Abstract
The proper function of the innate immune system depends on an intricate network of regulation that promotes effective responses to pathogens while avoiding autoimmunity. Circular RNAs (circRNAs), a class of RNAs that lack 5’ and 3’ ends, have emerged as key actors in these networks. Recent studies have demonstrated that endogenous circRNAs in eukaryotes regulate the activation of innate immune proteins and cells through diverse modes of action. Some DNA viruses also encode circRNAs, and foreign circRNAs have been found to stimulate an innate immune response. This review summarizes recent investigations that reveal the critical roles that circRNAs play in innate immunity and points to future areas of study in this emerging field.
Keywords: CircRNA, Viral RNA, N6-methyladenosine, self/non-self, innate immunity
Introduction
The precise balance between the proper eradication of pathogens and the avoidance of autoimmune recognition critically depends on the regulation of gene expression in the innate immune system. Most studies thus far have focused on the roles of innate immune proteins. Pattern recognition receptors, for example, detect products of microbial invasion to initiate innate immune signaling, while cytokines are the molecules through which this signaling occur [1,2]. However, relatively few studies have investigated the functions of RNA, particularly a new class of single-stranded RNAs that are covalently closed loops, circular RNAs (circRNAs). The discovery of non-linear RNAs has increased the diversity of the transcriptome as well as expanded our understanding of the regulatory roles of RNAs during innate immunity. In this review, we will present recent studies of the identification and characterization of circRNAs during pathogenic infection and the innate immune response and discuss the additional layer of regulation on circRNAs by RNA modifications.
Properties of CircRNAs
CircRNAs were initially thought to be products of aberrant splicing. However, in the past decade, advances in next-generation sequencing technologies and computational approaches have disrupted this presumption and established that the biogenesis of circRNAs is a carefully regulated process. CircRNA formation occurs via “back-splicing” in which the cellular spliceosomal machinery joins a downstream splice donor to an upstream splice acceptor [3–6]. Complementarity between introns that flank circularizing exons and RNA-binding proteins bring portions of the precursor mRNAs into close proximity to facilitate circRNA formation [3–6]. The site where the 3’ and 5’ ends join together forms the back-splice junction, the sole differentiating sequence between circRNAs and their linear counterparts. Back-splicing to generate circRNAs occurs in over 10% of transcripts in the human genome [7,8]. Some circRNAs are evolutionarily conserved between species, and others exhibit cell-type specific expression [7–9]. Most research has focused on endogenous circRNAs in eukaryotes, but circRNAs can also originate exogenously from viral genomes [10–14].
CircRNAs tend to share a set of general characteristics. They are predominantly exonic [7,8], though some consist of only introns or a mixture of both [15,16]. The lack of ends on circRNAs protects them from exonucleases, and thus, they exhibit much greater stability than linear RNAs [17]. Despite these defined physical properties, the functions of most circRNAs remain unclear, and many poorly expressed circRNAs are hypothesized to be transcriptional noise. However, numerous cellular and viral circRNAs have been shown to play significant roles in regulating gene expression and protein activity. As an additional layer of gene regulation, circRNAs can “sponge” microRNAs (miRNAs) through competitive binding to block miRNA interactions with mRNAs and long non-coding RNAs (lncRNAs) [18–20]. CircRNAs also directly regulate protein activation by docking in the active sites of RNA-binding proteins [21–24]. CircRNAs have additionally been shown to modulate mRNA stability [25], and several circRNAs are themselves translated to produce novel proteins [12,26]. Oftentimes, these diverse circRNA functions impact genes, proteins, and cells involved in innate immunity, implicating circRNAs as important actors in the innate immune response (Figure 1).
Figure 1:
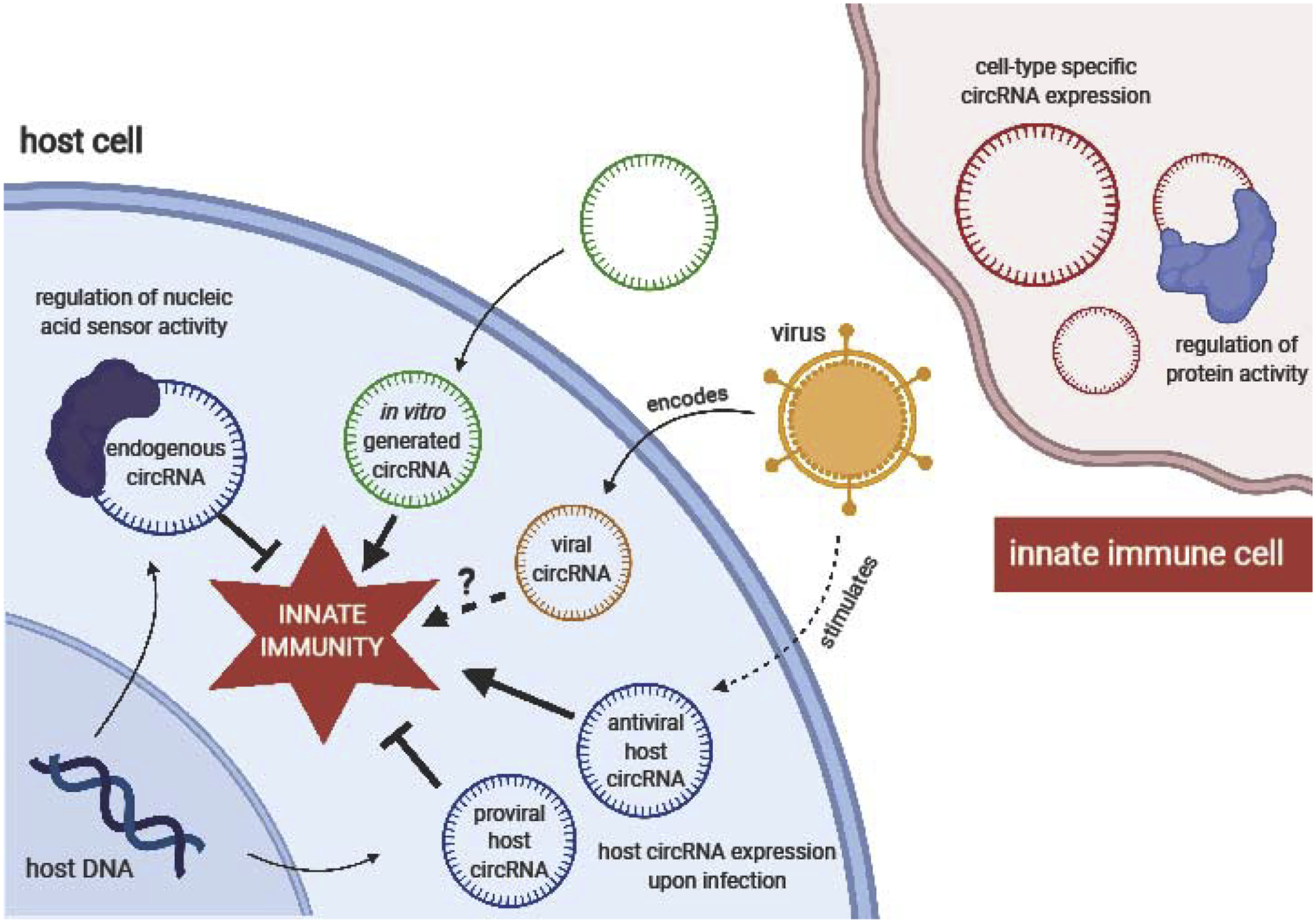
CircRNA functions in innate immunity. Innate immune cells exhibit cell-type specific circRNA expression, and some innate immune cell circRNAs have demonstrated roles in regulating protein function. In other cell types, endogenous circRNAs globally inhibit the activity of innate immune nucleic acid sensors, and specific endogenous circRNAs are expressed during viral infection with either inhibitory or promoting effects on innate immunity. Viruses themselves encode circRNAs, though their impacts on innate immunity are unclear. Foreign circRNAs generated by in vitro transcription stimulate an innate immune response.
Endogenous circRNAs in innate immune cells
CircRNAs drive innate immune cell differentiation and exhibit cell-type specific expression
Productive immunity requires the generation of specialized immune cell types from hematopoietic stem cells (HSCs). This process occurs during hematopoeisis, where tight regulation ensures that stem cells balance differentiation with self-renewal in order to maintain a continuous supply of innate immune cells throughout a lifetime. CircRNAs are an integral part of the regulatory system: a circular RNA antagonist for cGAS (cia-cGAS) sustains HSC dormancy by suppressing activity of the innate immune sensor cyclic GMPAMP synthase (cGAS) [22]. In the absence of cia-cGAS competitive binding, cGAS senses genomic DNA and induces type I interferon production that stimulates the differentiation of long-term HSCs into downstream cell types such as multipotent progenitor cells, which are precursors to innate immune cells [22] (Figure 2A).
Figure 2:
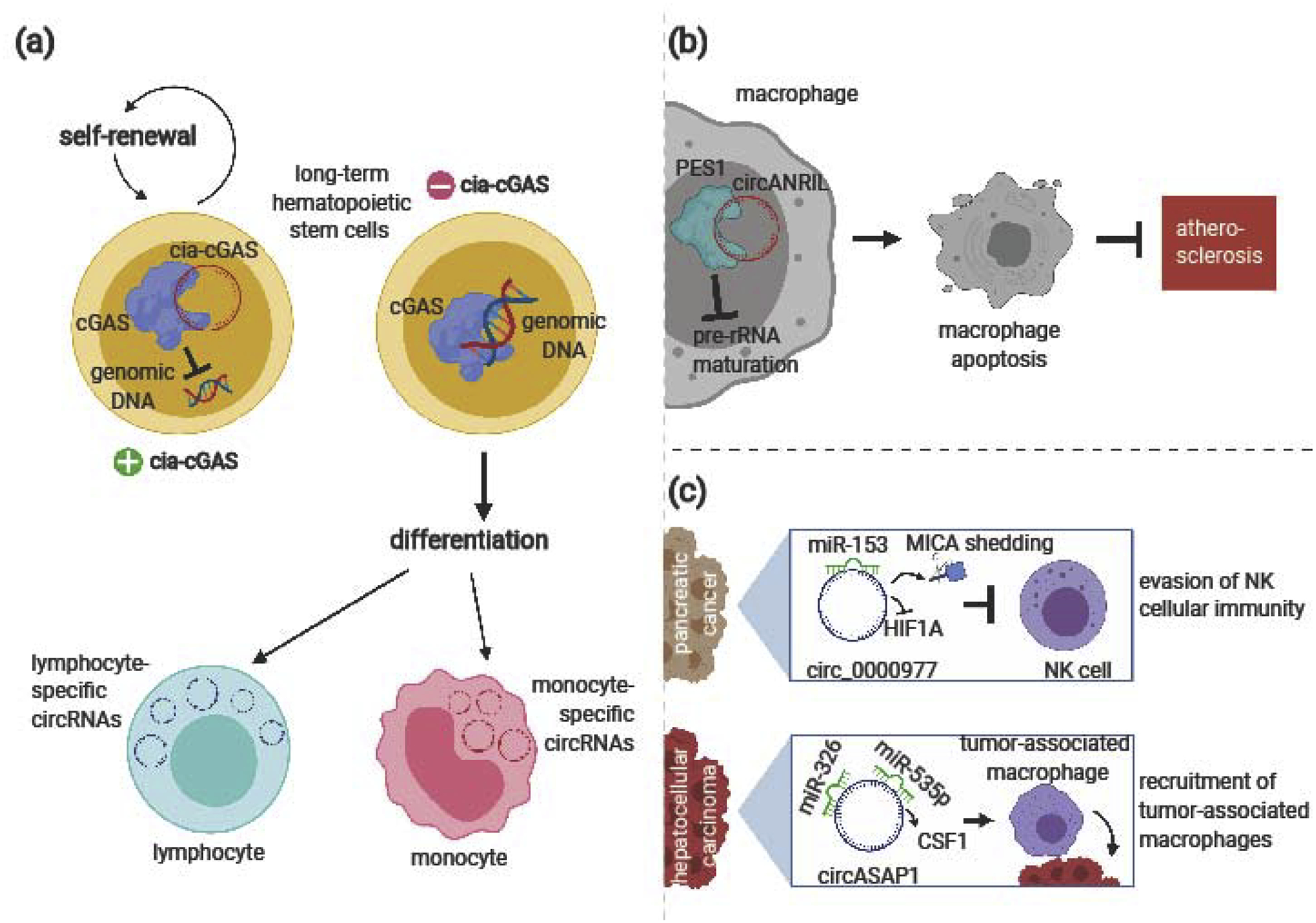
CircRNA roles in the development and regulation of innate immune cells. (a) CircRNAs control innate immune cell development and exhibit cell-type specific expression. In long-term hematopoietic stem cells, cia-cGAS maintains self-renewal to prevent exhaustion throughout a lifetime. Upon differentiation, immune cells exhibit cell-type specific circRNA expression profiles. (b) In macrophages, circANRIL binds PES1 to block pre-rRNA maturation, triggering apoptosis. This prevents atherosclerosis driven by excess macrophage proliferation. (c) Specific miRNA-sponging circRNAs can aid tumorigenesis by regulating innate immune cells in the tumor microenvironment. In pancreatic cancer cells, circ_0000977 blocks miR-153 activity, leading to HIF1A inhibition and MICA shedding that blocks NK cell activity. In hepatocellular carcinoma cells, circASAP1 sponges miR-326 and miR-535p, promoting CSF1 activity that recruits tumor-associated macrophages to the microenvironment.
When HSCs do differentiate, circRNAs exhibit cell-type specific expression in blood cell populations (Figure 2A). Computational analysis of RNA-seq data has revealed that undifferentiated hematopoietic progenitors, differentiated myeloid cells (such as monocytes), and differentiated lymphoid cells (such as T cells, B cells, and natural killer [NK] cells) exhibit distinctive circRNA transcriptional profiles [27]. Compared to B cells and T cells, monocytes are predicted to upregulate 438 circRNAs, many of which originate from genes associated with monocyte functions [28]. HSCs and monocyte and macrophage progenitors also differentially express circRNAs during each stage of osteoclast development, a process closely related to hematopoiesis [29]. CircRNAs have been further shown to directly impact this process. In bone marrow-derived macrophages, circRNA_28313 induces macrophage colony-stimulating factor 1 (CSF-1) to promote osteoclast differentiation [30]. CircRNA_28313 has a binding site for miR-195a, and the inhibition of miR-195a reverses the osteoclastogenic effects of circRNA_28313 silencing, suggesting that circRNA_29313 acts as an miRNA sponge [30]. While additional circRNAs have been linked to miRNAs through their correlated expression levels [29], the roles of most differentially expressed circRNAs remain an active area of investigation.
CircRNAs regulate innate immune cells that facilitate chronic disease development
In proper functioning immune systems, infection stimulates innate immune cells to initiate a cascade of cytotoxic and inflammatory activity. When dysregulated, this innate immune cell activity contributes to autoimmunity and chronic disease development. By modulating innate immune cell function and activation, individual circRNAs can therefore prevent or support the development of diseases such as atherosclerosis and cancer. Circular antisense non-coding RNA in the INK4 locus (circANRIL) is expressed in vascular tissue macrophages and may protect against atherosclerosis by preventing their uncontrolled proliferation [23]. Specifically, circANRIL binds to pescadillo ribosomal biogenesis factor 1 (PES1) to stop pre-rRNA maturation, leading to nucleolar stress, apoptosis, and decreased cell division [23] (Figure 2B). Compared to normal cells, cancerous cells aberrantly express specific circRNAs that modulate the activity of innate immune cells in the tumor microenvironment. Hypoxia-induced expression of circ_0000977 in pancreatic cancer cells enables the evasion of NK cell-mediated death [31]. Circ_0000977 suppresses the expression of hypoxia-inducible factor 1-alpha (HIF1A) and stimulates the shedding of cell surface MHC class I-related molecule A (MICA) to deactivate nearby NK cells [31]. Circ_0000977 likely acts as an miRNA sponge, as the circRNA contains one miR-153 binding site, and miR-153 inhibition attenuates the effects of circ_0000977 knockdown [31]. Moreover, in hepatocellular carcinoma cells, upregulated circASAP1 increases the expression of CSF-1 to promote the migration of tumor-associated macrophages into the microenvironment to support subsequent tumor metastasis [32]. CircASAP1 negatively regulates the activity of miR-326 and miR-535p, which otherwise reduces CSF-1 expression [32] (Figure 2C).
Endogenous circRNAs in innate immune response
Decrease in global circRNA levels stimulates innate immunity
One essential function of the innate immune system is to sense foreign pathogens and eradicate them, and numerous innate immunity proteins initiate the immune response by recognizing viral nucleic acids. Endogenous circRNAs bind to these proteins in the absence of viral infection; upon infection, endogenous circRNA levels globally decrease, freeing innate immune proteins to bind to viral RNA and therefore initiating host antiviral immunity. For example, the biogenesis of cellular circRNAs decreases during de novo viral infection due to the activity of nuclear factors 90 and 110 (NF90/NF110) [6]. In uninfected cells, the NF90/NF110 complex stabilizes the interactions between intronic complementary sequences that flank circularizing exons, facilitating nascent circRNA formation in the nucleus [6]. In the cytoplasm, NF90/NF110 also binds to mature circRNAs [6]. During vesicular stomatitis virus (VSV) or poly(I:C) treatment, nuclear NF90/NF110 translocates to the cytoplasm, reducing the global production of circRNAs [6]. Viral infection also triggers NF90/NF110 to release cytoplasmic circRNAs, freeing the protein complex to interact with and block replication of viral transcripts [6]. Accordingly, cells where individual cellular circRNAs are overexpressed experience increased VSV titer upon infection [6]. Thus, a decrease in global circRNA formation and an increase in unassociated circRNA, driven by reduced circRNA interaction with NF90/NF110, may aid NF90/NF110-mediated antiviral innate immunity [6]. Viral infection also stimulates global circRNA degradation, which induces the innate immune response. Endogenous circRNAs have secondary structures that bind to and inhibit antiviral protein kinase R (PKR), whose pathogen-associated molecular pattern is double-stranded RNA (dsRNA) [24]. In uninfected cells, the circRNA-PKR interaction prevents aberrant immune sensing by PKR. Encephalomyocarditis virus (EMCV) infection or poly(I:C) treatment activates endonuclease RNase L, which degrades circRNAs to free PKR, allowing it to sense and neutralize foreign dsRNA transcripts [24]. Thus, a decrease in global circRNA levels may represent an important step in activating NF90/NF110 and PKR binding to viral RNA in the innate immune response.
The immunosuppressive effect of circRNA interactions with innate immune proteins can be seen beyond antiviral immunity. Peripheral blood mononuclear cells (PBMCs) from patients with systemic lupus erythematosus (SLE), an autoimmune disorder, have globally decreased circRNA levels, and overexpression of endogenous circRNAs reduced the cells’ PKR activation and interferon gene expression levels [24]. Therefore, in both autoimmune disorders and viral infection, global circRNA expression is negatively associated with innate immune activation.
Individual host circRNA effects after viral and bacterial infection
In response to different environmental factors, including infection, gene expression often shifts to prime cells to respond to their changing conditions. As such, viral and bacterial infection stimulates the differential expression of host circRNAs that promote the innate immune response. The infection of human umbilical vein endothelial cells by Kaposi’s sarcoma herpesvirus (KSHV) leads to up- or downregulation of multiple circRNAs [14]. One upregulated host circRNA, hsa_circ_0001400, increases the production of a cytokine gene, tumor necrosis factor alpha (TNFα), and depletes expression of two KSHV-encoded genes, latency-associated nuclear antigen (LANA) and replication and transcription activator (RTA), which respectively regulate latent and lytic infection [14]. Hsa_circ_0001400 is computationally predicted to contain miRNA binding sites, but its mechanisms of action are unclear, and ectopic expression did not significantly alter viral genome copy number [14]. However, the ability of hsa_circ_0001400 to stimulate innate immune pathways and suppress viral genes suggests that it may possess potential antiviral functions. Host circRNAs also help initiate innate immune response to bacterial infection. Lipopolysaccharide (LPS) stimulation of macrophages upregulates circRasGEF1B, which stabilizes mature mRNA encoding intercellular adhesion molecule 1 (ICAM-1), a protein known to facilitate leukocyte recruitment to inflamed tissue [25]. By increasing its expression, circRasGEF1B may stimulate innate immunity to combat infection.
Conversely, viruses and bacteria often rely on host cell dependency factors to support pathogenesis. Altered expression of specific cellular circRNAs following infection promotes the pathogen’s life cycle by inhibiting innate immunity. Infection can upregulate circRNAs that suppress the innate immune response, as seen in the increased expression of pro-viral circPSD3 during de novo hepatitis C (HCV) infection of liver cells [33]. CircPSD3 inhibits the antiviral cellular nonsense-mediated decay (NMD) pathway, which degrades premature termination codon-containing mRNA transcripts that accumulate to promote pathogenesis [33]. Infection also leads to the suppression of circRNAs that promote innate immunity. For example, the suppression of circRNA cPWWP2A supports Mycobacterium tuberculosis (MTB) infection of primary human macrophages [34]. cPWWP2A has three binding sites for miR-579 [35], a suppressor of pro-survival genes Sirtuin 1 (SIRT1) and pyruvate dehydrogenase kinase (PDK) [34]. Knockdown of cPWWP2A led to increased miR-579 expression and triggered macrophage apoptosis [35]. Thus, depletion of cPWWP2A during MTB infection may aid pathogenesis by sponging miR-579 to reduce macrophage activity.
Many additional circRNAs are differentially expressed during viral and bacterial infection, though their impacts on immunity have not yet been elucidated. De novo MTB infection of human monocyte-derived macrophages leads to changes in the expression of numerous circRNAs beyond cPWWP2A [36]. Moreover, during infection of human immunodeficiency virus (HIV), Hantaan orthohantavirus (HTNV), or Middle East respiratory syndrome coronavirus (MERS-CoV), differentially expressed host circRNAs are associated with circRNA-miRNA-mRNA regulatory networks that are enriched in immune response and viral infection pathways [37–39]. The specific roles of these circRNAs during infection are continuing areas of investigation, but the documented effects of the previously described circRNAs suggest that some may support pathogenesis while others may stimulate innate immunity.
CircRNAs as potential biomarkers for immune diseases
CircRNAs are attractive biomarker candidates due to their high stability and high abundance in bodily fluids for non-invasive detection [40,41]. Microarray analysis of patient blood samples supports their potential application in the diagnosis of multiple immune-related diseases [40,41]. Two circRNAs implicated in the macrophage response to MTB infection were found to be differentially expressed—one upregulated and one downregulated—in PBMCs of MTB patients [36]. Similar analysis has identified putative circRNA biomarkers for autoimmune disorders: hsa_circ_0044235 was found to be downregulated in PBMCs of rheumatoid arthritis patients [42], while four differentially expressed circRNAs were identified and confirmed in the plasma of SLE patients [43]. CircRNAs may also be biomarkers for immunosenescence, as elderly patients with upregulated circular RNA100783 in their PBMCs experienced increased loss of CD28 expression, promoting CD8+ T-cell aging [44]. Though these studies on limited sample sizes require further validation, their results support a promising future application of circRNAs as biomarkers for immune-related disorders.
Foreign circRNAs in innate immunity
Viral genomes encode circRNAs
Viral genomes generate nucleic acids and proteins that facilitate viral replication and infection. The discovery of virally-encoded circRNAs adds to this landscape, though their overall impact on innate immunity remains unclear. From RNA-seq data of infected cell lines and tumors, circRNA detection software tools have identified circRNAs originating from a wide range of double-stranded DNA (dsDNA), single-stranded RNA (ssRNA), and retro-transcribing viruses [10–14,45]. The viral circRNA sequences tend to be flanked by complementary or short repeating sequences [45], which may promote their generation, and future investigations might seek to understand their use of cellular splicing and RNA binding proteins during viral circRNA formation. CircRNAs produced by dsDNA viruses such as the Epstein-Barr virus (EBV), KSHV, and human papillomaviruses (HPVs) have been experimentally validated, although the innate immune response to viral circRNAs has yet to be characterized [10–14]. Most viral circRNAs have ambiguous function, with a notable exception being HPV-encoded circE7, which is translated to produce E7, an oncoprotein [12]. Given the compactness of viral genomes, their circRNA products are expected to be significant in pathogenesis, particularly given the widespread prevalence of viral circRNAs.
In vitro-generated foreign circRNAs stimulate innate immunity
The low expression of viral circRNAs relative to their linear transcript counterparts poses barriers in the study and characterization of viral circRNAs’ specific effects. A useful tool for modeling viral circRNA functions and impacts on innate immunity may lie in engineered foreign circRNAs. Engineered foreign circRNAs are generated without cellular spliceosomal machinery through in vitro transcription of a permuted intron-exon template, derived from self-splicing phage introns [46]. Engineered foreign circRNAs stimulate innate immunity in vitro and in vivo, while circRNAs of the same sequence but generated from endogenous circularizing introns do not [46,47]. Transfection of engineered circRNAs into cells inhibited viral infection and stimulated expression of innate immunity genes such as PKR, retinoic-acid-inducible gene-I (RIG-I), and melanoma-differentiation-associated gene 5 (MDA5) [46]. RIG-I, an antiviral dsRNA sensor, detects unmodified foreign circRNAs, and undergoes a conformational change upon binding to trigger the filamentation of mitochondrial anti-viral signaling protein (MAVS) and downstream interferon production [47]. Of note, this mechanism is distinct from RIG-I’s typical activation pathway initiated upon binding with 5’-triphosphate dsRNA, its canonical pathogen-associated molecular pattern [47]. In mouse models, engineered foreign circRNAs also stimulated CD8+ T cell and B cell adaptive immunity, potentially due to signaling by circRNA-activated innate immune sentinels like dendritic cells [47]. The immunostimulatory effects of engineered foreign circRNAs suggest that native sources of foreign circRNAs, such as viral circRNAs, may generate a similar response.
Effect of m6A modification on foreign and endogenous circRNA innate immunity
Though circRNAs occupy numerous regulatory roles within cells, RNA modifications add another layer of control to determine the fates and functions of circRNAs. N6-methyladenosine (m6A) is an abundant post-transcriptional modification found on mRNAs and lncRNAs [48]. Numerous proteins work together to control m6A installation, removal, and detection, including the methyltransferase like 3 (METTL3) “writer,” the alkylation repair homolog protein 5 (ALKBH5) and fat mass and obesity-associated (FTO) “erasers,” and YTHDF1/2/3, members of the YTH (YT521-B homology) family of “readers” [49–51]. Endogenous circularizing introns undergo a protein-assisted splicing process that deposits an m6A modification on the resulting circRNA [47]. YTHDF2 binds m6A-modified circRNAs, preventing their detection by dsRNA sensor RIG-I and the subsequent stimulation of innate immunity [47]. Thus, m6A serves as a marker of “self” by labeling endogenous circRNAs to allow them to evade innate immune detection [47].
Over 1,400 endogenous circRNAs have been shown to possess m6A modifications, which are cell-type specific and often lie on exons that are not m6A-modified in their corresponding mRNAs [52]. Depletion of the m6A writer and reader proteins reveals that m6A modifications on endogenous circRNAs can regulate their cytoplasmic export, formation, and degradation [53–55] and promote translation [55,56]. Given this range of effects, it is likely that the m6A modification on endogenous circRNAs controls innate immunity beyond simply serving as a “self” label. Researchers seeking to understand this aspect of circRNA immunity may learn from the varying impacts of the m6A modification on linear RNAs during the antiviral innate immune response. Enriched m6A expression on endogenous RNAs due to ALKBH5 knockdown led to the cytoplasmic export of antiviral transcripts and increased type I interferon expression, implicating m6A in an immunostimulatory role [57]. Paradoxically, decreased m6A expression and detection following knockdowns of METTL3 and YTHDF2, respectively, stabilized mRNA of type I interferon gene IFNB and decreased viral propagation [58]. This suggests that m6A can also negatively regulate innate immunity [58]. Methylated RNA immunoprecipitation and sequencing (MeRIP-seq) revealed that upon Flaviviridae virus infection of human hepatoma cells, m6A patterns changed on 51 host genes, several of which were shown to either increase or decrease viral titer when knocked down [59]. These results indicate that m6A modifications on linear RNAs have diverse and emerging effects on innate immunity that may also apply to endogenous circRNAs.
Viral circRNAs also contain m6A modifications, which may be a mechanism to mimic cellular circRNAs, hide from host immune surveillance, or recruit required cellular proteins. The m6A modification on HPV-encoded circE7 is necessary for its translation to the E7 oncoprotein [12]. As is the case for endogenous circRNAs, studies of the innate immune response to m6A-modified viral circRNAs have been limited, and key insights may lie in other viral RNA classes with m6A modifications. For example, m6A-modified linear viral RNAs evade immune detection. Human metapneumovirus (HMPV) RNA contains m6A modifications, which decrease efficiency of RIG-I binding and reduce interferon production [60]. Given the widespread prevalence of m 6A modifications on both viral and endogenous circRNAs, the role of m6A-modified circRNAs in innate immunity is an important topic for further investigation.
Conclusions
While studies of the innate immune response have traditionally focused on cells and proteins, we now know that nucleic acids also play key regulatory roles. The significant influence of circRNAs of diverse origin and function exemplifies this. Endogenous circRNAs modulate the activation of innate immune proteins, as well as the differentiation and activity of innate immune cells. Viral and bacterial infection impacts the global and individual expression of host circRNAs, which vary in their antiviral or pro-viral effects. Viruses themselves encode circRNAs in their genomes, and in vitro-generated foreign circRNAs, which are potential models of viral infection, stimulate an innate immune response due to their lack of m6A modifications.
Many questions surrounding circRNAs in innate immunity remain, including the roles and biogenesis of virally-encoded circRNAs, the specific functions of endogenous circRNAs that are differentially expressed during the innate immune response, the impact of RNA modifications on endogenous and foreign circRNA-mediated innate immunity, and the potential for circRNAs to act as biomarkers. Future investigation into these questions will undoubtedly complicate and enrich our understanding of the layers of regulation that govern the innate immune response.
Figure 3:
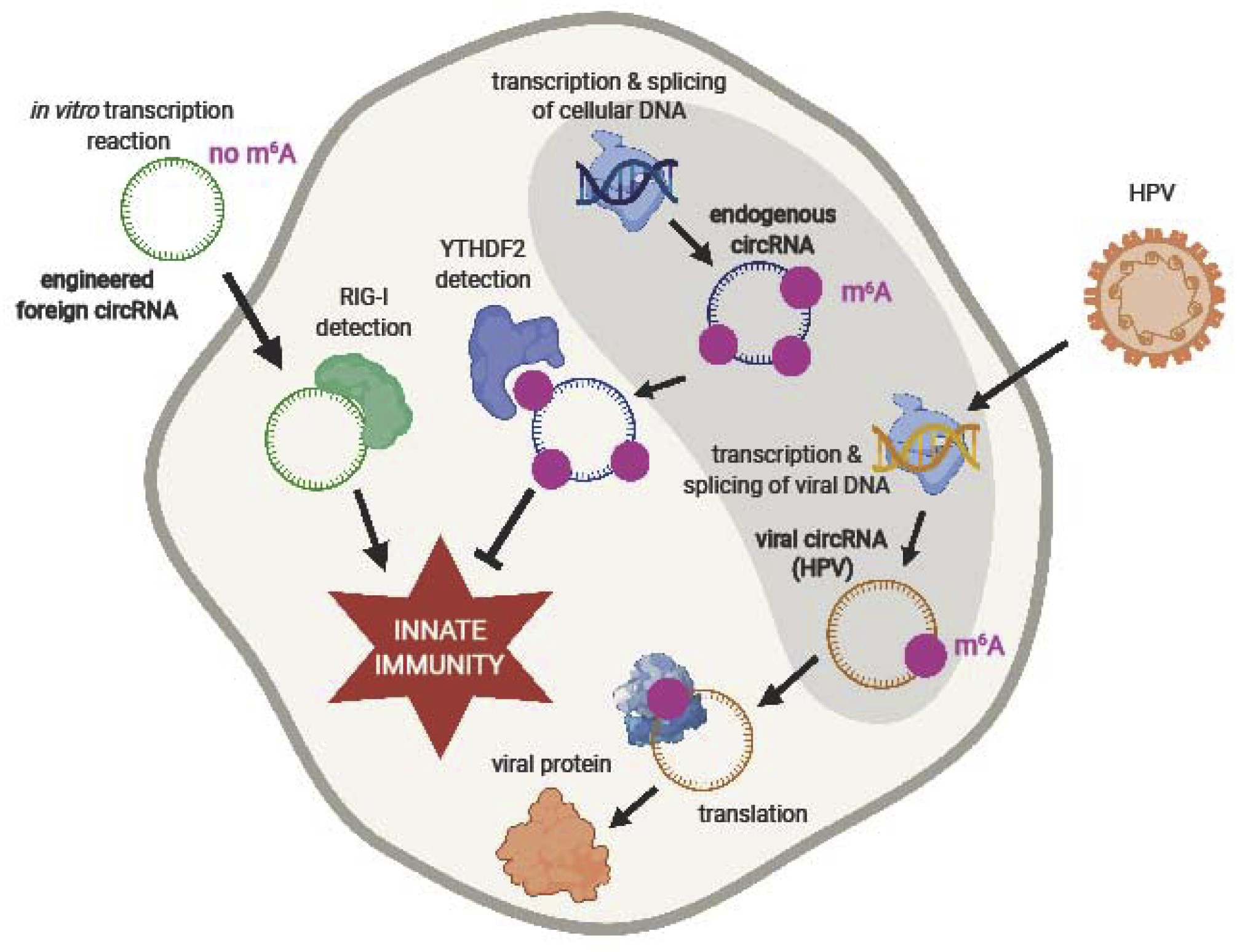
The m6A RNA modification regulates circRNA immunity. In vitro-transcribed foreign circRNAs that lack m6A modifications are detected by the nucleic acid sensor RIG-I, which then stimulate an innate immune response. Endogenous circRNAs, transcribed and spliced by cellular machinery, possess m6A modifications and do not stimulate innate immunity due to detection and sequestration by m6A “reader” YTHDF2. m6A is also required for the translation of specific viral circRNAs into protein, as seen in HPV-encoded circE7.
Highlights.
CircRNAs are a new class of RNA with diverse roles in innate immunity
Endogenous circRNAs regulate the activity of innate immune proteins and cells
Viruses encode circRNAs, though their roles require further investigation
Foreign circRNAs that lack the RNA modification N6-methyladenosine (m6A) “self” markers trigger an innate immune response
Acknowledgements
We thank our lab for helpful discussion and Dr. Peter Cresswell and Dr. Jie Jane Chen for critical reading of the manuscript. This work is supported by the National Institute of Health (5K12CA215110), the American Cancer Society (IRG17-172-57), and the Rita Allen Foundation to Y.G.C, and the Yale College Dean’s Research Fellowship to I.L. Figures were created with Biorender.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References:
- 1.Schlee M, Hartmann G: Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol 2016, 16:566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sokol CL, Luster AD: The chemokine system in innate immunity. Cold Spring Harb Perspect Biol 2015, 7:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XO, Wang H Bin, Zhang Y, Lu X, Chen LL, Yang L: Complementary sequence-mediated exon circularization. Cell 2014, 159:134–147. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Liu S, Zhang L, Issaian A, Hill RC, Espinosa S, Shi S, Cui Y, Kappel K, Das R, et al. : A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S: CircRNA Biogenesis competes with Pre-mRNA splicing. Mol Cell 2014, 56:5566. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL: Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell 2017, 67:214–227.e7. [DOI] [PubMed] [Google Scholar]
- 7.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO: Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS One 2012, 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE: Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, et al. : Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2014, 58:870–885. [DOI] [PubMed] [Google Scholar]
- 10.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M, et al. : The Epstein Barr virus circRNAome. PLoS Pathog 2018, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Chen J, Gong L, Bi Y, Liang J, Zhou L, He D, Shao C: Identification of virus-encoded circular RNA. Virology 2019, 529:144–151. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, et al. : Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun 2019, 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]; *From computational analysis of RNA-seq data, the authors identify circE7 as an HPV-encoded circRNA. Experiments in HPV-infected HEK293T cells and CaSki cervical carcinoma cells reveal that circE7 encodes E7, an oncoprotein, and that the cap-independent translation of circE7 depends on its m6A modification. This study provides an explicit role of a viral circRNA, as well as a direct impact of the m6A modification on viral circRNA function.
- 13.Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, Shair KH, Moore PS, Chang Y: Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A 2018, 115:E8737–E8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, Serquiña AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan R, et al. : Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci U S A 2018, 115:12805–12810. [DOI] [PMC free article] [PubMed] [Google Scholar]; *After de novo KSHV infection of primary umbilical vein endothelial cells, the authors computationally analyze circRNA microarray and RNA-seq data to identify KSHV-encoded circRNAs and differentially expressed host circRNAs. Ectopic expression of human circRNA hsa_circ_0001400 suppressed expression of LANA and RTA, two KSHV genes. This study is the first to report an antiviral host circRNA that is upregulated upon infection, and is one of the first to identify virally encoded circRNAs.
- 15.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL: Circular Intronic Long Noncoding RNAs. Mol Cell 2013, 51:792–806. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. : Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015, 22:256–264. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL: The Biogenesis of Nascent Circular RNAs. Cell Rep 2016, 15:611–624. [DOI] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. : Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495:333–338. [DOI] [PubMed] [Google Scholar]
- 19.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J: Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495:384–388. [DOI] [PubMed] [Google Scholar]
- 20.Kleaveland B, Shi CY, Stefano J, Bartel DP: A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018, 174:350–362.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, et al. : Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017, 14:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z: A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity 2018, 48:688–701.e7. [DOI] [PubMed] [Google Scholar]; **Using circRNA microarray analysis, the authors identify cia-cGAS to be upregulated in long-term hematopoeitic stem cells (LT-HSCs). Depletion experiments in mice and LT-HSCs reveal that cia-cGAS binds cGAS to inhibit IFN production, sustaining LT-HSC identity. Cia-cGAS is the first circRNA to have a clearly elucidated role in the differentiation of blood cells.
- 23.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. : Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016, 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H, et al. : Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177:865–880.e21. [DOI] [PubMed] [Google Scholar]; **The authors observe that poly(I:C) treatment or viral infection of HeLa cells leads to global reduction in circRNA levels, with RNase L revealed to be the degrading agent via depletion experiments. They find that in uninfected cells, circRNAs form duplex structures to suppress the activity of PKR, an innate immune nucleic acid sensor. The authors also show that blood cells of SLE patients have decreased circRNA levels, with overexpression of endogenous circRNAs leading to decreased PKR and interferon gene activation levels. This study provides evidence of a direct immunoregulatory role of global circRNA expression.
- 25.Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK: Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol 2016, 13:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. : Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017, 66:22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]; *The authors computationally analyze RNA-seq data from hematopoietic cells and validate their results in vivo with RT-qPCR. They determine that undifferentiated hematopoietic progenitors, differentiated myeloid cells, and differentiated lymphoid cells each express specific circRNA profiles, establishing type-specific expression of circRNAs in blood cells, including innate immune cells.
- 27.Nicolet BP, Engels S, Aglialoro F, Van Den Akker E, Von Lindern M, Wolkers MC: Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res 2018, 46:8168–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaffo E, Boldrin E, Dal Molin A, Bresolin S, Bonizzato A, Trentin L, Frasson C, Debatin KM, Meyer LH, te Kronnie G, et al. : Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci Rep 2019, 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dou C, Cao Z, Yang B, Ding N, Hou T, Luo F, Kang F, Li J, Yang X, Jiang H, et al. : Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep 2016, 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Ouyang Z, Shen Y, Liu B, Zhang Q, Wan L, Yin Z, Zhu W, Li S, Peng D: CircRNA_28313/miR-195a/CSF1 axis modulates osteoclast differentiation to affect OVX-induced bone absorption in mice. RNA Biol 2019, 16:1249–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou ZL, Luo Z, Wei W, Liang S, Gao TL, Lu Y Bin: Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA Biol 2019, 16:1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z, Zhou S, Li J, Zhou Z, Wang P, Xin H, Mao L, Luo C, Yu S, Huang X, et al. : Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2019, doi: 10.1002/hep.31068. [DOI] [PubMed] [Google Scholar]
- 33.Chen T-C, Tallo-Parra M, Kadener S, Böttcher R, Pérez-Vilaró G, Boonchuen P, Somboonwiwat K, Díez J, Sarnow P: Host-derived Circular RNAs Display Proviral Activities in Hepatitis C Virus - Infected Cells. bioRxiv 2020, doi: 10.1101/2020.01.24.917971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen X, Yao M Di, Li XM, Yao J, Zhou RM, et al. : Targeting pericyte–endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc Natl Acad Sci U S A 2019, 116:7455–7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Chen X li, Sun Q: microRNA-579 upregulation mediates death of human macrophages with mycobacterium tuberculosis infection. Biochem Biophys Res Commun 2019, 518:219–226. [DOI] [PubMed] [Google Scholar]
- 36.Huang Z, Su R, Deng Z, Xu J, Peng Y, Luo Q, Li J: Identification of differentially expressed circular RNAs in human monocyte derived macrophages response to Mycobacterium tuberculosis infection. Sci Rep 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Zhang H, An M, Zhao B, Ding H, Zhang Z, He Y, Shang H, Han X: Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection. J Transl Med 2018, 16:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu S, Zhu N, Guo W, Wang X, Li K, Yan J, Jiang C, Han S, Xiang H, Wu X, et al. : RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection. Front Cell Infect Microbiol 2020, 10:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Chu H, Wen L, Shuai H, Yang D, Wang Y, Hou Y, Zhu Z, Yuan S, Yin F, et al. : Competing endogenous RNA network profiling reveals novel host dependency factors required for MERSCoV propagation. Emerg Microbes Infect 2020, 9:733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S: Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res 2015, 25:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Memczak S, Papavasileiou P, Peters O, Rajewsky N: Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 2015, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing C, Su R, Xu J, Guo Y, Huang Z, et al. : Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin Exp Immunol 2018, 194:118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Li K, Lai W, Li X, Wang H, Yang J, Chu S, Wang H, Kang C, Qiu Y: Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta 2018, 480:17–25. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y hong, Yu X hui, Luo S shun, Han H: Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing 2015, 12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai Z, Fan Y, Zhang Z, Lu C, Zhu Z, Jiang T, Shan T, Peng Y: VirusCircBase: a database of virus circular RNAs. Brief Bioinform 2020, 2020:1–9. [DOI] [PubMed] [Google Scholar]
- 46.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY: Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell 2017, 67:228–238.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al. : N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell 2019, 76:96–109.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The authors demonstrate that in vitro-generated foreign circRNA, which lacks an m6A modification, triggers a conformational change in RIG-I and stimulates the expression of innate immunity genes in HeLa cells. They also show that endogenous circRNA is m6A-modified and binds to YTHDF2, an m6A “reader,” to circumvent immunity. Additionally, they deliver foreign circRNA as a vaccine adjuvant in vivo, which induced the sustained production of antigen-specific T cell and antibody responses. This study supports past work demonstrating that foreign circRNAs stimulate innate immunity and is the first to implicate m6A as a marker of self on endogenous circRNAs.
- 48.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR: Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. : Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485:201–206. [DOI] [PubMed] [Google Scholar]
- 50.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. : ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol Cell 2013, 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. : N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011, 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou C, Molinie B, Daneshvar K, Pondick JV., Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC: Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep 2017, 20:2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, et al. : N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun 2019, 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ok Hyun Park A, Ha H, Lee Y, Ho Boo S, Hoon Kwon D, Kyu Song H, Ki Kim Correspondence Y, Hyun Park O, Ki Kim Y: Endoribonucleolytic Cleavage of m 6 A-Containing RNAs by RNase P/MRP Complex. Mol Cell 2019, 74:494–507. [DOI] [PubMed] [Google Scholar]
- 55.Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, Incarnato D, Oliviero S, Fatica A, Morlando M, et al. : Modulation of circRNA Metabolism by m6A Modification. Cell Rep 2020, 31. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. : Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017, 27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Q, Hou J, Zhou Y, Li Z, Cao X: The RNA helicase DDX46 inhibits innate immunity by entrapping m 6 A-demethylated antiviral transcripts in the nucleus. Nat Immunol 2017, 18:1094–1103. [DOI] [PubMed] [Google Scholar]
- 58.Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, Nachshon A, Tai-Schmiedel J, Friedman N, Le-Trilling VTK, et al. : m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 2019, 20:173–182. [DOI] [PubMed] [Google Scholar]; **After deletion of METTL3 (the m6A “writer” protein) and YTHDF2 (one of several m6A “reader” proteins) in primary fibroblasts, the authors observe stabilization of IFNB mRNA, as well as increased expression of interferon-stimulated genes and decreased propagation of human cytomegalovirus (HCMV) upon infection. This study demonstrates that the m6A modification can serve to suppress innate immunity, particularly on linear RNAs, and raises further questions about m6A’s impacts on circRNAs involved in innate immunity.
- 59.Gokhale NS, McIntyre ABR, Mattocks MD, Holley CL, Lazear HM, Mason CE, Horner SM: Altered m6A Modification of Specific Cellular Transcripts Affects Flaviviridae Infection. Mol Cell 2020, 77:542–555.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, Liang X, Gao TZ, Xu Y, Zhou J, et al. : N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol 2020, 5:584–598. [DOI] [PMC free article] [PubMed] [Google Scholar]


