Characterizing the bat virome is important for understanding viral diversity and detecting viral spillover between animal species. Using an unbiased metagenomics method, we characterize the virome in guano collected from multiple roosts of common Northern California bat species. We describe several novel viral genomes and report the detection of viruses with close relatives reported in other bat species, likely reflecting cross-species transmissions. Viral sequences from well-known carnivore and rodent parvoviruses were also detected, whose presence are likely the result of contamination from defecation and urination atop guano and which reflect the close interaction of these mammals in the wild.
KEYWORDS: bat virome, emerging viruses, metagenomics
ABSTRACT
Bats are hosts to a large variety of viruses, including many capable of cross-species transmissions to other mammals, including humans. We characterized the virome in guano from five common bat species in 9 Northern California roosts and from a pool of 5 individual bats. Genomes belonging to 14 viral families known to infect mammals and 17 viral families infecting insects or of unknown tropism were detected. Nearly complete or complete genomes of a novel parvovirus, astrovirus, nodavirus, circular Rep-encoding single-stranded DNA (CRESS-DNA) viruses, and densoviruses, and more partial genomes of a novel alphacoronavirus and a bunyavirus were characterized. Lower numbers of reads with >90% amino acid identity to previously described calicivirus, circovirus, adenoviruses, hepatovirus, bocaparvoviruses, and polyomavirus in other bat species were also found, likely reflecting their wide distribution among different bats. Unexpectedly, a few sequence reads of canine parvovirus 2 and the recently described mouse kidney parvovirus were also detected and their presence confirmed by PCR; these possibly originated from guano contamination by carnivores and rodents. The majority of eukaryotic viral reads were highly divergent, indicating that numerous viruses still remain to be characterized, even from such a heavily investigated order as Chiroptera.
IMPORTANCE Characterizing the bat virome is important for understanding viral diversity and detecting viral spillover between animal species. Using an unbiased metagenomics method, we characterize the virome in guano collected from multiple roosts of common Northern California bat species. We describe several novel viral genomes and report the detection of viruses with close relatives reported in other bat species, likely reflecting cross-species transmissions. Viral sequences from well-known carnivore and rodent parvoviruses were also detected, whose presence are likely the result of contamination from defecation and urination atop guano and which reflect the close interaction of these mammals in the wild.
INTRODUCTION
Emerging infectious diseases are mostly of zoonotic origins and can pose great challenges to public health and the global economy. Bats, considered one of the most important natural reservoirs of a variety of zoonotic viruses, comprise more than 1,400 species that are widely distributed geographically (1–3). While the large number of species within the order Chiroptera may account for its high level of viral diversity, some unique ecological, behavioral, feeding, and genetic or immune characteristics may favor bats as a reservoir of viral diversity (4–6). In the past 20 years, several human viral outbreaks, including severe acute respiratory syndrome coronavirus (SARS-CoV) (7), Middle East respiratory syndrome coronavirus (MERS-CoV) (8), SARS-CoV-2 (9–11), Nipah virus (12), and possibly Ebola virus (13) have emerged from bats. The now-endemic human alphacoronavirus NL63 first described in 2004 (14) may also have originated in bats, possibly in the North American tricolored bat (Perimyotis subflavus) (15).
Such cross-species spillover events highlight the need to further characterize bat viruses to help identify viruses with the highest cross-species potential, namely, those with very close relatives in multiple host species. Previous studies of bat viruses detected in guano have described a wide range of viruses in families known to infect mammals, including astroviruses, adenoviruses, bunyaviruses, circoviruses, coronaviruses, flaviviruses, herpesviruses, nodaviruses, parvoviruses, picornaviruses, papillomaviruses, polyomaviruses, and rotaviruses (2, 5, 16–21). Although limited in their population sampling, these viromes differed between geographic locations as well as between bat species (2, 5, 16–21). The generation of such data will help identify which viruses are detected in multiple bat species and in other mammals (22–24).
Metagenomic sequencing-based methods targeting viral particle-associated nucleic acids from feces/respiratory swabs/tissues or total cellular RNA from tissues are accelerating virome characterizations (25–29). In order to characterize viruses in five bat species from Northern California and to investigate the possible presence of SARS-related coronaviruses, we analyzed viral sequences amplified from bat guano for similarities to all known eukaryotic viruses.
RESULTS
Overview of bat guano-associated viruses.
Guano samples were collected from 3 Northern California counties (Marin, Yolo, and Sacramento) between February and June 2020 (Fig. 1). Ten guano samples, including nine collected from different bat roosts and one mixed sample of five individual captured bats (Table 1), were processed to enrich viral particle-associated nucleic acids that were then randomly amplified and deep sequenced (see Materials and Methods). A total of 22 million paired-end sequence reads (median 2,233,723 sequences per sample) were generated. Following de novo assembly, both singlets and contigs were analyzed using BLASTx for virtually translated proteins sequences showing similarity to all currently known eukaryotic viral proteins. All 10 sequence libraries yielded eukaryotic viral reads (Fig. 2).
FIG 1.
Locations in northern California of the roosts where the bat guano samples were collected. Bottom left, Point Reyes in Marin County; bottom middle, Davis in Yolo County; bottom right, Sacramento in Sacramento County. (Courtesy of d-maps.com; https://d-maps.com/continent.php?num_con=25&lang=en.).
TABLE 1.
Summary of guano samples used in this study
| California county | Collection date | Namea | Primary bat speciesb | No. of samplesc | Estimated no. of animalsd |
|---|---|---|---|---|---|
| Bat roosts | |||||
| Marin | February 2020 | CR1 | Corynorhinus townsendii | 1* | ∼300 |
| February 2020 | CR2 | Corynorhinus townsendii | 1* | ∼500 | |
| February 2020 | MR1-A | Myotis yumanensis | 1* | >100 | |
| June 2020 | MR1-B | Tadarida brasiliensis | 10 | >100 | |
| June 2020 | UR | Myotis yumanensis | 10 | >100 | |
| Yolo | June 2020 | TR1 | Tadarida brasiliensis | 10 | >1,000 |
| Sacramento | June 2020 | TR2 | Tadarida brasiliensis | 10 | >100,000 |
| June 2020 | TR3 | Tadarida brasiliensis | 10 | >1,000 | |
| June 2020 | TR4 | Tadarida brasiliensis | 10 | >1,000 | |
| Individual batse | |||||
| Marin | February 2020 | MB | Myotis californicus and Myotis yumanensis | 5 | |
Roost name.
Species found in each roost.
No. of samples refers to the number of individual vials filled (an asterisk [*] indicates that many guano samples from the same roost were collected and mixed into one larger jar) from guano piles. For other roosts, 10 smaller guano samples were pooled prior to processing.
Estimated total size of the colony.
For individual bat samples, guano samples were collected from free-flying individual bats captured during a field study.
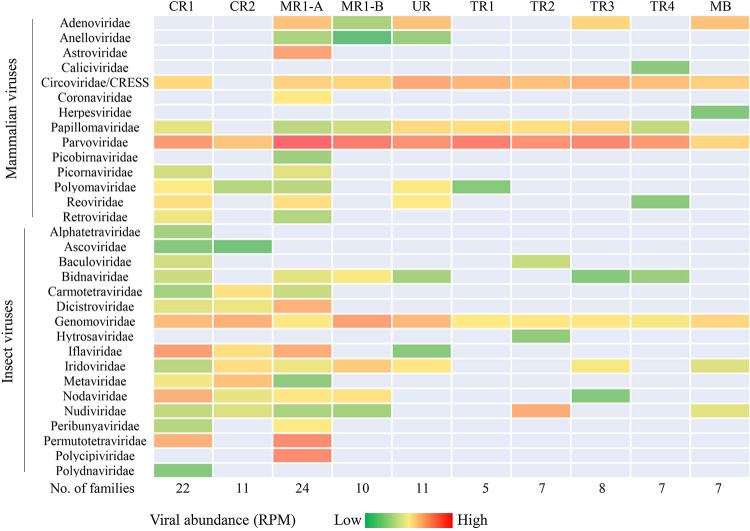
FIG 2.
Summary of the bat-associated viruses. All viral families identified from the 10 bat guano samples with E scores of <10−10. Only those eukaryotic viruses that could potentially infect mammals or insects are shown. Heat map was used to indicate the viral abundance (calculated as reads per million [RPM]), and RPM was displayed in log10 of each family. The numbers of viral families detected from each guano sample are listed at the bottom.
In total, we identified the presence of sequences related to 31 eukaryotic viral families. The most prevalent viruses, based on the overall number of sequence reads, belonged to the families Parvoviridae, Circoviridae, Genomoviridae, Papillomaviridae, Adenoviridae, Iridoviridae, Picornaviridae, Nudiviridae, Bidnaviridae, and Nodaviridae (Fig. 2). Viral sequences from the families Astroviridae, Caliciviridae, and Coronaviridae were found in only a single roost. The number of eukaryotic viral families detected from each roost ranged from 5 to 8 families in Tadarida brasiliensis (roosts TR1-4 from Yolo and Sacramento) and 11 to 24 families in Corynorhinus townsendii/Myotis yumanensis (roosts CR1-2 and MB1-2 from Marin). The majority of viral sequences showed limited protein identity to known viruses in the current database, indicating the detection of previously uncharacterized “new” viruses.
As in prior bat guano virome studies, sequences from bacterial viruses in the families Microviridae, Podoviridae, Siphoviridae, and Myoviridae, as well as viral families known to infect plants, algae, and protozoans were also detected (see File S1 in the supplemental material). These viral families, reflecting the presence of commensal gut prokaryotes, and/or that of consumed insects and their parasites, were not studied further (2, 17).
Identification of novel mammalian viruses.
Several novel complete or nearly complete viral genomes could be assembled, including those of an astrovirus, a chaphamaparvovirus, a nodavirus, 5 densoviruses, and 4 circular rep-expressing single-stranded DNA (CRESS-DNA) viruses. Fragments of an alphacoronavirus and a bunyavirus genome were also characterized. Phylogenetic analysis was used to compare these genomes to the most closely related and representative genomes in the same viral families.
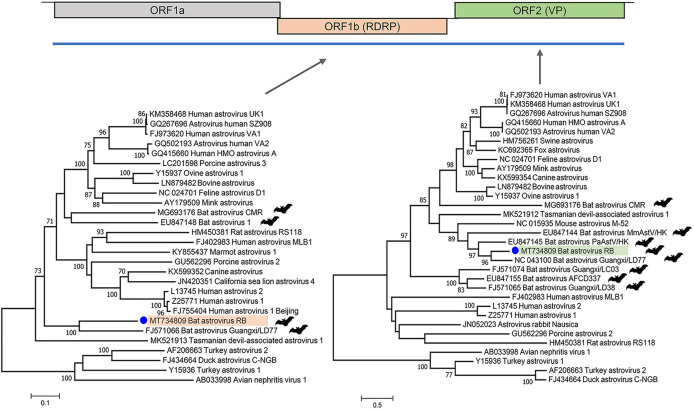
(i) Bat astrovirus. From the MR1-A roost, a nearly full-length astrovirus genome was generated (6,650 bases; GenBank accession number MT734809), encoding the three prototypical astrovirus open reading frames (ORFs; ORF1a/1b/2). Phylogenetic analyses of the full-length ORF1a (RNA-dependent RNA polymerase [RdRp]) and ORF2 (capsid protein VP) of this bat astrovirus RB (BAstV/RB) showed a close relationship with another bat (Taphozous melanopogon) astrovirus (Mamastrovirus 15; GenBank accession number FJ571066) described in 2019, China (30) (Fig. 3). These viruses share 66.2% and 57.0% identity in their RdRp and capsid proteins, respectively, and <50% identity to all other astroviruses in the capsid protein. Thus, bat astrovirus BAstV/RB is a new species under the genus Mamastrovirus based on the International Committee on the Taxonomy of Viruses (ICTV) criteria defining a new astrovirus species based on amino acid distance of their VP (31).
FIG 3.
Genome organization of astrovirus and phylogenetic analysis using the maximum likelihood method based on the complete amino acid sequence of the RNA-dependent RNA polymerase (RdRp) protein and capsid protein. The blue line indicates the genome coverage we got from this virus. Both Avastrovirus and Mamastrovirus reference genomes were included for phylogenetic analysis.
(ii) Bat parvovirus. From bat roost CR1, a 3,941-bases contig of a chaphamaparvovirus genome, named bat chaphamaparvovirus JR (GenBank accession number MT734803) could be assembled. Similarly to murine kidney parvovirus, ORFs that encode NS1, VP1, NP, and p15 were detected (Fig. 4). The 5′ ORF that encodes p10 fell outside the sequenced region. Phylogenetic analyses based on both NS1 and VP1 protein demonstrated that this virus clustered with chaphamaparvoviruses found in other bats, capuchins, Tasmanian devils, and mice (Fig. 4). NS1 and VP1 shared 64.4% to 67.0% and 66.1% to 69.2% identity, respectively, to those viruses in this cluster, with capuchin kidney parvovirus being the closest relative.
FIG 4.
Genome organization of chaphamaparvovirus and phylogenetic analysis using the maximum likelihood method based on the complete amino acid sequences of the NS1 and VP1 proteins. All currently known reference sequences from the Chaphamaparvovirus genus were included.
(iii) Bat coronavirus. Several reads of a coronavirus were identified from roost CR1. In order to check for the presence of other coronaviruses, including SARS-like genomes, we tested each library using previously described universal coronavirus PCR primers (23, 32). A conserved 440-bp RdRp region (GenBank accession number MT734810) was generated from the same CR1 roost. Phylogenetic trees based on the RdRp region and two contigs (447 bp) of the spike region indicated that it belongs to the Alphacoronavirus genus, with its sequenced RdRp region sharing ∼84% identity with a previously described coronavirus from free-tailed bats reported in both Brazil and Florida (33) (Fig. 5). The two spike gene contigs also showed closest identity (∼80%) to those found in Myotis lucifugus, in Colorado, USA (GenBank accession number KF430219). The multiple coronavirus consensus PCRs targeting both alpha- and betacoronaviruses were negative for all of the other roosts, consistent with metagenomic sequencing.
FIG 5.
Genome organization of coronavirus and phylogenetic analysis using the maximum likelihood method based on the 440-bp RdRp region and contigs from the spike gene. Both alphacoronaviruses and betacoronaviruses were included for the phylogenetic tree.
Distant relative of reported mammalian viruses.
Short regions of other divergent mammalian viruses in the families Adenoviridae, Papillomaviridae, Picornaviridae, Polyomaviridae, and Reoviridae were also detected, but limited numbers of sequencing reads precluded assembly of a large fraction of their genomes. These sequences were highly divergent from those of their closest relatives, sharing protein similarities ranging from 30% to 85%.
Close relatives of known mammalian viruses.
Also identified were reads and contigs that displayed a translated protein sequence identity of >90% to previously described viral proteins (Table 2; see also File S2 in the supplemental material). These viruses included calicivirus, circovirus, adenovirus, hepatovirus, bocavirus, and polyomavirus. The detection of closely related viruses in different bat species can be interpreted as reflecting cross-species transmissions.
TABLE 2.
Viral sequences that share high similarity with those of known viruses
| Virus hita | GenPept or GenBank accession no.b | Sample origin | Countryc | E value | Identity (%) | No. of contigs/reads | Total length (bp) | Roost |
|---|---|---|---|---|---|---|---|---|
| Bat calicivirus A10 | AWK23451 | P. subflavus | USA | 8E−58 | 100% | 1 | 284 | TR4 |
| Bat circovirus POA/V | AIX11629 | M. molossus/T. brasiliensis | Brazil | 2E−78 | 93.4% | 1 | 369 | TR1 |
| Bat hepatovirus | YP_009505614 | Coelura afra | Ghana | 1E−23 | 93.9% | 1 | 150 | CR1 |
| Bat mastadenovirus | AWT57880 | Myotis emarginatus | Spain | 8E−61 | 96.8% | 1 | 289 | MR1-A |
| Bat mastadenovirus G | YP_009325345 | Corynorhinus rafinesquii | USA | ∼1E−39 to 2E−68 | ∼93 to 97.2 | 2 | 757 | MR1-A |
| Bat bocaparvovirus | AIF74240 | Myotis pequinius | China | ∼1E−41 to 4E−51 | ∼92.8 to 97.6 | 2 | 462 | MR1-A |
| Bocaparvovirus sp. | AYG97822 | Rodents | China | ∼7E−47 to 9E−68 | ∼93.7 to 98.7 | 3 | 812 | MR1-B |
| Canine parvovirus 2 | – | Carnivores | * | 0 | 100 | 2 | 549 | TR4 |
| Bocaparvovirus 1 | AUD40074 | Himalayan marmot | China | ∼6E−57 to 3E−97 | ∼93.9 to 96.8 | 2 | 743 | MR1-B |
| Mouse kidney parvovirus | NC_040843 | Mus musculus | Australia/USA | ∼1E−177 to 0 | ∼97.5 to 98.6 | 4 | 2,063 | MR1-A |
| Myotis myotis bocavirus 1 | YP_009508788 | Myotis myotis | China | ∼8E−24 to 1E−53 | ∼91.4 to 93.7 | 2 | 452 | MR1-A |
| Porcine bocavirus 1 | AEM43610 | Pig | * | 1E−30 | 91.20 | 1 | 251 | MR1-B |
| Bat polyomavirus | AIF74282 | Rhinolophus ferrumequinum | China | 4E−42 | 94.50 | 1 | 221 | UR |
| Gammapapillomavirus 11 | ATQ38341 | Human | USA | 1E−46 | 100 | 1 | 225 | CR1 |
| Peromyscus papillomavirus 1 | YP_009508760 | Peromyscus (deer mouse) | USA | ∼6E−10 to 2E−101 | 91.6 to 100 | 3 | 834 | UR |
| Human rotavirus A | AIE45278 | Human | * | 1E−45 | 95.0 | 1 | 245 | MR1-A |
| Rotavirus H | – | Pig | * | ∼1E−18 to 4E−76 | ∼93.7 to 100 | 9 | 2,000 | UR |
Virus hits from NCBI database that shared high identity to the viral contigs/reads in this study.
A dash (–) indicates that the sequence shared the same identity (%) with multiple reference genomes.
An asterisk (*) indicates that the reference sequence could be found in multiple locations.
One contig and one read totaling 549 bases and showing 100% similarity to canine parvovirus 2 (CPV2) was found in guano from roost TR4. CPV2 tropism has been extensively studied as an example of a viral host jump from cats to dogs (34, 35). CPV2 has also been reported in multiple other carnivores, such as fox, raccoons, coyotes, puma, and minks (36). In MR1-A guano, we also detected 4 contigs totaling 2,063 bases (GenBank accession number MW151762) that shared more than 97.5% to 98.6% nucleotide similarity to over 46% of the mouse kidney parvovirus (MKPV) genome (GenBank accession number NC_040843.1). MKPV was only recently reported in wild New York City mice (37) and was shown to cause kidney failure in immunodeficient laboratory mice (38, 39). The presence of both of these parvovirus genomes in original guano samples TR4 and MR1-A was confirmed by reextraction, PCR, and amplicon Sanger sequencing (see Materials and Methods).
Viruses of unknown tropism—CRESS-DNA genomes.
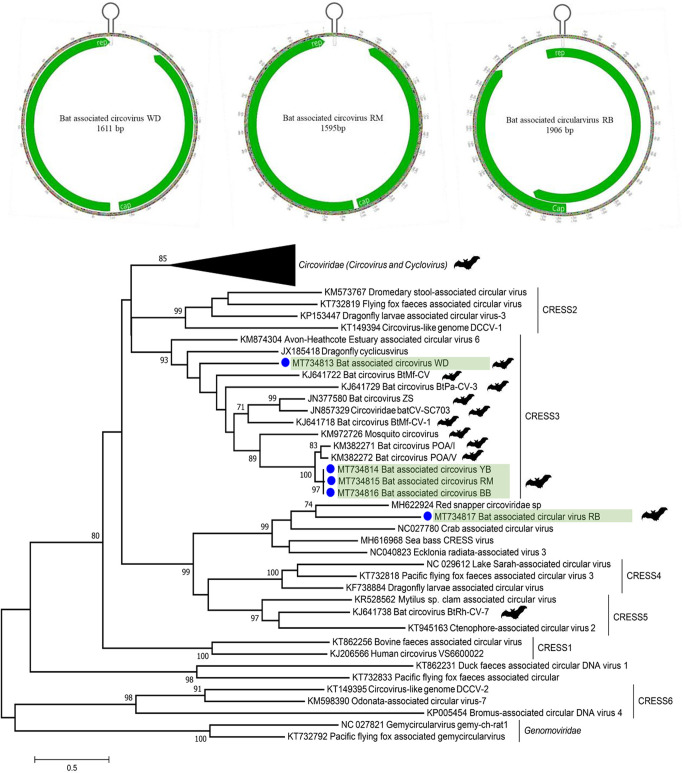
Circular Rep-encoding ssDNA (CRESS-DNA) genomes were detected in nearly all bat roosts (Fig. 2), and several complete circular genomes could be assembled. Genomes encoded ambisense or monosense Rep and Cap ORFs and a stem-loop structure with a conserved nonanucleotide motif (Fig. 6). Three genomes were 1,595 bases in length, sharing >99.5% nucleotide identity (bat-associated CRESS-DNA BB/YB/RM) with each other and 88.8% nucleotide identity with bat “circovirus” POV/I found in Brazil (40). Another genome (bat-associated circovirus WD; GenBank accession number MT734813) was 1,611 bases and showed 51% identity with its closest bat CRESS-DNA relative identified in China (41). Based on recent ICTV classification, these genomes clustered in the CRESS3 clade (42). Another 1,906-bp circular genome was assembled from bat roost MR1-A (bat-associated circular virus RB; GenBank accession number MT734817). The phylogenetic tree showed that its Rep protein mapped outside the six major CRESS1-6 clades (Fig. 6), sharing less than 40% identity with other, still unclassified, CRESS-DNA genomes.
FIG 6.
Genome organization of bat-associated CRESS-DNA WD/RM viruses and bat-associated circular virus RB. The phylogenetic tree was generated using the maximum likelihood method based on the complete amino acid sequence of the Rep protein. Reference sequences from cyclovirus, circovirus, and CRESS-DNA viruses in Circoviridae were included for the phylogenetic tree.
Insect viruses.
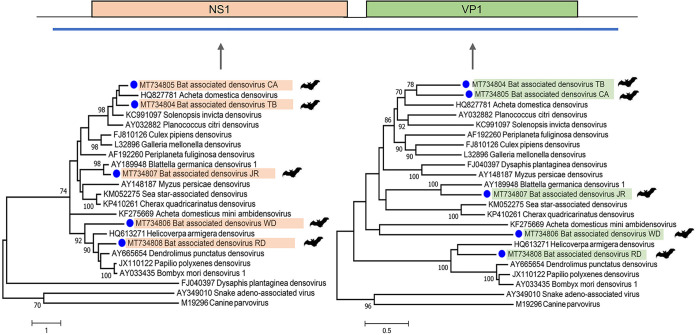
(i) Densovirus. In total, we could assemble five nearly full-length densovirus genomes from different bat roosts. Based on the complete NS1 and VP1 protein, all densoviruses were distinct and highly divergent from available genomes in GenBank (Fig. 7). Bat-associated densovirus CA and TB (GenBank accession numbers MT734804 and MT734805) clustered within the newly proposed Scindoambidensovirus genus, sharing ∼45% identity at NS1 and ∼36% at VP1 with Acheta domestica densovirus (GenBank accession number HQ827781). Bat-associated densovirus JR (GenBank accession number MT734807) clustered within the Blattambidensovirus genus, sharing ∼65% identity at NS1 and ∼42% at VP1 with Blattella germanica densovirus (GenBank accession number AY189948). Bat-associated densovirus RD (GenBank accession number MT734808) clustered with the Iteradensovirus genus, sharing ∼39% identity at NS1 with Dendrolimus punctatus densovirus (GenBank accession number NC_006555) and ∼41% at VP1 with Helicoverpa armigera densovirus (GenBank accession number NC_015718). Bat-associated densovirus WD (GenBank accession number MT734806) showed no clear clustering with any densovirus genera, sharing less than 35% identity at NS1 to any described densovirus. Therefore, based on ICTV criteria, it represents a possible member of a new genus in the Densovirinae subfamily.
FIG 7.
Genome organization of densovirus and phylogenetic analysis using the maximum likelihood method based on the complete amino acid sequences of the NS1 and VP1 proteins. Reference genomes from the Densovirinae subfamily were used for the phylogenetic tree, with adeno-associated virus and canine parvovirus as the outgroup.
(ii) Nodavirus. Five roosts were positive for nodavirus sequences, and from roost CR we were able to generate a nearly full-length genome of both genomic segments. Segment 1 of the bat-associated nodavirus JR (GenBank accession number MT734811) was 3,119 bases in length, and the closest relative with ∼40% identity over RdRp was an alphanodavirus (Nodamura virus) identified in Culex tritaeniorhynchus (GenBank accession number NC_002690) (Fig. 8). The genome of segment 2 was 1,799 bases in length (GenBank accession number MT734812), encoding a capsid protein with ∼32% identity with that of a Lutzomyia nodavirus identified in the sand fly Lutzomyia longipalpis (GenBank accession number KR003800).
FIG 8.
Genome organization of nodavirus and phylogenetic analysis using the maximum likelihood method based on the complete amino acid sequences of the RdRp protein (segment 1) and capsid protein (segment 2).
(iii) Bunyavirus. From our bat guano samples, we found two roosts positive for bunyaviruses, and we were able to assemble several contigs from both of the samples, all of which could be mapped to M and L segments (Fig. 9). Using the largest contig (838 bp from roost MR1-A) over the RdRp region, a phylogenetic tree indicated that this sequence clustered with bat bunyavirus JTM discovered in Rhinolophus ferrumequinum, sharing ∼88% identity at the amino acid level. The longest bunyavirus contig (594 bp) from roost CR1 was closest to a bunyavirus from an Australian flea (∼47% identity; GenBank accession number MN167501).
FIG 9.
Genome organization of bunyavirus and phylogenetic analysis using the maximum likelihood method based on the amino acid of the largest contig in the L segment.
DISCUSSION
Detection of novel viruses in bats is important for both surveillance and monitoring of bat populations, which provide important ecosystem functions, and for monitoring and understanding potential viral spillover between species. In order to characterize common enteric bat viruses, we analyzed guano samples of several Northern California bat roosts using viral metagenomics. Some eukaryotic viral genomes were completely or partially sequenced, while others were detected only in the form of one or a few viral reads. For parvoviruses, astrovirus, and coronavirus, their cellular hosts are likely enteric bat cells, while for other viruses, such as densoviruses and nodavirus, a dietary origin is expected, e.g., from ingested insects. For other viruses such as those with CRESS-DNA genomes, their cellular origin remains unknown but could conceivably be from ingested food or from parasites in their guts. Beside circoviruses, which are known to infect numerous birds, reptiles, and mammals, there is currently no evidence for replication of other CRESS-DNA viruses in mammalian cells.
Astroviruses can infect a wide range of hosts, including diverse birds and mammals (including humans), resulting often in asymptomatic infections but also diarrheal as well as occasional neurological infections (43, 44). We characterized a nearly complete genome of a novel mamastrovirus whose closest, although still considerably divergent, relative is from another bat (Taphozous melanopogon) found in South and South East Asia (30). A large number of astroviruses have been previously reported in bats and their genomes partially sequenced (45–51). Phylogenetic analysis has shown multiple bat astrovirus-containing clades separated by astroviruses from other mammals, likely reflecting multiple prior cross-species spillovers.
Members of the Parvoviridae family consist of nonenveloped icosahedral virions with single-stranded DNA genomes of 4 to 6 kb (52, 53). The Chaphamaparvovirus clade (previously known as chapparvoviruses) is a rapidly expanding genus whose members have been identified in numerous vertebrate animals, including rats and mice, bats, rhesus macaques, dogs, pigs, and Tasmanian devils, as well as birds and fish (54, 55). Murine chaphamaparvovirus (murine kidney parvovirus) was shown to be the cause of nephropathy in laboratory mice (38), and a recent study reported the detection of another chaphamaparvovirus genome in a plasma sample from a febrile individual (56). Here, we describe a new member of the Chaphamaparvovirus genus in bats, which joins the three previously reported bat relatives from Desmodus rotundus (common vampire bat) from Brazil (57) and from Eidolon helvum (straw-colored fruit bats) from Cameroon (58) and Ghana (59).
Also characterized were multiple genomes of CRESS-DNA viruses from different guano samples. These genomes are frequently detected in vertebrate fecal samples and diverse environmental samples and have recently been classified within a new phylum (42, 60). While the Cressdnaviricota phylum currently includes seven families, only one, the Circoviridae, includes viruses known to infect vertebrates (42, 60). Four of 5 Rep proteins in the CRESS-DNA genomes characterized here could be mapped to the CRESS3 family, while another may belong to a yet-to-be-described family. The host cells replicating these genomes may conceivably be enteric bat cells or parasites inside bat guts (61–63). An alternative source is from the largely insectivorous diets of these bats. We and others also reported that a CRESS-DNA genome (from a different genus) was released from silica-based nucleic acid purification columns (64, 65). Here, we used magnetic beads for nucleic acid extraction rather than the contaminated extraction columns (Materials and Methods). We also did not detect the same bat guano CRESS-DNA genomes in numerous prior studies using the same procedure. Lastly, the detection of closely related CRESS-DNA genomes in independent bat guano studies further supports the tentative conclusion that these CRESS-DNA genomes were genuinely present in guano.
Fragments of a novel alphacoronavirus could be derived from roost MR1-A, which consisted of Myotis yumanensis (Yuma myotis). That partial genome was distinct from previously reported bat alphacoronaviruses. The most closely related alphacoronaviruses in the RdRp region were from Tadarida brasiliensis (Mexican/Brazilian free-tailed bat) in Florida (GenBank accession number KX663833) and Brazil (GenBank accession number KC110781), and an undefined bat from Brazil (GenBank accession number MG266057). Alphacoronaviruses have been reported in multiple bat species from Colorado, USA (66), Trinidad (67), and Mexico (68), although none of them were closely related to SARS-CoV, SARS-CoV-2, or MERS, which are classified in the Betacoronavirus genus (69).
The viruses for which the largest numbers of reads could be identified were densoviruses. Densoviruses are known to infect insects and have been recently reclassified into seven genera (54, 55). Members of the Miniambidensovirus, Blattambidensovirus, Scindoambidensovirus, and Iteradensovirus genera were sequenced here (54, 55). While densoviruses have occasionally been reported in sterile mammalian samples (56, 70), their only currently known tropism consists of invertebrates, mainly insects (71). The detection of highly distinct densoviruses likely reflects the diversity of these bats’ insect diets. Nodavirus and bunyavirus genome segments were also detected. Members of these viral groups are capable of infecting insects as well as vertebrates (only fish in the case of nodaviruses), in which bunyaviruses can results in viremia (72, 73). Detection of these viral genomes in guano, rather than in plasma, indicates that their path was likely through ingestion of infected insects rather than by infection of bat cells.
Other viruses were detected with smaller numbers of reads but higher levels of similarity to previously reported bat viruses (Table 2). The detection of such closely related viruses in different bat species from different continents indicates that these viruses are likely to have a wide host range and are capable of infecting multiple bat species.
The detection of genome fragments of well-studied parvoviruses of carnivores (CPV2) and of mice (MKPV) in two guano samples was unexpected. The source of these genomes in bat guano remains unclear and may reflect either enteric infection of the sampled bats or, more likely given their currently known tropism and the low number of reads, as a result of contamination of guano by carnivores and rodents.
Characterizing bat virus diversity is important for understanding the ecological drivers of viral diversity in bats, and surveillance through guano samples collected at roosts allow for noninvasive virus monitoring and discovery of novel pathogens, including surveillance for viruses with zoonotic potential. Noninvasive screening of bat viruses also allows researchers to monitor for potential reverse zoonotic spillover of SARS-CoV-2 into North American bat populations, some of which may be susceptible to infection (74). The detection of SARS-CoV2 RNA in the feces of human (75) and other animals (76, 77) and the original detection of its closest relative (coronavirus RaTG13) from a fecal swab of a Rhinolophus affinis bat (9) does indicate that bat guano provide an readily accessible and appropriate material to screen for such viruses.
MATERIALS AND METHODS
Sample collection and virus enrichment.
Bat guano samples were collected from Marin, Yolo, and Sacramento counties in Northern California, USA (Table 1). Multiple guano samples were collected from the ground beneath each roost and pooled. Samples were initially stored at 4°C for several days and then transferred to −80°C until use. Two large maternity roosts from Marin County consisting of Corynorhinus townsendii and roosts containing Myotis yumanensis and Tadarida brasiliensis bats were sampled. Four roosts from Yolo and Sacramento counties, consisting exclusively of Tadarida brasiliensis, were also sampled. Fecal samples from individual bats were also collected from two Myotis californicus and three Myotis yumanensis bats during winter bat capture studies (CA Fish and Wildlife permit SC-10779).
Virus particle-associated nucleic acid enrichment was carried out based on our previously described methods (61). Briefly, 2 g of each guano sample was vigorously vortexed with 2 ml phosphate-buffered saline (PBS) and zirconia beads. The homogenate was centrifuged at 8,000 × g for 10 min at 4°C, and the supernatant was passed through a 0.45-μm filter (Merck Millipore, MA, USA). The filtrate was then digested with a cocktail of enzymes (Turbo DNase [Thermo Fisher Scientific, MA, USA]; Baseline Zero DNase [Epicentre, WI, USA]; Benzonase nuclease [Novagen, MA, USA]; and RNase A [Thermo Fisher Scientific]) at 37°C for 90 min to reduce the concentration of free nucleic acids (61). Residual RNA/DNA (protected from digestion within viral particles) was then extracted using magnetic beads covered with a proprietary silica-like coating (MagMax viral RNA isolation kit; Ambion, Inc., TX, USA).
Viral metagenomics analysis.
Nucleic acids were first amplified using random reverse transcription-PCR (RT-PCR) (78) using a PCR primer with a random nonamer at the 3′ end, followed by second-strand synthesis using Klenow polymerase (New England Biolabs, MA, USA) (61). Both cDNA and DNA were then amplified by AmpliTaq Gold DNA polymerase (Thermo Fisher Scientific), using the same PCR primer without its randomized 3′ end. An Illumina library was generated using the transposon-based Nextera XT sample preparation kit (Illumina, CA, USA) and sequenced on the MiSeq platform (2 × 250 bases, dual barcoding; Illumina). An inhouse pipeline was followed for the bioinformatic analysis (61); briefly, adaptor and primer sequences are trimmed using the default parameters of VecScreen (National Center for Biotechnology Information, MD, USA), duplicate reads were removed, and low-sequencing-quality tails were trimmed. Human and bacterial reads were subtracted by mapping to human reference genome hg38 and bacterial nucleotide sequences using Bowtie 2 v2.2.4 (79). De novo assembly was achieved by Ensemble Assembler program (v1.0) (80). Both contigs and singlets were then analyzed using BLASTx (v2.2.7) to search an in-house viral proteome database, and then candidate viral hits were aligned to the BLAST nonredundant (NR) universal proteome database using DIAMOND v0.9.15.116 (81).
Genome assembly and phylogenetic analysis.
Viral reads and contigs were aligned to reference viral genomes to generate full/partial genome sequences by Geneious R11 program (82). Sequences were first translated into amino acids and aligned using ClustalW. Phylogenetic trees were inferenced using the maximum likelihood method with MEGA v7.0 (83). The model test module of MEGA v7.0 was used to determine the best substitution model. Phylogenetic trees based on protein/nucleotide sequences were generated using the bootstrap method (1,000 times) under a GTR+I+G model.
PCR used to check for coronaviruses, canine parvovirus 2, and mouse kidney parvovirus.
Viral nucleci acids were directly extracted from the guano supernatant (not subjected to filtration and nuclease treatment) using a QIAamp virus minikit (Qiagen, Hilden, Germany). Reverse transcription was performed with by SuperScript III reverse transcriptase (Thermo Fisher Scientific). A 440-bp region of the RdRp of alpha- and betacoronaviruses was targeted for amplification using a published protocol with the first-round PCR primers 5′-CTTATGGGTTGGGATTATCCTAAGTGTGA-3′ and 5′-CTTATGGGTTGGGATTATCCCAAATGTGA-3′ and the second-round primers 5′-GGGTTGGGACTATCCTAAGTGTGA-3′ and 5′-CCATCATCAGATAGAATCATCATG-3′ (23, 32). The PCR primers used to confirm the presence of canine parvovirus 2 DNA were 5′-AAGACGTGCAAGCGAGTCC-3′ and 5′-GAGCGAAGATAAGCAGCGTAA-3′. The presence of MKPV DNA was confirmed using nested PCR with first round 5′-CAACATGGGGTCCACTCTCC-3′ and 5′-TAGGGCGCTGTCAAAGGAAG-3′ and second round 5′-TATGCACCAACATGGGGTCC-3′ and 5′-GGTGGCTTTACTGTCGGTGA-3′ primers. The PCR programs were as follows: 95°C for 3 min and 40 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 40 s, followed by an extension at 72°C for 10 min. PCR products were visualized on agarose gel and Sanger sequenced.
Data availability.
The short-read sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA565775 (BioSample accession numbers SAMN15468904 to SAMN15468913) and GenBank accession numbers MT734803 to MT734817 and MW151762).
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by Vitalant Research Institute and by the U.S. Geological Survey Ecosystems Mission Area for collection of guano samples.
Brian Reichert provided valuable comments on a draft of the manuscript.
Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Fenton M, Simmons N. 2015. Bats: a world of science and mystery, 1st ed. University of Chicago Press, Chicago, IL. [Google Scholar]
- 2.Bolatti EM, Zorec TM, Montani ME, Hošnjak L, Chouhy D, Viarengo G, Casal PE, Barquez RM, Poljak M, Giri AA. 2020. A preliminary study of the virome of the South American free-tailed bats (Tadarida brasiliensis) and identification of two novel mammalian viruses. Viruses 12:422. doi: 10.3390/v12040422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayman DT. 2016. Bats as viral reservoirs. Annu Rev Virol 3:77–99. doi: 10.1146/annurev-virology-110615-042203. [DOI] [PubMed] [Google Scholar]
- 4.Teeling EC, Springer MS, Madsen O, Bates P, O’Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 5.Mendenhall IH, Wen DLH, Jayakumar J, Gunalan V, Wang L, Mauer-Stroh S, Su YCF, Smith GJD. 2019. Diversity and evolution of viral pathogen community in cave nectar bats (Eonycteris spelaea). Viruses 11:250. doi: 10.3390/v11030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 8.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19:1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, Fung J, Chan TTY, Fung KSC, Woo PCY. 2020. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1542–1547. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC, Bi Y, Shi W. 2020. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30:2196–2203.e2193. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahman SA, Hassan SS, Olival KJ, Mohamed M, Chang LY, Hassan L, Saad NM, Shohaimi SA, Mamat ZC, Naim MS, Epstein JH, Suri AS, Field HE, Daszak P, Henipavirus Ecology Research Group. 2010. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerg Infect Dis 16:1990–1993. doi: 10.3201/eid1612.091790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 14.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat Med 10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, Frieman MB, Baric RS, Donaldson EF. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geldenhuys M, Mortlock M, Weyer J, Bezuidt O, Seamark ECJ, Kearney T, Gleasner C, Erkkila TH, Cui H, Markotter W. 2018. A metagenomic viral discovery approach identifies potential zoonotic and novel mammalian viruses in Neoromicia bats within South Africa. PLoS One 13:e0194527. doi: 10.1371/journal.pone.0194527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohl C, Brinkmann A, Radonić A, Dabrowski PW, Nitsche A, Mühldorfer K, Wibbelt G, Kurth A. 2020. Zwiesel bat banyangvirus, a potentially zoonotic Huaiyangshan banyangvirus (formerly known as SFTS)-like banyangvirus in northern bats from Germany. Sci Rep 10:1370. doi: 10.1038/s41598-020-58466-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmier A, Tirera S, de Thoisy B, Franc A, Darcissac E, Donato D, Bouchier C, Lacoste V, Lavergne A. 2017. Virome analysis of two sympatric bat species (Desmodus rotundus and Molossus molossus) in French Guiana. PLoS One 12:e0186943. doi: 10.1371/journal.pone.0186943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, Qian Z, Dong J, Sun L, Zhu Y, Du J, Yang F, Zhang S, Jin Q. 2016. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J 10:609–620. doi: 10.1038/ismej.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB. 2010. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J Virol 84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett AJ, Bushmaker T, Cameron K, Ondzie A, Niama FR, Parra HJ, Mombouli JV, Olson SH, Munster VJ, Goldberg TL. 2019. Diverse RNA viruses of arthropod origin in the blood of fruit bats suggest a link between bat and arthropod viromes. Virology 528:64–72. doi: 10.1016/j.virol.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS, Zheng XS, Wang MN, Daszak P, Wang LF, Cui J, Shi ZL. 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, Middleton D, Reid PA, McFarlane RA, Martin G, Tabor GM, Skerratt LF, Anderson DL, Crameri G, Quammen D, Jordan D, Freeman P, Wang LF, Epstein JH, Marsh GA, Kung NY, McCallum H. 2015. Ecological dynamics of emerging bat virus spillover. Proc Biol Sci 282:20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar A, Murthy S, Kapoor A. 2017. Evolution of selective-sequencing approaches for virus discovery and virome analysis. Virus Res 239:172–179. doi: 10.1016/j.virusres.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC. 2016. Uncovering Earth’s virome. Nature 536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 27.Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AM, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM. 2017. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 28.Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, Wang W, Eden JS, Shen JJ, Liu L, Holmes EC, Zhang YZ. 2018. The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 29.Shi M, Zhang YZ, Holmes EC. 2018. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res 243:83–90. doi: 10.1016/j.virusres.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu HC, Chu DKW, Liu W, Dong BQ, Zhang SY, Zhang JX, Li LF, Vijaykrishna D, Smith GJD, Chen HL, Poon LLM, Peiris JSM, Guan Y. 2009. Detection of diverse astroviruses from bats in China. J Gen Virol 90:883–887. doi: 10.1099/vir.0.007732-0. [DOI] [PubMed] [Google Scholar]
- 31.de Souza WM, Fumagalli MJ, de Araujo J, Ometto T, Modha S, Thomazelli LM, Durigon EL, Murcia PR, Figueiredo LTM. 2019. Discovery of novel astrovirus and calicivirus identified in ruddy turnstones in Brazil. Sci Rep 9:5556. doi: 10.1038/s41598-019-42110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Souza Luna LK, Heiser V, Regamey N, Panning M, Drexler JF, Mulangu S, Poon L, Baumgarte S, Haijema BJ, Kaiser L, Drosten C. 2007. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol 45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lima FE, Campos FS, Kunert Filho HC, Batista HB, Carnielli P, Jr, Cibulski SP, Spilki FR, Roehe PM, Franco AC. 2013. Detection of Alphacoronavirus in velvety free-tailed bats (Molossus molossus) and Brazilian free-tailed bats (Tadarida brasiliensis) from urban area of Southern Brazil. Virus Genes 47:164–167. doi: 10.1007/s11262-013-0899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Organtini LJ, Allison AB, Lukk T, Parrish CR, Hafenstein S. 2015. Global displacement of canine parvovirus by a host-adapted variant: structural comparison between pandemic viruses with distinct host ranges. J Virol 89:1909–1912. doi: 10.1128/JVI.02611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palermo LM, Hafenstein SL, Parrish CR. 2006. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J Virol 80:8482–8492. doi: 10.1128/JVI.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allison AB, Kohler DJ, Ortega A, Hoover EA, Grove DM, Holmes EC, Parrish CR. 2014. Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog 10:e1004475. doi: 10.1371/journal.ppat.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams SH, Che X, Garcia JA, Klena JD, Lee B, Muller D, Ulrich W, Corrigan RM, Nichol S, Jain K, Lipkin WI. 2018. Viral Diversity of House Mice in New York City. mBio 9:e01354-17. doi: 10.1128/mBio.01354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roediger B, Lee Q, Tikoo S, Cobbin JCA, Henderson JM, Jormakka M, O'Rourke MB, Padula MP, Pinello N, Henry M, Wynne M, Santagostino SF, Brayton CF, Rasmussen L, Lisowski L, Tay SS, Harris DC, Bertram JF, Dowling JP, Bertolino P, Lai JH, Wu W, Bachovchin WW, Wong JJ, Gorrell MD, Shaban B, Holmes EC, Jolly CJ, Monette S, Weninger W. 2018. An atypical parvovirus drives chronic tubulointerstitial nephropathy and kidney fibrosis. Cell 175:530–543.e524. doi: 10.1016/j.cell.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge Z, Carrasco SE, Feng Y, Bakthavatchalu V, Annamalai D, Kramer R, Muthupalani S, Fox JG. 2020. Identification of a new strain of mouse kidney parvovirus associated with inclusion body nephropathy in immunocompromised laboratory mice. Emerg Microbes Infect 9:1814–1823. doi: 10.1080/22221751.2020.1798288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lima FE, Cibulski SP, Dos Santos HF, Teixeira TF, Varela AP, Roehe PM, Delwart E, Franco AC. 2015. Genomic characterization of novel circular ssDNA viruses from insectivorous bats in Southern Brazil. PLoS One 10:e0118070. doi: 10.1371/journal.pone.0118070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y, Shi Z. 2012. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol 86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krupovic M, Varsani A, Kazlauskas D, Breitbart M, Delwart E, Rosario K, Yutin N, Wolf YI, Harrach B, Zerbini FM, Dolja VV, Kuhn JH, Koonin EV. 2020. Cressdnaviricota: a virus phylum unifying seven families of Rep-encoding viruses with single-stranded, circular DNA genomes. J Virol 94:e00582-20. doi: 10.1128/JVI.00582-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortez V, Meliopoulos VA, Karlsson EA, Hargest V, Johnson C, Schultz-Cherry S. 2017. Astrovirus biology and pathogenesis. Annu Rev Virol 4:327–348. doi: 10.1146/annurev-virology-101416-041742. [DOI] [PubMed] [Google Scholar]
- 44.Johnson C, Hargest V, Cortez V, Meliopoulos VA, Schultz-Cherry S. 2017. Astrovirus pathogenesis. Viruses 9:22. doi: 10.3390/v9010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afelt A, Lacroix A, Zawadzka-Pawlewska U, Pokojski W, Buchy P, Frutos R. 2018. Distribution of bat-borne viruses and environment patterns. Infect Genet Evol 58:181–191. doi: 10.1016/j.meegid.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu D, Zhu C, Wang Y, Ai L, Yang L, Ye F, Ding C, Chen J, He B, Zhu J, Qian H, Xu W, Feng Y, Tan W, Wang C. 2017. Virome analysis for identification of novel mammalian viruses in bats from Southeast China. Sci Rep 7:10917. doi: 10.1038/s41598-017-11384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seltmann A, Corman VM, Rasche A, Drosten C, Czirják G, Bernard H, Struebig MJ, Voigt CC. 2017. Seasonal fluctuations of astrovirus, but not coronavirus shedding in bats inhabiting human-modified tropical forests. Ecohealth 14:272–284. doi: 10.1007/s10393-017-1245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Taweel A, Kandeil A, Barakat A, Alfaroq Rabiee O, Kayali G, Ali MA. 2020. Diversity of astroviruses circulating in humans, bats, and wild birds in Egypt. Viruses 12:485. doi: 10.3390/v12050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu DK, Poon LL, Guan Y, Peiris JS. 2008. Novel astroviruses in insectivorous bats. J Virol 82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q. 2012. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol 86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drexler JF, Corman VM, Wegner T, Tateno AF, Zerbinati RM, Gloza-Rausch F, Seebens A, Müller MA, Drosten C. 2011. Amplification of emerging viruses in a bat colony. Emerg Infect Dis 17:449–456. doi: 10.3201/eid1703.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cotmore SF, Tattersall P. 2014. Parvoviruses: small does not mean simple. Annu Rev Virol 1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 53.Mietzsch M, Penzes JJ, Agbandje-McKenna M. 2019. Twenty-five years of structural parvovirology. Viruses 11:362. doi: 10.3390/v11040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Gordon E, Idle A, Altan E, Seguin MA, Estrada M, Deng X, Delwart E. 2020. Virome of a feline outbreak of diarrhea and vomiting includes bocaviruses and a novel chapparvovirus. Viruses 12:506. doi: 10.3390/v12050506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Penzes JJ, de Souza WM, Agbandje-McKenna M, Gifford RJ. 2019. An ancient lineage of highly divergent parvoviruses infects both vertebrate and invertebrate hosts. Viruses 11:525. doi: 10.3390/v11060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fahsbender E, Charlys da-Costa A, Elise Gill D, Augusto de Padua Milagres F, Brustulin R, Julio Costa Monteiro F, Octavio da Silva Rego M, Soares D'Athaide Ribeiro E, Cerdeira Sabino E, Delwart E. 2020. Plasma virome of 781 Brazilians with unexplained symptoms of arbovirus infection include a novel parvovirus and densovirus. PLoS One 15:e0229993. doi: 10.1371/journal.pone.0229993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Souza WM, Romeiro MF, Fumagalli MJ, Modha S, de Araujo J, Queiroz LH, Durigon EL, Figueiredo LTM, Murcia PR, Gifford RJ. 2017. Chapparvoviruses occur in at least three vertebrate classes and have a broad biogeographic distribution. J Gen Virol 98:225–229. doi: 10.1099/jgv.0.000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yinda CK, Ghogomu SM, Conceicao-Neto N, Beller L, Deboutte W, Vanhulle E, Maes P, Van Ranst M, Matthijnssens J. 2018. Cameroonian fruit bats harbor divergent viruses, including rotavirus H, bastroviruses, and picobirnaviruses using an alternative genetic code. Virus Evol 4:vey008. doi: 10.1093/ve/vey008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker KS, Leggett RM, Bexfield NH, Alston M, Daly G, Todd S, Tachedjian M, Holmes CE, Crameri S, Wang LF, Heeney JL, Suu-Ire R, Kellam P, Cunningham AA, Wood JL, Caccamo M, Murcia PR. 2013. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 441:95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breitbart M, Delwart E, Rosario K, Segalés J, Varsani A, Ictv Report C. 2017. ICTV virus taxonomy profile: Circoviridae. J Gen Virol 98:1997–1998. doi: 10.1099/jgv.0.000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siqueira JD, Dominguez-Bello MG, Contreras M, Lander O, Caballero-Arias H, Xutao D, Noya-Alarcon O, Delwart E. 2018. Complex virome in feces from Amerindian children in isolated Amazonian villages. Nat Commun 9:4270. doi: 10.1038/s41467-018-06502-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinsella CM, Bart A, Deijs M, Broekhuizen P, Kaczorowska J, Jebbink MF, van Gool T, Cotten M, van der Hoek L. 2020. Entamoeba and Giardia parasites implicated as hosts of CRESS viruses. Nat Commun 11:4620. doi: 10.1038/s41467-020-18474-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aiemjoy K, Altan E, Aragie S, Fry DM, Phan TG, Deng X, Chanyalew M, Tadesse Z, Callahan EK, Delwart E, Keenan JD. 2019. Viral species richness and composition in young children with loose or watery stool in Ethiopia. BMC Infect Dis 19:53. doi: 10.1186/s12879-019-3674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naccache SN, Greninger AL, Lee D, Coffey LL, Phan T, Rein-Weston A, Aronsohn A, Hackett J, Jr, Delwart EL, Chiu CY. 2013. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol 87:11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smuts H, Kew M, Khan A, Korsman S. 2014. Novel hybrid parvovirus-like virus, NIH-CQV/PHV, contaminants in silica column-based nucleic acid extraction kits. J Virol 88:1398. doi: 10.1128/JVI.03206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dominguez SR, O’Shea TJ, Oko LM, Holmes KV. 2007. Detection of group 1 coronaviruses in bats in North America. Emerg Infect Dis 13:1295–1300. doi: 10.3201/eid1309.070491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carrington CV, Foster JE, Zhu HC, Zhang JX, Smith GJ, Thompson N, Auguste AJ, Ramkissoon V, Adesiyun AA, Guan Y. 2008. Detection and phylogenetic analysis of group 1 coronaviruses in South American bats. Emerg Infect Dis 14:1890–1893. doi: 10.3201/eid1412.080642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pablo CS, Giovani HC. 2020. Bats and humans during the SARS-CoV-2 outbreak: the case of bat-coronavirus from Mexico. Transbound Emerg Dis doi: 10.1111/tbed.13751. [DOI] [PubMed] [Google Scholar]
- 69.Wong ACP, Li X, Lau SKP, Woo PCY. 2019. Global epidemiology of bat coronaviruses. Viruses 11:174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phan TG, Messacar K, Dominguez SR, da Costa AC, Deng X, Delwart E. 2016. A new densovirus in cerebrospinal fluid from a case of anti-NMDA-receptor encephalitis. Arch Virol 161:3231–3235. doi: 10.1007/s00705-016-3002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tijssen P, Pénzes JJ, Yu Q, Pham HT, Bergoin M. 2016. Diversity of small, single-stranded DNA viruses of invertebrates and their chaotic evolutionary past. J Invertebr Pathol 140:83–96. doi: 10.1016/j.jip.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Yong CY, Yeap SK, Omar AR, Tan WS. 2017. Advances in the study of nodavirus. PeerJ 5:e3841. doi: 10.7717/peerj.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshimatsu K, Arikawa J. 2012. [Bunyavirus and its ecology]. Uirusu 62:239–250. (In Japanese.) doi: 10.2222/jsv.62.239. [DOI] [PubMed] [Google Scholar]
- 74.Olival KJ, Cryan PM, Amman BR, Baric RS, Blehert DS, Brook CE, Calisher CH, Castle KT, Coleman JTH, Daszak P, Epstein JH, Field H, Frick WF, Gilbert AT, Hayman DTS, Ip HS, Karesh WB, Johnson CK, Kading RC, Kingston T, Lorch JM, Mendenhall IH, Peel AJ, Phelps KL, Plowright RK, Reeder DM, Reichard JD, Sleeman JM, Streicker DG, Towner JS, Wang LF. 2020. Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: a case study of bats. PLoS Pathog 16:e1008758. doi: 10.1371/journal.ppat.1008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bwire GM, Majigo MV, Njiro BJ, Mawazo A. 2020. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: a systematic review and meta-analysis. J Med Virol doi:10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salajegheh Tazerji S, Magalhães Duarte P, Rahimi P, Shahabinejad F, Dhakal S, Singh Malik Y, Shehata AA, Lama J, Klein J, Safdar M, Rahman MT, Filipiak KJ, Rodríguez-Morales AJ, Sobur MA, Kabir F, Vazir B, Mboera L, Caporale M, Islam MS, Amuasi JH, Gharieb R, Roncada P, Musaad S, Tilocca B, Koohi MK, Taghipour A, Sait A, Subbaram K, Jahandideh A, Mortazavi P, Abedini MA, Hokey DA, Hogan U, Shaheen MNF, Elaswad A, Elhaig MM, Fawzy M. 2020. Transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to animals: an updated review. J Transl Med 18:358. doi: 10.1186/s12967-020-02534-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haake C, Cook S, Pusterla N, Murphy B. 2020. Coronavirus infections in companion animals: virology, epidemiology, clinical and pathologic features. Viruses 12:1023. doi: 10.3390/v12091023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L, Deng X, Mee ET, Collot-Teixeira S, Anderson R, Schepelmann S, Minor PD, Delwart E. 2015. Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J Virol Methods 213:139–146. doi: 10.1016/j.jviromet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. 2015. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res 43:e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 82.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The short-read sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA565775 (BioSample accession numbers SAMN15468904 to SAMN15468913) and GenBank accession numbers MT734803 to MT734817 and MW151762).