Globally, respiratory syncytial virus (RSV) is a major cause of death in children under 1 year of age, yet no vaccine is available. We have generated a novel RSV live attenuated vaccine candidate containing mutations in the L and G proteins.
KEYWORDS: RSV, animal models, cotton rat, live attenuated vaccine, respiratory syncytial virus, rhesus macaque, vaccines
ABSTRACT
Respiratory syncytial virus (RSV) is the leading cause of acute lower respiratory tract infections in children of <5 years of age worldwide, infecting the majority of infants in their first year of life. Despite the widespread impact of this virus, no vaccine is currently available. For more than 50 years, live attenuated vaccines (LAVs) have been shown to protect against other childhood viral infections, offering the advantage of presenting all viral proteins to the immune system for stimulation of both B and T cell responses and memory. The RSV LAV candidate described here, rgRSV-L(G1857A)-G(L208A), contains two modifications: an attenuating mutation in the S-adenosylmethionine (SAM) binding site of the viral mRNA cap methyltransferase (MTase) within the large (L) polymerase protein and a mutation in the attachment (G) glycoprotein that inhibits its cleavage during production in Vero cells, resulting in virus with a “noncleaved G” (ncG). RSV virions containing the ncG have an increased ability to infect primary well-differentiated human bronchial epithelial (HBE) cultures which model the in vivo site of immunization, the ciliated airway epithelium. This RSV LAV candidate is produced efficiently in Vero cells, is highly attenuated in HBE cultures, efficiently induces neutralizing antibodies that are long lasting, and provides protection against an RSV challenge in the cotton rat, without causing enhanced disease. Similar results were obtained in a rhesus macaque.
IMPORTANCE Globally, respiratory syncytial virus (RSV) is a major cause of death in children under 1 year of age, yet no vaccine is available. We have generated a novel RSV live attenuated vaccine candidate containing mutations in the L and G proteins. The L polymerase mutation does not inhibit virus yield in Vero cells, the cell type required for vaccine production, but greatly reduces virus spread in human bronchial epithelial (HBE) cultures, a logical in vitro predictor of in vivo attenuation. The G attachment protein mutation reduces its cleavage in Vero cells, thereby increasing vaccine virus yield, making vaccine production more economical. In cotton rats, this RSV vaccine candidate is highly attenuated at a dose of 105 PFU and completely protective following immunization with 500 PFU, 200-fold less than the dose usually used in such studies. It also induced long-lasting antibodies in cotton rats and protected a rhesus macaque from RSV challenge. This mutant virus is an excellent RSV live attenuated vaccine candidate.
INTRODUCTION
Worldwide, respiratory syncytial virus (RSV) is the second leading cause of death in children between 1 month and 1 year of age and the most common cause of hospitalization in infants. Nearly all children have experienced RSV infection at least once by 2 years of age (1, 2). In infants, the elderly, and immunocompromised individuals, RSV infections often present as acute lower respiratory tract infection, which may progress to severe disease requiring intensive care unit management. In infants, many cases of advanced RSV infection manifest as a bronchiolitis associated clinically with coughing, wheezing, dyspnea, and hypoxia (3, 4). Currently, the only available, FDA-approved preventive therapy for RSV infection is a monoclonal antibody directed to the F protein, palivizumab (Synagis; MedImmune) (5, 6), which is costly and reserved for high-risk infants. In the early 1960s, two cohorts of RSV-naive children were immunized with a formalin-inactivated vaccine candidate (FI-RSV) or placebo in a clinical trial. Following natural exposure to RSV, 69% to 80% of the FI-RSV-immunized children developed vaccine-enhanced disease, resulting in numerous hospitalizations and two deaths (7, 8). The outcome of these clinical trials shifted the focus to a live attenuated vaccine (LAV) approach for RSV.
As a member of the Pneumoviridae family, RSV is a negative-sense nonsegmented (NNS) RNA virus that expresses three surface glycoproteins: the attachment (G), fusion (F), and small hydrophobic (SH) proteins. The G protein is responsible for attaching the virion to a target cell, and the F protein is responsible for fusing the virion membrane with the membrane of the target cell. The G protein is a type II membrane protein with extensive posttranslational modifications, including 4 N-linked glycans and an estimated 35 O-linked glycans, which increase the apparent weight of the protein from 33 kDa to 90 kDa when the virus is grown in immortalized cell cultures (9). Among the World Health Organization-approved cell lines for vaccine virus production, RSV grows to the highest titers in Vero, or African green monkey kidney, cells. This is likely due to the lack of type I interferon (IFN) production in these cultures (10). As such, all RSV vaccine candidates to date have been produced in Vero cultures (11–14).
The RSV large (L) polymerase protein is responsible for transcribing mRNA and replicating the viral genome, making it a major target for attenuating mutations. Modifications of mRNA include 5′ cap addition, methylation of the cap at guanine N-7 (G-N-7) and ribose 2′-O (2′-O) positions, and polyadenylation of the 3′ tail (15). The L protein includes a series of conserved regions (CR) I to VI. The KDKE motif in CR-VI is the methyltransferase (MTase) core, which catalyzes the addition of methyl groups to the mRNA cap at G-N-7 and 2′-O. The GxGxG motif in the same region binds S-adenosylmethionine (SAM), the methyl donor for mRNA cap MTase actions (16, 17). In NNS RNA viruses, a single MTase site is responsible for both G-N-7 and ribose 2′-O MTases, which share the same SAM-binding site (18, 19). Previous work in another NNS RNA virus, vesicular stomatitis virus (VSV), demonstrated that mutations in the KDKE motif impede both G-N-7 and 2′-O methylation, resulting in recombinant VSVs that are considerably more attenuated than those with mutations in the SAM-binding site. The KDKE mutant viruses were overattenuated and displayed poor growth rates in cell culture (16). However, mutations in the VSV SAM-binding site produced virus that grows to high titers in immortalized cell cultures despite significant attenuation in vivo, likely because mutations in the SAM-binding site reduced, but did not completely prevent, MTase activity (20, 21). Within the last decade, it was discovered that ribose 2′-O methylation of the mRNA cap protects the viral RNA from decapping and degradation while also acting as the molecular signature for discerning self versus non-self RNA by the host innate immune system (18). In the RSV LAV candidate evaluated here, we incorporated a point mutation in the L protein SAM-binding site (G1857A), which should reduce viral mRNA cap methylation, thereby reducing RNA stability, viral protein expression, and virus production.
RSV virions produced in HEp-2 cells contain the full-length, heavily glycosylated 90-kDa G protein, while the same virus grown in Vero cells contains a truncated (55 kDa) G protein (22). In Vero cells, the G protein is cleaved, likely by cathepsin L, between amino acids 209 and 210 (23). Vero-grown RSV, with its truncated G protein, is 5- to 10-fold less infectious in primary human bronchial epithelial (HBE) cell cultures than the same virus produced in HeLa or HEp-2 cells, where the virion contains the 90-kDa full-length G (23). Mutagenesis in and around the cleavage site identified a single residue whose substitution to alanine, G(L208A), prevents cleavage of G. This “noncleaved G” (ncG) mutation enhanced the infectivity of Vero-grown virus for HBE cultures by 5-fold (23). Increasing the initial infectivity of an RSV LAV candidate should reduce the number of virions needed for protection and, thus, reduce the cost of vaccine production.
Here, we have combined these two mutations in one recombinant green fluorescent protein (GFP)-expressing virus, rgRSV-G(L208A)-L(G1857A), and compared its immunogenicity to viruses with each mutation alone and with the parental virus. For simplicity, we will use the shortened name of “rgRSV-G/L” here. Although the L MTase mutation slows virus production in Vero cells, it reaches similar titers to the parental virus (rgRSV) by 72 h. In HBE cultures and in animals, the virus spread is dramatically reduced. Nevertheless, the immunogenicity and protection offered by rgRSV-G/L in vivo is retained. The ncG mutation increases the initial infectivity of the virus in HBE cultures. The result is a highly attenuated, highly immunogenic RSV LAV candidate that can be produced efficiently.
RESULTS
Properties of RSV with mutations in the L protein that attenuate and in the G protein that inhibit cleavage.
Recombinant GFP-expressing RSV (rgRSV) was modified by a single mutation in the L protein SAM-binding site, designated rgRSV-L(G1857A) and shortened to “rgRSV-L” for clarity. The rgRSV with an ncG mutation that prevents G protein cleavage during production in Vero cells (23), rgRSV-G(L208A), is shortened to “rgRSV-G.” The combination mutant, rgRSV-L(G1857A)-G(L208A) is shortened to “rgRSV-G/L.” The locations of these mutations in the RSV genome are shown in Fig. 1.
FIG 1.
Schematic illustrating the two mutations in rgRSV-G/L. (A) Within the second mucin-like domain in the attachment (G) protein, the L208 near the purported cleavage site of the G protein (between amino acids 209 and 210) is modified from leucine to alanine, G(L208A), which inhibits cleavage of the protein during its production in Vero cells, enhancing its ability to infect HBE cultures (23). In addition to the two mucin-like domains, the G protein contains a cytoplasmic tail (CT), transmembrane domain (TM), central conserved domain (CCD), and cysteine noose (CN). (B) Conserved region VI of the large polymerase (L) protein contains the methyltransferase (MTase) site that adds methyl groups to the viral mRNAs. By mutating G1857 in the L protein from glycine to alanine, L(G1857A), the S-adenosylmethionine (SAM)-binding site of the viral MTase is inhibited, reducing methylation of the viral mRNA caps.
An ideal RSV LAV candidate would grow well in Vero cells, since these are the cells approved for RSV vaccine production. To compare the growth rates of the three mutants and their parent virus, we inoculated Vero cells with a multiplicity of infection (MOI) of 1.0 and monitored viral yields daily for 5 days (Fig. 2). The viruses expressing the L mutation, rgRSV-L and rgRSV-G/L, grew more slowly for the first 48 h postinfection (hpi), reaching a 10-fold lower titer than the other two viruses expressing wild-type (wt) L, indicating that this L protein mutation caused slower growth under these conditions. However, by 72 hpi, both rgRSV-L and rgRSV-G/L had reached peak titers, which were similar to those of the viruses with wt L. By 96 hpi, cell death was apparent, and titers of all viruses decreased. Importantly, the ncG modification did not reduce virus yield in Vero cells, indicating that the ncG point mutation is not in a critical site for attachment to and infection of Vero cells or for production of maximal virus yield.
FIG 2.
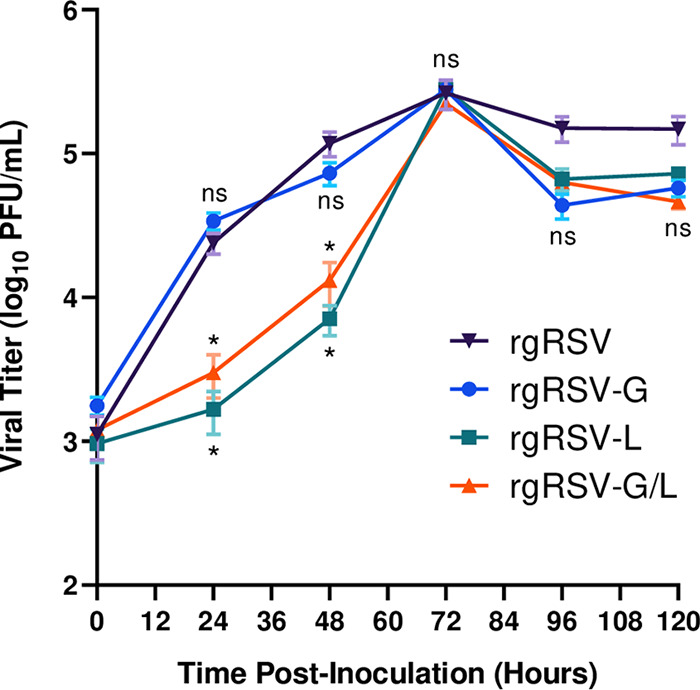
Virus growth kinetics in Vero cells. Vero cells in 24-well plates were inoculated with rgRSV, rgRSV-G, rgRSV-L, or rgRSV-G/L at an MOI of 1.0. Every 24 h, infected cells were scraped, vortexed to detach virions, and centrifuged to remove debris, and the supernatant was collected and aliquoted into cryovials. Cryovials were snap-frozen and stored at −80°C. Samples were titrated on HEp-2 cells and fluorescent foci were counted. Data points represent the means from four technical replicates of two biological replicates ± SDs. Statistical significance was determined by a one-way analysis of variance (ANOVA) with Dunnett’s post hoc test for individual comparisons to rgRSV. ns, not significant; *, P < 0.05.
The ncG mutation inhibits the cleavage of the RSV G protein when the virus is produced in Vero cells (23). To evaluate the size of the G protein in virions produced in Vero cells, all four viruses were purified by centrifugation to equilibrium on a linear sucrose gradient, and the size of the virion G protein in the virion fraction was evaluated by immunoblotting (Fig. 3). Truncated G was detected in rgRSV and rgRSV-L, both of which contained the wt G protein. As expected, the G protein was not cleaved in rgRSV-G or rgRSV-G/L virions, which express the ncG mutant protein, confirming that the G(L208A) mutation prevents G cleavage during virus production in Vero cells. Costaining the blot for N protein confirmed that similar amounts of virions had been loaded for each virus.
FIG 3.
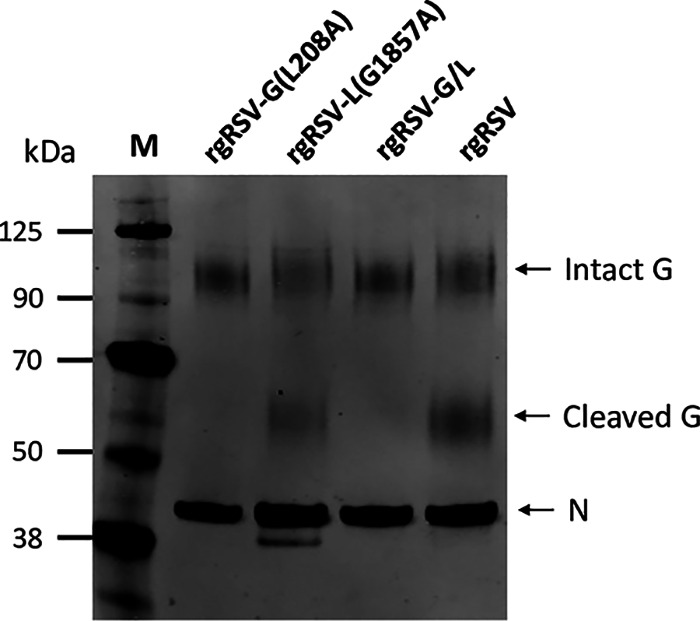
G protein in sucrose density gradient-purified RSV virions produced from Vero cells. Immunoblot of the G protein, intact (90 kDa) or cleaved (55 kDa). The RSV N protein was also immunostained to confirm that similar amounts of virions had been loaded in each well.
In HBE cultures, viruses with ncG display enhanced initial infectivity, and those with the L mutation are highly attenuated.
To confirm that full-length G enhances RSV infectivity of HBE cultures (23), we assessed the initial infectivity of the Vero-grown mutant viruses in HBE cultures and in HEp-2 cells. Both culture systems were inoculated with 1,000 PFU of each virus, as determined on HEp-2 cells, from the same virus dilution tube on the same day. Cells were inoculated with either rgRSV or rgRSV-G, and green cells were counted at 24 hpi. Cultures inoculated with rgRSV-L or rgRSV-G/L displayed very few GFP-expressing cells at 24 hpi but displayed more isolated green cells by 48 hpi and so were quantified at that time (Fig. 4A).
FIG 4.
Initial infectivity and spread in HBE cultures. Transwells of well-differentiated HBE cultures and HEp-2 cultures in 96-well plates, both in triplicates, were inoculated with 1,000 PFU of Vero-grown rgRSV, rgRSV-G, rgRSV-L, or rgRSV-G/L. (A) Fluorescence focus-forming units (FFUs) were counted at 24 hpi for viruses with the wt L protein and at 48 hpi for viruses expressing the L(G1857A) mutation. The ratio of infectious units in HBE cultures to the infectious units in HEp-2 cultures provides the relative infectivity of each virus for HBE cultures. Viral spread was measured every 24 h over a 5-day period and assessed by digital photography to quantify total well fluorescence (B) or by direct visualization of fluorescence (C). Levels of total well fluorescence in images were quantified with ImageJ software, and background fluorescence was subtracted to obtain a total well fluorescence. Data points represent the means from three transwells ± SDs. Statistical significance was determined by a one-way analysis of variance (ANOVA) with Dunnett’s post hoc test for individual comparisons. ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.001. Statistical significance of individual comparisons to rgRSV are plotted in panel B.
Compared to rgRSV, rgRSV-G was approximately 2-fold more infectious for HBE cultures, and rgRSV-G/L was 5-fold more infectious than rgRSV-L for HBE cultures, confirming the importance of ncG in the inoculum virus for infection of HBE cultures. Despite the additional 24 h of incubation, fewer infected cells were detected in the cultures inoculated with rgRSV-L and rgRSV-G/L viruses than in those inoculated with their wt L counterparts (10-fold and 4-fold, respectively). The lower levels of detectable infection are likely due to the less efficient translation of the viral mRNAs caused by the reduced cap methylation activity of the SAM-binding site mutant L protein.
In HBE cultures, RSV infects ciliated cells and is shed apically (24), analogous to being shed into the lumen of the airways. For F protein to be included in the virions, it must be in the apical membrane. Syncytia are not detected in HBE cultures (24), likely because the tight junctions between these columnar cells provide a barrier separating their apical and basolateral domains, thereby preventing the F protein in the apical membrane from contacting neighboring cells.
In most HBE cultures, apically shed virions infected neighboring cells in a distinctive “hurricane” pattern (Fig. 4C), driven by the coordinated beating of the cilia in the culture that moves the apical fluid in this pattern and the virions along with it. We did not sample the apical fluids to quantify virus yields at each time point, because sampling would require washing the culture with phosphate-buffered saline (PBS), spreading the virus artificially. Instead, we quantified the level of fluorescence as a measure of infection digitally and the increase as viral “spread.” Although rgRSV demonstrated a delay in spread relative to that for rgRSV-G (Fig. 4B), the difference did not reach statistical significance. The delay did correspond to the lower number of infected cells at 1 day postinfection (dpi) and the cleavage phenotype of its G protein. However, both viruses spread at similar rates thereafter (Fig. 4B), indicating that while the ncG mutation in rgRSV-G was initially more infectious for HBE cultures, the ncG mutation had no effect on viral spread in HBE cultures. This result was expected because the wt G protein is not cleaved when produced in HBE cultures the way it is in Vero cells (22).
The two viruses expressing wt L had spread to most areas of the cultures by 4 dpi, but the L mutant virus spread was not apparent until 4 dpi (Fig. 4C). Digital quantification demonstrated a major reduction in the spread of these two viruses with the same L mutation in the SAM-binding site (Fig. 4B) and therefore the level of attenuation caused by this mutation.
The rgRSV-G/L virus is highly attenuated in cotton rats.
Since HBE cultures are reflective of the in vivo target tissue for RSV, the dramatic attenuation observed for the mutant L viruses in HBE cultures should also be apparent in vivo. The cotton rat (Sigmodon hispidus) is semipermissive for human RSV infection (25–27), and as such, it is the best small-animal model for RSV LAV candidate assessment. To examine viral replication in cotton rats, we inoculated groups of 4 cotton rats intranasally with 104 PFU of one of the four Vero-grown viruses. At 4 dpi, viral titers in nasal turbinate and lung homogenates were quantified by a 50% tissue culture infective dose (TCID50) assay (Fig. 5).
FIG 5.
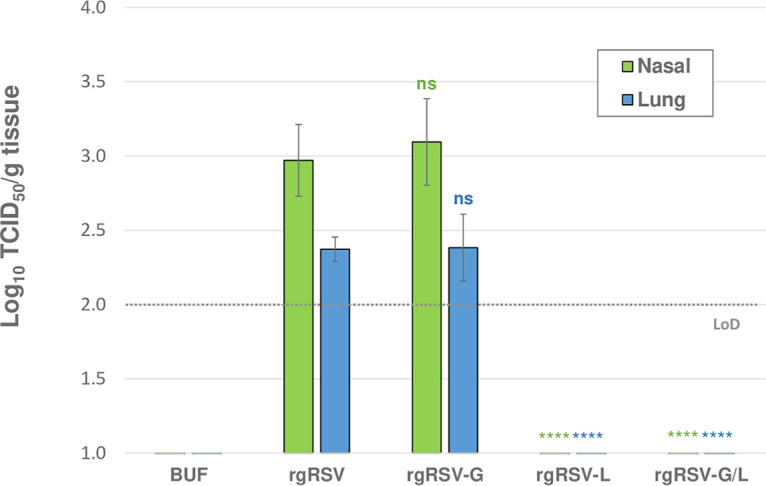
Viral replication in the cotton rat. Cohorts of 4 cotton rats were each immunized with 104 PFU of rgRSV, rgRSV-G, rgRSV-L, rgRSV-G/L, or PBS lacking infectious virus (BUF) and sacrificed at 4 dpi. Right lungs and nasal turbinates were homogenized, and virus was quantified in a TCID50 assay on HEp-2 cells. Viral replication was only detected in the rgRSV-G and rgRSV-inoculated groups, indicating that the SAM-binding modification (G1857A) attenuates viral replication below the limit of detection (LoD) for the TCID50 assay, log10 2. Mean viral titers for the four cotton rats ± SDs are plotted. Statistical significance was determined by a one-way analysis of variance (ANOVA) with Dunnett’s post hoc test for individual comparisons to rgRSV. ns, not significant; ****, P < 0.001.
Infectious virus was produced in the upper and lower respiratory tracts of animals inoculated with rgRSV and rgRSV-G, both of which express the wt L protein, with the highest viral replication present in the nasal tissue for both groups. However, no infectious virus was detected in the upper or lower respiratory tract for rgRSV-L or rgRSV-G/L, both expressing the mutant L protein. Overall, these results demonstrate that viruses containing the SAM-binding site mutation in the L protein are highly attenuated in vivo.
Calibration of the immunizing dose of RSV needed to protect cotton rats against challenge.
We aim to generate an RSV LAV candidate that requires the lowest possible inoculum that completely protects from RSV infection to minimize viral replication and reduce the change of symptoms caused by the vaccine in any recipient. In addition, to compare the relative potency of vaccine candidates and their protective efficacy to that of wt RSV, a minimal immunizing dose is essential. To identify the minimum immunizing dose required for protection, cotton rats were immunized with 10-fold serial dilutions of Vero-grown rgRSV, ranging from 105 PFU to 101 PFU (and a PBS-immunized group as an unimmunized control, labeled “BUF”), followed by challenge at 28 dpi with 105 PFU of RSV A2. The challenge virus is representative of the rgRSV parental virus, RSV A2, but lacks the GFP gene to avoid the possibility of a protective response targeting GFP rather than the virus. Four days after challenge, viral titers in nasal and lung homogenates were quantified in a TCID50 assay (Fig. 6).
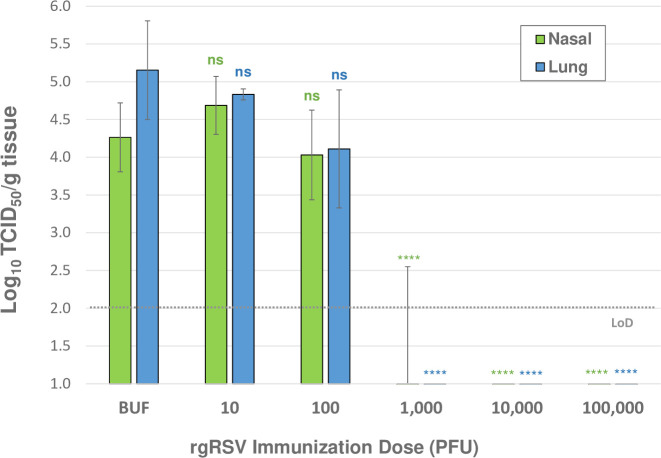
FIG 6.
Protection from challenge provided by immunization with decreasing concentrations of virus. Cohorts of 4 cotton rats were immunized with 105 PFU to 101 PFU of Vero-grown rgRSV and one control group immunized with PBS (labeled BUF). Animals were challenged with RSV A2 at 28 dpi and sacrificed 4 days later, and viral titers were quantified in lung and nasal tissue homogenates with a TCID50 assay. TCID50 assay limit of detection (LoD) is log10 2. The mean of viral titers from four cotton rats ± SDs are plotted. Statistical significance was determined by a one-way analysis of variance (ANOVA) with Dunnett’s post hoc test for individual comparisons to the buffer-immunized group. ns, not significant; ****, P < 0.001.
Immunization with 105 or 104 PFU of rgRSV completely protected cotton rats from challenge. Immunization with 103 PFU completely protected all but one cotton rat, in which virus was found in the nose only. Immunization with 102 PFU was not protective, yet there was a slight decrease in challenge virus replication compared to that in the buffer-immunized group, though not statistically significant. These results indicate that the minimum immunizing dose of rgRSV is between 102 and 103 PFU. A dose within this range should provide the most sensitive comparison of a potential vaccine candidate’s protective activity relative to its parental virus. Based on these data, we chose to immunize cotton rats with 500 PFU.
Low dose immunization of cotton rats with rgRSV mutants completely protects against challenge.
To assess the level of protection provided to cotton rats by immunization with 500 PFU of the mutant viruses, they were challenged at 28 dpi with 105 PFU of RSV A2. At 4 days postchallenge (dpc), virus in the nasal and lung homogenates was quantified in a TCID50 assay (Fig. 7A). Substantial viral replication was detected in the buffer-immunized group (BUF) after challenge, in both the nasal and lung tissues. Yet, no virus was detected in the TCID50 assay in any of the animals in any of the virus-immunized groups, indicating that the mutant viruses were all as effective at protecting cotton rats from RSV challenge as the parental rgRSV. This result is not surprising for rgRSV-G, whose infectivity for HBE cultures is twice that of rgRSV when both are produced in Vero cells (Fig. 4A). However, the ability of a low (500 PFU) immunizing dose of the highly attenuated L mutant viruses, rgRSV-L and rgRSV-G/L, to completely protect cotton rats against RSV challenge is striking and suggests that rgRSV-G/L is a strong vaccine candidate.
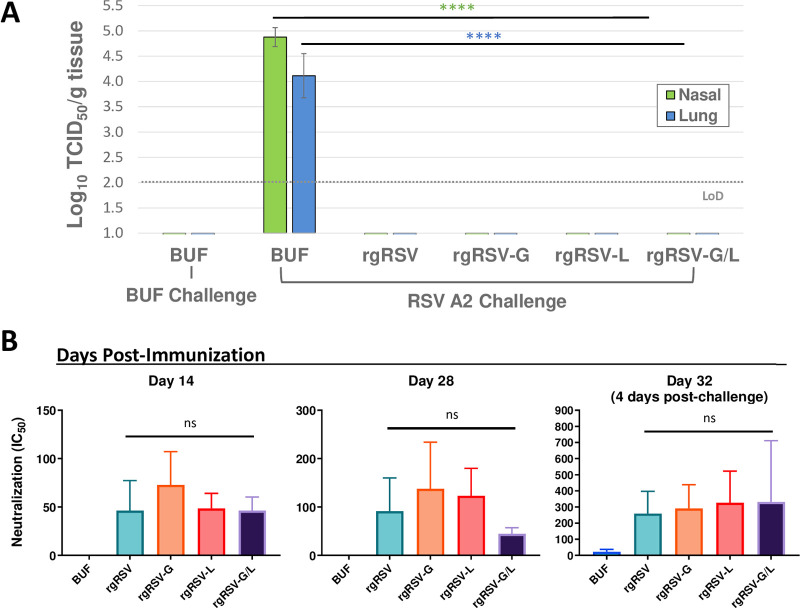
FIG 7.
Immunization with 500 PFU offers complete protection from RSV A2 challenge. Cohorts of 5 cotton rats were immunized with 500 PFU of rgRSV, rgRSV-G, rgRSV-L, rgRSV-G/L vaccine virus, or PBS lacking infectious virus (BUF). At 28 dpi, cotton rats were challenged with 105 PFU of RSV A2 and sacrificed 4 days later. (A) Viral replication in the nose and lungs was quantified by TCID50 assay. No viral replication is detected after RSV challenge in any of the rgRSV-immunized groups. (B) Neutralizing antibody titers at 14, 28, and 32 dpi were quantified in cotton rat sera, showing induction of nAbs in all virus-immunized groups and substantial increase in nAb titers at the 32-dpi time point, which was 4 days after RSV challenge. For TCID50 assays, the mean of viral titers from five cotton rats ± SDs are plotted. The limit of detection (LoD) for the TCID50 assay is log10 2. For nAb quantification, the half maximal inhibitory concentration (IC50) for each serum sample was determined and plotted as the geometric mean for the four cotton rats ± SEs. Statistical significance was determined by a one-way analysis of variance (ANOVA) with Dunnett’s post hoc test for individual comparisons. ns, not significant; ****, P < 0.001.
To assess the immunogenicity of the mutant viruses at the 500-PFU immunization dose, cotton rat sera were collected at 14 and 28 dpi. Cotton rats were challenged at 28 dpi with 105 PFU of RSV A2, and serum was also collected at 32 dpi (4 days postchallenge) at the time of termination. Neutralizing antibody (nAb) titers were quantified at each of these time points (Fig. 7B). nAbs were induced by all viruses at 14 and 28 dpi, and there was a significant boost in nAb titers at 32 dpi, 4 days after RSV challenge. Despite the rgRSV-G/L group showing the greatest increase in nAb titers after challenge, the nAb levels in this group remained constant from 14 to 28 dpi compared to an increase in the other groups, the reason for which is unclear. These findings demonstrate that even at a low immunization dose of 500 PFU, rgRSV-G/L still induces a neutralizing antibody response similar to that for the parental virus, resulting in complete protection from RSV challenge.
The rgRSV mutants do not elicit vaccine-enhanced disease in the cotton rat.
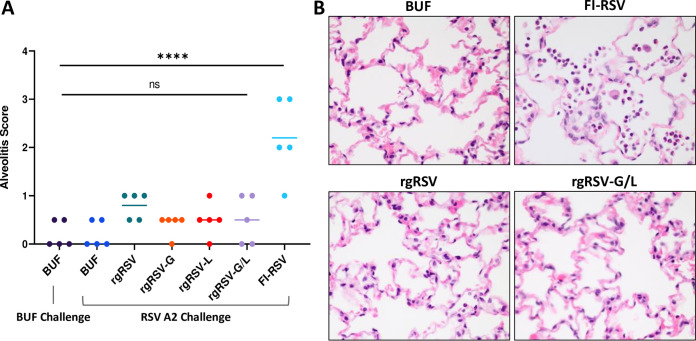
To determine if immunization with the rgRSV mutant viruses and subsequent RSV challenge leads to enhanced RSV disease (ERD), cotton rat lungs from the previous experiment were evaluated for “alveolitis,” or neutrophilic infiltrates in the alveolar sacs, as a primary indicator of ERD in cotton rats, paralleling ERD in humans (7, 27). As a positive control, cotton rats immunized with 200 µg of formalin-inactivated RSV (FI-RSV) from a previous study (28) were included for comparison. The FI-RSV-immunized group is a historical control in which cotton rats were sacrificed at 5 dpc rather than at 4 days.
The level of alveolitis was graded histologically, on a scale from 0 to 4 based on the percentage of the parenchyma affected. The median alveolitis score for each group was determined (Fig. 8A), and representative photomicrographs of groups immunized with buffer, FI-RSV, rgRSV, and rgRSV-G/L are shown in Fig. 8B. The alveolitis score in the FI-RSV group was significantly higher than that of the groups immunized with the rgRSV mutants, which is also reflected in the photomicrographs of the lung sections. These findings demonstrate that the rgRSV mutants do not elicit ERD in the cotton rat model after RSV challenge.
FIG 8.
Vaccine-enhanced disease is not observed with rgRSV mutants. Lung histopathology from the cotton rats immunized with 500 PFU of the rgRSV mutant viruses or PBS, denoted BUF, (followed by challenge at 28 dpi with RSV A2 and sacrifice 4 days later) was analyzed and compared with historical controls from a previous FI-RSV study. (A) The severity of alveolitis in each cotton rat was graded based on the extent of parenchyma affected (grade 0, 0% affected, within normal limits; grade 0.5, <5% affected; grade 1, <10% affected; grade 2, 10% to 25% affected; grade 3, 25% to 50% affected; grade 4, >50% affected) (51). (B) Representative photomicrographs depicting neutrophilic infiltrates in the alveolar spaces or lack thereof (magnification, ×60). rgRSV-immunized groups were compared to historical controls immunized with FI-RSV, challenged at 28 dpi, and sacrificed at 5 dpc. For the alveolitis scoring, the median alveolitis score from the five cotton rats is shown, with dots representing the individual scores. Statistical significance was determined by a one-way analysis of variance (ANOVA) with Dunnett’s post hoc test for individual comparisons. ns, not significant; ****, P < 0.001.
Immunization with rgRSV-G/L virus and subsequent RSV exposure induces long-lasting neutralizing antibodies in the cotton rat.
In addition to offering protection at 28 dpi, an ideal RSV LAV candidate should provide long-lasting protection. To assess serum antibody neutralizing activity over time, we immunized cohorts of 4 cotton rats with 105 PFU of rgRSV-G/L, rgRSV, or buffer and challenged all animals at 28 dpi with 105 PFU of RSV A2. This protocol was designed to mimic a child who was vaccinated and exposed naturally to RSV during their first winter season, an event that occurs in the majority of infants in their first year (1, 2). RSV-specific nAb titers were quantified once per week during the first 28 days of the study and approximately every 4 weeks thereafter for 7 months (Fig. 9). The rgRSV-G/L virus induced higher nAb titers than rgRSV following immunization and challenge at 28 dpi, and this trend continued up to 50 dpi. The nAb titers induced by both the rgRSV-G/L and rgRSV peaked at 50 dpi (22 dpc) followed by a “plateau phase” for approximately 4 months, extending until 179 dpi. During this period, nAb titers in the rgRSV-G/L-immunized group remained relatively constant but somewhat lower than in the rgRSV-immunized group, although not statically significant, and comparable to that in the group that only received RSV exposure at challenge. Titers of RSV-specific nAbs from all groups declined at 219 dpi, at which point the study was terminated.
FIG 9.
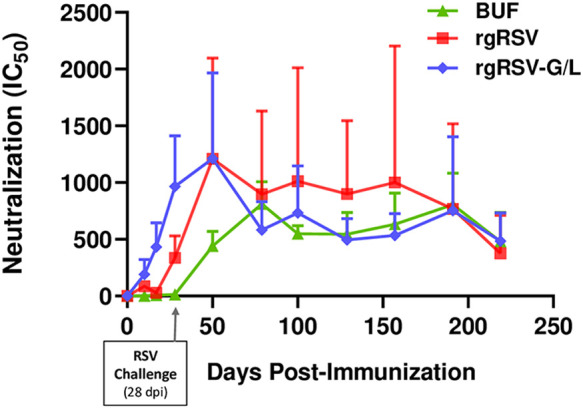
Longevity of the neutralizing antibody response in cotton rats immunized with rgRSV-G/L. Cotton rats were immunized with rgRSV, rgRSV-G/L, or PBS (lacking virus; denoted as BUF) and challenged at 28 dpi with 105 PFU of RSV A2 to mimic a child’s vaccination followed by a subsequent natural exposure to RSV during the following winter season. Peripheral blood was collected weekly for the first 28 days, and every 4 weeks for the remainder of the 7 months. The half maximal inhibitory concentration (IC50) for each serum sample was determined and plotted as the geometric mean from the four cotton rats ± SE.
During this phase, it is important to note that the large error bars for the rgRSV-immunized group are due to one outlier, representing a cotton rat with consistently high nAb titers during the plateau period. Without that animal, the rgRSV values would have tracked more closely with those for rgRSV-G/L. Regardless, both rgRSV and rgRSV-G/L induced substantial levels of nAbs that persisted for >6 months after immunization. These results demonstrate that the rgRSV-G/L virus induces long-lasting neutralizing antibody production despite its attenuation.
The rgRSV-G/L virus is highly attenuated and protective in a rhesus macaque.
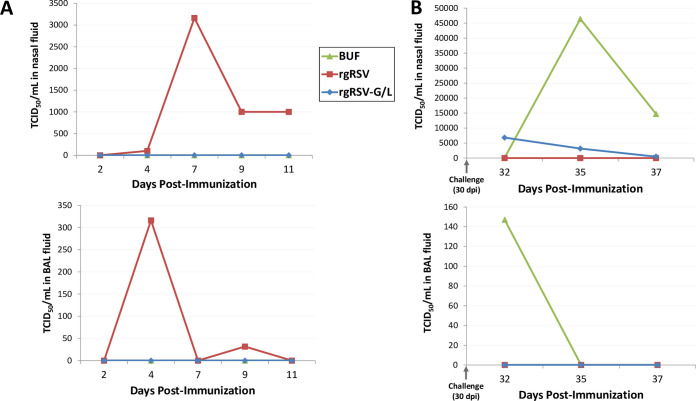
As an intermediate between the rodent model and eventual entrance into human clinical trials, we sought to corroborate the rgRSV-G/L virus infectivity, protection, and immunogenicity findings in the cotton rat in a nonhuman primate model, the rhesus macaque. Since this was a pilot study, only one macaque per group was included. Rhesus macaques were immunized with 106 PFU of either rgRSV-G/L, rgRSV, or buffer lacking infectious virus. Thirty days after immunization, macaques were challenged with 106 PFU of RSV A2, and terminated at 7 dpc. Bronchoalveolar lavage (BAL) and nasal rinse fluids were collected at various time points, and infectious virus was quantified. Postimmunization and postchallenge time points are shown in Fig. 10A and B, respectively. Findings from the upper and lower respiratory tracts were consistent throughout, with early replication of rgRSV peaking in the nose at 7 dpi and in the lung at 4 dpi. No detectable viral replication was apparent at any of the postimmunization time points in the rgRSV-G/L-immunized macaque. After challenge, viral replication was apparent in the buffer-immunized macaque, peaking at 5 and 2 dpc in the nose and lungs, respectively. In the rgRSV-G/L-immunized macaque, there were small amounts of virus in the nose at 2 dpc that decreased over the 7-day postchallenge period and no detectable viral replication in the lungs. These findings in the macaque corroborate our findings in the cotton rat model: there is minimal to nondetectable early viral replication with the rgRSV-G/L vaccine candidate and complete to nearly complete protection of the nose and lungs after RSV challenge.
FIG 10.
Viral replication in rhesus macaque nose and lungs after inoculation with rgRSV-G/L and after RSV challenge. Macaques were immunized intranasally and intratracheally with a total of 106 PFU of rgRSV, rgRSV-G/L, or PBS lacking virus (BUF) and challenged at 30 dpi with 106 PFU of RSV A2. Nasal rinsing and bronchoalveolar lavage (BAL) were performed on the indicated days and evaluated to quantify early viral replication after immunization (A) or viral replication after RSV challenge (B) in the nose (top) and lungs (bottom). Viral titers were quantified with a TCID50 assay.
The rgRSV-G/L virus is immunogenic in the macaque.
We assessed the induction of nAbs in rhesus macaques, comparing rgRSV-G/L with the parental rgRSV and the buffer control. Serum nAb was quantified every 7 days throughout the study (Fig. 11). All three macaques were challenged at 30 dpi with 106 PFU of RSV A2. The nAb titer in the rgRSV-immunized macaque was comparable to that of the rgRSV-G/L-immunized macaque on day 7 dpi, but by 14 dpi and beyond, the nAb titers in the rgRSV-immunized macaque were consistently 2-fold higher than those in the rgRSV-G/L-immunized macaque. Similar to the long-term immunogenicity findings in the cotton rat (Fig. 9), rgRSV induces approximately twice as much nAb as rgRSV-G/L in the rhesus macaque.
FIG 11.
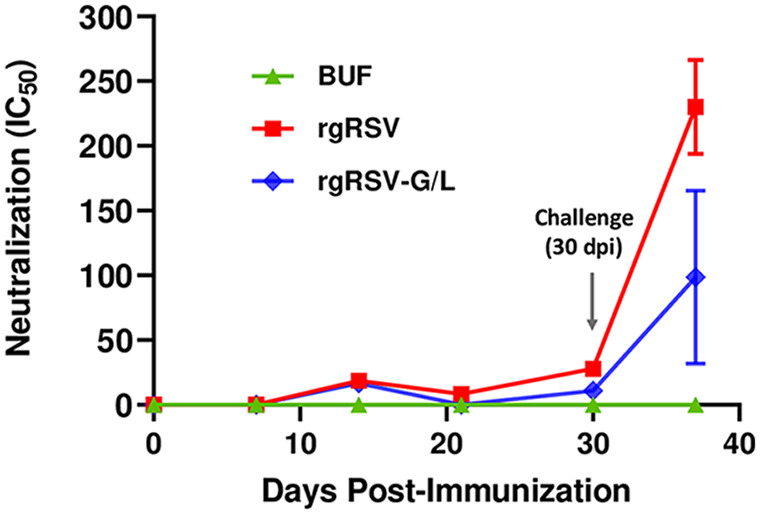
The rgRSV-G/L virus induced neutralizing antibodies in rhesus macaques. Peripheral blood was collected every 7 days throughout the study, and serum was isolated and analyzed. At 30 dpi, macaques were challenged with 106 PFU of RSV A2. Neutralizing antibodies (nAbs) were quantified with a viral neutralization assay utilizing a luciferase-expressing RSV (luc-RSV). The half maximal inhibitory concentration (IC50) for each serum sample was determined and plotted as the geometric mean. At the 37-dpi time point, the mean ± SD from three technical replicates was plotted.
DISCUSSION
We have generated and assessed an RSV LAV candidate with two mutations, rgRSV-G/L [rgRSV-G(L208A)-L(G1857A)], and compared it to viruses with each mutation separately and with the parental virus for its ability to induce a protective antibody response. This virus contains an attenuating L(G1857A) mutation in the SAM-binding site of the L protein methyltransferase region and an ncG mutation, G(L208A), in the G protein that prevents it from being cleaved during virus production in Vero cells, the cells that would be used to produce an RSV LAV for distribution. We confirm that the ncG mutation increases the initial infectivity of the vaccine virus produced in Vero cells by 2- to 5-fold for HBE cultures, as we have previously reported (22, 23). Although it is not clear what causes this variation, even a 2-fold increase in vaccine yield could be useful in reducing the cost of vaccine manufacturing. In addition, avoiding G protein cleavage would enhance the consistency of vaccine production.
HBE cultures are the ideal in vitro model for the airway epithelium and represent the intended site of vaccination in a child, since the vaccine will likely be delivered by nose drops or aerosol. While the enhanced infectivity caused by the G(L208A) mutation increases the functional yield of vaccine virus from Vero cells by 2- to 5-fold, this level of infectivity of RSV virions does not exceed that of virions produced by HEp-2 cells in which the native G protein is not cleaved. Furthermore, the initial advantage that the ncG gives the virus is not conferred to progeny produced by HBE cultures, which do not cleave the G protein (22, 23). Therefore, enhanced infectivity of the vaccine virus beyond the initially infected cells is not a concern.
The attenuating mutation in this RSV LAV candidate is in the SAM-binding site of the methyltransferase (MTase) region of the L protein CR-VI domain. The KDKE motif that defines this SAM-binding site activates both of the mRNA cap MTases, G-N-7 and ribose 2′-O. The mRNA cap G-N-7 methylation is essential for translation initiation (29), while the ribose 2′-O methylation is required for preventing the viral mRNA from being recognized as non-self RNA by the host immune system and for protection from RNA decapping and degradation (18). Disrupting the SAM-binding site of an NNS RNA virus inhibits both MTases, resulting in mRNA decapping and degradation, diminished viral protein translation, reduced viral replication, and attenuation of both viral infection and spread. In addition, modification of other viruses to abrogate ribose 2′-O cap methylation, namely, in human and murine coronavirus and West Nile virus, has been shown to initiate a type I interferon (IFN) response that could further inhibit viral replication (18, 19) and therefore contributes to the attenuated viral spread observed in HBE cultures and, perhaps, in the cotton rat. Enhancing type I IFN levels also boosts the adaptive immune response (30, 31) and may be the reason for roughly comparable neutralizing antibody titers induced by rgRSV-G/L and rgRSV in immunized cotton rats and macaques, despite no detectable virus production by rgRSV-G/L. We have found an enhanced type I IFN response (IFN-α2) to rgRSV-L(G1857A) in HBE cultures compared to that in cultures infected with rgRSV (R. Wang, M. Xue, M. Lu, O. Harder, Y. Zhang, X. Liang, P. Chen, S. Niewiesk, M. E. Peeples, J. Li, submitted for publication). The rgRSV-G/L vaccine virus is highly attenuated, similar to other RSV LAVs presently being evaluated in clinical trials, but appears to offer the added advantage of enhanced immunogenicity resulting from the SAM-binding site mutation.
For several NNS RNA viruses, recent studies have demonstrated that modifications in the SAM-binding site result in viruses that grow to high titers in Vero cell cultures, likely due to the lack of type I IFN in this cell line, but are significantly attenuated in HBE cultures and cotton rats compared to levels of wild-type virus and yet maintain their immunogenicity in vivo (20, 32–34). Our findings for the RSV LAV candidate presented here are consistent with those for other NNS RNA viruses, namely, VSV (20), measles virus (34), and human metapneumovirus (32). This is the first report of a SAM-binding site mutation in RSV and in an RSV LAV candidate. Most importantly, we demonstrated that the rgRSV-G/L vaccine candidate grows efficiently in Vero cell cultures but is highly attenuated in HBE cultures and yet immunogenic and protective in the cotton rat and rhesus macaque models. As with any attenuated LAV candidate, there is a risk of reversion to the wild type during serial passaging in vitro and replication in vivo. We demonstrated genetic stability of SAM-binding site mutants even after 12 passages in Vero cultures (Wang et al., submitted). Regardless, we will likely include more than one SAM-binding site or attenuating mutation in a future RSV LAV to further mitigate the risk of reversion after vaccination.
In the cotton rat model, animals are traditionally immunized and challenged with virus doses of between 104 and 106 PFU, corresponding to the dose range that has been used to immunize young seronegative children. For RSV, 105 PFU is the most commonly used immunization and challenge dose in the cotton rat (26, 27). Here, we have demonstrated that an immunizing dose of 105 PFU is more than 200 times the dose needed for a cotton rat to be completely protected from RSV challenge, even with the highly attenuated L mutant virus tested here. Immunizing with a higher viral dose may ensure protection from RSV challenge but limits the sensitivity of the challenge study in detecting differences in protection provided by different vaccine candidates and rgRSV.
There are limitations to the studies presented here. The single rhesus macaque in each arm of our primate study is, of course, not optimal and did not allow a determination of statistical significance. As such, we approached the primate study as a pilot study to corroborate our findings in the cotton rat model, where we found similar outcomes for undetectable virus replication and protection against challenge by the rgRSV-G/L virus. The induction of nAbs by the rgRSV-G/L vaccine candidate was consistent between the two animal models, showing a slight reduction in nAb titers in rgRSV-G/L groups compared to that for rgRSV. This finding may indicate that a less-attenuating SAM-binding site mutant might provide even better protection, and investigation of other less-damaging SAM-binding site mutants is warranted. Next, in both the cotton rat and rhesus macaque studies, we challenged with RSV A2, the same subgroup A virus as our vaccine virus, finding complete to near complete protection. We did not challenge with an RSV B subgroup virus. An RSV vaccine candidate must offer protection against both subgroups. In the cotton rat immunogenicity study, we had originally planned to challenge the cotton rats following the 4-month “plateau phase” to determine if the cotton rats were still protected at this time. Unfortunately, many of the cotton rats had developed age-related diseases by this time, such as geriatric dental malocclusion, which alters their ability to eat, and ophthalmic lesions (35). As such, it was best to terminate the study at 219 dpi and forgo the late-stage RSV challenge. Next, our RSV LAV candidate contained a GFP gene in the RSV genome, which facilitated viral visualization and quantification throughout this study. Once we have identified the best vaccine candidates, this gene will be removed prior to clinical trials in humans. Lastly, we did not include a control group that was immunized with heat or UV-inactivated virus serving to eliminate the possibility of antibody induction from the viral antigen alone. We consider this an unlikely source for the protective response given the very low viral inoculation dose (500 PFU) but may include this group in future studies to definitively rule out this possibility.
Many RSV LAV candidates have proceeded into clinical trials, and others are being developed; yet, no FDA-approved vaccine is available. A major challenge in the development of an RSV LAV is finding a mutant that is attenuated enough to avoid causing lung pathology and respiratory disease while producing enough viral antigens in the right context to be immunogenic (36, 37). Recently, a series of LAV candidates with deletions of the M2-2 gene (ΔM2-2) have shown promising results in nonhuman primates and recent phase I clinical trials in children. Deletion of the RSV M2-2 gene results in decreased genome replication but increases viral protein expression, resulting in a virus with attenuated replication that may retain immunogenicity in vivo. In the most recent study, children were immunized with D46/NS2/N/ΔM2-2-HindIII (38), which combines features of MEDIΔM2-2 (39) and LIDΔM2-2 (13), to generate a robust immune response, but levels of viral shedding and the development of mild clinical signs suggested this vaccine candidate may be underattenuated.
In addition to ΔM2-2 RSV LAV candidates, other promising candidates feature deletions of the NS1 or NS2 genes. The NS1 and NS2 proteins act to dampen the host type I and III IFN responses during infection, allowing for greater growth and spread of the virus in vivo (40, 41). By deleting either of these genes in a vaccine virus, the host IFN response is increased, which may enhance the adaptive immune response, a finding that was previously demonstrated in calves infected with bovine RSV with deletions in NS1 or NS2 (42). One human RSV LAV candidate with deletion of the NS2 gene, RSV/ΔNS2/Δ1313/I1314L (12), was recently found to be immunogenic, genetically stable, and protective in a phase 1 clinical trial.
Here, we have taken a novel approach to developing an RSV LAV candidate, rgRSV-G/L. In contrast to selecting random attenuating mutations or deleting entire viral genes, we have made targeted functional mutations in the G and L genes calculated to improve vaccine yield for its in vivo target cell and to strongly attenuate while enhancing interferon production, thereby maintaining a high level of immunogenicity. This is the first report of an RSV LAV vaccine candidate incorporating an ncG mutation or a SAM-binding site mutation. The rgRSV-G/L vaccine candidate displays increased initial infectivity in HBE cultures while attenuating viral replication and spread in HBE cultures and in vivo. In both primate and cotton rat models, the rgRSV-G/L vaccine candidate replicates to undetectable levels after inoculation with 105 PFU, while 500 PFU is enough to completely protect against RSV A2 challenge. The rgRSV-G/L is nearly as immunogenic as the parental rgRSV in both the cotton rat and macaque models. These findings in animal models demonstrate that the rgRSV-G/L [rgRSV-G(L208A)-L(G1857A)] vaccine is an excellent candidate RSV LAV.
MATERIALS AND METHODS
Cell culture.
HEp-2 (ATCC CCL-23), A549 (ATCC CCL-185), and Vero (ATCC CRL-CCL81) (ATCC, Manassas, VA) cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Corning Inc., Corning, NY) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA) and 1% of penicillin-streptomycin, HEPES buffer, and l-glutamine (Thermo Fisher Scientific), referred to as “medium.” All cell lines were maintained at 37°C in a 5% CO2 atmosphere. Primary, well-differentiated human bronchial epithelial (HBE) cultures were derived from human airway tissue, as previously described (43), and grown on collagen-coated Transwell inserts (Corning Inc.) and cultured with air-liquid interface (ALI) medium supplemented with Y-27632 ROCK inhibitor (BD Biosciences). Upon attaining confluence and forming tight junctions, the medium overlying the apical aspect of the cells was removed from the Transwells, and the basal medium was replaced with PneumaCult medium (Stem Cell Technologies, Inc., Vancouver, Canada) to promote differentiation of the epithelial cells at the air-liquid interface (ALI) over the following 3 to 4 weeks. Maturity of the cultures was confirmed by the microscopic presence of ciliary movement and mucus production on the apical surface. The basal PneumaCult medium was changed three times per week, with brief rinsing of the apical chamber with DMEM to remove accumulated mucus.
RSV stocks and purification.
Recombinant green fluorescent protein (GFP)-expressing RSV (rgRSV) was generated from a full-length cDNA, RW30 (22, 44). RW30 is a modified version of the original RSV genome cDNA and contains the GFP gene as the first gene at the 3′ end, prior to NS1 (Fig. 1). RW30 served as the backbone for mutagenesis of the G and L proteins. Synthetic double-stranded DNA (gBlocks; Integrated DNA Technologies, Coralville, IA) with the G(L208A) mutation in the G gene sequence was inserted into the RW30 plasmid using restriction sites flanking the G gene and the Gibson assembly kit (New England BioLabs, Ipswich, MA). Mutagenesis of the L protein within the SAM-binding site, L(G1857A), was achieved using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) for both the single and double G and L mutants. All three mutant viruses, rgRSV-G(L208A), rgRSV-L(G1857A), and rgRSV-G(L208A)/L(G1857A), were rescued from these plasmids as described previously (44). For simplicity, these mutants were described as rgRSV-G, rgRSV-L, and rgRSV-G/L throughout this report.
For virus propagation and collection, infected cells were inoculated with rgRSV or modified viruses diluted in medium lacking FBS, tipped at 37°C for 2 h, and the inoculum was replaced with fresh medium containing 2% to 5% FBS. After incubation at 37°C in a 5% CO2 incubator for 48 h postinoculation (hpi), the medium was replaced with fresh medium. At 72 hpi, the infected cells were manually scraped, the medium and cells were collected and vortexed, and the cells were pelleted by centrifugation at 1,550 × g for 15 min in a Sorvall Lynx 6000 centrifuge (Thermo Fisher Scientific). The supernatant was aliquoted into cryovials and snap-frozen on dry ice for 1 h before being stored at −80°C. Virus stocks were grown on either Vero or HEp-2 cultures and titrated on HEp-2 cells.
For infection experiments in HBE cultures and animal models, all RSV stocks were partially purified through a sucrose cushion. The 150-mm tissue culture dishes (Corning Inc., Corning, NY) containing Vero cells at approximately 30% confluence were infected with rgRSV or mutant viruses at a multiplicity of infection (MOI) of 0.1. At 72 hpi, infected cells were manually scraped and collected with the supernatant, briefly vortexed, and clarified by centrifugation at 1,550 × g for 15 min. The supernatant was then layered over a 35% sucrose solution in Dulbecco’s PBS (D-PBS) and centrifuged at 27,000 × g for approximately 5 h at 4°C in an F14-14x50cy rotor in a Sorvall Lynx 6000 centrifuge (Thermo Fisher Scientific). The pelleted partially purified virus was resuspended in 5 ml of DMEM containing 10% trehalose and titrated on HEp-2 cells. Every 24 h for 5 days, viral spread in HBE cultures was quantified with ImageJ software (https://imagej.nih.gov/ij/) (45) based on the fluorescence of Transwells photographed by an EVOS-2 image system.
Immunoblot analysis.
To maximize virion purification before assessing their G proteins, 1 ml of partially purified virus was layered on top of a sucrose gradient composed of 1-ml fractions of 25%, 35%, 45%, and 55% sucrose and centrifuged in a SW55 Ti swinging-bucket rotor in a Beckman ultracentrifuge at 287,000 × g for 17 h. The visible virus band between the 35% and 45% sucrose layers was collected, diluted in D-PBS, pelleted by centrifugation at 12,000 × g for 90 min in a tabletop centrifuge (model 5417R; Eppendorf, Hamburg, Germany), and resuspended in viral lysis buffer. Proteins were separated on NuPage Novex 4% to 12% bis-Tris protein gels (Life Technologies, Carlsbad, CA), transferred to a nitrocellulose membrane using an iBlot transfer stack (Thermo Fisher Scientific), and probed with mouse monoclonal antibody (MAb) L9 to the RSV G protein (46) and D14 to the RSV N protein (47) (Edward Walsh, University of Rochester) followed by IRDye goat anti-mouse IgG secondary antibody (LI-COR, Inc.) for 1 h at room temperature. Protein signals were visualized using an Odyssey CLx imaging system (LI-COR, Inc.) and Image Studio software.
Viral RNA sequencing.
Viral RNA was isolated from purified virions, animal tissue homogenates, or fluid samples using the Viral QIAamp RNA extraction kit (Qiagen, Valencia, CA). cDNA to the viral RNA was generated using a high-capacity cDNA reverse transcriptase kit (Applied Biosystems, Foster City, CA). A PCR step using PCR SuperMix (Invitrogen, Carlsbad, CA) with primers flanking the gene for either the G protein or the L protein amplified the cDNA prior to submission to Genewiz, Inc. (South Plainfield, NJ, USA) for Sanger sequencing.
Ethics statement.
The animal studies were coordinated and executed in accordance with the Animal Welfare Act and recommendations from the Guide for the care and use of laboratory animals of the National Research Council and the Weatherall report for “The use of nonhuman primates in research.” These studies were approved by The Ohio State University and Nationwide Children’s Hospital Research Institutional Animal Care and Use Committees (IACUC; animal protocol numbers 2009A0183 and AR17-00069, respectively). Cotton rats were housed in the University Laboratory Animal Resources (ULAR) facilities at The Ohio State University, and rhesus macaques were housed in the Nationwide Children’s Hospital Research Institute laboratory animal facilities. Animal care facilities at The Ohio State University and Nationwide Children’s Hospital Research Institute are AAALAC accredited. All procedures were performed according to the approved animal use protocols, and care was taken to minimize pain, distress, or discomfort to animals at all points throughout the study.
Cotton rat studies.
Male and female specific-pathogen-free (SPF) cotton rats (Sigmodon hispidus) (Envigo, Indianapolis, IN) ranging from 6 to 8 weeks of age were included in the studies, with 4 cotton rats per group. Cotton rats were anesthetized via isoflurane inhalation before being inoculated intranasally with a total inoculation volume of 100 µl containing one of the viruses under study, which was diluted in D-PBS. To assess viral replication after inoculation with 104 PFU, cotton rats were sacrificed at 4 dpi via inhalation of carbon dioxide. To assess the protection offered by the vaccine virus, cotton rats immunized with 500 PFU were challenged at 28 dpi with an intranasal administration of 105 PFU of parental RSV (A2 strain) lacking the GFP gene. Cotton rats in postchallenge studies were sacrificed 4 days after challenge (32 dpi) in a similar manner. At necropsy, the left lung and nasal turbinates were collected and homogenized for quantification of viral titer, and the right lung was fixed in 10% neutral buffered formalin for histologic assessment. Peripheral blood collection in cotton rats was performed every 2 to 4 weeks by retro-orbital blood sampling, collected in Microtainer (BD, Franklin Lakes, NJ) SST tubes (no. 365967) and centrifuged for 1 min to separate the serum from the erythrocytes before storing the serum samples at −20°C.
Rhesus macaque studies.
Three rhesus macaques (Macaca mulatta) (obtained from Children’s Hospital of Philadelphia, October 2013), specifically, two females and one male between the ages of 12 and 13 years, were enrolled in this study. Prior to handling, macaques were sedated with an intramuscular injection of tiletamine-zolazepam (Telazol) at 5 mg/kg body weight for procedures involving inoculation, nasal rinsing, and bronchoalveolar lavage (BAL) or at 2.5 mg/kg body weight for blood collection. On day 0, macaques were immunized with a total of 106 PFU of virus diluted into 4 ml, with 3 ml instilled intratracheally and 500 µl administered dropwise into each nostril. At 30 dpi, macaques were challenged with 106 PFU RSV A2. BAL and nasal rinsing were performed at 4, 7, 9, and 11 dpi to assess virus replication and at 32, 35, and 37 dpi to assess protection from RSV A2 challenge. Peripheral blood was collected from the femoral vein every 7 days throughout the study. Macaques were euthanized and necropsied at 37 dpi, or 7 days postchallenge.
Quantification of viral titer in lung and nasal tissues of animal models.
Tissue homogenates (cotton rat studies) or bronchoalveolar lavage/nasal rinse fluids (rhesus macaque studies) were assessed for virus replication. In the cotton rat, the left lung and nasal turbinates were removed, weighed, and homogenized in 2 ml and 3 ml of DMEM, respectively. The left lung was homogenized using a Precellys 24 tissue homogenizer (Bertin, MD). The mucosa from the nasal turbinates was homogenized by hand with a 0.90-mm mortar and pestle (CoorsTek, Golden, CO) and sterile sand. For macaque studies, bronchoalveolar lavage and nasal rinse fluids were collected in 3 ml of viral transport medium (number [no.] R12503; Thermo Fisher Scientific, Waltham, MA). The infectious virus present was quantified by TCID50 assay on HEp-2 cells, as previously described (48).
Serum RSV-neutralizing and protein-specific antibody quantification.
Neutralizing antibody titers were quantified with a virus neutralization assay, as previously described (49). Prior to assays, serum samples were heated at 65°C for 30 min to inactivate complement proteins. For neutralization assays, heat-inactivated serum was initially diluted 1:10 and then serially 4-fold, and each serum dilution was combined with 250 PFU of a luciferase-expressing RSV A virus (Luc-RSV) (50) and incubated at room temperature for 1 h. Previously plated A549 cells were inoculated with the mixture and incubated at 37°C for 2 h, and the medium was replaced with DMEM containing 2% FBS. After 12 to 18 h, cells were lysed and transferred to 96-well solid black plates, and luciferase assay substrate was added (Renilla luciferase assay system; Promega). Luminescence was quantified using a PE Wallac 1420 Victor2 microplate reader.
Pulmonary histology.
After sacrifice, lung sections from cotton rats were collected and preserved with 10% neutral buffered formalin. Fixed tissues were paraffin embedded and sectioned with a cryotome to generate 5-μm-thick tissue sections that were adhered to glass slides. Tissue slides were stained with hematoxylin and eosin (H&E) for standard assessment of pathology. For ERD evaluation studies, pulmonary histology from our cotton rat studies was compared to that of FI-RSV-immunized historical controls, as previously described (28). All histopathology slides were read in a blinded and randomized manner to eliminate bias.
Statistical analysis.
All statistical analyses were performed using Prism software, version 8 (GraphPad Software, San Diego, CA). Data are presented as means ± standard deviations (SDs) or standard errors (SEs), and statistical comparisons were performed using one-way analysis of variance (ANOVA) with post hoc multiple-comparison testing (Dunnett’s test). Two-tailed P values of less than 0.05 were deemed significant.
ACKNOWLEDGMENTS
We thank Cristina Capella, Bill Bremer, and Tiffany King for technical assistance, Amy Levinsky and Clara Wruck of the Cure CF Columbus Epithelial Cell Core (C3 RDP ECC) at Nationwide Children’s Hospital for the HBE cultures, Laurie Goodchild and her team for veterinary support in the macaque study, Krista La Perle for histopathology guidance, Edward Walsh for antibodies, and Peter Collins for the recombinant RSV system.
This work was supported by grants from the National Institutes of Health (P01 AI112524 to M.E.P., O.R., A.M., M.T., J.L., and S.N., 5T32AI112542 from the National Institute of Allergy and Infectious Diseases and administered by the OSU Infectious Disease Institute, and 5TL1TR002735 administered by the OSU Center for Clinical and Translational Science to T.J.) and by the Nationwide Children’s Hospital Technology Commercialization Fund. The Cure Cystic Fibrosis Columbus Research Development Program Epithelial Cell Core (C3 RDP ECC) is funded by a Research Development Program grant from Cystic Fibrosis Foundation Therapeutics, Inc. (MCCOY17R2). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Shi T, Mcallister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks A, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang D-A, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, Cecília De Freitas C, Emediato L, Fasce RA, Feikin DR, RSV Global Epidemiology Network, et al. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study for RSV Global Epidemiology Network. Lancet 390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glezen WP, Taber LH, Frank AL, Kasel JA. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 3.Simoes EAF. 1997. Respiratory syncytial virus infection: pathogenesis, treatment and prevention. Curr Opin Infect Dis 10:213–220. doi: 10.1097/00001432-199706000-00010. [DOI] [Google Scholar]
- 4.Falsey AR, Walsh EE. 2000. Respiratory syncytial virus infection in adults. Clin Microbiol Rev 13:371–384. doi: 10.1128/cmr.13.3.371-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sáez-Llorens X, Moreno M, Ramilo O, Sánchez P, Top F, Connor E, MEDI-493 Study Group . 2004. Safety and pharmacokinetics of palivizumab therapy in children hospitalized with respiratory syncytial virus infection. Pediatr Inf Dis J 23:707–712. doi: 10.1097/01.inf.0000133165.85909.08. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez WJ, Gruber WC, Groothuis JR, F Simoes EA, Rosas AJ, Lepow M, Kramer A, Hemming V, RSV-IGIV Study Group . 1997. Respiratory syncytial virus immune globulin treatment of RSV lower respiratory tract infection in previously healthy children. Pediatrics 100:937–942. doi: 10.1542/peds.100.6.937. [DOI] [PubMed] [Google Scholar]
- 7.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 8.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE, Kapikian AZ, Chanock M, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 9.Mclellan JS, Ray WC, Peeples ME. 2013. Structure and function of respiratory syncytial virus surface glycoproteins. Curr Top Microbiol Immunol 372:83–104. doi: 10.1007/978-3-642-38919-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desmyter J, Melnick JL, Rawls WE. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J Virol 2:955–961. doi: 10.1128/JVI.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malkin E, Yogev R, Abughali N, Sliman J, Wang CK, Zuo F, Yang CF, Eickhoff M, Esser MT, Tang RS, Dubovsky F. 2013. Safety and immunogenicity of a live attenuated RSV vaccine in healthy RSV-seronegative children 5 to 24 months of age. PLoS One 8:e77104. doi: 10.1371/journal.pone.0077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karron RA, Luongo C, Mateo JS, Wanionek K, Collins PL, Buchholz UJ. 2020. Safety and immunogenicity of the respiratory syncytial virus vaccine RSV/ΔNS2/Δ1313/I1314L in RSV-seronegative children. J Infect Dis 222:82–91. doi: 10.1093/infdis/jiz408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McFarland EJ, Karron RA, Muresan P, Cunningham CK, Valentine ME, Perlowski C, Thumar B, Gnanashanmugam D, Siberry GK, Schappell E, Barr E, Rexroad V, Yogev R, Spector SA, Aziz M, Patel N, Cielo M, Luongo C, Collins PL, Buchholz UJ, International Maternal Pediatric Adolescent AIDS Clinical Trials (IMPAACT) 2000 Study Team . 2018. Live-attenuated respiratory syncytial virus vaccine candidate with deletion of RNA synthesis regulatory protein M2-2 is highly immunogenic in children. J Infect Dis 217:1347–1355. doi: 10.1093/infdis/jiy040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchholz UJ, Cunningham CK, Muresan P, Gnanashanmugam D, Sato P, Siberry GK, Rexroad V, Valentine M, Perlowski C, Schappell E, Thumar B, Luongo C, Barr E, Aziz M, Yogev R, Spector SA, Collins PL, Mcfarland EJ, Karron RA, International Maternal Pediatric Adolescent AIDS Clinical Trials (IMPAACT) P1114 Study Team . 2018. Live respiratory syncytial virus (RSV) vaccine candidate containing stabilized temperature-sensitivity mutations is highly attenuated in RSV-seronegative infants and children. J Infect Dis 217:1338–1346. doi: 10.1093/infdis/jiy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whelan SPJ, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses, p 61–119. In Kawaoka Y (ed), Biology of negative strand RNA viruses: the power of reverse genetics. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Fontaine-Rodriguez EC, Whelan SPJ. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol 79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SPJ. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T-Y, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi P-Y, Diamond MS. 2010. 2′-O Methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Wei Y, Zhang X, Zhang Y, Cai H, Zhu Y, Shilo K, Oglesbee M, Krakowka S, Whelan SPJ, Li J, Lyles DS. 2014. mRNA cap methylation influences pathogenesis of vesicular stomatitis virus in vivo. J Virol 88:2913–2926. doi: 10.1128/JVI.03420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Wang JT, Whelan SPJ. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A 103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwilas S, Liesman RM, Zhang L, Walsh E, Pickles RJ, Peeples ME. 2009. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J Virol 83:10710–10718. doi: 10.1128/JVI.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corry J, Johnson SM, Cornwell J, Peeples ME. 2016. Preventing cleavage of the respiratory syncytial virus attachment protein in Vero cells rescues the infectivity of progeny virus for primary human airway cultures. J Virol 90:1311–1320. doi: 10.1128/JVI.02351-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol 76:5654–5666. doi: 10.1128/jvi.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boukhvalova MS, Prince GA, Blanco JCG. 2009. The cotton rat model of respiratory viral infections pathogenesis and immunity. Biologicals 37:152–159. doi: 10.1016/j.biologicals.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudd PA, Chen W, Mahalingam S. 2016. Mouse and cotton rat models of human respiratory syncytial virus. Methods Mol Biol 1442:209–217. doi: 10.1007/978-1-4939-3687-8_15. [DOI] [PubMed] [Google Scholar]
- 27.Boukhvalova M, Yim K, Blanco J. 2018. Cotton rat model for testing vaccines and antivirals against respiratory syncytial virus. Antivir Chem Chemother 26:2040206618770518. doi: 10.1177/2040206618770518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prince GA, Curtis SJ, Yim KC, Porter DD. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with lot 100 or a newly prepared reference vaccine. J General Virol 82:2881–2888. doi: 10.1099/0022-1317-82-12-2881. [DOI] [PubMed] [Google Scholar]
- 29.Both GW, Banerjee AK, Shatkin AJ. 1975. Methylation-dependent translation of viral messenger RNAs in vitro. Proc Natl Acad Sci U S A 72:1189–1193. doi: 10.1073/pnas.72.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo Y-J, Hahm B. 2010. Type I interferon modulates the battle of host immune system against viruses. Adv Appl Microbiol 73:83–101. doi: 10.1016/S0065-2164(10)73004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katze MG, Fornek JL, Palermo RE, Walters K-A, Korth MJ. 2008. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat Rev Immunol 8:644–654. doi: 10.1038/nri2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Wei Y, Zhang X, Cai H, Niewiesk S, Li J. 2014. Rational design of human metapneumovirus live attenuated vaccine candidates by inhibiting viral mRNA cap methyltransferase. J Virol 88:11411–11429. doi: 10.1128/JVI.00876-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Wei Y, Rauf A, Zhang Y, Ma Y, Zhang X, Shilo K, Yu Q, Saif YM, Lu X, Yu L, Li J. 2014. Methyltransferase-defective avian metapneumovirus vaccines provide complete protection against challenge with the homologous Colorado strain and the heterologous Minnesota strain. J Virol 88:12348–12363. doi: 10.1128/JVI.01095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Liu R, Lu M, Yang Y, Zhou D, Hao X, Zhou D, Wang B, Li J, Huang YW, Zhao Z. 2018. Enhancement of safety and immunogenicity of the Chinese Hu191 measles virus vaccine by alteration of the S-adenosylmethionine (SAM) binding site in the large polymerase protein. Virol 518:210–220. doi: 10.1016/j.virol.2018.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harder OE, Emmer KM, Sparks AE, Miller EJ, Gemensky-Metzler AJ, Coble DJ, Niewiesk S, la Perle KMD. 2020. Cause and treatment of exophthalmos in aged cotton rats (Sigmodon hispidus). Comp Med 70:291–299. doi: 10.30802/AALAS-CM-19-000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karron RA, Buchholz UJ, Collins PL. 2013. Live-attenuated respiratory syncytial virus vaccines, p 259–284. In Anderson LJ, Graham BS (ed), Challenges and opportunities for respiratory syncytial virus vaccines. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLellan JS, Ray WC, Peeples ME. 2013. Structure and function of respiratory syncytial virus surface glycoproteins, p 83–104. In Anderson LJ, Graham BS (ed), Challenges and opportunities for respiratory syncytial virus vaccines. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFarland EJ, Karron RA, Muresan P, Cunningham CK, Libous J, Perlowski C, Thumar B, Gnanashanmugam D, Moye J, Schappell E, Barr E, Rexroad V, Fearn L, Spector SA, Aziz M, Cielo M, Beneri C, Wiznia A, Luongo C, Collins P, Buchholz UJ. 2020. Live-attenuated respiratory syncytial virus vaccine with M2-2 deletion and with small hydrophobic noncoding region is highly immunogenic in children. J Infect Dis 221:534–543. doi: 10.1093/infdis/jiz603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karron RA, Luongo C, Thumar B, Loehr KM, Englund JA, Collins PL, Buchholz UJ. 2015. A gene deletion that up-regulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci Transl Med 7:312ra175. doi: 10.1126/scitranslmed.aac8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munir S, Le Nouen C, Luongo C, Buchholz UJ, Collins PL, Bukreyev A. 2008. Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J Virol 82:8780–8796. doi: 10.1128/JVI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hijano DR, Vu LD, Kauvar LM, Tripp RA, Polack FP, Cormier SA, Kalergis AM, Rehwinkel J, Kam-Lem Lay M, Cormier SA, Hijano DR, Vu LD, Kauvar LM, Tripp RA, Polack FP. 2019. Role of type I interferon (IFN) in the respiratory syncytial virus (RSV) immune response and disease severity. Front Immunol 10:566. doi: 10.3389/fimmu.2019.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valarcher J-F, Furze J, Wyld S, Cook R, Conzelmann K-K, Taylor G. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J Virol 77:8426–8439. doi: 10.1128/jvi.77.15.8426-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 44.Hallak LK, Spillmann D, Collins PL, Peeples ME. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J Virol 74:10508–10513. doi: 10.1128/jvi.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh EE, Falsey AR, Sullender WM. 1998. Monoclonal antibody neutralization escape mutants of respiratory syncytial virus with unique alterations in the attachment (G) protein. J Gen Virol 79:479–487. doi: 10.1099/0022-1317-79-3-479. [DOI] [PubMed] [Google Scholar]
- 47.Walsh EE, Hall CB, Schlesinger JJ, Brandriss MW, Hildreth S, Paradiso P. 1989. Comparison of antigenic sites of subtype-specific respiratory syncytial virus attachment proteins. J Gen Virol 70:2953–2961. doi: 10.1099/0022-1317-70-11-2953. [DOI] [PubMed] [Google Scholar]
- 48.Green MG, Petroff N, la Perle KMD, Niewiesk S. 2018. Characterization of cotton rat (Sigmodon hispidus) eosinophils, including their response to respiratory syncytial virus infection. Comp Med 68:31–40. [PMC free article] [PubMed] [Google Scholar]
- 49.Capella C, Chaiwatpongsakorn S, Gorrell E, Risch ZA, Ye F, Mertz SE, Johnson SM, Moore-Clingenpeel M, Ramilo O, Mejias A, Peeples ME. 2017. Prefusion F, postfusion F, G antibodies, and disease severity in infants and young children with acute respiratory syncytial virus infection. J Infect Dis 216:1398–1406. doi: 10.1093/infdis/jix489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuentes S, Crim RL, Beeler J, Teng MN, Golding H, Khurana S. 2013. Development of a simple, rapid, sensitive, high-throughput luciferase reporter based microneutralization test for measurement of virus neutralizing antibodies following respiratory syncytial virus vaccination and infection. Vaccine 31:3987–3994. doi: 10.1016/j.vaccine.2013.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider-Ohrum K, Cayatte C, Bennett AS, Rajani GM, McTamney P, Nacel K, Hostetler L, Cheng L, Ren K, O'Day T, Prince GA, McCarthy MP. 2017. Immunization with low doses of recombinant postfusion or prefusion respiratory syncytial virus F primes for vaccine-enhanced disease in the cotton rat model independently of the presence of a Th1-biasing (GLA-SE) or Th2-biasing (Alum) adjuvant. J Virol 91:e02180-16. doi: 10.1128/JVI.02180-16. [DOI] [PMC free article] [PubMed] [Google Scholar]