Human papillomavirus (HPV) is the causative agent of cervical and some other epithelial cancers. HPV vaccines generate functional (neutralizing) antibodies that target the virus particles (or capsids) of the most common HPV cancer-causing genotypes. Each genotype comprises variant forms that have arisen over millennia and which include changes within the capsid proteins. In this study, we explored the potential for these naturally occurring variant capsids to impact recognition by neutralizing monoclonal antibodies. All genotypes included at least one variant form that exhibited reduced recognition by at least one antibody, with some genotypes affected more than others. These data highlight the impact of naturally occurring variation on the structure of the HPV capsid proteins of vaccine-relevant oncogenic HPV genotypes.
KEYWORDS: human papillomavirus, variant, lineage, antigenicity, neutralization, monoclonal antibodies
ABSTRACT
Human papillomavirus (HPV) is the causative agent of cervical and other epithelial cancers. Naturally occurring variants of HPV have been classified into lineages and sublineages based on their whole-genome sequences, but little is known about the impact of this diversity on the structure and function of viral gene products. The HPV capsid is an icosahedral lattice comprising 72 pentamers of the major capsid protein (L1) and the associated minor capsid protein (L2). We investigated the potential impact of this genome variation on the capsid antigenicity of lineage and sublineage variants of seven vaccine-relevant, oncogenic HPV genotypes by using a large panel of monoclonal antibodies (MAbs) raised against the L1 proteins of lineage A antigens. Each genotype had at least one variant that displayed a ≥4-fold reduced neutralizing antibody sensitivity against at least one MAb, demonstrating that naturally occurring variation can affect one or more functional antigenic determinants on the HPV capsid. For HPV16, HPV18, HPV31, and HPV45, the overall impact was of a low magnitude. For HPV33 (sublineages A2 and A3 and lineages B and C), HPV52 (lineage D), and HPV58 (lineage C), however, variant residues in the indicated lineages and sublineages reduced their sensitivity to neutralization by all MAbs by up to 1,000-fold, suggesting the presence of key antigenic determinants on the surface of these capsids. These determinants were resolved further by site-directed mutagenesis. These data improve our understanding of the impact of naturally occurring variation on the antigenicity of the HPV capsid of vaccine-relevant oncogenic HPV genotypes.
IMPORTANCE Human papillomavirus (HPV) is the causative agent of cervical and some other epithelial cancers. HPV vaccines generate functional (neutralizing) antibodies that target the virus particles (or capsids) of the most common HPV cancer-causing genotypes. Each genotype comprises variant forms that have arisen over millennia and which include changes within the capsid proteins. In this study, we explored the potential for these naturally occurring variant capsids to impact recognition by neutralizing monoclonal antibodies. All genotypes included at least one variant form that exhibited reduced recognition by at least one antibody, with some genotypes affected more than others. These data highlight the impact of naturally occurring variation on the structure of the HPV capsid proteins of vaccine-relevant oncogenic HPV genotypes.
INTRODUCTION
Human papillomavirus (HPV) is the causative agent of cervical and other epithelial cancers and accounts for >600,000 cases globally per annum (1–3). Small double-stranded DNA (dsDNA) genomes typically exhibit a low evolutionary rate (4), although distinct HPV genotypes have arisen over time (5). Genotypes from the Alphapapillomavirus genus contribute to the development of cervical and other cancers (2), with HPV16 conferring the highest relative risk.
Whole-genome sequence analysis has led to the delineation of distinct HPV lineages and sublineages that exhibit both geographical bias in their distribution and differential disease risk (5–7). Efforts are also underway to understand the evolution of HPV variants from their prehistoric origins (6, 7). For example, HPV16 (6) and HPV58 (7) non-A lineages (B/C/D) are estimated to have split from their respective lineage A viruses approximately 400 to 600 thousand years ago (kya), followed by the further resolution of lineages B, C, and D by approximately 100 to 200 kya, coincident with the evolution and global migration of ancient hominins (6, 7).
The HPV capsid is an icosahedral lattice comprising 72 pentamers of the major capsid protein (L1) and includes the asymmetrical and/or stochastic distribution of the minor capsid protein (L2) (8, 9). Each L1 monomer consists of a core of β-strands and α-helices which support the five surface-exposed loop domains designated BC, DE, EF, FG, and HI. Type-specific neutralizing antibodies raised against the L1 capsid protein predominantly target these surface-exposed loops (10). The binding of a number of monoclonal antibodies (MAbs) to the capsid surface has been resolved to ca. 3 Å using pentameric crystals or ca. 10 Å by cryo-electron microscopy (11–16) and reveals the complexity of these interactions, including antibody footprints spanning multiple loops and adjacent monomers within a pentamer.
Neutralizing antibodies directed against the L1 capsid can passively protect in preclinical challenge models, leading to the development of highly efficacious L1 capsid-based prophylactic vaccines (17). Bivalent (Cervarix) and quadrivalent (Gardasil) vaccines target the most prevalent oncogenic genotypes (HPV16 and HPV18), while the nonavalent (Gardasil 9) vaccine targets five additional oncogenic genotypes (HPV31, HPV33, HPV45, HPV52, and HPV58). Quadrivalent and nonavalent vaccines also target nononcogenic genotypes, HPV6 and HPV11, which can cause genital warts. Other next-generation prophylactic vaccines are in various stages of development, with some attaining licensure (3, 18). The HPV vaccines are highly immunogenic (19) and have demonstrated remarkable efficacy in clinical trials (20), and vaccine effectiveness studies are beginning to confirm these experimental observations in target populations following introduction of national immunization programs (21).
The biological consequences of HPV genome variants are unclear. Studies examining the potential impact of natural variation on L1 antigenicity have examined variants of HPV16 (22, 23), HPV31 (24), HPV33 (25), HPV45 (26), HPV52 (27), and HPV58 (28) and demonstrated, in some cases, variant-specific differences in sensitivity to monovalent, bivalent, and quadrivalent virus-like particle (VLP) immune sera, animal immune sera, natural infection sera, and some MAbs. We recently examined the differential recognition of HPV lineage and sublineage variants by nonavalent vaccine sera (29). Here, we present a comprehensive evaluation of the antigenicity of lineage and sublineage variants of seven vaccine-relevant, oncogenic HPV genotypes using a large panel of MAbs raised against lineage A antigens.
RESULTS
Basic characterization of type-specific MAbs.
One hundred and one MAbs were assembled from four laboratories representing HPV16, HPV18, HPV31, HPV33, HPV45, HPV52, and HPV58 genotypes. Most (n = 87) MAbs were type-specific in their binding, with a small number (n = 14) demonstrating cross-reactivity to other genotypes. Approximately two-thirds of MAbs exhibited a strongly neutralizing phenotype and were represented by a range of isotypes: IgG1 (n = 18), IgG2a (n = 17), IgG2b (n = 21), and IgG3 (n = 3), along with two whose isotype could not be determined (data not shown).
Antigenic impact of natural variation in the capsid proteins.
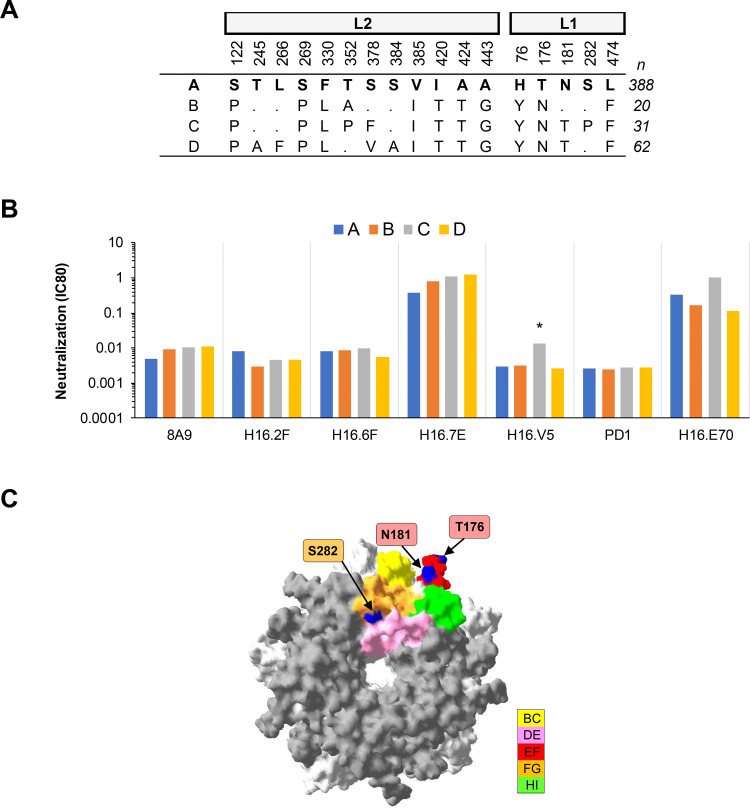
(i) HPV16. One MAb, H16.V5, exhibited a ≥4-fold increase in IC80 against HPV16 lineage C, while the remaining six MAbs exhibited similar (<4-fold difference) neutralizing antibody profiles against each HPV16 lineage (Fig. 1). Lineage C has an S282P polymorphism in the FG loop that distinguishes it from the other lineage variants. The footprint of MAb H16.V5 has been mapped (13, 14) and encompasses a number of residues in the FG loop, including S282. These data confirm residue S282 as being an important antigenic site in the H16.V5 footprint and corroborate previous site-directed mutagenesis data (23). H16.V5 is used in the competitive Luminex immunoassay (cLIA) to evaluate vaccine-induced and natural immunity to HPV (30–32). HPV16 lineage C is a minority variant found in Africa (5, 33).
FIG 1.
Neutralization of HPV16 lineage variants by MAbs. (A) Consensus lineage HPV16 L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of lineage L1 and L2 PsV variants to HPV16 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. The symbol * indicates a ≥4-fold reduced sensitivity in the magnitude of difference in MAb sensitivity (IC80) between the indicated variant and the consensus A/A1 PsV. (C) HPV16 lineage A homology model based upon the HPV16 L1 pentamer crystal (Protein Databank [PDB] accession number 2R5H.1; using DeepView Swiss-Pdb viewer v4.0) with top view shown. The quality of a predicted model was assessed by their qualitative model energy analysis (QMEAN4) and global model quality estimation (GMQ), which were −3.33 and 0.99, respectively. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC1; pink, DE2; red, EF2; orange, FG2; and green, HI1).
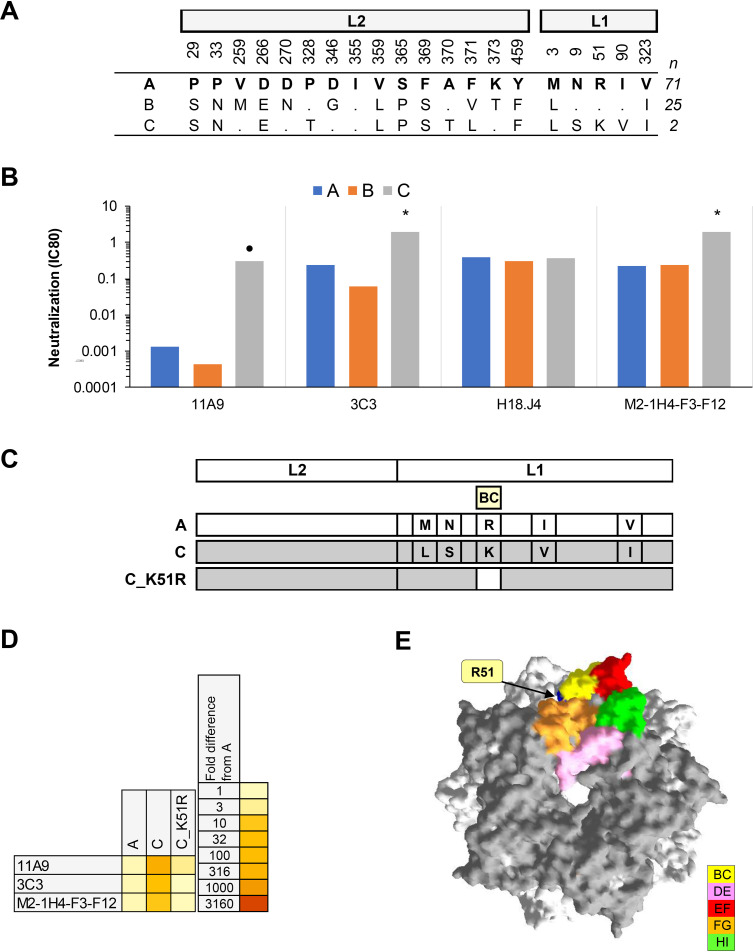
(ii) HPV18. Three MAbs exhibited a ≥4-fold increase in IC80 against HPV18 lineage C, including one that displayed a >100-fold increase in IC80. One MAb (H18.J4, a MAb also used in the cLIA [30]) displayed a similar neutralizing antibody profile against each HPV18 lineage (Fig. 2). Lineage C has an R51K polymorphism in the BC loop that distinguishes it from the other HPV18 lineages. A mutant pseudovirus (PsV) based upon lineage C with a K51R back mutation recapitulated the HPV18 lineage A phenotype, demonstrating the role of this single residue in the observed differential recognition of HPV18 lineage C by these MAbs. HPV18 lineage C is a minority variant found in Africa (34).
FIG 2.
Neutralization of HPV18 lineage variants by MAbs. (A) Consensus lineage HPV18 L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of lineage L1 and L2 PsV variants to HPV18 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. Symbols indicate magnitude of difference in MAb sensitivity (IC80) between indicated variant and consensus A/A1 PsV: *, ≥4-fold reduced sensitivity; ●, ≥100-fold reduced sensitivity. (C) Graphical representation of HPV18 L1 and L2 protein combinations from A (white) and C (gray). (D) Heatmap representation of fold difference in neutralization potency (IC80) against the A variant. (E) HPV18 lineage A homology model based upon the HPV18 L1 pentamer crystal (PDB accession number 2R5I.1) with top view shown. The quality of a predicted model was assessed by its QMEAN4 and GMQ, which were −2.20 and 0.99, respectively. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC1; pink, DE2; red, EF2; orange, FG2; and green, HI1).
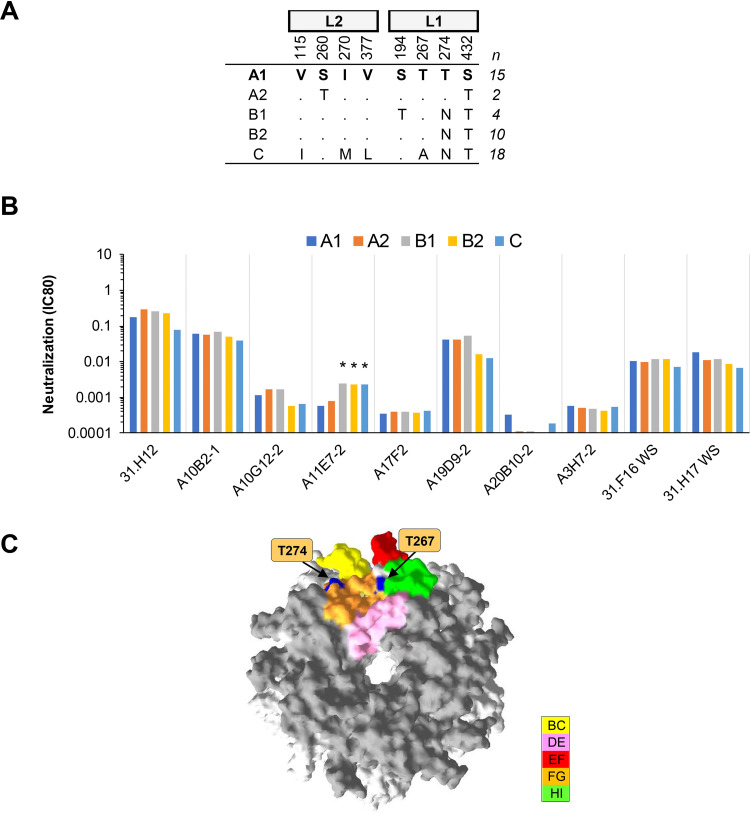
(iii) HPV31. One MAb, A11E7-2, exhibited a ≥4-fold increase in IC80 against HPV31 variants B1, B2, and C, while the remaining nine MAbs exhibited similar neutralizing antibody profiles against each HPV31 lineage (Fig. 3). The FG loop polymorphism T274N distinguishes variants B1, B2, and C from the rest. These data corroborate previous work (24, 35) suggesting that natural variation within the FG loop is unlikely to have significant implications for antibody recognition.
FIG 3.
Neutralization of HPV31 lineage and sublineage variants by MAbs. (A) Consensus lineage and sublineage HPV31 L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of lineage and sublineage L1 and L2 PsV variants to HPV31 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. Symbols indicate magnitude of difference in MAb sensitivity (IC80) between indicated variant and consensus A/A1 PsV: *, ≥4-fold reduced sensitivity. (C) HPV31 sublineage A1 homology model based upon the HPV16 L1 pentamer crystal (PDB accession number 2R5H.1) with top view shown. The quality of a predicted model was assessed by its QMEAN4 and GMQ, which were −3.35 and 0.96, respectively. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC1; pink, DE2; red, EF2; orange, FG2; and green, HI1).
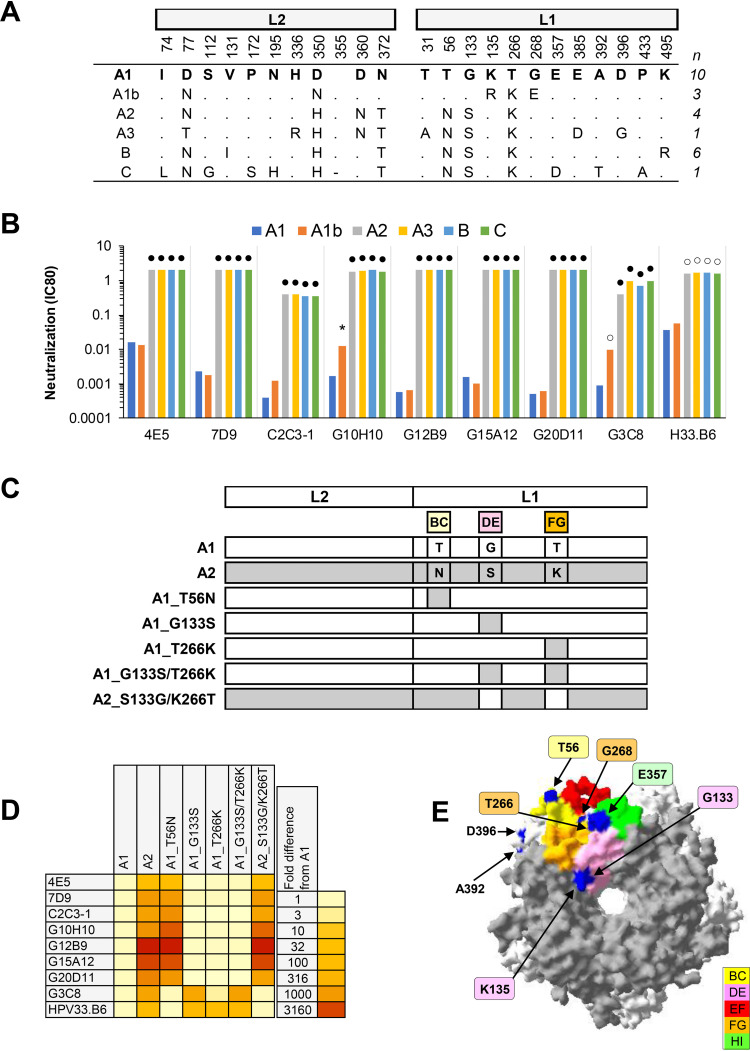
(iv) HPV33. All nine MAbs exhibited a median 1,046-fold increase in IC80 against HPV33 variants A2, A3, B, and C (Fig. 4). These variants share the motif of BC loop T56N, DE loop G133S, and FG loop T266K. The dependency on specific amino acid residues was further explored using mutant PsVs incorporating these T56N, G133S, and T266K polymorphisms. One MAb (G3C8) displayed a preference for the G133 residue, while another MAb (HPV33.B6) had a preference for residues in both the DE (G133) and FG (T266) loops (36). For the remaining MAbs, the BC loop residue (T56) alone appeared to be critical. These data corroborate observations on the differential recognition of these same variants by bivalent and quadrivalent vaccine-induced cross-neutralizing antibodies and nonavalent vaccine-induced type-specific antibodies (25, 29), suggesting that these variant residues are localized within a key antigenic determinant on the capsid surface. In addition, these data also highlight subtle differences in antibody specificity between vaccine-induced neutralizing antibodies that appear to target sites within the DE and FG loops (25) (analogous to MAb H33.B6) and the majority of type-specific MAbs tested here, which preferentially target one or more antigenic determinants that include the BC loop. One estimate suggests that variants A2, A3, B, and C account for ca. 50% of the global prevalence of HPV33 variants, though these appear to be distributed asymmetrically (37).
FIG 4.
Neutralization of HPV33 lineage and sublineage variants by MAbs. (A) Consensus lineage and sublineage HPV33 L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of lineage and sublineage L1 and L2 PsV variants to HPV33 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. Symbols indicate magnitude of difference in MAb sensitivity (IC80) between indicated variant and consensus A/A1 PsV: *, ≥4-fold reduced sensitivity; ○, ≥10-fold reduced sensitivity; ●, ≥100-fold reduced sensitivity. (C) Graphical representation of HPV33 L1 and L2 protein combinations from A1 (white) and A2 (gray). (D) Heatmap representation of fold difference in neutralization potency (IC80) against the A1 variant. (E) HPV33 sublineage A1 L1 pentamer crystal (PDB accession number: 6IGE.2) with top view shown. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC5; pink, DE4; red, EF4; orange, FG4; and green, HI5).
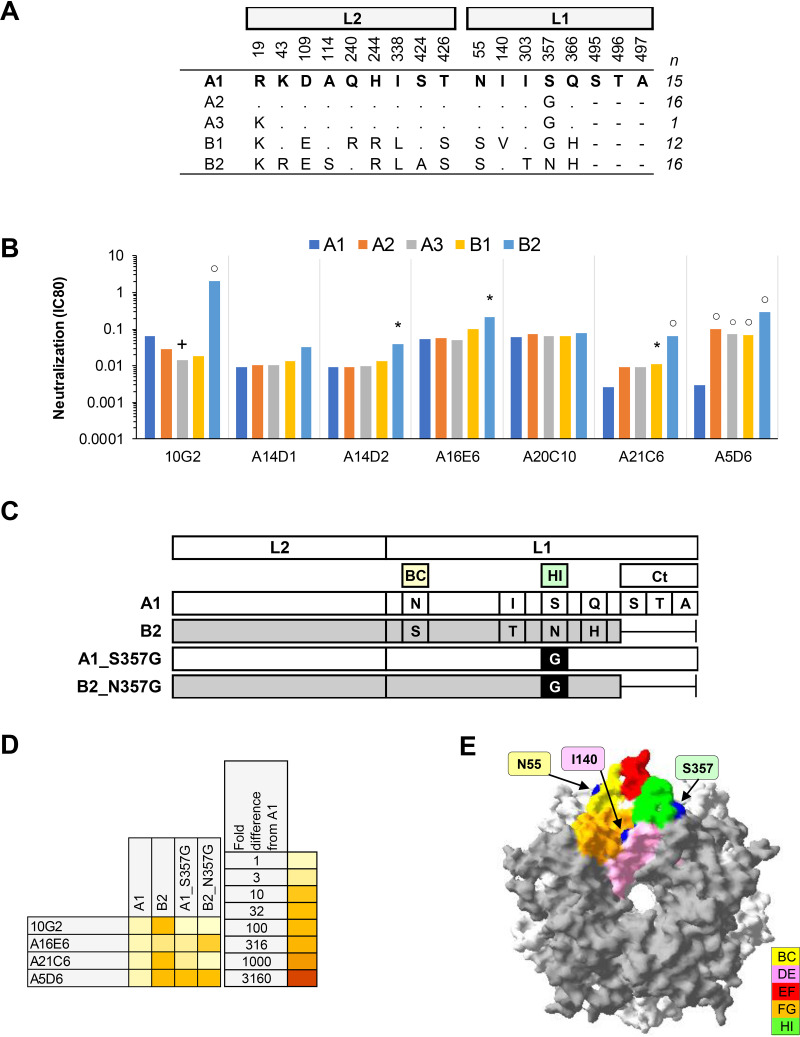
(v) HPV45. Five of seven MAbs exhibited a median 25-fold increase in IC80 against HPV45 lineage B2 (Fig. 5), while one MAb also demonstrated an increased IC80 against variants A2, A3, and B1. Two MAbs were unaffected by variant residue polymorphisms. Four MAbs were tested further against mutant PsV incorporating a G357 residue in a sublineage A1 or B2 background and demonstrated the importance of this residue in the epitopes of some of these MAbs. Nonavalent vaccine antibodies appear to recognize most HPV45 variants similarly but have a slightly enhanced recognition of sublineage B2 (29). Together with previously published data on bivalent and quadrivalent vaccine-induced cross-neutralizing antibodies (26), these data suggest that the HI loop residue 357 is located within an important antigenic site on the HPV45 capsid. HPV45 sublineages A2, A3, B1, and B2 dominate the distribution of HPV45 variants globally, with B2 constituting 20 to 30% of global variants (38).
FIG 5.
Neutralization of HPV45 sublineage variants by MAbs. (A) Consensus sublineage HPV45 L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of sublineage L1 and L2 PsV variants to HPV45 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. Symbols indicate magnitude of difference in MAb sensitivity (IC80) between indicated variant and consensus A/A1 PsV: +, ≥4-fold increased sensitivity; *, ≥4-fold reduced sensitivity; ○, ≥10-fold reduced sensitivity. (C) Graphical representation of HPV45 L1 and L2 protein combinations from A1 (white) and B2 (gray). (D) Heatmap representation of fold difference in neutralization potency (IC80) against the A1 variant. (E) HPV45 sublineage A1 homology model based upon the HPV18 L1 pentamer crystal (PDB accession number 2R5I.1) with top view shown. The quality of a predicted model was assessed by its QMEAN4 and GMQ, which were −2.81 and 0.99, respectively. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC1; pink, DE2; red, EF2; orange, FG2; and green, HI1).
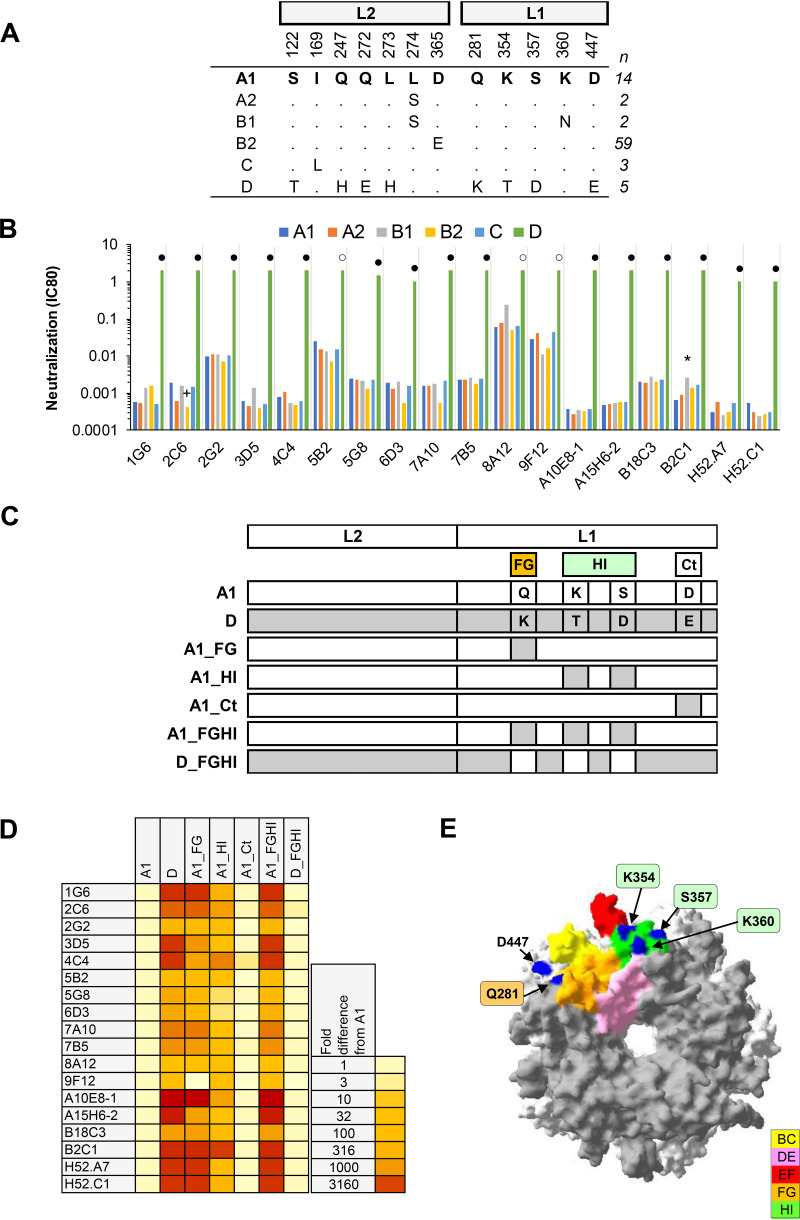
(vi) HPV52. Eighteen MAbs exhibited a median 1,184-fold increase in IC80 against HPV52 lineage D compared to A1, but neutralized sublineage A2, B1, and B2 and lineage C PsVs similarly (Fig. 6). HPV52 lineage D contains four variant residues in the FG (Q281K) and HI (K354T/S357D) loops and the C terminus (Ct) (D447E). MAbs were subsequently tested against sublineage A1 PsV constructs incorporating these variant residue mutations and demonstrated a lack of involvement of the Ct residue D447 and a dependency on residues in both the FG (Q281) and HI (K354/S357) loops. This dependency can be demonstrated whether the background is an A1 (A1_Q281K/K354T/S357D) or a D variant (D_K281Q/T354K/D357S). These data are consistent with the specificity of natural infection, preclinical immune sera (27), and nonavalent vaccine sera (29), and suggest that these residues are topographically located within a key antigenic site on the HPV52 capsid surface. HPV52 sublineage A1 appears to be the majority variant globally, apart from in Asia where sublineage B2 tends to predominate (39). HPV52 lineage D is a minority variant accounting for <2% of HPV52 infections globally but ca. 20% of HPV52 infections in Africa.
FIG 6.
Neutralization of HPV52 lineage and sublineage variants by MAbs. (A) Consensus lineage and sublineage L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of lineage and sublineage HPV52 L1 and L2 PsV variants to HPV52 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. Symbols indicate magnitude of difference in MAb sensitivity (IC80) between indicated variant and consensus A/A1 PsV: +, ≥4-fold increased sensitivity; *, ≥4-fold reduced sensitivity; ○, ≥10-fold reduced sensitivity; ●, ≥100-fold reduced sensitivity. (C) Graphical representation of HPV52 L1 and L2 protein combinations from A1 (white) and D (gray). (D) Heatmap representation of fold difference in neutralization potency (IC80) against the A1 variant. (E) HPV52 sublineage A1 L1 pentamer crystal (PDB accession number: 6IGF.1) with top view shown. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC2; pink, DE5; red, EF5; orange, FG5; and green, HI2).
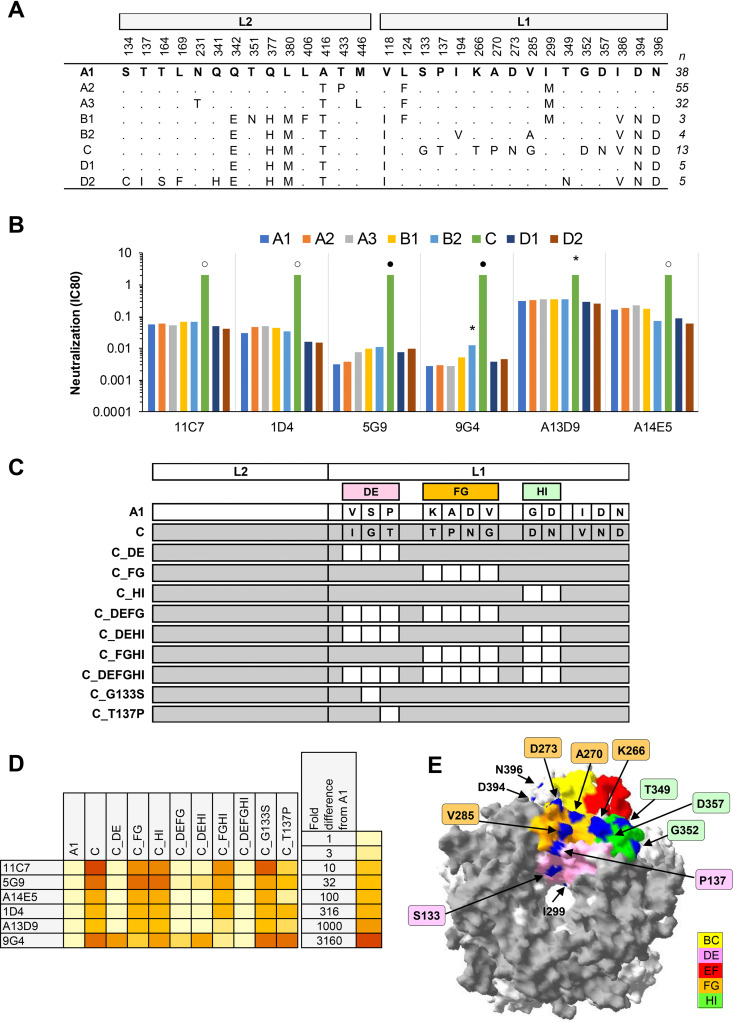
(vii) HPV58. Six MAbs exhibited a median 51-fold increase in IC80 against HPV58 lineage C but neutralized sublineage A2, A3, B1, B2, D1, and D2 PsVs similarly (Fig. 7). HPV58 lineage C incorporates 12 polymorphic residues compared to sublineage A1. The majority of these MAbs appeared to be dependent on residues in the DE loop, confirming previous data on the differential recognition of HPV58 capsid variants by HPV58 MAbs (28) and published L1 mapping data (16). We have extended these observations to demonstrate that the DE residue P137, but not S133, appears to be a key determinant of these epitope(s). One MAb (9G4) was also dependent on one or more residues in the FG loop. Together with previous observations on the differential recognition of the HPV58 lineage C variant by natural infection sera, animal immune sera (28), and nonavalent vaccine immune sera (29), these data suggest that residues in the DE, FG, and HI loops form part of a key antigenic determinant on the HPV58 capsid surface. It is noteworthy perhaps that while most MAbs were dependent on residues in the DE loop, the more complex antibody repertoire found within immune sera required residues from all three (DE, FG, and HI) loops (28). HPV58 sublineage A2 appears to be the most prevalent variant globally (7), while HPV58 lineage C constitutes ca. 10% of HPV58 infections in Europe and the Americas and ca. 30% of HPV58 infections in Africa.
FIG 7.
Neutralization of HPV58 lineage and sublineage variants by MAbs. (A) Consensus lineage and sublineage HPV58 L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of lineage and sublineage HPV58 L1 and L2 PsV variants to HPV58 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. Symbols indicate magnitude of difference in MAb sensitivity (IC80) between indicated variant and consensus A/A1 PsV: *, ≥4-fold reduced sensitivity; ○, ≥10-fold reduced sensitivity; ●, ≥100-fold reduced sensitivity. (C) Graphical representation of HPV58 L1 and L2 protein combinations from A1 (white) and C (gray). (D) Heatmap representation of fold difference in neutralization potency (IC80) against the A1 variant. (E) HPV58 sublineage A1 L1 pentamer crystal (PDB accession number: 5Y9E.1) with top view shown. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC2; pink, DE5; red, EF5; orange, FG5; and green, HI2).
DISCUSSION
Our understanding of the extent of HPV genome variation has improved recently due to the application of whole-genome sequencing, including definitive variant nomenclature (the designation of specific lineages and sublineages), improved resolution of variant disease association, and a better understanding of the global dispersal of HPV from its prehistorical origins (5–7). However, little is known about the biological consequences of HPV genome variation. The aim of the current study was to evaluate the extent to which naturally occurring variation in the sequence of the capsid genes of the most important oncogenic HPV genotypes impact functional antigenic determinants on the surface of these same capsid proteins.
Each genotype contained at least one lineage or sublineage variant that displayed a ≥4-fold reduced neutralizing antibody sensitivity against at least one MAb. For HPV16 and HPV31, the number of MAbs impacted and variants implicated was limited, and the differences when seen were of a low magnitude. These data suggest that for these two genotypes, natural variation does not appear to compromise important antigenic sites. For HPV18 and HPV45, the majority of neutralizing MAbs were affected by capsid variation in one or more variants that contained a specific polymorphic residue in the BC (HPV18) or HI (HPV45) loops, conferring an approximate 4- to 8-fold reduced sensitivity compared to the consensus A/A1 variant. For HPV33, HPV52, and HPV58, all neutralizing MAbs tested exhibited a reduced sensitivity against one or more variants by up to 1,000-fold. For HPV33, differential MAb reactivity identified a key antigenic determinant encompassing the BC loop residue T56, with secondary determinants that included DE (G133) and/or FG (T266) residues. For HPV52, MAb antibody specificities were mapped to a key antigenic determinant that encompassed the FG loop residue Q281 and HI residues K354/S357. For HPV58, the primary antigenic determinant for these MAbs included the DE loop (residue P137) with, in one case, contribution from variant residues in the FG.
These data delineate, for some genotypes, key neutralizing determinants on the L1 capsid surface and also suggest that these determinants are topographically distinct for each genotype. The spatial coincidence of these antigenic determinants with the limited number of polymorphic capsid residues that have arisen over millennia of natural virus evolution will require further study. It is unclear how relevant these MAbs are to the exact specificities of antibodies generated following vaccination or natural infection. Two of these MAbs (H16.V5 and H18.J4) are used in the cLIA to evaluate vaccine and natural immunity (30–32), while two HPV18 MAbs examined here were generated by the immunization of mice with the bivalent HPV vaccine. An elegant study of human MAbs isolated from naturally infected individuals before and after immunization with one dose of HPV vaccine (40) demonstrated improvements in magnitude and specificity. Furthermore, capsid recognition by natural infection and vaccine antibodies can also be compromised by the same variant residues identified here for some genotypes (25–29). Taken together, these observations suggest that naturally acquired and vaccine-induced antibody specificities at least overlap the specificities represented within this panel of MAbs.
One potential shortcoming of this study is that, despite the large number of MAbs used overall, there were a relatively small number of MAbs available for each type (<20), and even fewer that were neutralizing. Weakly neutralizing MAbs, including those that did not neutralize at 2 μg/ml, were not examined further though they may nevertheless be representative of antibodies elicited in response to HPV infection or vaccination. In addition, it should be noted that the process of MAb generation (including the mouse strain, the specific lineage A sequence used, the integrity of the VLP-based immunogen, and downstream MAb isolation methods) will inevitably introduce some selection bias. It is possible, therefore, that there exist antigenic determinants located on the capsid surface of some genotypes that would be recognized by other MAbs whose specificities are not represented here and that multiple MAbs examined here for a particular genotype may actually represent the same epitope. Nevertheless, this study represents the most comprehensive exploration of capsid antigenicity to date.
Another potential shortcoming is that the consensus lineage and sublineage sequences used here may not be sufficiently representative of all globally circulating HPV variants. The sequences used to construct the consensus lineage and sublineage PsVs were current as of 2018, but comprise less than 100 sequences per genotype in most cases (29). It is therefore possible that the deposition of additional whole-genome sequences over time will improve the resolution of lineage and sublineage consensus sequences. Similarly, data on the global dispersal of lineage and sublineage variants for each genotype, although extensive, are inevitably only estimates limited by the availability of data. In addition, although HPV PsVs have been used widely to monitor antibody responses to vaccines and natural infection, as well as elucidate steps in the entry process, differences between how PsVs behave in vitro and how authentic HPVs behave in vivo are uncertain, which is a limitation of all PsV-based systems.
It is unclear whether the differential antigenicity noted here for some variant lineages will have any consequence in a real-world setting, such as altered HPV vaccine effectiveness in some populations. Even so, these data should be seen in the context of low overall HPV33/52/58 disease burden (<10% of global burden) and estimated lineage prevalence (currently <50% of HPV33/52/58 infections). Together these data improve our understanding of the impact of naturally occurring variation on the antigenicity of lineage and sublineage variants of vaccine-relevant oncogenic HPV genotypes.
MATERIALS AND METHODS
Monoclonal antibodies.
A large panel of MAbs was assembled from several sources, including the National Institute of Diagnostics and Vaccine Development in Infectious Diseases (NIDVD), Xiamen University, Fujian, China (11, 27, 28, 41); the Jake Gittlen Laboratories for Cancer Research, The Pennsylvania State University College of Medicine, Hershey, PA, USA (42–46); and Groupe d'étude des interactions Hôte-Pathogène, Université d'Angers, Angers, France (47). HPV18 MAbs isolated at Public Health England were generated according to a published protocol (48) using Cervarix at a 1/10 human dose as the immunogen (49). MAb isotypes were assigned as previously described (11, 27, 28, 41–47) or ascertained using the Ig Isotyping mouse ELISA kit (Thermo Fisher Scientific).
L1L2 variant pseudoviruses.
Codon-optimized L1 and L2 genes representing consensus lineage and sublineage variant sequences were synthesized (GeneArt; Thermo Fisher Scientific) with additional site-directed mutagenesis (QuikChange site-directed mutagenesis kit, Agilent Technologies) as required (29). Inserts were confirmed by Sanger sequencing. Bicistronic psheLL vectors containing these inserts were assembled and, with a luciferase reporter (pGL4.51 [luc2/CMV/Neo]; Promega), used to transfect 293TT cells as previously described (29).
The PsV neutralization assay was performed as previously described (28, 29, 50). A standardized input of 300 50% tissue culture infective dose (TCID50) was used for all PsVs. MAbs were screened at 2 μg/ml for neutralization activity, defined as ≥80% reduction in the luciferase signal compared to control wells, against the reference PsV. Thereafter each neutralizing MAb was titrated against all relevant lineage and sublineage variants (and other mutant PsVs as appropriate) in parallel to generate an IC80: the concentration in μg/ml of MAb that resulted in an 80% reduction in the luciferase signal, compared to control wells (PsV and cells only) estimated by interpolation. The IC80 of each MAb against each variant PsV was compared to that generated against the consensus A/A1 PsV and a fold difference metric was generated to allow normalization and intra-/intergenotype comparisons (24–29). To accommodate a 4-fold difference between the consensus A/A1 PsV and the other variants, only MAbs with an IC80 of <0.5 μg/ml against the consensus A/A1 were considered strongly neutralizing and included in further analysis. Weak neutralizing MAbs and those that did not neutralize at 2 μg/ml were not analyzed further.
To demonstrate repeatability, a number of MAbs were retested against variant PsVs (HPV18 A, C; HPV33 A1, A2; HPV45 A1, B2; HPV52 A1, D; and HPV58 A1, C), resulting in a Pearson’s correlation coefficient (r2) of 0.989 and a median ratio of 1.00 (interquartile range [IQR], 0.85 to 1.00; n = 84) for the resulting paired IC80. The median of the log10 IC80 ratio of the test variant and the consensus A/A1 variant for each genotype indicated above for the initial and repeat tests was 1.01 (IQR, 0.95 to 1.03; n = 42) with an r2 of 0.949.
Multiplex VLP binding.
MAb binding was evaluated in a multiplex L1L2 VLP binding assay on the Bio-Plex 200 platform (Bio-Plex; Hercules, CA) using nonreporter containing L1L2 PsV as the target antigens. Briefly, L1L2 VLP representing reference sequences for HPV6, 11, 16, 18, 31, 33, 45, 52, 58, and BPV (50) were coupled to spectrally distinct microspheres using standard methodology and stored as individual coupled-microspheres in 0.1 M MES (2[N-morpholino]ethanesulfonic acid), 1% bovine serum albumin (BSA), and 0.2% Proclin 300 buffer. Antibody binding was resolved using biotinylated goat anti-mouse secondary antibody (Thermo Fisher; Rockford, IL) and streptavidin-PE (Prozyme; Hayward, CA). MAbs were titrated from 10 μg/ml and a midpoint binding concentration (μg/ml), defined as the concentration equivalent to 50% of the maximum binding (based upon median fluorescence intensity [MFI]) was estimated by interpolation. Type-specific antisera generated in BALB/c mice against each L1L2 VLP (27, 28) and preimmune sera were used as positive and negative controls, respectively.
Ethical statement.
All animal procedures were carried out according to appropriate local and national guidelines (11, 48). Production of mouse monoclonal antibodies followed PSU Institutional Animal Care and Use review and approval and followed NIH guidelines on the care and use of animals in research.
Statistical analysis.
Descriptive statistics were generated in Microsoft Excel 2016. Fold differences of equal to or greater than 4-fold between a variant lineage or sublineage and the reference A/A1 variant lineage or sublineage were considered significant.
ACKNOWLEDGMENTS
We are indebted to John T. Schiller and Christopher Buck (National Cancer Institute, Bethesda, MD, USA) for access to the psheLL backbone used for the pseudovirus clones. We thank Dhanraj Samuel, Lenesha Warrener, and Sara L. Bissett from Public Health England for generation of some of the HPV18-specific MAbs used in this study. Some of this work was conducted as part of an MSc Biomedical Science (Queen Mary University of London) project (D.B.).
This work was supported by Public Health England. This work did not receive a specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We declare no conflicts of interest.
REFERENCES
- 1.de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. 2020. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health 8:e180–e190. doi: 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
- 2.Schiffman M, Doorbar J, Wentzensen N, de Sanjose S, Fakhry C, Monk BJ, Stanley MA, Franceschi S. 2016. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2:16086. doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 3.Roden RBS, Stern PL. 2018. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat Rev Cancer 18:240–254. doi: 10.1038/nrc.2018.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 5.Burk RD, Harari A, Chen Z. 2013. Human papillomavirus genome variants. Virology 445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, DeSalle R, Schiffman M, Herrero R, Wood CE, Ruiz JC, Clifford GM, Chan PKS, Burk RD. 2018. Niche adaptation and viral transmission of human papillomaviruses from archaic hominins to modern humans. PLoS Pathog 14:e1007352. doi: 10.1371/journal.ppat.1007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Ho WCS, Boon SS, Law PTY, Chan MCW, DeSalle R, Burk RD, Chan PKS. 2017. Ancient evolution and dispersion of human papillomavirus 58 variants. J Virol 91:e01285-17. doi: 10.1128/JVI.01285-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck CB, Day PM, Trus BL. 2013. The papillomavirus major capsid protein L1. Virology 445:169–174. doi: 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JW, Roden RB. 2013. L2, the minor capsid protein of papillomavirus. Virology 445:175–186. doi: 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. 2007. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem 282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Wang D, Gu Y, Song S, He M, Shi J, Liu X, Wei S, Li J, Yu H, Zheng Q, Yan X, Baker TS, Zhang J, McLellan JS, Li S, Xia N. 2017. Crystal structures of two immune complexes identify determinants for viral infectivity and type-specific neutralization of human papillomavirus. mBio 8:e00787-17. doi: 10.1128/mBio.00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Christensen ND, Hafenstein S. 2015. The U4 antibody epitope on human papillomavirus 16 identified by cryo-electron microscopy. J Virol 89:12108–12117. doi: 10.1128/JVI.02020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. Structural comparison of four different antibodies interacting with human papillomavirus 16 and mechanisms of neutralization. Virology 483:253–263. doi: 10.1016/j.virol.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H, Brendle SA, Bywaters SM, Guan J, Ashley RE, Yoder JD, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. J Virol 89:1428–1438. doi: 10.1128/JVI.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Chen J, Wang Z, Wang D, He M, Qian C, Song S, Chi X, Kong Z, Zheng Q, Wang Y, Yu H, Zhao Q, Zhang J, Li S, Gu Y, Xia N. 2019. Neutralization sites of human papillomavirus-6 relate to virus attachment and entry phase in viral infection. Emerg Microbes Infect 8:1721–1733. doi: 10.1080/22221751.2019.1694396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Song S, He M, Wang D, Shi J, Liu X, Li Y, Chi X, Wei S, Yang Y, Wang Z, Li J, Qian H, Yu H, Zheng Q, Yan X, Zhao Q, Zhang J, Gu Y, Li S, Xia N. 2018. Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity. Nat Commun 9:5360. doi: 10.1038/s41467-018-07199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiller JT, Muller M. 2015. Next generation prophylactic human papillomavirus vaccines. Lancet Oncol 16:e217-25–e225. doi: 10.1016/S1470-2045(14)71179-9. [DOI] [PubMed] [Google Scholar]
- 18.Qiao YL, Wu T, Li RC, Hu YM, Wei LH, Li CG, Chen W, Huang SJ, Zhao FH, Li MQ, Pan QJ, Zhang X, Li Q, Hong Y, Zhao C, Zhang WH, Li YP, Chu K, Li M, Jiang YF, Li J, Zhao H, Lin ZJ, Cui XL, Liu WY, Li CH, Guo DP, Ke LD, Wu X, Tang J, Gao GQ, Li BY, Zhao B, Zheng FX, Dai CH, Guo M, Zhao J, Su YY, Wang JZ, Zhu FC, Li SW, Pan HR, Li YM, Zhang J, Xia NS. 2020. Efficacy, safety, and immunogenicity of an Escherichia coli-produced bivalent human papillomavirus vaccine: an interim analysis of a randomized clinical trial. J Natl Cancer Inst 112:145–153. doi: 10.1093/jnci/djz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiller J, Lowy D. 2018. Explanations for the high potency of HPV prophylactic vaccines. Vaccine 36:4768–4773. doi: 10.1016/j.vaccine.2017.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiller JT, Lowy DR. 2012. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat Rev Microbiol 10:681–692. doi: 10.1038/nrmicro2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drolet M, Benard E, Perez N, Brisson M, HPV Vaccination Impact Study Group . 2019. Population-level impact and herd effects following the introduction of human papillomavirus vaccination programmes: updated systematic review and meta-analysis. Lancet 394:497–509. doi: 10.1016/S0140-6736(19)30298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastrana DV, Vass WC, Lowy DR, Schiller JT. 2001. NHPV16 VLP vaccine induces human antibodies that neutralize divergent variants of HPV16. Virology 279:361–369. doi: 10.1006/viro.2000.0702. [DOI] [PubMed] [Google Scholar]
- 23.Ning T, Wolfe A, Nie J, Huang W, Chen XS, Wang Y. 2017. Naturally occurring single amino acid substitution in the L1 major capsid protein of human papillomavirus type 16: alteration of susceptibility to antibody-mediated neutralization. J Infect Dis 216:867–876. doi: 10.1093/infdis/jix274. [DOI] [PubMed] [Google Scholar]
- 24.Bissett SL, Godi A, Fleury MJ, Touze A, Cocuzza C, Beddows S. 2015. Naturally occurring capsid protein variants of human papillomavirus genotype 31 represent a single L1 serotype. J Virol 89:7748–7757. doi: 10.1128/JVI.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godi A, Bissett SL, Miller E, Beddows S. 2017. Impact of naturally occurring variation in the human papillomavirus (HPV) 33 capsid proteins on recognition by vaccine-induced cross-neutralizing antibodies. J Gen Virol 98:1755–1761. doi: 10.1099/jgv.0.000829. [DOI] [PubMed] [Google Scholar]
- 26.Godi A, Facchetti A, Bissett SL, Cocuzza C, Miller E, Beddows S. 2015. Naturally occurring major and minor capsid protein variants of human papillomavirus 45 (HPV45): differential recognition by cross-neutralizing antibodies generated by HPV vaccines. J Virol 90:3247–3252. doi: 10.1128/JVI.02859-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godi A, Bissett SL, Masloh S, Fleury M, Li S, Zhao Q, Xia N, Cocuzza CE, Beddows S. 2019. Impact of naturally occurring variation in the human papillomavirus 52 capsid proteins on recognition by type-specific neutralising antibodies. J Gen Virol 100:237–245. doi: 10.1099/jgv.0.001213. [DOI] [PubMed] [Google Scholar]
- 28.Godi A, Martinelli M, Haque M, Li S, Zhao Q, Xia N, Cocuzza CE, Beddows S. 2018. Impact of naturally occurring variation in the human papillomavirus (HPV) 58 capsid proteins on recognition by type-specific neutralizing antibodies. J Infect Dis 218:1611–1621. doi: 10.1093/infdis/jiy354. [DOI] [PubMed] [Google Scholar]
- 29.Godi A, Kemp TJ, Pinto LA, Beddows S. 2019. Sensitivity of human papillomavirus (HPV) lineage and sublineage variant pseudoviruses to neutralization by nonavalent vaccine antibodies. J Infect Dis 220:1940–1945. doi: 10.1093/infdis/jiz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dias D, Van Doren J, Schlottmann S, Kelly S, Puchalski D, Ruiz W, Boerckel P, Kessler J, Antonello JM, Green T, Brown M, Smith J, Chirmule N, Barr E, Jansen KU, Esser MT. 2005. Optimization and validation of a multiplexed Luminex assay to quantify antibodies to neutralizing epitopes on human papillomaviruses 6, 11, 16, and 18. Clin Diagn Lab Immunol 12:959–969. doi: 10.1128/CDLI.12.8.959-969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beachler DC, Jenkins G, Safaeian M, Kreimer AR, Wentzensen N. 2016. Natural acquired immunity against subsequent genital human papillomavirus infection: a systematic review and meta-analysis. J Infect Dis 213:1444–1454. doi: 10.1093/infdis/jiv753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferris D, Samakoses R, Block SL, Lazcano-Ponce E, Restrepo JA, Reisinger KS, Mehlsen J, Chatterjee A, Iversen OE, Sings HL, Shou Q, Sausser TA, Saah A. 2014. Long-term study of a quadrivalent human papillomavirus vaccine. Pediatrics 134:e657-65–e665. doi: 10.1542/peds.2013-4144. [DOI] [PubMed] [Google Scholar]
- 33.Clifford GM, Tenet V, Georges D, Alemany L, Pavon MA, Chen Z, Yeager M, Cullen M, Boland JF, Bass S, Steinberg M, Raine-Bennett T, Lorey T, Wentzensen N, Walker J, Zuna R, Schiffman M, Mirabello L. 2019. Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res 7:67–74. doi: 10.1016/j.pvr.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen AA, Gheit T, Franceschi S, Tommasino M, Clifford GM, IARC HPV Variant Study Group . 2015. Human papillomavirus 18 genetic variation and cervical cancer risk worldwide. J Virol 89:10680–10687. doi: 10.1128/JVI.01747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleury MJ, Touze A, Maurel MC, Moreau T, Coursaget P. 2009. Identification of neutralizing conformational epitopes on the human papillomavirus type 31 major capsid protein and functional implications. Protein Sci 18:1425–1438. doi: 10.1002/pro.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth SD, Sapp M, Streeck RE, Selinka HC. 2006. Characterization of neutralizing epitopes within the major capsid protein of human papillomavirus type 33. Virol J 3:83. doi: 10.1186/1743-422X-3-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen AA, Heideman DA, Boon D, Chen Z, Burk RD, De Vuyst H, Gheit T, Snijders PJ, Tommasino M, Franceschi S, Clifford GM. 2014. Human papillomavirus 33 worldwide genetic variation and associated risk of cervical cancer. Virology 448:356–362. doi: 10.1016/j.virol.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen AA, Heideman DA, Boon D, Gheit T, Snijders PJ, Tommasino M, Franceschi S, Clifford GM, IARC HPV Variant Study Group . 2014. Human papillomavirus 45 genetic variation and cervical cancer risk worldwide. J Virol 88:4514–4521. doi: 10.1128/JVI.03534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Park JS, Grce M, Hibbitts S, Palefsky JM, Konno R, Smith-McCune KK, Giovannelli L, Chu TY, Picconi MA, Pina-Sanchez P, Settheetham-Ishida W, Coutlee F, De Marco F, Woo YL, Ho WC, Wong MC, Chirenje MZ, Magure T, Moscicki AB, Sabol I, Fiander AN, Chen Z, Chan MC, Cheung TH, Burk RD, Chan PK. 2014. Geographical distribution and risk association of human papillomavirus genotype 52-variant lineages. J Infect Dis 210:1600–1604. doi: 10.1093/infdis/jiu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherer EM, Smith RA, Gallego DF, Carter JJ, Wipf GC, Hoyos M, Stern M, Thurston T, Trinklein ND, Wald A, Galloway DA. 2016. A single human papillomavirus vaccine dose improves B cell memory in previously infected subjects. EBioMedicine 10:55–64. doi: 10.1016/j.ebiom.2016.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Huang X, Chen S, Li Y, Li Y, Wang X, Tang J, Xia L, Lin Z, Luo W, Li T, Li S, Zhang J, Xia N, Zhao Q. 2018. Epitope clustering analysis for vaccine-induced human antibodies in relationship to a panel of murine monoclonal antibodies against HPV16 viral capsid. Vaccine 36:6761–6771. doi: 10.1016/j.vaccine.2018.09.035. [DOI] [PubMed] [Google Scholar]
- 42.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 43.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 44.Selinka HC, Giroglou T, Nowak T, Christensen ND, Sapp M. 2003. Further evidence that papillomavirus capsids exist in two distinct conformations. J Virol 77:12961–12967. doi: 10.1128/jvi.77.24.12961-12967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Combita AL, Touze A, Bousarghin L, Christensen ND, Coursaget P. 2002. Identification of two cross-neutralizing linear epitopes within the L1 major capsid protein of human papillomaviruses. J Virol 76:6480–6486. doi: 10.1128/jvi.76.13.6480-6486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizk RZ, Christensen ND, Michael KM, Muller M, Sehr P, Waterboer T, Pawlita M. 2008. Reactivity pattern of 92 monoclonal antibodies with 15 human papillomavirus types. J Gen Virol 89:117–129. doi: 10.1099/vir.0.83145-0. [DOI] [PubMed] [Google Scholar]
- 47.Fleury MJ, Touze A, Alvarez E, Carpentier G, Clavel C, Vautherot JF, Coursaget P. 2006. Identification of type-specific and cross-reactive neutralizing conformational epitopes on the major capsid protein of human papillomavirus type 31. Arch Virol 151:1511–1523. doi: 10.1007/s00705-006-0734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuel D, Warrener L, Hoschler K. 2011. Monoclonal antibodies to the haemagglutinin HA1 subunit of the pandemic influenza A/H1N1 2009 virus and potential application to serodiagnosis. J Med Virol 83:559–567. doi: 10.1002/jmv.21982. [DOI] [PubMed] [Google Scholar]
- 49.Bissett SL, Mattiuzzo G, Draper E, Godi A, Wilkinson DE, Minor P, Page M, Beddows S. 2014. Pre-clinical immunogenicity of human papillomavirus alpha-7 and alpha-9 major capsid proteins. Vaccine 32:6548–6555. doi: 10.1016/j.vaccine.2014.07.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Draper E, Bissett SL, Howell-Jones R, Edwards D, Munslow G, Soldan K, Beddows S. 2011. Neutralization of non-vaccine human papillomavirus pseudoviruses from the A7 and A9 species groups by bivalent HPV vaccine sera. Vaccine 29:8585–8590. doi: 10.1016/j.vaccine.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]