Abstract
Inflammatory bowel disease (IBD) is a chronic relapsing-remitting immune-mediated disorder affecting the gut. It is common in Westernized regions and is increasing in incidence in developing countries. At a molecular level, intrinsic deficiencies in epithelial integrity, mucosal barrier function, and mechanisms of immune response and resolution contribute to the development of IBD. Traditionally two platforms have been utilized for disease modeling of IBD; in-vitro monolayer cell culture and in-vivo animal models. Both models have limitations, including cost, lack of representative cell types, lack of complexity of cellular interactions in a living organism, and xenogeneity. Organoids, three-dimensional cellular structures which recapitulate the basic architecture and functional processes of the organ of origin, hold potential as a third platform with which to investigate the pathogenesis and molecular defects which give rise to IBD. Organoids retain the genetic and transcriptomic profile of the tissue of origin over time and unlike monolayer cell culture can be induced to differentiate into most adult intestinal cell types. They may be used to model intestinal host-microbe interactions occurring at the mucosal barrier, are amenable to genetic manipulation and can be co-cultured with other cell lines of interest. Bioengineering approaches may be applied to render a more faithful representation of the intestinal epithelial niche. In this review, we outline the concept of intestinal organoids, discuss the advantages and disadvantages of the platform comparative to alternative models, and describe the translational applications of organoids in IBD.
Keywords: organoids, inflammatory bowel disease, disease modeling, mucosal defense, epithelial barrier
Introduction
Inflammatory bowel disease (IBD) is an immune-mediated relapsing-remitting chronic disorder affecting the gut. Alterations in the intestinal microbiome, defects in mucosal barrier defense and aberrant innate and adaptive immune responses appear to be critical to the development of IBD (1–7). Clinically, two major phenotypes exist, Crohn's disease (CD) and ulcerative colitis (UC). CD features transmural inflammation in a “skip lesion” or discontinuous pattern. Although it may affect any part of the gut the terminal ileum is most frequently involved (1, 2, 8–10). UC affects the colon only, although a reactive “backwash ileitis” may occur. Inflammation is limited to the mucosa and submucosa and occurs in a continuous pattern, with rectal involvement extending proximally for a variable distance. Crypt abscesses due to accumulation of neutrophils are characteristic (5, 6, 11).
Pathogenesis
The underlying mechanisms which contribute to the etiology of IBD are highly complex and not yet fully elucidated. Genetic susceptibility, environmental factors, defects in mucosal barrier function, immune dysregulation, and dysbiosis have all been demonstrated to contribute to disease pathogenesis (12–18). Activation and recruitment of CD4+ T cells to the intestinal tissue and production of a proinflammatory cytokine cascade, particularly the Th1- and Th17-associated cytokines TNFα, IFNγ, IL-12, IL-21, and IL-23 in CD and Th2-associated cytokines IL-4 and IL-13 in UC are commonly observed (5, 19–21).
Loss of a functional epithelial barrier and increased permeability of the mucus gel layer, permits abnormal contact of luminal organisms with the epithelium, provoking an inflammatory response from the immune system located in the lamina propria (14, 22–25). Failure of inflammation to resolve along with lack of restoration of normal mucosal homeostasis results in progression to chronic inflammation, inadequate epithelial restitution, and ongoing tissue damage (26). This is accompanied by characteristic disturbances in the composition of the gut microbiome, with a reduction of obligate anaerobes such as Firmicutes, an increase in facultative anaerobes such as Enterobacteriaceae and the presence of invasive strains such as adherent-invasive E.coli (AIEC) (27–30).
Genome-wide association studies have thus far identified up to 250 susceptibility loci involved in IBD. The most well-known of these is the Crohn's susceptibility locus CARD15, formerly known as NOD2, which is responsible for sensing of luminal bacterial organisms; others include IL23R and ATG16L1, which play roles in IL23 signaling and autophagy, respectively (13, 31, 32). Many susceptibility loci are genes coding for components of the mucosal barrier. These include proteins responsible for assembly and maintenance of epithelial tight junctions, intercellular adhesion and polarity, mucin and glycoprotein synthesis, bacterial sensing mechanisms, and epithelial wound healing and restitution (13, 33, 34).
Current therapeutic strategies in IBD primarily function by modification of the immune response. Biologic therapies targeting the cytokines TNFα, IL-12 and IL-23, and integrin blockers which limit the migration of leukocytes to the GI tract have greatly expanded the repertoire of treatment options (35–37). However, up to 40% of patients fail to respond to biologic therapies, and up to 50% develop secondary treatment failure after an initial successful response (38). Although impaired barrier function is also a critical event in initiation and perpetuation of IBD no therapies directed at augmenting the barrier deficiency which occurs in IBD have successfully been developed for clinical practice. Due to the phenomenon of treatment-resistant IBD in a substantial proportion of patients, alternative strategies aimed at improving intestinal barrier function are warranted. Development of such therapies requires highly faithful modeling of the intestinal barrier in the preclinical setting.
Current Models of IBD
Traditional models for IBD comprise animal models and monolayer cell culture. Some animal models used to study IBD such as DSS-colitis and TNSB-colitis are extensively utilized and well-described (39, 40). In addition to chemically induced colitis, the creation of transgenic and knockout animal strains permit investigation of inflammation arising from specific defects in innate and adaptive immune responses (41–44). These models have the benefit of replicating the complex organization and simultaneous interactions that occur in the gut in a whole organism. Such models have been indispensable in unraveling the complex pathophysiology and molecular abnormalities that occur in IBD.
However, in-vivo disease modeling in animals does have some limitations. Chemical induction of colitis occurs by a heterogeneous mechanism to that by which inflammation occurs in human disease. While cell culture can be rapidly established, the length of animal reproductive cycles means that animal experiments are a slower process. Ethical considerations exist with the use of higher vertebrates which do not apply to cell culture. In addition, while the host-microbial interactions and inflammatory processes that occur in animal models are broadly applicable to humans, particular aspects of the microbiome, inflammatory response, and mucosal defense may be species-specific (45–47). Finally, animal models are poorly predictive of drug response and toxicity in humans (48, 49).
In-vitro immortalized intestinal human cell lines such as Caco-2, T84, and HT-29 cultures are excellent for investigating specific molecular interactions and signaling pathways under highly controlled conditions. They are derived from human tissue, are low-cost and can be rapidly established. However, monolayer cultures are reductive as a model and cannot replicate the complex interactions that occur in-vivo.
Organoids
Organoids are defined as 3-D structures derived from either pluripotent (embryonic or induced pluripotent), or adult tissue-resident stem cells, which spontaneously self-organize and undergo a degree of differentiation, producing functional cell types, and which have the capacity to undertake some functions of the relevant organ (50).
While systems for maintaining intestinal tissue explants ex-vivo had been described since 1992, (51, 52) it was Eiraku et al. (53) and Sato et al. (54), respectively, who first successfully developed a method of producing the stem-cell derived, constructs known today as organoids. Studies by Sato et al. derived these from Lgr5+ adult stem cells (ASCs), first from murine and subsequently human intestinal crypts. They self-organized into crypt-villus type architecture and had the potential to produce most mature cell lines of the gut (54–58). Since then organoid cultures have been successfully derived from other anatomical locations, including colonic, gastric and esophageal tissue (55, 56, 59–62).
Organoids derived from small bowel tissue are sometimes referred to as enteroids or simply small bowel organoids, while organoids derived from colonic tissue may similarly be referred to as colonoids. They can be expanded from small volumes of tissue, including from endoscopic biopsies. Lgr5+ ASCs can be induced to differentiate into organoids containing all cell lines propagated by the gut, including mature enterocytes, Paneth cells, goblet cells, enteroendocrine, and tuft cells (48, 54, 55, 63). PSC-derived organoids can additionally generate adjacent stromal cell types. They recapitulate the spatial organization and polarity observed in the intestinal mucosa. Gut organoids are also capable of many of the functions of the source tissue, including endocrine and paracrine secretion, filtration, molecular transport, absorption, and contraction (48).
By contrast, while cheap and rapidly established, immortalized monolayer cell lines cannot recapitulate the complex cell-cell interactions or interactions with the extracellular microenvironment which occur in whole organisms (64, 65). Typically only single cell types are represented (66). It is not possible to culture rarer intestinal cell types such as tuft cells, and it can be difficult to acquire immortalized cell lines which secrete mucus to mimic the mucosal barrier which exists in-vivo (48, 61). Further, as monolayer cell cultures are derived from malignant cells they intrinsically demonstrate different properties to those of non-malignant cells, particularly with respect to epithelial integrity, cell polarity, and adhesion. These cells are not fully differentiated, and cell division in monolayer cell culture does not respond to the usual cellular signaling mechanisms which regulate this process in-vivo (48, 61, 67) (Table 1).
Table 1.
Characteristics of different modeling platforms in IBD.
| Feature | 2D cell culture | Animal models | Organoids |
|---|---|---|---|
| Cost | + | ++ | ++ |
| Culture cycle length | + | +++ | ++ |
| Presence of all intestinal cell types | – | ++ | ++ |
| Presence of non-epithelial elements of intestinal niche | – | +++ | +/– |
| Genetic stability | + | +++ | ++ |
| Suitability for high-throughput studies | ++ | – | ++ |
| Suitability for drug toxicity screening | – | ++ | ++ |
Organoids theoretically have the potential to bridge the gap between monolayer cell culture and whole-organism environments. They are derived from human tissue and recapitulate the complex cellular organization seen in-vivo. However, they avoid the issues of xenogeneity which may be associated with animal models (68, 69). Organoids are also less costly and can be more rapidly established than animal models while retaining the potential for highly controlled molecular and genetic manipulation which is the salient attractive feature of monolayer cell culture (Table 1).
Translational Application of Organoid Models in IBD
Physiological Modeling of the Intestinal Niche
Differential protein expression, gene expression, cell migration, organization, survival, and cell signaling have been observed in organoid cultures comparative to monolayer cell culture (70–73). Defects in the function of multiple epithelial cell types have been demonstrated in IBD, underlining the need for a physiologically relevant model which includes multiple cell lineages (74–78). Gut organoids may also be co-cultured with non-epithelial cell lines of interest in order to more accurately represent the intestinal mucosal niche. Co-culture of gut epithelial organoids with cell lines such as macrophages and lymphocytes and with mesenchymal cells demonstrate promise in providing a more physiologically relevant model of the gastrointestinal mucosal environment (79–82).
Host-Microbe Interactions
Due to this ability to accurately simulate the intestinal microenvironment, intestinal organoids represent exciting models for investigating the host-microbial interactions which are key to the pathogenesis of IBD. Organoids have already been successfully utilized as a more accurate model for human virus infection. In a study by Saxena, fully differentiated cells present in organoid culture supported greater rotavirus viral load and replication than had been previously observed in monolayer culture; and infection of enteroendocrine cell types in addition to enterocytes with rotavirus was demonstrated (83). Organoids have also been used to investigate norovirus, which is difficult to cultivate in monolayer cell culture. Previously only successfully cultured in B cells, organoids permitted culture of norovirus in duodenal, jejunal, and ileal cell types with viral replication and growth occurring within (84). Current applications of organoids include disease modeling of SARS-CoV-2 in respiratory and small intestinal derived cell types, with viral infection, replication, and host viral response observed ex-vivo (85–87).
Organoid cultures have also been applied to simulate host-bacterial interactions. Salmonella, H. pylori, C. difficile, and pathogenic E. coli infection have all been modeled in organoid cultures (66, 88–91). In one study, gastric organoids which secrete mucous, include multiple epithelial cell types and retain the polarity of the in-vivo gastric epithelium have been successfully utilized as a model for host-microbe interactions in H.pylori infection (66). Interestingly, duodenal, ileal and colonic organoid cultures derived from different donors demonstrate a differential response to infection and differing patterns of bacterial adhesion, possibly due to the genetic variability based on the tissue of origin (89). A co-culture model developed to study the host-pathogen interactions of C. jejuni incorporates intestinal enterocytes, mucin-secreting goblet cells and dendritic cells, thus combining a mucus-secreting epithelial layer with cellular elements of the intestinal innate immune system (92).
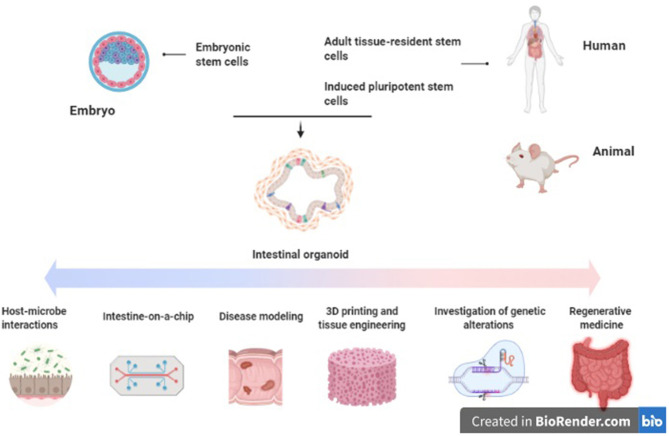
As well as modeling invasive microorganisms, organoids can also be used to study interactions between the gut and commensal microbiota. In one study, microbiota were found to play a role in epithelial regeneration in murine small bowel organoids. The pattern recognition receptor NOD2, single nucleotide polymorphisms (SNPs) of which are highly associated with Crohn's disease, is highly expressed in mouse intestinal stem cells (93–96). Stimulation of NOD2 by MDP (peptidoglycan muramyl-dipeptide), a bacterial cell wall constituent, enhanced organoid survival and protected them from oxidative-stress mediated cell death (96, 97). Organoids derived from adult and fetal murine tissues have also been utilized to determine developmental expression patterns of components of the innate immune system, including NOD2, TLR4, and TLR5 (98). Exposure of murine intestinal organoids to gut commensal bacteria including Akkermansia muciniphilia and Faecalibacterium prausnitzii has been shown to induce changes in gene expression and transcription, particularly of genes responsible for lipid metabolism (99). Similarly, exposure to the organism Bacteroides thetaiotaomicron and cytokine signaling via IL-22RA1 induces upregulation of Fut2 and increased fucosylation, which in turn inhibits colonization by opportunistic Enterococcus faecalis strains (97, 100, 101). Finally, alterations in the microbiome have been associated with colonic neoplasia; colonic organoid models have been used to demonstrate a mutational profile induced by exposure to colibactin synthesized by genotoxic E. coli which is also associated with colorectal cancer in-vivo (102). Thus, organoid systems may be utilized to explore activity of the gut microbiome on the epithelium and mechanisms of homeostasis (Figure 1).
Figure 1.
Applications of intestinal organoids in inflammatory bowel disease. Original figure (created with BioRender.com).
Disease Modeling
Unlike organoids derived from pluripotent stem cells, which rapidly accumulate mutations and epigenetic modifications, ASC-derived organoids are relatively genetically stable (103, 104). They retain the genetic profile and also the transcriptional and epigenetic landscape of the primary tissue from which they are derived (73, 105–107). While the majority of IBD is polygenic, some monogenic forms exist. These are mediated by specific genetic defects in epithelial dysfunction and stress response, defects in immune regulation of regulatory T cells or immunodeficiencies of phagocytic cells (108). Organoids represent useful models for studying these rare diseases, as well as other genetically determined intestinal disorders (109).
It is possible to culture intestinal organoids derived from patients with active IBD (110, 111). In one study, IBD colonic organoids demonstrated a distinct phenotype to those derived from control tissue, with a smaller size, increased cell death, abnormal cell polarization, and poorer budding capacity (110). Interestingly, they also expressed reduced quantities of the tight junction proteins ZO1, Occludin, and Claudin-1 as well as alterations in the expression of adherens junction and desmosomal proteins. These altered expression patterns persisted when the inflammatory stimulus was withdrawn (111). The phenotype and altered transcriptional profile noted in the IBD-derived organoids was inducible in the control organoids with administration of pro-inflammatory cytokines (TNFα, IL-1, and IL-6). Hibiya et al. demonstrated that murine colonic organoids which are exposed to chronic inflammatory stimuli (TNFα, IL-1β, IL-6, LPS, flagellin) underwent upregulation of the NFκB signaling pathway, which persisted after stimuli removal (112). These organoids also underwent transformation to an undifferentiated state, along with upregulation of genes related to oxidative stress and carcinogenesis (Smox and CD151), suggesting their potential utility as a model to study the epithelial changes which occur in colitis-induced carcinogenesis. Vermeire et al. also generated CD and UC-derived organoids which were subsequently exposed to TNF and flagellin, resulting in modulation of expression of the SARS-CoV-2 receptor ACE2. These changes were restored to baseline with anti-TNF treatments (113). Other studies utilizing patient-derived organoids from pediatric IBD patients demonstrated alterations in DNA methylation and transcriptional profiles, which correlated with treatment outcomes (114). Finally, a study by Jardine et al. successfully used colonic organoids generated from patients with TTC7A deficiency to perform high-throughput drug screening for candidate therapeutic agents (115). Loss of TTC7A causes intestinal epithelial apoptosis and immune defects which presents clinically as very early onset IBD. Thus, primary organoid cultures from inflamed tissue seem to represent an applicable model for investigation of the epithelial and mucosal abnormalities which occur in IBD (Figure 1).
Bioengineering and Gut-on-a-Chip
In the small intestine the mucosa of the gut is folded into villi and microvilli to maximize available surface area for absorption. Bioengineering techniques such as 3D printing and laser ablation allow the creation of scaffolds which recreate this intestinal topography. These can be directly seeded with epithelial organoids or used as molds to create hydrogel-based porous copies which reproduce the microanatomy of the gut (116). Alternatively, bioink comprising cell aggregates or organoids may be imprinted along with the desired biomaterials (hydrogels, matrix components) onto the scaffold via a computer-aided transfer process (117, 118). This allows for the creation of highly accurate and reproducible models with each component—organoids, biomaterial, and scaffold—spatially aligned at the desired patterns, gradients, and densities set by the modeling software. Such methods will help to address both reproducibility and scaling-up of organoid cultures into larger tissue constructs. Some 3D gut models aimed at investigating the pathophysiology of inflammatory bowel disease are already in use (119).
Aside from gut anatomy, intestinal motility and luminal flow are physiologic functions of the gut which are difficult to mimic ex-vivo. These can be simulated via epithelial cell-lined microfluidic platforms, sometimes referred to as a “gut-on-a-chip” (120–122). Such platforms permit recapitulation of flow patterns, mechanical deformation, shear stresses, and peristaltic activity with greater accuracy than has been possible previously (117, 123–125). Organoid-lined laser-ablated microchips with active perfusion of media components have also been developed, which permit simulation of intestinal homeostasis and cell turnover with a reduced need for passaging (126). These platforms are being utilized to further investigate the gut-microbiome relationship by inoculation with bacterial cultures and examining the effect of the physical environment on intestinal host-microbe interactions (127–132) (Figure 1).
Regenerative Medicine
The concept of mucosal healing as key to sustained remission of IBD has become increasingly prominent in recent years. This denotes absence of all mucosal ulceration at endoscopy, rather than resolution of clinical symptoms and serum biomarkers of inflammation alone (133, 134) Mucosal healing correlates with improved long-term clinical outcomes, including steroid use, hospital admissions and need for surgery in both CD and UC (135–139). The European Crohn's and Colitis Organization lists mucosal healing as a therapeutic target in its 2017 consensus guidelines for both UC and CD (140, 141). Local transplant of organoids to aid mucosal healing has been proposed as a potential therapy in IBD to aid epithelial regeneration and achieve mucosal healing (142). Studies using murine colitis models have demonstrated that human small bowel and colonic organoid cultures can engraft onto the ulcerated mucosa and reconstitute the normal crypt-villus architecture (58, 143, 144). More recently, patient-derived small intestinal organoids have been successfully expanded ex-vivo and engrafted into mice, with the ultimate aim of creating autologous small intestinal transplants to treat intestinal failure (145) (Figure 1).
Limitations of Organoids as a Model Platform
Despite the advantages described above, there are limitations associated with the use of organoids. Comparative to two-dimensional models they are more costly and less accessible, and require specialized medium to be maintained in culture. Matrigel and similar matrices in which they are typically cultured are expensive and increase the difficulty of manipulation. Particular studies such as transport and luminal exposure studies require injection of organoids which is a technically difficult and labor-intensive procedure; alternatives such as computer-assisted injection are again expensive and not readily available. Access to human tissue for generation of primary organoid cultures can be limited (78, 146). They are typically derived from epithelial tissues and so other components of the intestinal niche, including immune and mesenchymal elements, are underrepresented (146). As they are three-dimensional structures, this presents difficulty for investigations requiring access to the apical and basolateral surfaces. For this purpose they may be dissociated into 2D structures; however this disrupts their crypt-villus architecture and terminates their culture cycle (146). Finally, reproducibility of organoid cultures is challenging, as constructs of differing sizes and morphology result when they are grown in-vitro.
Conclusion
In summary, intestinal organoids represent a promising novel platform for further elucidating the host-microbe interactions, mucosal barrier deficiencies and genetic defects which underpin the pathogenesis of inflammatory bowel disease. Patient-derived organoids may have translational applications in the future as local therapy to aid mucosal healing. However, many limitations yet remain with this model. Some of these may be addressed by innovations such as computer-assisted bioprinting and 3D printed scaffolds to aid in reproducibility, and development of co-culture systems including immune and neuronal components to increase the physiological relevance of organoids as a platform for investigation of IBD.
Author Contributions
LO'C: preparation of manuscript. DCW and CMA: concept, review and editing of manuscript, and approval prior to submission. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by Crohn's and Colitis Foundation.
References
- 1.Baumgart DC, Sandborn WJ. Crohn's disease. Lancet. (2012) 380:1590–605. 10.1016/S0140-6736(12)60026-9 [DOI] [PubMed] [Google Scholar]
- 2.Torres J, Mehandru S, Colombel J-F, Peyrin-Biroulet L. Crohn's disease. Lancet. (2017) 389:1741–55. 10.1016/S0140-6736(16)31711-1 [DOI] [PubMed] [Google Scholar]
- 3.Ramos GP, Papadakis KA. Mechanisms of disease: inflammatory bowel diseases. Mayo Clin Proc. (2019) 94:155–65. 10.1016/j.mayocp.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. (2011) 474:307–17. 10.1038/nature10209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. (2007) 448:427–34. 10.1038/nature06005 [DOI] [PubMed] [Google Scholar]
- 6.Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. (2012) 380:1606–19. 10.1016/S0140-6736(12)60150-0 [DOI] [PubMed] [Google Scholar]
- 7.Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel J-F. Ulcerative colitis. Lancet. (2017) 389:1756–70. 10.1016/S0140-6736(16)32126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFilippis EM, Longman R, Harbus M, Dannenberg K, Scherl EJ. Crohn's disease: evolution, epigenetics, and the emerging role of microbiome-targeted therapies. Curr Gastroenterol Rep. (2016) 18:13. 10.1007/s11894-016-0487-z [DOI] [PubMed] [Google Scholar]
- 9.Kotze PG, Shen B, Lightner A, Yamamoto T, Spinelli A, Ghosh S, et al. Modern management of perianal fistulas in Crohn's disease: future directions. Gut. (2018) 67:1181–94. 10.1136/gutjnl-2017-314918 [DOI] [PubMed] [Google Scholar]
- 10.Gecse KB, Bemelman W, Kamm MA, Stoker J, Khanna R, Ng SC, et al. A global consensus on the classification, diagnosis and multidisciplinary treatment of perianal fistulising Crohn's disease. Gut. (2014) 63:1381–92. 10.1136/gutjnl-2013-306709 [DOI] [PubMed] [Google Scholar]
- 11.Feuerstein JD, Cheifetz AS. Ulcerative colitis: epidemiology, diagnosis, and management. Mayo Clin Proc. (2014) 89:1553–63. 10.1016/j.mayocp.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 12.Zeissig S, Bürgel N, Günzel D, Richter J, Mankertz J, Wahnschaffe U, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. (2007) 56:61–72. 10.1136/gut.2006.094375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCole DF. IBD candidate genes and intestinal barrier regulation. Inflamm Bowel Dis. (2014) 20:1829–49. 10.1097/MIB.0000000000000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson MEV, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. (2014) 63:281–91. 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, et al. mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis. (2016) 10:462–71. 10.1093/ecco-jcc/jjv223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Post S, Jabbar KS, Birchenough G, Arike L, Akhtar N, Sjovall H, et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. (2019) 68:2142–51. 10.1136/gutjnl-2018-317571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jostins L, Ripke S, Weersma RK, Duerr RH. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. (2012) 491:119–24. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange KM, Barrett JC. Understanding inflammatory bowel disease via immunogenetics. J Autoimmun. (2015) 64:91–100. 10.1016/j.jaut.2015.07.013 [DOI] [PubMed] [Google Scholar]
- 19.Parkes M. Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig Dis. (2012) 30:330–3. 10.1159/000338119 [DOI] [PubMed] [Google Scholar]
- 20.Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. (2014) 13:3–10. 10.1016/j.autrev.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 21.Xu X-R, Liu C-Q, Feng B-S, Liu Z-J. Dysregulation of mucosal immune response in pathogenesis of inflammatory bowel disease. World J Gastroenterol. (2014) 20:3255–64. 10.3748/wjg.v20.i12.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lidar M, Langevitz P, Shoenfeld Y. The role of infection in inflammatory bowel disease: initiation, exacerbation and protection. Isr Med Assoc J. (2009) 11:558–63. [PubMed] [Google Scholar]
- 23.Michielan A, D'Incà R. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm. (2015) 2015:628157. 10.1155/2015/628157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerova VA, Stoynov SG, Katsarov DS, Svinarov DA. Increased intestinal permeability in inflammatory bowel diseases assessed by iohexol test. World J Gastroenterol. (2011) 17:2211–5. 10.3748/wjg.v17.i17.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. (2007) 56:343–50. 10.1136/gut.2006.098160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. (2013) 7:1256–61. 10.1038/ismej.2013.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur N, Chen C-C, Luther J, Kao JY. Intestinal dysbiosis in inflammatory bowel disease. Gut Microbes. (2011) 2:211–6. 10.4161/gmic.2.4.17863 [DOI] [PubMed] [Google Scholar]
- 28.Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. (2018) 67:574–87. 10.1136/gutjnl-2017-314903 [DOI] [PubMed] [Google Scholar]
- 29.Barnich N, Carvalho FA, Glasser A-L, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. (2007) 11:1566–74. 10.1172/JCI30504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, et al. Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. (2013) 9:e1003141. 10.1371/journal.ppat.1003141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizoguchi A. Animal models of inflammatory bowel disease. Prog Mol Biol Transl Sci. (2012) 105:263–320. 10.1016/B978-0-12-394596-9.00009-3 [DOI] [PubMed] [Google Scholar]
- 32.Duerr RH. Update on the genetics of inflammatory bowel disease. J Clin Gastroenterol. (2003) 37:358–67. 10.1097/00004836-200311000-00003 [DOI] [PubMed] [Google Scholar]
- 33.Hong M, Ye BD, Yang S-K, Jung S, Lee H-S, Kim BM, et al. Immunochip meta-analysis of inflammatory owel disease identifies three novel loci and four novel associations in previously reported loci. J Crohns Colitis. (2018) 12:730–41. 10.1093/ecco-jcc/jjy002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. (2017) 49:256–61. 10.1038/ng.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong U, Cross RK. Expert opinion on interleukin-12/23 and interleukin-23 antagonists as potential therapeutic options for the treatment of inflammatory bowel disease. Expert Opin Investig Drugs. (2019) 28:473–9. 10.1080/13543784.2019.1597053 [DOI] [PubMed] [Google Scholar]
- 36.Verstockt B, Ferrante M, Vermeire S, Van Assche G. New treatment options for inflammatory bowel diseases. J Gastroenterol. (2018) 53:585–90. 10.1007/s00535-018-1449-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danese S, Vuitton L, Peyrin-Biroulet L. Biologic agents for IBD: practical insights. Nat Rev Gastroenterol Hepatol. (2015) 12:537–45. 10.1038/nrgastro.2015.135 [DOI] [PubMed] [Google Scholar]
- 38.Fine S, Papamichael K, Cheifetz AS. Etiology and management of lack or oss of Rresponse to anti-tumor necrosis factor therapy in patients with inflammatory bowel disease. Gastroenterol Hepatol. (2019) 15:656–65. [PMC free article] [PubMed] [Google Scholar]
- 39.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. (2014) 104:15.25.1–14. 10.1002/0471142735.im1525s104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neurath M, Fuss I, Strober W. TNBS-colitis. Int Rev Immunol. (2000) 19:51–62. 10.3109/08830180009048389 [DOI] [PubMed] [Google Scholar]
- 41.Rothemich A, Arthur JC. The Azoxymethane/Il10 -/- model of colitis-associated cancer (CAC). Methods Mol Biol. (2019) 1960:215–25. 10.1007/978-1-4939-9167-9_19 [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Wei W, Li X, Shen P, Ju D, Wang Z, et al. BMI1 and MEL18 promote colitis-associated cancer in mice via REG3B and STAT3. Gastroenterology. (2017) 153:1607–20. 10.1053/j.gastro.2017.07.044 [DOI] [PubMed] [Google Scholar]
- 43.Aherne CM, Collins CB, Masterson JC, Tizzano M, Boyle TA, Westrich JA, et al. Neuronal guidance molecule netrin-1 attenuates inflammatory cell trafficking during acute experimental colitis. Gut. (2012) 61:695–705. 10.1136/gutjnl-2011-300012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Sluis M, Bouma J, Vincent A, Velcich A, Carraway KL, Büller HA, et al. Combined defects in epithelial and immunoregulatory factors exacerbate the pathogenesis of inflammation: mucin 2-interleukin 10-deficient mice. Lab Invest. (2008) 88:634–42. 10.1038/labinvest.2008.28 [DOI] [PubMed] [Google Scholar]
- 45.Sollid LM, Johansen F-E. Animal models of inflammatory bowel disease at the dawn of the new genetics era. PLoS Med. (2008) 5:e198. 10.1371/journal.pmed.0050198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerquetella M, Spaterna A, Laus F, Tesei B, Rossi G, Antonelli E, et al. Inflammatory bowel disease in the dog: differences and similarities with humans. World J Gastroenterol. (2010) 16:1050–6. 10.3748/wjg.v16.i9.1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiminez JA, Uwiera TC, Douglas Inglis G, Uwiera RRE. Animal models to study acute and chronic intestinal inflammation in mammals. Gut Pathog. (2015) 7:29. 10.1186/s13099-015-0076-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.George MM, Rahman M, Connors J, Stadnyk AW. Opinion: are organoids the end of model evolution for studying host intestinal epithelium/microbe interactions? Microorganisms. (2019) 7:406 10.3390/microorganisms7100406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamb A. What's wrong with our cancer models? Nat Rev Drug Discov. (2005) 4:161–5. 10.1038/nrd1635 [DOI] [PubMed] [Google Scholar]
- 50.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. (2014) 345:1247125. 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- 51.Evans GS, Flint N, Somers AS, Eyden B, Potten CS. The development of a method for the preparation of rat intestinal epithelial cell primary cultures. J Cell Sci. (1992) 101:219–31. [DOI] [PubMed] [Google Scholar]
- 52.Fukamachi H. Proliferation and differentiation of fetal rat intestinal epithelial cells in primary serum-free culture. J Cell Sci. (1992) 103:511–9. [DOI] [PubMed] [Google Scholar]
- 53.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. (2008) 3:519–32. [DOI] [PubMed] [Google Scholar]
- 54.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. (2009) 459:262–5. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 55.Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, et al. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell. (2019) 24:829. 10.1016/j.stem.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. (2011) 141:1762–72. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- 57.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. (2011) 470:105–9. 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med. (2012) 18:618–23. 10.1038/nm.2695 [DOI] [PubMed] [Google Scholar]
- 59.Seidlitz T, Merker SR, Rothe A, Zakrzewski F, von Neubeck C, Grützmann K, et al. Human gastric cancer modelling using organoids. Gut. (2019) 68:207–17. 10.1136/gutjnl-2017-314549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, Francies H, Miremadi A, Grantham A, Grehan N, Devonshire G, et al. Su2042–derivation of oesophageal organoids to recapitulate the heterogeneity of primary umours and provide a model system for precision therapeutics. Gastroenterology. (2019) 156:S−696. 10.1016/S0016-5085(19)38664-0 [DOI] [Google Scholar]
- 61.Dotti I, Salas A. Potential use of human stem cell-derived intestinal organoids to study inflammatory bowel diseases. Inflamm Bowel Dis. (2018) 24:2501–9. 10.1093/ibd/izy275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. (2010) 6:25–36. 10.1016/j.stem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 63.Lau W de, de Lau W, Kujala P, Schneeberger K, Middendorp S, Li VSW, et al. Peyer's patch M cells derived from Lgr5 stem cellsrequire SpiB and are induced by RankL in cultured “Miniguts”. Mol Cell Biol. (2012) 32:3639–47. 10.1128/MCB.00434-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Connell L, Winter DC. Organoids: past learning and future directions. Stem Cells Dev. (2020) 29:281–9. 10.1089/scd.2019.0227 [DOI] [PubMed] [Google Scholar]
- 65.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. (2016) 539:560–4. 10.1038/nature20168 [DOI] [PubMed] [Google Scholar]
- 66.Boccellato F, Woelffling S, Imai-Matsushima A, Sanchez G, Goosmann C, Schmid M, et al. Polarised epithelial monolayers of the gastric mucosa reveal insights into mucosal homeostasis and defence against infection. Gut. (2018) 68:400–13. 10.1136/gutjnl-2017-314540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. (2010) 143:134–44. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- 68.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci USA. (2015) 112:1167–72. 10.1073/pnas.1401965111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. (2013) 110:3507–12. 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Williams S, Ho S, Loraine H, Hagan D, Whaley JM, et al. Quantitative PCR tissue expression profiling of the human SGLT2 gene and related family members. Diabetes Ther. (2010) 1:57–92. 10.1007/s13300-010-0006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zietek T, Rath E, Haller D, Daniel H. Intestinal organoids for assessing nutrient transport, sensing and incretin secretion. Sci Rep. (2015) 5:16831. 10.1038/srep16831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pelagalli A, Squillacioti C, Mirabella N, Meli R. Aquaporins in health and disease: an overview focusing on the gut of different species. Int J Mol Sci. (2016) 179(1):02419-17. 10.3390/ijms17081213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Middendorp S, Schneeberger K, Wiegerinck CL, Mokry M, Akkerman RDL, van Wijngaarden S, et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. (2014) 32:1083–91. 10.1002/stem.1655 [DOI] [PubMed] [Google Scholar]
- 74.Arijs I, De Hertogh G, Machiels K, Van Steen K, Lemaire K, Schraenen A, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol. (2011) 106:748–61. 10.1038/ajg.2011.27 [DOI] [PubMed] [Google Scholar]
- 75.Arijs I, De Hertogh G, Lemaire K, Quintens R. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE. (2009) 4:e7984. 10.1371/journal.pone.0007984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Surawicz CM, Haggitt RC, Husseman M, McFarland LV. Mucosal biopsy diagnosis of colitis: acute self-limited colitis and idiopathic inflammatory bowel disease. Gastroenterology. (1994) 107:755–63. 10.1016/0016-5085(94)90124-4 [DOI] [PubMed] [Google Scholar]
- 77.Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S. Bioprinting for cancer research. Trends Biotechnol. (2015) 33:504–13. 10.1016/j.tibtech.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 78.Noben M, Vanhove W, Arnauts K, Santo Ramalho A, Van Assche G, Vermeire S, et al. Human intestinal epithelium in a dish: current models for research into gastrointestinal pathophysiology. United European Gastroenterol J. (2017) 5:1073–81. 10.1177/2050640617722903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nozaki K, Mochizuki W, Matsumoto Y, Matsumoto T, Fukuda M, Mizutani T, et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J Gastroenterol. (2016) 51:206–13. 10.1007/s00535-016-1170-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF, et al. Erratum: a primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci Rep. (2017) 7:46790. 10.1038/srep46790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pastuła A, Middelhoff M, Brandtner A, Tobiasch M, Höhl B, Nuber AH, et al. Three-dimensional gastrointestinal organoid culture in combination with nerves or fibroblasts: a method to characterize the gastrointestinal stem cell niche. Stem Cells Int. (2016) 2016:3710836. 10.1155/2016/3710836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med. (2017) 23:49–59. 10.1038/nm.4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saxena K, Blutt SE, Ettayebi K, Zeng X-L, Broughman JR, Crawford SE, et al. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol. (2016) 90:43–56. 10.1128/JVI.01930-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. (2016) 353:1387–93. 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lamers MM, van der Vaart J, Knoops K, Riesebosch S, Breugem TI, Mykytyn AZ, et al. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human Alveolar-type II-like cells. EMBO J. (2020). 10.15252/embj.2020105912. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. (2020) 369:50–4. 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stanifer ML, Kee C, Cortese M, Zumaran CM, Triana S, Mukenhirn M, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. (2020) 32:107863. 10.1016/j.celrep.2020.107863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Forbester JL, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, et al. Interaction of salmonella enterica serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun. (2015) 83:2926–34. 10.1128/IAI.00161-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rajan A, Vela L, Zeng X-L, Yu X, Shroyer N, Blutt SE, et al. Novel segment- and host-specific patterns of enteroaggregative Escherichia coli adherence to human intestinal enteroids. mBio. (2018) 9:02419-17. 10.1128/mBio.02419-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engevik MA, Engevik KA, Yacyshyn MB, Wang J, Hassett DJ, Darien B, et al. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol. (2015) 308:G497–509. 10.1152/ajpgi.00090.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun. (2015) 83:138–45. 10.1128/IAI.02561-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zamora CY, Ward EM, Kester JC, Chen WLK, Velazquez JG, Griffith LG, et al. Application of a gut-immune co-culture system for the study of N-glycan-dependent host-pathogen interactions of Campylobacter jejuni. Glycobiology. (2020) 30:374–81. 10.1093/glycob/cwz105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adler J, Rangwalla SC, Dwamena BA, Higgins PDR. The prognostic power of the NOD2 genotype for complicated Crohn's disease: a meta-analysis. Am J Gastroenterol. (2011) 106:699–712. 10.1038/ajg.2011.19 [DOI] [PubMed] [Google Scholar]
- 94.Chen Y, Salem M, Boyd M, Bornholdt J, Li Y, Coskun M, et al. Relation between NOD2 genotype and changes in innate signaling in Crohn's disease on mRNA and miRNA levels. NPJ Genom Med. (2017) 2:3. 10.1038/s41525-016-0001-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Glas J, Seiderer J, Tillack C, Pfennig S, Beigel F, Jürgens M, et al. The NOD2 single nucleotide polymorphisms rs2066843 and rs2076756 are novel and common Crohn's disease susceptibility gene variants. PLoS ONE. (2010) 5:e14466. 10.1371/journal.pone.0014466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nigro G, Rossi R, Commere P-H, Jay P, Sansonetti PJ. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe. (2014) 15:792–8. 10.1016/j.chom.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 97.Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. (2017) 23:393–410. 10.1016/j.molmed.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 98.Kayisoglu O, Weiss F, Niklas C, Pierotti I, Pompaiah M, Wallaschek N, et al. Location-specific cell identity rather than exposure to GI microbiota defines many innate immune signalling cascades in the gut epithelium. Gut. (2020). 10.1136/gutjnl-2019-319919. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio. (2014) 12:01438–14. 10.1128/mBio.01438-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Engevik MA, Aihara E, Montrose MH, Shull GE, Hassett DJ, Worrell RT. Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth. Am J Physiol Gastrointest Liver Physiol. (2013) 305:G697–711. 10.1152/ajpgi.00184.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pham TAN, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. (2014) 16:504–16. 10.1016/j.chom.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature. (2020) 580:269–73. 10.1038/s41586-020-2080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. (2014) 513:422–5. 10.1038/nature13448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. (2013) 13:149–59. 10.1016/j.stem.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cramer JM, Thompson T, Geskin A, LaFramboise W, Lagasse E. Distinct human stem cell populations in small and large intestine. PLoS ONE. (2015) 10:e0118792. 10.1371/journal.pone.0118792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kraiczy J, Nayak KM, Howell KJ, Ross A, Forbester J, Salvestrini C, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. (2019) 68:49–61. 10.1136/gutjnl-2017-314817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yoo J-H, Donowitz M. Intestinal enteroids/organoids: a novel platform for drug discovery in inflammatory bowel diseases. World J Gastroenterol. (2019) 25:4125–47. 10.3748/wjg.v25.i30.4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Uhlig HH. Monogenic diseases associated with intestinal inflammation: implications for the understanding of inflammatory bowel disease. Gut. (2013) 62:1795–805. 10.1136/gutjnl-2012-303956 [DOI] [PubMed] [Google Scholar]
- 109.Bigorgne AE, Farin HF, Lemoine R, Mahlaoui N, Lambert N, Gil M, et al. TTC7A mutations disrupt intestinal epithelial apicobasal polarity. J Clin Invest. (2014) 124:328–37. 10.1172/JCI71471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.d'Aldebert E, Quaranta M, Sébert M, Bonnet D, Kirzin S, Portier G, et al. Characterization of human colon organoids from inflammatory bowel disease patients. Front Cell Dev Biol. (2020) 8:363. 10.3389/fcell.2020.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meir M, Salm J, Fey C, Schweinlin M, Kollmann C, Kannapin F, et al. Enteroids generated from patients with severe inflammation in Crohn's disease maintain alterations of junctional proteins. J Crohns Colitis. (2020) 14:1473–1487. 10.1093/ecco-jcc/jjaa085 [DOI] [PubMed] [Google Scholar]
- 112.Hibiya S, Tsuchiya K, Hayashi R, Fukushima K, Horita N, Watanabe S, et al. Long-term inflammation transforms intestinal epithelial cells of colonic organoids. J Crohns Colitis. (2017) 11:621–30. 10.1093/ecco-jcc/jjw186 [DOI] [PubMed] [Google Scholar]
- 113.Verstockt B, Verstockt S, Abdu Rahiman S, Ke B-J, Arnauts K, Cleynen I, et al. Intestinal receptor of SARS-CoV-2 in inflamed IBD tissue seems downregulated by HNF4A in ileum and upregulated by interferon regulating factors in colon. J Crohns Colitis. (2020). 10.1093/ecco-jcc/jjaa185. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Howell KJ, Kraiczy J, Nayak KM, Gasparetto M, Ross A, Lee C, et al. DNA Methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. (2018) 154:585–98. 10.1053/j.gastro.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jardine S, Anderson S, Babcock S, Leung G, Pan J, Dhingani N, et al. Drug screen identifies leflunomide for treatment of inflammatory bowel disease caused by TTC7A deficiency. Gastroenterology. (2020) 158:1000–15. 10.1053/j.gastro.2019.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim G-A, Spence JR, Takayama S. Bioengineering for intestinal organoid cultures. Curr Opin Biotechnol. (2017) 47:51–8. 10.1016/j.copbio.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fang Y, Eglen RM. Three-dimensional cell cultures in drug discovery and development. SLAS Discov. (2017) 22:456–72. 10.1177/1087057117696795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jin M-Z, Han R-R, Qiu G-Z, Ju X-C, Lou G, Jin W-L. Organoids: an intermediate modeling platform in precision oncology. Cancer Lett. (2018) 414:174–80. 10.1016/j.canlet.2017.11.021 [DOI] [PubMed] [Google Scholar]
- 119.Roh TT, Chen Y, Paul HT, Guo C, Kaplan DL. 3D bioengineered tissue model of the large intestine to study inflammatory bowel disease. Biomaterials. (2019) 225:119517. 10.1016/j.biomaterials.2019.119517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, et al. Microfluidic organ-on-a-hip models of human intestine. Cell Mol Gastroenterol Hepatol. (2018) 5:659–68. 10.1016/j.jcmgh.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beaurivage C, Naumovska E, Chang YX, Elstak ED, Nicolas A, Wouters H, et al. Development of a gut-on-A-chip model for high throughput disease modeling and drug discovery. Int J Mol Sci. (2019) 20:5661. 10.3390/ijms20225661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Poceviciute R, Ismagilov RF. Human-gut-microbiome on a chip. Nat Biomed Eng. (2019) 3:500–1. 10.1038/s41551-019-0425-0 [DOI] [PubMed] [Google Scholar]
- 123.Sidar B, Jenkins BR, Huang S, Spence JR, Walk ST, Wilking JN. Long-term flow through human intestinal organoids with the gut organoid flow chip (GOFlowChip). Lab Chip. (2019) 18:3552–62. 10.1039/C9LC00653B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Skardal A, Shupe T, Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today. (2016) 21:1399–411. 10.1016/j.drudis.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shin W, Kim HJ. Pathomimetic modeling of human intestinal diseases and underlying host-gut microbiome interactions in a gut-on-a-chip. Methods Cell Biol. (2018) 146:135–48. 10.1016/bs.mcb.2018.05.006 [DOI] [PubMed] [Google Scholar]
- 126.Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. (2020) 585:574–8. 10.1038/s41586-020-2724-8 [DOI] [PubMed] [Google Scholar]
- 127.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. (2012) 12:2165–74. 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 128.Kim HJ, Lee J, Choi J-H, Bahinski A, Ingber DE. Co-culture of living microbiome with microengineered human intestinal villi in a gut-on-a-chip icrofluidic device. J Vis Exp. (2016) 30:54344. 10.3791/54344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA. (2016) 113:E7–15. 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barrila J, Crabbé A, Yang J, Franco K, Nydam SD, Forsyth RJ, et al. Modeling host-pathogen interactions in the context of the microenvironment: three-dimensional cell culture comes of age. Infect Immun. (2018) 86:00282–18. 10.1128/IAI.00282-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grassart A, Malardé V, Gobaa S, Sartori-Rupp A, Kerns J, Karalis K, et al. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting shigella infection. Cell Host Microbe. (2019) 26:435–44.e4. 10.1016/j.chom.2019.08.007 [DOI] [PubMed] [Google Scholar]
- 132.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng. (2019) 3:520–31. 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lichtenstein GR, Rutgeerts P. Importance of mucosal healing in ulcerative colitis. Inflamm Bowel Dis. (2010) 16:338–46. 10.1002/ibd.20997 [DOI] [PubMed] [Google Scholar]
- 134.Bryant RV, Burger DC, Delo J, Walsh AJ, Thomas S, von Herbay A, et al. Beyond endoscopic mucosal healing in UC: histological remission better predicts corticosteroid use and hospitalisation over 6 years of follow-up. Gut. (2016) 65:408–14. 10.1136/gutjnl-2015-309598 [DOI] [PubMed] [Google Scholar]
- 135.Colombel JF, Rutgeerts P, Reinisch W, Esser D, Wang Y, Lang Y, et al. Early mucosal healing with infliximab is associated with improved long-term clinical outcomes in ulcerative colitis. Gastroenterology. (2011) 141:1194–201. 10.1053/j.gastro.2011.06.054 [DOI] [PubMed] [Google Scholar]
- 136.Rutgeerts P, Diamond RH, Bala M, Olson A, Lichtenstein GR, Bao W, et al. Scheduled maintenance treatment with infliximab is superior to episodic treatment for the healing of mucosal ulceration associated with Crohn's disease. Gastrointest Endosc. (2006) 63:433–42. 10.1016/j.gie.2005.08.011 [DOI] [PubMed] [Google Scholar]
- 137.Picco MF, Farraye FA. Targeting mucosal healing in Crohn's disease. Gastroenterol Hepatol. (2019) 15:529–38. [PMC free article] [PubMed] [Google Scholar]
- 138.Frøslie KF, Jahnsen J, Moum BA, Vatn MH, IBSEN Group . Mucosal healing in inflammatory bowel disease: results from a Norwegian population-based cohort. Gastroenterology. (2007) 133:412–22. 10.1053/j.gastro.2007.05.051 [DOI] [PubMed] [Google Scholar]
- 139.Baert F, Moortgat L, Van Assche G, Caenepeel P, Vergauwe P, De Vos M, et al. Mucosal healing predicts sustained clinical remission in patients with early-stage Crohn's disease. Gastroenterology. (2010) 138:463–8. e10–1. 10.1053/j.gastro.2009.09.056 [DOI] [PubMed] [Google Scholar]
- 140.Magro F, Gionchetti P, Eliakim R, Ardizzone S, Armuzzi A, Barreiro-de Acosta M, et al. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and Ileo-anal pouch disorders. J Crohns Colitis. (2017) 11:649–70. 10.1093/ecco-jcc/jjx008 [DOI] [PubMed] [Google Scholar]
- 141.Gomollón F, Dignass A, Annese V, Tilg H, Van Assche G, Lindsay JO, et al. 3rd European evidence-based consensus on the diagnosis and management of Crohn's disease 2016: part 1: diagnosis and medical management. J Crohns Colitis. (2017) 11:3–25. 10.1093/ecco-jcc/jjw168 [DOI] [PubMed] [Google Scholar]
- 142.Okamoto R, Shimizu H, Suzuki K, Kawamoto A, Takahashi J, Kawai M, et al. Organoid-based regenerative medicine for inflammatory bowel disease. Regen Ther. (2020) 13:1–6. 10.1016/j.reth.2019.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fukuda M, Mizutani T, Mochizuki W, Matsumoto T, Nozaki K, Sakamaki Y, et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev. (2014) 28:1752–7. 10.1101/gad.245233.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sugimoto S, Ohta Y, Fujii M, Matano M, Shimokawa M, Nanki K, et al. Reconstruction of the human colon epithelium in vivo. Cell Stem Cell. (2018) 22:171–6.e5. 10.1016/j.stem.2017.11.012 [DOI] [PubMed] [Google Scholar]
- 145.Meran L, Massie I, Campinoti S, Weston AE, Gaifulina R, Tullie L, et al. Engineering transplantable jejunal mucosal grafts using patient-derived organoids from children with intestinal failure. Nat Med. (2020) 26:1593–601. 10.1038/s41591-020-1024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fair KL, Colquhoun J, Hannan NRF. Intestinal organoids for modelling intestinal development and disease. Philos Trans R Soc Lond B Biol Sci. (2018) 373:2010217. 10.1098/rstb.2017.0217 [DOI] [PMC free article] [PubMed] [Google Scholar]