Differences in HIV pathogenesis between males and females, including immunity postinfection, have been well documented, as have steroid hormone effects on the microbiome, which is known to influence mucosal immune responses. Few studies have applied this knowledge to vaccine trials. We investigated two SIV vaccine regimens combining mucosal priming immunizations and systemic protein boosting. We again report a vaccine-induced sex bias, with female rhesus macaques but not males displaying significantly reduced acute viremia. The vaccine regimens, especially the mucosal primes, significantly altered the rectal microbiome. The greatest effects were in females. Striking differences between female and male macaques in correlations of prevalent rectal bacteria with viral loads and potentially protective immune responses were observed. Effects of the microbiome on vaccine-induced immunity and viremia control require further study by microbiome transfer. However, the findings presented highlight the critical importance of considering effects of sex and the microbiome in vaccine design and evaluation.
KEYWORDS: SIV vaccine, cellular immunity, humoral immunity, microbiome, mucosal and systemic immunization, rhesus macaque
ABSTRACT
An efficacious human immunodeficiency virus (HIV) vaccine will likely require induction of both mucosal and systemic immune responses. We compared the immunogenicity and protective efficacy of two mucosal/systemic vaccine regimens and investigated their effects on the rectal microbiome. Rhesus macaques were primed twice mucosally with replication-competent adenovirus type 5 host range mutant (Ad5hr)-simian immunodeficiency virus (SIV) recombinants and boosted twice intramuscularly with ALVAC-SIV recombinant plus SIV gp120 protein or with DNA for SIV genes and rhesus interleukin-12 plus SIV gp120 protein. Controls received empty Ad5hr vector and alum adjuvant only. Both regimens elicited strong, comparable mucosal and systemic cellular and humoral immunity. Prevaccination rectal microbiomes of males and females differed and significantly changed over the course of immunization, most strongly in females after Ad5hr immunizations. Following repeated low-dose intrarectal SIV challenges, both vaccine groups exhibited modestly but significantly reduced acute viremia. Male and female controls exhibited similar acute viral loads; however, vaccinated females, but not males, exhibited lower levels of acute viremia, compared to same-sex controls. Few differences in adaptive immune responses were observed between the sexes. Striking differences in correlations of the rectal microbiome of males and females with acute viremia and immune responses associated with protection were seen and point to effects of the microbiome on vaccine-induced immunity and viremia control. Our study clearly demonstrates direct effects of a mucosal SIV vaccine regimen on the rectal microbiome and validates our previously reported SIV vaccine-induced sex bias. Sex and the microbiome are critical factors that should not be overlooked in vaccine design and evaluation.
IMPORTANCE Differences in HIV pathogenesis between males and females, including immunity postinfection, have been well documented, as have steroid hormone effects on the microbiome, which is known to influence mucosal immune responses. Few studies have applied this knowledge to vaccine trials. We investigated two SIV vaccine regimens combining mucosal priming immunizations and systemic protein boosting. We again report a vaccine-induced sex bias, with female rhesus macaques but not males displaying significantly reduced acute viremia. The vaccine regimens, especially the mucosal primes, significantly altered the rectal microbiome. The greatest effects were in females. Striking differences between female and male macaques in correlations of prevalent rectal bacteria with viral loads and potentially protective immune responses were observed. Effects of the microbiome on vaccine-induced immunity and viremia control require further study by microbiome transfer. However, the findings presented highlight the critical importance of considering effects of sex and the microbiome in vaccine design and evaluation.
INTRODUCTION
The modest success of the RV144 clinical human immunodeficiency virus (HIV) vaccine trial in Thailand, resulting in 31.2% efficacy, provided hope that development of an efficacious HIV vaccine would be possible (1). The results also strengthened the interest of the vaccine field in humoral immunity, as IgG antibodies specific for HIV Env variable regions 1 and 2 (V1 and V2) were negatively correlated with the risk of HIV infection (2). Numerous approaches are under way, not only targeting the V1/V2 region but also aimed more generally at induction of broadly neutralizing antibodies. Strategies include improved design of native envelope trimers to serve as better immunogens (3, 4) and sequential vaccination strategies to steer the humoral immune response from germ line precursors eventually to the desired broadly neutralizing antibodies (5, 6). At the same time, while antibodies, including nonneutralizing antibodies that mediate protective functional activities, are of prime interest, development of vaccine strategies optimizing cellular immunity is ongoing. Polyvalent mosaic vaccines aim to induce immunity against the broad global spectrum of circulating HIV isolates (7) and have been shown to increase the breadth and depth of cellular immunity in nonhuman primates (NHPs) (8). Design of “conserved element” vaccines aims to circumvent dominant epitopes present in variable regions of the virus and to target a broad range of viral isolates by eliciting responses to subdominant epitopes present in conserved viral regions (9). A novel cytomegalovirus (CMV)-vectored vaccine was shown to be highly promising, resulting in viral clearance in approximately 50% of immunized and simian immunodeficiency virus (SIV)-challenged macaques (10). A recent attenuated version provided SIV control and progressive clearance in 59% of intravaginally challenged macaques (11). Persistent SIV-specific effector memory (EM) T cell responses restricted by major histocompatibility complex E (MHC-E) have been associated with this viral clearance (12).
The majority of these HIV vaccine approaches rely on systemic immunization; however, most HIV infections occur across the rectal/genital mucosa, suggesting a possible benefit of inducing strong mucosal immunity, as reviewed previously (13). In our preclinical vaccine trials, we have pursued a mucosal/systemic vaccine regimen, with priming by mucosal immunization to the upper respiratory tract with replication-competent adenovirus type 5 host range mutant (Ad5hr) recombinants able to replicate in rhesus macaques, followed by systemic intramuscular boosting with envelope proteins. This approach has led to partial protection against single high-dose HIV challenges in chimpanzees (14) and SIV (15, 16) and simian-human immunodeficiency virus (SHIV) (17, 18) challenges in rhesus macaques, as well as reduced risk of SIV acquisition following repetitive low-dose intrarectal challenges in rhesus macaques (19).
Recently, we evaluated the immunogenicity of several novel vaccine regimens that combined different mucosal priming and systemic booster immunizations (20–22). Among the booster immunizations, we evaluated an ALVAC/Env regimen mimicking the successful RV144 regimen except that the HIV immunogens of RV144 were changed to SIV. This regimen has since shown a vaccine efficacy in rhesus macaques of 44% (23). We also evaluated a DNA&Env regimen shown to elicit potent, long-lasting anti-Env antibodies (24, 25). The results of these combined strategies suggested that initial mucosal priming with replication-competent Ad5hr-SIV recombinants followed by boosting with either an ALVAC/Env or a DNA&Env regimen might induce strong, long-lasting, and functional systemic and mucosal SIV-specific antibodies. Here, we evaluated these two combined regimens in a preclinical vaccine study in rhesus macaques, comparing induction of cellular and humoral immune responses and subsequent protection against repeated low-dose intrarectal SIV challenges. Further, in view of our premise that mucosal priming is important for eventual vaccine protective efficacy, we investigated the microbiome over the course of immunization, as it is known to help shape mucosal immune responses (26).
RESULTS
Kinetics of cellular immune responses differ according to immunization route.
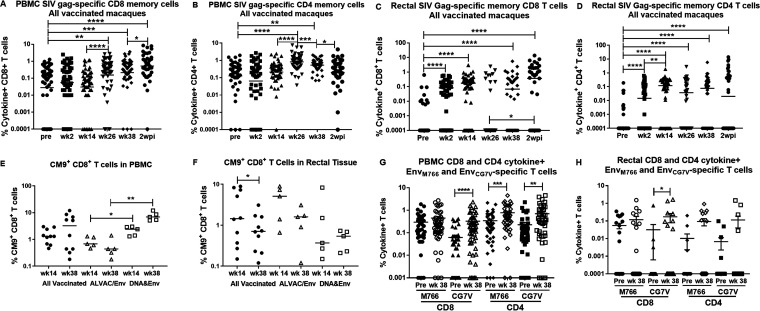
Induction of SIV Gag-specific CD8+ and CD4+ total memory (TM) T cells (defined in Materials and Methods) were assessed by intracellular cytokine staining and flow cytometry analysis prior to and 2 weeks following each immunization (see the vaccination schedule in Fig. 1) and 2 weeks postinfection (wpi). As shown in Fig. 2A and B, the frequencies of both CD8+ and CD4+ cytokine-positive TM T cells in blood increased significantly following the first intramuscular boost at week 24, with little change thereafter in the CD8+ T cell frequency and a slight decline in the CD4+ T cell frequency following the second boost at week 36. No changes in the CD8+ and CD4+ Gag-specific T cell frequencies in peripheral blood mononuclear cells (PBMCs) of the control animals were seen over the course of immunization (data not shown). In contrast to the PBMCs, SIV Gag-specific CD8+ and CD4+ TM T cell frequencies in rectal tissue were significantly induced by the first mucosal Ad5hr-SIV recombinant immunization (Fig. 2C and D). The rectal CD4+ T cells exhibited a further significant increase in frequency following the second Ad5hr-SIV recombinant immunization at week 12 (Fig. 2D). The rectal CD8+ and CD4+ TM T cell frequencies did not increase further following the booster immunizations. Similar to the PBMCs, no changes in the rectal CD8+ and CD4+ Gag-specific T cell frequencies of the control animals were observed over the course of immunization, although, compared to the preimmunization level, Gag-specific CD8+ TM T cells of the control macaques were increased marginally at 2 wpi (P = 0.041; data not shown). Overall, the sequential mucosal and systemic immunizations influenced induction of cellular immune responses in different compartments, with the mucosal primes eliciting responses in rectal tissue and the systemic boosts eliciting responses in PBMCs. This was also seen in analyses of CM9+ CD8+ T cells in a separate subset of 10 Mamu-A*01+ macaques. In PBMCs, CM9+ cells in all of the vaccinated macaques tended to increase at week 38 following the booster immunization (Fig. 2E). In contrast, in the same 10 macaques, CM9+ responses in rectal tissue significantly declined between week 14, following the mucosal Ad5hr-SIV recombinant immunization, and week 38 (Fig. 2F).
FIG 1.
Immunization and challenge scheme. As detailed in Materials and Methods, 50 macaques (24 females and 26 males) were primed mucosally at weeks 0 and 12 with replication-competent Ad5hr recombinants separately encoding SIVM766 gp120-TM and SIV239 Gag. Ten controls (4 females and 6 males) received empty Ad5hr vector. Subsequently, at weeks 24 and 36, macaques in the ALVAC/Env group (n = 25; 12 females and 13 males) were boosted systemically with ALVAC-SIVM766 Gag/pro/gp120-TM in one thigh and gD-SIVM766 gp120 and gD-SIVCG7V gp120, both in alum hydroxide, in the opposite thigh. Macaques in the DNA&Env group (n = 25; 12 females and 13 males) were boosted systemically with DNA encoding SIVM766 gp120-TM, SIV239 Gag, and macaque IL-12, followed by electroporation and immediate intramuscular administration of the same SIV gD-gp120 proteins in alum phosphate at the same anatomical sites. Control animals (5 in the ALVAC/Env group and 5 in the DNA&Env group) received alum hydroxide or alum phosphate only. At week 42, weekly repeated low-dose intrarectal challenges with SIVmac251 (1:500 dilution; 120 times the TCID50) were initiated. All macaques received up to 15 challenges until infection occurred. O, oral; IN, intranasal; IT, intratracheal; IM, intramuscular; IR, intrarectal.
FIG 2.
Kinetics of cellular immune responses differ according to immunization route. (A to D) Gag-specific total memory CD8+ (A and C) and CD4+ (B and D) T cells in PBMCs (A and B) and rectal tissue (C and D) are reported as percentages of cytokine-positive T cells (sum of IFN-γ+, IL-2+, and TNF-α+ cells) and are shown across the course of immunization for all vaccinated macaques. (E and F) The percentages of CM9+ CD8+ T cells in a group of vaccinated Mamu-A*01 macaques are shown for PBMCs (E) and rectal tissue (F). In both panels, results are shown for cells obtained at week 14 (after the second Ad5hr immunization) and at week 38 (after the second boost immunization) for all vaccinated macaques and for each vaccinated group separately. (G and H) EnvM766- and EnvCG7V-specific CD8+ and CD4+ cytokine-positive T cells prior to immunization and at week 38 in PBMCs (G) and rectal tissue (H) are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We focused here on the kinetics of Gag-specific T cell responses because they represented the most complete data sets for analyses of both PBMCs and rectal tissue. In some cases, we lacked sufficient rectal cells to assay more than one SIV antigen; in those cases, we selected Gag for analysis. However, we did evaluate induction of EnvM766- and EnvCG7V-specific CD8+ and CD4+ TM T cell responses. Significant increases in frequencies of EnvCG7V-specific but not EnvM766-specific CD8+ TM T cells in both blood and rectal tissue (Fig. 2G and H) were observed when preimmunization frequencies were compared to frequencies at the end of the immunization regimen (week 38). In contrast, the CD4+ Env-specific T cell frequencies increased significantly in blood for both the SIV Env strains (Fig. 2G), whereas no significant increases were seen for CD4+ T cells in rectal tissue (Fig. 2H).
A comparison of SIV Gag responses between the two vaccine regimens revealed few differences. The DNA&Env regimen exhibited a higher frequency of Gag-specific CD8+ TM T cells in blood at week 38, the end of the immunization regimen, compared to the ALVAC/Env group (mean percentages of 0.85 ± 0.19 and 0.31 ± 0.051, respectively; P = 0.011). No other differences in total Gag-specific CD4+ or CD8+ responses in blood and rectal tissue were observed. The CM9+ cell frequencies in blood were significantly elevated at both week 14 and week 38 in the DNA&Env group, compared to those in the ALVAC/Env group (Fig. 2E), whereas no differences between immunization groups were seen in rectal tissue (Fig. 2F). The higher CD8+ T cell responses in blood at week 38 elicited in the DNA&Env group are not surprising, as ALVAC-based vaccines poorly induce cellular responses (27). Moreover, interleukin 12 (IL-12) DNA present in the DNA immunogen has been shown to increase cytotoxic T cell responses in macaques immunized with SIV DNA vaccines (28).
Induction of binding and functional antibodies by the vaccine regimens.
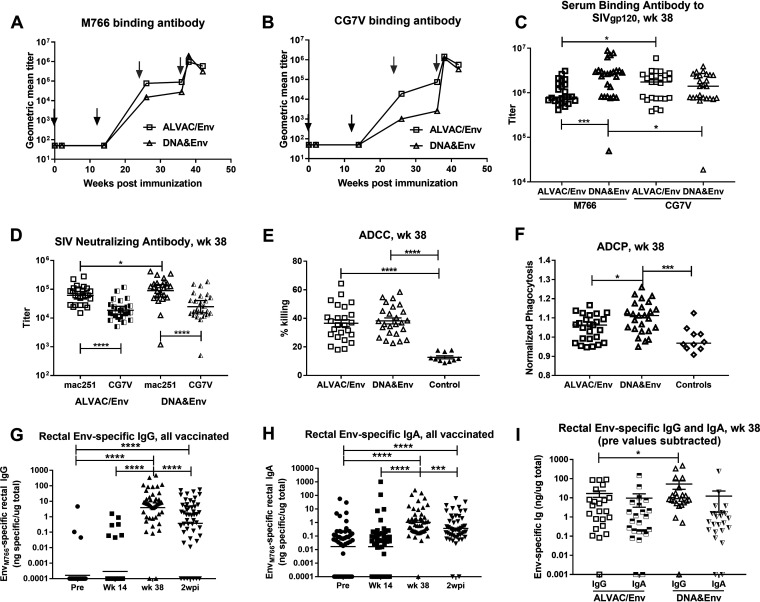
We next investigated the kinetics of serum SIV Env-specific antibody development, assessing reactivities to SIVM766 and SIVCG7V gp120s, as both were present in the booster immunogens. Significant binding antibodies did not develop until after the first systemic boost at week 24 (Fig. 3A and B). Comparisons of reactivities between the two vaccine arms gave mixed results. The DNA&Env group developed higher binding antibody titers to SIVM766 Env, compared to SIVCG7V Env, whereas the reverse was true for the ALVAC/Env group (Fig. 3C). Further, the DNA&Env group exhibited higher titers against SIVM766 Env than did the ALVAC/Env group. In keeping with its higher M766-specific binding titers, the DNA&Env group exhibited higher neutralizing antibody titers to tier 1A SIVmac251.6 than did the ALVAC/Env group (Fig. 3D), but no difference between groups in the ability to neutralize tier 1B SIVCG7V was seen. In both groups, the neutralizing titers against SIVmac251.6 were significantly higher than the titers against SIVCG7V (Fig. 3D). Similarly, no difference in antibody-dependent cellular cytotoxicity (ADCC) was seen between groups (Fig. 3E). However, the DNA&Env group showed elevated antibody-dependent cellular phagocytosis (ADCP) activity, compared to the ALVAC/Env group (Fig. 3F).
FIG 3.
Induction of binding and functional antibodies by the vaccine regimens. (A and B) Serum binding antibody titers (geometric mean) to SIV gp120 (M766 [A] and CG7V [B] strains) are shown over the course of immunization. Black arrows indicate mucosal priming immunizations at weeks 0 and 12; gray arrows indicate systemic booster immunizations at weeks 24 and 36. (C) A comparison of binding titers against both Env strains in the two vaccine arms is shown at week 38 (2 weeks after the second protein boost). (D) Serum neutralizing antibody titers against tier 1A SIVmac251.6 (mac251) and tier 1B SIVsmE660/BR-CG7V.IR1 (CG7V). (E and F) ADCC (percent killing) (E) and ADCP activity (F) are similarly shown at week 38 for both vaccine arms. (G and H) Rectal SIV Env-specific IgG (G) and IgA (H) binding antibodies are shown over the course of immunization for all vaccinated macaques. (I) Comparison of rectal Env-specific IgG and IgA at week 38 for the individual vaccine arms. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Mucosal Ad5hr-SIV recombinant immunization did not elicit detectable Env-specific IgG or IgA antibodies in rectal secretions; however, both developed following the booster immunizations (Fig. 3G and H). At week 38, 2 weeks after the last immunization, the DNA&Env group exhibited higher Env-specific IgG levels, compared to the ALVAC/Env group (Fig. 3I), consistent with results of the serum binding antibody titers. No difference between the groups was noted for rectal Env-specific IgA levels at week 38 (Fig. 3I).
Induction of TFH cells and B cells associated with antibody development.
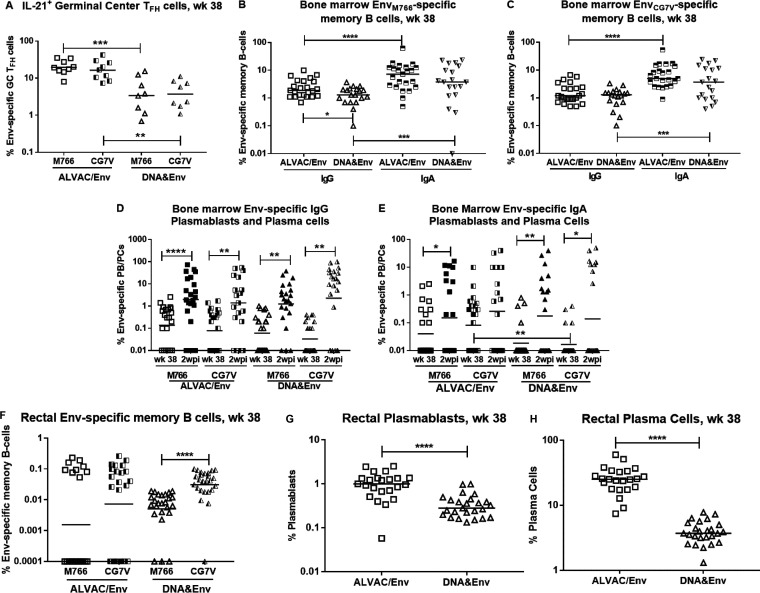
To investigate factors associated with antibody development, we assessed Env-specific T follicular helper (TFH) cells in germinal centers (GCs) of lymph node (LN) biopsy samples from a subset of the macaques by flow cytometry. The ALVAC/Env group exhibited significantly higher levels of both EnvM766- and EnvCG7V-specific TFH cells than did the DNA&Env group at week 38 following the last booster immunization (Fig. 4A), suggesting the potential to provide greater T cell help to B cells. This outcome was mirrored by moderately but significantly higher levels of IgG EnvM766-specific memory B cells in the bone marrow of the ALVAC/Env group, compared to the DNA&Env group (Fig. 4B). However, no differences were observed between immunization groups for EnvCG7V-specific IgG memory B cells or IgA memory B cells specific for either Env (Fig. 4B and C) or higher levels of bone marrow IgG plasmablasts (PBs) and plasma cells (PCs) at week 38, as assessed by enzyme-linked immunosorbent spot assay (ELISPOT) (Fig. 4D). As expected, levels of bone marrow Env-specific IgG and IgA antibody-secreting cells were increased in response to SIV Env antigen at 2 wpi (Fig. 4D and E). IgA bone marrow memory B cells specific for both Env proteins exhibited greater frequencies, compared to IgG memory B cells, for both immunization groups (Fig. 4B and C). We also observed significantly higher frequencies of bone marrow IgA EnvCG7V-specific PBs and PCs in the ALVAC/Env group, compared to the DNA&Env group, at week 38 (Fig. 4E). Flow cytometric analysis showed no significant differences between the vaccine groups in Env-specific memory B cells in rectal tissue of the immunized macaques (Fig. 4F). However, frequencies of total PBs and PCs in rectal tissue in the ALVAC/Env group were significantly higher than those in the DNA&Env group (Fig. 4G and H).
FIG 4.
Induction of TFH cells and B cells associated with antibody development. (A) TFH cells in LN GCs specific for both SIVM766 and SIVCG7V gp120 are shown at week 38 for each vaccine arm. (B and C) SIV Env-specific IgG and IgA bone marrow memory B cells are similarly shown for both Env strains (M766 [B] and CG7V [C]) and for both vaccine arms at week 38. (D and E) Bone marrow SIV Env-specific IgG (D) and IgA (E) PBs/PCs are shown for both Env strains and for each vaccine arm at week 38 and 2 wpi. (F to H) Rectal SIV Env-specific memory B cells for both vaccine arms and both Env strains (F) and PBs (G) and PCs (H) for both vaccine arms are shown at week 38. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Overall, both vaccine regimens elicited strong humoral immunity. While the DNA&Env group exhibited higher levels of binding antibodies in serum and rectal secretions and higher levels of circulating antibodies with functional activities, the ALVAC/Env group exhibited a greater potential to provide B cell help and elevated levels of PBs and PCs in bone marrow and rectal tissue. These contrasting results obtained at a single time point 2 weeks after the last immunization may reflect differences in how the vaccine boosts were administered and the kinetics of the responses. The ALVAC/Env group received ALVAC and Env boosts in opposite thighs, while the DNA&Env group received boosts of both immunogens in the same thigh (see Materials and Methods). Thus, B cell maturation and development at draining LNs might have experienced different kinetics of response, with the times of peak PB and PC development differing from that of serum antibody secretion; the single time point evaluated might not have captured the peak of one or the other. Future evaluation of vaccine-induced humoral immunity over a broader postimmunization time scale could assess this possibility.
The vaccine regimen strongly impacts the rectal microbiome.
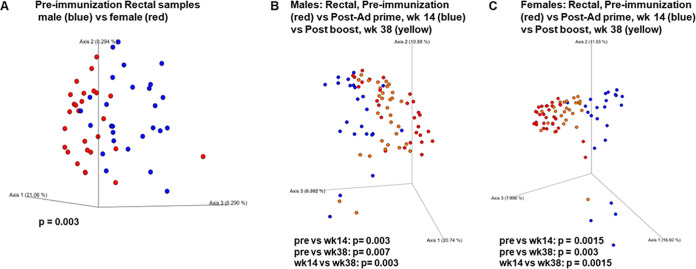
To investigate the effect of vaccination on the genital/rectal microbiome, DNA was purified from both vaginal and rectal swab samples collected prior to immunization and over the course of immunization, and 16S rRNA gene sequences were analyzed for identification and quantitation of the bacteria present. The rectal microbiota of females and males prior to immunization differed (Fig. 5A). Given this initial difference, we investigated the male and female rectal microbiota separately over the course of immunization. Both the mucosal priming immunizations and the systemic boosts resulted in distinct shifts in bacterial profiles. Comparison of the rectal bacterial populations prior to immunization with those present at week 14 after the mucosal Ad5hr-SIV recombinant immunizations revealed population changes, with significant differences in both male and female bacterial samples (Fig. 5B and C). Similarly, the bacterial populations present at week 14, compared to the postboost week 38 time point, exhibited significant differences in both males and females (Fig. 5B and C). The microbiota at week 38 remained significantly different from the populations observed prior to any immunization.
FIG 5.
The vaccine regimen strongly impacts the rectal microbiome. PCoA graphs were built using the Bray-Curtis dissimilarity matrix, a quantitative measure of community dissimilarity. (A) The difference in rectal microbiomes of male and female macaques prior to immunization. (B and C) Changes in rectal microbiomes of male (B) and female (C) macaques over the course of immunization. Combined PCoA plots include preimmunization, week 14, and week 38 time points. Statistical analysis was done using the PERMANOVA test for beta diversity.
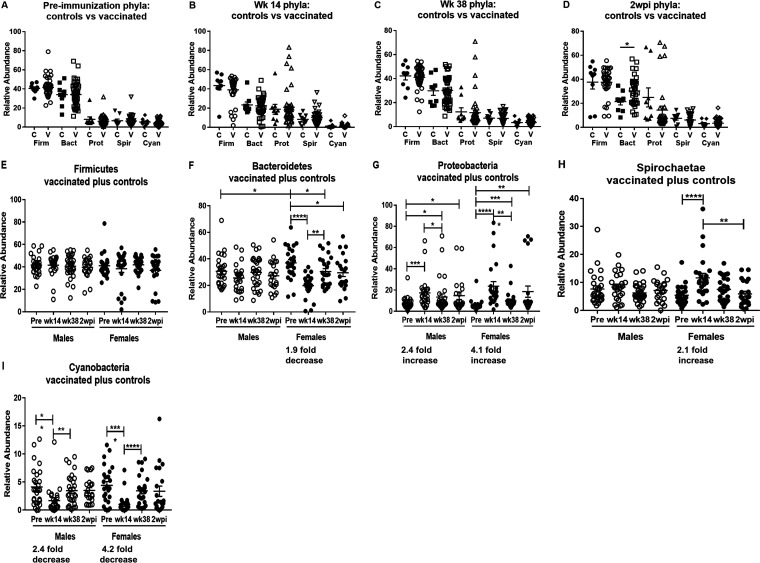
The five most prevalent bacterial phyla in the macaque rectal swabs were Firmicutes, Bacteroidetes, Proteobacteria, Spirochaetae, and Cyanobacteria (Fig. 6). There were no differences in the relative abundance of the five phyla between the vaccinated and control macaques (which also received empty Ad5hr vector and alum adjuvant) over the course of immunization at preimmunization to week 38 time points (Fig. 6A to D). A difference following infection (2 wpi) was only seen for Bacteroidetes (P = 0.020) (Fig. 6D). Therefore, here we plotted bacterial prevalences of the vaccinated and control macaques together.
FIG 6.
Analysis of the five most abundant bacterial phyla in the rectal microbiome of vaccinated macaques reveals the greatest changes in female macaques following Ad5hr priming. (A to D) Comparison of the relative abundances of the five most prevalent bacterial phyla for control and vaccinated macaques over the course of immunization and at 2 wpi. (E to I) The five most prevalent bacterial phyla found in the rectal microbiomes of the combined control plus vaccinated macaques, shown from most to least prevalent. In each panel, the relative abundance of each phylum is shown across the course of the vaccine regimen for males and females. Changes were most often detected between the preimmunization samples and post-Ad5hr immunization samples (week 14) and showed greatest statistical confidence for the female macaques, as indicated by the fold changes beneath the time points. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Prior to immunization, the prevalences of bacteria in these five phyla were very similar between males and females (Fig. 6E to I), with a significant difference seen only in the relative abundance of Bacteroidetes (Fig. 6F). The subsequent response to the two mucosal Ad5hr-SIV recombinant immunizations can be seen by comparing bacteria at the preimmunization and week 14 time points. At the phylum level, the greatest differences occurred in females, for which significant changes were seen in four of the five predominant phyla. The prevalences of Bacteroidetes and Cyanobacteria dropped significantly (Fig. 6F and I), while Proteobacteria and Spirochaetae prevalences increased (Fig. 6G and H). At the same preimmunization and week 14 time points, the male macaques exhibited changes in only two of the five phyla, exhibiting a more modest increase in Proteobacteria and a more modest decrease in Cyanobacteria (Fig. 6G and I). Shifts in bacterial prevalences in response to the two protein boosts can be seen by comparing populations present at week 14 with those at week 38. At the phylum level, bacterial frequencies indicated a reversion in females to levels close to those seen prior to immunization, with significant increases in Bacteroidetes and Cyanobacteria (Fig. 6F and I) and a significant decrease in Proteobacteria (Fig. 6G). At these same time points, males exhibited a small decrease in Proteobacteria and an increase in the abundance of Cyanobacteria (Fig. 6G and I). By 2 wpi, the rectal bacterial prevalences of these phyla in both males and females were similar to those at the preimmunization time points with the exception of a modestly decreased prevalence of Bacteroidetes in females and an increase in Proteobacteria in both males and females (Fig. 6F and G). Overall, the prime/boost vaccine regimen greatly impacted the rectal microbiome, with the Ad5hr-SIV recombinant and Ad5hr empty vector administrations having the greatest effect.
Repeated low-dose SIV intrarectal exposures reveal differences in viremia control between males and females.
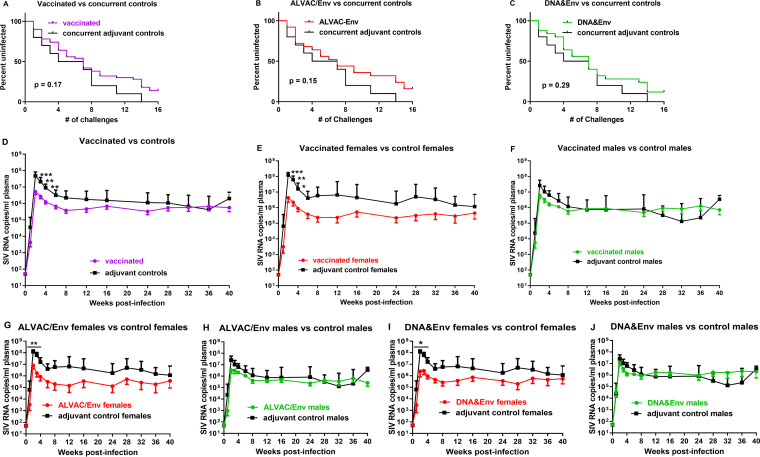
Having established that both vaccine regimens were immunogenic, eliciting cellular and humoral immune responses mucosally and systemically, we initiated repeated low-dose SIVmac251 intrarectal challenges to assess vaccine efficacy. After 15 weekly challenges, all 10 adjuvant-treated controls were infected, while 7 vaccinated macaques remained uninfected (4 in the ALVAC/Env group and 3 in the DNA&Env group) (Fig. 7A to C). However, we observed no significant reduction in the risk of SIV acquisition in all of the vaccinated macaques, compared to the adjuvant-treated controls (Fig. 7A). Further, neither immunization group exhibited significant acquisition risk reduction (Fig. 7B and C). Overall, however, the vaccinated macaques exhibited significantly, although modestly, decreased peak viral loads (Fig. 7D). Because we previously reported a sex bias in SIV vaccine challenge outcomes (19), we examined the results here in females and males separately. No significant difference in the peak viral loads of the control female and male macaques was observed (medians of 1.03 × 108 and 4.71 × 107 SIV RNA copies/ml, respectively; P = 0.24) indicating there was no difference in early pathogenesis between the sexes. However, the vaccinated female macaques exhibited significantly lower viral loads over the acute phase of infection, compared to the female controls (Fig. 7E), while the vaccinated males, compared to the male controls, did not (Fig. 7F). The result was retained when the two vaccination groups were evaluated separately (Fig. 7G to J), which validated our previous finding of a vaccine-induced sex bias in protection against SIV (19).
FIG 7.
Repeated low-dose SIV intrarectal exposures reveal differences in viremia control between males and females. (A to C) Survival curves for all vaccinated macaques (A) and the individual ALVAC/Env-vaccinated (B) and DNA&Env-vaccinated (C) groups, compared to adjuvant-treated controls, are shown. (D to F) Plasma viral loads for all vaccinated macaques. compared to adjuvant controls (D), all vaccinated females, compared to female controls (E), and all vaccinated males, compared to male controls (F). (G and H) Comparison of ALVAC/Env-vaccinated females (G) and males (H), compared to their respective controls. (I and J) Comparison of DNA&Env vaccinated females (I) and males (J), compared to their respective controls. Geometric mean viral loads are shown ± standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
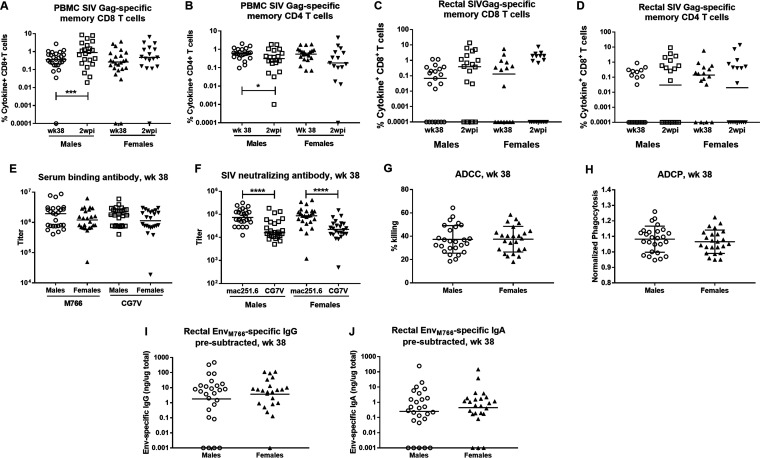
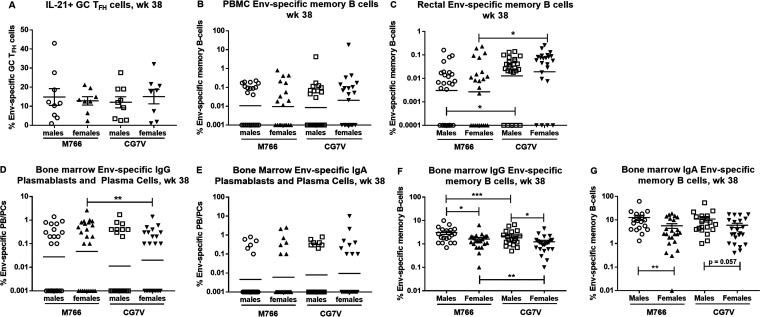
To determine the basis for this sex difference in control of viremia, we reanalyzed vaccine-induced immune responses, comparing outcomes in male versus female macaques at week 38, the last sampling time point prior to initiation of SIV challenge exposures. Overall, no differences between the sexes were seen with regard to vaccine-induced CD4+ or CD8+ SIV Gag-specific responses in PBMCs or rectal tissue (Fig. 8A to D), serum binding reactivities (Fig. 8E), and functional serum antibody activities (Fig. 8F to H). Neutralizing antibody activity was seen to be greater against the tier 1A SIVmac251.6 virus, compared to the tier 1B SIVsmE660/BR-CG7V.IR1 virus, but this was seen in both males and females (Fig. 8F). Differences between male and female vaccinated macaques were not observed with regard to rectal IgG or IgA Env-specific secretory antibodies (Fig. 8I and J). Sex differences were not observed in the frequencies of Env-specific GC TFH cells (Fig. 9A) or levels of Env-specific memory B cells in PBMCs or rectal tissue (Fig. 9B and C). We did see higher levels of rectal SIVCG7V Env-specific memory B cells in both males and females, compared to those specific for SIVM766 Env, presumably due to the close homology of the former Env to that in the priming Ad5hr recombinant. Bone marrow Env-specific IgG and IgA PB and PC responses also did not differ between males and females (Fig. 9D and E). The only differences in immune responses noted between male and female vaccinated macaques were in bone marrow IgG and IgA Env-specific memory B cell responses, with the males having slightly higher memory B cell frequencies than the females (Fig. 9F and G).
FIG 8.
No differences between vaccinated males and females in SIV Gag-specific T cells in blood or rectal tissue or in serum binding antibodies, antibodies with functional activity, or antibodies in rectal secretions. (A to D) SIV Gag-specific memory CD8+ (A and C) and CD4+ (B and D) T cells in PBMCs (A and B) or in rectal tissue (C and D) at week 38 and 2 wpi. (E) SIVM766 and SIVCG7V gp120-specific binding antibody titers at week 38 in males and females. (F) SIVmac251.6 and SIVCG7V neutralizing antibody titers at week 38 in vaccinated males and females. (G) ADCC activity (percent killing) in males and females at week 38. (H) ADCP activity in males and females at week 38. (I and J) EnvM766-specific IgG (I) and IgA (J) in rectal secretions of males and females at week 38. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
FIG 9.
Few differences in Env-specific GC TFH cells, Env-specific memory B cells in PBMCs and rectal tissue, or bone marrow antibody-secreting cells and Env-specific memory B cells between males and females. (A) EnvM766- and EnvCG7V-specific GC TFH cells in males and females at week 38. (B and C) SIVM766 and SIVCG7V Env-specific memory B cells in males and females in PBMCs (B) and rectal tissue (C) at week 38. (D and E) Bone marrow SIVM766 and SIVCG7V Env-specific IgG (D) and IgA (E) PBs/PCs in males and females at week 38. (F and G) Bone marrow IgG (F) and IgA (G) SIVM766 and SIVCG7V Env-specific memory B cells in males and females at week 38. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Because differences between the sexes in vaccine-induced immune responses were lacking except for the bone marrow memory B cell responses, we looked for immune responses that were directly associated with decreased peak viremia in female or male vaccinated macaques; however, none was observed. Therefore, we considered that the rectal bacterial microbiome, which was affected by the vaccine regimen and exhibited sex differences (Fig. 5 and 6), might have influenced the challenge results.
Differential effects of bacteria in rectal microbiomes of male and female macaques on control of viremia and vaccine-induced immune responses.
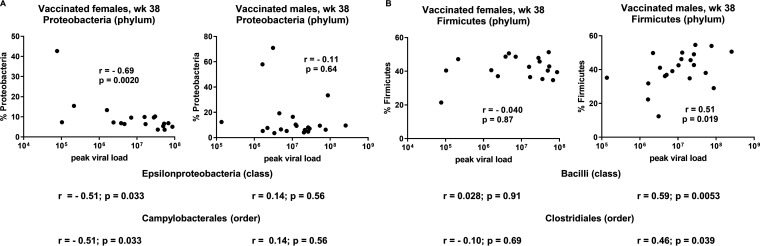
We first investigated relationships between the rectal microbiota and viremia control and observed strikingly different correlations in males and females. In female macaques, Proteobacteria, Epsilonproteobacteria, and Campylobacterales bacteria (phylum, class, and order levels, respectively) after the second boost (week 38) correlated with decreased peak viral load, while no such correlations were observed in vaccinated males (Fig. 10A). Similarly, Firmicutes, Bacilli, and Clostridiales bacteria (phylum, class, and order, respectively) at week 38 correlated with increased peak viral loads in vaccinated males, but no such relationships were seen in vaccinated females (Fig. 10B).
FIG 10.
Differential effects of bacteria in rectal microbiomes of male and female macaques on control of viremia and vaccine-induced immune responses. (A) Correlation of percentage of Proteobacteria at week 38 and peak viral loads of vaccinated female and male macaques. Results of similar analyses of Episilonproteobacteria and Campylobacterales are shown below without the actual plots. (B) Correlation of percentage of Firmicutes at week 38 versus peak viral loads of vaccinated female and male macaques, with results of correlations with Bacilli and Clostridiales shown below. Results were obtained by Spearman rank correlation analyses.
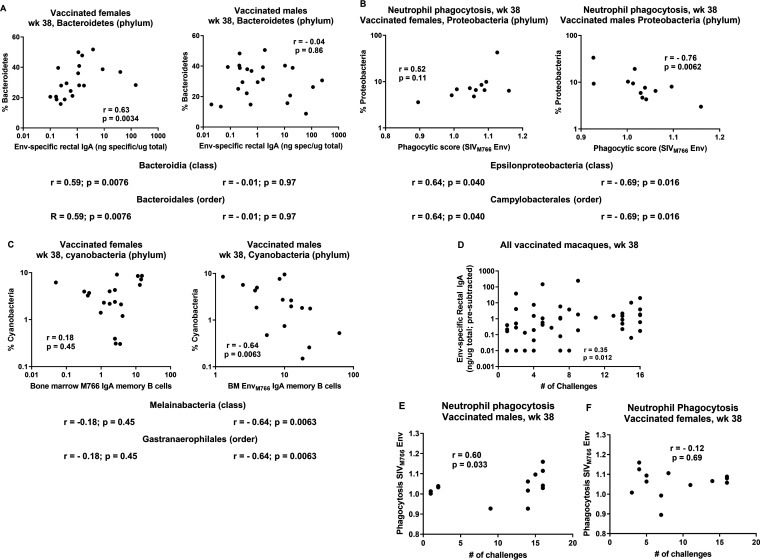
Further analysis of the rectal microbiota of the vaccinated macaques indicated that bacterial populations in males and females impacted several immune responses differently in males and females. Rectal Env-specific IgA was positively correlated with the frequency of Bacteroidetes (phylum), Bacteroidia (class), and Bacteroidales (order) in vaccinated females, but no such associations were seen in vaccinated males (Fig. 11A). In males, neutrophil phagocytosis was strongly negatively associated with Proteobacteria (phylum), Epsilonproteobacteria (class), and Campylobacteriales (order), whereas females tended to show positive correlations (Fig. 11B). Bone marrow IgA memory B cells of vaccinated male macaques correlated negatively with bacteria of the phylum Cyanobacteria, with identical correlation values seen for the class Melainabacteria and the order Gastranaerophilales, whereas vaccinated females displayed no such correlations (Fig. 11C). Importantly, some of these immune responses have been associated with protective efficacy. For example, levels of rectal Env-specific IgA were previously associated with reduced risk of SIV acquisition in vaccinated female macaques (19). Neutrophil phagocytic activity has also been associated with reduced risk of acquisition (30). While we did not observe overall SIV acquisition risk reduction here in our vaccinated macaques, we did see that both Env-specific rectal IgA in all the vaccinated animals (Fig. 11D) and neutrophil phagocytosis in vaccinated males but not females (Fig. 11E and F) were significantly correlated with a greater number of challenges needed for infection. These correlations suggest that the rectal microbiome in males and females differentially impacts both vaccine-induced immune responses potentially important for protective efficacy and viral load outcomes following the SIV intrarectal challenges.
FIG 11.
Differential effects of bacteria in rectal microbiomes of male and female macaques on vaccine-induced immune responses, including those associated with protection. (A) Correlation of percentages of Bacteroidetes at week 38 with Env-specific rectal IgA levels at the same time point in vaccinated female and male macaques. Results of similar analyses of Bacteroidia and Bacteroidales are shown below without the actual plots. (B) Correlation of percentages of Proteobacteria at week 38 with phagocytic scores at the same time point in female and male macaques. Results of correlations with Epsilonproteobacteria and Campylobacterales are shown below. (C) Correlation of percentages of Cyanobacteria at week 38 with bone marrow EnvM766 IgA memory B cells in female and male macaques at the same time point. Results of correlations with Melainabacteria and Gastranaerophilales are shown below. (D) Correlation of Env-specific rectal IgA levels at week 38 with number of challenges in all vaccinated macaques. (E) Positive correlation of neutrophil phagocytic activity at week 38 in vaccinated males with number of challenges. (F) Lack of correlation of neutrophil phagocytic activity in vaccinated females with number of challenges. Results were obtained by Spearman rank correlation analyses.
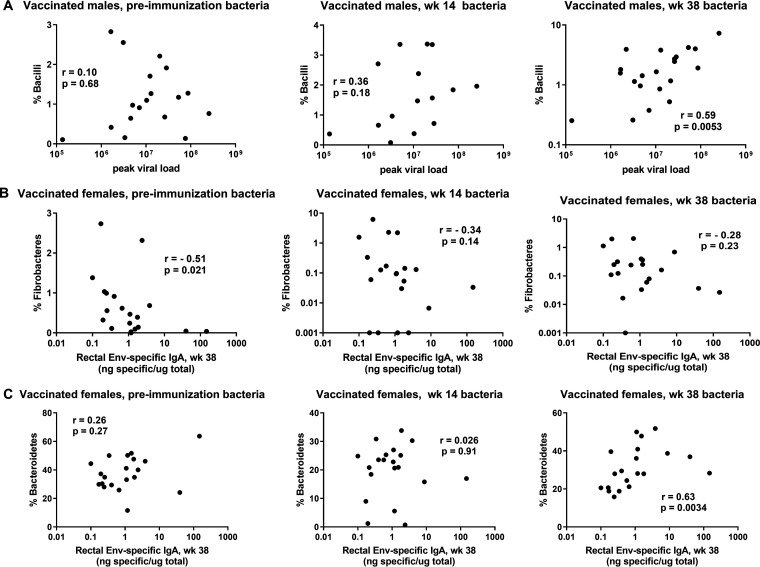
It is equally clear that the vaccine regimen resulted in alterations in the rectal microbiome potentially influencing viremia outcomes and vaccine-induced immunity. In addition to the principal-coordinate analysis (PCoA) plots (Fig. 5), which illustrate the impact of the mucosal and systemic elements of the vaccine regimen on the rectal microbiome, associations of peak viral loads with the frequency of particular bacteria over the course of vaccination fluctuated. For example, in vaccinated males, almost no correlation with the abundance of the bacterial class Bacilli was found prior to immunization and a weak correlation was found following the Ad5hr-SIV recombinant immunization at week 14; however, by week 38, following two booster immunizations, a strong positive correlation was seen between the frequency of the class Bacilli and peak viral loads (Fig. 12A). With regard to vaccine-induced immunity, we observed that, in vaccinated females, bacteria of the phylum Fibrobacteres prior to immunization were negatively correlated with week 38 rectal Env-specific IgA levels but this correlation was weakened over the course of immunization (Fig. 12B). In contrast, bacteria of the phylum Bacteroidetes in female macaques displayed no correlation with rectal Env-specific IgA levels at preimmunization and week 14 time points but week 38 bacteria showed a strong positive correlation (Fig. 12C). Overall, while our data do not point to a single bacterial species responsible for induced immune responses or lower peak viremia levels in vaccinated female macaques, they illustrate the potential impact of the rectal microbiota on induced immunity, outcomes of SIV exposures and infection, and differential effects on males and females. These possibilities should be confirmed in future microbiome transfer studies, which were not possible here.
FIG 12.
The rectal microbiome impacts immune responses; vaccine-induced alterations in the rectal microbiome impact viremia outcomes and induced immunity. (A) In vaccinated males, percentages of Bacilli do not correlate with peak viral loads prior to vaccination and following Ad5hr priming but do positively correlate following the booster immunization. (B) In vaccinated females, percentages of Fibrobacteres are negatively correlated with rectal Env-specific IgA levels prior to immunization but the correlation is lost following the Ad5hr prime and booster immunizations. (C) In vaccinated females, the percentages of Bacteroidetes prior to immunization and post-Ad5hr priming do not correlate with rectal Env-specific IgA levels but a positive correlation is seen following the booster immunization at week 38. Results were obtained by Spearman rank correlation analyses.
DISCUSSION
As expected, our combined mucosal-systemic immunization strategy elicited strong immunity, both cellular and humoral, at mucosal and systemic sites; however few differences overall were seen between the ALVAC/Env and DNA&Env regimens. Neither combined regimen resulted in reduced risk of SIV acquisition, compared to controls; however, significantly reduced acute viremia in female macaques, compared to female controls, was seen, a result not seen in males, confirming our earlier findings of a vaccine-induced sex bias (19). Further, the rectal microbiome was shown to vary with the vaccination regimen and differentially impact viremia control and immune response induction in male and female macaques. Both findings are significant with regard to the design and analysis of vaccine trials.
We previously documented a vaccine-induced sex bias in a preclinical study in which vaccinated females but not males exhibited significantly reduced SIV acquisition risk, compared to their corresponding controls (19). This was the first evidence of a sex bias in SIV vaccine efficacy, suggesting the importance of monitoring both males and females in preclinical and clinical vaccine trials. The reduced SIV acquisition risk in vaccinated females seen earlier was associated with local mucosal B cell immunity, including rectal IgA antibodies (19), but a variety of other responses were shown to impact protective outcomes differently between the sexes, including IgG subtype, Env-specific TFH cells, B regulatory cells, Fc glycosylation of antibodies, and antibody-dependent complement-mediated lysis (31–33). For that reason, here we evaluated a broad spectrum of vaccine-induced immune responses. We confirmed the previously observed sex bias, although the challenge outcomes differed. Significantly reduced acquisition risk in this study was not seen, perhaps due to changes in the vaccine regimen. Previously, the macaques were primed with Ad5hr SIVenvsmH4/rev, SIVgag239, and SIVnef239 recombinants and boosted with SIV gp120 or oligomeric gp140 in MF-59 adjuvant. Here, in contrast, the macaques received Ad5hr-SIVenvM766 and SIVgag239 recombinants and were boosted with an ALVAC recombinant or DNA for SIV genes, together with SIV gp120 proteins in alum. The change in adjuvant might have had the greatest impact. MF-59 has been shown to induce adaptive immune responses more strongly than alum (23), which might have influenced the challenge outcome. Nevertheless, a sex bias was again observed, although of a different character. Here, the vaccinated female macaques exhibited significantly lower acute viral loads, compared to the female controls, while the same outcome was not seen in the vaccinated males. Sex differences in susceptibility to viral infections and subsequent pathogenesis have been reported with regard to viral infections, including HIV; women initially exhibit lower viral loads than men but eventually progress more quickly to disease (34, 35). Female subjects also generally respond better to vaccines, developing greater immune responses (36). Overall, with regard to viral susceptibility and pathogenesis, the sex bias has been associated with differences in both innate and adaptive immunity, impacted by sex hormones, X-linked genes, and the microbiome (35, 37). Here, the male and female control macaques exhibited similar acute viral loads, indicating that the sex bias observed was not attributable to differences in early pathogenesis. However, we did not identify distinct immune responses associated with the decreased acute viremia in the vaccinated females, leading us to further explore effects of the vaccine-induced microbiome changes.
Because the microbiome helps shape mucosal immune responses (26) and steroid hormones modulate the microbiome (38), it was an obvious avenue to investigate concerning the observed sex difference in acute viremia levels. The predominant phyla identified in the rectal swabs of our rhesus macaques (Fig. 6) were not unexpected. Bacteroidetes and Firmicutes have been reported to be the two predominant phyla in the guts of healthy humans (39), and both phyla, along with Proteobacteria, have been reported as dominant in noncaptive Chinese rhesus macaques (40). Few studies, however, have investigated microbiome changes over the course of an immunization regimen. In humans, four studies (three conducted in infants and one conducted in adults) have examined the influence of the gut microbiome on induced vaccine responses (41). Actinobacteria and Firmicutes were found to be associated with higher humoral and cellular vaccine responses, while Proteobacteria and Bacteroidetes were associated with lower responses. However, intestinal microbiota were also found to induce a nonneutralizing dominant immune response reactive with HIV gp41, which is potentially confounding, as it was not associated with vaccine-induced protective efficacy (42). In rhesus macaques, microbial richness was shown to be reduced following vaccination and correlated inversely with the number of intrarectal challenges needed to achieve SHIV infection (43). Moreover, in that study, bacterial principal-component analysis component 1 at the genus level correlated inversely with the number of exposures needed for infection and with CD14+ DR− monocytes and correlated positively with TFH cells in mesenteric LNs. Further, in female rhesus macaques, DNA vaccination was shown to differentially impact microbiome profiles at vaginal and rectal sites and to correlate with induced humoral immune responses (44). A few studies have examined the microbiome with regard to SHIV transmission or effects following SIV infection. Sui et al. (45) reported that lower ratios of Bacteroides to Prevotella, as well as lower levels of Firmicutes, were associated with the susceptibility of naive macaques to SHIV infection and also inversely associated with greater immune activation in the rectal mucosa. Handley et al. (46) reported that successful vaccination against SIV, resulting in prevention of progression to AIDS, also prevented the emergence of viral and bacterial enteropathogens. Fecal microbial transplantation following antibiotic treatment of SIV-infected rhesus macaques on antiretroviral therapy resulted in increased peripheral Th17 and Th22 cells and decreased mucosal CD4+ T cell activation (47). These studies illustrate the potential effects of both vaccination and the microbiome on immune responses and vaccine efficacy. However, we think that the results reported here are the first regarding direct effects of vaccination on the rectal microbiome (Fig. 5 and 6), with associated changes in protective efficacy in male and female macaques.
Mucosal priming with replication-competent Ad5hr-SIV recombinants had the greatest impact on changes in the rectal microbiome (Fig. 6). This was not unexpected, as viral infections are known to induce changes in the composition of the microbiota (29). In fact, the majority of studies related to viruses and the microbiome have been conducted in the context of respiratory viral infections (48). The microbiome changes result from complex interactions arising from viral-induced inflammation and innate immunity and the interaction of pattern recognition receptors with viral pathogen-associated molecular patterns (49).
The basis for the difference in viral loads between males and females, the role of the microbiome, and the impact of the particular bacteria associated with the effect will require further experimentation. Microbial metabolites modulate host physiology in multiple ways, impacting metabolic and nutritional homeostasis, energy expenditure, and immunity (50, 51). Therefore, the specific effects of Proteobacteria in females and Firmicutes in males (Fig. 10) on viral load are not currently understood. That sex, however, influences the composition of the gut microbiota is not disputed (38). It may also be that the microbiome composition of males and females further influences sex differences in immune responses (52). The correlation of Bacteroidetes with rectal IgA levels in the vaccinated females (Fig. 11A) is better understood, as the genus Bacteroides has been shown to induce greater mucosal IgA production by increasing activation-induced cytidine deaminase expression in B cells of mice (53). Bacteroides acidifaciens has also been reported to promote IgA production in the large intestine by inducing GC formation and increasing the number of IgA+ B cells (54). Why the males do not respond similarly is not known, however. While our study highlights differential effects of the microbiome on viremia control and immune responses in males and females, it points to areas of research still to be explored.
HIV is primarily transmitted across mucosal surfaces. Compared to vaginal or oral transmission, intrarectal transmission has been shown to have the greatest relative risk, through unprotected receptive anal intercourse in humans (55) or through direct inoculation studies in rhesus macaques (56). Thus, our results here describing effects of the rectal microbiome on vaccine-induced immune responses and viremia control following intrarectal SIV transmission in male and female macaques are relevant for future vaccine design and development. However, it is well known that vaginal microbiota have profound effects on immune responses in females, as well as susceptibility to HIV infection (57). To our knowledge, studies regarding vaccine effects on the vaginal microbiome with subsequent impacts on protective efficacy against intravaginal exposures have not been reported but should be an important area for future investigation.
Overall, our study confirms that a combined mucosal priming/systemic boosting vaccination strategy can elicit strong humoral and cellular immunity in both compartments. Further design of such an approach should be valuable for an HIV vaccine, for which potent immune responses are necessary to prevent infection at rectal/genital sites of transmission and to control early systemic expansion of virus. Our study highlights the importance of assessing vaccine strategies in both males and females and the need to consider effects of microbiome differences in evaluating vaccine efficacy, especially as vaccines are tested in different parts of the world with different endemic microflora.
MATERIALS AND METHODS
Animals and ethics statement.
All animal experiments were approved by Institutional Animal Care and Use Committees (IACUCs) prior to study initiation, including the NCI Animal Care and Use Committee and the Advanced BioScience Laboratories, Inc. (ABL) Animal Care and Use Committee. Rhesus macaques were maintained at the NCI Animal Facility (Bethesda, MD) under protocol VB012. Following the challenge phase of the study, 24 macaques (10 from each vaccinated group and 4 controls evenly divided between males and females) were housed at ABL (Rockville, MD) under protocol AUP526. Each of these facilities is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. The standard practices closely follow recommendations made in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animals were housed in accordance with the recommendations of the AAALAC Standards and with the recommendations in the Guide for the Care and Use of Laboratory Animals. Details of animal welfare and steps taken to ameliorate suffering were in accordance with the Guide for the Care and Use of Laboratory Animals and the recommendations of the Weatherall report on the use of NHPs in research, as approved by the relevant IACUCs.
Immunization and challenge regimens.
Sixty Indian rhesus macaques (Macaca mulatta) that were 3 to 4 years of age and were negative for SIV, simian retrovirus, and simian T lymphotropic virus were used in this study. Macaques positive for Mamu-A*01 (n = 6) and Mamu-B*17 (n = 8) haplotypes were divided evenly among the three immunization groups. As illustrated in Fig. 1, 50 macaques were primed mucosally at weeks 0 and 12 with replication-competent Ad5hr recombinants (5 × 108 PFU/recombinant/dose/route) separately encoding SIVM766 gp120-TM and SIV239 Gag. Ten controls received empty Ad5hr vector (1 × 109 PFU/dose/route). Subsequently, at weeks 24 and 36, macaques in the ALVAC/Env group were boosted intramuscularly with ALVAC-SIVM766 Gag/pro/gp120-TM (108 PFU) in one thigh and 200 μg each of gD-SIVM766 gp120 and gD-SIVCG7V gp120 in alum hydroxide in the opposite thigh. The gD-gp120 proteins contain the 28-residue N terminus of the herpes simplex virus 1 glycoprotein D. Macaques in the DNA&Env group were boosted intramuscularly with DNA encoding SIVM766 gp120-TM (1 mg), SIV239 Gag (1 mg), and macaque IL-12 (0.2 mg) administered in the inner thigh, followed by electroporation (Elgen 1000; Inovio Pharmaceuticals, Inc., Plymouth Meeting, PA, USA) and then immediate intramuscular administration of the same gD-SIV gp120 proteins in alum phosphate (58) at the same anatomical site, as detailed previously (59). Control animals (5 in the ALVAC/Env group and 5 in the DNA&Env group) received alum hydroxide or alum phosphate only. At week 42, all macaques were challenged intrarectally using a low dose of SIVmac251 (1:500 dilution, 120 times the 50% tissue culture infective dose [TCID50]), a challenge stock developed by Ronald Desrosiers and provided by Nancy Miller, Division of AIDS, NIAID. Up to 15 challenges were continued weekly until the onset of infection, as determined by a plasma viral load of ≥50 SIV RNA copies/ml assessed by the nucleic acid sequence-based amplification method (60, 61). Macaques were monitored for at least 40 weeks after infection or until euthanasia criteria were met. An additional 12 macaques were added to this study later in order to evaluate additional immune responses in macaques vaccinated with the ALVAC/Env and DNA&Env vaccine regimens, as described above (6 macaques each). These macaques were not challenged. Five in each group were Mamu-A*01 positive.
Cellular immunity.
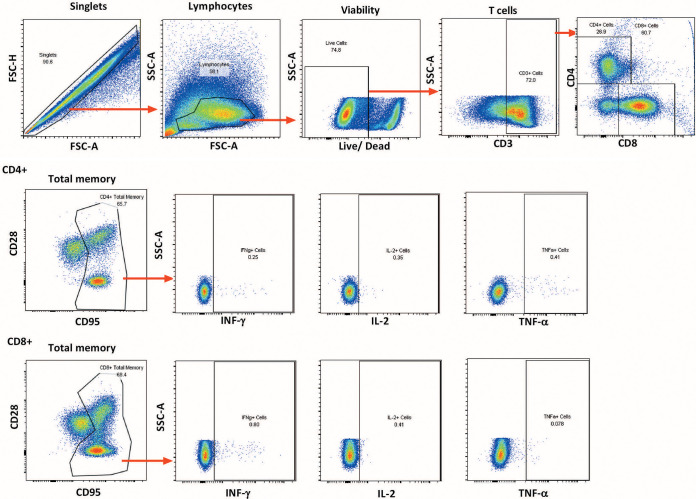
CD4+ and CD8+ T cell responses were assessed in PBMCs and rectal pinch biopsy samples over the course of immunization and 2 wpi. PBMCs were isolated from EDTA-treated blood by Ficoll gradient centrifugation and viably frozen. For assay, PBMCs were thawed and stimulated at a final concentration of 1 μg/ml with SIV EnvM766, EnvCG7V (ABL), or Gagmac239 (NIH AIDS Reagent Program) pooled peptides (15-mers overlapping by 11 amino acids). Controls included a nonstimulated negative control and a leukocyte activation cocktail (BD Biosciences)-stimulated positive control. Anti-CD28-phycoerythrin (PE)/Texas Red (clone CD28.2; Beckman Coulter) and anti-CD49d (clone 9F10; eBioscience) were added during stimulation, along with a protein transport inhibitor (monensin; BD Biosciences). After 6 h of incubation at 37°C in the dark, cells were washed with phosphate-buffered saline (PBS) and surface stained with the antibodies anti-CD95-PE/Cy5 (clone DX2; eBioscience) and anti-CD8-Qdot 655 (clone M-T807), both from the NHP Reagent Resource, as well as with a viability dye (Life Technologies). Following incubation for 20 min at room temperature in the dark, intracellular staining was performed. Cells were washed twice with PBS, resuspended in 300 μl Cytofix/Cytoperm solution (BD Biosciences), and incubated for 15 min at 4°C in the dark and then were washed twice with BD Perm/Wash buffer (BD Biosciences), resuspended in 100 μl BD Perm/Wash buffer, and stained with the antibodies anti-CD3-Qdot 800 (clone SP34-2; BD Biosciences), anti-CD4-Qdot 605 (clone 19Thy-5D7; NHP Reagent Resource), anti-tumor necrosis factor alpha (TNF-α)-PE (clone MAb11) and anti-IL-2-allophycocyanin (APC)/Cy7 (clone MQ1-17H12; both from Sony Biotechnologies Inc.), and anti-gamma interferon (IFN-γ)-Alexa 700 (clone B27; Life Technologies). Cells were incubated for 25 min at 4°C in the dark, washed twice with BD Perm/Wash buffer, and resuspended in PBS for data acquisition on an LSR II flow cytometer (BD Biosciences). The following consecutive gating strategy was used for flow cytometry analysis: singlets, lymphocytes, live cells, and then CD3+ T cells (a minimum of 50,000 CD3+ T cells were recorded). From the CD3+ T cell gate, CD4+and CD8+ T cells were gated separately and each population was further divided into CD28+ CD95+ central memory (CM) and CD28− CD95+ EM gates or combined (CD28+/− CD95+) for a TM gate. From those gates, the numbers and percentages of functional surface marker-positive and cytokine-positive cells in each memory cell subset were determined and background values from nonstimulated samples were subtracted. The sums of cytokine-positive cells (IL-2, TNF-α, and IFN-γ) in the CD4+ or CD8+ TM populations are reported. The data reported are overestimates, as doubly and triply cytokine-positive cells were not identified by Boolean analysis (62). Flow cytometric analyses were performed using FlowJo v9.8.1. A representative flow plot for PBMC T cell responses is shown in Fig. 13.
FIG 13.
Representative plot illustrating the gating strategy for memory CD8+ and CD4+ T cell responses in PBMCs. Details are provided in Materials and Methods.
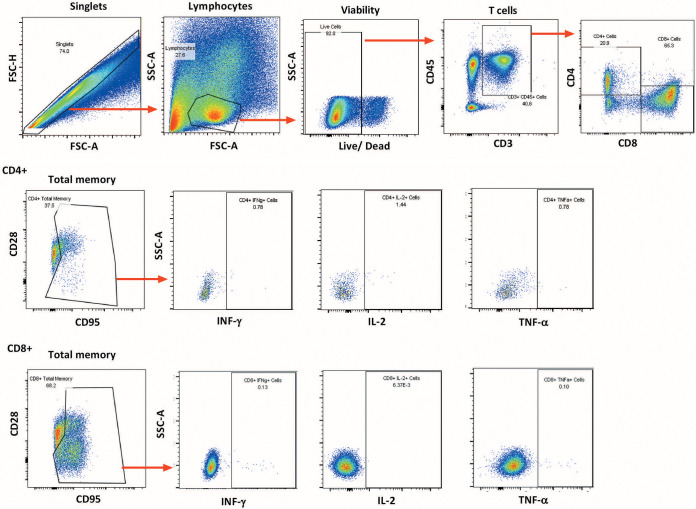
Rectal biopsy samples were rinsed with prewarmed RPMI 1640 medium (Life Technologies) containing 2× antibiotic-antimycotic solution, 2 mM l-glutamine (Life Technologies), and 2.5 mg/ml collagenase (Sigma-Aldrich). The pinch biopsy samples were minced using a scalpel and a 19 gauge needle, transferred in 10 ml of the same medium to a 50-ml tube, and incubated for 25 min at 37°C, with pulse vortex-mixing every 5 min. The digested tissue was passed five times through a blunt end cannula, and the liberated cells and tissue debris were passed through a 40-μm cell strainer and washed in R10 medium (RPMI 1640 medium containing 2× antibiotic-antimycotic solution, l-glutamine, and 10% fetal bovine serum [FBS]) prior to staining. Cells were stimulated with pooled SIV EnvCG7V, EnvM766, or Gagmac239 peptides at a final concentration of 1 μg/ml. Control tubes included a nonstimulated control and a leukocyte activation cocktail (BD Biosciences)-stimulated positive control. Anti-CD28-PE/Texas Red (clone CD28.2; Beckman Coulter) and anti-CD49d (clone 9F10; eBioscience) were also added during stimulation, along with a protein transport inhibitor (brefeldin A; BD Biosciences). After 6 h of incubation at 37°C in the dark, cells were washed with PBS and stained with the antibodies anti-CD95-PE/Cy7 (clone DX2; BD Biosciences), anti-CD45-brilliant violet 786 (BV786) (clone DO581283; BD Biosciences), and anti-CD8-PE (clone RPA-T8; BD Biosciences). A viability dye (Life Technologies) was also added to the antibody cocktail, to exclude dead cells in subsequent flow analyses. Following incubation for 20 min at room temperature in the dark, intracellular staining was performed. Cells were washed twice with PBS, resuspended in 300 μl Cytofix/Cytoperm solution (BD Biosciences), and incubated for 15 min at 4°C in the dark and then were washed twice with BD Perm/Wash buffer (BD Biosciences), resuspended in 100 μl BD Perm/Wash buffer, and stained with the antibodies anti-CD3-Alexa 700 (clone SP34-2; BD Biosciences), anti-CD4-peridinin chlorophyll protein (PerCP)/Cy5.5 (clone L200; BD Biosciences), anti-TNF-α-fluorescein isothiocyanate (FITC) (clone MAb11; BD Biosciences), anti-IFN-γ-APC (clone B27; BD Biosciences), and anti-IL-2-APC/Cy7 (clone MQ1-17H12; Sony Biotechnologies Inc.). Cells were incubated for 25 min at 4°C in the dark, washed twice with BD Perm/Wash buffer, and resuspended in PBS for data acquisition on an LSR II flow cytometer (BD Biosciences). The following consecutive gating strategy was used for flow cytometry analysis: singlets, lymphocytes, live cells, and then CD45+ CD3+ T cells (a minimum of 50,000 live cells were recorded during data acquisition). From the CD45+ CD3+ T cell gate, CD4+ and CD8+ T cells were gated separately and each population was further divided into CD28+ CD95+ CM and CD28− CD95+ EM gates or combined (CD28+/− CD95+) for a TM gate. From those gates, the numbers and percentages of functional surface marker-positive and cytokine-positive cells in each memory cell subset were determined and background values (from nonstimulated samples) were subtracted. Flow cytometric analyses were performed using FlowJo v9.8.1. A representative plot showing the gating strategy is shown in Fig. 14.
FIG 14.
Representative plot illustrating the gating strategy for memory CD8+ and CD4+ T cell responses in rectal tissue. Details are provided in Materials and Methods.
For processing and staining with CM9 dextramer, rectal pinch biopsy samples were incubated twice at room temperature for 15 min in Hanks’ balanced salt solution with 5 mM EDTA and 2 mM dithiothreitol. The supernatants were collected, pooled, and filtered through 100-μm cell strainers. Intraepithelial lymphocytes were collected after centrifugation. The tissues were then subjected to enzyme digestion with Liberase (Roche) at 37°C for 30 min. After digestion, the tissue chunks were mashed through a syringe end and filtered through a 100-μm cell strainer. Lamina propria cells were collected after centrifugation. Intraepithelial lymphocytes and lamina propria cells were mixed and washed in RPMI 1640 medium containing antibiotic-antimycotic solution and 10% FBS prior to staining. For staining, the rectal cells and PBMCs were incubated with CM9 dextramer (ImmuDex, Copenhagen, Denmark) for 15 min before the addition of other antibody mixtures (anti-CD45, anti-CD3, anti-CD4, and anti-CD8 from BD Biosciences and anti-CD28 and anti-CD95 from BioLegend). After 30 min of staining, cells were washed and subjected to flow cytometric analyses.
Humoral immunity.
Serum antibody binding titers for SIVM766 and SIVCG7V gp120 were assessed by enzyme-linked immunosorbent assay (ELISA), as described previously (63). The antibody titer was defined as the reciprocal of the serum dilution at which the optical density (OD) at 450 nm of the test serum was twice that of the negative control serum diluted 1:50.
Neutralizing antibody titers against SIVmac251.6 (tier 1A) and SIVsmE660/BR-CG7V.IR1 (tier 1B) were assayed in TZM-bl cells as described (64). Titers were defined as the serum dilution at which relative luminescence units were reduced 50%, compared to virus control wells with no test serum.
ADCC was assessed as described previously, using EGFP-CEM-NKr-CCR5-SNAP cells that constitutively express green fluorescent protein (GFP) as targets (65). Briefly, 1 million target cells were incubated with 50 μg of SIVmac251 gp120 protein for 2 h at 37°C, washed, and labeled for 30 min at room temperature with SNAP-Surface-Alexa 647 (product number S9136S; New England Biolabs), as recommended by the manufacturer. Plasma samples that had been heat inactivated at 56°C for 30 min were serially diluted 10-fold, and 100 μl was added to wells of a 96-well V-bottom plate (Millipore Sigma). A total of 5,000 target cells (50 μl) and 250,000 human PBMCs (50 μl) as effectors were added to each well to yield an effector/target ratio of 50:1. The plate was incubated at 37°C for 2 h, followed by two PBS washes. The cells were resuspended in 200 μl of a 2% PBS-paraformaldehyde (PFA) solution, and data were acquired on a LSR II flow cytometer equipped with a high-throughput system (BD Biosciences). Specific killing was measured by loss of GFP from the SNAP-Surface-Alexa 647+ target cells. Target and effector cells cultured in the presence of naive plasma were used as negative controls. The anti-SIVmac gp120 monoclonal antibody KK17 (NIH AIDS reagent program) was used as a positive control. Normalized percent killing was calculated as follows: (killing in the presence of plasma − background)/(killing in the presence of KK17 − background) × 100. The ADCC endpoint titer was defined as the reciprocal of the dilution at which the percent ADCC killing was greater than the mean percent killing of the negative control wells containing medium and target and effector cells, plus 3 standard deviations.
ADCP was assayed as described previously (66), with minor modifications. SIVM766 gp120 was biotinylated with a Biotin-XX microscale protein labeling kit (Thermo Fisher Scientific, Waltham, MA) and incubated with a 100-fold dilution of 1 μg yellow-green streptavidin fluorescent beads (Thermo Fisher Scientific) overnight at 4°C in the dark. A 1:100 dilution of serum from each macaque was added to 400,000 THP-1 cells plated in a U-bottom 96-well plate. The bead-gp120 mixture was further diluted 5-fold in R10 medium, and 50 μl was added to the cells and incubated for 3 h at 37°C. After incubation, 70 μl of 2% PFA was added for fixation. Fluorescent bead uptake by THP-1 cells was assessed on an LSR II flow cytometer (BD Biosciences). The phagocytic score of each sample was calculated by multiplying the percentage of bead-positive cells (frequency) by the degree of phagocytosis measured as mean fluorescence intensity (MFI) and dividing by 106. Values were normalized to background values (cells and beads without serum) by dividing the phagocytic score of the test sample by the phagocytic score of the background sample. Phagocytic scores postvaccination were obtained by dividing the phagocytic score of the test sample by the phagocytic score at the prevaccination time point.
Rectal Env-specific IgG and IgA binding antibodies.
Rectal secretions were collected using cotton swabs and stored at −70°C in 1 ml of PBS containing 0.1% bovine serum albumin, 0.01% thimerosal, and SigmaFAST protease inhibitor cocktail tablets (EDTA free; Sigma), used according to the manufacturer’s instructions, until analysis. Samples were tested for blood contamination using Chemstrip 5 test strips (Boehringer Mannheim) prior to assay. To remove fecal contaminants, samples were passed through 5-μm polyvinylidene difluoride microcentrifugal filter units (Millipore, Billerica, MA). Briefly, SIV gp120-specific IgA and IgG antibodies were measured by ELISA as described previously (67, 68). Env-specific IgA and IgG standards derived from IgG-depleted pooled serum samples or purified serum IgG, respectively, obtained from SIVmac251-infected macaques and quantified as described previously (69) were used to generate standard curves. Horseradish peroxidase-conjugated goat anti-monkey IgA and IgG (Alpha Diagnostics) and 3,3′,5,5′-tetramethylbenzidine (TMB) substrate were used in sequential steps, followed by the addition of phosphoric acid prior to reading of the OD450. Total IgA and IgG antibodies were measured in each sample and used to standardize gp120-specific IgA and IgG concentrations. Results are reported as Env-specific IgA or IgG levels per total IgA or IgG levels (nanograms specific per micrograms total).
GC TFH cells.
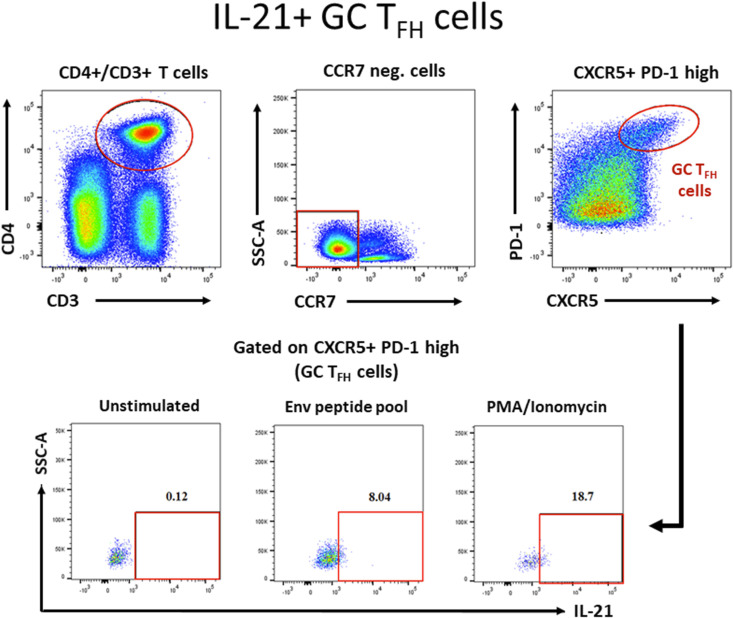
Inguinal LN biopsy samples were obtained from 9 macaques from each of the ALVAC/Env and DNA&Env groups and from 2 controls prior to immunization, 14 days following the second Ad5hr mucosal prime (week 14) and 14 days following the second intramuscular boost (week 38). Lymphocytes were isolated as described previously (70), by mincing and passing through a 70-μm cell strainer (BD Pharmingen). Cells were pelleted with 30 ml PBS at 550 × g for 7 min, incubated with 5 ml ammonium-chloride-potassium (ACK) lysing buffer (Lonza Bioscience) on ice for 10 min, and pelleted once again with PBS. Isolated LN cells were stored frozen in FBS-10% dimethyl sulfoxide solution. IL-21-producing TFH cells were detected by an intracellular staining assay as described previously (59). Briefly, frozen LN cells were thawed and resuspended in R10 medium. Aliquots (1.5 × 106 cells/500 μl) were stimulated with 1 μg/ml SIVM766 or SIVCG7V gp120 peptide pools or with 1× phorbol myristate acetate (PMA)-ionomycin cell stimulation cocktail (eBioscience) or were unstimulated. The lymphocytes received a mixture of 2 μg/ml anti-CD49d and anti-CD28 (BD Biosciences), BD GolgiPlug, and BD GolgiStop, as well as APC-eFluor 780-conjugated anti-CD197 (anti-CCR7) clone 2D12 (eBioscience) at a concentration recommended by the manufacturer, and were incubated at 37°C for 6 h. Cells were then washed with fluorescence-activated cell sorting (FACS) buffer and stained for surface markers for 25 min at room temperature with anti-PD-1-BV605 (EH12.2H7) and anti-CD25-BV785 (BC96) (BioLegend), anti-CXCR5-PerCP/eFluor 710 (MU5UBEE; eBioscience), anti-CD95-PE/CF594 (DX2), anti-CD3-Alexa Fluor 700 (SP34-2), and anti-CD4-BV711 (L200) (BD Biosciences), and Aqua Live/Dead viability dye (Invitrogen) for dead cell exclusion. Cells were washed with FACS buffer, treated with 1 ml of BD Cytofix/Cytoperm solution for 15 min at room temperature, and washed with BD Perm/Wash solution prepared at 1× dilution. Cells were then intracellularly stained with anti-IL-21-PE (3A3-N2.1), anti-Ki67-Alexa 647 (B56), and anti-CD154-PE/Cy5 (TRAP1) (BD Biosciences) and anti-Foxp3-eFluor 660 (236A/E7) (eBioscience) for 25 min at room temperature, washed with FACS buffer, and resuspended in PBS with 1% PFA. Cells were maintained at 4°C until data acquisition using an 18-parameter LSR II flow cytometer (BD Biosciences). A representative plot illustrating the gating strategy is shown in Fig. 15.
FIG 15.
Representative plot illustrating the gating strategy for Env-specific GC TFH cells. Details are provided in Materials and Methods.
Env-specific memory B cells.
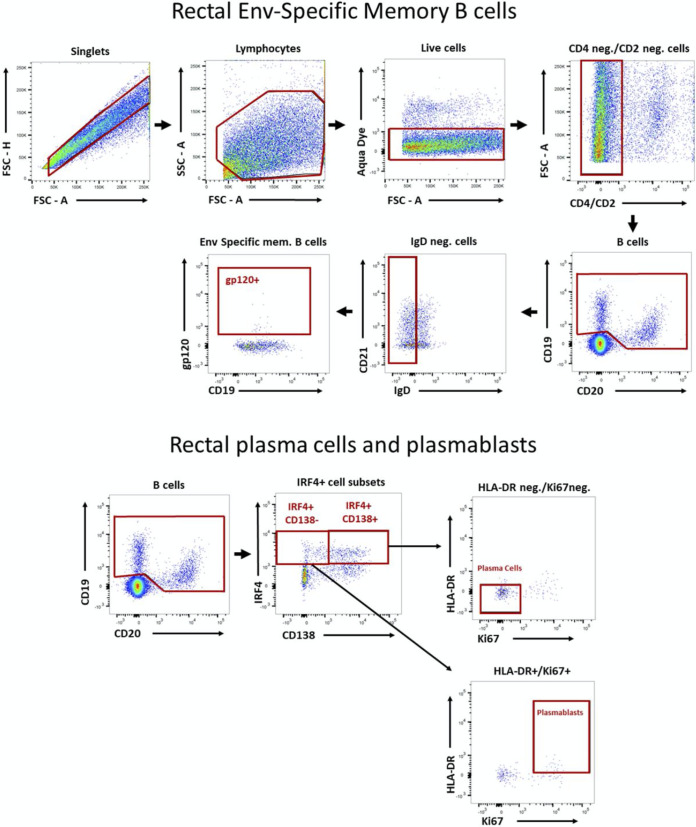
SIV Env-specific memory B cells in bone marrow and rectal biopsy samples and PBs and PCs in rectal tissue were assayed by flow cytometry as described previously (31, 63). Briefly, 1 × 106 to 2 × 106 cells were thawed, washed with R10 medium, and resuspended in PBS with 2% FBS (FACS buffer) for surface staining, which was carried out at room temperature for 25 min using the antibodies anti-CD2-Qdot 605 (S5.5), anti-CD14-Qdot 605 (Tu14), anti-CD20-eFluor 650NC (2H7), anti-HLA-DR-Pacific Blue (Tu36), and anti-CD27-PerCP/eFluor 710 (O323) (all from Thermo Fisher Scientific), anti-CD21-PE/Cy7 (B-ly4; BD Bioscience), anti-IgG-APC/Cy7 (G18-145; BD Biosciences), Texas Red-labeled anti-IgD polyclonal antibody (SouthernBiotech), anti-CD19-PE/Cy5 (J3-119; Beckman Coulter), and anti-CD138-PE (DL-101; BioLegend) (the latter only for rectal B cells). The Aqua Live/Dead viability dye (Invitrogen) was added during surface staining for dead cell exclusion. Unconjugated anti-CD4 antibodies (clones OKT4 and 19Thy5D7; NIH NHP Reagent Resource) were added to block reactivity to CD4. Envelope protein staining followed surface staining, with the addition of biotinylated gp120 proteins (2 μg/sample) and incubation for 25 min at 4°C. Cells were washed with FACS buffer, incubated for 25 min at 4°C with streptavidin conjugated to allophycocyanin, and washed with FACS buffer. Cells were incubated with 1 ml of 1× transcription factor Fix/Perm buffer (BD Pharmingen) and washed with 1× Perm/Wash buffer according to the manufacturer’s specifications. After permeabilization, intracellular staining was carried out at room temperature for 25 min with the antibodies anti-IRF4-FITC (3E4; e-Bioscience), anti-Ki67-Alexa 700 (B56), and anti-BCL2-PE (bcl2/100) (both BD Biosciences). Anti-BCL2 was used only for bone marrow B cells. Cells were finally washed with FACS buffer and resuspended in PBS with 1% PFA, and approximately 500,000 events were recorded on an 18-parameter LSR II flow cytometer (BD Biosciences), followed by analysis with FlowJo v10.2. Env-specific memory B cells were defined as described previously (71), as were PBs and PCs in rectal tissue (72). Representative plots illustrating the gating strategies are shown in Fig. 16.
FIG 16.
Representative plots illustrating gating strategies for rectal Env-specific memory B cells and rectal PCs and PBs. Details are provided in Materials and Methods.
Env-specific bone marrow PBs and PCs.
Bone marrow lymphocytes were purified as described previously (63) and frozen until analysis. Lymphocytes were thawed, and both total and EnvM766- or EnvCG7V-specific IgG and IgA antibody-secreting cells (combined as PBs/PCs) were quantified on unstimulated samples by ELISPOT as described previously (73). Env-specific IgA and IgG PBs/PCs were standardized to the total number of IgA and IgG PBs/PCs and are reported as the percentage of EnvM766 or EnvCG7V-specific PBs/PCs relative to the total number of PBs/PCs.
Rectal and vaginal microbiome analysis.
Rectal and vaginal swab samples (4N6 DNA swabs; Nova Biostorage) were collected over the course of immunization. Following collection, the swab tip was placed in 600 μl RLT Plus buffer (Qiagen), mixed briefly to release bacteria, and discarded. Vials were immediately frozen on dry ice and stored at −80°C. DNA was purified using an AllPrep DNA/RNA microkit (Qiagen) according to the manufacturer’s instructions. Swab samples were thawed and briefly vortex-mixed, and the RLT Plus lysate was pipetted into a QIAshredder spin column in a 2-ml collection tube and centrifuged for 2 min at 14,000 rpm. The lysate was again centrifuged for 3 min at 14,000 rpm, and the supernatant was transferred to an AllPrep DNA spin column in a 1.5-ml collection tube. Following centrifugation for 30 s at 10,000 rpm, the spin column was placed in a new 2-ml tube, 500 μl buffer AW1 was added, and the tube was centrifuged for 15 s at 10,000 rpm to wash the spin column membrane. The flow-through fraction was discarded, 500 μl buffer AW2 was added, and the tube was centrifuged for 2 min at 14,000 rpm to wash the membrane. The spin column was transferred to a new 1.5-ml collection tube, 50 μl buffer EB preheated to 70°C was added, the mixture was incubated for 2 to 3 min at room temperature, and the tube was centrifuged for 1 min at 10,000 rpm to elute the DNA. This step was repeated to elute additional DNA. The DNA concentration was measured using a NanoDrop spectrophotometer, and the samples were stored at −20°C prior to sequencing.
16S rRNA gene sequencing and analysis.
Paired-end, overlapping reads of the V4 region of the 16S rRNA gene (primers 515F and 806R) were generated for 306 rectal swab samples and 128 vaginal swab samples on an Illumina MiSeq platform (74). The demultiplexed paired-end fastq files were preprocessed and analyzed using QIIME2 v2-2018.2 (https://qiime2.org). The DADA2 algorithm (75), implemented in QIIME2, was used for error modeling and filtering of the raw fastq files. Following denoising, chimera removal, and rarefaction, a total of 9.27 million sequences were retained for 309 samples, with an average of 30,000 sequences per sample. Taxonomic classification was performed using the QIIME2 feature classifier (https://github.com/qiime2/q2-feature-classifier) plugin trained on the Silva database (release 132) (76). The alpha and beta diversity analyses were performed using the diversity plugin (https://github.com/qiime2/q2-diversity) at a sampling depth of 28,892×. Beta diversity using PCoA (77) was performed based on the Bray-Curtis distances, and permutational multivariate analysis of variance (PERMANOVA) was used to analyze statistical differences in beta diversity with QIIME2. The Benjamini-Hochberg false discovery rate correction was applied to all pairwise P values.
Statistical analysis.
Wilcoxon-Mann-Whitney and Wilcoxon signed rank tests were used for comparisons between groups and between times within groups. Logarithmic transformation of titers and arcsine transformation of percentage data were applied before analysis to reduce skewness in the raw data. Differences in viral acquisition during repeated challenge studies were assessed using the exact log rank test. Correlation coefficients and significance levels were estimated by the Spearman rank method, with Jonckheere-Terpstra test results reported for trends over discrete data distributions. Results are not corrected for multiple comparisons and are intended to provide individual assessments of null hypotheses, generally in an exploratory mode over microbiome taxa or in consistent, correlated combinations over times or between immunization groups. Analyses were performed using Prism (GraphPad Software, Inc.) and SAS/STAT software v9.4 (SAS Institute Inc.). All 16S rRNA gene sequencing data analysis was performed using QIIME2 v2-2018.2 (https://qiime2.org).
Data availability.
The microbiome data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA595360.
ACKNOWLEDGMENTS
We thank Josh Kramer, Matthew Breed, William Magnanelli, and Michelle Metrinko and their staff at the NCI Animal Facility for expert care of the rhesus macaques and performance of all animal procedures. Anti-CD4-Qdot 605, anti-CD8-Qdot 655, and anti-CD4 monoclonal antibodies OKT4 and 19Thy5D7 were obtained from the NIH NHP Reagent Resource. The complete set of SIV Gagmac239 peptides and the anti-SIVmac gp120 monoclonal antibody KK17 were obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID. The SIVmac251 challenge stock was obtained from Nancy Miller, Division of AIDS, NIAID, and was originally obtained from Ronald Desrosiers, University of Miami.
This work was funded by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting true views of the Department of the Army or the Department of Defense.
T.M., V.T., V.M., I.T., L.K.M.-N., S.H.H., M.A.R., R.H., E.H., W.Y., C.O., T.H., Y.S., C.L., J.B., M.R. and M.B. performed experiments; T.M., V.T., V.M., I.T., L.K.M.-N., S.H.H., M.A.R., R.H., E.H., C.O., Y.S., C.L., D.M., and M.R.-G. analyzed data; J.A.B., G.N.P., B.K.F., G.F., and M.R.-G. designed the study; D.J.V. performed statistical analysis; T.M. and M.R.-G. wrote the paper; and all authors reviewed and edited the manuscript.
REFERENCES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, Sok D, Zhou B, Burton DR, Ward AB, Wilson IA, Zhu J. 2016. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun 7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders RW, van Gils ML, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramírez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, Jacak R, Kalyuzhniy O, de Val N, Sesterhenn F, Le KM, Ramos A, Jones M, Saye-Francisco KL, Blane TR, Spencer S, Georgeson E, Hu X, Ozorowski G, Adachi Y, Kubitz M, Sarkar A, Wilson IA, Ward AB, Nemazee D, Burton DR, Schief WR. 2016. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 166:1459–1570.e11. doi: 10.1016/j.cell.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. 2012. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, Hahn BH, Korber BT. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med 13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 8.Barouch DH, O'Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, Sun Y-H, La Porte A, Riggs AM, Lynch DM, Clark SL, Backus K, Perry JR, Seaman MS, Carville A, Mansfield KG, Szinger JJ, Fischer W, Muldoon M, Korber B. 2010. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med 16:319–323. doi: 10.1038/nm.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X, Lu Z, Valentin A, Rosati M, Broderick KE, Sardesai NY, Marx PA, Mullins JI, Pavlakis GN, Felber BK. 2018. Gag and env conserved element CE DNA vaccines elicit broad cytotoxic T cell responses targeting subdominant epitopes of HIV and SIV able to recognize virus-infected cells in macaques. Hum Vaccin Immunother 14:2163–2177. doi: 10.1080/21645515.2018.1489949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen SG, Piatak M, Jr, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin G, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Früh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. 2013. Immune clearance of highly pathogenic SIV infection. Nature 502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen SG, Marshall EE, Malouli D, Ventura AB, Hughes CM, Ainslie E, Ford JC, Morrow D, Gilbride RM, Bae JY, Legasse AW, Oswald K, Shoemaker R, Berkemeier B, Bosche WJ, Hull M, Womack J, Shao J, Edlefsen PT, Reed JS, Burwitz BJ, Sacha JB, Axthelm MK, Früh K, Lifson JD, Picker LJ. 2019. A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci Transl Med 11:eaaw2607. doi: 10.1126/scitranslmed.aaw2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen SG, Wu HL, Burwitz BJ, Hughes CM, Hammond KB, Ventura AB, Reed JS, Gilbride RM, Ainslie E, Morrow DW, Ford JC, Selseth AN, Pathak R, Malouli D, Legasse AW, Axthelm MK, Nelson JA, Gillespie GM, Walters LC, Brackenridge S, Sharpe HR, López CA, Früh K, Korber BT, McMichael AJ, Gnanakaran S, Sacha JB, Picker LJ. 2016. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351:714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aldovini A. 2016. Mucosal vaccination for prevention of HIV infection and AIDS. Curr HIV Res 14:247–259. doi: 10.2174/1570162x14999160224103025. [DOI] [PubMed] [Google Scholar]
- 14.Lubeck MD, Natuk R, Myagkikh M, Kalyan N, Aldrich K, Sinangil F, Alipanah S, Murthy SC, Chanda PK, Nigida SM, Jr, Markham PD, Zolla-Pazner S, Steimer K, Wade M, Reitz MS, Jr, Arthur LO, Mizutani S, Davis A, Hung PP, Gallo RC, Eichberg J, Robert-Guroff M. 1997. Long-term protection of chimpanzees against high-dose HIV-1 challenge induced by immunization. Nat Med 3:651–658. doi: 10.1038/nm0697-651. [DOI] [PubMed] [Google Scholar]
- 15.Malkevitch NV, Patterson LJ, Aldrich MK, Wu Y, Venzon D, Florese RH, Kalyanaraman VS, Pal R, Lee EM, Zhao J, Cristillo A, Robert-Guroff M. 2006. Durable protection of rhesus macaques immunized with a replicating adenovirus-SIV multigene prime/protein boost vaccine regimen against a second SIVmac251 rectal challenge: role of SIV-specific CD8+ T cell responses. Virology 353:83–98. doi: 10.1016/j.virol.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Patterson LJ, Malkevitch N, Venzon D, Pinczewski J, Gomez-Roman VR, Wang L, Kalyanaraman VS, Markham PD, Robey FA, Robert-Guroff M. 2004. Protection against mucosal simian immunodeficiency virus SIVmac251 challenge by using replicating adenovirus-SIV multigene vaccine priming and subunit boosting. J Virol 78:2212–2221. doi: 10.1128/jvi.78.5.2212-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogers WM, Davis D, Baak I, Kan E, Hofman S, Sun Y, Mortier D, Lian Y, Oostermeijer H, Fagrouch Z, Dubbes R, van der Maas M, Mooij P, Koopman G, Verschoor E, Langedijk JPM, Zhao J, Brocca-Cofano E, Robert-Guroff M, Srivastava I, Barnett S, Heeney JL. 2008. Systemic neutralizing antibodies induced by long interval mucosally primed systemically boosted immunization correlate with protection from mucosal SHIV challenge. Virology 382:217–225. doi: 10.1016/j.virol.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demberg T, Florese RH, Heath MJ, Larsen K, Kalisz I, Kalyanaraman VS, Lee EM, Pal R, Venzon D, Grant R, Patterson LJ, Korioth-Schmitz B, Buzby A, Dombagoda D, Montefiori DC, Letvin NL, Cafaro A, Ensoli B, Robert-Guroff M. 2007. A replication-competent adenovirus-human immunodeficiency virus (Ad-HIV) tat and Ad-HIV env priming/Tat and envelope protein boosting regimen elicits enhanced protective efficacy against simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J Virol 81:3414–3427. doi: 10.1128/JVI.02453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuero I, Mohanram V, Musich T, Miller L, Vargas-Inchaustegui DA, Demberg T, Venzon D, Kalisz I, Kalyanaraman VS, Pal R, Ferrari MG, LaBranche C, Montefiori DC, Rao M, Vaccari M, Franchini G, Barnett SW, Robert-Guroff M. 2015. Mucosal B cells are associated with delayed SIV acquisition in vaccinated female but not male rhesus macaques following SIVmac251 rectal challenge. PLoS Pathog 11:e1005101. doi: 10.1371/journal.ppat.1005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon SN, Doster MN, Kines RC, Keele BF, Brocca-Cofano E, Guan Y, Pegu P, Liyanage NPM, Vaccari M, Cuburu N, Buck CB, Ferrari G, Montefiori D, Piatak M, Jr, Lifson JD, Xenophontos AM, Venzon D, Robert-Guroff M, Graham BS, Lowy DR, Schiller JT, Franchini G. 2014. Antibody to the gp120 V1/V2 loops and CD4+ and CD8+ T cell responses in protection from SIVmac251 vaginal acquisition and persistent viremia. J Immunol 193:6172–6183. doi: 10.4049/jimmunol.1401504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valentin A, McKinnon K, Li J, Rosati LJM, Kulkarni V, Pilkington GR, Bear J, Alicea C, Vargas-Inchaustegui DA, Patterson LJ, Pegu P, Liyanage NPM, Gordon SN, Vaccari M, Wang Y, Hogg AE, Frey B, Sui Y, Reed SG, Sardesai NY, Berzofsky JA, Franchini G, Robert-Guroff M, Felber BK, Pavlakis GN. 2014. Comparative analysis of SIV-specific cellular immune responses induced by different vaccine platforms in rhesus macaques. Clin Immunol 155:91–107. doi: 10.1016/j.clim.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vargas-Inchaustegui DA, Tuero I, Mohanram V, Musich T, Pegu P, Valentin A, Sui Y, Rosati M, Bear J, Venzon DJ, Kulkarni V, Alicea C, Pilkington GR, Liyanage NPM, Demberg T, Gordon SN, Wang Y, Hogg AE, Frey B, Patterson LJ, DiPasquale J, Montefiori DC, Sardesai NY, Reed SG, Berzofsky JA, Franchini G, Felber BK, Pavlakis GN, Robert-Guroff M. 2014. Humoral immunity induced by mucosal and/or systemic SIV-specific vaccine platforms suggests novel combinatorial approaches for enhancing responses. Clin Immunol 153:308–322. doi: 10.1016/j.clim.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NPM, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, Rao M, Chung AW, Dowell KG, Bailey-Kellogg C, Brown EP, Ackerman ME, Vargas-Inchaustegui DA, Whitney S, Doster MN, Binello N, Pegu P, Montefiori DC, Foulds K, Quinn DS, Donaldson M, Liang F, Loré K, Roederer M, Koup RA, McDermott A, Ma Z-M, Miller CJ, Phan TB, Forthal DN, Blackburn M, Caccuri F, Bissa M, Ferrari G, Kalyanaraman V, Ferrari MG, Thompson D, Robert-Guroff M, Ratto-Kim S, Kim JH, Michael NL, Phogat S, Barnett SW, Tartaglia J, Venzon D, Stablein DM, Alter G, Sekaly R-P, Franchini G. 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22:762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jalah R, Kulkarni V, Patel V, Rosati M, Alicea C, Bear J, Yu L, Guan Y, Shen X, Tomaras GD, LaBranche C, Montefiori DC, Prattipati R, Pinter A, Bess J, Jr, Lifson JD, Reed SG, Sardesai NY, Venzon JD, Valentin A, Pavlakis GN, Felber BK. 2014. DNA and protein co-immunization improves the magnitude and longevity of humoral immune responses in macaques. PLoS One 9:e91550. doi: 10.1371/journal.pone.0091550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel V, Jalah R, Kulkarni V, Valentin A, Rosati M, Alicea C, von Gegerfelt A, Huang W, Guan Y, Keele BF, Bess JW, Jr, Piatak M, Jr, Lifson JD, Williams WT, Shen X, Tomaras GD, Amara RR, Robinson HL, Johnson W, Broderick KE, Sardesai NY, Venzon DJ, Hirsch VM, Felber BK, Pavlakis GN. 2013. DNA and virus particle vaccination protects against acquisition and confers control of viremia upon heterologous simian immunodeficiency virus challenge. Proc Natl Acad Sci U S A 110:2975–2980. doi: 10.1073/pnas.1215393110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda K, Littman DR. 2012. The microbiome in infectious disease and inflammation. Annu Rev Immunol 30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. 2012. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum Vaccin Immunother 8:1192–1207. doi: 10.4161/hv.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jalah R, Patel V, Kulkarni V, Rosati M, Alicea C, Ganneru B, von Gegerfelt A, Huang W, Guan Y, Broderick KE, Sardesai NY, LaBranche C, Montefiori DC, Pavlakis GN, Felber BK. 2012. IL-12 DNA as molecular vaccine adjuvant increases the cytotoxic T cell responses and breadth of humoral immune responses in SIV DNA vaccinated macaques. Hum Vaccin Immunother 8:1620–1629. doi: 10.4161/hv.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazel-Sanchez B, Yildiz S, Schmolke M. 2019. Menage a trois: virus, host, and microbiota in experimental infection models. Trends Microbiol 27:440–452. doi: 10.1016/j.tim.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Ackerman ME, Das J, Pittala S, Broge T, Linde C, Suscovich TJ, Brown EP, Bradley T, Natarajan H, Lin S, Sassic JK, O'Keefe S, Mehta N, Goodman D, Sips M, Weiner JA, Tomaras GD, Haynes BF, Lauffenburger DA, Bailey-Kellogg C, Roederer M, Alter G. 2018. Route of immunization defines multiple mechanisms of vaccine-mediated protection against SIV. Nat Med 24:1590–1598. doi: 10.1038/s41591-018-0161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohanram V, Demberg T, Musich T, Tuero I, Vargas-Inchaustegui DA, Miller-Novak L, Venzon D, Robert-Guroff M. 2016. B cell responses associated with vaccine-induced delayed SIVmac251 acquisition in female rhesus macaques. J Immunol 197:2316–2324. doi: 10.4049/jimmunol.1600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller-Novak LK, Das J, Musich TA, Demberg T, Weiner JA, Venzon DJ, Mohanram V, Vargas-Inchaustegui DA, Tuero I, Ackerman ME, Alter G, Robert-Guroff M. 2018. Analysis of complement-mediated lysis of simian immunodeficiency virus (SIV) and SIV-infected cells reveals sex differences in vaccine-induced immune responses in rhesus macaques. J Virol 92:e00721-18. doi: 10.1128/JVI.00721-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vargas-Inchaustegui DA, Demers A, Shaw JM, Kang G, Ball D, Tuero I, Musich T, Mohanram V, Demberg T, Karpova TS, Li Q, Robert-Guroff M. 2016. Vaccine induction of lymph node-resident simian immunodeficiency virus Env-specific T follicular helper cells in rhesus macaques. J Immunol 196:1700–1710. doi: 10.4049/jimmunol.1502137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein SL. 2012. Sex influences immune responses to viruses, and efficacy of prophylaxis and treatments for viral diseases. Bioessays 34:1050–1059. doi: 10.1002/bies.201200099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rechtien A, Altfeld M. 2019. Sexual dimorphism in HIV-1 infection. Semin Immunopathol 41:195–202. doi: 10.1007/s00281-018-0704-y. [DOI] [PubMed] [Google Scholar]
- 36.Flanagan KL, Fink AL, Plebanski M, Klein SL. 2017. Sex and gender differences in the outcomes of vaccination over the life course. Annu Rev Cell Dev Biol 33:577–599. doi: 10.1146/annurev-cellbio-100616-060718. [DOI] [PubMed] [Google Scholar]
- 37.Klein SL, Flanagan KL. 2016. Sex differences in immune responses. Nat Rev Immunol 16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 38.Markle JG, Fish EN. 2014. SeXX matters in immunity. Trends Immunol 35:97–104. doi: 10.1016/j.it.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson K, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z, Yeoh YK, Hui M, Wong PY, Chan MCW, Ip M, Yu J, Burk RD, Chan FKL, Chan PKS. 2018. Diversity of macaque microbiota compared to the human counterparts. Sci Rep 8:15573. doi: 10.1038/s41598-018-33950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann P, Curtis N. 2018. The influence of the intestinal microbiome on vaccine responses. Vaccine 36:4433–4439. doi: 10.1016/j.vaccine.2018.04.066. [DOI] [PubMed] [Google Scholar]
- 42.Williams WB, Liao H-X, Moody MA, Kepler TB, Alam SM, Gao F, Wiehe K, Trama AM, Jones K, Zhang R, Song H, Marshall DJ, Whitesides JF, Sawatzki K, Hua A, Liu P, Tay MZ, Seaton KE, Shen X, Foulger A, Lloyd KE, Parks R, Pollara J, Ferrari G, Yu J-S, Vandergrift N, Montefiori DC, Sobieszczyk ME, Hammer S, Karuna S, Gilbert P, Grove D, Grunenberg N, McElrath MJ, Mascola JR, Koup RA, Corey L, Nabel GJ, Morgan C, Churchyard G, Maenza J, Keefer M, Graham BS, Bad En LR, Tomaras GD, Haynes BF. 2015. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science 349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sui Y, Lewis GK, Wang Y, Berckmueller K, Frey B, Dzutsev A, Vargas-Inchaustegui D, Mohanram V, Musich T, Shen X, DeVico A, Fouts T, Venzon D, Kirk J, Waters RC, Talton J, Klinman D, Clements J, Tomaras GD, Franchini G, Robert-Guroff M, Trinchieri G, Gallo RC, Berzofsky JA. 2019. Mucosal vaccine efficacy against intrarectal SHIV is independent of anti-Env antibody response. J Clin Invest 129:1314–1328. doi: 10.1172/JCI122110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elizaldi SR, Verma A, Walter KA, Rolston M, Dinasarapu AR, Durbin-Johnson BP, Settles M, Kozlowski PA, Raeman R, Iyer SS. 2019. Rectal microbiome composition correlates with humoral immunity to HIV-1 in vaccinated rhesus macaques. mSphere 4:e00824-19. doi: 10.1128/mSphere.00824-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sui Y, Dzutsev A, Venzon D, Frey B, Thovarai V, Trinchieri G, Berzofsky JA. 2018. Influence of gut microbiome on mucosal immune activation and SHIV viral transmission in naive macaques. Mucosal Immunol 11:1219–1229. doi: 10.1038/s41385-018-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, Nkolola JP, Norman ME, Miller AD, Wang D, Barouch DH, Virgin HW. 2016. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19:323–335. doi: 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hensley-McBain T, Zevin AS, Manuzak J, Smith E, Gile J, Miller C, Agricola B, Katze M, Reeves RK, Kraft CS, Langevin S, Klatt NR. 2016. Effects of fecal microbial transplantation on microbiome and immunity in simian immunodeficiency virus infected macaques. J Virol 90:4981–4989. doi: 10.1128/JVI.00099-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanada S, Pirzadeh M, Carver KY, Deng JC. 2018. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol 9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu C, Yang Y, Zhu W. 2015. Crosstalk between the immune receptors and gut microbiota. Curr Protein Pept Sci 16:622–631. doi: 10.2174/1389203716666150630134356. [DOI] [PubMed] [Google Scholar]
- 50.Levy M, Blacher E, Elinav E. 2017. Microbiome, metabolites and host immunity. Curr Opin Microbiol 35:8–15. doi: 10.1016/j.mib.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, Ritz T, Longerich T, Theriot CM, McCulloch JA, Roy S, Yuan W, Thovarai V, Sen SK, Ruchirawat M, Korangy F, Wang XW, Trinchieri G, Greten TF. 2018. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360:eaan5931. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fransen F, van Beek AA, Borghuis T, Meijer B, Hugenholtz F, van der Gaast-de Jongh C, Savelkoul HF, de Jonge MI, Faas MM, Boekschoten MV, Smidt H, El Aidy S, de Vos P. 2017. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol 8:754. doi: 10.3389/fimmu.2017.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanagibashi T, Hosono A, Oyama A, Tsuda M, Hachimura S, Takahashi Y, Itoh K, Hirayama K, Takahashi K, Kaminogawa S. 2009. Bacteroides induce higher IgA production than Lactobacillus by increasing activation-induced cytidine deaminase expression in B cells in murine Peyer's patches. Biosci Biotechnol Biochem 73:372–377. doi: 10.1271/bbb.80612. [DOI] [PubMed] [Google Scholar]
- 54.Yanagibashi T, Hosono A, Oyama A, Tsuda M, Suzuki A, Hachimura S, Takahashi Y, Momose Y, Itoh K, Hirayama K, Takahashi K, Kaminogawa S. 2013. IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA+ B cells. Immunobiology 218:645–651. doi: 10.1016/j.imbio.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 55.Hladik F, McElrath MJ. 2008. Setting the stage: host invasion by HIV. Nat Rev Immunol 8:447–457. doi: 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chenine AL, Siddappa NB, Kramer VG, Sciaranghella G, Rasmussen RA, Lee SJ, Santosuosso M, Poznansky MC, Velu V, Amara RR, Souder C, Anderson DC, Villinger F, Else JG, Novembre FJ, Strobert E, O’Neil SP, Evan Secor W, Ruprecht RM. 2010. Relative transmissibility of an R5 clade C simian-human immunodeficiency virus across different mucosae in macaques parallels the relative risks of sexual HIV-1 transmission in humans via different routes. J Infect Dis 201:1155–1163. doi: 10.1086/651274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wessels JM, Felker AM, Dupont HA, Kaushic C. 2018. The relationship between sex hormones, the vaginal microbiome and immunity in HIV-1 susceptibility in women. Dis Model Mech 11:dmm035147. doi: 10.1242/dmm.035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulmer JB, DeWitt CM, Chastain M, Friedman A, Donnelly JJ, McClements WL, Caulfield MJ, Bohannon KE, Volkin DB, Evans RK. 1999. Enhancement of DNA vaccine potency using conventional aluminum adjuvants. Vaccine 18:18–28. doi: 10.1016/s0264-410x(99)00151-6. [DOI] [PubMed] [Google Scholar]
- 59.Helmold Hait S, Vargas-Inchaustegui DA, Musich T, Mohanram V, Tuero I, Venzon DJ, Bear J, Rosati M, Vaccari M, Franchini G, Felber BK, Pavlakis GN, Robert-Guroff M. 2018. Early T follicular helper cell responses and germinal center reactions are associated with viremia control in immunized rhesus macaques. J Virol 93:e01687-18. doi: 10.1128/JVI.01687-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee EM, Chung HK, Livesay J, Suschak J, Finke L, Hudacik L, Galmin L, Bowen B, Markham P, Cristillo A, Pal R. 2010. Molecular methods for evaluation of virological status of nonhuman primates challenged with simian immunodeficiency or simian-human immunodeficiency viruses. J Virol Methods 163:287–294. doi: 10.1016/j.jviromet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 61.Romano JW, Shurtliff RN, Dobratz E, Gibson A, Hickman K, Markham PD, Pal R. 2000. Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J Virol Methods 86:61–70. doi: 10.1016/s0166-0934(99)00184-6. [DOI] [PubMed] [Google Scholar]
- 62.Sun Y, Santra S, Schmitz JE, Roederer M, Letvin NL. 2008. Magnitude and quality of vaccine-elicited T-cell responses in the control of immunodeficiency virus replication in rhesus monkeys. J Virol 82:8812–2219. doi: 10.1128/JVI.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Demberg T, Brocca-Cofano E, Xiao P, Venzon D, Vargas-Inchaustegui D, Lee EM, Kalisz I, Kalyanaraman VS, Dipasquale J, McKinnon K, Robert-Guroff M. 2012. Dynamics of memory B-cell populations in blood, lymph nodes, and bone marrow during antiretroviral therapy and envelope boosting in simian immunodeficiency virus SIVmac251-infected rhesus macaques. J Virol 86:12591–12604. doi: 10.1128/JVI.00298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12:Unit 12 1. [DOI] [PubMed] [Google Scholar]
- 65.Orlandi C, Flinko R, Lewis GK. 2016. A new cell line for high throughput HIV-specific antibody-dependent cellular cytotoxicity (ADCC) and cell-to-cell virus transmission studies. J Immunol Methods 433:51–58. doi: 10.1016/j.jim.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, Jost S, Berger CT, Sciaranghella G, Liu Q, Irvine DJ, Burton DR, Alter G. 2011. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods 366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertley FM, Kozlowski PA, Wang SW, Chappelle J, Patel J, Sonuyi O, Mazzara G, Montefiori D, Carville A, Mansfield KG, Aldovini A. 2004. Control of simian/human immunodeficiency virus viremia and disease progression after IL-2-augmented DNA-modified vaccinia virus Ankara nasal vaccination in nonhuman primates. J Immunol 172:3745–3757. doi: 10.4049/jimmunol.172.6.3745. [DOI] [PubMed] [Google Scholar]
- 68.Patterson LJ, Daltabuit-Test M, Xiao P, Zhao J, Hu W, Wille-Reece U, Brocca-Cofano E, Kalyanaraman VS, Kalisz I, Whitney S, Lee EM, Pal R, Montefiori DC, Dandekar S, Seder R, Roederer M, Wiseman RW, Hirsch V, Robert-Guroff M. 2011. Rapid SIV Env-specific mucosal and serum antibody induction augments cellular immunity in protecting immunized, elite-controller macaques against high dose heterologous SIV challenge. Virology 411:87–102. doi: 10.1016/j.virol.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manrique M, Kozlowski PA, Wang SW, Wilson RL, Micewicz E, Montefiori DC, Mansfield KG, Carville A, Aldovini A. 2009. Nasal DNA-MVA SIV vaccination provides more significant protection from progression to AIDS than a similar intramuscular vaccination. Mucosal Immunol 2:536–550. doi: 10.1038/mi.2009.103. [DOI] [PubMed] [Google Scholar]
- 70.Demberg T, Boyer JD, Malkevich N, Patterson LJ, Venzon D, Summers EL, Kalisz I, Kalyanaraman VS, Lee EM, D Weiner DB, Robert-Guroff M. 2008. Sequential priming with simian immunodeficiency virus (SIV) DNA vaccines, with or without encoded cytokines, and a replicating adenovirus-SIV recombinant followed by protein boosting does not control a pathogenic SIVmac251 mucosal challenge. J Virol 82:10911–10921. doi: 10.1128/JVI.01129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mohanram V, Demberg T, Tuero I, Vargas-Inchaustegui D, Pavlakis GN, Felber BK, Robert-Guroff M. 2014. Improved flow-based method for HIV/SIV envelope-specific memory B-cell evaluation in rhesus macaques. J Immunol Methods 412:78–84. doi: 10.1016/j.jim.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Demberg T, Mohanram V, Venzon D, Robert-Guroff M. 2014. Phenotypes and distribution of mucosal memory B-cell populations in the SIV/SHIV rhesus macaque model. Clin Immunol 153:264–276. doi: 10.1016/j.clim.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brocca-Cofano E, McKinnon K, Demberg T, Venzon D, Hidajat R, Xiao P, Daltabuit-Test M, Patterson LJ, Robert-Guroff M. 2011. Vaccine-elicited SIV and HIV envelope-specific IgA and IgG memory B cells in rhesus macaque peripheral blood correlate with functional antibody responses and reduced viremia. Vaccine 29:3310–3319. doi: 10.1016/j.vaccine.2011.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuur AF, Ieno EN, Smith GM. 2007. Analysing ecological data, p 259–264. Springer, New York, NY. doi: 10.1007/978-0-387-45972-1_15. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microbiome data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA595360.