Abstract
Background
Post-donation illness can be described as appearance of clinical symptoms in blood donors after donation. The consequent call back of the donor to report these symptoms to the blood collection institution is considered a post-donation illness report (PDIR). The most suitable way to examine whether PDIR is related to infection is to apply next-generation sequencing (NGS) and viral metagenomics. Investigation into a PDIR can reveal its importance for transfusion safety and help elaborate strategies for donor education in order to prevent the transfusion transmission of infections which are not routinely tested by the blood collection services.
Materials and methods
We applied NGS and viral metagenomics on blood donations which were deferred due to a PDIR. Thirty-three PDIR donations obtained in the Blood Center of Ribeirão Preto, Southeast Brazil, were evaluated. Sequencing was performed using Illumina NextSeq 550 (Illumina Inc, San Diego, CA, USA) equipment and the reads obtained for each sample were analysed by specific bioinformatic pipeline for the classification and discovery of emerging viruses. The identified viral agents by metagenomics were directly confirmed by molecular methods.
Results
In all PDIR donations, we found abundant reads of commensal viruses belonging to the Anelloviridae family as well as human pegivirus-1. However, we were also able to identify blood donations positive for clinically important viruses like dengue serotype-2 (DENV-2) of the Asian-American genotype and parvovirus B19 (B19V). Both viruses were also confirmed by real-time polymerase chain reaction, detecting DENV-2 RNA in a significant number of cases (7 samples, 21.2%), compared to B19V which was confirmed in 1 case (3.0%).
Discussion
Our study applies for the first time viral metagenomics to evaluate the significance of PDIRs. We confirm the crucial importance of the donor providing a timely PDIR for the prevention of transfusion transmission of viral infections which are not routinely tested in the blood banks worldwide.
Keywords: post-donation illness report, viral metagenomics, blood donors
INTRODUCTION
Correct information about the health status of the blood donor is obtained during the donation interview and is the first step towards increasing the transfusion safety of the applied blood components. Although the selection process of blood donors can be considered to be efficient, it is possible that some donors present a clinical manifestation of a post-donation event. Reporting any clinical symptoms which occur after donation by the blood donor is considered a post-donation illness report (PDIR). At the time of blood donation, the donor can be totally asymptomatic. If by this time there is a window period of a viral infection, the symptoms may appear some days after the donation. Therefore, a PDIR can lead to the deferral of the obtained blood components due to increased risk of transmission of transfusion-transmitted infection. The term "PDIR" is not exactly the same as the term "post-donation information" used by other research groups. Some define post-donation information as an error by which information known by the donor is not disclosed as part of the health history procedure but is rather provided subsequent to donation1, while for others, post-donation information includes not only information known to the donor but which is not disclosed, but also information received from a donor about a condition not known at the time of donation (e.g., symptoms of an infectious disease that developed after donation)2. However, all agree that the management of such reports is extremely complex and aggravated by the fact that most of the obtained blood components have already been distributed. Considering that most of the blood recipients are high-risk patients, probable transmission of an infectious disease may have serious clinical consequences. Therefore, careful examination of the respective PDIR with confirmation of its aetiology (presence of viral infection) can help improve transfusion safety (blood unit discharge) and blood donor education and counselling2.
Among US blood donors, the most important causes for post-donation information regard to travel-related information, medical reasons, high-risk behaviour, and exposure to infectious disease1. However, the causes for a PDIR remain unknown and up to now no such surveys have been performed. It is reasonable to suppose that in tropical countries PDIR may originate from endemic infectious diseases which are involved in explosive outbreaks with many asymptomatic cases which favour the transfusion transmission of these infections. Examples include but are not limited to viruses like zika3,4 and dengue5,6.
A PDIR related to the appearance of clinical symptoms with probable infectious aetiology needs to be examined in order to identify the cause; the vast number of viral infections which may affect blood donors makes this problematic. Next-generation sequencing (NGS) and viral metagenomics can help resolve this as these approaches can without bias identify virtually all genomes present in a clinical sample. Therefore, NGS can identify unexpected or unknown infectious agents which can compromise transfusion safety7. The application of NGS and viral metagenomics to plasma samples of blood donors who provide PDIRs may help evaluate the possible viral aetiology of some such donations.
The objective of this study was to evaluate the presence of viral agents in blood units with a PDIR related to symptoms compatible with infectious disease. Such a study may shed light on the variety of transfusion threats, and help establish strategies to prevent their transfusion transmission.
MATERIALS AND METHODS
Blood units with post-donation illness reports
The study was conducted between January and July, 2019, on 33 blood donations obtained from individual blood donors. The blood units were deferred from transfusion due to a PDIR based on the presence of one or more of the following clinical symptoms compatible with infectious disease: fever, retro-orbital pain, exanthema, headache, myalgia, diarrhoea, and jaundice. The analysis of the donor history forms demonstrated that, at the time of donation, none of the selected blood donors had any clinical symptoms or fever, were taking any type of medication, or had any at-risk behaviour. According to the Brazilian Ministry of Health, the time frame for PDIR is 7 days; however, our blood centre establishes a period of 14 days in order to promote transfusion safety. In addition, donors are counselled and receive information and a leaflet describing the symptoms of endemic infectious diseases which can be transmitted by blood transfusion (dengue, zika and chikungunya fevers). If such symptoms occur, the donors are advised to inform the blood collection institution as soon as possible.
We analysed 33 blood units with a PDIR which represented 44% of the total number of the PDIRs obtained for the study period. Due to ethical concerns, we were able to examine only blood units which had been discarded and had not been transfused.
The study was performed in the Blood Center of Ribeirão Preto, Faculty of Medicine of Ribeirão Preto, University of São Paulo, Southeast Brazil. To perform the study, we used plasma samples derived from the tube of the blood unit after its deferral. Approximately 1 mL of plasma was obtained from each blood unit and if not processed immediately it was stored at −80°C until use. Consent for this study was waived due to the type of collection and was approved by the Institutional Ethics Committee of the University Hospital at the Faculty of Medicine of Ribeirão Preto, University of São Paulo, Brazil (process n. 12196/2018).
Sample preparation and Illumina sequencing
Initially, 600 μL of plasma were pre-treated with Turbo DNAse (Turbo DNA-free kit, ThermoFisher Scientific, São Paulo, Brazil) in order to remove free host/bacterial DNA. After DNAse inactivation, the individual samples were mixed in 4 pools. The sample pools were extracted using the High Pure Viral Nucleic Acid Large Volume Kit (Roche, São Paulo, Brazil) following the manufacturer’s instructions with minor modifications: the polyA carrier RNA from the kit was replaced by the neutral carrier, GenElute Linear Polyacrylamide carrier (LPA) (Merck, São Paulo, Brazil), and Isopropanol was added to the binding buffer before the filtration step. After extraction, viral nucleic acid volume was recovered in nuclease-free water pre-heated to 70°C and submitted to reverse transcription using the Superscript III First-Strand Synthesis System (ThermoFisher Scientific, São Paulo, Brazil). cDNA amplification was performed using the QuantiTect Whole Transcriptome Kit (QIAGEN, São Paulo, Brazil). The sequence libraries were prepared using the Nextera DNA Flex Library Preparation Kit (Illumina, San Diego, CA, USA) and Nextera DNA CD Indexes following the manufacturer's instructions. Sequencing of the dual-indexed libraries was performed by Illumina NextSeq 500 sequencer using the NextSeq High Output Kit v 2.5, 300 cycles (Illumina, San Diego, CA, USA) following the manufacturer's instructions.
Bioinformatics analysis
The raw sequence data obtained were submitted to quality control analysis using FastaQC v.0.11.8 software8. Trimming was then performed using a Trimmomatic v.0.3.99 and Cutadapt v. 2.410 in order to select the best quality sequences. For the bioinformatic analysis, we used sequences with quality scores >30. Additional trimming to improve the overall quality was performed using AfterQCsoftware v.0.9.711 (Shenzhen Institutes of Advanced Technology, Shenzhen, China).
The Kraken2 v2.0.8 program12 (Johns Hopkins University, Baltimore, MD, USA) was used to determine the taxonomic classification. To examine the depth of coverage of the classified reads, we used BWA v.0.6 software13 (Wellcome Trust Sanger Institute, Hinxton, UK). “De novo” assembly was performed using SPAdes v.3.13.014 to generate contigs and scaffolds. Finally, to perform taxonomic classification based on protein identity we applied Blastx according to Diamond v.0.9.29 software15 (University of Tübingen, Tübingen, Germany).
Phylogenetic analysis
Phylogenetic analysis of the assembled contigs was performed using datasets of complete genomes which were obtained from viral databanks such as that of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) and the Virus Pathogen Resource (ViPR) (https://www.viprbrc.org/). Alignment was performed by MAFFT v.7.42916 (Kyoto University, Kyoto, Japan) and the phylogenetic signal was evaluated according to the Tree Puzzle v.5.2 programme17 (Max Planck Institute for Molecular Genetics, Berlin, Germany). To reconstruct the phylogenetic history of the identified viruses, we used the IQtree v.16.12 software18 (University of Vienna, Vienna, Austria) applying the maximum likelihood method with statistical support of ultrafast bootstrap with 10,000 replicates. The phylogenetic tree was visualised and edited by FigTree v.1.4.4 software19 (University of Edinburgh, Edinburgh, UK).
Molecular confirmation of the identified viruses
Important viral agents which had been identified by NGS were further confirmed in each individual blood donation using real-time and conventional polymerase chain reaction (PCR) methods. For DENV confirmation, we used a primer and probe system which was able to detect all DENV 1–4 serotypes, as described in the literature20. The confirmation of B19V DNA in plasma samples was performed by TaqMan® real-time PCR with a primer and probe set capable of detecting B19V 1–3 genotypes21. All TaqMan® (ThermoFisher Scientific, São Paulo, Brazil) real-time reactions were performed in an Applied Biosystems® 7500 real-time PCR system (ThermoFisher Scientific, São Paulo, Brazil) using the following protocol: initial hold at 50°C for 2 min and denaturation at 95°C for 10 min, followed by 40 cycles consisting of denaturation at 95°C for 10 sec, and a mixed annealing and elongation step at 60°C for 1 min.
In addition, we confirmed the presence of DENV-2 RNA by using an in-house designed specific primer set with the following sequences: forward primer DENV2F (5′-GCCTTTCAATATGCTGAAACGCG-3′) and reverse primer DENV2R (5′-GAAGGAGCGACAGCTGTCAGTA-3′). The amplification generated a fragment of 800 bp from the C-preM gene portion of the DENV-2 genome. The reaction was carried out in 50 μL of final volume with 400 nM of each primer, 200 μM of dNTPs, and 1.25 U of Taq polymerase. The cycling protocol included initial denaturation at 95°C for 5 min and 35 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min 30 secs. The reaction was finalised with a final elongation at 72°C for 10 min. Amplification was performed in a SimpliAmp Thermal Cycler (ThermoFisher Scientific, São Paulo, Brazil) and the amplified products were revealed using ChemiDocTM XRS+ equipment (Bio-Rad, Hercules, CA, USA). All of the procedures were performed in separate laboratory rooms in order to prevent contamination.
Data availability
Metagenomic datasets have been submitted to the NCBI SRA and are available under BioProject accession numbers PRJNA602694.
RESULTS
Blood donors' characteristics
Of the blood donors who provided a PDIR, 57.6% were male and 42.4% were female. The mean age of the tested group was 32.2 years of age (range 21–63 years of age). Most had completed higher education (78.8%); of the remaining 21.2%, 18.2% had completed secondary school education and 3% primary education.
Viral metagenomics on plasma samples with a post-donation information report
Next-generation sequencing was performed on 4 pools which were composed of 33 samples with post-donation information, and generated a total number of 605,249,682 reads. After removing low-quality reads, we obtained 599,045,507 reads with a quality score >30 to be used for taxonomic classification. After Kraken taxonomic classification2, 3,331,192 viral sequences were kept for assembly. The percentage of classified viral reads varied between 0.001% and 1.4% (medium 0.35%). The unclassified sequences corresponded to 20,850,054 reads. Complete or nearly complete viral genomes were assembled from 2 pools. The most abundant viruses identified were commensal. Among the non-pathogenic viruses found in the pools, the representatives of the Anelloviridae family were highly represented. We were able to identify a total number of 20,045 reads belonging to torque teno midi virus (TTMDV) types 2, 5–7, 11–15 and torque teno virus (TTV) types 12, 15, 16, 20, 22, 25 and 28. Another commensal virus found in our analysis was the human pegivirus-1 (HpgV-1), formerly known as GB virus C/hepatitis G virus belonging to the Flaviviridae family. We found 2,294 reads belonging to HPgV-1 in 2 pools (P10 and P22) (Table I).
Table I.
Viral agents identified in plasma samples obtained from blood donors who reported post-donation information characterised by symptoms compatible with infectious disease
| Pool number | Commensal viral agents | Number of reads | Pathogenic viruses | Reads |
|---|---|---|---|---|
| P10 | HPgV-11 TTV2-12,15,16, TTMDV3-5–7 |
2,291 68 |
Dengue serotype 2 | 23 |
| P11 | TTV-12,15,16,28, TTMDV-2,5–7 | 1,405 | Dengue serotype 2 | 159 |
| P12 | TTV-12,15,20,22,25,28, TTMDV-2,11–15 | 5,390 | Dengue serotype 2 | 2,084,960 |
| P22 | HPgV-1 TTV-12, 15, 16, 20, 22, 25,28, TTMDV-2, 5–7, 11–15 |
3 13,182 |
Parvovirus B19 | 1,198,765 |
HPgV-1: human pegivirus-1, GB virus C or hepatitis G virus;
TTV: torque teno virus;
TTMDV: torque teno midi virus.
We were also able to detect pathogenic viruses which may represent a transfusion threat. In 3 of the analysed pools, we were able to detect DENV-2 with a number of reads ranging from 31 to 2,084,960 (Table I). In particular, in one of the pools, we identified 2,084,960 DENV-2 reads giving us the opportunity to analyse the complete DENV-2 genome. In another pool, we obtained 1,198,765 reads belonging to B19V, which were sufficient to assemble almost the entire genome of 4,256 bp for phylogenetic analysis. The viral agents identified by viral metagenomics are shown in Table I.
The consensus assembly of the DENV-2 reads generated one contig with a length of 10,664 bp, with 45.7% GC content and an average coverage of 296,567 times; it demonstrated most hits with strain DENV-2 DR23/01 (GenBank number AB122020.1 from the Dominican Republic) with 98,41% similarity. The consensus assembly of B19V generated one contig with a length of 4,256 bp with 42.1% GC content and an average coverage of 242,236 time; it demonstrated most hits with the B19V isolate 09BRSP0883 from Brazil (GenBank number KC013331) with 97.24% similarity.
Phylogenetic analysis of DENV-2 and B19V
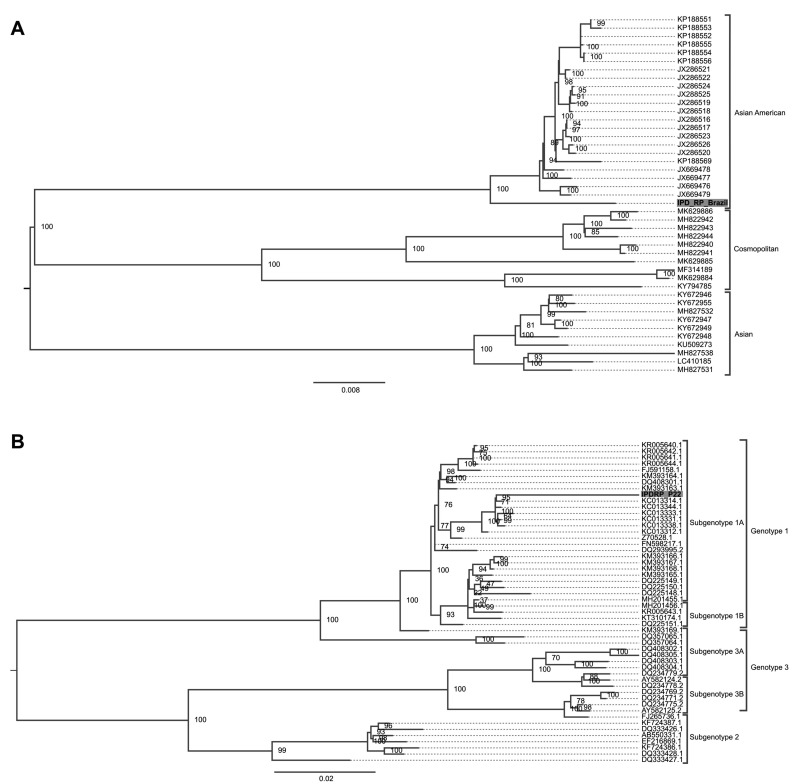
We performed phylogenetic analysis of the almost complete genomes of DENV-2 and B19V which were identified in our study. For phylogenetic analysis of DENV-2, we used the nucleotide substitution model TIM2+F+G4 chosen by the Bayesian Information Criterion (BIC) statistical model. The detected complete DENV-2 genome demonstrated that it belongs to the Southeast Asia-American genotype. The classification of DENV-2 is shown in Figure 1A. We also performed phylogenetic analysis of the partial genome of B19V. The nucleotide substitution model which was used for B19V analysis was TN+F+R2 chosen by the Bayesian Information Criterion (BIC) statistical model. The 4,256 bp B19V contig was classified as belonging to subgenotype 1A of the main genotype 1 (Figure 1B).
Figure 1.
Phylogenetic analysis of the viruses identified in blood donations with a post-donation illness report
(A) Maximum-likelihood tree of dengue 2 virus. The nucleotide substitution model applied for tree reconstruction was TIM2+F+G4 chosen by the Bayesian Information Criterion (BIC) model using 10,000 bootstrap replicates for the statistical support. Only values >75% are shown at the important tree branches. (B) Maximum likelihood tree of parvovirus B19. The nucleotide substitution model which was used was TN+F+R2 and was chosen by BIC using 10,000 bootstrap replicates for the statistical support of the branches. Only values of 75% were demonstrated at the important tree branches. The phylogenetics trees were constructed using IQtree v.16.12 software.
Individual confirmation of DENV-2 and B19V
We confirmed the direct presence of DENV-2 RNA and B19V DNA in all of the blood donor samples which made up the analysed pools. The real-time PCR for DENV-2 demonstrated the presence of viral RNA in 7 out of 33 blood donations with PDI (21.2%). This demonstrates that among the known pathogenic viruses, DENV-2 was most frequently identified in blood donations with a PDIR; this finding is consistent with the fact that this viral serotype was the cause of a large DENV-2 outbreak in the São Paulo State. The blood donor samples were characterised with different viral loads: Ct varied between 21.8 and 32.15. All the positive DENV-2 samples were also reconfirmed by conventional PCR. The DENV-2 positive donations were provided by blood donors who reported symptoms compatible with DENV infection-like fever, retro-orbital pain, myalgia, and headache 1–6 days (mean 2.42 days) after donation (Table II).
Table II.
Demographic and clinical characteristics of the blood donors who were positive for dengue serotype-2 virus (DENV-2) and parvovirus B19 (B19V) DNA and related post-donation illness
| Patient n. | Age | Gender | Education level | N. of days* | Symptoms | Virus |
|---|---|---|---|---|---|---|
| IPD-4 | 25 | Male | Primary | 2 | Fever | DENV-2 |
| IPD-6 | 56 | Male | Secondary | 1 | Fever, myalgia | DENV-2 |
| IPD-10 | 38 | Female | Secondary | 3 | Fever, myalgia | DENV-2 |
| IPD-14 | 36 | Male | Higher | 2 | Myalgia, afebrile | DENV-2 |
| IPD-18 | 35 | Female | Higher | 1 | Fever, malaise | DENV-2 |
| IPD-24 | 63 | Male | Higher | 4 | Fever, myalgia | DENV-2 |
| IPD-25 | 34 | Male | Higher | 6 | Fever, myalgia | DENV-2 |
| IPD-38 | 28 | Female | Secondary | 1 | High fever, retro orbital pain, headache | B19V |
Days after donation for the report of post donation illness.
All individual blood donations were also tested for B19V DNA. We were able to identify 1 sample out of 33 (3.0%) positive for B19V DNA with a viral load of 7.1x107 copies/mL; this belonged to a blood donor who provided a PDIR 4 days after donation.
The PDIR was characterised by symptoms like myalgia, headache, retro-orbital pain, and high fever.
Due to the fact that the obtained blood components were deferred from transfusion, the respective blood donors positive for DENV-2 and B19V were not followed-up through the evolution of the infections. In this study, the viruses identified by NGS were fully confirmed by molecular tests, which demonstrates the diagnostic applicability of NGS for the detection of clinically relevant viruses.
DISCUSSION
This is a pioneer study which evaluates the presence of viral infectious which can be implicated as causes for a PDIR in a tropical country like Brazil. The obtained data cannot be easily extrapolated to other studies examining post-donation information1,2 due to the different scope of the study. Nevertheless, the results obtained stress the importance of PDIRs in the prevention of transmission of viral infections which are not routinely tested in blood recipients.
Obtaining a PDIR issued by donors may depend on an investment in donor education during their counselling through which the benefits of PDIR in the prevention of transfusion-transmitted infections should be emphasised. In our institution, donors are invited to provide a PDIR in two ways: 1) during the pre-donation interview they are informed of the time-frame in which they must contact the collecting blood centre if symptoms appear (Ministry of Health of Brazil, Ordinance 158, Item 76, of February 4th, 2016); and 2) they are given a leaf let explaining the symptoms and conditions caused by endemic viral diseases (dengue, zika and chikungunya fevers) and their impact on transfusion safety. Such blood donor education practices reinforce the fact that provision of a PDIR is the simplest method to prevent the transfusion transmission of clinically important viral diseases which are not routinely tested by the blood transfusion services.
Tracing PDIR aetiology is problematic. On the one hand, we do not have enough information about the group of donors who provide a PDIR, a problem shared with other studies which examine post-donation information1. Our analysis showed that most of the donors who report PDI have a university education and were male. This is similar to studies that examined donors who report post-donation information1. In our study, due to ethical considerations, we only analysed blood units which had not been transfused. However, due to the detection of important viral agents which may be transmitted by blood transfusion like DENV-2 and B19V, we believe that systemic recording and the adequate management of PDIRs is of crucial importance for the prevention of transfusion transmission of important viral infections. A shortcoming of our study was that the blood units were obtained from a tropical region, endemic for arboviral infections like DENV. Therefore, more studies including analysis of PDIRs from other parts of the world are needed to compare the virome composition and evaluate the best strategies to help prevent transfusion transmission of the identified infectious agents.
However, in some cases, a PDIR cannot prevent the transfusion transmission of an infectious disease as the blood components have been distributed among the recipients. This is disappointing, and may involve specific follow up of the blood recipient and diagnosis of the infectious agent. In this respect, our study is important because it helps to identify the viruses that could result in the issue of a PDIR and alert hematologists who can then provide timely intervention. However, to definitively prevent the transfusion transmission of an infectious disease, the most effective way is the implementation of specific molecular tests for viral detection prior to transfusion. For the moment, it is not possible to implement routine diagnostic tests to all viruses which could compromise transfusion safety. For this reason, measures including donor education to emphasise the importance of the PDIR, establishing quarantine periods for the blood components, and specific universal physico-chemical treatments for pathogen reduction seem to be reasonable strategies of transfusion-transmission prevention of infections which are not routinely tested by the blood collection services. Nevertheless, implementation of any strategy must always be balanced against other health priorities.
Firstly, NGS identified many commensal viruses like TTV, TTMDV types (Anneloviridae) and HPgV-1 types (Flaviviridae). This finding was expected as all these viruses cause frequent infections in the human population and their prevalence may reach very high proportions. In a study performed among blood donors in Qatar, the detected prevalence for TTV and TTMDV were 83.4% and TTMDV, respectively22. HPgV-1 is also frequently found in healthy blood donors, with 3 out of 100 blood donations worldwide resulting positive23. Metagenomic analysis also identified two known pathogenic viruses as possible causes for PDIR: DENV-2 and B19V. DENV-2 was represented more frequently than B19V (21.2% vs. 3.0%). This is probably the result of a large DENV-2 outbreak in São Paulo State in 2019 with 437,047 cases reported up to September, 201924. During DENV outbreaks, a significant proportion of infected individuals remain asymptomatic, as reported in China25, Brazil26,27, Puerto Rico6,28,29, Honduras30, and Taiwan31, which enables transfusion transmission of this infection. In our study, phylogenetic analysis of the obtained complete DENV-2 genome demonstrated that it belongs to the American/Asian genotype (Subtype IIIb) (Figure 1A) but formed a separate lineage, probably due to intra-serotype genetic variations32. The introduction of a genetically divergent strain in the São Paulo State probably resulted in this large outbreak. The entry and dissemination of new DENV strains also have to be monitored by the transfusion services due to the large proportions of the outbreaks and the possibility of transfusion transmission of DENV33. The known viral pathogen detected in blood donations with PDI was B19V. Reports on the prevalence of B19V DNA in blood donations demonstrate that its detection is not a rare event due to the frequent B19V detection in blood donors34. B19V can be transmitted by blood transfusion and symptomatic cases due to treatment by red blood cell or concentrated plasma components have been reported35,36. A limitation of our study was that we detected only viral RNA/DNA and not a replication active DENV-2 and B19 virus. We believe that further studies including viral isolation and confirmation of infectious viral forms are important to better characterise the transfusion-transmission potential of the identified viral agents.
In conclusion, this study used NGS and metagenomic analysis to identify the viral causes for a PDIR and is the first to investigate the viruses leading to a PDIR. These findings underline the importance of efforts to educate donors and encourage the timely issue of a PDIR, and to provide appropriate clinical management of the report once the possible causes have been identified. Moreover, this study sheds light on viruses that might be transmitted by blood transfusion that are not routinely tested for. This can help in health policy decision-making in order to prevent their transfusion transmission.
ACKNOWLEDGEMENTS
The authors are grateful to the Blood Quality Department of the Blood Center of Ribeirão Preto for the separation of the post-donation information. We thank Dr. Kamila Chagas Peronni PhD for the support during the library preparations and the next-generation sequencing. We are also grateful to Sandra Navarro Bresciani for the artwork.
Footnotes
FUNDING
The research was supported by the São Paulo Research Foundation (FAPESP): project numbers17/23205-8, 19/07861-8, 19/08528-0, INCTC-FAPESP-2014/50947-7, and Conselho Nacional do Desenvolvimento Científico e Tecnológico, Brazil, project numbers INCTC-465539/2014-9; 455503/2014-1.
AUTHORSHIP CONTRIBUTIONS
RSB processed the experimental data, performed the analysis, drafted the manuscript, and designed the figures and the tables. LSO, ELM, and EMAU helped collect post-donation information and contributed to its interpretation. RMS contributed to sample preparations, library construction, molecular confirmation of the viral agents, and data analysis. WASJ, DTC and SK helped interpret the results and write the manuscript. SNS wrote the manuscript and was in charge of obtaining funding. All authors discussed the results and commented on the manuscript.
The Authors declare no conflicts of interest.
REFERENCES
- 1.Wilkinson SL, Steele WR, High PM, et al. NHLBI Retrovirus Epidemiology Donor Study-II. Characteristics of post donation information donors and comparison with appropriately deferred donors. Transfusion. 2011;51:1503–10. doi: 10.1111/j.1537-2995.2010.03043.x. [DOI] [PubMed] [Google Scholar]
- 2.Vuk T, Ljubièiæ J, Gulan Harcet J, et al. Post-donation information management - contribution to the safety of transfusion treatment. Transfus Clin Biol. 2019;26:353–4. doi: 10.1016/j.tracli.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Musso D, Nhan T, Robin E, et al. Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Euro Surveill. 2014;19:20761. doi: 10.2807/1560-7917.es2014.19.14.20761. [DOI] [PubMed] [Google Scholar]
- 4.Slavov SN, Hespanhol MR, Rodrigues ES, et al. Zika virus RNA detection in asymptomatic blood donors during an outbreak in the northeast region of São Paulo State, Brazil, 2016. Transfusion. 2017;57:2897–901. doi: 10.1111/trf.14322. [DOI] [PubMed] [Google Scholar]
- 5.Linnen JM, Vinelli E, Sabino EC, et al. Dengue viremia in blood donors from Honduras, Brazil, and Australia. Transfusion. 2008;48:1355–62. doi: 10.1111/j.1537-2995.2008.01772.x. [DOI] [PubMed] [Google Scholar]
- 6.Stramer SL, Linnen JM, Carrick JM, et al. Dengue viremia in blood donors identified by RNA and detection of dengue transfusion transmission during the 2007 dengue outbreak in Puerto Rico. Transfusion. 2012;52:1657–66. doi: 10.1111/j.1537-2995.2012.03566.x. [DOI] [PubMed] [Google Scholar]
- 7.Sauvage V, Gomez J, Boizeau L, et al. The potential of viral metagenomics in blood transfusion safety. Transfus Clin Biol. 2017;24:218–222. doi: 10.1016/j.tracli.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. [Accessed on 07/01/2020.]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 9.Bolger AM, Marc L, Bjoern U. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. [Google Scholar]
- 11.Chen S, Huang T, Zhou Y, et al. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics. 2017;18:80. doi: 10.1186/s12859-017-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood DE, Steven LS. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. [Accessed on 14/01/2020]. arXiv:1303.3997 [q-bio.GN]. Available at: https://arxiv.org/abs/1303.3997.
- 14.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Compt Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchfink B, Chao X, Daniel HH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 16.Katoh K, Misawa K, Kuma KI, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt HA, Strimmer K, Vingron M, et al. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–4. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen LT, Schmidt HA, Von HA, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2014;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambaut A. FigTree v1. 4. Molecular evolution, phylogenetics and epidemiology. 2012. [Accessed on 17/01/2020.]. Available at: http://tree.bio.ed.ac.uk/software/figtree.
- 20.Huhtamo E, Hasu E, Uzcátegui NY, et al. Early diagnosis of dengue in travelers: comparison of a novel real-time RT-PCR, NS1 antigen detection and serology. J Clin Virol. 2010;47:49–53. doi: 10.1016/j.jcv.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Slavov SN, Haddad SK, Silva-Pinto AC, et al. Molecular and phylogenetic analyses of human Parvovirus B19 isolated from Brazilian patients with sickle cell disease and â-thalassemia major and healthy blood donors. J Med Virol. 2012;84:1652–65. doi: 10.1002/jmv.23358. [DOI] [PubMed] [Google Scholar]
- 22.Al-Qahtani AA, Alabsi ES, AbuOdeh R, et al. Prevalence of anelloviruses (TTV, TTMDV, and TTMV) in healthy blood donors and in patients infected with HBV or HCV in Qatar. Virol J. 2016;13:208. doi: 10.1186/s12985-016-0664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang N, Dai R, Zhang X. Global prevalence of human pegivirus-1 in healthy volunteer blood donors: a systematic review and meta-analysis. Vox Sang. 2020;115:107–19. doi: 10.1111/vox.12876. [DOI] [PubMed] [Google Scholar]
- 24.Boletim epidemiológico. Secretaria de vigilância em saúde, Ministério da Saúde. [Accessed on 21/01/2020]. Available at: http://portalarquivos2.saude.gov.br/images/pdf/2019/setembro/11/BE-arbovirose-22.pdf. [In Portuguese.]
- 25.Liao Q, Shan Z, Wang M, et al. An evaluation of asymptomatic Dengue infections among blood donors during the 2014 Dengue outbreak in Guangzhou. China. J Med Virol. 2017;89:2037–40. doi: 10.1002/jmv.24883. [DOI] [PubMed] [Google Scholar]
- 26.Busch MP, Sabino EC, Brambilla D, et al. International Component of the NHLBI Recipient Epidemiology and Donor Evaluation Study-III (REDS-III). Duration of Dengue viremia in blood donors and relationships between donor viremia, infection incidence and clinical case reports during a large epidemic. J Infect Dis. 2016;214:49–54. doi: 10.1093/infdis/jiw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabino EC, Loureiro P, Lopes ME, et al. International Component of the NHLBI Recipient Epidemiology and Donor Evaluation Study-III. Transfusion-transmitted Dengue and associated clinical symptoms during the 2012 epidemic in Brazil. J Infect Dis. 2016;13:694–702. doi: 10.1093/infdis/jiv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen LR, Tomashek KM, Biggerstaff BJ. Estimated prevalence of dengue viremia in Puerto Rican blood donations, 1995 through 2010. Transfusion. 2012;52:1647–51. doi: 10.1111/j.1537-2995.2011.03529.x. [DOI] [PubMed] [Google Scholar]
- 29.Mohammed H, Linnen JM, Muñoz-Jordán JL, et al. Dengue virus in blood donations, Puerto Rico, 2005. Transfusion. 2008;48:1348–54. doi: 10.1111/j.1537-2995.2008.01771.x. [DOI] [PubMed] [Google Scholar]
- 30.Linnen JM, Vinelli E, Sabino EC, et al. Dengue viremia in blood donors from Honduras, Brazil, and Australia. Transfusion. 2008;48:1355–62. doi: 10.1111/j.1537-2995.2008.01772.x. [DOI] [PubMed] [Google Scholar]
- 31.Tsai JJ, Lin PC, Tsai CY, et al. Low frequency of asymptomatic dengue virus-infected donors in blood donor centers during the largest dengue outbreak in Taiwan. PLoS One. 2018;13:e0205248. doi: 10.1371/journal.pone.0205248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–41. doi: 10.1016/s0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabino EC, Loureiro P, Lopes ME, et al. Transfusion-transmitted dengue and associated clinical symptoms during the 2012 epidemic in Brazil. J Infect Dis. 2016;213:694–702. doi: 10.1093/infdis/jiv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juhl D, Hennig H. Parvovirus B19: What Is the Relevance in Transfusion Medicine? Front Med. 2018;1:4. doi: 10.3389/fmed.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagaharu K, Sugimoto Y, Hoshi Y, et al. Persistent symptomatic parvovirus B19 infection with severe thrombocytopenia transmitted by red blood cell transfusion containing low parvovirus B19 DNA levels. Transfusion. 2017;57:1414–8. doi: 10.1111/trf.14088. [DOI] [PubMed] [Google Scholar]
- 36.Satake M, Hoshi Y, Taira R, et al. Symptomatic parvovirus B19 infection caused by blood component transfusion. Transfusion. 2011;51:1887–95. doi: 10.1111/j.1537-2995.2010.03047.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Metagenomic datasets have been submitted to the NCBI SRA and are available under BioProject accession numbers PRJNA602694.