Dengue virus (DENV) is a mosquito-borne pathogen that threatens 2.5 billion people in more than 100 countries annually. Dengue infection induces a spectrum of clinical symptoms, ranging from classical dengue fever to severe dengue hemorrhagic fever or dengue shock syndrome; however, the complexities of DENV immunopathogenesis remain controversial. Previous studies have reported the importance of the transcription factor Nrf2 in the control of redox homeostasis and antiviral/inflammatory or death responses to DENV. Importantly, the production of reactive oxygen species and the subsequent stress response have been linked to the development of inflammation and progression toward the more severe forms of the disease. Here, we demonstrate that DENV uses the NS2B3 protease complex to strategically target Nrf2 for degradation, leading to a progressive increase in oxidative stress, inflammation, and cell death in infected cells. This study underlines the pivotal role of the Nrf2 regulatory network in the context of DENV infection.
KEYWORDS: DENV, oxidative stress, Nrf2, interferon, inflammation, NS2B3, dengue, immunometabolism
ABSTRACT
Dengue virus (DENV) is a mosquito-borne virus that infects upward of 300 million people annually and has the potential to cause fatal hemorrhagic fever and shock. While the parameters contributing to dengue immunopathogenesis remain unclear, the collapse of redox homeostasis and the damage induced by oxidative stress have been correlated with the development of inflammation and progression toward the more severe forms of disease. In the present study, we demonstrate that the accumulation of reactive oxygen species (ROS) late after DENV infection (>24 hpi) resulted from a disruption in the balance between oxidative stress and the nuclear factor erythroid 2-related factor 2 (Nrf2)-dependent antioxidant response. The DENV NS2B3 protease complex strategically targeted Nrf2 for degradation in a proteolysis-independent manner; NS2B3 licensed Nrf2 for lysosomal degradation. Impairment of the Nrf2 regulator by the NS2B3 complex inhibited the antioxidant gene network and contributed to the progressive increase in ROS levels, along with increased virus replication and inflammatory or apoptotic gene expression. By 24 hpi, when increased levels of ROS and antiviral proteins were observed, it appeared that the proviral effect of ROS overcame the antiviral effects of the interferon (IFN) response. Overall, these studies demonstrate that DENV infection disrupts the regulatory interplay between DENV-induced stress responses, Nrf2 antioxidant signaling, and the host antiviral immune response, thus exacerbating oxidative stress and inflammation in DENV infection.
IMPORTANCE Dengue virus (DENV) is a mosquito-borne pathogen that threatens 2.5 billion people in more than 100 countries annually. Dengue infection induces a spectrum of clinical symptoms, ranging from classical dengue fever to severe dengue hemorrhagic fever or dengue shock syndrome; however, the complexities of DENV immunopathogenesis remain controversial. Previous studies have reported the importance of the transcription factor Nrf2 in the control of redox homeostasis and antiviral/inflammatory or death responses to DENV. Importantly, the production of reactive oxygen species and the subsequent stress response have been linked to the development of inflammation and progression toward the more severe forms of the disease. Here, we demonstrate that DENV uses the NS2B3 protease complex to strategically target Nrf2 for degradation, leading to a progressive increase in oxidative stress, inflammation, and cell death in infected cells. This study underlines the pivotal role of the Nrf2 regulatory network in the context of DENV infection.
INTRODUCTION
Dengue virus (DENV), the leading arthropod-borne virus infection in the world, represents a major global human health concern. DENV is endemic in more than 100 countries, with as many as 3 billion people in tropical regions of the world at risk of infection (1–4). Among the estimated 300 million annual cases, the majority of infected individuals develop a self-limiting febrile illness, but approximately 500,000 clinical cases result in more-severe manifestations, such as DENV-induced hemorrhagic fever (DHF) and DENV-induced shock syndrome (DSS) (4), leading to 25,000 to 50,000 deaths per year (1).
Four antigenically distinct DENV serotypes (DENV-1 to -4) can be transmitted to humans via the bite of mosquitoes of the genus Aedes; infection with one strain confers durable protection against the same strain of the virus, whereas a secondary infection with a different serotype can provoke a pathological phenomenon known as antibody-dependent enhancement (ADE) (5). The antibodies generated during the first infection bind, but fail to neutralize, the heterologous DENV strain; coated with nonneutralizing antibodies, the DENV particles are facilitated to enter phagocytic cells expressing Fcγ receptor on the surface, resulting in increased viral replication (6, 7). ADE is associated with the most-severe manifestations of dengue fever, although many other factors contribute to the progression of the disease; aberrant production of proinflammatory cytokines increases the risk of vascular leakage, and high levels of circulating interleukin 1β (IL-1β) and tumor necrosis factor alpha (TNF-α) in DENV-infected patients correlate with severe dengue fever (8). Another validated prognostic marker is NS1; high levels of the circulating nonstructural DENV protein 1 have been correlated with increased vascular permeability and severe disease (9). NS1 differentially modulates dengue pathogenesis by directly activating monocytes/macrophages to secrete cytokines that disturb endothelial integrity, while also promoting platelet aggregation, which can cause thrombocytopenia and hemorrhage (10–12).
DENV alters the homeostatic redox conditions within infected cells (13–15). Oxidative stress induces endothelial dysfunction and cell death and hence alters vascular homeostasis. Evidence for oxidative injury comes predominantly from the measurement of biochemical markers of lipid peroxidation and protein oxidation, including malondialdehyde (MDA) and protein carbonyls, the by-products of oxidation of lipids and proteins, respectively. In DENV-infected patients, the level of oxidative stress was found to be maximally increased in DSS, followed by DHF, and minimal in dengue fever (DF) (16). However, there is no clear explanation as to how DENV infection induces oxidative stress, although previous studies have reported lower levels of antioxidant markers such as catalase (CAT), superoxide dismutase (SOD), glutathione peroxidase (GPx), reduced glutathione (GSH), and paraoxonase in the peripheral circulation of dengue study subjects (17, 18). Thus, one possible mechanism of oxidative stress could be the downregulation of endogenous antioxidant enzymes during dengue infection.
Many of these antioxidant enzymes are controlled by the Nrf2 (nuclear factor erythroid 2-related factor 2)–Keap1 (kelch-like ECH-associated protein 1) pathway (19–21). Under resting conditions, Nrf2 and Keap1 form an inactive cytosolic complex, which is targeted for constitutive degradation by the ubiquitin-proteasome pathway (22). Oxidative stress alters the conformation of Keap1, resulting in the release and stabilization of Nrf2; following translocation to the nucleus, Nrf2 forms a heterodimer with small Maf (sMaf) proteins and activates a network of genes containing an antioxidant response element (ARE) (23, 24). This network of antioxidant and cytoprotective proteins includes antioxidant enzymes and anti-inflammatory mediators, as well as proteins involved in the regulation of autophagy and the clearance of damaged proteins (25–27). Activation of Nrf2 also leads to the upregulation of metabolic proteins involved in the synthesis of glutathione, the main intracellular small-molecule antioxidant (28, 29), and NADPH, which provides reducing equivalents for the regeneration of GSH from its oxidized form, glutathione disulfide (GSSG) (30–33).
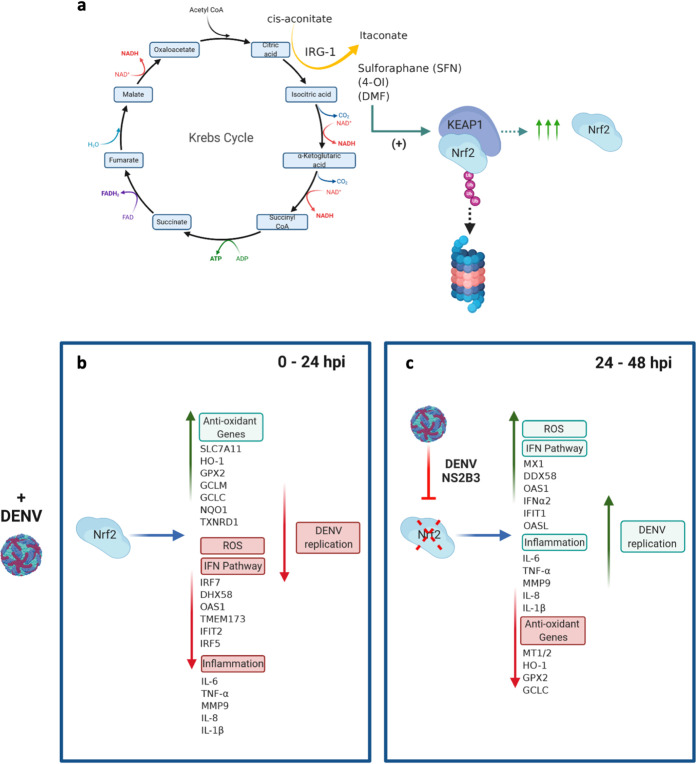
In our previous studies, DENV infection of monocyte-derived dendritic cells (Mo-DC) triggered an NADPH-oxidase (NOX)-dependent oxidative stress response that was required for the activation of interferon regulatory factor 3 (IRF3)/signal transducer and activator of transcription 1 (STAT1)- and NF-κB-mediated antiviral responses and for mitochondrion-dependent apoptosis (34). Furthermore, transcription factor Nrf2 and the antioxidant gene network were shown to regulate both antiviral and inflammatory gene responses by feedback modulation of oxidative stress. Expression of the Nrf2-mediated antioxidant gene network suppressed both antiviral and inflammatory gene expression, highlighting the interplay between oxidative stress and innate immune responses (34). Recently, the regulatory importance of Nrf2 was highlighted with observations that Nrf2 also functioned as a link between cellular metabolism and innate immunity (see Fig. 8a) (23, 33, 35). The endogenous metabolite itaconate, a by-product of Krebs cycle metabolism, directly modified Keap1 via alkylation of cysteine residues, thus enabling Nrf2 release and the induction of genes with antioxidant and anti-inflammatory capacities (35). Furthermore, type I interferon (IFN) increased the expression of IFN-regulated gene 1 (Irg1), also known as cis-aconitate decarboxylase (ACOD1), whose proximal enzymatic activity is required to convert cis-aconitate to itaconate (36). Conversely, itaconate production limited type I IFN responses, as well as IL-1β transcription, thus characterizing a negative-feedback loop that involved IFN and itaconate (35). Subsequently, it was shown that Nrf2 activation decreased the expression of the cytosolic DNA sensor STING (stimulator of interferon genes) and increased the susceptibility of cells to infection with DNA viruses (37). Mechanistically, Nrf2 regulated STING expression by decreasing STING mRNA stability, thus positioning Nrf2 at the interface between metabolic reprogramming and the control of innate immune responses.
FIG 8.
Nrf2 as a link between host metabolism and antiviral immunity. The Nrf2 transcription factor and the antioxidant network sit at the interface between metabolism and immunity. (a) Schematic of the tricarboxylic acid cycle (also called the Krebs cycle), illustrating the production of itaconate from cis-aconitate, mediated by Irg1. Increased itaconate production can also be mimicked by the isothiocyanate sulforaphane (SFN), the water-soluble compound 4-octyl itaconate (4-OI), or the FDA-approved compound dimethyl fumarate (DMF). Treatment with these compounds leads to the release of Nrf2 from KEAP1 and to Nrf2 stabilization, followed by transcriptional upregulation of the antioxidant gene network. (b) DENV infection likewise triggered Nrf2 release during the first 24 hpi; in turn, the Nrf2 response restricted the type I IFN response, decreased inflammatory gene transcription, and reduced ROS accumulation (34–37). (c) At later times after infection (24 to 48 hpi), the virally encoded NS2B3 protease complex inhibited Nrf2, resulting in decreased antioxidant gene expression, increased antiviral and inflammatory gene expression, and ROS accumulation, accompanied by increased dengue virus replication.
In the present study, we asked how dengue infection might alter the balance between oxidative stress and innate immune responses. By monitoring the kinetics of the antiviral, inflammatory, metabolic, and antioxidant responses in DENV-infected Mo-DC, we identified increases in the levels of reactive oxygen species (ROS) and in antiviral and inflammatory gene expression, coupled with DENV NS2B3-mediated degradation of the Nrf2 antioxidant transcription factor. As a consequence, DENV-mediated turnover of Nrf2 inhibited the expression of antioxidant response genes, including the antiviral heme oxygenase 1 (HO-1) gene, and provoked elevated levels of ROS-associated inflammatory and cell death responses.
RESULTS
Kinetic analysis of the innate immune and cellular stress responses to DENV infection.
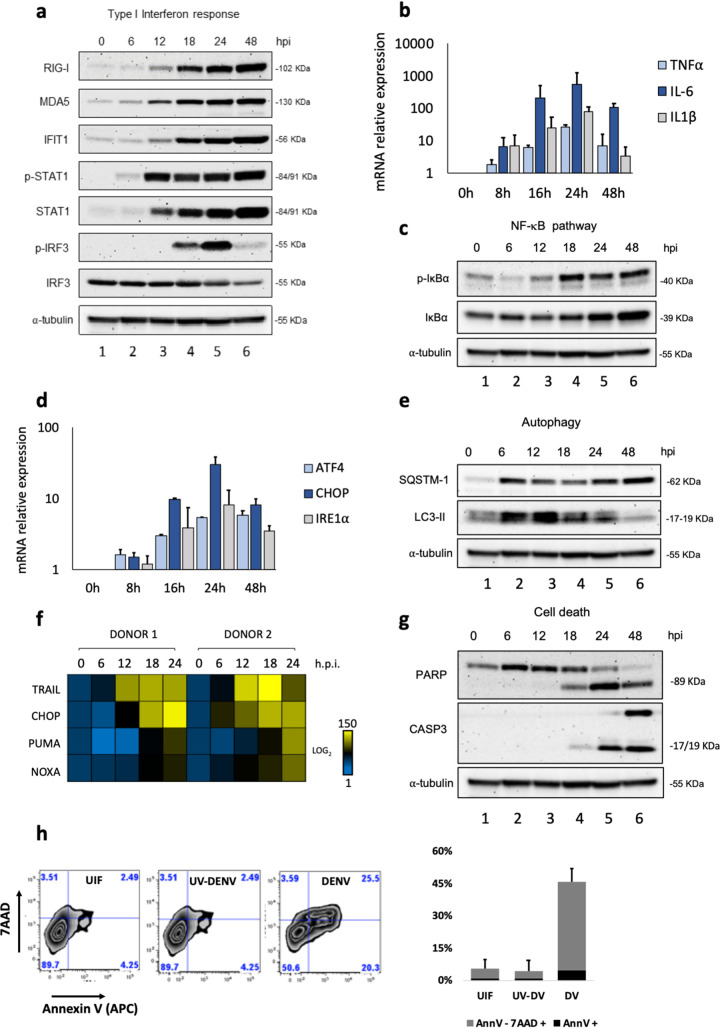
Dendritic cells (DC) in the blood and Langerhans cells in the dermal skin layers are among the first cellular targets of DENV infection and produce several inflammatory cytokines that contribute to endothelial dysfunction and vascular leakage (38). As described previously, we used primary human monocyte-derived dendritic cells (Mo-DC), differentiated in vitro with granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 to immature DC, defined by the expression of characteristic surface markers—DC-SIGNhigh, CD1ahigh, CD14very low—as a model with which to study the host cellular response to infection with the attenuated strain DENV-2 NGC (34). To assess the ability of Mo-DC to sense and respond to DENV infection, the kinetics of type I interferon signaling were evaluated; multiple components of the antiviral response were differentially stimulated as early as 6 h postinfection (hpi), including STAT1 phosphorylation and induction of the cytosolic RNA sensors retinoic acid-inducible gene 1 protein (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) (Fig. 1a). In parallel, activation of NF-κB-dependent inflammatory genes, including the TNF-α, IL-6, and IL-1β genes, was detected within 12 to 24 h (Fig. 1b), and as a reflection of NF-κB signaling, phosphorylation of the regulatory inhibitor IκBα was detected at 12 hpi (Fig. 1c).
FIG 1.
DENV-associated host innate immune and stress responses in Mo-DC. Human Mo-DC were infected with DENV at an MOI of 1, and samples of cells were collected at 0, 6, 12, 18, 24, and 48 hpi. (a) Protein analysis by Western blotting of primary Mo-DC treated with DENV-2 (MOI, 1). Detection of endogenous human RIG-I, MDA5, IFIT1, STAT1, and IRF3 proteins was used to address the activation of the type I interferon pathway. An anti-α-tubulin antibody was used to measure total loaded protein in the Western blot. Results for all Western blots are representative of two independent experiments. (b) TNF-α, IL-6, and IL-1β gene expression levels were evaluated by qPCR analysis; results are representative of three independent experiments and are expressed as means ± SD from three biological replicates. (c) Antibodies against phosphorylated IκBα or total IκBα (Cell Signaling) were used to determine the levels of IκBα and phospho-IκBα, as a measure of NF-κB activation. (d) ATF4, CHOP, and IRE1α gene expression levels were evaluated by qPCR analysis. (e) Antibodies against SQSTM-1 and LC3-II were used as markers for the induction of autophagy. (f) High-throughput qPCR Biomark analysis of the expression of selected apoptotic genes. Gene expression levels were calculated using the ΔΔCT method. The scale represents the log2 values, where yellow shows upregulation, and blue shows downregulation, in gene expression. Data are representative of one experiment performed on two individual donors (donor 1 and donor 2). (g) PARP and caspase-3 antibodies were used as measures of cell death. (h) Apoptosis was quantified by combined staining with annexin V and 7-AAD, and fluorescence was analyzed at 48 hpi using flow cytometry. Percentages are shown for early apoptotic cells (annexin V positive) and late apoptotic/necrotic cells (positive for both annexin V and 7-AAD). UIF, uninfected. Data are representative of the results of three independent experiments.
DENV replication in endoplasmic reticulum (ER)–mitochondrial-membrane junctions caused new membrane synthesis and rearrangement through the induction of unfolded protein response (UPR) genes (39). Mo-DC rapidly upregulated transcription of the UPR genes—ATF4, C/EBP-homologous protein (CHOP), and IRE1α (Fig. 1d)—with temporal kinetics similar to those of inflammatory genes. In addition to the antiviral and inflammatory responses and the UPR, increased autophagy stimulation (Fig. 1e) and transcriptional upregulation of several markers of intrinsic apoptosis, including the death receptor TRAIL (TNF-related apoptosis-inducing ligand) and the BH3-only proapoptotic genes NOXA and PUMA (p53 upregulated modulator of apoptosis), were detected in response to DENV infection (Fig. 1f). Likewise, cleaved forms of poly(ADP-ribose) polymerase (PARP) and caspase-3 (Fig. 1g), markers of ongoing apoptosis, were detected in DENV-infected Mo-DC. Fluorescence-activated cell sorter (FACS) analysis confirmed the cytotoxic effects of DENV: ∼50% of the Mo-DC were actively undergoing early or late stages of apoptosis at 48 hpi, as indicated by the increase in the number of annexin V-positive or annexin V- and 7-aminoactinomycin D (7-AAD)-positive cells, respectively (Fig. 1h, left); no cytotoxicity was observed after infection with UV-inactivated DENV (Fig. 1h, right). Clearly, Mo-DC induced multiple host response pathways in response to DENV infection, including antiviral, inflammatory, ER stress, and cell death programs.
DENV increases oxidative stress in primary human DC.
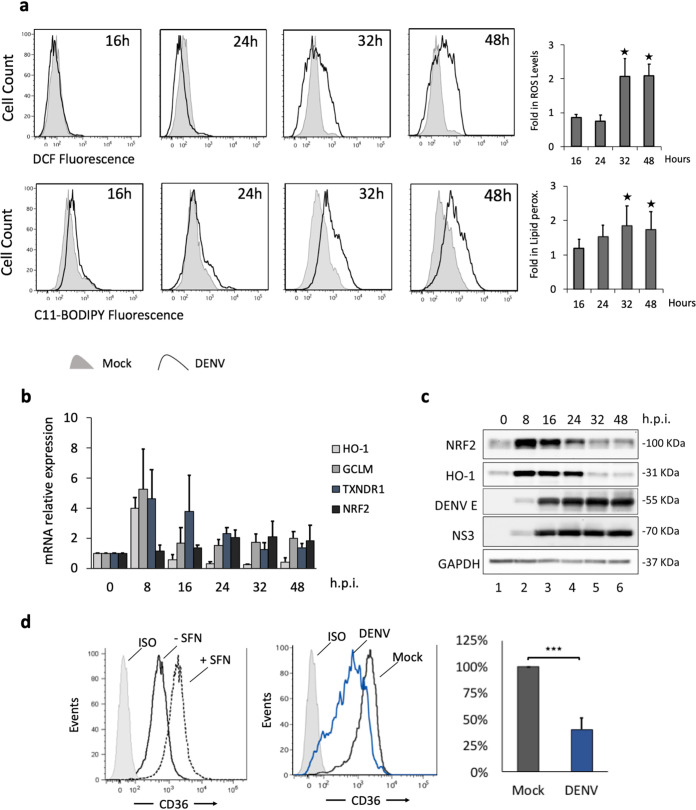
DENV infection also stimulated a 2-fold increase in the production of reactive oxygen species, as evaluated using the fluorescent detection probe CM-H2DCFDA (Fig. 2a, top); given that dead cells also produce ROS, this population was excluded by 7-AAD staining. ROS induction was observed mainly during the late phase of infection (after the initiation of viral replication) between 32 and 48 hpi, indicating that DENV infection effectively perturbed redox homeostasis. To further examine the consequences of ROS elevation in Mo-DC, the fluorescence emission of the Bodipy 581/591-C11 probe was used to detect lipid peroxidation; because Bodipy 581/591-C11 contains alkene groups that mimic unsaturated fatty acids, this probe serves a substrate for peroxidation (40). By 48 hpi, lipid peroxidation was increased >2-fold (Fig. 2a, bottom), further illustrating the induction of oxidative stress.
FIG 2.
DENV increases oxidative stress in primary human DC. Human Mo-DC were either infected with DENV-2 NGC at an MOI of 1 or mock infected, and cells were collected at 16, 24, 32, and 48 hpi. (a) (Top) The accumulation of ROS was measured by flow cytometry using CM-H2DCFDA (1 μM) at the indicated times. (Bottom) Lipid peroxidation was measured by using C11-Bodipy (1 μM). Data represent means from at least three different donors. P values and fold increases were determined based on comparison with uninfected cells. (b) HO-1, GCLM, TXNDR1, and NRF2 gene expression levels were evaluated by qPCR analysis, normalized to GAPDH expression, and described relative to control levels (0 hpi). Results are representative of three different donors and are expressed as means ± SD. (c) Protein analysis by Western blotting of primary Mo-DC treated with DENV-2 (MOI, 1). Endogenous human NRF2 and HO-1 proteins were detected after 0, 8, 16, 24, 32, and 48 hpi. DENV infection was detected using a polyclonal rabbit antibody raised against the DENV-2 E protein or the DENV-2 NS3 protein. An anti-GAPDH antibody was used to measure total loaded protein in the Western blot. Results are representative of two different donors. (d) (Left) Mo-DC were examined for CD36 expression by flow cytometry after 24 h of treatment with sulforaphane (10 μM). ISO, •••••••. (Center) Mo-DC were infected with DENV at an MOI of 1 or were mock infected, and samples were collected at 40 hpi prior to FACS analysis. (Right) Downregulation of CD36 was measured as a percentage determined based on comparison with levels in mock-infected cells. Data are means of results from three independent donors.
To determine how ROS production in DENV-infected Mo-DC altered the expression of the antioxidant defense pathways, the transcription of the NFE2L2 gene, encoding the Nrf2 antioxidant transcription factor, and of genes encoding a subset of Nrf2 downstream effectors (the glutamate-cysteine ligase modifier subunit [GCLM], involved in glutathione synthesis; thioredoxin reductase 1 [TXNRD1], a member of the thioredoxin system; and heme oxygenase 1 [HO-1], involved in the catabolism of heme) was measured by real-time PCR. By 8 h after DENV infection, the GCLM, TXNRD1, and HO-1 antioxidant genes were induced 4- to 6-fold (Fig. 2b), while by 16 h and thereafter, antioxidant response gene expression decreased to near-baseline levels. In contrast to differential regulation at the RNA level, both the Nrf2 and HO-1 protein levels were induced by DENV infection between 8 and 24 h (Fig. 2c, lanes 2 to 4) but decreased thereafter to near-baseline levels (Fig. 2c, lanes 5 and 6). Both the DENV envelope (E) and DENV NS3 proteins were detected at 8 hpi and accumulated thereafter in DENV-infected Mo-DC (Fig. 2c, lanes 2 to 6).
The expression of CD36, an Nrf2-regulated anti-inflammatory scavenger receptor for oxidized low- density lipoprotein and malaria-infected erythrocytes (41–43), was analyzed at 40 h, and the effect of DENV on CD36 expression was determined when Nrf2 and HO-1 protein levels were low (Fig. 2d). DENV infection reduced CD36 expression by almost 60%, confirming the inhibition of Nrf2 activity. On the other hand, treating Mo-DC with sulforaphane (SFN), a natural isothiocyanate compound known to increase Nrf2 levels via Keap1 cysteine modification (44), increased CD36 expression, in agreement with the specific involvement of Nrf2 in CD36 regulation (43).
Nrf2 KO increased DENV replication and inflammatory gene expression but suppressed host metabolic genes.
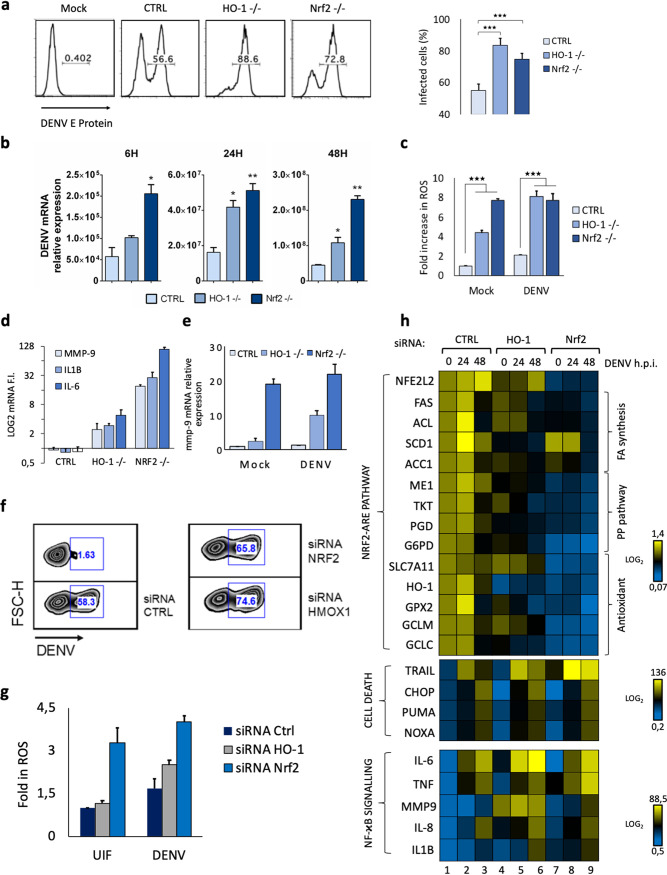
To evaluate the impact of Nrf2 signaling on DENV replication, cell metabolism, and the host innate response, we generated A549 cells in which Nrf2 or the Nrf2-responsive HO-1 gene was stably knocked out. Knockout (KO) of either Nrf2 or HO-1 increased the number of infected cells at 24 hpi from ∼50% to >80%, as determined by DENV E protein accumulation (Fig. 3a); measurement of viral RNA expression at 6, 24, and 48 hpi further confirmed this observation, demonstrating that knockout of either Nrf2 or HO-1 increased viral RNA levels ∼2- to 4-fold (Fig. 3b). Similar results were also observed in Nrf2 or HO-1 knockdown cells (Fig. 3f). In the absence of HO-1, the basal mRNA levels of IL-1β, IL-6, and matrix metalloproteinase 9 (MMP-9) genes were markedly increased, by ∼6- to ∼30-fold (Fig. 3d), while Nrf2 KO presented a >100-fold increase in basal levels of these inflammatory markers, indicating that the Nrf2-related pathway controlled the basal expression of these inflammatory genes. As seen in Fig. 3e, DENV infection further increased MMP-9 mRNA expression by ∼10-fold in HO-1 KO cells, but not in Nrf2 KO cells, where MMP-9 was highly expressed in the absence of DENV infection (Fig. 3d and e). In terms of ROS production, HO-1 KO increased basal ROS levels ∼4-fold, and with DENV infection, ROS levels increased ∼8-fold in HO-1–/– cells. In contrast, Nrf2 KO completely deregulated ROS production, resulting in an ∼8-fold increase in basal ROS levels that were not further increased by DENV infection (Fig. 3c). Similar results were observed in cells silenced for Nrf2 and HO-1 (Fig. 3g).
FIG 3.
Nrf2 knockout increased DENV replication and inflammatory gene expression but suppressed host metabolic genes. (a) The percentage of infected cells among A549 cells with HO-1 or Nrf2 knockout was determined by flow cytometry at 24 hpi using a mouse IgG2a MAb specific for DENV E protein (clone 4G2). (b) DENV RNA expression levels were evaluated by qPCR analysis in A549 wild-type and knockout cells after 6, 24, and 48 h of infection. Gene expression was normalized to GAPDH expression and is presented as RNA levels relative to those in uninfected cells, set by default at 1. Results are representative of three different donors and are expressed as means ± SD. (c) The mean fluorescence intensities, measured by flow cytometry of A549 knockout cells after 48 h following infection with DENV at an MOI of 1, were used to calculate the fold increase in ROS levels over those in mock-infected cells. The frequencies of ROS-positive cells were determined by gating against stained live cells (7-AAD negative). Results are representative of three independent experiments and are expressed as means ± SD from three biological replicates. (d) Gene expression levels of MMP-9, IL-1β, and IL-6 were evaluated in A549 knockout cells by qPCR analysis. (e) Comparison of MMP-9 gene expression between mock-infected and DENV-infected (at an MOI of 1) A549 knockout cells. Results were normalized to GAPDH levels and are expressed as RNA levels relative to those for the mock-infected control, set by default at 1. (f) The percentage of infected cells in Nrf2- or HO-1 silenced A549 cells was determined by flow cytometry using a mouse IgG2a MAb specific for DENV E protein (clone 4G2). (g) The mean fluorescence intensities, measured by flow cytometry in Nrf2- or HO-1-silenced A549 cells 48 h after infection with DENV at an MOI of 10, were used to calculate the fold increase in ROS levels over those in mock-infected cells. The frequencies of ROS-positive cells were determined by gating against stained live cells (7-AAD negative). Results are representative of three independent experiments and are expressed as means ± SD from three biological replicates. (h) High-throughput analysis of gene expression evaluated by qPCR Biomark analysis. mRNA analysis was performed using a customized Biomark chip encompassing selected Nrf2 target genes, cell death-related genes, and NF-κB-dependent genes. Gene expression levels were calculated using the ΔΔCT method. The scale represents log2 values, where yellow shows an upregulation and blue a downregulation in gene expression. Data are means for two independent experiments.
Considering that Nrf2 regulates aspects of host metabolism such as lipogenesis and the pentose phosphate pathway (PPP), we investigated how Nrf2 or HO-1 ablation would impact the expression of metabolic, inflammatory, and apoptotic markers following DENV infection. For this purpose, a nanoscale Biomark assay was used to measure selected genes at 24 h and 48 h after DENV infection (Fig. 3h). The assay contained representative genes of the pentose phosphate pathway (ME1, ACC1, phosphogluconate dehydrogenase [PGD], transketolase isoform 1 [TKT], glucose-6-phosphate dehydrogenase [G6PD]), fatty acid synthesis genes (repressed by Nrf2) (fatty acid synthase [FAS], stearoyl coenzyme A desaturase [SCD1], ATP-citrate lyase [ACL]), and antioxidant response genes (glutamate–cysteine ligase catalytic [GCLC], GCLM, solute carrier family 7 member 11 [SLC7A11], glutathione peroxidase 2 [GPX2], HO-1). Genes associated with cell death (TRAIL, CHOP, PUMA, NOXA) and inflammation (IL-6, TNF, MMP-9, IL-8, IL-1β) were upregulated in both HO-1 (Fig. 3h, lanes 4 to 6) and Nrf2 (Fig. 3h, lanes 7 to 9) knocked-down A549 cells, particularly at 48 h. In cells lacking Nrf2, the expression of genes associated with cell metabolism and the antioxidant response was silent and was not induced by DENV infection (Fig. 3h, lanes 7 to 9). At the same time, the expression of inflammatory and death response networks predominated in DENV-infected HO-1 and Nrf2 knockdown cells (Fig. 3h, lanes 4 to 9).
Nrf2 or HO-1 expression interferes with DENV infection.
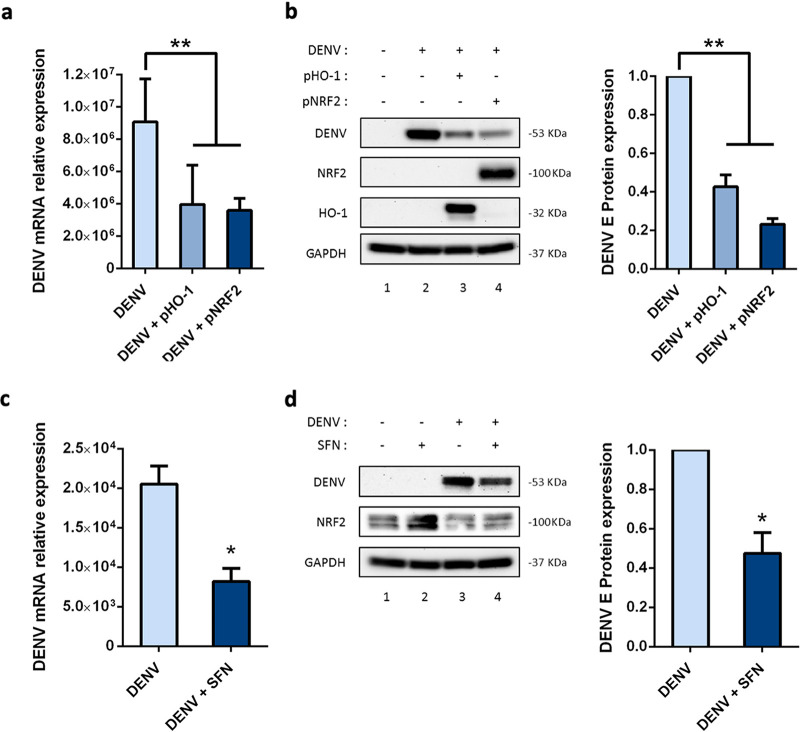
We next evaluated the effects of Nrf2 and HO-1 overexpression on DENV replication by comparing Huh-7 cells transfected with Nrf2 or HO-1 expression plasmids to Huh-7 cells transfected with an empty plasmid vector. Both Nrf2 and HO-1 reduced DENV infection, as demonstrated by a 2- to 3-fold reduction in viral RNA levels (Fig. 4a) and a higher reduction in the expression of DENV E protein (Fig. 4b), with Nrf2 overexpression leading to greater inhibition than HO-1 (Fig. 4b, right). The pharmacological activation of endogenous Nrf2 prior to DENV infection recapitulated the effects of exogenous Nrf2 expression; SFN activation of Nrf2 moderately decreased viral RNA and E protein when administered to Huh-7 cells (Fig. 4c and d).
FIG 4.
Nrf2 overexpression limited DENV infection. (a) Huh-7 cells were transfected for 24 h with an empty vector (0.5 μg) or an Nrf2 (0.5 μg) or HO-1 (0.5 μg) expression plasmid, infected for 24 h with DENV (MOI, 5), and then evaluated by qPCR analysis for DENV RNA expression levels, which were normalized to GAPDH levels and presented as RNA levels relative to those in uninfected cells, set by default at 1. (b) Cells were evaluated for NRF2, HO-1, and DENV-2 E protein expression by Western blotting (left panel) and were evaluated as described for panel a. DENV infection was detected using an antibody against DENV-2 E protein, and total protein was normalized with an anti-GAPDH antibody; the right panel provides a quantification of DENV-2 E protein levels, expressed as the fold difference over the control. Results are representative of two independent experiments and are expressed as means ± SD. (c) Analysis of DENV RNA expression in Huh-7 cells after pharmacological stimulation of Nrf2 with SFN (20 μM). Cells were treated for 8 h with SFN or DMSO (control) and were then infected for 24 h with DENV (MOI, 5). Viral RNA expression levels were measured by qPCR analysis, normalized to GAPDH levels, and expressed as RNA levels relative to those in uninfected cells, set by default at 1. (d) Quantification of NRF2 and DENV-2 E protein expression by Western blotting (left panel). DENV infection was detected using an antibody against DENV-2 E protein, and total protein was normalized with an anti-GAPDH antibody; the right panel provides a quantification of DENV-2 E protein levels, expressed as the fold difference over the control. Results are representative of two independent experiments and are expressed as means ± SD.
DENV infection targets Nrf2 for degradation.
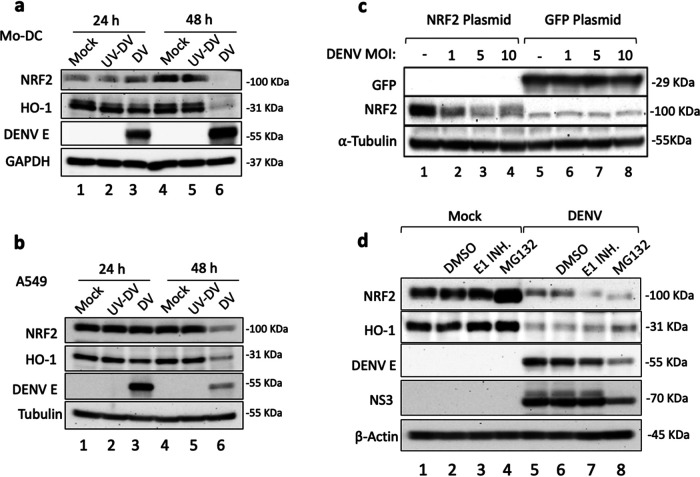
We next addressed the possibility that DENV infection specifically altered Nrf2 expression; in both Mo-DC and A549 cells, de novo DENV infection reduced Nrf2 and HO-1 protein levels between 24 and 48 h (Fig. 5a and b, lanes 6), whereas UV-inactivated DENV failed to reduce Nrf2 levels (Fig. 5a and b, lanes 2 and 5), indicating that active virus replication was required to interfere with Nrf2 and HO-1 expression. To test DENV-induced turnover of Nrf2, HEK293T cells expressing either Nrf2 or green fluorescent protein (GFP) as a control were infected with DENV at different multiplicities of infection (MOI); a dose-dependent reduction in Nrf2 protein levels (Fig. 5c, lanes 1 to 4), but not in GFP (Fig. 5c, lanes 5 to 8), was observed in infected cells, supporting the idea that DENV specifically interfered with Nrf2 expression. Finally, using pharmacological inhibitors—the proteasome inhibitor MG132 or an E1 ubiquitin inhibitor (PYR41)—we tested whether blockade of ubiquitination or proteasome activity reversed Nrf2 degradation (Fig. 5d); although MG132 appeared to stabilize Nrf2 turnover in control cells (Fig. 5d, compare lanes 1 and 4), inhibitor treatment did not reverse Nrf2 downregulation (Fig. 5d, lanes 7 and 8), arguing that ubiquitination and proteasome-mediated degradation were not involved in DENV-induced Nrf2 turnover.
FIG 5.
DENV infection targets Nrf2 for degradation. (a and b) Mo-DC (a) and A549 cells (b) were either left uninfected or infected with UV-DENV or DENV at an MOI of 10. Cells were collected at 24 and 48 hpi for Western blot analysis of endogenous human NRF2 and HO-1 proteins. DENV infection was detected using an antibody against DENV-2 E protein, and total protein was normalized with an anti-GAPDH or an anti-α-tubulin antibody. (c) HEK293T cells were transfected with 500 ng of a plasmid expressing human Nrf2 or GFP. After 4 h, cells were either mock infected or infected with DENV at the indicated MOI. Nrf2 protein and GFP were analyzed by Western blotting 48 hpi using anti-Nrf2 and GFP antibodies; an anti-α-tubulin antibody was used to normalize total protein loaded in the blot. Results are representative of two independent experiments. (d) A549 cells were either mock infected or infected with DENV at an MOI of 1, and at 24 h, cells were treated with DMSO (vehicle control), the E1 ubiquitin enzyme PYR41 (1 μM), or the proteasome inhibitor MG132 (5 μM). At 48 h, whole-cell extracts were prepared from infected cultures, and Nrf2 and HO-1 proteins were detected by SDS-PAGE and Western blotting. Results are representative of two independent experiments.
DENV NS2B3 protease complex promotes Nrf2 degradation.
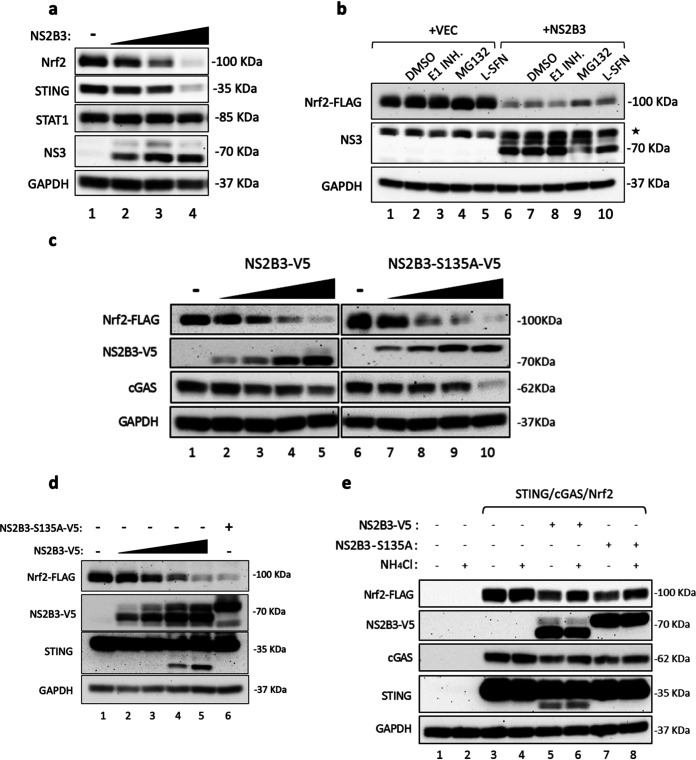
To test the idea that specific viral activities may be involved in Nrf2 turnover, particularly given the observation that the DENV protease cleaved both the ER-localized proteins STING and FAM134B (45, 46), the effects of NS2B3 protease on Nrf2 and STING were evaluated; both proteins displayed concentration-dependent degradation in the presence of increasing concentrations of NS2B3, whereas STAT1 was not subject to DENV NS2B3-dependent degradation (Fig. 6a). Cells expressing Flag-Nrf2 and DENV-2 NS2B3 were treated with an E1 ubiquitin inhibitor or the proteasome inhibitor MG132, or with SFN, a well-known activator of Nrf2 (Fig. 6b). Again, NS2B3 expression alone induced Nrf2 degradation, which was not prevented by blockade of ubiquitination or proteasome activity (Fig. 6b, lanes 6 to 10), indicating that Nrf2 degradation was NS2B3 dependent. Additionally, SFN stimulation was not sufficient to recover Nrf2 from NS2B3-mediated degradation (Fig. 6b, lane 10).
FIG 6.
The DENV NS2B3 protease complex mediates Nrf2 degradation. (a) 293T cells were cotransfected with human Nrf2, STING, and different amounts of an NS2B3 plasmid (0.5, 1, and 2 μg). Protein expression was analyzed at 24 h by Western blotting using anti-Nrf2, anti-STING, and anti-NS3 antibodies. An anti-STAT1 antibody was used to detect endogenous STAT1 protein. GAPDH was used as a loading control. (b) Coexpression of a human Nrf2-FLAG plasmid (1 μg) and WT DENV NS2B3 (2 μg) or an empty vector (VEC). At 24 h after transfection, cells were treated for 24 h with either DMSO, the E1 ubiquitin enzyme PYR41 (1 μM), or the proteasome inhibitor MG132 (5 μM). Protein expression was analyzed by Western blotting using anti- FLAG, anti-NS3, and anti-GAPDH antibodies. The asterisk indicates a nonspecific band. (c) HEK293T cells were cotransfected with human Nrf2-FLAG (1 μg), cGAS (150 ng), and different amounts of the wild-type DENV NS2B3-V5 plasmid or the protease-dead mutant DENV NS2B3-S135A-V5 plasmid (0, 1, 2, 4, and 10 μg). Cells were harvested 24 h after transfection, and protein expression was analyzed by Western blotting using anti-FLAG, anti-V5, anti-cGAS, and anti-GAPDH antibodies. (d) Coexpression of Nrf2-FLAG and different amounts of WT DENV NS2B3-V5 (0.5, 1, 2, and 5 μg) or NS2B3-S135A-V5 (5 μg) analyzed at 24 h by Western blotting using anti-FLAG, anti-V5, and anti-GAPDH antibodies. (e) 293T cells were cotransfected with human Nrf2 (1 μg), STING (150 ng), cGAS (150 ng), and either WT DENV NS2B3-V5 (2 μg) or NS2B3–S135A-V5 (2 μg). At 16 h after transfection, cells were either left untreated or treated for 8 h with NH4Cl (10 mM). Analysis by Western blotting was performed using anti-FLAG, anti-V5, anti-STING, anti-cGAS, and anti-GAPDH antibodies. Results are representative of three independent experiments.
To determine if Nrf2 degradation was de facto proteolysis, dependent on NS2B3 activity, wild-type (WT) NS2B3 and an equivalent construct expressing the Ser135Ala point mutation of the protease (NS2B3-S135A) (47) were expressed in HEK293T and A549 cells. Again, Nrf2 was degraded in a dose-dependent manner by NS2B3 (Fig. 6c and d, lanes 2 to 5), and surprisingly, Nrf2 degradation also occurred with the protease-dead mutant (Fig. 6c, compare lanes 6 and 10; Fig. 6d, compare lanes 1 and 6). As a control for protease activity, STING was cleaved by NS2B3 (Fig. 6d, lanes 1, 4, and 5), whereas cleavage of STING and autocleavage of NS2B3 were blocked in the presence of NS2B3-S135A (Fig. 6d, compare lanes 5 and 6), demonstrating that Nrf2 degradation was protease activity independent. Subsequently, wild-type NS2B3 and NS2B3-S135A were expressed in A549 cells, and Nrf2 turnover was compared with the degradation of cyclic GMP-AMP synthase (cGAS) by NS2B3 protease (Fig. 6c and e). Nrf2 turnover was stimulated by either the wild-type (Fig. 6c, lanes 2 to 5) or the inactive (Fig. 6c, lanes 7 to 10) form of NS2B3.
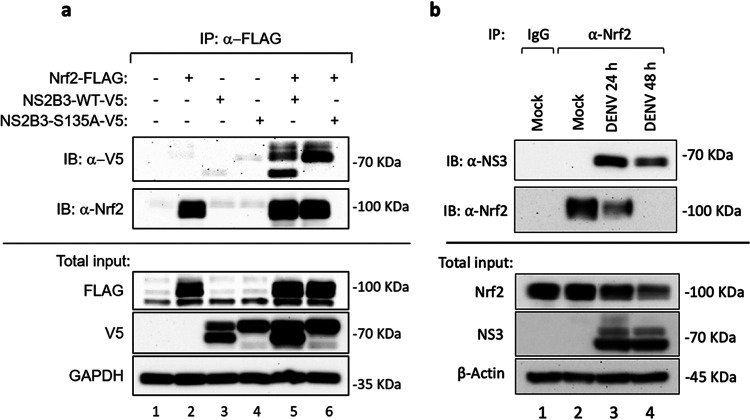
Interestingly, Nrf2 turnover shared similarities with NS2B3-dependent degradation of the DNA sensor cGAS, which was degraded by NS2B3 in a proteolysis-independent manner (Fig. 6c) but was dependent on lysosomal degradation/autophagy (48). In agreement with this observation, addition of the autophagy inhibitor ammonium chloride (NH4Cl), which alters lysosomal pH and blocks phagosome-lysosome fusion (49), resulted in prominent (up to 85%) restoration of Nrf2 expression (Fig. 6e, compare lane 5 with lane 6 and lane 7 with lane 8). Finally, the physical interaction between Nrf2 and NS2B3 was analyzed by immunoprecipitation: Nrf2-FLAG was able to interact with either WT NS2B3 or the S135A mutant (Fig. 7a, lanes 5 and 6). Moreover, Nrf2 was immunoprecipitated at 24 and 48 hpi using an anti-Nrf2 antibody (Fig. 7b, lanes 3 and 4), and subsequent immunoblot analysis revealed a 70-kDa band corresponding to DENV NS3 (Fig. 7b, lanes 3 and 4). Taken together, these results demonstrate that DENV infection antagonized the Nrf2 antioxidant pathway and that DENV NS2B3 protease physically interacted with Nrf2 to induce degradation in a proteolysis-independent manner.
FIG 7.
The DENV NS2B3 protease complex associates with Nrf2. (a) 293T cells were transfected with human Nrf2-FLAG (2 μg), NS2B3-WT-V5 (1 μg), NS2B3-S135A-V5 (1 μg), or an empty vector, alone or in combination. After 16 h, cells were collected and lysed. (Top) Immunoprecipitation (IP) with anti-FLAG was performed, and the amount of coprecipitated NS2B3 (immunoblot [IB]: anti-V5) (upper panel) or Nrf2 (IB: anti-Nrf2) (lower panel) was analyzed by Western blotting. (Bottom) Total lysates were also analyzed for the expression of Nrf2-FLAG by using an anti-FLAG Ab and for the expression of NS2B3-V5 (WT or S135A) by using an anti-V5 Ab. GAPDH was used as a loading control. (b) A549 cells were infected with DENV (MOI, 1), collected after 24 h and 48 h of infection, and lysed. (Top) Immunoprecipitations with anti-Nrf2 or an isotype-matched control IgG Ab were performed, and the amount of coprecipitated DENV NS3 (upper panel) or Nrf2 (lower panel) was analyzed by Western blotting. (Bottom) Total lysates were also analyzed for the expression of Nrf2 and DENV NS3. β-Actin was used as a loading control. Results are representative of two independent experiments.
DISCUSSION
The mechanisms contributing to DENV immunopathogenesis and the development of severe disease, including DHF and DSS, are complex and multifactorial. One factor contributing to DENV pathogenesis is the interplay between oxidative stress damage and DENV infection. In this study, we demonstrated that (i) DENV infection generated a strong intrinsic response to infection in Mo-DC, with the induction of the type I IFN response, induction of NF-κB-directed inflammatory pathways, and upregulation of the unfolded protein response and apoptotic markers; (ii) DENV infection stimulated reactive oxygen species (ROS) production; (iii) expression of both Nrf2 and HO-1 was ablated between 24 and 48 h after infection; and (iv) the viral protease complex NS2B3 targeted Nrf2 protein for degradation between 24 and 48 hpi. As a consequence of the loss of Nrf2, ROS production increased, and the IFN antiviral response and NF-κB-dependent inflammatory responses were no longer inhibited, but the expression of several Nrf2-regulated metabolic enzymes was limited (Fig. 3). These studies suggest that a negative regulatory loop controlled by Nrf2 (50), and the concomitant changes in host metabolic activity, limited the antiviral response during the early stages of DENV infection to facilitate replication (Fig. 8b). At later times (>24 hpi), Nrf2 degradation mediated by the NS2B3 protease complex dissociated the effects of metabolic activity, potentially mediated endogenously through the Krebs cycle metabolite itaconate (34, 35, 50), and the type I IFN response, leading to a rapid increase in ROS production in infected cells as well as increased inflammation, and generally supporting the development of an intracellular environment favoring DENV replication (Fig. 8c). From these observations, we speculate that at 24 to 48 hpi, the proviral effect of ROS overcame the antiviral effects of the IFN response.
Dengue virus, like other members of the Flaviviridae family, uses nonstructural viral proteins, such as NS2A, NS4A, NS4B, and NS5, to interfere with or degrade crucial signaling components in order to circumvent the IFN antiviral response. Common targets of these proteins are the signal transducer and activator of transcription 1 (STAT1) and STAT2, which are required for type I IFN signaling and for induction of the hundreds of antiviral interferon-stimulated genes (ISGs) (51). In addition, DENV actively inhibited the host intracellular sensing pathways that lead to IFN induction (52). An original, and initially enigmatic, observation was that DENV protease complex NS2B3, in addition to its role in processing the DENV polyprotein, also proteolytically cleaved the endoplasmic reticulum (ER)-resident STING protein, which is responsible for triggering the antiviral response to DNA viruses (48, 53). Subsequently, the cyclic GMP-AMP synthase (cGAS) was shown to be degraded by the NS2B3 complex during DENV infection, although in a proteolysis-independent manner (46). cGAS recognized and bound DNA in a sequence-independent manner and synthesized the second messenger cGAMP, which, in turn, bound to and activated STING, leading to type I IFN production. Overall, the autophagy-lysosome-dependent mechanism of cGAS degradation, coupled with the proteolysis-dependent cleavage of STING, appears to be an effective strategy for evading cGAS-STING activation by mitochondrial DNA released during DENV infection (46). As seen in Fig. 6, both Nrf2 and cGAS levels decreased with increasing NS2B3 expression, suggesting that degradation of Nrf2 by NS2B3 may occur by an NS2B-dependent mechanism similar to that identified for cGAS (48).
What, then, is the functional significance of Nrf2 degradation during DENV infection? The effect appears to be twofold: to dissociate host metabolism from innate immune regulation and to inhibit the IFN-independent antiviral activity associated with HO-1 activity. Although HO-1 is well characterized as an Nrf2-inducible protein with antiapoptotic, antioxidant, and anti-inflammatory activities, a number of studies have also demonstrated that HO-1 has antiviral activity and can interfere with the replication of several viruses, such as the human immunodeficiency virus (HIV) (54), influenza virus (55), human respiratory syncytial virus (HRSV) (56), hepatitis B virus (57), hepatitis C virus (HCV) (58), enterovirus 71 (59), dengue virus (60), and Ebola virus (61).
While the mechanisms underlying the antiviral effects of HO-1 have remained elusive for some viruses, in other cases, its antiviral effects have been identified. Among Flaviviridae family members, bilirubin derived from HO-1 has been reported to inhibit the activity of the nonstructural 3/4A protease of HCV (62) and to noncompetitively inhibit the protease of DENV (60). Here, we demonstrated that cells lacking either HO-1 or its transcriptional regulator Nrf2 were more susceptible to DENV infection than their WT counterparts (Fig. 3). Thus, strategic targeting of Nrf2 degradation by DENV may represent an evasion strategy to limit HO-1 antiviral activity during DENV infection.
Oxidative stress is a characteristic of several viral infections, including influenza and HIV (63); for example, generation of ROS and reactive nitrogen species (RNS) contributed to the development of influenza virus-induced pathogenesis in the lung (64–66). In addition, decreased expression of Nrf2 and its downstream effectors, NQO1 and GCLC, has been reported in HIV-1-infected human monocyte-derived macrophages (67). In the Flaviviridae family, HCV downregulated Nrf2 activation by impairing its nuclear import through delocalization of Maf proteins, leading to chronic infection, ROS production, and inflammation (68). Similarly, HRSV induced Nrf2 deacetylation and subsequent proteasomal degradation, which led to the downregulation of antioxidant enzyme expression (69). In the case of DENV, the process is independent of proteasome-mediated degradation but seems to rely on lysosomal degradation, as demonstrated by Nrf2 recovery in the presence of the autophagy inhibitor NH4Cl (Fig. 6e).
A recent study showed that DENV infection selectively activated Nrf2 following nuclear translocation, and after ER stress, protein kinase R-like ER kinase (PERK) facilitated Nrf2-mediated transcriptional activation of C-type lectin domain family 5, member A (CLEC5A), to increase CLEC5A expression. Signaling downstream of the Nrf2–CLEC5A interaction enhanced tumor necrosis factor alpha (TNF-α) production following DENV infection, while forced expression of the NS2B3 viral protein induced Nrf2 nuclear translocation/activation and CLEC5A expression, which increased DENV-induced TNF-α production. These results demonstrate that DENV infection causes Nrf2-regulated TNF-α production by increasing levels of CLEC5A (70).
Enhanced susceptibility to DENV infection was also reported in individuals with a deficiency of glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the pentose phosphate pathway (PPP) that catalyzes the synthesis of NADPH, an important cofactor used for the activity of some Nrf2-dependent antioxidant enzymes (71). G6PD deletion enhanced ROS production, thus linking the cellular oxidative state and susceptibility to DENV infection (71).
Nrf2-dependent protection against oxidative stress relies, in fact, on an intact PPP. Upon Nrf2 activation, glucose is redirected into the PPP through the upregulation of metabolic genes, including the G6PD gene, resulting in an increase in NADPH synthesis. However, interference with the supply of glucose or the PPP and NADPH generation inhibited both Nrf2-mediated ROS detoxification by antioxidant enzymes and Nrf2-dependent gene expression (72). Thus, the Nrf2 antioxidant network provides an interface between redox and intermediary metabolism.
A deficiency in G6PD, the enzyme required to maintain glutathione in the reduced state, led to the production of nitric oxide, superoxide radicals, and viral persistence in DENV-2-infected monocytes (54). Similarly, genetic silencing of Nrf2 in DENV-infected Mo-DC perturbed intracellular redox homeostasis and provoked increased inflammatory and cell death responses (34). These findings suggest that temporal modulation of redox balance is conducive to enhanced virus replication. Thus, induction of Nrf2 activity in DENV-infected cells limited development of the full antiviral response early after infection (<24 hpi), while subsequent cleavage of Nrf2 by NS2B3 created an intracellular environment at later times (24 to 48 hpi) where ROS accumulation and inflammatory gene expression supported viral replication and essentially overcame the inhibitory effects of the IFN response. In conclusion, this study suggests a pivotal role of the Nrf2 regulatory network in the progression of DENV infection (Fig. 8), highlighting the metabolic changes derived from Nrf2 suppression and its effects on the antiviral, inflammatory, and oxidative stress responses.
MATERIALS AND METHODS
Cell lines and culture conditions.
Nrf2 KO A549 cells were a kind gift of Christian K. Holm (Aarhus University) and have been reported previously (37). Using the same parental A549 cells, HO-1 was also knocked out by CRISPR-Cas9 technology. Huh-7 cells were a kind gift of Marco Tripodi (Sapienza University of Rome). HEK293T cells, A549 cells, C6/36 Aedes albopictus mosquito cells, and Vero monkey kidney cells were obtained from the American Type Culture Collection (ATCC). Huh-7 cells, HEK293T cells, C6/36 mosquito cells, and Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) in the presence of 10% fetal bovine serum (FBS; Gibco), 1% l-glutamine (Life Technologies), and 1% antibiotics (Sigma-Aldrich). A549 cells were cultured in F12K medium (Invitrogen) supplemented with 10% FBS.
Monocyte isolation and differentiation into Mo-DC.
Briefly, human peripheral blood mononuclear cells (PBMC) were isolated from freshly collected blood using Ficoll-Paque PLUS medium (GE Healthcare Bio) as per manufacturer’s instructions. CD14+ monocytes were isolated by positive selection using CD14 microbeads and a magnetic cell separator as per kit instructions (Miltenyi Biotec). Purified CD14+ monocytes were seeded in 6-well plates at a concentration of 1 × 106 cells/ml in complete RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum (HyClone), 1% antibiotics, 100 ng/ml GM-CSF, and 50 ng/ml IL-4 at 37°C (under 5% CO2). After 5 days, the phenotype of these cells was concordant with an immature DC population: DC-SIGNhigh, CD1ahigh, CD14very low.
Reagents and antibodies.
MG132, PYR41, and SFN were purchased from Cayman Chemicals. Primary antibodies—anti-pIRF3 at Ser 396, anti-IRF3, anti-RIG-I, anti-MDA5, anti-cGAS, anti-STING, anti-pSTAT1 at Tyr701, anti-STAT1, anti–p-IkBa at Ser32, anti-IkBa, anti-pEIF2α, anti-PARP, anti-cleaved (CL) caspase-3, anti-α-tubulin, anti-β-actin, and anti-V5 epitope—were purchased from Cell Signaling. Ammonium chloride, anti-FLAG, and anti-interferon-induced protein with tetratricopeptide repeats 1 (anti-IFIT1) were purchased from Sigma. Anti-Nrf2 (H10), anti-HO-1, anti-hemagglutinin (anti-HA), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH), and anti-GFP were purchased from Santa Cruz Biotechnology.
Plasmids.
The HO-1 overexpression plasmid was provided by Hyman Schipper (Lady Davis Institute) and has been described previously (73). The STING expression plasmid was obtained from John Cambier, National Jewish Health, Albany Medical College (74). V5-tagged WT DENV NS2B3 and its active-site S135A mutant were provided by Carolyn B. Coyne and are described in reference 46. The NC16 pCDNA3.1 FLAG Nrf2 plasmid and pCDNA3.1 empty vector were bought from Addgene.
Virus production, quantification, and infection.
Confluent monolayers of C6/36 Aedes albopictus mosquito cells were infected with DENV serotype 2 strain New Guinea C (DENV-2 NGC) at a multiplicity of infection (MOI) of 0.05. Virus was allowed to adsorb for 1 h at 28°C in a minimal volume of serum-free DMEM. After adsorption, the monolayer was washed once with serum-free medium and covered with DMEM containing 2% FBS. After 7 days of infection, the medium was harvested, cleared by centrifugation (500 × g, 5 min), and concentrated by centrifugation (2,000 × g, 8 min) through a 15-ml Millipore Amicon centrifugal filter unit (Millipore, Billerica, MA). The virus was concentrated by ultracentrifugation on a sucrose density gradient (20% sucrose cushion) using a Sorvall WX 100 ultracentrifuge (Thermo Scientific, Rockford, IL) for 2 h at 134,000 × g and 10°C with the brake turned off. The concentrated virus was then washed to remove sucrose using a 15-ml Amicon tube. After two washes, the virus was resuspended in DMEM plus 0.1% bovine serum albumin (BSA) and stored at −80°C. Titers of DENV stocks were determined by fluorescence-activated cell sorting (FACS), infection of Vero monkey kidney cells with 10-fold serial dilutions of the stock, and then immunofluorescence staining of intracellular DENV E protein at 24 h postinfection as described by Lambeth et al. in 2005 (75). Titers were expressed as infectious units per milliliter. In infection experiments, A549 cells, Huh-7 cells, HEK293T cells, or Mo-DC were first infected in a small volume of medium without FBS for 1 h at 37°C and then incubated with complete medium for 24 to 48 h prior to analysis.
Genome editing by CRISPR-Cas9.
The CRISPR design tool (http://crispr.mit.edu) was used to identify candidate guide RNA (gRNA) sequences in the Hmox-1 gene. Forward and reverse oligonucleotides targeting exon 3 of HO-1 (listed in Table 1) were phosphorylated and annealed according to Zhang’s protocol (76) and then subcloned within plasmid pSpCas9(BB)-2A-GFP (pX458) (kindly provided by Søren Riis Paludan, Aarhus University) using the BbsI restriction enzyme. The engineered pX458-XO-1 vector was subsequently amplified in chemically competent Escherichia coli DH5α cells and then extracted by using a QIAprep spin miniprep kit, and its sequence was validated by sequencing starting from the U6 promoter.
TABLE 1.
List of primers
| Sequence ID | Target gene | Primer name | Sequence |
|---|---|---|---|
| gi_158976983 | Dengue virus NS4A | DENV2_UPL5_F | ATCCTCCTATGGTACGCACAAA |
| DENV2_UPL5_R | CTCCAGTATTATTGAAGCTGCTATCC | ||
| NM_002083.3 | Glutathione peroxidase 2 | GPX2_UPL_2F | GTCCTTGGCTTCCCTTGC |
| GPX2_UPL_2R | TGTTCAGGATCTCCTCATTCTG | ||
| NM_014331.3 | Cystine/glutamate transporter | SLC7A11_UPL_80F | CCATGAACGGTGGTGTGTT |
| SLC7A11_UPL_80R | GACCCTCTCGAGACGCAAC | ||
| NM_002395.5 | Malic enzyme 1 | ME1_UPL_59F | GCAGTGCTACAAAATAACCAAGG |
| ME1_UPL_59R | TGGAAGAGTGACTGGATCAAAA | ||
| NM_002631.3 | Phosphogluconate dehydrogenase | PGD_UPL_32F | ATTGCTGCAAAAGTGGGAAC |
| PGD_UPL_32R | GTTGTGCACCATCTTCACGA | ||
| NM_001064.3 | Transketolase | TKT_UPL_23F | GGATGACCAGGTGACCGTTA |
| TKT_UPL_23R | CGCGGATGTTGATCTTTTCT | ||
| NM_001096.2 | ATP citrate lyase | ACL_UPL_49F | AACCCCAAAGGGAGGATCT |
| ACL_UPL_49R | TTGACACCCCCTAGATCACAG | ||
| NM_198834.2 | Acetyl-CoA carboxylase α | ACC1_UPL_73F | GATGTGGATGATGGGCTACA |
| ACC1_UPL_73R | TGAGGCCTTGATCATTACTGG | ||
| NM_004104.4 | Fatty acid synthase | FAS_UPL_11F | CAGGCACACACGATGGAC |
| FAS_UPL_11R | CGGAGTGAATCTGGGTTGAT | ||
| NM_005063.4 | Stearoyl-CoA desaturase | SCD1_UPL_82F | CCTAGAAGCTGAGAAACTGGTGA |
| SCD1_UPL_82R | ACATCATCAGCAAGCCAGGT | ||
| NM_000594.4 | Tumor necrosis factor alpha | TNF_UPL40F | CAGCCTCTTCTCCTTCCTGAT |
| TNF_UPL40R | GCCAGAGGGCTGATTAGAGA | ||
| NM_000600.5 | Interleukin 6 | IL6_UPL40F | GATGAGTACAAAAGTCCTGATCCA |
| IL6_UPL40R | CTGCAGCCACTGGTTCTGT | ||
| NM_000576.3 | Interleukin 1β | IL1β_UPL78F | TACCTGTCCTGCGTGTTGAA |
| IL1β_UPL78R | TCTTTGGGTAATTTTTGGGATCT | ||
| NM_001675.4 | Activating transcription factor 4 | ATF4_UPL88F | GGTCAGTCCCTCCAACAACA |
| ATF4_UPL88R | CTATACCCAACAGGGCATCC | ||
| NM_001195053.1 | C/EBP-homologous protein | CHOP_UPL21F | AAGGCACTGAGCGTATCATGT |
| CHOP_UPL21R | TGAAGATACACTTCCTTCTTGAACA | ||
| NM_001433.5 | Inositol-requiring enzyme 1α | IRE1a_UPL50F | GAAGCATGTGCTCAAACACC |
| IRE1a_UPL50R | TCTGTCGCTCACGTCCTG | ||
| NM_004994.3 | Matrix metallopeptidase 9 | MMP9_UPL27F | CCTGGAGACCTGAGAACCAA |
| MMP9_UPL27R | GAGTGTAACCATAGCGGTACAGG | ||
| NM_002046.7 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH_UPL60_F | AGCCACATCGCTCAGACAC |
| GAPDH_UPL60_R | GCCCAATACGACCAAATCC | ||
| NM_006164 | Nuclear factor erythroid 2-related factor 2 | NFE2L2_UPL9F | CCGGCATTTCACTAAACACA |
| NFE2L2_UPL9R | TGTGTCTGGATAGCTGGAAGATT | ||
| NM_002133.2 | Heme oxygenase 1 | HMOX1_UPL15F | GGCAGAGGGTGATAGAAGAGG |
| HMOX1_UPL15R | AGCTCCTGCAACTCCTCAAA | ||
| NM_001498.3 | Glutamate-cysteine ligase catalytic subunit | GCLC_UPL80F | ATGCCATGGGATTTGGAAT |
| GCLC_UPL80R | GATCATAAAGGTATCTGGCCTCA | ||
| NM_002061.4 | Glutamate-cysteine ligase modifier subunit | GCLM_UPL18F | GTTGGAACAGCTGTATCAGTGG |
| GCLM_UPL18R | CAGTCAAATCTGGTGGCATC | ||
| NM_182729.3 | Thioredoxin reductase 1 | TXNRD1_UPL60F | TGAGGAGAAAGCTGTGGAGAA |
| TXNRD1_UPL60R | CCATTCCAATGGCCAAAA | ||
| NM_000584.4 | Interleukin 8 | IL8_UPL72F | AGACAGCAGAGCACACAAGC |
| IL8_UPL72R | TGGTTCCTTCCGGTGGT | ||
| NM_001256067.2 | NADPH oxidase activator 1 | NOXA_UPL5F | GTGGATTTCCTGGGCAAG |
| NOXA_UPL5R | TCATGGTTCGCTCCTGGT | ||
| HMOX1_CrisprCas9_F | CACCGTCACCCAGGTAGCGGGTGT | ||
| HMOX1_CrisprCas9_R | AAACACACCCGCTACCTGGGTGAC |
The pX458-XO-1 plasmid was transfected in A549 cells by Lipofectamine 2000. At 24 h after transfection, GFP-positive cells were isolated by FACS, and colonies were grown up from single cells in 96-well plates. The medium was replaced every 3 to 5 days, and after 3 weeks, all clonal populations were screened for HO-1 expression by Western blotting.
DENV E protein staining.
The percentage of cells infected with DENV was determined by intracellular staining using a mouse IgG2a monoclonal antibody (MAb) specific for DENV E protein (clone 4G2). Harvested cells were washed with phosphate-buffered saline (PBS), fixed with 1% formaldehyde, and incubated for 10 min with a permeabilized solution (1× PBS, 0.1% saponin, and 1% FBS) together with a primary DENV E antibody. Cells were then washed twice with PBS and incubated another 10 min in a permeabilized solution containing a phycoerythrin (PE)-conjugated goat anti-mouse IgG MAb (BioLegend). Data were acquired by a FACSCanto II flow cytometer (BD Biosciences).
ROS staining.
Total ROS production was evaluated by flow cytometry using the CM-H2DCFDA probe (1 μM; Life Technologies). C11-Bodipy (1 μM; Life Technologies) was used to measure lipid peroxidation in live cells. Following DENV infection, cells were washed in PBS before incubation with the probes for 30 min at 37°C. After incubation, cells were washed twice in PBS before FACS analysis. The frequency of ROS-positive cells was determined by gating against stained live cells (7-AAD negative). Fluorescence data were further analyzed by FlowJo software using the mean fluorescence intensity (MFI) data for each condition.
Plasmid transfection and RNA interference (RNAi).
HEK293T and Huh-7 cells were transiently transfected with expression vectors by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For small interfering RNA (siRNA) experiments, A549 cells were transfected with 80 pmol of human Nrf2 (sc-37030), human heme oxygenase-1 (sc-35554), or control siRNA (sc-37007) diluted in Opti-MEM (Gibco) by using Lipofectamine RNAiMax according to the manufacturer’s instructions. Cells were incubated for 60 h and then infected with DENV.
Protein extraction and immunoblot analysis.
DENV-infected cells were washed twice in ice-cold PBS and lysed in lysis buffer (50 mM Tris-HCl [pH 8], 1% sodium deoxycholate, 1% NP-40, 5 mM EDTA, 150 mM NaCl, 0.1% sodium dodecyl sulfate) in the presence of protease and phosphatase inhibitors, and the insoluble fraction was removed by centrifugation at 17,000 × g for 15 min (4°C). Protein concentrations were determined using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). Proteins were resolved by SDS-PAGE on 4% to 20% precast Novex Tris-glycine gradient gels (Thermo Fisher Scientific) and blotted onto nitrocellulose membranes (0.22 μM; GE Healthcare). Blots were incubated with primary antibodies, extensively washed with Tris-buffered saline with Tween 20 (TBST), and, after incubation with horseradish peroxidase (HRP)-labeled goat anti-rabbit or goat anti-mouse antibodies (Abs) (GE Healthcare), developed with the enhanced chemiluminescence (ECL) detection system according to the manufacturer’s instructions (Thermo Fisher Scientific).
Coimmunoprecipitation.
Primary antibodies (5 μg) were added to 300 μl of cell lysate (500 μg) and incubated with gentle rocking overnight at 4°C. Protein A or G agarose beads (Sigma-Aldrich) were washed with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40) for 30 min at 4°C. The beads were then incubated with the cell lysate at 4°C for at least 4 h. Each lysate was washed four times in radioimmunoprecipitation assay (RIPA) lysis buffer. Samples were subsequently neutralized, mixed with SDS-loading buffer and a reducing agent, heated, and loaded onto SDS-PAGE gels.
Nrf2 degradation by DENV NS2B3.
293T cells were transfected with plasmids encoding Nrf2-FLAG and DENV NS2B3-V5 or NS2B3-S135A-V5 and were infected with DENV-2 (MOI, 1). After 24 h, cells were treated with dimethyl sulfoxide (DMSO), MG-132 (5 μM; Cayman Chemicals), or the E1 inhibitor PYR41 (1 μM; Cayman Chemicals); cells were lysed 18 h later, and cell extracts were used to quantify the expression of Nrf2 and DENV NS2B using anti-FLAG and anti-V5 antibodies.
Cell viability analysis.
Cell surface expression of phosphatidylserine was measured using an allophycocyanin (APC)-conjugated annexin V antibody, as recommended by the manufacturer (BioLegend, San Diego, CA). Briefly, specific annexin V binding was achieved by incubating A549 cells in annexin V binding buffer (10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2 [pH 7.4]) containing a saturating concentration of APC-annexin V antibody and 7-aminoactinomycin D (7-AAD) for 15 min in the dark. APC-annexin V and 7-AAD binding to the cells was analyzed by flow cytometry.
mRNA extraction and quantitative real-time PCR (qRT-PCR).
RNA was isolated by column separation using the RNeasy kit (Qiagen) according to the manufacturer’s instructions, and the concentration was measured with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). A 350-ng portion of RNA was used for cDNA synthesis using the High-Capacity cDNA reverse transcription kit (Thermo Fisher Scientific). Quantitative PCR (qPCR) was then performed using the TaqMan Fast Advanced master mix with TaqMan probes or Universal ProbeLibrary probes (Roche) with specific primers designed using the Roche Life Science Assay Design Center on a StepOnePlus real-time PCR system (Thermo Fisher Scientific). A relative quantification method was used, with GAPDH as the housekeeper gene.
Fluidigm Biomark assay.
Total RNA and cDNA were prepared as described above, and a Biomark assay on Nrf2-inducible, apoptotic, antiviral, and inflammatory genes was performed (77). cDNAs along with the entire pool of primers were preamplified for 14 cycles using the TaqMan PreAmp master mix according to the manufacturer’s protocol (Applied Biosystems). cDNA was treated with exonuclease I (Thermo Fisher Scientific). cDNA samples were prepared with 2× FastStart TaqMan Probe Master (Roche), GE sample loading buffer (Fluidigm), and Taq polymerase (Invitrogen). Assays were prepared with 2× assay loading reagent (Fluidigm), primers (Roche), and probes (Roche). Samples and assay components were loaded in their appropriate inlets on a 48.48 Biomark chip. The chip was run on the Biomark HD system (Fluidigm), which enabled quantitative measurement of as many as 48 different mRNAs in 48 samples under identical reaction conditions. Runs were 40 cycles. Raw threshold cycle (CT) values were calculated by real-time PCR analysis software (Fluidigm), and software-designated failed reactions were discarded from analysis. A relative quantification method was used, with GAPDH as the housekeeping gene. The n-fold differential expression of mRNA gene samples was expressed as 2–ΔΔCT. The heat maps were produced with Microsoft Excel. Gene-level expression is shown as log2 (2–ΔΔCT).
Statistical analysis.
Values were expressed as means (standard deviations [SD]). Graphs and statistics were computed using Microsoft Excel. An unpaired, two-tailed Student t test was used to determine the significance of the differences between the control and each experimental condition. P values of <0.05 were considered statistically significant (***, P < 0.001; **, P < 0.01; *, P < 0.05).
ACKNOWLEDGMENTS
This research project was supported by funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 813343 for the Marie Curie ITN INITIATE program. Support was also provided to J.H. by Istituto Pasteur Italia-Fondazione Cenci Bolognetti and by the Italian Association for Cancer Research (IG22891). D.O. was supported by a Carlsberg Foundation International Research Fellowship and by a Weiman Foundation Associate Professor Fellowship.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halstead SB. 2008. Dengue virus–mosquito interactions. Annu Rev Entomol 53:273–291. doi: 10.1146/annurev.ento.53.103106.093326. [DOI] [PubMed] [Google Scholar]
- 3.Wilder-Smith A, Ooi E-E, Vasudevan SG, Gubler DJ. 2010. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr Infect Dis Rep 12:157–164. doi: 10.1007/s11908-010-0102-7. [DOI] [PubMed] [Google Scholar]
- 4.Rothman AL. 2011. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol 11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 5.Ubol S, Halstead SB. 2010. How innate immune mechanisms contribute to antibody-enhanced viral infections. Clin Vaccine Immunol 17:1829–1835. doi: 10.1128/CVI.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flipse J, Wilschut J, Smit JM. 2013. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic 14:25–35. doi: 10.1111/tra.12012. [DOI] [PubMed] [Google Scholar]
- 7.Halstead SB. 2003. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res 60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 8.Jaiyen Y, Masrinoul P, Kalayanarooj S, Pulmanausahakul R, Ubol S. 2009. Characteristics of dengue virus-infected peripheral blood mononuclear cell death that correlates with the severity of illness. Microbiol Immunol 53:442–450. doi: 10.1111/j.1348-0421.2009.00148.x. [DOI] [PubMed] [Google Scholar]
- 9.Libraty DH, Young PR, Pickering D, Endy TP, Kalayanarooj S, Green S, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis 186:1165–1168. doi: 10.1086/343813. [DOI] [PubMed] [Google Scholar]
- 10.Halstead SB, Russell PK, Brandt WE. 2020. NS1, dengue’s dagger. J Infect Dis 221:857–860. doi: 10.1093/infdis/jiz083. [DOI] [PubMed] [Google Scholar]
- 11.Glasner DR, Puerta-Guardo H, Beatty PR, Harris E. 2018. The good, the bad, and the shocking: the multiple roles of dengue virus nonstructural protein 1 in protection and pathogenesis. Annu Rev Virol 5:227–253. doi: 10.1146/annurev-virology-101416-041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glasner DR, Ratnasiri K, Puerta-Guardo H, Espinosa DA, Beatty PR, Harris E. 2017. Dengue virus NS1 cytokine-independent vascular leak is dependent on endothelial glycocalyx components. PLoS Pathog 13:e1006673. doi: 10.1371/journal.ppat.1006673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seet RCS, Lee C-YJ, Lim ECH, Quek AML, Yeo LLL, Huang S-H, Halliwell B. 2009. Oxidative damage in dengue fever. Free Radic Biol Med 47:375–380. doi: 10.1016/j.freeradbiomed.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 14.Gil L, Martínez G, Tápanes R, Castro O, González D, Bernardo L, Vázquez S, Kourí G, Guzmán MG. 2004. Oxidative stress in adult dengue patients. Am J Trop Med Hyg 71:652–657. doi: 10.4269/ajtmh.2004.71.652. [DOI] [PubMed] [Google Scholar]
- 15.Soundravally R, Hoti SL, Patil SA, Cleetus CC, Zachariah B, Kadhiravan T, Narayanan P, Kumar BA. 2014. Association between proinflammatory cytokines and lipid peroxidation in patients with severe dengue disease around defervescence. Int J Infect Dis 18:68–72. doi: 10.1016/j.ijid.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 16.Soundravally R, Sankar P, Hoti SL, Selvaraj N, Bobby Z, Sridhar MG. 2008. Oxidative stress induced changes in plasma protein can be a predictor of imminent severe dengue infection. Acta Trop 106:156–161. doi: 10.1016/j.actatropica.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Chandrasena LG, Peiris H, Kamani J, Wanigasuriya P, Jayaratne SD, Wijayasiri WAA, Wijesekara GUS. 2014. Antioxidants in patients with dengue viral infection. Southeast Asian J Trop Med Public Health 45:1015–1022. [PubMed] [Google Scholar]
- 18.Rasool M, Malik A, Khan KM, Qureshi MS, Shabbir B, Zahid S, Asif M, Manan A, Rashid S, Khan SR, Arsalan HM, Alam R, Arooj M, Qazi MH, Chaudhary AGA, Abuzenadah AM, Al-Qahtani MH, Karim S. 2015. Assessment of biochemical and antioxidative status in patients suffering from dengue fever. J Huazhong Univ Sci Technol Med Sci 35:411–418. doi: 10.1007/s11596-015-1446-x. [DOI] [PubMed] [Google Scholar]
- 19.Magesh S, Chen Y, Hu L. 2012. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev 32:687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungvari Z, Bagi Z, Feher A, Recchia FA, Sonntag WE, Pearson K, de Cabo R, Csiszar A. 2010. Resveratrol confers endothelial protection via activation of the antioxidant transcription factor Nrf2. Am J Physiol Heart Circ Physiol 299:H18–H24. doi: 10.1152/ajpheart.00260.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta RK, Patel AK, Shah N, Chaudhary AK, Jha UK, Yadav UC, Gupta PK, Pakuwal U. 2014. Oxidative stress and antioxidants in disease and cancer: a review. Asian Pacific J Cancer Prev 15:4405–4409. doi: 10.7314/apjcp.2014.15.11.4405. [DOI] [PubMed] [Google Scholar]
- 22.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes JD, Dinkova-Kostova AT. 2014. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. 2005. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem 280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 25.Kwak M-K, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. 2003. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. J Biol Chem 278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 26.Pajares M, Jiménez-Moreno N, García-Yagüe ÁJ, Escoll M, de Ceballos ML, Van Leuven F, Rábano A, Yamamoto M, Rojo AI, Cuadrado A. 2016. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou Y-S, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S-i, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 28.Wild AC, Moinova HR, Mulcahy RT. 1999. Regulation of γ-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J Biol Chem 274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 29.Moinova HR, Mulcahy RT. 1999. Up-regulation of the human γ-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem Biophys Res Commun 261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 30.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. 2002. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res 62:5196–5203. [PubMed] [Google Scholar]
- 31.Lee J-M, Calkins MJ, Chan K, Kan YW, Johnson JA. 2003. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem 278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 32.Wu KC, Cui JY, Klaassen CD. 2011. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci 123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. 2012. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Olagnier D, Peri S, Steel C, van Montfoort N, Chiang C, Beljanski V, Slifker M, He Z, Nichols CN, Lin R, Balachandran S, Hiscott J. 2014. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 10:e1004566. doi: 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, Szpyt J, Runtsch MC, King MS, McGouran JF, Fischer R, Kessler BM, McGettrick AF, Hughes MM, Carroll RG, Booty LM, Knatko EV, Meakin PJ, Ashford MLJ, Modis LK, Brunori G, Sévin DC, Fallon PG, Caldwell ST, Kunji ERS, Chouchani ET, Frezza C, Dinkova-Kostova AT, Hartley RC, Murphy MP, O’Neill LA. 2018. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordes T, Wallace M, Michelucci A, Divakaruni AS, Sapcariu SC, Sousa C, Koseki H, Cabrales P, Murphy AN, Hiller K, Metallo CM. 2016. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J Biol Chem 291:14274–14284. doi: 10.1074/jbc.M115.685792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olagnier D, Brandtoft AM, Gunderstofte C, Villadsen NL, Krapp C, Thielke AL, Laustsen A, Peri S, Hansen AL, Bonefeld L, Thyrsted J, Bruun V, Iversen MB, Lin L, Artegoitia VM, Su C, Yang L, Lin R, Balachandran S, Luo Y, Nyegaard M, Marrero B, Goldbach-Mansky R, Motwani M, Ryan DG, Fitzgerald KA, O’Neill LA, Hollensen AK, Damgaard CK, de Paoli FV, Bertram HC, Jakobsen MR, Poulsen TB, Holm CK. 2018. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat Commun 9:3506. doi: 10.1038/s41467-018-05861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luplertlop N, Missé D, Bray D, Deleuze V, Gonzalez J‐P, Leardkamolkarn V, Yssel H, Veas F. 2006. Dengue-virus-infected dendritic cells trigger vascular leakage through metalloproteinase overproduction. EMBO Rep 7:1176–1181. doi: 10.1038/sj.embor.7400814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peña J, Harris E. 2011. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem 286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drummen GP, van Liebergen LC, Op den Kamp JA, Post JA. 2002. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic Biol Med 33:473–490. doi: 10.1016/S0891-5849(02)00848-1. [DOI] [PubMed] [Google Scholar]
- 41.Harvey CJ, Thimmulappa RK, Sethi S, Kong X, Yarmus L, Brown RH, Feller-Kopman D, Wise R, Biswal S. 2011. Targeting Nrf2 signaling improves bacterial clearance by alveolar macrophages in patients with COPD and in a mouse model. Sci Transl Med 3:78ra32. doi: 10.1126/scitranslmed.3002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishii T, Mann GE. 2014. Redox status in mammalian cells and stem cells during culture in vitro: critical roles of Nrf2 and cystine transporter activity in the maintenance of redox balance. Redox Biol 2:786–794. doi: 10.1016/j.redox.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olagnier D, Lavergne R-A, Meunier E, Lefèvre L, Dardenne C, Aubouy A, Benoit-Vical F, Ryffel B, Coste A, Berry A, Pipy B. 2011. Nrf2, a PPARγ alternative pathway to promote CD36 expression on inflammatory macrophages: implication for malaria. PLoS Pathog 7:e1002254. doi: 10.1371/journal.ppat.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu C, Eggler AL, Mesecar AD, van Breemen RB. 2011. Modification of Keap1 cysteine residues by sulforaphane. Chem Res Toxicol 24:515–521. doi: 10.1021/tx100389r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LCF, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lennemann NJ, Coyne CB. 2017. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 13:322–332. doi: 10.1080/15548627.2016.1265192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khumthong R, Angsuthanasombat C, Panyim S, Katzenmeier G. 2002. In vitro determination of dengue virus type 2 NS2B-NS3 protease activity with fluorescent peptide substrates. J Biochem Mol Biol 35:206–212. doi: 10.5483/bmbrep.2002.35.2.206. [DOI] [PubMed] [Google Scholar]
- 48.Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, Webb LG, Bernal-Rubio D, Solovyov A, Greenbaum B, Simon V, Basler CF, Mulder LCF, García-Sastre A, Fernandez-Sesma A. 2017. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2:17037. doi: 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hart PD, Young MR, Jordan MM, Perkins WJ, Geisow MJ. 1983. Chemical inhibitors of phagosome-lysosome fusion in cultured macrophages also inhibit saltatory lysosomal movements. A combined microscopic and computer study. J Exp Med 158:477–492. doi: 10.1084/jem.158.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hooftman A, O’Neill LAJ. 2019. The immunomodulatory potential of the metabolite itaconate. Trends Immunol 40:687–698. doi: 10.1016/j.it.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Miorin L, Maestre AM, Fernandez-Sesma A, García-Sastre A. 2017. Antagonism of type I interferon by flaviviruses. Biochem Biophys Res Commun 492:587–596. doi: 10.1016/j.bbrc.2017.05.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suthar MS, Aguirre S, Fernandez-Sesma A. 2013. Innate immune sensing of flaviviruses. PLoS Pathog 9:e1003541. doi: 10.1371/journal.ppat.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu C-Y, Chang T-H, Liang J-J, Chiang R-L, Lee Y-L, Liao C-L, Lin Y-L. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devadas K, Dhawan S. 2006. Hemin activation ameliorates HIV-1 infection via heme oxygenase-1 induction. J Immunol 176:4252–4257. doi: 10.4049/jimmunol.176.7.4252. [DOI] [PubMed] [Google Scholar]
- 55.Hashiba T, Suzuki M, Nagashima Y, Suzuki S, Inoue S, Tsuburai T, Matsuse T, Ishigatubo Y. 2001. Adenovirus-mediated transfer of heme oxygenase-1 cDNA attenuates severe lung injury induced by the influenza virus in mice. Gene Ther 8:1499–1507. doi: 10.1038/sj.gt.3301540. [DOI] [PubMed] [Google Scholar]
- 56.Espinoza JA, León MA, Céspedes PF, Gómez RS, Canedo-Marroquín G, Riquelme SA, Salazar-Echegarai FJ, Blancou P, Simon T, Anegon I, Lay MK, González PA, Riedel CA, Bueno SM, Kalergis AM. 2017. Heme oxygenase-1 modulates human respiratory syncytial virus replication and lung pathogenesis during infection. J Immunol 199:212–223. doi: 10.4049/jimmunol.1601414. [DOI] [PubMed] [Google Scholar]
- 57.Protzer U, Seyfried S, Quasdorff M, Sass G, Svorcova M, Webb D, Bohne F, Hösel M, Schirmacher P, Tiegs G. 2007. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology 133:1156–1165. doi: 10.1053/j.gastro.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 58.Lehmann E, El-Tantawy WH, Ocker M, Bartenschlager R, Lohmann V, Hashemolhosseini S, Tiegs G, Sass G. 2010. The heme oxygenase 1 product biliverdin interferes with hepatitis C virus replication by increasing antiviral interferon response. Hepatology 51:398–404. doi: 10.1002/hep.23339. [DOI] [PubMed] [Google Scholar]
- 59.Tung W-H, Hsieh H-L, Lee IT, Yang C-M. 2011. Enterovirus 71 induces integrin β1/EGFR-Rac1-dependent oxidative stress in SK-N-SH cells: role of HO-1/CO in viral replication. J Cell Physiol 226:3316–3329. doi: 10.1002/jcp.22677. [DOI] [PubMed] [Google Scholar]
- 60.Tseng C-K, Lin C-K, Wu Y-H, Chen Y-H, Chen W-C, Young K-C, Lee J-C. 2016. Human heme oxygenase 1 is a potential host cell factor against dengue virus replication. Sci Rep 6:32176. doi: 10.1038/srep32176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill-Batorski L, Halfmann P, Neumann G, Kawaoka Y. 2013. The cytoprotective enzyme heme oxygenase-1 suppresses Ebola virus replication. J Virol 87:13795–13802. doi: 10.1128/JVI.02422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Z, Wilson AT, Luxon BA, Brown KE, Mathahs MM, Bandyopadhyay S, McCaffrey AP, Schmidt WN. 2010. Biliverdin inhibits hepatitis C virus nonstructural 3/4A protease activity: mechanism for the antiviral effects of heme oxygenase? Hepatology 52:1897–1905. doi: 10.1002/hep.23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarz KB. 1996. Oxidative stress during viral infection: a review. Free Radic Biol Med 21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- 64.Akaike T, Noguchi Y, Ijiri S, Setoguchi K, Suga M, Zheng YM, Dietzschold B, Maeda H. 1996. Pathogenesis of influenza virus-induced pneumonia: involvement of both nitric oxide and oxygen radicals. Proc Natl Acad Sci U S A 93:2448–2453. doi: 10.1073/pnas.93.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akaike T, Maeda H. 2000. Nitric oxide and virus infection. Immunology 101:300–308. doi: 10.1046/j.1365-2567.2000.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Vliet A, Eiserich JP, Cross CE. 2000. Nitric oxide: a pro-inflammatory mediator in lung disease? Respir Res 1:67–72. doi: 10.1186/rr14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staitieh BS, Ding L, Neveu WA, Spearman P, Guidot DM, Fan X. 2017. HIV-1 decreases Nrf2/ARE activity and phagocytic function in alveolar macrophages. J Leukoc Biol 102:517–525. doi: 10.1189/jlb.4A0616-282RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carvajal-Yepes M, Himmelsbach K, Schaedler S, Ploen D, Krause J, Ludwig L, Weiss T, Klingel K, Hildt E. 2011. Hepatitis C virus impairs the induction of cytoprotective Nrf2 target genes by delocalization of small Maf proteins. J Biol Chem 286:8941–8951. doi: 10.1074/jbc.M110.186684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komaravelli N, Tian B, Ivanciuc T, Mautemps N, Brasier AR, Garofalo RP, Casola A. 2015. Respiratory syncytial virus infection down-regulates antioxidant enzyme expression by triggering deacetylation-proteasomal degradation of Nrf2. Free Radic Biol Med 88:391–403. doi: 10.1016/j.freeradbiomed.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng YL, Lin YS, Chen CL, Tsai TT, Tsai CC, Wu YW, Ou YD, Chu YY, Wang JM, Yu CY, Lin CF. 2016. Activation of Nrf2 by the dengue virus causes an increase in CLEC5A, which enhances TNF-α production by mononuclear phagocytes. Sci Rep 6:32000. doi: 10.1038/srep32000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Alimi AA, Ali SA, Al-Hassan FM, Idris FM, Teow S-Y, Mohd Yusoff N. 2014. Dengue virus type 2 (DENV2)-induced oxidative responses in monocytes from glucose-6-phosphate dehydrogenase (G6PD)-deficient and G6PD normal subjects. PLoS Negl Trop Dis 8:e2711. doi: 10.1371/journal.pntd.0002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heiss EH, Schachner D, Zimmermann K, Dirsch VM. 2013. Glucose availability is a decisive factor for Nrf2-mediated gene expression. Redox Biol 1:359–365. doi: 10.1016/j.redox.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song W, Su H, Song S, Paudel HK, Schipper HM. 2006. Over-expression of heme oxygenase-1 promotes oxidative mitochondrial damage in rat astroglia. J Cell Physiol 206:655–663. doi: 10.1002/jcp.20509. [DOI] [PubMed] [Google Scholar]
- 74.Jin L, Lenz LL, Cambier JC. 2010. Cellular reactive oxygen species inhibit MPYS induction of IFNβ. PLoS One 5:e15142. doi: 10.1371/journal.pone.0015142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lambeth CR, White LJ, Johnston RE, de Silva AM. 2005. Flow cytometry-based assay for titrating dengue virus. J Clin Microbiol 43:3267–3272. doi: 10.1128/JCM.43.7.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olagnier D, Chiang C, Hiscott J. 2017. Evaluation of innate immune gene expression following HDAC inhibitor treatment by high throughput qPCR and PhosFlow cytometry. Methods Mol Biol 1510:245–255. doi: 10.1007/978-1-4939-6527-4_18. [DOI] [PubMed] [Google Scholar]