Abstract
Background:
Metaplastic carcinoma is an aggressive, triple-negative breast cancer (TNBC) with differentiation towards squamous, spindle, or mesenchymal cell types. The molecular underpinnings of the histological subtypes are unclear. Our lab discovered a cytoplasmic function of EZH2, a transcriptional repressor, whereby pEZH2 T367 binds to cytoplasmic proteins in TNBC cells and enhances invasion and metastasis. Here, we investigated the expression and subcellular localization of pEZH2 T367 protein in metaplastic carcinomas.
Methods:
Thirty-five metaplastic carcinomas (17 squamous, 10 mesenchymal, and 8 spindle) were evaluated and immunostained with anti-pEZH2 T367. We analyzed staining intensity (score 1–4), subcellular localization (nuclear/cytoplasmic), and localization within the tumor (center/invasive edge). Protein expression of pEZH2 T367-binding partners was measured from a quantitative multiplex proteomics analysis performed in our lab.
Results:
Cytoplasmic pEZH2 T367 was significantly upregulated in squamous (14 of 17, 82%) compared to mesenchymal (4 of 10, 40%) and spindle (2 of 6, 33%) subtypes (p=0.011). Twenty-five of 34 (73%) tumors with available tumor–normal interface showed accentuated cytoplasmic pEZH2 T367 at the infiltrative edge. Cytoplasmic pEZH2 T367 was upregulated in 9 of 10 (90%) tumors with lymph node metastasis (p=0.05). Bioinformatics analyses identified an EZH2 protein network in metaplastic carcinomas (p value: <1.0e-16). Using quantitative proteomics, we found significantly increased expression of cytoplasmic EZH2-binding partners in squamous compared to spindle and mesenchymal subtypes.
Conclusions:
pEZH2 T367 expression and subcellular localization may be useful to distinguish metaplastic carcinoma subtypes. pEZH2 T367 may play a role in the histological diversity and behavior of these tumors.
Keywords: Metaplastic carcinoma, pEZH2, Metastasis, Epithelial-to-mesenchymal transition, Claudin low
Introduction
Metaplastic breast carcinoma is an aggressive subtype of triple-negative breast cancer (TNBC) in which part or all of the carcinomatous epithelium exhibits differentiation towards metaplastic, non-glandular, component(s) [1, 2]. The metaplastic component(s) may be spindled, squamous, or mesenchymal which includes chondroid and/or other heterologous components such as osseous differentiation [3]. Metaplastic carcinomas are rare, accounting for approximately 1% of breast carcinomas. However, they may constitute up to 14% of tumors in women of African descent [4, 5]. Compared to non-metaplastic TNBC, metaplastic tumors have poorer prognosis, greater chemoresistance, and increased propensity for metastasis [5–7]. Studies suggest that histological subtype may influence prognosis [8–11]. The molecular underpinnings for the aggressive clinical features of metaplastic carcinomas are not fully understood.
The Polycomb Group Protein Enhancer of Zeste Homologue 2, EZH2, maintains heritable gene expression profiles. EZH2 binds to members of the Polycomb Repressive Complex 2 (PRC2) including EED and SUZ12, and deposits trimethyl marks on histone tails of lysine 27 of histone H3 (H3K27me3) leading to transcriptional repression of target genes [12]. In this manner, EZH2 regulates cell-type identity and differentiation. EZH2 overexpression occurs in solid and hematologic malignancies including breast cancer [4, 13–16]. Our lab and others have demonstrated that EZH2 overexpression in invasive breast carcinomas is associated with aggressive behavior, ER negative status, and worse clinical outcome compared to tumors with low EZH2 expression [13, 17].
Recent studies have shown that the oncogenic function of EZH2 in breast cancer may be regulated by phosphorylation and/or may involve H3K27me3-independent mechanisms. For example, CDK1 phosphorylates EZH2 at T487 in TNBC cells leading to disruption of EZH2 binding to PRC2 and inhibition of EZH2 histone methyltransferase activity [18]. CDK2 also phosphorylates EZH2 at T416 which induces differentiation towards a TNBC basal-like phenotype by changing the transcriptomic profiles [19]. Our laboratory has discovered that in a subset of TNBC tumors p38 Mitogen-Activated Kinase (MAPK) phosphorylates EZH2 at T367 leading to pEZH2 T367 cytoplasmic localization and binding to cytoplasmic proteins with roles in actin-binding, cell adhesion, and cytosolic functions in an H3K27me3-independent manner. We found that EZH2 phosphorylation at T367 is necessary for the metastasis-inducing function of EZH2 [20]. In this study, we hypothesized that metaplastic carcinomas may exhibit pEZH2 T367 expression in the cytoplasm and that the subcellular localization of pEZH2 T367 may be associated with the tumor histopathology and prognosis.
Materials and methods
Tissue collection and immunohistochemistry
Thirty-five cases of metaplastic breast carcinomas with available blocks from 1988 to 2015 were identified in the surgical pathology files at the University of Michigan Pathology Department with IRB approval (HUM0005330). All slides were reviewed and tumors classified according to the predominant metaplastic component into spindle, squamous, or mesenchymal (chondroid and/or osseous) independently reviewed by three pathologists (SLS, ERM, and CGK). Five micron-thick paraffin-embedded sections were de-paraffinized in xylene and rehydrated through graded alcohols to water. Heat-Induced Epitope Retrieval (HIER) was performed in the Decloaking Chamber (Biocare Medical) with FLEX FTRS Low pH Retrieval buffer, pH 6.1 (Dako, North America). Slides were incubated in 3% hydrogen peroxide for 5 min to quench endogenous peroxidases. Rabbit polyclonal anti-pEZH2 T367 generated and validated in our lab[20] was incubated (1:8000) with the sections for 1 h at room temperature. Antibodies were detected with Envision+HRP Labeled Polymer (DakoCytomation) for 30 minutes at room temperature. HRP staining was visualized with the DAB+Kit (DakoCytomation). Negative control slides were run. Slides were counterstained in hematoxylin, blued in running tap water, dehydrated through graded alcohols, cleared in xylene and then mounted with Permount.
pEZH2 T367 staining was evaluated at least twice by three pathologists (SLS, ERM, and CGK) blinded to tumor stage and clinical information. pEZH2 T367 was evaluated for intensity, subcellular localization (nuclear and/or cytoplasmic), and localization within the tumor (central and/or peripheral at the edge of the tumor). We employed a 4 tiered scoring system: score 0 no staining, score 1 weak staining, score 2 moderate staining, and score 3 strong staining, with 0–1 classified as low, and 2–3 as high based on our previous studies [20]. Statistical analyses using chi-square and two-tailed t test were performed, and a p value of ≤0.05.
Bioinformatics analyses of metaplastic breast carcinoma proteomics data
We analyzed the expression of EZH2 protein and reported pEZH2 T367-binding protein partners by analyzing a comprehensive proteomics dataset of metaplastic breast carcinomas recently generated by our group [21]. We employed gene ontology (GO) over-representation tests (GO annotations: biological process, molecular function, cellular compartment, protein domain) in PANTHER (v14.1) and STRING (v11.0) databases to perform enrichment analyses, identify protein-protein networks, and to elucidate the top GO biological processes associated with the EZH2 pathway proteins expressed in metaplastic breast carcinomas.
For patient-stratified analyses of protein expression in individual metaplastic tumors in the proteomics dataset, we employed one-way ANOVA statistical analysis between multiple groups. For all tests, a p value of <0.05 was considered statistically significant.
TCGA analysis
Correlation analysis between EZH2 and EZH2 cytoplasmic-binding proteins that we identified in our previous studies [17, 20, 22, 23] (SYNE2, APS8, MLPH, MYO1F, VCL,DBNL, EPS8L2, MAPK13, MAPK14, and AKT1) was performed by DSC and SV using RNA-seq data from The Cancer Genome Atlas (TCGA). TCGA assembler [24] was used to download clinical patient data and Level 3 RNA-seq data (scaled estimate for each sample) related to primary invasive breast carcinomas (BRCA). Transcript per million (TPM) value for each gene was obtained by multiplying the scaled estimate by 1×106. TPM values related to metaplastic carcinoma of the breast (n=9) were considered for correlation analysis. Pearson correlation analyses was performed using R. The scatter plots were generated using “ggpubr” (https://cran.r-project.org/web/packages/ggpubr/index.html) R package.
Results
pEZH2 T367 protein is differentially upregulated in the cytoplasm of squamous metaplastic carcinomas compared to the spindle and mesenchymal subtypes
The 35 metaplastic carcinomas in this study consisted of 17 squamous, 10 mesenchymal, and 8 spindle subtypes. All patients were female, with a median age of 54 years old, 10 patients developed lymph node metastasis, and 7 distant metastases, most frequent to the lung and pleura (4/7). Clinical and pathological features are in Table 1.
Table 1.
Summary of clinical and pathological information on the 35 patients with metaplastic carcinoma of the breast included in this study
| Characteristics | |
|---|---|
| No. of patients | 35 |
| Median age | 54 |
| Pathologic stage, n (%) | |
| I/II | 22 (63%) |
| III/IV | 8 (23%) |
| Unknown | 5 (14%) |
| Median tumor size, cm, (range) | 2.5 (0.5–10.5) |
| Estrogen Receptor, n (%) | |
| Negative | 30 (86%) |
| Positive | 1 (3%)* |
| Unknown | 4 (11%) |
| Progesterone Receptor n (%) | |
| Negative | 31 (89%) |
| Positive | 0 (0%) |
| Unknown | 4 (11%) |
| HER2/neu overexpression, n (%) | |
| Negative | 25 (71%) |
| Positive | 0 (0%) |
| Unknown | 10 (29%) |
| Lymph node metastasis, n(%) and subtype | |
| Negative | 20 (57%) |
| Positive | 10 (29%), 9 squamous, 1 chondroid |
| Unknown | 5 (14%) |
| Site of distant metastasis, n and subtype | |
| Lung and pleura | 4 – 2 squamous, 2 spindled |
| Vertebra | 1 - Squamous |
| CNS | 1 - Squamous |
| Soft tissue (neck) | 1 - Squamous |
| Predominant metaplastic component, n(%) | |
| Squamous | 17 (49%) |
| Spindle | 8 (23%) |
| Mesenchymal | 10 (28%) |
| Osseous | 1 (3%) |
| Chondroid | 9 (26%) |
| Histologic grade, n (%) | |
| 1 | 0 (0) |
| 2 | 6 (17) |
| 3 | 26 (74) |
| Unknown | 3 (9) |
ER positive cells were approximately 5% tumor cells
pEZH2 T367 protein is upregulated in 25 (71.4%) and low in 10 (28.6%) of the metaplastic carcinomas. Of the 25 tumors with upregulated pEZH2 T367 expression, 17 (68%) tumors display low nuclear and high cytoplasmic expression, 5 (29%) exhibit high nuclear and low cytoplasmic, and 3 (12%) have high expression in the nucleus and in the cytoplasm.
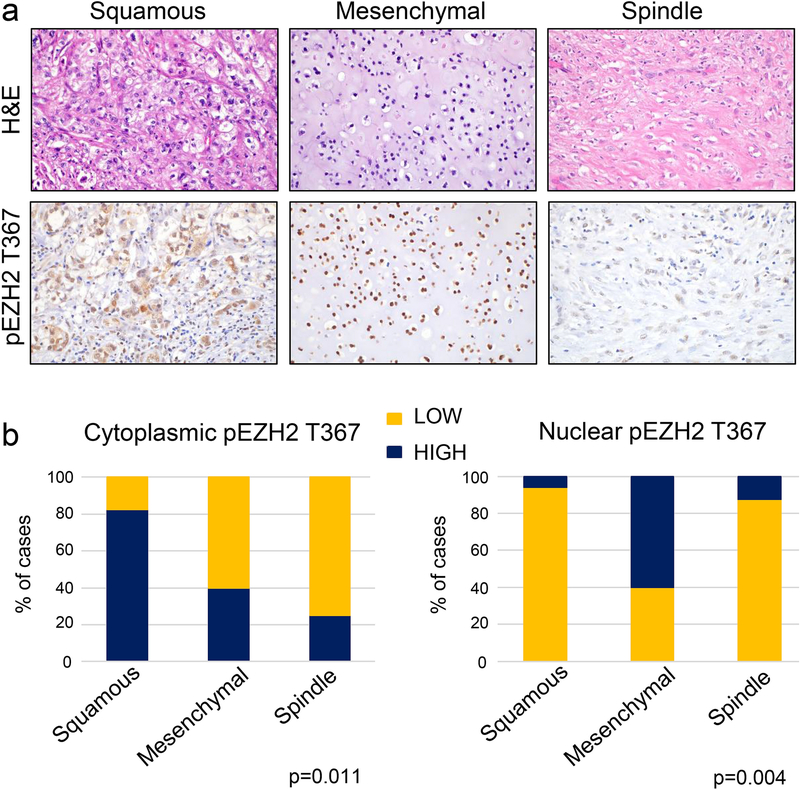
We found that high cytoplasmic pEZH2 T367 is significantly associated with squamous (14 of 17, 82%) compared to mesenchymal (4 of 10, 40%) and spindle (2 of 6, 33%) histological subtypes (p=0.011). In contrast, high nuclear pEZH2 T367 is significantly associated with mesenchymal histology (6 of 10, 60%), compared with spindle (1 of 8, 12.5%) and squamous tumors (1 of 17, 6%) (p=0.004). The majority of spindle metaplastic carcinomas showed low pEZH2 T367 in the nucleus and in the cytoplasm (Fig. 1a–b). The relationship between pEZH2 T367 expression in the nucleus and the cytoplasm with the histological subtype of metaplastic carcinoma is summarized in Table 2. Taken together, these data show that the histological subtypes of metaplastic carcinoma exhibit distinct patterns of pEZH2 T367 expression and subcellular localization.
Figure 1. pEZH2 T367 is differentially expressed in the nucleus and/or the cytoplasm of metaplastic carcinoma histopathological subtypes.
a. Representative images of metaplastic carcinomas including squamous subtype with cytoplasmic pEZH2 T367, mesenchymal subtype with chondroid differentiation and nuclear expression of pEZH2 T367, and spindle subtype with low nuclear and cytoplasmic pEZH2 T367. b. The bar graphs show quantification of the immunostaining results of 35 metaplastic carcinomas, revealing statistically significant differences in pEZH2 T367 subcellular localization according to the histological subtype.
Table 2.
Nuclear and cytoplasmic pEZH2 T367 expression in relationship to histological subtype of metaplastic carcinoma
| Histological Subtype (n) | NHigh /Chigh | NHigh /CLow | NLow /CHigh | NLow /CLow |
|---|---|---|---|---|
| Squamous (17) | 1 (5.9%) | 0 | 13 (76.4%) | 3 (17.6%) |
| Sarcomatoid (10) | 2 (20%) | 4 (40%) | 2 (20%) | 2 (20%) |
| Spindle (8) | 0 | 1 (12.5%) | 2 (25%) | 5 (62.5%) |
p=0.002
Cytoplasmic pEZH2 T367 is increased at the invasive front of metaplastic carcinomas and is associated with the presence of lymph node metastasis
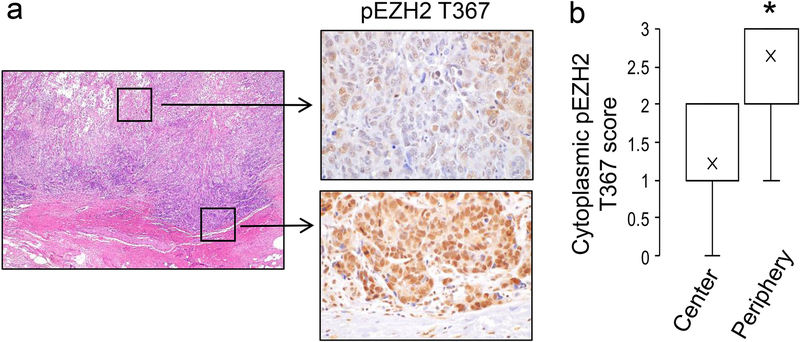
Our previous studies showing that pEZH2 T367 expression in the cytoplasm enhances breast cancer migration and invasion [20], led us to hypothesize that the phosphorylation event may be more frequent in the cancer cells at the invasive front of the tumor. Towards this, we analyzed pEZH2 T367 staining at the leading edge of each metaplastic carcinoma in whole tissue sections, focusing mainly in the cytoplasmic pEZH2 T367 expression. Of the 34 sections with available tumor-normal interface, 25 (73%) showed accentuated cytoplasmic pEZH2 T367 at the infiltrative edge. These included 12 squamous, 9 mesenchymal, and 4 spindle subtypes. The average intensity score of cytoplasmic pEZH2 T367 was significantly higher compared to the center of the tumor (average intensity score of 2.7 vs. 1.2 at the periphery and center, respectively, p value <0.001) (Fig. 2a–b). Although 25 of 34 metaplastic carcinomas demonstrated peripheral accentuation, 8 still demonstrated overall low cytoplasmic expression of pEZH2 T367 (value <2).
Figure 2. Metaplastic carcinomas show accentuated cytoplasmic pEZH2 T367 at the infiltrative edge compared to the center of the tumor.
a. Representative images of a metaplastic carcinoma with squamous differentiation demonstrating upregulated expression of cytoplasmic and nuclear pEZH2 T367 at the infiltrative edge, compared to the low expression in the center of the tumor (H&E 2x, pEZH2 T367 40x). b. Bar graph shows quantification of the staining intensity for cytoplasmic pEZH2 T367 at the center and the periphery of whole sections of 25 tumors with available tumor-normal interface (average intensity score 2.7 vs. 1.2 at the periphery and center, respectively, p<0.001).
Lymph node metastasis developed in 10 of 30 cases (33%) with available clinical information. We found that metaplastic carcinomas with squamous differentiation had a significantly higher frequency compared to spindle and mesenchymal (9/16, 56% compared to 0/6 (0%) and 1/8 (12.5%), p=0.016). High cytoplasmic pEZH2 T367 was significantly associated with lymph node metastasis, detected in 9 of 10 tumors with lymph node disease (p=0.05). Distant metastases developed in 5 of 15 (33.3%) squamous and in 2 of 8 (25%) spindled metaplastic carcinomas. No distant metastases were recorded in the mesenchymal tumors for the available follow up period. Although high cytoplasmic pEZH2 T367 was observed in 5 of 7 (71%) of tumors that metastasized to distant sites, this association did not reach statistical significance.
Metaplastic carcinomas display a predicted EZH2 protein–protein interaction network of nuclear and cytoplasmic partners
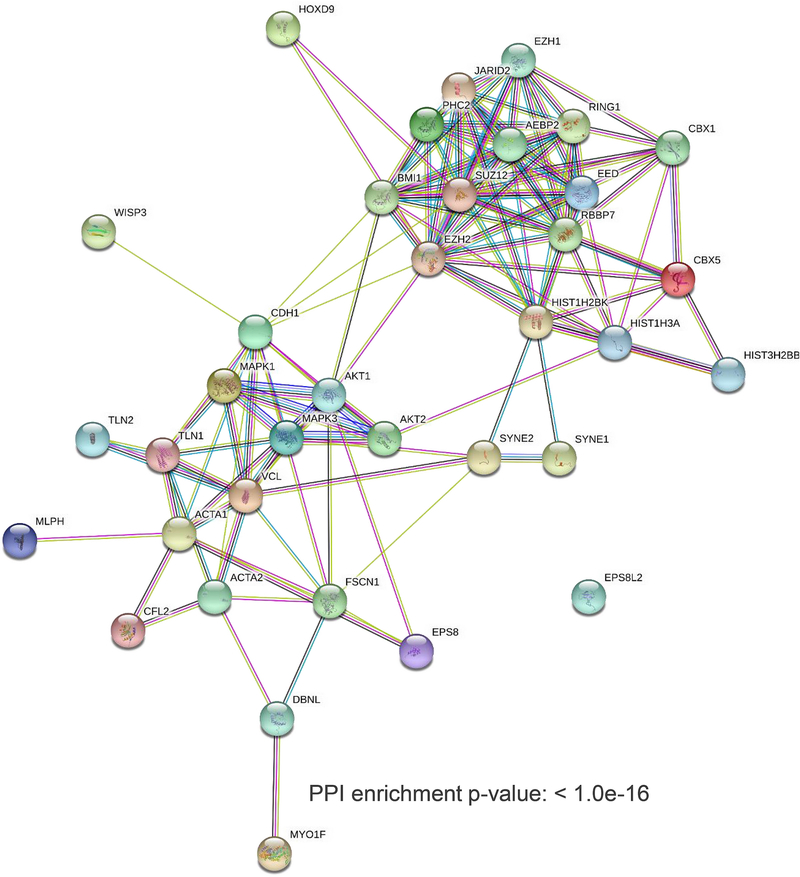
Based on the above results, we set out to investigate whether EZH2 is predicted to interact with cytoplasmic proteins in metaplastic carcinomas. Leveraging a robust bioinformatics approach, we interrogated our recently generated quantitative proteomics profile of metaplastic carcinomas consisting of 5798 individual proteins [21] for expression of well-established EZH2 nuclear and cytoplasmic interacting proteins. The latter include the pEZH2 T367 cytoplasmic binding proteins that we have reported using MDA-MB-231 TNBC cells, which recapitulate the basal-like and EMT features of metaplastic carcinomas [20], as well as those reported in other studies[22, 23, 25]. As shown in Fig. 3, metaplastic carcinomas display a predicted enriched EZH2 protein network with high significance that includes nuclear as well cytoplasmic proteins (PPI enrichment p value<0.1e-16). Enrichment analysis demonstrated that the top 15 altered pathways associated with the network proteins include nuclear processes such as epigenetic regulation, chromatin organization, and transcription, as well as cytoplasmic pathways including cell-cell junction organization assembly, cytoskeletal organization, and caveolin-mediated endocytosis (Table 3). Supporting these findings, analysis of the nine metaplastic carcinomas available in the TCGA databases show a significant positive correlation between EZH2 mRNA and mRNA expression of MAPK13 (p38δ isoform), which exhibits a predominant cytoplasmic localization in breast cancer (Pearson CC=0.94, P value=0.00012), as well as positive but non-statistically significant correlations with EPS8, EPS8L2, MAPK14, TLN1, SYNE2, MLPH, and AKT1 (Supplementary Figure 1).
Figure 3. Predicted interaction network of EZH2 nuclear and cytoplasmic interacting proteins in metaplastic carcinomas.
Enrichment analysis in STRING database (v11.0) of a recently reported quantitative proteomics landscape of metaplastic carcinomas [21] predicts the presence of EZH2 nuclear and cytoplasmic networks of EZH2 based on topological features from protein-protein interactions (p value < 1.0e-16). Selection of nuclear and cytoplasmic proteins were based on the literature [31, 32] and our recent study [20], respectively. Average clustering coefficients for canonical (0.84), non-canonical (0.63) and both clusters (0.66) represent a correlation representative of distinct EZH2 mechanisms.
Table 3.
Enrichment analyses with biological functions of top 15 altered biological pathways (REACTOME).
| Top GO Biological Process | P value |
|---|---|
| Regulation of cell cycle process, G0 to G1 transition | 3.8E-09 |
| Epigenetic regulation | 3.18E-08 |
| Chromatin organization | 5.75E-07 |
| Organelle and cellular component organization | 5.12E-07 |
| Gland development | 7.61E-05 |
| Regulation of transcription, DNA-templated | 7.9E-05 |
| Regulation of RNA metabolic process | 0.00019 |
| Cell-cell junction organization and assembly | 0.00073 |
| Mesenchyme migration | 0.0019 |
| Caveolin-mediated endocytosis | 0.0019 |
| Cytoskeletal organization and anchoring at nuclear membrane | 0.0028 |
| Actin filament organization and support | 0.0039 |
| Mammary gland epithelial cell differentiation | 0.0064 |
| Regulation of cell migration | 0.0155 |
| Stem cell differentiation | 0.014 |
Patient-stratified analysis of quantitative proteomics reveals differential expression of cytoplasmic EZH2-binding partners in metaplastic carcinoma subtypes
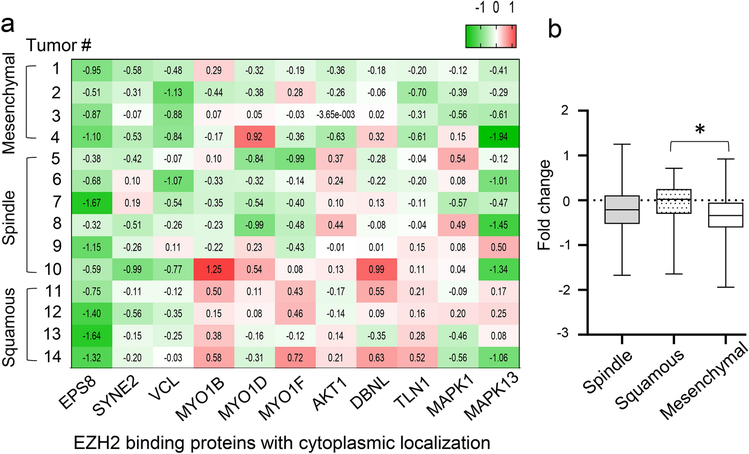
Analysis of the expression of cytoplasmic EZH2 interacting proteins in the 14 individual tumors subjected to quantitative proteomics showed upregulation of cytoplasmic pEZH2 T367 binding partners in metaplastic carcinomas with squamous differentiation compared to tumors with mesenchymal elements (p value=0.002) (Figure 4a–b). Among the upregulated EZH2 cytoplasmic interactors are the actin binding proteins MYO1B, MYO1D, MYO1F, DBNL, and TLN1 involved in cytoskeletal organization, cell migration and adhesion [26], and the oncogenic signal transduction proteins AKT1 and MAPK13 (p38δ).
Figure 4. Expression of EZH2 cytoplasmic interacting proteins in metaplastic carcinomas using unsupervised protein expression analysis.
a. Heatmap showing increased expression of reported EZH2 cytoplasmic interacting proteins in squamous compared to mesenchymal subtypes of metaplastic carcinoma. The heatmaps shows fold change values of each patient relative to the average of normal breast tissues. Key markers for each hallmark signature have been previously reported [20, 25, 33–38]. b. Bar graph shows quantification of fold change in protein expression according to histological metaplastic carcinoma subtype. One-way ANOVA statistical analyses were performed between each subtype, where p<0.05 was considered significant (*).
Discussion
Metaplastic breast carcinomas exhibit defining histopathological features, but the underlying molecular alterations are far from understood. At present, these tumors remain a significant clinical challenge with unfavorable prognosis and no targeted therapies [5, 7, 27]. Our study provides evidence that pEZH2 T367 is upregulated in the nucleus and cytoplasm of metaplastic carcinomas, and that cytoplasmic pEZH2 T367 upregulation is a feature of tumors with squamous differentiation compared to the other subtypes, and is associated with lymph node metastasis.
While the cell of origin of metaplastic carcinomas of the breast is still unclear, their differentiation along different cell types (e.g. spindle, squamous, chondroid, osseous) and aggressive clinical course suggest that alterations in cell type identity and of pathways that regulate cell migration and invasion may be important in these tumors. Our laboratory has recently reported a quantitative proteomic landscape of metaplastic breast carcinomas, with an important observation being that compared to non-metaplastic TNBCs, metaplastic carcinoma subtypes share deregulated profiles including epithelial-to-mesenchymal transition, extracellular matrix, and metabolic pathways. We identified that the subtypes exhibit distinct upregulated profiles, including translation and ribosomal proteins in spindle, inflammation and apical junction-related proteins in squamous, and extracellular matrix proteins in mesenchymal tumors [21]. These data suggest that while metaplastic carcinomas may share initial molecular events, each subtype may have a unique and active differentiation program.
One of the main regulators of cell differentiation across species is the Polycomb group protein EZH2. EZH2 possesses the enzymatic activity of the Polycomb Repressive Complex 2 (PRC2) leading to trimethylation of histone H3 to induce transcriptional repression of target genes. Studies have demonstrated that upregulation of EZH2 has tumorigenic functions in breast cancer, especially in TNBC by enhancing initiation, proliferation, invasion and metastasis [13, 22, 23]. Recently, our lab has reported that EZH2 may undergo T367 phosphorylation by p38 MAPK in a subset of TNBC tumors, which initiates a non-canonical cytoplasmic pathway by which pEZH2 T367 binds to cytoplasmic proteins and mediates migration, invasion, and metastasis in mouse models [20]. In this study, we found that 71.4% of metaplastic carcinomas display upregulation of pEZH2 T367 in the nucleus and/or in the cytoplasm, and that the subcellular localization of pEZH2 T367 expression varies according to the differentiation of the metaplastic component. Specifically, tumors with squamous differentiation are significantly more likely to have the combination of high cytoplasmic/low nuclear pEZH2 T367, while mesenchymal tumors exhibit frequent low cytoplasmic/high nuclear, and spindle tumors show low cytoplasmic/low nuclear pEZH2 T367.
Studies have suggested that the histological subtypes of metaplastic breast carcinomas may have different clinical behavior, with high-grade spindle tumors having a worse prognosis than those with mesenchymal differentiation or production of cartilaginous and or osseous matrix [2, 8, 9, 11, 28, 29]. Leyrer and colleagues reported that the majority of metaplastic carcinomas with lymph node metastasis had squamous differentiation [30]. Despite the small number of cases, of the 25 patients with available follow-up information, 10 had lymph node metastasis, 9 of which were squamous and 1 mesenchymal. We found that high cytoplasmic pEZH2 T367 was significantly associated with lymph node metastasis. Consistent with the function of pEZH2 T367 in migration and invasion that we identified in laboratory studies [20], we found that pEZH2 T367 expression is accentuated at the advancing front of the metaplastic carcinomas compared to the center of the tumors.
The relevance of cytoplasmic EZH2 expression and function is further supported by analyses of our proteomics landscape data in this study. Using an independent quantitative proteomics dataset generated in our lab using tandem mass tag (TMT) based proteomics on 14 human metaplastic carcinomas, we identified that EZH2 and a network of known interacting proteins are enriched in metaplastic carcinomas, and that cytoplasmic binding partners are specially upregulated in tumors with squamous differentiation. Pathway analyses revealed that metaplastic carcinomas exhibit deregulation of pathways involved in cell differentiation and in cytoskeletal organization and migration.
A limitation of this study is the small sample size, reflecting the infrequent presentation of metaplastic carcinomas. However, our cohort of 35 tumors allowed investigation of the association between pEZH2 T367 and clinical and pathological features. The independent group of 14 metaplastic carcinomas subjected to quantitative proteomics provided additional support for the presence of pEZH2 T367 cytoplasmic interacting proteins in squamous metaplastic carcinomas compared to mesenchymal tumors. In summary, our data suggest that expression and localization of pEZH2 T367 differs according to the differentiation of the metaplastic component and is associated with lymph node metastasis. These findings suggest a subtype specific role for pEZH2 T367 with diagnostic and clinical implications.
Supplementary Material
Supplementary Figure 1. Pearson correlation analysis between EZH2 and EZH2 cytoplasmic binding proteins in TCGA metaplastic carcinoma of breast. Gene expression values (TPM) in nine metaplastic carcinoma samples was used for the analysis. “ggpubr” R package was used to generate scatter plots showing Pearson correlation coefficient and p value from correlation test.
Acknowledgements.
We thank members of the Kleer lab for helpful discussions during the execution of the study.
Funding: This work was supported by National Institute of Health (NIH) grants R01CA125577 and R01CA107469 (C.G.K.), and the University of Michigan Rogel Cancer Center support grant P30CA046592.
Footnotes
Conflict of interest: Emily McMullen declares that she has no conflict of interest. Stephanie Skala declares that she has no conflict of interest. Maria Gonzalez declares that she has no conflict to interest. Sabra Djomehri declares that she has no conflict of interest, Celina Kleer declares that she has no conflict of interest.
Compliance with ethical standards:
Ethical approval: This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent: This article does not involve studies in human participants.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Oberman HA. Metaplastic carcinoma of the breast. A clinicopathologic study of 29 patients. Am J Surg Pathol. 1987;11(12):918–29. doi: 10.1097/00000478-198712000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Rakha EA, Tan PH, Varga Z, Tse GM, Shaaban AM, Climent F, et al. Prognostic factors in metaplastic carcinoma of the breast: a multi-institutional study. Br J Cancer. 2015;112(2):283–9. doi: 10.1038/bjc.2014.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinnon E, Xiao P. Metaplastic carcinoma of the breast. Arch Pathol Lab Med. 2015;139(6):819–22. doi: 10.5858/arpa.2013-0358-RS. [DOI] [PubMed] [Google Scholar]
- 4.Pang J, Toy KA, Griffith KA, Awuah B, Quayson S, Newman LA, et al. Invasive breast carcinomas in Ghana: high frequency of high grade, basal-like histology and high EZH2 expression. Breast Cancer Res Tr. 2012;135(1):59–66. doi: 10.1007/s10549-012-2055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pezzi CM, Patel-Parekh L, Cole K, Franko J, Klimberg VS, Bland K. Characteristics and treatment of metaplastic breast cancer: analysis of 892 cases from the National Cancer Data Base. Ann Surg Oncol. 2007;14(1):166–73. doi: 10.1245/s10434-006-9124-7. [DOI] [PubMed] [Google Scholar]
- 6.Song Y, Liu X, Zhang G, Song H, Ren Y, He X, et al. Unique clinicopathological features of metaplastic breast carcinoma compared with invasive ductal carcinoma and poor prognostic indicators. World J Surg Oncol. 2013;11:129. doi: 10.1186/1477-7819-11-1291477-7819-11-129 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hilli Z, Choong G, Keeney MG, Visscher DW, Ingle JN, Goetz MP, et al. Metaplastic breast cancer has a poor response to neoadjuvant systemic therapy. Breast Cancer Res Treat. 2019;176(3):709–16. doi: 10.1007/s10549-019-05264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Zein D, Hughes M, Kumar S, Peng X, Oyasiji T, Jabbour H, et al. Metaplastic Carcinoma of the Breast Is More Aggressive Than Triple-negative Breast Cancer: A Study From a Single Institution and Review of Literature. Clin Breast Cancer. 2017;17(5):382–91. doi: S1526–8209(16)30556–0 [pii] 10.1016/j.clbc.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han M, Salamat A, Zhu L, Zhang H, Clark BZ, Dabbs DJ, et al. Metaplastic breast carcinoma: a clinical-pathologic study of 97 cases with subset analysis of response to neoadjuvant chemotherapy. Mod Pathol. 2019;32(6):807–16. doi: 10.1038/s41379-019-0208-x. [DOI] [PubMed] [Google Scholar]
- 10.Lee H, Jung SY, Ro JY, Kwon Y, Sohn JH, Park IH, et al. Metaplastic breast cancer: clinicopathological features and its prognosis. J Clin Pathol. 2012;65(5):441–6. doi: 10.1136/jclinpath-2011-200586 jclinpath-2011–200586 [pii]. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi R, Horii R, Maeda I, Suga S, Makita M, Iwase T, et al. Clinicopathologic study of 53 metaplastic breast carcinomas: their elements and prognostic implications. Hum Pathol. 2010;41(5):679–85. doi: 10.1016/j.humpath.2009.10.009 S0046–8177(09)00374–8 [pii]. [DOI] [PubMed] [Google Scholar]
- 12.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. Embo J. 1997;16(11):3219–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100(20):11606–11. doi: 10.1073/pnas.1933744100 1933744100 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–9. [DOI] [PubMed] [Google Scholar]
- 15.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–34. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez ME, Li X, Toy K, DuPrie M, Ventura AC, Banerjee M, et al. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28(6):843–53. doi: 10.1038/onc.2008.433onc2008433 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13(1):87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie L, Wei Y, Zhang F, Hsu YH, Chan LC, Xia W, et al. CDK2-mediated site-specific phosphorylation of EZH2 drives and maintains triple-negative breast cancer. Nat Commun. 2019;10(1):5114. doi: 10.1038/s41467-019-13105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anwar T, Arellano-Garcia C, Ropa J, Chen YC, Kim HS, Yoon E, et al. p38-mediated phosphorylation at T367 induces EZH2 cytoplasmic localization to promote breast cancer metastasis. Nat Commun. 2018;9(1):2801. doi: 10.1038/s41467-018-05078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djomehri SI, Gonzalez ME, da Veiga Leprevost F, Tekula SR, Chang HY, White MJ, et al. Quantitative proteomic landscape of metaplastic breast carcinoma pathological subtypes and their relationship to triple-negative tumors. Nat Commun. 2020;11(1):1723. doi: 10.1038/s41467-020-15283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore HM, Gonzalez ME, Toy KA, Cimino-Mathews A, Argani P, Kleer CG. EZH2 inhibition decreases p38 signaling and suppresses breast cancer motility and metastasis. Breast Cancer Res Treat. 2013;138(3):741–52. doi: 10.1007/s10549-013-2498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez ME, DuPrie ML, Krueger H, Merajver SD, Ventura AC, Toy KA, et al. Histone methyltransferase EZH2 induces Akt-dependent genomic instability and BRCA1 inhibition in breast cancer. Cancer Res. 2011;71(6):2360–70. doi: 10.1158/0008-5472.CAN-10-1933 71/6/2360 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11(6):599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunawan M, Venkatesan N, Loh JT, Wong JF, Berger H, Neo WH, et al. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol. 2015;16(5):505–16. doi: 10.1038/ni.3125ni.3125 [pii]. [DOI] [PubMed] [Google Scholar]
- 26.Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133–55. doi: 10.1146/annurev-cellbio-100809-151502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMullen ER, Zoumberos NA, Kleer CG. Metaplastic Breast Carcinoma: Update on Histopathology and Molecular Alterations. Arch Pathol Lab Med. 2019;143(12):1492–6. doi: 10.5858/arpa.2019-0396-RA. [DOI] [PubMed] [Google Scholar]
- 28.Carter MR, Hornick JL, Lester S, Fletcher CD. Spindle cell (sarcomatoid) carcinoma of the breast: a clinicopathologic and immunohistochemical analysis of 29 cases. Am J Surg Pathol. 2006;30(3):300–9. doi: 10.1097/01.pas.0000184809.27735.a1 00000478–200603000-00002 [pii]. [DOI] [PubMed] [Google Scholar]
- 29.Chuthapisith S, Warnnissorn M, Amornpinyokiat N, Pradniwat K, Angsusinha T. Metaplastic carcinoma of the breast with transformation from adenosquamous carcinoma to osteosarcomatoid and spindle cell morphology. Oncol Lett. 2013;6(3):728–32. doi: 10.3892/ol.2013.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leyrer CM, Berriochoa CA, Agrawal S, Donaldson A, Calhoun BC, Shah C, et al. Predictive factors on outcomes in metaplastic breast cancer. Breast Cancer Res Treat. 2017;165(3):499–504. doi: 10.1007/s10549-017-4367-5. [DOI] [PubMed] [Google Scholar]
- 31.Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. 2018;6:10. doi: 10.1186/s40364-018-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raaphorst FM, Meijer CJ, Fieret E, Blokzijl T, Mommers E, Buerger H, et al. Poorly differentiated breast carcinoma is associated with increased expression of the human polycomb group EZH2 gene. Neoplasia. 2003;5(6):481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Cruz CC, Kirmizis A, Simon MD, Isono K, Koseki H, Panning B. The polycomb group protein SUZ12 regulates histone H3 lysine 9 methylation and HP1 alpha distribution. Chromosome Res. 2007;15(3):299–314. doi: 10.1007/s10577-007-1126-1. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Li B, Dong C, Wang Y, Li Q. 20(S)-Ginsenoside Rh2 suppresses proliferation and migration of hepatocellular carcinoma cells by targeting EZH2 to regulate CDKN2A-2B gene cluster transcription. Eur J Pharmacol. 2017;815:173–80. doi: 10.1016/j.ejphar.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21(4):1360–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8(2):195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Warden C, Zou Z, Neman J, Krueger JS, Jain A, et al. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One. 2013;8(11):e80970. doi: 10.1371/journal.pone.0080970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian X, Pelton A, Shahsafaei A, Dorfman DM. Differential expression of enhancer of zeste homolog 2 (EZH2) protein in small cell and aggressive B-cell non-Hodgkin lymphomas and differential regulation of EZH2 expression by p-ERK1/2 and MYC in aggressive B-cell lymphomas. Mod Pathol. 2016;29(9):1050–7. doi: 10.1038/modpathol.2016.114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Pearson correlation analysis between EZH2 and EZH2 cytoplasmic binding proteins in TCGA metaplastic carcinoma of breast. Gene expression values (TPM) in nine metaplastic carcinoma samples was used for the analysis. “ggpubr” R package was used to generate scatter plots showing Pearson correlation coefficient and p value from correlation test.