Abstract
Intermediate filaments (IFs) are key players in multiple cellular processes throughout human tissues. Their biochemical and structural properties are important for understanding filament assembly mechanisms, interactions between IFs and binding partners, and for developing pharmacological agents that target IFs. IF proteins share a conserved coiled-coil central rod domain flanked by variable N-terminal “head” and C-terminal “tail” domains. There have been several recent advances in our understanding of IF structure from the study of keratins, glial fibrillary acidic protein, and lamin. These include discoveries of: (i) a knob-pocket tetramer assembly mechanism in coil 1B; (ii) a lamin-specific coil 1B insert providing one-half superhelix turn; (iii) helical yet flexible linkers within the rod domain; and (iv) the identification of coil 2B residues required for mature filament assembly. Furthermore, the head and tail domains of some IFs contain low complexity aromatic rich kinked segments, and structures of IFs with binding partners show electrostatic surfaces are a major contributor to complex formation. These new data advance the connection between IF structure, pathologic mutations, and clinical diseases in humans.
Keywords: intermediate filament, assembly, structure, keratin, vimentin, lamin, crystallography, binding
Graphical Abstract
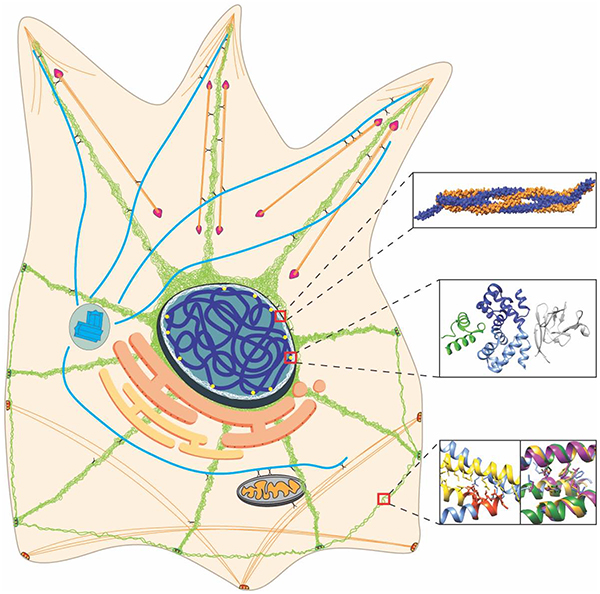
Introduction
Intermediate filaments (IFs) are a group of fibrous proteins that have a variety of biological functions important to human health. Multiple human diseases result from mutations in IFs or aberrant regulation of their expression [1–5]. Unfortunately, despite progress in biochemical, biophysical, and cellular studies of IFs [6], our molecular understanding of how IFs function at atomic resolution remains elusive due to intrinsic properties of IFs, such as high flexibility, insolubility, and aggregation-prone behavior [7]. The basic structure of an IF protein comprises a flexible N-terminal domain (head), coiled-coil central part (rod domain), and an unstructured C-terminal domain (tail). Atomic-resolution knowledge of the rod domain comes from crystallization of IF fragments [8, 9], while the heads and tails are less characterized (Figure 1). In this review, we highlight recent IF structures that advance our understanding of IF assembly, cellular function, and the role of IFs in human disease pathogenesis.
Figure 1. Organization of intermediate filament proteins and their corresponding atomic resolution structures.
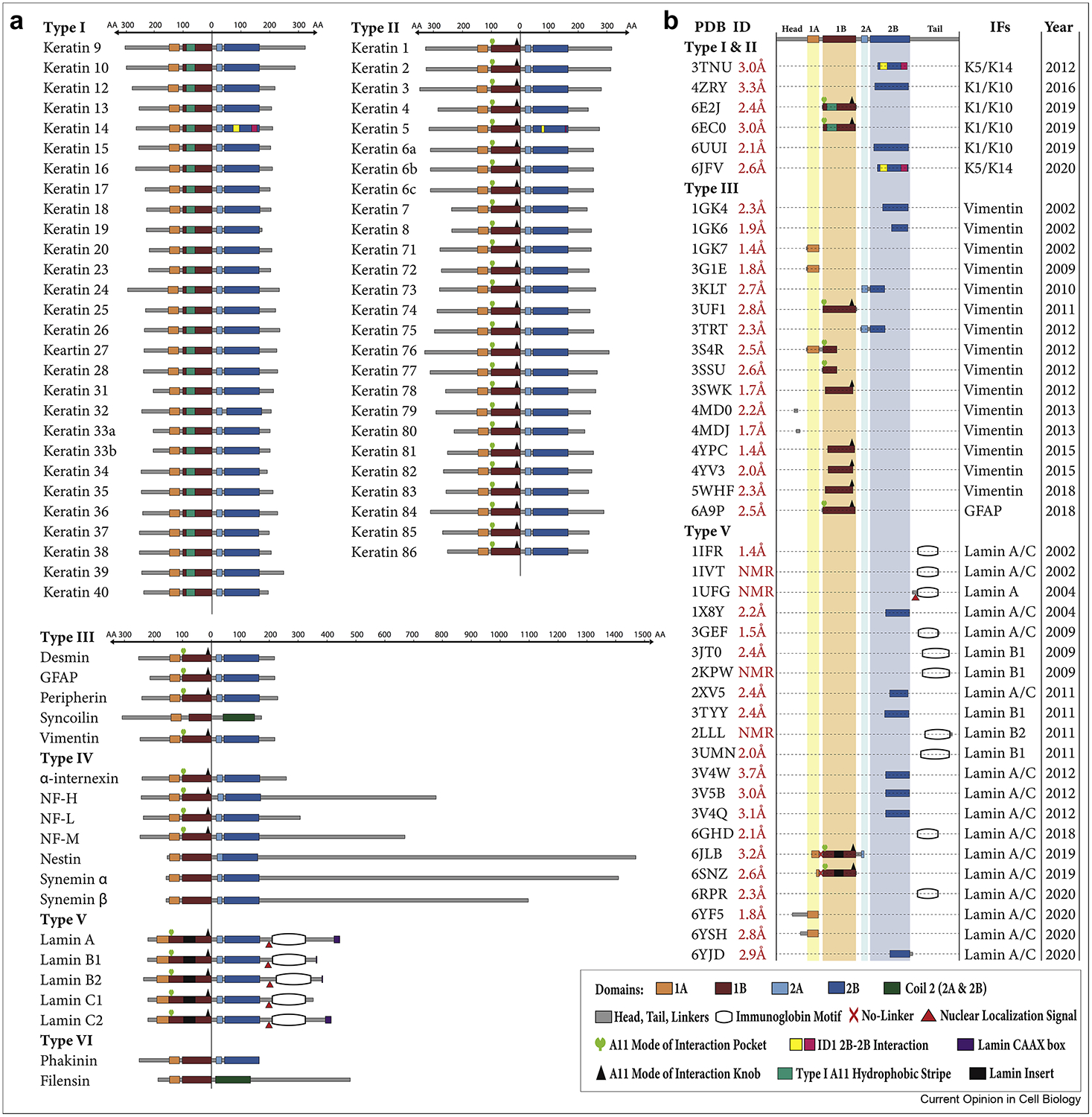
(A) Diagram of all intermediate filament (IF) proteins, separated by type, showing precise, to-scale mapping of the amino acid length of the various IF domains. All IF proteins are aligned with the “0” reference point at the end of the 1B domain. Specific structural elements identified to date, such as the A11 knob, A11 pocket, A11 hydrophobic stripe, nuclear localization signal, and immunoglobulin-fold domain, are annotated on the appropriate IFs. The variability in the length of the head and tail regions is evident across IF types. The 1B domain insert and immunoglobulin fold domain distinguish lamins from all other IFs. (B) The 40 current atomic resolution crystal or NMR structures of IF proteins deposited in the Protein Data Bank (PDB). Indicated in the panel are the PDB ID number, the resolution of all crystal structures, to-scale mapping of the IF fragment whose structure was determined, and the year of release in the PDB. Of these 40 structures, only 4 capture a physiological mutation known to cause human disease: 6E2J (K1S233L/K10 mutant complex associated with epidermolytic palmoplantar keratoderma), 3V4W (E347K lamin A/C mutant associated with dilated cardiomyopathy), 3V4Q (R335W lamin A/C mutant associated with dilated cardiomyopathy), and 3GEF (R482W lamin A/C immunoglobulin domain mutant associated with dominant familial partial lipodystrophy).
Advances in Rod Domain Structure
The rod domain historically was divided into four helical domains (1A, 1B, 2A, 2B) separated by linkers (L1, L12, L2). Identification of hendecad repeats in 2A and L2, and determination of a fully helical vimentin coil 2 structure led to calling 2A, L2, and 2B simply “coil 2” [10, 11]. We use here the historical nomenclature because it enhances direct correlation between newly determined structural mechanisms and prior biochemical and clinicopathologic studies.
Numerous biophysical studies over the years have defined a basic assembly process for IFs. Using the heteromeric keratins as an example, IF assembly is believed to occur as follows: a type I and type II keratin pairs to form a parallel heterodimer; two heterodimers bind to form an anti-parallel tetramer; eight tetramers merge into a protofibril/unit-length filament (ULF); then four protofibrils assemble into the complete KIF [7, 12, 13]. In contrast to keratins, homomeric IFs self-assemble into parallel homodimers. Nearest-neighbor cross-linking studies defined four ways that IF dimers align to form a tetramer [14, 15]. These tetramer alignments are denoted A11, A22, A12, and ACN, and are defined in the “Fundamental IF Principles” box.
Recent structures of coils 1B and 2B provided insight into higher-order IF assembly by capturing molecular interactions above the dimer level. Structural properties of the IF tetramer, considered the building block for mature filaments [7, 16–19], were elucidated through coil 1B crystal structures of keratin 1-keratin 10 (K1/K10) (PDB 6EC0) [19], lamin A (PDB ID 6JLB and 6SNZ) [20, 21], and GFAP (PDB 6A9P) [22]. These structures agree with an earlier tetrameric vimentin structure (PDB 3UF1) [23], and confirmed that the 1B tetramer is an anti-parallel dimer of dimers in an A11 mode of alignment [15]. The tetrameric assembly relies on a symmetric, highly conserved, “anchoring knob-hydrophobic pocket” interaction at each end of the 1B domain (Figure 2) [19]. Disruption of the knob-pocket interaction drastically inhibited assembly of both heteromeric (K1/K10, K8/K18) and homomeric (vimentin) filaments in vitro [19].
Figure 2. Molecular interactions contributing to intermediate filament assembly.
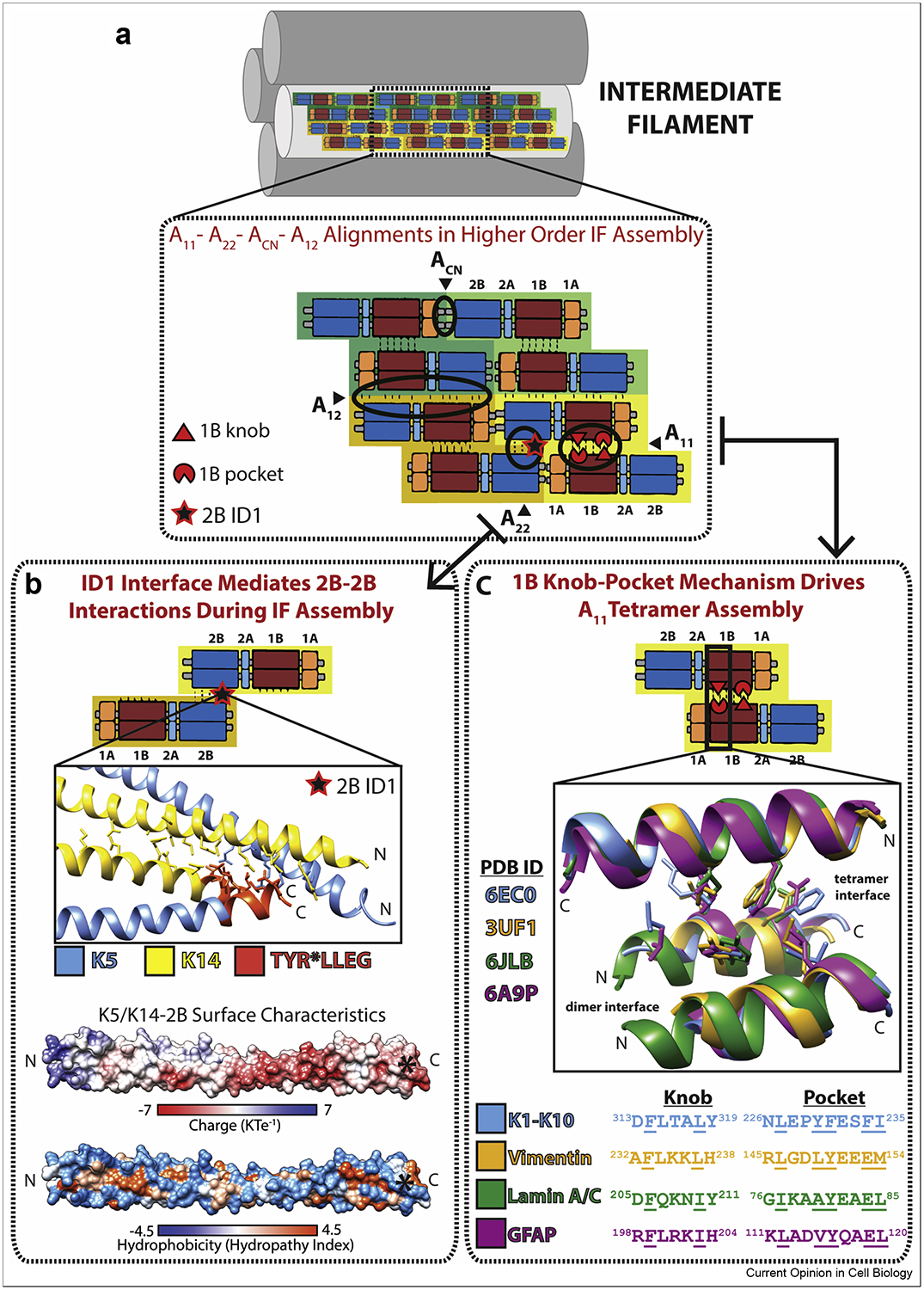
(A) Schematic of an intermediate filament (IF) depicting the four major alignments of anti-parallel tetramers believed to occur within a unit length filament (ULF) and mature IF. The model is based on anti-parallel tetramers being the building block of IFs because of their stability, which comes largely from A11 knob-pocket interactions. Octamers form longitudinally via ACN and A22 interactions, the latter of which may be stabilized in part by contacts identified in the “ID1” interface. Octamers are joined in an anti-parallel orientation via A12 interactions, providing lateral filament growth, and further association of octameric units leads to ULF formation. (B) The ID1 interaction involves many K14 residues (yellow) and a few K5 residues (blue) at the N and C termini of the K5/K14 2B domain (PDB ID 6JFV). The highly conserved 2B residues TYR*LLEG (orange) are present at the C-terminus of the structure. Electrostatic and hydrophobic surfaces of K5/K14–2B show that the TYR*LLEG motif (asterisk) occupies an acidic portion of the molecule. The electrostatic and hydrophobic surfaces were calculated using APBS/PDB2PQR and UCSF Chimera software. (C) The 1B anchoring knob-hydrophobic pocket mechanism that stabilizes A11 tetramer formation is structurally homologous in four coil 1B structures: heteromeric K1/K10 and homomeric vimentin, lamin A/C, and GFAP.
The structure of the A11 lamin A tetramer from 1A to the beginning of the 2B domain (residues 27–282) (PDB ID 6JLB) depicted symmetric “knob-pocket” interactions in the 1B domain [20]. A shorter tetrameric lamin A structure of linker L1 and 1B (residues 63–222) (PDB ID 6SNZ) did not capture a tight knob-pocket interaction [21], suggesting that crystal lattice contacts displaced the physiological orientation of the dimers. Both lamin A structures demonstrated that the 42 residue lamin-specific 1B insert contributes ~1/2 superhelix turn in the middle of the 1B domain. The implications of this turn are that it preserves heptad periodicity for proper dimer formation, conserves the positions of the distal coiled-coil regions for proper tetramer formation, and provides additional inter-dimer molecular contacts [21]. The lamin A structures also show that linkers L1, L12, and L2 adopt an untwisted helical conformation, but retain significant capacity to bend (Figure 3A). Advances in understanding lamin structure enabled a 3.5 nm-wide model for lamin assembly consistent with recent cryo-electron tomography (ET) data [20, 24].
Figure 3. Structural basis for linker bending, LARK formation in IF heads and tails, and interactions with IF-associated proteins.
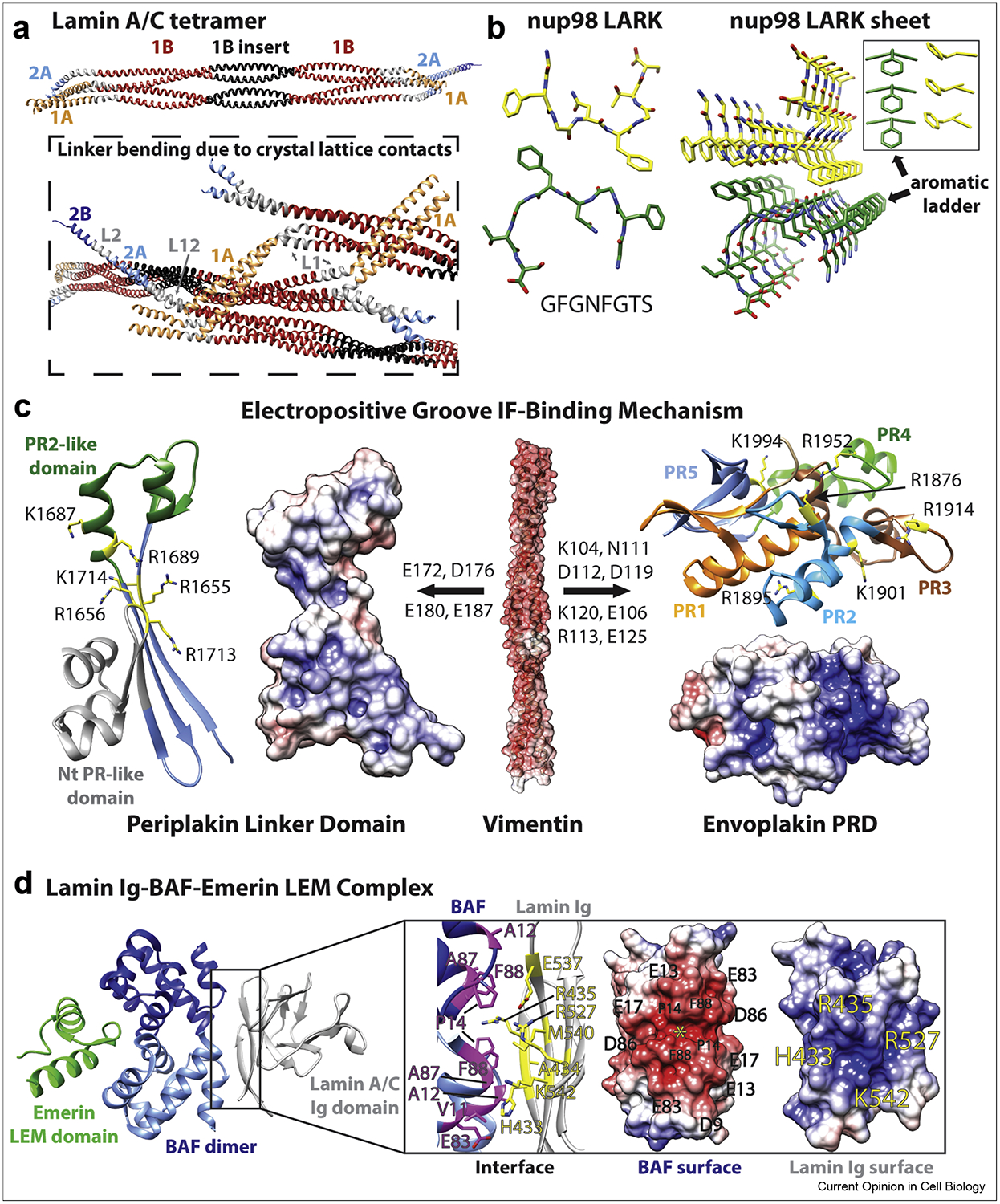
(A) The largest IF structure published to date is the lamin A/C 1A-1B-2A tetramer (PDB ID 6JLB), shown here (top) as a ribbon diagram (colored according to same scheme in Figure 1a). Three lamin A/C tetramers from the crystal lattice are depicted in the box (bottom). Molecular forces caused by crystal lattice contacts resulted in significant linker L1 and L12 bending in this structure. (B) Crystal structure of the nup98 low-complexity aromatic-rich kinked segment (LARKS) (left, PDB ID 6BZM). LARKS are present in many IF heads and/or tails. Crystal lattice packing revealed that LARKS can form sheets (right) stabilized by “aromatic ladders.” (C) With its acidic molecular surface, vimentin (PDB ID 3ULF) binds at basic grooves on the surfaces of periplakin linker domain (PDB ID 4Q28) and envoplakin plakin repeat domain (PRD) (PDB ID 4QMD). Key residues in the basic groove mediating the interaction with vimentin are identified in the ribbon diagrams. Key residues in vimentin involved in plakin binding are identified above the arrows. Modeling suggested that vimentin residues E172, D176 and E180 stabilize the binding interface by ion pair interaction. (D) The structure of the lamin A/C Ig domain-BAF-emerin complex (left, PDB ID 6GHD) revealed an interaction between the acidic BAF dimer and the basic lamin Ig domain (right). Lamin A/C Ig domain’s strands β1 and β9 contact the first BAF monomer, whereas strand β1 and loop β8β9 interact with the second one. The yellow star marks the acidic groove on the BAF dimer that lamin Ig residue R435 binds into. (C, D) Electrostatic surfaces were calculated using APBS/PDB2PQR and charge is displayed from −7 KTe−1 (red) to 7 KTe−1 (blue).
2B domain interactions involved in higher-order IF assembly were revealed by crystal structures of the K1/K10 (PDB 6UUI) [25] and K5/K14 (PDB 6JFV) [26] 2B domains. These structures contain a cysteine-to-alanine substitution to prevent disulfide bonding [27, 28], resulting in denser crystal packing and lattices containing many intermolecular contacts, particularly involving the 2B C-terminus. Physiologically relevant mutations in this region, along with the presence of a highly conserved TYR*LLEGE motif [29], indicate that the 2B C-terminus contributes critical intermolecular interactions to the mature IF. A specific interaction in the K5/K14 2B crystal lattice between the N- and C- termini of adjacent 2B dimers (termed “ID1”) was used to identify residues biologically relevant to IF assembly (Figure 2B). Mutating residues in this ID1 interface disrupted K5/K14 assembly in cells and in vitro by electron microscopy [26, 30].
The crystallographically captured (and biochemically confirmed) 1B-1B and 2B-2B interactions advance our understanding of contacts important to the A11 and A22 modes of alignment previously determined by chemical cross-linking studies [15]. Models of unit-length filament (ULF) configuration incorporating these interactions have been described [19, 20, 26]. Notably, these models highlight the lack of structural data describing A12 interactions, perhaps due to the difficulty of crystallizing an IF construct large enough to capture the multiple interacting subunits in this alignment. Recent attempts to model A12 interactions in vimentin used a crystal lattice substitution approach, but lacked experimental validation [31]. The process of IF assembly has been observed via stopped-flow spectroscopy and static light scattering to involve ULFs of varying numbers of tetramers per cross-section [32]. Evidence of polymorphic ULFs, along with differences in width between mature lamins (~3.5 nm) and the other IF types (~10 nm), suggests that ULF formation is a complex process and that the molecular mechanisms by which ULFs facilitate IF assembly likely vary across the IF types.
At the super-structural level beyond the ULF, advances in IF structure have been made in hair and nail keratins (“hard keratins”) using electron tomography (ET) and small-angle neutron scattering (SANS). An analysis of existing ET data showed that keratin IFs in the hair of humans and sheep form exclusively left-handed macrofibrils, contrary to previous reports of a mixture of left- and right-handed macrofibrils [33]. Furthermore, analyzing the diffusion of water into the hair KIF assembly by SANS supported a hexagonal model of IF packing and suggested that the IF structural order in hair is decreased by the presence of a cuticle and the reduction of disulfide bonds [34].
IF Head and Tail Structure
The N-terminal (head) and C-terminal (tail) regions of IFs are low-complexity protein domains (LCDs) important to mature filament assembly and cellular functions. They are the most poorly characterized parts of IF proteins and vary significantly in sequence and size across IF types (Figure 1A). Some IFs, particularly keratins and nuclear lamins, have glycine-rich quasi-repeating peptides in the head and tail regions that form a “glycine loop” structural motif [35]. The glycine loop follows the form x(y)n, where x is an aromatic residue or occasionally a long-chain aliphatic one, y is glycine or sometimes other polar residue, and n is the variable number of tandem glycine loop motifs. The potential for glycine loops to contain β-turns was noted, but elucidating the structural contribution of glycine loops and their aromatic residues to IF assembly has proven difficult. One modeling analysis proposed an aromatic zipper mechanism that brings together two tetramers by a tetrameric terminal domain complex [36].
Recent investigations of membraneless organelles, like keratohyalin granules, demonstrate LCDs help drive macromolecular phase separation [37]. Crystal structures of several segments from LCDs identified a new structural motif termed LARKS (low-complexity aromatic rich kinked segments) [38]. LARKS adopt a kinked β-sheet structure and pair into protofilaments in which the aromatic residues provide intra- (aromatic “ladders”) and inter-sheet stabilization (Figure 3B). Since keratins are highly enriched in LARKS, these findings are relevant to understanding IF head and tail structure, including the biochemical basis for ACN head-to-tail interactions in filament elongation. Kinks at glycine or aromatic residues restrict the side chains from interdigitating across the β-sheet, distinguishing LARKS from steric zippers [38]. Crystallography of LARKS provides experimental evidence that heads and tails of certain IF types may utilize β-sheet architecture to promote macromolecular interactions involved in IF assembly and cellular processes.
For IFs that lack or have fewer LARKS or glycine-loop motifs, different ACN mechanisms may exist to enable successful head to tail interactions during the longitudinal annealing phase of IF assembly. Molecular modeling of vimentin ACN based on prior 1A and 2B vimentin structures suggested a parallel, dimeric coiled-coil interaction between two IF census motifs (the conserved N- and C-termini of the 1A and 2B coils, respectively) with three heptad repeat overlap [39]. Progress has also been made in elucidating the molecular basis for the ACN interaction in lamins. Three cleverly designed chimeric proteins fusing capping motifs onto N- or C-terminal lamin A rod domain fragments enabled the determination of three lamin A crystal structures [40]. Together, these structures produced a molecular model for the lamin A ACN tetramer which contains a 6.5 nm overlap of the rod domain ends.
Structural basis of IF-Protein Complexes
IFs perform many tissue-specific functions including mechanical support of cells and regulation of intracellular organization, stress response, cell growth, proliferation, migration and death. It is not surprising then that IFs interact with multiple cellular proteins and these complexes play an important role in human health [6, 41]. For example, the interactions of keratins 1/10 with desmoplakin in the desmosome and filaggrin in the stratum corneum are important for establishing integrity of the epidermal skin barrier [37, 42–45]. Our molecular understanding of IF interactions with IF-associated proteins (IFAPs) remains limited. Flexibility, insolubility of the mature filament, and relatively weak binding of IFs to IFAPs in vitro are several reasons for this limitation. Recent research, however, has elucidated some of the mechanisms used by IF proteins to bind target proteins [46–48].
Mechanism of IF protein binding to plakins
Cornified envelope formation, an essential component of the epidermal permeability barrier, is initiated during keratinocyte terminal epidermal differentiation when envoplakin dimerizes with periplakin [49, 50]. Both proteins belong to the plakin family, a group of proteins which shares a common domain organization: an N-terminal plakin domain, a central coiled-coil domain, a C-terminal tail with a linker domain, and at least one plakin repeat domain (PRD) (except in the case of periplakin, which lacks a PRD domain) [51]. Atomic resolution structures were determined for the three types of PRDs (denoted A, B, C) [52–54]. These structures revealed a conserved basic groove in PRDs which may act as an IF-binding site by accommodating acidic patches that exist on the surface of IF proteins. PRD residues important for the interaction of envoplakin with vimentin and keratin were identified by mutational analysis and nuclear magnetic resonance (NMR) [53]. Similarly, the crystal structure of the periplakin C-terminal linker domain (PDB ID 4Q28) was used to study vimentin-periplakin interactions [48]. The periplakin linker is sufficient for binding to keratin 8 and vimentin [55]. Molecular modeling of the periplakin, desmoplakin, and plectin linkers demonstrated, like the linker crystal structure, that two plakin repeat (PR)-like folds exist and flank a basic groove–a predicted IF binding site [48, 56] (Figure 3C). Several positively charged arginine and lysine residues exist in the basic groove of periplakin, suggesting that electrostatic forces are the primary driver of its interaction with IF proteins. Mutations of these basic residues were made and periplakin-vimentin (coils 1A and 1B) binding analyzed using an in vitro microscale thermophoresis (MST)-based assay. The binding is weak: the KD ranges from 70 to 31 μM in 150 to 10 mM NaCl, respectively. Though desmoplakin’s linker domain is structurally similar to periplakin’s, no binding was detected by MST between the desmoplakin linker and vimentin. Linker domains include only two PRs, which may explain why vimentin binding is weaker if compared to the PRDs, which have 4.5 PRs forming a positively charged groove [53, 54]. However, even in the case of the envoplakin PRD, the KD for vimentin (coils 1A and 1B) binding is about 19 μM. More studies are needed to understand how these weak binding affinities are strengthened through elevations in intracellular concentrations of the interacting proteins and/or multimerization of binding interfaces.
These studies showed that PRD and linker domains in plakin family proteins can connect plakins with IFs; however, they did not reveal the molecular specificity of the interactions. For example, one isoform of microtubule actin cross-linking factor (MACF1b) contains a plakin region comprised of multiple PRDs which targets MACF1b to the Golgi complex [57]. Improved biochemical understanding of how PRDs discriminate between binding partners could explain why the PRDs of MACF1b, but not other plakins, target it to the Golgi complex [51, 57].
Mechanism of Lamin Ig domain binding to BAF and emerin
The tail of human lamin A/C is important for its function. High numbers of mutations in the tail, specifically in its Ig-fold domain, are linked to muscle diseases, lipodystrophies, or progeroid syndromes [58]. The mutations primarily localize to the hydrophobic core where they destabilize the Ig domain structure, or to two solvent-exposed sites of the Ig domain where they disrupt interactions with a binding partner [58–60]. Emerin is a multifunctional nuclear envelope protein involved in regulation of gene expression and genome organization. EmN contains a LAP2-emerin-MAN1 (LEM) domain that is important for the ability of emerin to bind lamin A/C tail and to tether repressive chromatin at the nuclear periphery in a barrier-to-autointegration (BAF)-dependent manner [61, 62]. Recently two crystal structures of the ternary complex (TC) comprising the human lamin A/C Ig-fold domain, emerin N-terminal domain (EmN), and DNA-binding protein BAF were determined [46, 47]. The first structure focused on the TC [46], while the second provided insights into the effect of the emerin LEM-domain mutations [47].
The TC structure (PDB ID 6GHD) revealed that BAF dimerization is needed for emerin and lamin A/C binding [46]. Both monomers of the BAF dimer bind emerin LEM domain on one side, while lamin A/C Ig domain binds at the opposite side (Figure 3D). Analysis of the binding interface suggested that progeroid syndrome mutations in the lamin A/C Ig-fold domain decreased binding affinity for BAF [46]. NMR analysis also showed that lamin Ig-fold domain may directly bind EmN, but only if EmN is oligomerized. This structural and biochemical data led to the first model of the interface between LEM-domain proteins, the nucleoskeleton, and the chromatin-associated protein BAF [46].
Genotype-Structurotype-Phenotype Correlation
A common paradigm used to discuss and comprehend human diseases is the correlation between a person’s genotype and their clinical phenotype. While this paradigm has its merits, it ignores the wealth of structural biology information that bridges genotype and phenotype. Hence, it was proposed that scientific explanation of human disease should strive for a more inclusive paradigm: genotype-structurotype-phenotype correlation [25, 30]. “Structurotype” refers to the macromolecular structure dictated by a person’s genotype and how that structure drives biochemical and functional processes in humans to cause a specific clinical phenotype (Figure 4). IF proteins are excellent models for illustrating genotype-structurotype-phenotype correlation, since dozens of IF-pathies exist [1].
Figure 4. Genotype-structurotype-phenotype correlation emphasizes the role structural mechanisms play in the pathogenesis of human diseases.
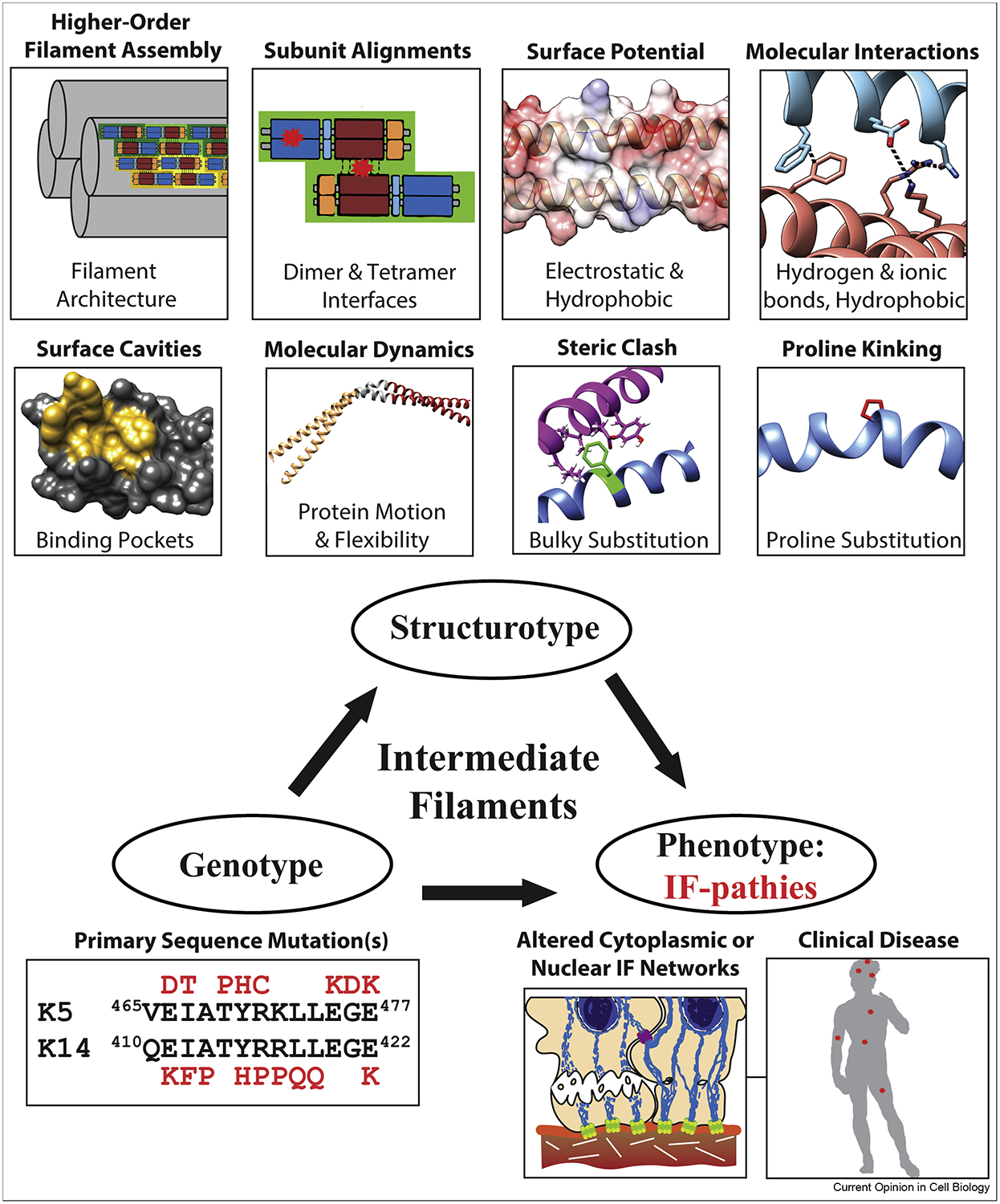
Mutations in IFs can disrupt IF structure through a variety of mechanisms that ultimately inhibit or distort IF assembly (top). Common outcomes from IF mutations are the formation of cytoplasmic (or nuclear) aggregates, misfolded filaments, re-organized IF networks, and decreased mechanical support to the cell (often leading to cell fragility).
Recent structural characterization of several physiologically relevant IF mutations reinforces the structurotype concept. The crystal structure of the K1/K10 1B heterocomplex containing a S233L mutation in K1, which is known to cause epidermolytic palmoplantar keratoderma through tonotubule formation [63, 64], demonstrated increased hydrophobicity on the K1/K10 1B tetramer surface [19]. This extra hydrophobicity drove higher-order aggregation (octamer in the structure) that was not seen in the wild-type structure. A three-residue (Q113E114L115) deletion in the proximal 1A domain of desmin is associated with left ventricular noncompaction cardiomyopathy and leads to cytoplasmic accumulation of desmin and degraded sarcomeres [65]. A molecular model of the desmin 1A domain suggests this deletion causes a local twisting of the helical backbone. A combination of NMR, circular dichroism, and fluorescence spectroscopy along with molecular dynamics simulation were used to characterize the lamin A Ig domain containing mutation W514R, which is associated with skeletal muscle dystrophy [66]. The mutant W514R lamin A Ig domain demonstrated enhanced oligomerization, likely from increased positive electrostatic surface potential, resulting in misshapen laminar networks. Lamin A mutations Y45C (muscular dystrophy) and L59R (dilated cardiomyopathy) in coil 1A had opposite effects on the binding of a coil 2B fragment: Y45C decreased binding, whereas L59R strengthened binding suggesting that enhanced filament strength can also be pathogenic by preventing proper dynamic remodeling [20]. The GFAP 1B structure was used to analyze structural perturbations caused by 14 mutations associated with Alexander disease, a fatal neurodegenerative disease [22]. Like prior analyses of K1/K10 mutations associated with keratinopathies [27, 67] and K5/K14 mutations associated with epidermolysis bullosa simplex [28], the GFAP 1B mutations altered the IF structure in a variety of ways. These include altered interfaces (dimer vs. tetramer), surface potential (electrostatic vs. hydrophobic), molecular interactions (hydrogen bond, ionic, hydrophobic), and IF protein packing (steric clashes, helix kinking).
The Future of IF Structural Biology
IFs accomplish numerous functions throughout the human body, making their study relevant to many organ systems [2–5, 68]. Given the ubiquity of IFs in human biology, it is surprising that the structural basis of IF function remains poorly understood; only a few interactions contributing to IF assembly have been captured and the assembly processes among the different IF types remain to be fully elucidated. It has been 18 years since the first IF protein structures were determined. Today, the field stands at 43 atomic resolution structures, the majority belonging to homomeric vimentin (16) and lamin (21). In contrast, only 6 heteromeric keratin structures have been determined, accounting for 4 out of 54 keratins (7.4%). Type IV neurofilaments have zero structures. While it is true that many structural elements are conserved across IFs, dissimilarities among IFs in oligomizeration properties and head and tail length and sequence indicate that new assembly mechanisms discovered in IF structures may not be directly applicable across all IF types. Thus, the IF structure field needs to greatly expand its investigation of these important differences that may account for the varied cellular functions of IFs and plethora of IF-pathies.
Current knowledge of IF structural biology lags behind that of actin microfilaments and microtubules, both of which are actively targeted for pharmacological purposes [69, 70]. Our inability to manipulate IF systems for human therapeutic benefit has been lamented [71]. The association of IFs with cancer, as just one example, justifies the need for heightened attention to IF structure and the pharmacological manipulation of IF biology [68, 72–79].
Continued integration of solution biophysics, structure determination techniques, and cell biology to rigorously validate discoveries is essential for the advancement of the IF field. The increasing use and capabilities of cryo-EM has the potential to capture higher-order filament interactions which x-ray crystallography, to date, has been unable to accomplish. Ultimately, we expect that the body of structural IF data will grow over the coming years and provide valuable advances in our understanding of disease mechanisms and the eventual development of IF-targeting drugs.
Highlights.
Intermediate filament domain structures provide insight into assembly mechanisms
Heads and tails may contain low complexity aromatic rich kinked segments
Electrostatic surfaces help intermediate filaments bind partner proteins
Genotype and structurotype contribute to the clinic phenotype in human diseases
Acknowledgements
We thank Professor Leonard Milstone for constructive feedback. This work was supported by the Richard K. Gershon, M.D., Student Research Fellowship at Yale University School of Medicine (to SAE), and NIH/NIAMS Awards K08AR070290 and R03AR076484 (to CGB).
List of abbreviations:
- BAF
Binding protein barrier to autointegration
- EM
electron microscopy
- EmN
Emerin N-terminal domain
- ET
electron tomography
- GFAP
glial fibrillary acidic protein
- IF
intermediate filament
- IFAP
intermediate filament-associated protein
- KIF
keratin intermediate filament
- LARKS
low-complexity aromatic rich kinked segments
- LCD
low-complexity protein domain
- LEM
LAP2-emerin-MAN1
- MACF1b
microtubule actin cross-linking factor 1b
- MST
microscale thermophoresis
- NMR
Nuclear magnetic resonance
- PDB
Protein Data Bank
- PR
plakin repeat
- PRD
plakin repeat domain
- SANS
small-angle neutron scattering
- TC
ternary complex
- ULF
unit length filament
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: SAE, MH, and CGB are inventors on pending patent PCT/US19/55115.
References
- 1.Omary MB, “IF-pathies”: a broad spectrum of intermediate filament-associated diseases. J Clin Invest, 2009. 119(7): p. 1756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danielsson F, et al. , Vimentin Diversity in Health and Disease. Cells, 2018. 7(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan A, et al. , Neurofilaments and Neurofilament Proteins in Health and Disease. Cold Spring Harb Perspect Biol, 2017. 9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toivola DM, et al. , Keratins in health and disease. Curr Opin Cell Biol, 2015. 32: p. 73–81. [DOI] [PubMed] [Google Scholar]
- 5.Jones LN and Steinert PM, Hair keratinization in health and disease. Dermatol Clin, 1996. 14(4): p. 633–50. [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S, Cytoplasmic Intermediate Filaments in Cell Biology. Annu Rev Cell Dev Biol, 2018. 34: p. 1–28. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann H and Aebi U, Intermediate Filaments: Structure and Assembly. Cold Spring Harb Perspect Biol, 2016. 8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strelkov SV, et al. , Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. J Mol Biol, 2001. 306(4): p. 773–81. [DOI] [PubMed] [Google Scholar]
- 9.Guzenko D, Chernyatina AA, and Strelkov SV, Crystallographic Studies of Intermediate Filament Proteins. Subcell Biochem, 2017. 82: p. 151–170. [DOI] [PubMed] [Google Scholar]
- 10.Nicolet S, et al. , Atomic structure of vimentin coil 2. J Struct Biol, 2010. 170(2): p. 369–76. [DOI] [PubMed] [Google Scholar]
- 11.Parry DA, Hendecad repeat in segment 2A and linker L2 of intermediate filament chains implies the possibility of a right-handed coiled-coil structure. J Struct Biol, 2006. 155(2): p. 370–4. [DOI] [PubMed] [Google Scholar]
- 12.Parry DA, Marekov LN, and Steinert PM, Subfilamentous protofibril structures in fibrous proteins: cross-linking evidence for protofibrils in intermediate filaments. J Biol Chem, 2001. 276(42): p. 39253–8. [DOI] [PubMed] [Google Scholar]
- 13.Aebi U, et al. , The fibrillar substructure of keratin filaments unraveled. J Cell Biol, 1983. 97(4): p. 1131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinert PM, Marekov LN, and Parry DA, Diversity of intermediate filament structure. Evidence that the alignment of coiled-coil molecules in vimentin is different from that in keratin intermediate filaments. J Biol Chem, 1993. 268(33): p. 24916–25. [PubMed] [Google Scholar]
- 15.Steinert PM, et al. , Keratin intermediate filament structure. Crosslinking studies yield quantitative information on molecular dimensions and mechanism of assembly. J Mol Biol, 1993. 230(2): p. 436–52. [DOI] [PubMed] [Google Scholar]
- 16.Quinlan RA, et al. , Heterotypic tetramer (A2D2) complexes of non-epidermal keratins isolated from cytoskeletons of rat hepatocytes and hepatoma cells. J Mol Biol, 1984. 178(2): p. 365–88. [DOI] [PubMed] [Google Scholar]
- 17.Mücke N, et al. , Molecular and biophysical characterization of assembly-starter units of human vimentin. J Mol Biol, 2004. 340(1): p. 97–114. [DOI] [PubMed] [Google Scholar]
- 18.Geisler N, Schünemann J, and Weber K, Chemical cross-linking indicates a staggered and antiparallel protofilament of desmin intermediate filaments and characterizes one higher-level complex between protofilaments. Eur J Biochem, 1992. 206(3): p. 841–52. [DOI] [PubMed] [Google Scholar]
- 19.**.Eldirany SA, et al. , Human keratin 1/10–1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly. EMBO J, 2019. 38(11). [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies highly conserved knob-pocket mechanism as key to the A11 tetramer based on the crystal structure of the K1/K10–1B tetramer and electron microscopy of knob-mutated filaments. The crystal structure of K1/K10–1B harboring the disease-causing mutation S233L in Keratin 1 also captures aberrant hydrophobic interactions thought to be the mechanism of the disease epidermolytic palmoplantar keratoderma.
- 20.**.Ahn J, et al. , Structural basis for lamin assembly at the molecular level. Nat Commun, 2019. 10(1): p. 3757. [DOI] [PMC free article] [PubMed] [Google Scholar]; Crystal structure of a lamin A/C construct spanning coils 1A, 1B, and half of coil 2 captures an A11 tetramer. Binding between a stabilized tetramer and coil 2 is demonstrated, and the effect of disease-causing mutations on the tetramer-coil 2 interaction is characterized.
- 21.**.Lilina AV, et al. , Lateral A11 type tetramerization in lamins. J Struct Biol, 2020. 209(1): p. 107404. [DOI] [PubMed] [Google Scholar]; Crystal structure of lamin A/C coil 1B and part of 1A captures the A11 tetramer. Detailed comparison to keratin and vimentin 1B structures.
- 22.*.Kim B, Kim S, and Jin MS, Crystal structure of the human glial fibrillary acidic protein 1B domain. Biochem Biophys Res Commun, 2018. 503(4): p. 2899–2905. [DOI] [PubMed] [Google Scholar]; Crystal structure of GFAP 1B domain represents the only non-vimentin Type III IF structure to date. Molecular modeling of mutations related to Alexander disease.
- 23.Aziz A, et al. , The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography. J Biol Chem, 2012. 287(34): p. 28349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turgay Y, et al. , The molecular architecture of lamins in somatic cells. Nature, 2017. 543(7644): p. 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.**.Lomakin IB, et al. , Crystal structure of keratin 1/10(C401A) 2B heterodimer demonstrates a proclivity for the C-terminus of helix 2B to form higher order molecular contacts. Yale J Biol Med, 2020. 93(1). [PMC free article] [PubMed] [Google Scholar]; Crystal structure of K1/K10C401A 2B heterocomplex demonstrated a propensity for the 2B C-terminus to make higher-order inter-filament contacts.
- 26.**.Lee CH, et al. , Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly. Structure, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies a 2B-2B interface, termed ID1, based on the crystal structure of the K5/K14C367A 2B domain and demonstrates the importance of the interaction to filament assembly via electron microscopy of mutated filaments and imaging of cells transfected with mutated filaments.
- 27.Bunick CG and Milstone LM, The X-Ray Crystal Structure of the Keratin 1-Keratin 10 Helix 2B Heterodimer Reveals Molecular Surface Properties and Biochemical Insights into Human Skin Disease. J Invest Dermatol, 2017. 137(1): p. 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee CH, et al. , Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol, 2012. 19(7): p. 707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson AK, Coulombe PA, and Fuchs E, The roles of K5 and K14 head, tail, and R/K L L E G E domains in keratin filament assembly in vitro. J Cell Biol, 1992. 119(2): p. 401–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eldirany SA, Ho M, and Bunick CG, The Interface between Keratin Structurotype and Human Disease. Structure, 2020. 28(3): p. 271–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang AH, et al. , A crystal structure of coil 1B of vimentin in the filamentous form provides a model of a high-order assembly of a vimentin filament. FEBS J, 2018. 285(15): p. 2888–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mücke N, et al. , Assembly Kinetics of Vimentin Tetramers to Unit-Length Filaments: A Stopped-Flow Study. Biophys J, 2018. 114(10): p. 2408–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.*.Harland DP, et al. , Helical twist direction in the macrofibrils of keratin fibres is left handed. J Struct Biol, 2019. 206(3): p. 345–348. [DOI] [PubMed] [Google Scholar]; Analyzes 41 electron tomograms from previously published studies of hair and wool and reimaged some samples using a handedness marker to conclude that all keratin macrofibrils from humans and sheep (and likely all mammals) have a left handed helical twist.
- 34.*.Murthy NS, Wang W, and Kamath Y, Structure of intermediate filament assembly in hair deduced from hydration studies using small-angle neutron scattering. J Struct Biol, 2019. 206(3): p. 295–304. [DOI] [PubMed] [Google Scholar]; Uses small-angle neutron scattering to investigate the water signal from hair under various conditions. A hexagonal lattice of keratin assembly in hair is proposed based on the observed water distribution, and the effects of humidity, disulfide bonds, hair cuticles, and oils on hair structure and hydration are investigated.
- 35.Steinert PM, et al. , Glycine loops in proteins: their occurrence in certain intermediate filament chains, loricrins and single-stranded RNA binding proteins. Int J Biol Macromol, 1991. 13(3): p. 130–9. [DOI] [PubMed] [Google Scholar]
- 36.Badowski C, et al. , Modeling the Structure of Keratin 1 and 10 Terminal Domains and their Misassembly in Keratoderma. J Invest Dermatol, 2017. 137(9): p. 1914–1923. [DOI] [PubMed] [Google Scholar]
- 37.Quiroz FG, et al. , Liquid-liquid phase separation drives skin barrier formation. Science, 2020. 367(6483). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.**.Hughes MP, et al. , Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science, 2018. 359(6376): p. 698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]; Crystal structures of several low complexity protein domains reveal a common kinked B-sheet structure termed a “low complexity aromatic rich kinked segment” or LARK. Analysis of the human proteome shows that LARKS are present in a variety of proteins including keratins and keratin-associated proteins.
- 39.Dey R and Burkhard P, A proposed atomic model of the head-to-tail interaction in the filament structure of vimentin. J Biomol Struct Dyn, 2019: p. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.**.Stalmans G, et al. , Addressing the Molecular Mechanism of Longitudinal Lamin Assembly Using Chimeric Fusions. Cells, 2020. 9(7). [DOI] [PMC free article] [PubMed] [Google Scholar]; Determines the structures of three lamin A fragments fused to special capping motifs to uncover molecular insights into the ACN tetrameric alignment in lamin filaments.
- 41.Bott CJ and Winckler B, Intermediate filaments in developing neurons: Beyond structure. Cytoskeleton (Hoboken), 2020. [DOI] [PubMed] [Google Scholar]
- 42.Sumigray KD and Lechler T, Cell adhesion in epidermal development and barrier formation. Curr Top Dev Biol, 2015. 112: p. 383–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinbest AJ, et al. , Structural properties of target binding by profilaggrin A and B domains and other S100 fused-type calcium-binding proteins. J Dermatol Sci, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown SJ and McLean WH, One remarkable molecule: filaggrin. J Invest Dermatol, 2012. 132(3 Pt 2): p. 751–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandilands A, et al. , Filaggrin in the frontline: role in skin barrier function and disease. J Cell Sci, 2009. 122(Pt 9): p. 1285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.**.Samson C, et al. , Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases. Nucleic Acids Res, 2018. 46(19): p. 10460–10473. [DOI] [PMC free article] [PubMed] [Google Scholar]; Crystal structure of the Lamin A/C Ig fold domain-BAF-emerin complex reveals BAF dimerization is important for Lamin A/C-emerin interaction and lamin A/C mutations that cause progeroid syndromes decrease binding affinity for BAF.
- 47.Essawy N, et al. , An Emerin LEM-Domain Mutation Impairs Cell Response to Mechanical Stress. Cells, 2019. 8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.*.Odintsova E, et al. , Binding of the periplakin linker requires vimentin acidic residues D176 and E187. Commun Biol, 2020. 3(1): p. 83. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mutational affinity and cell transfection studies and docking analysis demonstrate binding of acidic vimentin residues by a basic groove at the periplakin linker surface.
- 49.Al-Jassar C, et al. , Hinged plakin domains provide specialized degrees of articulation in envoplakin, periplakin and desmoplakin. PLoS One, 2013. 8(7): p. e69767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Candi E, Schmidt R, and Melino G, The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol, 2005. 6(4): p. 328–40. [DOI] [PubMed] [Google Scholar]
- 51.Hu L, et al. , Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer. Int J Mol Sci, 2018. 19(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi HJ, et al. , Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure. Nat Struct Biol, 2002. 9(8): p. 612–20. [DOI] [PubMed] [Google Scholar]
- 53.Fogl C, et al. , Mechanism of intermediate filament recognition by plakin repeat domains revealed by envoplakin targeting of vimentin. Nat Commun, 2016. 7: p. 10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang H, et al. , Structure of the Intermediate Filament-Binding Region of Desmoplakin. PLoS One, 2016. 11(1): p. e0147641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kazerounian S, Uitto J, and Aho S, Unique role for the periplakin tail in intermediate filament association: specific binding to keratin 8 and vimentin. Exp Dermatol, 2002. 11(5): p. 428–38. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 2008. 9: p. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin CM, et al. , Microtubule actin crosslinking factor 1b: a novel plakin that localizes to the Golgi complex. J Cell Sci, 2005. 118(Pt 16): p. 3727–38. [DOI] [PubMed] [Google Scholar]
- 58.Krimm I, et al. , The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure, 2002. 10(6): p. 811–23. [DOI] [PubMed] [Google Scholar]
- 59.Dhe-Paganon S, et al. , Structure of the globular tail of nuclear lamin. J Biol Chem, 2002. 277(20): p. 17381–4. [DOI] [PubMed] [Google Scholar]
- 60.Verstraeten VL, et al. , Compound heterozygosity for mutations in LMNA causes a progeria syndrome without prelamin A accumulation. Hum Mol Genet, 2006. 15(16): p. 2509–22. [DOI] [PubMed] [Google Scholar]
- 61.Berk JM, Tifft KE, and Wilson KL, The nuclear envelope LEM-domain protein emerin. Nucleus, 2013. 4(4): p. 298–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koch AJ and Holaska JM, Emerin in health and disease. Semin Cell Dev Biol, 2014. 29: p. 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terron-Kwiatkowski A, et al. , Mutation S233L in the 1B domain of keratin 1 causes epidermolytic palmoplantar keratoderma with “tonotubular” keratin. J Invest Dermatol, 2006. 126(3): p. 607–13. [DOI] [PubMed] [Google Scholar]
- 64.Wevers A, Kuhn A, and Mahrle G, Palmoplantar keratoderma with tonotubular keratin. J Am Acad Dermatol, 1991. 24(4): p. 638–42. [DOI] [PubMed] [Google Scholar]
- 65.Marakhonov AV, et al. , Noncompaction cardiomyopathy is caused by a novel in-frame desmin (DES) deletion mutation within the 1A coiled-coil rod segment leading to a severe filament assembly defect. Hum Mutat, 2019. 40(6): p. 734–741. [DOI] [PubMed] [Google Scholar]
- 66.Dutta S, et al. , Skeletal Muscle Dystrophy mutant of lamin A alters the structure and dynamics of the Ig fold domain. Sci Rep, 2018. 8(1): p. 13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hinbest AJ, et al. , Molecular Modeling of Pathogenic Mutations in the Keratin 1B Domain. Int J Mol Sci, 2020. 21(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karantza V, Keratins in health and cancer: more than mere epithelial cell markers. Oncogene, 2011. 30(2): p. 127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nepali K, et al. , Tubulin inhibitors: a patent survey. Recent Pat Anticancer Drug Discov, 2014. 9(2): p. 176–220. [DOI] [PubMed] [Google Scholar]
- 70.Gandalovičová A, et al. , Migrastatics-Anti-metastatic and Anti-invasion Drugs: Promises and Challenges. Trends Cancer, 2017. 3(6): p. 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun J, et al. , High-Throughput Screening for Drugs that Modulate Intermediate Filament Proteins. Methods Enzymol, 2016. 568: p. 163–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma P, et al. , Intermediate Filaments as Effectors of Cancer Development and Metastasis: A Focus on Keratins, Vimentin, and Nestin. Cells, 2019. 8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trogden KP, et al. , An image-based small-molecule screen identifies vimentin as a pharmacologically relevant target of simvastatin in cancer cells. FASEB J, 2018. 32(5): p. 2841–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bollong MJ, et al. , A vimentin binding small molecule leads to mitotic disruption in mesenchymal cancers. Proc Natl Acad Sci U S A, 2017. 114(46): p. E9903–E9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sutoh Yoneyama M, et al. , Vimentin intermediate filament and plectin provide a scaffold for invadopodia, facilitating cancer cell invasion and extravasation for metastasis. Eur J Cell Biol, 2014. 93(4): p. 157–69. [DOI] [PubMed] [Google Scholar]
- 76.Grin B, et al. , Withaferin a alters intermediate filament organization, cell shape and behavior. PLoS One, 2012. 7(6): p. e39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satelli A and Li S, Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci, 2011. 68(18): p. 3033–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schoumacher M, et al. , Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol, 2010. 189(3): p. 541–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hendrix MJ, et al. , Role of intermediate filaments in migration, invasion and metastasis. Cancer Metastasis Rev, 1996. 15(4): p. 507–25. [DOI] [PubMed] [Google Scholar]


