Abstract
The microtubule cytoskeleton is assembled from a finite pool of α,β-tubulin, the size of which is controlled by an autoregulation mechanism. Cells also tightly regulate the architecture and dynamic behavior of microtubule arrays. Here, we discuss progress in our understanding of how tubulin autoregulation is achieved, and highlight work showing that tubulin, in its unassembled state, is relevant for regulating the formation and organization of microtubules. Emerging evidence suggests that tubulin regulates microtubule-associated proteins and kinesin motors that are critical for microtubule nucleation, dynamics and function. These relationships create feedback loops that connect the tubulin assembly cycle to the organization and dynamics of microtubule networks. We term this concept the “tubulin economy”, which emphasizes the idea that tubulin is a resource that can be deployed for the immediate purpose of creating polymers, or alternatively as a signaling molecule that has more far-reaching consequences for the organization of microtubule arrays.
INTRODUCTION
The cell uses microtubules, polymers assembled from αβ-tubulin, to carry out a dizzying range of functions. Nanometer-sized tubulin subunits assemble into typically micron-length microtubule polymers which can function individually, e.g., as tracks for molecular motor proteins that organize contents of the cell, or collectively, as higher order structures that are tasked for specific cellular roles. For example, microtubules assemble into structures such as the mitotic spindle, which segregates chromosomes during cell division; axonemes within primary and motile cilia, which form the structural core of these fingerlike projections; and the marginal band in red blood cells and platelets, which stabilizes the biconcave shape of these cells. A key property of microtubules is that they are dynamic, switching between growth and shrinkage as subunits are added or removed from their ends [1]. Microtubules retain their dynamicity in higher order arrays, allowing these assemblies to carry out work, such as powering chromosome movement during mitosis. Importantly, the continuous remodeling of microtubule networks produces a pool of soluble, unpolymerized tubulin subunits, and the size of this pool dictates the assembly properties of microtubules, e.g., the growth rate and the frequency at which microtubules switch from growth to shrinkage (catastrophe).
Because microtubules and the assemblies that they form are essential for life, it is not surprising that most research on tubulin focuses on the properties and functions of their polymeric forms. By comparison, much less attention is paid to unassembled subunits. However, tubulin – in its unpolymerized form – has the potential to impact cell physiology in significant ways. For example, many microtubule-associated proteins (MAPs) have multiple sites that interact with microtubules and tubulin. Indeed, TOG-domain containing proteins (e.g. XMAP215) use such multivalency to promote microtubule assembly [2–4]. In principle, tubulin thus has the capacity to govern the activity of MAPs, creating powerful feedback loops that connect microtubule dynamics to the architecture of microtubule-based arrays. The focus of this review is to discuss the concept that unassembled tubulin subunits critically impact the formation and function of microtubule networks. We discuss evidence showing that tubulin can be highly concentrated at the centrosome, providing an example of how tubulin distribution within the cell can be heterogeneous, and that unpolymerized tubulin can act in a regulatory manner by controlling a wide range of processes, from production of tubulin itself to the formation of microtubule-based arrays. We suggest that the cell employs a “tubulin economy”, judiciously using tubulin subunits as either building blocks for microtubules or as signaling molecules to regulate the overall properties and function of microtubule networks.
CONTROL OF TUBULIN PROTEIN LEVELS BY AUTOREGULATION
One of the first indications that tubulin has a significant function outside of its polymeric form came from the observation that an acute increase in the levels of unpolymerized tubulin, e.g., by treatment of cells with microtubule depolymerizing agents, causes degradation of α- and β-tubulin mRNAs [5] and a subsequent decrease in α,β-tubulin protein synthesis [6]. Treatment of cells with taxol, a microtubule-stabilizing drug, on the other hand, causes an increase in tubulin mRNA and protein levels [5, 7]. These cellular responses, which are thought to affect the expression of all α- and β-tubulin isotypes in all higher eukaryotic cells, are referred to as tubulin autoregulation [7, 8]. An important aspect of tubulin autoregulation mechanism is that it appears to monitor tubulin protein levels, rather than polymer mass. If the objective of tubulin autoregulation was to maintain a precise mass of tubulin polymer, cells containing low polymer mass should increase the levels of α- and β-tubulin mRNAs and protein, and do the opposite in the presence of high levels of polymer, contrary to what has been observed. This conundrum has given rise to the intriguing hypothesis that cells are sensitive to intracellular concentrations of α,β-tubulin dimers because they have a function(s) outside of building microtubules [9].
Seminal work revealed core features of the tubulin autoregulation pathway. Translation of the first 13 nucleotides of β-tubulin mRNA, which encode the sequence Met-Arg-Glu-Ile (MREI), is required for β-tubulin mRNA degradation [10–14]. Moreover, occlusion of the MREI sequence with a β-tubulin monoclonal antibody prevents autoregulation. For this reason, it had long-been hypothesized that mRNA degradation was triggered by a factor that binds the MREI motif as nascent tubulin is in the process of being translated [15].
Using a photo-crosslinking approach, Lin et al. recently identified tetratricopeptide protein 5 (TTC5) as a protein that fits all of the criteria to be the key mediator of tubulin autoregulation [16]. TTC5 forms protein-protein contacts between the nascent N-termini of both α- and β-tubulin and the ribosome to form a “ribosome-nascent chain” complex (RNC). Knockout of TTC5 in multiple cell lines eliminated the decay of α- and β-tubulin mRNAs in response to drug-induced microtubule destabilization. In addition, the ability of cells to specifically reduce the synthesis of both α- and β-tubulin proteins is also lost in the absence of TTC5. Interestingly, cells appear to produce an inhibitor of TTC5 that is inactivated by elevated levels of unpolymerized tubulin; the identity of this inhibitor is not currently known.
Although most research on tubulin autoregulation has focused on α- and β-tubulin, Gasic et al. recently demonstrated that γ-tubulin mRNA levels are similarly controlled by microtubule damaging agents [9]. Whether this autoregulatory mechanism impacts protein levels of γ-tubulin and/or microtubule nucleation rates is not known. Interestingly, the authors did not observe co-regulation of MAPs or microtubule-associated motor proteins which is striking because proteins involved in a common biological process are often coordinately regulated [17, 18].
SOLUBLE TUBULIN AS A REGULATOR OF MICROTUBULE NUCLEATION
The distribution of soluble tubulin within the cell is not homogeneous. An excellent example of local enrichment in the soluble tubulin pool is the centrosome, a cellular organelle with the primary responsibility of nucleating microtubule polymers. A recent study [19] directly measured the amounts of polymer and soluble tubulin within centrosomes of C. elegans one-cell embryos. By segmenting individual microtubules using serial-section EM tomography, this study reported that the metaphase centrosome contains ~10,000 short (1.1 μm average length) microtubules. Combining this EM characterization with quantitative fluorescence light microscopy measurements of total tubulin concentration allowed the authors to determine the concentration of soluble tubulin fraction throughout the centrosome. Their measurements revealed that centrosomes concentrate soluble tubulin by an order of magnitude – while the overall tubulin concentration in the cell was 47 μM, the soluble tubulin fraction reached values of 470 μM within centrosomes. Combined with the polymer fraction, the overall tubulin concentration in one-cell stage C. elegans centrosomes was up to a staggering 660 μM.
How centrosomes maintain such a high concentration of unpolymerized tubulin remains a puzzle. With purified tubulin in vitro, the critical concentration for spontaneous microtubule nucleation is typically estimated to be in the tens of μM range [20]. However, the conditions in vitro are dramatically different than those within centrosomes, where assembly inhibitors or tubulin sequestering proteins may suppress microtubule nucleation. Importantly, Baumgart et al. [19] used FRAP measurements to demonstrate that the vast majority of unpolymerized tubulin in centrosomes is not immobilized by centrosome scaffolding proteins, albeit displaying a lower effective diffusion than soluble tubulin in the cytoplasm.
Although the critical concentration for microtubule nucleation may depend on specific local conditions, the significant enrichment of soluble tubulin within mitotic centrosomes is likely to promote nucleation. Thus, the finding that tubulin is super-concentrated at centrosomes can explain why gamma-tubulin, an important nucleation template, is not necessary for microtubule nucleation in C. elegans [21]. Along those lines, a previous study [22] demonstrated that artificial C. elegans centrosomes, reconstituted from a minimal system of recombinant components not including gamma-tubulin, have potent microtubule nucleation capacity. While the mechanism(s) by which tubulin concentrates at centrosomes are not fully understood, the findings by Woodruff et al. provided important insight. Namely, the authors showed that condensates of pericentriolar material (PCM)-scaffold protein SPD-5 in vitro recruited TPXL-1 and ZYG-9 (C. elegans homologs of MAPs TPX2 and ch-TOG/XMAP215), which in turn further promoted tubulin recruitment and microtubule nucleation. Thus, the authors concluded that centrosomes may be condensates that promote microtubule nucleation by selectively gating specific client proteins such as tubulin.
Aside from selective condensation within centrosomes, soluble tubulin can control acentrosomal microtubule nucleation through its regulation of the MAP TPX2. Previous in vitro studies showed that TPX2 can serve as a microtubule nucleating factor, particularly in concert with the microtubule polymerase ch-TOG/XMAP215 [23, 24]. This activity is likely to be important in the context of branching microtubule nucleation, wherein microtubules are nucleated from the wall of a pre-existing microtubule lattice [25]. Earlier work showed that TPX2 promotes the formation of tubulin aggregates or “stubs” [23, 26], suggesting that TPX2 can increase the local concentration of tubulin and/or drive the assembly of a microtubule seed. Recent work sheds further light on the ability of TPX2 to nucleate microtubules by showing that TPX2 can form a condensate, and that this behavior is stimulated by unpolymerized tubulin [27]. Significantly, TPX2 and tubulin co-condense on the surface of microtubules, and these condensates promote branching microtubule nucleation. Unpolymerized tubulin thus regulates the spatial distribution and microtubule nucleation activity of TPX2.
SOLUBLE TUBULIN AS A REGULATOR OF MICROTUBULE DYNAMICS
Global levels of soluble tubulin in the cytosol have been linked to the size of microtubule structures. For example, an increase in soluble tubulin availability led to longer cilia, while limiting tubulin supply resulted in shorter cilia [28–31]. Reconstituting mitotic spindles in decreasing cytoplasmic volumes resulted in smaller spindle sizes, correlated with lower concentrations of soluble tubulin [32, 33]. A new study reveals that even local spatiotemporal control of soluble tubulin levels may induce microtubule cytoskeleton perturbations [34]. Here, the authors developed a FRET-based sensor compatible with optogenetic manipulation, which allowed them to directly visualize interactions between soluble tubulin and a well-known tubulin-sequestering protein Op18/stathmin in HEK293T cells. They found that a local release of tubulin promoted microtubule extensions into lamellipodia, demonstrating the local feedback between polymer and soluble tubulin pools.
In the unperturbed physiological context, the process of tubulin assembly into microtubules could, in principle, on its own lead to transient local inhomogeneities in the pool of soluble tubulin and/or MAPs. Given that the parameters of dynamic instability, including growth and catastrophe rates, are acutely sensitive to concentrations of tubulin and MAPs, any such perturbations may result in a negative feedback, where microtubule assembly locally depletes the protein components, leading to a slow-down in further assembly or even triggering microtubule disassembly. A new study by Geisterfer et al. [35] devised a highly controlled in vitro setup to measure the extent to which an increased density of polymer assembly affects local microtubule dynamics.
Using hydrogel photolithography approaches to confine precise volumes of Xenopus egg extracts in a manner compatible with high spatiotemporal resolution imaging of the dynamics of individual microtubules, Geisterfer et al. found a direct negative correlation between the microtubule growth rate and the density of microtubule plus ends in this cell-free system. With a clever setup, in which Xenopus egg extracts were contained within an hourglass-shaped device with an artificial microtubule organizing center restricted to one half of the device while the cytoplasm was continuous across the entire device, the authors concluded that the mechanism of local growth-rate regulation is likely diffusion-based. Namely, local regions with smaller density of EB-comets, which mark growing microtubule ends, displayed faster microtubule growth rates than those in the regions of higher EB-comet density. Their findings are thus consistent with the limiting component model [36], in which a local depletion of protein components limits microtubule assembly.
Could soluble tubulin itself be the protein component that is limiting microtubule growth in relation to microtubule end density? Based on the classic measurements of the tubulin diffusion coefficient in the cytoplasm of sea urchin embryos (D ≈ 6 μm2/s) [37], the estimates of overall concentration of tubulin dimers in the cytosol (typically in the high micromolar range) [38], and theoretical models [39], the diffusion of tubulin dimers alone is not expected to produce diffusion-limited effects on the large scales observed by Geisterfer et al. In addition, not only the growth rate, but also microtubule catastrophe is expected to be sensitive to tubulin concentration. However, Geisterfer et al. observed no concurrent effects on catastrophe rates. Rather, the authors speculate that a local depletion of a larger and less abundant regulator, such as the microtubule polymerase XMAP215, may play a dominant role in the observed variability of microtubule growth.
Importantly, the activity of XMAP215 and other microtubule regulating MAPs itself can be regulated by soluble tubulin. MAPs that contain TOG (Tumor Overexpressed Gene) domains, e.g. ch-TOG/XMAP215 proteins and CLASPs, are among the microtubule plus end-binding proteins (+TIPs) that regulate microtubule dynamics [40, 41]. TOG proteins contain multiple copies of TOG domains, eg., ch-TOG contains 5 whereas human CLASP1 contains 3. The ability of these domains to bind either soluble tubulin or microtubules enables them to act as microtubule polymerases [4, 42–45] or as rescue and anti-catastrophe factors [46–50]. Interestingly, soluble tubulin affects the microtubule assembly promoting role of Stu2, the yeast orthologue of ch-TOG. In the absence of soluble tubulin, Stu2 decorates the microtubule lattice. However, tubulin causes Stu2 to decorate the microtubule end, and therefore facilitates its function as a microtubule polymerase [3]. The ability of Stu2 to be regulated in this fashion depends on a basic stretch of amino acids; whether other ch-TOG family members use basic regions in a similar fashion is unknown. Additionally, the TOG5 domain of Msps, the Drosophila melanogaster member of the family, has been shown to mediate microtubule lattice association [51]. These findings raise the possibility that regulation by unpolymerized tubulin may be a general feature of TOG-domain proteins.
Tubulin has the potential to also affect kinesin motors which regulate microtubule dynamics [52–54]. In the case of kinesin-13s, it is well established that they deform protofilaments, thus destabilizing lateral contacts that preserve microtubule architecture, resulting in microtubule depolymerization [52, 55]. Tubulin subunits that are removed from microtubules re-enter the soluble pool, fueling microtubule assembly. Initial work [52] on kinesin-13s showed that a motor-tubulin complex forms as a byproduct of microtubule disassembly. ATP hydrolysis by kinesin-13 is thought to trigger dissociation of the complex, and is thus an important step in the microtubule depolymerization cycle [52, 56]. Work from the Hirokawa lab extends this observation, showing that KIF2 forms a 1:2 (KIF2:tubulin) complex with tubulin [57]. An important implication of this result is that kinesin-13 has the potential to be product-inhibited. Key parameters that can impact the extent to which kinesin-13 is product-inhibited include: 1) the rate at which tubulin-bound motor hydrolyzes ATP; and 2) the likelihood that kinesin-13 will form a complex with microtubules versus unpolymerized tubulin. High concentrations of soluble tubulin may reduce kinesin-13 activity via motor sequestration, which is logical in the context of a cell: kinesin-13-dependent catastrophes may need to be minimized in circumstances where the mass of polymeric tubulin is low or when tubulin is in a greater state of flux between soluble and polymerized states, e.g. during the G2-M transition.
Kip3, a kinesin-8 from yeast is a microtubule depolymerase that processively walks along the microtubule lattice, but also associates with soluble tubulin as a part of its depolymerase activity [58, 59]. Therefore, like for kinesin-13s, the interaction of kinesin-8s with tubulin indicates that kinesin-8s may also be subject to product-inhibition. While kinesins that promote microtubule assembly, including polymerases Kip2 and Kinesin-5, may also be able to bind soluble tubulin, the extent to which these interactions could regulate their activity is less well understood [60, 61].
Finally, katanin, a AAA ATPase that severs microtubules, has been reported to be inhibited by the acidic C-terminal tail (CTT) of tubulin [62], and also by free tubulin [63]. These observations are consistent with work showing that the hexamerization of katanin, and its severing activity are triggered by katanin’s interaction with the CTT of β-tubulin [64]. The interaction of katanin with the CTT of tubulin that is incorporated into the microtubule lattice can thus be competed away by the CTT of free, unpolymerized tubulin. In cells, katanin is key for a number of processes that require microtubule disassembly, including spindle formation and length homeostasis [65–67] and disassembly of cilia [68–70]. Therefore, concentrations of unpolymerized tubulin are likely to affect the ability of katanin in numerous biological contexts. Interestingly, spastin, another prominent microtubule severing enzyme, does not appear to be similarly affected by soluble tubulin [71].
THE ROLE OF SOLUBLE TUBULIN IN MICROTUBULE CYTOSKELETON ORGANIZATION
Though microtubule motor proteins are primarily known for their roles in intracellular transport, many motors also impact microtubule dynamics and organization. These activities depend on their catalytic motor heads, and in some cases, additional non-motor microtubule-binding sites [72]. The kinesin-14 HSET, for example, contains an N-terminal microtubule-binding domain as well as a motor domain at its C-terminus. Recent work has demonstrated that the N-terminal microtubule-binding site of HSET can also bind unpolymerized tubulin [73]. In vitro, this interaction drives oligomerization of the motor through a mechanism that is not yet well-defined. A curious feature of HSET-tubulin assemblies is that they are composed, on average, of 3–4 HSET molecules and 12 tubulin dimers. This result was obtained even in the presence of agents that prevent microtubule polymerization (colchicine, GDP), implying that a small fraction of tubulin exists in oligomeric form.
Tubulin-dependent formation of multi-motor HSET teams results in a large increase in the distance that the motor translocates along microtubules; like other Ncd-class kinesin-14s, HSET is only weakly processive as a single molecule [74]. The ability of tubulin to physically link multiple HSET molecules together is key for the motor to organize microtubules into asters, potentially by allowing HSET to transport microtubules as cargo for long distances. Binding of tubulin to the tail of HSET may also block its interactions with neighboring microtubules, thus preventing static crosslinking of microtubules into bundles. The physiological significance of HSET’s ability to form multi-motor teams via an interaction with soluble tubulin remains to be investigated, although Norris et al. were able to demonstrate that high levels of unpolymerized tubulin and HSET promote aster formation during mitosis.
One question that has not been addressed concerns the fate of tubulin that becomes shuttled to microtubule minus ends by HSET. In principle, minus end-directed transport of tubulin would be one way to concentrate tubulin at a pole, thus promoting nucleation or regulation of microtubule dynamics. Consistent with this, Strothman et al. showed that HSET is a potent suppressor of microtubule minus-end catastrophe when present under conditions conducive to microtubule assembly [75]. Collectively, these studies show that HSET’s activities may depend critically on interactions with unpolymerized tubulin in cells.
CONCLUDING REMARKS
The roles of tubulin in its polymerized form are vast, but we know little about how tubulin in its soluble, unpolymerized form affects cellular processes. We propose that tubulin is a resource within the cell that can be deployed either to build microtubule-based structures, or as a signaling molecule that can modulate other reactions. Here, we use the term signaling to describe molecular interactions that produce a biochemical output as a result of changes to the microtubule network. Interestingly, the recent identification of a cryptic tubulin-binding domain in a MAP kinase family member (MEKK1; [76]) suggests that signaling in its more classical sense may rely on the microtubule cytoskeleton. Other more exotic functions of tubulin have yet to be discovered, like the recent suggestion that tubulin, via its C-terminal tail, may regulate the closure of VDAC channels on the cytoplasmic surface of mitochondria [77]. We suggest that future work should consider the impact of the “tubulin economy” on cellular processes that critically depend on microtubule-based assemblies, and perhaps beyond.
Figure 1. Impact of the tubulin economy on the microtubule cytoskeleton.
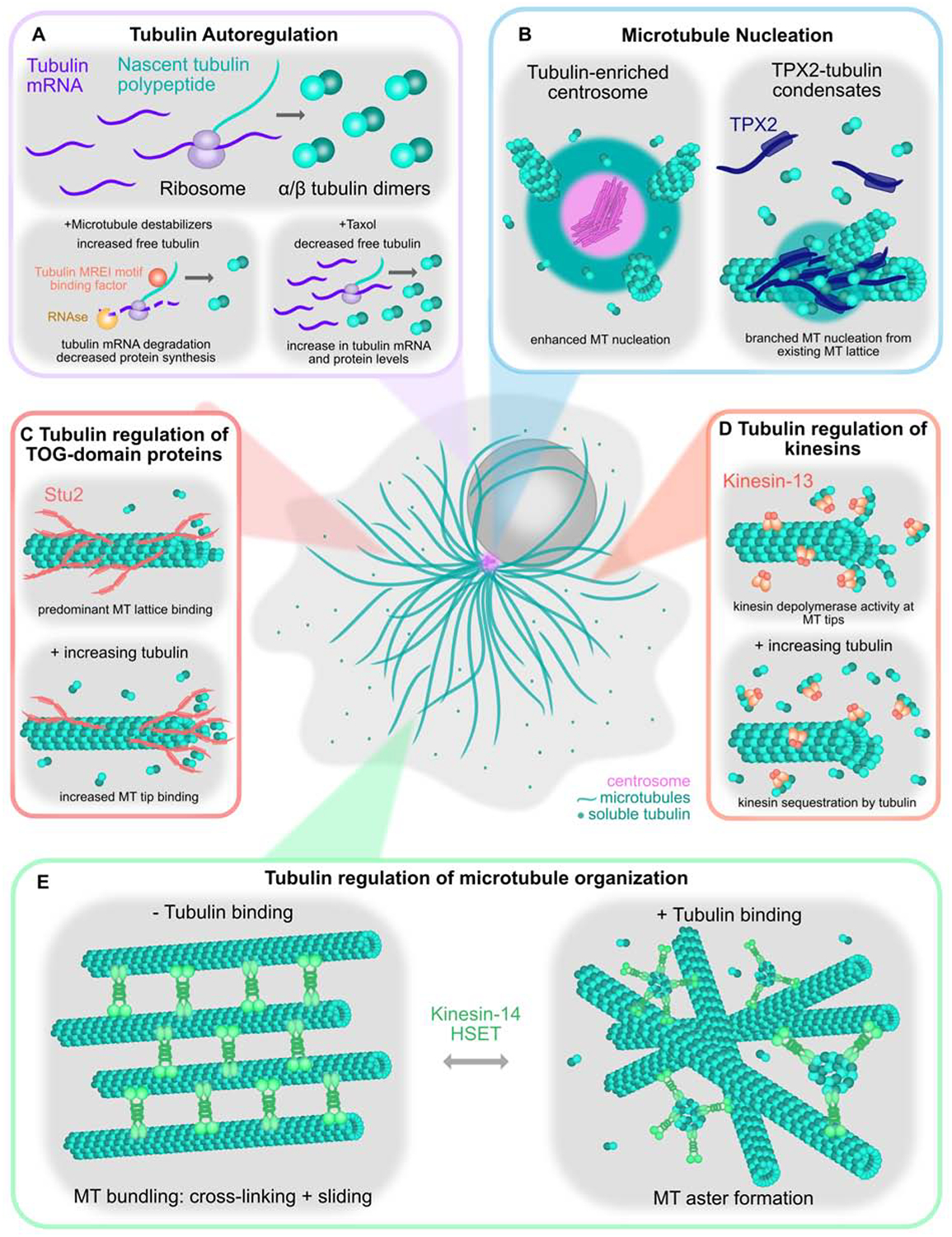
A) Soluble (unpolymerized) tubulin controls the stability of tubulin mRNAs, and therefore controls the cellular levels of tubulin proteins. Microtubule destabilizing agents cause an increase in the available pool of soluble tubulin, which triggers degradation of tubulin mRNA, resulting in a decrease in tubulin protein synthesis. Tubulin autoregulation occurs by recognition of a ribosome-associated nascent tubulin peptide by TTC5, which leads to RNAse digestion of tubulin mRNA transcripts. Conversely, treatment of cells by taxol decreases the available pool of soluble tubulin and causes an increase in tubulin mRNA levels and increased tubulin protein synthesis. B) Pockets of highly concentrated tubulin within the cell create sites of enhanced microtubule nucleation. The centrosome, for example, concentrates tubulin ~10-fold. TPX2 forms co-condensates with tubulin, leading to enhanced microtubule nucleation both in solution, and along existing microtubule polymers. C) Tubulin can regulate microtubule dynamics indirectly through regulation of microtubule-associated proteins. Soluble tubulin affects the localization of TOG-domain protein Stu2; in the absence of tubulin, Stu2 has a higher affinity for the microtubule lattice, however, Stu2 shifts to the microtubule tip in the presence of tubulin, where it acts as a microtubule polymerase. D) Kinesin-13 family motors promote the dissociation of terminal tubulin dimers, which increases the frequency of microtubule catastrophes. A by-product of this reaction is the formation of a tubulin-motor complex, which occurs following dissociation of the motor from the microtubule end. In principle, kinesin-13 motors may therefore be subject to product-inhibition, ultimately decreasing the depolymerase activity of the enzyme. E) Soluble tubulin controls the ability of kinesin-14 HSET to organize microtubules into different types of architectural arrays. HSET as a single molecule may bind across multiple microtubules through both its motor and tail domains, therefore promoting microtubule bundling. HSET will crosslink parallel bundles, but slide antiparallel bundles through its minus-end directed motility. Binding of soluble tubulin enables HSET form multi-motor clusters, which are then capable of organizing microtubules into aster-like formations.
ANNOTATIONS FOR HIGHLIGHTED REFERENCES.
Of Outstanding Interest
Baumgart et al. JCB 2019. This study determined the amounts of dimeric and polymeric forms of tubulin within C. elegans mitotic centrosomes using a combination of light and electron microscopy.
Geisterfer et al. Curr Bio 2020. This study established a negative feedback between the local density of growing microtubule ends and the microtubule growth rate.
Lin et al. Science 2020. Using a photocrosslinking mass spectrometry approach, the authors identify TTC5 as a long sought after protein factor that mediates auto-regulation of tubulin protein production. Loss of TTC5 results in an inability of cells to downregulate translation of tubulin in the presence of microtubule depolymerizing agents.
Norris et al. Nat. Comm. 2018. In this work, the authors show that HSET can use its N-terminal non-motor microtubule-binding domain to also bind tubulin. Tubulin-binding triggers the formation of multi-motor ensembles, dramatically increasing HSET’s ability to traverse long distances on the microtubule lattice and its ability to promote the formation of microtubule asters in vitro.
Of Special Interest:
King and Petry. Nat. Comm. 2020. TPX2, a microtubule-associated protein that can use its ability to bind tubulin to nucleate microtubules, is shown here to undergo a liquid-liquid phase transition in a manner that is stimulated by tubulin. The formation of TPX2-tubulin condensates stimulates microtubule nucleation.
Gasic and Mitchison PLoSBiology (2019) Using gene expression analysis, the authors show that mRNAs encoding TUBAs, TUBBs, and TUBGs show strong responses to microtubule damaging agents and other perturbations (e.g., drugs that target signaling and metabolic pathways). mRNAs for MAPs do not show a similar response, suggesting a critical role for tubulin proteins in many biological responses.
Geyer et al. eLife (2018) Through studying the roles of the TOG domains and a basic stretch of amino acids in Stu2, the authors observe that the binding of tubulin by Stu2 modulates the ability of this microtubule polymerase to localize at the microtubule plus end. In the absence of tubulin, Stu2 decorates the lattice, but in the presence of tubulin, Stu2 tracks growing microtubule plus ends.
Strothman et al. JCB (2019) In this study, the authors determine that microtubule minus ends undergo catastrophe at a lower frequency than plus ends because the off-rate of tubulin at minus ends is lower than at plus ends. HSET further suppresses the off-rate, causing minus ends to grow for prolonged periods of time in a manner that is less sensitive to the action of MCAK.
Van Geel et al. Sci Rep 2020. The authors developed a new FRET sensor compatible with optogenic approaches, allowing imaging of local perturbations in tubulin levels.
ACKNOWLEDGMENTS
We apologize to those colleagues whose work we have not cited due to space constraints. We thank Will Hancock, Beth Lawrence, Luke Rice, Dan Sackett and Kristen Verhey for insightful discussions and critical reading of the manuscript.
FUNDING
R.O. is funded by R01 GM086610 and also by start-up funds from the University of Michigan. M.Z. acknowledges support from NIH R35GM119552 and NSF grant number 2018661.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES AND RECOMMENDED READING
- 1.Mitchison T, and Kirschner M (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- 2.Ayaz P, Munyoki S, Geyer EA, Piedra FA, Vu ES, Bromberg R, Otwinowski Z, Grishin NV, Brautigam CA, and Rice LM (2014). A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife 3, e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geyer EA, Miller MP, Brautigam CA, Biggins S, and Rice LM (2018). Design principles of a microtubule polymerase. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, and Howard J (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci U S A 108, 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleveland DW, Lopata MA, Sherline P, and Kirschner MW (1981). Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell 25, 537–546. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Ze’ev A, Farmer SR, and Penman S (1979). Mechanisms of regulating tubulin synthesis in cultured mammalian cells. Cell 17, 319–325. [DOI] [PubMed] [Google Scholar]
- 7.Cleveland DW (1989). Autoregulated control of tubulin synthesis in animal cells. Curr Opin Cell Biol 1, 10–14. [DOI] [PubMed] [Google Scholar]
- 8.Gasic I, and Mitchison TJ (2019). Autoregulation and repair in microtubule homeostasis. Curr Opin Cell Biol 56, 80–87. [DOI] [PubMed] [Google Scholar]
- 9.Gasic I, Boswell SA, and Mitchison TJ (2019). Tubulin mRNA stability is sensitive to change in microtubule dynamics caused by multiple physiological and toxic cues. PLoS Biol 17, e3000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachurski CJ, Theodorakis NG, Coulson RM, and Cleveland DW (1994). An amino-terminal tetrapeptide specifies cotranslational degradation of beta-tubulin but not alpha-tubulin mRNAs. Mol Cell Biol 14, 4076–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gay DA, Sisodia SS, and Cleveland DW (1989). Autoregulatory control of beta-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci U S A 86, 5763–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pachter JS, Yen TJ, and Cleveland DW (1987). Autoregulation of tubulin expression is achieved through specific degradation of polysomal tubulin mRNAs. Cell 51, 283–292. [DOI] [PubMed] [Google Scholar]
- 13.Yen TJ, Gay DA, Pachter JS, and Cleveland DW (1988). Autoregulated changes in stability of polyribosome-bound beta-tubulin mRNAs are specified by the first 13 translated nucleotides. Mol Cell Biol 8, 1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yen TJ, Machlin PS, and Cleveland DW (1988). Autoregulated instability of beta-tubulin mRNAs by recognition of the nascent amino terminus of beta-tubulin. Nature 334, 580–585. [DOI] [PubMed] [Google Scholar]
- 15.Theodorakis NG, and Cleveland DW (1992). Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol 12, 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Z, Gasic I, Chandrasekaran V, Peters N, Shao S, Mitchison TJ, and Hegde RS (2020). TTC5 mediates autoregulation of tubulin via mRNA degradation. Science 367, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- 18.Scarpulla RC, Vega RB, and Kelly DP (2012). Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab 23, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumgart J, Kirchner M, Redemann S, Bond A, Woodruff J, Verbavatz JM, Julicher F, Muller-Reichert T, Hyman AA, and Brugues J (2019). Soluble tubulin is significantly enriched at mitotic centrosomes. J Cell Biol 218, 3977–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fygenson DK, Braun E, and Libchaber A (1994). Phase diagram of microtubules. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 50, 1579–1588. [DOI] [PubMed] [Google Scholar]
- 21.Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, and Saxton W (2001). Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell 12, 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077 e1010. [DOI] [PubMed] [Google Scholar]
- 23.Roostalu J, Cade NI, and Surrey T (2015). Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol 17, 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieczorek M, Bechstedt S, Chaaban S, and Brouhard GJ (2015). Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat Cell Biol 17, 907–916. [DOI] [PubMed] [Google Scholar]
- 25.Petry S, Groen AC, Ishihara K, Mitchison TJ, and Vale RD (2013). Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, and Carazo-Salas RE (2003). Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J 22, 2060–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King MR, and Petry S (2020). Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun 11, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keeling J, Tsiokas L, and Maskey D (2016). Cellular Mechanisms of Ciliary Length Control. Cells 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirvis M, Stearns T, and James Nelson W (2018). Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem J 475, 2329–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma N, Kosan ZA, Stallworth JE, Berbari NF, and Yoder BK (2011). Soluble levels of cytosolic tubulin regulate ciliary length control. Mol Biol Cell 22, 806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Piao T, Cao M, Qin T, Huang L, Deng H, Mao T, and Pan J (2013). Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J Cell Sci 126, 1531–1540. [DOI] [PubMed] [Google Scholar]
- 32.Good MC, Vahey MD, Skandarajah A, Fletcher DA, and Heald R (2013). Cytoplasmic volume modulates spindle size during embryogenesis. Science 342, 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, and Gatlin JC (2013). Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342, 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Geel O, Cheung S, and Gadella TWJ (2020). Combining optogenetics with sensitive FRET imaging to monitor local microtubule manipulations. Sci Rep 10, 6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geisterfer ZM, Zhu DY, Mitchison TJ, Oakey J, and Gatlin JC (2020). Microtubule Growth Rates Are Sensitive to Global and Local Changes in Microtubule Plus-End Density. Curr Biol 30, 3016–3023 e3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goehring NW, and Hyman AA (2012). Organelle growth control through limiting pools of cytoplasmic components. Curr Biol 22, R330–339. [DOI] [PubMed] [Google Scholar]
- 37.Salmon ED, Saxton WM, Leslie RJ, Karow ML, and McIntosh JR (1984). Diffusion coefficient of fluorescein-labeled tubulin in the cytoplasm of embryonic cells of a sea urchin: video image analysis of fluorescence redistribution after photobleaching. J Cell Biol 99, 2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons SF, and Salmon ED (1997). Microtubule assembly in clarified Xenopus egg extracts. Cell Motil Cytoskeleton 36, 1–11. [DOI] [PubMed] [Google Scholar]
- 39.Odde DJ (1997). Estimation of the diffusion-limited rate of microtubule assembly. Biophys J 73, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Bassam J, and Chang F (2011). Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol 21, 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slep KC (2009). The role of TOG domains in microtubule plus end dynamics. Biochem Soc Trans 37, 1002–1006. [DOI] [PubMed] [Google Scholar]
- 42.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, and Hyman AA (2008). XMAP215 is a processive microtubule polymerase. Cell 132, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gard DL, and Kirschner MW (1987). A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol 105, 2203–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podolski M, Mahamdeh M, and Howard J (2014). Stu2, the budding yeast XMAP215/Dis1 homolog, promotes assembly of yeast microtubules by increasing growth rate and decreasing catastrophe frequency. J Biol Chem 289, 28087–28093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang PJ, and Huffaker TC (1997). Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol 139, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aher A, Kok M, Sharma A, Rai A, Olieric N, Rodriguez-Garcia R, Katrukha EA, Weinert T, Olieric V, Kapitein LC, et al. (2018). CLASP Suppresses Microtubule Catastrophes through a Single TOG Domain. Dev Cell 46, 40–58 e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence EJ, Arpag G, Norris SR, and Zanic M (2018). Human CLASP2 specifically regulates microtubule catastrophe and rescue. Mol Biol Cell 29, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence EJ, and Zanic M (2019). Rescuing microtubules from the brink of catastrophe: CLASPs lead the way. Curr Opin Cell Biol 56, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence EJ, Zanic M, and Rice LM (2020). CLASPs at a glance. J Cell Sci 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Majumdar S, Kim T, Chen Z, Munyoki S, Tso SC, Brautigam CA, and Rice LM (2018). An isolated CLASP TOG domain suppresses microtubule catastrophe and promotes rescue. Mol Biol Cell 29, 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byrnes AE, and Slep KC (2017). TOG-tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. J Cell Biol 216, 1641–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desai A, Verma S, Mitchison TJ, and Walczak CE (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69–78. [DOI] [PubMed] [Google Scholar]
- 53.Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, and Howard J (2006). Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol 8, 957–962. [DOI] [PubMed] [Google Scholar]
- 54.Walczak CE, Gayek S, and Ohi R (2013). Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol 29, 417–441. [DOI] [PubMed] [Google Scholar]
- 55.Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, and Milligan RA (2002). A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell 9, 903–909. [DOI] [PubMed] [Google Scholar]
- 56.Friel CT, and Howard J (2011). The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J 30, 3928–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogawa T, Saijo S, Shimizu N, Jiang X, and Hirokawa N (2017). Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation. Cell Rep 20, 2626–2638. [DOI] [PubMed] [Google Scholar]
- 58.Arellano-Santoyo H, Geyer EA, Stokasimov E, Chen GY, Su X, Hancock W, Rice LM, and Pellman D (2017). A Tubulin Binding Switch Underlies Kip3/Kinesin-8 Depolymerase Activity. Dev Cell 42, 37–51 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varga V, Leduc C, Bormuth V, Diez S, and Howard J (2009). Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138, 1174–1183. [DOI] [PubMed] [Google Scholar]
- 60.Chen GY, Cleary JM, Asenjo AB, Chen Y, Mascaro JA, Arginteanu DFJ, Sosa H, and Hancock WO (2019). Kinesin-5 Promotes Microtubule Nucleation and Assembly by Stabilizing a Lattice-Competent Conformation of Tubulin. Curr Biol 29, 2259–2269 e2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hibbel A, Bogdanova A, Mahamdeh M, Jannasch A, Storch M, Schaffer E, Liakopoulos D, and Howard J (2015). Kinesin Kip2 enhances microtubule growth in vitro through length-dependent feedback on polymerization and catastrophe. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roll-Mecak A, and Vale RD (2008). Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 451, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey ME, Sackett DL, and Ross JL (2015). Katanin Severing and Binding Microtubules Are Inhibited by Tubulin Carboxy Tails. Biophys J 109, 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zehr EA, Szyk A, Szczesna E, and Roll-Mecak A (2020). Katanin Grips the beta-Tubulin Tail through an Electropositive Double Spiral to Sever Microtubules. Dev Cell 52, 118–131 e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, and Heald R (2011). Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McNally K, Berg E, Cortes DB, Hernandez V, Mains PE, and McNally FJ (2014). Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol Biol Cell 25, 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mishra-Gorur K, Caglayan AO, Schaffer AE, Chabu C, Henegariu O, Vonhoff F, Akgumus GT, Nishimura S, Han W, Tu S, et al. (2014). Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron 84, 1226–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Casanova M, Crobu L, Blaineau C, Bourgeois N, Bastien P, and Pages M (2009). Microtubule-severing proteins are involved in flagellar length control and mitosis in Trypanosomatids. Mol Microbiol 71, 1353–1370. [DOI] [PubMed] [Google Scholar]
- 69.Hu WF, Pomp O, Ben-Omran T, Kodani A, Henke K, Mochida GH, Yu TW, Woodworth MB, Bonnard C, Raj GS, et al. (2014). Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron 84, 1240–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, and Gaertig J (2007). Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol 178, 1065–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vemu A, Szczesna E, Zehr EA, Spector JO, Grigorieff N, Deaconescu AM, and Roll-Mecak A (2018). Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Welburn JP (2013). The molecular basis for kinesin functional specificity during mitosis. Cytoskeleton (Hoboken) 70, 476–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Norris SR, Jung S, Singh P, Strothman CE, Erwin AL, Ohi MD, Zanic M, and Ohi R (2018). Microtubule minus-end aster organization is driven by processive HSET-tubulin clusters. Nat Commun 9, 2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reinemann DN, Norris SR, Ohi R, and Lang MJ (2018). Processive Kinesin-14 HSET Exhibits Directional Flexibility Depending on Motor Traffic. Curr Biol 28, 2356–2362 e2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strothman C, Farmer V, Arpag G, Rodgers N, Podolski M, Norris S, Ohi R, and Zanic M (2019). Microtubule minus-end stability is dictated by the tubulin off-rate. J Cell Biol 218, 2841–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filipcik P, Latham SL, Cadell AL, Day CL, Croucher DR, and Mace PD (2020). A cryptic tubulin-binding domain links MEKK1 to curved tubulin protomers. Proc Natl Acad Sci U S A 117, 21308–21318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sheldon KL, Gurnev PA, Bezrukov SM, and Sackett DL (2015). Tubulin tail sequences and post-translational modifications regulate closure of mitochondrial voltage-dependent anion channel (VDAC). J Biol Chem 290, 26784–26789. [DOI] [PMC free article] [PubMed] [Google Scholar]


