Abstract
Heat shock proteins (HSPs) are a family of cellular proteins involved in a variety of biological functions including chaperone activity. HSPs are classified based on their molecular weight and each family has several isoforms in eukaryotes. HSP40 is the most diverse family acting as a co-chaperone for the highly conserved HSP70 family. Some of the isoforms are reported to be induced during heat stress. Few studies have also highlighted the diverse role of some isoforms in different stress conditions including viral infections. But till date, no study has comprehensively examined the expression profile of different HSP40 and 70 isoforms in either heat stress or HIV-1 infection, a virus that is responsible for the pandemic of AIDS. In the present study, we have compared the mRNA expression profile of HSP40 and HSP70 isoforms during heat stress and HIV-1 infection in a T-cell line and also validated the HIV-1 stress results in peripheral blood mononuclear cells. In case of HSP70, we observed that three isoforms (HSPA1A, HSPA1B, and HSPA6) are highly upregulated during heat stress, but these isoforms were found to be downregulated during the peak of HIV-1 infection. While in case of HSP40, we found that only DNAJA4, DNAJB1, and DNAJB4 showed significant upregulation during heat stress, whereas in HIV-1 infection, majority of the isoforms were induced significantly. Stress-dependent differential expression observed here indicates that different HSP40 and HSP70 isoforms may have specific roles during HIV-1 infection and thus could be important for future studies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12192-020-01185-y.
Keywords: HIV-1, HSP70, HSP40, HSPA, DNAJ, Isoforms, Real-time PCR
Introduction
Heat shock proteins (HSP) are present in cellular or extracellular environment of the cell and are known to be induced due to various stress conditions such as heat, cold, microbial infection, UV radiation, etc. (Ritossa 1962, 1996; Trautinger et al. 1996; Colinet et al. 2010). They are highly conserved proteins, many of which are referred to as molecular chaperones as they help in folding, transport, assembly, and degradation of other proteins (Sørensen et al. 2003; Arrigo 2005). They have been classified into different groups based on their molecular weights: small HSPs, HSP40, HSP60, HSP70, HSP90, and HSP110. In 2009, a new nomenclature was proposed for the heat shock protein family members: HSPA (HSP70), HSPH (HSP110), HSPC (HSP90), DNAJ (HSP40), HSPB (small HSP), and HSPD/E (HSP60/HSP10) (Kampinga et al. 2009). It is more consistent with the HUGO Gene Nomenclature Committee and NCBI Entrez Gene database. In the present report, we will use the new nomenclature while referring to different isoforms of HSP40 (DNAJ) and HSP70 (HSPA).
HSP70 is one of the most conserved proteins among the HSP family, showing its presence from bacteria to higher mammals (Lindquist and Craig 1988). This family comprises of thirteen different isoforms in humans as proposed by Kampinga and others in 2009 and listed in Table 1 (Kampinga et al. 2009). They have a similar structure comprising of a N-terminal nucleotide-binding domain or ATPase domain and a C-terminal substrate-binding domain composed of two subdomains (Mayer and Bukau 2005). Of the thirteen isoforms, HSPA1A, HSPA1B, and HSPA6 are reported to be highly induced upon heat stress, whereas HSPA8 is a constitutively expressed isoform (Kampinga et al. 2009). On the other hand, the HSP40 protein family is the most diverse family which specifies the function of other chaperone proteins like HSP70, HSP90, and HSP104 acting as a co-chaperone (Reidy et al. 2014), but the molecular function of different isoforms of the family, if any, remains to be elucidated. The human genome encodes about 49 isoforms of HSP40 protein as listed in Table 1 (Kampinga et al. 2009). Isoforms of HSP40 are very diverse at the primary sequence level. They have combinations of four typical domains: a highly conserved N-terminal sequence of about 70 amino acids (signature J domain); followed by a Gly/Phe-rich region (G/F-rich domain); four repeats of the CxxCxGxG-type zinc finger domain; and a less well-conserved C-terminal substrate-binding domain (Cheetham and Caplan 1998). Based on the differences in these regions, HSP40 isoforms can be categorized into three groups: type I (group A) proteins comprising of all four domains, while type II (group B) proteins lack the zinc finger domain, and type III (group C) proteins retain only the signature J domain, which can be located at any position in the protein sequence. Multiple HSP40 protein isoforms also exist in both prokaryotic and eukaryotic cells, e.g., 6 DNAJ/HSP40 homologues have been identified in Escherichia coli (Qiu et al. 2006), 22 in Saccharomyces cerevisiae (Walsh et al. 2004), and 43 in Plasmodium falciparum (Botha et al. 2007).
Table 1.
List of different HSP40 and HSP70 isoforms expressed in human cells (Kampinga et al. 2009)
| HSP40 Isoforms | ||
| Class A | DNAJB14 | DNAJC15 |
| DNAJA1 | Class C | DNAJC16 |
| DNAJA2 | DNAJC1 | DNAJC17 |
| DNAJA3 | DNAJC2 | DNAJC18 |
| DNAJA4 | DNAJC3 | DNAJC19 |
| Class B | DNAJC4 | DNAJC20 |
| DNAJB1 | DNAJC5 | DNAJC21 |
| DNAJB2 | DNAJC5B | DNAJC22 |
| DNAJB3 | DNAJC5G | DNAJC23 |
| DNAJB4 | DNAJC6 | DNAJC24 |
| DNAJB5 | DNAJC7 | DNAJC25 |
| DNAJB6 | DNAJC8 | DNAJC26 |
| DNAJB7 | DNAJC9 | DNAJC27 |
| DNAJB8 | DNAJC10 | DNAJC28 |
| DNAJB9 | DNAJC11 | DNAJC29 |
| DNAJB11 | DNAJC12 | DNAJC30 |
| DNAJB12 | DNAJC13 | |
| DNAJB13 | DNAJC14 | |
| HSP70 Isoforms | ||
| HSPA1A | HSPA6 | HSPA13 |
| HSPA1B | HSPA8 | HSPA14 |
| HSPA1L | HSPA9 | |
| HSPA2 | HSPA12a | |
| HSPA5 | HSPA12b | |
HSP70 and HSP40 work together to facilitate the folding of proteins. The function of HSP70 is dependent on its ATPase cycle. HSP70 is present in two states. One form is the ATP-bound state that has low affinity for client proteins and high association and dissociation rate. The other form is the ADP-bound state that has a high affinity for client proteins but low association and dissociation rate. The catalysis of ATP to ADP occurs through the low, intrinsic ATPase activity of HSP70 (Laufen et al. 1999; Lopez-Buesa et al. 1998). HSP40 physically interacts with HSP70 through the DNAJ domain and activates the ATPase activity of HSP70 (Minami et al. 1996). HSP40 provides the client specificity required by HSP70 to perform its function. The exchange of ADP with ATP is catalyzed by nucleotide exchange factors such as HSP110 and HSPBP1 (Bracher and Verghese 2015).
Human immunodeficiency virus (HIV-1) is the causative agent of acquired immune deficiency syndrome (AIDS) (Barré-Sinoussi et al. 1983; Gallo et al. 1984; Levy et al. 1984). With just a 9.7-kb genome and 15 different proteins, the virus hijacks the host machinery to complete its replication cycle and propagate. Several genome wide screens have shown that HIV-1 requires and utilizes a large number of cellular proteins during its life cycle including some heat shock proteins (Zhou et al. 2008; Zhu et al. 2014; Park et al. 2017). A study conducted two decades ago showed that HSP70 mRNA is upregulated in CD4+ T-cells infected with HIV-1 (Wainberg et al. 1997). A few studies since then have looked at the expression and role of heat shock proteins in HIV-1 infection although most of these reports have not looked at isoform-specific expression profile and functions. Our lab has previously shown that HSP40 and HSP70 reciprocally regulate HIV-1 gene expression (Kumar et al. 2011). We have also shown that HSP40 is required for the Nef-mediated increase in viral gene expression and replication (Kumar and Mitra 2005).
Recent studies have highlighted the role of few of the isoforms of HSP40 and HSP70 in other viral infections. For example, certain cytosolic HSP70 isoforms helped by distinct HSP40 isoforms are needed during the dengue virus life cycle (Taguwa et al. 2015). The isoform-specific functional implication of both HSP70 and HSP40 is also highlighted in Kaposi’s sarcoma-associated herpes virus infection, wherein there is an increased production or nuclear translocation of some of the isoforms (Baquero-Pérez and Whitehouse 2015). DNAJA1, an isoform of HSP40, acts as a positive regulator for influenza virus replication (Cao et al. 2014; Batra et al. 2016). Earlier, it was also shown that HSPA8, an isoform of HSP70, is associated with the influenza virus matrix protein 1, which is important for virus replication, assembly, and budding (Watanabe et al. 2006). The role of individual isoforms in HIV-1 infection is being studied as well in a fragmented manner and some reports indicate differences in function of isoforms based on HIV-1 subtypes or clades. HIV-1 clade B gp120 induces HSPA5 and other ER stress markers as compared to clade C gp120 in astrocytoma cells (López et al. 2017). A protective response is induced in clade B gp120-treated cells, while an apoptotic response is activated by clade C gp120. HIV-1 Tat is also known to induce HSPA5 and other ER stress markers (Norman et al. 2008). A report in 2013 showed that four HSP40 protein family isoforms DNAJB1, DNAJB6, DNAJA1, and DNAJC5 negatively regulate the HIV-1 replication by downregulating viral Rev protein expression (Urano et al. 2013).
However, till date, researchers are yet to comprehensively analyze the expression of all the different isoforms of HSP40 and HSP70 either during heat stress or during HIV-1 infection. It is unclear if response of these isoforms could be same or different in HIV-1 infection from that of heat stress. HIV-1 might specifically activate a specific pattern of heat shock protein response through these isoforms, which could be either beneficial or detrimental for virus replication. In addition, the HSP40-HSP70 interaction is poorly understood in case of HIV-1 infection. Thus, it becomes particularly important to look into this aspect as future drugs can target the interaction itself. Also, while drug targets for HSP70 isoforms may be very nonspecific due to the high similarity between the isoforms, the huge diversity in HSP40 isoforms can provide ideal drug targets (Wang et al. 2020). As a first step towards answering these questions, we have analyzed the expression of HSP40 and HSP70 isoforms at mRNA level upon heat stress and HIV-1 infection in CEM-GFP T-cell line. The heat stress was given at 42 °C with or without recovery time as previous studies have reported that cellular functions are extremely sensitive to temperature changes between 42 and 42.5 °C (Sapareto et al. 1978) and the actual heat shock protein synthesis occur after cells are returned to 37 °C (Slater et al. 1981). The expressions of the isoforms were also analyzed at mRNA level in CEM-GFP cells infected with HIV-1 and harvested at different times post-infection. Our results clearly show differential expression of various isoforms in these two different stress conditions indicating thereby stress-specific regulation of these HSP isoforms.
Materials and methods
Cell lines and primary cells
CEM-GFP (#3655) (Gervaix et al. 1997) and TZM-bl (#8129) (Platt et al. 1998) cell lines were obtained from NIH AIDS reagent program, Division of AIDS, NIAID, USA. These are reporter cell lines expressing GFP and β-galactosidase/luciferase, respectively, as reporters upon transactivation by HIV-1 infection. Buffy coat of seronegative healthy blood donors was procured from ISI blood bank, Navi Peth, Pune, and PBMCs were isolated using Histopaque 1077 (Sigma, USA) by gradient centrifugation (Panda and Ravindran 2013). TZM-bl cells were grown in DMEM (Invitrogen, USA), whereas PBMCs and CEM-GFP cells were grown in RPMI 1640 medium (Invitrogen, USA) containing 10% fetal bovine serum (FBS) and penicillin-streptomycin (Invitrogen, USA) at 37 °C in a humidified 5% CO2 incubator. For CEM-GFP cells, media were supplemented with 500 μg/ml G418 (Invitrogen, USA).
Heat stress of cells
CEM-GFP cells were given heat stress at 42 °C for 30 minutes followed by either immediate harvesting of cells or harvesting after a recovery period of 2 hours at 37 °C post-heat stress (Slater et al. 1981). RNA was isolated from harvested cells using TRIzol reagent (Invitrogen, USA) following manufacturer’s instructions.
Virus stock preparation
HIV-1 pNL4-3 molecular clone (#114), obtained from the National Institutes of Health AIDS repository, was used for preparing virus stock as described earlier (Adachi et al. 1986). Briefly, HEK-293 T-cells were transfected with the molecular clone using the CalPhos Mammalian Transfection Kit (Clontech, Takara, USA) as per the manufacturer’s instructions. Cell culture media was collected at 24 hours post-media change, clarified at 1800 g for 5 min, and concentrated by ultracentrifugation at 141000 × g for 2.5 hours at 4 °C. The viral pellet was resuspended in RPMI 1640 containing a final concentration of 50-mM HEPES in small aliquots and was stored at −70 °C. Antigen capture ELISA for p24 antigen (Advanced Biosciences Laboratories, USA) was used to calculate the concentration of virus in the stock as per the instruction of manufacturer.
Viral infectivity assay
For calculation of infectious virus particles generated, different concentrations of virus stock based on p24 antigen (analyzed by ELISA) were used to infect TZM-bl reporter cells at a confluence of 50–60%. Infectivity was calculated at 36 hours post-infection by β-gal staining after fixing the cells using 0.25% glutaraldehyde. The cells that stained positive were counted in random fields and the infectious virion per nanogram virus (p24) was calculated (Augustine et al. 2017).
HIV-1 infection
CEM-GFP cells were infected with 0.1 MOI of HIV-1 NL4-3 virus for 4 hours at 37 °C in the presence of polybrene (1 μg/ml) with intermittent mixing. 0.1 MOI indicates that the number of virus particles used in infection was 1/10th of the total number of cells used in the experiment. Cells were then washed and resuspended in complete medium and were kept in humidified CO2 incubator post-infection. Cells were harvested on different days post-infection for RNA extraction with TRIzol (Invitrogen, USA) reagent using manufacturer’s protocol. cDNA synthesis from RNA was carried out using MMLV Reverse Transcriptase (Invitrogen, USA) as per manufacturer’s protocol. PBMCs were isolated from Buffy coat using Histopaque by gradient centrifugation as mentioned earlier. Cells were grown in RPMI medium (Invitrogen, USA) supplemented with 10% fetal bovine serum (Invitrogen, USA) and incubated at 37 °C in a humidified CO2 incubator. The PBMCs were activated with phytohemagglutinin (5 μg/ml) for 36 hours. After 36 hours, cells were then infected with 0.1 MOI of HIV-1 NL4.3 virus for 4 hours at 37 °C in the presence of polybrene (1 μg/ml) with intermittent mixing. The cells were then washed, resuspended in complete medium supplemented with recombinant human IL-2 (Roche Applied Science, Germany) at 20 units/ml, and incubated until harvested (Augustine et al. 2017). Cells were harvested at different days post-infection for RNA isolation and cDNA synthesis as described above. Culture supernatants from infected cells were used to determine virus production and peak of infection by p24 antigen capture ELISA (Advanced Bioscience Laboratories, USA).
Quantitative real-time PCR
Expression of various HSP40 and 70 isoforms was analyzed by quantitative real-time RT-PCR in a reaction mixture containing SYBR Green iTaq supermix (Bio-Rad, USA) and 10-pmol concentrations of hGAPDH or isoform-specific oligonucleotide primer pairs and run on a Realplex4 Mastercycler (Eppendorf, Germany) using the following program: initial denaturation at 95 °C for 2 min and 40 cycles of 95 °C for 30s, 55 °C for 30s, and 72 °C for 30s followed by the melt curve analysis. All the error bars represent mean ± standard error of mean of at least three independent experiments performed in duplicate wells of the plate for each sample. The sequence of all the HSP40 and HSP70 isoform-specific PCR primers used in this study is listed in Supplementary Tables S1 and S2.
The fold change values were calculated as:
Fold difference = 2−∆∆CT
where ΔCT = CT (target) − CT (GAPDH) and ΔΔCT = ΔCT (treated) – ΔCT (control)
Generation of heat maps
Heat maps for the gene expression results were built using Latex software. Each grid in the heat map shows the log of the fold change value for the mRNA expression of that gene at that time point averaged over three replicates. A value of zero is onefold regulation, i.e., no regulation, and is represented in black. Gradations of green represent upregulation and gradations of red represent downregulation.
Statistical analysis
All the error bars represent mean ± standard error of mean of at least three independent experiments. The statistical significance of difference between two groups was evaluated by Student’s t test using SigmaPlot 12.5. The significance is represented as ns = p≧0.05, * = p≦0.05, ** = p≦0.001, and *** = p≦0.0001.
Results
Heat stress modulates expression of several HSP40 isoforms in T-cells
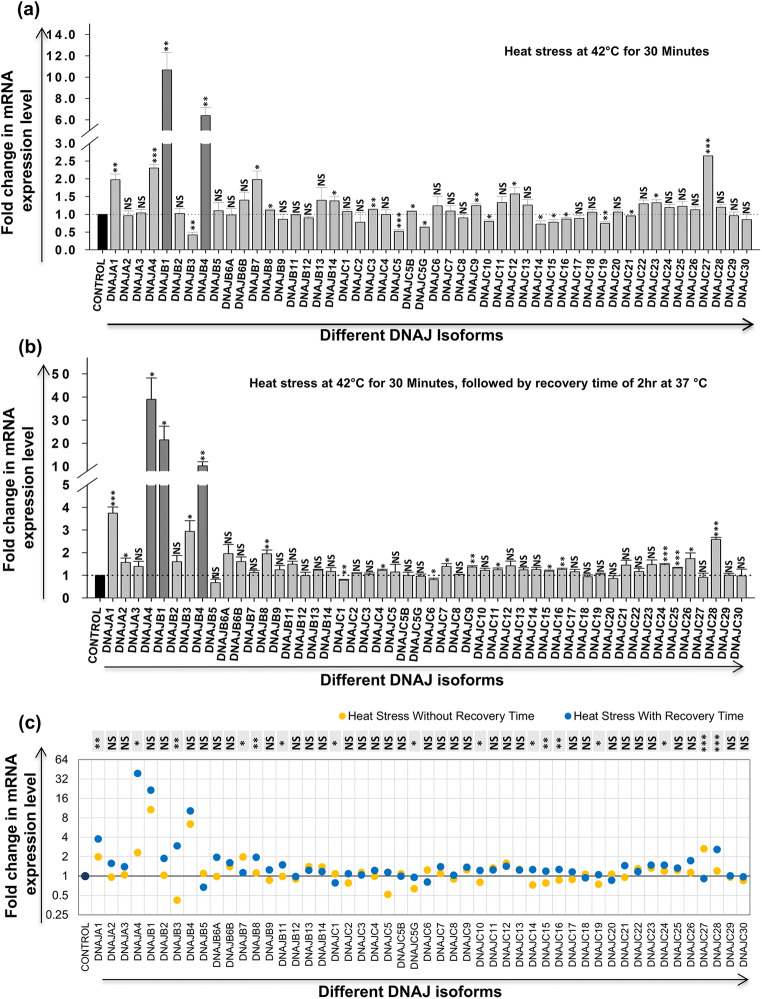
HSPs are proteins that are thought to be induced during heat stress or any other kind of stress; however, gene expression profile of all HSP40 isoforms during heat stress has not been studied till date. So it is important to know whether all the DNAJ isoforms are heat stress inducible or only specific isoform/s show modulation during heat stress. CEM-GFP is a reporter T-cell line that is used in HIV-1 infection studies as a model cell line and was thus used in the present work for comparative analysis. First, heat stress at 42 °C for 30 minutes was given to CEM-GFP cells followed by harvesting of cells for RNA extraction either with or without a recovery period of 2 h at 37 °C. Heat stress without recovery period caused significant upregulation (≥ 1.5-fold) in the expression of 7 isoforms (DNAJA1, DNAJA4, DNAJB1, DNAJB4, DNAJB7, DNAJC12, and DNAJC27) (Fig. 1a). However, very significant increase (≥ fivefold) was seen in DNAJB1 (~10.68-fold) and DNAJB4 (~6.39-fold). Interestingly, significant decrease (≤ 1.5-fold) was observed only in 3 isoforms: DNAJB3, DNAJC5, and DNAJC5G. Heat stress with recovery period on the other hand resulted in significant upregulation (≥ 1.5-fold) of 9 isoforms (DNAJA1, DNAJA2, DNAJA4, DNAJB1, DNAJB3, DNAJB4, DNAJB8, DNAJC26, and DNAJC28) (Fig. 1b). Among these, 3 isoforms were very highly upregulated (≥ fivefold): DNAJA4 (~38.99-fold), DNAJB1 (~21.47-fold), and DNAJB4 (~10.23-fold). On the other hand, none of the isoforms showed significant downregulation in heat stress with recovery period. Results obtained from these experiments show that all the isoforms of DNAJ are not heat inducible. Upon comparison of these two datasets, we find that some of the isoforms which show significant modulation during heat stress without recovery time does not show any significant change in expression in cells kept for recovery after heat stress (DNAJB7, DNAJC5, DNAJC5G, DNAJC12, and DNAJC27). Certain isoforms that did not show any change in expression during heat stress without recovery time showed a significant upregulation during heat stress with recovery time (DNAJA2, DNAJB8, DNAJC26, and DNAJC28) (Fig. 1c). These results suggest that some of the isoforms show immediate response to heat stress while others get activated later on to probably maintain homeostasis in the cell. DNAJB1 and DNAJB4 showed very significant upregulation in expression in both the conditions and therefore can be probably used as inducible markers of heat stress.
Fig. 1.
Effect of heat stress on expression of different HSP40 isoforms in CEM-GFP cells. CEM-GFP cells were given heat shock at 42 °C for 30 minutes, and the cells were harvested immediately without any recovery time or after a recovery time of 2 hours at 37 °C. Modulation in mRNA expression of different HSP40 isoforms was determined by using qRT-PCR (a) without recovery time and (b) with recovery time of 2 hours at 37 °C. The results represent mean ± S.E. from n = 3 independent experiments, and statistical significance is determined by using Student’s t test, as *p < 0.05, **p ≤ 0.01, and ***p ≤ 0.001. (c) Comparative analysis of variation in expression of different HSP70 isoforms between the above conditions (a and b). Statistical significance is determined by using the two-tailed Student’s t test, as *p < 0.05, **p ≤ 0.01, and ***p ≤ 0.001
Several isoforms of HSP70 family are induced during heat stress in T-cells
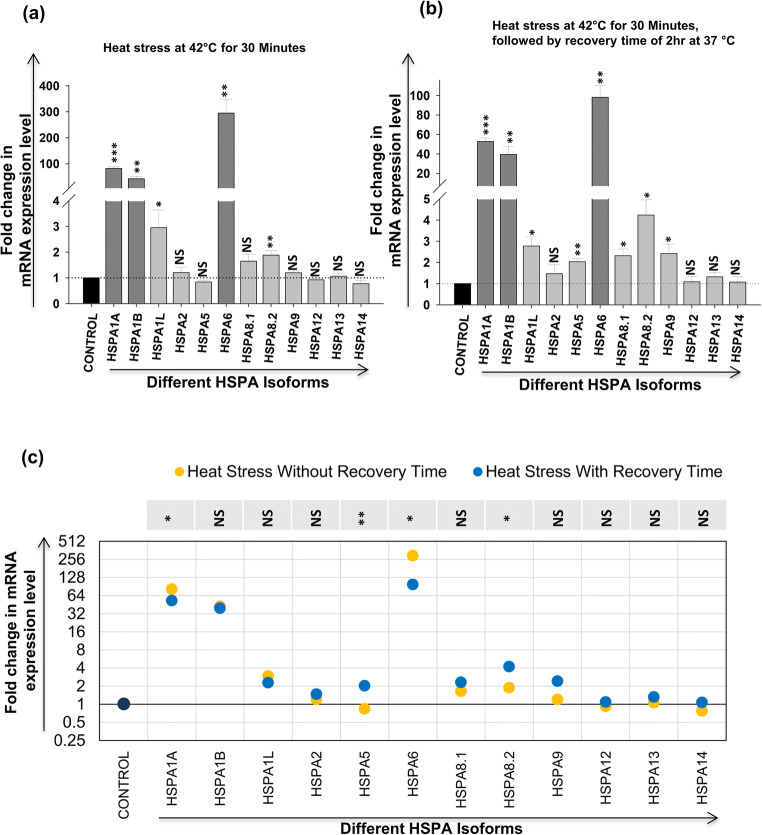
In case of HSP70, upon heat stress of CEM-GFP cells without recovery time, 5 isoforms (HSPA1A, HSPA1B, HSPA1L, HSPA6, and HSP8.2) showed upregulation (≥ 1.5-fold) in expression. There was very significant upregulation in expression of HSPA1A (~82-fold), HSPA1B (~40-fold), and HSPA6 (~295-fold) (Fig. 2a). On the other hand, when cells were given heat stress followed by a recovery period of 2 h, 8 isoforms (HSPA1A, HSPA1B, HSPA1L, HSPA5, HSPA6, HSPA8.1, HSP8.2, and HSPA9) were upregulated (≥ 1.5-fold) but again the same three isoforms HSPA1A (~53-fold), HSPA1B (~40-fold), and HSPA6 (~98-fold) were very significantly induced (Fig. 2b). In both the conditions, these three isoforms HSPA1A, HSPA1B, and HSPA6 were consistently highly upregulated (≥ fivefold). However, it needs to be mentioned that there was no decrease in the expression of any isoform in both the conditions. Moreover, pattern of expression in both conditions remained almost similar except HSPA5, HSPA8.1, and HSPA9, which showed upregulation in the heat stress with recovery time (Fig. 2c). The upregulations in HSPA1A, HSPA1B, and HSPA6 have been well documented in literature and they are thus called heat inducible isoforms of the HSP70 family. The above results show that the same three isoforms are highly heat stress inducible even in T-cells, an important host for HIV-1.
Fig. 2.
Effect of heat stress on expression of different HSP70 isoforms in CEM-GFP cells. CEM-GFP cells were given heat shock at 42 °C for 30 minutes and the cells were harvested immediately without any recovery time or after a recovery time of 2 hours at 37 °C. Modulation in mRNA expression of different HSP70 isoforms was determined by using qRT-PCR (a) without recovery time and (b) with recovery time of 2 hours at 37 °C. The results represent mean ± S.E. from n = 3 independent experiments and statistical significance is determined by using Student’s t test, as *p < 0.05, **p ≤ 0.01, and ***p ≤ 0.001. (c) Comparative analysis of variation in expression of different HSP70 isoforms between the above conditions (a and b). Statistical significance is determined by using the two-tailed Student’s t test, as *p < 0.05, **p ≤ 0.01, and ***p ≤ 0.001
HSP40 isoforms are differentially modulated during HIV-1 infection in T-cells
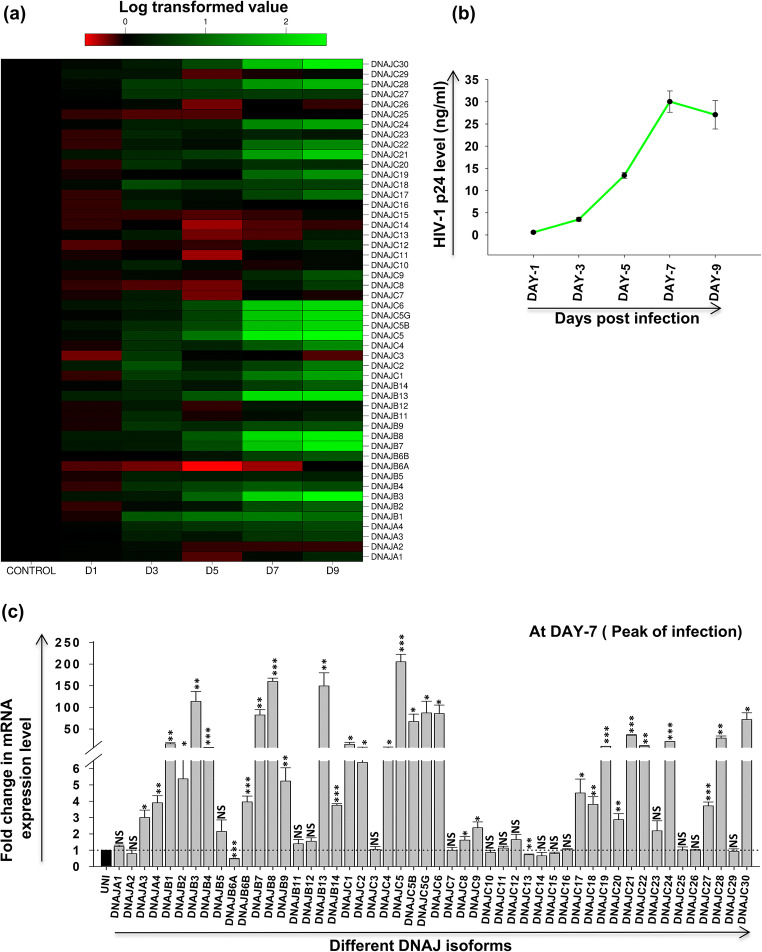
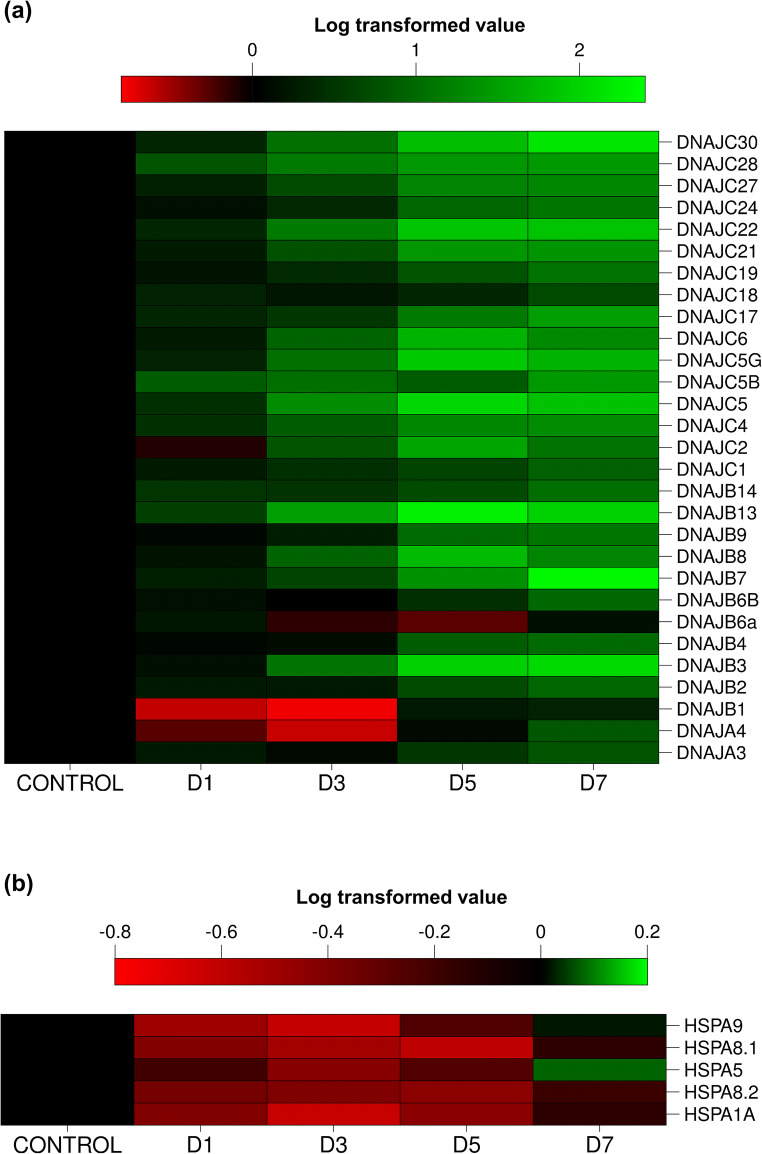
CEM-GFP cells infected with HIV-1 at 0.1 MOI were harvested at different days post-infection. RNA was isolated from these cells followed by cDNA preparation and qRT-PCR for different isoforms of DNAJ family, where mock-infected cells were taken as control. A heat map for the mRNA expression level of various isoforms at different days post-infection is presented in Fig. 3a. Initially, there is no significant change in expression but over the course of time post-infection, significant changes are observed (either up- or downregulation) in expression of several isoforms with infection progression, as observed in the heat map. Further, to find the day of the peak of HIV-1 infection in the specific experimental condition, we performed p24 antigen capture ELISA from the supernatant collected at different days post-infection. We find the peak of infection to be at day 7 post-infection based on p24 antigen present in the supernatant (Fig. 3b). Thus, further comparative analysis was performed at day 7 post-infection as shown in Fig. 3c. Our results clearly indicate that almost 28 isoforms of DNAJ family showed more than threefold upregulations, while only one isoform DNAJB6a showed significant downregulation during the peak of infection. Moreover, some isoforms exhibited very significant increase in expression like DNAJB3 (~114-fold), DNAJB7 (~82-fold), DNAJB8 (~160-fold), DNAJB13 (~150-fold), DNAJC5 (~205-fold), DNAJC5B (~67-fold), DNAJC5G (~87-fold), DNAJC6 (~86-fold), and DNAJC30 (~72-fold). Such differential expression results from these experiments indicate that some of these DNAJ family isoforms might play an important role in the life cycle of HIV-1 in T-cells.
Fig. 3.
Expression profile of different HSP40 isoforms during HIV-1 infection. CEM-GFP cells were infected with 0.1 MOI of HIV-1NL4.3 virus and harvested at different days post-infection (days 1, 3, 5, 7, and 9) and followed by qRT-PCR to determine mRNA expression level of different HSP40 isoforms. (a) Expression kinetics of different HSP40 isoforms at different days post-infection presented as heat map using logarithmic-transformed fold change mean values (n = 3). The abscissa represents the number of days post-infection in the experiment and the ordinate represents different isoforms of DNAJ. (b) Determination of peak day of infection by p24 antigen capture ELISA of supernatants collected from HIV-1 NL4.3 virus-infected CEM-GFP cells as described in materials and methods. (c) Fold change in mRNA expression level of different HSP40 isoforms at peak of HIV-1 infection (day 7 post-infection). The results represent mean ± S.E. from n = 3 independent experiments and statistical significance is determined by using Student’s t test, *p < 0.05, **p ≤ 0.01, and ***p ≤ 0.001
HSP70 isoforms are significantly modulated during HIV-1 infection in T-cells
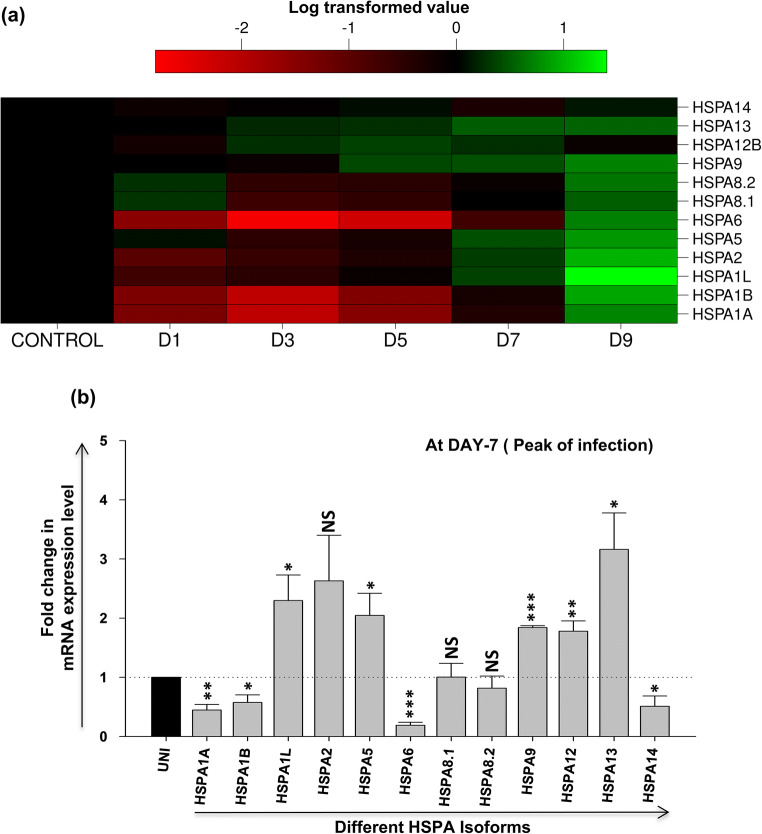
A heat map for the expression of various isoforms of HSP70 family at RNA level for different days post-HIV-1 infection is shown in Fig. 4a, indicating changes in the expression during the course of HIV-1 infection. Overall, we observed that all isoforms are initially downregulated but expression of some of them is induced later in the infection as shown in the heat map. Notably, the heat inducible isoforms such as HSPA1A, HSPA1B, and HSPA6 are highly and significantly downregulated between days 1 and 5. From day 7 onwards, upregulation is observed in several isoforms, most significantly in HSPA1L, HSPA2, HSPA5, HSPA9, HSPA12, and HSPA13. As the peak of infection was at day 7 based on our results in Fig. 3b, the expression of all the HSP70 isoforms was then compared at day 7 as presented in Fig. 4b. Here, we observe that four isoforms were modestly downregulated: HSPA1A (~0.45-fold), HSPA1B (~0.6-fold), HSPA6 (~0.20-fold), and HSPA14 (~0.5-fold) at the peak of infection. When compared to HSP40 isoforms, none of the HSP70 isoforms showed a very significant (≥ threefold) increase in expression at the peak of infection except HSPA13 (~threefold). The results suggest that some of the HSP70 isoforms which are significantly downregulated may potentially be involved in HIV-1 life cycle in T-cells.
Fig. 4.
Expression profile of different HSP70 isoforms during HIV-1 infection. CEM-GFP cells were infected with 0.1 MOI of HIV-1NL4.3 virus and harvested at different days post-infection (days 1, 3, 5, 7, and 9) and was followed by qRT-PCR to determine mRNA expression level of different HSP70 isoforms. (a) expression kinetics of different HSP70 isoforms at different days post-infection presented as heat map using logarithmic-transformed fold change mean values (n = 3). The abscissa represents the number of days post-infection in the experiment and the ordinate represents different isoforms of HSP70. (b) Fold change in mRNA expression level of different HSP70 isoforms at peak of HIV-1 infection (day 7 post-infection). The results represent mean ± S.E. from n = 3 independent experiments and statistical significance is determined by using Student’s t test, *p < 0.05, **p ≤ 0.01, and ***p ≤ 0.001
Validation of expression profile of selected HSP40 and HSP70 isoforms in HIV-1-infected human peripheral blood mononuclear cells (hPBMCs)
Further to validate our results obtained for various isoforms during HIV-1 infection in CEM-GFP T-cell line also in primary cells, peripheral blood mononuclear cells were obtained from a local blood bank. PBMCs were infected with HIV-1 at 0.1 MOI and cells were harvested for RNA isolation followed by qRT-PCR at different days post-infection. Mock-infected cells were used as control. Further, in case of DNAJ family, we selected 29 isoforms that were showing significant modulation (≥ threefold upregulations or ≤ twofold downregulation) in expression in HIV-1-infected CEM-GFP cells. Our results show that all the selected isoforms showed same kind of expression profile in infected hPBMCs as observed earlier in CEM-GFP cells (Fig. 5a). In case of HSP70, we selected the common cytoplasmic isoforms for expression profiling in hPBMCs—HSPA1A, HSPA5, HSPA8.1, HSPA8.2, and HSPA9 (Fig. 5b). We found that while there was not a significant downregulation, a similar trend was observed in three of the isoforms, HSPA1A, HSPA8.1, and HSPA8.2. HSPA5 showed modest increase during late infection in PBMCs as was observed in CEM-GFP cells in peak of HIV-1 infection.
Fig. 5.
Differential expression of selected HSP40 and 70 isoforms in HIV-1-infected human PBMCs. Human PBMCs activated with PHA were infected with HIV-1NL4.3 virus at 0.1 MOI and cells were harvested at different days post-infection (day 1, 3, 5, and 7). Further, RNA was isolated and qRT-PCR was performed for selected HSP40 and HSP70 isoforms as described in the text. Differential expression of selected HSP40 (a) and HSP70 (b) isoforms presented as heat map using logarithmic-transformed fold change mean values (n = 3), respectively. The abscissa represents the number of days post-infection in the experiment and the ordinate represents different isoforms of HSP40 and HSP70, respectively
Comparative analysis of expression profile of HSP40 and HSP70 isoforms during heat stress and HIV-1 infection in T-cells
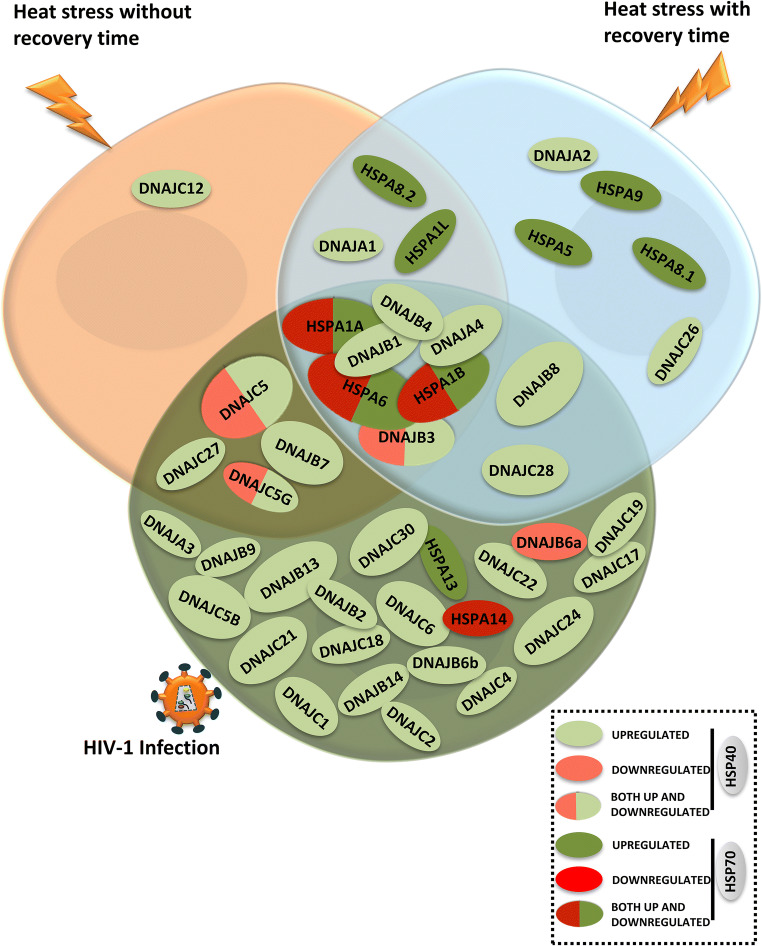
Our results clearly suggest that all the isoforms of HSP40 or HSP70 family are not heat inducible. Moreover, these isoforms behave differently during heat stress and HIV-1 infection. Comparative analysis of the expression profile indicates that there are some isoforms in these two families that are specifically modulated either due to heat stress (DNAJA1, DNAJA2, DNAJC12, DNAJC26, HSPA1L, HSPA8.1, HSPA8.2, HSPA5, and HSPA9) or during HIV-1 infection (DNAJA3, DNAJB2, DNAJB6a, DNAJB6b, DNAJB9, DNAJB13, DNJAB14, DNJAC1, DNAJC2, DNAJC4, DNAJC5B, DNAJC6, DNAJC17, DNAJC18, DNAJC19, DNAJC21, DNAJC22, DNAJC24, DNAJC30, HSPA13, and HSPA14). Also few isoforms showed modulation in both heat stress and HIV-1 infection (DNAJA4, DNAJB1, DNAJB3, DNAJB4, HSPA1A, HSPA1B, and HSPA6) as shown in Table 2. We have also tried to graphically represent this differential modulation of HSP40/HSP70 isoforms in T-cells during heat stress or HIV-1 infection conditions as a Venn diagram (Fig. 6). In case of HSP40 family, during heat stress conditions, only few isoforms showed modulation but during HIV-1 infection, most of the isoforms were induced (oval shape with light green denotes upregulation, light red denotes downregulation, while ovals with both color denote opposite regulation in both the stress conditions). In case of HSP70, 3 isoforms showed huge upregulation during heat stress but there was significant downregulation during HIV-1 infection (oval shape with dark green denotes upregulation, dark red denotes downregulation, while ovals with both color denote opposite regulation in the two stress conditions). Differential expression of various isoforms of HSP40 and HSP70 family observed during HIV-1 infection in T-cells or heat stress suggests that the presence of multiple isoforms may not be merely redundant but could have potentially important roles in different stress conditions.
Table 2.
List of significantly differentially regulated isoforms of HSP40 and HSP70 during heat stress and HIV-1 infection in CEM-GFP T-cells
| Stress-induced isoforms | |||||
|---|---|---|---|---|---|
| Heat stress specific | Common in both | HIV-1 infection specific | |||
| DNAJA1 | HSPA5 | DNAJA4 | DNAJA3 | DNAJC1 | DNAJC19 |
| DNAJA2 | HSPA9 | DNAJB1 | DNAJB2 | DNAJC2 | DNAJC21 |
| DNAJC12 | DNAJB3 | DNAJB6a | DNAJC4 | DNAJC22 | |
| DNAJC26 | DNAJB4 | DNAJB6b | DNAJC5B | DNAJC24 | |
| HSPA1L | HSPA1A | DNAJB9 | DNAJC6 | DNAJC30 | |
| HSPA8.1 | HSPA1B | DNAJB13 | DNAJC17 | HSPA13 | |
| HSPA8.2 | HSPA6 | DNAJB14 | DNAJC18 | HSPA14 | |
Fig. 6.
Graphical representation for differential modulation of HSP40 and HSP70 isoforms during heat stress (with and without recovery period) and HIV-1 infection in CEM-GFP cells at peak of infection (day 7) highlighting modulated common and unique isoforms. In case of HSP40 isoforms, oval shape with light green denotes upregulation, light red denotes downregulation, while oval with both color denotes opposite modulation in both the stress conditions. In case of HSP70 isoforms, oval shape with dark green denotes upregulation, dark red denotes downregulation, while oval with both color denotes opposite modulation in the two stress conditions)
Discussion
HSP70 is one of the most conserved heat shock protein family, whereas HSP40 is the most diverse. Together, they form an association that maintains homeostasis inside the cells by facilitating folding, transport, assembly, and degradation of proteins. Human HSP70 family comprises of 13 different isoforms, whereas human HSP40 family has 49 different isoforms (Kampinga et al. 2009). The presence of so many isoforms raises the question as to whether all these isoforms have specific roles or are they redundant. While research on the role of both HSP40 and HSP70 isoforms in viral infection is nascent, some recent studies do highlight the same. For example, in dengue virus infection, among different HSP70 and HSP40 isoforms, HSPA8 and DNAJB6b seem to play a vital role in virion production (Taguwa et al. 2015). The same group showed that HSPA1A and HSPA8 are also required for Zika virus production (Taguwa et al. 2019). Six isoforms of HSP70 and HSP40, HSPA1A, HSPA8, HSPA5, DNAJA1, DNAJB1, and DNAJB6, were found to be associated with the replication and transcription complexes of Kaposi’s sarcoma-associated Herpes virus (Baquero-Pérez and Whitehouse 2015). In another interesting report, it was shown that DNAJB1-HSP70 complex regulates innate antiviral response generated by Melanoma differentiation-associated gene-5 and mitochondrial antiviral signaling protein (MDA5-MAVS) pathway, which is known to play an important role in type I interferon induction in dsRNA virus-infected cells (Takashima et al. 2018). Another HSP40 isoform, DNAJB6, negatively regulates the replication of Japanese encephalitis virus (JEV) replication by interacting with NS3 viral protein (Cao et al. 2019), whereas DNAJA3 was reported to play an imperative antiviral role against foot-and-mouth disease virus (FMDV) by promoting VP1 degradation and restoring of IFN-β signaling pathway (Zhang et al. 2019). The above studies focus on the role of HSP40 or HSP70 isoforms on viral infection. Even in case of HIV-1 infection, one study in the past has indicated that few isoforms of HSP40 restrict HIV-1 infection (Urano et al. 2013). From our mRNA expression results in HIV-1 infection, we find that although all the HSP40 isoforms studied by Urano et al. (DNAJA1, DNAJB1, DNAJB6, DNAJC5, and DNAJC3) were modulated throughout the time course, only DNAJB1, DNAJB6 (B6a and B6b), and DNAJC5 are significantly induced at the peak of infection (day 7). On the other hand, DNAJA1 and DNAJC3 did not show significant change in the level of mRNA expression on day 7 post-infection. Interestingly, that study also reported that DNAJC3 was unable to inhibit viral production. Such a comparison strengthens the idea that isoforms which are found to be very significantly modulated during infection can be taken up for further studies to understand their role in the virus life cycle. Furthermore as mentioned earlier, our lab has previously shown that HSP70 tends to inhibit HIV-1 replication whereas HSP40 enhances virus replication, thus reciprocally regulating HIV-1 infection (Kumar et al. 2011). However, till date, a comprehensive study on the role of different stress on all isoforms of HSP70 and HSP40 has not been published.
Therefore in this study, we have focused on the regulation of mRNA expression of different isoforms of HSP40 and HSP70 during two stress conditions wherein we looked into changes due to heat stress and HIV-1 infection. Three HSP70 isoforms, HSPA1A, HSPA1B, and HSPA6, were previously reported to be highly induced during heat stress (Kampinga et al. 2009). The same results were obtained in our analysis when T-cells were subjected to heat stress both with and without a recovery period. The literature in case of modulation of HSP40 isoforms during heat stress is less studied. In the present investigation, our results clearly show that DNAJA4, DNAJB1, and DNAJB4 were highly upregulated at mRNA level during heat stress. Interestingly, during heat stress without recovery time, three HSP40 isoforms were downregulated, DNAJB3, DNAJC5, and DNAJC5G while there was no downregulation in any of the HSP70 isoforms. The upregulation of HSPs during heat stress is to confer the cell thermotolerant, protecting it from harmful effects of the heat (Parsell et al. 1993). Thus the downregulation of three HSP40 isoforms during heat stress needs to be further studied to understand their role.
Thereafter, we looked into the regulation of HSP70 and HSP40 isoforms during HIV-1 infection. We find that HSP40 isoforms are differentially induced whereas many HSP70 isoforms were significantly downregulated during the first 5 days post-infection. At the peak of infection, when viral load is highest, our results show that 28 isoforms of the HSP40 family were significantly upregulated out of which some isoforms were very highly upregulated. Only one isoform DNAJB6a was downregulated. On the other hand, 3 isoforms of HSP70 were downregulated at the peak of infection, in contrast to their expression during heat stress. These results were further validated in HIV-1-infected human peripheral blood mononuclear cells (PBMCs) showing broadly similar pattern of modulation. Collectively, various members of HSP40-HSP70 family are differentially regulated at mRNA level during heat stress or HIV-1 infection in T-cells. Our results also indicate that certain isoforms are modulated both in HIV-1 infection and heat stress (both with and without recovery) yet some other isoforms are modulated only in one or the other stress condition. Thus, we believe that those isoforms that are modulated in both kinds of stresses can be considered as general stress markers. Conversely, isoforms modulated in only one or the other kind of stress can be stress-specific markers.
Isoforms that are differentially expressed during heat stress and HIV-1 infection could be involved in maintaining cellular homeostasis during adverse conditions either in early or late phase. Although there is very little literature on gene expression profile of HSP40 and 70 isoforms during any kind of stress but based on the changes in mRNA expression level observed during the two different stress conditions in our study, we are tempted to hypothesize that those isoforms, which are modulated in heat stress without recovery time and early stage of HIV-1 infection could be involved in acute response to stress, while those isoforms that are modulated during heat stress with recovery time and late stage of HIV-1 infection could be functional at a later stage of the stress. For example, in our study, we find that DNAJC27 is upregulated during heat stress without recovery time and early stage of HIV-1 infection indicating its possible role in acute response during stress condition. DNAJB3, DNAJB8, and DNAJC28 are upregulated in heat stress with recovery time and in late stage of HIV-1 infection which suggests possibly their role in chronic stress. Furthermore, reports show that DNAJB8 has anti-aggregation activity and helps in the clearance of polyQ protein aggregates in chronic neurodegenerative diseases protecting cells from cell death (Hageman et al. 2010; Gillis et al. 2013). In our study, DNAJB8 was found to be upregulated during heat stress with recovery time and in late stage of HIV-1 infection which suggests its possible role in late stage of HIV-1 infection. In another report, DNAJA4 expression increases during hyperthermia, which reduces the expression of F-actin, leading to destabilizing the actin cytoskeleton, and shows antiviral activity for human papilloma virus infection (Liu et al. 2020). HIV-1 utilizes the cytoskeleton for entry, internal transport, and budding out from host cells (Stolp and Fackler 2011). In our study, we found an increased mRNA level of DNAJA4 in both conditions of heat stress and HIV-1 infection indicating its possible role in stress. An isoform of HSP70, HSPA5, is upregulated only in heat stress with recovery time, possibly as a late response. Even in case of HIV-1 infection, our result shows its upregulation in late infection. As a part of the unfolded protein response in cells, HSPA5 initially tries to reduce the unfolded protein load, thereby maintaining cell viability (Hetz 2012). With chronic stress, it directs the cells towards apoptosis. Chronic stress is seen in late HIV-1 infection. The markers of the unfolded protein response are also reported to be upregulated with time due to HIV-1 Tat protein (Norman et al. 2008). Other HSP70 isoforms such as HSPA1A, HSPA1B, and HSPA6 are modulated in both stress conditions and throughout HIV-1 infection, possibly because they play a role in maintaining cellular homeostasis throughout the stress.
Such comparison of expression profile between heat stress and HIV-1 infection and their further characterization might help us in elucidation of their specific roles in different stresses. While our work shows changes in expression of different HSP40 and HSP70 isoforms at mRNA level during two different stress conditions, a further study of their protein level expression will provide a better understanding about their impact in regulation of heat stress and HIV-1 virus life cycle. It will also be conducive to investigate the HSPA-DNAJ isoform pairs that are involved in different stress conditions such as heat and viral infection stress.
Supplementary Information
(PDF 179 kb)
Acknowledgments
This work was supported by a COE grant of Department of Biotechnology, Govt. of India, and JC Bose fellowship of SERB to DM. KC and KI is grateful to University Grants Commission, India, for research fellowship. Authors also acknowledge the intramural support of National Centre for Cell Science.
Author’s contributions
Debashis Mitra contributed to the study concept, whereas all the authors contributed in study design. Experimental work and data collection were performed by Kailash Chand and Kruthika Iyer. Data analysis was done by all the authors. The initial draft of the manuscript was written by Kailash Chand and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the Department of Biotechnology, Government of India, COE project no. BT/PR/15450/COE/34/46/2016 and the JC Bose Fellowship of SERB to DM. The work was also supported by intramural funding from National Centre for Cell Science, Pune, India.
Data Availability
The data that supports the findings of this study are available from the corresponding author upon reasonable request.
Compliance with ethical standards
Conflicts of interests/competing interests
The authors declare that they have no competing interests.
Consent for publication
Consent is taken from all the coauthors.
Code availability
We used Sigma Plot, Microsoft Excel, and Latex to generate the figures in this manuscript.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59(2):284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo AP. Heat shock proteins as molecular chaperones. Medecine/Sciences. 2005;21(6-7):619–625. doi: 10.1051/medsci/2005216-7619. [DOI] [PubMed] [Google Scholar]
- Augustine T, Chaudhary P, Gupta K, Islam S, Ghosh P, Santra MK, Mitra D. Cyclin F/FBXO1 interacts with HIV-1 viral infectivity factor (Vif) and restricts progeny virion infectivity by ubiquitination and proteasomal degradation of vif protein through SCFcyclin F E3 ligase machinery. J Biol Chem. 2017;292(13):5349–5363. doi: 10.1074/jbc.M116.765842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero-Pérez B, Whitehouse A. Hsp70 isoforms are essential for the formation of Kaposi’s sarcoma-associated herpesvirus replication and transcription compartments. PLoS Pathog. 2015;11(11):e1005274. doi: 10.1371/journal.ppat.1005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vézinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- Batra J, Tripathi S, Kumar A, Katz JM, Cox NJ, Lal RB, Sambhara S, Lal SK. Human Heat shock protein 40 (Hsp40/DnaJB1) promotes influenza A virus replication by assisting nuclear import of viral ribonucleoproteins. Sci Rep. 2016;6(1):19063. doi: 10.1038/srep19063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha M, Pesce ER, Blatch GL (2007) The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: Regulating chaperone power in the parasite and the host. Int. J. Biochem. Cell Biol 39(10):1781–1803. 10.1016/j.biocel.2007.02.011 [DOI] [PubMed]
- Bracher A, Verghese J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2015;2:10. doi: 10.3389/fmolb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Wei C, Zhao L, Wang J, Jia Q, Wang X, Jin Q, Deng T. DnaJA1/Hsp40 is co-opted by influenza A virus to enhance its viral RNA polymerase activity. J Virol. 2014;88(24):14078–14089. doi: 10.1128/jvi.02475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YQ, Yuan L, Zhao Q, Yuan JL, Miao C, Chang YF, Wen XT, Wu R, Huang XB, Wen YP, Yan QG, Huang Y, Han XF, Ma XP, Cao SJ. Hsp40 protein DNAJB6 interacts with viral NS3 and inhibits the replication of the Japanese encephalitis virus. Int J Mol Sci. 2019;20(22):5719–5736. doi: 10.3390/ijms20225719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3(1):28–36. 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2 [DOI] [PMC free article] [PubMed]
- Colinet H, Lee SF, Hoffmann A (2010) Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. FEBS J 277(1):174–185. 10.1111/j.1742-4658.2009.07470.x [DOI] [PubMed]
- Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, White G, Foster P, Markham PD. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224(4648):500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- Gervaix A, West D, Leoni LM, Richman DD, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci U S A. 1997;94(9):4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J, Schipper-Krom S, Juenemann K, Gruber A, Coolen S, Van Den Nieuwendijk R, Van Veen H, Overkleeft H, Goedhart J, Kampinga HH, Reits EA. The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem. 2013;288(24):17225–17237. doi: 10.1074/jbc.M112.421685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman J, Rujano MA, van Waarde MAWH, Kakkar V, Dirks RP, Govorukhina N, Oosterveld-Hut HMJ, Lubsen NH, Kampinga HH. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell. 2010;37(3):355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14(1):105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Mitra D. Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J Biol Chem. 2005;280(48):40041–40050. doi: 10.1074/jbc.M508904200. [DOI] [PubMed] [Google Scholar]
- Kumar M, Rawat P, Khan SZ, Dhamija N, Chaudhary P, Ravi DS, Mitra D. Reciprocal regulation of human immunodeficiency virus-1 gene expression and replication by heat shock proteins 40 and 70. J Mol Biol. 2011;410(5):944–958. doi: 10.1016/j.jmb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B (1999) Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proceedings of the National Academy of Sciences 96. 10.1073/pnas.96.10.5452 [DOI] [PMC free article] [PubMed]
- Levy JA, Hoffman AD, Kramer SM, Landis JA, Shimabukuro JM, Oshiro LS (1984) Isolation of lymphocytopathic retroviruses from San Francisco patients with AIDS. Science 225(4664):840-842. 10.1126/science.6206563 [DOI] [PubMed]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22(1):631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Liu RJ, Niu XL, Yuan JP, Chen HD, Gao XH, Qi RQ (2020) DnaJA4 is involved in responses to hyperthermia by regulating the expression of F-actin in HaCaT cells. Chin Med J (Engl) Publish Ahead of Print. 10.1097/CM9.0000000000001064 [DOI] [PMC free article] [PubMed]
- López SN, Rodríguez-Valentín M, Rivera M, Rodríguez M, Babu M, Cubano LA, Xiong H, Wang G, Kucheryavykh L, Boukli NM (2017) HIV-1 Gp120 clade B/C induces a GRP78 driven cytoprotective mechanism in astrocytoma. Oncotarget 8(40):68415–68438. 10.18632/oncotarget.1947410 [DOI] [PMC free article] [PubMed]
- Lopez-Buesa P, Pfund C, Craig EA. The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15253–15258. doi: 10.1073/pnas.95.26.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Höhfeld J, Ohtsuka K, Hartl FU. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J Biol Chem. 1996;271(32):19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- Norman JP, Perry SW, Reynolds HM, Kiebala M, De Mesy Bentley KL, Trejo M, Volsky DJ, Maggirwar SB, Dewhurst S, Masliah E, Gelbard HA. HIV-1 Tat activates neuronal ryanodine receptors with rapid induction of the unfolded protein response and mitochondrial hyperpolarization. PLoS One. 2008;3(11):e3731. doi: 10.1371/journal.pone.0003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Ravindran B (2013) Isolation of human PBMCs. BIO-PROTOCOL 3(3):e323. 10.21769/bioprotoc.323
- Park RJ, Wang T, Koundakjian D, Hultquist JF, Lamothe-Molina P, Monel B, Schumann K, Yu H, Krupzcak KM, Garcia-Beltran W, Piechocka-Trocha A, Krogan NJ, Marson A, Sabatini DM, Lander ES, Hacohen N, Walker BD. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet. 2017;49(2):193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Taulien J, Lindquist S. The role of heat-shock proteins in thermotolerance. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1993;339(1289):23–30. doi: 10.1007/978-94-011-2108-8_4. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(4):2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M, Sharma R, Shastry S, Roberts BL, Albino-Flores I, Wickner S, Masison DC. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS genetics. 2014;10(10):e1004720. doi: 10.1371/journal.pgen.1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia. 1962;18(12):571–573. doi: 10.1007/BF02172188. [DOI] [Google Scholar]
- Ritossa F (1996) Discovery of the heat shock response. Cell Stress Chaperones 1(2):97–98. 10.1379/1466-1268(1996)001<0097:DOTHSR>2.3.CO;2 [DOI] [PMC free article] [PubMed]
- Sapareto SA, Hopwood LE, Dewey WC. Effects of Hyperthermia on survival and progression of Chinese hamster ovary cells. Cancer Res. 1978;38(2):393–400. [PubMed] [Google Scholar]
- Slater A, Cato ACB, Sillar GM, Kioussis J, Burdon RH. The pattern of protein synthesis induced by heat shock of HeLa cells. Eur J Biochem. 1981;117(2):341–346. doi: 10.1111/j.1432-1033.1981.tb06343.x. [DOI] [PubMed] [Google Scholar]
- Sørensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003;6(11):1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x. [DOI] [Google Scholar]
- Stolp B, Fackler OT. How HIV takes advantage of the cytoskeleton in entry and replication. Viruses. 2011;3(4):293–311. doi: 10.3390/v3040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, Andino R, Fernandez-Sesma A, Frydman J. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell. 2015;163(5):1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguwa S, Yeh MT, Rainbolt TK, Nayak A, Shao H, Gestwicki JE, Andino R, Frydman J (2019) Zika virus dependence on host Hsp70 provides a protective strategy against infection and disease. Cell Rep 26(4):906-920.e3. 10.1016/j.celrep.2018.12.095 [DOI] [PMC free article] [PubMed]
- Takashima K, Oshiumi H, Matsumoto M, Seya T. DNAJB1/HSP40 suppresses melanoma differentiation-associated gene 5-mitochondrial antiviral signaling protein function in conjunction with HSP70. Journal of innate immunity. 2018;10(1):44–55. doi: 10.1159/000480740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger F, Kindås-Mügge I, Knobler RM, Hönigsmann H. Stress proteins in the cellular response to ultraviolet radiation. J Photochem Photobiol B. 1996;35(3):141–148. doi: 10.1016/S1011-1344(96)07344-7. [DOI] [PubMed] [Google Scholar]
- Urano E, Morikawa Y, Komano J. Novel role of HSP40/DNAJ in the regulation of HIV-1 replication. J Acquir Immune Defic Syndr. 2013;64(2):154–162. doi: 10.1097/QAI.0b013e31829a2ef8. [DOI] [PubMed] [Google Scholar]
- Wainberg Z, Oliveira M, Lerner S, Tao Y, Brenner BG. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology. 1997;233(2):364–373. doi: 10.1006/viro.1997.8618. [DOI] [PubMed] [Google Scholar]
- Walsh P, Bursać D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5(6):567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu X, Jiang Z, You Q. Modulation of protein fate decision by small molecules: Targeting molecular chaperone machinery. Acta Pharm. Sin. B. 2020;10:1904–1925. doi: 10.1016/j.apsb.2020.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Fuse T, Asano I, Tsukahara F, Maru Y, Nagata K, Kitazato K, Kobayashi N. Identification of Hsc70 as an influenza virus matrix protein (M1) binding factor involved in the virus life cycle. FEBS Lett. 2006;580(24):5785–5790. doi: 10.1016/j.febslet.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yang F, Zhu Z, Yang Y, Wang Z, Cao W, Dang W, Li L, Mao R, Liu Y, Tian H, Zhang K, Liu X, Ma J, Zheng H. Cellular DNAJA3, a novel VP1-interacting protein, inhibits foot-and-mouth disease virus replication by inducing lysosomal degradation of VP1 and attenuating its antagonistic role in the beta interferon signaling pathway. J Virol. 2019;93(13):e00588–e00519. doi: 10.1128/JVI.00588-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4(5):495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zhu J, Davoli T, Perriera JM, Chin CR, Gaiha GD, John SP, Sigiollot FD, Gao G, Xu Q, Qu H, Pertel T, Sims JS, Smith JA, Baker RE, Maranda L, Ng A, Elledge SJ, Brass AL. Comprehensive identification of host modulators of HIV-1 replication using multiple orthologous RNAi reagents. Cell Rep. 2014;9(2):752–766. doi: 10.1016/j.celrep.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 179 kb)
Data Availability Statement
The data that supports the findings of this study are available from the corresponding author upon reasonable request.