Abstract
Cancer is generally characterized by acquired genomic aberrations in a broad spectrum of types and sizes, ranging from single nucleotide variants to structural variants (SVs). At least 30% of cancers have a known pathogenic SV used in diagnosis or treatment stratification. However, research into the role of SVs in cancer has been limited due to difficulties in detection. Biological and computational challenges confound SV detection in cancer samples, including intratumor heterogeneity, polyploidy, and distinguishing tumor-specific SVs from germline and somatic variants present in healthy cells. Classification of tumor-specific SVs is challenging due to inconsistencies in detected breakpoints, derived variant types and biological complexity of some rearrangements. Full-spectrum SV detection with high recall and precision requires integration of multiple algorithms and sequencing technologies to rescue variants that are difficult to resolve through individual methods. Here, we explore current strategies for integrating SV callsets and to enable the use of tumor-specific SVs in precision oncology.
Subject terms: High-throughput screening, Genomic analysis
The importance of structural variant detection in cancer
Genomic aberrations acquired in cancer genomes encompass a broad spectrum of types and sizes. These range from single nucleotide variants (SNVs) to larger structural variants (SVs) that impact genome organization (Fig. 1, Table 1)1,2. SVs are a major contributor to genomic variation, they affect more base pairs in the genome than SNVs3 and can have serious phenotypic impact4,5. Some SVs are known to drive carcinogenesis and SVs resulting in gene fusions were the first recurrent mutations observed in many pediatric cancers6,7. With at least 30% of cancer genomes affected by a pathogenic SV, detection of SVs is essential for both diagnosis and treatment stratification6–11. In addition, discovering new oncogenic SV driver events is beneficial for understanding cancer etiology. However, research into the role of SVs in cancer has been limited due to difficulties in their detection which has partially resulted from co-opting sequencing technologies designed for SNV detection.
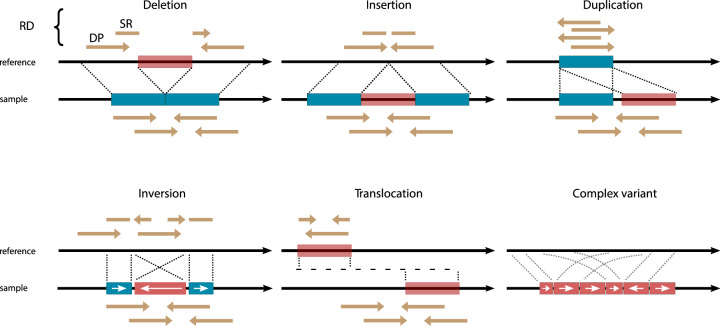
Fig. 1. Major SV types and their characteristic read-alignment patterns.
Alignment of paired-end sequencing reads to a reference genome is used to infer sites of discontinuity or breakpoints. Structural variants (SVs) are generally defined as larger than 50 base pairs and further classified in five major SV types: deletions, insertions of non-reference sequence or mobile elements, duplications, inversions and translocations. Clusters of breakpoints in a genomic region which cannot be classified are considered “complex SVs” and likely result from either progressive rearrangements or a major genomic disturbance. SVs (red blocks) are characterized by patterns in breakpoints and reads aligned to flanking reference sequences (blue blocks). The reads directly below the sample DNA strand represent the distance and orientation at which they are generated during sequencing. If the reads align differently than expected to the reference strand this is indicative of an SV. Changes in read depth (RD) or coverage indicate mostly larger duplications or deletions and are useful for detecting copy number variants (CNVs). Discordant pairs (DP) align to the reference at a different relative distance or orientation than expected. DPs are best suited for detecting large SVs such as inter-chromosomal translocations or inversions. Split reads (SR) span breakpoints and can only be partially aligned. SR can detect small variants with base-pair resolution, especially those smaller than the size of the read.
Table 1.
Glossary of key terms.
| Breakpoint | The location at which a structural variant differs from the reference genome, and forms a novel junction between two previously unconnected segments. |
| Chimeric transcript | A transcript consisting of exons from two different genes, resulting from a genomic mutation or transcriptional process like intergenic splicing or read-through. |
| Complex rearrangement | Structural variant consisting of multiple breakpoints that can not be traced back to a basic type. |
| Differential analysis of tumor-normal data | Also known as “somatic analysis”. By using paired sequencing data, the aim is to classify detected variants as either tumor-specific or also occurring in the matching normal sample. |
| Discordant read pairs | Sequencing reads which have an abnormal insert size when mapped to the reference genome, either larger or smaller than expected, but also mapping to two different chromosomes. |
| Haplotyping/phasing variants | Determining if detected variants occur on the same homologous chromosome and potentially affect the same allele. |
| Long-read sequencing technologies | Single molecule sequencing technologies are actively developed by Pacific Biosciences and Oxford Nanopore Technologies. Reads are ~10 kb+ with a nucleotide accuracy of ~85% depending on the platform version and base calling algorithm (Table 3). |
| Polyploid | Cells which contain more than two chromosomes of each pair. |
| Read alignment patterns | Alignment of read pairs to a reference genome which behave differently than expected. Specific patterns can indicate a structural variant is present. Patterns include changes in read-depth, discordantly paired reads, split reads, soft-clipped reads and one-end mapped reads (Fig. 1). |
| Short-read sequencing technologies | Often used synonymously with sequencing-by-synthesis technology from Illumina. Generates paired-end reads of 150–250 bp with 99% nucleotide accuracy (Table 3). |
| Split reads | Sequencing reads that span breakpoints and therefore map to two locations (split reads) or can only be partially mapped to a single location (soft-clipped reads). Since the default aligner BWA-MEM soft-clips also split reads, they are often used synonymously. |
| Structural variant (SV) | Genomic variant larger than 50 bp in size. Five major SV types are distinguished: deletions, duplications, inversions, translocations and insertions of non-reference sequence or mobile elements. |
| Tumor purity | The proportion of cancer cells within a tumor sample. |
| Variant allele frequency | The relative abundance of a variant allele versus the unchanged reference allele based on read support. |
Advances in sequencing technologies have increased the number of SVs identified per genome from ~2, 1–2, 5k in the 1000 genomes project to more than 27k in recent multi-platform sequencing efforts3,4,12. Specifically for the cancer genomics community, recent contributions of the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium have provided an extensive resource of paired tumor-normal genomes13. The insights obtained from multi-platform analyses also highlight current SV blindspots in cancer variant databases like COSMIC. Despite technological innovations, confident SV detection in cancer genomes remains challenging due to biological factors including contamination from healthy tissue, intratumor heterogeneity and polyploidy. Identification of variants acquired in tumor cells requires discerning tumor-specific somatic SVs (TSSVs) from variants in the germline and mosaic variants present in unaffected cells14. This is often done by differential analysis between paired tumor-normal samples15. The classification of SVs as tumor-specific or normal is confounded by inconsistencies in detected breakpoints and derived variant types, as well as the biological complexity of some rearrangements.
Confident SV detection and subsequent classification of variants as either germline, tumor-specific or mosaic variation in healthy tissue is not only important for diagnostics and cancer etiology but also for research into cancer predisposition and genetic interactions. In addition, the genetic context of somatic variants and interplay with germline variants may influence their tumorigenic potential16. Here, we focus on the detection of TSSVs from paired tumor-normal WGS data. First, we explore current approaches for SV detection and their integration, whilst accounting for challenges specific to cancer samples. Second, we address different approaches aimed at distinguishing TSSVs from normal SVs. Third, we highlight the impact that long-read sequencing can have on somatic SV detection. Last, we explore how orthogonal sequencing technologies can be combined to improve TSSV detection.
Detection of somatic SVs in short-read WGS data
SVs can be detected using short-read sequencing data based on patterns in aligned reads (Fig. 1). These reads are sequenced as paired ends of 150–250 bp long. Changes in read-depth (RD) are used to derive copy-number variants (CNVs). Discordant read-pairs (DP) that align with an abnormal distance and/or orientation to the reference genome are suited for detecting large SVs. Split or soft-clipped reads (SR) are partially mapped reads and can indicate breakpoints with base-pair resolution17. Both the alignment method and reference genome used, influence the performance of SV detection algorithms17,18. BWA-MEM is predominantly used for alignment prior to SV detection, as it provides secondary alignments to reads mapping to multiple locations rather than placing the reads randomly19,20. However, alignment uncertainty is inherent to short-read sequencing data. In parallel, the reference genome continues to evolve, resulting in improved alignments and fewer false-positive variants in studies which adopted GRCh38 (hg38) compared to GRCh37 (hg19)8,21–23.
Combinatorial algorithms integrate multiple read-alignment patterns
The latest generation of SV detection algorithms that combine multiple read-alignment patterns can detect SVs across a broad range of types and sizes. At present, many different strategies and methods exist (Table 2). How these combinatorial algorithms integrate read-alignment patterns influences their ability to detect specific variant classes (Fig. 2A)24,25. As a result, no single algorithm performs best across the full spectrum of SVs, implying that integration of multiple algorithms is beneficial25. Although most studies comparing SV algorithms focus on germline SVs, these findings were recently also confirmed for somatic SV detection26. The methodology used by DELLY, LUMPY, Manta, SvABA, and GRIDSS for detecting SVs (Box 1) achieves high performance in detecting both germline and somatic SVs25,26.
Table 2.
SV detection algorithms.
| Tool1 | Platform | Method | Reference | Used in study |
|---|---|---|---|---|
| DELLY | IL | DP, SR | 34 | 8,13,89,93 |
| LUMPY | IL | DP, SR | 35 | 8,81 |
| Manta | IL | DP, SR, AS/l | 36 | 89,94 |
| GRIDSS | IL | DP, SR, AS/l | 37,39 | |
| SvABA | IL, 10× | DP, SR, AS/l | 38 | 13,83 |
| Varlociraptor | IL | Post-processing | 31 | |
| Lancet | IL | 40 | ||
| GROC-SVS | 10× | 95 | 81,83 | |
| Longranger | 10× | 96 | 81,83 |
| Long read tools | Platform | Method | Reference | Cited by/remarks |
|---|---|---|---|---|
| HySA | PacBio and IL | Assembly and alignment | 76 | Hybrid assembly of IL and PacBio reads |
| SVIM | ONT, PacBio | Alignment | 69 | 67,93,97 |
| Sniffles | ONT, PacBio | Alignment | 55 | 25,32,56,67,89,93,94,97,98 |
| pbhoney | PacBio | Alignment | 99 | 25,76 |
| pbsv | PacBio | Alignment | 100 | 25,97 |
| NanoSV | ONT | Alignment | 98 | 32,56,93 |
| Picky | ONT | Alignment | 56 | 32,93 |
| NanoVar | ONT, PacBio | Alignment | 93 | Applied to a leukemia sample |
| cuteSV | ONT, PacBio | Alignment | 97 | |
| nanomonsv | ONT | Alignment | 68 | Detects tumor-specific SVs |
1Inclusion criteria: published tool focussed on tumor-specific SV detection in cancer from tumor-normal paired WGS data, used in key studies addressed in this review. Long-read alignment based SV detection tools that are commonly used are included regardless of their ability to detect tumor-specific SVs.
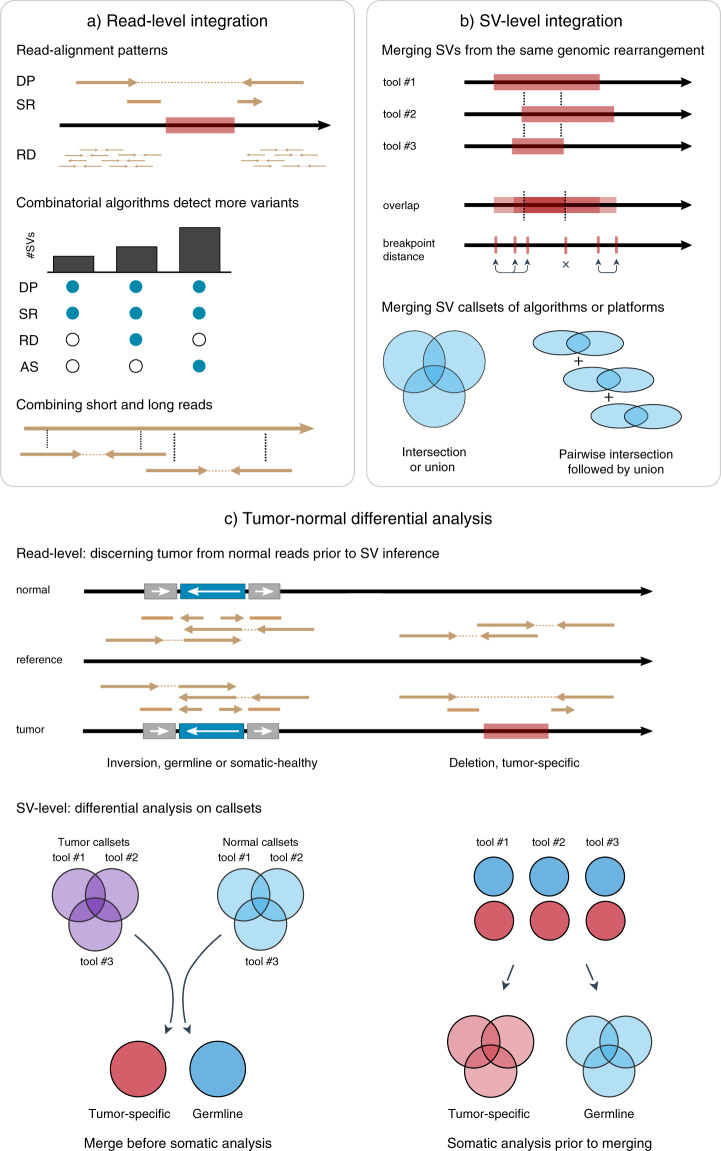
Fig. 2. Data integration to improve tumor-specific SV detection.
a Alignment of sequencing data against a reference is used to infer SVs by detecting aberrant patterns of read-alignment: discordant pairs (DP), split reads (SR), read depth (RD) and (local) assembly (top, see also Fig. 1). Algorithms that combine multiple read-alignment patterns can resolve more SVs (middle). Likewise, read-level integration of technologies can aid SV detection, i.e., combining short and long reads (bottom). b Comparison of SV callsets requires merging variants from the same genomic rearrangement based on e.g., reciprocal overlap or breakpoint distance (top). These merging approaches can yield different outcomes as shown by how only a small segment of the deletion overlaps between tools and not all breakpoints could be matched. Intersection of callsets identifies the SVs with support from multiple algorithms or technologies. Alternatively, sensitivity can be increased by taking the union of callsets or their pairwise intersections (bottom). c Identification of tumor-specific SVs (red) requires tumor-normal differential analysis of reads or events. A tumor sample (purple) is expected to contain tumor-specific variants (red, bottom stand), as well as germline variants (blue, top strand). Tumor/normal reads can be distinguished prior to SV inference or afterwards by comparison of the variants or breakpoints as in b. If multiple SV tools are used, differential analysis can be done after merging tumor and normal callsets (bottom left) or first by using each algorithm’s somatic filtering feature (bottom right).
Box 1: Integration of read-alignment patterns by combinatorial algorithms.
Integration of read-alignment patterns by SV detection algorithms influence which SVs can be confidently detected. DELLY, LUMPY, GRIDSS, Manta, and SvABA are state-of-the-art algorithms and have amongst the best performance for germline SV detection25. They can detect all the major SV types at base-pair resolution using SR or assembly and also perform somatic classification.
DELLY uses DP and SR in a stepwise manner to detect ~200 bp–5 kbp SVs34. Since DELLY analyses SV types separately, it can detect nested SVs and infer complex events which is useful for somatic SV detection. LUMPY has a probabilistic model that combines parallel analyses of DP and SR such that both contribute independently to the detection of breakpoints35. Overlapping breakpoints are clustered to identify SVs, except for insertions. GRIDSS can detect SVs and indels regardless of size using a combination of assembly, SR and DP-support37. Break-end contigs spanning SV breakpoints are assembled from SR, DP, one-end anchored, gapped, and unmapped reads. Variants are inferred with a probabilistic model combining evidence from realignment of these break-end contigs, SR and DP. GRIDSS can rescue un/misaligned reads, detect novel non-reference sequence insertions, and resolve micro-homology surrounding breakpoints. Manta uses a graph-based approach to generate candidate SVs from DP, SR and gapped reads, followed by local assembly and realignment of contigs to the genome. SVs are scored by a model that integrates evidence from discordant reads and the assembly. SvABA performs genome-wide local assembly in 25 kb windows based on SR, DP, gapped, and unmapped reads38. Variants are inferred from alignment of contigs to the reference and subsequently scored by realignment of reads to the contigs.
Despite their differences in approach, for overlapping/shared SVs these tools agree on breakpoints within ~2 bp based on simulations in optimal detection conditions26.
SV-level integration of multiple algorithms improves precision
Since the optimal detection algorithm differs between SV type and size range, full-spectrum SV detection with high recall and precision currently requires multiple algorithms25,27. The optimal method to combine the resulting callsets remains a largely unanswered question and a variety of tools and in-house pipelines are currently used4,13,25,28. To compare and combine SV callsets, variants from the same genomic rearrangement need to be merged first, this is complicated by diversity in breakpoint resolution and SV typing (Fig. 2B). The recent review by Ho et al. addresses different “ensemble” integration approaches currently in use in germline SV research4. In general, simple integration strategies use (reciprocal) overlap or breakpoint distance to merge SVs whilst more complex solutions combine this with read-evidence integration, local assembly or machine learning29–32.
After overlapping variants are merged, integration of SV callsets from multiple algorithms can either be performed by taking the union or intersection (Fig. 2B). Since achieving high precision takes priority in most cancer research and clinical applications, an intersection strategy is often preferred but reduces recall. The precision/recall trade-off can be optimized by carefully selecting which tools to intersect25 and by taking the union of pairwise intersections26.
Distinguishing somatic from germline SVs
TSSV detection aims to identify variants that uniquely occur in a patient’s tumor cells. Typically paired tumor-normal samples are used to classify SVs as either germline, mosaic-normal or tumor-specific variants15. Detection of TSSVs is a two-step process that involves the detection of SVs in both samples, followed by differential analysis of the callsets (Fig. 2C). Also, cancer genomes can have highly complex rearrangements. Alternatively, if patient-derived healthy material is not available, SVs can be filtered using a panel-of-normals. A sufficiently large panel-of-normals can provide more statistical power for filtering recurrent germline variants, but is less effective than a patient-derived normal sample when filtering rare or private germline variants4. Also, strictly filtering out regions with germline CNVs excludes potentially interesting genomic regions from SV analysis, which are susceptible to rearrangements because of their architecture33.
Tools for somatic SV detection in WGS data
Somatic SV detection algorithms differ in their approach to identify TSSVs from paired tumor-normal samples and as a result can classify the same event differently26. Despite their differences, DELLY, LUMPY, SvABA, Manta, and GRIDSS have successfully been used to report somatic SVs in various studies34–37. DELLY and LUMPY use ad hoc filtering whereby SVs supported by at least one read from the normal sample are removed from the tumor SV callset34,35, which is highly sensitive contamination. In contrast, Manta uses a probabilistic scoring system for somatic SVs integrating evidence from tumor and normal reads36. SvABA uses both the tumor and normal data during assembly before distinguishing somatic variants38. GRIDSS has yet another approach and applies extensive rule-based filtering to both single break-ends and breakpoints37,39.
Specialized somatic SV detection tools such as Lancet and Varlociraptor account for challenges specific to the identification of TSSVs (Box 2)31,40. The first challenge in comparing tumor and normal SV callsets are differences in SV breakpoints and types, analogous to the issues with overlapping SV callsets of different algorithms25. Second, somatic SVs are often complex which can be problematic for algorithms that are not equipped to resolve these complex SV signatures and instead infer (false-positive) small indels41. As an alternative to ad-hoc filtering of SV callsets, Varlociraptor and Lancet, respectively, compare breakpoints and aberrant reads between tumor-normal samples at an earlier stage of the analysis (Fig. 2C). Specifically, Varlociraptor compares the statistical support for an altered reference with simulated variant versus an unadjusted reference (Box 2)31. Using read-level or breakpoint-level comparison can account for the subsequent mutations at germline variant locations, as these mutations may convolute somatic-germline comparisons. Third, issues inherent to analyzing tumor samples such as contamination, polyploidy, and heterogeneity are accounted for by Varlociraptor and Lancet (Box 2).
Box 2: SV detection algorithms specialized in differential analysis.
Lancet and Varlociraptor address challenges specific to tumor-normal analysis, e.g., contamination, polyploidy, intratumor heterogeneity (subclonality) and thus aid in identification of tumor-specific SVs.
Lancet is specialized in the detection of somatic SNVs, insertions (<200 bp) and deletions (<400 bp) from short-read WGS data using local (micro-)assembly and re-alignment to the reference40. By using a graph-based approach, Lancet can resolve haplotypes and use the origin of supporting reads to distinguish TSSVs from germline variants. Sample contamination can be accounted for by adjusting the number of allowed supporting normal-reads. Lancet can detect rare variants (>5% AF) in a virtual tumor whilst preventing false-positives in short-tandem repeat regions, achieving higher precision than other algorithms but at cost of sensitivity.
Varlociraptor is a post-processing tool which uses a Bayesian framework to differentiate between somatic and germline breakpoints by calculating false discovery rate (FDR) values from unfiltered callsets31. During FDR calculation it quantifies uncertainties due to ambiguous read alignments, how reads support SVs (typing uncertainty), gap-placement bias and strand bias30,31. This is done by simulating the variant into the reference, re-aligning reads and comparing the statistical support for the adjusted versus unadjusted reference. Challenges specific to tumor samples are taken into account, as additional uncertainties e.g., mosaic-normal variants, contamination, intratumor heterogeneity and aneuploidy. By doing so, it is able to control the FDR of SNVs and small insertions/deletions (30–250 bp) and achieves better precision/recall on callsets of DELLY, Manta, and Lancet compared to the filtering of the tools themselves31.
Challenges for accurate SV detection in cancer genomes
The analysis of tumor-normal paired samples is confounded by challenges inherent to cancer samples, including polyploidy, heterogeneity and contamination17. First, potential aneuploidy of tumor cells complicates haplotype reconstruction and phasing reads12,42. Second, intratumor heterogeneity can result in multiple subclonal variants which have low allele frequency (AF) and few supporting reads, making them difficult to detect. Third, contamination of the tumor sample with healthy material and vice versa complicates differential analysis between paired samples due to mislabelled reads. This can result in algorithms falsely discarding somatic variants with one or more supporting reads from the control sample. Adjusting the filtering threshold based on an estimated contamination fraction is a balance between precision and sensitivity for detecting low-AF variants.
The detection of rare TSSVs is limited by sequencing depth and AF. In practice, a minimum of 20% AF is required for reliable variant detection from tumor-normal pairs26,31. Increasing sequencing depth to 75x-90x for tumor samples improves the sensitivity of detection, especially for variants below 20% AF, whilst maintaining precision26. In addition, interpretation of TSSV allele frequencies is not straightforward since they can reflect intratumor heterogeneity and/or multiple alleles within a polyploid tumor genome. Note that the SV type should be considered during AF interpretation43. For diploid normal cells, variants are expected to have an AF of 0%, 50%, 100%, or 33% in case of a heterozygous duplication. However, mosaic-normal variants can occur at varying AF and be difficult to distinguish from TSSVs14. Computational modeling with AF can provide insight into intratumor heterogeneity and clonal architecture, both of which are important for therapeutic resistance and relapse44. The majority of SV tools operate under a diploid genome assumption. A multitude of tools independently quantify purity and ploidy of tumor samples however benchmarking studies show little consensus39,45. These tools can rely solely on CNV deletion events to model the cell purity and ploidy, and/or incorporate heterozygous known SNPs into their probabilistic models. At present, only SVclone uses SVs to estimate intra-tumor heterogeneity due to the complexities of calculating variant AF for SVs43.
Computational challenges of complex variant detection
Genomic instability in cancer genomes results in more breakpoints and more complex SVs compared to germline variation46. Complex SVs are characterized by signatures of many breakpoints clustering together and are hypothesized to be caused by a single catastrophic process followed by repair or progressive rearrangements47. The presence of breakpoint clusters complicates the inference of the underlying genomic rearrangements and therefore also the identification of tumor-specific events. Alternatively, when breakpoint clusters confound confident SV calling, breakpoint-level differential analysis can be used to identify tumor-specific events. In addition, unsupervised clustering can discern complex from simple SVs and help to study both events more accurately41.
Technical limitations of short-read WGS influence SV detection
The detection of SVs is also influenced by technical limitations of the sequencing platform; most notably genome coverage bias and alignment uncertainty. Illumina (IL) is currently the most commonly used short-read sequencing platform since it’s relatively affordable, fast and has a high nucleotide accuracy (>99%)48. However, IL sequencing has inherent biases in genome coverage with regions that have a high, or low GC content (<10% and >85% GC) or long homopolymers49. Although PCR-free library preparation does reduce GC biases it does require a large amount of input DNA (Table 3)49.
Table 3.
Comparison of long-read and short-read sequencing technologies.
| Illumina | 10× linked-read | Short-read RNA-seq | ||
|---|---|---|---|---|
| Avg read length | 2× 150–250 bp | IL platform dependent | 2× 75 bp | |
| Max read length | 2× 250 bp | (~100 kb span) | 2× 100 bp | |
| Accuracy per nucleotide1 | >99% | (see Illumina) | (see Illumina) | |
| Error bias | Substitutions in high/low GC regions | (see Illumina) | (see Illumina) | |
| Coverage bias | Low coverage of high/low GC regions. Mapping issues with highly homologous regions | (see Illumina) | Illumina biases and additional: ligation bias due to the reverse transcriptase enzyme101, protocol differences poly-A only versus ribodepletion | |
| Accuracy after error correction2 | N/A | N/A | N/A | |
|
Sample requirements PCR-free3 |
1–2 μg/(default > 500 ng) | IL + controller chip 1 ng | 0.1–1 μg | |
|
Low-throughput sample requirements |
100 ng3 | – | 25 ng | |
| Base modifications | Bisulfite sequencing required | – | – | |
| Nanopore | PacBio | BNG | Hi–C | |
|---|---|---|---|---|
| Avg read length | 15–20 kb | 10–15 kb | ~100 kb resolution, variants >500 bp | ~1 kb–1 Mb resolution102 |
| Max read length | >800 kb103,104 | >60 kb103–105 | ||
| Accuracy per nucleotide1 | 60–85%103,104,106 | >85%105 | no base pair resolution | no base pair resolution |
| Error bias | Small indels, mostly deletions49,107 | Small indels, mostly insertions49,107 | N/A | N/A |
| Coverage bias | Truncation of homopolymers and low-complexity regions103 | Homopolymers | Fragile sites, nick enzyme-dependent108 | Biases depend on protocol used: restriction enzymes, PCR, and IL seq102,109 |
|
Accuracy after error correction2 |
After 1D2 97% After hybrid correction: >99% |
After CCS: 95–99%60. | N/A | N/A |
| Sample requirements3 |
1 μg–400 ng HMW 0.4–1 μg HMW |
10 μg HMW | 0.3–0.9 μg HMW | 1–10 million cells102,109 |
|
Low-throughput sample requirements |
10–100 ng | 400–800 ng | 300 ng | 1000 cells110 |
| Base modifications | Theoretically all |
Methylation, Mostly bacterial |
– | – |
Comparison of Pacific Biosciences (PacBio), Oxford Nanopore Technologies (ONT), Illumina (IL), 10× Genomics linked-read sequencing on the Illumina platform (10×), RNA sequencing on the Illumina platform (RNA-seq), BioNano Genomics (BNG) and the genome-wide chromatin conformation capture protocol Hi–C (Hi–C). Many characteristics of 10× and RNA-seq are shared with IL, since the same sequencing platform is used. Also note that Hi–C protocols are under active development, they vary in biases, sample requirements and use of IL sequencing102,109,110. Unreferenced values are derived from the manufacturer’s websites last accessed in October 2020 [Oxford Nanopore Technologies (https://nanoporetech.com/), PacBio (https://www.pacb.com/), Illumina (https://www.illumina.com/), 10x Genomics (https://www.10xgenomics.com/), Illumina Stranded mRNA Prep (https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/stranded-mrna-prep.html), Bionano Genomics (https://bionanogenomics.com/)].
1Reported accuracy for PacBio and ONT strongly depends on the sequencing platform version and polishing steps. Using regular single-pass sequencing without self-correction, both PacBio and ONT theoretically have per-nucleotide error rates of ~15%103–105 but previous versions of the MinION up to ~40%106.
2The latest ONT and PacBio technologies attain >99% accuracy for de novo human assemblies. PacBio achieves >99.8% accuracy using circular consensus sequencing (CSS) where the same read is sequenced many times and averaged, although this limits read length to ~13 kb60. ONT reports >99% after polishing with short reads (hybrid correction) which is necessary due to truncation of homopolymers and low-complexity regions103. ONT 1D2 technology sequences both DNA strands and uses consensus to attain >97% whilst maintaining read lengths, although only ~60% of the molecules can be sequenced using this approach [Oxford Nanopore Technologies (https://nanoporetech.com/)].
3Sample requirements as listed by the manufacturer and dependent on the library preparation method used, e.g., insert size and use of PCR, as well as the exact version of the machine. High molecular weight (HMW) DNA is required to attain long read lengths, but the read length of PacBio is limited by the polymerase and for ONT by the length of the DNA molecules hence it can report ultra-long reads >800 kb103. The minimum sample amount of 10 ng listed by ONT is likely insufficient for a human genome. Whilst for IL in practice smaller amounts e.g., 50 ng are used as low-throughput minimum.
The detection of SVs relies on identifying aberrant read alignment patterns (Fig. 1). Reads derived from highly homologous regions, such as pseudogenes and segmental duplications, are often not long enough to uniquely map to the reference genome50. Yet repeat-rich regions comprise about half of the human genome and are vulnerable to SVs due to homologous recombination errors and replication slippage33,51. Depending on the alignment algorithm, uncertainty usually results in either random placement of reads or multi-mapping to all possible locations52. Multi-mapping, for example as done by BWA-MEM, causes unequal genome coverage altering the signal-to-noise ratio52. Hence, alignment uncertainty is problematic for accurate SV detection and should be addressed with a sound statistical model30,31,52. Current estimates suggest ~55 Mb of GRCh38 are “dark regions” inaccessible to IL sequencing due to alignment ambiguity (i.e., repeat-rich regions) or the sequencing chemistry (i.e., GC content)53. The over 4000 affected gene bodies53 also include disease-related genes, such as the TERT promoter which was found to be mutated in 9% of tumors in the PCAWG study but mutations can be missed due to its high GC content13.
Impact of long-read sequencing
Single-molecule long-read sequencing technologies by Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) are valuable for SV detection54. PacBio and ONT generate reads of ~10+ kb versus ~250 bp from IL; the longer reads reduce alignment ambiguity and do not have a GC bias, resulting in improved coverage of “dark” regions in the genome55. In addition, long reads allow for haplotype phasing of variants and de novo assembly of complex rearrangements56. For example, sequencing lung cancer cell lines with PromethION detected both known cancer-driver SNVs and revealed large previously unknown genomic rearrangements, including an 8 Mb amplification of MYC57. Similarly, direct comparison of a PacBio assembly with IL sequencing shows ~2.5× more uniquely identified SVs (~48k and ~20k, respectively), in particular more inversions and 50 bp–2 kb insertions/deletions located in repeat-rich regions12.
Limitations of long-read sequencing
The disadvantages of PacBio and ONT platforms include costs and sample requirements, which are substantial compared to IL sequencing and can be problematic for tumor samples (Table 3)55. In addition, they have a lower nucleotide accuracy of ~85% for single molecule sequencing and up to 99% using consensus sequencing of the same DNA molecule58–61. Continuous improvements in algorithms for base calling and error correction have increased the accuracy of these platforms58,59. Since low nucleotide accuracy can impede read-alignment, error correction potentially improves SV detection by increasing the fraction of aligned reads62. However, error-correction strategies come with trade-offs for SV detection. Long reads can be aligned to each other as a self-correction strategy when sufficient coverage (~50×) is available55. However, haplotyping information is lost as a result of using the consensus of reads with mixed molecular origin. This makes the consensus sequence unsuitable for variant phasing or for studying intra-tumor heterogeneity or polyploidy. Alternatively, short reads can be used for error correction by aligning them to the long reads, but this approach only improves accuracy of genomic regions accessible to IL sequencing55,61.
Long-read data requires specialized algorithms
Long-read SV detection algorithms are either based on de novo assembly or read-alignment to a reference genome. Assembly-based strategies have a higher sensitivity for detecting non-template insertions and homozygous SVs. During assembly, contigs are compared to the reference genome and can provide more evidence than individual reads32,55. However, variant calling using alignment requires less coverage than assembly (~20× versus ~50×) and statistical significance when identifying SVs is achieved relatively easily due to the low alignment uncertainty of long reads32,50,55. Compared to assembly methods, alignment-based approaches are more suited to identify heterozygous SVs and more robust to amplifications in highly homologous regions such as low-complexity regions12,55. Within clinical applications, often insufficient resources are available to perform long-read sequencing of tumor-normal pairs to depths required for de novo assembly (Table 3). Therefore, we focus on using alignment-based strategies (Table 2).
Alignment of long reads differs from short reads due to the increase in base pairs to align and different errors profiles55. Although BWA-MEM offers support for long reads, it often infers many small gaps during alignment and misses large indels63,64. Specialized long-read alignment algorithms have been developed to overcome these issues. In contrast to short-read data, there is no best practise for which aligner should be used when performing SV detection63–66. Preliminary comparisons suggest that NGMLR and minimap2 perform well and both algorithms are designed to handle the higher error rates and adjust for the 1 bp indels in long-reads12.
Alignment-based SV detection algorithms for long-read data
Currently, many tools are actively developed to detect SVs from alignment of ONT and PacBio data (Table 2). However, studies comparing long-read SV detection tools have been scarce and predominantly show the limitations of available truth sets by identifying many novel variants12,67. At present only nanomonsv reports somatic SVs from long-read data68. The commonly used tools SVIM and Sniffles have shown good precision and sensitivity in multiple performance assessments63,67,69. They were among the first to process both ONT and PacBio data despite their different error profiles and have been followed by additional tools like NanoVar and CuteSV (Table 2). Similar to short-read SV detection tools, long-read tools combine multiple read-alignment patterns to detect SVs. They infer patterns similar to split reads and discordant pairs using intra-alignment and inter-alignment signatures, despite long reads not being paired-end. Similar to short-read tools, using a consensus callset created by intersecting multiple long-read SV detection algorithms can increase precision32,67. Alternatively, machine learning approaches can attain greater improvements in precision and sensitivity than ad hoc intersection, given a truth set is available for training32.
Multi-platform data integration to improve detection of somatic SVs in cancer
Limitations in both short-read and long-read WGS can potentially be overcome by using a multi-platform approach and as such improve the identification of TSSVs. Integration can improve both precision and sensitivity by combining read-alignment patterns (Fig. 2A) and integrating SV callsets from multiple algorithms or technologies (Fig. 2B).
Gene fusion detection by combined analysis of RNA and WGS
Integration of genomic and transcriptomic data can further improve variant detection and provide insight into the phenotypic effect of SVs; specifically resolving gene fusions, splice variants and linking SVs to altered gene expression70. RNA sequencing of tumor samples offers unique advantages such as tissue specificity and time specificity, but obtaining high-quality RNA can be problematic. In addition, sufficient expression is necessary to detect events, which may impede detection of low AF variants.
RNA-seq is especially suitable for detecting gene fusion events through their chimeric transcripts. Gene fusions have high clinical relevance since they are often cancer drivers and otherwise occur rarely in the general population6,70. Specialized gene fusion algorithms predict gene fusions from chimeric transcripts by using read-alignment patterns such as SR crossing exonic junctions and DP mapping to both gene partners71. However, these algorithms can suffer from a high false positive rate which requires extensive filtering72. Chimeric transcripts can occur without genomic rearrangement, for example through intergenic splicing (trans-splicing and cis-splicing) or transcriptional slippage on short homologous sequences73. Since these chimeric transcripts are also present in healthy cells, this advocates for tissue matched RNA-seq of paired tumor-normal samples to allow the identification of tumor-specific events.
Combining RNA-seq with WGS data could resolve specificity issues and improve gene fusion detection. By itself, WGS can detect gene fusions, but not the occurrence of functional transcripts. Although sometimes used for validation purposes74, there are no established algorithms which integrate WGS and RNA-seq such that they both contribute to detection. The advantages of combining WGS, RNA-seq and exome sequencing has been demonstrated for detecting SVs in heterogeneous pediatric cancers75. Similarly, joint analysis of RNA-seq and short-read WGS in the PCAWG study identified the underlying SV for 82% of gene fusions. The remaining fusions were either the result of RNA-only alterations such as transcriptional read-through or underdetection of SVs5.
Integration of short-read and long-read WGS
Short-read and long-read data can complement each platform’s strengths and overcome individual limitations12. Combining SV callsets after detection can increase sensitivity and requiring orthogonal support for variants across platforms can increase their confidence. However, the union or intersection of callsets is still affected by platform-specific technical biases. Read-level integration can overcome some of these issues as illustrated by error correction approaches which use IL reads to improve the accuracy of PacBio/ONT reads55. Likewise, hybrid assembly of short and long reads benefits from their respective high accuracy and scaffolding properties. Localized hybrid assembly tailored to SV detection as implemented by HySA shows that problematic SVs can be detected that have too little support in either PacBio or IL76. However, HySA cannot infer somatic SVs and some variants were missed due to few supporting aberrant IL reads and PacBio alignment issues. Hybrid assembly can also reduce coverage requirements for de novo assembly77.
As an alternative to long-read technologies, linked-read sequencing from 10× Genomics (10×) performs well for haplotype construction and variant phasing12. A read-barcode is added during library preparation to trace the molecule of origin at costs similar to IL sequencing78 (Table 3). In addition, 10× can report variants in repeat-rich regions not accessible by standard short-read IL sequencing79,80. Integration of short-read WGS and 10× enabled chromosome-scale haplotyping and phasing of detected variants of the polyploid cancer cell line HepG281,82. Variant phasing can help to gain biological insights, as shown for associated regulatory and coding mutations in treatment-resistant prostate cancer83 and identification of SVs as potential cancer drivers by altering cis-regulation of genes84.
Discovery of large, complex variants by chromatin assays
Combining sequencing data with technologies that provide insight into genomic organization can elucidatie large complex rearrangements. Technologies such as Bionano Genomics (BNG) and Hi–C have shown limitations of SV detection using sequencing. The combination of short-read WGS, BNG, and Hi–C on a cancer cell line showed most of the large (>1 Mb) intra-chromosomal and inter-chromosomal SV events were uniquely detected by a single technology with only ~20–35% validated by multiple platforms8. Each platform has its own scope of variant detection. Short-read WGS detected the largest number of variants across a broad range, whilst BNG and Hi–C lack base-pair resolution but can detect >1 kb deletions in repeat rich regions unlike short-read WGS8. BNG has promising diagnostic applications as it can confidently detect large variants with low input requirements (Table 3). Also, BNG had full concordance with standard diagnostic assays in pediatric ALL and identified additional variants85.
Incorporating pre-existing technologies in ongoing studies
Continuous technological improvements provide exciting new data and SV discoveries, but this does not make existing datasets obsolete. The phenotypic effect of CNVs is often better understood than for SVs and established technologies have had more opportunity to collect samples, including rare cancer types. Currently many samples are available in repositories that profile genomic imbalances either via SNV array or exome sequencing technologies13,86. Challenges in integrating these datasets result from differences between technologies, such as breakpoint resolution and platform-specific biases, and systematic solutions are rare87. The widely varying detection resolution of different technologies invalidates callset intersection strategies, as smaller events are below the detection limits for lower resolution arrays, and exome sequencing is limited to events involving multiple exons. The absence of an event in a callset should not be considered proof that the event does not exist. Gene-centric approaches based on unions seem the most applicable. Although integration of pre-existing datasets assayed with different technologies with recently acquired datasets provides a complex computational challenge and is often ignored, it is likely to be an ongoing issue as technologies and platforms continue to evolve.
Challenges in using sequencing for precision oncology
In clinical practice, next-generation sequencing (NGS) is increasingly used to replace targeted assays subject to budgetary and sample requirements. NGS can simultaneously detect different variant types and discover new biomarkers, and is more cost-effective than a series of single-gene assays. Although turn-around times are often longer, sensitivity and precision are maintained88 provided sufficient sequencing depth is achieved26,31. As a result, NGS makes pan-cancer biomarker testing feasible, leading to the approval of drugs based on molecular alterations shared by different cancer types like the use of TRK inhibitors for all solid tumors with a NTRK fusion88. However, the distribution of NGS data over multiple repositories and lack of data harmonization complicates clinical decision-making and prevents precision medicine from reaching its full potential.
Variant interpretation is a major challenge in precision oncology often done by expert panels such as interdisciplinary molecular tumor boards88. Despite its challenges, integration of multi-omics data is increasingly being used to improve variant interpretation and increase the number of identified drivers or actionable targets5,88,89. However, standards on variant interpretation and prioritization are still emerging90. As a result, there is low concordance between the recommendations of different molecular tumor boards when given identical case studies, especially for complex genomic alterations90.
Recent initiatives have attempted to resolve this need for standardization in variant assessment and clinical decision through the Molecular Tumor Board Portal91 and Somatic Working Group of the Clinical Genome92. Both harmonize different variant repositories, curated knowledge bases and computational predictions to acquire insights into variant-gene-drug-disease relationships with the focus on clinical use Although extremely valuable, these efforts focus only on SNVs and to a limited extent gene fusions. Similar initiatives for SVs and complex genomic alterations are currently lacking. Largely due to tumor-specific SVs not yet commonly being used as molecular targets or biomarkers to guide patient-specific treatment. We anticipate that improved confidence of TSSV detection will enable the subsequent research necessary for the use of the full spectrum of variants in precision oncology.
Conclusion
The field of SV detection is continuously improving through advancements in sequencing technologies and tools. These advancements will contribute to discoveries into the role of SVs in cancer, as well as the incorporation of SVs in precision oncology programs. Nevertheless, SV detection and interpretation in tumor samples is complicated by unique biological and technical challenges, i.e., contamination, intra-tumor heterogeneity and aneuploidy. These challenges are addressed by algorithms specialized in identifying TSSVs from tumor-normal paired sequencing data, which requires both SV detection and distinguishing tumor-specific variants.
Based on studies of normal genomic variation, a multi-platform approach is necessary to detect the full spectrum of variants and reduce false positives. Truth sets and procedures developed for SV detection from short-read data show that combining multiple tools improves precision and recall. Despite this, short-read sequencing has inherent limitations such as GC coverage bias and mapping ambiguities leading to inaccessible genomic regions. Long-read sequencing technologies can resolve large, complex SVs and improve coverage, but have lower per-nucleotide accuracy, higher costs and sample requirements. SV detection tools for long-read data have yet to mature with performance assessments and truth sets lacking.
Integration of long-read and short-read data is likely required for complete characterization of tumor genomes. However, adopting sequencing technologies in clinical laboratories requires a clear added value compared to the standardized assays, as well as being fast and affordable. Considering IL and 10× provide high accuracy WGS at low sample requirements, they are most feasible for tumor-normal sequencing in a clinical setting. Supplementary low-coverage sequencing with ONT can cover regions inaccessible to short-read WGS and aid in variant phasing. Alternatively, RNA sequencing has proven to be highly beneficial in a clinical setting for the detection of gene fusion events.
In conclusion, improving detection of TSSVs by integrating data derived from multiple platforms and detection tools enables the use of TSSVs in precision oncology and research into their role in cancer. With accurate TSSV datasets becoming more available, previously unchartered territories of variant types can be explored to potentially discover novel SV cancer driver events.
Acknowledgements
This work was financially supported by KiKa.
Author contributions
A.S. and P.K. substantially contributed to the conception and design of the article. I.A.E.M.B. and J.H.K. drafted the article. All authors discussed the concepts and contributed to the final manuscript.
Data availability
No datasets were generated or analyzed during this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Aplan PD. Causes of oncogenic chromosomal translocation. Trends Genet. 2006;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho SS, Urban AE, Mills RE. Structural variation in the sequencing era. Nat. Rev. Genet. 2019;21:1–19. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calabrese C, et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578:129–136. doi: 10.1038/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Wu N, Liu D, Jin Y. Recurrent fusion genes in leukemia: an attractive target for diagnosis and treatment. Curr. Genomics. 2017;18:378–384. doi: 10.2174/1389202918666170329110349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon JR, et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 2018;50:1388–1398. doi: 10.1038/s41588-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupain C, et al. Discovery of new fusion transcripts in a cohort of pediatric solid cancers at relapse and relevance for personalized medicine. Mol. Ther. 2019;27:200–218. doi: 10.1016/j.ymthe.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairncross JG, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 11.Cohen MH, et al. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin. Cancer Res. 2002;8:935–942. [PubMed] [Google Scholar]
- 12.Chaisson MJP, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pleasance ED, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Horebeek L, Dubois B, Goris A. Somatic variants: new kids on the block in human immunogenetics. Trends Genet. 2019;35:935–947. doi: 10.1016/j.tig.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Mandelker D, Ceyhan-Birsoy O. Evolving significance of tumor-normal sequencing in cancer care. Trends Cancer Res. 2020;6:31–39. doi: 10.1016/j.trecan.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramroop JR, Gerber MM, Toland AE. Germline variants impact somatic events during tumorigenesis. Trends Genet. 2019;35:515–526. doi: 10.1016/j.tig.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, et al. Structural variation discovery in the cancer genome using next generation sequencing: computational solutions and perspectives. Oncotarget. 2015;6:5477–5489. doi: 10.18632/oncotarget.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruffalo M, LaFramboise T, Koyuturk M. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics. 2011;27:2790–2796. doi: 10.1093/bioinformatics/btr477. [DOI] [PubMed] [Google Scholar]
- 19.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv [q-bio.GN] (2013).
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan B, et al. Similarities and differences between variants called with human reference genome HG19 or HG38. BMC Bioinforma. 2019;20:17–29. doi: 10.1186/s12859-018-2573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisfeldt J, Mårtensson G, Ameur, Nilsson D, Lindstrand A. Discovery of Novel Sequences in 1,000 Swedish Genomes. Mol. Biol. Evol. 2019;37:18–30. doi: 10.1093/molbev/msz176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, et al. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics. 2017;109:83–90. doi: 10.1016/j.ygeno.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Lin K, Smit S, Bonnema G, Sanchez-Perez G, de Ridder D. Making the difference: integrating structural variation detection tools. Brief. Bioinform. 2015;16:852–864. doi: 10.1093/bib/bbu047. [DOI] [PubMed] [Google Scholar]
- 25.Kosugi S, et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong, T., Hayes, V. M. & Chan, E. K. F. Detection of somatic structural variants from short-read next-generation sequencing data. Brief. Bioinform. bbaa056 (2020). [DOI] [PMC free article] [PubMed]
- 27.Pabinger S, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief. Bioinforma. 2014;15:256–278. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarate S, et al. Parliament2: Accurate structural variant calling at scale. GigaScience. 2020;9:giaa145. doi: 10.1093/gigascience/giaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohiyuddin M, et al. MetaSV: an accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics. 2015;31:2741. doi: 10.1093/bioinformatics/btv204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittler R, Marschall T, Schönhuth A, Mäkinen V. Repeat- and error-aware comparison of deletions. Bioinformatics. 2015;31:2947–2954. doi: 10.1093/bioinformatics/btv304. [DOI] [PubMed] [Google Scholar]
- 31.Köster J, Dijkstra LJ, Marschall T, Schönhuth A. Varlociraptor: enhancing sensitivity and controlling false discovery rate in somatic indel discovery. Genome Biol. 2020;21:1–25. doi: 10.1186/s13059-020-01993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou A, Lin T, Xing J. Evaluating nanopore sequencing data processing pipelines for structural variation identification. Genome Biol. 2019;20:1–13. doi: 10.1186/s13059-018-1612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvalho CMB, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 37.Cameron, D. L. et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. (2017). [DOI] [PMC free article] [PubMed]
- 38.Wala JA, et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018;28:581–591. doi: 10.1101/gr.221028.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron, D. L. et al. GRIDSS, PURPLE, LINX: unscrambling the tumor genome via integrated analysis of structural variation and copy number. Preprint at bioRxiv 10.1101/781013. (2019).
- 40.Narzisi G, et al. Genome-wide somatic variant calling using localized colored de Bruijn graphs. Commun. Biol. 2018;1:20. doi: 10.1038/s42003-018-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, et al. Patterns of structural variation in human cancer. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huddleston J, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677–685. doi: 10.1101/gr.214007.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cmero M, et al. Inferring structural variant cancer cell fraction. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-020-14351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffith M, et al. Optimizing cancer genome sequencing and analysis. Cell Syst. 2015;1:210. doi: 10.1016/j.cels.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Z, Fan X, Su Y, Huang YS. Accurity: accurate tumor purity and ploidy inference from tumor-normal WGS data by jointly modelling somatic copy number alterations and heterozygous germline single-nucleotide-variants. Bioinformatics. 2018;34:2004–2011. doi: 10.1093/bioinformatics/bty043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi K, Ju YS. Patterns and mechanisms of structural variations in human cancer. Exp. Mol. Med. 2018;50:98. doi: 10.1038/s12276-018-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinsella M, Patel A, Bafna V. The elusive evidence for chromothripsis. Nucleic Acids Res. 2014;42:8231–8242. doi: 10.1093/nar/gku525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodwin S, McPherson JD, Richard McCombie W. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross MG, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Freudenberg J. Mappability and read length. Front. Genet. 2014;5:381. doi: 10.3389/fgene.2014.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 52.Oloomi, S. M. H. The Impact of Multi-mappings in Short Read Mapping. Doctoral dissertation (2018).
- 53.Ebbert MTW, et al. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biol. 2019;20:97. doi: 10.1186/s13059-019-1707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Coster W, Van Broeckhoven C. Newest methods for detecting structural variations. Trends Biotechnol. 2019;37:973–982. doi: 10.1016/j.tibtech.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Sedlazeck FJ, Lee H, Darby CA, Schatz MC. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 2018;19:329–346. doi: 10.1038/s41576-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 56.Gong L, et al. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat. Methods. 2018;15:455–460. doi: 10.1038/s41592-018-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakamoto Y, et al. Long-read sequencing for non-small-cell lung cancer genomes. Genome Res. 2020;30:1243–1257. doi: 10.1101/gr.261941.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rang FJ, Kloosterman WP, de Ridder J. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:90. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Travers KJ, Chin C-S, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wenger AM, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu S, Wang A, Au KF. A comparative evaluation of hybrid error correction methods for error-prone long reads. Genome Biol. 2019;20:1–17. doi: 10.1186/s13059-018-1605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakamoto Y, Sereewattanawoot S, Suzuki A. A new era of long-read sequencing for cancer genomics. J. Hum. Genet. 2019;65:3–10. doi: 10.1038/s10038-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sedlazeck FJ, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinforma. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Coster W, et al. Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. Genome Res. 2019;29:1178–1187. doi: 10.1101/gr.244939.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiraishi, Y. et al. Precise characterization of somatic structural variations and mobile element insertions from paired long-read sequencing data with nanomonsv. Preprint at bioRxiv 10.1101/2020.07.22.214262. (2020).
- 69.Heller D, Vingron M. SVIM: structural variant identification using mapped long reads. Bioinformatics. 2019;35:2907–2915. doi: 10.1093/bioinformatics/btz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reisle C, et al. MAVIS: merging, annotation, validation, and illustration of structural variants. Bioinformatics. 2019;35:515–517. doi: 10.1093/bioinformatics/bty621. [DOI] [PubMed] [Google Scholar]
- 71.Haas BJ, et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20:1–16. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng Z, et al. Hypothesis: artifacts, including spurious chimeric RNAs with a short homologous sequence, caused by consecutive reverse transcriptions and endogenous random primers. J. Cancer. 2015;6:555–567. doi: 10.7150/jca.11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chwalenia K, Facemire L, Li H. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip. Rev. 2017;8:e1427. doi: 10.1002/wrna.1427. [DOI] [PubMed] [Google Scholar]
- 74.Gao Q, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23:227–238.e3. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rusch M, et al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-06485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan X, Chaisson M, Nakhleh L, Chen K. HySA: a Hybrid Structural variant Assembly approach using next-generation and single-molecule sequencing technologies. Genome Res. 2017;27:793–800. doi: 10.1101/gr.214767.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma ZS, Li L, Ye C, Peng M, Zhang Y-P. Hybrid assembly of ultra-long Nanopore reads augmented with 10x-Genomics contigs: Demonstrated with a human genome. Genomics. 2019;111:1896–1901. doi: 10.1016/j.ygeno.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 78.Zheng GXY, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marks P, et al. Resolving the full spectrum of human genome variation using Linked-Reads. Genome Res. 2019;29:635–645. doi: 10.1101/gr.234443.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mostovoy Y, et al. A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods. 2016;13:587–590. doi: 10.1038/nmeth.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou B, et al. Haplotype-resolved and integrated genome analysis of the cancer cell line HepG2. Nucleic Acids Res. 2019;47:3846. doi: 10.1093/nar/gkz169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bell JM, et al. Chromosome-scale mega-haplotypes enable digital karyotyping of cancer aneuploidy. Nucleic Acids Res. 2017;45:e162–e162. doi: 10.1093/nar/gkx712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Viswanathan SR, et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell. 2018;174:433–447.e19. doi: 10.1016/j.cell.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, et al. High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis -regulatory alterations. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13885-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neveling, K. et al. Next generation cytogenetics: comprehensive assessment of 48 leukemia genomes by genome imaging. Preprint at bioRxiv 10.1101/2020.02.06.935742. (2020).
- 86.Barrett T, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou Z, Wang W, Wang L-S, Zhang NR. Integrative DNA copy number detection and genotyping from sequencing and array-based platforms. Bioinformatics. 2018;34:2349–2355. doi: 10.1093/bioinformatics/bty104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:1–19. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nattestad M, et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 2018;28:1126–1135. doi: 10.1101/gr.231100.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rieke DT, et al. Comparison of treatment recommendations by molecular tumor boards worldwide. JCO Precis. Oncol. 2018;2:1–14. doi: 10.1200/PO.18.00098. [DOI] [PubMed] [Google Scholar]
- 91.Tamborero D, et al. Support systems to guide clinical decision-making in precision oncology: The Cancer Core Europe Molecular Tumor Board Portal. Nat. Med. 2020;26:992–994. doi: 10.1038/s41591-020-0969-2. [DOI] [PubMed] [Google Scholar]
- 92.Yu Y, et al. PreMedKB: an integrated precision medicine knowledgebase for interpreting relationships between diseases, genes, variants and drugs. Nucleic Acids Res. 2018;47:D1090–D1101. doi: 10.1093/nar/gky1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tham CY, et al. NanoVar: accurate characterization of patients’ genomic structural variants using low-depth nanopore sequencing. Genome Biol. 2020;21:1–15. doi: 10.1186/s13059-020-01968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roberts, H. E. et al. Short and long-read genome sequencing methodologies for somatic variant detection; genomic analysis of a patient with diffuse large B-cell lymphoma. Preprint at bioRxiv 10.1101/2020.03.24.999870. (2020). [DOI] [PMC free article] [PubMed]
- 95.Spies N, et al. Genome-wide reconstruction of complex structural variants using read clouds. Nat. Methods. 2017;14:915–920. doi: 10.1038/nmeth.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Genomics, 10x. Whole Genome Phasing and SV Calling. 10x Genomics Support https://support.10xgenomics.com/genome-exome/software/pipelines/latest/using/wgs. (2020)
- 97.Jiang T, et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020;21:1–24. doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stancu MC, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017;8:1–13. doi: 10.1038/s41467-016-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.English AC, Salerno WJ, Reid JG. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinforma. 2014;15:1–7. doi: 10.1186/1471-2105-15-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pacific Biosciences. pbsv. https://github.com/PacificBiosciences/pbsv. (2020)
- 101.Boivin V, et al. Reducing the structure bias of RNA-Seq reveals a large number of non-annotated non-coding RNA. Nucleic Acids Res. 2020;48:2271–2286. doi: 10.1093/nar/gkaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sati S, Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 2016;126:33–44. doi: 10.1007/s00412-016-0593-6. [DOI] [PubMed] [Google Scholar]
- 103.Jain M, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tyson JR, et al. MinION-based long-read sequencing and assembly extends the Caenorhabditis elegans reference genome. Genome Res. 2018;28:266–274. doi: 10.1101/gr.221184.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rhoads A, Au KF. PacBio sequencing and its applications. Genom. Proteom. Bioinforma. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laver T, et al. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detection Quant. 2015;3:1. doi: 10.1016/j.bdq.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jain M, et al. Improved data analysis for the MinION nanopore sequencer. Nat. Methods. 2015;12:351–356. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen P, et al. Modelling BioNano optical data and simulation study of genome map assembly. Bioinformatics. 2018;34:3966. doi: 10.1093/bioinformatics/bty456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Niu L, et al. Amplification-free library preparation with SAFE Hi-C uses ligation products for deep sequencing to improve traditional Hi-C analysis. Commun Biol. 2019;2:1–8. doi: 10.1038/s42003-019-0519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Díaz N, et al. Chromatin conformation analysis of primary patient tissue using a low input Hi-C method. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analyzed during this study.