Abstract
Chromosomes are selectively organized within the nuclei of interphase cells reflecting the current fate of each cell and are reorganized in response to various physiological cues to maintain homeostasis. While substantial progress is being made to establish the various patterns of genome architecture, less is understood on how chromosome folding/positioning is achieved. Here we discuss recent insights into the cellular mechanisms dictating chromatin movements including the use of epigenetic modifications and allosterically regulated transcription factors as well as a nucleoskeleton system comprised of actin, myosin, and actin-binding proteins. Together, these nuclear factors help coordinate the positioning of both general and cell-specific genomic architectural features.
Introduction
A eukaryotic cell houses its genetic material within a nucleus. Here, the longest and heaviest biomolecules, chromosomes, are condensed to fit within this central organelle. In humans ~2 m of nucleic acid polymer totaling ~2.15 X 1012 Da is fit into nuclei having diameters of ~5–10 μM. This challenge is compounded since DNA is not stored naked but is an amalgam with proteins and RNA to form chromatin, which minimally doubles its mass [1]. Notably, chromatin is not simply compacted into a tangled mass within interphase cells but rather genomes are spatially arranged reflecting both the developmental type and physiological status of each cell [2]. Likely, methodical chromosome folding fosters access to the necessary tracts of DNA to support homeostasis. Despite the conserved three-dimensional (3D) configuration of genomes [3], the mechanisms mediating genome packaging are still unclear. As chromosome reorganization is a common biological event reoccurring after each cell division and also in response to ever fluctuating environmental and internal cues, a better understanding of this process is imperative. Here, we review recent work supporting the model that genomes are packaged using a hierarchical architecture within interphase cells and discuss insights into the cellular pathways mediating the organization process.
Chromosome Architectural Features
Classic work argued that chromosomes are arranged into territories within interphase nuclei [4]. Eventually the preferential placement of every chromosome within a nucleus, relative to the periphery and interior, was shown using fluorescent in situ hybridization (FISH) and chromosome paints [5]. The resolution of chromosome arrangements was greatly enhanced by the development of chromatin conformation capture (3C)-based assays (ie, 3C to Hi-C) that identify statistically significant contact frequencies between DNA sites across a genome [2]. A seminal Hi-C study demonstrated that the human genome partitions into two compartments that are composed of open (active; A) and closed (inactive; B) chromatin [6]. Notably, the A/B compartments correspond to the classic euchromatin and heterochromatin areas originally described by cytological staining [7]. The compartments further divide into smaller units such as topologically associated domains (TADs) or lamin associated domains (LADs) [2]. Of note, these domains dissipate during prometaphase and must be recurrently formed after each cell division [8]. In general, the domain boundaries are constant across cell types and species and the DNA within a subdomain displays a higher frequency of internal chromosomal interactions and limited associations with the adjacent domains [2]. Yet, the genetic connections within a TAD (sub-TADs) and interactions between domains (loops) can differ as well as the relative nuclear positions of the domains [9]. Hence, differences in these chromosomal arrangements likely contribute to cell (eg, tissue type) and physiological (eg, aging and disease) fates. Although it is now evident that cells have both common and specific 3D chromosomal features, how either is achieved is unclear. Minimally, building any genomic structure will rely on the same principles that are central to all organizational systems: 1) the ability to selectively recognize the items to be moved; 2) a mechanism to mobilize the marked units; and 3) the instructions on where to place the objects.
Genome Organization Tags (GOTs)
The most apparent sources of information to initiate chromosome architecture are the DNA elements within a genome sequence as well as epigenetic marks on the histone octamers integrated into the chromatin. We refer to these factors as well as any other physical features that mark chromosome regions for select 3D packaging as Genome Organization Tags (GOTs). Notably, DNA elements and histone modifications can serve to assemble both common or cell-specific structures, since the epigenetic landscape as well as the DNA binding factors expressed in any cell can vary. Likely, these GOTs are key guides in building an appropriate 3D genome structure on a cell-to-cell basis.
Early work on TADs and LADs demonstrated that the conserved border regions are marked by CCCTC-binding factor (CTCF) and cohesin binding sites [2]. CTCF and cohesin anchor loops that can connect gene promoters and distal enhancers [10]. The loops are thought to form through an extrusion mechanism in which a cohesin ring translocates along the DNA until it encounters DNA-bound CTCF [11]. Hence, the CTCF DNA elements deliver critical information to the system, as these are recurrently used to reform TADS following each cell division [12]. Overall, CTCF governs ~80% of all TADs, yet loss of CTCF does not impact partitioning of the A/B compartments [13]. Hence, CTCF serves as a major determinant of TAD boundaries, but additional mechanisms must contribute to genome organization including the formation of some TADs.
The initial discovery of the A/B compartments by Hi-C noted that the partitioned regions correlate with the relative DNase sensitivities of the associated chromatin [6]. A higher resolution Hi-C map further divided the 2 compartments into 6 sub-compartments (A1–2 and B1–4) based on refined DNA interactions and, importantly, select histone modifications (H3K36me3, H3K27me3, H3K4me1, H3K4me2, H3K4me3, H3K9me3, H3K79me2, and H4K20me1) [11] (Figure 1a). As expected the gene-rich A1 and A2 sections favor activating chromatin marks such as H3K36me3, H3K79me2, H3K27ac and H3K4me1 whereas B1–3 correlate with various heterochromatin characteristics such as H3K27me3 (B1; facultative heterochromatin), nuclear lamina/pericentromeric heterochromatin (B2), or lacking an enrichment in any particular mark suggesting common heterochromatin (B3). The B4 region displays a distinctive pattern favoring both activating (H3K36me3) and repressing (H3K9me3 and H4K20me3) modifications [10]. Significantly, using just the epigenetic modification patterns it is possible to predict the coarse 3D structure of a genome de novo leading to the concept that genome architecture self-assembles through a phase separation mechanism that is based upon epigenetic signatures [11,14]. Although the contributions of phase separation to genome folding is yet to be empirically shown, the predictive powers of the epigenetic code suggest that the intrinsic information present along the chromatin polymers is sufficient to guide folding under ideal conditions. In certain respects, this concept is akin to protein folding where the primary amino acid sequence is sufficient to attain a final native protein structure under ideal in vitro conditions yet all polypeptides rely on molecular chaperones for proper folding in vivo [15]. Importantly, the cell interior rarely, if ever, provides ideal conditions and therefore chromosome organization is likely facilitated by extrinsic factors in a manner comparable to molecular chaperones promoting the protein folding quality control process. Despite the correlation between histone code patterns and chromosome compartments, it is still unclear how the epigenetic histone marks functionally contribute to genome organization.
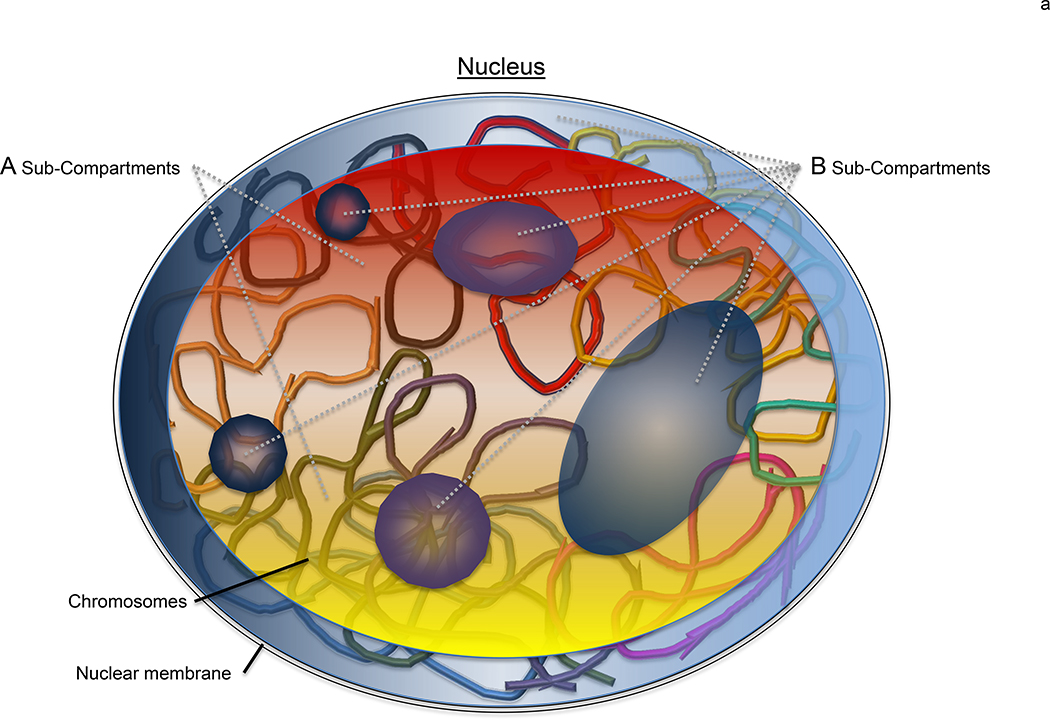
Figure 1. Chromosome regions are clustered based upon epigenetic signatures.
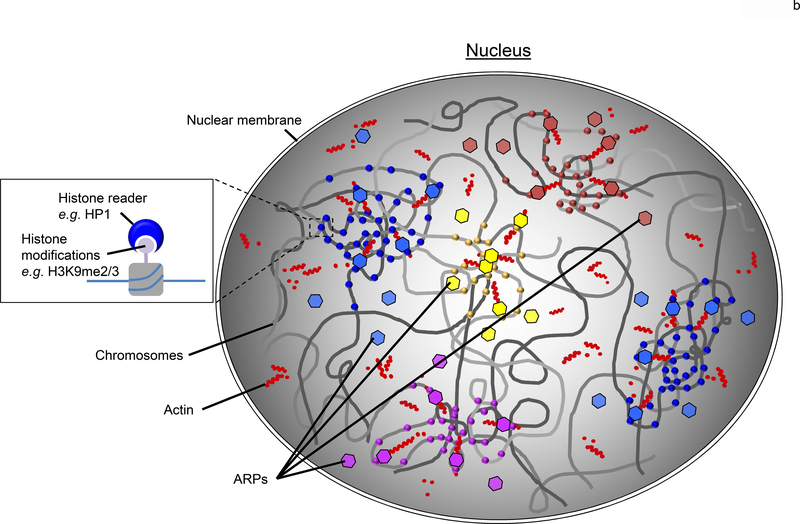
(a) In a nucleus, eukaryotic genomes are partitioned into two main compartments (A/B), as detected by Hi-C [6], that correspond to the classic heterochromatin and euchromatin regions [7]. The main compartments are further sub-divided (compartments A1-A2 and B1-B4) based upon varying epigenetic signatures. Here, the general locale of A1 and A2 sub-compartments are represented by the red and yellow regions primarily found in the nuclear interior whereas the B1-B4 sub-compartments are shown by the blue and purple areas that are minimally enriched at the nuclear periphery and around the nucleolus. While it is clear that the various regions correlate with select epigenetic modifications [11,16], how this information is used to physically cluster the chromatin is yet to be revealed. (b) A plausible model to account for the nuclear organization of large-scale genomic features such as the A/B compartments and sub-compartments (A1-B4) is the coordinated use of a dynamic nuclear actin network to effectively move the large chromosome regions, epigenetic readers (eg, HP1) to selectively distinguish the various histone marks (circles), and ARPs (hexagons) to bridge the readers to the actin (red dots) network. The various colors of the circles and hexagons, represent the various readers and ARPs found with eukaryotic nuclei. Together, this system would provide an effective means to reorganize a genome after each cell division within a biologically useful timescale (<hour).
One proposed mechanism to account for an epigenetic-dependent clustering of select genomic regions through a self-assembly mode is the use of histone “reader” proteins that can both selectively recognize histone marks and self-associate [11,16,17]. The prototypical molecule for this concept is Heterochromatin Protein 1 (HP1), which selectively binds H3K9me2/3 and can self-associate [18]. HP1 was originally identified as a heterochromatin-associated protein in Drosophila melanogaster and mammalian HP1 maintains a compacted heterochromatic state that is dependent upon HP1 dimerization [18]. It has been suggested that HP1 undergoes a phase separation to form constitutive heterochromatin compartments [16,17]. However, it is unclear whether the phase separation drives heterochromatin formation or if a phase shift happens once the domain is established. We favor the latter since phase separation typically requires high concentrations of the shifting protein, which are unlikely to exist while chromosomes are being actively organized [19]. Minimally, HP1 maintains a heterochromatin state once the chromatin has been clustered by spreading across adjacent nucleosomes using its distance-limited H3K9me3-induced self-association bridging activity [20,21]. Nevertheless, it is still plausible that HP1 fosters the organization of constitutive heterochromatin compartments independent of its phase separation behavior.
The ability of HP1 to specifically bind H3K9me2/3 satisfies the first step of an organization system—selective recognition of the item to be moved. Missing is how the binding of HP1 to these marks would allow sizable regions of chromatinized chromosomes to be coerced together into an organized unit—what nuclear system might drive the motion of these large chromatin polymers? We suspect that other protein partners connect HP1 to such a system. In particular, we believe that the Actin related protein 6 (Arp6), which associates with HP1 from flies to humans [22,23], links chromatin-bound HP1 to an actin-based system in the nucleus (Figure 1b), which is described below. While a direct role for HP1-Arp6 in this process is yet to be shown, it is notable that Arp6 colocalizes with HP1 at pericentric heterochromatin [23,24]. Regardless of the exact role(s) of HP1 with heterochromatin, additional information beyond broadly used epigenetic marks will be required to properly organize genomes.
The most apparent source of detailed information to coordinate chromosome architecture is the genome sequence itself. Yet, few examples of DNA elements capable of directing chromatin organization in interphase cells, besides CTCF elements, have been reported. A notable exception is the pioneering work exploiting the INO1 gene in budding yeast as a model to understand directed chromosome motion [25]. In brief, the INO1 locus is inactive and localized predominantly to the inner nucleoplasm in the presence of inositol but colocalizes with the nuclear membrane following inositol starvation and gene activation [25]. Significantly, movement of INO1 to the nuclear periphery is dependent upon a Gene Recruitment Sequence I (GRS I) upstream of the transcription start site along with the Put3 transcription factor that binds to GRS I [26]. Loss of Put3 or ablation of GRS I reveals a second site, GRS II, capable of mobilizing INO1 to the nuclear membrane in conjunction with the Cbf1 transcription factor [26]. Intriguingly, the DNA sequences of either GRS do not resemble the known consensus elements of the associated transcription factors. This divergence in sequence of the element dictating nuclear positioning might help explain why it has been generally difficult to identify DNA elements associated with genome architecture. Significantly, the INO1 system was instrumental in discovering a potential nuclear pathway that satisifies the second step of an organization system—a mechanism to mobilize a marked unit.
Actin-based Transport in the Nucleus
Cargo transport in a cell is often associated with a cytoskeleton system where actin, actin-binding proteins (ABPs), and myosin motors move large biomolecules precisely from one locale to another [27]. Although actin was discovered in the nucleus over half a century ago by subcellular fractionation [28], a role for actin-based transport in the nucleus had been met with skepticism based, in part, on a limited capacity to detect filamentous actin in the nucleus using the common actin-stain phalloidin [29]. Fortunately, the development of new detection reagents (eg, ABPs or actin-nanobodies fused to GFP) have yielded significant new insights in the existence and relevance of a nucleoskeletal system incorporating various actin species including monomers, polymers, and rods [30]. Perhaps one of the more exciting concepts to emerge is a seemingly general use of the nuclear actin system in genome organization events.
In normal eukaryotic cells from yeast to flies to mammals visualization of nuclear actin with an actin-nanobody reveals a punctate pattern [31–34]. At least in yeast, the puncta are lost following treatment with an actin polymerization inhibitor [34]. Further interrogation of the actin-nanobody signal in yeast using an Airyscan confocal microscope revealed a dynamic polymerization behavior for nuclear actin [34]. Intriguingly, various cell perturbations including cell division, environmental cues, or DNA damage readily stabilize actin filaments in the nucleus.
The functional relevance of nuclear f-actin appears broad including for the expansion of the nuclear volume and decondensing chromosomes following mitosis or during embryonic development [35]. In the course of development nuclear actin shifts from puncta near the DNA to a filamentous cage surrounding the chromosomes within oocytes [36]. More subtle physiological events also correlate with f-actin formation such as activation of G protein-coupled receptors that trigger polymerization of f-actin starting at the nuclear membrane and moving into the nucleoplasm to alter chromatin organization [37]. Conversely, DNA damage events produce actin filaments to promote the repair process by minimally transporting double-stranded DNA breaks (DSB) from the inner nucleoplasm to the nuclear periphery [32,33]. Our group has shown that actin polymerization is required to transport activated genes to the nuclear membrane although stable actin polymers were not part of this process [34]. In several cases it has been noted that the association between actin and chromatin requires different histone marks including H3K27me3 and H3K9me3 during oogenesis [38] or decreased H3S10p and increased H4K16ac during the mitotic exit [37], which might explain site-specific nucleation of actin at certain chromatin sites.
At heterochromatic DSBs actin is selectively recruited to the damaged site by the combined efforts of Mre11, which is a subunit of the MRN complex that recognizes the damaged DNA site, and HP1 [32,33]. In brief, the Mre11 and HP1 nucleate the Arp2/3 complex, Arp2/3 fosters f-actin polymerization towards the nuclear periphery, and the Hsp90 co-chaperone Unc45 activates the nuclear myosins (Myo1A, Myo1B, and MyoV) to transport the DSBs to the nuclear membrane (Figure 2a). In some respects, the activated INO1 gene uses a parallel mechanism to move to the nuclear periphery.
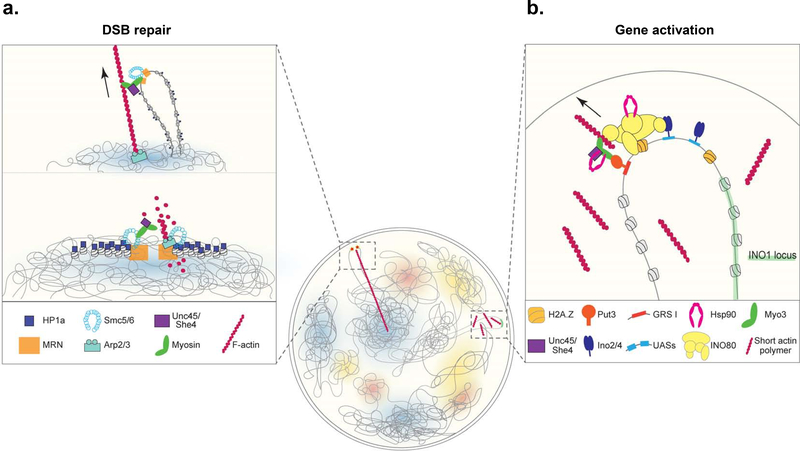
Figure 2. Chromatin sites are reorganized using a myosin motor-driven nucleoskeleton system selectively targeted to a genomic locus.
(a) Double stranded DNA breaks (DSBs) within heterochromatin regions are recognized by the Mre11/Rad50/Nbs1 (MRN) complex, in conjunction with HP1, Arp2/3 is recruited to site to polymerize F-actin towards the nuclear periphery (lower panel). Nuclear myosins along with the Hsp90 co-chaperone Unc45 actively transport the break sites to the nuclear periphery along the actin tracks where the DNA will be effectively repaired (upper panel) [32,33]. (b) Upon inositol starvation in budding yeast the INO1 locus moves from the inner nucleoplasm to the nuclear periphery [25]. The promoter bound transcription factors Ino2/4 at the UASs and Put3 at the GRS I DNA elements nucleate the proteins mediating the transport including the INO80 nucleosome remodeler and Myo3 myosin motor. Together with Unc45 and the Hsp90 chaperone, INO1 is mobilized to the nuclear periphery utilizing dynamic actin polymers [34].
At the INO1 locus inositol starvation triggers the recruitment of several transcription factors including Ino2, Ino4, and Put3 [26, 34]. As mentioned, Put3 binds to the DNA element, GRS I, required to move INO1 to the nuclear periphery whereas Ino2 and Ino4 bind the upstream activation sequences (UASs) of the INO1 promoter to recruit the factors driving gene transcription including the INO80 and SWR-C chromatin remodelers that mediate the exchange of H2A/H2A.Z. In addition to gene promoters, the non-canonical H2A.Z histone is enriched at most TAD boundaries [12,39]. Both of these remodelers contain an actin monomer along with several Arp subunits, which facilitate an interaction with f-actin [40]. At INO1 this function stabilizes a Put3-mediated nucleation of the unconvention myosin Myo3 to the chromatin that allows the single-headed myosin to transport INO1 to the nuclear periphery [34]. As with DSBs, motion of this chromatin site requires an Unc45 homolog along with the Hsp90 chaperone itself (Figure 2b). In contrast to DSBs, Arp2/3 is not used for actin polymerization nor are stable actin filaments apparent during the motion but rather Formin homologs are required to drive INO1 transport [34]. Besides INO1, Formins have proven important for the transcriptional response to serum stimulation as well as mammalian cell spreading [31,41,42].
A common thread in many of the actin-dependent genome organization events is the inclusion of one or more ABP. ABPs were first reported in the nucleus in 1987 when cofilin was found to interact with nuclear actin rods [43]. Of note, cofilin1 is vital for f-actin assembly during mitotic exit to increase the nuclear volume of daughter cells [35]. In general, ABPs serve to control the behavior of actin [27]. A critical class of ABPs are the Actin-related proteins (ARPs), which are conserved from yeast to human. ARPs share structural features with conventional actin despite varied primary sequences [44]. Initial characterization of the ARPs in budding yeast identified 10 homologs with 4 primarily found in the cytoplasm (Arp1, 2, 3, and 10) and 6 enriched in the nucleus (Arp4, 5, 6, 7, 8, and 9) [45]. ARP nomenclature is based on an increasing diversity relative to actin with Arp1 sharing 52% identity and Arp10 17% [44]. Overall, actin and the ARPs belong to a larger protein family that includes the 70 kDa heat shock proteins (Hsp70s), hexokinases, and glycerol kinases where the actin-fold is minimally used as a conformational switch point [46].
As the best studied ARPs, Arp2 and Arp3 form a complex enriched in the cytoplasm that controls actin polymerization through actin nucleation and branching [27], it was anticipated that most ARPs would govern actin dynamics. Yet, nuclear ARPs are primarily known as components of large, multi-subunit chromatin modifying complexes including nucleosome remodelers (INO80, SWR-C, SWI/SNF, RSC, p400) and histone acetylases (NuA4) where an ARP effectively caps the barbed end of a monomeric actin co-subunit [40]. However, chromatin modifiers often contain additional ARP-subunits that are not in contact with the actin subunit. Moreover, the majority of the nuclear ARPs are not part of large protein structures (i.e. >150 kDa), which would be comparable to most chromatin modifying complexes [46]. Hence, the functional relevance of most nuclear ARPs is not clear. Yet, the capacity of the ARPs to contribute to f-actin binding, even when part of a large chromatin modifier, suggests that mediating actin interactions is a major ARP contribution within the nucleus. Might the interaction between HP1 and Arp6 serve a similar role to coordinate an initial clustering of chromosome domains after cell division in conjunction with a dynamic nuclear actin network (Figure 1b)? Intriguingly, in Arabidopsis the different nuclear Arps display distinct nuclear localization patterns [44], which might be useful in either establishing or maintaining different chromosome domains.
Once a chromosome site/domain has been targeted for transport, a force-generating activity, such as a myosin motor, will be engaged at least for physiological events that require immediate action such as the response to nutrient deprivation (INO1) or DNA damage (DSBs). For example, homology-dependent pathways directing either DNA repair or allele-paired transcription rely on nuclear myosin I (NMI) or myosin VI, respectively [47,48]. In general, it seems that most rapid long distance chromatin movements depend on nuclear myosin and actin [49,50]. A recent report from Christof Gebhardt’s group following single molecules of nuclear myosin VI found that transcription-dependent long-range chromatin rearrangements occurred along nuclear actin filaments [A Große-Berkenbusch et al., unpublished]. Whether all chromosome movements will require a myosin motor or if other avenues are utilized is an open question. For instance, it is possible that large-scale partitioning events such as the formation of the A/B compartments might just rely on the rapid actin polymerization/depolymerization found within interphase cells [34]. In this case, the Arp-dependent connections between histone code readers and the dynamic actin network would facilitate local concentration gradients formed by the weak interactions between the same reader proteins (eg, HP1 dimers) thereby clustering the chromosome regions with the same epigenetic signatures (Figure 1b). As each large-scale domain coalesces, the finer details of cell-specific chromosome architecture would be further guided by the DNA elements along the genome in conjunction with the expressed cognate transcription factors. Once a suitable concentration of the reader is achieved, a phase transition could then occur to enforce the maintenance of the newly formed domain. Persistence of such a dynamic actin-based system throughout interphase also provides a plausible explanation for variances in genome organization that have been noted in single cell experiments [51].
Where does it end?
While a variety of reports are providing exciting insights into the first two steps governing an effective genome organizing system (ie, marking items to be moved and a mechanism to move the items), less is understood about the parameters determining the final step—instructions on where to place the objects. What signals and/or mechanisms are in place to guide chromosome domains and/or chromatin sites to select areas of the nuclear space. For instance, placement of a GRS I sequence within an unrelated gene is sufficient to guide that site to same spot along the nuclear periphery as the inositol-dependent reorganized INO1 locus [26]—what cellular signaling network provides the necessary long-distance information to the nuclear transport system to guide it from the inner nucleoplasm to a select site along the nuclear membrane? Recent super-resolution microscopy work as well as proximity techniques (i.e. DamID and TSA-Seq) might be revealing these details by showing how various nuclear bodies contribute to genome organization [52,53] as well as how the packaging fluctuates in response to transcription activation including interactions with the nuclear envelope [54] and splicing speckles [55]. Clearly, there is much left to be discovered on how cells make critical decision on where and when to position their genomic information.
Acknowledgements
The laboratory of BCF was supported by NIH Public Service grant GM118306.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the past 2 years, have been highlighted as:
* of special interest
** of outstanding interest
- [1].Kornberg RD: Chromatin Structure: A Repeating Unit of Histones and DNA. Science 1974, 184:868–871. [DOI] [PubMed] [Google Scholar]
- [2].Dekker J, Misteli T: Long-Range Chromatin Interactions. Cold Spring Harb Perspect Biol 2015, doi: 10.1101/cshperespect.a0193567:a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cremer M, Cremer T: Nuclear compartmentalization, dynamics, and function of regulatory DNA sequences. Genes Chromo Cancer 2019, 58:427–436. [DOI] [PubMed] [Google Scholar]
- [4].Rabl C: Über Zelltheilung. Morph Jb 1885, 10:214–330. [Google Scholar]
- [5].Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA: The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 2001, 10:211–220. [DOI] [PubMed] [Google Scholar]
- [6].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. : Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science 2009, 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heitz E: Das Heterochromatin der Moose. Jahrb Wiss Botanik 1928, 69:762–818. [Google Scholar]
- [8].Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J: Organization of the Mitotic Chromosome. Science 2013, 342:948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Collas P, Liyakat Ali TM, Brunet A, Germier T.: Finding Friends in the Crowd: Three-Dimensional Cliques of Topological Genomic Domains. Front Genet, 2019, 10:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn A, Machol I, Omer AD, Lander ES, et al. : A three-dimensional map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hildebrand EM, Dekker J: Mechanisms and Functions of Chromosome Compartmentalization. Trends Biochem Sci 2020, 45:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Oomen ME, Hansen AS, Liu Y, Darzacq X, Dekker J: CTCF sites display cell cycle–dependent dynamics in factor binding and nucleosome positioning. Genome Res 2019, 29:236–249.*This report uses a 4-pronged strategy to confirm that CTCF dynamically associates with its DNA elements at TAD boundaries in a cell cycle-dependent manner that correlates with loss and gain of TADs as well as the positioning of nearby nucleosomes.
- [13].Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG: Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 2017, 169:930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pierro MD, Cheng RR, Aiden EL, Wolynes PG, Onuchic JN: De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc Natl Acad Sci USA 2017, 114:12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Balchin D, Hayer-Hartl M, Hartl FU: In vivo aspects of protein folding and quality control. Science 2016, 353. [DOI] [PubMed] [Google Scholar]
- [16].Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ: Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH: Phase separation drives heterochromatin domain formation. Nature 2017, 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kumar A, Kono H: Heterochromatin protein 1 (HP1): interactions with itself and chromatin components. Biophys Rev 2020, 12:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McSwiggen DT, Mir M, Darzacq X, Tjian R: Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 2019, 33:23–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Danzer JR, Wallrath LL: Mechanisms of HP1-mediated gene silencing in Drosophila. Development 2004, 131:3571–3580. [DOI] [PubMed] [Google Scholar]
- [21].Canzio D, Liao M, Naber N, Pate E, Larson A, Wu S, Marina DB, Garcia JF, Madhani HD, Cooke R, et al. : A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature 2013, 496:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kato M, Sasaki M, Mizuno S, Harata M: Novel actin-related proteins in vertebrates: similarities of structure and expression pattern to Arp6 localized on Drosophila heterochromatin. Gene 2001, 268:133–140. [DOI] [PubMed] [Google Scholar]
- [23].Ohfuchi E, Kato M, Sasaki M, Sugimoto K, Oma Y, Harata M: Vertebrate Arp6, a novel nuclear actin-related protein, interacts with heterochromatin protein 1. Eur J Cell Biol 2006, 85:411–421. [DOI] [PubMed] [Google Scholar]
- [24].Frankel S, Sigel EA, Craig C, Elgin SC, Mooseker MS, Artavanis-Tsakonas S: An actin-related protein in Drosophila colocalizes with heterochromatin protein 1 in pericentric heterochromatin. J Cell Sci 1997, 110:1999–2012. [DOI] [PubMed] [Google Scholar]
- [25].Brickner JH, Walter P: Gene Recruitment of the Activated INO1 Locus to the Nuclear Membrane. PLoS Biol 2004, doi: 10.1371/journal.pbio.0020342 2:e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH: Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell 2012, 22:1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pollard TD: Actin and Actin-Binding Proteins. Cold Spring Harb Perspect Biol 2016, doi: 10.1101/cshperspect.a018226 8:p.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ohnishi T, Kawamura H, Yamamoto T: [EXTRACTION OF A PROTEIN RESEMBLING ACTIN FROM THE CELL NUCLEUS OF THE CALF THYMUS]. J Biochem 1963, 54:298–300. [DOI] [PubMed] [Google Scholar]
- [29].Belin BJ, Mullins RD: What we talk about when we talk about nuclear actin. Nucleus 2013, 4:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kyheröinen S, Vartiainen MK: Nuclear actin dynamics in gene expression and genome organization. Sem Cell Dev Biol 2020, 102:105–112. [DOI] [PubMed] [Google Scholar]
- [31].Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R: Nuclear F-actin formation and reorganization upon cell spreading. J Biol Chem 2015, 290:11209–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Caridi CP, D’Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, et al. : Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559:54–60.**The active movement of heterochromatic double-stranded DNA breaks from the inner nucleus to the periphery is shown to use an Unc45-dependent nuclear myosin motor activity where the DSB-cargo is mobilized along Arp2/3-dependent actin filaments.
- [33].Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J: Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 2018, 559:61–66.**Nuclear actin, Arp2/3, and the Wiskott-Aldrich syndrome protein (WASp) are recruited to DSBs to mobilize and cluster damage DNA at the nuclear periphery by a transport mechanism that is dependent upon actin polymerization.
- [34].Wang A, Kolhe JA, Gioacchini N, Baade I, Brieher WM, Peterson CL, Freeman BC: Mechanism of Long-Range Chromosome Motion Triggered by Gene Activation. Dev Cell 2020, 52:309–320.**Transport of the activated INO1 gene locus is shown to be dependent upon nuclear actin polymerization. The gene locus is connected to the actin network by Arp-containing chromatin remodelers and moved by nuclear myosin supported by the Hsp90/Unc45 chaperones and nucleated at the gene by an allosterically regulated transcription factor.
- [35].Baarlink C, Plessner M, Sherrard A, Morita K, Misu S, Virant D, Kleinschnitz E-M, Harniman R, Alibhai D, Baumeister S, et al. : A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat Cell Biol 2017, 19:1389–1399. [DOI] [PubMed] [Google Scholar]
- [36].Wineland DM, Kelpsch DJ, Tootle TL: Multiple Pools of Nuclear Actin. Anatom Record 2018, 301:2014–2036.*Using various actin probes the existence of multiple forms of actin pools in the nuclei of different cell types of developing Drosophila embryos is demonstrated.
- [37].Wang Y, Sherrard A, Zhao B, Melak M, Trautwein J, Kleinschnitz E-M, Tsopoulidis N, Fackler OT, Schwan C, Grosse R: GPCR-induced calcium transients trigger nuclear actin assembly for chromatin dynamics. Nat Comm 2019, 10:5271.*The ability of G protein-coupled receptors to trigger f-actin polymerization using the ER-associated formin INF2 is shown. Correlating with the f-actin are changes in the chromatin organization.
- [38].Clough E, Tedeschi T, Hazelrigg T: Epigenetic regulation of oogenesis and germ stem cell maintenance by the Drosophila histone methyltransferase Eggless/dSetDB1. Dev Biol 2014, 388:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, Huttley GA, Tremethick DJ: Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol 2012, 19:1076–1083. [DOI] [PubMed] [Google Scholar]
- [40].Kapoor P, Shen X: Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol 2014, 24:238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Baarlink C, Wang H, Grosse R: Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 2013, 340:864–867. [DOI] [PubMed] [Google Scholar]
- [42].Plessner M, Grosse R: Extracellular signaling cues for nuclear actin polymerization. Eur J Cell Biol 2015, 94:359–362. [DOI] [PubMed] [Google Scholar]
- [43].Nishida E, Iida K, Yonezawa N, Koyasu S, Yahara I, Sakai H: Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc Natl Acad Sci USA 1987, 84:5262–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Meagher RB, Kandasamy MK, McKinney EC, Roy E: Nuclear Actin-Related Proteins in Epigenetic Control. Int Rev Cell Mol Biol 2009, 277:157–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Harata M, Oma Y, Tabuchi T, Zhang Y, Stillman DJ, Mizuno S: Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J Biochem 2000, 128:665–671. [DOI] [PubMed] [Google Scholar]
- [46].Hurley JH: The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct 1996, 25:137–162. [DOI] [PubMed] [Google Scholar]
- [47].Zorca CE, Kim LK, Kim YJ, Krause MR, Zenklusen D, Spilianakis CG, Flavell RA: Myosin VI regulates gene pairing and transcriptional pause release in T cells. Proc Natl Acad Sci USA 2015, 112:E1587–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Evdokimova VN, Gandhi M, Nikitski AV, Bakkenist CJ, Nikiforov YE: Nuclear myosin/actin-motored contact between homologous chromosomes is initiated by ATM kinase and homology-directed repair proteins at double-strand DNA breaks to suppress chromosome rearrangements. Oncotarget 2018, 9:13612–13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS: Long-range directional movement of an interphase chromosome site. Curr Biol 2006, 16:825–831. [DOI] [PubMed] [Google Scholar]
- [50].Dundr M, Ospina JK, Sung M-H, John S, Upender M, Ried T, Hager GL, Matera AG: Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 2007, 179:1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, Dekker J, Mirny L, Misteli T: Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell 2019, 176:1502–1515.*The inherent flexibility of genome architecture is shown by a combination of Hi-C and hiFISH to map the 3D chromosome organizations of single cells within the same cell population.
- [52].Bintu B, Mateo LJJ, Su JHJ-H, Sinnott-Armstrong NAA, Parker M, Kinrot S, et al. : Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 2018, 362:eaau1783.*Direct imaging of chromatin positioning in single cells visualizes the cell-to-cell varability in genome organization.
- [53].Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. : Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174:744–757.*A novel method, split-pool recognition of interactions by tag extension (SPRITE), is described that permits higher-order identification of genomic interactions and demonstrates that chromosome positioning correlates with nuclear bodies including the nucleolus and nuclear speckles.
- [54].Brueckner L, Zhao PA, van Schaik T, Leemans C, Sima J, Peric-Hupkes D, Gilbert DM, van Steensel B: Local rewiring of genome-nuclear lamina interactions by transcription. EMBO J 2020, doi: 10.15252/embj.2019103159 39:e103159.*The influence of gene activation or inactivation on the attachment of the association chromatin with the nuclear lamina is explored. In brief, gene activation tends to detach the chromatin from the lamina whereas inactivation strengthens the association.
- [55].Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, van Steensel B, Ma J, Belmont AS: Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 2018, 217:4025–4048.*A novel method of interrogating genome organization is described, TSA-Seq, that is capable of serving as a cytological ruler by exploiting proximity labeling as a means of determing 3D positioning of chromatin near specific nuclear targets.