Abstract
Group 1 innate lymphoid cells (ILCs) comprise the natural killer (NK) cells and ILC1s. Both cells co-exist in peripheral tissues and despite effort to characterise the molecular identity and developmental pathways of ILC1s however their relationship with NK cells remains elusive. ILC1s and NK cells share many common features and analysis of ILC1s in tissues revealed a great heterogeneity and distinct transcriptional requirement of each ILC1 subsets complexifying the organisation of this group. Here, we discuss whether ILC1 and NK cells can be considered as distinct lineages based on their origin, location, phenotype or transcriptional regulation. Discrimination of NK cells and ILC1s represent an important challenge to unravel the individual functions of these cells during infection and tumour immunosurveillance.
Keywords: innate lymphoid cells, ILC1, NK cells, phenotype, transcriptional regulation, development, function, lineage
1. Introduction
Innate lymphoid cells (ILCs) provide the first line of defense against invading pathogens and are also involved in tissue repair [1]. Unlike T and B lymphocytes, ILCs develop independently of the recombinant activating gene (RAG), and their activity is not, therefore, regulated by antigen-specific receptors. Instead, their activity is dependent on cytokines, the engagement of activating and inhibitory receptors, and physiological signals from their microenvironment. They were initially classified into three subsets on the basis of the cytokines and transcription factors they express: group 1 ILCs (ILC1s), group 2 ILCs (ILC2s) and group 3 ILCs (ILC3s), which mirror the functions of T helper (h) 1, Th2 and Th17 cells, respectively. The classification of ILCs has recently been expanded to five groups, with the addition of NK cells and lymphoid tissue-inducer cells (LTi), previously classified as ILC1s and ILC3s, respectively, as distinct subsets [2]. This new classification is based on the evidence for distinctive developmental pathways for each of these subsets [3–8].
However, within group 1 ILCs, the definition of ILC1s and NK cells has been challenged by comparisons of these populations in different tissues, at steady state or in inflammatory conditions. Analyses of these populations at single-cell level have revealed considerable heterogeneity in the ILC1 subsets residing within tissues. It therefore remains unclear whether ILC1s and NK cells correspond to different developmental stages, activation states, functional plasticity or the imprinting of particular tissue microenvironments on the same lineage.
2. Do ILC1s and NK cells have different origins?
Not really
The developmental relationship between ILC1s and NK cells remains a matter of debate. All ILCs develop from common lymphoid progenitors (CLPs) in the fetal liver and adult bone marrow. The induction of the transcription factors Nfil3 and ID2 leads to the emergence of the innate lineage, which is characterized by the induction of α4β7 on a subset of CLPs [8,9]. The resulting common innate lymphoid progenitors (CILPs) do not generate B and T cells, but can generate all ILC subsets in vivo. The identification a common helper-ILC progenitor (CHILP) suggested the existence of a branch between the ILC and NK cell lineages, as no NK cells were generated following the adoptive transfer of CHILPs. However, the distinction between CILPs and CHILPs is not clear, as these two types of progenitor seem to have very similar patterns of surface marker and transcription factor expression. The expression of PLZF in the ILC precursor (ILCp) marks the bifurcation of LTi and NK cells from the other ILC1, 2 and 3 subsets [4]. However, a fate mapping experiment showed that 25% of NK cells had expressed PLZF, suggesting NK cells and ILC1 share a progenitor expressing PLZF or that ILC1s might acquire an NK cell phenotype. Indeed, a recent analysis of the progenitor in triple reporter (ID2, PZLF and Bcl11b) mice revealed that Id2+Zbtb16+ CILPs retained the potential to generate NK cells [10**]. These data suggest that ILC progenitors retain the potential to generate ILC1s and NK cells, as currently defined, and that the type of cell actually generated depends on the microenvironment and the signals received by progenitor.
In humans, CD117+ cells in the bloodstream or tissues can give rise to all ILC subsets, including NK cells, in vitro and in vivo [11]. Unbiased hierarchical stochastic neighbor-embedding (HSNE) analyses of blood CD117+ ILCs revealed that the CD117+CRTH2−NKp44− ILC population could be split into different subsets on the basis of KLRG1 and NKp46 expression. KLRG1-expressing ILCs correspond to a transitional stage of ILC2s, but they retain the potential to give rise to other ILC subsets if stimulated with appropriate signals. By contrast, NKp46-expressing ILCs give rise exclusively to ILC3s and ILC1s [12*]. CD200r, which can be used to distinguish between ILC1s and NK cells in mice [13], is also expressed on all human ILCs, but not on NK cells [12*]. This receptor can, therefore, be used to distinguish between NK cells and ILC1s in both humans and mice.
3. Can we define ILC1s and NK cells on the basis of their location and phenotype?
Yes, but essentially only at steady state.
One key characteristic distinguishing ILC1s from NK cells is their location: ILC1s reside in tissues, whereas NK cells recirculate in the bloodstream [14]. A careful analysis of NKp46+ cells in diverse tissues revealed that several ILC1 subsets co-exist with NK cells (Figure 1).
Figure 1: Tissue-specific markers and transcriptional requirements of the group 1 ILC in mouse.
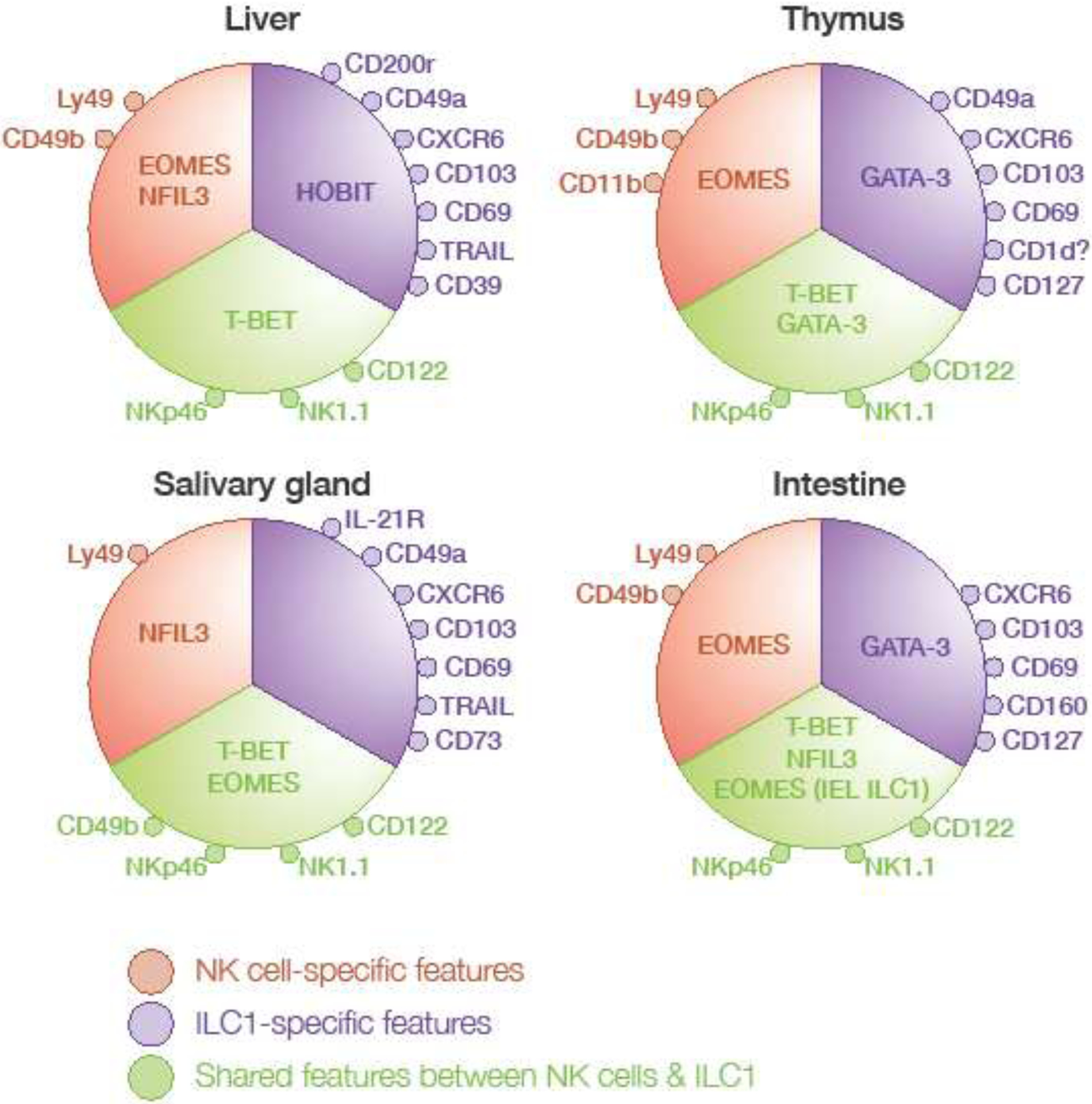
Characteristic markers and transcription factors of ILC1 and NK cells in the liver, thymus, salivary gland and intestine are shown. Common features of ILC1 and NK cells are depicted in green, while ILC1 and NK cell-specific receptors and transcription factors are shown in purple and red respectively (IEL, intraepithelial lymphocytes).
Intrahepatic ILC1s were the first ILC1 subset distinctly different from NK cells to be discovered [15]. This unique subset was initially thought to correspond to immature NK cells, as the cells lack expression of most of the Ly49 molecules, CD49b and the transcription factor Eomesodermin (Eomes), which control the maturation of the NK cells, but express the death-inducing ligand TRAIL [15,16]. Transcriptional analyses of hepatic TRAIL+ ILC1s and CD49b+ NK cells showed that liver ILC1s displayed a unique expression patterns for chemokine receptors and adhesion molecules, including CXCR6, CXCR3, CD103, CD49a, CD69, and CD39; cytokine receptors, such as IL-7Rα, IL-17RD, IL-21R, and TGF-βR; and regulatory molecules, such as CD200R, PD1-L, ICOSL, and Lag3 [6,17,18]. Single-cell multiplex transcriptional analyses of liver NK1.1+ NKp46+ cells identified four clusters and revealed the presence of more than two subsets in the liver [19]. Eomes was detected in one of the hepatic ILC1 subgroups and was differentially expressed between NK subsets. These findings suggest that the use of Eomes expression to distinguish between ILC1 and NK cells may not be appropriate.
Thymic NK cells or ILC1s?
There is also some debate about the identity of NKp46+ cells in the thymus. These cells were first described as NK cells, but their particular phenotype, including low levels of Ly49 and high levels of IL-7Rαand CD69 expression, and their requirement for the transcription factor GATA-3 for development, as reported for ILC1s [20], suggested that they might actually correspond to an ILC1 subset [21]. However, they also express Eomes and CD49b, which are generally associated with the NK cell lineage. Recent studies have shown that the NK1.1-expressing cells in the thymus are heterogeneous, with the majority of these cells having an ILC1-like phenotype (CD122+NK1.1+CD127+CD49b−), and a minority presenting an NK-cell phenotype (CD122+NK1.1+CD127+CD49b+CD11blow). The cells of the CD49b− population express CD49a and CD103, but not Eomes, consistent with currently used definitions of ILC1s. However, the authors found that this population of CD49b− cells was absent from Rag1−/− mice and tested positive for CD1d tetramers, suggesting that they may belonged to the NKT lineage rather than the ILC1 lineage [22*] (Figure 1).
Salivary gland ILC1s or TGF-β-imprinted NK cells?
As in the liver, NKp46+ cells in the salivary gland were originally described as unconventional NK cells [23], however more recent analysis revealed that both NK cells and ILC1 co-exist [24]. ILC1 in the salivary gland are phenotypically close to those found in the liver, expressing CD49a, CD103, CXCR6, IL21R and CD69 [25]. However, salivary gland ILC1s also display some transcriptional features common to NK cells, such as the expression of Eomes and Ly49H, and a lack of CD127 (Figure 1). Salivary gland ILCs therefore provide another example of ILCs producing transcripts considered characteristic of both ILC1s and NK cells. The lack of TGF-β signaling triggers a loss of ILC1-associated markers, including CD49a, CD103 and CD69, on salivary gland NKp46+ cells, but not on liver and intestinal ILC1s [25]. Moreover, recent studies have shown that tumor microenvironments enriched in TGF-β can modulate the features of NK cells, resulting in the acquisition of features characteristic of ILC1s and the downregulation of EOMES expression [26**]. These findings raise questions as to whether the microenvironment leaves an imprint on NK cells and induces the expression of ILC 1-associated markers. For a formal definition distinguishing between ILC 1 and NK cells, improvements are required in our understanding of the transcriptional requirements and expression profiles of these cells.
Intestinal ILC1s
In the small intestine, single-cell RNA-seq on the entire CD127+ cell population highlighted the complexity and diversity of ILC states [27]. As only CD127+ cells were analyzed, this analysis excluded a large proportion of NK cells and did not, therefore, fully capture all the diversity of group 1 ILCs [27]. However, it nevertheless revealed the existence of four clusters within the ILC1 group, with a gradient of Tbx21 expression among ILC1s. The ILC1a cluster expressed Gata3, suggesting possible involvement in the plasticity between ILC1s and ILC2s reported in previous studies. Another subset expressed high levels of NKp46 and RORγt [5,28], potentially corresponding to a transient or plastic ILC state, as reported for ex-ILC3 cells acquiring an ILC1-like expression profile. In the presence of IL-12 produced by DCs, ILC3s lose RORγt and acquire Tbet expression, leading to the production of IFN-γ and the loss of IL-22 production [28]. Conversely, CD127+ ILC1s can differentiate into Rorγt+ ILC3s when exposed to IL-23 and IL-1β, whereas CD103+ ILC1s and NK cells cannot [29].
In humans, two populations of ILC1s can be identified on the basis of CD127+ and CD103+ expression [29,30]. CD103+ ILC1s also express NKp44, CD161 and, like NK cells, they express CD56, CD94 and Eomes. These cells are present in the tonsils and the ileal epithelium, but not in mesenteric lymph nodes [30], whereas CD127+ ILC1s are present in the lamina propria [29].
So, can surface markers reliably distinguish ILC1s from NK cells?
Some markers, such as CD49a, CD103, CD69 and CXCR6, appear to be preferentially expressed on ILC1s, but a population of Lin−NK1.1+NKp46+ cells also express these markers, together with EOMES or other NK cell markers, such as CD49b or Ly49H (Figure 1). This mixed phenotype in a particular tissue can be explained by imprinting effects of the microenvironment, such as the TGF-β in the salivary gland ILC1s, but it does not occur in the liver [25]. Furthermore, these markers are not stable during activation, as the expression of CD49a and CD69 can be upregulated during MCMV infection or following exposure to cytokines, such as IL-2, IFN-γ, or IL-15 [25,31,32], suggesting that these key markers in Lin−NK1.1+NKp46+ cells are not sufficient to distinguish ILC1s reliably from NK cells at different developmental or activation stages. Finally, enzymatic digestion used to isolate ILCs from tissues can disrupt the expression of some markers and potentially explain some phenotypical differences observed.
4. Do ILC1s and NK cells have different regulatory programs?
Yes, but this is not the case for all ILC1s.
No transcription factor controlling the development of ILC1 and NK cells differentially has yet been identified, at least if all ILCs are considered together. The transcriptional control of ILC1 development seems to depend on the tissue in which the cell resides. Both ILC1 and NK cells express Tbet, a defining characteristic of the ILC1 family, but EOMES controls the maturation of NK cells [16]. Tbet deficiency leads to the total loss of ILC1s, whereas NK cells persist but in smaller numbers [33] and with impaired functions [16,34,35].
Nfil3 was one of the first transcription factors identified as exerting differential control over ILC1s and NK cells in the liver [6] and, subsequently, in the salivary gland [24,36,37] and uterus [33]. However, ILC1s in other tissues require Nfil3 for their development [9,38,39]. The transcription factor Hobit is required for hepatic ILC1s, but not for NK cells or ILC1s in the small intestine [40] (Figure 1).
Gata3 regulates the development of ILC progenitors [20] and thymic ILC1s [21], but does not affect the development of NK cells. The deletion of Gata3 in hematopoietic stem cells impairs ILC1 development in the small intestine, but it remains unknown whether this deletion affects liver or salivary gland ILC1s [20].
The long non-coding RNA Rroid, which controls Id2 transcription in ILC1s and NK cells but not in other ILCs, has different effects on ILC1s in different tissues [41]. Its deletion results in low levels of NK cells and ILC1s in the liver, lung, and spleen, but ILC1s from the small intestine and the salivary gland are not affected.
These particular transcriptional requirements for ILC1s increase the number of subsets and complexify the classification of the ILC1 family. They also make it more difficult to develop an ILC1-deficient model for studies of the role of these cells in immune responses.
5. Do ILC1s and NK cells have different roles in immunity?
Yes, at different times.
One crucial reason for identifying ILC1s and NK cells as different lineages relates to the possibility of these cells having different immunological functions. Early re-investigations of the role of NKp46-expressing cells in the control of infection were performed with specific markers found on liver ILC1s, such as CD49a. During infection with Toxoplasma gondii or Clostridium difficile, ILC1s are the major source of IFN-γ and TNF-α, producing much more of these cytokines than NK cells or NKp46 ILC3s [5,42]. However, in the absence of an ILCl-deficient model, these studies used Tbet−/− mice, which have relatively normal numbers of cells [5,42], but impaired NK cell function [34,35,43]. It therefore remains difficult to draw any firm conclusions concerning the specific and non-redundant roles of ILC1s. Interestingly, Rroid−/− mice, which have no ILC1s or NK cells in any organ other than ILC1s in the small intestine and salivary glands, can clear Salmonella enterica infection, supporting the hypothesis that intestinal ILC1s rather than NK cells control bacterial infection [41].
A temporal analysis of ILC1s during viral infection showed that these cells were required for optimal viral control, as they served as an early source of IFNγ [13**]. The elimination of liver ILC1s in Hobit-deficient mice leads to an increase in viral load after mouse cytomegalovirus (MCMV) clearance [13**]. CD49a+Eomes−CD200rl+ ILC1s remained Eomes− after adoptive transfer, confirming that endogenous ILC1s constitute a stable lineage during inflammation. By contrast, CD49b+Eomes+ NK cells in MCMV-infected mice displayed an increase in the expression of markers associated with an ILCl phenotype, such as CD39a and CD69, and no upregulation of CD200r, suggesting that this marker may be more reliable for tracking ILC1s in an inflammatory context [13**]. However, CD200r can be upregulated on liver NK cells in obese individuals [44]. This conversion to an ILCl phenotype is also mediated by TGF-β in the salivary gland [25] and in the tumor microenvironment [26**].
The conversion of NK cells into ILC1s is accompanied by a loss of antitumor properties. CD49a acquisition is followed by the upregulation of CTLA-4, Lag3 and CD96, and the downregulation of IFN-γ, contributing to tumor growth [26**]. It is, therefore, important to unravel the roles of bona fide ILC1s and TGF-β-imprinted NK cells that have acquired an ILC1 phenotype.
Acknowledgments:
C.S is supported by National Health and Medical Research Council (NHMRC) of Australia grants (APP1165443) and fellowships (APP1123000 C.S). L.B. is supported by National Institutes of Health Research Grants AI46709 and AI122217. The E.V. laboratory at CIML and Assistance-Publique des Hôpitaux de Marseille is supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (TILC, grant agreement No. 694502 and MInfla-TILC, grant agreement No. 875102 - MInfla-Tilc), the Agence Nationale de la Recherche including the PIONEER Project (ANR-17-RHUS-0007), MSDAvenir, Innate Pharma and institutional grants to the CIML (INSERM, CNRS, and Aix-Marseille University) and to Marseille Immunopole.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
E.V. is a cofounder and employee of Innate Pharma.
B. References
- 1.Artis D, Spits H: The biology of innate lymphoid cells. Nature 2015, 517:293–301. [DOI] [PubMed] [Google Scholar]
- 2.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. : Innate Lymphoid Cells: 10 Years On. Cell 2018, 174:1054–1066. [DOI] [PubMed] [Google Scholar]
- 3.Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, Golub R: Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat Immunol 2016, 17:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A: A committed precursor to innate lymphoid cells. Nature 2014, 508:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klose CSN, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca Pereira D, et al. : Differentiation of Type 1 ILCs from a Common Progenitor to All Helper-like Innate Lymphoid Cell Lineages. Cell 2014, 157:340–356. [DOI] [PubMed] [Google Scholar]
- 6.Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJM, Busslinger M, Smyth MJ, Belz GT, Carotta S: Differential Requirement for Nfil3 during NK Cell Development. The Journal of Immunology 2014, 192:2667–2676. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, Brandt C, Kane L, Campos LS, Lu L, et al. : Single-cell RNA-seq identifies a PD-1(hi) ILC progenitor and defines its development pathway. Nature 2016, 539:102–106. [DOI] [PubMed] [Google Scholar]
- 8.Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, Almeida FF, Shi W, Ritchie ME, Naik SH, Huntington ND, et al. : Deciphering the Innate Lymphoid Cell Transcriptional Program. Cell Rep 2016, 17:436–447. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, Hooper LV: The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, Liu P, Golub R, Di Santo JP: An Id2(RFP)-Reporter Mouse Redefines Innate Lymphoid Cell Precursor Potentials. Immunity 2019, 50:1054–1068 e1053. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper shows that Zbtb16 (PLZF) expressing ILCp retains NK cells potential. This study used triple gene reporter mouse that may explain contradicting results from previous reports.
- 11.Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, et al. : Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell 2017, 168:1086–1100 e1010. [DOI] [PubMed] [Google Scholar]
- 12.Nagasawa M, Heesters BA, Kradolfer CMA, Krabbendam L, Martinez-Gonzalez I, de Bruijn MJW, Golebski K, Hendriks RW, Stadhouders R, Spits H, et al. : KLRG1 and NKp46 discriminate subpopulations of human CD117(+)CRTH2(−) ILCs biased toward ILC2 or ILC3. J Exp Med 2019, 216:1762–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This papers shows that blood CD117+ ILC population can be segregated by the expression of KLRG1 and NKp46. KLRG1+ ILCs are transitional cells that are biased toward the ILC2 lineage. NKp46+ ILCs are biased toward the ILC3 lineage but they retain the capacity to differentiate into ILC1s under IL-12 stimulation.
- 13.Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, Lau C, Degli-Esposti MA, Leslie CS, Sun JC, O’Sullivan TE: ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 2017, 171:795–808 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper shows that tissue-resident ILC1s represent an early source of IFN-γduring MCMV infection and the deletion of ILC1 using Hobit-deficient mice leads to increased viral load. It also reports that CD200r is a stable marker to track ILC1 during the course of infection.
- 14.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY: Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 2015, 350:981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, Yagita H, Kinoshita K, Okumura K, Smyth MJ: TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 2005, 105:2082–2089. [DOI] [PubMed] [Google Scholar]
- 16.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL: The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 2012, 36:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z: Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. The Journal of clinical investigation 2013, 123:1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, et al. : T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. Journal of Experimental Medicine 2014, 11:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perchet T, Petit M, Banchi EG, Meunier S, Cumano A, Golub R: The Notch Signaling Pathway Is Balancing Type 1 Innate Lymphoid Cell Immune Functions. Front Immunol 2018, 9:1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, et al. : The Transcription Factor GATA3 Is Critical for the Development of All IL-7Rα-Expressing Innate Lymphoid Cells. Immunity 2014, 40:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vosshenrich CAJ, García-Ojeda ME, Samson-Villéger SI, Pasqualetto V, Enault L, Richard-Le Goff O, Corcuff E, Guy-Grand D, Rocha B, Cumano A, et al. : A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nature Immunology 2006, 7:1217–1224. [DOI] [PubMed] [Google Scholar]
- 22.Gabrielli S, Sun M, Bell A, Zook EC, de Pooter RF, Zamai L, Kee BL: Murine thymic NK cells are distinct from ILC1s and have unique transcription factor requirements. Eur J Immunol 2017, 47:800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study concludes that thymic NK1.1+ cells are more related NK cells than ILC1 as they dependent on Nfil3 to develop. However these thymic NK cells express CD49a and CD103, therefore more investigations will be requiered to determine whether thymic NK1.1+ cells in the thymus are an heterogeneous population of NK cells and ILC1.
- 23.Tessmer MS, Reilly EC, Brossay L: Salivary gland NK cells are phenotypically and functionally unique. PLoS Pathog 2011, 7:e1001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erick TK, Anderson CK, Reilly EC, Wands JR, Brossay L: NFIL3 Expression Distinguishes Tissue-Resident NK Cells and Conventional NK-like Cells in the Mouse Submandibular Glands. J Immunol 2016, 197:2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortez VS, Cervantes-Barragan L, Robinette ML, Bando JK, Wang Y, Geiger TL, Gilfillan S, Fuchs A, Vivier E, Sun JC, et al. : Transforming Growth Factor-beta Signaling Guides the Differentiation of Innate Lymphoid Cells in Salivary Glands. Immunity 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, Rautela J, Straube J, Waddell N, Blake SJ, et al. : Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol 2017, 18:1004–1015. [DOI] [PubMed] [Google Scholar]; ** This paper shows how TGF-0 induce the conversion of NK cells into intermediate type 1 innate lymphoid cell (CD49a+Eomes+) and ILC1 populations in the tumor microenvironment. Unlike NK cells, these converted ILC1s are unable to control tumor growth.
- 27.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. : The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell 2016, 166:1231–1246 e1213. [DOI] [PubMed] [Google Scholar]
- 28.Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. : Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol 2013, 14:221–229. [DOI] [PubMed] [Google Scholar]
- 29.Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, Munneke JM, Hazenberg MD, Villaudy J, Buskens CJ, et al. : Interleukin-12 and −23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity 2015, 43:146–160. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M: Intraepithelial Type 1 Innate Lymphoid Cells Are a Unique Subset of IL-12- and IL-15-Responsive IFN-γ-Producing Cells. Immunity 2013:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth MJ, Takeda K, Hayakawa Y, Peschon JJ, van den Brink MR, Yagita H: Nature’s TRAIL-on a path to cancer immunotherapy. Immunity 2003, 18:1–6. [DOI] [PubMed] [Google Scholar]
- 32.Schuster IS, Wikstrom ME, Brizard G, Coudert JD, Estcourt MJ, Manzur M, O'Reilly LA, Smyth MJ, Trapani JA, Hill GR, et al. : TRAIL+ NK Cells Control CD4+ T Cell Responses during Chronic Viral Infection to Limit Autoimmunity. Immunity 2014, 41:646–656. [DOI] [PubMed] [Google Scholar]
- 33.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, et al. : Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 2014, 3:e01659–e01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenne CN, Enders A, Rivera R, Watson SR, Bankovich AJ, Pereira JP, Xu Y, Roots CM, Beilke JN, Banerjee A, et al. : T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. The Journal of experimental medicine 2009, 206:2469–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH: T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 2004, 20:477–494. [DOI] [PubMed] [Google Scholar]
- 36.Crotta S, Gkioka A, Male V, Duarte JH, Davidson S, Nisoli I, Brady HJM, Wack A: The Transcription Factor E4BP4 Is Not Required for Extramedullary Pathways of NK Cell Development. The Journal of Immunology 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M: Cutting Edge: Salivary Gland NK Cells Develop Independently of Nfil3 in Steady-State. The Journal of Immunology 2014. [DOI] [PubMed] [Google Scholar]
- 38.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S: Nfil3 is required for the development of all innate lymphoid cell subsets. Journal of Experimental Medicine 2014, 211:1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, Lopez-Lastra S, Motomura Y, Moreira-Santos L, Bihl F, Braud V, et al. : NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep 2015, 10:2043–2054. [DOI] [PubMed] [Google Scholar]
- 40.Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. : Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352:459–463. [DOI] [PubMed] [Google Scholar]
- 41.Mowel WK, McCright SJ, Kotzin JJ, Collet MA, Uyar A, Chen X, DeLaney A, Spencer SP, Virtue AT, Yang E, et al. : Group 1 Innate Lymphoid Cell Lineage Identity Is Determined by a cis-Regulatory Element Marked by a Long Non-coding RNA. Immunity 2017, 47:435–449 e438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B, Ling L, Leiner I, Pamer EG: Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell Host Microbe 2015, 18:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Sullivan TE, Rapp M, Fan X, Weizman OE, Bhardwaj P, Adams NM, Walzer T, Dannenberg AJ, Sun JC: Adipose-Resident Group 1 Innate Lymphoid Cells Promote Obesity-Associated Insulin Resistance. Immunity 2016, 45:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuff AO, Sillito F, Dertschnig S, Hall A, Luong TV, Chakraverty R, Male V: The Obese Liver Environment Mediates Conversion of NK Cells to a Less Cytotoxic ILC1-Like Phenotype. Front Immunol 2019, 10:2180. [DOI] [PMC free article] [PubMed] [Google Scholar]


