Astroviruses cause gastroenteritis and encephalitis in human, and nephritis, hepatitis, and gout disease in poultry. However, the host immune response activated by astrovirus is mostly unknown. Here, we found that a novel goose astrovirus, GAstV-GD, and its ORF2 protein could efficiently induce a high level of OASL in vitro and in vivo, which could feed back to restrict the replication of GAstV-GD, revealing novel innate molecules triggered by astroviruses and highlighting that the ORF2 of GAstV-GD and OASL can be potential antiviral targets for astroviruses.
KEYWORDS: goose astrovirus, ORF2, innate immunity, OASL, antiviral activity
ABSTRACT
Although astroviruses causes enteric diseases and encephalitis in humans and nephritis and hepatitis in poultry, astrovirus infection is thought to be self-limiting. However, little is known about its molecular mechanism. In this study, we found that a novel goose astrovirus (GAstV), GAstV-GD, and its open reading frame 2 (ORF2) could efficiently activate the innate immune response and induce a high level of OASL in vitro and in vivo. The truncation assay for ORF2 further revealed that the P2 domain of ORF2 contributed to stimulating OASL, whereas the acidic C terminus of ORF2 attenuated such activation. Moreover, the overexpression and knockdown of OASL could efficiently restrict and promote the viral replication of GAstV-GD, respectively. Our data not only give novel insights for elucidating self-limiting infection by astrovirus but also provide virus and host targets for fighting against astroviruses.
IMPORTANCE Astroviruses cause gastroenteritis and encephalitis in human, and nephritis, hepatitis, and gout disease in poultry. However, the host immune response activated by astrovirus is mostly unknown. Here, we found that a novel goose astrovirus, GAstV-GD, and its ORF2 protein could efficiently induce a high level of OASL in vitro and in vivo, which could feed back to restrict the replication of GAstV-GD, revealing novel innate molecules triggered by astroviruses and highlighting that the ORF2 of GAstV-GD and OASL can be potential antiviral targets for astroviruses.
INTRODUCTION
Astroviruses are currently divided into two genera, Mamastrovirus and Avastrovirus (1). Mamastroviruses infect mammals, including humans, whereas avastroviruses generally infect different avian species (1, 2). The genome of astroviruses is about 6.8 to 7.9 kb in length, which includes a 5′ untranslated region (UTR), three open reading frames (ORFs) (1a, 1b, and 2), a 3′ UTR, and a poly(A) tail (3). ORF2 is thought to be the major structural protein encoded by astroviruses. Although astrovirus infection generally leads to mild diseases such as diarrhea and gastroenteritis in human and animals, as initially reported, astroviruses can also cause nephritis in chickens and pigeons, hepatitis in ducklings, and encephalitis in humans, cattle, and sheep (2, 4–6). Recently, a novel goose astrovirus (GAstV) related to the outbreak of gosling gout disease in China has been reported (7). The genetic variation and multiple hosts of astroviruses pose risks for cross-transmission and zoonotic infection (2, 3). Notably, unlike other viruses, most of the astroviruses both from Mamastrovirus and Avastrovirus show low replication in cell culture systems and do not cause a cytopathic effect (CPE) in infected cells, which is why infection is termed self-limiting (8). However, the molecular mechanism for self-limiting infection by astroviruses is unknown.
Although infection by many viruses shuts down the cellular antiviral responses or hijacks the antiviral molecules to escape innate immunity and cause severe CPE in the infected cells, cellular innate immunity plays vital roles in inhibiting or restricting virus replication or spread (9–12). To investigate whether the astrovirus activates the cellular innate immunity to restrict the viral replication itself, a novel goose astrovirus, GAstV-GD, which emerged in 2015 and caused gosling gout disease in China, was tested in vitro and in vivo. Interestingly, we found that not only GAstV-GD but also its ORF2 could efficiently activate innate immunity and induce a high level of OASL in vitro and in vivo, which could feed back to inhibit the replication of GAstV-GD.
RESULTS
Self-limiting replication of GAstV-GD in LMH cells.
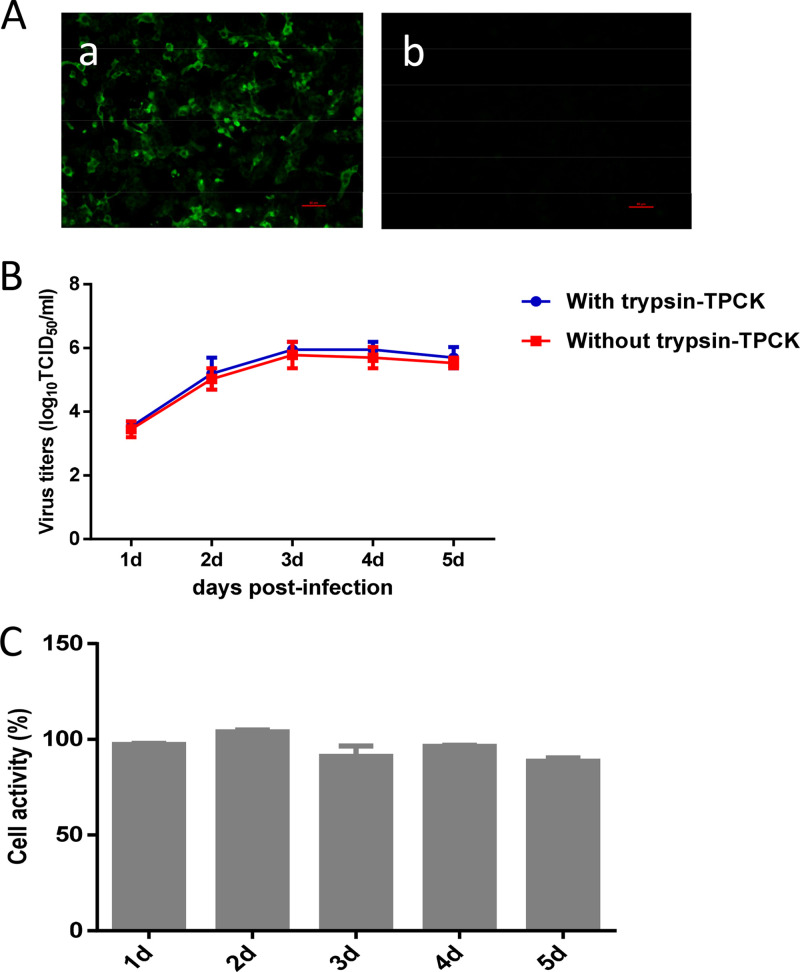
As we all know, the astroviruses are difficult to isolate and culture in vitro (13). In our previous study, a novel goose astrovirus, GAstV-GD, was efficiently isolated using the LMH cell line. Although the novel goose astrovirus GAstV-GD could infect the LMH cells and replicated in the LMH cells, as shown in Fig. 1A, GAstV-GD showed diffuse rather than clustered distributions in the LMH cells at different time points. Moreover, GAstV-GD did not cause CPE in the infected LMH cells. The viral growth curve in LMH cells further showed that the GAstV-GD virus grows slowly, and the peak titer of GAstV-GD in LMH cells was only 1.58 × 106 50% tissue culture infective dose (TCID50)/ml at 3 days postinfection (dpi), as described in Fig. 1B. Notably, although no apparent CPE was found in the infected LMH cells, the titer of GAstV-GD was slightly decreased at 4 to 5 dpi. A cell activity assay using the cell-counting kit-8 (CCK-8) approach further demonstrated that the cell activity of the LMH cells infected with GAstV-GD was very similar to that of the wild-type LMH cells as described in Fig. 1C. These findings demonstrate that the novel goose astrovirus GAstV-GD shows self-limiting infection in LMH cells, possibly due to the activation of antiviral innate immunity.
FIG 1.
Self-limiting replication of GAstV-GD in LMH cells. (A) LMH cells infected with GAstV-GD (a) or not infected (b) were reacted with the mouse sera against ORF2 of GAstV-GD by indirect immunofluorescence assay. (B) The growth curve of GAstV-GD in LMH cells. LMH cells were infected with GAstV-GD at a multiplicity of infection (MOI) of 0.1 and cultured with or without trypsin-tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK). At the indicated number of days postinfection, the titer of the supernatant of the infected cells was determined by 50% tissue culture infective dose (TCID50). (C) Cell activity assay of GAstV in LMH cells. At different time points postinfection, the activity of the infected cells was determined by a CCK-8 approach comparing with activity of the wild-type LMH cells. All experiments were done in triplicate and repeated twice.
GAstV-GD induced high-level OASL in vitro and in vivo.
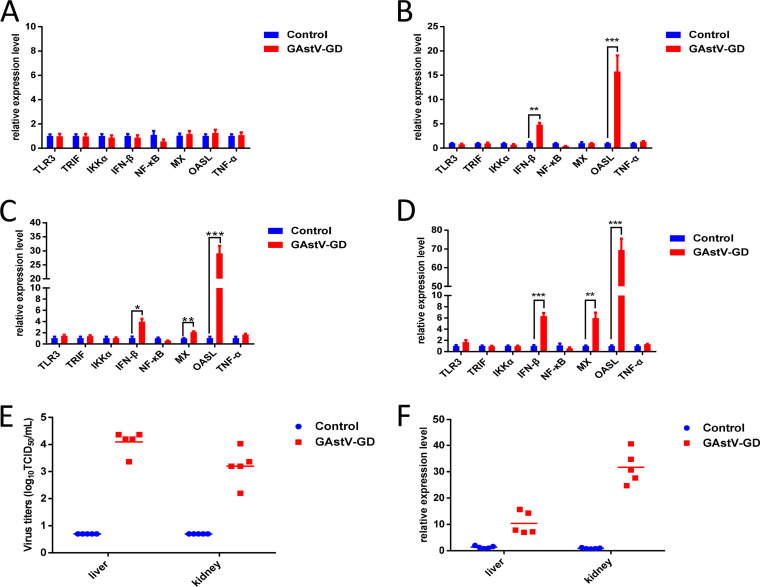
To elucidate the underlying mechanism for the self-limiting infection of the astrovirus, LMH cells were infected with GAstV-GD, and the expression levels of several cytokines associated with innate immunity were analyzed in the infected cells by real-time PCR. As described in Fig. 2, the levels of beta interferon (IFN-β) and OASL were significantly increased in LMH cells infected with GAstV-GD at 48 hours postinfection (hpi), 72 hpi, and 96 hpi compared with those in the uninfected LMH cells. At 72 hpi (Fig. 2C) and 96 hpi (Fig. 2D), the expression level of Mx was also sharply increased in the infected cells. However, the levels of other cytokines or molecules such as Toll-like receptor 3 (TLR3), TRIF, IκB kinase α (IKKα), NF-κB, and tumor necrosis factor alpha (TNF-α) detected were not increased in either the infected cells or the control cells. Since the level of OASL in the infected cells was the highest in these molecules tested, to confirm this finding in vivo, the expression levels of OASL in the livers and kidneys from the goslings infected with GAstV-GD were analyzed by real-time PCR. The viral load in the livers and kidneys confirmed the infection of goslings inoculated with GAstV-GD, as described in Fig. 2E. As shown in Fig. 2F, the levels of OASL in the livers and kidneys from the infected goslings were significantly higher than those from the control goslings. These data demonstrate that infection by GAstV-GD in vitro or in vivo can induce a high level of OASL, which might contribute to the self-limiting replication of the astrovirus.
FIG 2.
GAstV-GD activated innate immunity in vitro and in vivo. Transcriptional levels of TLR3, TRIF, IKKα, IFN-β, NF-κB, Mx1, OASL and TNF-α associated with innate immunity in GAstV-GD infected LMH cells and control cells were analyzed at 24 hpi (A), 48 hpi (B), 72 hpi (C) and 96 hpi (D); (E) Viral titers in the liver and kidney from the GAstV-GD infected goslings (n = 5) and control goslings (n = 5); (F) Transcriptional level of OASL in the liver and kidney from the GAstV-GD infected goslings (n = 5) and control goslings (n = 5). All experiments were done in triplicates and repeated twice.
ORF2 played vital roles in activating innate immunity.
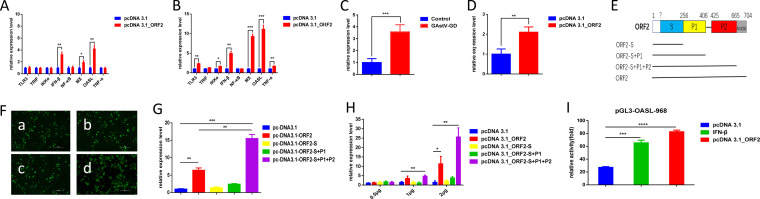
The LMH cells were transfected with pcDNA 3.1_ORF2, encoding the major structural protein of the astrovirus, to investigate whether ORF2 of GAstV-GD activates innate immunity. The mRNA level of several cytokines in the transfected cells were analyzed. As shown in Fig. 3A and B, the levels of IFN-β, OASL, and Mx were substantially increased in LMH cells transfected with pcDNA 3.1_ORF2 at 24 hpi and 48 hpi. Although the levels of TLR3, IKKα, and TNF-α were not increased in LMH cells transfected with ORF2 at 24 hpi, those in LMH cells transfected with pcDNA 3.1_ORF2 were significantly increased at 48 hpi in comparison with those in the control LMH cells transfected with pcDNA 3.1 vector. In addition, we also found that GAstV-GD and its ORF2 could efficiently activate the chicken lambda interferon (IFN-λ) as described in Fig. 3C and D. A series of truncations of the ORF2 were generated to map which domains of the ORF2 play vital roles in activating these cytokines, and the levels of these cytokines in the transfected LMH cells were analyzed. As described in Fig. 3E and F, three truncations of the ORF2 were constructed and could be efficiently expressed in the LMH cells with very similar transfection efficacy. Interestingly, the truncated ORF2-S+P1+P2 could induce a higher level of OASL than that induced by the full-length ORF2, whereas other truncated ORF2 tested, such as ORF2-S+P1 and ORF2-S, could not efficiently activate the higher levels of OASL, as shown in Fig. 3G. Notably, the activity for inducing OASL by ORF2 and ORF2-S+P1+P2 showed efficient dose dependence, described in Fig. 3H. To further test whether ORF2 can activate OASL through its promoter, the promoter region (−968) of OASL was cloned into a luciferase (Luci) reporter vector and evaluated by ORF2. As shown in Fig. 3I, ORF2 could significantly activate the luciferase activity under the control of the OASL promoter in comparison with that in the control. These data demonstrate that the ORF2 and its P2 domain efficiently activate innate immunity, whereas the acidic C terminus of ORF2 attenuates such activation.
FIG 3.
ORF2 played vital roles in activating innate immunity. Transcriptional levels of TLR3, TRIF, IKKα, IFN-β, NF-κB, Mx1, OASL, and TNF-α associated with innate immunity in LMH cells transfected with pcDNA3.1_ORF2 and control cells were analyzed at 24 hpi (A) and at 48 hpi (B). (C) Transcriptional levels of IFN-λ in the LMH cells infected with GAstV-GD were analyzed at 72 hpi. (D) Transcriptional levels of IFN-λ in the LMH cells transfected with pcDNA3.1_ORF2 or pcDNA3.1 vector were analyzed at 48 hpi. (E) Pattern diagrams of the recombinant plasmid for different truncated ORF2 of GD. (F) Expressions of pcDNA 3.1_ORF2 (a), pcDNA 3.1_ORF2-S (b), pcDNA 3.1_ORF2-S+P1 (c), and pcDNA3.1_ORF2-S+P1+P2 (d) were measured by indirect immunofluorescence assay/ (G) Transcriptional level of OASL at 48 hpi in the LMH cells transfected with different truncations of ORF2. (H) Transcriptional level of OASL at 48 hpi in the LMH cells transfected with different doses of different truncations of ORF2. All experiments were done in triplicate and repeated twice; (I) ORF2 efficiently activated the promoter of OASL. The LMH cells were cotransfected with pGL3-OASL-968 and pRL-TK. After 24 h, these LMH cells were transfected with pcDNA 3.1_ORF2 and pcDNA 3.1, while IFN-β was added as a positive control. Luciferase activity in these LMH cells was then detected using a luciferase reporter gene assay kit at 24 h posttransfection.
Overexpression and knockdown of OASL in LMH cells.
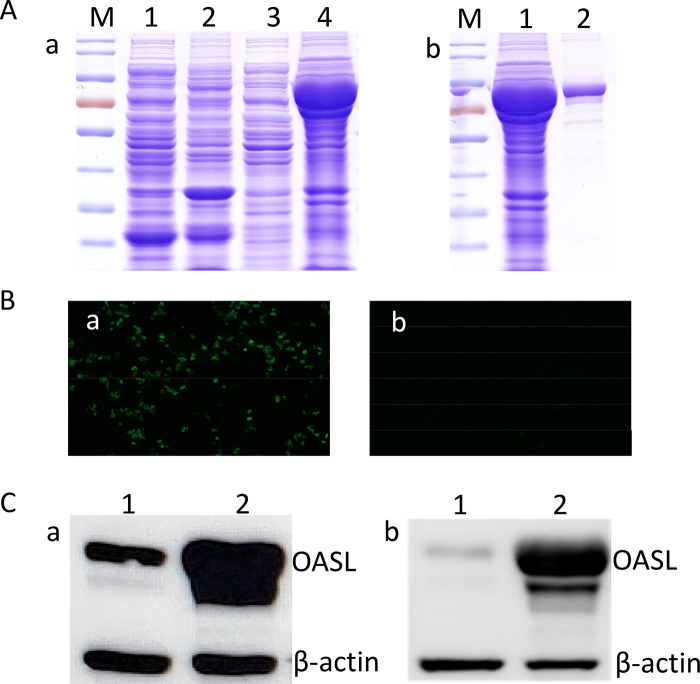
Since the expression vectors for chicken OASL and its specific antibodies were not available, the chicken OASL gene was first cloned into prokaryotic and eukaryotic expression vectors, and then the LMH cells with knockdown of OASL were generated using the CRISPR-Cas9 technique as described in Materials and Methods. The responding recombinants for expression were designated pGEX-6P-1_OASL and pcDNA 3.1_OASL, respectively. The expression of the glutathione S-transferase (GST)-OASL fusion protein in Escherichia coli transformed with pGEX-6P-1_OASL was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) and analyzed by SDS-PAGE as described in Fig. 4A. Then, the mouse sera against OASL were generated by immunizing BALB/c mice with the purified GST-OASL. Immunofluorescence assay (IFA) showed that the mouse sera generated against GST-OASL could efficiently react with the OASL expressed in LMH cells transfected with the recombinant plasmid pcDNA 3.1_OASL, as shown in Fig. 4B and C. In the Western blot analysis, the mouse sera against GST-OASL could efficiently recognize the endogenous chicken OASL in LMH cells (Fig. 4C). As described in Fig. 4C, panel b, the LMH cells with knockdown of OASL (LMH_KD-OASL) by CRISPR-Cas9 showed significantly lower expression of OASL than that in the wild-type LMH cells. All these data demonstrate that the constructed expression vectors for OASL, the prepared mouse sera against OASL, and the generated LMH cells with knockdown of OASL could provide efficient tools and reagents for further studying the function of chicken OASL.
FIG 4.
Overexpression and knockdown of OASL in LMH cells. (A) SDS-PAGE analysis for expression and purification of GST-OASL. (a) Lanes 1, 2, 3, and 4 show the supernatant and the precipitate of GST and GST-OASL, respectively; (b) lanes 1 and 2 show the unpurified and purified GST-OASL, respectively. (B) The expression of pcDNA 3.1_OASL (a) was measured by indirect immunofluorescence assay compared with wild-type LMH cells (b). (C) Western blot analysis for the expression of pcDNA 3.1_OASL. (a) Lanes 1 and 2 show LMH cells transfected with pcDNA3.1 and pcDNA3.1_OASL, respectively. Western blot analysis for the expression of OASL in LMH_KD-OASL cells and wild-type LMH cells. (b) Lanes 1 and 2 show LMH_KD-OASL cells and wild-type LMH cells, respectively. All experiments were performed three times with comparable results.
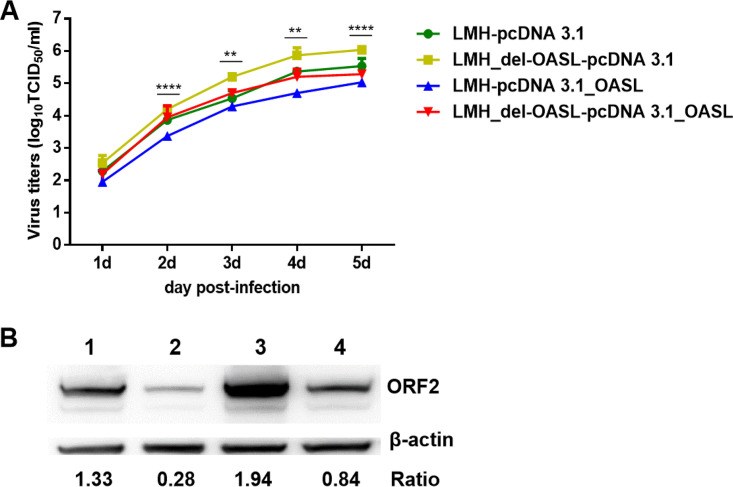
OASL could efficiently restrict the replication of GAstV-GD.
Since GAstV-GD and its ORF2 could induce OASL, to further investigate the effect of OASL on the viral replication of GAstV-GD, the wild-type LMH cells and the LMH_KD-OASL cells were transfected with pcDNA 3.1_OASL and pcDNA 3.1, respectively, and then infected with GAstV-GD. The viral titer in these infected cells was determined by TCID50 and analyzed by Western blotting. As described in Fig. 5A, the GAstV-GD showed the highest virus titers in the LMH_KD-OASL cells transfected with pcDNA 3.1 vector and the lowest virus titers in LMH cells transfected with pcDNA 3.1_OASL, compared with that in the wild-type LMH cells transfected with pcDNA 3.1 vector. The viral titers in LMH_KD-OASL cells were significantly lower than those in LMH cells, with overexpression of OASL at 2 to 6 dpi (P < 0.01). Notably, the virus titers in LMH_KD-OASL cells transfected with pcDNA 3.1_OASL were very similar to those in the LMH cells transfected with pcDNA 3.1 vector. This finding was further confirmed by Western blotting, as described in Fig. 5B. The expression level of ORF2 in the LMH_KD-OASL cells transfected with pcDNA 3.1 vector was the highest, whereas that in LMH cells transfected with pcDNA 3.1_OASL was the lowest compared with that in the wild-type LMH cells transfected with pcDNA 3.1 vector. The ratios of the expression of ORF2 in LMH cells and in LMH_KD-OASL cells were 4.75 and 6.93 times of that in LMH cells with the overexpression of OASL, respectively. That ratio in LMH_KD-OASL cells was 2.31 times of that in LMH_KD-OASL cells with the overexpression of OASL. The expression level of ORF2 in the LMH_KD-OASL cells transfected with pcDNA 3.1_OASL was very similar to that in LMH cells transfected with pcDNA 3.1 vector. All these data demonstrate that OASL can be an essential host factor to efficiently restrict the viral replication of GAstV-GD.
FIG 5.
OASL could efficiently inhibit the replication of GAstV-GD. Growth curves of GAstV-GD at 0.01 MOI in the wild-type LMH cells and the LMH_KD-OASL cells which were transfected with pcDNA 3.1_OASL and pcDNA 3.1 before infection. At the indicated number of days postinfection, titer of the supernatant of the infected cells was determined by TCID50 (A), and the infected cells were analyzed at 5 dpi by Western blotting (B). Lanes 1 and 2 show the wild-type LMH cells transfected with pcDNA 3.1 and pcDNA 3.1_OASL, respectively; lanes 3 and 4 show the LMH_KD-OASL cells transfected with pcDNA 3.1 and pcDNA 3.1_OASL, respectively. All experiments were done in triplicate and repeated twice.
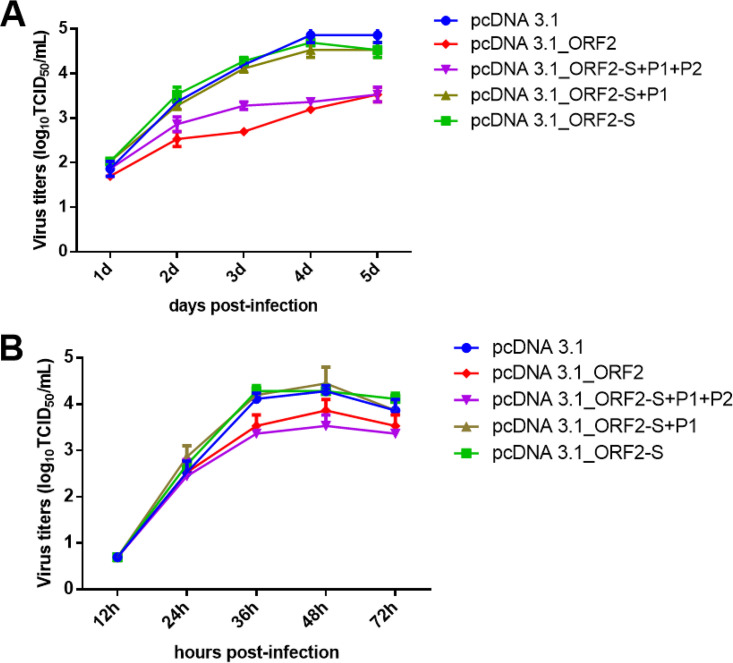
ORF2 could efficiently restrict the replication of GAstV-GD.
Since OASL plays vital roles in blocking the infection and replication of viruses, to evaluate further whether ORF2 can efficiently inhibit the viral replication of GAstV-GD, the LMH cells were first transfected with pcDNA 3.1_ORF2 and then infected with GAstV-GD or H9N2 influenza virus. The viral titers in the transfected cells were analyzed in comparison with those of the control cells. As described in Fig. 6, the ORF2 could significantly restrict the viral replication of GAstV-GD and efficiently inhibit the viral replication of H9N2 in LMH cells. The viral titers of GAstV-GD in LMH cells transfected with ORF2 and ORF2-S+P1+P2 were 10 times lower than those in LMH cells transfected with ORF2-S+P1, ORF2-S, or pcDNA3.1 vector at 3 to 5 dpi (Fig. 6A). ORF2 and ORF2-S+P1+P2 also restricted the replication of H9N2. The viral titers of H9N2 in LMH cells transfected with ORF2 and ORF2-S+P1+P2 were about 5 times lower than those in LMH cells transfected with ORF2-S+P1, ORF2-S, or pcDNA3.1 vector at 36 hpi (Fig. 6B). These data showed that the antiviral activity of ORF2 was not specific to GAstV-GD, indicating the innate antiviral activity induced by ORF2 might contribute to this activity for restricting the viral replication of GAstV-GD.
FIG 6.
ORF-2 restricted the replication of GAstV-GD via its P2 domain. (A) Viral growth curves of GAstV-GD. The LMH cells that were transfected with pcDNA 3.1_ORF2, pcDNA 3.1_ORF2-S+P1+P2, pcDNA 3.1_ORF2-S+P1, pcDNA 3.1_ORF2-S, and pcDNA 3.1 were infected with GAstV-GD at 0.01 MOI, and the supernatants of the infected cells were collected at the indicated time points and titer was determined by TCID50; (B) Viral growth curves of H9N2. The LMH cells that were transfected with pcDNA 3.1_ORF2, pcDNA 3.1_ORF2-S+P1+P2, pcDNA 3.1_ORF2-S+P1, pcDNA 3.1_ORF2-S, and pcDNA 3.1 were infected with H9N2 at 0.001 MOI, and the supernatants of the infected cells were collected at the indicated time points and titer was determined by TCID50.
DISCUSSION
It is well known that astroviruses from Mamastrovirus or Avastrovirus are difficult to isolate and culture in vitro (13). The infection and replication of the astroviruses generally show self-limitation in cell culture, and astroviruses do not cause CPE in the infected cells (14–16). However, the factors from either the virus or the host that contribute to this phenotype of the astroviruses are mostly unknown, and the roles of the innate and adaptive immune responses in astrovirus infection have also not been extensively studied. A few reports of human astrovirus (HAstV) demonstrate that astroviruses can elicit a weak type I interferon response, and the exogenous type I interferon efficiently limits astrovirus infection in vitro (16, 17). Notably, the deficiency of the IFN-α/IFN-β receptor increases the replication of murine astrovirus (18). Moser et al. reported that the astrovirus could activate the ERK 1/2 pathway early in the infection, independently of viral replication (19). In this study, we found that a novel goose astrovirus, GAstV-GD, which is associated with gout disease endemic in goslings in China, also showed self-limiting replication in the LMH cell line.
Moreover, we found that GAstV-GD could activate the high levels of OASL in the LMH cells via the P2 domain of the ORF2. Notably, the OASL could also be efficiently activated in the liver and kidney from the geese infected with GAstV-GD. The overexpression and knockdown assay revealed that OASL could be a vital host factor to restrict the viral replication of GAstV-GD in LMH cells. Therefore, OASL induced in the infected LMH cells may contribute to the self-limiting replication of GAstV-GD. It also should be noted that the acidic domain at the C terminus of the ORF2 could attenuate the activation of OASL compared with that of the intact ORF2 and the truncated ORF2 without the acidic domain. The ORF2 of HAstV has been reported to efficiently bind to complement 1q and mannose-binding lectin to inhibit complement activity (20, 21). However, the molecular mechanism for the ORF2 of GAstV-GD to activate the OASL needs to be further elucidated. To demonstrate whether ORF2 can activate OASL through its promoter, we cloned the promoter region (−968) of OASL into Luci reporter vector and evaluated the activity of ORF2 on the promoter of OASL. As shown in Fig. 3I, ORF2 could significantly activate the luciferase activity under the control of the OASL promoter in comparison with that in the control. However, whether ORF2 can directly bind to the OASL promoter or indirectly activate luciferase activity needs to be further investigated.
Since OASL is a critical host antiviral molecule that can efficiently inhibit the replication of RNA viruses, possibly through a RIG-I-dependent manner (22–24), the potential antiviral activity of the ORF2 of GAstV-GD was also investigated. Interestingly, our data showed that ORF2 and ORF2-S+P1+P2 could efficiently restrict the viral replication of both GAstV-GD and H9N2 avian influenza virus in vitro. Notably, the acidic domain of ORF2 is thought to be a binding site for cell receptors during astrovirus infection (25). ORF2-S+P1+P2 in this study lacks the acidic domain present in the full-length ORF2. Therefore, the antiviral activity of ORF2 on GAstV-GD was mainly dependent on the innate antiviral responses induced by ORF2 via its S+P1+P2 region but was not due to the blocking of the cell receptor by ORF2 via its acidic domain or by impairing normal virus formation and/or budding.
It should be noted that Ingle et al. recently reported that the murine astrovirus and its ORF2 could complement primary immunodeficiency to protect against murine norovirus and rotavirus infections through cytokine IFN-λ (26). We also found that GAstV-GD and its ORF2 could efficiently activate the chicken IFN-λ as described in Fig. 3C and D. The effects of chicken IFN-λ triggered by GAstV-GD and ORF2 on other pathogens need to be further elucidated. All these findings demonstrate that some astroviruses and their ORF2, both from Mamastrovirus and Avastrovirus, can efficiently activate the host innate immunity to limit itself replication and provide protection against other pathogens. These also indicate the possibility that the infection of some low pathogenic microbes might assist the host to fight against other highly pathogenic pathogens.
In summary, this is the first demonstration that a novel goose astrovirus, GAstV-GD, and its ORF2 efficiently activate OASL in vitro and in vivo and that OASL can be an essential host factor to restrict the viral replication of GAstV. Besides, the ORF2 of GAstV-GD can be a potential antiviral agonist to restrict the replication of GAstV-GD and H9N2 in vitro. We hypothesized that the high level of OASL induced by the ORF2 of GAstV contributes to the self-limiting replication of the astrovirus and plays a vital role in restricting the viral replication of GAstV-GD and H9N2. However, whether the high level of OASL induced in the infected geese contributes to the gout disease caused by GAstV-GD need to be further investigated.
MATERIALS AND METHODS
Cell line and virus.
Chicken liver cell line (LMH; ATCC) was cultured in Dulbecco’s modified Eagle medium (DMEM)/F-12 with 10% fetal bovine serum (FBS) and 1% penicillin (100 U/ml)-streptomycin (10 mg/ml) at 37°C with 5% CO2. A strain of goose astrovirus, GAstV-GD, was isolated from a goose in Guangdong, China, and was propagated in LMH cells with DMEM/F-12 medium and 1 μg/ml trypsin-tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) (7). The strain of the H9N2 avian influenza virus [A/Chicken/Jiangsu/WS1/2012 (H9N2)] was kept in our laboratory.
Construction of different ORF2 truncations.
Linear pcDNA3.1 was amplified using the pcDNA3.1 plasmid as a template and pc-F and pc-R as primers. For amplifying ORF2 of GAstV-GD, the cDNA of GAstV-GD was used as a template, and pcDNA 3.1_ORF2-flag-F and pcDNA 3.1_ORF2-flag-R were used as primers. The different truncated ORF2 were also amplified using the corresponding primers listed in Table 1. The linearized vector and the corresponding fragment of PCR products were then ligated using a ClonExpress II kit (Vazyme, Nanjing, China). The recombinant plasmids were further confirmed by sequencing.
TABLE 1.
Primer list for truncation of GAstV ORF2
| Primer name | Direction | Sequence (5′–3′) |
|---|---|---|
| pc | Forward | GAATTCTGCAGATATCCAGCACAGTG |
| pc | Reverse | GCTCGGTACCAAGCTTAAGTTTAAACG |
| pcORF2-flag | Forward | AGCTTGGTACCGAGCATGGATTACAAGGATGACGACGATAAGATGGCAGACAGGGCGGTG |
| pcORF2-flag | Reverse | ATATCTGCAGAATTCTTATCACTCATGTCCGCCCTTCTC |
| pcORF2-S-flag | Reverse | ATATCTGCAGAATTCTTATGGCTTTGGACCATAGTTCGAG |
| pcORF2-S+P1-flag | Reverse | ATATCTGCAGAATTCTTACATTGGTGCCACTGGCAGAGGC |
| pcORF2-S+P1+P2-flag | Reverse | ATATCTGCAGAATTCTTAGGTCTTGAGCGAGACTGCTAGGTG |
Virus replication kinetics.
LMH cells were infected with GAstV-GD at a multiplicity of infection (MOI) of 0.1. At 2 h postinfection, the supernatant of the infected cells was discarded, and Opti-MEM medium containing 1 μg/ml TPCK-trypsin was added to the cells. Then, the supernatants from the infected cells were collected at 1, 2, 3, 4, and 5 dpi, and titer was determined in LMH cells by TCID50 as previously described (7). In brief, the supernatants were subjected to 10-fold serial dilutions using DMEM/F-12 medium with 1 μg/ml trypsin-TPCK, which resulted in 8 concentrations. Aliquots (200 μl) of supernatants in each concentration were dispensed into each well in 96-well plates with four repeats, and the infected LMH cells were cultured for 5 days. At 5 dpi, the infected LMH cells were fixed and analyzed by IFA for the detection of the virus, and the viral titers were calculated using the Reed-Muench method.
RNA extraction and quantitative real-time PCR.
The total RNAs of the cells were extracted using an AxyPre total RNA miniprep kit, and cDNA was synthesized using HiScript Q RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme) according to the manufacturer’s protocol. Real-time PCR was performed using AceQ Universal SYBR quantitative PCR (qPCR) Mastermix (Vazyme, Nanjing, China) in an ABI 7500 real-time PCR system (Applied Biosystems, CA, USA) according to the manufacturer’s protocol (27). The genes tested and the primers used are listed in Table 2. Gene expression levels were normalized to the 18S rRNA housekeeping gene. The changes in gene expression were calculated using the threshold cycle (2−ΔΔCT) method. Statistical analysis was conducted using GraphPad Prism 6 software.
TABLE 2.
Primers used for real-time PCR
| Target gene | Product size (bp) | GenBank accession no. | Primer sequencea |
|---|---|---|---|
| ch18S | 154 | AF173612 | F: 5′-TCAGATACCGTCGTAGTTCC-3′ |
| R: 5′-TTCCGTCAATTCCTTTAAGTT-3′ | |||
| chTLR3 | 99 | NM_001011691.3 | F: 5′-GCACCTGTGAAAGCATTG-3′ |
| R: 5′-TAGGCGGGGTGTTACAAATG-3′ | |||
| chTRIF | 134 | NM_001081506.1 | F: 5′-CACAGACCTTGCAATCCTCA-3′ |
| R: 5′-ATCACTGGTGCTCACTTCAC-3′ | |||
| chIKKα | 244 | NM_001012904.1 | F: 5′-CTTTCATCTATGGCAACTCCTG-3′ |
| R: 5′-ATGTCCAAACCAAGACGTGAT-3′ | |||
| chNF-κB | 181 | NM_205129.1 | F: 5′-GCCAGGTTGCCATCGTGTTCC-3′ |
| R: 5′-CGCGTGCGTTTGCGCTTCTC-3′ | |||
| chIFN-β | 151 | NM_001024836.1 | F: 5′-GCCCACACACTCCAAAACACTG-3′ |
| R: 5′-TTGATGCTGAGGTGAGCGTTG-3′ | |||
| chTNF-α | 229 | NM_204267.1 | F: 5′-TGTGTATGTGCAGCAACCCGTAGT-3′ |
| R: 5′-GGCATTGCAATTTGCACAGAAGT-3′ | |||
| chMx1 | 137 | NM_204609.1 | F: 5′-AATAAGGCTACTATCCCACA-3′ |
| R: 5′-GTGTACTTTTGGAGTTCCTT-3′ | |||
| chOASL | 105 | NM_205041.1 | F: 5′-GAGATAGAGAAGGAGTGGTG-3′ |
| R: 5′-GTAGACTGTGGTCTTGTTAC-3′ | |||
| chIFN-λ | 195 | KF680103.1 | F: 5′-CTTTGGAGTTGAAGGCAGTGTGG-3′ |
| R: 5′-TCTGGGTTGTGGGGTTTGTGAG-3′ | |||
| GoOASL | 133 | KU058695.1 | F: 5′-CAGCGTGTGGTGGTTCTC-3′ |
| R: 5'-AACCAGACGATGACATACAC-3' |
F, forward; R, reverse.
Expression of OASL and preparation of its polyclonal antibodies.
Linearized pcDNA3.1 and pGEX-6P-1 were amplified using pcDNA3.1 vector and pGEX-6P-1 vector as a template, and pc-F plus pc-R and pGEX-6P-1-F plus pGEX-6P-1-R as primers, respectively. For amplifying OASL, the cDNA of HD11 cells was used as a template, and pcDNA 3.1_OASL-F, pcDNA 3.1_OASL-R, pGEX-6P-1_OASL-F, and pGEX-6P-1_OASL-R were used as primers (Table 3). The linearized vector and the corresponding fragment of PCR products were then ligated using the ClonExpress II kit (Vazyme, Nanjing, China). The recombinant plasmids, pcDNA 3.1_OASL, and pGEX-6P-1_OASL were further confirmed by sequencing. The expression of GST-OASL fusion protein in E. coli transformed with pGEX-6P-1_OASL was induced by IPTG and analyzed by SDS-PAGE. The mouse sera against OASL were generated by immunizing BALB/c mice with the purified GST-OASL.
TABLE 3.
Primers for construction of expression plasmids for OASL
| Plasmid | Primer name | Direction | Sequence (5′–3′) |
|---|---|---|---|
| pc-DNA3.1-OASL | pc | Forward | GAATTCTGCAGATATCCAGCACAGTG |
| Reverse | GCTCGGTACCAAGCTTAAGTTTAAACG | ||
| pc-OASL | Forward | AGCTTGGTACCGAGCATGGAGATGGGGTTGGAGAGCGTGAG | |
| Reverse | ATATCTGCAGAATTCTTACAGAGTTCACAGCTTTTATTCCCAGGG | ||
| GST-OASL | pGEX-6P-1 | Forward | TAATGACGGTGAAAACCTCTGACACATGC |
| Reverse | CATGGGCCCCTGGAACAGAACTTCCAGAT | ||
| pG-OASL | Forward | GTTCCAGGGGCCCATG GAGATGGGGTTGGAGAGCGTGAG | |
| Reverse | GTTTTCACCGTCATTACAGAGTTCACAGCTTTTATTCCCAGGGGGAC |
Establishment of LMH cells with the knockdown of OASL.
Two single guide RNAs (sgRNAs) listed in Table 4 were first cloned into lentiCRISPRv2 vector, and the recombinant plasmids were designated lentiCRISPRv2-OASL1 and lentiCRISPRv2-OASL2, respectively. The sequences for the two sgRNAs targeting OASL were referred to the reference (28). The LMH cells were transfected with the two lentiCRISPRv2-OASL plasmids containing Cas9 nuclease and sgRNA to generate the LMH cells with the knockdown of OASL. After 24 h, the LMH cells were treated with 8 μg/ml puromycin for 10 days to kill nontransfected cells. The remaining cells (designated LMH-KD-OASL) were cultured, and a part of them were used to determine the expression level of OASL protein by Western blotting.
TABLE 4.
sgRNA sequences targeting chicken OASL
| Target gene | Direction | Sequence (5′–3′) |
|---|---|---|
| OASL | Forward | CACCGTACCCGGGGGAAATAGCGGG |
| Reverse | AAACCCCGCTATTTCCCCCGGGTAC | |
| OASL | Forward | CACCGCCGGCGATGACCGCAACCCG |
| Reverse | AAACCGGGTTGCGGTCATCGCCGGC |
Construction of OASL promoter-luciferase reporter gene.
For amplifying OASL promoter, the cDNA of HD11 cells was used as a template, and pGL3-OASL-968-F (5′-CCGCTCGAGCTTGATGGCATGAGTCCTCCTGCC-3′) and pGL3-OASL-968-R (5′-GGGAAGCTTGCTCCGCCGCCGCTCGCTGCAG-3′) were used as primers. The pGL3-Basic vector (Promega, Madison, WI) and the purified fragment of OASL promoter were digested by Confine digestion enzymes XhoI and HindIII, respectively. Then, they were ligated with T4 DNA ligase (Vazyme). The recombinant plasmids were further confirmed by sequencing.
Luciferase reporter assay.
The LMH cells were cotransfected with pGL3-OASL-968, constructed in this study, and pRL-TK (Promega). After 24 h, these LMH cells were transfected with pcDNA 3.1_ORF2 and pcDNA 3.1, while IFN-β was added as a positive control. Luciferase activity in these LMH cells was then detected using the luciferase reporter gene assay kit (Vazyme) at 24 h posttransfection.
Indirect immunofluorescence assay.
The LMH cells infected with GAstV-GD were fixed with a stationary liquid composed of acetone and ethanol (3:2) for 5 min. After washing with phosphate-buffered saline (PBS), the fixed cells were incubated with the specific mouse serum as the primary antibody, such as polyclonal antibodies against ORF2 of the GAstV-GD (prepared in our lab), polyclonal antibodies against OASL (generated in this study), and the mouse monoclonal anti-FLAG antibody (Abclonal, Wuhan, China). Then, fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Sigma-Aldrich, USA) as a secondary antibody. Finally, the stained cells were examined under a fluorescence microscope.
Western blot analysis.
The LMH cells infected with viruses or transfected with different truncated ORF2 were lysed with radioimmunoprecipitation assay (RIPA) buffer (CWBio, Beijing, China) with protease inhibitors (CST, MA, USA). The lysates were loaded with 5× denaturing sample buffer and separated in 10% SDS-PAGE gel electrophoresis, and they were transferred onto 0.2-μm nitrocellulose (NC) membranes (GE Healthcare Life Sciences, Freiburg, Germany). In the Western blot analysis, the mouse monoclonal anti-GAPDH antibody (ABclonal, Wuhan, China), mouse polyclonal anti-ORF2 of GAstV antibody, mouse anti-H9N2 monoclonal antibody 3A11 (29), and polyclonal antibodies against OASL (generated in this study) were used as primary antibodies, and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG was served as the secondary antibody. The signals of proteins were detected with enhanced chemiluminescence.
Infection study in geese.
Ten 5-day-old goslings were assigned equally into two groups. The infection group was infected with GAstV-GD at a dose of 105 TCID50 through muscle injection routes, whereas the control goslings were inoculated with 0.2 ml of phosphate-buffered saline (PBS). The goslings’ livers and kidneys were collected at 5 dpi and used to analyze viral titer and the level of cytokine expression.
Statistical analysis.
The data were analyzed using Prism 6 software (GraphPad Software, La Jolla, CA). The Student’s t test was used to calculate P values. All experiments were done in triplicates and repeated twice, and all data are shown with the means ± standard error of the mean (SEM). Significance levels were set as follow: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Ethics approval and consent to participate.
All animal experiments complied with institutional animal care guidelines and were approved by the Animal Care Committee of Yangzhou University. At the end of the experiment, all the geese were euthanized by CO2.
Data availability.
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENTS
We thank Jianjun Zhang (Sinopharm Yangzhou VAC Biological Engineering Co. Ltd.) for kindly providing us goslings.
We declare that we have no competing interests.
This study was supported by the National Key Research & Development (R&D) Plan (2018YFD0500106) and by the Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality (grant 26116120), the Research Foundation for Talented Scholars in Yangzhou University, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
D.R., J.Y., and A.Q. designed the project. D.R., T.L., X.Z., X.Y., and Q.X. carried out the experiments. D.R., W.G., J.Y., H.X., and Z.W. analyzed the data. D.R., T.L., and J.Y. drafted the manuscript. J.Y. supervised all the experiments and participated in the data analysis. J.Z., X.Z., and A.Q. discussed and prepared the final report. All of the authors have read and approved the final manuscript.
REFERENCES
- 1.Qin Y, Fang Q, Liu H, Ji C, Chen Y, Ouyang K, Wei Z, Huang W. 2018. Construction of a reverse genetic system for porcine astrovirus. Arch Virol 163:1511–1518. doi: 10.1007/s00705-018-3771-4. [DOI] [PubMed] [Google Scholar]
- 2.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. 2011. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol 11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Wang F, Liu N, Yang L, Zhang D. 2017. Complete genome sequence of a novel avastrovirus in goose. Arch Virol 162:2135–2139. doi: 10.1007/s00705-017-3297-1. [DOI] [PubMed] [Google Scholar]
- 4.Vu DL, Cordey S, Brito F, Kaiser L. 2016. Novel human astroviruses: novel human diseases? J Clin Virol 82:56–63. doi: 10.1016/j.jcv.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, Talbot T, Blanchard P, Rimoldi G, Fahsbender E, Page B, Phan TG, Wang C, Deng X, Pesavento P, Delwart E. 2013. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis 19:1385–1392. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boujon CL, Koch MC, Wuthrich D, Werder S, Jakupovic D, Bruggmann R, Seuberlich T. 2017. Indication of cross-species transmission of astrovirus associated with encephalitis in sheep and cattle. Emerg Infect Dis 23:1604–1608. doi: 10.3201/eid2309.170168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Ren D, Li T, Zhou H, Liu X, Wang X, Lu H, Gao W, Wang Y, Zou X, Sun H, Ye J. 2018. An emerging novel goose astrovirus associated with gosling gout disease. China Emerg Microbes Infect 7:1–8. doi: 10.1038/s41426-018-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch A, Pintó RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng M, Zhang X. 2016. Immunity to avian leukosis virus: where are we now and what should we do? Front Immunol 7:624. doi: 10.3389/fimmu.2016.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goraya MU, Wang S, Munir M, Chen JL. 2015. Induction of innate immunity and its perturbation by influenza viruses. Protein Cell 6:712–721. doi: 10.1007/s13238-015-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heim MH. 2013. Innate immunity and HCV. J Hepatol 58:564–574. doi: 10.1016/j.jhep.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Domingo-Calap P, Segredo-Otero E, Duran-Moreno M, Sanjuan R. 2019. Social evolution of innate immunity evasion in a virus. Nat Microbiol 4:1006–1013. doi: 10.1038/s41564-019-0379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson C, Hargest V, Cortez V, Meliopoulos VA, Schultz-Cherry S. 2017. Astrovirus pathogenesis. Viruses 9:22. doi: 10.3390/v9010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koci MD, Moser LA, Kelley LA, Larsen D, Brown CC, Schultz-Cherry S. 2003. Astrovirus induces diarrhea in the absence of inflammation and cell death. J Virol 77:11798–11808. doi: 10.1128/jvi.77.21.11798-11808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woode GN, Pohlenz JF, Gourley NE, Fagerland JA. 1984. Astrovirus and Breda virus infections of dome cell epithelium of bovine ileum. J Clin Microbiol 19:623–630. doi: 10.1128/JCM.19.5.623-630.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebire NJ, Malone M, Shah N, Anderson G, Gaspar HB, Cubitt WD. 2004. Pathology of astrovirus associated diarrhoea in a paediatric bone marrow transplant recipient. J Clin Pathol 57:1001–1003. doi: 10.1136/jcp.2004.017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guix S, Perez-Bosque A, Miro L, Moreto M, Bosch A, Pinto RM. 2015. Type I interferon response is delayed in human astrovirus infections. PLoS One 10:e0123087. doi: 10.1371/journal.pone.0123087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marvin SA, Huerta CT, Sharp B, Freiden P, Cline TD, Schultz-Cherry S. 2016. Type I interferon response limits astrovirus replication and protects against increased barrier permeability in vitro and in vivo. J Virol 90:1988–1996. doi: 10.1128/JVI.02367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser LA, Schultz-Cherry S. 2008. Suppression of astrovirus replication by an ERK1/2 inhibitor. J Virol 82:7475–7482. doi: 10.1128/JVI.02193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hair PS, Gronemus JQ, Crawford KB, Salvi VP, Cunnion KM, Thielens NM, Arlaud GJ, Rawal N, Krishna NK. 2010. Human astrovirus coat protein binds C1q and MBL and inhibits the classical and lectin pathways of complement activation. Mol Immunol 47:792–798. doi: 10.1016/j.molimm.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Gronemus JQ, Hair PS, Crawford KB, Nyalwidhe JO, Cunnion KM, Krishna NK. 2010. Potent inhibition of the classical pathway of complement by a novel C1q-binding peptide derived from the human astrovirus coat protein. Mol Immunol 48:305–313. doi: 10.1016/j.molimm.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, Ibsen MS, Schmid-Burgk JL, Schmidt T, Ganapathiraju MK, Fujita T, Hartmann R, Barik S, Hornung V, Coyne CB, Sarkar SN. 2014. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi UY, Kang JS, Hwang YS, Kim YJ. 2015. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp Mol Med 47:e144. doi: 10.1038/emm.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eskildsen S, Justesen J, Schierup MH, Hartmann R. 2003. Characterization of the 2′-5′-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res 31:3166–3173. doi: 10.1093/nar/gkg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Méndez E, Aguirre-Crespo G, Zavala G, Arias CF. 2007. Association of the astrovirus structural protein VP90 with membranes plays a role in virus morphogenesis. J Virol 81:10649–10658. doi: 10.1128/JVI.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingle H, Lee S, Ai T, Orvedahl A, Rodgers R, Zhao G, Sullender M, Peterson ST, Locke M, Liu TC, Yokoyama CC, Sharp B, Schultz-Cherry S, Miner JJ, Baldridge MT. 2019. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-lambda. Nat Microbiol 4:1120–1128. doi: 10.1038/s41564-019-0416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santhakumar D, Iqbal M, Nair V, Munir M. 2017. Chicken IFN kappa: a novel cytokine with antiviral activities. Sci Rep 7:2719. doi: 10.1038/s41598-017-02951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong E, Wang X, Chen H, Yang C, Hu J, Liu W, Wang Z, Chen X, Zheng H, Pu J, Sun H, Smith J, Burt DW, Liu J, Li N, Huang Y. 2018. Molecular mechanisms for the adaptive switching between the OAS/RNase L and OASL/RIG-I pathways in birds and mammals. Front Immunol 9:1398. doi: 10.3389/fimmu.2018.01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Wan Z, Li T, Xie Q, Sun H, Chen H, Liang G, Shao H, Qin A, Ye J. 2019. A novel linear epitope crossing group 1 and group 2 influenza A viruses located in the helix A of HA2 derived from H7N9. Vet Microbiol 228:39–44. doi: 10.1016/j.vetmic.2018.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.