Swine serve as a mixing vessel for generating pandemic strains of human influenza virus. All hemagglutinin subtypes of IAVs can infect swine; however, only sporadic cases of infection with avian IAVs are reported in domestic swine. The molecular mechanisms affecting the ability of avian IAVs to infect swine are still not fully understood. From the findings of phenotypic analyses, this study suggests that the tissue tropisms (i.e., in swine upper respiratory tracts) of avian IAVs affect their spillovers from wild birds to pigs. It was found that this phenotype is determined not by receptor binding preference but is determined by other markers across genomic segments, such as those in the ribonucleoprotein complex. In addition, our results show that such a phenotypic trait was sporadically and randomly distributed among IAVs across multiple avian species and geographic and temporal orders. This study suggests an efficient way for assessment of the risk posed by avian IAVs, such as in evaluating their potentials to be transmitted from birds to pigs.
KEYWORDS: influenza A viruses, avian influenza virus, swine influenza virus, mixing vessel, H4N6, spillover, tissue tropism, host tropism, swine upper respiratory tract, transmission, avian viruses, risk assessment
ABSTRACT
Wild aquatic birds maintain a large, genetically diverse pool of influenza A viruses (IAVs), which can be transmitted to lower mammals and, ultimately, humans. Through phenotypic analyses of viral replication efficiency, only a small set of avian IAVs were found to replicate well in epithelial cells of the swine upper respiratory tract, and these viruses were shown to infect and cause virus shedding in pigs. Such a phenotypic trait of the viral replication efficiency appears to emerge randomly and is distributed among IAVs across multiple avian species and geographic and temporal orders. It is not determined by receptor binding preference but is determined by other markers across genomic segments, such as those in the ribonucleoprotein complex. This study demonstrates that phenotypic variants of viral replication efficiency exist among avian IAVs but that only a few of these may result in viral shedding in pigs upon infection, providing opportunities for these viruses to become adapted to pigs, thus posing a higher potential risk for creating novel variants or detrimental reassortants within pig populations.
IMPORTANCE Swine serve as a mixing vessel for generating pandemic strains of human influenza virus. All hemagglutinin subtypes of IAVs can infect swine; however, only sporadic cases of infection with avian IAVs are reported in domestic swine. The molecular mechanisms affecting the ability of avian IAVs to infect swine are still not fully understood. From the findings of phenotypic analyses, this study suggests that the tissue tropisms (i.e., in swine upper respiratory tracts) of avian IAVs affect their spillovers from wild birds to pigs. It was found that this phenotype is determined not by receptor binding preference but is determined by other markers across genomic segments, such as those in the ribonucleoprotein complex. In addition, our results show that such a phenotypic trait was sporadically and randomly distributed among IAVs across multiple avian species and geographic and temporal orders. This study suggests an efficient way for assessment of the risk posed by avian IAVs, such as in evaluating their potentials to be transmitted from birds to pigs.
INTRODUCTION
Influenza A viruses (IAVs) are members of the Orthomyxoviridae family and are negative single-stranded RNA viruses with eight gene segments encoding at least 11 proteins. The nomenclature for IAVs is determined on the basis of the antigenic properties of two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). To date, 18 HA subtypes and 11 NA subtypes have been identified (1, 2). Besides humans, pigs, dogs, and horses, IAVs have been recovered from a variety of bird species, including at least 105 wild bird species of 26 different families (3). Among wild birds, birds of wetlands and aquatic environments, such as the Anseriformes (particularly ducks, geese, and swans) and Charadriiformes (particularly gulls, terns, and waders), constitute the major natural IAV reservoir (4). They maintain a large IAV genetic pool, which contributes to the appearance of new IAVs in other birds, lower mammals, and, ultimately, humans. To date, 16 HA subtypes (H1 to H16) and 9 NA subtypes (N1 to N9) of IAVs have been identified in these wild aquatic birds (4–6).
Like avian species, swine are one of the most important natural hosts for IAVs. To date, only three predominant subtypes of IAVs (i.e., H1N1, H1N2, and H3N2) are enzootic in pigs (7, 8). Having both avian-like receptors (α2,3-linked sialic acid [SA2,3Gal]) and human-like receptors (α2,6-linked sialic acid [SA2,6Gal]), swine may serve as an intermediate host, serving as mixing vessels for generating human pandemic influenza virus strains (9). For example, the 2009 H1N1 pandemic IAV is a reassortant with HA, NP, and NS from the classical swine (North American) lineage, basic polymerase 2 (PB2) and PA from the avian (North American) lineage, PB1 from human seasonal H3N2 strains, and NA and M from the Eurasian swine lineage (10). Although these pandemic viruses have avian-origin genetic segments, how and when these avian genes are introduced into swine are not clear. Under laboratory conditions, all HA subtypes of IAVs can infect swine (11). However, only sporadic cases of avian IAVs are detected in pigs, including subtypes H1, H3, H4, H5, H6, H7, and H9, and most of these spillovers are transient (12–14). Thus, as a key component of preparedness for influenza pandemics, it will be critical to understand which avian IAVs from among a large IAV genetic pool maintained by wild aquatic birds can be potentially transmitted to and subsequently spread in pigs.
Prior studies have demonstrated that receptor binding preference is one of the key factors affecting the transmissibility of avian IAVs in mammals (4). Acquired mutations in the HA receptor binding sites can switch the receptor binding properties of avian IAVs and enhance virus transmissibility in mammals. For example, four amino acid substitutions in the HA receptor binding sites and one in the polymerase complex protein basic polymerase 2 (PB2) enabled H5N1 avian IAV to transmit in ferrets through airborne droplets (15). Mutations at HA residues 222 (alanine to valine) and 228 (glycine to serine) increased the receptor binding preference of H6N6 avian IAVs to SA2,6Gal (16). In addition to the glycan receptor binding preference, the genetic constellation of the ribonucleoprotein (RNP) complex and adaptative mutations in RNP genes, including PB1, have been shown to affect the host and tissue tropisms of IAVs (17–20). It has been suggested that the avian IAV RNP complex has defects in its replication ability in human cells, and mutations that occur across PB2, PB1, PA, and NP of the RNP complex are demonstrated to facilitate the adaptation of avian IAVs in humans and other mammalian species (reviewed by Mänz et al. [21]). Nevertheless, the prerequisite for an avian IAV to be transmitted in mammals, such as pigs, is that the virus should be able to cause infection and viral shedding, thereby allowing further infection and allowing the virus opportunities to acquire adaptive mutations and/or genetic reassortment, which may further facilitate virus circulation in pigs.
This study aimed to understand the molecular mechanism(s) by which avian IAVs infect pigs and further to determine which avian IAVs among a large IAV genetic pool maintained by wild aquatic birds are more likely to be transmitted and further adapted to pigs. Our previous study suggested that the tissue tropism of an IAV, especially the efficiency of replication in swine upper respiratory tract epithelial cells (e.g., swine nasal epithelial [SNE] cells), is an important factor selecting for a transmissible reassortant in pigs (22). Here, we hypothesize that a large diversity in replication phenotypes on SNE cells is present in avian IAVs but that there is no evolutionary pressure and that the viruses with a high replication ability on SNE cells would be more likely to be transmitted in pigs. To test these hypotheses, we focused on subtype H4N6 avian IAVs, which have been detected twice in domestic swine populations in North America (23, 24).
RESULTS
H4N6 avian viruses have a large extent of diversity in replication efficiency on respiratory tract epithelial cells.
In 2015, A/swine/Missouri/A01727926/2015 (abbreviated MO/15) was recovered from a sick pig with influenza-like clinical signs due to a spillover of H4N6 avian viruses from wild birds to domestic pigs (23). Phylogenetic analyses showed that, similar to the isolate from a prior spillover, A/Swine/Ontario/01911/99 (H4N6) (abbreviated ON/99) (24), MO/15 is also an isolate of avian origin of the North America lineage. However, the HA gene of MO/15 belongs to genetic lineage IV, whereas that of ON/99 belongs to genetic lineage I, demonstrating that these viruses were from two independent spillover incidences (Fig. 1). We hypothesize that H4N6 avian IAVs circulating in wild birds possess a large range of replication efficiencies in mammalian respiratory tract epithelial cells and that only a small portion of avian IAVs have phenotypes similar to those of the spillover viruses detected in pigs (i.e., MO/15 and ON/99). To test this hypothesis, a total of 115 H4N6 isolates (Table 1) from wild birds in North America were selected for phenotype analyses, and these viruses were selected to represent the diversity in genomic sequences, avian host species, sampling times, and sampling locations to maximize the possibility of covering the phenotypic diversity of H4N6 viruses in wild birds.
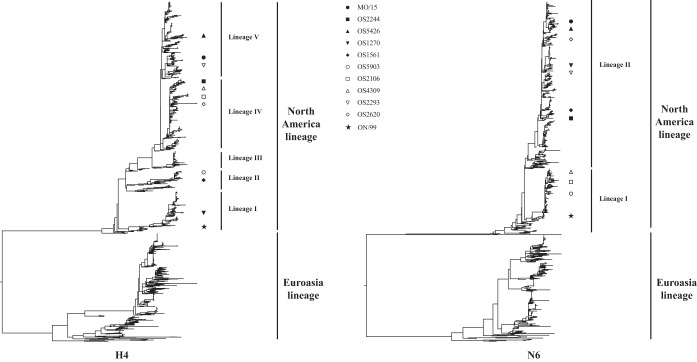
FIG 1.
Phylogenetic analysis of the HA and NA genes of H4N6 AIVs. The phylogenetic trees for each gene segment (HA and NA) were inferred using a maximum likelihood method implemented in the RAxML (version 8.2.9) program. A gamma model of rate heterogeneity and a generalized time-reversible (GTR) substitution model were applied in the analysis. Phylogenetic trees were then visualized by use of the ggtree (version 1.6.11) program. The representative isolates are marked on the trees with symbols. Abbreviations: MO/15, A/swine/Missouri/A01727926/2015 (H4N6); OS2244, A/blue-winged teal/Ohio/12OS2244/2012 (H4N6); OS5426, A/blue-winged teal/Ohio/15OS5426/2015 (H4N6); OS1270, A/mallard/Ohio/08OS1270/2008 (H4N6); OS1561, A/blue-winged teal/Illinois/10OS1561/2010 (H4N6); OS5903, A/American green-winged teal/Mississippi/11OS5903/2011 (H4N6); OS2106, A/blue-winged teal/Ohio/12OS2106/2012 (H4N6); OS4309, A/mallard/Wisconsin/11OS4309/2011 (H4N6); OS2293, A/blue-winged teal/Ohio/12OS2293/2012; OS2620, A/blue-winged teal/Missouri/11OS2620/2011 (H4N6); ON/99, A/Swine/Ontario/01911/99 (H4N6). Three isolates, MO/15, OS2244, and OS5426, were used in the animal study.
TABLE 1.
H4N6 viruses used in this study
| Virus name | Subtype | Passagea | HA titer (log2)b | Log10 TCID50/mlc |
|---|---|---|---|---|
| A/swine/Missouri/A01727926/2015 | H4N6 | MDCK3 | 7 | 6.67 |
| A/blue-winged teal/Ohio/15OS5426/2015 | H4N6 | EGG1 | 9 | 5.5 |
| A/mallard/Ohio/08OS1270/2008 | H4N6 | EGG1 | 9 | 7.33 |
| A/blue-winged teal/Illinois/10OS1561/2010 | H4N6 | EGG1 | 9 | 7 |
| A/American green-winged teal/Mississippi/11OS5903/2011 | H4N6 | EGG1 | 8 | 6.33 |
| A/blue-winged teal/Ohio/12OS2106/2012 | H4N6 | EGG1 | 8 | 7.33 |
| A/mallard/Wisconsin/11OS4309/2011 | H4N6 | EGG1 | 10 | 6.67 |
| A/blue-winged teal/Ohio/12OS2244/2012 | H4N6 | EGG1 | 9 | 7.5 |
| A/blue-winged teal/Ohio/12OS2293/2012 | H4N6 | EGG1 | 9 | 6.5 |
| A/blue-winged teal/Missouri/11OS2620/2011 | H4N6 | EGG1 | 8 | 6.67 |
| A/environment/Maryland/1655/2006 | H4N6 | EGG1 | 8 | 7.67 |
| A/environment/Maryland/09OS1315/2009 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Maryland/10OS1040/2010 | H4N6 | EGG1 | 9 | 8.5 |
| A/mallard/Wisconsin/1538/2009 | H4N6 | EGG1 | 9 | 7.5 |
| A/American green-winged teal/Ohio/13OS1429/2013 | H4N6 | EGG1 | 9 | 7.5 |
| A/blue-winged teal/Ohio/12OS2295/2012 | H4N6 | EGG1 | 9 | 7.67 |
| A/mallard/Maryland/09OS1134/2009 | H4N6 | EGG1 | 9 | 7.67 |
| A/mallard/Maryland/11OS951/2011 | H4N6 | EGG1 | 9 | 8.33 |
| A/blue-winged teal/Missouri/10MO003/2010 | H4N6 | EGG1 | 9 | 8 |
| A/blue-winged teal/Iowa/13OS2359/2013 | H4N6 | EGG1 | 9 | 7 |
| A/mallard/Wisconsin/11OS3481/2011 | H4N6 | EGG1 | 8 | 8 |
| A/mallard/Maryland/08OS1421/2008 | H4N6 | EGG1 | 8 | 7.5 |
| A/mallard/Wisconsin/2653/2009 | H4N6 | EGG1 | 8 | 8 |
| A/American green-winged teal/Ohio/13OS2050/2013 | H4N6 | EGG1 | 8 | 7.67 |
| A/environment/Maryland/09OS1301/2009 | H4N6 | EGG1 | 8 | 7.67 |
| A/northern shoveler/Illinois/11OS4707/2011 | H4N6 | EGG1 | 9 | 8 |
| A/mallard/Maryland/11OS964/2011 | H4N6 | EGG1 | 9 | 8.5 |
| A/mallard/Maryland/14OS1477/2014 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Maryland/09OS1170/2009 | H4N6 | EGG1 | 6 | 6.67 |
| A/environment/Maryland/09OS1302/2009 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Maryland/14OS1322/2014 | H4N6 | EGG1 | 7 | 6.33 |
| A/mallard/Iowa/10OS2420/2010 | H4N6 | EGG1 | 8 | 7.67 |
| A/blue-winged teal/Ohio/12OS2321/2012 | H4N6 | EGG1 | 8 | 7 |
| A/blue-winged teal/Ohio/12OS3128/2012 | H4N6 | EGG1 | 8 | 7.5 |
| A/green-winged teal/Ohio/1292/2005 | H4N6 | EGG1 | 9 | 8 |
| A/mallard/Maryland/08OS1414/2008 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Ohio/1695/2009 | H4N6 | EGG1 | 8 | 7.67 |
| A/northern pintail/Wisconsin/11OS3299/2011 | H4N6 | EGG1 | 7 | 7.5 |
| A/northern pintail/Ohio/15OS5731/2015 | H4N6 | EGG1 | 8 | 8 |
| A/environment/Maryland/14OS1379/2014 | H4N6 | EGG1 | 9 | 6.67 |
| A/American green-winged teal/Ohio/13OS2063/2013 | H4N6 | EGG1 | 8 | 8.5 |
| A/blue-winged teal/Wisconsin/11OS3027/2011 | H4N6 | EGG1 | 8 | 8.33 |
| A/redhead/Ohio/13OS0376/2013 | H4N6 | EGG1 | 8 | 8.33 |
| A/blue-winged teal/Ohio/15OS5173/2015 | H4N6 | EGG1 | 6 | 6.67 |
| A/mallard/Wisconsin/10OS2918/2010 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Ohio/660/2002 | H4N6 | EGG1 | 6 | 6 |
| A/mallard/Maryland/07OS1669/2007 | H4N6 | EGG1 | 8 | 7.5 |
| A/mallard/Ohio/657/2002 | H4N6 | EGG1 | 8 | 8 |
| A/mallard/Ohio/15OS4260/2015 | H4N6 | EGG1 | 9 | 7.67 |
| A/blue-winged teal/Ohio/12OS4593/2012 | H4N6 | EGG1 | 8 | 7.5 |
| A/mallard/Ohio/14OS3201/2014 | H4N6 | EGG1 | 8 | 7.5 |
| A/environment/Maryland/09OS1293/2009 | H4N6 | EGG1 | 8 | 6.67 |
| A/environment/Maryland/09OS1314/2009 | H4N6 | EGG1 | 8 | 7.67 |
| A/environment/Maryland/2115/2006 | H4N6 | EGG1 | 5 | 6.33 |
| A/blue-winged teal/Ohio/1850/2006 | H4N6 | EGG1 | 9 | 7.67 |
| A/American green-winged teal/Ohio/13OS2021/2013 | H4N6 | EGG1 | 9 | 8 |
| A/mallard/Ohio/671/2002 | H4N6 | EGG1 | 8 | 7.33 |
| A/mallard/Ohio/10OS1324/2010 | H4N6 | EGG1 | 8 | 7 |
| A/environment/Maryland/09OS1291/2009 | H4N6 | EGG1 | 8 | 7.5 |
| A/green-winged teal/Ohio/1324/2005 | H4N6 | EGG1 | 8 | 8 |
| A/mallard duck/Ohio/15OS5358/2015 | H4N6 | EGG1 | 7 | 5.5 |
| A/mallard/Maryland/14OS1312/2014 | H4N6 | EGG1 | 8 | 6.33 |
| A/mallard/Maryland/14OS1310/2014 | H4N6 | EGG1 | 8 | 6.5 |
| A/mallard/Illinois/08OS2315/2008 | H4N6 | EGG1 | 8 | 8 |
| A/blue-winged teal/Missouri/15OS4858/2015 | H4N6 | EGG1 | 8 | 7.5 |
| A/mallard/Ohio/11OS2085/2011 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Ohio/15OS3997/2015 | H4N6 | EGG1 | 8 | 8.33 |
| A/American green-winged teal/Illinois/10OS1551/2010 | H4N6 | EGG1 | 9 | 7.5 |
| A/environment/Maryland/09OS1292/2009 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Ohio/298/1987 | H4N6 | EGG1 | 9 | 7.5 |
| A/mallard/Ohio/1501/2006 | H4N6 | EGG1 | 6 | 7.33 |
| A/mallard/Ohio/15OS4235/2015 | H4N6 | EGG1 | 9 | 7.5 |
| A/blue-winged teal/Missouri/10MO0407/2010 | H4N6 | EGG1 | 7 | 7.5 |
| A/blue-winged teal/Wisconsin/2753/2009 | H4N6 | EGG1 | 8 | 7 |
| A/American green-winged teal/Ohio/12OS2137/2012 | H4N6 | EGG1 | 8 | 7.33 |
| A/mallard/Ohio/15OS4181/2015 | H4N6 | EGG1 | 7 | 4.5 |
| A/mallard/Ohio/15OS3981/2015 | H4N6 | EGG1 | 8 | 4.5 |
| A/blue-winged teal/Missouri/10MO030/2010 | H4N6 | EGG1 | 8 | 8 |
| A/environment/Maryland/09OS1290/2009 | H4N6 | EGG1 | 8 | 7.67 |
| A/blue-winged teal/Missouri/15OS4861/2015 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Maryland/09OS1359/2009 | H4N6 | EGG1 | 7 | 6.5 |
| A/mallard/Maryland/1241/2005 | H4N6 | EGG1 | 8 | 7.67 |
| A/mallard/Maryland/11OS952/2011 | H4N6 | EGG1 | 8 | 8.5 |
| A/northern shoveler/Wisconsin/11OS4110/2011 | H4N6 | EGG1 | 8 | 8 |
| A/mallard/Maryland/14OS1321/2014 | H4N6 | EGG1 | 8 | 7.5 |
| A/mallard/Wisconsin/08OS2261/2008 | H4N6 | EGG1 | 8 | 8 |
| A/mallard/Maryland/09OS1361/2009 | H4N6 | EGG1 | 8 | 8 |
| A/blue-winged teal/Wisconsin/2741/2009 | H4N6 | EGG1 | 9 | 8 |
| A/green-winged teal/Ohio/15OS5497/2015 | H4N6 | EGG1 | 9 | 8 |
| A/American green-winged teal/Wisconsin/11OS3592/2011 | H4N6 | EGG1 | 6 | 8 |
| A/American green-winged teal/Wisconsin/12OS2997/2012 | H4N6 | EGG1 | 9 | 8 |
| A/blue-winged teal/Missouri/15OS5134/2015 | H4N6 | EGG1 | 9 | 5.5 |
| A/environment/Maryland/1101/2006 | H4N6 | EGG1 | 6 | 6.5 |
| A/blue-winged teal/Missouri/15OS4803/2015 | H4N6 | EGG1 | 8 | 7.33 |
| A/blue-winged teal/Illinois/10OS1562/2010 | H4N6 | EGG1 | 10 | 9 |
| A/mallard/Maryland/09OS1355/2009 | H4N6 | EGG1 | 10 | 8.5 |
| A/gadwall/Wisconsin/11OS3415/2011 | H4N6 | EGG1 | 10 | 8 |
| A/blue-winged teal/Ohio/15OS5174/2015 | H4N6 | EGG1 | 10 | 8.67 |
| A/environment/Maryland/09OS1303/2009 | H4N6 | EGG1 | 9 | 8 |
| A/environment/Maryland/09OS1308/2009 | H4N6 | EGG1 | 9 | 8.33 |
| A/blue-winged teal/Wisconsin/11OS2657/2011 | H4N6 | EGG1 | 8 | 7.67 |
| A/green-winged teal/Ohio/15OS4704/2015 | H4N6 | EGG1 | 9 | 8.5 |
| A/environment/Maryland/14OS1486/2014 | H4N6 | EGG1 | 9 | 8 |
| A/blue-winged teal/Missouri/15OS4830/2015 | H4N6 | EGG1 | 6 | 7 |
| A/mallard/Ohio/15OS4308/2015 | H4N6 | EGG1 | 10 | 8.5 |
| A/blue-winged teal/Ohio/13OS2016/2013 | H4N6 | EGG1 | 8 | 8 |
| A/environment/Maryland/14OS1364/2014 | H4N6 | EGG1 | 8 | 7 |
| A/mallard/Ohio/655/2002 | H4N6 | EGG1 | 9 | 8 |
| A/mallard/Maryland/06OS1110/2006 | H4N6 | EGG1 | 9 | 6.5 |
| A/blue-winged teal/Iowa/13OS2338/2013 | H4N6 | EGG1 | 9 | 8 |
| A/mallard/Ohio/12OS5547/2012 | H4N6 | EGG1 | 9 | 8 |
| A/environment/Maryland/09OS1295/2009 | H4N6 | EGG1 | 7 | 6.67 |
| A/blue-winged teal/Missouri/15OS4786/2015 | H4N6 | EGG1 | 9 | 8 |
| A/mallard/Ohio/15OS4302/2015 | H4N6 | EGG1 | 9 | 8 |
| A/American green-winged teal/Missouri/15OS6530/2015 | H4N6 | EGG1 | 9 | 6.67 |
| A/blue-winged teal/Missouri/14OS1868/2014 | H4N6 | EGG1 | 9 | 7.67 |
MDCK3, passage in MDCK cells; EGG1, passage in specific-pathogen-free embryonated chicken eggs.
HA titers were determined using 0.5% turkey red blood cells.
The TCID50 was determined in MDCK cells.
The replication phenotypic analyses were carried out on SNE cells, swine trachea epithelial (STE) cells, and human A549 cells. Testing temperatures of 33, 37, and 39°C were used for SNE, STE, and A549 cells, respectively, to model the temperatures across the upper, middle, and lower respiratory tracts of the pig, as performed previously (22). Due to a lack of available swine alveolar epithelial cells and the limited availability of swine lung primary cells, human A549 cells were used to model swine alveolar epithelial cells from the swine lower respiratory tract. Human A549 cells are characterized as a type II pulmonary epithelial cell to model IAV infection (25), and both human and swine type II pneumocytes express α-2,3- and α-2,6-linked sialic acid receptors (26, 27) and are targets of IAV infection (26, 28–30).
The 115 avian viruses showed a large diversity of replication efficiencies within and across the three cell models. These avian viruses replicated the best on A549 cells at 39°C (5.875 ± 1.426 [standard deviation] log10 50% tissue culture infective doses [TCID50]/ml), followed by STE cells at 37°C (4.314 ± 1.699 log10 TCID50/ml) and SNE cells at 33°C (2.422 ± 1.398 log10 TCID50/ml) (Fig. 2). There was a large diversity of the replication efficiencies across the three cell model systems: using SNE cells, only 43 test viruses reached a viral titer of 2 log10 TCID50/ml or higher; using STE cells, 102 H4N6 test viruses reached a viral titer of 2 log10 TCID50/ml or higher; and using A549 cells, 110 H4N6 test IAVs reached a viral titer of 2 log10 TCID50/ml or higher.
FIG 2.
Growth dynamics of H4N6 IAVs in swine nasal epithelial (SNE) cells, swine trachea epithelial (STE) cells, and human alveolar basal epithelial (A549) cells. (A) Growth kinetics of nine representative H4N6 avian isolates and an H4N6 swine isolate, MO/15. (B) Summary of growth phenotypes on SNE, STE, and A549 cells at 72 h postinoculation. Cells were infected with the indicated viruses at a multiplicity of infection of 0.001 TCID50/cell. Infected cells were incubated at 33°C, 37°C, or 39°C. Growth curves were determined by using the viral titers in the supernatants of infected cells obtained at 12, 24, 48, and 72 h postinoculation. The experiments were performed in triplicate, and the data shown represent the mean titers ± standard errors (n = 3 cultures). The abbreviations are defined in the legend to Fig. 1.
The results showed that the swine isolate MO/15 replicated efficiently in all three cell models: on SNE cells at 33°C with a titer of 5.613 ± 0.098 log10 TCID50/ml, on STE cells at 37°C with a titer of 3.500 ± 0.000 log10 TCID50/ml, and on A549 cells at 39°C with a titer of 5.223 ± 0.479 log10 TCID50/ml. Of the 115 viruses tested, only 4 had a growth titer greater than or equal to that of MO/15 in SNE cells at 33°C. Twenty viruses replicated well across all three cell types with a titer of ≥3.5 log10 TCID50/ml and showed a replication pattern similar to that of MO/15.
Growth kinetics analyses support the distinct replication patterns of H4N6 avian viruses on respiratory tract epithelial cells.
To further differentiate the phenotypic diversity of these viruses, we determined the growth kinetics of MO/15 and nine other representative avian isolates that demonstrated distinct growth phenotypes in the above-described analyses. In addition to the three cell models mentioned above, various temperatures (i.e., 33, 37, and 39°C) were used to evaluate the effects of temperature on the growth of the IAVs. The results demonstrated that there were significant variations in the growth kinetic patterns among these 10 isolates within and across the three cell models, and these were consistent with those described above in the large-scale endpoint replication efficiency analyses. Of interest, the results showed that temperature had an effect on the viral growth kinetics in all three cell models. SNE cells supported virus replication at all three temperatures tested, STE cells supported virus replication at 37°C better than at the other two temperatures, and A549 cells supported high rates of replication of H4N6 viruses at 37°C and 39°C but not at 33°C. In general, 33°C, 37°C, and 39°C were shown to be the optimal temperatures for virus replication on SNE, STE, and A549 cells, respectively (Fig. 2).
The results showed that MO/15 replicated efficiently in all three cell models (Fig. 2). Among the other nine avian viruses tested, A/blue-winged teal/Ohio/15OS5426/2015 (H4N6) (abbreviated OS5426) replicated poorly in all three cell models, whereas A/blue-winged teal/Ohio/12OS2244/2012 (H4N6) (abbreviated OS2244) replicated well in all three cell models, with peak viral titers of 5.943 ± 0.418, 4.890 ± 0.191, and 6.610 ± 0.348 log10 TCID50/ml on SNE (33°C), STE (37°C), and A549 (39°C) cells, respectively.
Based on the phenotypic analyses, MO/15, OS5426, and OS2244 were selected for further phenotypic analysis of virus binding preference, polymerase activities, and infectivity and transmission in animals.
The growth phenotypes of H4N6 avian viruses are likely affected by genetic diversity but are genetic clade independent.
To evaluate whether the replication phenotypes of H4N6 viruses are associated with certain genetic clades of H4N6 viruses, we constructed phylogenetic trees of the selected H4N6 viruses and colored the viruses by their growth efficiency on SNE cells at 33°C (Fig. 3A). The results suggested that for each of the eight gene segments, the high-growth strains were sporadically located across the phylogenetic tree, and no clear patterns between viral growth efficiency and the genetic clades of any of the genes were identified. The two spillover strains, MO/15 and ON/99, were not genetically associated with the high-growth H4N6 avian strains identified in our phenotypic analyses. No patterns between virus growth phenotypes on STE cells and A549 cells and the genetic clades of any of the genes were identified either (Fig. 3B and C).
FIG 3.
Distribution of growth phenotypic variants in phylogenetic trees. The phenotypes used in this figure were the TCID50 titers at 72 h and 33°C on SNE cells (A), STE cells (B), and A549 cells (C). The phylogenetic trees for each gene segment were inferred using a maximum likelihood method implemented in the RAxML (version 8.2.9) program. A gamma model of rate heterogeneity and a generalized time-reversible (GTR) substitution model were applied in the analysis. Phylogenetic trees were then visualized by the use of ggtree (version 1.6.11) software. Three viruses, MO/15, OS2244, and OS5425, used in the animal study are annotated. The abbreviations are defined in the legend to Fig. 1.
H4N6 avian viruses showed similar receptor binding properties.
To evaluate whether the receptor binding properties affect growth phenotypes, we performed molecular analyses of receptor binding sites and receptor binding preference analyses. Sequence analyses suggested that the HA receptor binding sites were conserved across all 115 test viruses, including OS2244 and OS5426, two prototype viruses that we selected in this study. However, compared to avian viruses, both swine isolates, MO/15 and ON/99, had two mutations, Q226L and G228S, at the HA receptor binding sites (Fig. 4A).
FIG 4.
Receptor binding preference of H4N6 IAVs. (A) Sequence logo of the HA receptor binding sites of avian and swine H4N6 IAVs. Both the H3 and H4 numbering are indicated, and the mutations at residues 226 and 228 on the HA protein are highlighted. (B) Receptor binding analyses using biolayer interferometry assays. 3′-Sialyl-N-acetyllactosamine (3′SLN; avian-like IAV receptor analog) and 6′-sialyl-N-acetyllactosamine (6′SLN; human-like IAV receptor analog) were used in these analyses. Streptavidin-coated biosensors were immobilized with biotinylated glycans at different concentrations. Sugar loading-dependent binding signals were captured in the association step and normalized to the same background. Binding curves were fitted by using the binding-saturation method in GraphPad Prism (version 7) software. The horizontal dashed lines indicate half of the fractional saturation (f = 0.5), and the vertical dashed line indicates the relative sugar loading at f equal to 0.5 (RSL0.5); the higher that the RSL0.5 value is, the smaller that the binding affinity is. The abbreviations are defined in the legend to Fig. 1.
The receptor binding specificities were determined through biolayer interferometer assays. The 50% relative sugar loadings (RSL0.5) were quantified; the smaller that the RSL0.5 value was, the stronger that binding affinity for a test virus was (Fig. 4B). The avian viruses bound to both 3′-sialyl-N-acetyllactosamine (3′SLN; avian-like IAV receptor analog) and 6′-sialyl-N-acetyllactosamine (6′SLN; human-like IAV receptor analog). The avian viruses showed very similar binding patterns, with both viruses preferentially binding to 3′SLN rather than to 6′SLN: for 3′SLN, RSL0.5 was 0.007 for OS2244 and <0.001 for OS5426; for 6′SLN, RSL0.5 was 0.293 for OS2244 and 0.303 for OS5426 (Fig. 4B). The swine-origin virus (MO/15) also bound to both 3′SLN and 6′SLN but had a higher affinity of binding to 6′SLN (RSL0.5 = 0.228) than to 3′SLN (RSL0.5 = 0.448).
The RNP complex of H4N6 avian viruses showed variations in polymerase activities in mammalian cells.
To further identify the molecular mechanisms affecting the diversity of the replication phenotypes of the H4N6 avian viruses, we determined the polymerase activities of the ribonucleoprotein (RNP) complex of H4N6 viruses. We hypothesized that the constellation of the RNP complex would affect the polymerase activity of H4N6 IAVs and, thus, the viral replication efficiency. To test this hypothesis, the PB2, PB1, PA, and NP genes of MO/15, OS2244, and OS5246 were cloned and the polymerase activities were quantified using a minigenome assay in human embryonic kidney (HEK) 293T cells (Fig. 5). Before the analyses by the minigenome assay, each gene that we cloned was validated to be functional by generating a live reassortant virus with 7 other segments from A/Puerto Rico/8/1934 (H1N1) (abbreviated PR8) using reverse genetics (data not shown).
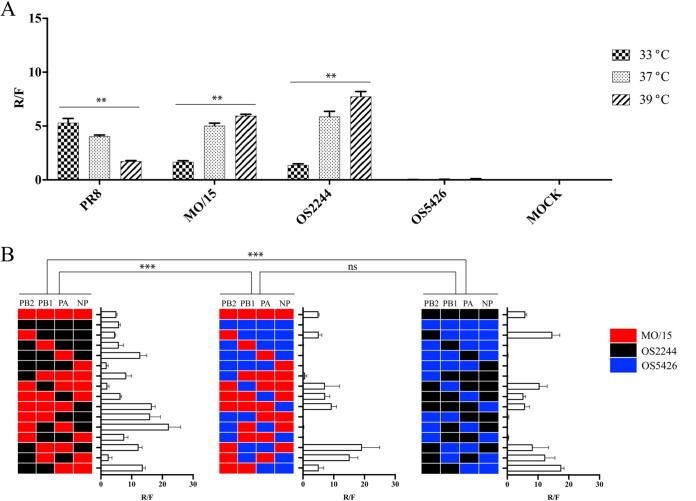
FIG 5.
Polymerase activity of the RNP complex. (A) Polymerase activities of the RNP complexes from wild-type viruses on human embryonic kidneys 293T cells at 33, 37, and 39°C; (B) polymerase activities of the RNP reassortant complex on human embryonic kidney 293T cells at 37°C. The polymerase activities were determined using minigenome luciferase assays. The mean and standard deviation of the R/F value for each RNP complex were derived from the luciferase assay data in triplicate. ns, no statistically significant difference; **, P < 0.01; ***, P < 0.001.
The results showed that the polymerase activities of each RNP were consistent with the replication patterns of the corresponding isolates described above. For example, the RNP complexes of MO/15 and OS2244 had Renilla/firefly (R/F) values of 1.665 and 1.377 (33°C), respectively, whereas the RNP complex of OS5246 had an R/F value of <0.1 (33°C) (Fig. 5A). In addition, our results showed that the polymerase activities can be temperature dependent (Fig. 5A). For example, the MO/15 and OS2244 complexes significantly increased the polymerase activity as the temperature increased (P = 0.0036). As a positive control, the polymerase activities of PR8, a laboratory-adapted strain, were also quantified. The polymerase activities of PR8 decreased at the higher temperature, indicating a difference in the optimal temperature conditions between the RNP complexes from laboratory-adapted strains and those from H4N6 wild-type viruses.
In order to investigate the effects of the genomic constellation on the polymerase activities of the RNP complex, we determined the polymerase activities of all possible RNP reassortants between MO/15 and OS2244 (n = 16), between MO/15 and OS5426 (n = 16), and between OS2244 and OS5426 (n = 16) (Fig. 5B). The results showed a large extent of diversity among these 48 RNP reassortants (Fig. 5). In general, the polymerase activities of the reassortants between MO/15 and OS2244 were relatively higher than those between OS2244 and OS5426 (P < 0.0001) and those between MO/15 and OS5426 (P < 0.0001). The polymerase activities between OS2244 and OS5426 and those between MO/15 and OS5426 were not statistically significantly different (P = 0.3315). Further analyses suggested that the PB2 gene is important for the high polymerase activity of the RNP complex. All 16 RNP complexes tested with PB2 of OS5246 showed a very low or minimal polymerase activity, whereas the others tested with either the PB2 of MO/15 or the PB2 of OS2244 did not (Fig. 5B).
The H4N6 swine virus infected pigs and caused limited transmission through direct contacts in pigs.
To evaluate virus infectivity and transmissibility, four pigs (referred to as inoculation pigs) were inoculated with MO/15, and three influenza virus-seronegative pigs (contact pigs) were paired with either one or two inoculation pigs in the same pen. The results showed that all four inoculation pigs shed virus in nasal wash samples at 2, 4, and 6 days postinoculation (dpi), with the viral loads ranging from 3.23 ± 0.35 to 6.28 ± 0.47 log10 copies/ml (Fig. 6 and Table 2), and the results were further confirmed by reverse transcription-droplet digital PCR (RT-ddPCR), which produced similar viral loads ranging from 2.79 to 5.89 log10 copies/ml at 2, 4, and 6 dpi (Table 3). To confirm viral infection in the respiratory tracts of pigs, one inoculation pig was euthanized and necropsied on 5 dpi, and another one was euthanized and necropsied on 7 dpi. Viral titration by quantitative reverse transcription-PCR (qRT-PCR) showed that the upper and middle respiratory tract tissues had viral loads with titers ranging from 4.10 ± 0.17 to 5.11 ± 0.06 log10 copies/g (Table 4), and the viral loads were further confirmed by RT-ddPCR, with titers ranging from 4.29 to 5.21 log10 copies/g (Table 5). Serological analyses by hemagglutination inhibition (HI) assays showed that two remaining inoculation pigs seroconverted as early as 10 dpi with the same HI titers of 1:160 and remained seroconverted on 21 dpi with HI titers of 1:40 and 1:80, respectively (Table 6).
FIG 6.
Viral shedding in the animal experiments. Seven pigs were assigned to three groups, consisting of one group for each isolate, and each group contained two or three animals. In each group, one or two animals were used as the inoculation animal and one animal was used as the contact animal. Each inoculation animal was inoculated nasally with 106 TCID50 of each indicated virus on day 0; the contact animal was brought into the pen 24 h later. Nasal washes were collected at 2-day intervals, and one inoculated pig was euthanized and necropsied on day 5 and/or day 7. Bars of the same color indicate that the pigs were in the same pen. Open bars represent inoculation pigs, while filled bars represent contact pigs. The viral titer in each nasal wash sample was determined by qRT-PCR, and the dashed lines indicate the limit of detection (3.574 log10 copies/ml) by qRT-PCR. No viral shedding was detected in any of the pigs in the negative-control groups (data not shown).
TABLE 2.
Viral titers for the nasal wash samples from feral swine determined from qRT-PCR data
| Virus | Pen | Treatment | Pig | Time of necropsy (dpia ) | Viral shedding in nasal wash samples (mean ± SD log10 copies/ml) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 dpi | 2 dpi | 4 dpi | 6 dpi | 8 dpi | 10 dpi | |||||
| MO/15 | 3 | Inoculation | 143 | 5 | NAb | 4.68 ± 0.38 | 3.28 ± 0.37 | —c | — | — |
| MO/15 | 3 | Contact | 150 | 21 | NA | NA | NA | NA | NA | NA |
| MO/15 | 3 | Inoculation | 163 | 21 | NA | 5.09 ± 0.37 | 6.28 ± 0.47 | 3.61 ± 0.37 | NA | NA |
| MO/15 | 5 | Contact | 142 | 21 | NA | NA | NA | NA | NA | NA |
| MO/15 | 5 | Inoculation | 144 | 7 | NA | 4.90 ± 0.38 | 5.55 ± 0.42 | 3.23 ± 0.35 | — | — |
| MO/15 | 7 | Contact | 147 | 21 | NA | NA | NA | 4.74 ± 0.35 | 5.39 ± 0.29 | NA |
| MO/15 | 7 | Inoculation | 148 | 21 | NA | 4.70 ± 0.42 | 4.60 ± 0.36 | NA | NA | NA |
| OS2244 | A | Inoculation | 155 | 5 | NA | NA | NA | — | — | — |
| OS2244 | A | Contact | 140 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | A | Inoculation | 162 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | C | Contact | 153 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | C | Inoculation | 156 | 7 | NA | 4.56 ± 0.26 | 3.52 ± 0.31 | NA | — | — |
| OS2244 | B | Contact | 149 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | B | Inoculation | 151 | 21 | NA | NA | 3.38 ± 0.97 | NA | NA | NA |
| OS5426 | C | Inoculation | 159 | 5 | NA | NA | NA | — | — | — |
| OS5426 | C | Contact | 141 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | C | Inoculation | 139 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | A | Contact | 146 | 7 | NA | NA | NA | NA | — | — |
| OS5426 | A | Inoculation | 145 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | B | Contact | 154 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | B | Inoculation | 152 | 21 | NA | NA | NA | NA | NA | NA |
| PBS | 1 | Control | 160 | 5 | NA | NA | NA | — | — | — |
| PBS | 3 | Control | 164 | 5 | NA | NA | NA | — | — | — |
| PBS | 1 | Control | 158 | 7 | NA | NA | NA | NA | — | — |
| PBS | 3 | Control | 165 | 7 | NA | NA | NA | NA | — | — |
| PBS | 1 | Control | 164 | 21 | NA | NA | NA | NA | NA | NA |
| PBS | 3 | Control | 161 | 21 | NA | NA | NA | NA | NA | NA |
dpi, days postinoculation.
NA, not detected.
—, sample not available.
TABLE 3.
Viral titers for the nasal wash samples from feral swine determined from RT-ddPCR data
| Virus | Pen | Treatment | Pig | Time of necropsy (dpia ) | Viral shedding in nasal wash samples (mean ± SD log10 copies/ml) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 dpi | 2 dpi | 4 dpi | 6 dpi | 8 dpi | 10 dpi | |||||
| MO/15 | 3 | Inoculation | 143 | 5 | NAb | 4.49 | 2.79 | —c | — | — |
| MO/15 | 3 | Contact | 150 | 21 | NA | NA | NA | NA | NA | NA |
| MO/15 | 3 | Inoculation | 163 | 21 | NA | 4.75 | 5.89 | 3.19 | NA | NA |
| MO/15 | 5 | Contact | 142 | 21 | NA | NA | NA | NA | NA | NA |
| MO/15 | 5 | Inoculation | 144 | 7 | NA | 4.72 | 5.00 | 3.00 | — | — |
| MO/15 | 7 | Contact | 147 | 21 | NA | NA | NA | 4.40 | 5.06 | NA |
| MO/15 | 7 | Inoculation | 148 | 21 | NA | 4.33 | 4.24 | NA | NA | NA |
| OS2244 | A | Inoculation | 155 | 5 | NA | NA | NA | — | — | — |
| OS2244 | A | Contact | 140 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | A | Inoculation | 162 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | C | Contact | 153 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | C | Inoculation | 156 | 7 | NA | 4.62 | 3.13 | NA | — | — |
| OS2244 | B | Contact | 149 | 21 | NA | NA | NA | NA | NA | NA |
| OS2244 | B | Inoculation | 151 | 21 | NA | NA | 2.57 | NA | NA | NA |
| OS5426 | C | Inoculation | 159 | 5 | NA | NA | NA | — | — | — |
| OS5426 | C | Contact | 141 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | C | Inoculation | 139 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | A | Contact | 146 | 7 | NA | NA | NA | NA | — | — |
| OS5426 | A | Inoculation | 145 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | B | Contact | 154 | 21 | NA | NA | NA | NA | NA | NA |
| OS5426 | B | Inoculation | 152 | 21 | NA | NA | NA | NA | NA | NA |
| PBS | 1 | Control | 160 | 5 | NA | NA | NA | — | — | — |
| PBS | 3 | Control | 164 | 5 | NA | NA | NA | — | — | — |
| PBS | 1 | Control | 158 | 7 | NA | NA | NA | NA | — | — |
| PBS | 3 | Control | 165 | 7 | NA | NA | NA | NA | — | — |
| PBS | 1 | Control | 164 | 21 | NA | NA | NA | NA | NA | NA |
| PBS | 3 | Control | 161 | 21 | NA | NA | NA | NA | NA | NA |
dpi, days postinoculation.
NA, not detected.
—, sample not available.
TABLE 4.
Viral titers for respiratory tract tissue samples from feral swine determined from qRT-PCR data
| Sample | Viral load in respiratory tract tissues (mean ± SD log10 no. of copies/g) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MO/15 (inoculation) |
OS2244 (inoculation) |
OS5426 |
PBS (control) |
|||||||
| Pig 143 (5 dpia ) | Pig 144 (7 dpi) | Pig 155 (5 dpi) | Pig 156 (7 dpi) | Inoculation pig 159 (5 dpi) | Contact pig 146 (7 dpi) | Pig 160 (5 dpi) | Pig 164 (5 dpi) | Pig 158 (7 dpi) | Pig 165 (7 dpi) | |
| RT | 4.10 ± 0.17 | 4.51 ± 0.36 | NAb | 3.85 ± 0.40 | 4.86 ± 0.06 | NA | NA | NA | NA | NA |
| MT | 4.26 ± 0.05 | NA | NA | NA | 6.63 ± 0.02 | NA | NA | NA | NA | NA |
| ET | NA | NA | 3.77 ± 0.04 | 4.18 ± 0.20 | 6.64 ± 0.01 | NA | NA | NA | NA | NA |
| SP | NA | NA | NA | NA | 4.84 ± 0.01 | NA | NA | NA | NA | NA |
| TR-U | 4.74 ± 0.05 | NA | 3.83 ± 0.13 | NA | 3.91 ± 0.29 | NA | NA | NA | NA | NA |
| TR-M | 4.72 ± 0.03 | NA | 4.21 ± 0.06 | NA | 4.25 ± 0.02 | NA | NA | NA | NA | NA |
| TR-D | 4.67 ± 0.03 | NA | 4.60 ± 0.10 | NA | NA | NA | NA | NA | NA | NA |
| BR | 5.11 ± 0.06 | NA | 7.59 ± 0.04 | NA | 4.24 ± 0.04 | NA | NA | NA | NA | NA |
| LCR | NA | NA | 5.98 ± 0.05 | 3.90 ± 0.63 | 4.43 ± 0.04 | NA | NA | NA | NA | NA |
| LCD | NA | NA | 4.30 ± 0.09 | 3.81 ± 0.11 | 3.68 ± 0.26 | NA | NA | NA | NA | NA |
| RCR | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| RMD | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| RCD | NA | NA | 5.18 ± 0.02 | NA | NA | NA | NA | NA | NA | NA |
| RA | NA | NA | 5.70 ± 0.02 | NA | NA | NA | NA | NA | NA | NA |
dpi, day postinoculation on which necropsy was performed.
NA, not detected.
TABLE 5.
Viral titers for respiratory tract tissue samples from feral swine determined from RT-ddPCR data
| Sample | Viral load in respiratory tract tissues (mean log10 no. of copies/g) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MO/15 (inoculation) |
OS2244 (inoculation) |
OS5426 |
PBS (control) |
|||||||
| Pig 143 (5 dpia ) | Pig 144 (7 dpi) | Pig 155 (5 dpi) | Pig 156 (7 dpi) | Inoculation pig 159 (5 dpi) | Contact pig 146 (7 dpi) | Pig 160 (5 dpi) | Pig 164 (5 dpi) | Pig 158 (7 dpi) | Pig 165 (7 dpi) | |
| RT | 4.33 | NAb | NA | 3.49 | 4.85 | NA | NA | NA | NA | NA |
| MT | 4.29 | NA | NA | NA | 6.64 | NA | NA | NA | NA | NA |
| ET | NA | NA | 4.13 | 4.27 | 6.69 | NA | NA | NA | NA | NA |
| SP | NA | NA | NA | NA | 4.83 | NA | NA | NA | NA | NA |
| TR-U | 4.74 | NA | 4.21 | NA | 4.07 | NA | NA | NA | NA | NA |
| TR-M | 4.68 | NA | 4.54 | NA | 4.17 | NA | NA | NA | NA | NA |
| TR-D | 4.77 | NA | 4.84 | NA | NA | NA | NA | NA | NA | NA |
| BR | 5.21 | NA | 8.06 | NA | 4.38 | NA | NA | NA | NA | NA |
| LCR | NA | NA | 6.28 | 3.17 | 4.56 | NA | NA | NA | NA | NA |
| LCD | NA | NA | 4.39 | 3.83 | 3.89 | NA | NA | NA | NA | NA |
| RCR | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| RMD | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| RCD | NA | NA | 5.48 | NA | NA | NA | NA | NA | NA | NA |
| RA | NA | NA | 6.15 | NA | NA | NA | NA | NA | NA | NA |
dpi, day postinoculation on which necropsy was performed.
NA, not detected.
TABLE 6.
HI titers for sera collected from feral swine
| Virus | Pen | Treatment | Pig | Time of necropsy (dpia ) | HI titer against MO/15 in serum samples |
||||
|---|---|---|---|---|---|---|---|---|---|
| 0 dpi | 6 dpi | 10 dpi | 14 dpi | 21 dpi | |||||
| MO/15 | 3 | Inoculation | 143 | 5 | <1:10 | —b | — | — | — |
| MO/15 | 3 | Contact | 150 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| MO/15 | 3 | Inoculation | 163 | 21 | <1:10 | <1:10 | 1:160 | 1:80 | 1:40 |
| MO/15 | 5 | Contact | 142 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| MO/15 | 5 | Inoculation | 144 | 7 | <1:10 | <1:10 | — | — | — |
| MO/15 | 7 | Contact | 147 | 21 | <1:10 | <1:10 | 1:40 | 1:640 | 1:160 |
| MO/15 | 7 | Inoculation | 148 | 21 | <1:10 | <1:10 | 1:160 | 1:160 | 1:80 |
| OS2244 | A | Inoculation | 155 | 5 | <1:10 | — | — | — | — |
| OS2244 | A | Contact | 140 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| OS2244 | A | Inoculation | 162c | 21 | <1:10 | <1:10 | 1:80 | — | — |
| OS2244 | C | Contact | 153 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| OS2244 | C | Inoculation | 156 | 7 | <1:10 | <1:10 | — | — | — |
| OS2244 | B | Contact | 149 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| OS2244 | B | Inoculation | 151 | 21 | <1:10 | <1:10 | 1:320 | 1:320 | 1:160 |
| OS5426 | C | Inoculation | 159 | 5 | <1:10 | — | — | — | — |
| OS5426 | C | Contact | 141 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| OS5426 | C | Inoculation | 139 | 21 | <1:10 | <1:10 | <1:10 | 1:80 | 1:40 |
| OS5426 | A | Contact | 146 | 7 | <1:10 | <1:10 | — | — | — |
| OS5426 | A | Inoculation | 145 | 21 | <1:10 | <1:10 | 1:10 | 1:40 | 1:20 |
| OS5426 | B | Contact | 154 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| OS5426 | B | Inoculation | 152 | 21 | <1:10 | <1:10 | 1:80 | 1:40 | 1:20 |
| PBS | 1 | Control | 160 | 5 | <1:10 | — | — | — | — |
| PBS | 3 | Control | 164 | 5 | <1:10 | — | — | — | — |
| PBS | 1 | Control | 158 | 7 | <1:10 | <1:10 | — | — | — |
| PBS | 3 | Control | 165 | 7 | <1:10 | <1:10 | — | — | — |
| PBS | 1 | Control | 164 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
| PBS | 3 | Control | 161 | 21 | <1:10 | <1:10 | <1:10 | <1:10 | <1:10 |
dpi, day postinoculation.
—, not available.
The pig was euthanized on 12 dpi due to a leg injury.
Among three paired contact pigs, one had virus detected in the nasal wash samples at 4.74 ± 0.35 and 5.39 ± 0.29 log10 copies/ml on 6 and 8 dpi, respectively (Fig. 5 and Table 2). The quantification was also confirmed by RT-ddPCR, which produced similar viral copy numbers, showing 4.40 log10 copies/ml on 6 dpi and 5.06 log10 copies/ml on 8 dpi (Table 3). This shedding contact pig seroconverted on 21 dpi with an HI titer of 1:160, whereas the other two contact pigs remained seronegative against IAV (Table 6).
In summary, our results showed that MO/15 infected feral swine with a high efficiency (4/4; 100%) and that MO/15 caused transmission among pigs through direct contact, though with a low transmission efficiency (1/3; 33.33%).
H4N6 avian IAVs infected pigs but showed distinct phenotypes of virus shedding.
We hypothesize that a high replication efficacy in swine respiratory tract epithelial cells, especially upper respiratory tract epithelial cells, such as SNE cells, could enhance viral infectivity and shedding in pigs. To test this hypothesis, we compared the infectivity, virus shedding, and transmission ability in pigs for two prototype avian viruses, OS2244 and OS5426, which showed high- and low-growth phenotypes, respectively, in SNE cells (Fig. 2B). The design of the animal experiments was the same as that for MO/15 described above.
The results showed that two of four pigs inoculated with OS2244 shed viruses in nasal wash samples, with peak titers of 4.56 ± 0.26 log10 copies/ml being obtained for pig 156 and 3.38 ± 0.97 log10 copies/ml being obtained for pig 151 (Fig. 6 and Table 2), and the results were further confirmed by RT-ddPCR, which produced viral copy numbers ranging from 2.57 to 4.62 log10 copies/ml (Table 3). Viral titrations suggested that viruses were detected in the respiratory tract tissues of both inoculation pigs euthanized on 5 and 7 dpi, with viral copy numbers ranging from 3.77 ± 0.04 to 7.59 ± 0.04 log10 copies/g (Table 4), and the results were further confirmed by RT-ddPCR, which produced titers ranging from 3.17 to 8.06 log10 copies/g (Table 5). Both of the two remaining inoculation pigs seroconverted, with HI titers being at least 1:80 (Table 6). These data show that all four pigs inoculated with OS2244 were infected with the virus and that two of these four pigs shed viruses.
In contrast, none of four pigs inoculated with OS5426 had detectable viral loads in any of the nasal washes collected from 0 through 14 dpi. On the other hand, viral titrations demonstrated that viruses were detected in the respiratory tract tissues of the inoculation pig euthanized on 5 dpi (Tables 4 and 5). Three remaining pigs inoculated with OS5426 seroconverted, with HI titers ranging from 1:10 to 1:80 (Table 6). These data show that all four pigs inoculated with OS5426 were infected with the virus but that none of them shed viruses.
No viruses were detected in nasal washes from the pigs in contact with either OS2244 or OS5466 or those from the negative-control pigs; none of the pigs in contact with either OS2244 or OS5466 and none of the negative-control pigs seroconverted. In addition, none of inoculation pigs, contact pigs, and negative-control pigs showed any clinical signs.
In summary, the animal study showed that animals inoculated with OS2244, which replicated efficiently in SNE cells, shed viruses in their nasal wash samples, whereas animals inoculated with OS5426, which replicated poorly in SNE cells, did not shed any viruses in their nasal wash samples, validating our hypotheses, although both avian viruses effectively infected pigs through nasal inoculation.
DISCUSSION
In this study, through phenotypic analyses of 115 genetically diverse H4N6 avian IAVs, we identified only a small set of isolates with a high growth efficiency on epithelial cells of the upper swine respiratory tract (i.e., SNE cells). A similar high-growth phenotype was also identified for an H4N6 isolate that was detected in a spillover event from wild birds to domestic swine. Genetic analyses suggested that the genetic constellation of the RNP complex, but not the receptor binding properties, is a major factor contributing to the observed phenotypic diversity. Animal studies suggested that both of the viruses with a high-growth phenotype on SNE cells as well as the virus with a low-growth phenotype on SNE cells infected pigs through nasal inoculation. Only the pigs infected with viruses with high-growth phenotypes had detectable viral shedding. Those viruses with high-growth phenotypes on SNE cells could potentially enable virus to be transmitted to other pigs, thus helping the virus acquire adaptive mutations or a gene from other cocirculating viruses through genetic reassortment. For example, Q226L and G228S were detected in the HA protein of both H4N6 swine isolates but rarely in avian isolates, and these mutations increased the human-like receptor binding ability and could have further enhanced virus transmission ability in pigs (Fig. 4) (31, 32). In summary, this study suggested that the tissue tropisms of H4N6 avian IAVs affect their spillover from wild birds to pigs. Thus, characterization of the tissue tropism of avian IAVs could be an efficient way for risk assessment, such as in evaluating their potentials to be transmitted from avians to pigs.
In wild birds, there is a lack of selection pressure for a mammal-based replication efficiency. While wild birds maintain a large, genetically diverse pool of IAVs, the majority of wild-type viruses would not be expected to bear a high-growth phenotype in mammalian cells. Our results from the screening of 115 H4N6 avian viruses showed that only 4 reached the same growth titers as MO/15 in SNE cells and that the high-growth viruses were scattered across the phylogenetic trees of eight genetic segments (Fig. 2). No particular genetic constellations correlated with the various growth phenotypes that we determined (Fig. 3). Our study suggests that assessment of the risk of avian IAVs infecting pigs should include phenotypic analyses of a broad range of genetically diverse isolates rather than phenotypic analyses of only the isolates from the most commonly found genotypes. In prior studies, a few genetic markers, such as residue 627 in PB2 (33–35) and residue 701 in PB1 (34), were shown to increase virus replication efficiencies in mammals, such as mice and ferrets. Thus, it would be useful to develop a genomic sequence-based risk assessment tool by identifying synergistic genetic markers through the use of a large set of phenotypic data.
Among all subtypes of avian IAVs, subtype H4 IAVs are enzootic in both wild birds and domestic poultry, and both pigs and humans may be exposed to these viruses (3, 13, 23, 24, 36–39). In addition to two spillovers of H4N6 IAVs mentioned above in Ontario, Canada (1999) (24), and in Missouri, USA (2015) (23), H4N1 and H4N8 isolates were recovered from pigs in China (40, 41). Serological surveillance suggested that transmission of these H4 IAVs from pig to pig occurred during the outbreaks (23, 24). A serological surveillance in southeastern China showed that swine had seropositivity rates of 2.4% (n = 127) from 1978 to 1982 and 8.9% (n = 101) in 1982 (13). However, in a feral swine population in the United States, among 406 IAV-positive samples from a total of 8,239 serum samples collected from 2010 to 2013, none was seropositive for the H4 virus (42).
Because of the long-term cocirculation of multiple subtypes of IAVs in wild aquatic birds, H4 avian IAVs have had frequent reassortments and show a large extent of genotypic diversity (37). Prior studies have also shown that these H4 avian viruses maintain a large pool of phenotypic diversity. For example, some H4 isolates exhibited infectivity in pigs and mice without prior adaptation and were efficiently transmitted in guinea pigs (11, 37). Similar to the receptor binding properties of two H4N6 avian viruses that we demonstrated in this study (Fig. 4), these avian H4 IAVs bind to both avian-like and human-like receptors, showing a stronger affinity to avian-like receptors (37). On the other hand, similar to our results (Fig. 5), another study suggested that the RNP complex of H4 IAVs contributes to higher virulence in mice (43). These data support our findings that, in addition to glycan receptor binding, the genomic makeup, such as that in the RNP complex, can affect virus infectivity and the transmissibility of avian IAVs in mammals.
The animal study demonstrated that the swine inoculated with MO/15 shed virus and also that the virus showed limited transmission (33%) in pigs through direct contact (Fig. 6). This result contradicts the results reported in a prior study that showed that the same isolate was not transmitted in pigs (23). To validate our analyses, we performed viral titration using the TCID50 of the virus shed from those four inoculation pigs and one contact pig in the animal experiment with MO/15 (Tables 2 and 3). Our results confirmed that 3 of 4 pigs inoculated with MO/15 had viral titers ranging from 2.50 to 3.67 log10 TCID50/ml and that the contact pig had a viral titer of 4.5 log10 TCID50/ml at 8 dpi (data not shown). The nasal washes of one of the four inoculation pigs did not have a detectable TCID50, possibly due to the relatively smaller viral loads and the limited growth of avian viruses on Madin-Darby canine kidney (MDCK) cells. The feral swine used in this study were less likely to be primed with IAVs than the domestic pigs used in the other study (42), although the pigs in both studies were shown to be seronegative for IAVs. On the other hand, the inoculation routes and doses were different in these two studies. In this study, 106 TCID50 in 2 ml of inoculum (1 ml per nostril) was intranasally inoculated for each pig; in the other study, a relatively lower concentration (3 × 105 TCID50 in 3 ml) of virus was inoculated into each pig intranasally (1 ml of virus) and intratracheally (2 ml of virus) (23). It has been shown that viral loads, the inoculation volume, and inoculation routes can affect virus infection ability (44). Nevertheless, the H4N6 swine isolate still seems to have a limited transmission ability in pigs, which is consistent with the low seroprevalence among the pigs at the outbreak farm and the other pigs in the production system (23).
Of interest, none of four pigs inoculated with OS5426 had detectable viral titers in their nasal washes, although the turbinate tissues of the pig euthanized on 5 dpi had viral loads of >4.85 log10 copies/g (Tables 4 and 5). There was a poor replication efficiency of OS5426 in SNE cells. These results suggest that the low viral shedding in the nasal washes was associated with inefficient virus replication in the nasal epithelial cells of the upper respiratory tract tissues. Further studies need to evaluate whether OS5426 has a high replication efficiency in other types of cells other than SNE cells in upper respiratory tract tissues.
In summary, this study suggests that a large pool of phenotypic variants exists among avian IAVs but that only a few may cause virus shedding in swine upon infection, providing more opportunities for these avian viruses to become adapted to pigs, thus posing a higher potential risk for creating novel variants or detrimental reassortants within swine populations.
MATERIALS AND METHODS
Cells.
Madin-Darby canine kidney (MDCK) cells, human embryonic kidney (HEK) 293T cells, and human alveolar adenocarcinoma (A549) cells (American Type Culture Collection, Manassas, VA, USA) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA, USA) at 37°C under 5% CO2. Swine nasal epithelial (SNE) cells and swine trachea epithelial (STE) cells were kindly provided by Stacey Schultz-Cherry, St. Jude Children’s Research Hospital, and grown at 37°C with 5% CO2 in DMEM–Ham’s F-12 medium (Thermo Fisher Scientific, Asheville, CA) supplemented with FBS (10%).
Viruses.
A/swine/Missouri/A01727926/2015 (H4N6) (abbreviated MO/15), an isolate recovered from a sick domestic swine, was kindly provided by the U.S. Department of Agriculture (USDA) Swine Influenza Surveillance Program and propagated for one passage on MDCK cells at 37°C with 5% CO2 in Opti-MEM I reduced serum medium (Thermo Fisher Scientific, Asheville, NC, USA) supplemented with 1 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Gibco, Gran Island, NY, USA), before being used for the molecular studies, phenotypic determination, and the animal study.
A total of 115 H4N6 isolates originating from wild birds in North America were selected to represent genomic diversity, in addition to the diversity in avian hosts, by sampling location, and by sampling time (Table 1). These avian isolates were propagated for one passage in specific-pathogen-free (SPF) 10-day-old chicken embryonated eggs (Charles River Laboratories, Inc., Norwich, CT), before being used in this study for the molecular studies, phenotypic determination, and the animal study.
Hemagglutination and hemagglutination inhibition assays.
Hemagglutination and hemagglutination inhibition (HI) assays were carried out by using 0.5% turkey erythrocytes, as previously described (45).
Viral titration.
For viral titration, the 50% tissue culture infective dose (TCID50) was determined on MDCK cells. Briefly, MDCK cells were seeded in a 96-well plate at 2 × 104 cells/well with Opti-MEM I reduced serum medium. The cells were incubated at 37°C with 5% CO2 for 18 to 20 h before virus inoculation. Viral samples were serial diluted in Opti-MEM I reduced serum medium supplemented with 1 μg/ml of TPCK-trypsin. The cell medium was removed, and 100 μl of each virus dilution was inoculated onto MDCK cells in quadruplicate. Infected cells were incubated at 37°C with 5% CO2 for 72 h, and then the number of positive and negative wells for each dilution were recorded for TCID50 calculation. The TCID50 was calculated as previously described by Reed and Muench (46).
Molecular cloning and genomic sequencing.
Viral RNA was extracted using a GeneJet viral DNA/RNA extraction kit (Thermo Fisher Scientific, Asheville, NC, USA), and cDNA was transcribed with the viral RNA as the template by using SuperScript III reverse transcriptase (Invitrogen, Thermo Fisher Scientific, Asheville, NC, USA). The PB2, PB1, PA, and NPs gene of MO/15, OS2244, and OS5426 were cloned into the pHW2000 vector with universal primers (47). After cloning, the plasmids were prepared for sequencing to further confirm that the sequences were correct and contained no additional mutations.
Minigenome.
For the luciferase assay, 293T cells were plated in a 96-well plate at 4 × 104 cells/well for transfection on the next day, when the cell density reached 80% confluence. The 293T cells were transfected with 40 ng of each plasmid harboring an RNP gene (pHW2000-PB2, pHW2000-PB1, pHW2000-PA, and pHW2000-NP), 40 ng of human Renilla luciferase reporter plasmid, and 4 ng of the pGL4.13 firefly luciferase reporter plasmid using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) in Opti-MEM medium (Gibco, Carlsbad, CA) for 48 h. Transfection was performed in triplicate. The luciferase activity was developed using a dual-luciferase reporter system (Promega, Madison, WI) according to the manufacturer’s instructions and measured on a cell imaging multimode reader (Cytation 5; BioTek, Winooski, VT).
Replication efficiency.
To determine the replication efficiency of the H4N6 viruses, we determined the titers of these viruses after they were amplified for 72 h in SNE cells at 33°C, STE cells at 37°C, and A549 cells at 39°C.
Growth kinetics.
The growth kinetics were determined in SNE, STE, and A549 cells. Briefly, cells were seeded in 6-well plates at a density of 5 × 105 cells/well. Twenty-four hours later, the cells were washed once with phosphate-buffered saline (PBS) and then infected with a test virus at a multiplicity of infection (MOI) of 10−3 TCID50/cell. After absorption for 1 h at 37°C, the inoculum was removed. The cells were washed once with PBS, and then 3 ml of Opti-MEM medium supplemented with 1 μg/ml of TPCK-trypsin was added. The cultures were incubated at 33°C, 37°C, or 39°C for the duration of the experiment. At 12, 24, 48, and 72 h postinfection, the supernatants were collected, and their TCID50 were then determined. In addition to MO/15, nine representative avian isolates (Table 1) with distinct growth phenotypes across the three testing temperatures were used for the growth kinetics analyses.
RNA extraction and viral copy number determination.
Viral RNA was extracted from nasal wash and tissue samples by using a MagMAX pathogen RNA/DNA kit (Thermo Fisher Scientific, Asheville, NC, USA) according to the manufacturer’s instructions. Quantitative reverse transcription-PCR (qRT-PCR) was used to determine the copy numbers of IAVs in nasal wash samples and homogenized tissue samples as previously described (22). Briefly, qRT-PCR was performed in triplicate by using TaqMan Fast virus 1-step master mix (Life Technology, Carlsbad, CA) with M gene-specific primers 5′-GACCRATCCTGTCACCTCTGAC-3′ (forward primer) and 5′-AGGGCATTYTGGACAAAKCGTCTA-3′ (reverse primer) and 6-carboxyfluorescein-labeled probe 5′-TGCAGTCCTCGCTCACTGGGCACG-3′. The viral copy numbers in the samples were determined with the standard curve, which was generated by a plasmid containing the M gene of PR8. The copy number determinations by qRT-PCR were performed in triplicate.
The quantities of the viral copies in both the nasal wash and homogenized tissue samples were confirmed by using reverse transcription-droplet digital PCR (RT-ddPCR) on a QX200TM droplet digital PCR system (Bio-Rad, Hercules, CA) with a one-step RT-ddPCR advanced kit for probes (Bio-Rad, Hercules, CA). The influenza A virus-specific primers and probe set used in the RT-ddPCR were the same as those used in the qRT-PCR. The RT-ddPCR experiment was performed in duplicate, and the data are shown as the averages of the two values.
Viral purification and quantification of virus particles.
Viruses were purified by sucrose gradient centrifugation as described elsewhere (22). The purified viruses were dissolved in PBS and dialyzed against PBS at 4°C overnight. The concentrations of the virus particles were determined using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as described elsewhere (48).
Virus-glycan receptor binding assay and data analyses.
Two biotinylated glycan analogs, the carbohydrates 3′-sialyl-N-acetyllactosamine (3′SLN), representing SA2,3Gal, and 6′-sialyl-N-acetyllactosamine (6′SLN), representing SA2,6Gal, were purchased from GlycoTech (Gaithersburg, MD). The glycan stocks were reconstituted at 1 mg/ml in a 50% (vol/vol) glycerol–PBS solution according to the manufacturer’s instructions and were stored at 4°C until use. Binding of viruses (at a concentration of 5 nM) to the biotinylated glycan analogs was performed as previously described in an Octet RED96 biolayer interferometer equipped with streptavidin biosensor tips (Pall FortéBio, Menlo Park, CA, USA). The glycan concentrations ranged from 0.007 to 1.5 μg/ml.
Responses were normalized by the highest value obtained during the experiment, and binding curves were fitted by using the binding-saturation method in GraphPad Prism (version 8) software. The normalized response curves report the fractional saturation (f) of the sensor surface, as described in a previous study (49). RSL0.5 (relative sugar loading at an f value of 0.5) was used to quantitate the affinity of binding of two selected viruses to the two glycan analogs. The higher that the RSL0.5 is, the weaker that the binding affinity is.
Feral swine.
For the animal experiments, a total of 26 feral swine (body weight, 16 to 22 kg) were trapped in a rural area of Starkville, MS, USA, by using corral traps similar to those described previously (50). Animals were transported to the National Wildlife Research Center, Mississippi Field Station, in Mississippi State, MS, USA, where they were quarantined for 1 week. Before the animals were included in the experiments, we confirmed that these feral swine had not been exposed to brucellosis, pseudorabies virus, or IAV, using enzyme-linked immunosorbent assays (Idexx, Westbrook, ME) as previously described (50), and that all HI assay results were negative for the three test H4N6 IAVs (MO/15, OS2244, and OS5426) and three endemic human IAVs [i.e., A/California/04/2009 (H1N1), A/Switzerland/9715293/2013 (H3N2), and A/Hong Kong/4801/2014 (H3N2)]. The swine were then housed and fed according to a standard protocol, as described elsewhere (50).
Animal experiment for evaluating the pathogenesis and transmission of H4N6 IAVs.
Twenty-six feral swine were randomly assigned to four experimental groups, including three treatment groups and one control group. The treatment groups included seven pigs each, and the swine were exposed to one of three test H4N6 isolates, MO/15, OS2244, and OS5426.
The pigs in each treatment group were housed in three pens (two or three pigs per pen), and the pigs in the control groups were housed in two pens (two or three pigs per pen). One or two pigs in each pen were experimentally inoculated with virus or sterile PBS, while the remaining cohoused pigs served as the contact. The control group was housed in a building separate from the building housing the experimental groups. Prior to virus inoculation and sample collection, the pigs were anesthetized using a method described previously (50). After being anesthetized, four pigs within each treatment group were intranasally inoculated with 106 TCID50 of virus in a volume of 2 ml (1 ml/nostril). Three IAV-seronegative contact pigs were cohoused at 24 h after virus inoculation. Two pigs in the control group were intranasally inoculated with 2 ml of PBS.
On 1 to 10 dpi, nasal wash fluids were collected from both nostrils of all pigs, placed into 3 ml of PBS with 100 units/ml of penicillin-streptomycin, and then subjected to viral copy number determination and titration by TCID50. The body temperature of each pig was measured before samples were taken. Serum from each pig was also collected for seroconversion analysis using an HI assay with 0.5% turkey red blood cells, as described previously (45). At 5 and 7 dpi, one pig from each treatment group and one control pig were euthanized, and a necropsy was performed according to a previously described procedure (50).
Tissues of the respiratory tract were collected for viral titration: left cranial lung (LCR), left caudal lung (LCD), right cranial lung (RCR), right caudal lung (RCD), right middle lung (RMD), right accessory (RA), upper trachea (TR-U), middle trachea (TR-M), distal trachea (TR-D), bronchus (BR), soft palate (SP), ethmoid turbinate (ET), rostral turbinate (RT), and middle turbinate (MT). To quantify IAV, each tissue specimen was homogenized at a ratio of 1:4 (wt/vol; 1 g tissue in 4 ml of buffer) in PBS containing 100 units/ml of penicillin-streptomycin, and then the solutions were subjected to three freeze-thaw cycles prior to performing RNA extraction and virus titrations.
Genomic sequences.
A total of 1,497 H4N6 genomic sequences were downloaded from the Influenza Research Database (IRD), the database of the Global Initiative on Sharing All Influenza Data (GISAID), and the database of the National Center for Biotechnology Information (NCBI) on 2 May 2017, and the genomic sequences of 35 H4N6 isolates were obtained from USDA. The repeated sequence records were removed, and data from the three databases were merged. A total of 115 avian H4N6 isolates from North America were selected to represent the wide genomic diversity of H4N6 IAVs in North America as well as a diversity of avian host species, sampling times, and sampling locations (Table 1).
MSA and phylogenetic analysis.
Multiple-sequence alignments (MSA) were generated by using MAFFT (version 7.273) software (51). Phylogenetic analysis was performed using a maximum likelihood tree to represent the evolutionary relationship among different isolates, as described elsewhere (52, 53). The phylogenetic trees for all gene segments (PB2, PB1, PA, HA, NP, NA, MP, and NS) were inferred using a maximum likelihood method implemented in the RAxML (version 8.2.9) program (54). A gamma model of rate heterogeneity and a generalized time-reversible (GTR) substitution model were applied in the analysis. Phylogenetic trees were then visualized by use of the ggtree (version 1.6.11) program (55).
Statistical analyses.
A nonparametric method, the Kruskal-Wallis test (GraphPad Prism software), was used to test the hypothesis that the polymerase activities of influenza viruses is affected by temperature. This analysis was performed on the polymerase activities of the RNP of MO/15, the RNP of OS2244, and the RNP of PR8 under three testing temperatures, 33, 37, and 39°C. To test the hypothesis that the genomic constellation would affect the polymerase activities of the RNP complex, a 2-way analysis of variance was used to compare the difference in polymerase activity between the RNP reassortants: between the MO/15-OS2244 RNP reassortant and the MO/15-OS5426 RNP reassortant, the MO/15-OS2244 RNP reassortant and the OS2244-OS5426 RNP reassortant, and the MO/15-OS5426 RNP reassortant and the OS2244-OS5426 RNP reassortant. A P value of 0.05 was determined to be statistically significant for all analyses.
Biosafety protocol for laboratory and animal experiments.
Virus titration and purification and virus inoculation in feral swine were conducted under biosafety level 2 conditions, in compliance with the protocols approved by the U.S. Department of Agriculture Institutional Biosafety Committee.
Ethics statement.
The animal experiments were performed under protocol number QA2625, titled Potential of Avian Influenza A Virus To Infect Feral Swine, which was approved by the Institutional Animal Care and Use Committee of the National Wildlife Research Center (NWRC), in accordance with USDA animal welfare regulations.
ACKNOWLEDGMENTS
We acknowledge the USDA Swine Influenza Surveillance Program for providing influenza virus isolates and Stacey Schultz-Cherry at St. Jude Children’s Research Hospital for providing swine primary cells.
This project was partially supported by grant R21AI135820 from the National Institutes of Health.
REFERENCES
- 1.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 4.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. doi: 10.1128/MMBR.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph U, Su YC, Vijaykrishna D, Smith GJ. 2017. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir Viruses 11:74–84. doi: 10.1111/irv.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Short KR, Richard M, Verhagen JH, van Riel D, Schrauwen EJ, van den Brand JM, Manz B, Bodewes R, Herfst S. 2015. One Health, multiple challenges: the inter-species transmission of influenza A virus. One Health 1:1–13. doi: 10.1016/j.onehlt.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown IH. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol 74:29–46. doi: 10.1016/s0378-1135(00)00164-4. [DOI] [PubMed] [Google Scholar]
- 8.Webby RJ, Swenson SL, Krauss SL, Goyal SM, Rossow KD, Webster RG. 2001. Evolving H3N2 and emerging H1N2 swine influenza viruses in the United States. Int Congr Ser 1219:241–249. doi: 10.1016/S0531-5131(01)00408-3. [DOI] [Google Scholar]
- 9.Scholtissek C. 1990. Pigs as the ‘mixing vessel’ for the creation of new pandemic influenza A viruses. Med Princ Pract 2:65–71. doi: 10.1159/000157337. [DOI] [Google Scholar]
- 10.Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med 360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 11.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, Kawaoka Y, Webster RG. 1994. Potential for transmission of avian influenza viruses to pigs. J Gen Virol 75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 12.Su S, Qi W, Chen J, Zhu W, Huang Z, Xie J, Zhang G. 2013. Seroepidemiological evidence of avian influenza A virus transmission to pigs in southern China. J Clin Microbiol 51:601–602. doi: 10.1128/JCM.02625-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ninomiya A, Takada A, Okazaki K, Shortridge KF, Kida H. 2002. Seroepidemiological evidence of avian H4, H5, and H9 influenza A virus transmission to pigs in southeastern China. Vet Microbiol 88:107–114. doi: 10.1016/s0378-1135(02)00105-0. [DOI] [PubMed] [Google Scholar]
- 14.Jung K, Song D-S, Kang B-K, Oh J-S, Park B-K. 2007. Serologic surveillance of swine H1 and H3 and avian H5 and H9 influenza A virus infections in swine population in Korea. Prev Vet Med 79:294–303. doi: 10.1016/j.prevetmed.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun H, Kaplan BS, Guan M, Zhang G, Ye J, Long L-P, Blackmon S, Yang C-K, Chiang M-J, Xie H, Zhao N, Cooley J, Smith DF, Liao M, Cardona C, Li L, Wang GP, Webby R, Wan X-F. 2017. Pathogenicity and transmission of a swine influenza A(H6N6) virus. Emerg Microbes Infect 6:e17. doi: 10.1038/emi.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snyder MH, Betts RF, DeBorde D, Tierney EL, Clements ML, Herrington D, Sears SD, Dolin R, Maassab HF, Murphy BR. 1988. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol 62:488–495. doi: 10.1128/JVI.62.2.488-495.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin H, Zhou H, Lu B, Kemble G. 2004. Imparting temperature sensitivity and attenuation in ferrets to A/Puerto Rico/8/34 influenza virus by transferring the genetic signature for temperature sensitivity from cold-adapted A/Ann Arbor/6/60. J Virol 78:995–998. doi: 10.1128/jvi.78.2.995-998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin H, Lu B, Zhou H, Ma C, Zhao J, Yang CF, Kemble G, Greenberg H. 2003. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (FluMist) derived from cold-adapted A/Ann Arbor/6/60. Virology 306:18–24. doi: 10.1016/s0042-6822(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 20.Herlocher ML, Clavo AC, Maassab HF. 1996. Sequence comparisons of A/AA/6/60 influenza viruses: mutations which may contribute to attenuation. Virus Res 42:11–25. doi: 10.1016/0168-1702(96)01292-0. [DOI] [PubMed] [Google Scholar]
- 21.Mänz B, Schwemmle M, Brunotte L. 2013. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol 87:7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Sun H, Cunningham FL, Li L, Hanson-Dorr K, Hopken MW, Cooley J, Long LP, Baroch JA, Li T, Schmit BS, Lin X, Olivier AK, Jarman RG, DeLiberto TJ, Wan XF. 2018. Tissue tropisms opt for transmissible reassortants during avian and swine influenza A virus co-infection in swine. PLoS Pathog 14:e1007417. doi: 10.1371/journal.ppat.1007417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abente EJ, Gauger PC, Walia RR, Rajao DS, Zhang J, Harmon KM, Killian ML, Vincent AL. 2017. Detection and characterization of an H4N6 avian-lineage influenza A virus in pigs in the midwestern United States. Virology 511:56–65. doi: 10.1016/j.virol.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karasin AI, Brown IH, Carman S, Olsen CW. 2000. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol 74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li IW, Chan KH, To KW, Wong SS, Ho PL, Lau SK, Woo PC, Tsoi HW, Chan JF, Cheng VC, Zheng BJ, Chen H, Yuen KY. 2009. Differential susceptibility of different cell lines to swine-origin influenza A H1N1, seasonal human influenza A H1N1, and avian influenza A H5N1 viruses. J Clin Virol 46:325–330. doi: 10.1016/j.jcv.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Khatri M, Goyal SM, Saif YM. 2012. Oct4+ stem/progenitor swine lung epithelial cells are targets for influenza virus replication. J Virol 86:6427–6433. doi: 10.1128/JVI.00341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hui CF, Chan RW, Fung K, Yu WC, Tsao S, Chan MC, Nicholls JM. 2011. The regional distribution of different types of influenza receptors in cultured human alveolar epithelial cells and correlation with in vitro infection. Influenza Other Respir Viruses 5(Suppl 1):436–437. [PMC free article] [PubMed] [Google Scholar]
- 28.Trebbien R, Larsen LE, Viuff BM. 2011. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol J 8:434. doi: 10.1186/1743-422X-8-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujino N, Kubo H, Ota C, Suzuki T, Takahashi T, Yamada M, Suzuki S, Kondo T, Nagatomi R, Tando Y, Yamaya M. 2013. Increased severity of 2009 pandemic influenza A virus subtype H1N1 infection in alveolar type II cells from patients with pulmonary fibrosis. J Infect Dis 207:692–693. doi: 10.1093/infdis/jis739. [DOI] [PubMed] [Google Scholar]
- 30.Weinheimer VK, Becher A, Tonnies M, Holland G, Knepper J, Bauer TT, Schneider P, Neudecker J, Ruckert JC, Szymanski K, Temmesfeld-Wollbrueck B, Gruber AD, Bannert N, Suttorp N, Hippenstiel S, Wolff T, Hocke AC. 2012. Influenza A viruses target type II pneumocytes in the human lung. J Infect Dis 206:1685–1694. doi: 10.1093/infdis/jis455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bateman AC, Busch MG, Karasin AI, Bovin N, Olsen CW. 2008. Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for α2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. J Virol 82:8204–8209. doi: 10.1128/JVI.00718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song H, Qi J, Xiao H, Bi Y, Zhang W, Xu Y, Wang F, Shi Y, Gao GF. 2017. Avian-to-human receptor-binding adaptation by influenza A virus hemagglutinin H4. Cell Rep 20:1201–1214. doi: 10.1016/j.celrep.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 33.Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67:1761–1764. doi: 10.1128/JVI.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. 2007. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog 3:e133. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kayali G, Barbour E, Dbaibo G, Tabet C, Saade M, Shaib HA, Debeauchamp J, Webby RJ. 2011. Evidence of infection with H4 and H11 avian influenza viruses among Lebanese chicken growers. PLoS One 6:e26818. doi: 10.1371/journal.pone.0026818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang L, Deng G, Shi J, Wang S, Zhang Q, Kong H, Gu C, Guan Y, Suzuki Y, Li Y, Jiang Y, Tian G, Liu L, Li C, Chen H. 2016. Genetics, receptor binding, replication, and mammalian transmission of H4 avian influenza viruses isolated from live poultry markets in China. J Virol 90:1455–1469. doi: 10.1128/JVI.02692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slavec B, Krapez U, Racnik AJ, Hari A, Wernig JM, Dovc A, Zadravec M, Lindtner-Knific R, Marhold C, Zorman-Rojs O. 2012. Surveillance of influenza A viruses in wild birds in Slovenia from 2006 to 2010. Avian Dis 56:999–1005. doi: 10.1637/10175-041012-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Q, Li Q, Zhong L, Gu M, Zhu J, Zhao G, Chen C, Wang X, Liu X, Liu X. 2013. Complete genomic sequence of a novel reassortant H4N2 avian influenza virus isolated from domestic ducks in Jiangsu, China. Genome Announc 1:e00091-13. doi: 10.1128/genomeA.00091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su S, Qi W-B, Chen J-D, Cao N, Zhu W-J, Yuan L-G, Wang H, Zhang G-H. 2012. Complete genome sequence of an avian-like H4N8 swine influenza virus discovered in southern China. J Virol 86:9542. doi: 10.1128/JVI.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y, Liu X, Li S, Guo X, Yang Y, Jin M. 2012. Complete genome sequence of a novel H4N1 influenza virus isolated from a pig in central China. J Virol 86:13879. doi: 10.1128/JVI.02726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin BE, Sun H, Carrel M, Cunningham FL, Baroch JA, Hanson-Dorr KC, Young SG, Schmit B, Nolting JM, Yoon KJ, Lutman MW, Pedersen K, Lager K, Bowman AS, Slemons RD, Smith DR, DeLiberto T, Wan XF. 2017. Feral swine in the United States have been exposed to both avian and swine influenza A viruses. Appl Environ Microbiol 83:e01346-17. doi: 10.1128/AEM.01346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bui VN, Ogawa H, Xininigen Karibe K, Matsuo K, Awad SS, Minoungou GL, Yoden S, Haneda H, Ngo LH, Tamaki S, Yamamoto Y, Nakamura K, Saito K, Watanabe Y, Runstadler J, Huettmann F, Happ GM, Imai K. 2012. H4N8 subtype avian influenza virus isolated from shorebirds contains a unique PB1 gene and causes severe respiratory disease in mice. Virology 423:77–88. doi: 10.1016/j.virol.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belser JA, Barclay W, Barr I, Fouchier RAM, Matsuyama R, Nishiura H, Peiris M, Russell CJ, Subbarao K, Zhu H, Yen HL. 2018. Ferrets as models for influenza virus transmission studies and pandemic risk assessments. Emerg Infect Dis 24:965–971. doi: 10.3201/eid2406.172114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun H, Yang J, Zhang T, Long LP, Jia K, Yang G, Webby RJ, Wan XF. 2013. Using sequence data to infer the antigenicity of influenza virus. mBio 4:e00230-13. doi: 10.1128/mBio.00230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 47.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ, Gamblin SJ, Hay AJ, Daniels RS, McCauley JW. 2012. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A 109:21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. 2013. Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 50.Sun H, Cunningham FL, Harris J, Xu Y, Long LP, Hanson-Dorr K, Baroch JA, Fioranelli P, Lutman MW, Li T, Pedersen K, Schmit BS, Cooley J, Lin X, Jarman RG, DeLiberto TJ, Wan XF. 2015. Dynamics of virus shedding and antibody responses in influenza A virus-infected feral swine. J Gen Virol 96:2569–2578. doi: 10.1099/jgv.0.000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, Bowman AS, DeLiberto TJ, Killian ML, Krauss S, Nolting JM, Torchetti MK, Ramey AM, Reeves AB, Stallknecht DE, Webby RJ, Wan X-F. 2018. Genetic evidence supports sporadic and independent introductions of subtype H5 low-pathogenic avian influenza A viruses from wild birds to domestic poultry in North America. J Virol 92:e00913-18. doi: 10.1128/JVI.00913-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, DeLiberto TJ, Killian ML, Torchetti MK, Wan X-F. 2018. Evolutionary pathway for the 2017 emergence of a novel highly pathogenic avian influenza A (H7N9) virus among domestic poultry in Tennessee, United States. Virology 525:32–39. doi: 10.1016/j.virol.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]