Avian influenza H5N1 and H7N9 viruses infected humans with high mortality rates. A highly safe and effective vaccine is needed to protect against a potential pandemic.
KEYWORDS: influenza A virus, replication-defective virus vaccine, H5N1 and H7N9 virus, H5N1 and H7N9 influenza virus
ABSTRACT
The recent highly pathogenic avian influenza (HPAI) H5N1 and H7N9 viruses have caused hundreds of human infections with high mortality rates. Although H5N1 and H7N9 viruses have been limited mainly to avian species, there is high potential for these viruses to acquire human-to-human transmission and initiate a pandemic. A highly safe and effective vaccine is needed to protect against a potential H5N1 or H7N9 influenza pandemic. Here, we report the generation and evaluation of two reassortant influenza viruses, PR8-H5-H7NA and PR8-H7-H5NA. These viruses contain six internal segments from A/Puerto Rico/8/1934 (PR8), the HA segment from either A/Alberta/01/2014 (H5N1) [AB14 (H5N1)] or A/British Columbia/01/2015 (H7N9) [BC15 (H7N9)], and a chimeric NA segment with either the BC15 (H7N9) HA gene or the AB14 (H5N1) HA gene flanked by the NA packaging signals of PR8. These viruses expressed both H5 and H7 HAs in infected cells, replicated to high titers when exogenous NA was added to the culture medium in vitro, and were replication defective and nonvirulent when administered intranasally in mice. Moreover, intranasal vaccination with PR8-H5-H7NA elicited robust immune responses to both H5 and H7 viruses, conferring complete protection against both AB14 (H5N1) and BC15 (H7N9) challenges in mice. Conversely, vaccination with PR8-H7-H5NA only elicited robust immune responses toward the H7 virus, which conferred complete protection against BC15 (H7N9) but not against AB14 (H5N1) in mice. Therefore, PR8-H5-H7NA has strong potential to serve as a vaccine candidate against both H5 and H7 subtypes of influenza viruses.
IMPORTANCE Avian influenza H5N1 and H7N9 viruses infected humans with high mortality rates. A highly safe and effective vaccine is needed to protect against a potential pandemic. We generated and evaluated two reassortant influenza viruses, PR8-H5-H7NA and PR8-H7-H5NA, as vaccine candidates. Each virus contains one type of HA in segment 4 and the other subtype of HA in segment 6, thereby expressing both H5 and H7 subtypes of the HA molecule. The replication of viruses is dependent on the addition of exogenous NA in cell culture and is replication defective in vivo. Vaccination of PR8-H5-H7NA virus confers protection to both H5N1 and H7N9 virus challenge; conversely, vaccination of PR8-H7-H5NA provides protection only to H7N9 virus challenge. Our data revealed that when engineering such a virus, the H5 or H7 HA in segment 6 affects the immunogenicity. PR8-H5-H7NA has strong potential to serve as a vaccine candidate against both H5 and H7 subtypes of influenza viruses.
INTRODUCTION
Influenza A viruses (IAV), members of the Orthomyxoviridae family, are composed of a segmented, negative-sense, single-stranded RNA genome (1). While wild aquatic birds are known to be the natural reservoir for IAV, other permissive species consist of humans and domestic animals, including pigs and poultry (2, 3). Zoonotic IAVs also have the potential to spill over into naive species, posing a pandemic risk to the susceptible population due to their lack of preexisting immunity. Throughout the last hundred years, there have been four IAV strains that crossed species from avian or porcine to humans, resulting in pandemics with sustained human-to-human transmission: the 1918 Spanish A/H1N1 virus, the 1957 Asian A/H2N2 virus, the 1968 Hong Kong A/H3N2 virus, and the 2009 A/H1N1 virus (4). Moreover, since 1997, several human infections with avian IAV of the H5, H7, H9, H6, and H10 subtypes have been reported (5). Of these avian IAVs, H5N1 and H7N9 viruses stand out due to their high mortality rates in humans, which poses great concerns for public health (6). The highly pathogenic avian influenza (HPAI) H5N1 virus emerged in 1997 and has resulted in 861 laboratory-confirmed human cases and 455 fatalities from 2003 to 16 April 2020 (7, 8). Conversely, ever since the low-pathogenicity avian influenza (LPAI) H7N9 was first reported in humans in 2013 in China (9), it has caused six epidemic waves from 2013 to 2018, resulting in a total of 1,568 confirmed human infections, including 616 deaths as of 4 March 2020 (10).
Vaccination is the most effective public health intervention strategy in the event of a pandemic. Readily available and effective vaccines are critical for preventing potential influenza epidemics and pandemics. Unfortunately, there are some challenges to the development of prepandemic avian IAV vaccines. Some of these challenges include the extended time period required for vaccine production and the low-immunogenicity conventional subunit and inactivated vaccines generated against zoonotic avian IAVs compared to the seasonal influenza vaccines (4). Moreover, the inactivated influenza vaccines frequently administered intramuscularly predominantly induce neutralizing antibodies directed against the hemagglutinin (HA) protein, which frequently mutates to escape the host’s immune response. In contrast, live attenuated influenza vaccines (LAIV) are intranasally administered and mimic the natural route of infection (11). This ability to resemble a natural infection aids in LAIVs inducing broader, longer-lasting, and robust cellular and humoral immune responses in the mucosal system compared to the inactivated influenza vaccines (12–14). However, one disadvantage of their use is the possibility of virulence reversion, limiting their widespread use (15). Replication-defective virus vaccines are composed of viruses that lack one or more vital components to their replication, synthesis, or assembly of the virion. These viruses, therefore, cannot replicate in vivo but can be propagated in vitro when provided the appropriate component (16). Thus, replication-defective IAVs function similarly to LAIVs, which may elicit robust host immune responses while avoiding influenza-induced pathogenesis (13, 17).
The genome of IAV is comprised of eight viral RNA (vRNA) segments (PB2, PB1, PA, HA, NP, NA, M, and NS), which contain the central coding regions that are flanked at both termini by noncoding regions (NCRs) (18). For successful packaging, the eight vRNA segments are incorporated into the nascent virions by segment-specific packaging signals, which are located at the distal 3′ and 5′ ends of each vRNA segment, along with the NCRs and the adjacent terminal coding regions (19–22). IAVs are classified into 18 HA (H1 to H18) and 11 NA (N1 to N11) subtypes based on the antigenic properties of the viral surface glycoproteins, HA and NA (23). HA is the major protein that the host produces, with neutralizing antibodies against and functions in viral uptake by binding to sialic acid receptors on the cell surface to initiate internalization by endocytosis (24). Conversely, the sialidase activity of NA is critical for the complete release of newly assembled viral particles to infect neighboring cells (23). The distinct NA packaging signal has been identified to allow the insertion of a gene encoding a foreign protein antigen (25, 26). Interestingly, previous research has reported that IAV with a large deletion in the NA segment can replicate by the addition of exogenous bacterial NA into the culture media. In a study conducted by Masic et al., a chimeric swine influenza virus (SIV) generated by fusing the SIV H3 HA ectodomain to the NA packaging signal from H1N1 SIV had growth kinetics identical to those of its wild-type counterpart and was replication defective in pigs (27).
In this study, we generated two reassortant IAVs, PR8-H5-H7NA and PR8-H7-H5NA. Specifically, the reassortant viruses contain six internal segments from A/Puerto Rico/8/1934 [PR8 (H1N1)], the HA segment from either A/Alberta/01/2014 (H5N1) [AB14 (H5N1)] or A/British Columbia/01/2015 (H7N9) [BC15 (H7N9], and a chimeric NA segment with either the BC15 (H7N9) HA gene or the AB14 (H5N1) HA gene flanked by the NA packaging signals of PR8 (H1N1). The missing function of NA in the virus is provided through the addition of bacterial sialidase (NA) in the culture medium. These viruses grew well in vitro through the addition of exogenous NA and were replication defective and immunogenic in mice. Of them, PR8-H5-H7NA conferred complete protection against both BC15 (H7N9) and AB14 (H5N1) challenges in mice, demonstrating its great potential as a replication-defective bivalent vaccine candidate against H5 and H7 IAVs.
RESULTS
Generation and characterization of reassortant viruses expressing H5 and H7 HAs.
In this study, we generated four reassortant influenza viruses. The first two viruses, AB14-HA/NA (PR8) and BC15-HA/NA (PR8), were constructed as the control viruses, composed of six internal genomic segments from PR8 (H1N1) and the HA and NA from either AB14 (H5N1) or BC15 (H7N9), respectively. The other two viruses, PR8-H5-H7NA and PR8-H7-H5NA, possess six internal segments from PR8 (H1N1), the HA from either AB14 (H5N1) or BC15 (H7N9), and the modified NA segment containing the HA ectodomain from either BC15 (H7N9) or AB14 (H5N1) flanked by the PR8 (H1N1) NA segment packaging sequences (Fig. 1A and B). The NA packaging sequences are defined as the 3′-NCR (20 nucleotides [nt]), followed by a terminal coding region (186 nt), and the 5′ NCR (28 nt), preceded by a terminal coding region (156 nt). These reassortant viruses, PR8-H7-H5NA and PR8-H5-H7NA, were rescued in the presence of exogenous bacterial NA.
FIG 1.
Schematic diagram of the PR8-H5-H7NA and PR8-H7-H5NA viruses expressing both H5 and H7 HAs. The six genomic internal segments (PB2, PB1, PA, NP, M, and NS) of both viruses are from PR8 (H1N1) (in black). (A) The HA of PR8-H5-H7NA is from AB14 (H5N1). (B) The HA of PR8-H7-H5NA is from BC15 (H7N9). The NA of PR8-H5-H7NA and PR8-H7-H5NA are both composed of the ORF of the HA ectodomain (ED) from BC15 (H7N9) or AB14 (H5N1) flanked by the NA segment-specific packaging sequences derived from PR8 (H1N1). The 186-nt region derived from NA consists of the cytoplasmic tail (CT), transmembrane domain (TMD), and stalk region. The length of the genes is not to scale.
To examine whether both H5 and H7 HAs were expressed, the lysates of virus-infected cells were subjected to Western blotting using anti-NP, anti-M1, anti-H5 HA, and anti-H7 HA antibodies. The viral NP and M1 proteins were detected in all virus-infected cells (Fig. 2A); the H7 HA was present in PR8-H7-H5NA-, PR8-H5-H7NA-, and BC15-HA/NA (PR8)-infected cells; and the H5 HA was present in PR8-H7-H5NA-, PR8-H5-H7NA-, and AB14-HA/NA (PR8)-infected cells (Fig. 2B). These results demonstrated that both H5 and H7 HAs were expressed in cells infected with PR8-H7-H5NA or PR8-H5-H7NA. When purified virions were subjected to Western blotting, H7 HA was present in the PR8-H7-H5NA-, PR8-H5-H7NA-, and BC15-HA/NA (PR8)-purified virions. However, H5 HA was only present in PR8-H5-H7NA and AB14-HA/NA (PR8), not in PR8-H7-H5NA virions (Fig. 2C). These results suggested that both H5 and H7 HAs were incorporated in the PR8-H5-H7NA virions, while only a few H5 HAs were incorporated in the PR8-H7-H5NA virions.
FIG 2.
Detection of H5 and H7 HAs by Western blotting. MDCK cells were mock infected (M) or infected with PR8 (H1N1), PR8-H7-H5NA, PR8-H5-H7NA, BC15-HA/NA (PR8), or AB14-HA/NA (PR8) at an MOI of 0.01, and the whole-cell lysates were harvested at 8 hpi. (A) Detection of nucleoprotein (NP) and matrix (M1) proteins in infected cell lysates by Western blotting. (B) Detection of H5 and H7 HAs in infected cell lysates by Western blotting. (C) Detection of H5 and H7 HAs in the purified virions.
To determine the growth potential of PR8-H7-H5NA and PR8-H5-H7NA in cell culture, we infected MDCK cells with the respective virus at a multiplicity of infection (MOI) of 0.001 and determined the viral titers at the indicated time points in the presence of the appropriate enzyme, NA or trypsin. The reassortant virus PR8-H5-H7NA and its counterpart, wild-type virus AB14-HA/NA (PR8), grew at a one-log lower rate than the PR8 virus. At 72 h postinfection (hpi), the PR8 virus titer is plateaued (2.3 × 107 PFU/ml), while the PR8-H5-H7NA and AB14-HA/NA (PR8) viruses grew to a titer of 5.5 × 106 PFU/ml and 6.3 × 106 PFU/ml, respectively. Conversely, PR8-H7-H5NA and its wild-type counterpart, BC15-HA/NA (PR8), grew at a higher rate than PR8-H5-H7NA and AB14-HA/NA (PR8), albeit slightly lower than that of the PR8 virus. At 72 hpi, the titers of PR8-H5-H7NA and AB14-HA/NA (PR8) reached 1.3 × 107 PFU/ml and 1.6 × 107 PFU/ml, respectively, which are similar to the titer of the PR8 virus (Fig. 3A). It is noted that in the absence of exogenous bacterial NA, no viral replication was detected at all time points for PR8-H7-H5NA and PR8-H5-H7NA.
FIG 3.
Growth properties of PR8-H5-H7NA and PR8-H7-H5NA. (A) The growth curve of PR8-H5-H7NA, PR8-H7-H5NA, PR8 (H1N1), AB14-HA/NA (PR8), and BC15-HA/NA (PR8) on MDCK cells. The cells were infected with the respective virus at an MOI of 0.001. PR8 (H1N1), AB14-HA/NA (PR8), and BC15-HA/NA (PR8) were propagated with TPCK-trypsin, while PR8-H5-H7NA and PR8-H7-H5NA were grown in the presence of exogenous bacterial NA. The viral supernatants were collected at the indicated time points, and the titers were determined by plaque assay on MDCK cells. (B) Plaques formed by PR8 (H1N1) (control), PR8-H5-H7NA, and PR8-H7-H5NA on MDCK cells in the presence or absence of exogenous bacterial NA.
We also observed that in the presence of exogenous bacterial NA (2.5 to 5 mU/ml), PR8-H7-H5NA formed relatively large plaques that were slightly smaller than those formed by the PR8 virus. In contrast, PR8-H5-H7NA formed small plaques (Fig. 3B). In the absence of exogenous bacterial NA, PR8-H7-H5NA and PR8-H5-H7NA were unable to form any plaques. These results suggested the propagation of PR8-H5-H7NA and PR8-H7-H5NA are highly dependent on the presence of exogenous NA and can grow to high titers in cell culture.
PR8-H5-H7NA and PR8-H7-H5NA are replication defective in mice.
To evaluate the virulence of PR8-H5-H7NA and PR8-H7-H5NA, 60 6-week-old BALB/c mice were randomly divided into five groups (n = 12 per group, equal numbers of males and females) (Table 1). Two groups of mice were intranasally infected with 1 × 103 PFU of either PR8-H5-H7NA or PR8-H7-H5NA; two groups of mice were infected with the two wild-type counterpart viruses, AB14-HA/NA (PR8) and BC15-HA/NA (PR8), at 1 × 103 PFU; and one group of mice was infected with medium only (minimal essential medium [MEM]). After infection, the mice were evaluated daily for survival rate and body weight change. At 3 and 14 days postinfection (dpi), lung tissue samples were collected for viral titration (Fig. 4A). At 3 dpi, low viral load was detected in two out of four mice that were infected with PR8-H5-H7NA (mean titer of 2.3 × 102 PFU/g) as well as in all four mice that were infected with PR8-H7-H5NA (mean titer of 2.37 × 103 PFU/g). In contrast, the mice that were infected with AB14-HA/NA (PR8) and BC15-HA/NA (PR8) had high viral loads in their lungs (mean titer of 1.43 × 106 PFU/g and 1.79 × 106 PFU/g, respectively) (Fig. 4B). At 14 dpi, there was no virus detected in any mouse from any of the groups infected with PR8-H5-H7NA and PR8-H7-H5NA. Since a low viral titer in mice infected with PR8-H5-H7NA and PR8-H7-H5NA was observed, we wanted to clarify whether this amount of virus was due to viral replication in mice respiratory tract owing to NA activity resulted from commensal bacteria or due to reactivation of the unreleased virus owing to higher NA activity (2.5 mU/ml) provided in the plaque assay. Based on a report that screening of 34 strains of 13 species of bacteria isolated from human respiratory tract found the highest NA activity was 0.13 mU/ml (28), we conducted an in vitro experiment to simulate this scenario. MDCK cells were infected with PR8-H5-H7NA or PR8-H7-H5NA at an MOI of 0.001 PFU/ml in the presence of 0.2 mU/ml NA, and supernatant harvested at 72 hpi was subjected to the plaque assay in the presence of either 0.2 mU/ml or 2.5 mU/ml NA. While no virus was detected when 0.2 mU/ml NA was provided in the plaque assay, fewer than 10 plaques were observed when 2.5 mM/ml NA was added in the plaque assay.
TABLE 1.
Assignment of mice for the virulence study of the reassortant viruses, PR8-H5-H7NA and PR8-H7-H5NA (trial no. 1)
| Group (n) | Challenge virus | Amt (PFU) | Route |
|---|---|---|---|
| A (12) | MEM | Intranasally | |
| B (12) | AB14-HA/NA (PR8) | 103 | Intranasally |
| C (12) | BC15-HA/NA (PR8) | 103 | Intranasally |
| D (12) | PR8-H5-H7NA | 103 | Intranasally |
| E (12) | PR8-H7-H5NA | 103 | Intranasally |
FIG 4.
PR8-H5-H7NA and PR8-H7-H5NA viruses are replication-defective in mice. (A) The schedule of the mouse trial to evaluate the virulence of PR8-H5-H7NA and PR8-H7-H5NA. Male and female BALB/c mice (n = 12 per group, six males and six females) were intranasally inoculated with either MEM (control), PR8-H5-H7NA (1 × 103 PFU), or PR8-H7-H5NA (1 × 103 PFU). The mice were evaluated daily for both survival rate and body weight change. In addition, any mouse that reached a humane intervention point was humanely euthanized and their lung samples harvested for viral titration. (B) Viral titration of lung tissues collected at 3 dpi. Viral titers are shown as the number of PFU per gram. The samples were analyzed in duplicate. For PR8-H5-H7NA and PR8-H7-H5NA, plaque assays were performed in the presence of exogenous bacterial NA. Each symbol represents an individual mouse, and the horizontal lines indicate the mean values. (C) The survival rate. (D) The body weight change. The values represent the means ± standard deviations (SD).
In agreement with the lung viral titers, all mice in the MEM group as well as all mice infected with either PR8-H7-H5NA or PR8-H5-H7NA survived the duration of the trial and gained weight as the days proceeded (Fig. 4C and D). Conversely, the mice that were infected with AB14-HA/NA (PR8) began to lose weight from 2 dpi, with the lowest weights happening at 7 dpi. In this group, two mice succumbed to infection at 6 dpi, two mice at 8 dpi, and one mouse at 9 dpi. Similarly, the mice that were infected with BC15-HA/NA (PR8) began to lose weight from 3 dpi, with the lowest weights happening at 7 dpi. In this group, two mice succumbed to infection at 8 and 9 dpi, respectively (Fig. 4C and D). These results indicate that the reassortant viruses PR8-H5-H7NA and PR8-H7-H5NA are replication defective and nonvirulent in mice.
PR8-H5-H7NA and PR8-H7-H5NA are immunogenic in mice.
To determine whether PR8-H5-H7NA and PR8-H7-H5NA are immunogenic and able to provide protection against AB14 (H5N1) and BC15 (H7N9) challenges in mice, two animal trials were performed (Table 2). The schedule of the vaccination, sample collection, and viral challenge is illustrated in Fig. 5. Serum samples were collected prior to each vaccination and before viral challenge (on days 0, 21, and 30). The serum from vaccinated mice was tested against AB14 (H5N1) and BC15 (H7N9) by hemagglutinin inhibition (HAI) and serum viral neutralization (SVN) assays.
TABLE 2.
Assignment of mice for the immune protection study of the reassortant chimeric viruses, PR8-H5-H7NA and PR8-H7-H5NA
| Trial no. and group (n) | Vaccination (day 0) | Boost (day 21) | Challenge (day 31) |
|---|---|---|---|
| 2-1 | |||
| A (12) | MEM | MEM | MEM |
| B (14) | PR8-H5-H7NA | PR8-H5-H7NA | AB14 (H5N1) |
| C (14) | PR8-H7-H5NA | PR8-H7-H5NA | AB14 (H5N1) |
| D (12) | MEM | MEM | AB14 (H5N1) |
| 2-2 | |||
| A (6) | MEM | MEM | MEM |
| B (14) | PR8-H5-H7NA | PR8-H5-H7NA | BC15 (H7N9) |
| C (14) | PR8-H7-H5NA | PR8-H7-H5NA | BC15 (H7N9) |
| D (12) | MEM | MEM | BC15 (H7N9) |
FIG 5.
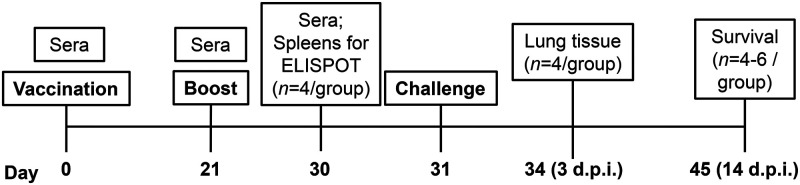
Schedule of mouse immunization, sample collection, and challenge. For the mouse trials no. 2-1 and no. 2-2, BALB/c mice (n = 12 or 14, equal males and females) were intranasally vaccinated on day 0 and boosted on day 21 with 1 × 103 PFU of PR8-H5-H7NA, PR8-H7-H5NA, or MEM. On day 30, four mice per group (two males and two females) were humanely euthanized for splenocyte isolation to detect IFN-γ-secreting cells by the ELISpot assay. On day 31, the remaining mice were challenged with MEM (control), 1 × 103 PFU of AB14 (H5N1) (trial 2-1), or 1 × 103 of PFU BC15 (H7N9) (trial 2-2). Three days postchallenge, four mice per group (two males and two females) were euthanized for sampling. On day 45 (14 days postchallenge), the remaining mice were euthanized and sampled.
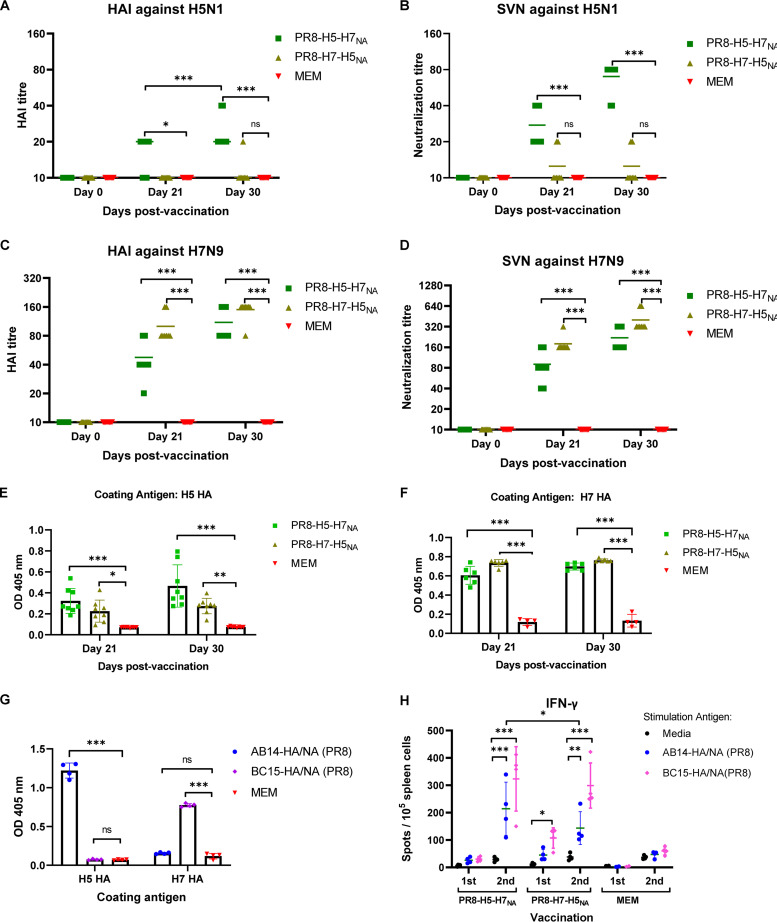
All mice were negative on day 0 for both AB14 (H5N1) and BC15 (H7N9) antibodies in both the HAI assay (HAI, ≤10) and the SVN assay (SVN, ≤10) (Fig. 6). The first vaccination (day 21) with PR8-H5-H7NA resulted in a slight increase in HAI titers against the AB14 (H5N1) antigen, with six out of eight mice having an HAI titer of 20 (Fig. 6A). This contrasts with the mice that were vaccinated with PR8-H7-H5NA that did not have an increase in HAI titers against AB14 (H5N1) antigen after the first vaccination (HAI, ≤10). After the second vaccination (day 30), while the mice that were vaccinated with PR8-H5-H7NA maintained HAI titers against AB14 (H5N1) antigen comparable to those after the first vaccination (HAI of 20, with the exception of one mouse that had an HAI titer of 40), the mice that were vaccinated with PR8-H7-H5NA did not have an increase in HAI titers against the AB14 (H5N1) antigen (HAI, ≤20). In agreement with the HAI assay, the SVN assay also showed that PR8-H5-H7NA stimulated moderate neutralizing antibodies against the AB14 (H5N1) antigen after the first vaccination (SVN, ≤40) and a significant increase after the second vaccination (SVN, ≤80) (Fig. 6B). Conversely, PR8-H7-H5NA induced little production of the neutralizing antibodies against the AB14 (H5N1) antigen after the first (SVN, ≤20) and second (SVN, ≤20) vaccinations.
FIG 6.
Immune responses generated after intranasal vaccination with the replication-defective PR8-H5-H7NA and PR8-H7-H5NA viruses in mice. BALB/c mice were intranasally vaccinated on days 0 and 21 with either MEM (control), PR8-H5-H7NA (1 × 103 PFU), or PR8-H7-H5NA (1 × 103 PFU). The mouse serum was collected on days 0, 21, and 30 for analysis by the HAI assay, SVN assay, and ELISA to detect H5 and H7-HA specific IgG. (A) HAI titers to AB14 (H5N1). (B) SVN titers to AB14 (H5N1). (C) HAI titers to BC15 (H7N9). (D) SVN titers to BC15 (H7N9). (E) H5 HA-specific IgG antibody responses. (F) H7 HA-specific IgG antibody responses. (G) The cross-reactions between H5 HA- and H7 HA-specific IgG antibodies in the mouse serum against both AB14-HA/NA (PR8) and BC15-HA/NA (PR8). The samples were tested in duplicate. (H) ELISpot assay for antigen-specific splenocytes secreting IFN-γ. Each sample was analyzed in triplicate. The splenocytes from mice after one or two vaccinations with PR8-H5-H7NA or PR8-H7-H5NA were harvested and stimulated with purified BC15-HA/NA (PR8), AB14-HA/NA (PR8), or medium only as the antigen control. The graphs represent the means ± SD. The dots represent the individual mice. The differences between two groups were analyzed by a two-way ANOVA with Tukey’s multiple-comparison test. A probability (P) value of <0.05 was considered statistically significant. Significant differences between groups are denoted by * (P < 0.05), ** (P < 0.01), *** (P < 0.001), or ns (not significant; P > 0.05).
In contrast to the antibody levels against the AB14 (H5N1) virus, vaccination with PR8-H5-H7NA and PR8-H7-H5NA resulted in a significantly higher induction of antibodies against the BC15 (H7N9) virus (Fig. 6C and D). Specifically, after the first vaccination (day 21) with either PR8-H5-H7NA or PR8-H7-H5NA, mice developed sufficient antibody levels against the BC15 (H7N9) antigen. Specifically, seven out of eight mice in the PR8-H5-H7NA-vaccinated group had an HAI titer of ≥40, and all of the mice in the PR8-H7-H5NA-vaccinated group had an HAI titer of ≥80. These antibody levels meet or exceed the gold standard HAI titer cutoff of 40, which is associated with reducing the probability of contracting influenza by 50% (29). After the second vaccination (day 30), the HAI titer against the BC15 (H7N9) antigen continued to rise, with all the mice from both vaccination groups (PR8-H5-H7NA and PR8-H7-H5NA) having HAI titers of ≥80 (Fig. 6C). Similarly, for PR8-H5-H7NA and PR8-H7-H5NA, the SVN titers against the BC15 (H7N9) antigen increased after the first vaccination (PR8-H5-H7NA SVN titers of ≥40; PR8-H7-H5NA SVN titers of ≥160) and the second vaccination (PR8-H5-H7NA SVN titers of ≥160; PR8-H7-H5NA SVN titers of ≥320) (Fig. 6D).
Influenza-specific IgG in the mouse serum was determined for days 0, 21, and 30 by enzyme-linked immunosorbent assay (ELISA) using purified H5 or H7 HA protein as the capture antigen. Concerning the IgG levels against the H5 HA, after the first vaccination (day 21) with PR8-H5-H7NA, the IgG optical density (OD) levels were significantly elevated compared to those for the MEM-vaccinated control group (PR8-H5-H7NA, mean OD of 0.323; MEM, mean OD of 0.075) (P < 0.001) (Fig. 6E). Similarly, after the second vaccination (day 30), the IgG levels were further upregulated compared to those of the MEM-vaccinated control group (PR8-H5-H7NA, mean OD of 0.465; MEM, mean OD of 0.078) (P < 0.001). In contrast, the IgG levels against the H5 HA antigen after the first and second vaccinations with PR8-H7-H5NA were only moderately elevated compared to those of the MEM-vaccinated control group (day 21 PR8-H7-H5NA, mean OD of 0.226; day 30 PR8-H7-H5NA, mean OD of 0.274). Conversely, after the first vaccination with either PR8-H5-H7NA or PR8-H7-H5NA, there was a robust production of IgG specific for the H7 HA antigen compared to that induced by MEM vaccination (Fig. 6F). The PR8-H5-H7NA group had a mean OD of 0.605, the PR8-H7-H5NA group had a mean OD of 0.739, and the MEM-vaccinated control group had a mean OD of 0.118. After the second vaccination, the IgG levels remained at levels similar to those induced after the first vaccination.
To understand whether AB14 (H5N1) viral infection would induce the cross-reactive antibodies against BC15 (H7N9) virus or vice versa, serum from mice that survived infection with either AB14-HA/NA (PR8) or BC15-HA/NA (PR8) were subjected to ELISA using purified H5 or H7 HA proteins as the capture antigens. Interestingly, there were no cross-reactive antibodies detected against either the AB14 (H5N1) or the BC15 (H7N9) virus (Fig. 6G).
To evaluate the antigen-specific gamma interferon (IFN-γ)-secreting cells induced by PR8-H5-H7NA and PR8-H7-H5NA vaccination, the mouse spleens were harvested after the first (day 21) and the second (day 30) vaccination from the MEM control group and the PR8-H5-H7NA- and PR8-H7-H5NA-vaccinated mice (two males and two females per group on both days). The splenocytes were isolated and the antigen-specific responses were measured by the IFN-γ enzyme-linked immunosorbent spot (ELISpot) assay (Fig. 6H). After the first vaccination with either PR8-H5-H7NA or PR8-H7-H5NA, the levels of antigen-specific IFN-γ remained low compared to that of the MEM control group. However, after the second vaccination with either PR8-H5-H7NA or PR8-H7-H5NA, the frequencies of both H5- and H7-specific IFN-γ-secreting cells significantly increased compared to those of the control mice (P < 0.001). Of note, the frequency of H5-specific IFN-γ-secreting cells induced by PR8-H5-H7NA is significantly higher than that induced by PR8-H7-H5NA (P = 0.0199).
Vaccination with PR8-H5-H7NA completely protects mice from a lethal challenge of both AB14 (H5N1) and BC15 (H7N9) viruses, whereas vaccination with PR8-H7-H5NA only protects mice from a lethal challenge of BC15 (H7N9) virus.
After the mice were intranasally challenged with either AB14 (H5N1) or BC15 (H7N9), they were monitored daily for 14 days for survival rate and body weight (Fig. 7). As expected, all of the mice that were mock vaccinated and mock challenged with MEM survived the duration of the trial, remaining at normal body weights. In agreement with our previous observations (30), all mice that were mock vaccinated with MEM and challenged with a lethal dose of either AB14 (H5N1) virus or BC15 (H7N9) virus rapidly lost body weight, reaching a humane endpoint of weight loss greater than 20% of their initial body weight within 6 and 7 days postchallenge, respectively. Similarly, all the mice that were vaccinated with PR8-H7-H5NA and challenged with AB14 (H5N1) rapidly lost body weight starting at 2 days postchallenge, and they all succumbed to infection by 7 days postchallenge (Fig. 7A and B). In sharp contrast, all the mice that were vaccinated with PR8-H5-H7NA survived AB14 (H5N1) lethal challenge over the duration of the trial, gradually gaining weight to a level comparable to that of the MEM control group. Interestingly, vaccination with either PR8-H5-H7NA or PR8-H7-H5NA provided complete protection to mice against lethal challenge with the BC15 (H7N9) virus (Fig. 7C and D). These mice exhibited gradual body weight gain at a rate similar to that of the mice mock vaccinated and mock challenged with MEM.
FIG 7.
Survival rate, body weight change, and viral titration of PR8-H5-H7NA- and PR8-H7-H5NA-vaccinated mice after challenge with either AB14 (H5N1) or BC15 (H7N9). BALB/c mice were intranasally vaccinated on days 0 and 21 with MEM, PR8-H5-H7NA (1 × 103 PFU), or PR8-H7-H5NA (1 × 103 PFU). On day 31, these mice were intranasally challenged with MEM (control), AB14 (H5N1) (1 × 103 PFU), or BC15 (H7N9) (1 × 103 PFU). (A and B) For the AB14 (H5N1)-challenged mice, the survival rates (A) and body weight changes (B) are shown. (C and D) For the BC15 (H7N9)-challenged mice, the survival rates (C) and body weight changes (D) are shown. (E and F) The viral titers in the lung tissues of AB14 (H5N1) (E)- and BC15 (H7N9) (F)-challenged mice at 3 dpi. The samples were analyzed in quadruplicate. Each symbol represents an individual mouse, and the horizontal lines indicate mean values. The differences between two groups were analyzed by a two-way ANOVA with Tukey’s multiple-comparison test. A probability (P) value of <0.05 was considered statistically significant. Significant differences between groups are denoted by * (P < 0.05), ** (P < 0.01), *** (P < 0.001), or ns (P > 0.05). The individual mouse values are represented as the means ± SD.
On day 3 postchallenge with AB14 (H5N1) or BC15 (H7N9), four mice per group were euthanized (two males and two females), and the lungs were collected for virus isolation and titration. With regard to the AB14 (H5N1)-challenged mice, high viral titers could be detected in the MEM mock-vaccinated and PR8-H7-H5NA-vaccinated mice (mean titers of 106.63 50% tissue culture infective dose [TCID50]/g and 106.5 TCID50/g, respectively). Conversely, in the PR8-H5-H7NA-vaccinated mice, only one mouse had a detectable viral titer (102.65 TCID50/g) (Fig. 7E). This PR8-H5-H7NA-vaccinated group had significantly lower viral titers than the mice mock vaccinated with MEM and challenged with AB14 (H5N1) (P < 0.001). In agreement with the survival and body weight data, while the mice that were mock vaccinated with MEM and challenged with BC15 (H7N9) displayed high lung viral loads 3 days postchallenge (mean titer of 107.35 TCID50/g), no infectious virus could be detected from the lungs of PR8-H5-H7NA- or PR8-H7-H5NA-vaccinated and BC15 (H7N9)-challenged mice (Fig. 7F). These results indicate that intranasal vaccination with PR8-H5-H7NA provided complete protection against both AB14 (H5N1) and BC15 (H7N9) viral challenge in mice, whereas vaccination with PR8-H7-H5NA only provided complete protection against BC15 (H7N9).
DISCUSSION
Effective vaccines against H5N1 and H7N9 IAVs are needed due to their ability to cause severe diseases in humans, their high mortality rates, and their pandemic potential (4, 6). Unfortunately, the commonly used inactivated vaccines based on wild-type H5N1 or H7N9 virus cannot be produced on a large scale due to the requirements of working under biosafety containment level 3 conditions and their high virulence in embryonated chicken eggs (31). In this study, we evaluated the immunogenicity and protective efficacy of two replication-defective bivalent IAV vaccines against AB14 (H5N1) and BC15 (H7N9). These vaccines were generated by reverse-genetic technology that expressed both the H5 and H7 HAs in the genetic background of the high-yield strain PR8 (H1N1). The eight packaging signals were retained while replacing the ectodomain of NA with the ectodomain of either the AB14 (H5N1) HA or the BC15 (H7N9) HA. The reassortant chimeric viruses, PR8-H5-H7NA and PR8-H7-H5NA, lost the natural viral NA enzymatic activity, capable of replicating and forming plaques only when exogenous NA was provided (Fig. 3A). Although the viral kinetics of the reassortant chimeric viruses were slightly lower than that of the parental virus PR8 (H1N1), PR8-H5-H7NA and PR8-H7-H5NA reached titers equivalent to those of PR8 (H1N1) by 72 hpi in the presence of exogenous bacterial NA (Fig. 3B). Therefore, the virus could be produced in high yield, meeting one of the essential criteria to be considered a vaccine candidate.
The reassortant chimeric viruses, PR8-H5-H7NA and PR8-H7-H5NA, have the potential to serve as replication-defective IAVs in vivo. Replication-defective viruses are defined by the feature of lacking an essential component in the viral replication cycle that renders it incapable of properly replicating in vivo but capable of replicating in vitro if the appropriate component is provided (16). After intranasal infection, these replication-defective viruses will infect the epithelial cells, replicate genomic RNA, and express the viral antigen H5 and H7 HAs. However, without NA, the progeny virions will remain attached to the cellular membrane, restricting budding of the progeny virion to infect neighboring cells. We detect low viral titers on day 3 p.i. in PR8-H5-H7NA- and PR8-H7-H5NA-infected mice (Fig. 4B), and this may raise the concern that host NA-like function or NA activity resulting from commensal bacterial could complement the missing NA function from the virus. However, we also showed that growing the reassortant chimeric viruses in the presence of NA at concentrations slightly higher than the physiological concentration did not result in any viral detection in plaque assay when lower NA activity is provided, yet a few plaques could be detected when higher NA activity is provided in plaque assay. These results suggested that the lower virus titer seen in mouse lung was due to the reactivation of the unreleased replication-defective virus over the plaque assay procedure; the NA activity in the respiratory tract is not sufficient to support efficient productive replication of PR8-H5-H7NA and PR8-H7-H5NA in vivo.
In this study, intranasal vaccination with PR8-H5-H7NA provided complete protection against both AB14 (H5N1) and BC15 (H7N9) infection in mice, inducing neutralizing antibodies against both AB14 (H5N1) and BC15 (H7N9). In contrast, intranasal vaccination with PR8-H7-H5NA only provided protection against BC15 (H7N9) infection. It was noted that PR8-H7-H5NA induced neutralizing antibodies against BC15 (H7N9) but not against AB14 (H5N1). There are several possible explanations for the lack of protection of PR8-H7-H5NA against AB14 (H5N1) infection. First, while Western blotting showed both H5 and H7 HAs were incorporated in the PR8-H5-H7NA virions, only the H7 HA was detected in PR8-H7-H5NA virions, although there was expression of both H5 and H7 HAs in PR8-H7-H5NA-infected cells (Fig. 2). Second, the H5 HA-ectodomain (HA-ED) was introduced by replacing the NA open reading frame (ORF) in PR8-H7-H5NA, and the H7 HA-ED was introduced by replacing the NA ORF in PR8-H5-H7NA. Theoretically, the ratio of HA to NA on influenza virus particles ranges from 4:1 to 10:1 (32). Therefore, the H5 HA possibly was presented at a lower number than the H7 HA in PR8-H7-H5NA. Moreover, previous studies have shown that H5N1 vaccines appeared to be poorly immunogenic in mammalian models (4). Thus, even though the H5 HA is present on the virion surface in the proper conformation, it is tempting to speculate that the smaller amount and poor antigenicity of H5 HA did not grant PR8-H7-H5NA the ability to induce sufficient H5-specific immune responses, providing protection to AB14 (H5N1) viral challenge. Interestingly, after two vaccinations, PR8-H7-H5NA induced a moderate number of IFN-γ-secreting splenocytes upon AB14 (H5N1) stimulation; however, this number was significantly lower than that induced by PR8-H5-H7NA. The lack of protection by PR8-H7-H5NA against AB14 (H5N1) is attributable to lower titer of HAI as well as cell-mediated immune response.
Immune response after vaccination is affected and is different between biological sexes. In C57BL/6 mice, vaccination of inactivated influenza vaccine resulted in the generation of higher antibody titers in female mice than in male mice, which conferred a better protection in female mice against drifted variant viral infection than in male mice (33). In our study, we used both male and female mice; however, we did not observe significant differences concerning the immune responses or the protection effect between different sexes. The genetic background of mice, type of vaccine, virus strains used in the challenge study, and sample size all could contribute to the outcome of the results. This direction warrants further investigation.
In conclusion, we generated a replication-defective IAV, PR8-H5-H7NA, in the genetic background of PR8 (H1N1) virus. PR8-H5-H7NA vaccination elicited robust immune responses, which conferred complete immune protection against both AB14 (H5N1) and BC15 (H7N9) challenges in mice, demonstrating its great potential as a replication-defective bivalent vaccine candidate.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney (MDCK) (ATCC; number CRL-2936) cells were grown in minimal essential medium (MEM) (Sigma-Aldrich, St. Louis, MO, USA) that was supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich) and gentamicin (50 μg/ml; Bio Basic, Markham, ON, Canada). Human embryonic kidney (HEK-293T) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. MDCK and HEK-293T cells were maintained at 37°C in a humidified 5% CO2 incubator. Influenza virus strain isolates A/Alberta/01/2014 (H5N1) [AB14 (H5N1)] and A/British Columbia/01/2015 (H7N9) [BC15 (H7N9)] were kind gifts from Yan Li at the National Microbiology Laboratory, Public Health Agency of Canada (PHAC). The viruses were propagated in MDCK cells and titrated by plaque assay. All infectious experiments were conducted in a biosafety containment level 3 facility at the Vaccine and Infectious Disease Organization-International Vaccine Centre (VIDO-InterVac, Saskatoon, SK, Canada) at the University of Saskatoon, Canada, under the guidelines of PHAC and the Canadian Food Inspection Agency (CFIA; Saskatoon, SK, Canada).
The reassortant wild-type viruses, AB14-HA/NA (PR8) and BC15-HA/NA (PR8), which are composed of six internal genes from PR8 (H1N1) and the wild-type HA and NA genes from either AB14 (H5N1) or BC15 (H7N9), respectively, were grown in the presence of 0.2% bovine serum albumin (BSA) (A7030; Sigma-Aldrich, St. Louis, MO, USA) and 2 μg/ml of l-[(toluene-4-sulfonamido)-2-phenyl] ethyl chloromethyl ketone (TPCK)-trypsin. The reassortant chimeric viruses, PR8-H5-H7NA and PR8-H7-H5NA, composed of six internal genes from PR8 (H1N1) and the HA and NA plasmids as described below, were rescued by the influenza virus reverse genetics technique in the presence of 1 ml of Opti-MEM containing 0.2% BSA, 2 μg/ml of TPCK-trypsin, and 20 mU/ml Vibrio cholerae neuraminidase (N6514; Sigma-Aldrich, St. Louis, MO, USA). After viral rescue, the viruses were grown in MDCK cells in the presence of 0.2% BSA, 1 μg/ml TPCK-trypsin, and 2.5 to 5 mU/ml Vibrio cholerae neuraminidase, unless otherwise stated.
Plasmids and the generation of the reassortant H5 and H7 viruses.
The wild-type HA and NA genes derived from AB14 (H5N1) and BC15 (H7N9) viruses were cloned into pHW2000 to result in the plasmids pHW-AB14-HA, pHW-AB14-NA, pHW-BC15-HA, and pHW-BC15-NA as previously described (34–36). The plasmid pHW-BC15/H7NA was generated by modifying pHW186-NA to include the H7 HA ectodomain flanked by the PR8 (H1N1) NA packaging signals. Specifically, the PR8 (H1N1) NA segment-specific packaging signals at the 3′ and 5′ ends (206 nucleotides [nt] and 184 nt, respectively) were amplified by PCR using pHW186-NA as the template. The primers used for amplifying the 3′-NA packaging signal were 5′-TAA CGC TAG CAG TTA ACC GGA GTA C-3′ and 5′-CGA GGC AGA TTT TGT CTA CCC AGG TGC TAT TTT TAT AGG-3′. The primers used for amplifying the 5′-NA packaging signal were 5′-CAG CGG CTA CAA AGA TTG AGG CCT TGC TTC TGG GTT GAA T-3′ and 5′-GAA ATG CTG CTC GCA CTA GTC-3′. The H7 HA ectodomain (excluding the signal peptide sequence, transmembrane domain, and cytoplasmic tail) from BC15 (H7N9) was amplified by PCR using pHW-BC15-HA as the template with the following primers: 5′-AAA TAG CAC CTG GGT AGA CAA AAT CTG CCT CGG ACA TC-3′ and 5′-CCC AGA AGC AAG GCC TCA ATC TTT GTA GCC GCT GCT TAG T-3′. The three PCR products (3′-NA packaging signal, 5′-NA packaging signal, and the H7 HA ectodomain) were joined by overlapping PCR to obtain the plasmid pHW-BC15/H7NA. The plasmid pHW-AB14/H5NA, encoding the H5 HA ectodomain of AB14 (H5N1) flanked by the PR8 (H1N1) NA packaging signals, was constructed using the same method as that for the plasmid pHW-BC15/H7NA. The following primers were used for amplifying the 3′-NA packaging signal: 5′-TAA CGC TAG CAG TTA ACC GGA GTA C-3′ and 5′-CAA TGC AAA TAT GAT CCA CCC AAG TAT TGT TTT CGT AG-3′. The H5 HA ectodomain from AB14 (H5N1) was amplified by PCR using pHW-AB14-HA as the template with the primers 5′-AAA CAA TAC TTG GGT GGA TCA TAT TTG CAT TGG TTA TCA-3′ and 5′-AGC AAG GCC TCA TTG GTA GAT TCC TAT TGA TTC C-3′. The PCR products (3′-NA packaging signal, 5′-NA packaging signal, and the H5 HA ectodomain) were joined by overlapping PCR to obtain the plasmid pHW-AB14/H5NA. To generate the PR8-H5-H7NA virus, the following plasmids were used: pHW181-PB2, pHW182-PB1, pHW183-PA, pHW-AB14-HA, pHW185-NP, pHW-BC15/H7NA, pHW187-M, and pHW188-NS. To generate PR8-H7-H5NA virus, the following plasmids were used: pHW181-PB2, pHW182-PB1, pHW183-PA, pHW-BC15-HA, pHW185-NP, pHW-AB14/H5NA, pHW187-M, and pHW188-NS. Viral titers were determined on MDCK cells by plaque assay as previously described (37). The viruses for the animal experiments were purified and prepared as described previously (38).
Western blotting.
Virus-infected MDCK cell lysates or purified virions were prepared as described previously (38), after which they were resolved on SDS-PAGE and transferred onto nitrocellulose membranes (0.45 μm; Bio-Rad, Hercules, CA, USA). The membranes were blocked with 5% skim milk in TBST (0.1 M Tris, 0.17 M NaCl, and 1% Tween 20) for 1 h and then washed twice with TBST. After washing, they were probed overnight at 4°C with either rabbit anti-HA (A/Anhui/1/2005 [H5N1]) polyclonal antibody (11048-T62; Sinobiological), rabbit anti-HA (A/Anhui/1/2013 [H7N9]) polyclonal antibody (40103-RP02; Sinobiological), rabbit polyclonal anti-M1 (made in-house), or a rabbit polyclonal anti-NP (made in-house) (39). After overnight incubation, the membranes were washed four times with TBST, followed by incubation with donkey anti-rabbit IgG (IR Dye 680RD; Li-Cor, Lincoln, NE, USA) for 1 h at room temperature. The membranes were visualized using an Odyssey infrared imager (Li-Cor Biosciences).
Ethics statement.
All animal procedures were approved by the University Animal Care Committee (UACC) and Animal Research Ethics Board (AREB) of the University of Saskatchewan. The approval for the virulence trial (animal use protocol number 20190041) and for the immune protection trial (animal use protocol number 20190079) was granted on 12 April 2019 and 18 June 2019, respectively, in accordance with the standards stipulated by the Canadian Council on Animal Care (CCAC).
Animal experiments.
For this study, three animal trials were designed and executed to assess (i) the virulence of the reassortant chimeric viruses, PR8-H5-H7NA and PR8-H7-H5NA, in mice, and (ii) the immune protection of PR8-H5-H7NA and PR8-H7-H5NA against challenge of AB14 (H5N1) and BC15 (H7N9) in mice. For the first animal trial (Table 1), 60 6-week-old male and female BALB/c mice (30 males and 30 females) (Charles River Laboratories, Saint-Constant, QC, Canada) were randomly separated into five groups with 12 mice per group (six males and six females) and allowed to acclimatize for 1 week. After 1 week, the mice were intranasally inoculated with 50 μl (25 μl/nostril) of either MEM or the following reassortant viruses at a dose of 1 × 103 PFU: PR8-H5-H7NA, PR8-H7-H5NA, AB14-HA/NA (PR8), and BC15-HA/NA (PR8). Four mice from each group were humanely euthanized at 3 days postinfection (dpi), and their lung tissues were collected for virus isolation and titration. The remaining mice were monitored daily for 14 days for survival rate and body weight change. Any mouse that dropped below 20% of its initial body weight was humanely euthanized.
The second and the third animal trials were designed to evaluate the immune protection of PR8-H5-H7NA and PR8-H7-H5NA against challenge of AB14 (H5N1) and BC15 (H7N9) in mice. For the trial to evaluate the immune protection against challenge with AB14 (H5N1) (trial no. 2-1), 52 6-week-old male and female BALB/c mice (Charles River Laboratories, Saint-Constant, QC, Canada) were randomly assigned into four groups with either 12 or 14 mice in each group (six or seven males and females, respectively) (Table 2). For the trial to evaluate the immune protection against challenge with BC15 (H7N9) (trial no. 2-2), 46 6-week-old male and female BALB/c mice (Charles River Laboratories, Saint-Constant, QC, Canada) were randomly assigned into four groups with either 6, 12, or 14 mice in each group (equal number of males and females in each group) (Table 2). The males and females were housed in separate cages within groups and allowed to acclimatize for 1 week prior to vaccination. After acclimatization, the mice in groups A and D for both trials were intranasally mock vaccinated with 50 μl of MEM. The mice in groups B and C were intranasally vaccinated with 50 μl of 1 × 103 PFU of either PR8-H7-H5NA or PR8-H5-H7NA. On day 21, the mice received a booster vaccination identical to the first vaccination. On day 30 (9 days postboost), four mice from each group (two males and two females) were euthanized and their spleens were collected for splenocyte isolation to detect IFN-γ-secreting cells. Ten days after the second vaccination (day 31), the mice were intranasally challenged with a 100% lethal dose (LD100) at 1 × 103 PFU of AB14 (H5N1) (trial no. 2-1) or BC15 (H7N9) (trial no. 2-2). After viral challenge, four mice from each group (two males and two females) were humanely euthanized at 3 days postchallenge, and their serum and lungs were collected. The other mice were monitored daily for 14 days for survival rate and body weight change. Serum samples were collected before each vaccination and at euthanization. Mice that lost over 20% of their total body weight were humanely euthanized. All infectious experiments were conducted in a biosafety containment level 3 facility at VIDO-InterVac, University of Saskatchewan, under the guidelines of PHAC, CFIA, the University of Saskatchewan, and the CCAC.
HAI and SVN assays.
The HAI and SVN assays were performed using chicken red blood cells as described previously (35, 36, 40). Four HA units of both AB14 (H5N1) and BC15 (H7N9) were used for the HAI assay.
ELISA to determine antigen-specific IgG levels.
Ninety-six-well Immulon-2 plates (Dynex Technology, Inc., Chantilly, VA, USA) were coated with influenza A H7N9 (A/Anhui/1/2013) HA protein (1 μg/ml) (40103-V08H4; Sino Biological) or influenza A H5N1 (A/Alberta/01/2014) HA protein (1 μg/ml) (homemade; expressed in 293T cells [36]) using a carbonate buffer and incubated overnight at 4°C. The plates were then washed twice with phosphate-buffered saline (PBS) and blocked with 2% BSA in PBS overnight at 4°C. The plates were washed twice and incubated with serially diluted mouse serum in 1% BSA–PBST (PBS plus 0.1% Tween 20). After incubation, plates were washed twice with PBST and were incubated with goat anti-mouse IgG (H+L) (B2763; Invitrogen, Carlsbad, CA, USA) in 1% BSA–PBST for 1 h at room temperature. The plates were washed with PBST, and color development was initiated through the addition of alkaline phosphatase (AP)-conjugated streptavidin (Jackson ImmunoResearch) and p-nitrophenyl phosphate (pNPP) substrate [10 mg/ml p-nitrophenyl phosphate di(Tris) salt crystalline (Sigma-Aldrich, St. Louis, MO, USA), 1% diethanolamine (Sigma-Aldrich, St. Louis, MO, USA), 0.5 mg/ml MgCl2, pH 9.8]. The optical density (OD) was measured at 405 nm on a microplate reader (SpectraMax Plus 384; Molecular Devices, San Jose, CA, USA). The significant differences between groups are denoted by one (P < 0.05), two (P < 0.01), or three (P < 0.001) asterisks or were not significant (ns; P > 0.05).
ELISpot assay.
The frequency of cytokine-secreting cells was measured by the ELISpot assay. Multiscreen-HA plates, 0.45 μm, sterile (MAHAS4510; Millipore) were first coated and incubated overnight at 4°C with purified rat anti-mouse IFN-γ (551216; BD). The next day, mouse splenocytes were seeded at 5 × 105 cells/well, followed by stimulation with 50 μg/ml of either purified β-propiolactone-inactivated BC15-HA/NA (PR8), AB14-HA/NA (PR8), or medium only for 20 h at 37°C. After incubation, plates were blocked and incubated, and the spots developed and counted with the ELISpot reader (AID, Strassberg, Germany) as previously described (36). The data are expressed as the number of spots per 105 spleen cells with the medium only spots subtracted as the background.
Virus isolation and titration.
Directly after tissue collection, the lungs were weighed and homogenized in MEM supplemented with penicillin-streptomycin (Gibco, Thermo Fisher, ON, Canada) in a TissueLyser II (Qiagen, Hilden, Germany) at 25 Hz for 5 min. The samples were clarified by centrifugation at 5,000 × g for 10 min at 4°C, and the supernatants were collected in screw-cap tubes and stored at −80°C for further titration. For the virulence animal trial (Table 1), lung tissues were analyzed by plaque assay in the presence of NA and TPCK-trypsin, with the viral titers expressed as the number of PFU per gram of tissue. For the immune protection trials (Table 2, trial no. 2-1 and 2-2), lung tissues were subjected to the 50% tissue culture infective dose (TCID50) assay as previously described (35, 36). The TCID50 titer was calculated by the Spearman-Kärber algorithm and expressed as TCID50 per gram of tissue (41, 42).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 8.3.0 software (GraphPad Software Inc., San Diego, CA, USA). The differences between two groups were analyzed by a two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test. The data are expressed as the means ± standard deviations (SD). A probability (P) value of <0.05 was considered statistically significant. Significant differences between groups are denoted by one (P < 0.05), two (P < 0.01), or three (P < 0.001) asterisks or were considered not significant (ns; P > 0.05).
ACKNOWLEDGMENTS
We are thankful to Y. Li from the National Microbiology Laboratory, Public Health Agency of Canada, for sharing the AB14 (H5N1) and BC15 (H7N9) strain isolates with us. We are grateful to the animal care staff at VIDO-InterVac for the enormous support in housing, monitoring, infecting, and processing the mice. We also appreciate Tracey Thue’s assistance in her coordination with CFIA and PHAC for the biosafety regulation guidelines.
This work was supported by a grant from the Public Health Agency of Canada to Y.Z. X.T. was supported by a Saskatchewan Health Research Foundation Postdoctoral Fellowship.
We have no conflict of interests to declare.
Conceptualization, Y.Z.; Methodology, X.T., S.L., and Y.Z.; Formal Analysis, X.T. and Y.Z.; Investigation, X.T., S.L., Y.L., and K.P.; Resources, X.T., S.L., Y.L., K.P., and Y.Z.; Writing – Original Draft Preparation, X.T. and Y.Z.; Writing – Review and Editing, S.L. and Y.Z.; Supervision, Y.Z.; Funding Acquisition, Y.Z.
Footnotes
The manuscript was approved for publication by the director of VIDO-InterVac and was assigned manuscript serial number 908.
REFERENCES
- 1.Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26:D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, Garcia-Sastre A. 2018. Influenza. Nat Rev Dis Primers 4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Short KR, Richard M, Verhagen JH, van Riel D, Schrauwen EJ, van den Brand JM, Manz B, Bodewes R, Herfst S. 2015. One health, multiple challenges: the inter-species transmission of influenza A virus. One Health 1:1–13. doi: 10.1016/j.onehlt.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vries RD, Herfst S, Richard M. 2018. Avian influenza A virus pandemic preparedness and vaccine development. Vaccines 6:46. doi: 10.3390/vaccines6030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard M, de Graaf M, Herfst S. 2014. Avian influenza A viruses: from zoonosis to pandemic. Future Virol 9:513–524. doi: 10.2217/fvl.14.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutton TC. 2018. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses 10:461. doi: 10.3390/v10090461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 8.WHO. 2020. Human infection with avian influenza A(H5) viruses. World Health Organization, Geneva, Switzerland. https://wwwwhoint/docs/default-source/wpro—documents/emergency/surveillance/avian-influenza/ai-20200417pdf?sfvrsn=30d65594_56. [Google Scholar]
- 9.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 10.Food and Agriculture Organization of the United Nations. 2020. H7N9 situation update. Food and Agriculture Organization of the United Nations, Rome, Italy. http://www.fao.org/ag/againfo/programmes/en/empres/H7N9/situation_update.html. Accessed 15 March 2020. [Google Scholar]
- 11.Calzas C, Chevalier C. 2019. Innovative mucosal vaccine formulations against influenza A virus infections. Front Immunol 10:1605. doi: 10.3389/fimmu.2019.01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sridhar S, Brokstad KA, Cox RJ. 2015. Influenza vaccination strategies: comparing inactivated and live attenuated influenza vaccines. Vaccines 3:373–389. doi: 10.3390/vaccines3020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanco-Lobo P, Nogales A, Rodriguez L, Martinez-Sobrido L. 2019. Novel approaches for the development of live attenuated influenza vaccines. Viruses 11:190. doi: 10.3390/v11020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abente EJ, Rajao DS, Santos J, Kaplan BS, Nicholson TL, Brockmeier SL, Gauger PC, Perez DR, Vincent AL. 2018. Comparison of adjuvanted-whole inactivated virus and live-attenuated virus vaccines against challenge with contemporary, antigenically distinct H3N2 influenza A viruses. J Virol 92:e01323-18. doi: 10.1128/JVI.01323-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe T, Watanabe S, Neumann G, Kida H, Kawaoka Y. 2002. Immunogenicity and protective efficacy of replication incompetent influenza virus-like particles. J Virol 76:767–773. doi: 10.1128/jvi.76.2.767-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudek T, Knipe DM. 2006. Replication-defective viruses as vaccines and vaccine vectors. Virology 344:230–239. doi: 10.1016/j.virol.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Nogales A, Baker SF, Domm W, Martinez-Sobrido L. 2016. Development and applications of single-cycle infectious influenza A virus (sciIAV). Virus Res 216:26–40. doi: 10.1016/j.virusres.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann G, Brownlee GG, Fodor E, Kawaoka Y. 2004. Orthomyxovirus replication, transcription, and polyadenylation. Curr Top Microbiol Immunol 283:121–143. doi: 10.1007/978-3-662-06099-5_4. [DOI] [PubMed] [Google Scholar]
- 19.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J Gen Virol 91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 20.Noda T, Kawaoka Y. 2012. Packaging of influenza virus genome: robustness of selection. Proc Natl Acad Sci U S A 109:8797–8798. doi: 10.1073/pnas.1206736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto H, Muramoto Y, Noda T, Kawaoka Y. 2013. The genome-packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome-bundling signal. J Virol 87:11316–11322. doi: 10.1128/JVI.01301-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shafiuddin M, Boon ACM. 2019. RNA sequence features are at the core of influenza A virus genome packaging. J Mol Biol 431:4217–4228. doi: 10.1016/j.jmb.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gamblin SJ, Skehel JJ. 2010. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem 285:28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krammer F. 2019. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol 19:383–397. doi: 10.1038/s41577-019-0143-6. [DOI] [PubMed] [Google Scholar]
- 25.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc Natl Acad Sci U S A 100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinya K, Fujii Y, Ito H, Ito T, Kawaoka Y. 2004. Characterization of a neuraminidase-deficient influenza a virus as a potential gene delivery vector and a live vaccine. J Virol 78:3083–3088. doi: 10.1128/jvi.78.6.3083-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masic A, Pyo HM, Babiuk S, Zhou Y. 2013. An eight-segment swine influenza virus harboring H1 and H3 hemagglutinins is attenuated and protective against H1N1 and H3N2 subtypes in pigs. J Virol 87:10114–10125. doi: 10.1128/JVI.01348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikawa T, Shimizu K, Tanaka T, Kuroda K, Takayama T, Yamamoto T, Hanada N, Hamada Y. 2012. Bacterial neuraminidase rescues influenza virus replication from inhibition by a neuraminidase inhibitor. PLoS One 7:e45371. doi: 10.1371/journal.pone.0045371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trombetta CM, Remarque EJ, Mortier D, Montomoli E. 2018. Comparison of hemagglutination inhibition, single radial hemolysis, virus neutralization assays, and ELISA to detect antibody levels against seasonal influenza viruses. Influenza Other Respir Viruses 12:675–686. doi: 10.1111/irv.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Landreth S, Gaba A, Hlasny M, Liu G, Huang Y, Zhou Y. 2019. In vivo characterization of avian influenza A (H5N1) and (H7N9) viruses isolated from Canadian travelers. Viruses 11:193. doi: 10.3390/v11020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood JM, Robertson JS. 2004. From lethal virus to life-saving vaccine: developing inactivated vaccines for pandemic influenza. Nat Rev Microbiol 2:842–847. doi: 10.1038/nrmicro979. [DOI] [PubMed] [Google Scholar]
- 32.Mitnaul LJ, Castrucci MR, Murti KG, Kawaoka Y. 1996. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J Virol 70:873–879. doi: 10.1128/JVI.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fink AL, Engle K, Ursin RL, Tang W-Y, Klein SL. 2018. Biological sex affects vaccine efficacy and protection against influenza in mice. Proc Natl Acad Sci U S A 115:12477–12482. doi: 10.1073/pnas.1805268115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landreth S, Lu Y, Pandey K, Zhou Y. 2020. A replication-defective influenza virus vaccine confers complete protection against H7N9 viral infection in mice. Vaccines 8:207. doi: 10.3390/vaccines8020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Landreth S, Liu G, Brownlie R, Gaba A, Littel-van den Hurk SD, Gerdts V, Zhou Y. 2020. Innate immune modulator containing adjuvant formulated HA based vaccine protects mice from lethal infection of highly pathogenic avian influenza H5N1 virus. Vaccine 38:2387–2395. doi: 10.1016/j.vaccine.2020.01.051. [DOI] [PubMed] [Google Scholar]
- 37.Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. 2007. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J Gen Virol 88:13–18. doi: 10.1099/vir.0.82419-0. [DOI] [PubMed] [Google Scholar]
- 38.Masic A, Babiuk LA, Zhou Y. 2009. Reverse genetics-generated elastase-dependent swine influenza viruses are attenuated in pigs. J Gen Virol 90:375–385. doi: 10.1099/vir.0.005447-0. [DOI] [PubMed] [Google Scholar]
- 39.Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. 2007. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. J Gen Virol 88:942–950. doi: 10.1099/vir.0.82483-0. [DOI] [PubMed] [Google Scholar]
- 40.WHO. 2002. WHO manual on animal influenza diagnosis and surveillance, 2nd ed. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 41.Karber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-Schmiedebergs Arch Exp Pathol Pharmakol 162:480–483. doi: 10.1007/BF01863914. [DOI] [Google Scholar]
- 42.Spearman C. 1908. The method of “right and wrong cases” (“constant stimuli”) without Gauss’s formulae. Br J Psychol 2:227–242. [Google Scholar]