Abstract
Mitochondria form networks that continually remodel and adapt to carry out their cellular function. The mitochondrial network is remodeled by changes in mitochondrial morphology, number, and distribution within the cell. Mitochondrial dynamics depend directly on fission, fusion, shape transition, and transport or tethering along the cytoskeleton. Over the past several years, many of the mechanisms underlying these processes have been uncovered. It has become clear that each process is precisely and contextually regulated within the cell. Here, we discuss the mechanisms regulating each aspect of mitochondrial dynamics, which together shape the network as a whole.
Keywords: mitochondria, morphology, fission, fusion, cytoskeleton, transport
Introduction
Mitochondria are highly dynamic organelles that morphologically adapt to fit cellular needs. The mitochondrial network changes in response to diverse cellular pathways, such as metabolism, intracellular calcium signaling, apoptosis, mitosis, and mitochondrial DNA replication. Despite the diversity of contexts that alter mitochondrial dynamics, the resultant effects on the mitochondrial network are dependent on four distinct processes. Fission, the division of a single mitochondrion into two mitochondria by cleavage of the Inner Mitochondrial Membrane (IMM) and Outer Mitochondrial Membrane (OMM), and fusion, the joining of the OMM and IMM, are in equilibrium to determine network connectivity. Network morphology is simultaneously determined by mitochondrial shape transitions independent of fission/fusion and precise positioning along the cytoskeleton (Fig. 1). As our understanding of each process develops, we must also establish a holistic understanding of how mitochondrial dynamics are orchestrated to control network properties. Here, we highlight recent progress that provides new insights into the complexity of each aspect of mitochondrial dynamics.
Figure 1. Overview of mitochondrial dynamics.
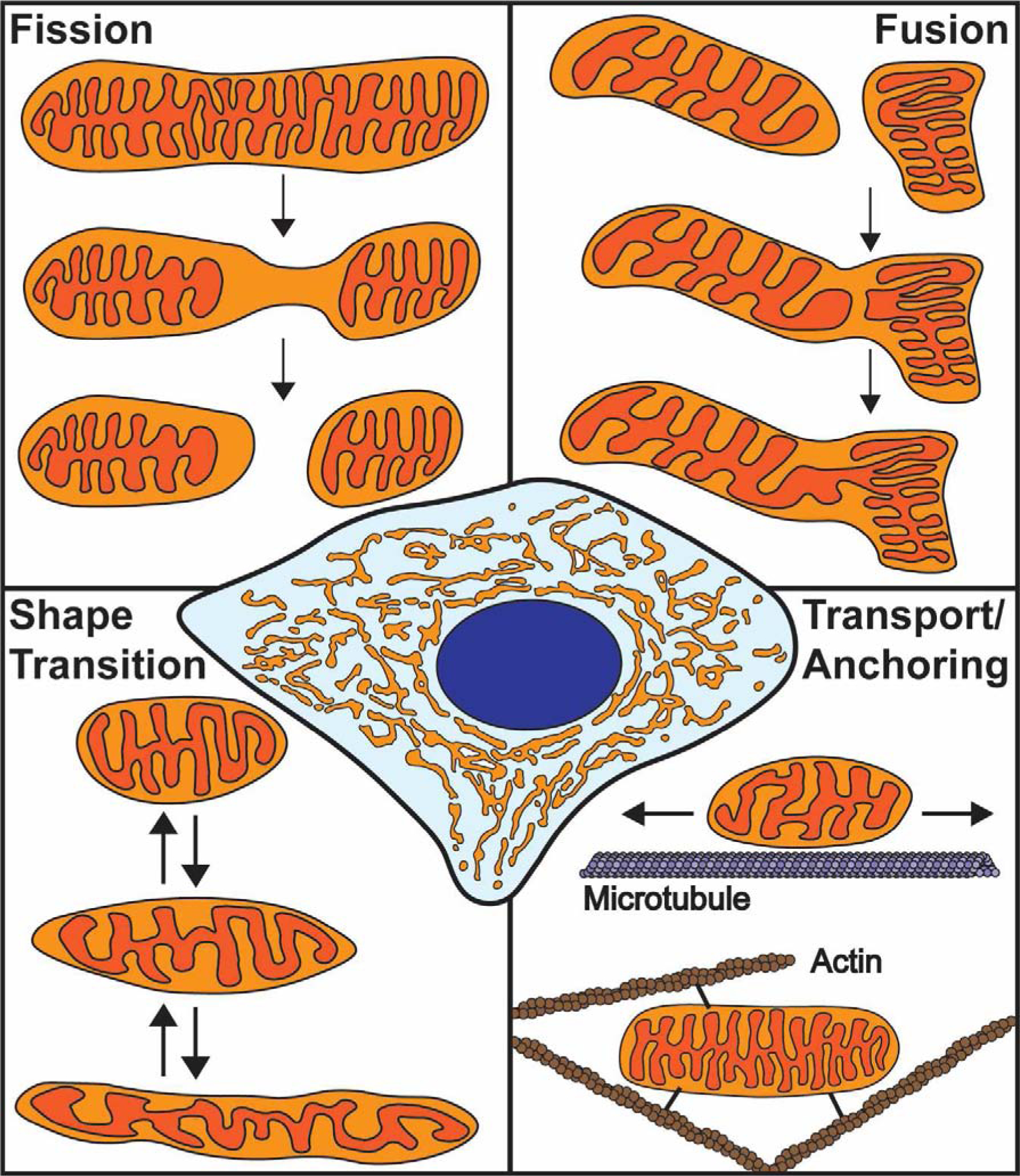
Mitochondria form a complex, interconnected network within the cell (center). The morphology of this network is determined by fission, fusion, mitochondrial shape transition, and positioning along the cytoskeleton. Fission begins with IMM division followed by OMM scission. Fusion involves merging of two outer membranes followed by joining of the inner membranes. Mitochondrial shape transition is a process independent of fission/fusion that controls the transition between rounded and elongated mitochondrial morphologies. Mitochondrial positioning involves transport and tethering along the microtubule and actin cytoskeletons.
Dividing mitochondria with Drp1 and actin
The key events of mitochondrial fission are constriction and scission of both the OMM and IMM. Outer membrane constriction is driven by Drp1, a GTPase that dynamically associates with the endoplasmic reticulum (ER) and mitochondria (Fig. 2A)1,2. Drp1 is recruited to mitochondria via interactions with receptors in the OMM: mitochondrial fission factor (MFF) and mitochondrial dynamics proteins 49 and 51 (MID49/51)3–6. Some Drp1 is transferred to the OMM following MFF-dependent oligomerization on the ER; this transfer likely occurs at mitochondria-ER contact sites, which mark sites of mitochondrial division (Fig. 2B)2,7. Drp1-dependent fission at mitochondria-ER contacts is facilitated by actin assembly, as inhibiting actin polymerization reduces fission frequency and Drp1 recruitment to mitochondria1,2,8,9. Actin assembly at mitochondria-ER contacts depends on two actin nucleating proteins, the formin INF2 and Spire1C, which reside on the ER and mitochondria, respectively. These proteins interact to promote actin assembly and mitochondrial constriction (Fig. 2B)8,10. Actin filaments locally assemble in a wave-like manner around mitochondrial subpopulations to induce fission11. Following actin disassembly, these mitochondrial subpopulations undergo fusion to locally remodel the mitochondrial network.
Figure 2. Mechanism of mitochondrial fission.
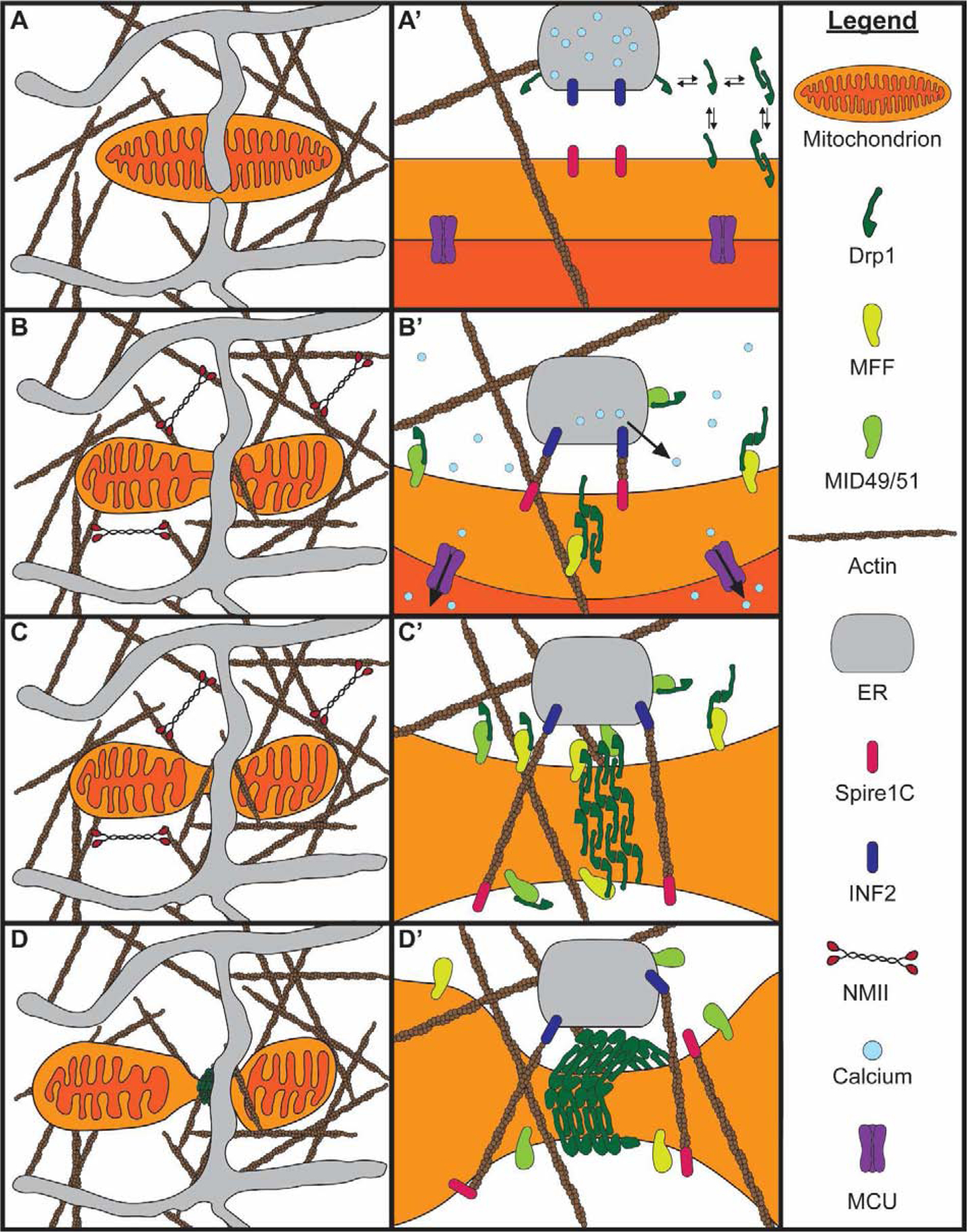
A) A mitochondrion is embedded in the interstitial actin network and closely associated with the ER. A closer view of the mitochondria-ER contact is shown in A’. Drp1 dynamically associates with the cytosol, mitochondria, and ER prior to fission. B) Peripheral NMII pulls on actin filaments to deform the mitochondrial membrane. Increased cytosolic calcium induces actin polymerization at mitochondria-ER contacts by INF2 on the ER and Spire1C on mitochondria. Mff and Mid49/51 begin recruiting Drp1 to the mitochondria-ER contact. Calcium is released from the ER and enters the mitochondria through the MCU, causing IMM constriction. C) Elevated mitochondrial matrix calcium causes IMM division prior to OMM division. Mff and Mid49/51 continue recruiting Drp1 to the mitochondria-ER contact, with some Drp1 coming from the ER. Drp1 oligomerizes along the constricted OMM. D) The Drp1 oligomer fully assembles around the OMM. Drp1 GTP hydrolysis dissociates Mid49/51, constricting the Drp1 ring. The Drp1 ring constricts the OMM and completes the process of fission.
Recent advances in electron microscopy have revealed the three-dimensional ultrastructure of the actin cytoskeleton during mitochondrial constriction. Yang and Svitkina (2019) found dense arrays of filamentous actin with criss-cross orientation at mitochondrial constrictions12. Many of these actin filaments extend from the nearby ER. This study also examined the positioning of non-muscle myosin II (NMII), as this motor has been implicated along with actin and INF2 in fission1,13. NMII is located near mitochondrial constrictions, primarily along the interstitial actin network (Fig. 2B–C), and is proposed to pull on the interstitial actin network to deform mitochondria upstream of Drp1, consistent with findings that NMII promotes Drp1 recruitment to mitochondria13.
Once recruited, Drp1 oligomerizes to wrap around the outer membrane (Fig. 2C). Upon GTP hydrolysis, Drp1 changes conformation, dissociating MID49/51 to shrink the oligomeric ring (Fig. 2D)14. While the Drp1 ring constricts the OMM, there is debate as to whether Drp1 carries out membrane scission. Initial studies found no evidence that Drp1 could drive membrane scission. Dynamin-2 (Dnm2), another dynamin GTPase, was found at fission sites following Drp1 recruitment; Dnm2 knockdown was also found to inhibit mitochondrial fission15. However, several recent studies implicate Drp1 as the protein responsible for membrane scission. Fibroblasts lacking Dnm2 or all three dynamin proteins display normal mitochondrial division, suggesting dynamins 1–3 are dispensable for fission16,17; in contrast, Drp1 is required for fission9,16,17. Further, purified Drp1 can induce the fission of membrane tubules up to 250 nm in radius17. While these results implicate Drp1 as the protein responsible for scission, more work is required to confirm whether Drp1 drives the final step in fission.
Whereas outer membrane scission depends on Drp1 oligomerization and GTP hydrolysis, the mechanism of inner membrane scission is less clear. Recent studies have shown that the IMM constricts and divides at mitochondria-ER contacts prior to Drp1-dependent OMM fission18,19. IMM constriction depends on INF2-mediated actin polymerization and NMII, similar to outer membrane constriction. Actin assembly at mitochondria-ER contacts stimulates calcium release from the ER and subsequent mitochondrial uptake through the mitochondrial calcium uniporter (MCU; Fig. 2B)18. Elevated mitochondrial calcium then stimulates IMM constriction in a Drp1-independent manner, but the subsequent mechanism of IMM scission is a black box.
While it is clear that mitochondrial fission is largely coordinated by the ER, several other factors determine sites of fission. Fission relies on the dynamic recruitment of lysosomes and the lysosomal GTPase RAB7. GTP-bound RAB7 is recruited to mitochondria by the mitochondrial fission protein 1 (Fis1), an OMM protein with two tetratricopeptide repeat domains exposed to the cytosol20. Once recruited, GTP-bound RAB7 promotes mitochondria-lysosome contact formation21. Mitochondria-lysosome contacts restrict mitochondrial motility, regulate inter-mitochondrial tethering, and mark sites of fission22. Fission is also modulated by the dynamic recruitment of the trans-Golgi network (TGN). The small GTPase ADP-ribosylation factor 1 (Arf1) and its effector, phosphatidylinositol 4-kinase-III-b [PI(4)KIIIb] are recruited to fission sites on TGN vesicles after Drp1 recruitment23. Loss of Arf1 or PI(4)KIIIb produces a hyperfused and branched network, suggesting these proteins affect mitochondrial branching in addition to fission. Intriguingly, TGN vesicles converged with lysosomes and ER at fission sites. Each of these organelles is present at most, but not all mitochondrial fission sites. Further analysis of the temporal and spatial dynamics of these organelles and their effector proteins is necessary to understand how they are coordinated to promote fission.
Promoting fusion or inhibiting fission: a balancing act
Mitochondrial fusion is mediated by the dynamin family GTPases mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), and Opa1. Fusion begins with Mfn1/2-mediated OMM tethering and merging followed by Opa1-mediated joining of the IMM (Fig. 3A)24,25. Opa1 has two isoforms: a long isoform (L-Opa1) containing a transmembrane domain, and a short isoform (S-Opa1) lacking the transmembrane domain. S-Opa1 is produced via proteolytic cleavage of L-Opa1 by one of two proteases, Yme1L or Oma126. Yme1L knockdown produces a fragmented mitochondrial network, suggesting that Opa1 processing promotes fusion26. A separate study found that L-Opa1 was sufficient for fusion in cells lacking Yme1L and Oma1; conversely, S-Opa1 overexpression in these cells resulted in mitochondrial fragmentation27. These contrasting results raised the question of whether Opa1 processing promotes fission or fusion. Two recent studies used in vitro membrane fusion assays to gain mechanistic insight into Opa1-mediated fusion28,29. Both studies tested the sufficiency of Opa1 to facilitate membrane fusion of liposomes. In these assays, L-Opa1 is sufficient to drive fusion through a heterotypic interaction with cardiolipin (CL), a mitochondrial phospholipid, whereas S-Opa1 is unable to drive fusion28,29. However, these studies found that S-Opa1 and L-Opa1 work synergistically to catalyze fusion. Ge et al. (2020) show that fusion efficiency peaks at an equimolar ratio of S-Opa1 to L-Opa1 (Fig. 3B)28. Thus, Opa1 processing tightly regulates fusion, with insufficient or excess processing inhibiting fusion.
Figure 3. Mitochondrial fusion and shape transition.
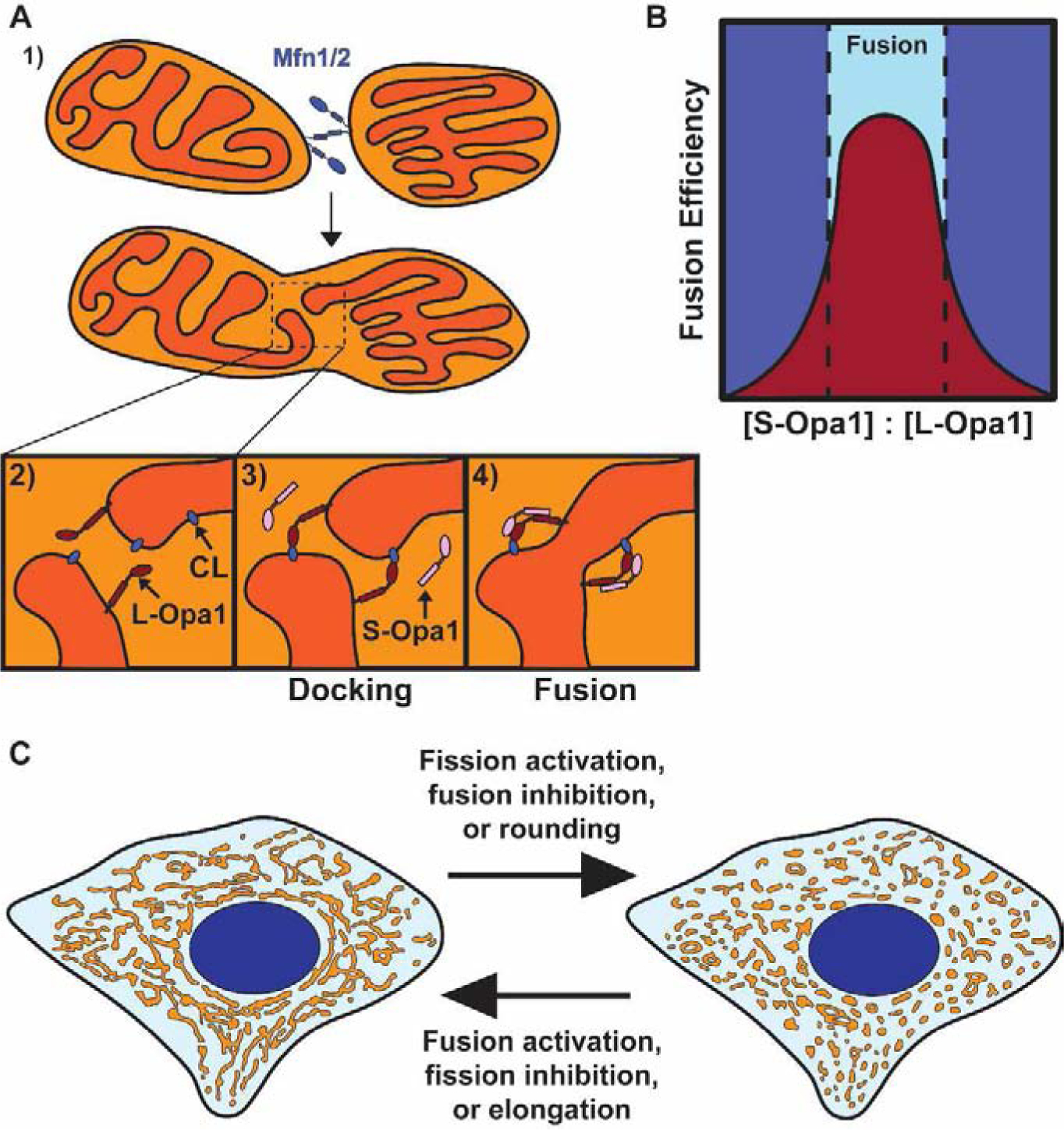
A) Proposed model of mitochondrial fusion. 1) Mitochondrial fusion begins with Mfn1/2-mediated tethering of two mitochondrial outer membranes. 2) The inner membranes are positioned for fusion upon outer membrane fusion. 3) Interactions between L-Opa1 and cardiolipin (CL) dock the inner membranes, bringing them closer together. 4) S-Opa1 functions with L-Opa1 and cardiolipin to promote efficient inner membrane fusion. B) Fusion efficiency at different S-Opa1:L-Opa1 ratios. Fusion efficiency peaks at an equimolar ratio of S-Opa1 to L-Opa1, with higher and lower ratios inhibiting fusion. C) Schematic of a connected mitochondrial network (left) and fragmented mitochondrial network (right). The transition between these networks can occur through direct regulation of fission, fusion, or mitochondrial shape transition.
Additional insight into the control of fusion has come from reexamination of Fis1 and mitochondria-ER contact sites. Mammalian Fis1 was initially thought to promote fission since its yeast homolog recruits Drp1 to mitochondria and because Fis1 overexpression induces mitochondrial fragmentation30. However, human Fis1 does not function through Drp1 and is dispensable for fission3,5,6. Fis1 has recently been shown to inhibit the activity of the fusion GTPases Opa1 and Mfn1/231, suggesting that fusion inhibition is sufficient to fragment the mitochondrial network, mirroring fission activation (Fig. 3C). Mitochondrial fusion also occurs at mitochondria-ER contact sites, similar to fission22,32,33. Fission and fusion proteins colocalize at mitochondria-ER contacts to form hotspots for membrane dynamics32; these ER-associated dynamics also include contact untethering between mitochondria22. Thus, the ER regulates multiple aspects of mitochondrial dynamics at contact sites. The next challenge is to determine how these separate machineries are coordinated to promote a single process.
Mitochondrial shape transition independent of fission/fusion
Mitochondrial shape varies depending on a variety of cellular signals. Two stimuli commonly used to alter mitochondrial network morphology are increased intracellular calcium and mitochondrial depolarization. Both produce a mitochondrial network comprised of small, rounded mitochondria, leading to speculation that both induce fission. However, Fung et al. (2019) revealed that calcium-induced and depolarization-induced mitochondrial fragmentation are distinct34. Calcium-induced actin assembly on mitochondria requires INF2, and thus undergoes canonical INF2-dependent fission (Fig. 2). Mitochondrial depolarization induced with the mitochondrial uncoupler carbonyl cyanide 3-chlorophenylhydrazone (CCCP) has a different effect on actin and mitochondria. CCCP-induced actin dynamics are INF2-independent, relying instead on the Arp2/3 complex to form transient actin clouds around depolarized mitochondria34. Depolarized mitochondria then undergo inner membrane rounding, resulting in shape deformation.
CCCP-induced mitochondrial deformations appear as rings. However, a recent study combining live imaging and volume electron microscopy found that mitochondrial rings are actually three-dimensional discs with central invaginations that only appear as rings in cross sections35. Most CCCP-induced shape changes were generated from rounding of the mitochondrion, rather than fission or fusion34,35. Consistently, CCCP-induced shape change is Drp1-independent and occurs due to IMM rearrangement, while the OMM remains intact34. Thus, CCCP-induced mitochondrial shape transition regulates the switch between a connected and fragmented mitochondrial network independent of fission/fusion (Fig. 3C).
Other mitochondrial inhibitors, such as inhibition of ATP synthesis with oligomycin, have distinct effects on mitochondrial morphology9. Thus, the relationship between mitochondrial function and morphology depends on multiple factors; metabolic function affects mitochondrial dynamics through fission, fusion, transport, and more36. However, mitochondrial form does not always match function. In Drosophila neurons, normal mitochondrial function is required for organism viability, independent of mitochondrial distribution, indicating that these processes are separable37.
Increasing cytosolic calcium has multiple effects on mitochondrial morphology. While increasing intracellular calcium promotes fission through canonical Drp1 oligomerization and actin polymerization1,2,18, calcium separately affects mitochondrial morphology through Miro1, an OMM transmembrane protein with two GTPase domains and two calcium-binding EF hands. Calcium induces mitochondrial shortening by binding to a single EF-hand of Miro138. Miro1-dependent mitochondrial shortening produces small, rounded mitochondria independent of Drp1, indicating this transition is distinct from fission38. Thus, calcium affects multiple aspects of mitochondrial morphology by promoting fission and shape transition through separate mechanisms. Additional work is needed to understand how these pathways intersect and cooperate to remodel the mitochondrial network in response to cytosolic calcium levels.
Mitochondrial transport and anchoring: stop and go on two cytoskeletons
Mitochondrial network morphology is also controlled by many additional interactions with the cytoskeleton. Mitochondrial transport is critical in highly polarized cells, such as neurons, where mitochondria undergo long-distance transport39. Most mitochondrial transport is microtubule-based, with transport toward the microtubule plus-end mediated by kinesin-1 and transport toward the minus-end mediated by cytoplasmic dynein 1 (dynein) and its partner complex, dynactin40,41. In the canonical model of mitochondrial transport, these opposing motors are tethered to mitochondria through the TRAK/Miro motor adaptor complex (Fig. 4A)39,41. TRAK1 and TRAK2, the mammalian orthologs of Drosophila Milton, interact with kinesin-1 and dynein-dynactin41, while Miro1 and Miro2 function as calcium-sensitive adaptors that link the motor/TRAK complexes to mitochondria42.
Figure 4. Mitochondrial transport and anchoring on the cytoskeleton.
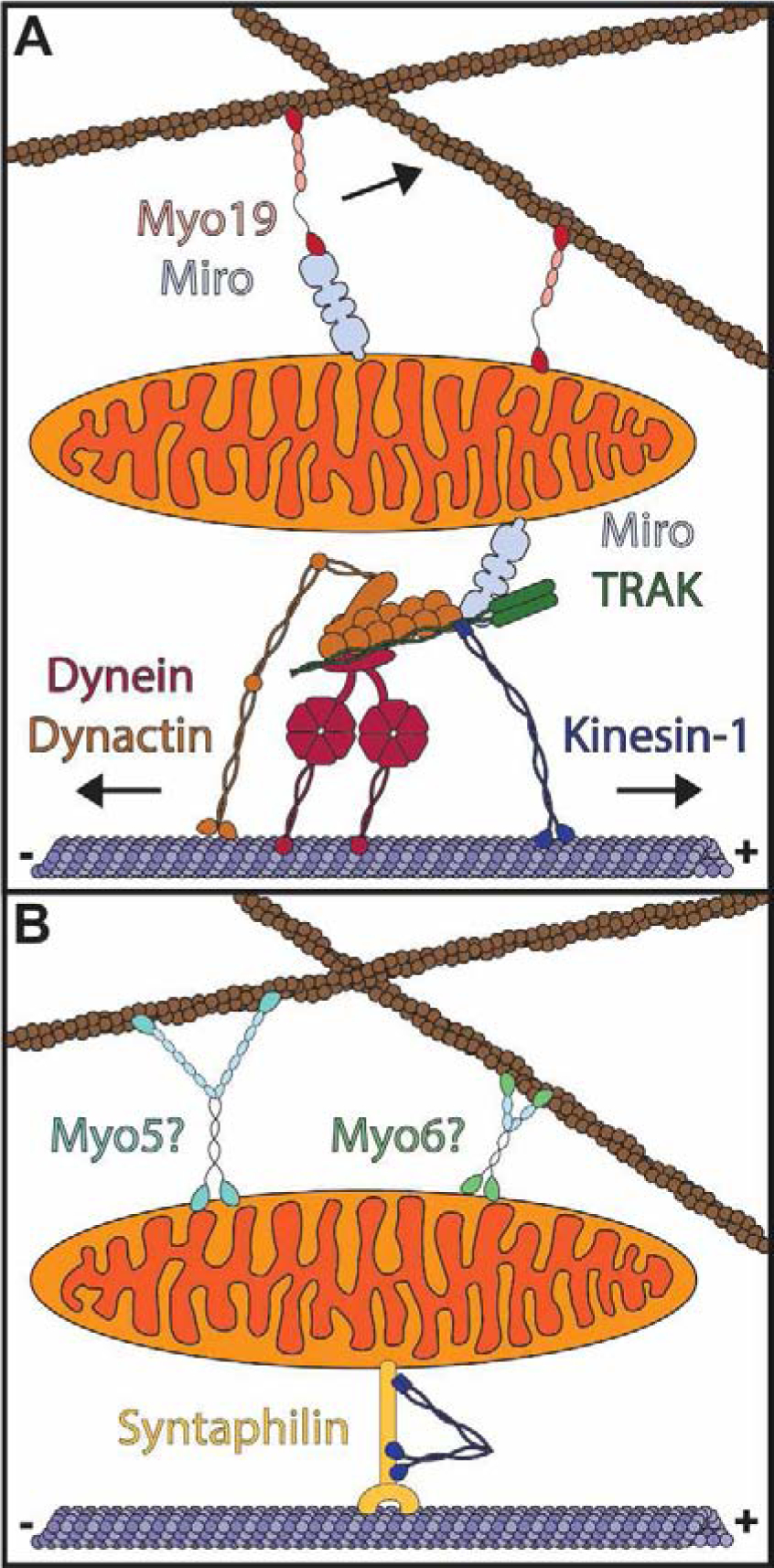
A) TRAK and Miro proteins serve as adaptors for microtubule-based mitochondrial transport (below). Kinesin-1 drives transport to the microtubule plus-end while transport to the microtubule minus end is mediated by dynein/dynactin. Myo19 associates with Miro proteins and directly with the mitochondrial outer membrane to drive mitochondrial transport along the actin cytoskeleton. B) Mitochondria are anchored to the actin and microtubule cytoskeletons. Syntaphilin anchors mitochondria to microtubules while myosin V (Myo5), myosin VI (Myo6), or another tether may anchor mitochondria to actin.
Motors, TRAKs and Miro proteins are required for mitochondrial transport, but the functional interactions among these components remain largely untested, and the molecular basis by which opposing kinesin and dynein motors are coordinated to produce directional transport of mitochondria is not understood. Motor regulation may be adaptor-specific. For instance, TRAK2 has been proposed to predominantly interact with dynein-dynactin whereas TRAK1 interacts with both kinesin-1 and dynein-dynactin41. TRAK1 overexpression promotes plus-end directed mitochondrial transport in mouse embryonic fibroblasts (MEFs) while TRAK2 overexpression promotes minus-end directed mitochondrial transport. However, TRAK2 requires Miro1, but not Miro2, to promote dynein-dependent transport43. Combined, these results suggest that the direction of mitochondrial transport is determined by specific associations between TRAK and Miro isoforms. Further studies are required to determine how individual TRAK and Miro proteins interact with microtubule motors to selectively promote transport toward either microtubule end.
A recent study found that TRAKs localize to mitochondria and drive transport in MEFs lacking Miro1 and Miro243. While transport is reduced in Miro1/2 knockout cells, this finding suggests that TRAKs can function independently of Miro. Since TRAKs interact with other OMM proteins, such as Mfn144,45, another OMM protein may function as an alternate adaptor for TRAK1/2. Miro proteins also serve as adaptors for myosin XIX (Myo19), a mitochondria-associated myosin motor, though Myo19 can also associate with the OMM independent of Miro (Fig. 4A)43,46,47. Myo19 overexpression increases mitochondrial motility in an actin-dependent manner48. Furthermore, TRAK overexpression reduces the association of Myo19 with mitochondria46, suggesting that Myo19 and TRAKs compete for Miro binding to induce actin- or microtubule-based mitochondrial motility. Given the nature of Miro proteins, it will be interesting to see how calcium binding and GTP hydrolysis affect the interaction of Miro with TRAKs and Myo19.
Mitochondria are also anchored to the cytoskeleton at specific cellular locations. Mitochondrial anchoring is particularly important in neurons, where mitochondria are tethered at presynaptic sites in axons to supply energy for neurotransmission. In mammalian neurons, actin stabilizes mitochondria at presynaptic terminals49,50. A recent study found that stationary mitochondria at presynaptic sites are more firmly anchored in place than other mitochondria49. The tethering of these presynaptic mitochondria is partially dependent on actin. Given that Myosin V and VI oppose mitochondrial motility in Drosophila neurons51, and Myosin VI can form actin cages around mitochondria52, it will be interesting to determine whether these myosins or a separate tether link mitochondria to actin for anchoring at presynaptic sites (Fig. 4B). Mitochondrial anchoring at presynaptic sites is also facilitated by syntaphilin, a microtubule-binding protein that has been proposed to dock mitochondria by binding kinesin-1, preventing motor activation by the Miro-TRAK complex (Fig. 4B)53. Thus, similar to mitochondrial transport, mitochondrial anchoring relies on both the actin and microtubule cytoskeletons and further investigations of mitochondrial anchoring must account for the effects of each.
Conclusion
As new imaging techniques have uncovered the precise shaping and remodeling of the mitochondrial network, we have increased our understanding of this dynamic organelle. Recent work has helped define the molecular dynamics of fission, fusion, shape transition, transport, and tethering. However, the mechanistic details of each process and their interplay with each other have not been worked out. The intersection of these pathways, with varied effects on mitochondrial morphology, gives rise to the morphological complexity found in this dynamic organelle. Future endeavors accounting for each aspect of mitochondrial dynamics will more fully uncover the nature of this organelle network and determine how it remodels and reshapes to fit cellular needs.
Supplementary Material
Acknowledgements
We would like to thank Stephen Coscia, Elizabeth Gallagher, Andrew Moore, and Chantell Evans for helpful discussion. We apologize that we could not mention many important works and articles on mitochondrial dynamics due to space limitations. This work has been supported by National Institutes of Health Grants T32 GM008216 to A.R.F., AG064618 to T.A.J., and R35 GM126950 and RM1 GM136511 to E.L.F.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Ji W, Hatch AL, Merrill RA, Strack S & Higgs HN Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji W-K et al. Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J. Cell Biol 216, 4123–4139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otera H et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol 191, 1141–1158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu R et al. MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff. Sci. Rep 7, 880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer CS et al. Adaptor Proteins MiD49 and MiD51 Can Act Independently of Mff and Fis1 in Drp1 Recruitment and Are Specific for Mitochondrial Fission. J. Biol. Chem 288, 27584–27593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osellame LD et al. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci 129, 2170–2181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman JR et al. ER Tubules Mark Sites of Mitochondrial Division. Science 334, 358–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korobova F, Ramabhadran V & Higgs HN An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 339, 464–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Vos KJ, Allan VJ, Grierson AJ & Sheetz MP Mitochondrial Function and Actin Regulate Dynamin-Related Protein 1-Dependent Mitochondrial Fission. Curr. Biol 15, 678–683 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Manor U et al. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 4, e08828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore AS, Wong YC, Simpson CL & Holzbaur ELF Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission–fusion balance within mitochondrial networks. Nat. Commun 7, 12886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C & Svitkina TM Ultrastructure and dynamics of the actin–myosin II cytoskeleton during mitochondrial fission. Nat. Cell Biol 21, 603–613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; *The authors use platinum replica electron microscopy to reveal the structure of actin filaments around mitochondria during fission. They find criss-cross arrays of actin filaments at mitochondrial constriction sites, with NMII located nearby, but not directly at sites of constriction.
- 13.Korobova F, Gauvin TJ & Higgs HN A Role for Myosin II in Mammalian Mitochondrial Fission. Curr. Biol 24, 409–414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalia R et al. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 558, 401–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JE, Westrate LM, Wu H, Page C & Voeltz GK Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540, 139–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonseca TB, Sánchez-Guerrero Á, Milosevic I & Raimundo N Mitochondrial fission requires DRP1 but not dynamins. Nature 570, E34–E42 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kamerkar SC, Kraus F, Sharpe AJ, Pucadyil TJ & Ryan MT Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat. Commun 9, 5239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study follows up on previous reports suggesting that Drp1 does not sever mitochondrial membranes, but rather constricts them for Dnm2 to carry out scission. The authors find that fission occurs normally in Dnm2 knockout cells, but not in Drp1 knockout cells. Drp1 is also able to constrict and sever membrane tubules, similar in size and composition to that of mitochondria in vitro. These findings indicate that Drp1 is capable of constricting and severing the mitochondrial membrane for the final step of fission.
- 18.Chakrabarti R et al. INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol 217, 251–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho B et al. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun 8, 15754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki M, Jeong S-Y, Karbowski M, Youle RJ & Tjandra N The Solution Structure of Human Mitochondria Fission Protein Fis1 Reveals a Novel TPR-like Helix Bundle. J. Mol. Biol 334, 445–458 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Wong YC, Ysselstein D & Krainc D Mitochondria–lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong YC, Peng W & Krainc D Lysosomal Regulation of Inter-mitochondrial Contact Fate and Motility in Charcot-Marie-Tooth Type 2. Dev. Cell 50, 339–354.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagashima S et al. Golgi-derived PI (4) P-containing vesicles drive late steps of mitochondrial division. Science 367, 1366–1371 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Ishihara N Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci 117, 6535–6546 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Cipolat S, de Brito OM, Dal Zilio B & Scorrano L OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci 101, 15927–15932 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra P, Carelli V, Manfredi G & Chan DC Proteolytic Cleavage of Opa1 Stimulates Mitochondrial Inner Membrane Fusion and Couples Fusion to Oxidative Phosphorylation. Cell Metab. 19, 630–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anand R et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol 204, 919–929 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge Y et al. Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. eLife 9, e50973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study uses in vitro membrane fusion assays with purified Opa1 to demonstrate that L-Opa1 and S-Opa1 cooperate to efficiently promote membrane fusion at an equimolar ratio. This finding resolves conflicting reports of insufficient and excessive Opa1 processing inhibiting fusion.
- 29.Ban T et al. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol 19, 856–863 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Yoon Y, Krueger EW, Oswald BJ & McNiven MA The Mitochondrial Protein hFis1 Regulates Mitochondrial Fission in Mammalian Cells through an Interaction with the Dynamin-Like Protein DLP1. Mol. Cell. Biol 23, 5409–5420 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu R, Jin S, Lendahl U, Nistér M & Zhao J Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 38, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abrisch RG, Gumbin SC, Wisniewski BT, Lackner LL & Voeltz GK Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J. Cell Biol 219, e201911122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y et al. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 175, 1430–1442.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Fung TS, Ji W-K, Higgs HN & Chakrabarti R Two distinct actin filament populations have effects on mitochondria, with differences in stimuli and assembly factors. J. Cell Sci 132, jcs234435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; **This article uses live imaging of actin and mitochondrial dynamics to show that calcium- and depolarization-induced mitochondrial fragmentation are distinct processes. Calcium-induced actin assembly on mitochondria requires INF2, whereas CCCP-induced actin assembly is Arp2/3-dependent and INF2-independent. CCCP-induced mitochondrial dynamics are mediated by Opa1-dependent rearrangement and circularization of the inner membrane. These dynamics are Drp1-indpendent and keep the outer membrane intact, indicating this process is distinct from fission.
- 35.Miyazono Y et al. Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fission-independent manner. Sci. Rep 8, 350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishra P & Chan DC Metabolic regulation of mitochondrial dynamics. J. Cell Biol 212, 379–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trevisan T et al. Manipulation of Mitochondria Dynamics Reveals Separate Roles for Form and Function in Mitochondria Distribution. Cell Rep. 23, 1742–1753 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Nemani N et al. MIRO-1 Determines Mitochondrial Shape Transition upon GPCR Activation and Ca2+ Stress. Cell Rep. 23, 1005–1019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misgeld T & Schwarz TL Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron 96, 651–666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilling AD, Horiuchi D, Lively CM & Saxton WM Kinesin-1 and Dynein Are the Primary Motors for Fast Transport of Mitochondria in Drosophila Motor Axons. Mol. Biol. Cell 17, 12 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Spronsen M et al. TRAK/Milton Motor-Adaptor Proteins Steer Mitochondrial Trafficking to Axons and Dendrites. Neuron 77, 485–502 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Eberhardt EL, Ludlam AV, Tan Z & Cianfrocco MA Miro: A molecular switch at the center of mitochondrial regulation. Protein Sci. 29, 1269–1284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Doménech G et al. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 37, 321–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; **Using Miro1/2 single and double knockout fibroblasts, this elegant study demonstrates non-canonical and non-redundant roles for Miro proteins in mitochondrial transport. TRAK2 functions with Miro1 to promote retrograde mitochondrial transport whereas Miro2 functions with TRAK2 to promote anterograde transport. However, TRAK1/2, kinesin-1, and dynein-dynactin can associate with mitochondria independent of Miro. Miro proteins also recruit Myo19 to mitochondria, showing that Miro coordinates actin- and microtubule-based mitochondrial motility.
- 44.Lee CA, Chin L-S & Li L Hypertonia-linked protein Trak1 functions with mitofusins to promote mitochondrial tethering and fusion. Protein Cell 9, 693–716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misko A, Jiang S, Wegorzewska I, Milbrandt J & Baloh RH Mitofusin 2 Is Necessary for Transport of Axonal Mitochondria and Interacts with the Miro/Milton Complex. J. Neurosci 30, 4232–4240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oeding SJ et al. Identification of Miro1 and Miro2 as mitochondrial receptors for myosin XIX. J. Cell Sci 131, jcs219469 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Bocanegra JL et al. The MyMOMA domain of MYO19 encodes for distinct Miro-dependent and Miro-independent mechanisms of interaction with mitochondrial membranes. Cytoskeleton 77, 149–166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quintero OA et al. Human Myo19 Is a Novel Myosin that Associates with Mitochondria. Curr. Biol 19, 2008–2013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutnick A, Banghart MR, West ER & Schwarz TL The light-sensitive dimerizer zapalog reveals distinct modes of immobilization for axonal mitochondria. Nat. Cell Biol 21, 768–777 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; *The authors used an inducible dimerizer to tether constitutively active kinesin-3 to mitochondria in mammalian neurons. Stationary mitochondria at presynaptic sites resist this inducible motility whereas other stationary mitochondria show inducible movement. This presynaptic mitochondrial tethering is partially dependent on actin, as inhibiting actin polymerization reduced the number of synaptic mitochondria resisting motility. How mitochondria are tethered to the actin cytoskeleton at presynaptic sites remains an open question.
- 50.Chada SR & Hollenbeck PJ Nerve Growth Factor Signaling Regulates Motility and Docking of Axonal Mitochondria. Curr. Biol 14, 1272–1276 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Pathak D, Sepp KJ & Hollenbeck PJ Evidence That Myosin Activity Opposes Microtubule-Based Axonal Transport of Mitochondria. J. Neurosci 30, 8984–8992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kruppa AJ et al. Myosin VI-Dependent Actin Cages Encapsulate Parkin-Positive Damaged Mitochondria. Dev. Cell 44, 484–499.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y & Sheng Z-H Kinesin-1–syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. J. Cell Biol 202, 351–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


