Abstract
AXL, a TAM family receptor tyrosine kinase, is increasingly being recognized as a key determinant of resistance to targeted therapies as well as chemotherapy and radiation in non-small cell lung cancer (NSCLC) and other cancers. We further show here that high levels of AXL and EMT were frequently expressed in subsets of both treatment-naïve and treatment-relapsed NSCLC. Previously, we and others have demonstrated a role for AXL in mediating DNA damage repair (DDR) as well as resistance to inhibition of WEE1, a replication stress response kinase. Here, we show that BGB324 (bemcentinib), a selective small-molecule AXL inhibitor, caused DNA damage and induced replication stress, indicated by ATR/CHK1 phosphorylation, more significantly in TP53-deficient NSCLC cell lines. Similar effects were also observed in large cell neuroendocrine carcinoma (LCNEC) cell lines. High AXL protein levels were also associated with resistance to ATR inhibition. Combined inhibition of AXL and ATR significantly decreased cell proliferation of NSCLC and LCNEC cell lines. Mechanistically, combined inhibition of AXL and ATR significantly increased RPA32 hyper-phosphorylation and DNA double strand breaks and induced markers of mitotic catastrophe. Notably, NSCLC cell lines with low levels of SLFN11, a known predictive biomarker for platinum and PARP inhibitor sensitivity, were more sensitive to AXL/ATR co-targeting. These findings demonstrate a novel and unexpected role for AXL in replication stress tolerance, with potential therapeutic implications.
Implications:
These findings demonstrate that the combination of AXL and ATR inhibitors could be a promising therapeutic combination for NSCLC, LCNEC and other cancers.
Keywords: Non-small cell lung cancer, Large cell neuroendocrine carcinoma, AXL, ATR, Replication stress, DNA damage
Introduction
AXL, a TAM (Tyro3, AXL, Mer) family receptor tyrosine kinase, has emerged as a key determinant of therapeutic resistance in multiple cancers, including non-small cell lung cancer (NSCLC).1 AXL overexpression has been shown by our group, as well as others, to confer acquired resistance to targeted therapies such EGFR, ALK and BRAF inhibitors.2–6 AXL promotes cell proliferation, and survival via the RAS/MEK/ERK and the PI3K/AKT/mTOR pathways. AXL signaling has also been shown to induce invasion and migration1, is strongly associated with a mesenchymal phenotype, and has been shown to drive epithelial to mesenchymal transition (EMT), which is also associated with therapeutic resistance.3, 7, 8 Several small-molecule AXL inhibitors and anti-AXL biologics are currently being investigated as monotherapies and in combination with targeted agents and chemotherapy in clinical trials for NSCLC (e.g. NCT02922777, NCT02729298, NCT02988817, NCT03425279).1, 9–11 AXL inhibitors have also demonstrated anti-viral effects and are being investigated in clinical trials for SARS-CoV-2 infection.
Replication stress (RS), which often manifests by the slowing or stalling of replication fork progression, is considered an important driver of genomic instability, a hallmark of cancer.12 As a result of aberrant oncogenic stimulation and/or loss of cell cycle checkpoints, cancer cells undergo increased replication of unrepaired DNA, which results in elevated RS. Several oncogenic alterations prevalent in NSCLC tumors, such as KRAS mutations, STK11/LKB1 loss and MYC amplifications have been shown to induce RS.13, 14 Specifically, about 40% of lung adenocarcinomas and 10% of squamous cell lung carcinomas, the two predominant histological subtypes of NSCLC, have alterations in one or more of these genes.15–17 To alleviate RS, cancer cells rely on an ATR/CHK1-mediated RS response, which prevents collapse of stalled replication forks and premature restart of aberrant replication, both of which result in mitotic catastrophe and cell death. Given that more than 50% of NSCLC tumors have alterations in TP53 or a DNA damage response (DDR) gene (e.g. ATM, PRKDC, FANCM, POLE, BRCA),17 NSCLC tumors are highly reliant on the ATR-CHK1 axis. Similarly, another lung cancer subtype, large cell neuroendocrine carcinoma (LCNEC), also has a high prevalence of TP53 inactivation (92%)18, and frequent co-occurring alterations in genes associated with RS such as KRAS, NRAS, STK11 or RB118, suggesting that, LCNEC tumors may also be dependent on the ATR-CHK1 pathway. Elevating intrinsic RS levels, as well as targeting the RS response pathways, have emerged as potential therapeutic approaches for specific subsets of NSCLC. RS response inhibitors such as ATR and CHK1 inhibitors in combination with radiation therapy and chemotherapeutic agents or other DNA damaging agents are in early clinical investigations in NSCLC (e.g. NCT02589522, NCT02264678, NCT01139775, NCT02797964). Standard cancer treatments such as chemotherapy and radiation therapy have also been shown to induce RS by causing DNA damage.12, 19
Increased AXL expression has been observed in response to both chemotherapy and radiation therapy, and is associated with resistance to these treatments.20–22 Conversely, AXL inhibition promotes sensitivity to chemotherapy and radiation.22 Based on these findings, we hypothesize that elevated AXL expression may play a role in tolerance of RS inherent to lung cancer cells or in response to treatment. Previously, our group and others have demonstrated an unexpected role for AXL in DDR8, 23 where AXL knockdown and inhibition induced DNA damage and impaired the efficiency of homologous recombination (HR)-mediated DNA repair.8 AXL inhibition in combination with PARP inhibitor (olaparib) and CHK1/2 inhibitor (AZD7762) have also been reported to result in synergistic cell death.8, 24 However, the effects of AXL inhibition on the RS response pathway are not well understood.
In this study, we found that AXL, which is associated with resistance to therapy, is expressed at higher levels in subsets of treatment-naïve NSCLC and LCNEC tumors as well as treatment-resistant, relapsed NSCLC. We also examined the effects of AXL inhibition with the selective small-molecule AXL inhibitor BGB324 (bemcentinib) and AXL knockdown on DNA damage and RS in NSCLC and LCNEC cells. BGB324 induced DNA damage and activated the ATR/CHK1 axis, an effect that was more pronounced in a TP53-deficient background. Furthermore, high AXL levels predicted resistance to ATR inhibitors while the combination of BGB324 and ATR inhibitor (VX-970 or AZD6738) was synergistic. Together, these findings demonstrate a novel effect of BGB324 on the ATR/CHK1 axis and indicate the combination of AXL and ATR inhibitors for treatment of TP53-deficient NSCLC and LCNEC.
Materials and methods
Reagents.
Antibodies for western blotting purchased from Cell signaling (Danvers, MA): phospho-CHK1 (S345) (2348, 1:1000), CHK1 (2360, 1:1000), γH2Ax (9718, 1:1000), AXL (8661, 1:1000), Cyclin B1 (4138), phospho-cdc2 (Y15) (4539), phospho-Histone H3 (S10) (53348), phospho-AKT (S473) (4060), AKT (9272), phospho-S6 (S240/244) (2215), S6 (2217), p21 (2946); Bethyl laboratories : RPA32 (A300–244, 1:1000), phospho-RPA32 (S4/S8) (A300–245, 1:1000), phospho-RPA32 (S33) (A300–246, 1:1000) and phospho-KAP1 (S824) (A300–246, 1:1000); Santa Cruz Biotechnology (Dallas, TX) : β-actin (sc-47778, 1:2000), GAPDH (sc-20357, 1:2000). BGB324 was generously provided by BergenBio and AZD6738 by AstraZeneca. VX-970 was purchased from Selleck Chemicals (Houston, TX). Vinculin (V9131), Propidium iodide and Hydroxyurea were purchased from and Sigma-Aldrich (St. Louis, MO), respectively.
Cell lines.
Human NSCLC, LCNEC, and mesothelioma cell lines were obtained from The University of Texas MD Anderson Lung Cancer Moon Shot Program. GEMM-derived NSCLC cell lines 344SQ, 344SQpLKO.1shControl, 344SQshAXL−10.1 and 344SQshAXL−12.1 were provided by Dr. Don Gibbons. Cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 100 IU/mL penicillin and 100 μg/mL streptomycin at 37 °C in a humidified atmosphere of 5% CO2. All cell lines were in early passages and maintained in culture for less than two months, human cell lines were authenticated by STR profiling and tested regularly for Mycoplasma contamination using MycoAlert Plus (Lonza).
Single agent and combination viability assays.
NSCLC and LCNEC cell lines (2000 cells/well) were seeded in 96-well white bottom microtiter plates. After overnight attachment, cells were treated with BGB324, ATR inhibitors or DMSO control at indicated concentrations in triplicate for 120 h. Cell viability was measured using CellTiter-Glo (Promega) and luminescence was read on a Synergy HT microplate reader (BioTek). For single drug treatments, dose response curves were modeled using non-linear curve fitting and the drug concentration that produces 50% inhibition of relative cell proliferation (IC50) was estimated using our previously published method (drexplorer software).25 Replicate reproducibility was determined by concordance correlation coefficient and goodness of fit by residual standard error. For drug combination experiments, area under the curve (AUC) for the observed effect of the combination was compared to the AUC for the additive effect predicted by the BLISS model. The difference between the two AUCs, denoted by ΔAUC, was computed. ΔAUC<−0.1 was considered to be a greater than additive; −0.1<ΔAUC<0.1, an additive effect of the drug combination, based on an estimated 10% margin of experimental variability.26 Combination index (C.I.) at 50% fraction affected was computed using the Chou-Talalay model.27
RPPA.
Reverse phase proteomic array (RPPA) was performed as described previously.26
Western blotting.
Cells were seeded in 10 cm dishes and treated as indicated. Cells were washed with ice-cold PBS and lysed with RPPA lysis buffer supplemented with protease and phosphatase inhibitor cocktail. The lysate was centrifuged at 14,000 rpm for 10 min to remove cell debris. Total protein concentration of the supernatant was measured using DC protein assay reagent (Biorad). 30μg of cell lysate was boiled for 5 min at 100°C with 2X laemmli buffer, resolved on a 15% polyacrylamide gel and electroblotted on a nitrocellulose membrane. Membranes were blocked in 1X Caesin blocking solution (Biorad) for 1 hour at room temperature and incubated overnight with primary antibodies at specified dilutions at 4°C. Membranes were washed with Tris-buffered saline with 0.1% Tween-20 (TBST) and incubated with appropriate horseradish peroxidase-linked secondary antibodies for 1 h at room temperature. The immunoblots were visualized using the SuperSignal West Pico Plus chemiluminescent substrate (Thermo Scientific) on a Biorad ChemiDoc™ Touch imaging system. Relative band intensities were quantified using Image J software and normalized to loading control.
Knockdown experiments –
Human AXL Stealth siRNA (#HSS100897, #HSS100898, #HSS183343) and negative control siRNA (#12935112) were purchased from Thermo Fisher Scientific. Cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s instructions for 48 h. For stable AXL knockdown, mouse Axl shRNA constructs (TRCN0000023310 and TRCN0000023312) were purchased from Horizon/GE-Dharmacon. shRNA used were expressed in the pLKO.1 puro vector with a scramble sequence as the control. Lentivirus were generated by co-transfection of Axl shRNA constructs with psPAX2/pMD2.G into 293T cells using lipofectamine LTX (Thermo Fisher). Following transduction into 344SQ cells and puromycin selection, Axl knockdown was confirmed by quantitative reverse transcriptase polymerase chain reaction and western blotting.
Clonogenic survival assay –
Calu-1 and H1299 cells were seeded in a 6-well plate at 250 cells/well. After overnight attachment, cells were treated with DMSO, BGB324 (1μM), VX-970 (1μM) or their combination for 48 h. Colonies were allowed to grow in drug-free media for two weeks and stained with 0.25% crystal violet.
Immunofluorescence –
Calu-1 and H1299 cells (200 cells/well) in 24-well plates were treated with DMSO, BGB324 (1μM), VX-970 (1μM) or the combination for 24 h. Cells were fixed with 4% formaldehyde and permeabilized with 0.03% triton X-100. After blocking with 5% normal goat serum, cells were incubated overnight with γH2Ax antibody (Cell signaling #9718, 1:500) at 4°C, followed by addition of the secondary goat anti-rabbit AlexaFluor-546-conjugated antibody (4μg/mL) (A11010, Thermo Fisher). Nuclei were stained with DAPI (1μg/mL) (Sigma). Fluorescence microscopy was performed with an Olympus IX73 microscope system (Waltham, MA).
Cell cycle analysis –
0.25×106 cells (Calu-1, H1299) were plated in a 10 cm dish and after overnight attachment, treated with DMSO, BGB324 (1μM), VX-970 (1μM) or the combination for 72 h. For mitotic release assay, cells were treated with nocodazole (200 ng/mL; Sigma) and the inhibitors for 24 h and released into fresh media. Cells were harvested at indicated time points, fixed in 70% ethanol overnight at 4°C and stained with 50μg/mL propidium iodide and 250μg/mL RNAase A for 1 h at 37°C. Cells analyzed on a LSR Fortessa flow cytometer (BD Biosciences) and data was analyzed using Flow Jo software (Treestar, San Carlos, CA).
Intracellular flow cytometry staining of phospho Histone H3 –
Calu-1 cells were plated and treated as described above. Cells were harvested, fixed overnight with fixation buffer (Biolegend, San Diego, CA) and permeabilized with intracellular staining perm wash buffer (Biolegend). Cells were then stained using Alexa Fluor® 488 anti-phospho Histone H3 (Ser10) antibody (Biolegend #613408) and propidium iodide (50μg/mL) and analyzed on an LSR Fortessa flow cytometer.
Apoptosis assay –
H1299 cells were treated with indicated concentrations of BGB324, VX-970 or their combination for 48 h. Apoptosis was measured using Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA), per the manufacturer’s instructions.
Statistical Analysis –
Data statistics and bioinformatics analyses were performed using R (version 3.3.0, https://www.r-project.org/) and Bioconductor packages (https://www.bioconductor.org/). Mutation and frequency analysis was done using Fisher’s exact or Chi-squared test, as indicated. ANOVA followed by Tukey’s post hoc test was used to compare across treatment groups. To identify proteins most highly correlated with drug sensitivity or EMT score, we used Spearman’s rank correlation test. For mRNA and RPPA expression data analyses, Benjamini-Hochberg method was used to control false discovery rate (FDR).28
Results
AXL is overexpressed in subsets of treatment-naïve and relapsed NSCLC tumors
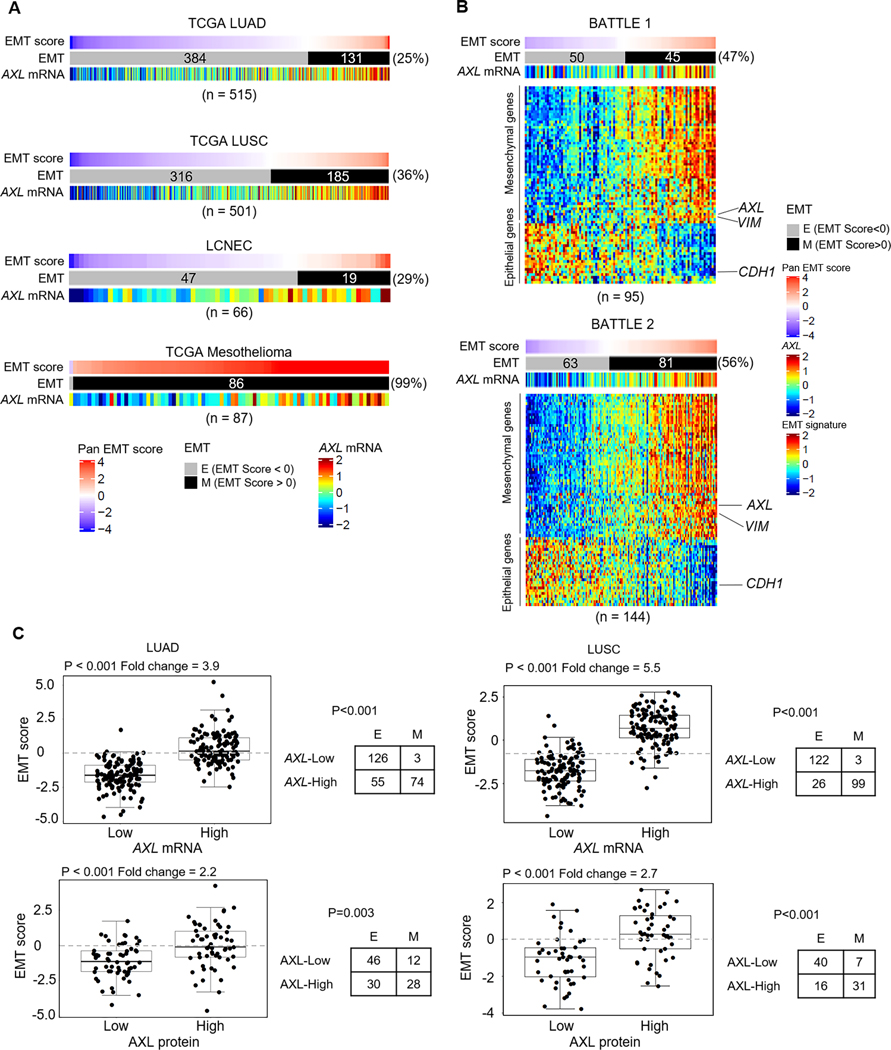
EMT has been implicated as a mechanism of resistance to multiple therapies.29 To determine if EMT was increased following treatment in NSCLC clinical samples, we examined EMT scores of patient tumors using our previously established 77-gene Pancancer EMT signature.30 Using the EMT signature, tumors expressing epithelial genes at higher levels relative to mesenchymal genes in the EMT signature have an EMT score<0 and exhibit an epithelial phenotype. Conversely, tumors with a mesenchymal phenotype, have an EMT score>0.3, 30 In the two treatment-naïve NSCLC clinical cohorts - TCGA lung adenocarcinoma (LUAD),15 and TCGA lung squamous cell carcinoma (LUSC),16 (total n=1016 tumors), an average of 31% of the tumors had a mesenchymal phenotype (Fig. 1A). In tumors from treatment-resistant advanced NSCLC patients who had received prior systemic treatment including chemotherapy, radiation and EGFR inhibitors, but subsequently relapsed (BATTLE-131 and BATTLE-232) (total n=239 tumors), approximately 53% of tumors exhibit a mesenchymal phenotype (Fig. 1B). This statistically significant enrichment of mesenchymal tumors in the treatment-refractory cohorts (p<0.001 by Chi-squared test) suggests that prior therapy may induce EMT in NSCLC.
Figure 1. AXL is a potential therapeutic target in a subset of treatment-naïve and treatment-resistant lung tumors.
A. Range of EMT scores and AXL mRNA expression in treatment-naïve patient cohorts - lung adenocarcinoma (TCGA LUAD), lung squamous cell carcinoma (TCGA LUSC), large cell neuroendocrine carcinoma (LCNEC) and mesothelioma. B. EMT scores, EMT gene signature and AXL mRNA expression in tumor samples from treatment-refractory advanced NSCLC patients, obtained post-progression (BATTLE-1, BATTLE-2). C. Using a quartile cutoff for AXL expression (mRNA (Top) and protein (Bottom)), EMT scores of LUAD and LUSC tumors with AXL-high (4th quantile) and AXL-low (1st quantile) expression were compared by Welch’s t-test. Fold change in EMT score between AXL-high and AXL-low tumors are indicated. Frequency of epithelial (E; EMT score < 0) and mesenchymal (M; EMT score > 0) tumors in the two expression groups was analyzed by Pearson’s Chi-squared test with Yates’ continuity correction.
As we have shown previously AXL to be both a marker of a mesenchymal phenotype, and a driver of EMT3, 8, we next directly examined AXL expression in the NSCLC tumor cohorts. In the treatment-naïve NSCLC patient cohorts (LUAD and LUSC), we observed a subset of tumors that expressed higher levels of AXL mRNA (Fig. 1A) and AXL protein (Supplementary Fig. S1A-B). Furthermore, 48% of LUAD tumors and 66% of LUSC tumors expressing high levels of AXL were mesenchymal (EMT score > 0) (Fig. 1C). This suggests that early co-targeting of AXL along with standard therapies could be useful in preventing resistance and improving treatment outcomes in treatment-naïve NSCLC patients. Similarly, consistent with AXL’s role in therapeutic resistance, a larger subset of relapsed NSCLC tumors from the BATTLE-1 and BATTLE-2 cohorts also expressed high AXL (Fig. 1B). Together, these findings underscore the therapeutic utility of targeting AXL, particularly in treatment-resistant NSCLC.
To further assess the spectrum of EMT and AXL expression across multiple cancers, we examined EMT scores and TCGA gene expression data of 32 different thoracic and extra-thoracic malignancies (Supplementary Fig. S2). Among other cancers, some of the highest AXL mRNA expression was seen in mesothelioma, which was also one of the most mesenchymal tumors (Fig. 1A and Supplementary Fig. S2). In the recently published cohort of treatment-naïve large cell neuroendocrine carcinoma (LCNEC) tumors,18 29% of tumors had a mesenchymal EMT score, along with high levels of AXL expression in many of these tumors (Fig. 1A and Supplementary Fig. S1C). LCNEC are rare pulmonary tumors, accounting for about 3% of lung cancer diagnoses, with genomic similarities to NSCLC (STK11, NRAS mutations) as well as neuroendocrine features like small cell lung cancer (SCLC).18 LCNECs, also similar to SCLC, are primarily treated with platinum-based chemotherapy and lack any specific targeted therapy options.33 Overall, these observations identify sub-populations of patients with higher AXL expression in both treatment-naïve and relapsed NSCLC tumors, and across multiple cancer types, that may benefit from co-treatment with AXL inhibitors.
AXL inhibition by selective small-molecule inhibitor BGB324 results in DNA damage and ATR/CHK1 activation.
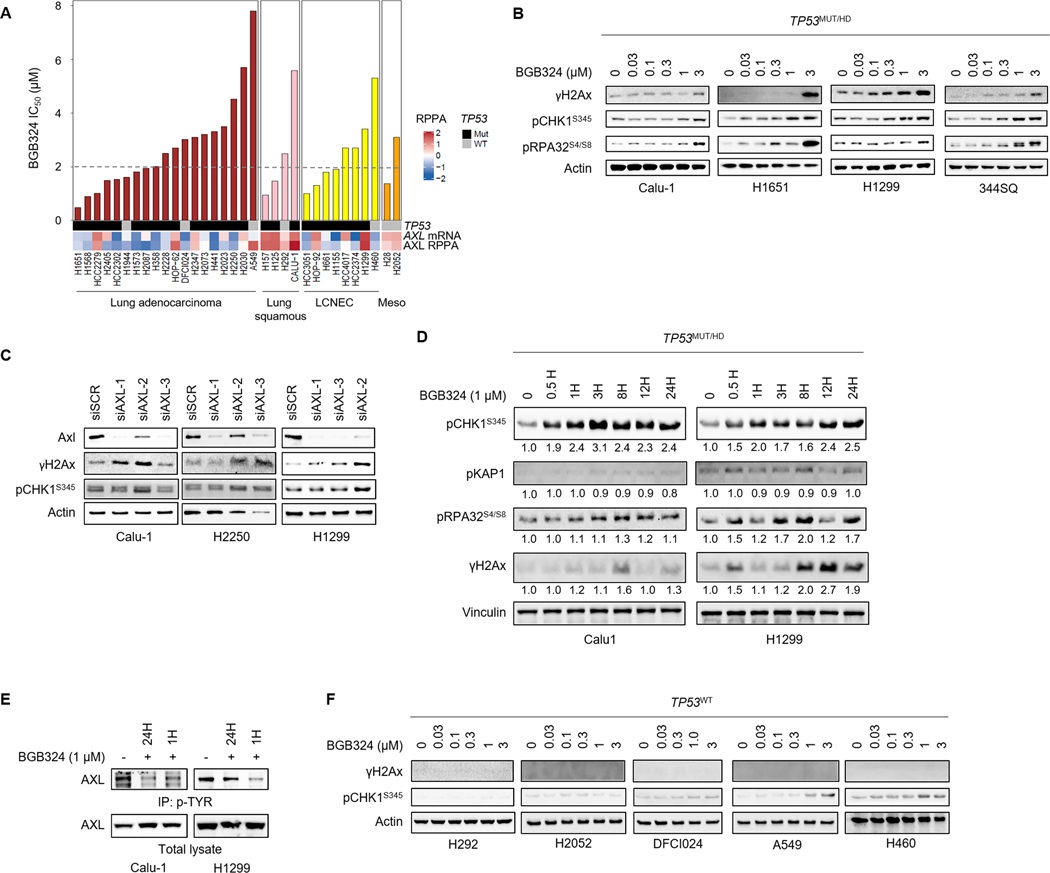
BGB324 (bemcentinib, R428) is a selective AXL inhibitor that is currently in Phase I and II clinical trials in combination with EGFR inhibition, chemotherapy and immunotherapy in previously treated advanced NSCLC (NCT02424617, NCT02922777, NCT03184571).9, 34 To assess the effect of BGB324 on growth inhibition, we screened a panel of 23 NSCLC cell lines with varying AXL levels, as determined by RPPA, and genetic background in 5 day cell proliferation assays. Both NSCLC adenocarcinoma and squamous cell carcinoma cell lines showed a range of sensitivities to BGB324 (IC50 values range from 0.67 to >9.61μM, median 2μM) (Fig. 2A). In addition, we tested BGB324 in a panel of additional lung cancer types including LCNEC and mesothelioma. Several LCNEC cell lines also showed a similar sensitivity to BGB324 (median IC50 2.3 μM) (Fig. 2A). Next, as oncogenic drivers of NSCLC have differential sensitivities to treatments35, we looked for associations between these genes and sensitivity to BGB324. However, in our panel of cell lines (Supplementary Table S1), no association between these oncogenes and BGB324 sensitivity was observed.
Figure 2. AXL inhibition and knockdown induces DNA damage and activates ATR/CHK1 signaling.
A. Sensitivity of a panel of lung cancer cell lines to BGB324 (ranked by IC50). Dashed line indicates median IC50 (2μM). AXL mRNA levels, AXL protein expression determined by RPPA, and TP53 mutation status are indicated below. B. Expression of markers of DNA damage (γH2Ax) and RS (pCHK1, pRA32) by western blotting, following treatment with indicated concentrations of BGB324 for 24 h, in TP53-deficient lung cancer cell lines. β-Actin was used as loading control. Representative immunoblots of at least 2 independent experiments are shown. C. AXL was silenced in Calu-1, H2250 and H1299 cells using 3 different siRNA sequences. Increase in CHK1 phosphorylation and γH2Ax accumulation 48 h post-AXL depletion was observed by western blotting. D. Calu-1 and H1299 cells were treated with 1μM BGB324 for indicated durations. Expression of the different DDR pathway mediators and Vinculin (loading control) was analyzed by western blotting. Relative band intensities, quantified by Image J and normalized to loading control, are indicated below the blot. E. Inhibition of AXL phosphorylation in Calu-1 and H1299 cells by BGB324. Phosphorylated proteins were immunoprecipitated using Phospho tyrosine antibody and immunoblotted for AXL. F. Immunoblots show limited γH2Ax induction and CHK1 phosphorylation in TP53-wildtype lung cancer cell lines.
Next, to test our hypothesis that AXL promotes tolerance of RS and DNA damage, we examined markers of RS and DNA damage following AXL inhibition by BGB324. In a panel of NSCLC and LCNEC cell lines treated for 24 hours (h) with BGB324, changes in protein expression were analyzed by western blotting. In TP53-deficient NSCLC cell lines (H1651 and Calu-1), and the NRAS-mutant/TP53-deficient LCNEC cell line H1299, treatment with BGB324 caused a dose-dependent accumulation of γH2Ax, a marker of double stranded DNA breaks (Fig. 2B). We next examined the effects in a mesenchymal murine lung cancer cell line (344SQ) derived from a KrasLA1/+Trp53R172HΔG genetically engineered mouse model (GEMM)36 and observed a similar increase in γH2Ax accumulation following BGB324 treatment. As with AXL inhibition by BGB324, siRNA-mediated depletion of AXL also resulted in DNA damage (Fig. 2C). This is consistent with our previous findings8 and further supports a role for AXL in DDR.
Beyond DNA damage, we also observed that BGB324 induced a strong increase in CHK1 phosphorylation at serine 345 in these TP53-deficient cell lines. ATR/CHK1 activation commonly occurs in response to exposed single stranded DNA, such as during RS. Furthermore, hyperphosphorylation of the 32 kDa single stranded DNA binding replication protein A 2 (RPA2 or RPA32) at serine 4 and 8 (S4/8) was also detected at higher concentrations, (Fig. 2B). AXL knockdown also similarly activated CHK1 phosphorylation, although the effect appeared to be variable (Fig. 2C).
To assess the temporal dynamics of early DNA damage response to AXL inhibition by BGB324, we examined the time course of activation of key DDR pathway proteins in the TP53-deficient Calu-1 and H1299 cells following BGB324 treatment. CHK1 phosphorylation was detected as early as 0.5 h post-treatment, signaling the onset of RS, and was sustained up to 24 h (Fig. 2D). Phosphorylation of RPA32 (S4/8), however, was observed only at 8 h following BGB324 treatment and was co-incident with increasing γH2Ax accumulation, suggesting possible collapse of stalled replication forks and formation of double-stranded DNA breaks (Fig. 2D).37 No significant increases in phosphorylation of KAP1, an ATM substrate, were observed (Fig. 2D). AXL phosphorylation, in basal condition as well as upon ligand (GAS6)-stimulation, measured by immunoprecipitation, was also confirmed to be inhibited both at 1 h and 24 h post BGB324 treatment (Fig. 2E, Supplementary Fig. S3A,B). As described previously38, we also noted a dose-dependent increase in total AXL levels following 24 h treatment with BGB324 (Supplementary Fig. S3C).
Compared to the TP53-deficient cell lines, CHK1 phosphorylation in response to BGB324 was less pronounced in cell lines with intact TP53 (Fig. 2F) with no detectable increase in γH2Ax. In a TP53 wild type background, treatment with BGB324, instead, resulted in the accumulation of p21, a cyclin-dependent kinase inhibitor, indicating the onset of senescence (Supplementary Fig. S3D). Overall, these findings suggest that TP53-deficient lung cancer cells could be more susceptible to the RS and DNA damage induced by BGB324.
AXL inhibition sensitizes cancer cells to ATR inhibitors
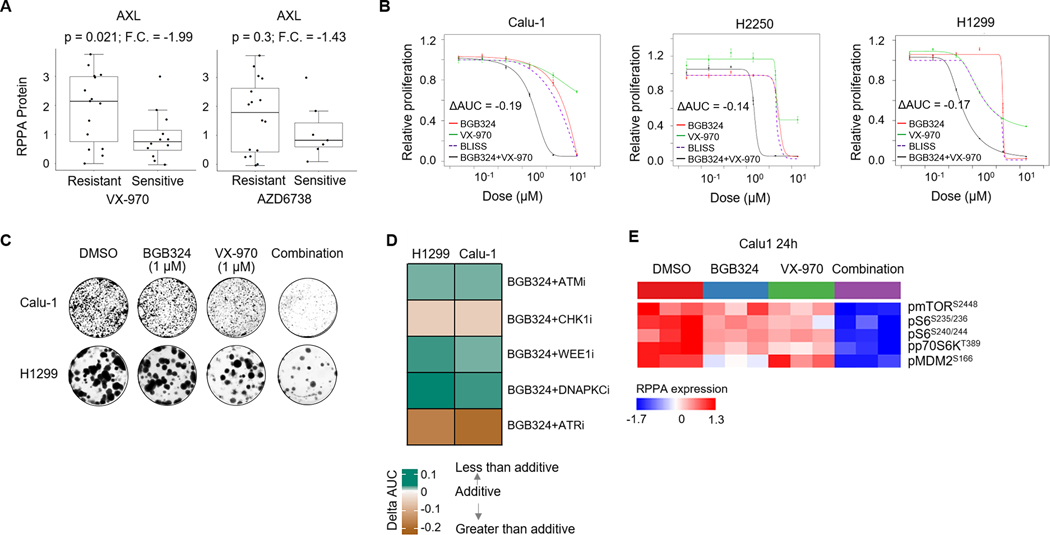
As BGB324 treatment results in DNA damage and RS (and thus activating the ATR-CHK1 checkpoint), we hypothesized that AXL inhibition-induced RS would sensitize cancer cells to ATR inhibition. To test this hypothesis, we first determined the single agent activities of two selective ATR inhibitors currently in clinical trials, VX-970 (berzosertib) and AZD6738 (ceralasertib), in a panel of 25 NSCLC and LCNEC cell lines. Both VX-970 and AZD6738 showed potent cytotoxicity in a subset of cell lines (Supplementary Fig. S4A-B). However, despite potent inhibition of ATR-mediated CHK1 phosphorylation, several cell lines showed inherent resistance to the ATR inhibitors. Interestingly, AXL expression was higher in cell lines that were resistant to the ATR inhibitors VX-970 (Fold change, F.C., between sensitive versus resistant =−1.9, p=0.04) (Fig. 3A) and AZD6738 (F.C.= - 1.4, p=0.3).
Figure 3. AXL inhibition increases sensitivity of lung cancer cells to ATR inhibitors.
A. Comparison of AXL protein expression between lung cancer cell lines sensitive (IC50<3μM) and resistant (IC50>3μM) to the ATR inhibitors, VX-970 and AZD6738. Fold change, F.C. and p value by Welch’s t-test are indicated. B. Relative proliferation of Calu-1, H2250 and H1299 cell lines following 4–5 day treatment with BGB324, VX-970 or their combination at indicated concentrations, as measured by CellTiter-Glo viability assay. Data are mean±s.e.m. ΔAUC value denoting shift in dose response curve of the drug combination beyond the predicted additive effect of the single agents was calculated using the BLISS independence model. ΔAUC<−0.1 indicate a greater than additive effect of the combination. C. Clonogenic survival of Calu-1 and H1299 cells following treatment with DMSO, BGB324, VX-970 or their combination. Colonies stained with 025% crystal violet after 14 days. Representative image from two independent experiments shown. D. Heatmap depicts the effect of combination (calculated as ΔAUC by BLISS model) of BGB324 with various DDR inhibitors – ATM inhibitor (AZD0156), CHK1 inhibitor (LY2606368), WEE1 inhibitor (AZD1775) and DNAPKC inhibitor (NU7441) in Calu-1 and H1299 cells. E. Heatmap shows proteins in the AXL/PI3K/AKT/mTOR signaling pathway, determined by RPPA, following 24 h treatment of Calu-1 cells with DMSO, BGB324, VX-970 or their combination (1μM), significantly altered by ANOVA comparison at FDR=0.01 cutoff.
To determine whether AXL inhibition could enhance sensitivity of lung cancer cells to ATR inhibitors, NSCLC and LCNEC cells were treated with fixed ratio concentrations of BGB324 and VX-970 in 5-day cell proliferation assays. In the ATR inhibitor-resistant cell lines, Calu-1 and H2250, the combination of BGB324 and VX-970 had a greater than predicted additive effect on decreasing cell viability (i.e. ΔAUC<−0.1) (Fig. 3B). Chou-Talalay combination indices (C.I.) of 0.24 and 0.24 and 0.58, in Calu-1, H2250 and H1299 cell lines, respectively, calculated at 50% fraction affected for the drug combination, also suggested a synergistic interaction. Even at a non-growth inhibitory concentration, BGB324 potentiated the cytotoxic effect of VX-970 (Supplementary Fig. S4C). This synergistic effect was further confirmed in a clonogenic assay, wherein the clonogenic survival of Calu-1 and H1299 cells treated with the drug combination was more effectively decreased as compared to either single agent (Fig. 3C). To confirm if the observed effects were AXL-dependent, we knocked down AXL in the Kras/Trp53-mutant GEMM-derived 344SQ cells (Supplementary Fig. S4D) and assessed its effect on sensitivity to ATR inhibition. While the parental 344SQ cells were inherently highly sensitive to VX-970 (Supplementary Fig. S4A), AXL knockdown further increased the sensitivity of 344SQ cells to ATR inhibition (Supplementary Fig. S4E). Together, these data show that AXL inhibition by BGB324 strongly sensitizes inherently resistant AXL-high lung cancer cells to ATR inhibitors.
To test if the observed effects of the BGB324/VX-970 combination were specific to ATR inhibition or were more broadly applicable to DDR inhibitors, we next tested the combination of BGB324 with other DDR inhibitors. In addition to ATR inhibitors, BGB324 also modestly sensitized cancer cells to CHK1 inhibition (LY2606368) (Fig. 3D, Supplementary Fig. S5A). In contrast, combinations with other DDR inhibitors including an ATM inhibitor (AZD0156), a DNAPKC inhibitor (NU7441) or a WEE1 inhibitor (AZD1775) showed no effect (Fig. 3D, Supplementary Fig. S5A). These results further support the idea that the sensitization of NSCLC and LCNEC cells to VX-970 was due to the specific increase in RS and activation of ATR/CHK1 axis by BGB324. Next, we observed by RPPA analysis, that phosphorylation of mTOR and its downstream mediators such as S6 and MDM2 was significantly inhibited following treatment of Calu-1 cells with BGB324/VX-970 combination (Fig. 3E). AXL exerts its oncogenic pro-survival effects through PI3K/AKT/mTOR signaling.1 Furthermore, mTOR has also been shown to mediate DDR and RS.39, 40 To determine if the observed synergistic effect of BGB324/VX-970 combination was mediated by the mTOR pathway, we tested the combination of VX-970 with AZD2014, a potent and selective inhibitor of mTOR complexes mTORC1 and mTORC2. AZD2014 partially recapitulated the effects seen with BGB324, suggesting observed synergistic interaction maybe mediated in part by inhibition of AXL/mTOR signaling (Supplementary Fig. S5B).
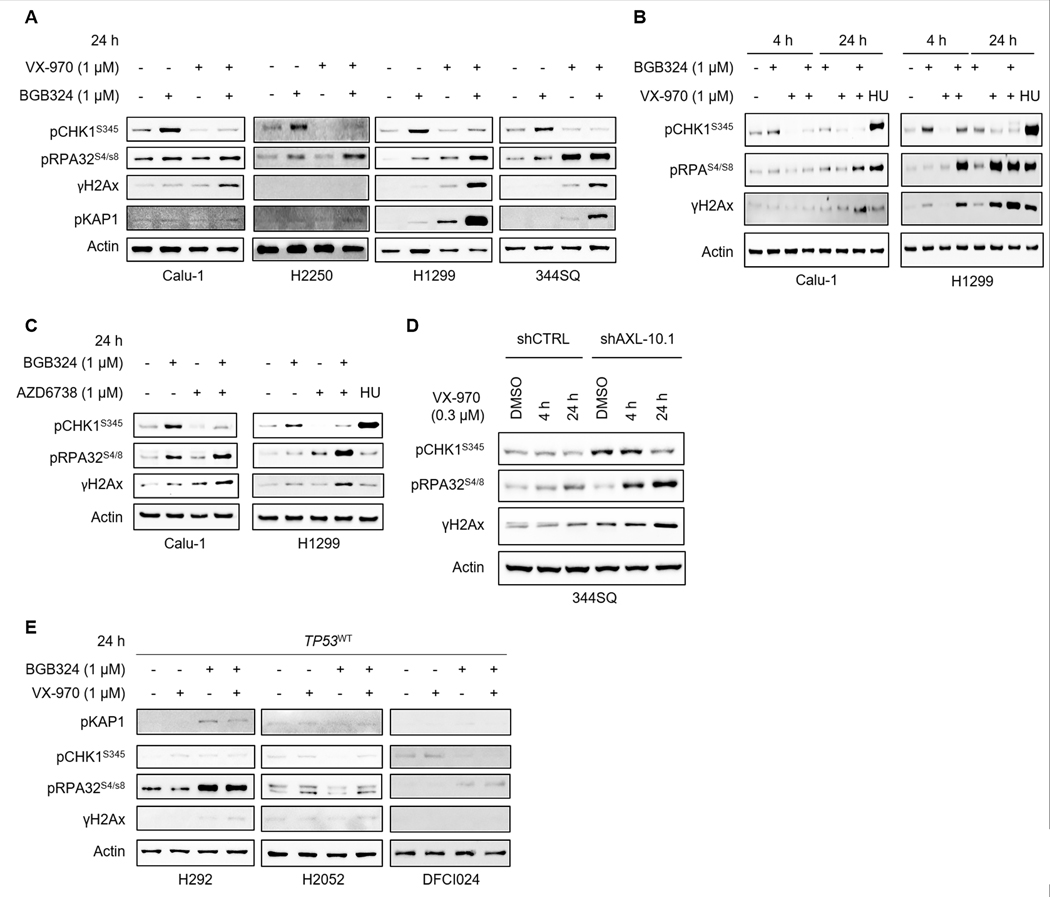
Combination of AXL and ATR inhibitors caused significant DNA damage
Having seen increased γH2Ax accumulation (DNA damage), CHK1 phosphorylation (RS) and RPA32 hyperphosphorylation (Fig. 2B) in TP53-deficient cell lines (Calu-1, H2250, H1299, and 344SQ) following 24 h treatment with BGB324, we were interested to understand the effect of AXL/ATR co-targeting on these markers. BGB324-induced increases in pCHK1 were abrogated by the ATR inhibitor VX-970 (Fig. 4A). Greater induction of phospho-RPA32 (S4/8) was detected as early as 4 h in H1299 cells (Fig. 4B) and was pronounced at 24 h in most cell lines (Fig. 4A) when both drugs were combined as compared to treatment with the single agents, suggesting fork collapse and progression to double strand breaks. Consequently, γH2Ax levels (DNA damage) were significantly increased upon combined AXL/ATR inhibition (Fig. 4A). Appreciable γH2Ax levels in H2250 could not be detected by western blotting. Induction of pKAP1 at 24 h was also evident in H1299 and 344SQ cells, when both drugs were combined. Prolonged treatment with hydroxyurea (HU), a ribonucleotide reductase inhibitor that induces replication fork stalling and RS, also showed a similar increase in RPA32 phosphorylation and γH2Ax (Fig. 4B). The effects of combined AXL/ATR inhibition were further confirmed using another ATR inhibitor, AZD6738, which likewise increased γH2Ax and phospho-RPA32 levels (Fig. 4C).
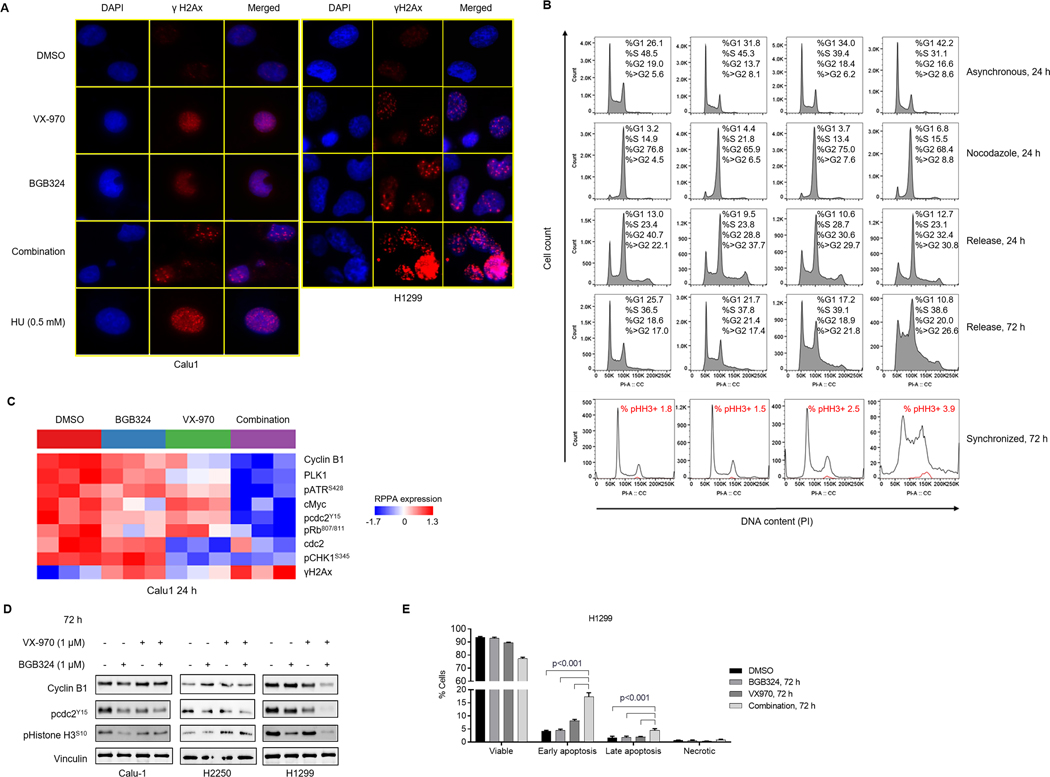
Figure 4. Combination of AXL and ATR inhibitors causes significant DNA damage and induces markers of mitotic catastrophe.
A. Expression of key DDR and RS markers following treatment of TP53-deficient lung cancer cell lines Calu-1, H2250, H1299 and 344SQ with BGB324, VX-970 or their combination (1μM) for 24 h analyzed by western blotting. β-Actin was used as loading control. B. Immunoblots of Calu-1 and H1299 cells treated with BGB324, VX-970, or their combination for indicated durations. Hydroxyurea (0.5mM) was used as a positive control for RS. Changes in RS markers (pCHK1, pRPA32) and DNA damage (γH2Ax) seen as early as 4 h post treatment with BGB324/VX-970 combination. C. Similar increases in DNA damage and RS markers were observed in Calu-1 and H1299 cells treated with BGB324 and another ATR inhibitor, AZD6738. D. 344SQ shCTRL and 344SQ AXL knockdown cells were treated with 0.5μM VX-970 for 4 h or 24 h. Expression of DNA damage and RS markers were analyzed by western blotting. E. Increases in DNA damage and RS markers were not as pronounced in TP53-wildtype cell lines treated with BGB324/VX-970 combination.
To further confirm if these increases were AXL-dependent, Kras/Trp53-mutant 344SQ AXL knockdown cells were treated with VX-970. In cells lacking AXL, addition of VX-970 resulted in enhanced DNA damage and RPA32 hyperphosphorylation, as compared to the parental cells (Fig. 4D). As expected, the effect of the drug combination on DNA damage was less pronounced in TP53-intact cell lines (Fig. 4E). The effect of VX-970 and BGB324/VX-970 was also unexpectedly more pronounced in TP53 WT cell lines harboring STK11 co-mutations (Supplementary Fig. S5C), suggesting that there may be other molecular contexts in which the combination could be effective. Together, these data suggest that the combined inhibition of AXL and ATR induces severe RS and DNA damage, resulting in cell death. In general, this effect appeared to be more pronounced in TP53-deficient lung cancer cells.
We next analyzed the nuclear distribution of γH2Ax using immunofluorescence. In asynchronous H1299 and Calu-1 cells treated with BGB324 for 24 h, a heterogenous pattern of diffuse pan-nuclear γH2Ax staining along with a few distinct punctate foci was detected (Fig. 5A). γH2Ax foci formation is typically seen with DNA damage, indicating double stranded DNA breaks, while uniform pan-nuclear staining has been observed during lethal RS.41, 42 As expected, treatment with the ATR inhibitor or HU showed a pan-nuclear γH2Ax staining, characteristic of RS (Fig. 5A, Supplementary Fig. S6). Further, in response to BGB324/VX-970 combination, most of the cells showed widespread γH2Ax foci formation. In a fraction of cells, intense pan-nuclear γH2Ax staining, likely a result of severe RS and DNA-PK hyperactivation at stalled replication forks41, was also observed following treatment with the combination (Fig. 5A, Supplementary Fig. S6).
Figure 5. Combined inhibition of AXL and AXL increases γH2Ax foci and induces markers of mitotic catastrophe, resulting in S/G2-M arrest and apoptosis.
A. Immunofluorescence analysis in Calu-1 and H1299 cells treated as indicated for 24 h. Representative images of 2 independent experiments show γH2Ax (Red) and DAPI (Blue) staining. B. Calu-1 cells were treated with DMSO, BGB324, VX-970 or the combination for 24 h with and without nocodazole (200 ng/mL) followed by washout. Cells were harvested at indicated times and fixed for PI staining. Representative profiles from two independent experiments are shown. Comparison of DNA content (propidium iodide staining) in all single cells (Black) and phospho Histone H3 positive cells (Red), following treatment of Calu-1 cells synchronized with nocodazole (100 ng/mL) for 24 h followed by treatments as indicated. Percentage of total γH2Ax-positive cells indicated. C. Heatmap shows expression of proteins, determined by RPPA following 24 h treatment of Calu-1 cells with DMSO, BGB324, VX-970 or their combination (1μM), significantly altered by ANOVA comparison at FDR=0.01 cutoff. D. Western blotting analysis of Calu-1, H2250 and H1299 cells treated as indicated for 72 h shows inactivation of G2/M checkpoint (phospho cdc2) and increase in mitotic progression (phospho Histone H3). Vinculin was used as loading control. Representative images of at least 2 independent experiments are shown. E. Induction of apoptosis in H1299 cells by 72 h treatment with BGB324/VX-970 combination, assessed by flow cytometric analysis of Annexin-V FITC staining.
BGB324 and VX-970 combination induced mitotic catastrophe through premature cdc2 activation
In response to DNA damage, TP53-deficient cell lines fail to activate the G1 checkpoint, instead, relying on the ATR/CHK1-mediated G2/M checkpoint to halt cell cycle progression and repair DNA damage. ATR/CHK1 activation causes the inhibitory phosphorylation of the mitotic cyclin-dependent kinase cdc2 at Tyr 15, resulting in G2/M arrest, while ATR inhibition abrogates this checkpoint, allowing cells to progress prematurely into mitosis. Since the BGB324/VX-970 combination induced significant DNA damage in TP53-deficient cell lines, we next examined its effect on cell cycle progression. In asynchronous Calu-1 cells, treatment with the BGB324/VX-970 combination for 24 h strongly increased the number of cells in G1 phase, as compared to either single agent alone (Fig. 5B). To test whether if this increase was due to restoration of the G1 checkpoint, cells were treated with BGB324 and/or VX-970, followed by addition of nocodazole, a microtubule inhibitor. Cell cycle profiles post treatment showed that majority of the cells progressed through the cell cycle and were arrested at M phase by nocodazole, with no significant induction of G1 checkpoint. These results indicate that the increase in G1 population was likely as a result of downregulation of cdc2 phosphorylation and abrogation of the G2 checkpoint, which was seen here (Fig. 5C,D). A previous study reported a similar effect of BGB324 on cdc2 dephosphorylation, as well as a synergistic combination with anti-mitotic agents.43 Of note, treatment with either BGB324 or the BGB324/VX-970 combination appeared to induce a small fraction of cells to arrest at the G1 phase, suggesting a partial but incomplete G1 checkpoint restoration. This was further supported by a significant decrease in RB phosphorylation in cells treated with the inhibitor combination (Fig. 5C). To further examine mitotic exit following treatment with the inhibitors, Calu-1 cells were treated with DMSO, BGB324, VX-970 or the combination in the presence of nocodazole. Upon release from the mitotic arrest, the control cells returned to normal cell cycle kinetics by 72 h. On the other hand, cells treated with the ATR inhibitor or the drug combination failed to re-enter cell cycle efficiently with an exacerbation of the endoreduplication and polyploidy induced by nocodazole44 (Fig. 5B). Consistent with this, expression of phospho-Histone H3, a mitosis marker, was increased in Calu-1 and H2250 cells (Fig. 5D). A similar increase in Histone H3-positive cells was also observed by flow cytometry (Fig. 5B, Supplementary Fig. S7A). These findings, together with cdc2 dephosphorylation by BGB324/VX-970 combination (Fig. 5C, 5D), suggest an aberrant mitotic exit in the presence of significant DNA damage. In the LCNEC cell line H1299, despite cdc2 reactivation, expression of phospho-Histone H3 was significantly inhibited at 72 h and the cells persisted in prolonged G2/M arrest (Fig. 5D, Supplementary Fig. S7B). In this same model, an increase in proportion of cells undergoing early and late apoptosis was detected by Annexin-V/propidium iodide staining at 72 h following treatment of H1299 cells with the drug combination (Fig. 5E). Together, these data suggest that DNA damage induced by BGB324/VX-970 combination results in cancer cell death through multiple mechanisms, including mitotic catastrophe and apoptosis.
Biomarkers of response to BGB324 and VX-970 combination
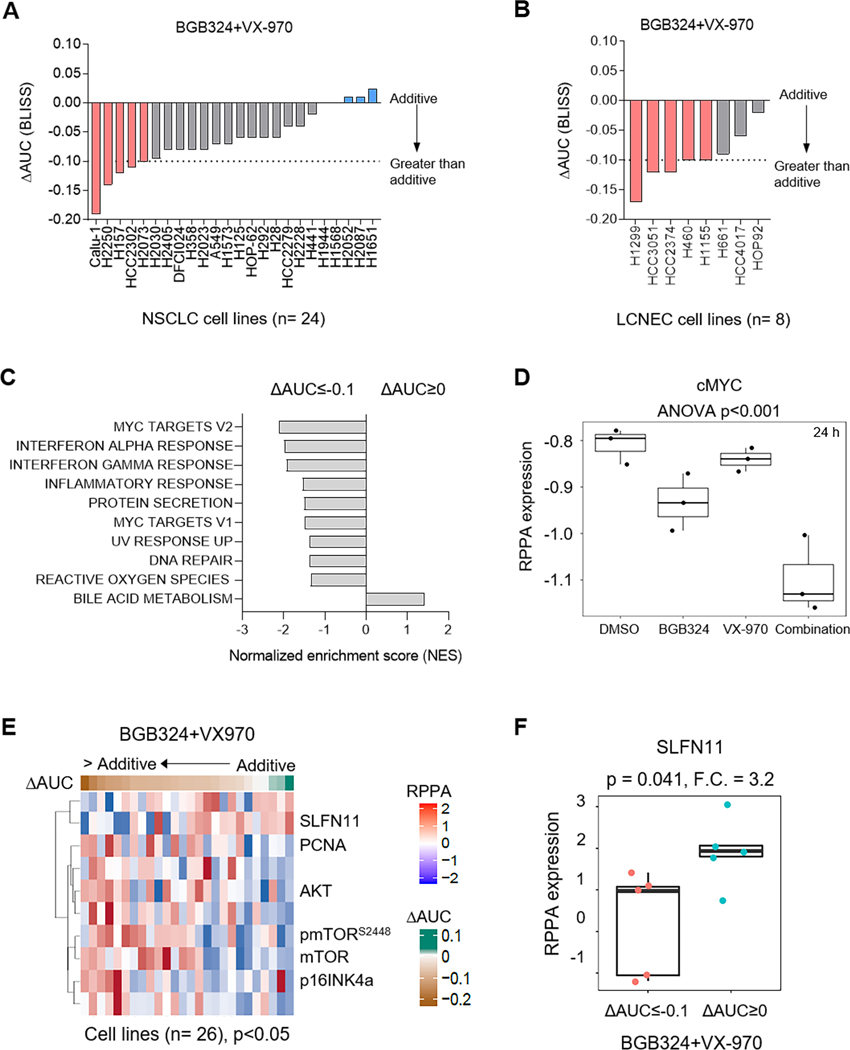
In order to identify markers that predict sensitivity to AXL/ATR co-targeting, we screened the AXL/ATR inhibitor combinations in a panel of 24 lung cancer cell lines (Fig. 6A, Supplementary Fig. S7C). In 4 NSCLC cell lines with primary resistance to ATR inhibition, the combination of BGB324 and VX-970 was synergistic (ΔAUC<−0.1). Similarly, in 5 out of 8 LCNEC cell lines, a greater than additive combinatorial effect on inhibiting cell proliferation was observed (Fig. 6B).
Figure 6. Lung cancer cells with low SLFN11 expression and enriched MYC targets are more sensitive to combined AXL and ATR inhibition.
A. Range of sensitivities of lung cancer cell lines to BGB324 and VX-970 combination. BLISS ΔAUC values were calculated from 5 day proliferation assay, as described previously. B. Bar plots show sensitivities of LCNEC cell lines to BGB324/VX-970 combination treatment. BLISS ΔAUC values indicated. C. GSEA analysis of differential mRNA expression between cell lines with a greater than additive response (ΔAUC<−0.1) and additive response (ΔAUC>0) to BGB324/VX-970 combination showed enrichment of gene sets regulated by MYC among others. NES, normalized enrichment scores of the gene sets are indicated. p<0.05 cutoff was used. D. Expression of cMYC, determined by RPPA, in Calu-1 cells treated with DMSO, BGB324, VX-970 or the combination for 24 h. p value by ANOVA indicated. E. Correlation of RPPA proteomic expression profiles with ΔAUC values of BGB324/VX-970 combination across the panel of 24 lung cancer cell lines tested. Heatmap shows significant biomarkers at a p<0.05 value cutoff. F. Comparison of SLFN11 expression between cell lines with a greater than additive response (ΔAUC<−0.1) and additive response (ΔAUC>0) to BGB324/VX-970 combination. Fold change, F.C. and p values by t-test are indicated.
Next, to identify gene expression signatures associated with sensitivity to the AXL/ATR inhibitor combination, we performed an exploratory analysis of differential gene expression, between cell lines that showed a greater than additive response (ΔAUC<−0.1) and those that showed the least additive response (0<ΔAUC<0.1) to the drug combination, using gene set enrichment analysis. Gene sets associated with MYC-regulated genes, interferon response as well as DNA repair were enriched in the cell lines most susceptible to AXL/ATR co-targeting (Fig. 6C). In line with this, cMYC levels were significantly decreased following treatment with BGB324/VX-970 combination (Fig. 6D). To identify proteomic biomarkers, we correlated proteomic profiles of cell lines to the observed combinatorial effect (ΔAUC). NSCLC cell lines that exhibited a greater than additive effect to BGB324/VX-970 combination (ΔAUC<−0.1) expressed higher mTOR levels and pathway activation, consistent with data above that mTOR signaling was turned off by the combination (Fig. 6D). p16INK4a expression was also higher in these cell lines, which also expressed low RB protein levels (rho = −0.35; p=0.08, data not shown). In addition to its role in promoting G1 arrest, p16INK4a impairs homologous DNA repair and sensitizes to DNA damaging agents.45, 46 Interestingly, cell lines that showed a greater than additive response to BGB324/VX-970 combination had low SLFN11 protein levels (rho=0.64, p<0.001) (Fig. 6D, 6E). In response to DNA damage and RS, SLFN11, a DNA/RNA helicase, is recruited to stalled replication forks by RPA1 and induces an irreversible replication arrest through chromatin unwinding.47 As a result, high SLFN11 levels have been associated with sensitivity to DNA damaging agents and DDR inhibitors.48–50 In the absence of SLFN11, resistance to drugs that induce DNA damage or RS has been overcome by targeting the ATR/CHK1 axis.47, 51 Consistent with these previous findings, many cell lines with high SLFN11 expression were sensitive to single agent BGB324 or VX-970 (without significant further enhancement of response when combined, indicated by an additive response). While, in SLFN11-low cell lines, a relatively greater in vitro effect of the combination (as compared to single agents), indicated by a greater than additive response (ΔAUC≤−0.1), was observed. Together, these results suggest that lung cancer cells with elevated cMYC-induced RS and a compromised RS and DDR response (as indicated by lower SLFN11 expression), were more susceptible to AXL/ATR co-targeting.
Discussion
AXL upregulation is emerging as a mechanism of resistance to treatments targeting RS and DNA damage (e.g. chemotherapy21, 52, radiation20, 22), and the RS response (WEE1 inhibitors53) in various cancers. Similar to our previous study8, we show here that inhibition of AXL using BGB324 (bemcentinib) resulted in DNA damage accumulation. In addition, we found increased RS and ATR/CHK1 axis activation following treatment with BGB324. Together, these findings support a protective role for AXL in suppressing DNA damage and tolerance of RS. Although the precise mechanism of how AXL inhibition results in RS is unclear, we speculate that this may be mediated in part via RAD51 depletion. We have shown previously that AXL inhibition and knockdown causes RAD51 depletion8, which has been linked to defective replication fork protection and ATR/CHK1 activation.8, 54 Furthermore, AXL-mediated mTOR signaling could also have an effect on RS as mTOR complexes 1 and 2 (mTORC 1/2) are known to regulate CHK1 and promote the transcriptional abundance of DNA replication licensing factors such as CDC6.39, 40
We observe elevated levels of DNA damage and RS induced by AXL inhibition in NSCLC cell lines with a TP53-deficient background as compared to cells with an intact TP53 axis. In the absence of a p53-dependent G1 checkpoint, cancer cells are likely to be more susceptible to RS and reliant ATR/CHK1 signaling.55 This could explain the striking induction of γH2Ax and pCHK1 in response to BGB324 treatment in these cells. On the other hand, consistent with recent reports56, we find that TP53-deficiency alone did not increase sensitivity of NSCLC cell lines to ATR inhibitors. Our data here shows that NSCLC cell lines with high AXL levels were more resistant to ATR inhibitors. In line with this, we demonstrate greater than additive effects of AXL and ATR inhibitor combinations in many NSCLC cell lines. These findings also extend to LCNEC cell lines that frequently harbor TP53 mutations. Particularly, LCNEC cell lines respond to both BGB324 as a single agent and its combination with ATR inhibitors. Given the rarity of targetable mutations and shared characteristics, LCNEC tumors are treated similar to SCLC, predominantly with chemotherapy.18 In light of our findings that a significant subset of mesenchymal LCNEC tumors express high levels of AXL, AXL inhibition and AXL/ATR co-targeting could be a therapeutically useful strategy to treat these aggressive tumors that otherwise lack effective targeted therapies. Mechanistically, we show here that combined inhibition of AXL and ATR induced significant DNA damage and abrogated the G2M checkpoint. In contrast to the results we observed in NSCLC and LCNEC cell lines, a recent study, which examined the combination of BGB324 and a CHK1/2 inhibitor (AZD7762), found no increase in DNA damage or CHK1 phosphorylation following BGB324 treatment in melanoma cell lines.24 Differences in genetic background and tumor type likely contribute to these differential effects. Similarly, our group has previously shown in SCLC that AXL overexpression confers primary and acquired resistance to inhibition of WEE1, a DDR kinase in the RS response pathway.53 However, unlike AXL/ATR combination, the combination of BGB324 and WEE1 inhibitor AZD1775 showed mostly an additive effect in the NSCLC cell lines tested. SCLC has a higher basal RS and frequency of DDR mutations, which could explain its greater susceptibility to AXL/WEE1 inhibitor combination. Of note, we observe that while loss of AXL, by siRNA knockdown, induced DNA damage, some residual AXL appeared to be required for CHK1 phosphorylation. Additional studies are needed to better understand AXL’s direct role in RS.
Finally, we identify that cell lines with low levels of SLFN11 were sensitive to AXL/ATR co-targeting. SLFN11, a DNA/RNA helicase, induces an irreversible replication arrest in response to DNA damage and RS.47 Previously, we and others have reported SLFN11 as a predictive marker of sensitivity to DNA damaging agents such as cisplatin and PARP inhibitors in SCLC49, 57. High SLFN11 expression has also been associated with platinum response in LCNEC tumors.58 Conversely, SLFN11 inactivation, such as by promoter hypermethylation, conferred resistance to chemotherapy in NSCLC and other cancers.49, 59 Consistent with our observations, in the absence of SLFN11, resistance to drugs that induce DNA damage or RS have been overcome by targeting ATR (such as with ATR/CHK1 inhibitor combinations).48, 51 Therefore, our findings suggest the AXL and ATR inhibitor combination could be effective in platinum-resistant SLFN11-deficient NSCLC and LCNEC.
In conclusion, these findings underscore a critical role for AXL in tolerance of RS and DNA damage in lung cancer, which can shed light on the implication of its upregulation in response to multiple anticancer treatments. In addition, our results show that TP53-deficient NSCLC and LCNEC are specifically susceptible to the novel combination of AXL and ATR inhibitors. We further identify SLFN11, a clinically used biomarker, to be a strong predictor of response to AXL/ATR co-targeting.
Supplementary Material
Acknowledgements
Support for this research was provided by the NIH/NCI CCSG P30-CA016672 grant (Bioinformatics Shared Resource), NIH/NCI T32 CA009666 to C.M. Gay, NIH/NCI R01-CA207295 to L.A. Byers, the University of Texas-Southwestern and MD Anderson Cancer Center Lung SPORE (5 P50 CA070907), ASCO Young Investigator Award to C.M. Gay, the LUNGevity foundation to D.L. Gibbons and L.A. Byers, MD Anderson Cancer Center Physician Scientist Award to L.A. Byers, Andrew Sabin Family Fellowship to L.A. Byers, the Rexanna Foundation for Fighting Lung Cancer to J.V. Heymach and L.A. Byers, and through generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shot Program (J.V. Heymach, J. Wang, L.A. Byers). LCNEC mRNA expression data was generously provided by Dr. Julie George, University of Cologne. BGB324 was provided by BerGenBio ASA, Norway. AZD6738, AZD2014 and AZD1775 were provided by AstraZeneca, UK.
L.A. Byers serves on advisory committees for AstraZeneca, AbbVie, GenMab, BergenBio, Pharma Mar SA, Sierra Oncology, Merck, Bristol Myers Squibb, Genentech, and Pfizer and has research support from AbbVie, AstraZeneca, GenMab, Sierra Oncology, Tolero Pharmaceuticals. D.L. Gibbons serves on scientific advisory committees for AstraZeneca, Sanofi, Alethia Biotherapeutics and Janssen and has received research support from Janssen, Takeda, Ribon Therapeutics and AstraZeneca. J.V. Heymach serves on advisory committees for AstraZeneca, Boehringer Ingelheim, Exelixis, Genentech, GlaxoSmithKline, Guardant Health, Hengrui, Lilly, Novartis, Spectrum, EMD Serono and Synta, and has research support from AstraZeneca, Bayer, GlaxoSmithKline and Spectrum and royalties and licensing fees from Spectrum. C.M. Gay reports research funding from AstraZeneca.
Footnotes
Conflict of interest: All other authors have no pertinent financial or non-financial conflicts of interest to report.
References
- 1.Gay CM, Balaji K, Byers LA. Giving AXL the axe: targeting AXL in human malignancy. Br J Cancer 2017;116:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi H, Yamada T, Wang R, Tanimura K, Adachi Y, Nishiyama A, et al. AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat Commun 2019;10:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamichi S, Seike M, Miyanaga A, Chiba M, Zou F, Takahashi A, et al. Overcoming drug-tolerant cancer cell subpopulations showing AXL activation and epithelial-mesenchymal transition is critical in conquering ALK-positive lung cancer. Oncotarget 2018;9:27242–27255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuo Q, Liu J, Huang L, Qin Y, Hawley T, Seo C, et al. AXL/AKT axis mediated-resistance to BRAF inhibitor depends on PTEN status in melanoma. Oncogene 2018;37:3275–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asiedu MK, Beauchamp-Perez FD, Ingle JN, Behrens MD, Radisky DC, Knutson KL. AXL induces epithelial-to-mesenchymal transition and regulates the function of breast cancer stem cells. Oncogene 2014;33:1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaji K, Vijayaraghavan S, Diao L, Tong P, Fan Y, Carey JP, et al. AXL Inhibition Suppresses the DNA Damage Response and Sensitizes Cells to PARP Inhibition in Multiple Cancers. Mol Cancer Res 2017;15:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbons D, Byers L, Gerber D, Peguero J, Micklem D, Yule M, et al. MA 02.09 A Ph I/II Study of BGB324, a Selective AXL Inhibitor as Monotherapy and in Combination with Erlotinib in Advanced NSCLC. J Thorac Oncol 2017;12:S1805. [Google Scholar]
- 10.Myers SH, Brunton VG, Unciti-Broceta A. AXL Inhibitors in Cancer: A Medicinal Chemistry Perspective. J Med Chem 2016;59:3593–3608. [DOI] [PubMed] [Google Scholar]
- 11.Boshuizen J, Koopman LA, Krijgsman O, Shahrabi A, van den Heuvel EG, Ligtenberg MA, et al. Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat Med 2018;24:203–212. [DOI] [PubMed] [Google Scholar]
- 12.Forment JV, O’Connor MJ. Targeting the replication stress response in cancer. Pharmacol Ther 2018;188:155–167. [DOI] [PubMed] [Google Scholar]
- 13.Kotsantis P, Petermann E, Boulton SJ. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov 2018;8:537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science 2008;319:1352–1355. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George J, Walter V, Peifer M, Alexandrov LB, Seidel D, Leenders F, et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat Commun 2018;9:1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickoloff JA, Boss MK, Allen CP, LaRue SM. Translational research in radiation-induced DNA damage signaling and repair. Transl Cancer Res 2017;6:S875-S891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skinner HD, Giri U, Yang LP, Kumar M, Liu Y, Story MD, et al. Integrative Analysis Identifies a Novel AXL-PI3 Kinase-PD-L1 Signaling Axis Associated with Radiation Resistance in Head and Neck Cancer. Clin Cancer Res 2017;23:2713–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y, et al. Gas6/Axl Axis Contributes to Chemoresistance and Metastasis in Breast Cancer through Akt/GSK-3beta/beta-catenin Signaling. Theranostics 2016;6:1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand TM, Iida M, Stein AP, Corrigan KL, Braverman CM, Coan JP, et al. AXL Is a Logical Molecular Target in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res 2015;21:2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kariolis MS, Miao YR, Diep A, Nash SE, Olcina MM, Jiang D, et al. Inhibition of the GAS6/AXL pathway augments the efficacy of chemotherapies. J Clin Invest 2017;127:183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flem-Karlsen K, McFadden E, Omar N, Haugen MH, Oy GF, Ryder T, et al. Targeting AXL and the DNA Damage Response Pathway as a Novel Therapeutic Strategy in Melanoma. Mol Cancer Ther 2020;19:895–905. [DOI] [PubMed] [Google Scholar]
- 25.Tong P, Coombes KR, Johnson FM, Byers LA, Diao L, Liu DD, et al. drexplorer: A tool to explore dose-response relationships and drug-drug interactions. Bioinformatics 2015;31:1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardnell RJ, Feng Y, Mukherjee S, Diao L, Tong P, Stewart CA, et al. Activation of the PI3K/mTOR Pathway following PARP Inhibition in Small Cell Lung Cancer. PLoS One 2016;11:e0152584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300. [Google Scholar]
- 29.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010;29:4741–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res 2016;22:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR Jr., Tsao A, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov 2011;1:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadimitrakopoulou V, Lee JJ, Wistuba II, Tsao AS, Fossella FV, Kalhor N, et al. The BATTLE-2 Study: A Biomarker-Integrated Targeted Therapy Study in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naidoo J, Santos-Zabala ML, Iyriboz T, Woo KM, Sima CS, Fiore JJ, et al. Large Cell Neuroendocrine Carcinoma of the Lung: Clinico-Pathologic Features, Treatment, and Outcomes. Clin Lung Cancer 2016;17:e121-e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res 2010;70:1544–1554. [DOI] [PubMed] [Google Scholar]
- 35.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 2009;23:2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res 2012;40:10780–10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauter M, Weber A, Torka R. Targeting of the AXL receptor tyrosine kinase by small molecule inhibitor leads to AXL cell surface accumulation by impairing the ubiquitin-dependent receptor degradation. Cell Commun Signal 2019;17:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silvera D, Ernlund A, Arju R, Connolly E, Volta V, Wang J, et al. mTORC1 and −2 Coordinate Transcriptional and Translational Reprogramming in Resistance to DNA Damage and Replicative Stress in Breast Cancer Cells. Mol Cell Biol 2017;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Li S, Hu X, Xiang X, Halloran M, Yang L, et al. mTOR Signaling Upregulates CDC6 via Suppressing miR-3178 and Promotes the Loading of DNA Replication Helicase. Sci Rep 2019;9:9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moeglin E, Desplancq D, Conic S, Oulad-Abdelghani M, Stoessel A, Chiper M, et al. Uniform Widespread Nuclear Phosphorylation of Histone H2AX Is an Indicator of Lethal DNA Replication Stress. Cancers (Basel) 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 2001;276:47759–47762. [DOI] [PubMed] [Google Scholar]
- 43.Wilson C, Ye X, Pham T, Lin E, Chan S, McNamara E, et al. AXL inhibition sensitizes mesenchymal cancer cells to antimitotic drugs. Cancer Res 2014;74:5878–5890. [DOI] [PubMed] [Google Scholar]
- 44.Vogel C, Kienitz A, Hofmann I, Muller R, Bastians H. Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene 2004;23:6845–6853. [DOI] [PubMed] [Google Scholar]
- 45.Dok R, Kalev P, Van Limbergen EJ, Asbagh LA, Vazquez I, Hauben E, et al. p16INK4a impairs homologous recombination-mediated DNA repair in human papillomavirus-positive head and neck tumors. Cancer Res 2014;74:1739–1751. [DOI] [PubMed] [Google Scholar]
- 46.Duan J, Chen Z, Liu P, Zhang Z, Tong T. Wild-type p16INK4a suppresses cell growth, telomerase activity and DNA repair in human breast cancer MCF-7 cells. Int J Oncol 2004;24:1597–1605. [PubMed] [Google Scholar]
- 47.Murai J, Tang SW, Leo E, Baechler SA, Redon CE, Zhang H, et al. SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol Cell 2018;69:371–384 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murai J, Feng Y, Yu GK, Ru Y, Tang SW, Shen Y, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 2016;7:76534–76550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart CA, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 2017;8:28575–28587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lok BH, Gardner EE, Schneeberger VE, Ni A, Desmeules P, Rekhtman N, et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin Cancer Res 2017;23:523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coussy F, El-Botty R, Chateau-Joubert S, Dahmani A, Montaudon E, Leboucher S, et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci Transl Med 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinn JM, Greenwade MM, Palisoul ML, Opara G, Massad K, Guo L, et al. Therapeutic Inhibition of the Receptor Tyrosine Kinase AXL Improves Sensitivity to Platinum and Taxane in Ovarian Cancer. Mol Cancer Ther 2019;18:389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sen T, Tong P, Diao L, Li L, Fan Y, Hoff J, et al. Targeting AXL and mTOR Pathway Overcomes Primary and Acquired Resistance to WEE1 Inhibition in Small-Cell Lung Cancer. Clin Cancer Res 2017;23:6239–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krajewska M, Fehrmann RS, Schoonen PM, Labib S, de Vries EG, Franke L, et al. ATR inhibition preferentially targets homologous recombination-deficient tumor cells. Oncogene 2015;34:3474–3481. [DOI] [PubMed] [Google Scholar]
- 55.Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci U S A 2001;98:9092–9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Middleton FK, Pollard JR, Curtin NJ. The Impact of p53 Dysfunction in ATR Inhibitor Cytotoxicity and Chemo- and Radiosensitisation. Cancers (Basel) 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pietanza MC, Waqar SN, Krug LM, Dowlati A, Hann CL, Chiappori A, et al. Randomized, Double-Blind, Phase II Study of Temozolomide in Combination With Either Veliparib or Placebo in Patients With Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. J Clin Oncol 2018;36:2386–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabari JK, Julian RA, Ni A, Halpenny D, Hellmann MD, Drilon AE, et al. Outcomes of advanced pulmonary large cell neuroendocrine carcinoma stratified by RB1 loss, SLFN11 expression, and tumor mutational burden. J Clin Oncol 2018;36:e20568-e20568. [Google Scholar]
- 59.Nogales V, Reinhold WC, Varma S, Martinez-Cardus A, Moutinho C, Moran S, et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 2016;7:3084–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.