Despite tremendous efforts, there are no FDA-approved vaccines for human respiratory syncytial virus (RSV). A live attenuated RSV vaccine is one of the most promising vaccine strategies for RSV. However, it has been a challenge to identify an RSV vaccine strain that has an optimal balance between attenuation and immunogenicity. In this study, we generated a panel of recombinant RSVs carrying a single and double deletion or a single alanine insertion in the large (L) polymerase protein that are genetically stable, sufficiently attenuated, and grow to high titer in cultured cells, while retaining high immunogenicity. Thus, these recombinant viruses may be promising vaccine candidates for RSV.
KEYWORDS: vaccine, polymerases, respiratory syncytial virus
ABSTRACT
Human respiratory syncytial virus (RSV) is the leading viral cause of lower respiratory tract disease in infants and children worldwide. Currently, there are no FDA-approved vaccines to combat this virus. The large (L) polymerase protein of RSV replicates the viral genome and transcribes viral mRNAs. The L protein is organized as a core ring-like domain containing the RNA-dependent RNA polymerase and an appendage of globular domains containing an mRNA capping region and a cap methyltransferase region, which are linked by a flexible hinge region. Here, we found that the flexible hinge region of RSV L protein is tolerant to amino acid deletion or insertion. Recombinant RSVs carrying a single or double deletion or a single alanine insertion were genetically stable, highly attenuated in immortalized cells, had defects in replication and spread, and had a delay in innate immune cytokine responses in primary, well-differentiated, human bronchial epithelial (HBE) cultures. The replication of these recombinant viruses was highly attenuated in the upper and lower respiratory tracts of cotton rats. Importantly, these recombinant viruses elicited high levels of neutralizing antibody and provided complete protection against RSV replication. Taken together, amino acid deletions or insertions in the hinge region of the L protein can serve as a novel approach to rationally design genetically stable, highly attenuated, and immunogenic live virus vaccine candidates for RSV.
IMPORTANCE Despite tremendous efforts, there are no FDA-approved vaccines for human respiratory syncytial virus (RSV). A live attenuated RSV vaccine is one of the most promising vaccine strategies for RSV. However, it has been a challenge to identify an RSV vaccine strain that has an optimal balance between attenuation and immunogenicity. In this study, we generated a panel of recombinant RSVs carrying a single and double deletion or a single alanine insertion in the large (L) polymerase protein that are genetically stable, sufficiently attenuated, and grow to high titer in cultured cells, while retaining high immunogenicity. Thus, these recombinant viruses may be promising vaccine candidates for RSV.
INTRODUCTION
Human respiratory syncytial virus (RSV) is the leading causative agent of acute viral respiratory tract infections (1). RSV is a globally prevalent pathogen infecting individuals of all ages, with high morbidity and mortality seen in infants, children, the elderly, and immunocompromised individuals. Epidemiological studies suggest that 69% of infants are infected by RSV during the first year of life and 83% during the second year (2, 3). By the age of 5 years, nearly all children have been infected by RSV. The clinical signs and symptoms associated with RSV range from mild respiratory problems to severe cough, bronchiolitis, pneumonia, and death (1). Worldwide it is estimated that RSV causes 3.4 million hospitalizations and between 66,000 and 199,000 deaths in young children less than 5 years of age (4, 5). The only FDA-approved therapy for RSV infection is palivizumab (Synagis; MedImmune), a monoclonal antibody targeting the viral fusion protein, which is reserved for high-risk infants (such as cases of prematurity and congenital heart diseases) (6). Currently, there is no FDA-approved RSV vaccine, although several vaccine candidates, including live attenuated, subunit, and virus-vectored vaccines are in preclinical or clinical trials (reviewed in reference 7).
Generally, inactivated and live attenuated vaccines are the two most common strategies used in vaccine development. For safety reasons, an inactivated vaccine is often preferred. However, a formalin-inactivated RSV vaccine developed and tested in the 1960s not only failed to induce a protective immune response in humans, but induced enhanced respiratory disease upon natural infection with RSV (8, 9). Eighty percent of the vaccinated children were hospitalized following natural RSV infection, and two children died. In contrast to inactivated vaccines, enhanced lung disease has not been observed for candidate live attenuated RSV vaccines (1, 10). Therefore, live attenuated vaccines are one of the most promising vaccine candidates for RSV. However, it has been a challenge to identify an RSV vaccine strain that is genetically stable, sufficiently attenuated, and grows to high titer in cultured cells while retaining high immunogenicity (1, 10, 11). From the early 1960s to 1990, a panel of cold-passaged (cp) and temperature-sensitive (ts) RSVs were identified and tested clinically (12–17), but none was found to have a satisfactory balance between attenuation and immunogenicity (15–17).
In 1995, reverse genetics for RSV was established, which provided a powerful tool to rationally design attenuated strains by manipulating the viral genome (18). Attenuation approaches that have been attempted include engineering mutations in the two major glycoproteins (F or G), deleting a nonessential gene (encoding nonstructural proteins NS1 and NS2, the small hydrophobic [SH] protein, or the M2-2 protein), and deoptimizing codon usage of the genes expressing viral proteins (10, 19–22). Many of these approaches either affect genetic stability, virus growth in vitro, or immunogenicity or lead to insufficient attenuation or overattenuation. Recently, several new candidates (such as RSV with deleted M2-2, RSV with thermostabilized F protein, and RSV with mutations in NS1 or NS2 protein) have been developed and show good attenuation and immunogenicity in animal models (23–25). Although some of these vaccine candidates are promising, the exploration of new approaches for the attenuation of RSV for vaccine development are needed.
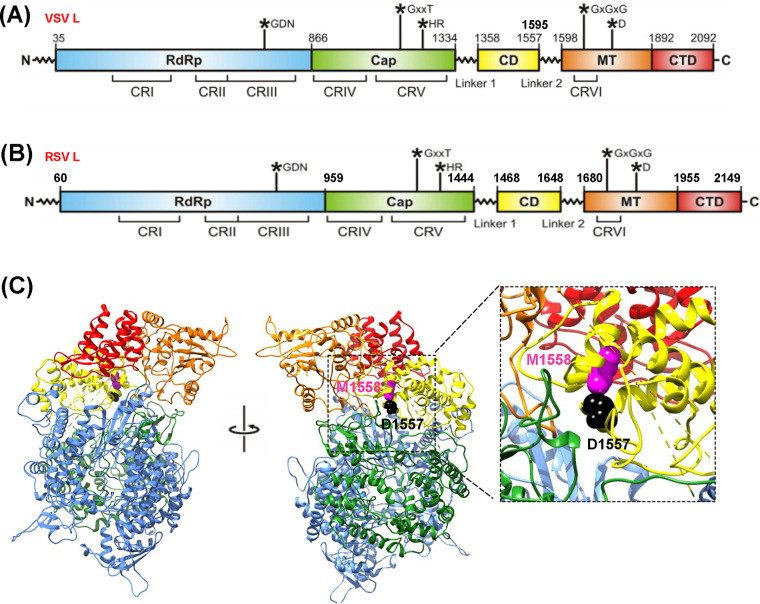
RSV belongs to the family Pneumoviridae in the order Mononegavirales. Similar to all other nonsegmented negative-sense (NNS) RNA viruses, RSV encodes a large (L) polymerase protein with molecular weight of 230 kDa that possesses all of the enzymatic activities for genome replication, mRNA transcription, and mRNA modifications (26, 27). Amino acid sequence alignment of the L proteins of NNS RNA viruses has identified six conserved regions (CR) numbered I to VI (Fig. 1A). Functions of CR III, CR V, and CR VI have been defined (28–33). CR III contains a GDN motif that is essential for polymerase activity, whereas CR V and CR VI function as the mRNA capping enzyme and the cap-modifying methyltransferases (MTases), respectively (28–33). A recent structural study of the L protein of vesicular stomatitis virus (VSV) within the related family Rhabdoviridae revealed that the L protein organizes as a core ring-like domain containing the RNA-dependent RNA polymerase and an appendage of globular domains containing a capping region (CR V) and a cap methyltransferase region (CR VI), which are linked by a flexible hinge region (34). Interestingly, morbilliviruses (measles virus, MeV [35]; rinderpest virus, RPV [36]; and canine distemper virus, CDV [37]) within the family Paramyxoviridae, as well as VSV within the Rhabdoviridae (38), were shown to tolerate in-frame insertion of the entire enhanced GFP (EGFP) at the region between CR V and CR VI in the L protein, which resulted in viable recombinant viruses. In addition, constructs with insertion of a hemagglutinin (HA) tag in this flexible region in Nipah virus and RSV L proteins were found to be functional in a minigenome assay, retaining 40 to 60% and 10 to 60% polymerase activity compared to the wild-type L protein, respectively (39). However, it is unknown whether the HA insertion will lead to recovery of viable recombinant viruses.
FIG 1.
Design of RSV L deletion and insertion mutants. (A) Conserved regions (CRs) in the VSV L protein. The nucleotide polymerization motif (GDN) in CR III, mRNA capping motif (GxxT[n]HR) in CR V, and mRNA cap methyltrasferase motif (SAM binding GxGxG…D) in CR VI are indicated. (B) CRs in the RSV L protein. Based on the sequence alignment of RSV L (YP009518860.1) and VSV L (Q98776.1), the CRs and their predicted amino acid positions are assigned. CD, connector domain; MT, methyltransferase; CTD, C-terminal domain; and EGFP, enhanced green fluorescent protein gene. (C) Structure modeling of RSV L protein. The cryo-EM structure of the VSV L protein (PDB no. 5A22) was used as the template for RSV L protein structure prediction using MODELLER program (version 9.20). Amino acids D1557 and M1558 are indicated.
In this study, we found that the flexible hinge region between CR V and CR VI of RSV L protein not only was tolerant to amino acid insertion but also tolerates amino acid deletion. Recombinant RSVs (rRSVs) carrying a single or double deletion, or an alanine insertion in this flexible region, grew to a high titer, and were genetically stable and sufficiently attenuated, yet retained high immunogenicity in cotton rats. Therefore, these rRSVs’ deletion and insertion mutants are highly promising vaccine candidates for RSV.
RESULTS
Recovery of rgRSVs carrying a deletion or insertion in the L protein.
Recent structural studies of VSV L protein showed that CR V and CR VI are connected by two linkers (linkers 1 and 2) separated by a connector domain (CD), suggesting that the region between CR V and CR VI is flexible (34). More recently, the structure of RSV L has been solved (40, 41). Unfortunately, the RSV L structure does not include the CD and CR VI. Thus, we performed amino acid sequence alignment and structural homology analysis between VSV and RSV L proteins. The analysis showed that CR I to VI, linkers 1 and 2, and CD are conserved between RSV and VSV L proteins (Fig. 1A and B). Previously, in-frame insertion of EGFP into linker 2 of VSV L resulted in a functional L protein and a viable recombinant VSV (38). Based on the sequence alignment between VSV and RSV L protein, we hypothesized that the homologous positions at amino acids D1557 and M1558 in RSV L will tolerate amino acid insertions and deletions (Fig. 1C). To generate mutants, we first deleted single amino acid D1557 or M1558 from the RSV L gene, resulting in L-ΔD1557 and L-ΔM1558, respectively. Both D1557 and M1558 were also deleted in one virus to generate the double deletion mutant, L-ΔD1557-ΔM1558. To generate an insertion mutant, a single alanine residue was inserted between D1557 and M1558, resulting in L-1557-A-1558.
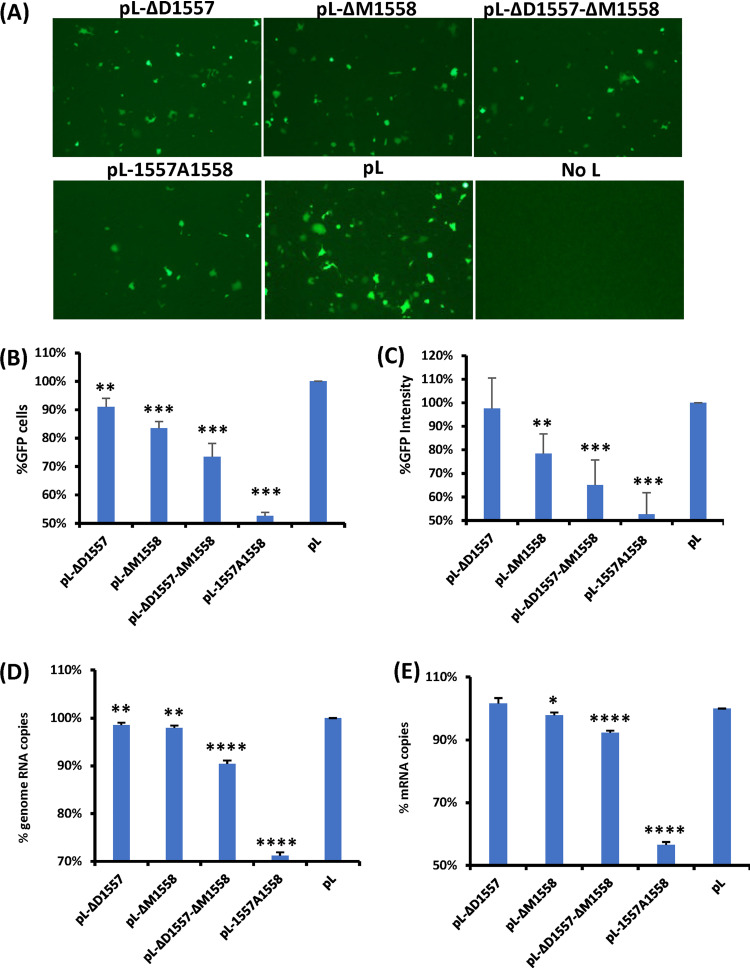
These RSV L mutants were tested in an RSV minigenome that expresses GFP and found to be functional (Fig. 2A), although these L mutants expressed less GFP relative to the wild-type (wt) L, suggesting that the L deletion and L insertion mutants are capable of replicating genome and transcribing mRNA and thus may not be lethal to the virus. Quantitative analysis showed that L-ΔD1557 expressed 90% as many GFP-positive cells compared to wt L (P < 0.01) but had no significant difference in GFP intensity from wt L (P > 0.05) (Fig. 2B and C). L-ΔM1558, L-ΔD1557-ΔM1558, and L-1557A1558 produced approximately 83, 73, and 52% as much GFP-positive cells as the wt minigenome expression, respectively (Fig. 2B). In addition, the GFP intensities of L-ΔM1558, L-ΔD1557-ΔM1558, and L-1557A1558 were 78, 65, and 53% relative to that of wt L, respectively. The total RNA was extracted from the cells transfected with the minigenome, and the RNA copies of the replication product (minigenome) and transcription product (GFP mRNA) were quantified by real-time reverse transcription-PCR (RT-PCR). All of the L mutants had a significant reduction in minigenome replication (Fig. 2D). With the exception of L-ΔD1557, all of the L mutants had a significant reduction in GFP mRNA transcription (Fig. 2E). Therefore, these results showed that the L deletion and L insertion mutants reduced both replication and transcription, leading to reduced gene expression.
FIG 2.
Examination of the function of L deletion and insertion mutants using a minigenome assay. (A) L deletion and insertion mutants diminished GFP expression. Confluent HEp-2 cells were transfected with the minigenome plasmid together with pTM1-N, pTM1-P, pTM1-M2.1, pTM1-L, or a pTM1-L mutant, using Lipofectamine 2000. GFP expression was visualized by fluorescence microscopy at 48 h posttransfection. (B and C) Quantification of GFP-positive cells (B) and GFP intensity (C) by flow cytometry. HEp-2 cells were transfected with the minigenome and support plasmids. At 48 h posttransfection, cells were trypsinized, and the percentages of GFP-positive cells (B) and GFP intensity (C) were counted by flow cytometry. (D and E) Quantification of genome (D) and mRNA (E) of minigenome assay by real-time RT-PCR. HEp-2 cells were transfected with the minigenome and support plasmids. At 48 h posttransfection, cells were lysed by TRIzol reagent, and total RNA was extracted. Purified RNA was further treated by DNase to remove potential contamination of plasmid DNA. Then the genome (D) and mRNA (E) of the minigenome were quantified by real-time RT-PCR. Data for each L mutant were normalized by wild-type pL. The average from three independent experiments ± standard deviation is shown. NS, no significant difference (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We next built these deletion and insertion mutations into an infectious cDNA clone of RSV A2 strain in which the GFP gene had been inserted between the leader and the NS1 gene. GFP expression allowed us to monitor replication and gene expression of the recombinant virus. All of the mutant viruses were successfully recovered and were named rgRSV-ΔD1557, -ΔM1558, -ΔD1557-ΔM1558, and -1557A1558. All recombinant viruses were plaque purified and further passaged 4 or 5 times in Vero cells. The entire genome was amplified by RT-PCR and sequenced. All retained the desired deletion or insertion in the L gene. No additional mutations were found elsewhere in their genomes.
rgRSV deletion and insertion mutants are attenuated in cell culture.
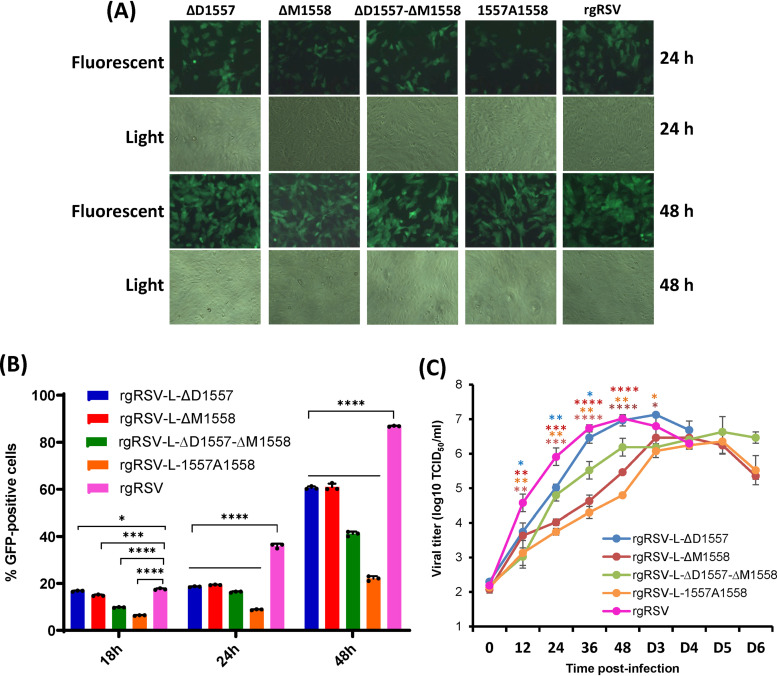
We next determined whether these rgRSV mutants were attenuated in HEp-2 and Vero cells. HEp-2 and Vero cells were infected with each recombinant virus, and the expression of GFP and virus production kinetics were monitored. The parental rgRSV had a maximal level of GFP expression at day 3 postinfection, developing extensive cytopathic effect (CPE) by day 4. However, all rgRSV mutants had delayed GFP expression and CPE in HEp-2 cells, reaching maximal GFP expression at days 4 to 5 postinoculation. Extensive CPE was not observed for these rgRSV mutants until days 5 to 6 postinoculation.
Quantification by flow cytometry showed that significantly fewer GFP-positive cells were detected in HEp-2 cells that were infected by each of the mutants at 24 and 48 h postinfection, compared to rgRSV (P < 0.05) (Fig. 3A and B). We also compared the growth kinetics of these rgRSV mutants with the parental rgRSV (Fig. 3C). The parental rgRSV reached a peak titer of 107.02 50% tissue culture infective doses (TCID50) at 48 h postinoculation. The rgRSV-ΔD1557 mutant grew to a higher titer than rgRSV, reaching a peak titer of 107.13 TCID50 at day 3 postinoculation. The growth kinetics of rgRSV-ΔM1558, -ΔD1557-ΔM1558, and -1557A1558 had a significant delay. Those rgRSV mutants reached a peak titer at days 4 to 5 postinoculation and had a 0.4- to 0.6-log reduced peak titer compared to rgRSV. These results demonstrated that all of these rgRSV mutants had a significant delay in viral replication in HEp-2 cells.
FIG 3.
Characterization of rgRSVs carrying deletion and insertion in the flexible hinge region of the L protein. (A) Delayed GFP expression by rgRSV mutants in Vero cells. Confluent Vero cells were infected by each rgRSV at an MOI of 0.1, and GFP expression was monitored at the indicated time by fluorescence microscope. (B) Quantification of GFP-positive cells by flow cytometry. Confluent HEp-2 cells were infected by each rgRSV (MOI of 1.0), and at the indicated time points, cells were trypsinized and GFP-positive cells were quantified by flow cytometry. Data are the average from three independent experiments ± standard deviation. (C) Single-step growth curve of rgRSV mutants. HEp-2 cells in 12-well-plates were infected with each rgRSV at an MOI of 1.0. After adsorption for 1 h, the inocula were removed and the infected cells were washed 3 times with Opti-MEM medium. Fresh DMEM containing 2% FBS was added, and the cells were incubated at 37°C for various times. The supernatant and cells were harvested by three freeze-thaw cycles, followed by centrifugation at 1,500 × g at 4°C for 15 min at the indicated intervals. The viral titer was determined by TCID50 assay in HEp-2 cells. The viral titers shown are the geometric mean titer (GMT) from three independent experiments ± standard deviation. NS, no significant difference (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Antigen expression of rgRSV deletion and insertion mutants in A549 cells.
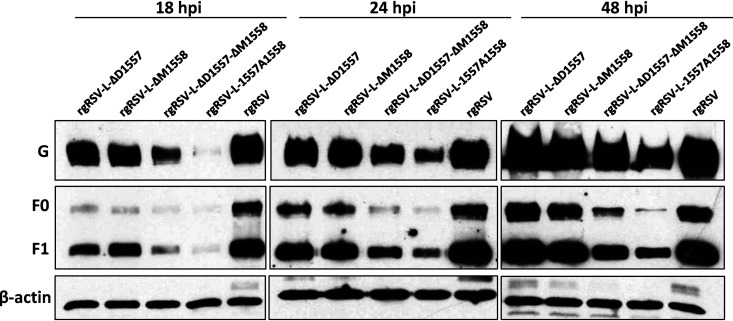
An ideal attenuated strain for vaccine purposes should generate sufficient amounts of viral antigens in order to trigger a strong immune response. Since F and G proteins are the two major surface glycoproteins responsible for inducing neutralizing antibody, we monitored the dynamics of F and G protein expression in virus-infected cells (Fig. 4). At 18 h postinfection, all rgRSV mutants had significant defects in both F and G protein expression compared to rgRSV. However, rgRSV-ΔD1557 and rgRSV-ΔD1557-ΔM1558 reached a similar level of F and G expression to rgRSV at 24 and 48 h postinoculation. rgRSV-1557A1558 and rgRSV-ΔM1558 had less F and G expression at all three time points compared to rgRSV. Consistent with the attenuation phenotype in cell culture, all rgRSV mutants had a delay in antigen expression, but two mutants eventually reached similar levels of antigen expression, relative to rgRSV.
FIG 4.
F and G protein expression. A549 cells were infected with the parental rgRSV or rgRSV mutants at an MOI of 0.1. At 18, 24, and 48 h postinoculation, total cell lysates were harvested and subjected to Western blotting using a monoclonal antibody against RSV F or G protein.
Genetic stability of rgRSV deletion and insertion mutants in Vero cells.
We hypothesize that amino acid deletion and insertion in the L gene will enhance genetic stability of rgRSV mutants. To do this, we successively passed these recombinant viruses in Vero cells 15 times. At each passage, a 1.0-kb fragment spanning the deletion or insertion mutations was amplified by RT-PCR and sequenced. The result showed that all recombinant viruses retained the desired deletions or insertion at each passage. At passage 15, the entire genome was amplified by RT-PCR and sequenced. No additional mutations were found in the genome (data not shown). This result suggests that rgRSV deletion and insertion mutants were genetically stable in cell culture.
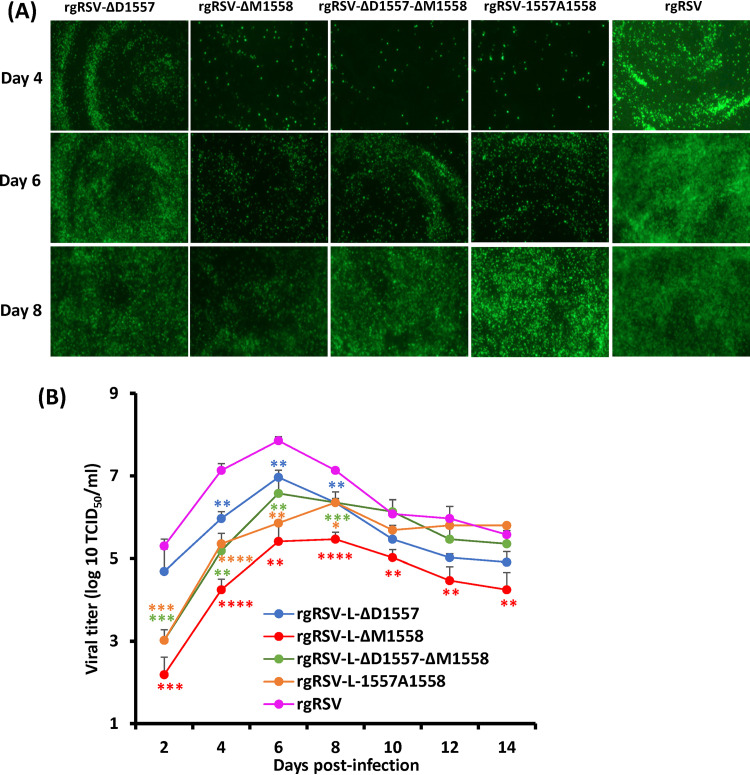
Replication of rgRSVs in HBE culture.
RSV infection has been studied mainly in immortalized cell lines. However, virus replication in vitro in immortalized cell lines may differ from replication in vivo in the human airway epithelium in many aspects, such as receptors and entry and spread mechanisms. Primary, well-differentiated human bronchial epithelial (HBE) cultures have been shown to accurately represent the human airway epithelium, in both appearance and function. Similar to RSV infection in the airways of human lungs, RSV infects HBE cultures via the apical surface of the ciliated cells. Therefore, we tested replication and spread of rgRSV deletion and insertion mutants in HBE cultures at an inoculation dose of 400 TCID50. The parental rgRSV produced visible GFP-positive cells at day 1, rapidly spreading at days 2 to 4, and reaching a peak that involved most of the culture by day 6 (Fig. 5A). All rgRSV mutants had delays in spreading in HBE cultures (Fig. 5A). At the indicated times, mucus was washed off, supernatants were collected, and the dynamics of virus release from HBE cultures was determined by TCID50 (Fig. 5B). The parental rgRSV reached a peak titer of 107.86 TCID50/ml at day 6 postinoculation and decreased by day 8. All rgRSV mutants had a significant delay in virus released from HBE cultures. rgRSV-ΔD1557 and rgRSV-ΔD1557-ΔM1558 reached peak titers of 106.97 TCID50/ml and 106.58 TCID50/ml, respectively, at day 6, whereas rgRSV-ΔM1558 and rgRSV-1557A1558 peaked at day 8, with titers of 105.40 TCID50/ml and 106.36 TCID50/ml, respectively, before receding.
FIG 5.
rgRSV production in HBE cultures. (A) rgRSVs in HBE culture. HBE cultures were inoculated with 400 TCID50 of each rgRSV (equivalent to an MOI of 0.001). At the indicated times, virus spread was monitored by fluorescence microscopy. Representative images at each time point are shown. (B) Virus release from rgRSV-infected HBE culture. Apical washes were collected every 2 days until day 14 postinoculation. Infectious virus in washes was quantified by the TCID50 assay. The viral titers shown are the geometric mean titer (GMT) from three Transwells ± standard deviation. NS, no significant difference (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
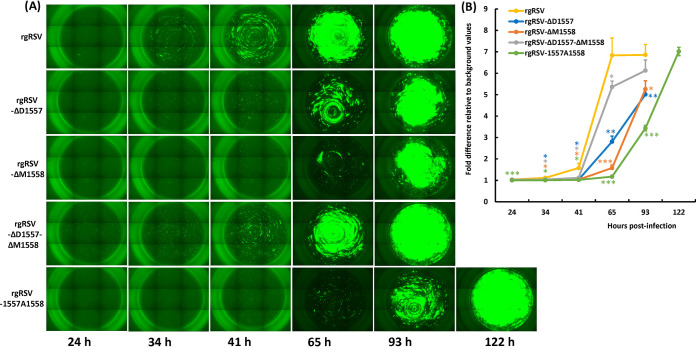
We also tested the replication of rgRSV mutants in HBE culture derived from a different lung donor at a higher inoculation dose (2,000 TCID50). In this experiment, we did not disturb and wash the mucus off HBE culture until 65 h postinoculation. Significantly less GFP signal was observed in these cultures for the rgRSV mutants (Fig. 6A). Quantification of GFP by ImageJ software shows significant delays in viral spreading for all rgRSV mutants, in some more than others (Fig. 6B). Based on the GFP expression and virus titer, the robustness of spread and virus replication in HBE culture can be ranked as follows: rgRSV > rgRSV-ΔD1557 > rgRSV-ΔD1557-ΔM1557 > rgRSV-1557A1558 > rgRSV-ΔM1558.
FIG 6.
rgRSV mutants spread more slowly in HBE culture. (A) Spreading of rgRSVs in HBE culture. HBE cultures were inoculated with 2,000 TCID50 of each rgRSV (equivalent to an MOI of 0.005). At the indicated times, the entire Transwell was imaged by fluorescence microscopy. At 65 h postinoculation, 100 μl of PBS was added to the apical surface of the HBE cultures. Representative images of three Transwells at each time point for each rgRSV are shown. (B) GFP expression in infected HBE cultures. The GFP signal was quantified from a digital image by ImageJ software for each rgRSV-infected culture on the day they reached maximum infection. Fold differences in GFP intensity of each rgRSV mutant-infected HBE culture relative to mock-infected HBE were calculated. The data are expressed as the mean from three Transwells ± standard deviation. NS, no significant difference (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Cytokine production in HBE cultures infected by rgRSV.
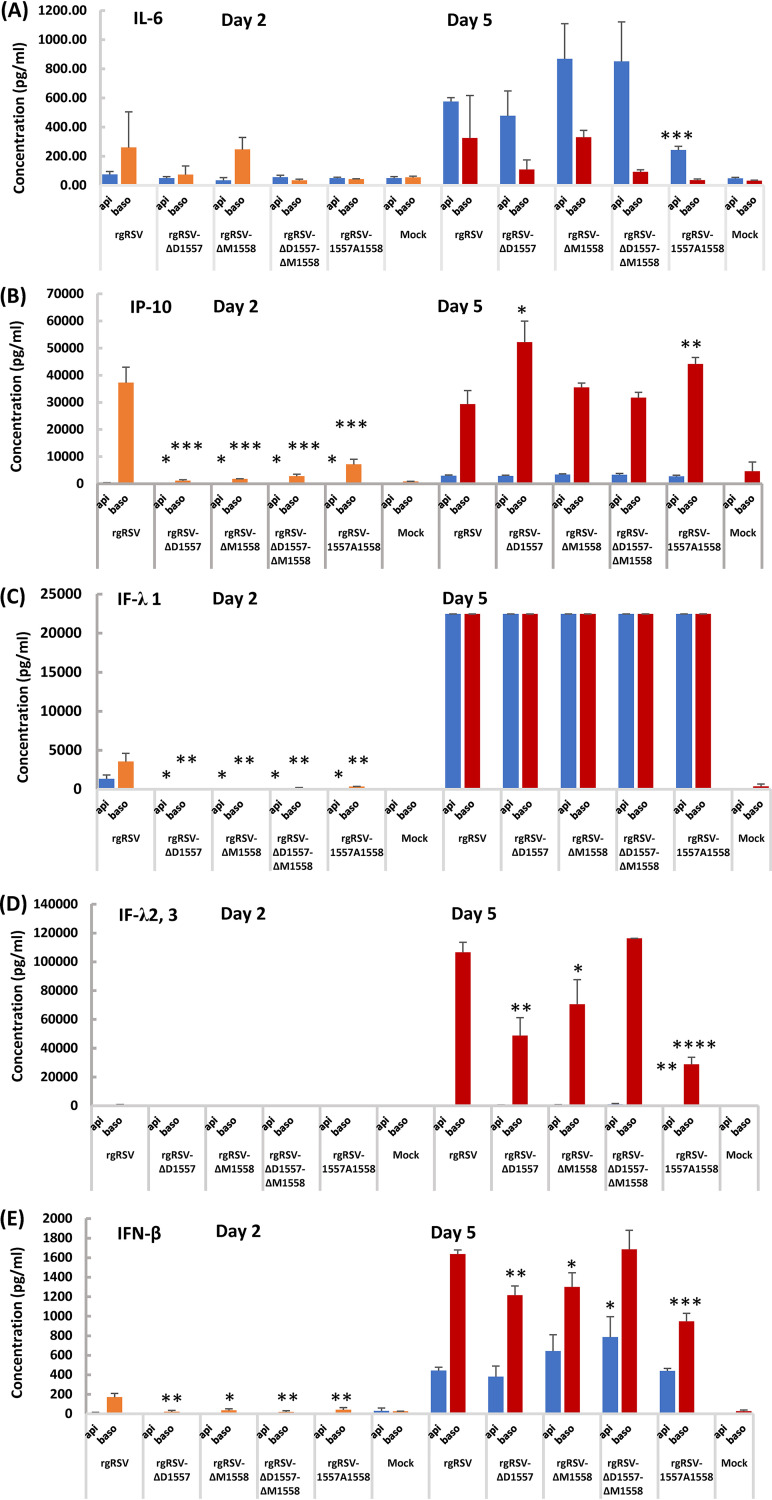
A robust innate immune response induced by live attenuated RSV vaccine candidates is critical for inducing the subsequent adaptive immunity (42). Thus, we analyzed the production of cytokines in HBE cultures. HBE cultures are a good model for RSV infection, likely mimicking RSV infection in human lungs. We are particularly interested in those cytokines involved in innate immunity, which include beta interferon (IFN-β [type I IFN response]), IFN-λ1, -2, and -3 (type III IFN response), IP-10 (interferon gamma-inducible protein 10 kDa, also known as CXCL10 chemokine), and interleukin-6 (IL-6 [a signature cytokine for inflammatory response]). Briefly, HBE cultures were inoculated with 2,000 TCID50 of rgRSV or each mutant, and apical and basolateral fluids were sampled at days 2 and 5 postinoculation for cytokines. The amount of each cytokine was normalized by the degree of viral spreading (GFP intensity). As shown in Fig. 7A, there was no significant difference in IL-6 production at day 2 for medium harvested from the apical or basolateral surface (P > 0.05). However, rgRSV-1557A1558 had significantly lower IL-6 production in the apical surface compared to rgRSV at day 5 (P > 0.001). All rgRSV mutants had less IP-10 at day 2 compared to rgRSV (Fig. 7B) (P < 0.05 or P < 0.001). However, rgRSV-ΔD1557 and rgRSV-1557A1558 had higher IP-10 in basolateral samples (P < 0.05 or 0.01), and rgRSV-ΔD1557-ΔM1558 and rgRSV-ΔM1558 had a similar level of IP-10 compared to rgRSV at day 5 (P > 0.05). All rgRSV mutants had significantly less IFN-λ1 at day 2 (P < 0.05 or 0.01) but reached a similar level at day 5 compared to rgRSV (Fig. 7C) (P > 0.05). Interestingly, the majority of IFN-λ2 and -3 was found in basolateral samples. rgRSV-ΔD1557-ΔM1558 had a similar level of IFN-λ2 and -3 (P > 0.05), whereas other mutants had less IFN-λ2 and -3 at day 5 (Fig. 7D) (P < 0.05, 0.01, or 0.0001). All rgRSV mutants had less IFN-β in basolateral samples at day 2 relative to rgRSV (Fig. 7E) (P < 0.05 or 0.01). Interestingly, rgRSV-ΔD1557-ΔM1558 had significantly higher IFN-β in apical samples compared to rgRSV (Fig. 7E) (P < 0.05). In addition, rgRSV-ΔD1557-ΔM1558 had a similar level of IFN-β (P > 0.05), whereas all other mutants had less IFN-β in basolateral samples at day 5 (P < 0.05). These results demonstrated that rgRSV mutants had a delay in cytokine production at an early time point (day 2) but were capable of inducing a high level of the cytokines involved in innate immunity at day 5 in HBE culture.
FIG 7.
Production of interferons, IP-10, and IL-6 in RSV mutant-infected HBE cultures. The apical andbasolateral fluid from the infected HBE cultures described in the legend to Fig. 6 were sampled as described in the legend to Fig. 5 at the time that each mutant-infected culture reached its maximum GFP expression (indicated below each pair of bars). IL-6 (A), IP-10 (B), IFN-λ1 (C), IFN-λ2/3 (D), and IFN-β (E) levels were assayed using the Legendplex human antivirus response panel (Biolegend) bead assay. Basolateral values were multiplied by 5 to account for the dilution in 5 times more liquid. The data from rgRSV mutants were compared to those from rgRSV. Statistical analysis is indicated. Data with no significant difference (P > 0.05) are not labeled in the figure panels. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Replication of rgRSVs in cotton rats.
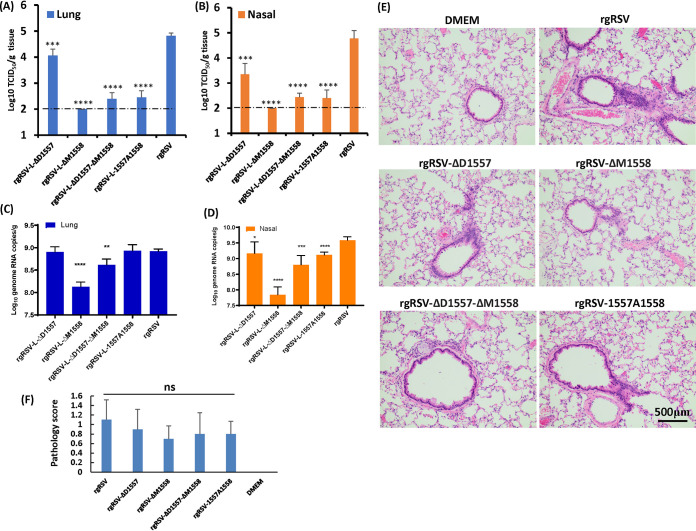
We next determined whether rgRSV deletion and insertion mutants are attenuated in vivo. To do this, 6-week-old female cotton rats were intranasally inoculated with 2 × 105 TCID50 of parental rgRSV or rgRSV deletion and insertion viruses. At day 4 postinfection, cotton rats were sacrificed, and viral replication in nasal turbinates and lungs and pulmonary histology were examined. Under our experimental conditions, the detection limit of rgRSV in lung and nasal turbinate tissues was 2 log TCID50/g tissue. The parental rgRSV replicated efficiently in the nasal turbinates and lungs of all five cotton rats. Average viral titers of 4.82 ± 0.11 and 4.77 ± 0.31 log10 TCID50/g were found in the lungs (Fig. 8A) and nasal turbinates (Fig. 8B), respectively. The rgRSV deletion and insertion mutants had significantly reduced viral replication in the nasal turbinates and lungs (Fig. 8A and B). For rgRSV-ΔD1557, average viral titers of 3.35 ± 0.43 and 4.06 ± 0.25 log10 TCID50/g were found in the nasal turbinates and lung, respectively. For rgRSV-ΔD1557-ΔM1558, an average titer of 2.44 ± 0.16 log10 PFU/g was detected in the nasal turbinates (P < 0.0001), and 4 out of 5 rats had detectable virus in lung tissue, with a titer of 2.39 ± 0.24 log10 PFU/g (P < 0.0001). For rgRSV-1557A1558, 4 out of 5 cotton rats had detectable virus in the nasal turbinates, with an average titer of 2.40 ± 0.32 log10 PFU/g (P < 0.0001), and 4 out of 5 rats had detectable virus in lung tissue, with a titer of 2.45 ± 0.26 log10 PFU/g (P < 0.0001). rgRSV-ΔM1558 was the most attenuated in cotton rats, and the infectious virus in nasal turbinate and lung tissue was below the detection limit.
FIG 8.
Replication of rgRSV mutants in cotton rats. (A and B) RSV titer in lungs (A) and in nasal turbinates (B). Four-week-old SPF cotton rats were inoculated intranasally with 2.0 × 105 TCID50 of each rgRSV. At day 4 postinfection, the cotton rats were sacrificed, and lungs and nasal turbinates were collected for virus titration by TCID50 assay. The viral titers shown are the geometric mean titer (GMT) from 5 animals ± standard deviation. The detection limit is 2.0 log TCID50/g tissue. (C and D) RSV genome RNA copies in lungs (C) and in nasal turbinates (D). Total RNA was extracted from the homogenized tissue using TRIzol reagent. RSV genome copies were quantified by real-time RT-PCR. (E) Representative histologic changes from each group are shown. Hematoxylin-eosin (H&E) staining of lung tissue is shown. Histological images were taken under light microscopy. Micrographs with ×20 magnification (scale bar of 500 μm) are shown. (F) Histology score. Each slide was scored based on severity of peribronchiolitis, perivasculitis, bronchiolitis, alveolitis, and interstitial pneumonia. Scores: 0, no lesion; 1, mild; 2, moderate; 3, severe. NS, no significant difference (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, total RNA was extracted from lung and nasal turbinate tissues, and the RSV genome copies were quantified by real-time PCR, which is more sensitive than the TCID50 assay. Interestingly, lungs from the rgRSV-1557A1558 group had no significant reduction in RNA copies compared to the lungs from the rgRSV group (P > 0.05) (Fig. 8C), despite the fact that infectious RSV titer had 2.5-log TCID50/g tissue reductions (Fig. 8A). The RNA copies in lung samples from rgRSV-ΔM1558 and rgRSV-ΔD1557-ΔM1558 had only 0.8- and 0.3-log reductions compared to rgRSV, respectively (Fig. 8C) (P < 0.0001 or 0.01). All rgRSV mutants had various degrees of reductions in RSV genome RNA copies in nasal turbinates compared to rgRSV (P < 0.05, 0.001, or 0.0001) (Fig. 8D). Histopathologic changes were comparably mild in lungs from all RSV-infected groups and were generally characterized by few leukocytes surrounding bronchioles and blood vessels, evidence of occasional bronchiolar goblet cell hyperplasia with granulocyte exocytosis, and small numbers of discrete nodules of alveolar macrophages (Fig. 8E). In addition, there were no significant differences among the RSV-infected group (P > 0.05) (Fig. 8F). These results demonstrated that the rgRSV-ΔM1558, -ΔD1557-ΔM1558, and -1557A1558 mutants were highly attenuated in viral replication in both the upper and lower respiratory tracts in cotton rats, whereas rgRSV-ΔD1557 was moderately defective in replication in cotton rats.
Deletion and insertion mutants of rgRSV provide complete protection against RSV infection.
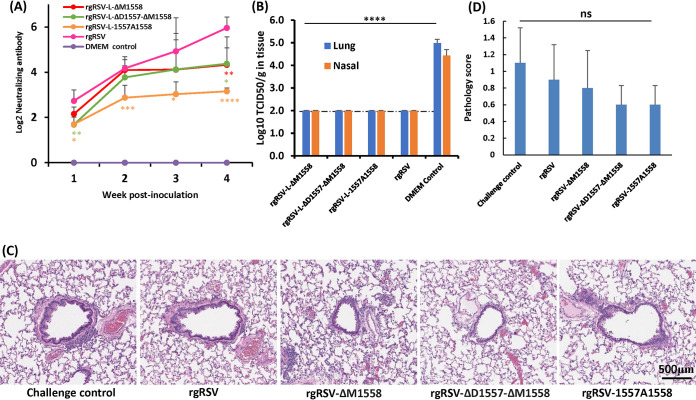
Since rgRSV-ΔM1558, -ΔD1557-ΔM1558, and -1557A1558 were highly attenuated in vivo, we next determined whether they are immunogenic. Briefly, 6-week-old female cotton rats were intranasally immunized with 2 × 105 TCID50 of parental rgRSV, or rgRSV deletion or insertion viruses. The parental rgRSV was used as a control. An ideal attenuated vaccine candidate should retain a level of immunogenicity similar to or higher than that of the parental rgRSV. After immunization, serum antibody levels were determined weekly by a plaque reduction neutralization assay. Figure 9A shows the dynamics of neutralizing antibody responses following immunization. rgRSV-ΔM1558 and -ΔD1557-ΔM1558 triggered levels of neutralizing antibody comparable to those induced by rgRSV immunization (P > 0.05). However, rgRSV-1557A1558 induced relatively lower antibody responses at weeks 3 and 4 compared to rgRSV (P < 0.05). No RSV-specific neutralizing antibody was detected in the unvaccinated control. At week 4 postimmunization, cotton rats were challenged intranasally with 2 × 105 TCID50 of rgRSV. At day 4 postchallenge, cotton rats were sacrificed, and viral replication in nasal turbinates and lungs and pulmonary histology were examined. Infectious RSV was below the detection limit in all nasal turbinate and lung samples from the animals immunized with rgRSV or rgRSV-ΔM1558, -ΔD1557-ΔM1558, or -1557A1558 followed by challenge with rgRSV (Fig. 9B). Microscopically, all five groups had variable but mild pulmonary changes (Fig. 9C and D). The rgRSV- or rgRSV mutant-immunized group had fewer pathological changes compared to the unimmunized challenged control. However, there were no statistically significant differences among the groups (P > 0.05) (Fig. 9D). No enhanced lung damage was found for the rgRSV mutants. Therefore, these results showed that cotton rats immunized with rgRSV deletion and insertion mutants were protected from RSV challenge.
FIG 9.
Immunogenicity of rgRSV mutants in cotton rats. Four-week-old SPF cotton rats were inoculated intranasally with 2.0 × 105 TCID50 of each rgRSV. Blood samples were collected from each rat weekly by retro-orbital bleeding. (A) Neutralizing antibody production. The RSV neutralizing antibody titer was determined using a plaque reduction neutralization assay, as described in Materials and Methods. (B) Protection from RSV challenge. At week 4 postimmunization, cotton rats were challenged with 2.0 × 105 TCID50 of rgRSV. At day 4 postchallenge, the cotton rats were sacrificed, and lungs and nasal turbinates were collected for virus titration by TCID50 assay. The viral titers shown are the geometric mean titer (GMT) from 5 animals ± standard deviation. The detection limit is 2.0 log TCID50/g tissue. (C) Representative histologic changes from each group are shown. Hematoxylin-eosin (H&E) staining of lung tissue is shown. Micrographs with ×20 magnification (scale bar of 500 μm) are shown. (D) Histology score. Each slide was scored based on severity of peribronchiolitis, perivasculitis, bronchiolitis, alveolitis, and interstitial pneumonia. Scores: 0, no lesion; 1, mild; 2, moderate; 3, severe. NS, no significant difference (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
Despite major efforts, there is no FDA-approved vaccine for RSV. In this study, we generated three RSV live attenuated vaccine candidates by deleting and inserting single or double amino acid residues in the flexible hinge region between CR V and CR VI of the L protein. We showed that these vaccine candidates were genetically stable, highly attenuated in immortalized cells, HBE cultures, and an animal model, and capable of inducing high levels of innate immune cytokines in HBE culture and triggering a high level of neutralizing antibody and providing complete protection against RSV infection in cotton rats. To the best of our knowledge, this is first time that the flexible hinge region of L protein has been shown to tolerate amino acid deletion, although it has been shown that this region is able to tolerate an in-frame GFP insertion for several NNS RNA viruses.
The L protein is an important target for designing live attenuated vaccine candidates for RSV, since the L protein is a multifunctional protein containing domains that perform at least 10 functions, including nucleotide polymerization, replication, transcription, mRNA capping, cap methylation, and polyadenylation (26, 27, 34). Suppression or partial suppression of these enzymatic activities will lead to diminished viral replication and gene expression, which potentially results in virus attenuation. Between 1960 and 1990, several RSV attenuated strains were developed by classic methods such as cold or warm passage (cp or ts) and chemical mutagenesis (12–17, 43). Interestingly, the amino acid residue(s) responsible for the temperature-sensitive (ts) phenotype of cp ts RSV mutants often mapped to the CRI-IV region of L protein (44, 45), which is essential for polymerase activity. Consistent with these findings, random substitutions of a cluster of charged amino acids to alanine in the L protein resulted in recombinant RSVs that also exhibited a ts phenotype (46). Interestingly, mutations responsible for the host range and ts phenotype of other NNS RNA viruses (such as VSV and Sendai virus) map to the flexible region or CR VI in the L protein, which affected the mRNA cap MTase activity (33, 47, 48).
Genetic stability is one of the major challenges for developing a live attenuated RSV vaccine. The primary advantage of using amino acid deletion and insertion is that this strategy can permanently retain the genetic stability, as it would not be possible for these deletion or insertion mutants to revert to wild-type virus. In our study, no reversions or additional mutations were observed when these RSV mutants were repeatedly passaged in Vero cells. In addition, RT-PCR fragments amplified from nasal turbinate and lung tissues retained the desired deletion and insertion mutations. These results suggest that these RSV mutants are genetically stable.
We found for the first time that the flexible hinge region of RSV L protein tolerates amino acid deletion. Notably, a single or double deletion was sufficient to achieve attenuation. rgRSV-ΔD1557 was moderately attenuated in replication in immortalized cells, HBE cultures, and cotton rats. However, rgRSV-ΔM1558 and rgRSV-ΔD1557-ΔM1558 were highly attenuated in vitro and in vivo. For these two mutants, the infectious RSV titer was near or below the detection limit in lungs and nasal turbinates of cotton rats. However, significant amounts of RSV genome RNA were detected in lung and nasal tissues, suggesting that these RSV mutants have some levels of replication in vivo, which may be sufficient to trigger an RSV-specific immune response. In addition, no enhanced lung damage was observed after reinfection with rgRSV. Another important characteristic of these two RSV mutants is that they grow to a high titer in cell culture despite high attenuation, making vaccine production economically feasible. Thus, L deletion is a novel approach for rational design of live attenuated vaccines for RSV and perhaps for other NNS RNA viruses.
We also found that a single alanine insertion at the flexible hinge region was sufficient to attenuate RSV. The resultant recombinant virus, rgRSV-1557A1558, was highly attenuated in cell culture, HBE culture, and cotton rats. Although it triggered a relatively lower neutralizing antibody response compared to the two RSV deletion mutants, it provided complete protection against RSV replication. In the case of MeV, c-Myc tag or EGFP tag was inserted in the flexible region (35). Recombinant MeV (rMeV) containing an EGFP insertion (240 amino acids) was more attenuated in cell culture than rMeV with a c-Myc tag (6 amino acids) at the same position, suggesting that the size of the insert correlates with the degree of attenuation (35). For CDV, it was shown that insertion of EGFP at this position resulted in overattenuation of the virus, only providing partial protection against CDV challenge (37). Insertion of GFP into this flexible region of VSV L resulted in a virus displaying unusual properties, including a temperature-sensitive growth phenotype and a lack of virion-associated polymerase activity in vitro (38).
Although the exact mechanism(s) behind the attenuation of these RSV deletion and insertion mutants is not clear, one possibility is that these insertion and deletion RSV L proteins reduced the polymerase activity, which leads to the synthesis of less genomic and antigenomic RNA as well as mRNAs. In a minigenome assay, we found significantly fewer GFP-positive cells and lower GFP intensity for these L protein deletion and insertion mutants. Analysis of minigenome RNA and GFP mRNA showed these L mutants affected both replication and transcription. In RSV-infected cells, we found that synthesis of viral F and G proteins was significantly delayed at early time points compared to the parental RSV. Previously, it was shown that insertion of HA tag into amino acid positions 1749 and 1695 in RSV L resulted in 60% and 10% bioactivity in a minigenome assay (39). These results suggest that multiple locations in the flexible hinge region in RSV L are tolerant to amino acid insertions, although it is unknown whether viable recombinant virus can be recovered. Similarly, the EGFP inserted into the PRV and CDV L proteins significantly reduced polymerase activity, retaining 1 to 85% and 30 to 60% of wt polymerase activity, respectively (37). For VSV, insertion of EGFP into the flexible region of L (LEGFP) resulted in a ts phenotype for polymerase activity (38). Another possible mechanism of viral attenuation by these L deletion and insertion mutants is that they affect mRNA capping and/or methylase activities since they are located in the region between the mRNA capping and methylase domains. In fact, it was previously shown that single amino acid substitutions in the flexible region of VSV L significantly reduced both G-N-7 and ribose 2′-O MTase activities (47).
RSV encodes two nonstructural proteins (NS1 and NS2) that strongly inhibit innate immunity (25, 42, 49). Generally, a robust innate immune response is critical for induction of strong adaptive immunity (25, 42, 50). Thus, we examined cytokine production in HBE culture, a near in vivo airway model for RSV infection. Production of innate immune cytokines, including IFN-β, IFN-λ1, -2, and -3, and IP-10, of rgRSV mutants had a significant delay compared to rgRSV. However, all rgRSV mutants had induced high levels of these cytokines at day 6. The production of these cytokines will likely provide adjuvant effects for these vaccine candidates. IL-6 is a signature cytokine for inflammatory response and correlates with disease severity upon pneumovirus infection (51, 52). All rgRSV deletion mutants produced a similar level of IL-6 (P > 0.05), whereas rgRSV-1557A1558 had a significant reduction in IL-6 (P < 0.001), suggesting that some of these rgRSV mutants may not induce more pathogenesis than rgRSV. We also observed differential secretion of cytokines/chemokines from the apical and basolateral surfaces in HBE, which is probably due to the difference in cell types in these two sites. Similarly, it was recently found that the secretome of HBE is distinct at the apical and basolateral surfaces and this profile is altered during RSV infection (53).
In summary, we found that amino acid deletions or insertions in the hinge region of the L protein can serve as a novel approach to rationally design genetically stable, highly attenuated, and immunogenic live vaccine candidates for RSV.
MATERIALS AND METHODS
Ethics statement.
The animal study was conducted in strict accordance with USDA regulations and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (54) and was approved by The Ohio State University Institutional Animal Care and Use Committee (IACUC; animal protocol no. 2009A0221). Every effort was made to minimize potential distress, pain, or discomfort to the animals throughout all experiments.
Cell lines.
The HeLa (ATCC CCL-2), A549 (ATCC CCL-185), Vero (ATCC CRL-CCL81), and HEp-2 (ATCC CCL-23) cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and were grown in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS). Primary, well-differentiated human bronchial epithelial (HBE) cultures were grown on collagen-coated Transwell inserts (Corning, Inc., Corning, NY) at an air-liquid interface, as previously described (55). Upon reaching confluence and forming tight junctions, the apical medium was removed, and cultures were maintained at the air-liquid interface for 4 to 6 weeks to generate well-differentiated, polarized cultures. All cell lines used in this study were free of mycoplasma, as confirmed by the LookOut Mycoplasma PCR detection kit (Sigma, St. Louis, MO).
Virus stocks and purification.
Recombinant RSV containing a green fluorescence protein (GFP) gene between the leader sequence and NS1 gene (rgRSV) (55) was propagated and its titer was determined in HeLa or HEp-2 cells. Virus was purified through a sucrose cushion by ultracentrifugation and was resuspended in DMEM with 10% trehalose.
Plasmids and site-directed mutagenesis.
Plasmid (RW30) encoding the full-length antigenomic cDNA of RSV strain A2 with GFP inserted between the leader and the NS1 gene and support plasmids expressing RSV A2 strain N (pTM1-N), P (pTM1-P), L (pTM1-L), and M2-1 (pTM1-M2-1) proteins were generously provided by Peter Collins, NIAID, Bethesda, MD (18). The L deletions (ΔD1557, ΔM1558, and ΔD1557-ΔM1558) and insertion (1557A1558) were introduced into the pTM1-L and RW30 plasmids using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All plasmids and mutations were confirmed by DNA sequencing.
Minigenome assay.
Confluent HEp-2 cells were transfected with the minigenome plasmid together with pTM1-N, pTM1-P, pTM1-M2.1, pTM1-L, or a pTM1-L mutant, using Lipofectamine 2000. GFP expression was visualized by a fluorescence microscope at 48 h posttransfection. In addition, total RNA was extracted and purified from transfected cells, and the minigenome RNA and GFP mRNA were quantified by real-time RT-PCR.
Recovery of RSV from the full-length cDNA clones.
rgRSV mutants were rescued from the full-length cDNA of the RSV A2 strain (18). Briefly, HEp-2 cells were infected with MVA-T7 at a multiplicity of infection (MOI) of 0.1 and transfected with 1.2 μg of plasmid RW30 or RW30 mutant, 0.4 μg of pTM1-N, 0.2 μg of pTM1-P, 0.1 μg of pTM1-M2-1, and 0.1 μg of pTM1-L using the Lipofectamine 3000 reagent (Life Technologies). The successful recovery of the rgRSV was confirmed by the presence of green fluorescent cells, followed by RT-PCR and sequencing. All rgRSV mutants were plaque purified. rgRSV mutants carrying L deletion and insertion mutations were designated rgRSV-ΔD1557, -ΔM1558, -ΔD1557-ΔM1558, and -1557A1558.
RT-PCR and sequencing.
All plasmids, viral mutants and stocks, and virus isolates from the nasal turbinates and lungs of cotton rats were sequenced to confirm virus identity. Viral RNA was extracted from 100 μl of each recombinant virus using an RNeasy minikit (Qiagen, Valencia, CA). A 1.5-kb DNA fragment spanning the flexible region between CR V and CR VI of the L gene was amplified by RT-PCR. Also, the entire RSV genome was amplified using six overlapping fragments by RT-PCR. The PCR products were purified and sequenced using a sequencing primer at The Ohio State University Plant Microbe Genetics Facility to confirm the presence of the designed mutations.
Viral replication kinetics.
Confluent HEp-2 and Vero cells in a 12-well plate were infected with wild-type or mutant rgRSV at an MOI of 1.0. At different time points postinoculation, the supernatants were harvested and kept on ice. Cell pellets were subjected to three freeze-thaw cycles in 0.2 ml of fresh DMEM with 10% trehalose. The two portions of supernatants were combined, and the virus titer was determined by TCID50 assay in HEp-2 cells (55).
Genetic stability of rgRSV mutants in cell culture.
Confluent Vero cells in T25 flasks were infected with each rgRSV mutant at an MOI of 0.1. At day 3 postinoculation, the cell culture supernatant was harvested and used for the next passage in Vero cells. Using this method, each rgRSV mutant was repeatedly passaged 15 times in Vero cells. At each passage, the flexible region between CR V and CV VI of the L gene was amplified by RT-PCR and sequenced. At passage 15, the entire genome of each recombinant virus was amplified by RT-PCR and sequenced.
Analysis of RSV protein expression by Western blotting.
Confluent A549 cells were infected with rgRSV and rgRSV mutants at an MOI of 0.1. At 18, 24, and 48 h postinfection, the cell culture supernatant was removed and the cells were lysed in 150 μl of radioimmunoprecipitation assay (RIPA) buffer (Abcam) supplemented with protease inhibitor cocktail (Sigma-Aldrich). RSV F and G proteins were detected by Western blotting using anti-RSV F (Abcam) or serum (Virostat) antibody. β-Actin was used as a loading control.
Examination of GFP expression by microscopy and flow cytometry.
Vero or HEp-2 cells were infected with rgRSV or mutants at an MOI of 1.0, and GFP expression was monitored at the indicated times by fluorescence microscopy. At the indicated time points, cells were trypsinized and fixed in 0.01% of paraformaldehyde solution, and then the number of GFP-positive cells and GFP density were quantified by flow cytometry.
Replication, spreading, and cytokine production in HBE culture.
Purified virions were titrated on HEp-2 cells and were diluted in HBE cell medium. The apical surface of well-differentiated HBE cells in Transwells was washed with phosphate-buffered saline (PBS) for 2 h, and the basal medium was changed before the virus was added (at 400 or 2,000 TCID50) to the apical chamber of the Transwell. Fluorescent cells were visualized with an EVOS2 fl inverted fluorescence microscope (Life Technologies). At indicated times, 100 μl of PBS was added to the apical surface of HBE culture, gently rocked for 30 min, and collected for virus titration. The amount of the HBE culture expressing GFP-positive cells was determined from a digital image using ImageJ software. In addition, the medium at the apical wash and the basolateral medium from HBE cultures were collected at days 2 and 5 postinoculation for detection of cytokines (IFN-β, IFN-λ1, IFN-λ2/3, IP-10, and IL-6) by a flow cytometer bead assay (LEGENDplex; Biolegend, San Diego, CA).
Replication and pathogenesis of rgRSV mutants in cotton rats.
Thirty 4-week-old specific-pathogen-free (SPF) male cotton rats (Envigo, Indianapolis, IN) were randomly divided into 6 groups (5 cotton rats per group). Prior to virus inoculation, the cotton rats were anesthetized with isoflurane. The cotton rats in group 1 were inoculated with 2.0 × 105 TCID50 of parental rgRSV and served as positive controls. The cotton rats in groups 2 to 5 were inoculated with 2.0 × 105 TCID50 of four rgRSV mutants: rgRSV-ΔD1557, -ΔM1558, -ΔD1557-ΔM1558, and 1557A1558. The cotton rats in group 6 were inoculated with DMEM and served as mock-infected controls. Each cotton rat was inoculated intranasally with a volume of 100 μl. At day 4 postinfection, the cotton rats were sacrificed via carbon dioxide inhalation. The left lung and nasal turbinates were collected for virus titration, and the right lung was collected for histological analysis. In addition, total RNA was extracted from lung and nasal turbinate tissues, and RSV genome RNA copies were determined by real-time RT-PCR.
Immunogenicity of rgRSV in cotton rats.
For the immunogenicity study, 25 4-week-old female cotton rats (Envigo) were randomly divided into five groups (5 cotton rats per group). Cotton rats in groups 1 to 4 were intranasally inoculated with 2.0 × 105 TCID50 of three rgRSV mutants (rgRSV-ΔM1558, -ΔD1557-ΔM1558, and -1557A1558) and rgRSV, respectively. Cotton rats in group 5 were mock infected with DMEM and served as the unvaccinated challenged controls. After immunization, the cotton rats were evaluated daily for any possible abnormal reaction, blood samples were collected from each cotton rat weekly by orbital sinus blood sampling, and serum was used for detection of neutralizing antibodies. At 4 weeks postimmunization, the cotton rats in all groups were challenged with 2.0 × 105 TCID50 of parental rgRSV via the intranasal route and evaluated twice daily for the presence of any clinical symptoms. At 4 days postchallenge, all cotton rats were euthanized by CO2 asphyxiation, and their lungs and nasal turbinates were collected for virus titration. The immunogenicity of rgRSV mutants was assessed based on their ability to trigger neutralizing antibody, the ability to prevent rgRSV replication in lungs and the nose, and the ability to protect the lung from pathological changes.
Pulmonary histology.
After sacrifice, the right lung of each animal was removed, inflated, and fixed with 4% paraformaldehyde. Fixed tissues were routinely processed, embedded in paraffin, and sectioned at 4 μm. Slides were then stained with hematoxylin and eosin (H&E) and evaluated by light microscopy by veterinary anatomic pathologists board certified by the American College of Veterinary Pathologists. Sections were evaluated for peribronchiolitis, perivasculitis, bronchiolitis, alveolitis, and interstitial pneumonia using a modification of adapted criteria (56, 57).
Determination of viral titer in lung and nasal turbinates.
The nasal turbinates and the left lung from each cotton rat were removed, weighed, and homogenized in either 3 or 2 ml of DMEM. The lung was homogenized using a Precellys 24 tissue homogenizer (Bertin Instruments, Rockville, MD) by following the manufacturer's recommendations. The nasal turbinates were homogenized by hand with a 15-ml-capacity Pyrex homogenizer (Corning, Inc., Corning, NY). The presence of infectious virus was determined by TCID50 assay in HEp-2 cells. The detection limit of rgRSV in lung and nasal turbinates was 2 logTCID50/g tissue. Titer below the detection limit was reported as 2 log TCID50/g tissue.
Determination of RSV neutralizing antibody.
RSV-specific neutralizing antibody titers were determined using a plaque reduction neutralization assay as described previously (58, 59). The virus-serum mixtures were incubated at 37°C for 1 h, added to confluent HEp-2 cells for 1 h, and overlaid with 0.75% methylcellulose in overlay medium (1× minimal essential medium [MEM], 2% FBS, sodium bicarbonate, 25 mM HEPES, 1% l-glutamine, 1% penicillin-streptomycin [Pen-Strep]) and incubated for 3 days before counting the fluorescent foci. The numbers of foci at each serum dilution were plotted, and the 50% plaque reduction titer was used as the RSV-specific neutralizing antibody titer.
Sequence alignment and analysis.
L protein sequences were aligned by ClustalW2. The RSV-L (YP009518860.1), VSV-L (Q98776.1), RPV-L (NC_006296.2), CDV-L (NP_047207.1), and MV-L (Z66517.1) were included.
Structure modeling of RSV L protein.
The cryo-electron microscopy (cryo-EM) structure of the L protein of vesicular stomatitis virus (PDB no. 5A22) was chosen as the template for RSV L protein structure prediction using the MODELLER program (version 9.20).
Statistical analysis.
Quantitative analysis was performed by either densitometric scanning of autoradiographs or by using a phosphorimager (Typhoon; GE Healthcare, Piscataway, NJ) and ImageQuant TL software (GE Healthcare, Piscataway, NJ). Statistical analysis was performed by one-way multiple comparisons using SPSS (version 8.0) statistical analysis software (SPSS, Inc., Chicago, IL). A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH/NIAID (P01AI112524).
We thank Peter Collins for the recombinant RSV and minigenome systems. We are grateful to members of the J. Li laboratory for critically reading the manuscript.
REFERENCES
- 1.Collins PL, Crowe JEJ. 2007. Respiratory syncytial virus and metapneumovirus, p 1601–1646. Knipe PM, Howley DE (ed), Fields virology, 5th ed, vol 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Glezen WP, Taber LH, Frank AL, Kasel JA. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 3.Chang J. 2011. Current progress on development of respiratory syncytial virus vaccine. BMB Rep 44:232–237. doi: 10.5483/BMBRep.2011.44.4.232. [DOI] [PubMed] [Google Scholar]
- 4.Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang DA, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, Emediato CCDL, Fasce RA, Feikin DR, Feng LZ, RSV Global Epidemiology Network, et al. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EAF, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saez-Llorens X, Moreno MT, Ramilo O, Sanchez PJ, Top FH, Jr, Connor EM, MEDI-493 Study Group . 2004. Safety and pharmacokinetics of palivizumab therapy in children hospitalized with respiratory syncytial virus infection. Pediatr Infect Dis J 23:707–712. doi: 10.1097/01.inf.0000133165.85909.08. [DOI] [PubMed] [Google Scholar]
- 7.Mejias A, Rodriguez-Fernandez R, Peeples ME, Ramilo O. 2019. Respiratory syncytial virus vaccines are we making progress? Pediatr Infect Dis J 38:E266–E269. doi: 10.1097/INF.0000000000002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 9.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 10.Buchholz UJ, Nagashima K, Murphy BR, Collins PL. 2006. Live vaccines for human metapneumovirus designed by reverse genetics. Expert Rev Vaccines 5:695–706. doi: 10.1586/14760584.5.5.695. [DOI] [PubMed] [Google Scholar]
- 11.Collins PL, Graham BS. 2008. Viral and host factors in human respiratory syncytial virus pathogenesis. J Virol 82:2040–2055. doi: 10.1128/JVI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowe JE, Bui PT, Davis AR, Chanock RM, Murphy BR. 1994. A further attenuated derivative of a cold-passaged temperature-sensitive mutant of human respiratory syncytial virus retains immunogenicity and protective efficacy against wild-type challenge in seronegative chimpanzees. Vaccine 12:783–790. doi: 10.1016/0264-410x(94)90286-0. [DOI] [PubMed] [Google Scholar]
- 13.Crowe JE, Bui PT, London WT, Davis AR, Hung PP, Chanock RM, Murphy BR. 1994. Satisfactorily attenuated and protective mutants derived from a partially attenuated cold-passaged respiratory syncytial virus mutant by introduction of additional attenuating mutations during chemical mutagenesis. Vaccine 12:691–699. doi: 10.1016/0264-410x(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 14.Crowe JE, Bui PT, Siber GR, Elkins WR, Chanock RM, Murphy BR. 1995. Cold passaged, temperature-sensitive mutants of human respiratory syncytial virus (Rsv) are highly attenuated, immunogenic, and protective in seronegative chimpanzees, even when Rsv antibodies are infused shortly before immunization. Vaccine 13:847–855. doi: 10.1016/0264-410x(94)00074-w. [DOI] [PubMed] [Google Scholar]
- 15.Karron RA, Wright PF, Crowe JE, Jr, Clements-Mann ML, Thompson J, Makhene M, Casey R, Murphy BR. 1997. Evaluation of two live, cold-passaged, temperature-sensitive respiratory syncytial virus vaccines in chimpanzees and in human adults, infants, and children. J Infect Dis 176:1428–1436. doi: 10.1086/514138. [DOI] [PubMed] [Google Scholar]
- 16.Wright PF, Karron RA, Belshe RB, Thompson J, Crowe JE, Boyce TG, Halburnt LL, Reed GW, Whitehead SS, Anderson EL, Wittek AE, Casey R, Eichelberger M, Thumar B, Randolph VB, Udem SA, Chanock RM, Murphy BR. 2000. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis 182:1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- 17.Wright PF, Shinozaki T, Fleet W, Sell SH, Thompson J, Karzon DT. 1976. Evaluation of a live, attenuated respiratory syncytial virus-vaccine in infants. J Pediatr 88:931–936. doi: 10.1016/s0022-3476(76)81044-x. [DOI] [PubMed] [Google Scholar]
- 18.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5' proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci U S A 92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchholz UJ, Biacchesi S, Pham QN, Tran KC, Yang L, Luongo CL, Skiadopoulos MH, Murphy BR, Collins PL. 2005. Deletion of M2 gene open reading frames 1 and 2 of human metapneumovirus: effects on RNA synthesis, attenuation, and immunogenicity. J Virol 79:6588–6597. doi: 10.1128/JVI.79.11.6588-6597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, Murphy BR. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol 73:3438–3442. doi: 10.1128/JVI.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng MN, Whitehead SS, Bermingham A, St Claire M, Elkins WR, Murphy BR, Collins PL. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J Virol 74:9317–9321. doi: 10.1128/jvi.74.19.9317-9321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin H, Cheng X, Traina-Dorge VL, Park HJ, Zhou H, Soike K, Kemble G. 2003. Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine 21:3647–3652. doi: 10.1016/s0264-410x(03)00426-2. [DOI] [PubMed] [Google Scholar]
- 23.Stobart CC, Rostad CA, Ke Z, Dillard RS, Hampton CM, Strauss JD, Yi H, Hotard AL, Meng J, Pickles RJ, Sakamoto K, Lee S, Currier MG, Moin SM, Graham BS, Boukhvalova MS, Gilbert BE, Blanco JC, Piedra PA, Wright ER, Moore ML. 2016. A live RSV vaccine with engineered thermostability is immunogenic in cotton rats despite high attenuation. Nat Commun 7:13916. doi: 10.1038/ncomms13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rostad CA, Stobart CC, Todd SO, Molina SA, Lee S, Blanco JCG, Moore ML. 2017. Enhancing the thermostability and immunogenicity of a respiratory syncytial virus (RSV) live-attenuated vaccine by incorporating unique RSV line19F protein residues. J Virol 92:e01568-17. doi: 10.1128/JVI.01568-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee S, Luthra P, Esaulova E, Agapov E, Yen BC, Borek DM, Edwards MR, Mittal A, Jordan DS, Ramanan P, Moore ML, Pappu RV, Holtzman MJ, Artyomov MN, Basler CF, Amarasinghe GK, Leung DW. 2017. Structural basis for human respiratory syncytial virus NS1-mediated modulation of host responses. Nat Microbiol 2:17101. doi: 10.1038/nmicrobiol.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowton VM, McGivern DR, Fearns R. 2006. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J Gen Virol 87:1805–1821. doi: 10.1099/vir.0.81786-0. [DOI] [PubMed] [Google Scholar]
- 27.Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 28.Sleat DE, Banerjee AK. 1993. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol 67:1334–1339. doi: 10.1128/JVI.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Rahmeh A, Morelli M, Whelan SPJ. 2008. A conserved motif in region V of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol 82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JR, Fontaine-Rodriguez EC, Whelan SPJ. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol 79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li JR, Wang JT, Whelan SPJ. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A 103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino T, Yadav SP, Banerjee AK. 2010. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci U S A 107:3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grdzelishvili VZ, Smallwood S, Tower D, Hall RL, Hunt DM, Moyer SA. 2005. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J Virol 79:7327–7337. doi: 10.1128/JVI.79.12.7327-7337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SPJ. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duprex WP, Collins FM, Rima BK. 2002. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J Virol 76:7322–7328. doi: 10.1128/jvi.76.14.7322-7328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown DD, Rima BK, Allen IV, Baron MD, Banyard AC, Barrett T, Duprex WP. 2005. Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase. J Virol 79:14330–14338. doi: 10.1128/JVI.79.22.14330-14338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silin D, Lyubomska O, Ludlow M, Duprex WP, Rima BK. 2007. Development of a challenge-protective vaccine concept by modification of the viral RNA-dependent RNA polymerase of canine distemper virus. J Virol 81:13649–13658. doi: 10.1128/JVI.01385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruedas JB, Perrault J. 2009. Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro. J Virol 83:12241–12252. doi: 10.1128/JVI.01273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dochow M, Krumm SA, Crowe JE, Jr, Moore ML, Plemper RK. 2012. Independent structural domains in paramyxovirus polymerase protein. J Biol Chem 287:6878–6891. doi: 10.1074/jbc.M111.325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilman MSA, Liu C, Fung A, Behera I, Jordan P, Rigaux P, Ysebaert N, Tcherniuk S, Sourimant J, Eleouet JF, Sutto-Ortiz P, Decroly E, Roymans D, Jin Z, McLellan JS. 2019. Structure of the respiratory syncytial virus polymerase complex. Cell 179:193–204.e14. doi: 10.1016/j.cell.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao D, Gao Y, Roesler C, Rice S, D'Cunha P, Zhuang L, Slack J, Domke M, Antonova A, Romanelli S, Keating S, Forero G, Juneja P, Liang B. 2020. Cryo-EM structure of the respiratory syncytial virus RNA polymerase. Nat Commun 11:368. doi: 10.1038/s41467-019-14246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karron RA, Buchholz UJ, Collins PL. 2013. Live-attenuated respiratory syncytial virus vaccines. Curr Top Microbiol Immunol 372:259–284. doi: 10.1007/978-3-642-38919-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HW, Arrobio JO, Pyles G, Brandt CD, Camargo E, Chanock RM, Parrott RH. 1971. Clinical and immunological response of infants and children to administration of low-temperature adapted respiratory syncytial virus. Pediatrics 48:745–755. [PubMed] [Google Scholar]
- 44.Connors M, Crowe JE, Jr, Firestone CY, Murphy BR, Collins PL. 1995. A cold-passaged, attenuated strain of human respiratory syncytial virus contains mutations in the F and L genes. Virology 208:478–484. doi: 10.1006/viro.1995.1178. [DOI] [PubMed] [Google Scholar]
- 45.Luongo C, Winter CC, Collins PL, Buchholz UJ. 2013. Respiratory syncytial virus modified by deletions of the NS2 gene and amino acid S1313 of the L polymerase protein is a temperature-sensitive, live-attenuated vaccine candidate that is phenotypically stable at physiological temperature. J Virol 87:1985–1996. doi: 10.1128/JVI.02769-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang RS, Nguyen N, Zhou H, Jin H. 2002. Clustered charge-to-alanine mutagenesis of human respiratory syncytial virus L polymerase generates temperature-sensitive viruses. Virology 302:207–216. doi: 10.1006/viro.2002.1596. [DOI] [PubMed] [Google Scholar]
- 47.Grdzelishvili VZ, Smallwood S, Tower D, Hall RL, Hunt DM, Moyer SA. 2006. Identification of a new region in the vesicular stomatitis virus L polymerase protein which is essential for mRNA cap methylation. Virology 350:394–405. doi: 10.1016/j.virol.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 48.Murphy AM, Grdzelishvili VZ. 2009. Identification of Sendai virus L protein amino acid residues affecting viral mRNA cap methylation. J Virol 83:1669–1681. doi: 10.1128/JVI.01438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. 2004. Suppression of the induction of alpha, beta, and gamma interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J Virol 78:4363–4369. doi: 10.1128/jvi.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hijano DR, Vu LD, Kauvar LM, Tripp RA, Polack FP, Cormier SA. 2019. Role of type I interferon (IFN) in the respiratory syncytial virus (RSV) immune response and disease severity. Front Immunol 10:566. doi: 10.3389/fimmu.2019.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka T, Narazaki M, Kishimoto T. 2014. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol 6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell CD, Unger SA, Walton M, Schwarze J. 2017. The human immune response to respiratory syncytial virus infection. Clin Microbiol Rev 30:481–502. doi: 10.1128/CMR.00090-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Touzelet O, Broadbent L, Armstrong SD, Aljabr W, Cloutman-Green E, Power UF, Hiscox JA. 2020. The secretome profiling of a pediatric airway epithelium infected with hRSV identified aberrant apical/basolateral trafficking and novel immune modulating (CXCL6, CXCL16, CSF3) and antiviral (CEACAM1) proteins. Mol Cell Proteomics 19:793–807. doi: 10.1074/mcp.RA119.001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 55.Johnson SM, McNally BA, Ioannidis I, Flano E, Teng MN, Oomens AG, Walsh EE, Peeples ME. 2015. Respiratory syncytial virus uses CX3CR1 as a receptor on primary human airway epithelial cultures. PLoS Pathog 11:e1005318. doi: 10.1371/journal.ppat.1005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delgado MF, Coviello S, Monsalvo AC, Melendi GA, Hernandez JZ, Batalle JP, Diaz L, Trento A, Chang HY, Mitzner W, Ravetch J, Melero JA, Irusta PM, Polack FP. 2009. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat Med 15:34–41. doi: 10.1038/nm.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw CA, Galarneau JR, Bowenkamp KE, Swanson KA, Palmer GA, Palladino G, Markovits JE, Valiante NM, Dormitzer PR, Otten GR. 2013. The role of non-viral antigens in the cotton rat model of respiratory syncytial virus vaccine-enhanced disease. Vaccine 31:306–312. doi: 10.1016/j.vaccine.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, Liang X, Gao TZ, Xu Y, Zhou J, Feng Z, Niewiesk S, Peeples ME, He C, Li J. 2020. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol 5:584–598. doi: 10.1038/s41564-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue M, Zhao BS, Zhang Z, Lu M, Harder O, Chen P, Lu Z, Li A, Ma Y, Xu Y, Liang X, Zhou J, Niewiesk S, Peeples ME, He C, Li J. 2019. Viral N(6)-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat Commun 10:4595. doi: 10.1038/s41467-019-12504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]