Abstract
The nucleoskeleton has been associated with partitioning the genome into active and inactive compartments that dictate local transcription factor (TF) activity. However, recent data indicates that the nucleoskeleton and TFs reciprocally influence each other in dynamic TF trafficking pathways through the functions of LEM proteins. While the conserved peripheral recruitment of TFs by LEM proteins has been viewed as a mechanism of repressing transcription, a diversity of release mechanisms from the lamina suggest this compartment serves as a refuge for nuclear TF accumulation for rapid mobilization and signal stability. Detailed mechanisms suggest that TFs toggle between nuclear lamina refuge and nuclear matrix lamin-LEM protein complexes at sites of active transcription. In this review we will highlight emerging LEM functions acting at the interface of chromatin and nucleoskeleton to create TF trafficking networks.
Introduction
The 1952 electron micrographs of the Amoeba proteus nuclear membrane demonstrated a fibrous lamina supporting the inner leaflet, an observation which “necessitate(d) a reinterpretation of the structure of this membrane”[1]. This observation launched seven decades of reinterpretation of the astonishingly complex nucleoskeleton that extends from the periphery (nuclear lamina) into the nucleoplasm (nuclear matrix). The nucleoskeleton profoundly influences transcription via the regulation of transcription factors (TFs) and reciprocally relies on TFs in order to achieve specificity[2]. At the nuclear periphery A and B-type lamins form tetrameric 3.5nm filaments which assemble into distinct but interacting semiregular meshworks[3]. This lamin framework delineates the transcriptionally repressive nuclear lamina compartment which is characterized by dense peripheralized heterochromatin interacting with several classes of lamina-associated proteins[4]. By contrast, the transcriptionally active nuclear matrix consists of A-type lamins solubilized by post-translational modifications working with lamin-associated proteins[5,6].
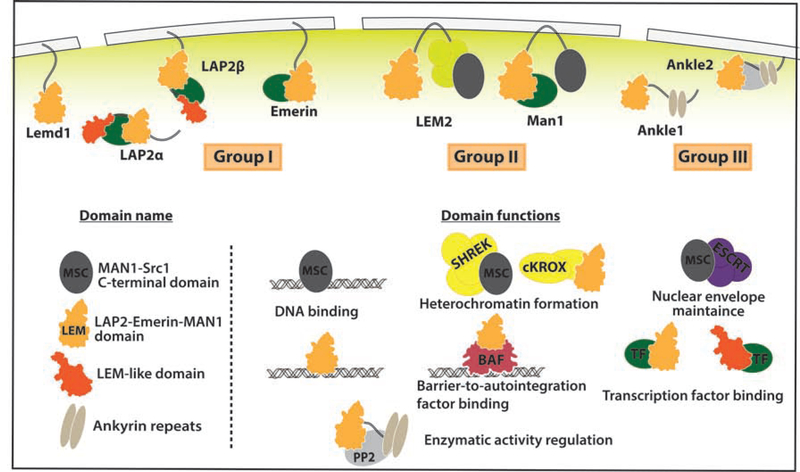
A key link between the nucleoskeleton and transcription is a unique family of seven genes encoding integral inner nuclear membrane proteins sharing a LEM domain (named after the founding members LAP2, Emerin, Man1)(Figure 1)[7]. Abnormalities associated with LEM protein function range from aging, cardiovascular disease, and cancer resistance, highlighting their critical role in signal transduction[8]. Three LEM subgroups reflect their unique domain organization. Group I LEM proteins contain a N-terminal LEM-domain and a C-terminal single-pass transmembrane domain (apart from certain LAP2 splice forms). Group II LEM-proteins contain two transmembrane domains, a DNA-binding MSC domain, and a N-terminal LEM domain. Group III LEM-proteins are highly divergent, lacking transmembrane domains and containing ankyrin repeats[9]. The ability of LEM proteins to directly bind lamins and DNA make them prime candidates for Lamina-Associated Domain (LAD) tethers, regions of heterochromatin that interact with the lamina[10]. However, numerous recent studies suggest a reinterpretation of LEM protein function that highlights new aspects of transcriptional regulation and disease relevance. Here we highlight recent work revealing the ancestral functions of these LEM proteins and how they have been co-opted to generate complex regulatory circuits on nucleoskeletal elements for localized TF function.
Figure 1. LEM family of nuclear lamina proteins.
LEM proteins are classified into three groups: group I includes LAP2, Emerin and Lemd1; group II includes LEM2 and Man1; group III includes Ankle1 and Ankle2 (also named Lem4). Apart from their shared LEM domain, each group contains unique domains with distinct known functions. See text for details.
Ancestral LEM proteins tether chromatin and participate in nuclear structure
Group II LEM proteins are well conserved throughout eukaryotic evolution[9,11], and recent studies deepen our understanding of their ancestral functions. In protozoa, the LEM domains demonstrates weaker sequence conservation than the MSC domain, indicating that the DNA-binding capacity of the MSC domain predated the development of the LEM-domain with its own indirect DNA-binding function. Functional studies in the binucleate protist Tetrahymena thermophila identify two LEM2-like proteins that localize to the nuclear envelope with one accumulating in the macronuclei designated for degradation and the other localizing to the nuclear pore complexes of the micronucleus and inhibiting its degradation, reinforcing the LEM protein role in nuclear structural maintenance[12].
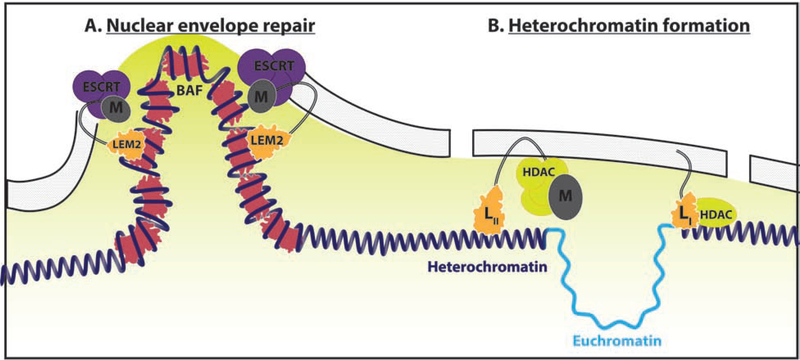
A key nuclear maintenance partner emerging with Group II LEM proteins is the Barrier-to-Autointegration (BAF) protein, a DNA binding protein conserved in metazoa and first identified for its role in blocking viral integration into the mouse genome[13]. Vertebrate LEM2 and BAF, working with Charged Multivesicular Body Protein 7(CHMP7), and ESCRT-III complex, enforce nuclear-cytoplasmic segregation in the settings of mitosis and envelope rupture[14–17]. BAF ranked as the top hit in a siRNA depletion screen of 1,295 genes to induce micronuclei formation when the mitotic spindle was disrupted. BAF cross-bridges low-complexity distal regions of chromatin by forming dimers coating the surface of chromosomes, condensing the chromatin locally to generate a diffusion barrier. In a telling experiment, dimerization-deficient BAF mutant fails to rescue knockdown of endogenous BAF, indicating the DNA compaction by BAF is a major mechanism of structuring the nucleus[18*]. In a fascinating recent follow-up, LEM2 uses phase-separation onto microtubules via its low complexity region to coordinate the actions of BAF in chromatin cross-bridging with CHMP7 and ESCRT machinery in nuclear membrane repair during envelope reformation[19**] (Figure 2A). Together, it appears BAF co-evolved with the LEM domain to enforce appropriate nuclear-cytoplasmic compartmentalization, with BAF creating a diffusion barrier by compacting chromatin.
Figure 2. Ancestral LEM proteins tether chromatin and participate in nuclear structure.
(A) Nuclear envelope repair coordinated by LEM2-CHMP7-ESCRTIII recruitment to BAF coated DNA. LEM2: orange, ESCRT: purple, BAF: pink. (B) Group I and II LEM proteins (LI/II) tether and silence (yellow gradient) peripheral heterochromatin via the recruitment of HDAC complexes (yellow). LEM domain: orange, MSC domain: dark grey.
An intriguing question is what takes the place of BAF chromatin compaction and LAD-like formation in yeast where the LEM2 amino-terminal LEM domain acts to tether chromatin and centromeres to the nuclear periphery without lamins or BAF. The carboxy-terminal MSC domain anchors telomeres and epigenetically silences peripheral chromatin via recruitment of the HDAC-containing SHREC complex[20](Figure 2B). These LEM2 associated LADs appear to be pruned by the same ESCRT machinery LEM2 associates with for nuclear envelope repair[21]. Studies suggest lamina proteins Nur1 and Bqt4, which form complexes with LEM2, are vital to this process[20,22]. Super resolution microscopy reveals that Bqt4, LEM2, and Nur1 aggregate into distinct microdomains within the nuclear lamina, with LEM2 distributing both diffusely across the lamina and forming distinct puncta. Deletion of Bqt4 results in loss of the diffusely perinuclear LEM2 population with the Bqt4-dependent diffuse LEM2 population functioning to maintain heterochromatin silencing[23*]. This intriguing finding suggests that subpopulations of LEM-associated complexes exist within the nuclear lamina, allowing for greater compartmentalization and functional specialization.
In higher eukaryotes, lamina sequestration and repression of cardiac genes occurs in part through LAP2β and HDAC3[24]. HDAC3, which associates with LAP2β on the nuclear lamina, functions through a mechanism independent of its catalytic activity. Knockout of HDAC3 results in the mislocalization of cardiac genes into the nucleoplasm prematurely, leading to precocious differentiation that can be rescued by exogenous catalytically-dead HDAC3 mutants or HDAC3-LAP2β fusions, but not a LAP2β-binding deficient HDAC3 mutant. Given the diversification of the LEM protein family in these organisms, an open question is how Group I and Group II LEM proteins differ in LAD anchorage functionality, how the loss of the direct DNA-binding capacity of the MSC domain changes the functionality of Group I LEM proteins, and how LEM proteins in general work in concert with other lamina components to mediate a diversity of long-term and short-term lamina-chromatins interactions[25–28].
Nuclear Lamina: LEM proteins make a TF refuge, not a graveyard
While LEM proteins help form LADs that repress transcription, neither direct nor indirect LEM protein binding to chromatin provides the sequence specificity required for tight transcriptional regulation, directing attention to LEM protein interaction with sequence-specific TFs. Analysis of Igh, Ikzf1, and Bcl11a loci, which are peripherally localized in LADs in fibroblasts but nucleoplasmic and active in pro-B cells, reveals that flanking LAD chromatin sequences are sufficient for peripheral targeting of ectopic loci. The LAD flanking sequences were enriched in YY1 and CTCF binding sites, and knockdown of YY1 and lamin A/C (but not lamin A alone or CTCF) were sufficient to disrupt LAD formation[29]. Notably lamin A/C disruption has previously been reported to disrupt LEM protein recruitment to the nuclear lamina[30]. These data suggest that transcription factors act in concert with the nuclear lamina, likely through a LEM protein bridge, to sequester genes to the nuclear periphery as a mechanism of transcriptional silencing. Similarly, the peripheral sequestration of non-DNA-bound TFs by LEM proteins exerts a predictably repressive function on signal transduction[31]. This relationship is typified by the repressive interaction between Man1 and SMAD transcription factors, as MAN1 scaffolds SMAD2/3 dephosphorylation by PPM1A[32–34].
However recent studies suggest LEM interactions with TFs are more than just repressive, but also serve as a dynamic nuclear refuge for the accumulation of TFs without the risk of inadvertent activation or degradation. For example, acetylated GLI1 is sequestered to the nuclear lamina by the LEM protein LAP2β[35**] (Figure 3). While this interaction represses GLI1 transcriptional activity, it also antagonizes GLI1 nuclear export. Without LAP2β expression, GLI1 fails to accumulate in the nucleus and cannot execute its transcriptional program. Inhibition of the proteasome with MG132 in LAP2β−/− cell results in the cytoplasmic accumulation of GLI1, while nuclear export inhibition by leptomycin B rescues the nuclear accumulation of GLI1. Therefore, LAP2β generates a nuclear reserve of inactivated GLI1 poised for subsequent activation by the inhibition of its nuclear export.
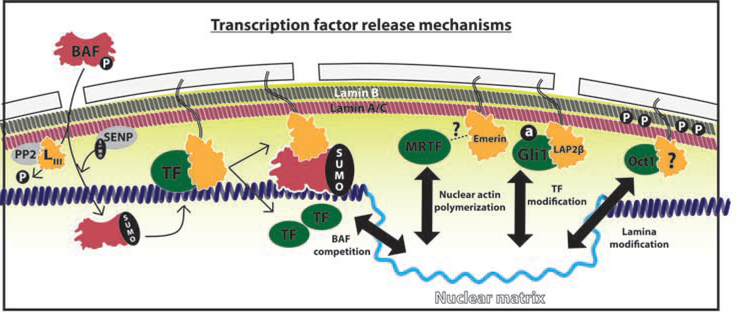
Figure 3. Nuclear Lamina: LEM proteins make a TF refuge, not a graveyard.
LEM proteins create a transcription factor (TF, green) refuge at the nuclear lamina (yellow gradient) which release TFs through diverse mechanisms. Left: Proposed model of BAF (pink) competition with TFs (green) for LEM protein (orange) and DNA interaction. BAF phosphorylation (P) is regulated by phosphatases (PP2) directed by Group III LEM proteins (LIII) while SUMOylation (SUMO) requires lamina-bound SENP1/2 (SENP). Right: Mechanisms of TF release from the nuclear lamina include nuclear actin polymerization by Emerin, TF modification such as GLI1 acetylation (a), and lamina modification such as B-type lamin phosphorylation (P).
Dynamic LEM-lamina-TF interactions appear to hinge on post-translational modifications. Over a decade ago the release of c-Fos from the nuclear lamina was found to be dependent on its phosphorylation by lamina-associated and serum-stimulated Erk1/2[36,37]. More recently de-acetylation of GLI1was found to liberate it from LAP2β and allow accumulation on the nucleoplasmic LAP2 isoform LAP2ɑ[35**]. These LAP2 splice-forms compete for a common zinc-finger binding site on GLI1, with LAP2β outcompeting LAP2ɑ to bind acetylated GLI1 by a secondary interaction site at the acetyl moiety. Countering this repression, LAP2ɑ catalyzes the deacetylation of GLI1 by the recruitment of HDAC1, facilitating the LAP2 hand-off mechanism. Overexpression of LAP2β, which represses GLI1 transcriptional activity at steady-state but generates a larger acetylated/inactivated lamina-bound reserve, allows more rapid recovery from deacetylation blockade compared to wild-type. These results illustrate how LEM proteins act as a refuge for TFs waiting to gain access to chromatin and can rapidly mobilize nuclear reserves[35**].
In addition to TF modifications, emerging data suggest post-translational modifications of the nucleoskeleton may be another mechanism controlling the egress of TFs from the lamina. The direct phosphorylation of lamin B1 by JNK at T575 has been suggested as a mechanism of Oct-1 release from the nuclear lamina[38,39] (Figure 3). Meanwhile, Emerin regulates the rapid mechano-responsiveness of the MRTF/SRF transcriptional complex in part by helping to induce nuclear actin polymerization. Disruption of Emerin localization by lamin A depletion resulted in a failure of MRTF to accumulate in the nucleus[40–42] (Figure 3). Finally, β-catenin uses the cytoplasmic Nesprins, a member of the mechano-sensing LINC complex for nuclear entry[43]. Wnt pathway activation induces the transient accumulation of β-catenin at the nuclear lamina prior transitioning to the nucleoplasm[44]. Emerin, known to act in association with the LINC complex, also induces β-catenin nuclear export[45,46]. Yet to be elucidated in each of these cases is the mechanism and identity of the nuclear matrix recipient of these key TFs.
A further blind spot in our understanding is how the DNA compaction functions of BAF control LEM-TF interactions. LEM-bound BAF-dimers form femtomolar range DNA interactions and in principle should simply disengage all associated transcription factors[13]. Indeed all reported BAF-TF interactions are inhibitory[13,47,48]. Providing a potential solution, phosphorylation of BAF by Vaccinia-related kinases VRK1 and VRK2a at S4, with minor sites at T2 and T3, controls BAF nuclear-cytoplasmic distribution by disrupting its interaction with LEM-proteins, DNA, and TFs[49,50]. The Group III LEM-protein ANKLE2 balances VRK1/2 phosphorylation of BAF and PP2/PP4 dephosphorylation[50–52]. Recently the phosphorylation of BAF was reported to control interaction with E2F1 in Drosophila, with BAF antagonizing E2F1 nuclear accumulation [13,48]. In addition, BAF has been recently found to be SUMOylated at K6 by lamina associated SENP1/2 which promotes its DNA, Lamin A, and PCNA interactions without changing its LEM-protein interaction[53]. These data suggest that LEM-associated, stimulus-specific modifications of BAF provide a mechanism to drive TFs from the lamina refuge (Figure 3).
Nuclear Matrix: Nucleoplasmic structure enforces transcription
In addition to their established role in enforcing heterochromatin, recent studies support a vital role for LEM proteins in the establishment of the nuclear matrix. Solubilization of lamins depends upon both the phosphorylation of lamin A/C at S22 and S392 and the function of the nucleoplasmic LEM protein LAP2ɑ[5,6,54]. LAP2ɑ, a splice form of the LAP2 proteins, appears to have evolved in mammals following a retrotransposon domestication event, replacing its transmembrane domain with a unique coiled-coil domain[55] that binds and facilitates solubilization of lamins[54]. Genomic binding studies find that LAP2ɑ and lamin A bind overlapping regions of chromatin in the nuclear interior[56] and depletion of lamin A results in increased nucleoplasmic genomic loci mobility[57], consistent with the formation of nucleoplasmic structural scaffolds for transcription. How lamin phosphorylation, LAP2ɑ, and TFs influence each other remains an active area of investigation.
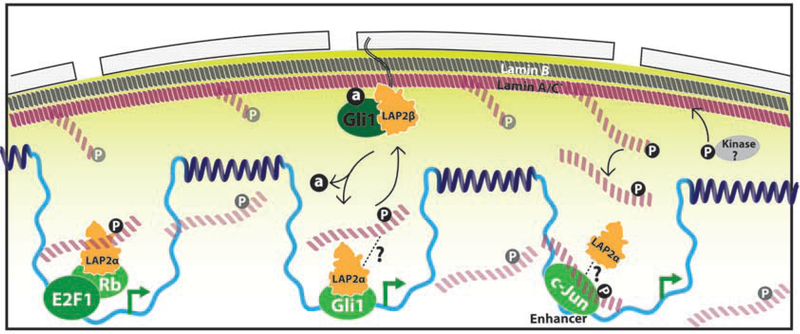
In addition to previous work showing the cell cycle regulatory retinoblastoma protein anchorage by LAP2ɑ[5,58], recent work adds GLI1 to the list of LAP2ɑ nuclear matrix transcriptional partners[35**] (Figure 4). Vicinal DNA-labeling via DamID indicates LAP2ɑ and GLI1 co-bind hedgehog target genes, with FRAP measurements of GLI1 showing reduced mobility when associated with LAP2ɑ. Loss of nucleoplasmic anchorage is correlated with a decrease in GLI1 transcriptional output and an increase in CRM1-binding (and subsequent nuclear export). The relative contributions of nuclear matrix anchorage to nucleoplasmic positioning versus scaffolding nucleoplasmic interaction surfaces remains to be determined.
Figure 4. Nuclear Matrix: Nucleoplasmic structure enforces transcription.
Transcription factors (green) dock with nuclear matrix structures composed of depolymerized phosphorylated A-type lamins (P, pink) and LAP2 (orange) on euchromatin (light blue) to activate transcription (green arrow).
An exciting recent report indicates that lamin A/C phosphorylation and LAP2ɑ play a direct role in the regulation of transcription. The authors find that nucleoplasmic and soluble S22 phosphorylated lamin A/C co-binds binds active enhancers with the c-Jun transcription factor[59**] (Figure 4). This confirms that the euchromatin regions previously found to bind lamin A/C and LAP2ɑ are indeed bound by a subpopulation of phosphorylated lamin, with global run-on sequencing experiments confirming these are regions of active transcription. An intriguing finding was that the pathological lamin A/C mutant progerin itself does not show detectable levels of pS22, yet the regions bound by wildtype pS22 lamin A/C were altered in HGPS patient derived fibroblasts. This result supports the importance of lamin post-translational modifications and reinforces how LEM-lamina-matrix interactions are critical for disease prevention. Similar to GLI1, it is unclear whether c-Jun directs the nucleation of the nuclear matrix onto these regions or if the lamins bind preexisting sites that stabilize c-Jun chromatin accumulation. Studies into how c-Jun interacts with the nuclear matrix lamins, either through LAP2ɑ or other scaffolding mechanisms, will likely shed light on how this dynamic structure influences transcription.
Concluding Remarks
LEM proteins have evolved at the interface of chromatin and nucleoskeleton to control chromatin structure and modulate the activity of sequence specific transcription factors, providing the local dynamism needed to adapt to changing cellular environments. The ability to epigenetically silence peripheralized chromatin in LADs has led to the development of a transcriptionally inert compartment which TF nuclear trafficking pathways use to store inactive TFs which can be rapidly deployed by post-translational modifications. Rather than being a graveyard for TFs, LEM proteins enable the lamina to serve as a vital refuge that modifies transcriptional dynamics. Future questions include determining if LEM-based TF peripheral sequestration contributes to LAD specificity, how kinases/deacetylases act with specificity to release TFs from the lamina, and how BAF contributes to TF antagonism. Progress in the field will depend on overcoming some of the major technical challenges associated with studying the dynamic nature of the compartment. These include identifying the significance of transient and changing physical chemical properties of TF-LEM interactions, the functional redundancy of LEM proteins, the possible existence of sub-populations of LEM proteins, and the pleiotropic effects associated with the perturbation of these core nuclear structural entities. However, the introduction of proximal DNA and protein labeling technologies, generation of state-specific antibodies, and the ability to assay local mobility by rapidly improving imaging technologies make these questions trackable for the first time. Similarly, the recent appreciation of phase-separated compartments within the nucleoplasm makes their interactions with the nuclear matrix all the more intriguing. While phase-separation efficiently segregates factors based on solubility, lamin-based structures in the nucleoplasm promise an unprecedented mechanism of structural stability while allowing for dynamic changes. The sky is the limit for how LEM proteins contribute to a panoply of biological processes and human diseases.
Supplementary Material
Acknowledgements
This work was funded by NIH R01 2R37-ARO54780 (AEO), Cancer Biology Training Grant (FG) and Stanford MSTP (ANM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Greider MH, Kostir WJ, Frajola WJ: ELECTRON :MICROSCOPY OF THE NUCLEAR M E M B R A N E OF AMOEBA PROTEUS *. 1956. [PubMed]
- 2.Adam SA: The Nucleoskeleton. Cold Spring Harb Perspect Biol 2017, 9:a023556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O: The molecular architecture of lamins in somatic cells. Nat Publ Gr 2017, doi: 10.1038/nature21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briand N, Collas P: Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol 2020, 21:1–25.A useful review on the heterogeneity of Lamin-associated domains from recent cell biology and single-cell sequencing studies. It provides a backdrop for future studies on lamin-chromatin interactions and transcriptional regulation.
- 5.Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, Kral R, Gotic I, Fuchs P, Cohen TV, Bittner R, et al. : Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat Cell Biol 2008, 10:1341–8. [DOI] [PubMed] [Google Scholar]
- 6.Kochin V, Shimi T, Torvaldson E, Adam SA, Goldman A, Pack C-G, Melo-Cardenas J, Imanishi SY, Goldman RD, Eriksson JE: Interphase phosphorylation of lamin A. J Cell Sci 2014, 127:2683–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun S, Barrales RR: Beyond Tethering and the LEM domain: MSCellaneous functions of the inner nuclear membrane Lem2. Nucleus 2016, doi: 10.1080/19491034.2016.1252892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brachner A, Foisner R: Lamina-associated polypeptide (LAP)2α and other LEM proteins in cancer biology. Adv Exp Med Biol 2014, 773:143–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brachner A, Foisner R: Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem Soc Trans 2011, 39:1735–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Steensel B, Furlong EEM: The role of transcription in shaping the spatial organization of the genome. Nat Rev Mol Cell Biol 2019, 20:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koreny L, Field MC: Ancient Eukaryotic Origin and Evolutionary Plasticity of Nuclear Lamina. Genome Biol Evol 2016, 8:2663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwamoto M, Fukuda Y, Osakada H, Mori C, Hiraoka Y, Haraguchi T: Identification of the evolutionarily conserved nuclear envelope proteins Lem2 and MicLem2 in Tetrahymena thermophila. Gene X 2019, 1:100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margalit A, Brachner A, Gotzmann J, Foisner R, Gruenbaum Y: Barrier-to-autointegration factor - a BAFfling little protein. Trends Cell Biol 2007, 17:202–208. [DOI] [PubMed] [Google Scholar]
- 14.Lusk CP, Ader NR: CHMPions of repair: Emerging perspectives on sensing and repairing the nuclear envelope barrier. Curr Opin Cell Biol 2020, 64:25–33.This work reviews homeostatic mechanisms of nuclear-cytoplasmic compartmentalization, emphazing interactions of the LEM proteins with ESCRT machinery.
- 15.Gu M, LaJoie D, Chen OS, Von Appen A, Ladinsky MS, Redd MJ, Nikolova L, Bjorkman PJ, Sundquist WI, Ullman KS, et al. : LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc Natl Acad Sci U S A 2017, 114:E2166–E2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olmos Y, Perdrix-Rosell A, Carlton JG: Membrane Binding by CHMP7 Coordinates ESCRT-III-Dependent Nuclear Envelope Reformation. Curr Biol 2016, 26:2635–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaller DJ, Allegretti M, Borah S, Ronchi P, Beck M, Lusk CP: An escrt-lem protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, Gerlich DW: DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 2017, 170:956–972.e23.* BAF dimers function to cross-bridge distal segments of DNA during mitotic exit to create a diffusion-limiting surface at the developing nuclear periphery, ensuring nucleocytoplasmic segregation. This study brings together the known DNA bridging functions of BAF to create a model for its indispensable role in nuclear structural maintenance.
- 19.von Appen A, LaJoie D, Johnson IE, Trnka MJ, Pick SM, Burlingame AL, Ullman KS, Frost A: LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 2020, 582:115–118.** LEM2 phase separates onto microtubes at the nuclear periphery via the self-association of its low complexity domain to coordinate BAF, CHMP7, and ESCRTIII. This is the first demonstration of LEM proteins participating in phase separation and explains how LEM2 coordinates its LEM-BAF and MSC-CHMP7/ESCRTIII functions in nuclear envelope reformation.
- 20.Barrales RR, Forn M, Georgescu PR, Sarkadi Z, Braun S: Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev 2016, 30:133–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pieper GH, Sprenger S, Teis D, Oliferenko S: ESCRT-III/Vps4 Controls Heterochromatin-Nuclear Envelope Attachments. Dev Cell 2020, 53:27–41.e6.Investigating how fission yeast Schizosaccharomyces japonicus break and reform nuclear envelope-chromatin interactions during mitosis, this work documents the importance of the ESCRT-III/Vps4 release from INM -associated Lem2-Nur1 at mitosis. Failure leads to persistent association of chromosomes with the INM throughout mitosis and and diffuclty re-establishing the nucleocytoplasmic compartmentalization after division.
- 22.Hirano Y, Kinugasa Y, Asakawa H, Chikashige Y, Obuse C, Haraguchi T, Hiraoka Y: Lem2 is retained at the nuclear envelope through its interaction with Bqt4 in fission yeast. Genes to Cells 2018, 23:122–135. [DOI] [PubMed] [Google Scholar]
- 23.Ebrahimi H, Masuda H, Jain D, Cooper JP: Distinct ‘safe zones’ at the nuclear envelope ensure robust replication of heterochromatic chromosome regions. Elife 2018, 7.* Subpopulations of Lem2 segregated by Bqt4 are diffusely perinuclear or accumulate into distinct microdomains with unique functions such as pericentric heterochromatin maintenance. This is the first report of subpopulations of LEM proteins in the nuclear lamina participating in distinct activities.
- 24.Poleshko A, Shah PP, Gupta M, Babu A, Morley MP, Manderfield LJ, Ifkovits JL, Calderon D, Aghajanian H, Sierra-Pagán JE, et al. : Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction. Cell 2017, 171:573–587.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. : LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation. 2013, doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Amendola M, Van Steensel B: Mechanisms and dynamics of nuclear lamina-genome interactions. Curr Opin Cell Biol 2014, 28:61–68. [DOI] [PubMed] [Google Scholar]
- 27.Holla S, Dhakshnamoorthy J, Folco HD, Balachandran V, Xiao H, Sun L ling, Wheeler D, Zofall M, Grewal SIS: Positioning Heterochromatin at the Nuclear Periphery Suppresses Histone Turnover to Promote Epigenetic Inheritance. Cell 2020, 180:150–164.e15.A S. cerevisiae screen for heterochromatic silencers identifies the Amo1-RixC-HP1 complex and demonstrates how it creates a peripheral subdomain that enforces stable gene repression and maintains heterochromatin in a heritable manner.
- 28.Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, et al. : Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 2019, 570:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL: Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol 2015, 208:33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaughan OA, Alvarez-Reyes M, Bridger JM, Broers JLV, Ramaekers FCS, Wehnert M, Morris GE, Whitfield WGF, Hutchison CJ: Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J Cell Sci 2001, 114 (14): 2577–90. [DOI] [PubMed] [Google Scholar]
- 31.Heessen S, Fornerod M: The inner nuclear envelope as a transcription factor resting place. EMBO Rep 2007, 8:914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan D, Estévez-Salmerón LD, Stroschein SL, Zhu X, He J, Zhou S, Luo K: The integral inner nuclear membrane protein MAN1 physically interacts with the R-smad proteins to repress signaling by the transforming growth factor-β superfamily of cytokines. J Biol Chem 2005, 280:15992–16001. [DOI] [PubMed] [Google Scholar]
- 33.Lin F, Morrison JM, Wu W, Worman HJ: MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-β signaling. Hum Mol Genet 2005, 14:437–445. [DOI] [PubMed] [Google Scholar]
- 34.Bourgeois B, Gilquin B, Tellier-Lebègue C, Östlund C, Wu W, Pérez J, Hage P El, Lallemand F, Worman HJ, Zinn-Justin S: Inhibition of TGF-β signaling at the nuclear envelope: Characterization of interactions between MAN1, Smad2 and Smad3, and PPM1A. Sci Signal 2013, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirza AN, McKellar SA, Urman NM, Brown AS, Hollmig T, Aasi SZ, Oro AE: LAP2 Proteins Chaperone GLI1 Movement between the Lamina and Chromatin to Regulate Transcription. Cell 2019, 176:198–212.e15.** Identifies a LAP2 isoform-dependent nuclear chaperoning system that involves accumulation of acetylated GLI1 on the nuclear lamina and mobilization of GLI1 to the nuclear matrix by deacetylation. This study is the first detailed mechanism of how modification of a TF controls shuttling between lamina and matrix.
- 36.Ivorra C, Kubicek M, González JM, Sanz-González SM, Alvarez-Barrientos A, O’Connor J-E, Burke B, Andrés V: A mechanism of AP-1 suppression through interaction of c-Fos with lamin A/C. Genes Dev 2006, 20:307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzàlez JM, Navarro-Puche A, Casar B, Crespo P, Andrès V: Fast regulation of AP-1 activity through interaction of lamin A/C, ERK1/2, and c-Fos at the nuclear envelope. J Cell Biol 2008, 183:653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boubriak II, Malhas AN, Drozdz MM, Pytowski L, Vaux DJ: Stress-induced release of Oct-1 from the nuclear envelope is mediated by JNK phosphorylation of lamin B1. PLoS One 2017, 12:e0177990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malhas AN, Lee CF, Vaux DJ: Lamin B1 controls oxidative stress responses via Oct-1. J Cell Biol 2009, 184:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J: Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 2013, 497:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K: Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol 2014, 16:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willer MK, Carroll CW: Substrate stiffness-dependent regulation of the SRF−Mkl1 co-activator complex requires the inner nuclear membrane protein Emerin. [date unknown], doi: 10.1242/jcs.197517. [DOI] [PMC free article] [PubMed]
- 43.Neumann S, Schneider M, Daugherty RL, Gottardi CJ, Eming SA, Beijer A, Noegel AA, Karakesisoglou I: Nesprin-2 interacts with α-catenin and regulates Wnt signaling at the nuclear envelope. J Biol Chem 2010, 285:34932–34938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uzer G, Bas G, Sen B, Xie Z, Birks S, Olcum M, McGrath C, Styner M, Rubin J: Sun-mediated mechanical LINC between nucleus and cytoskeleton regulates βcatenin nuclear access. J Biomech 2018, 74:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markiewicz E, Tilgner K, Barker N, van de Wetering M, Clevers H, Dorobek M, Hausmanowa-Petrusewicz I, Ramaekers FCS, Broers JLV, Blankesteijn WM, et al. : The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J 2006, 25:3275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tilgner K, Wojciechowicz K, Jahoda C, Hutchison C, Markiewicz E: Dynamic complexes of A-type lamins and emerin influence adipogenic capacity of the cell via nucleocytoplasmic distribution of β-catenin. J Cell Sci 2009, 122:401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Xu S, Rivolta C, Li LY, Peng GH, Swain PK, Sung CH, Swaroop A, Berson EL, Dryja TP, et al. : Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem 2002, 277:43288–43300. [DOI] [PubMed] [Google Scholar]
- 48.Unnikannan CP, Reuveny A, Grunberg DT, Volk T: Mechanosensitive recruitment of BAF to the nuclear membrane inhibits nuclear E2F1 and Yap levels. bioRxiv 2019, doi: 10.1101/803932. [DOI] [Google Scholar]
- 49.Nichols RJ, Wiebe MS, Traktman P: The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell 2006, 17:2451–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birendra KC, May DG, Benson BV., Kim DI, Shivega WG, Ali MH, Faustino RS, Campos AR, Roux KJ: VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol Biol Cell 2017, 28:2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TBN, Mall M, Wallenfang MR, Mattaj IW, Gorjánácz M: Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell 2012, 150:122–135. [DOI] [PubMed] [Google Scholar]
- 52.Zhuang X, Semenova E, Maric D, Craigie R: Dephosphorylation of barrier-to-autointegration factor by protein phosphatase 4 and its role in cell mitosis. J Biol Chem 2014, 289:1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin Q, Yu B, Wang X, Zhu S, Zhao G, Jia M, Huang F, Xu N, Ren H, Jiang Q, et al. : K6-linked SUMOylation of BAF regulates nuclear integrity and DNA replication in mammalian cells. Proc Natl Acad Sci U S A 2020, 117:10378–10387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R, Ashery-Padan R, Weiss AM, Feinstein N, Gruenbaum Y, et al. : Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J Cell Sci 2000, 113 Pt 19:3473–84. [DOI] [PubMed] [Google Scholar]
- 55.Abascal F, Tress ML, Valencia A: Alternative splicing and co-option of transposable elements: The case of TMPO/LAP2alpha and ZNF451 in mammals. Bioinformatics 2015, 31:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonhoure N, Bernasconi D, Lammers F, Canella D, Willis IM, Herr W: A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2alpha. Genome Biol 2016, doi: 10.1111/2049-632X.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bronshtein I, Kepten E, Kanter I, Berezin S, Lindner M, Redwood AB, Mai S, Gonzalo S, Foisner R, Shav-Tal Y, et al. : Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat Commun 2015, 6:8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markiewicz E, Dechat T, Foisner R, Quinlan RA, Hutchison CJ: Lamin A/C binding protein LAP2alpha is required for nuclear anchorage of retinoblastoma protein. Mol Biol Cell 2002, 13:4401–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ikegami K, Secchia S, Almakki O, Lieb JD, Moskowitz IP: Phosphorylated Lamin A/C in the Nuclear Interior Binds Active Enhancers Associated with Abnormal Transcription in Progeria. Dev Cell 2020, 52:699–713.e11.** Serine 22 phosphorylated lamin A/C co-binds nucleoplasmic open chromatin regions with c-Jun and is associated with active transcription that can be modified by the presence of lamina-associated progerin. This study is the first demonstration of interphase phosphorylated lamins co-binding with a transcription factor onto nucleoplasmic sites of active transcription.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.