMCs (from Fig. 5c) is plotted against the replication of these viruses in tissue and MC+PM1 cocultures derived from the multivirus competitions in penile tissue (from Fig. 5a and b). Previous studies revealed a direct correlation between the replicative fitness of the infecting HIV-1 (as determined in PBMCs and primary T cells) and subsequent disease progression (26, 31–33). We observed no correlation between replicative fitness of Envacute HIV-1 with the replicative fitness in tissue or with the ability of the virus to be transmitted by MC to replicate in T cells (i.e., transmission fitness) (Fig. 5d).
FIG 5.
Replication fitness does not predict transmission fitness. Multivirus infection assays in penile tissue were performed, and the average replication of each Envchronic and each Envacute virus in tissue (a) and in MC+PM1 cocultures (b) was determined by sequencing. (c) Each of the Envacute viruses was competed against the Envchronic controls in PHA- and IL-2-activated PBMCs to assess the pathogenic fitness (21). The average pathogenic fitness versus the three Envchronic viruses is shown. (d) The percent replication of Envacute viruses in competitions in tissue and in MC + PM1 cocultures was plotted against the average percent replication in PBMC competitions. ND, not done.
Collectively, these data suggest that replication fitness as measured in PBMCs is different from the transmission fitness assessed in the ex vivo human penile explant model. As discussed below, replicative fitness of HIV-1 in free T cells was unlikely to predict the efficiency of viruses to penetrate the mucosa and be bound by MC to trans-infect T cells, which we refer to as transmission fitness.
Transmission fitness is linked to C-type lectin binding affinity.
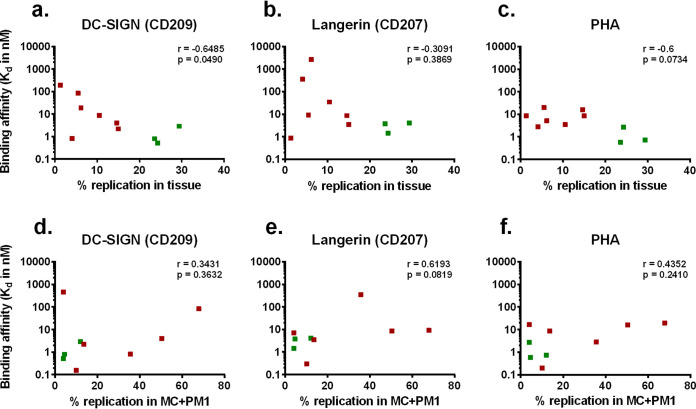
Based on previous studies, these analyses of Envacute versus Envchronic could not define a specific amino acid sequence, altered function, or phenotype (12, 13). The only unique feature appeared to be fewer conserved N-linked sites for some of the Envacute HIV-1 than Envchronic HIV-1 viruses. Based on these observations, we proposed that the Env glycoproteins of acute HIV-1 may have differential binding affinity to C-type lectins. Binding affinities of Envacute and Envchronic viruses for DC-SIGN and langerin were measured on a RapID4 acoustic biosensor. In this system, DC-SIGN and langerin were covalently bound to the surface of a quartz crystal chip. Inactivated Envacute and Envchronic virus stocks were passed over the chip surface, and kinetic binding parameters were measured by the variation in resonant frequency. The binding affinity of each virus for DC-SIGN, langerin, and PHA was then compared to the replication efficiency in both tissue and MC+PM1 cocultures (Fig. 6). The ability of the viruses to replicate in the tissue was related to DC-SIGN binding affinity (Fig. 6a). There was also a trend for viruses that replicated in tissue to bind with higher affinity to langerin (Fig. 6b). While we observed a 1,000-fold variation in langerin and DC-SIGN binding affinity, all viruses had similar binding affinity to PHA (<50-fold variation in dissociation constants) (Fig. 6c). There were, however, no correlations between binding affinity to these C-type lectins and transmission fitness, i.e., the ability to bind MC, pass through the tissue, and infect free CD4+ T cells (Fig. 6d to f).
FIG 6.
Binding profile of Envacute and Envchronic viruses to C-type lectins. Binding to DC-SIGN and langerin was determined using an acoustic biosensor. Binding affinity (dissociation constant [Kd], in nanomolar units) was determined by binding the lectins of interest to the chip and allowing viruses to flow over the surface. Average replication in tissue of Envacute (red squares) and Envchronic (green squares) viruses was correlated with the binding affinity to DC-SIGN (a), langerin (b), and PHA (c). Average transmission fitness in MC+PM1 cocultures of Envacute (red squares) and Envchronic (green squares) viruses was plotted against the binding affinity to DC-SIGN (d), langerin (e), and PHA (f). PHA was tested as the nonspecific control. Each symbol represents one virus. Statistical analysis was performed using a Spearman rank correlation analysis.
C-type lectins trap HIV-1 replication in the tissue.
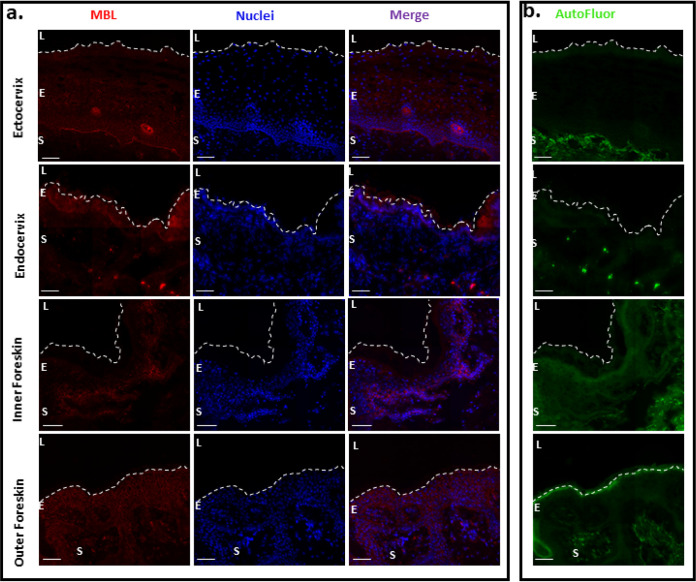
Previous studies have described high levels of soluble C-type lectins, like mannose-binding lectins (MBLs), in mucosal tissue as well as Dectin-1, a C-type lectin on epithelial and endothelial cells. MBLs are thought to trap pathogens with high-mannose glycans in the mucosa and prevent translocation (34). Our fluorescent deconvolution microscopy confirms the presence of MBL in different mucosal tissues (Fig. 7). These analyses serve only to illustrate that DC-SIGN and langerin on the myeloid cell lineage in mucosal tissue provide relatively few binding sites for high-mannose glycans compared to the abundant MBL in these same tissues. Previous studies have shown that the HIV-1 Env gp120/gp41 found on the virus surface retains N-linked high-mannose glycans and somehow avoids late Golgi complex (GC) modification responsible for trimming and complex glycan content (35, 36).
FIG 7.
Presence of MBL in different mucosal tissue types. (a) Fluorescent deconvolution microscopy images of ectocervix, endocervix, inner foreskin, and outer foreskin stained for MBL (red) and DAPI (blue). (b) Autofluorescence (green) of the corresponding tissues. Magnification, ×40. Bar, 40 μm. Dashed lines indicate the luminal surface. L, lumen; E, epithelium; S, stroma.
To evaluate the influence of glycans on the transmission fitness, penile and cervical explants were pretreated with mannan for 12 h and washed prior to addition of one acute B4 and one chronic Q0 virus. Saccharomyces cerevisiae mannan, derived from N linkage, has a conserved mannose(×8)–N-acetylglucosamine(×2) core structure with high affinity to all C-type lectins. Replication of virus in both tissue and MC+PM1 was evaluated by a Gag-specific quantitative PCR (qPCR). Analysis of the total viral copy number revealed that cervical tissue soaked in mannan prior to virus exposure exhibited 6.7× reduced viral replication compared to untreated tissue (Fig. 8a). In penile tissue, no differences in viral replication were observed between mannan-treated and untreated tissue (Fig. 8b). In contrast, 3.2× and 3.9× increased HIV-1 copy numbers were found in MC+PM1 cocultures derived from penile and cervical tissue, respectively, when tissue was treated with mannan (Fig. 8a and b). These findings suggest that fewer viruses were trapped in tissue treated with mannan and that more viruses were able to cross the mucosa with MC to infect T cells.
FIG 8.
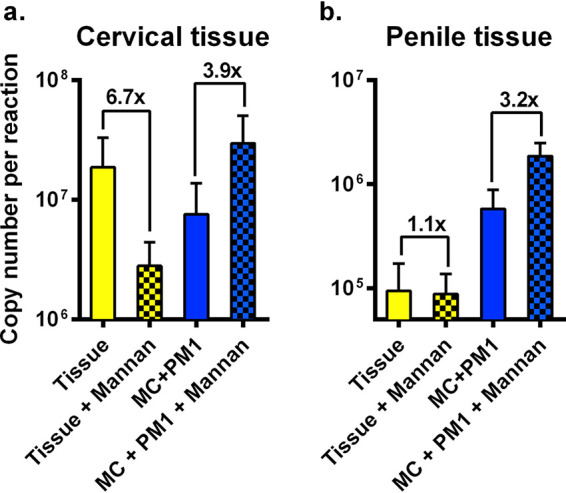
Mannan alters the transmission of HIV-1 across mucosal tissue. (a) Cervical and (b) penile tissues were incubated in medium alone or medium containing mannan for 12 h before exposure to a mix of one Envacute and one Envchronic virus. Migratory cells were harvested after 24 h and cocultured with PM1 T cells. After 10 days in culture, the total HIV-1 copy number in tissue and MC+PM1 cells with and without mannan was assessed using a gag-specific qPCR. Each assay was performed with tissue from three donors. The values are means and standard errors of the means.
DISCUSSION
In the vast majority of new HIV-1 infections, a stringent transmission bottleneck leads to the establishment of a systemic infection by a single viral founder (incidence estimated to be between 76 and 90%) (3). A recent study suggests that this dramatic decline in the HIV-1 heterogeneity from donor to recipient during transmission may be related to a genetic bottleneck during the virus inoculation in the recipient as well as during the translocation of the HIV-1 from the recipient genital mucosa to the blood (6). It remains poorly understood if the bottleneck leading to systemic infection by a single HIV-1 variant is caused by a stochastic event or if selective factors influence the efficiency of HIV-1 transmission through the genital mucosa. During transmission, the HIV-1 genetic bottleneck and any selection (as opposed to stochastic effects) would likely have multifactorial origins. For example, the genetic bottleneck maybe the result of a lower activation state of CD4 T cells (37) and/or the fact that in any transmission event, the majority (>93%) of HIV proviruses are defective (38).
In this study, we evaluated the transmission fitness of HIV-1 harboring subtype B chronic and acute Env glycoproteins by employing a multivirus competition infection of primary human penile and cervical tissue. Env chimeric clones were chosen to specifically determine the role different Env phenotypes play in the mucosal transmission event. In previous studies, replicative fitness in primary CD4+ T cells and macrophages was dominated by the efficiency of host cell entry and mapped to the env gene (39, 40). By using chimeric clones that share the same backbone, we limited transmission efficiency to Env derived from HIV-infected patients during acute or chronic infection. Although Env activity may be dominant or an important trait for transmission, we are not discounting other HIV-1 proteins/factors that may impact transmission efficiency (9, 11, 17). Our data show that Envchronic viruses replicate mainly in the mucosal tissue, while Envacute viruses pass through the tissue with migratory cells and initiate infection in CD4+ T cell lines. Replication of HIV-1 trapped in tissue directly correlated with binding affinity to C-type lectins. Treating the tissue with a saturating concentration of mannan resulted in higher efficiency of HIV-1 transmission to T cells.
Based on our tissue model, HIV-1 harboring Env with the highest binding affinity to C-type lectins showed increased virus replication in the mucosal tissue but was not efficiently transmitted through to CD4+ T cells. Our studies suggest that C-type lectins may “trap” HIV-1 replication in the mucosa and even prevent translocation for systemic infection. Mucosal biologists/immunologists have known for decades that C-type lectins are highly abundant in foreskin, penile, vaginal, and cervical tissues (41, 42) and are key innate barriers preventing pathogen translocation. Understanding the role of C-type lectins in HIV-1 transmission has a long and convoluted history. As early as 1997, Garred et al. (43) described protection from HIV-1 in high-risk cohorts with a specific promoter polymorphism in mannose-binding lectin loci which led to reduced expression of mannose-binding lectin (MBL). However, the protective role of C-type lectins in HIV-1 infection was soon eclipsed by studies describing a role of DC-SIGN in capturing and transmitting HIV-1 (44), which remains a controversial topic (45, 46). Langerhans cells (LCs) expressing langerin or DCs expressing DC-SIGN are probably the first immune cells to come into contact with infecting virus in these mucosal tissues. The proposed role of LCs in HIV-1 transmission was questioned in a study describing the capture, endocytosis, and degradation of HIV-1 by Birkbeck granules following binding to langerin on mucosal LCs (47).
Our findings suggest that replicating HIV-1 in the genital mucosa may be trapped by soluble MBL, shown to have binding specificity for mannose similar to that of the less abundant DC-SIGN and langerin found on DCs and LCs, respectively (48, 49). Addition of mannan to genital mucosal tissue would saturate C-type lectin binding and as a result, reduce trapped HIV-1 replication in the tissue. Surprisingly, addition of mannan increased HIV-1 transmission through the tissue to infect T cells via tissue migratory cells. These findings suggest competing roles of C-type lectins in mucosal tissue to prevent HIV-1 translocation for systemic infection versus the role of DC-SIGN and langerin in enhancing heterosexual HIV-1 transmission. Again, C-type lectins in mucosal tissue are a primary innate defense mechanism preventing pathogen translocation (50, 51), but this process has not been well defined for HIV-1. There is of course the added complication that unlike pathogenic microbes that harbor surface glycoproteins with high-mannose glycans (e.g., Chlamydia and Candida albicans), HIV-1 glycoproteins are subject to the same posttranslational machinery as cellular glycoproteins, typically resulting in greater glycan diversity and reduced branching. However, Doores et al. (36) described high-mannose glycans on Env directly from virus particles, whereas recombinant Env, produced from the same cell types, had more complex glycans, similar to that observed on cellular glycoproteins. These findings suggest that HIV-1 Env may escape the medial or trans GC processing in the context of virus replication, possibly through the coexpression of HIV-1 Nef or Vpu accessory proteins.
In summary, we determined that Envacute HIV-1 viruses were more readily transmitted in penile or cervical tissue than Envchronic viruses. Trapped replication in these mucosal tissues was observed with Envchronic viruses and was associated with higher C-type lectin binding affinity. As described herein and as previously reported, the soluble MBL is omnipresent in genital mucosal tissues and serves as an innate defense mechanism to prevent microbial translocation. We propose that HIV-1 particles with high mannose content may be retained in the mucosa and that HIV-1 with Env with reduced glycan/mannose content may break this trap for systemic infection. These findings are consistent with our recent observation that a more diverse HIV-1 population infected the female genital tract while a single HIV-1 variant was found in the blood at early infection (6). Future studies will focus on determining the actual glycan composition on the Env glycoprotein of the transmitter/founder HIV-1 clone that is responsible for new infections and avoids the lectin trap in genital mucosa.
MATERIALS AND METHODS
Acute and chronic Env chimeric viruses.
Envacute and Envchronic were constructed from 11 acute subtype B envelopes obtained from the Center for HIV-1/AIDS Vaccine Immunology (CHAVI) acute infection studies (3) and three chronic envelopes from patient samples from a Belgian cohort (12, 26). Env chimeric viruses were constructed using a yeast-based recombination cloning method as previously described (52). Briefly, the envelope genes from acute and chronic clones were recombined into pREC_nfl_NL4-3_Δenv/URA3 following transfection into Saccharomyces cerevisiae. Virus was produced by transfection of the pREC construct with the supplementary vector pCMV_cplt into 293T cells using the Effectene lipid system (Qiagen, USA) according to the manufacturer’s instructions. All chimeric viruses were tested for coreceptor usage on U87.CD4 cells expressing either CCR5 or CXCR4 (12, 21) (all were CCR5 using) and then propagated, and titers were determined on U87.CD4.CCR5 cells using the 50% tissue culture infective dose (TCID50) assay. Subsequent experiments on transmission fitness involved tissue explants that harbor a variety of lymphoid and myeloid cells that are susceptible to HIV-1 infection. Previous studies have shown that these HIV-1 stocks differ by 1 log in absolute TCID50 titers derived from PBMCs and primary T cells from different donors and from T cell lines, but the relative difference in titers remained similar. The acute viruses are termed B1, B2, B3, B4, B7, B8, B9, B14, B17, B19, and B20, and the three chronic control Env chimeric viruses are named I10, K44, and Q0 (12). Neighbor-joining phylogenetic tree analysis was performed to compare sequences of the original acute HIV-1 env genes derived from single genome amplification (SGA) to those of the acute chimeric env viruses and to highlight the diversity within the chronic env chimeric viruses. Neighbor-joining trees were constructed with SEAVIEW 4 and visualized with FigTree 1.4.2.
Ethics statement.
Human penile and cervical tissue was collected with written consent according to local research committee (LRC) guidelines. All tissues were obtained under protocols approved by the Imperial College NHS Trust Tissue Bank and the National Research Ethics Committee in accordance with the Human Tissue Act 2004.
HIV-1 transmission fitness assay in human explant tissue.
For this study, for lack of a better term, we describe ex vivo transmission fitness as the ability of the virus to enter the tissue, to be carried by and/or infect migratory cells, and then to be transferred to free T cells.
Deidentified penile tissue was obtained from men undergoing gender reassignment surgery at Charing Cross Hospital, London, United Kingdom. Deidentified endo- and ectocervical tissue was collected from women undergoing therapeutic hysterectomy at St. Mary’s Hospital, London, United Kingdom. Penile and cervical tissue, including both epithelium and stroma, were cut into 3-mm3 explants, and four tissue pieces were added per well in a 24-well tissue culture plate. Multivirus competitions were set up such that each of the 11 Envacute viruses was competed against the other and always against the three chronic viruses. Tissue blocks were then exposed to a mixture of four or five Envacute viruses (B1, B2, B3, B4, B7, B8, B9, B14, B17, B19, and B20) and the same three Envchronic viruses (I10, K44, and Q0) at 150 IU per virus for 3 h at 37°C. The tissue was then washed three times to remove free virus and cultured in complete RPMI 1640 medium (Sigma, United Kingdom) at 37°C.
Migratory cells (MC) migrating out of the tissue were harvested at 24 h and washed 3 times with phosphate-buffered saline (PBS) by centrifugation for 3 min at 350 × g. MC were then cocultured with 105 PM1 CD4+ T cells in complete RPMI for 10 days at 37°C. The explant tissues were washed 3 times with PBS, resuspended in complete RPMI, and cultured for 10 days at 37°C. For MC–T cell cocultures and tissue cultures, half of the supernatant was harvested for analysis and replaced with fresh medium every 3 days. On day 10, the tissue was washed 3 times with PBS and lysed with 300 μl of 1% Triton X-100 supplemented with 1 mg/ml proteinase K overnight at 56°C. Cells from MC+PM1 T cell cocultures were washed 3 times with PBS and lysed with 300 μl of 1% Triton X-100. Each multivirus competition was performed in tissues from two donors in triplicate. DNA was extracted from lysates using the PureLink Pro 96 genomic DNA kit (Invitrogen, USA) according to the manufacturer’s instructions. The extraction procedure was performed using the epMotion 5075 automated liquid handling and pipetting system (Eppendorf, USA).
HIV-1 transmission fitness assay in mannan-treated human explant tissue.
Transmission fitness assays were performed as described above with penile and cervical explant tissues incubated in medium alone or medium containing 1 mg/ml mannan (Sigma, Canada) for 12 h before infection with a mixture of one Envacute (B4) and one Envchronic (Q0) virus. Each condition was tested in tissues from three donors in triplicate. The total HIV-1 copy numbers in tissue and MC+PM1 cocultures were assessed using a Gag-specific qPCR.
Flow cytometry.
For phenotypic analysis of DCs migrating from penile and cervical tissue, a multicolored flow cytometry antibody cocktail was used, consisting of CD14 Qdot605 (TüK4) (Invitrogen), CD11c AF700 (B-ly6), CD123 PECy5 (9F5), CD209 V450 (DCN46), Lin-1 fluorescein isothiocyanate (FITC), CD1c PECy7 (L161) (BioLegend, United Kingdom), HLA-DR allophycocyanin (APC) H7 (G46-6), CD207 APC (10E2) (BioLegend, United Kingdom), and CD83 phycoerythrin (PE) (HB15e). Unless otherwise specified, all antibodies were obtained from BD Biosciences, United Kingdom. Dead cells were excluded from analysis through staining with Aqua viability dye (Invitrogen, United Kingdom). Samples were acquired using an LSRIIFortessa fluorescence-activated cell sorter (FACS; BD Biosciences, United Kingdom) and analyzed using FlowJo (Tree Star, USA). Compensation matrices were created on FlowJo using single stained anti-mouse Ig(κ) negative-control compensation beads (BD Biosciences, United Kingdom).
PCR and NGS analysis of multivirus infections.
HIV-1 DNA was PCR amplified from cellular DNA extracts. It is important to stress that the HIV-1 DNA and not the viral RNA was PCR amplified from cellular DNA extracts to ensure some level of virus infection rather than just residual inoculating virus after extensive washing. At a minimum, this viral DNA results from initial reverse transcription, but it likely represents a spreading HIV-1 infection due to the increasing viral DNA load over the 10-day incubation. PCR amplification of the C2-V3 envelope region and 454 sequencing were performed as previously described (6). The C2-V3 region of the envelope gene was amplified by an external-nested PCR amplification. Primers used for the external PCR were forward ENVB (5′-AGAAAGAGCAGAAGACAGTGGCAATGA-3′) (HXB2 positions 6202 to 6228) and reverse ED14 (5′-TCTTGCCTGGAGCTGCTTGATGCCCCAGAC-3′) (HXB2 positions 7932 to 7961), and those for nested PCR were forward E80 (5′-CCAATTCCCATACATTATTGTG-3′) (HXB2 positions 6858 to 6879) and reverse E125 (5′-CAATTTCTGGGTCCCCTCCTGAGG-3′) (HXB2 positions 7315 to 7338) (6). PCR mixtures contained a 0.2 μM concentration of each primer, 1.5 mM MgCl2, 1× Platinum Taq PCR buffer, 0.2 mM deoxynucleotide triphosphates (dNTPs), and 2 U Platinum Taq DNA polymerase. PCR cycle conditions were 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min (external) or 45 s (nested), and a final extension of 72°C for 10 min.
To prepare the amplicon library for 454 sequencing, fusion primers including the Roche 454 Titanium key sequence, a multiplex identifier (MID) sequence for forward and reverse primers followed by the template-specific sequences forward E110 (5′-CTGTTAAATGGCAGTCTAGCAGAA-3′) (HXB2 positions 7002 to 7025) and reverse E125 (5′-CAATTTCTGGGTCCCCTCCTGAGG-3′) (HXB2 positions 7315 to 7338) were used. PCR cycle conditions were 95°C for 2 min, followed by 35 cycles of 95°C for 30s, 55°C for 30 s, and 72°C for 45 s, and a final extension of 72°C for 10 min. PCR products were run on a 1% agarose gel to verify the product size of 406 bp and purified using the Agencourt AMPure XP bead system with a bead-to-DNA ratio of 0.7:1 according to the Roche manual. Sample libraries were quantified using the Quant-iT PicoGreen double-stranded-DNA assay kit (Invitrogen, Canada), diluted, and pooled at 106 molecules/μl for pyrosequencing. Emulsion PCR (emPCR) was performed with a ratio of 0.5 molecules of sample library per bead. Enriched beads (5 × 105) were then loaded onto titanium picotiter plates according to the Roche 454 instructions. The sequencing run was performed on the Roche 454 GS Junior sequencer as previously described (6). Next-generation sequencing (NGS) analyses was employed as a sophisticated and sensitive counting tool, and the higher error rate seen with Roche 454 has no bearing on these results.
Sequence data analysis.
A custom analysis pipeline (a revision of Segminator II [53] and SeekDeep [27]) was used to quantify each virus within a competition. First, raw sequence data were extracted by the MID tag. Next-generation sequences were analyzed for quality and length, and then the relative amount of each virus was determined via maximum-likelihood phylogenetic tree alignments, SeekDeep alignments, groups, and count sequences within a cluster aligning to each of the input viral sequences.
Lectin binding assay.
The relative binding affinity of Envacute and Envchronic HIV-1 to C-type lectins was assessed using an Akubio RapID4 acoustic biosensor (TTP Labtech). DC-SIGN and langerin were prepared in sodium acetate buffer and covalently bound to a quartz crystal using an ethyl carbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (NHS) amine linkage. Free ester groups were neutralized by ethanolamine blocking. Viral stocks normalized for p24 concentration were allowed to flow over the immobilized lectins for 3 min at a flow rate of 25 μl min−1, followed by a 5-min dissociation step. The quartz crystal surface was regenerated using a 100 mM glycine buffer (pH 2.5) containing 0.05% Tween 20 for 3 min. PHA was used as a nonspecific lectin control.
Immunofluorescence and imaging.
Frozen tissue sections (12 μm) were fixed in 3.7% formaldehyde in PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer for 5 to 10 min, followed by washing in cold PBS. Samples were blocked with normal donkey serum for 10 min and washed again. For mannan-binding lectin (MBL) identification, samples were stained (1:100; clone 3B6; Abcam) for 1 h at 37°C and then washed with cold PBS. Next, a Cy5-labeled donkey anti-mouse immunoglobulin secondary antibody (Jackson ImmunoResearch, USA) was added for ∼20 to 30 min at room temperature. Samples were washed, and Hoechst DAPI (4′,6-diamidino-2-phenylindole; Invitrogen, USA) was applied for 10 min to stain nuclei before a final wash with PBS. Mounting medium (DakoCytomation) and coverslips were applied to sections, sealed with clear nail polish, and stored at 4°C. Images were obtained by deconvolution microscopy on a DeltaVision RT system collected on a digital camera (CoolSNAP HQ; Photometrics) using a 40× oil objective. Panel images at a magnification of ×40 were acquired to include the epithelium and the underlying connective tissue in all ex vivo human samples. After image acquisition, SoftWoRx analysis software (Applied Precision) was used to stitch and quickly project up to 30 z-stacks per panel set.
Quantitative PCR.
Samples were run using the QuantStudio5 real-time PCR system (Applied Biosystems, Canada) using gag-specific primers as previously described (54). First, an external HIV-1 gag-specific PCR was performed in a 50-μl reaction mixture containing 1× PCR buffer, 1 mM MgCl2, 0.2 μM dNTPs, 2 U of Platinum Taq DNA polymerase, and 0.2 μM concentrations of forward primer gag-cons1 (5′-GAGAGAGATGGGTGCGAGAGCG-3′) (HXB2 positions 783 to 804) and reverse primer UNIV-GA4 (5′-TTGCCAAAGAGTGACCTGAGGGAA-3′) (HXB2 positions 2250 to 2273). PCR was performed using the Simpliamp thermocycler (Applied Biosystems, Canada) under the following reaction conditions: 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min 50 s, and a final extension of 72°C for 10 min.
Following PCR, 20 μl PCR product was added to a qPCR mixture which included 0.5 μM concentrations of forward primer gSCA F (5′-CATGTTTTCAGCATTATCAGAAGGA-3′) (HXB2 positions 1299 to 1323) and reverse primer gSCA R (5′-TGCTTGATGTCCCCCCACT-3′) (HXB2 positions 1359 to 1377), 0.2 μM gSCA probe (5′-FAM [6-carboxyfluorescein]-TACTGGGACAGCTACAACCATCCCTT-BHQ [black hole quencher]-3′) (HXB2 positions 968 to 993), and TaqMan Fast advanced master mix. Samples were run under the following reaction conditions: 60°C for 2 min, 95°C for 20 s, and 40 cycles of 95°C for 1 s and 60°C for 43 s. All samples were run in duplicate. Samples were considered negative when cycle threshold (CT) values were above 35.
Statistical analysis.
Statistical analysis was performed on GraphPad Prism 6, using the Holm-Šidák test to compare the average replication of Envchronic with Envacute in each competition, a two-tailed Mann-Whitney test to compare the total average replication of Envchronic and Envacute in tissue (with and without mannan) with that in MC+PM1 cocultures, and a Spearman rank correlation analysis for comparison of lectin binding affinity and replication in tissue or MC+PM1 cocultures. P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
We thank all tissue donors from Charing Cross and St. Mary’s Hospital in London, United Kingdom.
This study was funded by NIAID/NIH grant AI49170 and the CWRU/UH Center for AIDS Research (P30 AI036219). H.M.C. and R.J.S. were funded by the European Union’s Horizon 2020 for Research & Innovation program under grant agreement no. 681137.
Conceptualization and Methodology N.H., J.A.B., Y.G., J.F.S.M., T.J.H., R.J.S. and E.J.A.; Investigation K.K., A.N.R, G.N., I.N., A.A.J.M., D.J.S., H.M.C, A.M.C., M.A.L.; Writing and Visualization K.K. and E.J.A.; Funding Acquisition E.J.A.
We declare no competing interest.
REFERENCES
- 1.Patel P, Borkowf CB, Brooks JT, Lasry A, Lansky A, Mermin J. 2014. Estimating per-act HIV transmission risk: a systematic review. AIDS 28:1509–1519. doi: 10.1097/QAD.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison A, Colvin CJ, Kuo C, Swartz A, Lurie M. 2015. Sustained high HIV incidence in young women in southern Africa: social, behavioral, and structural factors and emerging intervention approaches. Curr HIV/AIDS Rep 12:207–215. doi: 10.1007/s11904-015-0261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 5.Joseph SB, Swanstrom R, Kashuba AD, Cohen MS. 2015. Bottlenecks in HIV-1 transmission: insights from the study of founder viruses. Nat Rev Microbiol 13:414–425. doi: 10.1038/nrmicro3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein K, Nickel G, Nankya I, Kyeyune F, Demers K, Ndashimye E, Kwok C, Chen PL, Rwambuya S, Poon A, Munjoma M, Chipato T, Byamugisha J, Mugyenyi P, Salata RA, Morrison CS, Arts EJ. 2018. Higher sequence diversity in the vaginal tract than in blood at early HIV-1 infection. PLoS Pathog 14:e1006754. doi: 10.1371/journal.ppat.1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgener A, McGowan I, Klatt NR. 2015. HIV and mucosal barrier interactions: consequences for transmission and pathogenesis. Curr Opin Immunol 36:22–30. doi: 10.1016/j.coi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Dupont HA, Lam J, Woods MW, Zahoor MA, Kaushic C. 2018. Hormonal influence on HIV-1 transmission in the female genital tract: new insights from systems biology. Am J Reprod Immunol 80:e13019. doi: 10.1111/aji.13019. [DOI] [PubMed] [Google Scholar]
- 9.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P. 2013. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinnon LR, Achilles SL, Bradshaw CS, Burgener A, Crucitti T, Fredricks DN, Jaspan HB, Kaul R, Kaushic C, Klatt N, Kwon DS, Marrazzo JM, Masson L, McClelland RS, Ravel J, van de Wijgert J, Vodstrcil LA, Tachedjian G. 2019. The evolving facets of bacterial vaginosis: implications for HIV transmission. AIDS Res Hum Retroviruses 35:219–228. doi: 10.1089/aid.2018.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deymier MJ, Ende Z, Fenton-May AE, Dilernia DA, Kilembe W, Allen SA, Borrow P, Hunter E. 2015. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-alpha resistance. PLoS Pathog 11:e1005154. doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King DF, Siddiqui AA, Buffa V, Fischetti L, Gao Y, Stieh D, McKay PF, Rogers P, Ochsenbauer C, Kappes JC, Arts EJ, Shattock RJ. 2013. Mucosal tissue tropism and dissemination of HIV-1 subtype B acute envelope-expressing chimeric virus. J Virol 87:890–899. doi: 10.1128/JVI.02216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lassen KG, Lobritz MA, Bailey JR, Johnston S, Nguyen S, Lee B, Chou T, Siliciano RF, Markowitz M, Arts EJ. 2009. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog 5:e1000377. doi: 10.1371/journal.ppat.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abraha A, Nankya IL, Gibson R, Demers K, Tebit DM, Johnston E, Katzenstein D, Siddiqui A, Herrera C, Fischetti L, Shattock RJ, Arts EJ. 2009. CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic. J Virol 83:5592–5605. doi: 10.1128/JVI.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, Lapedes AS, Shen T, Gaschen B, Krishnamoorthy M, Li H, Decker JM, Salazar-Gonzalez JF, Wang S, Jiang C, Gao F, Swanstrom R, Anderson JA, Ping LH, Cohen MS, Markowitz M, Goepfert PA, Saag MS, Eron JJ, Hicks CB, Blattner WA, Tomaras GD, Asmal M, Letvin NL, Gilbert PB, Decamp AC, Magaret CA, Schief WR, Ban YE, Zhang M, Soderberg KA, Sodroski JG, Haynes BF, Shaw GM, Hahn BH, Korber B. 2011. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog 7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol 79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathys L, Balzarini J. 2015. Several N-glycans on the HIV envelope glycoprotein gp120 preferentially locate near disulphide bridges and are required for efficient infectivity and virus transmission. PLoS One 10:e0130621. doi: 10.1371/journal.pone.0130621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollakis G, Baan E, van Werkhoven MB, Berkhout B, Bakker M, Jurriaans S, Paxton WA. 2015. Association between gp120 envelope V1V2 and V4V5 variable loop profiles in a defined HIV-1 transmission cluster. AIDS 29:1161–1171. doi: 10.1097/QAD.0000000000000692. [DOI] [PubMed] [Google Scholar]
- 21.Krebs KC, Tian M, Asmal M, Ling B, Nelson K, Henry K, Gibson R, Li Y, Han W, Shattock RJ, Veazey RS, Letvin N, Arts EJ, Gao Y. 2016. Infection of rhesus macaques with a pool of simian immunodeficiency virus with the envelope genes from acute HIV-1 infections. AIDS Res Ther 13:41. doi: 10.1186/s12981-016-0125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith SA, Burton SL, Kilembe W, Lakhi S, Karita E, Price M, Allen S, Hunter E, Derdeyn CA. 2016. Diversification in the HIV-1 envelope hyper-variable domains V2, V4, and V5 and higher probability of transmitted/founder envelope glycosylation favor the development of heterologous neutralization breadth. PLoS Pathog 12:e1005989. doi: 10.1371/journal.ppat.1005989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen R, Kappes JC, Smythies LE, Richter HE, Novak L, Smith PD. 2014. Vaginal myeloid dendritic cells transmit founder HIV-1. J Virol 88:7683–7688. doi: 10.1128/JVI.00766-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiffoleau E. 2018. C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front Immunol 9:227. doi: 10.3389/fimmu.2018.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geijtenbeek TB, van Kooyk Y. 2003. DC-SIGN: a novel HIV receptor on DCs that mediates HIV-1 transmission. Curr Top Microbiol Immunol 276:31–54. doi: 10.1007/978-3-662-06508-2_2. [DOI] [PubMed] [Google Scholar]
- 26.Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, Toossi Z, Colebunders RL, Jensen MA, Mullins JI, Vanham G, Arts EJ. 2005. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol 79:9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA. 2018. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res 46:e21. doi: 10.1093/nar/gkx1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheeseman HM, Carias AM, Evans AB, Olejniczak NJ, Ziprin P, King DF, Hope TJ, Shattock RJ. 2016. Expression profile of human Fc receptors in mucosal tissue: implications for antibody-dependent cellular effector functions targeting HIV-1 transmission. PLoS One 11:e0154656. doi: 10.1371/journal.pone.0154656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Q, Frank I, Williams V, Santos JJ, Watts P, Griffin GE, Moore JP, Pope M, Shattock RJ. 2004. Blockade of attachment and fusion receptors inhibits HIV-1 infection of human cervical tissue. J Exp Med 199:1065–1075. doi: 10.1084/jem.20022212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trifonova RT, Lieberman J, van Baarle D. 2014. Distribution of immune cells in the human cervix and implications for HIV transmission. Am J Reprod Immunol 71:252–264. doi: 10.1111/aji.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quiñones-Mateu ME, Ball SC, Marozsan AJ, Torre VS, Albright JL, Vanham G, van der Groen G, Colebunders RL, Arts EJ. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol 74:9222–9233. doi: 10.1128/JVI.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura T, Brumme ZL, Brockman MA, Rosato P, Sela J, Brumme CJ, Pereyra F, Kaufmann DE, Trocha A, Block BL, Daar ES, Connick E, Jessen H, Kelleher AD, Rosenberg E, Markowitz M, Schafer K, Vaida F, Iwamoto A, Little S, Walker BD. 2010. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. JVI 84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selhorst P, Combrinck C, Ndabambi N, Ismail SD, Abrahams MR, Lacerda M, Samsunder N, Garrett N, Abdool Karim Q, Abdool Karim SS, Williamson C. 2017. Replication capacity of viruses from acute infection drives HIV-1 disease progression. J Virol 91. doi: 10.1128/JVI.01806-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auriti C, Prencipe G, Moriondo M, Bersani I, Bertaina C, Mondi V, Inglese R. 2017. Mannose-binding lectin: biologic characteristics and role in the susceptibility to infections and ischemia-reperfusion related injury in critically ill neonates. J Immunol Res 2017:7045630. doi: 10.1155/2017/7045630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2011. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One 6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A 107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz-Arias I, Doitsh G, Yang Z, Sowinski S, Ruelas D, Greene WC. 2015. Blood-derived CD4 T cells naturally resist pyroptosis during abortive HIV-1 infection. Cell Host Microbe 18:463–470. doi: 10.1016/j.chom.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ball SC, Abraha A, Collins KR, Marozsan AJ, Baird H, Quiñones-Mateu ME, Penn-Nicholson A, Murray M, Richard N, Lobritz M, Zimmerman PA, Kawamura T, Blauvelt A, Arts EJ. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J Virol 77:1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rangel HR, Weber J, Chakraborty B, Gutierrez A, Marotta ML, Mirza M, Kiser P, Martinez MA, Este JA, Quinones-Mateu ME. 2003. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J Virol 77:9069–9073. doi: 10.1128/JVI.77.16.9069-9073.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulla R, De Seta F, Radillo O, Agostinis C, Durigutto P, Pellis V, De Santo D, Crovella S, Tedesco F. 2010. Mannose-binding lectin is produced by vaginal epithelial cells and its level in the vaginal fluid is influenced by progesterone. Mol Immunol 48:281–286. doi: 10.1016/j.molimm.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Hirbod T, Bailey RC, Agot K, Moses S, Ndinya-Achola J, Murugu R, Andersson J, Nilsson J, Broliden K. 2010. Abundant expression of HIV target cells and C-type lectin receptors in the foreskin tissue of young Kenyan men. Am J Pathol 176:2798–2805. doi: 10.2353/ajpath.2010.090926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garred P, Madsen HO, Balslev U, Hofmann B, Pedersen C, Gerstoft J, Svejgaard A. 1997. Susceptibility to HIV infection and progression of AIDS in relation to variant alleles of mannose-binding lectin. Lancet 349:236–240. doi: 10.1016/S0140-6736(96)08440-1. [DOI] [PubMed] [Google Scholar]
- 44.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, Figdor CG, van Kooyk Y. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587–597. doi: 10.1016/S0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 45.Lin G, Simmons G, Pohlmann S, Baribaud F, Ni H, Leslie GJ, Haggarty BS, Bates P, Weissman D, Hoxie JA, Doms RW. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J Virol 77:1337–1346. doi: 10.1128/JVI.77.2.1337-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turville SG, Cameron PU, Handley A, Lin G, Pohlmann S, Doms RW, Cunningham AL. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat Immunol 3:975–983. doi: 10.1038/ni841. [DOI] [PubMed] [Google Scholar]
- 47.Ballweber L, Robinson B, Kreger A, Fialkow M, Lentz G, McElrath MJ, Hladik F. 2011. Vaginal Langerhans cells nonproductively transporting HIV-1 mediate infection of T cells. J Virol 85:13443–13447. doi: 10.1128/JVI.05615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feinberg H, Mitchell DA, Drickamer K, Weis WI. 2001. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 49.Ji X, Gewurz H, Spear GT. 2005. Mannose binding lectin (MBL) and HIV. Mol Immunol 42:145–152. doi: 10.1016/j.molimm.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 50.Clemons KV, Martinez M, Axelsen M, Thiel S, Stevens DA. 2011. Efficacy of recombinant human mannose binding lectin alone and in combination with itraconazole against murine Candida albicans vaginitis. Immunol Invest 40:553–568. doi: 10.3109/08820139.2011.569627. [DOI] [PubMed] [Google Scholar]
- 51.Pellis V, De Seta F, Crovella S, Bossi F, Bulla R, Guaschino S, Radillo O, Garred P, Tedesco F. 2005. Mannose binding lectin and C3 act as recognition molecules for infectious agents in the vagina. Clin Exp Immunol 139:120–126. doi: 10.1111/j.1365-2249.2005.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore DM, Arts EJ, Gao Y, Marozsan AJ. 2005. A yeast recombination-based cloning system to produce chimeric HIV-1 viruses and express HIV-1 genes. Methods Mol Biol 304:369–385. doi: 10.1385/1-59259-907-9:369. [DOI] [PubMed] [Google Scholar]
- 53.Archer J, Rambaut A, Taillon BE, Harrigan PR, Lewis M, Robertson DL. 2010. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time—an ultra-deep approach. PLoS Comput Biol 6:e1001022. doi: 10.1371/journal.pcbi.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]