The massive release of circulating mediators of inflammation is responsible for lung injury during influenza A virus infection. This phenomenon is referred to as the “cytokine storm.” However, the mechanism by which influenza induces the cytokine storm is not fully understood.
KEYWORDS: influenza virus, H9N2, CD83, cytokines, neuraminidase, dendritic cells
ABSTRACT
Influenza A viruses cause severe respiratory illnesses in humans and animals. Overreaction of the innate immune response to influenza virus infection results in hypercytokinemia, which is responsible for mortality and morbidity. However, the mechanism by which influenza induces hypercytokinemia is not fully understood. In this study, we established a mouse-adapted H9N2 virus, MA01, to evaluate the innate immune response to influenza in the lung. MA01 infection caused high levels of cytokine release, enhanced pulmonary injury in mice, and upregulated CD83 protein in dendritic cells and macrophages in the lung. Influenza virus neuraminidase (NA) unmasked CD83 protein and contributed to high cytokine levels. Furthermore, we provide evidence that CD83 is a sialylated glycoprotein. Neuraminidase treatment enhanced lipopolysaccharide (LPS)-stimulated NF-κB activation in RAW264.7 cells. Anti-CD83 treatment alleviated influenza virus-induced lung injury in mice. Our study indicates that influenza virus neuraminidase modulates CD83 status and contributes to the “cytokine storm,” which may suggest a new approach to curb this immune injury.
IMPORTANCE The massive release of circulating mediators of inflammation is responsible for lung injury during influenza A virus infection. This phenomenon is referred to as the “cytokine storm.” However, the mechanism by which influenza induces the cytokine storm is not fully understood. In this study, we have shown that neuraminidase unmasked CD83 protein in the lung and contributed to high cytokine levels. Anti-CD83 treatment could diminish immune damage to lung tissue. The NA-CD83 axis may represent a target for an interruption of influenza-induced lung damage.
INTRODUCTION
Influenza viruses are enveloped and embedded with hemagglutinin (HA) and neuraminidase (NA) on the surface (1). The genome of influenza virus is composed of eight segments of negative-sense single-stranded RNA (ssRNA). Influenza viruses cause severe pulmonary infection and represent a major pathogen for both humans and animals. The induction of high levels of inflammatory cytokines, including tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), alpha interferon (IFN-α), IFN-β, IL-1β, and monocyte chemoattractant protein 1 (MCP-1), during influenza virus infection has been demonstrated to be the principal factor contributing to mortality and morbidity (2, 3). This phenomenon of hypercytokinemia, or the so-called “cytokine storm,” was suggested to be responsible for pulmonary damage during influenza virus infection (4). An excessive innate immune response, enhanced inflammasome activation, and overreaction of interferon regulatory factor 5 (IRF5) are related to high inflammatory cytokine levels (5–7). However, the mechanisms by which influenza virus induces hypercytokinemia remain to be determined.
The influenza A virus surface glycoprotein NA plays an important role in viral attachment and entry as well as virion release from infected cells. NA acts as a sialidase enzyme, which enzymatically cleaves sialic acids from cell surface proteins and carbohydrate side chains on nascent virions (8). There is evidence that both influenza virus and bacterial NA could modulate the innate immune response in dendritic cells (DCs) and macrophages (9, 10). Moreover, the removal of sialic acid from glycoconjugates on the surface of monocytes and DCs enhances their response to innate stimulators (11, 12). NA is known to contribute to lung disease by activating IL-1β and transforming growth factor β (TGF-β) (13–15). However, the immunostimulatory role of NA still needs to be further determined.
CD83 is a type I transmembrane glycoprotein that is highly and stably expressed by mature DCs; it exists in two forms: membrane-bound CD83 is immunostimulatory, while soluble CD83 (sCD83) is immunosuppressive (16). Administration of sCD83 has been suggested to inhibit monocyte differentiation into DCs, alter the DC cytoskeleton, prevent DC maturation, and reduce DC-mediated T-cell proliferation (17–21). sCD83 can also inhibit autoimmune diseases and allergic responses in mice (22, 23). Furthermore, varicella-zoster virus and cytomegalovirus can modulate CD83 expression on DCs (24, 25). Inhibition of CD83 alleviates systemic inflammation induced by herpes simplex virus infection in mice (26). However, whether influenza A virus contributes to hypercytokinemia via CD83 modulation has not been determined yet.
The cytokine storm is the main cause of the mortality of influenza virus infection. However, mice deficient in the hallmark inflammatory cytokines TNF-α, IL-6, and CC chemokine ligand 2 are not protected from lethal H5N1 influenza virus infection (27). The combination of an NA inhibitor with the immunomodulators celecoxib and mesalazine reduces mortality in mice infected by influenza H5N1 virus (28). Moreover, sphingosine analogs protect against the pathogenic influenza virus through hypercytokinemia inhibition in mice (29). However, new approaches that could curb hypercytokinemia during influenza A virus infection still need to be identified.
In this study, we provide evidence that influenza A virus infection upregulates CD83 on the cell surface of DCs and macrophages in the lungs of mice. NA is involved in this modulation of CD83, which contributes to hypercytokinemia. Anti-CD83 treatment alleviates influenza virus-induced lung injury in mice. Our research suggests a new approach for preventing and treating influenza A virus-induced lung injury.
RESULTS
Mouse-adapted influenza virus exhibits enhanced virulence in mice.
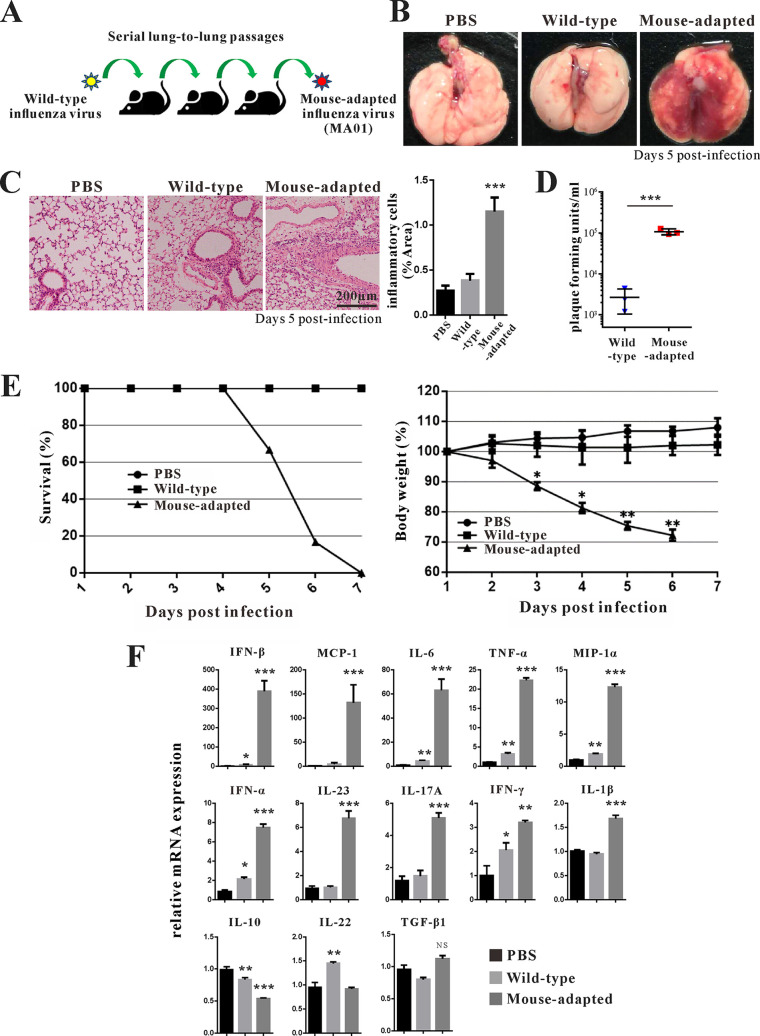
Previously, we and other groups have shown that avian influenza virus H9N2 can establish infection in mice after adaptation (30–32); this could serve as a model to evaluate the host immune response to influenza virus infection. To obtain mouse-adapted influenza virus H9N2, we performed a series of lung passages of wild-type (wt) influenza virus H9N2 in mice (Fig. 1A). After 10 passages, we obtained mouse-adapted influenza virus H9N2, referred to as MA01. Mice suffered much more severe lung pathology after infection with MA01 (Fig. 1B and C). Furthermore, compared with wild-type influenza virus H9N2, the lung virus titers of MA01 were significantly enhanced (Fig. 1D). After infection with MA01, the mortality rate was higher, and progressive weight loss was observed in mice (Fig. 1E). In contrast, infection with wt avian influenza virus H9N2 did not cause obvious lung pathology in mice, was not lethal, and resulted in only slight weight loss. To determine the molecular basis for the enhanced virulence of the H9N2 virus during mouse adaptation, the complete genomes of MA01 were sequenced. Compared with wild-type H9N2, we observed the PB2 mutation 627(E→K), which is characterized as a virulence determinant (Table 1). Next, we evaluated cytokine levels in mouse lung after virus infection by quantitative PCR (Q-PCR). Both wt and MA01 infection upregulated IFN-α, IFN-β, IFN-γ, MCP-1, macrophage inflammatory protein 1α (MIP-1α), TNF-α, IL-6, and IL-17A levels in the lung; however, these cytokine levels were much higher in mice infected with MA01 (Fig. 1F). MA01 infection upregulated IL-23 and IL-1β; in contrast, wt influenza virus infection upregulated IL-22 (Fig. 1F). TGF-β1 was not changed, whereas the regulatory cytokine IL-10 was decreased after infection (Fig. 1F). Collectively, these data indicate that mouse adaptation to low avian influenza virus H9N2 virulence could change its pathogenicity, with a higher mortality rate and a stronger lung inflammatory response.
FIG 1.
Mouse-adapted influenza virus with enhanced virulence. (A) Scheme of mouse-adapted avian influenza H9N2 virus generation. After 10 serial lung-to-lung passages, avian influenza H9N2 virus acquired profoundly affected virulence in wild-type (wt) C57BL/6 mice. Mice were infected with the avianinfluenza H9N2 virus as described in Materials and Methods. (B and C) Appearance of mouse lung (B) and H&E staining analysis of mouse lung tissue (C) 5 days after infection with the control (PBS) and wt and mouse-adapted H9N2 viruses (n = 3 mice per group). The percentages of inflammatory cells per area were measured with the ImageJ program. ***, P < 0.001 (compared with PBS). (D) The lung virus titers of each group were analyzed by PFU. ***, P < 0.001 (compared with PBS). (E) Survival (left) and body weight (right) of mice were recorded daily for 10 days after infection (n = 7 mice per group). *, P < 0.05; **, P < 0.01 (compared with the wt). (F) Q-PCR analysis of cytokine IFN-α, IFN-β, IFN-γ, MCP-1, MIP-1α, TNF-α, IL-6, IL-1β, IL-17A, IL-22, IL-23, IL-10, and TGF-β expression levels in the lungs of wt and mouse-adapted H9N2-infected mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (compared with PBS). Data are presented as means ± SD; P values were calculated using two-tailed Student’s t test. The experiments depicted in this figure have been reproduced three times.
TABLE 1.
Nucleotide and amino acid substitutions during mouse adaptation
| Protein | Nucleotide alteration | Amino acid substitution |
|---|---|---|
| PB2 | 1059(A→G) | |
| 1236(G→A) | ||
| 1329(G→A) | ||
| 1332(T→G) | ||
| 1335(G→A) | ||
| 1879(G→A) | 627(E→K) | |
| 2173(T→C) | 725(S→L) | |
| 2174(C→T) | 725(S→L) | |
| 2175(G→C) | 725(S→L) | |
| 2193(G→A) | ||
| PB1 | 2007(A→G) | |
| PA | 1040(A→G) | 347(D→G) |
| 1062(T→C) | ||
| 1065(C→A) | ||
| 1122(T→G) | 374(I→M) | |
| 1126(T→C) | 376(S→P) | |
| 2119(G→T) | 707(V→F) | |
| HA | 27(A→G) | 9(I→M) |
| 700(T→A) | 234(L→M) | |
| 761(G→T) | 254(R→I) | |
| 808(G→C) | ||
| 1641(T→G) | 547(C→W) | |
| 1648(A→T) | 550(T→S) | |
| 1652(G→A) | 551(S→N) | |
| NP | 974(G→A) | 325(R→K) |
| 1486(G→T) | 496(D→Y) | |
| NA | 1128(C→G) | |
| 1347(T→A) | ||
| 1352(T→G) | 451(V→G) | |
| NS | 63(G→A) | |
| M | ||
Influenza virus infection upregulates CD83 protein levels in the lung.
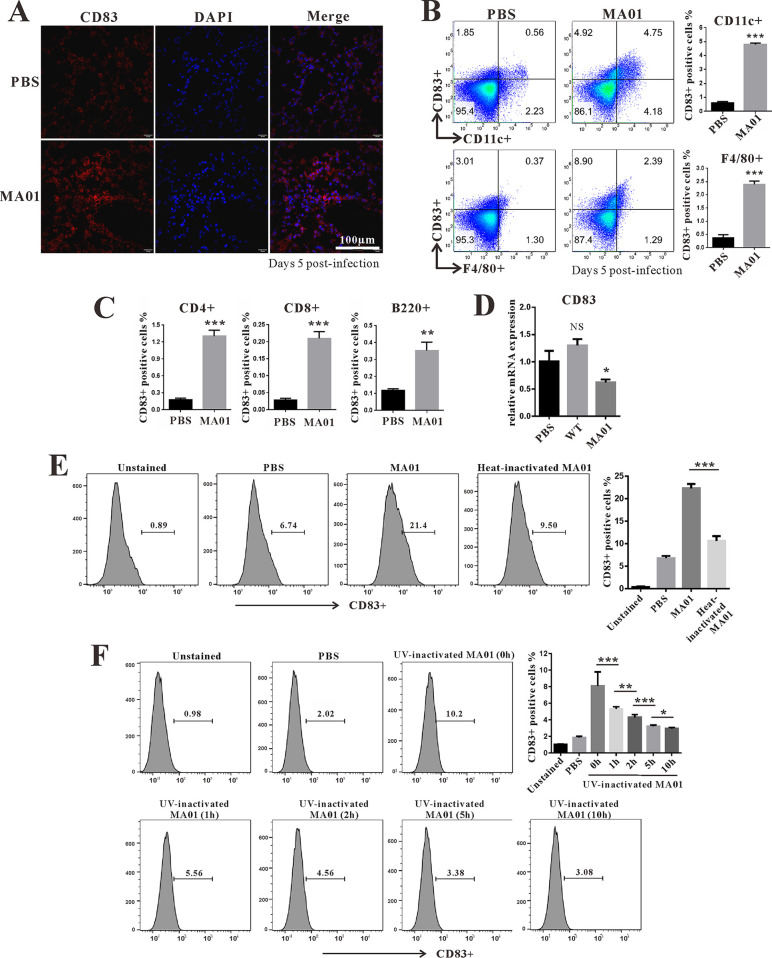
To determine whether MA01 infection induces innate immune cell activation in the lung, we analyzed immune cell surface markers by flow cytometry. First, we tested whether MA01 infection could promote innate immune cell infiltration. MA01 infection significantly increased DC (CD11c+) and macrophage(F4/80+) infiltration in the lung (see Fig. S1A in the supplemental material). Next, we analyzed immune cell surface markers during MA01 infection. CD40 and CD86 were both upregulated on DCs and macrophages in the lung during MA01 infection (Fig. S1B and C). Furthermore, we found that CD83 levels were upregulated (Fig. 2A) not only on DCs and macrophages but also on CD4+, CD8+ T, and B220+ B cells (Fig. 2B and C). However, CD83 levels were more prominent on DCs, consistent with previous reports that CD83 is mainly expressed on DCs (33). Moreover, we demonstrated that MA01 infection-induced CD83 upregulation was not due to mRNA levels, as CD83 mRNA was decreased in the lung during MA01 infection (Fig. 2D). Collectively, these data indicate that influenza virus infection increases CD83 levels in the lung.
FIG 2.
Influenza virus modulates CD83 in the lung. (A) Wild-type C57BL/6 mice were infected with PBS and mouse-adapted H9N2 (MA01) for 5 days (n = 5 mice per group). Immunofluorescence analysis of CD83 (red) and nuclear (DAPI) (blue) staining in mouse lung tissue of each group was performed. (B and C) The percentages of CD83-positive cells in lung dendritic cells (CD11c+) and macrophages (F4/80+) (B) as well as T cells (CD4+ and CD8+) and B cells (B220+) (C) were analyzed by flow cytometry. (D) Q-PCR analysis of CD83 mRNA levels in the lungs of PBS- and MA01-infected mice. (E) RAW264.7 cells were stimulated with MA01 (50 μl) or heat-inactivated MA01 (50 μl) for 3 h. (F) RAW264.7 cells were stimulated with MA01 (50 μl) or MA01 treated with UV for 0 h, 1 h, 2 h, 5 h, and 10 h (50 μl). The percentages of CD83-positive cells were analyzed by flow cytometry. *, P < 0.05; **, P < 0.01; ***, P < 0.001. These data are representative of results from three independent experiments and presented as means ± SD; P values were calculated using two-tailed Student’s t test. The experiments depicted in this figure have been reproduced two to three times.
NA unmasks CD83 on the cell surface and enhances cytokine production.
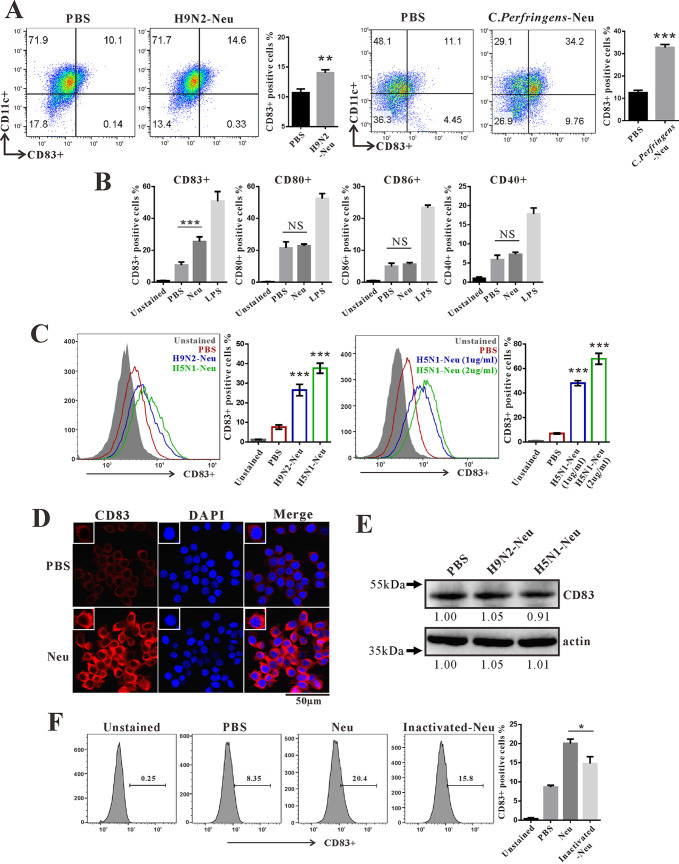
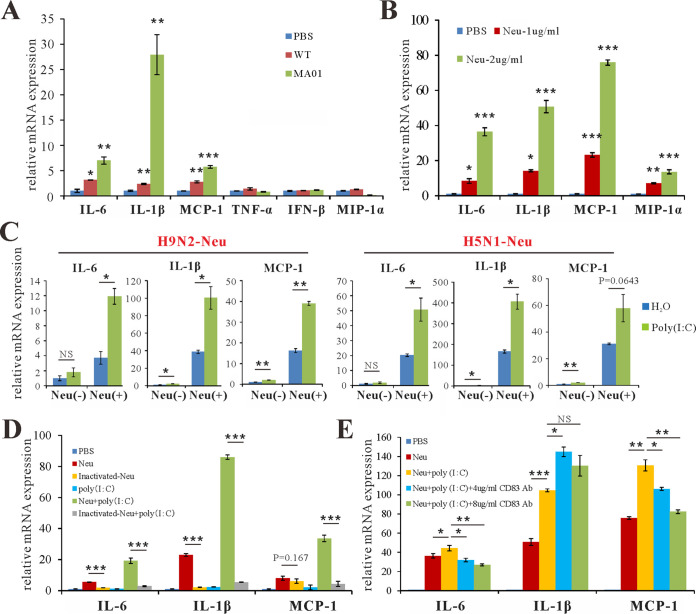
Next, we investigated the mechanism by which influenza virus infection increases CD83 levels on DCs and macrophages. First, we tested whether influenza virus could upregulate CD83 in macrophages in vitro. Infection of RAW264.7 cells with MA01 increased CD83 levels (Fig. 2E). However, both heat-inactivated and UV-inactivated MA01 lost the ability to upregulate CD83. These data implied that a heat-sensitive component other than viral RNA from influenza virus exerts its effects on CD83 (Fig. 2E and F). CD83 is a type I transmembrane glycoprotein and has been suggested to be a sialic acid-binding Ig-like lectin adhesion receptor (34). As CD83 mRNA was decreased and the CD83 protein signal was detected more strongly by flow cytometry in the lung during MA01 infection, we postulated that NA from influenza virus may be involved in this process. To test this, purified NA protein from influenza virus H9N2 was added to cultured bone marrow dendric cells (BMDCs), which increased the CD83 signal on BMDCs (Fig. 3A) and RAW264.7 cells (Fig. 3D). Bacterial NA from Clostridium perfringens also possessed the ability to increase the CD83 signal on BMDCs (Fig. 3A). C. perfringens NA treatment also upregulated the CD83 signal on CD4+, CD8+, B220+, and CD11c+ cells from mouse spleen (Fig. S2). However, CD40, CD80, and CD86 were not upregulated on RAW264.7 cells after influenza virus NA treatment (Fig. 3B). Furthermore, NA from influenza virus H5N1 also increased the CD83 signal on macrophages, and this effect was enhanced with higher concentrations of NA (Fig. 3C). However, the influenza virus NA-induced CD83 signal increase was not due to changed total CD83 protein levels in cells, as Western blot quantifications of CD83 levels were similar between groups (Fig. 3E). Heat combined with repeated freezing and thawing inactivated influenza virus NA and diminished its ability to improve the CD83 signal on macrophage cells (Fig. 3F). These data indicate that NA specifically targets CD83 and modifies its status on the cell surface. Next, we tested the cytokines and chemokines affected by NA. Infection of RAW264.7 cells by both wt and MA01 influenza virus stimulates the production of IL-6, IL-1β, and MCP-1; however, MA01 exhibited a stronger effect on these cytokines (Fig. 4A). Incubation of BMDCs with H9N2 NA upregulated the expression of IL-6, IL-1β, MIP-1α, and MCP-1 (Fig. 4B), consistent with a previous report that NA is an inducer of inflammation (12). Next, we tested whether NA can enhance cytokine production after stimulation with poly(I·C), considering that the influenza virus genome is composed of RNA. Both H5N1 and H9N2 NA treatments enhanced the poly(I·C)-induced production of the cytokines IL-6, IL-1β, and MCP-1 on RAW264.7 cells. (Fig. 4C). Freeze-thaw-inactivated NA lost the ability to stimulate these cytokines (Fig. 4D). As we have shown that NA can upregulate the CD83 signal on the cell surface, we then postulated whether inhibiting CD83 could inhibit the cytokine response of RAW264.7 cells in the presence of NA. The anti-CD83 antibody was added during RAW264.7 cell stimulation with poly(I·C) and NA. CD83 inhibition decreased IL-6 and MCP-1 levels in cells in vitro (Fig. 4E). Collectively, these data indicate that NA targets CD83 on the cell surface and enhances cytokine production in vitro.
FIG 3.
Neuraminidase unmasks CD83 on the cell surface. (A) BMDCs (5 to 7 days) isolated from wild-type C57BL/6 mice were stimulated with 1 μg/ml H9N2-Neu (NA protein from H9N2) and 1 μg/ml C. perfringens-Neu (NA from Clostridium perfringens) for 3 h. The percentage of CD83-positive cells wasanalyzed by flow cytometry. (B) RAW264.7 cells were incubated with H9N2-Neu (1 μg/ml) and LPS (100 ng/ml) for 3 h. The percentages of CD83-, CD80-, CD40-, and CD86-positive cells were analyzed by flow cytometry. (C) RAW264.7 cells were stimulated with 1 μg/ml H9N2-Neu, 1 μg/ml H5N1-Neu, and 2 μg/ml H5N1-Neu (NA protein from H5N1) for 3 h. The percentage of CD83-positive cells was analyzed by flow cytometry. **, P < 0.01; ***, P < 0.001; NS, not significant (compared with PBS). (D) Immunofluorescence analysis of CD83 (red) and nuclear (DAPI) (blue) staining in RAW264.7 cells stimulated with H5N1-Neu (2 μg/ml) for 3 h. (E) Western blot analysis of CD83 protein levels in RAW264.7 cells that were treated with H9N2-Neu (1 μg/ml) and H5N1-Neu (1 μg/ml) for 3 h. (F) RAW264.7 cells were incubated with H9N2-Neu (1 μg/ml) and inactivated H9N2-Neu (1 μg/ml) for 3 h. The percentage of CD83-positive cells was analyzed by flow cytometry. *, P < 0.05 (compared with H9N2-Neu). Data are representative of results from three independent experiments and presented as means ± SD; P values were calculated using two-tailed Student’s t test. Data presented in this figure have been reproduced at least three times.
FIG 4.
Neuraminidase enhances cytokine production. (A) Q-PCR analysis of cytokine MCP-1, MIP-1α, TNF-α, IL-6, IL-1β, and IFN-β expression levels in RAW264.7 cells treated with PBS (50 μl), wild-type H9N2 (WT) (50 μl), and mouse-adapted H9N2 (MA01) (50 μl) for 12 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with PBS). (B) Q-PCR analysis of cytokine MCP-1, MIP-1α, IL-6, and IL-1β expression levels in RAW264.7 cells treated with 1 μg/ml and 2 μg/ml H9N2-Neu for 12 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with PBS). (C) Q-PCR analysis of cytokine MCP-1, IL-6, and IL-1β expression levels in RAW264.7 cells treated with 2.5 μg/ml H5N1-Neu, 2.5 μg/ml H9N2-Neu, and 2.5 μg/ml Neu plus 20 μg/ml poly(I·C) for 12 h. *, P < 0.05; **, P < 0.01; NS, not significant (compared with H2O). (D) Q-PCR analysis of cytokine MCP-1, IL-6, and IL-1β expression levels in RAW264.7 cells treated with poly(I·C), H9N2-Neu, inactivated H9N2-Neu, H9N2-Neu plus poly(I·C), and inactivated H9N2-Neu plus poly(I·C) for 12 h. The concentration for each stimulation is described above. ***, P < 0.001 (compared with H9N2-Neu). (E) Q-PCR analysis of cytokine MCP-1, IL-6, and IL-1β expression levels in RAW264.7 cells treated with H9N2-Neu, poly(I·C), H9N2-Neu plus poly(I·C), H9N2-Neu plus poly(I·C), and anti-CD83 antibody (Ab) (4 or 8 μg/ml) for 12 h. The concentration for each stimulation is described above. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant [compared with Neu plus poly(I·C)]. Data are presented as means ± SD; P values were calculated using two-tailed Student’s t test. The experiments depicted in this figure have been reproduced three times.
CD83 is a sialylated glycoprotein.
Next, we examined whether CD83 is a sialylated glycoprotein. We used biotinylated Sambucus nigra lectin (SNA) and Maackia amurensis lectin I (MAL) to probe sialic acid modification on the CD83 protein. SNA binds preferentially to sialic acid attached to terminal galactose in α-2,6 and, to a lesser degree, α-2,3. However, MAL appears to bind sialic acid via an α-2,3 linkage (35). Both SNA and MAL bind to purified mouse CD83 protein (Fig. 5A). However, the binding activity of SNA and MAL was severely reduced in the presence of MA01 (Fig. 5B). In contrast, heat-inactivated MA01 lost the ability to quench the binding activity (Fig. 5B). Next, we tested the influence of NA and MA01 on the SNA-binding activity of RAW264.7 and spleen cells. SNA binding was decreased and the CD83 signal was increased in the presence of NA and MA01 on RAW264.7, spleen, and spleen CD11c+ DCs (Fig. 5C and D). This phenomenon was also observed in immune and F4/80+ cells from the lung after MA01 infection (Fig. 5E). These data hint at the possibility that CD83 is sialylated and sequestered on the surface of cells.
FIG 5.
CD83 is a sialylated glycoprotein. (A) SNA (20 μg/ml) and MAL (20 μg/ml) were immobilized on ELISA plates. After blocking the plate with binding buffer, CD83-Fc protein (10 μg/ml) was added to the plate in triplicates, and the plate was incubated for 3 h. Binding was detected as described in Materials and Methods. ***, P < 0.001 (compared with no CD83). (B) After SNA and MAL were coincubated with CD83 protein, MA01 (50 μl) or heat-inactivated MA01 (50 μl) was added to the plate, and the plate was incubated for an additional 3 h. ***, P < 0.001 (compared with MA01). (C) RAW264.7 cells were stimulated with H9N2-Neu (1 μg/ml) and MA01 (50 μl) for 3 h. The percentages of CD83-positive cells and SNA-binding cells were analyzed by flow cytometry. ***, P < 0.001 (compared with PBS). (D) Wild-type C57BL/6 mice were infected with PBS and MA01 for 5 days (n = 5 mice per group). Flow cytometry analyses of the percentages of CD83-positive cells and SNA-binding cells in infected mouse spleen cells and spleen CD11c+ dendritic cells were performed. *, P < 0.05; ***, P < 0.001 (compared with PBS). (E) Flow cytometry analysis of the percentages of CD83-positive cells and SNA-binding cells in infected mouse lung cells and lung F4/80+ macrophages. *, P < 0.05; ***, P < 0.001 (compared with PBS). These data are presented as means ± SD; P values were calculated using two-tailed Student’s t test. Data presented in this figure have been reproduced at least three times.
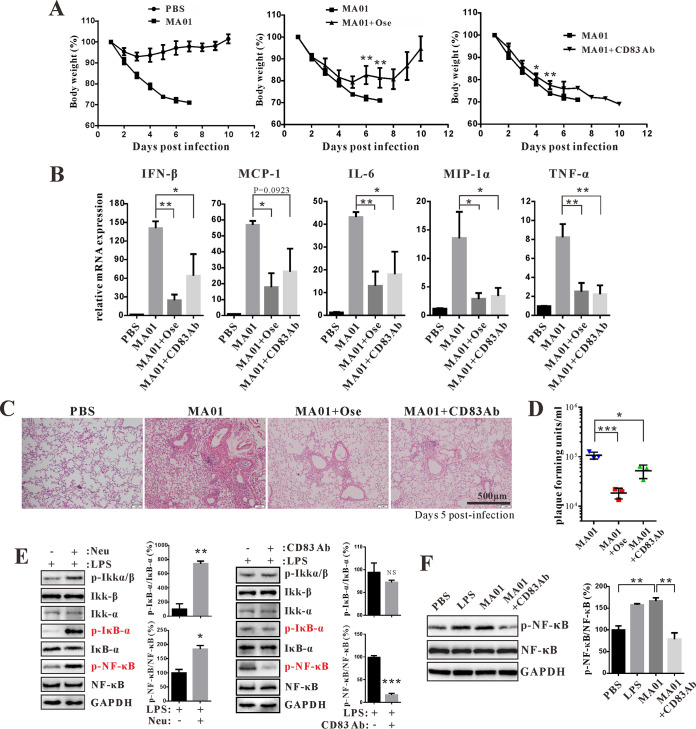
Anti-CD83 treatment alleviates influenza virus-induced lung injury in mice.
Next, we tested whether inhibiting CD83 could reduce influenza virus-induced lung injury. Injection of anti-CD83 antibody and the NA inhibitor oseltamivir decreased weight loss in mice during MA01 infection (Fig. 6A). Furthermore, we found that after MA01 infection, CD83 levels were downregulated by oseltamivir treatment on DCs (CD11c+) and macrophages (F4/80+) in the lung (Fig. S3). Next, cytokine levels of IFN-β, MCP-1, IL-6, MIP-1α, and TNF-α were decreased by anti-CD83 antibody and oseltamivir in mice (Fig. 6B). Consistent with the cytokine levels, influenza-induced lung injury was alleviated by anti-CD83 antibody and oseltamivir (Fig. 6C). Treatment with anti-CD83 antibody and oseltamivir also reduced lung viral titers in MA01-infected mice (Fig. 6D). Next, we tested whether NA could alter NF-κB activation. NA enhanced lipopolysaccharide (LPS)-stimulated NF-κB activation on RAW264.7 cells, as the phosphorylation of IκB-α and NF-κB P56(S536) was increased (Fig. 6E). However, the anti-CD83 antibody inhibited the phosphorylation of NF-κB stimulated by LPS and MA01 (Fig. 6E and F). These data indicate that CD83 inhibition can alleviate influenza virus-induced lung injury in mice.
FIG 6.
CD83 inhibition alleviates influenza virus-induced lung injury in mice. (A) Mice were injected with PBS and MA01 for 5 days (n = 5 mice per group). Infected mice were intraperitoneally injected with PBS, oseltamivir phosphate (Ose) (3 mg/mouse), and anti-CD83 antibody (CD83 Ab) (40 μg/mouse) twice daily for 5 days, starting at 12 h postinfection. Body weight was monitored daily for 10 days. *, P < 0.05; **, P < 0.01 (compared with MA01). (B) Q-PCR analysis of cytokine IFN-β, MCP-1, MIP-1α, TNF-α, and IL-6 expression levels in the lungs of each group as described above. *, P < 0.05; **, P < 0.01 (compared with MA01). (C) Histopathological analysis of mouse lung tissue in each group as described above. (D) The lung virus titers of each group were analyzed by PFU. *, P < 0.05; ***, P < 0.001 (compared with PBS). (E) RAW264.7 cells were incubated with LPS (100 ng/ml), LPS plus H9N2-Neu (1 μg/ml), and LPS plus anti-CD83 antibody (5 μg/ml) for 30 min, and the activation of the NF-κB pathway was determined by Western blotting. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with LPS). p-Ikkα/β, phosphorylated IκB kinase α/β. (F) Western blot analysis of NF-κB phosphorylation in RAW264.7 cells stimulated with LPS (100 ng/ml), MA01 (50 μl), and MA01 plus anti-CD83 antibody (5 μg/ml) for 30 min. **, P < 0.01 (compared with MA01). Data are presented as means ± SD; P values were calculated using two-tailed Student’s t test. The experiments depicted in this figure have been reproduced two to three times.
DISCUSSION
Influenza A virus infection causes the hyperrelease of inflammatory mediators, which leads to lung injury. However, the mechanisms by which influenza induces hypercytokinemia are not fully understood. In this study, we obtained a highly pathogenic H9N2 virus (MA01) through serial lung-to-lung passages of wt influenza virus H9N2 in mice. MA01 infection upregulated cytokine, chemokine, and CD83 levels in the lung. Furthermore, we demonstrated that CD83 is a sialylated glycoprotein and that NA can unmask CD83 on the cell surface and enhance cytokine production. Anti-CD83 treatment decreased cytokine levels and alleviated influenza virus-induced lung injury in mice. This research may suggest a new approach to reduce influenza virus-induced lung injury.
The influenza A virus subtypes H5N1 and H1N1 cause severe infection in humans and animals and are recognized as highly pathogenic influenza viruses (36). These subtypes easily infect mice with high pathogenicity and could serve as a good model to evaluate influenza-induced hypercytokinemia and lung injury (37, 38). However, experimental studies of these subtypes have been restricted to laboratories with high biosafety levels, which limits potential research. In contrast, the H9N2 subtype normally causes only slight pathogenicity in humans and animals and is thus recognized as a low-pathogenicity influenza virus (39, 40). However, after adaptation, H9N2 can gain the ability to cause severe infection in mice (30–32). In this study, we generated the MA01 virus by serial lung-to-lung passages of wt influenza virus H9N2 in mice. MA01 infection caused higher mortality rates and stronger lung inflammatory responses in mice. As such, the MA01 virus could serve as a model virus to investigate influenza-induced hypercytokinemia and lung injury in mice under broader laboratory conditions.
Hypercytokinemia, the so-called “cytokine storm,” is the key factor of pulmonary damage during influenza virus infection. In this study, we have demonstrated that both wt and MA01 infections upregulate IFN-α, IFN-β, IFN-γ, MCP-1, MIP-1α, TNF-α, IL-6, and IL-17A levels in the lung; however, this cytokine storm was much more prominent in a mouse infected with MA01. The stronger MA01-induced cytokine storm correlated with reduced survival rates and excess lung pathologies in mice. IL-10 is a pleiotropic cytokine with important immunoregulatory functions and is produced by a variety of cell types, including macrophages, monocytes, T cells, and B cells (41). IL-10 inhibits the expression of proinflammatory cytokines such as IL-1 and TNF-α. During influenza virus infection, IL-10 produced in the lung regulates pulmonary inflammation and acts as an anti-inflammatory cytokine (42). In this study, we have shown that both wt H9N2 and MA01 infections reduce IL-10 levels in the lung. Moreover, infection with wt H9N2 but not MA01 induces IL-22. IL-22 belongs to the IL-10 family that is produced by T helper 17 (Th17), γδ T, NKT, and innate lymphoid cells (43). IL-22 is essential for epithelial regeneration, repair, and inflammatory protection during influenza virus infection (44, 45). In this study, we have demonstrated that the influenza virus NA protein unmasks CD83 on the cell surface and contributes to an enhanced cytokine response. However, whether NA and CD83 can modulate IL-10 and IL-22 production warrants further investigation.
Influenza virus-induced hypercytokinemia may involve multiple mechanisms. The genome of influenza virus is composed of ssRNA and acts via pathogen-associated molecular patterns. The replication of the viral genome generates Z-RNA inside cells that is detected by Z-DNA/RNA-binding protein 1 in the nucleus, which triggers receptor-interacting serine/threonine protein kinase 3-mediated pathways of apoptosis and mixed-lineage kinase domain-like pseudokinase-dependent necroptosis in infected cells (46). Both of these pathways could activate innate host immune responses, contributing to hypercytokinemia. The NA protein exerts several actions on influenza virus-induced pathogenicity. NA inhibitors (i.e., oseltamivir and zanamivir) represent the most effective therapy for patients (47, 48). Influenza virus infection induces broadly cross-reactive and protective NA-specific antibodies in humans (49). These antibodies robustly inhibit the enzymatic activity of NA, protecting mice from lethal influenza virus infection. In this study, we have demonstrated that the sialidase activity of NA unmasks CD83 on the cell surface and enhances cytokine production. NA treatment could enhance LPS-stimulated NF-κB activation.
CD83 is expressed by a great variety of cell types but mainly by activated immune cells such as dendritic, B, and T cells. The expression of CD83 is important for DC phenotype and function (50). The homotypic interaction of CD83 regulates DC activation via mitogen-activated protein kinase (MAPK) by inhibiting p38α phosphorylation (21). Anti-CD83 antibody would presumably block this homotypic interaction. NA treatment may induce this homotypic interaction of CD83. A plethora of distinct viruses could regulate CD83 expression levels and thus change the DC function. Herpesviruses are capable of establishing latency and have acquired immune evasion mechanisms through targeting CD83 (51). Varicella-zoster virus can selectively inhibit CD83 expression on DCs upon infection (24). In contrast, Epstein-Barr virus was found to promote the expression of CD83 on B cells (52). In this study, we have found that the NA protein from influenza virus could unmask CD83 on DCs and macrophages, promoting the cytokine response.
Collectively, our research reveals a new mechanism of the influenza-induced cytokine storm. NA acts as a stimulator of the innate immune response induced by influenza virus infection. NA induced an immune overreaction by unmasking CD83 protein on the cell surface of DCs and macrophages. Therefore, inhibiting CD83 could alleviate influenza virus-induced hypercytokinemia and pulmonary injury in mice.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed according to the recommendations outlined in the Guide for the Care and Use of Laboratory Animals (53). All research animal protocols were approved by the Institutional Animal Care and Use Committee at the Model Animal Research Center, Nanjing University (animal protocol number AP#ZZD01).
Reagents.
The following reagents were purchased: oseltamivir phosphate (Aladdin); mouse CD83/HB15 protein (Fc tag), Clostridium perfringens neuraminidase (NA) (His tag), influenza virus A H9N2 (A/Hong Kong/1073/99) neuraminidase (His tag), and influenza virus A H5N1 [A/Thailand/1(KAN-1)/2004] neuraminidase (His tag) (Sino Biological); poly(I·C) and LPS (InvivoGen); and Maackia amurensis lectin I (MAL) and Sambucus nigra (elderberry bark) lectin (SNA) (Vector Laboratories).
Virus and mice.
H9N2 influenza virus A/chicken/China/AV1534/2011(H9N2) (54) with pandemic potential was purchased from the China Veterinary Culture Collection Center (CVCC). Virus stocks (wt) were propagated in the chorioallantoic cavity of 10-day-old pathogen-free embryonated chicken eggs for 48 h at 37°C. The lyophilized powder of the H9N2 influenza virus from the CVCC was dissolved in 1 ml sterile phosphate-buffered saline (PBS) and stored at −80°C. Mouse-adapted H9N2 virus (MA01) was propagated in the mouse lung for 48 h to 72 h. The titers of virus fluid were tested using PFU. C57BL/6 mice were purchased from the Nanjing Biomedical Research Institute of Nanjing University.
Mouse-adapted H9N2 virus (MA01).
Female 6-week-old C57BL/6 mice were intranasally inoculated with 50 μl wt H9N2 virus fluid (∼2.67 × 103 PFU/ml) under light anesthesia with an intraperitoneal injection of pentobarbital sodium. After 2 days, the mouse lungs were harvested and homogenized with 1 ml of PBS containing 100 U/ml penicillin and 100 μg/ml streptomycin; 50 μl of the centrifuged supernatant was used as the inoculum for the next passage. After 10 passages, a mouse-adapted variant of avian influenza virus H9N2 acquired mutations with profoundly affected virulence.
RNA isolation, RT-PCR amplification, and sequencing.
Viral RNA was extracted from the lungs of virus-infected mice using the TIANamp virus RNA kit (catalog number U8106; Tiangen) according to the manufacturer’s protocol. Reverse transcription (RT) was performed with the PrimeScript RT reagent kit with gDNA Eraser (catalog number RR047A; TaKaRa), using the Uni12 primer (AGC AAA AGC AGG). After reverse transcription, PCR amplification of cDNA was directed by Platinum Hot Start PCR 2× master mix (catalog number 13000-012; Invitrogen). After agarose gel electrophoresis, the PCR products were excised and purified with a FastPure gel DNA extraction minikit (catalog number DC301-01; Vazyme). After purification, The PCR products were subjected to sequence analysis by Sangon Biotechnology Co., Ltd. (Shanghai, China).
Virus titer analysis.
MDCK cells were seeded at 5 × 105 cells per well in 6-well plates. After 12 h of culture, the cells were washed twice with 1 ml of PBS, and 500 μl of the virus dilution was added to each well (before infection, 10-fold serial dilutions of the virus were prepared for each group). The virus was adsorbed to the cells for 1 h at 37°C. After adsorption, the supernatant was removed, and each well was washed with 2 ml of PBS. Two milliliters of agarose plaque assay medium was added, and the plates were incubated at 37°C for 72 h. After incubation, stationary liquid (acetic acid-methanol-water, 1:4:5) was added at 2 ml per well, and the plates were incubated at 37°C for 1 h. The agarose overlay was removed, crystal violet (1%, wt/vol) was added at 1 ml per well, and the plates were incubated for 10 min. The plates were washed by submersion in a water bath until all excess crystal violet was gone. Plaques were counted, and the PFU per milliliter were calculated. MDCK cells were kindly provided by the Stem Cell Bank, Chinese Academy of Sciences.
Experimental infection of mice.
To test the different pathogenicities of wt H9N2 virus and MA01 in vivo, mice were inoculated with 50 μl (∼2.67 × 103 PFU/ml) of virus fluid and the same amount of PBS (the mock control). Body weight and survival were recorded for 10 days after inoculation. Tissue specimens of the lung were collected from mice for subsequent analysis on day 5 postinoculation. The possibility that CD83 inhibition could reduce influenza virus-induced lung injury was tested. The virus fluid of MA01 was inoculated as described above. Inoculated mice were treated with oseltamivir phosphate (3 mg/mouse), soluble CD83 (40 μg/mouse), or anti-CD83 antibody (40 μg/mouse) twice daily for 5 days by intraperitoneal injection. Oseltamivir was used as a positive control.
BMDC culture.
DCs were generated from bone marrow (BM) cells obtained from 5- to 7-week-old wt C57BL/6 mice. Briefly, BM cells were flushed out from femurs and tibias. BM cells were cultured in complete culture medium (RPMI 1640 supplemented with 10% fetal bovine serum [FBS], 25 mM HEPES, 5 mM β-mercaptoethanol [β-ME], and antibiotics; HyClone) containing 10 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) and 10 ng/ml IL-4 (Peprotech).
FACS analysis and antibodies.
Cultured RAW264.7 and BMDCs were washed and suspended in fluorescence-activated cell sorter (FACS) buffer (PBS, 2% FBS), incubated with fluorochrome-coupled agents for 30 min at 4°C, and then washed in FACS buffer. Data were obtained with the Acea NovoCyte series flow cytometer and analyzed using FlowJo software v9.6 (FlowJo, Ashland, OR, USA). The following antimurine antibodies were purchased from eBioscience and used for flow cytometric analysis: CD83 (clone Michel-19), CD11c (N418), F4/80 (BM8), CD4 (RM4-5), CD8 (53-6.7), B220 (RA3-6B2), CD40 (HM40-3), CD80 (16-10A1), CD86 (GL1), CD3 (17A2), and major histocompatibility complex class II (MHCII) (M5/114.15.2).
Enzyme-linked immunosorbent assay.
The lectin of sialic acids SNA (20 μg/ml) and MAL (20 μg/ml) was immobilized on enzyme-linked immunosorbent assay (ELISA) plates at 4°C overnight. After blocking the plate with binding buffer, CD83-Fc protein (10 μg/ml) was added to the plate well in triplicate and incubated with SNA and MAL for 3 h at 37°C. Subsequently, MA01 (50 μl) and heat-inactivated MA01 (50 μl) were added to the plate, and the plate was incubated for an additional 3 h. The plate was washed twice with PBS and then incubated with the primary antibody CD83 (catalog number PA5-47138; Invitrogen) (1:100) at 37°C for 1 h. After washing, horseradish peroxidase (HRP)-labeled secondary antibodies (Southern Biotech) were added. TMB (3,3′,5,5′-tetramethyl benzidine) reagent (BioLegend) was used for color development, and the optical density was read at 450 nm.
Histological analysis.
The lung tissues of mice inoculated with wt H9N2 and MA01 were harvested on day 5 postinoculation, fixed in 10% neutral buffered formalin for 24 h at room temperature, and then embedded in paraffin. The tissue samples were then cut into 5-μm sections and subjected to standard hematoxylin and eosin (H&E) staining. The percentages of inflammatory cells per area were measured with the ImageJ program.
Immunofluorescence staining.
Next, we analyzed CD83 expression in NA-treated RAW264.7 cells and avian influenza virus-infected mouse lung. RAW264.7 cells were cultured in 24-well plates with glass coverslips and stimulated with H5N1 neuraminidase (H5N1-Neu) for 3 h. Subsequently, RAW264.7 cells and lung tissue sections from infected mice were fixed with 4% formaldehyde at room temperature. After blocking with 1% bovine serum albumin (BSA), the samples were incubated with the CD83 primary antibody (catalog number PA5-47138; Invitrogen) (1:100) at 4°C overnight. The following day, the samples were incubated with the secondary antibody Alexa Fluor 555 donkey anti-goat IgG (catalog number A-21432; Life Technologies) (1:200). 4′,6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining. Images were obtained with an Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan) with a constant exposure time. RAW264.7 cells were kindly provided by the Stem Cell Bank, Chinese Academy of Sciences.
Western blotting.
RAW264.7 cells were collected and lysed on ice with radioimmunoprecipitation assay (RIPA) buffer (catalog number 87787; Thermo Fisher Scientific) containing a protease/phosphatase inhibitor cocktail (catalog number 78420; Thermo Fisher Scientific) and 1 mM phenylmethylsulfonyl fluoride (PMSF) (catalog number ST506; Beyotime) for 10 min. Cell lysates were then centrifuged at 12,000 × g (4°C) for 15 min, and the supernatant was collected to determine the protein concentration using a bicinchoninic acid assay kit (catalog number P0012; Beyotime). Samples were then mixed with 5× SDS-PAGE sample buffer (catalog number P0015L; Beyotime) and heated to 95°C for 5 min. Subsequently, samples were loaded onto a 10% gel, separated by electrophoresis, and transferred onto polyvinylidene difluoride membranes (catalog number ISEQ00010; Merck Millipore). After blocking with 5% nonfat dry milk in Tris-buffered saline and Tween 20 for 1 h at room temperature, the membranes were incubated with CD83 primary antibody (catalog number SC-55535; Santa Cruz Biotechnology) (1:1,000), an NF-κB pathway sampler kit (catalog number 9936T; CST) (1:1,000), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog number AF0006; Beyotime) (1:1,000) at 4°C overnight. After washing with Tris-buffered saline and Tween 20 three times, the secondary antibodies were applied to the membranes for 2 h. Membranes were imaged using the ChemiDoc Touch system with the Clarity Western ECL substrate (catalog number 170-5061; Bio-Rad).
Quantitative real-time PCR.
Total RNA was extracted using the RNA simple total RNA kit (catalog number DP419; Tiangen) and reverse transcribed using the QuantScript RT kit (catalog number KR103; Tiangen). RT-PCR was performed on a LightCycler 480 instrument, using SuperReal PreMix Plus (SYBR green) (catalog number FP205; Tiangen). PCR primers of mouse genes were purchased from OriGene. GAPDH was used as an internal control. Measurements for all reactions were performed in triplicate. The results were analyzed using comparative critical threshold (ΔΔCT) methods in the Q-PCR software program.
Statistical analysis.
The statistical analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA). All graphs show means with standard deviations (SD). Statistical significance was determined by two-tailed Student’s t test. A P value of less than 0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant).
Data availability.
Gene sequences (MA01) determined in this study have been submitted to GenBank. The GenBank accession numbers are MT939243, MT939296, MT939317, MT939432, MT939437, MT939439, MT939462, and MT939481.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (number 82000016); the Fundamental Research Funds for the Central Universities (090314380026); PI start-up funding from the Model Animal Research Center, Nanjing University; open funding from the Key Laboratory of Allergy and Immunology, Shenzhen University, and the Shenzhen Science and Technology Peacock Team Project (number KQTD20170331145453160); the Foundation of Luzhou Science and Technology Program and Southwest Medical University (number 00022690); the Natural Science Foundation of Southwest Medical University (numbers 00031586 and 00031294); and the Ph.D. Research Startup Foundation of The Affiliated Hospital of Southwest Medical University (numbers 18053 and 18076).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, Garcia-Sastre A. 2018. Influenza. Nat Rev Dis Primers 4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukuyama S, Kawaoka Y. 2011. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol 23:481–486. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog 4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiore-Gartland A, Panoskaltsis-Mortari A, Agan AA, Mistry AJ, Thomas PG, Matthay MA, PALISI PICFlu Investigators, Hertz T, Randolph AG. 2017. Cytokine profiles of severe influenza virus-related complications in children. Front Immunol 8:1423. doi: 10.3389/fimmu.2017.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbester JL, Clement M, Wellington D, Yeung A, Dimonte S, Marsden M, Chapman L, Coomber EL, Tolley C, Lees E, Hale C, Clare S, Udalova I, Dong T, Dougan G, Humphreys IR. 2020. IRF5 promotes influenza virus-induced inflammatory responses in human induced pluripotent stem cell-derived myeloid cells and murine models. J Virol 94:e00121-20. doi: 10.1128/JVI.00121-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAuley JL, Gilbertson BP, Trifkovic S, Brown LE, McKimm-Breschkin JL. 2019. Influenza virus neuraminidase structure and functions. Front Microbiol 10:39. doi: 10.3389/fmicb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamatos NM, Carubelli I, van de Vlekkert D, Bonten EJ, Papini N, Feng C, Venerando B, d’Azzo A, Cross AS, Wang LX, Gomatos PJ. 2010. LPS-induced cytokine production in human dendritic cells is regulated by sialidase activity. J Leukoc Biol 88:1227–1239. doi: 10.1189/jlb.1209776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirayama Y, Inaba K, Inaba M, Kato T, Kitaura M, Hosokawa T, Ikehara S, Muramatsu S. 1988. Neuraminidase-treated macrophages stimulate allogenic CD8+ T cells in the presence of exogenous interleukin 2. J Exp Med 168:1443–1456. doi: 10.1084/jem.168.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nita-Lazar M, Banerjee A, Feng C, Vasta GR. 2015. Galectins regulate the inflammatory response in airway epithelial cells exposed to microbial neuraminidase by modulating the expression of SOCS1 and RIG1. Mol Immunol 68:194–202. doi: 10.1016/j.molimm.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crespo HJ, Cabral MG, Teixeira AV, Lau JTY, Trindade H, Videira PA. 2009. Effect of sialic acid loss on dendritic cell maturation. Immunology 128:e621–e631. doi: 10.1111/j.1365-2567.2009.03047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz-Cherry S, Hinshaw VS. 1996. Influenza virus neuraminidase activates latent transforming growth factor beta. J Virol 70:8624–8629. doi: 10.1128/JVI.70.12.8624-8629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson CM, Turpin EA, Moser LA, O’Brien KB, Cline TD, Jones JC, Tumpey TM, Katz JM, Kelley LA, Gauldie J, Schultz-Cherry S. 2010. Transforming growth factor-beta: activation by neuraminidase and role in highly pathogenic H5N1 influenza pathogenesis. PLoS Pathog 6:e1001136. doi: 10.1371/journal.ppat.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng C, Zhang L, Nguyen C, Vogel SN, Goldblum SE, Blackwelder WC, Cross AS. 2013. Neuraminidase reprograms lung tissue and potentiates lipopolysaccharide-induced acute lung injury in mice. J Immunol 191:4828–4837. doi: 10.4049/jimmunol.1202673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Ju X, Silveira PA, Abadir E, Hsu WH, Hart DNJ, Clark GJ. 2019. CD83: activation marker for antigen presenting cells and its therapeutic potential. Front Immunol 10:1312. doi: 10.3389/fimmu.2019.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin H, Liang S, Zhong Z, Wen J, Li W, Wang L, Xu J, Zhong F, Li X. 2014. Soluble CD83 inhibits human monocyte differentiation into dendritic cells in vitro. Cell Immunol 292:25–31. doi: 10.1016/j.cellimm.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Zhu Y, Zhang G, Gao C, Zhong W, Zhang X. 2011. CD83-stimulated monocytes suppress T-cell immune responses through production of prostaglandin E2. Proc Natl Acad Sci U S A 108:18778–18783. doi: 10.1073/pnas.1018994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotzor N, Lechmann M, Zinser E, Steinkasserer A. 2004. The soluble form of CD83 dramatically changes the cytoskeleton of dendritic cells. Immunobiology 209:129–140. doi: 10.1016/j.imbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Senechal B, Boruchov AM, Reagan JL, Hart DN, Young JW. 2004. Infection of mature monocyte-derived dendritic cells with human cytomegalovirus inhibits stimulation of T-cell proliferation via the release of soluble CD83. Blood 103:4207–4215. doi: 10.1182/blood-2003-12-4350. [DOI] [PubMed] [Google Scholar]
- 21.Bates JM, Flanagan K, Mo L, Ota N, Ding J, Ho S, Liu S, Roose-Girma M, Warming S, Diehl L. 2015. Dendritic cell CD83 homotypic interactions regulate inflammation and promote mucosal homeostasis. Mucosal Immunol 8:414–428. doi: 10.1038/mi.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu YJ, Song YN, Geng XR, Ma F, Mo LH, Zhang XW, Liu DB, Liu ZG, Yang PC. 2020. Soluble CD83 alleviates experimental allergic rhinitis through modulating antigen-specific Th2 cell property. Int J Biol Sci 16:216–227. doi: 10.7150/ijbs.38722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W, Buscher K, Wang B, Fan Z, Song N, Li P, Yue Y, Li B, Li C, Bi H. 2018. Soluble CD83 alleviates experimental autoimmune uveitis by inhibiting filamentous actin-dependent calcium release in dendritic cells. Front Immunol 9:1567. doi: 10.3389/fimmu.2018.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrow G, Slobedman B, Cunningham AL, Abendroth A. 2003. Varicella-zoster virus productively infects mature dendritic cells and alters their immune function. J Virol 77:4950–4959. doi: 10.1128/jvi.77.8.4950-4959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilingloh CS, Grosche L, Kummer M, Muhl-Zurbes P, Kamm L, Scherer M, Latzko M, Stamminger T, Steinkasserer A. 2017. The major immediate-early protein IE2 of human cytomegalovirus is sufficient to induce proteasomal degradation of CD83 on mature dendritic cells. Front Microbiol 8:119. doi: 10.3389/fmicb.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam SMS, Byun HO, Choi B, Sohn S. 2019. Inhibition of CD83 alleviates systemic inflammation in herpes simplex virus type 1-induced Behcet’s disease model mouse. Mediators Inflamm 2019:5761392. doi: 10.1155/2019/5761392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomon R, Hoffmann E, Webster RG. 2007. Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc Natl Acad Sci U S A 104:12479–12481. doi: 10.1073/pnas.0705289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng BJ, Chan KW, Lin YP, Zhao GY, Chan C, Zhang HJ, Chen HL, Wong SS, Lau SK, Woo PC, Chan KH, Jin DY, Yuen KY. 2008. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc Natl Acad Sci U S A 105:8091–8096. doi: 10.1073/pnas.0711942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh KB, Teijaro JR, Wilker PR, Jatzek A, Fremgen DM, Das SC, Watanabe T, Hatta M, Shinya K, Suresh M, Kawaoka Y, Rosen H, Oldstone MB. 2011. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc Natl Acad Sci U S A 108:12018–12023. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Hu S, Li Z, Wang X, Liu M, Guo Z, Li S, Xiao Y, Bi D, Jin H. 2011. Multiple amino acid substitutions involved in enhanced pathogenicity of LPAI H9N2 in mice. Infect Genet Evol 11:1790–1797. doi: 10.1016/j.meegid.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 31.Wu R, Sui Z, Liu Z, Liang W, Yang K, Xiong Z, Xu D. 2010. Transmission of avian H9N2 influenza viruses in a murine model. Vet Microbiol 142:211–216. doi: 10.1016/j.vetmic.2009.09.068. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Sun Y, Xu Q, Tan Y, Pu J, Yang H, Brown EG, Liu J. 2012. Mouse-adapted H9N2 influenza A virus PB2 protein M147L and E627K mutations are critical for high virulence. PLoS One 7:e40752. doi: 10.1371/journal.pone.0040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lechmann M, Berchtold S, Hauber J, Steinkasserer A. 2002. CD83 on dendritic cells: more than just a marker for maturation. Trends Immunol 23:273–275. doi: 10.1016/s1471-4906(02)02214-7. [DOI] [PubMed] [Google Scholar]
- 34.Scholler N, Hayden-Ledbetter M, Hellstrom KE, Hellstrom I, Ledbetter JA. 2001. CD83 is an I-type lectin adhesion receptor that binds monocytes and a subset of activated CD8+ T cells [corrected]. J Immunol 166:3865–3872. doi: 10.4049/jimmunol.166.6.3865. [DOI] [PubMed] [Google Scholar]
- 35.Heiskanen A, Hirvonen T, Salo H, Impola U, Olonen A, Laitinen A, Tiitinen S, Natunen S, Aitio O, Miller-Podraza H, Wuhrer M, Deelder AM, Natunen J, Laine J, Lehenkari P, Saarinen J, Satomaa T, Valmu L. 2009. Glycomics of bone marrow-derived mesenchymal stem cells can be used to evaluate their cellular differentiation stage. Glycoconj J 26:367–384. doi: 10.1007/s10719-008-9217-6. [DOI] [PubMed] [Google Scholar]
- 36.Webster RG, Govorkova EA. 2014. Continuing challenges in influenza. Ann N Y Acad Sci 1323:115–139. doi: 10.1111/nyas.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taubenberger JK, Morens DM. 2008. The pathology of influenza virus infections. Annu Rev Pathol 3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nachbagauer R, Shore D, Yang H, Johnson SK, Gabbard JD, Tompkins SM, Wrammert J, Wilson PC, Stevens J, Ahmed R, Krammer F, Ellebedy AH. 2018. Broadly reactive human monoclonal antibodies elicited following pandemic H1N1 influenza virus exposure protect mice against highly pathogenic H5N1 challenge. J Virol 92:e00949-18. doi: 10.1128/JVI.00949-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A 97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cameron KR, Gregory V, Banks J, Brown IH, Alexander DJ, Hay AJ, Lin YP. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:36–41. doi: 10.1006/viro.2000.0585. [DOI] [PubMed] [Google Scholar]
- 41.Saraiva M, O’Garra A. 2010. The regulation of IL-10 production by immune cells. Nat Rev Immunol 10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 42.Sun J, Madan R, Karp CL, Braciale TJ. 2009. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med 15:277–284. doi: 10.1038/nm.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudakov JA, Hanash AM, van den Brink MR. 2015. Interleukin-22: immunobiology and pathology. Annu Rev Immunol 33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar P, Thakar MS, Ouyang W, Malarkannan S. 2013. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal Immunol 6:69–82. doi: 10.1038/mi.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov S, Renneson J, Fontaine J, Barthelemy A, Paget C, Fernandez EM, Blanc F, De Trez C, Van Maele L, Dumoutier L, Huerre MR, Eberl G, Si-Tahar M, Gosset P, Renauld JC, Sirard JC, Faveeuw C, Trottein F. 2013. Interleukin-22 reduces lung inflammation during influenza A virus infection and protects against secondary bacterial infection. J Virol 87:6911–6924. doi: 10.1128/JVI.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T, Yin C, Boyd DF, Quarato G, Ingram JP, Shubina M, Ragan KB, Ishizuka T, Crawford JC, Tummers B, Rodriguez DA, Xue J, Peri S, Kaiser WJ, Lopez CB, Xu Y, Upton JW, Thomas PG, Green DR, Balachandran S. 2020. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180:1115–1129.e13. doi: 10.1016/j.cell.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNicholl IR, McNicholl JJ. 2001. Neuraminidase inhibitors: zanamivir and oseltamivir. Ann Pharmacother 35:57–70. doi: 10.1345/aph.10118. [DOI] [PubMed] [Google Scholar]
- 48.Moscona A. 2005. Neuraminidase inhibitors for influenza. N Engl J Med 353:1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- 49.Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm AE, Stamper CT, Lan LY, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173:417–429.e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grosche L, Knippertz I, Konig C, Royzman D, Wild AB, Zinser E, Sticht H, Muller YA, Steinkasserer A, Lechmann M. 2020. The CD83 molecule—an important immune checkpoint. Front Immunol 11:721. doi: 10.3389/fimmu.2020.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salio M, Cella M, Suter M, Lanzavecchia A. 1999. Inhibition of dendritic cell maturation by herpes simplex virus. Eur J Immunol 29:3245–3253. doi:. [DOI] [PubMed] [Google Scholar]
- 52.Dudziak D, Kieser A, Dirmeier U, Nimmerjahn F, Berchtold S, Steinkasserer A, Marschall G, Hammerschmidt W, Laux G, Bornkamm GW. 2003. Latent membrane protein 1 of Epstein-Barr virus induces CD83 by the NF-kappaB signaling pathway. J Virol 77:8290–8298. doi: 10.1128/jvi.77.15.8290-8298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 54.Song Y, Zhang Y, Chen L, Zhang B, Zhang M, Wang J, Jiang Y, Yang C, Jiang T. 2019. Genetic characteristics and pathogenicity analysis in chickens and mice of three H9N2 avian influenza viruses. Viruses 11:1127. doi: 10.3390/v11121127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene sequences (MA01) determined in this study have been submitted to GenBank. The GenBank accession numbers are MT939243, MT939296, MT939317, MT939432, MT939437, MT939439, MT939462, and MT939481.