Noroviruses are a major cause of acute gastroenteritis in humans. New norovirus variants and recombinants (re)emerge regularly in the human population. From animal experiments and surveillance studies, it has become clear that at least seven animal models are susceptible to infection with human strains and that domesticated and wild animals shed human noroviruses in their feces. As virus attachment is an important first step for infection, we used a novel method utilizing FITC-labeled VLPs to test for norovirus attachment to intestinal tissues of potential animal hosts. We further characterized these tissues with regard to their HBGA expression, a well-studied norovirus susceptibility factor in humans. We found attachment of several human strains to a variety of animal species independent of their HBGA phenotype. This supports the hypothesis that human strains could reside in an animal reservoir.
KEYWORDS: animal reservoir, histo-blood group antigens, host tropism, noroviruses, virus attachment, virus histochemistry, virus-like particles
ABSTRACT
Human noroviruses are the most common nonbacterial cause of gastroenteritis outbreaks, with new variants and genotypes frequently emerging. The origin of these new viruses is unknown; however, animals have been proposed as a potential source, as human noroviruses have been detected in animal species. Here, we investigated the potential of animals to serve as a reservoir of human noroviruses by testing norovirus attachment to formalin-fixed intestinal tissues of a range of potential reservoir animals. We set up a novel method to study norovirus binding using fluorescein isothiocyanate (FITC)-labeled virus-like particles (VLPs). In humans, noroviruses interact with histo-blood group antigens (HBGAs), carbohydrates that are expressed, among others, on the epithelial lining of the gastrointestinal tract. In animals, this interaction is not well understood. To test if virus binding depends on HBGAs, we characterized the HBGA phenotype in animal tissues by immunohistochemistry. With the exception of the black-headed gull and the straw-colored fruitbat, we observed the attachment of several human norovirus genotypes to the intestinal epithelium of all tested animal species. However, we did not find an association between the expression of a specific HBGA phenotype and virus-like particle (VLP) attachment. We show that selected human noroviruses can attach to small-intestinal tissues across species, supporting the hypothesis that human noroviruses can reside in an animal reservoir. However, whether this attachment can subsequently lead to infection needs to be further assessed.
IMPORTANCE Noroviruses are a major cause of acute gastroenteritis in humans. New norovirus variants and recombinants (re)emerge regularly in the human population. From animal experiments and surveillance studies, it has become clear that at least seven animal models are susceptible to infection with human strains and that domesticated and wild animals shed human noroviruses in their feces. As virus attachment is an important first step for infection, we used a novel method utilizing FITC-labeled VLPs to test for norovirus attachment to intestinal tissues of potential animal hosts. We further characterized these tissues with regard to their HBGA expression, a well-studied norovirus susceptibility factor in humans. We found attachment of several human strains to a variety of animal species independent of their HBGA phenotype. This supports the hypothesis that human strains could reside in an animal reservoir.
INTRODUCTION
Noroviruses are an important cause of gastroenteritis in humans and animals. To date, 10 genogroups (G) have been identified (GI to GX), which are further divided into 49 genotypes (1). Viruses within genogroups GI, GII, GIV.1, GVIII, and GIX are known to infect humans, while viruses from other genogroups have been found in a range of animals: pigs (GII.11, GII.18, and GII.19), cattle (GIII.1 and GIII.2), sheep (GIII.3), rodents (GV.1 and GV.2), cats (GIV.2, GVI.1, and GVI.2), lions (GIV.2), dogs (GVI.1, GVI.2, and GVII), harbor porpoises (GNA1), sea lions (GNA2), and bats (GX). Based on the whole capsid protein, viruses of different genogroups share <50% amino acid identity, while genotypes within the same genogroup share >60% amino acid identity. Therefore, porcine and feline/canine genotypes are of special interest with regard to their zoonotic potential, as they share ∼70% amino acid identity with human genotypes.
New variants, genotypes, and recombinants frequently emerge in the human population, yet their origin is unknown. It is assumed that these viruses emerge either from an unsampled population (e.g., asymptomatic or immunocompromised patients or demographic regions from which surveillance data are lacking) or from an animal reservoir. Anti-bovine and -canine norovirus antibodies have been reported in humans, and, conversely, various species of animals have tested positive for antibodies to human noroviruses (2). Furthermore, viral RNA of human GI and GII strains has been found in fecal material of calves, pigs, birds, captive macaques, dogs, and rodents (3–14; reviewed in reference 2). With the exception of birds and dogs (which have not been used for inoculation experiments), these species are also susceptible to human noroviruses under experimental conditions (15). This implies that animals can be a reservoir for human noroviruses.
The best-studied susceptibility factors for human noroviruses are histo-blood group antigens (HBGAs) (16–18). These terminal sugars of carbohydrate chains are linked to glycoproteins or glycolipids on red blood cells and tissues, including the epithelial cells of the gastrointestinal tract (19, 20). Moreover, HBGAs are secreted by these cells into bodily fluids, including mucosa and saliva (21). In the intestine, HBGAs are derived from precursor structures to which an α1,2-fucosyltransferase 2 (FUT2) adds a fucose group, resulting in the H1, H2, or H3 antigens. The addition of an α1,3- or α1,4-linked fucose group to the H1 or H2 antigen or their precursor structure results in the Lewis a, b, x, and y antigens. These steps are carried out by either the FUT3 to -7 or FUT9 enzyme. The A and B enzymes, encoded by the ABO locus, add either an N-acetylgalactosamine or a galactose in a α1,3 linkage to the H antigen, resulting in the A and B antigens, respectively (21).
Most human, canine, and bat noroviruses bind to synthetic HBGAs in a strain-dependent manner (22–24). In contrast, bovine (GIII) and murine (GV) noroviruses recognize receptors that are not expressed in humans; GIII.2 (Newbury agent 2) attaches to the alpha-galactosidase (Galα1,3), and the GV receptor is the transmembrane protein CD300lf, which is expressed on murine tuft cells in the intestine, but the main expression of this molecule is on hematopoietic cells (25–28). For other animal noroviruses, including the viruses identified in harbor porpoises (GNA1), sheep (GIII.2), and cats (GIV.2), no HBGA ligand or alternative attachment factors have been identified.
Norovirus attachment to HBGAs in vitro is assumed to be the primary step for virus uptake into the target cell. However, HBGA expression alone is not sufficient to enable infection of cells in culture. Therefore, it has been hypothesized that HBGAs are necessary for infection but not sufficient by themselves to initiate a full infectious cycle (29). The important role that HBGAs play in norovirus susceptibility has been confirmed by volunteer studies and epidemiological data (reviewed in reference 30). They indicated that a subset of the human population, which does not express HBGA in the mucosa or saliva, is resistant to certain norovirus strains. Dependent on geographical location, these nonsecretors make up between 5% and 20% of the human population and do not express a functional FUT2, thereby lacking the H-antigen-based structures on their intestinal epithelium and in their saliva.
Binding studies using fluorescein isothiocyanate (FITC)-labeled viruses and formalin-fixed paraffin-embedded (FFPE) tissues, or virus histochemistry, have been shown to be a valuable tool for studying host and cell tropism for viruses such as avian influenza virus and Middle East respiratory syndrome coronavirus (MERS-CoV) (31, 32), and we have now set up this technique for norovirus. The aim of the study was to assess the potential susceptibility of different animals to human noroviruses. As attachment is the first crucial step for a virus to initiate infection of a host cell, we tested attachment of a diverse range of human noroviruses using FITC-labeled virus-like particles (VLPs) on human (for validation) and animal FFPE tissues. The expression of the recombinant major capsid protein (VP1) results in self-assembly of empty capsids that are morphologically similar to the infective norovirus virions and therefore are commonly used as surrogates to study norovirus-HBGA interactions (33). The FITC label allowed us to study attachment of all genotypes by eliminating the need for secondary anti-norovirus antibodies that are currently only available for a limited selection of genotypes. To test if norovirus attachment is associated with a host HBGA profile, we defined the HBGA phenotype of these tissues using immunohistochemistry. Studying virus attachment to tissues from potential hosts will lead to a better understanding of which animal species are more likely to be susceptible to infection with human noroviruses and, therefore, focus efforts in the search for a reservoir for human noroviruses.
RESULTS
FITC-labeled human norovirus VLPs attach to human intestinal epithelium.
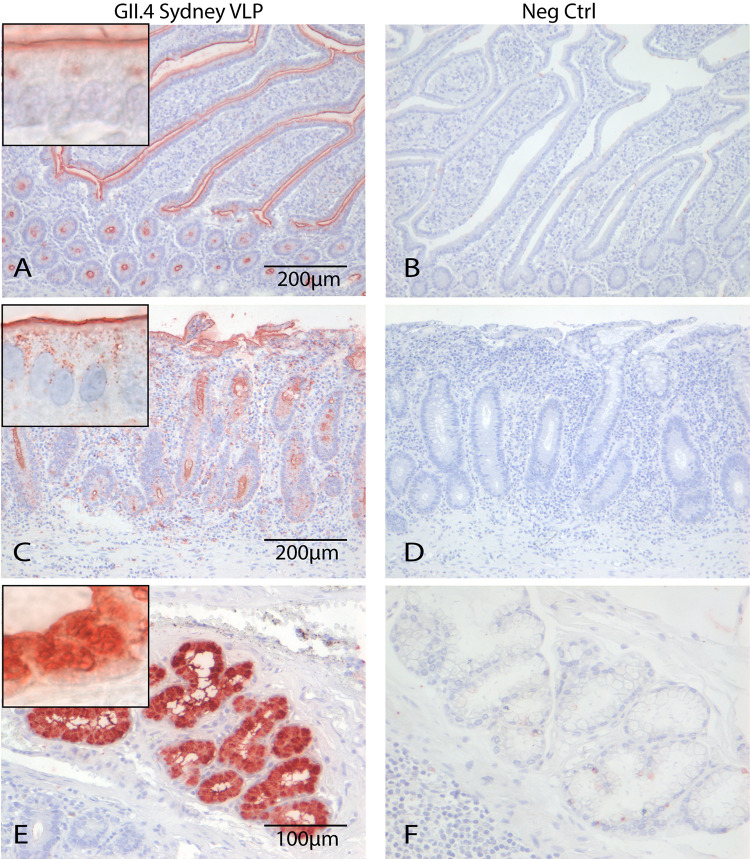
To validate binding of the FITC-labeled VLPs, we tested attachment of GII.4 Sydney 2012 on 6 human tissues, originating from duodenum, jejunum, or ileum. To exclude any effect of the FITC label on the VLP binding, we also tested the attachment of unlabeled GII.4 that was detected with an anti-GII.4 antibody. FITC-labeled and unlabeled GII.4 attached with similar efficiency to the epithelium of the villi and the crypts of tissues derived from duodenum, jejunum, and ileum of 3/6 human tissues (Fig. 1A and C and Table 1), while the negative control did not attach to any of the tissues (Fig. 1B and D). In one sample, additional staining was detected in the Brunner glands (Fig. 1E and F). GII.4 attachment depends on the α1,2-fucose group (34) that is added by FUT2, and we enzymatically cleaved this group with a 1,2α-fucosidase, which was confirmed by Ulex europeus (UEA-I) lectin staining (Fig. 2B). Upon α1,2-fucose removal, GII.4 binding was completely lost (Fig. 2C and D). FITC-labeled and unlabeled GII.4 VLPs showed identical attachment patterns, indicating that FITC-labeled particles can be used to study attachment.
FIG 1.
GII.4 VLPs attached to epithelium of villi and crypts in human small intestinal tissues (red). No difference was seen between FITC-labeled (A) and unlabeled (C) VLPs that were detected with an anti-FITC and an anti-GII.4 antibody, respectively. (E) In some tissues, VLPs additionally attached to the Brunner glands. (B, D, and F) No staining was seen in the negative controls (Neg Ctrl). Magnifications, 20× (A, B, C, and D) and 20× (E and F). Picture insets of attachment signal are 100×.
TABLE 1.
Summary of the virus histochemistry and immunohistochemistry results
| Tissue source | Histochemistry results fora: |
HBGA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI.3 | GI.6 | GII.1 | GII.3 | GII.4 | GII.6 | GII.17 2005 | GII.17 2014 | GII.17 2015 | GIV.1 | GIX | GNA1 | GIII.2, GII.18, GIV.2 | Total | ||
| Humanb | |||||||||||||||
| Human1 (ile) | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | A, B, Lea,x,y |
| Human2 (jej) | − | + | − | + | + | + | − | + | + | + | + | − | − | 8/15 | A, H2, Leb |
| Human3 (duo) | − | + | − | + | + | + | − | + | + | + | + | − | − | 8/15 | A, H2, Leb |
| Human4 (ile) | + | + | − | + | + | + | + | + | + | − | + | − | − | 9/15 | H2, Lea,b,y |
| Human5 (duo) | + | + | − | + | − | + | + | + | + | + | + | + | − | 10/15 | H2, Lea,b,x |
| Human6 (duo) | + | + | − | − | − | + | + | + | + | + | + | + | − | 9/15 | Lea,b,x |
| Animalc | |||||||||||||||
| Pig1h ,i | − | − | − | − | + | + | + | + | + | − | + | − | − | 6/15 | H1, H2 |
| Pig2h | − | + | − | − | − | − | − | + | + | + | + | − | − | 5/15 | A, H2 |
| Pig3h | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | H2 |
| Pig4h | − | + | − | − | − | − | − | − | − | + | − | − | − | 2/15 | A, H1, H2 |
| Dog1h | − | − | − | + | + | + | + | − | − | − | − | + | − | 5/15 | H1, H2 |
| Dog2h ,i | + | + | − | + | + | + | − | + | + | + | + | − | − | 9/15 | A, Lea,x,y |
| Dog3h | − | − | − | − | + | + | + | + | − | − | + | + | − | 6/15 | H1, H2 |
| Cat1h | + | + | − | − | + | − | − | − | − | − | + | + | − | 5/15 | A, H2, Leb,x,y |
| Rat1h | − | + | − | − | + | + | − | + | − | + | + | − | − | 6/15 | A, H1, H2 |
| Rat2h | − | − | − | − | + | − | − | − | − | − | + | − | − | 2/15 | H1, H2 |
| Rat3h | − | + | − | − | + | + | − | + | − | + | + | − | − | 6/15 | A, B, H1, H2 |
| Chimpanzee1 | − | + | − | + | + | + | − | + | + | + | + | − | − | 8/15 | A, H2, Leb |
| Chimpanzee2 | − | + | − | − | + | + | − | + | + | + | + | − | − | 7/15 | A, H2Leb |
| Porpoise1h | + | + | − | − | + | − | − | − | − | − | − | + | − | 4/15 | H2, Lex,y |
| Porpoise2h | + | + | − | − | − | − | − | − | − | − | − | − | − | 2/15 | Lex,y |
| Porpoise3h | + | + | − | − | − | − | − | − | − | − | − | + | − | 3/15 | Lex,y |
| Pipistrelle1d ,h | + | + | − | − | + | + | + | + | − | − | + | + | − | 8/15 | A, H1, H2 |
| Pipistrelle2d ,h | + | + | − | − | + | + | − | − | − | + | + | − | − | 6/15 | A, B, H2, Ley |
| Pipistrelle3d ,h | + | + | − | − | + | + | − | + | + | + | + | − | − | 8/15 | A, B, H1, H2 |
| Pipistrelle4d ,h | + | + | − | + | + | + | + | + | + | + | + | + | − | 11/15 | A, H1, H2, Lex,y |
| Straw-colored FB1 | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | B, Lea |
| Straw-colored FB2g | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | |
| Straw-colored FB3 | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | B, Ley |
| Egyptian FB1 | + | + | − | − | − | − | − | − | − | + | − | − | − | 3/15 | B, Lea,x,y |
| Egyptian FB2g | + | + | − | − | − | − | − | − | − | − | − | + | − | 3/15 | Lea,x |
| Egyptian FB3 | + | + | − | − | − | − | − | − | − | − | − | + | − | 3/15 | Lea,x,y |
| Black-headed gull1e ,g | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | |
| Black-headed gull2e ,g | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | |
| Black-headed gull3e ,g | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | |
| Black-headed gull4e ,g | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/15 | |
| Mallard1g | + | + | − | − | − | − | − | − | − | + | − | + | − | 4/15 | Lex |
| Mallard2i | + | + | − | + | − | + | − | + | − | + | + | − | − | 7/15 | Lea,b,x |
| Mallard3 | + | + | − | − | − | + | − | − | − | + | − | − | − | 4/15 | Lea,b,x |
| Turkey1 | + | + | − | − | − | − | − | − | − | − | − | − | − | 2/15 | Leb,x,y |
| Chicken1 | + | + | − | − | − | − | − | − | − | + | − | − | − | 3/15 | Leb,x,y |
| Chicken2g | + | + | − | − | − | − | − | − | − | − | − | − | − | 2/15 | Lex |
| Oyster1 | − | − | − | − | + | − | − | − | − | − | − | − | − | 1/15 | A |
| Oyster2 | − | + | − | − | + | − | − | + | − | − | + | − | − | 4/15 | A, H1 |
| Oyster3 | + | − | − | − | + | − | − | − | − | − | + | − | − | 3/15 | A |
| Oyster4 | − | − | − | − | + | − | − | − | − | − | − | − | − | 1/15 | A, H1 |
| No. of species positive/no. of species testedf | 10/15 | 13/15 | 0/15 | 5/15 | 9/15 | 7/15 | 4/15 | 8/15 | 5/15 | 9/15 | 9/15 | 7/15 | 0/15 | ||
| Individuals positive/Individuals testedf | 22/46 | 30/46 | 0/46 | 9/46 | 22/46 | 19/46 | 8/46 | 18/46 | 12/46 | 19/46 | 22/46 | 12/46 | 0/46 | ||
Binding, +; no binding, −.
ile, ileum; jej, jejunum; duo, duodenum.
Underlined are species in which human strains have been detected or which have been susceptible to human noroviruses in the laboratory.
GX was found in species belonging to another microbat genus, the horseshoe bat (Rhinolophus).
Human strains were found in feces of gulls within the Larus genus.
Including humans.
Nonsecretor animals.
Species with own norovirus.
Duodenum, jejunum and ileum were tested (Table 2).
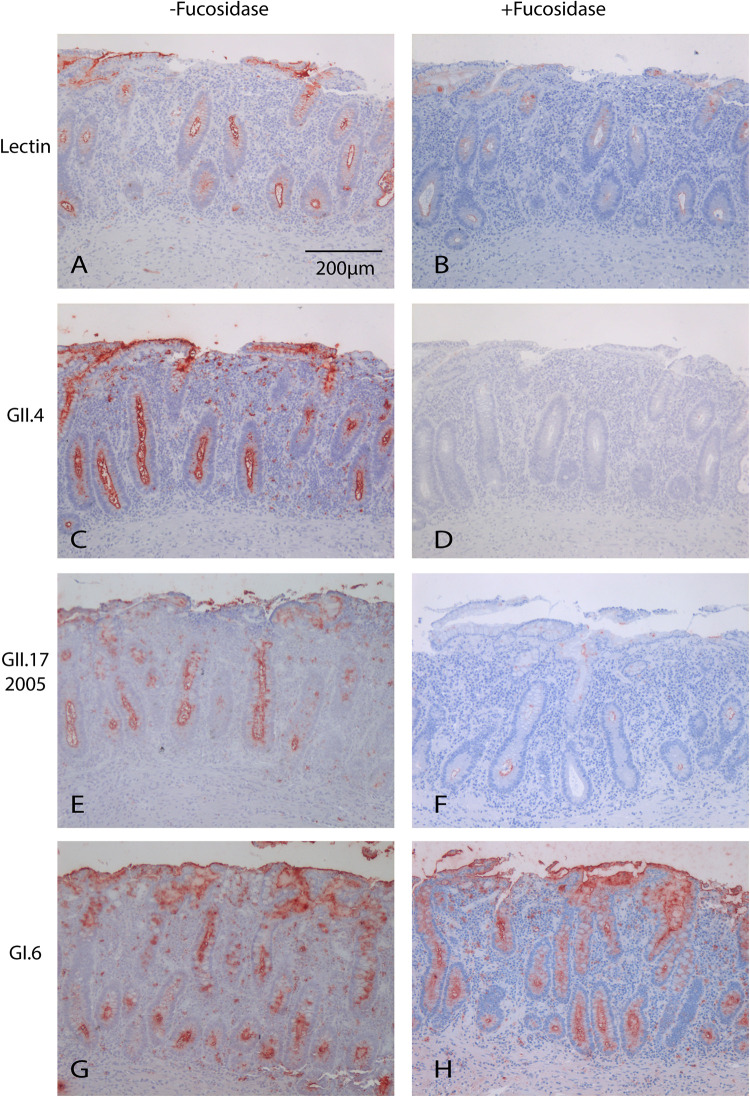
FIG 2.
Attachment of VLPs to human intestinal tissue after enzymatic removal of the α1,2 fucose group. UEA-I lectin staining without (A) and with (B) fucosidase treatment was performed as a control to confirm cleavage of the α1,2 fucose group. Upon 1,2α-fucosidase treatment, no attachment of GII.4 was observed (C and D). Attachment of GII.3, GII.6, GII.17 (2005, 2014, and 2015), and GIX was reduced (represented by GII.17 2005 staining [E and F]). Attachment of GI.3, GI.6, and GIV.1 was unchanged (represented by GI.6 staining [G and H]). Magnification, 10×.
To further validate our assay and compare the attachment pattern of different human norovirus genotypes, the VLPs were first tested on human tissues. We included VLPs representing a wide range of human norovirus genotypes: GI (GI.3, GI.6), GII (GII.1, GII.3, GII.4, GII.6, and GII.17 [2005, 2014, and 2015]), GIV.1, and GIX. All strains, except GII.1, attached to at least one of the human tissues. While all genotypes attached to the epithelium, variation was found in genotype binding between individuals, as not all strains attached to the same individuals (Table 1). Of all the human tissues, there was only one, a human ileum tissue, to which none of the VLPs attached. GI.6, GII.6, GII.17 (2014 and 2015), and GIX attached to 5/6 tissues, showing the broadest range of attachment. When we removed the α1,2-fucose group, thereby changing the HBGA to a nonsecretor phenotype, attachment of GII.3, GII.17, GII.6, and GIX was clearly reduced (Fig. 2E and F), while that of GI and GIV.1 was unaffected (Fig. 2G and H). Thus, VLPs from different genotypes attached to the epithelium of human tissue. The finding that not all VLPs attached to all tissues suggests the role of a host factor.
FITC-labeled VLPs of genogroups GI, GII, GIV, and GIX attach to animal intestinal tissue.
To investigate the potential susceptibility of different animal species, we tested the attachment of the human norovirus VLPs on a variety of animal tissues. Tested were tissues of species or families in which human noroviruses have previously been detected or have been susceptible to human norovirus infection in the laboratory (Table 1). We further tested tissues of species that have developed and sustained their own noroviruses (Table 1) as well as oysters, which are known bioaccumulators of human noroviruses and in which a few animal noroviruses have been detected (35, 36).
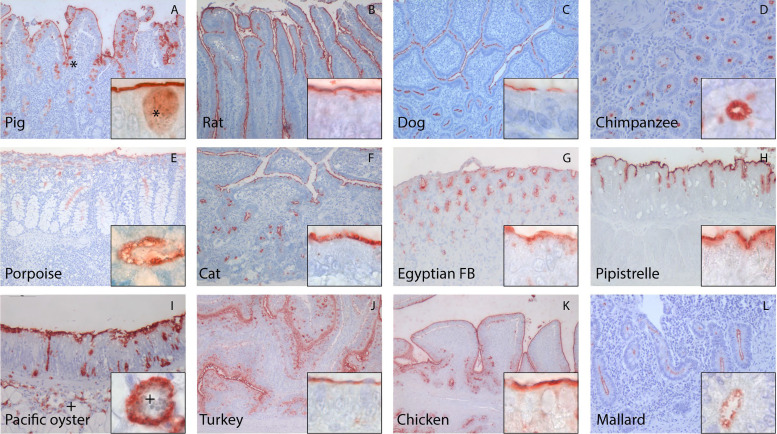
Like on human tissue, GI, GII, GIV.1, and GIX VLPs also attached to intestinal epithelium of animals (Fig. 3 and Table 1). Out of the 14 species tested, tissue samples from the black-headed gull and the straw-colored fruit bat were the only samples to which none of the VLPs attached. The highest diversity of genotype attachment was seen for the dog and the common pipistrelle bat samples, to which VLPs from all tested genotypes attached. On the contrary, only a few genotypes attached to tissue samples from harbor porpoise (GI.3, GI.6, and GII.4), Egyptian fruit bat (GI.3, GI.6, and GIV.1), turkey (GI.3 and GI.6), and chicken (GI.3, GI.6, and GIV.1). The ability to attach to several species differed between VLPs of different genogroups and genotypes. For instance, GI.6 VLP attached to intestinal tissue from 13/15 species (including humans), whereas GII.17 (2005) VLP only attached to tissues from 4/15 species. Thus, although most genotypes were capable of attaching to animal tissues, we did observe genotype- and species-specific differences.
FIG 3.
Human norovirus VLPs attached to the epithelium of intestinal tissue originating from various animals. A representation of different VLPs is shown here. In pigs and oysters, attachment was also detected intracellularly in goblet cells (*) and unidentified cells (+), respectively. Magnifications, 10× (A, B, C, E, F, J, K and L), 20× (D, G, H and I), and 100× (insets).
To test if attachment varies between different parts of the GI tract within one individual, we tested VLP attachment to duodenum, jejunum, and ileum for one pig, one dog, and one mallard specimen for which these additional samples were available (Table 2). In the pig samples, VLPs attached only to the ileum and not the duodenum and jejunum. Conversely, in the dog, most genotypes attached to the ileum and jejunum and fewer to the duodenum. In the mallard, GII.17 (2014) and GIX VLPs attached exclusively to the jejunum and the cecum, GIV.1 VLPs to the duodenum, the jejunum, and the cecum, and GII.3 and GII.6 only to the cecum. GI.3 and GI.6 attached to all parts except the cecum. For some species, additional non-small-intestinal tissues were available, to which we noticed unexpected attachment, i.e., in the common pipistrelle (stomach, colon, and bladder), Egyptian fruit bat (colon), mallard (cecum), and cat (esophagus and stomach).
TABLE 2.
Attachment of human noroviruses to different sections of the small intestine of one pig, one dog, and one mallard
| Animala | Attachment in: |
HBGA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI.3 | GI.6 | GII.1 | GII.3 | GII.4 | GII.6 | GII.17 2005 | GII.17 2014 | GII.17 2015 | GIV.1 | GIX | GNA1 | GIII.2, GII.18, GIV.2 | ||
| Pig1 (duo) | − | − | − | − | − | − | − | − | − | − | − | − | − | H1, H2 |
| Pig1 (jej) | − | − | − | − | − | − | − | − | − | − | − | − | − | H1, H2 |
| Pig1 (ile) | − | − | − | − | + | + | + | + | + | − | + | − | − | H1, H2 |
| Dog2 (duo) | + | + | − | − | − | − | − | − | − | + | + | − | − | A, Lea,x,y |
| Dog2 (jej) | + | + | − | + | + | + | − | + | + | + | + | − | − | A, Lea,x,y |
| Dog2 (ile) | + | + | − | + | + | + | − | + | + | + | + | − | − | A, Le,x,y |
| Mallard2 (duo) | + | + | − | − | − | − | − | − | − | + | − | − | − | Lea,b |
| Mallard2 (jej) | + | + | − | − | − | − | − | + | − | + | + | − | − | Lea,b |
| Mallard2 (ile) | + | + | − | − | − | − | − | − | − | − | − | − | − | Lea,b |
| Mallard2 (cae) | − | − | − | + | − | + | − | + | − | + | + | − | − | Lea,b,x |
ile, ileum; jej, jejunum; duo, duodenum; cae, cecum.
In conclusion, we detected the attachment of FITC-labeled VLPs of genogroups GI, GII, GIV, and GIX to intestinal epithelium of all animal species, except for the black-headed gull and the straw-colored fruit bat, and with the broadest range of VLPs attaching to dog and common pipistrelle tissues.
Porpoise norovirus attaches to animal and human tissues.
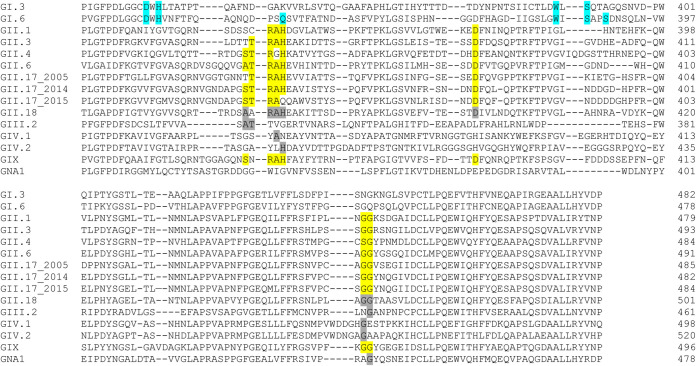
In addition to human strains, we also tested animal norovirus genotypes, GII.18 (porcine), GIII.2 (bovine), GIV.2 (feline), and GNA1 (harbor porpoise), on human and animal tissues. For GII.18, we experienced problems expressing VP1, and we used p-particles instead. The structural features and critical residues for HBGA binding are known for several GI and GII genotypes, while these are largely unknown for GIV.1 and some of the genogroups infecting animal species. Sequence alignment of all VLPs tested (Fig. 4) showed that there are large differences for the animal noroviruses compared to the known residues of the HBGA binding sites of GI and GII, indicating that these have different ligand binding sites.
FIG 4.
Sequence alignment of the HBGA-binding interfaces and the surrounding regions of human and animal norovirus genotypes used in this study. All VP1 sequences of VLPs (accession numbers are in Table 4) were aligned with MUSCLE. Amino acids that have experimentally been shown to be required for GII (yellow) and GI (blue) interaction with HBGAs are marked (53). Amino acids that are identical to known HBGA interaction residues are shaded gray. The number indicates the amino acid position counted from the start of VP1.
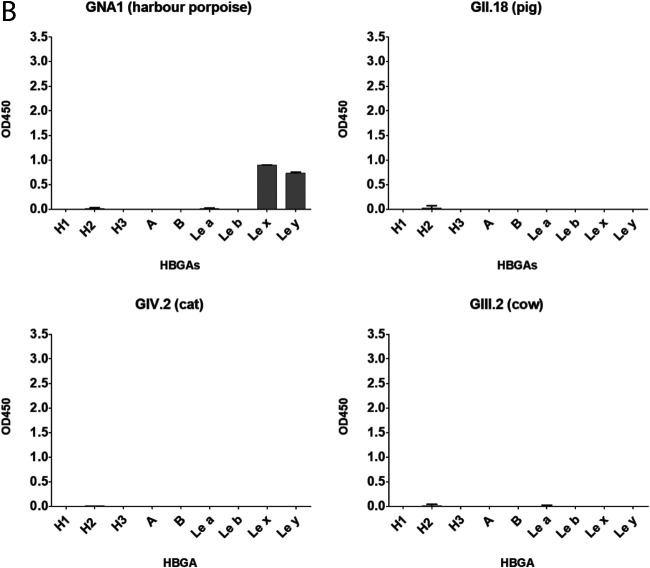
The harbor porpoise strain GNA1 attached to epithelium of 2/3 porpoise specimens as well as to intestinal tissues of humans, dogs, cats, common pipistrelles, Egyptian fruit bat, and a mallard (Table 1). VLPs from none of the other animal strains attached to any of the tissues. Neither chemical (citric acid) nor enzymatic (protease) antigen retrieval treatment resulted in binding. For canine and bat noroviruses, an effect of temperature on binding has been reported (23, 24). Therefore, we tested binding at various temperatures (4°C, 25°C, and 37°C) as well as increased VLP concentration, but we did not observe any binding. Thus, perhaps virus histochemistry is not a suitable method to study attachment for all animal norovirus strains.
Animals express HBGAs similar to those of humans.
To determine whether VLP attachment can be associated with the expression of specific HBGAs, we characterized the HBGA profile of humans and potential animal hosts by immunohistochemistry (IHC) on the same FFPE tissues that had been used for virus histochemistry. Humans expressed H and Lewis antigens on the epithelium of the villi and crypts, and in some individuals additional expression was detected in the mucin-producing goblet cells and Brunner glands (Tables 1 and 3). In most animal tissues, HBGAs were expressed on the epithelium and sometimes in goblet cells and Brunner glands. Samples of humans, dogs, and the common pipistrelle expressed the widest diversity of HBGAs, being positive for H, A, and Lewis antigens. On the contrary, the bird species showed the lowest diversity, expressing only Lewis antigens. Tissues of the black-headed gull as well as one straw-colored fruit bat were the only tissues in which we did not detect any HBGA expression. It is known that the detection of the H1 and H2 antigens can be hindered by the presence of the A, B, Leb, and Ley antigens (23, 37, 38). Therefore, secretors were defined as being positive for H1, H2, A, B, Leb, or Ley. Of note, although defined as nonsecretor antigens, Lea and Lex were also detected in secretors, indicative of FUT3 activity. In total, only eight nonsecretors were found, namely, within the chicken, the mallards, the black-headed gulls, the Egyptian fruit bats, and the straw-colored fruit bats (Table 1). Due to the limited number of nonsecretor individuals, attachment data on these individuals were scarce, but GI.3, GI.6, GIV.1, and GNA1 were the only genotypes attaching to nonsecretor tissues. For the tissue regions where VLPs attach to overlap HBGA expression, we further observed differences in HBGA phenotype between species and between individuals. However, we did not find a one-to-one correlation between VLP attachment and specific ABH or Lewis phenotypes (Table 1). Similarly, differences in VLP binding in different sections of the intestine were not correlated with differences in HBGA expression (Table 2). We did, however, note that the dog and pipistrelle tissues to which the broadest diversity of VLP attached were also the species where HBGA expression most closely resembled that in humans.
TABLE 3.
Summary of HBGA expression in intestinal epithelium of humans and different animal species
| Hostb | Result for HBGAc
: |
Summary per species |
HBGA from prev studiesd | Fut2(47) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | Lea | Leb | Lex | Ley | H1 | H2 | Secretor | ||||
| Human | 4/6 | 1/6 | 4/6 | 5/6 | 3/6 | 2/6 | 0/6 | 4/6 | 6/6 | A, B, H2, Lea,b,x,y | A, B, H1, H2, Lea,b,x,y (19, 20, 38, 48) | + |
| Pig | 2/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 2/4 | 4/4 | 4/4 | A, H1, H2 | A, H1, Leb (70) | + |
| Dog | 1/3 | 0/3 | 1/3 | 0/3 | 1/3 | 1/3 | 2/3 | 2/3 | 3/3 | A, H1, H2, Lea,x,y | A, H, Lea,b,y (23, 71) | + |
| Cat | 1/1 | 0/1 | 0/1 | 1/1 | 1/1 | 1/1 | 0/1 | 0/1 | 1/1 | A, H2, Leb,x,y | A, H | + |
| Rat | 2/3 | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 3/3 | 3/3 | 3/3 | A, B, H1, H2 | A, B, H (72–74) | + |
| Chimpanzee | 2/2 | 0/2 | 0/2 | 2/2 | 0/2 | 0/2 | 0/2 | 2/2 | 2/2 | A, H2, Leb | A, H, lewis (75) | + |
| Porpoise | 0/3 | 0/3 | 0/3 | 0/3 | 3/3 | 3/3 | 0/3 | 1/3 | 3/3 | H2, Lex,y | NA | +a |
| Pipistrelle | 4/4 | 2/4 | 0/4 | 0/4 | 1/4 | 2/4 | 3/4 | 4/4 | 4/4 | A, B, H1, H2, Lex,y | NA | + |
| Straw-colored FB | 0/3 | 2/3 | 1/3 | 0/3 | 0/3 | 1/3 | 0/3 | 0/3 | 2/3 | B, Lea,y | NA | |
| Egyptian FB | 0/3 | 1/3 | 2/3 | 0/3 | 2/3 | 2/3 | 0/3 | 0/3 | 2/3 | B, Lea,x,y | NA | |
| Black-headed gull | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | NA | NA | |
| Mallard | 0/3 | 0/3 | 2/3 | 2/3 | 1/3 | 0/3 | 0/3 | 0/3 | 2/3 | Lea,b,x | NA | |
| Turkey | 0/1 | 0/1 | 0/1 | 1/1 | 1/1 | 1/1 | 0/1 | 0/1 | 1/1 | Leb,x,y | NA | |
| Chicken | 0/2 | 0/2 | 0/2 | 1/2 | 2/2 | 1/2 | 0/2 | 0/2 | 1/2 | Leb,x,y | NA | |
| Oyster | 4/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 2/4 | 0/4 | 4/4 | A, H1 | A, H1 (76, 77) | NA |
| No. of species positive/no. of species tested | 8/15 | 5/15 | 5/15 | 7/15 | 9/15 | 9/15 | 5/15 | 7/15 | 14/15 | |||
Dolphins.
FB, fruitbat.
Shown are number of positive individuals/individuals tested.
NA, not applicable.
FITC-labeled VLPs attach to synthetic HBGAs.
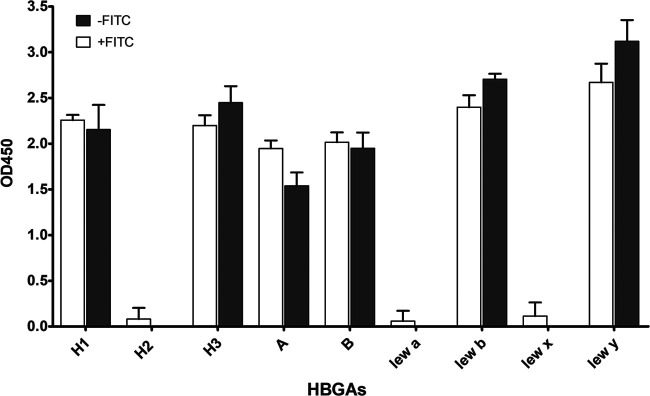
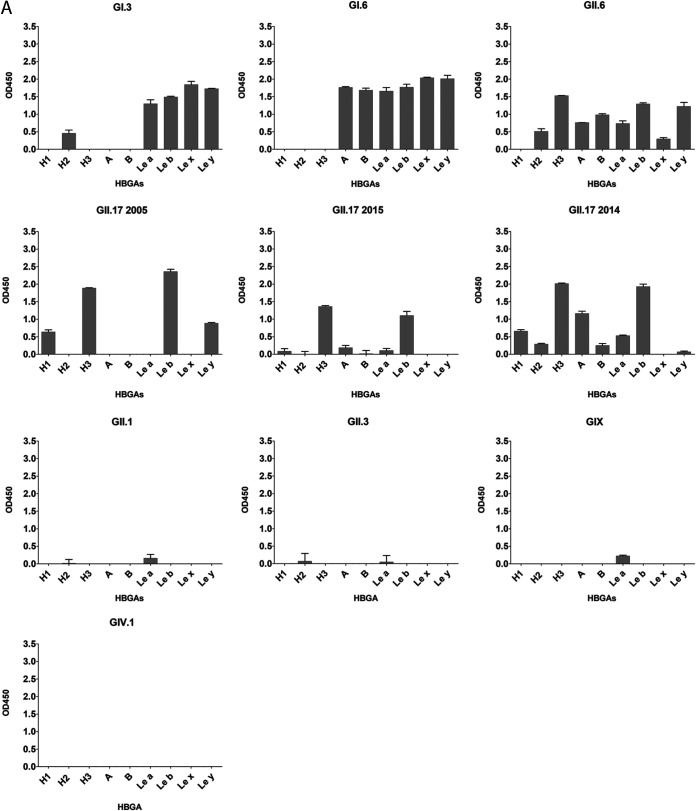
To characterize specific VLP-HBGA interaction, we further investigated their binding to synthetic HBGAs. To exclude the interference of FITC, labeled and unlabeled GII.4 samples were tested for HBGA binding using either an anti-FITC or an anti-GII.4 antibody. GII.4 attached to H1, H3, A, B, Leb, and Ley independently of the FITC label, indicating that the FITC labeling does not impact the VLP-HBGA interaction (Fig. 5). We further tested the same panel of VLPs that had been used for virus histochemistry. As described by others, VLP-HBGA interaction varied between strains. GII.6 had the broadest binding pattern, attaching to A, B, H, and Lewis antigens (Fig. 6A). In contrast, GII.1, GII.3, GII.15, and GIV.1 VLPs did not attach to any of the tested HBGAs (data not shown). GI.3, GI.6, and GII.6 attached to both secretor (Leb and Ley) and nonsecretor (Lea and Lex) antigens, while GII.4 and GII.17 2005 and 2015 exclusively bound to secretor antigens. The three GII.17 strains showed differences in their HBGA interactions. While all three bound strongest to H3 and Leb, GII.17 2005 additionally bound to H1 and Ley and GII.17 2014 to H1, H2, A, B, and Lea. Of the animal VLPs, only the porpoise GNA1 attached to Lex and Ley, while the others did not bind to any of the HBGAs (Fig. 6B).
FIG 5.
Binding of unlabeled (white) and labeled (gray) GII.4 VLPs to synthetic carbohydrates. Streptavidin plates were coated overnight at 4°C with 10 μg/ml PAA-conjugated HBGAs, and 100 ng of VLPs was incubated overnight at 4°C. Bound labeled and unlabeled VLPs were detected with an anti-FITC and an anti-GII.4 antibody, respectively. Each reaction was done in duplicates, and the error bars represent the standard deviations.
FIG 6.
Binding of human (A) and animal (B) FITC-labeled VLPs to synthetic carbohydrates. Plates were coated with 10 μg/ml PAA-conjugated HBGAs. Attachment of 100 ng FITC-labeled VLPs was detected with anti-FITC antibody. Shown are the mean absorbance values, OD450, from which the negative-control values (d-mannose) have been subtracted. Each reaction was done in duplicate, and the error bars represent the standard deviations.
These data showed that GII.4 and GII.17 attached to secretor antigens only, while GI and GII.6 attached to secretor and nonsecretor antigens. Interestingly, GII.3 and GIX, which contain the conserved residues within the HBGA binding site and attach to tissues, did not recognize any of the synthetic carbohydrates.
DISCUSSION
This study demonstrates that a broad range of mammalian and bird species could be susceptible to human noroviruses based on their carbohydrate expression and attachment of human norovirus VLPs to intestinal tissue. Human norovirus VLPs attached to intestinal tissues that originated from species that have been found positive for human norovirus RNA, including pigs, dogs, rats, primates, and bird species. Dogs are the only animals for which human-to-animal transmission has been reported (14, 39). Interestingly, we also detected broad attachment to species in which no human norovirus has been found to date. These include cats, the common pipistrelle, mallards, and, to a lesser extent, harbor porpoises, Egyptian fruit bats, turkeys, and chickens. Bats and birds are of special interest, as they are important reservoirs for many known human pathogens (40). In bats, no human noroviruses have been found to date, although bat norovirus GX has been isolated from Rhinolophus sinicus and Rhinolophus affinis (also microbats) without obvious clinical signs. In wild birds in Finland and Brazil, RNA of human noroviruses (GII.4, GII.3, GI.2, and GII.p31) has been detected (8, 41). The identified birds in Finland were mostly gull species from the genus Larus. The gulls that we tested, however, were from a different genus, Chroicocephalus, possibly explaining the lack of attachment of VLPs in this species.
In intestinal biopsy specimens of chronically infected patients, the dominant cell type found to be infected were enterocytes, which are the most prevalent cell type in the epithelium of the villi (42). Similarly, in most animal species as well as the human samples, we detected VLPs that attach to the epithelium of villi. We also observed attachment to crypts, which have not been found to be positive in human biopsy specimens (42). In vivo, the crypts have a different environment than the villi, consisting of mostly nonenterocytes that secrete antimicrobial peptides and hormones, possibly hindering the ability of the virus to infect. VP1 antigen has also been detected in human biopsy specimens in macrophages in the lamina propria, and whole-virus staining with inactivated GII.4 Sakai showed binding exclusively to the lamina propria and Brunner glands (42, 43). In contrast, we detected no attachment to immune cells in the lamina propria or Peyer’s patches, supporting the hypothesis that staining in these cells could be due to phagocytosed infected enterocytes (42).
Depending on the animal model and genotype, viral antigen was previously detected in the duodenum, the jejunum, or the ileum without a tendency toward one location. Interestingly, we did see differences in attachment to different parts of the intestine in pigs, dogs, and mallards. For some species, additional non-small-intestinal tissues were available for which we noticed unexpected attachment. This indicates that there are differences between norovirus strains in their preference to infect certain parts of the intestine and that noroviruses, in some cases, are able to infect sites outside the small intestine. The latter has been observed in animal infection experiments with human norovirus strains where virus has also been detected in lymphatic organs (44–46).
There are intrahost and intra- and interspecies differences in the attachment of noroviruses. To investigate what underlies this, we compared VLP attachment to HBGA expression, which, in humans, is the best understood host susceptibility factor. Genetic analysis has shown that genes encoding enzymes that are involved in HBGA synthesis are found in a broad range of vertebrates and invertebrates (47). As in humans, we found HBGAs expressed on the intestinal epithelium in a broad variety of animal species. Our findings of HBGA expression in the mallard, chicken, turkey, and megabat species is in contrast to the previous findings described by Yamamoto et al., in which the genetic foundation of FUT and ABO genes across the animal kingdom was analyzed (47). Megabats and bird species, including chickens, mallards, and turkeys, were identified as lacking the ABO- and FUT-related genes. An explanation for this discrepancy could be that unrecognized homologs of the enzymes needed for the addition of the different antigens exist or, alternatively, that our antibodies are cross-reacting with similar carbohydrate structures.
VLP attachment did not correlate one-to-one with any specific ABH or Lewis phenotype, which has also been described in other studies (19, 48). We did, however, notice that those specimens with a wider range of HBGA expression also showed attachment of more noroviruses. Dogs and the common pipistrelle expressed almost the same diversity of HBGA as humans, and, in accordance with this, all VLPs attached to tissues from these species. This indicates that dogs as well as some microbat species are susceptible to a broader range of noroviruses. The close contact between humans and dogs also makes them interesting candidates for human-to-animal transmission. Rather than specific HBGA expression, the secretor status might be more important for susceptibility, especially for GII genotypes. The dependence on the α1,2-fucose group is documented for GII.4 Sydney 2012 and GI.1 (19), and, accordingly, GII.4 only attached to secretor-positive animals. The limited number of eight nonsecretor individuals does not allow us to draw conclusions, but GI and GIV.1 were the only VLPs attaching to these tissues. When treated with a 1,2α-fucosidase, GII.4 binding was completely diminished, while attachment of the other GIIs was reduced and that of GI and GIV.1 remained unchanged. This fits into the notion that attachment of GII strains depends on the α1,2-fucose group, while GI strains bind to a terminal Gal group (49). Our data support the hypothesis that HBGAs are necessary but insufficient for norovirus attachment and infection. This was evidenced by the fact that HBGA expression alone did not automatically lead to VLP attachment and that, although within the same tissue VLP attachment was always colocated with HBGA expression, HBGA expression was also detected in locations where VLPs did not attach.
GII.4 is the genotype most frequently found in human outbreaks and the most commonly found human genotype in animals (2, 50). Therefore, it is surprising that not GII.4 but GI.3 and GI.6 were the strains that attached to the broadest range of species (10 and 13/15 species, respectively). Similarly, the broad attachment of GIV.1 and GIX was unexpected, as the GIX genogroup consists of only a few reported strains, and GIV.1 strains, although found in sewage, are only sporadically detected in humans (51). On the one hand, this indicates that attachment alone is not an indicator of how likely a strain is causing symptomatic outbreaks. On the other hand, attachment can also be affected by differences in stability. For example, in oysters, GI.1 has been shown to bioaccumulate more efficiently than GII.4, because the latter had been degraded in salt water (52). We detected attachment of GII.4 to all oysters, which would likely be impacted by the conditions in salt water.
We observed a lack of binding of some human and most animal strains to synthetic glycans. We did not detect attachment the human GII.1, GII.3, GIV.1, and GIX strains to synthetic carbohydrates, even though HBGA ligands have been described and included for all but GIV.1 (53). GNA1 was the only animal strain that attached to intestinal tissue as well as to synthetic glycans, Lex and Ley, which were also the only glycans expressed on harbor porpoise tissue. Some of the other animal genotypes have been tested before, and the lack of attachment was expected. These include GIV.2 and GII.11 and GII.19, which did not attach to HBGAs or human saliva, respectively (54–56). Interestingly, the porcine genotypes have the conserved binding site, but no attachment factor has been identified yet. The change of two amino acids adjacent to GII.11 and GII.19 HBGA binding led to their attachment to type A and B antigens (56). Similarly, GIII.2 attaches to α-galactosidase, which was not included in our glycan panel (25). These variations in VLP-HBGA interaction data could be a result of several factors. They include the nature of the carbohydrates themselves (i.e., linker, mono- versus polysaccharides, and manufacturer) as well as the condition under which the interaction is tested (i.e., pH, temperature, and salt content). Differences in binding specificity have been shown for different strains of the same genotype. The labeling of the particles with FITC occurs at pH 9.6, although subsequent steps are performed at pH 7.5. Noroviruses and VLPs are stable at a pH range of 3 to 7 (57); however, conformational changes have been reported, which could have implications for HBGA binding. Therefore, the lack of binding of some human as well as animal genotypes could be a result of the restricted selection of glycans that we tested as well as variation in stabilities of genotypes at high pH. For some genotypes and variants, no HBGA binding partner has been identified to date (including some GII.1 variants), and alternative or additional attachment molecules have been proposed, such as heparan sulfate (58, 59) and sialic acids (60). Further, it cannot be excluded that a protein receptor plays a role for some strains, similar to what is observed for murine norovirus.
In summary, using FITC-labeled VLPs is a promising method to investigate potential tissue and cell tropism of different genotypes and strains, as it eliminates the need for specific antibodies. We have shown that many animals express HBGAs on their intestinal tissues, and additional factors are likely important for norovirus attachment and host susceptibility. Nevertheless, this is a first approximation to identify potential norovirus hosts and reservoirs. The broad attachment of many VLPs to these tissues should be followed up by investigating whether attachment subsequently leads to infection and if these are isolated or frequently occurring transmissions.
MATERIALS AND METHODS
Plasmid constructs.
VP1 sequences of human and animal noroviruses were custom synthesized (IDT, Coralville, IA) and cloned into pCAGGS with EcoRI and XhoI restriction sites added to the 5′ and 3′ ends, respectively (Table 4). VP1s of GIV.1, GII.4, and GII.17 2015 were amplified from stool samples, and sequences were submitted to GenBank (accession numbers are listed in Table 4). RNA was isolated with the high-pure RNA isolation kit (Roche, Basel, Switzerland) and 5 μl of RNA, 1 μl (2 pmol) of random primers, 1 μl deoxynucleotide triphosphates (dNTPs; 10 mM each), 0.5 μl (20 U) RNase inhibitor. Samples were incubated for 5 min at 65°C and cooled down on ice for 5 min. 1× SuperScript IV RT buffer, 1 μl 0.1 M dithiothreitol, 0.5 μl (20 U) RNase inhibitor, and 1 μl (200 U) SuperScript IV RT buffer were added and incubated for 5 min at 25°C, 10 min at 50°C, and 10 min at 80°C. Five microliters of the resulting cDNA was used for specific VP1 amplification with 1.25 μl dNTPs, 1× Pfu buffer, 1 μl Pfu, and 10 pmol primers containing EcoRI and XhoI restriction sites (underlined): GIV.1, AAAGAATTCATGAAGATGGCGTCGAGTGA/TAGCTCGAGTTATTGAAACCTCACTCTAC; GII.4, GGAGAATTCTGAAGATGGCGTCGAGTGAC/GTTCTCGAGTTATAGTGCACGTCTACGCCCCGTTC; GII.17 2015, GGAGAATTCATGAAGATGGCGTCGAATGAC/GTTCTCGAGTTACTGAGCCCTCCTTCGCCCATT. The PCR protocol consisted of 2 min at 95°C and then 39 cycles of 30 s at 95°C, 1 min at 55°C, and 2.5 min at 72°C, followed by an elongation step of 6 min at 72°C. The PCR product was purified and ligated into the pCAGGs plasmid. The accuracy of the plasmids was checked by Sanger sequencing.
TABLE 4.
VP1 sequences used to produce VLPs
| Norovirus genotype (ORF2) | Accession no. |
|---|---|
| Human | |
| GI.3 | JQ911594 |
| GI.6 | LN854564 |
| GII.1 | LN854570 |
| GII.3 | LN854569 |
| GII.4 (Sydney 2012) | MT232050 |
| GII.6 | KJ407072 |
| GII.17 2005 | DQ438972 |
| GII.17 2014 (Kawasaki323) | AB983218 |
| GII.17 2015 | KX424646 |
| GIX (previously GII.15) | KJ196290 |
| GIV.1 | MT232232 |
| Animal | |
| GNA1 (porpoise) | KP987888 |
| GIV.2 (cat) | JF781268 |
| GIII.2 (cow) | AF320625 |
| GII.18 (pig) | AY823305 |
VLP production and FITC labeling.
VLPs were produced by adapting a previously published protocol (61, 62). Twenty-four hours prior to transfection, 3 × 106 293T cells were seeded in gelatinized 10-cm plates in Dulbecco’s modified Eagle’s medium (DMEM; Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO, USA), 1× nonessential amino acids (Lonza), 1× PenStrep (Lonza), 1× l-glutamine (Lonza), and 1× sodium pyruvate (Gibco, Waltham, MA, USA). Cells were kept at 37°C with 5% CO2. The pCAGGs-VP1 construct was transfected using calcium phosphate transfection. As a negative control, the pCAGGS construct without insert was transfected. For transfection, 6.2 μl CaCl2, 40 μl plasmid, and 400 μl sterile water were mixed, and 500 μl HEPES buffered saline solution (8.18% NaCl, 5.94% HEPES, and 0.2% Na2HPO4 [all wt/vol]) was added. After 5 min, the transfection mix was added to the cells and incubated at 37°C for 16 h. Cells were then washed with phosphate-buffered saline (PBS) and the medium was refreshed. After another 48 h, the cells were harvested and centrifuged at 1,000 rpm for 10 min. The pellet was resolved in lysis buffer (10% Triton X-100 with protease inhibitor [cOmplete Mini EDTA-free protease inhibitor cocktail; Roche]) and left for 5 min at room temperature. All subsequent centrifugation steps were done at 4°C. The lysate and supernatant were cleared by centrifugation at 3,000 rpm for 15 min. The supernatant was then centrifuged through a 20% (wt/wt) sucrose cushion for 2 h at 27,000 rpm (SW32 rotor). The pellet was dissolved in 1 ml PBS for 20 min at 4°C and subsequently centrifuged overnight through a 20% to 60% sucrose gradient at 30,000 rpm (SW41 rotor). Fractions corresponding to 35% to 45% (wt/wt) sucrose were concentrated and washed through a 100-kDa Amicon filter at 4,000 × g for 20 min at 4°C. The presence of VP1 was confirmed by SDS-PAGE. For GII.18, we experienced problems expressing VP1; therefore, we produced p-particles as described before (63). The presence of VLPs and p-particles was confirmed by electron microscopy. VLPs in PBS were stored at –80°C until further used. VLPs and p-particles were FITC labeled as previously described (31). Equal amounts of VLPs and 0.1 mg/ml FITC (Sigma) in 0.5 mol/liter bicarbonate buffer (pH 9.5) were mixed under constant stirring in the dark for 1 h. To lose excessive unbound FITC, the samples were dialyzed against PBS overnight in dialysis cassettes (GeBaFlex-Midi tubes, 8-kDa cutoff). VLPs were aliquoted and stored at –80°C until used. For quantification, FITC-labeled VLPs were run on a 12.5% acrylamide gel together with a bovine serum albumin (BSA) concentration marker. Silver staining was done with the silver staining kit (Pierce, Waltham, MA, USA) according to the manufacturer’s instructions.
Intestinal tissues.
Human intestinal tissues were obtained from the Pathology Research and Trial Service (PARTS) at Erasmus MC. The following archival FFPE sections were obtained from the Department of Virology, Erasmus MC: harbor porpoises (Phocoena phocoena, n = 3) that had been part of an unrelated study (64), chimpanzees (Pan troglodytes, n = 2), domestic cat (Felis catus, n = 1), which had been the negative-control animal in an unrelated animal study (65), chicken (Gallus gallus domesticus, n = 2), which were the negative-control animals in an unrelated study (66), wild mallards (Anas platyrhynchos, n = 3), black-headed gulls (Chroicocephalus ridibundus, n = 4) that were from experiments with wild animals that had been published previously (67), common pipistrelles (Pipistrellus pipistrellus, microbat, n = 4), Egyptian fruit bats (Rousettus aegyptiacus, megabat, n = 3), and straw-colored fruit bats (Eidolon Helvum, megabat, n = 3). The bat samples were used in an unrelated study (68). The rats (Rattus norvegicus n = 3) were published in an unrelated study (69). The pacific oysters (Crassostrea gigas, n = 4) were collected from a market. Domestic pigs (Sus scrofa domesticus, n = 4), dogs (Canis lupus familiaris, n = 3), and turkey (Meleagris gallopavo, n = 1) had been used for unrelated purposes. If available, we included three healthy individuals per species.
HBGA typing by immunohistochemistry.
FFPE tissues were obtained from the Department of Pathology at Viroscience Erasmus MC. Three-micrometer FFPE tissue slides were deparaffinized with xylene and hydrated using a graded ethanol series (100 > 100 > 95 > 90 > 70%). HBGA expression was assessed by immunohistochemistry (IHC). To block endogenous peroxidase, slides were incubated with 3% H2O2 diluted in PBS at room temperature (RT) for 10 min. All antibody incubation steps were conducted in 0.1% BSA for 1 h at RT with two washing steps (PBS plus 0.01% Tween) in between. HBGAs were detected with primary antibodies against antigen A (1:1; 9113D10; Diagast, Loos, France), B (1:1; 9621A8; Diagast), AB (1:1; 9113D10 + 152D12; Diagast), Neg (Diagast), Ley (1:50; H18A; Absolute Antibody), Lea (1:50; 7-LE; Sigma), Leb (1:50; 2-25LE; Sigma), Lex (1:100; MC480; Thermo Fisher, Waltham, MA), and H1 (1:100; 17-206; Thermo Fisher). A secondary biotinylated rabbit anti-mouse (1:100; Dako, Glostrup, Denmark) was used, followed by Streptavidin-horseradish peroxidase (HRP)-conjugated antibody (1:300; Dako). Peroxidase was revealed with 3-amino-9-ethyl-carbazole (Sigma). Tissues were counterstained with hematoxylin and embedded in Meyer’s glycerol-gelatin (Merck, Darmstadt, Germany).
H type 2 was detected by lectin staining. Slides were blocked with 1% BSA in Tris-buffered saline (TBS; 50 mM Tris-HCl, pH 7.5), and biotin-labeled Ulex europeus lectin (1:200 UEA-I; Sigma) was added in TBS with 1 mM MgCl2, 1 mM MnCl2, 1 mM CaCl2, pH 7.5, and incubated overnight at 4°C. Slides were washed with TBS, and Streptavidin-HRP diluted in 1% BSA in TBS was added for 1 h at RT. The rest was done as described above.
Virus histochemistry on tissue sections.
Tissue slides were prepared as for IHC (described above). For all blocking and antibody steps, TNB (0.1 M Tris, 0.15 M NaCl, pH 7.5, with 0.5% blocking reagent [Perkin Elmer, Boston MA]) was used. Slides were blocked for 30 min at RT, and 25 ng of VLP or negative control was added and incubated overnight at 4°C. Between all subsequent steps, slides were washed twice with 0.01% Tween 20 in PBS, and all incubation steps were done at RT. Virus was detected by peroxidase-labeled rabbit anti-FITC (1:100; Dako) for 1 h. The signal was amplified using a tyramide signal amplification system (Perkin Elmer, Boston, MA) according to the manufacturer’s instructions. Streptavidin-HRP was added at 1:300 and incubated for 30 min. The HRP revelation was done as described above for IHC. As a control, unlabeled VLPs were used and stained with a primary anti-GII.4 antibody (1:100; SMV59; Maine Biotechnology, Portland, ME, USA). The rest of the steps were done as described above for the IHC. The treatment with the 1,2α-fucosidase (kindly provided by Takane Katayama, Kyoto Unversity, Japan) was done prior to incubation with VLPs or the lectin staining. Twenty micrograms was added per slide in 100 mM sodium phosphate buffer, pH 6.5, overnight at 37°C.
Enzyme-linked immunosorbent assay (ELISA)-based carbohydrate microtiter plate assays.
Streptavidin-coated high-capacity plates (Pierce, Invitrogen) were coated overnight at 4°C with 10 μg/ml biotinylated synthetic oligosaccharides that were polyacrylamide (PAA) conjugated and that were eluted in 1× TBS buffer (20 mM Tris, 150 mM NaCl, pH 7.2). The carbohydrates were ordered at GlycoTech (Gaithersburg, MD, USA), Carbosynth (Berkshire, UK), and GlycoNZ (Auckland, New Zealand) and are listed in Table 5. Plates were washed five times with cold PBS and blocked at RT with 5% BSA in PBS. After blocking and between all subsequent incubation steps, plates were washed five times with 0.01% Tween 20 in PBS (PBS-T). One hundred nanograms of FITC-labeled or unlabeled VLPs in 0.1% BSA in PBS-T was added and incubated overnight at 4°C. FITC-labeled VLPs were detected with an HRP-conjugated anti-FITC antibody (1:1,000; Dako) for 1 h at 4°C. The unlabeled VLPs were detected with a mouse anti-GII.4 (1:100; SMV59; Maine Biotechnology, Portland, ME), followed by an anti-mouse-HRP antibody (1:100; Dako). The peroxidase signal was detected using the TMB 2-component microwell peroxidase substrate kit (SeraCare, Milford, MA, USA). The reaction was stopped after 10 min with 3 M H2SO4, and the mean absorbance values, i.e., the optical densities at 450 nm (OD450), were measured, followed by subtraction of the negative-control values (d-mannose). Each VLP was tested in duplicate.
TABLE 5.
Synthetic HBGA structures used for the binding assay
| Name | Structurea | Category | Manufacturer |
|---|---|---|---|
| Led (H type 1)-PAA-biotin | Fucα1-2Galβ1-3GlcNAcβ1 | Trisaccharide | Glycosynth |
| H (type 2)-PAA-biotin | Fuca1-2Galβ1-4GlcNAcβ1 | Trisaccharide | Glycotech |
| H (type 3)-PAA-biotin | Fucα1-2Galβ1-3GalNAcα1 | Disaccharide | Glycotech |
| Blood type A (tri)-PAA biotin | GalNAcα1-3(Fucα1-2)Galβ1 | Trisaccharide | Glycotech |
| Blood type B (tri)-PAA biotin | Galα1-3(Fucα1-2)Galβ1 | Trisaccharide | Glycotech |
| Lea-PAA-biotin | Galβ1-3(Fucα1-4)GlcNAcβ | Trisaccharide | Glycotech |
| Leb-PAA-biotin | Fucα1-2Galβ1-3(Fucα1-4)GlcNAcβ | Tetrasaccharide | Glycotech |
| Lex-PAA-biotin | Galβ1-4(Fucα1-3)GlcNAcβ1 | Trisaccharide | Glycotech |
| Ley-PAA-biotin | Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1 | Tetrasaccharide | Glycotech |
| α-d-Mannose-PAA-biotin | α-d-Man | GlycoNZ |
Glc, glucose; Fuc, fucose; Gal, galactose; GlcNac, N-acetylglucosamine; Lac, lactose; GalNac, N-acetylgalactosamine; Man, mannose.
Data availability.
Sequences determined in the course of this work were submitted to GenBank (accession numbers are listed in Table 4).
ACKNOWLEDGMENTS
We thank Thijs Kuiken, Lineke Begeman, Josanne Verhagen, Judith van den Brand, Peter van Run, and Niels van Elk for providing the formalin-fixed intestinal tissues and for their technical assistance. We thank the Pathology Research and Trial Service (PARTS) at Erasmus MC for providing the human intestinal tissues. For the R. aegyptiacus and E. helvum tissues, we thank Andrew A. Cunningham from the Institute of Zoology, Zoological Society of London. We thank Stichting Vleermuisopvang Oss for providing the tissues of P. pipistrellus. We thank Sofia Strubbia and Soizick Le Guyader for providing excellent technical assistance with the oyster samples, Jacques Le Pendu for advice with the HBGA antibodies, and lectins and Takane Katayama for providing the fucosidase. We also thank Monique Spronken for her technical help with the ELISA-based assay.
This work was supported by European Union’s Horizon 2020 research and innovation program under grant agreement no. 643476 (COMPARE) and ZonMW TOP project 91213058.
REFERENCES
- 1.Chhabra P, de Graaf M, Parra GI, Chan MC-W, Green K, Martella V, Wang Q, White PA, Katayama K, Vennema H, Koopmans MPG, Vinje J. 2019. Updated classification of norovirus genogroups and genotypes. J Gen Virol 100:jgv001318. doi: 10.1099/jgv.0.001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villabruna N, Koopmans M, De Graaf M. 2019. Animals as reservoir for human norovirus. Viruses 11:478. doi: 10.3390/v11050478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattison K, Shukla A, Cook A, Pollari F, Friendship R, Kelton D, Bidawid S, Farber JM. 2007. Human noroviruses in swine and cattle. Emerg Infect Dis 13:1184–1188. doi: 10.3201/eid1308.070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sisay Z, Djikeng A, Berhe N, Belay G, Abegaz WE, Wang QH, Saif LJ. 2016. First detection and molecular characterization of sapoviruses and noroviruses with zoonotic potential in swine in Ethiopia. Arch Virol 161:2739–2747. doi: 10.1007/s00705-016-2974-9. [DOI] [PubMed] [Google Scholar]
- 5.Taku O, Iweriebor BC, Nwodo UU, Obi LC, Okoh AI, SAMRC Microbial Water Quality Monitoring Centre. 2017. Occurrence of norovirus in pig faecal samples in the Eastern Cape, South Africa. Asian Pacific J Trop Dis 7:151–155. doi: 10.12980/apjtd.7.2017D6-393. [DOI] [Google Scholar]
- 6.Nakamura K, Saga Y, Iwai M, Obara M, Horimoto E, Hasegawa S, Kurata T, Okumura H, Nagoshi M, Takizawa T. 2010. Frequent detection of noroviruses and sapoviruses in swine and high genetic diversity of porcine sapovirus in Japan during fiscal year 2008. J Clin Microbiol 48:1215–1222. doi: 10.1128/JCM.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao DY, Wei JY, Chang WF, Wang J, Wang LC. 2012. Detection of multiple genotypes of calicivirus infection in asymptomatic swine in Taiwan. Zoonoses Public Health 59:434–444. doi: 10.1111/j.1863-2378.2012.01483.x. [DOI] [PubMed] [Google Scholar]
- 8.Summa M, Henttonen H, Maunula L. 2018. Human noroviruses in the faeces of wild birds and rodents-new potential transmission routes. Zoonoses Public Health 65:512–518. doi: 10.1111/zph.12461. [DOI] [PubMed] [Google Scholar]
- 9.Farkas T. 2016. Natural norovirus infections in rhesus macaques. Emerg Infect Dis 22:1272–1274. doi: 10.3201/eid2207.151740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Z, Liu B, Tao Y, Li C, Xia M, Zhong W, Jiang X, Liu H, Tan M. 2017. Norovirus GII.17 natural infections in Rhesus monkeys, China. Emerg Infect Dis 23:316–319. doi: 10.3201/eid2302.161077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farkas T, Cross RW, Hargitt IE, Lerche NW, Morrow AL, Sestak K. 2010. Genetic diversity and histo-blood group antigen interactions of rhesus enteric caliciviruses. J Virol 84:8617–8625. doi: 10.1128/JVI.00630-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Tao Y, Li C, Li X, Liu J, He Z, Xia M, Jiang X, Tan M, Liu H. 2016. Complete genome sequence of a GII.17 norovirus isolated from a rhesus monkey in China. Genome Announc 4:e00904-16. doi: 10.1128/genomeA.00904-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolf S, Reetz J, Johne R, Heiberg AC, Petri S, Kanig H, Ulrich RG. 2013. The simultaneous occurrence of human norovirus and hepatitis E virus in a Norway rat (Rattus norvegicus). Arch Virol 158:1575–1578. doi: 10.1007/s00705-013-1646-2. [DOI] [PubMed] [Google Scholar]
- 14.Summa M, von Bonsdorff CH, Maunula L. 2012. Pet dogs–a transmission route for human noroviruses? J Clin Virol 53:244–247. doi: 10.1016/j.jcv.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Todd KV, Tripp RA. 2019. Human norovirus: experimental models of infection. Viruses 11:151. doi: 10.3390/v11020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang XI, Lindblad L, Stewart P, LePendu J, Baric R. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat Med 9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 17.Hutson AM, Atmar RL, Graham DY, Estes MK. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis 185:1335–1337. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- 18.Thorne L, Nalwoga A, Mentzer AJ, de Rougemont A, Hosmillo M, Webb E, Nampiija M, Muhwezi A, Carstensen T, Gurdasani D, Hill AV, Sandhu MS, Elliott A, Goodfellow I. 2018. The first norovirus longitudinal seroepidemiological study from sub-Saharan Africa reveals high seroprevalence of diverse genotypes associated with host susceptibility factors. J Infect Dis 218:716–725. doi: 10.1093/infdis/jiy219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marionneau S, Ruvoën N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravn V, Dabelsteen E. 2000. Tissue distribution of histo-blood group antigens. APMIS 108:1–28. doi: 10.1034/j.1600-0463.2000.d01-1.x. [DOI] [PubMed] [Google Scholar]
- 21.Marionneau S, Cailleau-Thomas A, Rocher J, Le Moullac-Vaidye B, Ruvoën N, Monique C, Le Pendu J. 2001. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83:565–573. doi: 10.1016/S0300-9084(01)01321-9. [DOI] [PubMed] [Google Scholar]
- 22.Tan M, Jiang X. 2005. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol 13:285–293. doi: 10.1016/j.tim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Caddy S, Breiman A, Le Pendu J, Goodfellow I. 2014. Genogroup IV and VI canine noroviruses interact with histo-blood group antigens. J Virol 88:10377–10391. doi: 10.1128/JVI.01008-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kocher JF, Lindesmith LC, Debbink K, Beall A, Mallory ML, Yount BL, Graham RL, Huynh J, Gates JE, Donaldson EF, Baric RS. 2018. Bat caliciviruses and human noroviruses are antigenically similar and have overlapping histo-blood group antigen binding profile. mBio 9:e00869-18. doi: 10.1128/mBio.00869-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zakhour M, Ruvoën-Clouet N, Charpilienne A, Langpap B, Poncet D, Peters T, Bovin N, Le Pendu J. 2009. The αGal epitope of the histo-blood group antigen family is a ligand for bovine norovirus Newbury2 expected to prevent cross-species transmission. PLoS Pathog 5:e1000504. doi: 10.1371/journal.ppat.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haga K, Fujimoto A, Takai-Todaka R, Miki M, Doan YH, Murakami K, Yokoyama M, Murata K, Nakanishi A, Katayama K. 2016. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc Natl Acad Sci U S A 113:E6248–E6255. doi: 10.1073/pnas.1605575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilic T, Koromyslova A, Malak V, Hansman GS. 2018. Atomic structure of the murine norovirus protruding domain and sCD300lf receptor complex. J Virol 92:e00413-18. doi: 10.1128/JVI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orchard RC, Wilen CB, Doench JG, Baldridge MT, McCune BT, Lee YCJ, Lee S, Pruett-Miller SM, Nelson CA, Fremont DH, Virgin HW. 2016. Discovery of a proteinaceous cellular receptor for a norovirus. Science 353:933–936. doi: 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guix S, Asanaka M, Katayama K, Crawford SE, Neill FH, Atmar RL, Estes MK. 2007. Norwalk virus RNA is infectious in mammalian cells. J Virol 81:12238–12248. doi: 10.1128/JVI.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordgren J, Svensson L. 2019. Genetic susceptibility to human norovirus infection: an update. Viruses 11:226. doi: 10.3390/v11030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Riel D, Munster VJ, De Wit E, Rimmelzwaan GF, Fouchier RAM, Osterhaus ADME, Kuiken T. 2006. H5N1 virus attachment to lower respiratory tract. Science 312:399–399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 32.Widagdo W, Okba NMA, Li W, de Jong A, de Swart RL, Begeman L, van den Brand JMA, Bosch B-J, Haagmans BL. 2019. Species-specific colocalization of Middle East respiratory syndrome coronavirus attachment and entry receptors. J Virol 93:e00107-19. doi: 10.1128/JVI.00107-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang XI, Wang MIN, Graham DY, Estes MK. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol 66:6527–6532. doi: 10.1128/JVI.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol 81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costantini V, Loisy F, Joens L, Le Guyader FS, Saif LJ. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Appl Environ Microbiol 72:1800–1809. doi: 10.1128/AEM.72.3.1800-1809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zakhour M, Maalouf H, Di Bartolo I, Haugarreau L, Le Guyader FS, Ruvoën-Clouet N, Le Saux JC, Ruggeri FM, Pommepuy M, Le Pendu J. 2010. Bovine norovirus: carbohydrate ligand, environmental contamination, and potential cross-species transmission via oysters. Appl Environ Microbiol 76:6404–6411. doi: 10.1128/AEM.00671-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyström K, Le Gall-Reculé G, Grassi P, Abrantes J, Ruvoën-Clouet N, Le Moullac-Vaidye B, Lopes AM, Esteves PJ, Strive T, Marchandeau S, Dell A, Haslam SM, Le Pendu J. 2011. Histo-blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain-dependent manner. PLoS Pathog 7:e1002188. doi: 10.1371/journal.ppat.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marionneau S, Airaud F, Bovin NV, Le Pendu J, Ruvoën-Clouet N. 2005. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J Infect Dis 192:1071–1077. doi: 10.1086/432546. [DOI] [PubMed] [Google Scholar]
- 39.Charoenkul K, Nasamran C, Janetanakit T, Tangwangvivat R, Bunpapong N, Boonyapisitsopa S, Suwannakarn K, Theamboonler A, Chuchaona W, Poovorawan Y, Amonsin A. 2020. Human norovirus infection in dogs, Thailand. Emerg Infect Dis 26:350–353. doi: 10.3201/eid2602.191151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A Duarte M, Silva F, João M, R Brito C, S Teixeira D, L Melo F, M Ribeiro B, Nagata T, S Campos F. 2019. Faecal virome analysis of wild animals from Brazil. Viruses 11:803. doi: 10.3390/v11090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karandikar UC, Crawford SE, Ajami NJ, Murakami K, Kou B, Ettayebi K, Papanicolaou GA, Jongwutiwes U, Perales MA, Shia J, Mercer D, Finegold MJ, Vinje J, Atmar RL, Estes MK. 2016. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J Gen Virol 97:2291–2300. doi: 10.1099/jgv.0.000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan MCW, Ho WS, Sung JJY. 2011. In vitro whole-virus binding of a norovirus genogroup II genotype 4 strain to cells of the lamina propria and Brunner's glands in the human duodenum. J Virol 85:8427–8430. doi: 10.1128/JVI.05016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bok K, Parra GI, Mitra T, Abente E, Shaver CK, Boon D, Engle R, Yu C, Kapikian AZ, Sosnovtsev SV, Purcell RH, Green KY. 2011. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc Natl Acad Sci U S A 108:325–330. doi: 10.1073/pnas.1014577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo DJ, Jung D, Jung S, Ha SK, Ha SD, Choi IS, Myoung J, Choi C. 2018. Experimental miniature piglet model for the infection of human norovirus GII. J Med Virol 90:655–662. doi: 10.1002/jmv.24991. [DOI] [PubMed] [Google Scholar]
- 46.Park BJ, Jung ST, Choi CS, Myoung J, Ahn HS, Han SH, Kim YH, Go HJ, Lee JB, Park SY, Song CS, Lee SW, Choi IS. 2018. Pathogenesis of human norovirus genogroup II genotype 4 in postweaning gnotobiotic pigs. J Microbiol Biotechnol 28:2133–2140. doi: 10.4014/jmb.1810.09061. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto F, Cid E, Yamamoto M, Saitou N, Bertranpetit J, Blancher A. 2014. An integrative evolution theory of histo-blood group ABO and related genes. Sci Rep 4:6601. doi: 10.1038/srep06601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang P, Farkas T, Marionneau S, Zhong W, Ruvoën-Clouet N, Morrow AL, Altaye M, Pickering LK, Newburg DS, LePendu J, Jiang X. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo–blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis 188:19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- 49.Choi JM, Hutson AM, Estes MK, Prasad BVV. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc Natl Acad Sci U S A 105:9175–9180. doi: 10.1073/pnas.0803275105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Beek J, de Graaf M, Al-Hello H, Allen DJ, Ambert-Balay K, Botteldoorn N, Brytting M, Buesa J, Cabrerizo M, Chan M, Cloak F, Di Bartolo I, Guix S, Hewitt J, Iritani N, Jin M, Johne R, Lederer I, Mans J, Martella V, Maunula L, McAllister G, Niendorf S, Niesters HG, Podkolzin AT, Poljsak-Prijatelj M, Rasmussen LD, Reuter G, Tuite G, Kroneman A, Vennema H, Koopmans MPG. 2018. NoroNet. Molecular surveillance of norovirus, 2005–16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis 18:545–553. doi: 10.1016/S1473-3099(18)30059-8. [DOI] [PubMed] [Google Scholar]
- 51.La Rosa G, Iaconelli M, Pourshaban M, Fratini M, Muscillo M. 2010. Molecular detection and genetic diversity of norovirus genogroup IV: a yearlong monitoring of sewage throughout Italy. Arch Virol 155:589–593. doi: 10.1007/s00705-010-0619-y. [DOI] [PubMed] [Google Scholar]
- 52.Maalouf H, Schaeffer J, Parnaudeau S, Le Pendu J, Atmar RL, Crawford SE, Le Guyader FS. 2011. Strain-dependent norovirus bioaccumulation in oysters. Appl Environ Microbiol 77:3189–3196. doi: 10.1128/AEM.03010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan M, Xia M, Chen Y, Bu W, Hegde RS, Meller J, Li X, Jiang X. 2009. Conservation of carbohydrate binding interfaces–evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. doi: 10.1371/journal.pone.0005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farkas T, Nakajima S, Sugieda M, Deng X, Zhong W, Jiang X. 2005. Seroprevalence of noroviruses in swine. J Clin Microbiol 43:657–661. doi: 10.1128/JCM.43.2.657-661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh BK, Glatt S, Ferrer JL, A DK, Leuthold MM, Dunder J, Hansman GS. 2015. Structural analysis of a feline norovirus protruding domain. Virology 474:181–185. doi: 10.1016/j.virol.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y, Xia M, Wang L, Arumugam S, Wang Y, Ou X, Wang C, Jiang X, Tan M. 2019. Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses. Emerg Microbes Infect 8:1642–1657. doi: 10.1080/22221751.2019.1686335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pogan R, Dülfer J, Uetrecht C. 2018. Norovirus assembly and stability. Curr Opin Virol 31:59–65. doi: 10.1016/j.coviro.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Tamura M, Natori K, Kobayashi M, Miyamura T, Takeda N. 2004. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J Virol 78:3817–3826. doi: 10.1128/jvi.78.8.3817-3826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almand EA, Moore DM, Jaykus LA. 2017. Norovirus binding to ligands beyond histo-blood group antigens. Front Microbiol 8:2549. doi: 10.3389/fmicb.2017.02549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rydell GE, Nilsson J, Rodriguez-Diaz J, Ruvoen-Clouet N, Svensson L, Le Pendu J, Larson G. 2009. Human noroviruses recognize sialyl Lewis x neoglycoprotein. Glycobiology 19:309–320. doi: 10.1093/glycob/cwn139. [DOI] [PubMed] [Google Scholar]
- 61.de Graaf M, Bodewes R, van Elk CE, van de Bildt M, Getu S, Aron GI, Verjans GMGM, Osterhaus ADME, van den Brand JMA, Kuiken T, Koopmans MPG. 2017. Norovirus infection in harbor porpoises. Emerg Infect Dis 23:87–91. doi: 10.3201/eid2301.161081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taube S, Kurth A, Schreier E. 2005. Generation of recombinant norovirus-like particles (VLP) in the human endothelial kidney cell line 293T. Arch Virol 150:1425–1431. doi: 10.1007/s00705-005-0517-x. [DOI] [PubMed] [Google Scholar]
- 63.Tan M, Hegde RS, Jiang X. 2004. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J Virol 78:6233–6242. doi: 10.1128/JVI.78.12.6233-6242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Elk CE, van de Bildt MWG, van Run PRWA, Bunskoek P, Meerbeek J, Foster G, Osterhaus ADME, Kuiken T. 2019. Clinical, pathological, and laboratory diagnoses of diseases of harbour porpoises (Phocoena phocoena), live stranded on the Dutch and adjacent coasts from 2003 to 2016. Vet Res 50:88. doi: 10.1186/s13567-019-0706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reperant LA, van de Bildt MWG, van Amerongen G, Leijten LME, Watson S, Palser A, Kellam P, Eissens AC, Frijlink HW, Osterhaus ADME, Kuiken T. 2012. Marked endotheliotropism of highly pathogenic avian influenza virus H5N1 following intestinal inoculation in cats. J Virol 86:1158–1165. doi: 10.1128/JVI.06375-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richard M, Herfst S, van den Brand JMA, de Meulder D, Lexmond P, Bestebroer TM, Fouchier RAM. 2017. Mutations driving airborne transmission of A/H5N1 virus in mammals cause substantial attenuation in chickens only when combined. Sci Rep 7:1–13. doi: 10.1038/s41598-017-07000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verhagen JH, Höfle U, van Amerongen G, van de Bildt M, Majoor F, Fouchier RAM, Kuiken T. 2015. Long-term effect of serial infections with H13 and H16 low-pathogenic avian influenza viruses in black-headed gulls. J Virol 89:11507–11522. doi: 10.1128/JVI.01765-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Widagdo W, Begeman L, Schipper D, van Run PR, Cunningham AA, Kley N, Reusken CB, Haagmans BL, van den Brand JMA. 2017. Tissue distribution of the MERS-coronavirus receptor in bats. Sci Rep 7:1–8. doi: 10.1038/s41598-017-01290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maas M, van Heteren M, de Vries A, Kuiken T, Hoornweg T, Veldhuis Kroeze E, Rockx B. 2019. Seoul virus tropism and pathology in naturally infected feeder rats. Viruses 11:531. doi: 10.3390/v11060531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian P, Jiang X, Zhong W, Jensen HM, Brandl M, Bates AH, Engelbrektson AL, Mandrell R. 2007. Binding of recombinant norovirus like particle to histo-blood group antigen on cells in the lumen of pig duodenum. Res Vet Sci 83:410–418. doi: 10.1016/j.rvsc.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 71.McKibbin JM, Spencer WA, Smith EL, Mansson JE, Karlsson KA, Samuelsson BE, Li YT, Li SC. 1982. Lewis blood group fucolipids and their isomers from human and canine intestine. J Biol Chem 257:755–760. [PubMed] [Google Scholar]
- 72.Iwamoto S, Kumada M, Kamesaki T, Okuda H, Kajii E, Inagaki T, Saikawa D, Takeuchi K, Ohkawara S, Takahashi R, Ueda S, Inoue S, Tahara K, Hakamata Y, Kobayashi E. 2002. Rat encodes the paralogous gene equivalent of the human histo-blood group ABO gene association with antigen expression by overexpression of human ABO transferase. J Biol Chem 277:46463–46469. doi: 10.1074/jbc.M206439200. [DOI] [PubMed] [Google Scholar]
- 73.Oriol R, Le Pendu J, Mollicone R. 1986. Genetics of ABO, H, Lewis, X and related antigens. Vox Sang 51:161–171. doi: 10.1111/j.1423-0410.1986.tb01946.x. [DOI] [PubMed] [Google Scholar]
- 74.Kim YS, Perdomo J. 1972. Glycoprotein biosynthesis in small intestine. III. Enzymatic basis for the difference in the antigenicity of mucins. J Clin Investig 51:1135–1145. doi: 10.1172/JCI106906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiener AS, Moor‐Jankowski J, Gordon EB. 1963. Blood groups of apes and monkeys. II. The A‐B‐O blood groups, secretor and Lewis types of apes. Am J Phys Anthropol 21:271–281. doi: 10.1002/ajpa.1330210303. [DOI] [PubMed] [Google Scholar]
- 76.Morozov V, Hanisch FG, Wegner KM, Schroten H. 2018. Pandemic GII.4 Sydney and epidemic GII.17 Kawasaki308 noroviruses display distinct specificities for histo-blood group antigens leading to different transmission vector dynamics in Pacific oysters. Front Microbiol 9:2826. doi: 10.3389/fmicb.2018.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian P, Engelbrektson AL, Jiang X, Zhong W, Mandrell RE. 2007. Norovirus recognizes histo-blood group antigens on gastrointestinal cells of clams, mussels, and oysters: a possible mechanism of bioaccumulation. J Food Prot 70:2140–2147. doi: 10.4315/0362-028x-70.9.2140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences determined in the course of this work were submitted to GenBank (accession numbers are listed in Table 4).