Abstract
The current study focuses on the isolation and in vitro characterization of bioactive metabolites produced by endophytic fungi isolated from the Himalayan yew (Taxus wallichiana Zucc.). The endophytic fungi were isolated on artificial media from inner tissues of bark and needles. Antimicrobial and antioxidant activity, along with total phenolic- and flavonoid-content assays were used in the evaluation of bioactivity of the fermented crude extracts. The ability of the endophytes to produce the anticancer compound Taxol was also analyzed using thin-layer chromatography (TLC) and reverse-phase high-performance liquid chromatography (RP-HPLC). A total of 16 fungal morphotypes were obtained from asymptomatic inner tissues of the bark and needles of T. wallichiana. Among the 16 isolates, the ethyl acetate (EA) fraction of isolate MUS1, showed antibacterial and antifungal activity against all test-pathogens used (Streptococcus faecalis ATCC 19433, Staphylococcus aureus ATCC 12600, Bacillus subtilis ATCC 6633, Escherichia coli ATCC 25922, Salmonella enterica ATCC 13076, Pseudomonas aeruginosa ATCC 27853, and Candida albicans). MUS1 showed significant inhibition against Pseudomonas aeruginosa ATCC 27853 (minimum inhibitory concentration (MIC): 250 µg/ml) and the pathogenic yeast, Candida albicans (MIC: 125 µg/ml). Antioxidant activity, total phenolic, and total flavonoid content as well as in vitro Taxol production were evaluated for EA fraction of isolate MUS1. Antioxidant activity was evaluated using 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay. At a concentration of 100 µg/ml, the % DPPH radical scavenging activity was 83.15 ± 0.40, 81.62 ± 0.11, and 62.36 ± 0.29, for ascorbic acid, butylated hydroxytoluene (BHT), and the EA fraction of MUS1, respectively. The DPPH-Half maximal inhibitory concentration (DPPH-IC50) value for the EA fraction was 81.52 ± 0.23 µg/ml, compared to BHT (62.87 ± 0.08 µg/ml) and ascorbic acid (56.15 ± 0.19 µg/ml). The total phenolic and flavonoid content in the EA fraction were 16.90 ± 0.075 µg gallic acid equivalent (GAE) and 11.59 ± 0.148 µg rutin equivalent (RE), per mg of dry crude extract, respectively. TLC and RP-HPLC analysis showed that the isolate MUS1 also produces Taxol (282.05 µg/l of fermentation broth). Isolate MUS1 was identified as Annulohypoxylon sp. by internal transcribed spacer (ITS) sequencing. Having the ability to produce antimicrobial and antioxidant metabolites, as well as the anticancer compound Taxol, makes Annulohypoxylon sp. strain MUS1, a promising candidate for further study of naturally occurring bioactive metabolites.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02693-z.
Keywords: Annulohypoxylon sp., Bioactive metabolites, Endophytic fungi, Taxol, Taxus wallichiana
Introduction
Taxus wallichiana Zucc. (Taxaceae), also known as Himalayan yew (Vernacular name: Lauth-salla), is a medicinal plant found in hilly regions of Nepal. T. wallichiana grows on steep mountain slopes of altitude 1500–3000 m, and mostly in areas not disturbed by man-made practices (Strobel et al. 1996). Taxus species are the source of a highly functionalized diterpene molecule called Taxol, first isolated from the stem bark of Western yew, Taxus brevifolia (Wani et al. 1971), and used in cancer treatment. This has led to the overharvesting of needles from the Yew trees and consequently, Taxus forests in the Himalayan regions are on the verge of extinction and now on the International Union for Conservation of Nature (IUCN) Red List. This has compelled an immediate search for alternative sources of Taxol to save these trees and maintain their biodiversity for future generations.
Endophytes are a group of microbial colonizers that reside in the inner tissue of plants (Bascom-Slack et al. 2009). These microorganisms form a symbiotic association with the host that can range from commensalistic to slightly pathogenic (Strobel and Daisy, 2003). Some endophytes also can produce the same or similar secondary metabolites as their plant-host (Venieraki et al. 2017). For example, the discovery that some endophytic fungi could produce Taxol opened the possibilities of using them as an alternative source to the Taxus plant (Stierle et al. 1993; Venieraki et al. 2017). Since the initial report, more than 200 endophytic fungi isolated from plants have been reported to produce Taxol in culture media, albeit in small quantities or requiring optimization in some cases (Flores-Bustamante et al. 2010; Gupta et al. 2020; Kusari et al. 2014). Different biological classes of natural products, including antibacterial, antifungal, anticancer, antiviral as well as plant protective agents, have been reported to be produced by endophytes (Singh et al. 2011; Strobel and Daisy 2003). However, the endophytic microbiota in plants still represents a largely untapped resource of natural products (Wijeratne et al. 2010). Therefore, the current study focuses on the isolation and in vitro characterization of bioactive metabolites produced by endophytic fungi isolated from the Himalayan yew (T. wallichiana) collected from the Mustang district of Nepal.
Materials and methods
Location of the study area and sample collection
The Taxus plant materials were collected from Taglung forest of the Lower-Mustang region of Mustang district (Fig. S1, suppl. file), at similar altitudes (2722 m to 2729 m) and geographic locations (latitude 28.62 to 28.63°N and longitude 83.66 to 83.67°E) in October 2016. The plant samples (i.e., healthy and asymptomatic bark and needles) from T. wallichiana, were packed in polythene bags and brought to the laboratory in an icebox. The samples were processed within 48 h of collection and stored at 4 °C until isolation procedures were finished. The specimen of each sample was deposited at the Department of Biotechnology, Kathmandu University in Dhulikhel, Kavre, Nepal.
Media and microorganisms used
Potato dextrose agar (PDA, HiMedia) or Potato dextrose broth (PDB, HiMedia) was used for fungal isolation and routine growth of fungal endophytes, for genomic DNA (gDNA) extraction and secondary metabolites production. Gram-positive bacteria (Streptococcus faecalis ATCC 19433, Staphylococcus aureus ATCC 12600, and Bacillus subtilis ATCC 6633), Gram-negative bacteria (Escherichia coli ATCC 25922, Salmonella enterica ATCC 13076, and Pseudomonas aeruginosa ATCC 27853), and the yeast, Candida albicans (isolated from oral thrush of HIV positive patient and verified as C. albicans), were used as test-pathogens. The bacterial strains were obtained directly from the American Type Culture Collection (ATCC) and, Candida albicans was obtained from the Department of Microbiology, Dhulikhel Hospital of Kathmandu University, Dhulikhel, Nepal. All the pathogens were grown and maintained in Muller-Hinton Agar (MHA) or Muller-Hinton broth (MHB) (Schwalbe et al. 2007).
Sample processing and isolation of endophytic fungi
The isolation of fungi from inner tissues of needles and bark of T. wallichiana was carried out in two methods as previously described (Huang et al. 2001). In the first one, the bark and needle samples were washed thoroughly with running tap water to remove the dust and external debris. The bark was cut into 1 cm × 1 cm × 0.5 cm pieces, while the needles were separated into groups of 4–6 needles and washed with sterile water before surface sterilization. Surface sterilization was carried out with 70% (v/v) ethyl alcohol for 2 min, followed by 0.2% mercuric chloride for 1 min. The sterilized pieces of bark and needles were washed with sterile distilled water three times using separate sterile containers. After washing with sterile water, bark and needles were dried using sterile blotting paper. A single needle was excised and was cut into 0.5 cm long pieces for each sample. The cut needles were placed over the PDA media (pH 5.6) horizontally—freshly exposed tissue facing down—or vertically, slightly dipping the breadth side of the needle into the media. The outer bark was removed using a sterile blade and inner sterile tissue was excised. The inner bark-tissue was cut into 0.5 cm × 0.5 cm pieces for each sample and placed over PDA media (pH 5.6) in a Petri-plate. The plates were sealed with parafilm and incubated at 25 ± 2 °C for at least 10 days.
In the second method and as above, 0.5 cm × 0.5 × 0.5 cm sterile tissues were excised by removing the outer bark using a sharp blade. One to two bark pieces (0.5 cm × 0.5 cm) of each sample were ground into a paste with 2 ml of sterile-distilled water using a mortar and pestle (sterile). Similarly, two to three surface-sterilized leaf-needles were excised, blotted dry, and ground. The maceration of bark and needle tissues was carried out in two steps. First, one ml of sterile distilled water was added to the sample and grounded with gentle force. In the second step, an additional one ml of sterile-distilled water was added and the sample was grounded well to make a paste/solution. About one ml of the grounded paste/solution was transferred to a Petri-plate and approximately 20 ml of molten PDA media was poured into the plate. The mixture was homogenized by gently moving the plate in the shape of the number eight. Once solid, the plates were sealed with parafilm and incubated at 25 ± 2 °C for at least 10 days. No antibiotics were added to the media. Every single sample was processed by both methods. Hence, the processing of a single sample comprised four plates, two for intact tissue (one for bark and one for needle) and two for ground tissue (one for bark and one for needle). Fungal isolates obtained from both methods were checked for contamination. Finally, the pure fungal isolates were obtained by transferring the tissue-extruded hyphae filaments to another PDA media plate (pH 5.6) using the hyphal tip method (Strobel et al. 1996).
Characterization of endophytic fungi
Morphotypes were assigned based on the phenotype of the fungal colony (e.g., color, shape, and size), mycelial mat characteristics in the growth media, as well as the presence of sexual or asexual structures seen under the microscope (Barnett and Hunter 1998; Watanabe 2010). Tape-lift mounts stained with Lactophenol cotton blue were visualized under the compound microscope (Li 2013).
Genomic DNA (gDNA) from the isolated fungal endophytes was extracted for molecular characterization using the Quick-DNA™ Fungal/Bacterial Miniprep Kit (Zymo Research Company) with little modification. For this, agar-plugs of isolated fungi grown in PDA were used to seed Erlenmeyer flasks containing 50 ml of Potato dextrose broth (PDB) and incubated at 25 ± 2 °C, shaking at 150 rounds-per-minute (rpm). After 3 days, mycelia were harvested by filtration and washed thoroughly with distilled water to remove any media and extracellular metabolites. The harvested mycelia (≈ 300 mg) were pulverized with liquid nitrogen using mortar and pestle and transferred to 2 ml Eppendorf tubes. Lysis Solution, 750 µl, was added to the tube containing the pulverized fungal mycelium powder and the samples were incubated at 65 °C for 1 h in a water-bath. The remaining steps were followed as prescribed in the instruction manual provided with the kit.
The gDNA obtained was used for polymerase chain reaction (PCR) to amplify the internal transcribed spacer (ITS) using primers ITS1 (F-5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (R-5′-TCCTCCGCTTATTGATATGC-3′) to identify the endophytic fungi at the molecular level as previously described (Raja et al. 2017). The 25 µl PCR-mixture consisted of 13.5 µl of PCR grade water, 1.25 µl of ITS1-F (10 µM), 1.25 µl of ITS4-R (10 µM), 2.5 µl of amplification buffer (10X), 0.75 µl of MgCl2 (50 mM), 0.5 µl of dNTP mix (10 mM), 0.25 µl of Taq DNA polymerase (5 U/µl) and 5 µl of Template DNA. PCR was carried out in a thermocycler (BIO-RAD T100) under the following amplification conditions: an initial denaturing step at 94 °C for 5 min, followed by 35 amplification cycles of 94 °C for 1 min, 55 °C for the 40 s, and 72 °C for 1 min and a final extension step at 72 °C for 10 min. The PCR products were visualized on 1% (w/v) agarose gel and aliquots were sent to Macrogen-Europe for sequencing. The obtained sequences were compared to existing DNA sequences in NCBI GenBank using BLASTn (megablast). The phylogenetic relations of the fungal endophytes were analyzed by MEGAX software (Kumar et al. 2018).
Production, extraction, and analysis of secondary metabolites
Production and extraction
To test the production of intracellular and secreted extracellular secondary metabolites, a small-scale fermentation process was carried out as previously described (Yashavantha Rao et al. 2015). Briefly, fungi were grown in 250 ml Erlenmeyer flasks containing 100 ml of Potato dextrose broth (PDB) at 25 ± 2 °C and shaken at 150 rounds per minute (rpm). After 10 days, mycelia and liquid fermented culture media were separated by filtration. The mycelia were homogenized with mortar and pestle and extracted with 10 ml each of either Methanol (MH), Ethyl acetate (EA), or Chloroform (CH). Similarly, the liquid fermented filtrate was extracted with equal volumes (100 ml) of either EA, CH, or Hexane (HX), using a separatory funnel. The solvent-extracts obtained from both mycelia (10 ml) and culture-filtrates (100 ml) were filtered using Whatman filter paper No.1 and evaporated to dryness at room temperature under an exhaust fan. The dried-fractions extracted with each solvent (i.e., EA, MH, CH, or HX) were weighed and stored at 4 °C until further analysis.
TLC analysis of secondary metabolites
Thin-layer Chromatography (TLC) analysis of the crude extracts was done according to Garyali et al. 2013, with little modifications (Garyali et al. 2013). The crude fractions were dissolved in ethyl acetate (EA) (10 mg/ml) and 10 µl from each were spotted onto 20 × 20 cm TLC plates (Silica gel 60 F254, Merck) with at least one cm between the spots and placed in a binary mobile phase consisting of Chloroform: Methanol (9:1 v/v). Once the mobile phase reached the top, the TLC plate was removed, the solvent evaporated, and the plate was observed under short- and long-wave ultraviolet (UV) light. Furthermore, the TLC plate was sprayed with 2% AlCl3 and again observed under short- and long-wave UV. A second TLC plate was made in the same manner and was sprayed with 0.04 mg/ml DPPH solution, incubated in the dark for 10 min, and observed under visible light.
In vitro bioactivity analysis
Antimicrobial activity
Altogether, six human pathogenic-bacteria and one human yeast-pathogen were used in an agar-well diffusion (AWD) assay, to determine the antimicrobial activity of the crude extracts obtained from the fungal endophytes (Schwalbe et al. 2007; Sharma et al. 2016; Yashavantha Rao et al. 2015). Stocks (10 mg/ml) for both the intracellular and extracellular-extracts were made using dimethyl sulfoxide (DMSO) as a solvent. Antibiotic discs of gentamicin (10 µg/disc) and fluconazole (25 µg/disc), were used as positive controls against bacteria and yeast, respectively. Gram-positive bacteria (ATCC strains of Streptococcus faecalis, Staphylococcus aureus, and Bacillus subtilis), Gram-negative bacteria (ATCC strains of Escherichia coli, Salmonella enterica, and Pseudomonas aeruginosa), and the ascomycete yeast (a clinical strain of Candida albicans) were grown overnight in Muller-Hinton broth (MHB) and adjusted to a McFarland turbidity standard of 0.5 (1 × 108 Colony Forming Units (CFU)/ml). The test-pathogens were spread on Muller-Hinton agar (MHA) plates using an L-shaped spreader and allowed to dry for a few minutes. Once dried, for each treatment, agar-wells were made with a sterile borer (6 mm inner diameter) and 50 μl of extract (10 mg/ml) were loaded into the agar-wells. A standard antibiotic disc of gentamicin or fluconazole (6 mm in diameter) was placed on top of the media. DMSO-only (50 µl) was also included as a negative control. The diameter (mm) of the zone of inhibition (ZOI) was measured for each treatment and compared to the controls. The assay was carried out in triplicate for each test-pathogen.
Minimum inhibitory concentration assay
The minimum inhibitory concentration (MIC) was calculated using a macro-broth dilution method with little modifications (Schwalbe et al. 2007). P. aeruginosa ATCC 27853 and clinical strain of C. albicans were chosen as test-pathogens based on the greater ZOI values obtained in the AWD assay. The test-pathogens (0.5 McFarland turbidity standard, 1 × 108 CFU/ml) were diluted with sterile-distilled water in the ratio of 1:100 to a concentration of approx. 106 CFU/ml. Gentamicin sulfate (HiMedia), a broad-spectrum bactericidal agent, was used as a standard antibiotic for MIC assay. The antibiotic stock was filtered-sterilized (0.45 µm filter) and stored at 4 °C until the use.
TLC-bioautography assay
The antimicrobial activity of the extracts was also evaluated using the TLC-bioautography agar overlay method, which is a combination of contact and direct bioautography (Dewanjee et al. 2015). After the TLC plate was prepared as described above, the plate was placed in a 90 mm Petri-plate and covered with molten Muller-Hinton agar (MHA) medium to a thickness of 2–3 mm. Once solid, the agar was overlaid with 106 CFU/ml of P. aeruginosa ATCC 27853. The plates were incubated at 37 ± 2 °C for 18–24 h. After incubation, the ZOI, which can be seen as clear spots against a purple background, was visualized by staining with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) solution (0.5 mg ml−1).
Antioxidant activity
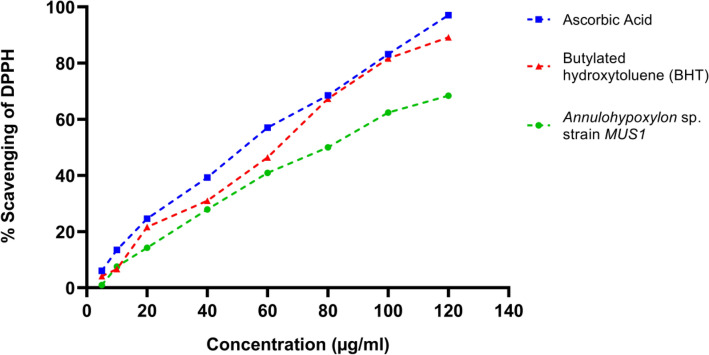
A DPPH-radical scavenging assay using 2,2-diphenyl-1-picrylhydrazyl (DPPH; Sigma-Aldrich) was carried out as previously described (Pan et al. 2017) with some modifications to determine the antioxidant activity of the endophytic fungal crude extracts. The DPPH reagent was freshly prepared in Methanol (0.04 mg/ml). Ascorbic acid and Butylated hydroxytoluene (BHT) (used as controls), and the ethyl acetate (EA) fraction were prepared in methanol in the concentration range of 5, 10, 20, 40, 60, 80, 100, and 120 µg/ml. One ml of the standards, or the EA fraction, was mixed with three ml of the DPPH solution (1:3) in a glass test-tube. After mixing, the test tubes were incubated in the dark at 27 °C for 10 min. The absorbance at 517 nm was recorded using a UV–Vis Spectrophotometer (Shimadzu, UV-1800) and Methanol was used to blank the instrument. The assay was repeated three times. The percent scavenging ability for DPPH-radicals was calculated as described in Pan et al. 2017. Additionally, a measure of half-maximal inhibitory concentration (IC50) for 50% DPPH scavenging activity was carried out for standards and EA fraction of MUS1 by fitting a linear regression to DPPH scavenging curves, respectively.
Total phenol and flavonoid assays
The total phenolic content (TPC) and total flavonoid content (TFC) of the fungal crude extracts obtained using the solvents Ethyl acetate (EA), Chloroform (CH), and Hexane (HX), were determined as described (Pan et al. 2017). However, the assay volumes were halved (total volume = 12.5 ml) and methanol was used to dissolve the standards i.e., gallic acid (HiMedia), rutin (HiMedia), and the crude extracts (1 mg/ml). For the TPC assay, the absorbance was measured at 733 nm, while the TFC assay was measured at 507 nm using a UV–Vis Spectrophotometer (Shimadzu, UV-1800). The TPC from the extracts was determined based on the calibration curve obtained from the gallic acid standards (i.e., 1, 2, 4, 6, 8, 10, 12, and 16 µg/ml), and expressed as µg of gallic acid equivalents (GAEs) per mg of dry fungal extract. Similarly, the calibration curve from the rutin standards (i.e., 5, 10, 20, 40, 60, 80, 100, and 120 µg/ml), was used to calculate the TFC and expressed as µg of rutin equivalents (REs) per mg of dry fungal extract.
Taxol analysis
Preparative TLC
TLC was used to test for the presence of Taxol in the EA fraction obtained from crude extracts of the endophytic fungal isolates (Głowniak and Mroczek 1999). 20 µl of the EA fraction (10 mg/ml) were spotted onto 20 × 20 cm TLC plates (Silica gel 60 F254, Merck) with at least one cm between the spots and placed in a quaternary mobile phase consisting of Benzene: Chloroform: Acetone: Methanol (20:92.5:15:7.5). Once the mobile phase reached the top, the TLC plate was removed. The standard anticancer compound, Taxol (Paclitaxel; Sigma-Aldrich) was also included and used for comparison. The bands horizontally in-line with the Taxol standard were visualized under short-wave UV light and marked with a pencil. The pencil-marked spots were scraped off the plates using a sharp blade and eluted using acetone-methanol (1:1) mixture followed by centrifugation at 10,000 rpm for 10 min. After centrifugation, the supernatant was taken and evaporated to dryness under reduced pressure and dissolved in HPLC grade methanol for the HPLC analysis of Taxol.
RP-HPLC analysis
Reverse-phase High-performance liquid chromatography (RP-HPLC) analysis and quantification of Taxol was done using a reverse-phase C18-column (Shim-pack GIST C18, 5 µm, 150 × 4.6 mm I.D.) in a Shimadzu LC-2030 instrument with UV absorbance at 227 nm (detector) as described elsewhere (Li et al. 2017; Liu et al. 2009) with some modifications. For this, 15 µl of the sample was injected into the column at a column temperature of 30 °C. The separation was carried out at a flow rate of 1 ml min−1 with gradient acetonitrile–water (v/v) system (50% of acetonitrile for 0–10 min, 90% of acetonitrile for 10–12 min, again 50% of acetonitrile for 12–15 min and stop after 15 min). A standard curve (R2 = 0.99) was generated using authentic Taxol (Paclitaxel; Sigma-Aldrich) solutions (i.e., 5, 10, 15, 25, 50, 100 µg/ml) and used to calculate the amount of Taxol present in fermentation cultures as described in Liu et al. (2009).
Statistical analysis
Statistical analysis was carried out using GraphPad Prism version 8.0.0 for Windows, GraphPad Software, San Diego, USA, for t-test, ANOVA, and Post Hoc tests. The t-test and one-way ANOVA followed by Tukey’s Post Hoc test with P < 0.05 were used to analyze the significant differences between the results obtained.
Results
Endophytic fungi from Himalayan Yew
A total of 16 fungal morphotypes were obtained from asymptomatic inner tissues of bark and needles of T. wallichiana using PDA media.
Identification of isolate MUS1 from Himalayan Yew
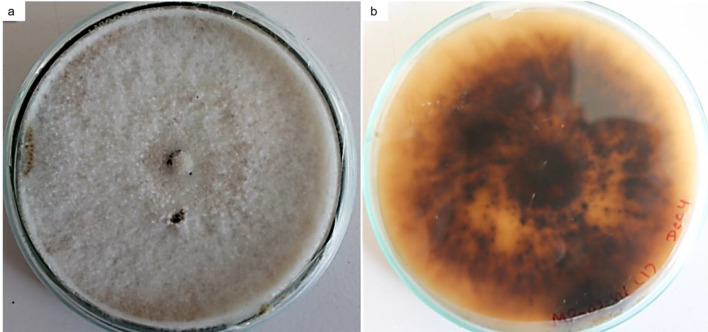
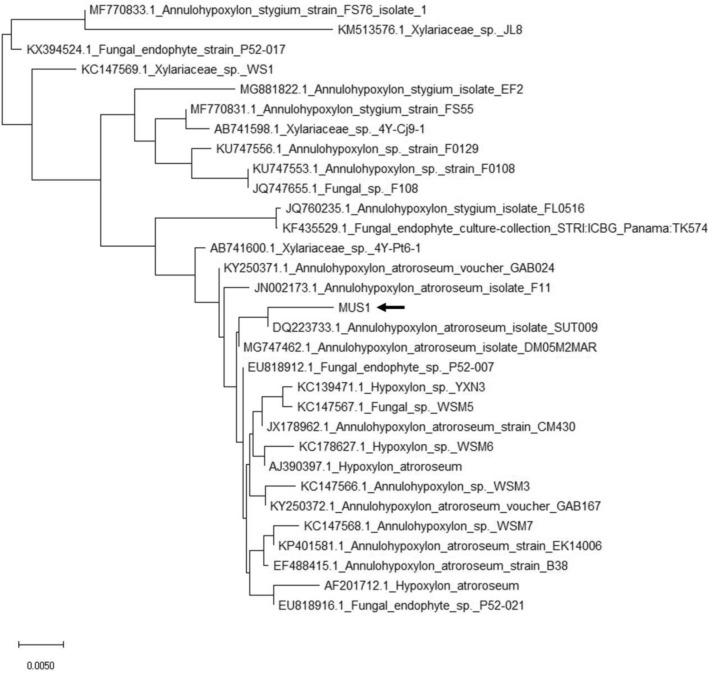
Fungal genera identified by morphological characterization included Alternaria, Aspergillus, Fusarium, Mucor, Penicillium, and Trichoderma spp. Some of the endophytic isolates were unidentified (Fig. 1). An endophytic fungus isolated among the 16 different morphotypes, initially named isolate MUS1, exhibited bioactive-metabolite activity and was chosen for further study. The isolate showed a creamy-white flocculent mycelium in the dorsal-side of the PDA plate and yellowish-brownish-black taint on the ventral-side of the PDA plate after 10 days of incubation (Fig. 2). No signs of sporulation or conidia were seen (Fig. S3). MUS1 was identified as Annulohypoxylon sp. (Fig. 2) and the ITS sequence has been deposited in GenBank under accession number MN699475 (Fig. 3).
Fig. 1.
Morphological characteristics of 10-day-old cultures of fungal endophytic isolates from Taxus wallichiana. Dorsal view of fungal isolates in Potato dextrose agar media (a–h), and respective microscopic structures (same letter), seen under the compound microscope (400X). Plate a, c, and h are identified as Aspergillus sp., Mucor sp., and Trichoderma sp., respectively, while others remained unidentified by phenotypic characterization
Fig. 2.
Ten-day-old culture of fungal isolate, MUS1, later identified as Annulohypoxylon sp. by internal transcribed spacer (ITS) sequencing, grown on Potato dextrose agar (PDA) media; a dorsal side—a creamy-white flocculent mycelium, and b ventral side—yellowish-brownish-blackish color. No signs of sporulation or conidia were seen under the microscope (Fig. S3)
Fig. 3.
Phylogenetic analysis of the ITS1-5.8S-ITS2 sequence from the endophytic fungal isolate MUS1 using the Neighbor-joining (NJ) method. The sequence of endophytic fungi under study marked with symbol ‘MUS1’ (pointed with arrow) with bootstrapping of 1000 replicates (conducted in MEGAX software). Scale bar represents the number of differences between sequences. In this case, there is a 0.5% difference between two sequences. MUS1 was identified as Annulohypoxylon sp. (Fig. 2) and the ITS sequence has been deposited in GenBank under accession number MN699475
TLC analysis of secondary metabolites
TLC analysis of crude extracts from isolate MUS1 extracted using various solvents showed a diversity of extracellular metabolites. However, the number of metabolites was higher for the EA fraction (Rf = 0.25 to 0.98) in comparison to the CH and HX fractions (Fig. 4). Furthermore, when the TLC plate was sprayed with a 2% AlCl3 solution, a bluish-green fluorescence spot could be seen under short-UV suggesting the presence of flavonoid metabolites. The second TLC plate made in the same manner but sprayed with the DPPH solution also showed yellow spots on the fluorescent regions (Fig. 4). This suggests that extracellular metabolites from isolate MUS1 are potentially rich in bioactive metabolites including antioxidants.
Fig. 4.
Thin-layer chromatography (TLC) plates of crude fractions from the fungal isolate MUS1. The crude extract was extracted from fermented broth using Potato dextrose broth (PDB) as a culture medium using three different solvents in decreasing polarity, Ethyl acetate (EA), Chloroform (CH), and Hexane (HX). The three different fractions, MP1 (extracted with EA), MP2 (extracted with CH), and MP3 (extracted with HX), in each plate, were run in a solvent mixture of chloroform: methanol (9:1, v/v) and visualized using different methods: a under long-wave UV light, b under short-wave UV light, c TLC chromatogram after spraying with 2% AlCl3 under short-wave UV light, and d TLC chromatogram after spraying with 0.04 mg/ml DPPH under visible light. RU denotes the standard flavonoid compound, rutin. The TLC chromatograms in plates a and b show that the number of metabolite bands is decreasing with decreasing polarity of the solvents used [i.e., higher for EA (MP1) in comparison to CH (MP2) and HX (MP3)]. Similarly, fluorescence, after spraying with 2% AlCl3, is higher in MP1 than MP2 and MP3 (plate c), and Plate d represents the DPPH-radical scavenging activity of the metabolites
Antimicrobial activity
The EA fraction from the 16 fungal endophytes isolated, was analyzed for antimicrobial activities against seven human pathogens using the Agar well diffusion (AWD) assay. Only the extracellular EA-fraction of isolate MUS1 showed antimicrobial activity against all the pathogens tested. Two isolates showed activity against the bacterial pathogens but had less activity as compared to MUS1. Other isolates were unable to produce any antimicrobial activity (results not shown).
Out of the three solvents used in the extraction (i.e., HX, EA, or CH), the EA fraction (500 µg/well) had significant antimicrobial activity compared to the CH or HX fractions (Tukey's post-hoc test, P < 0.05; results not shown), suggesting that the metabolite or metabolites responsible for the antimicrobial activity were soluble in EA. However, no activity was seen for the intracellular fractions for any of the solvents used including EA in the extraction (results not shown). Interestingly, the yield of the extracellular metabolites was less in comparison to intracellular metabolites from isolate MUS1 for all the solvents used (Table 1).
Table 1.
Metabolites extraction from isolate MUS1
| Solvent fraction | Intracellular metabolites (mg)a | Extracellular metabolites (mg)a |
|---|---|---|
| EA | 29.5 [7.04] | 40.1 [0.0401] |
| CH | 17.2 [4.11] | 9.8 [0.0098] |
| HX | N/a | 8.6 [0.0086] |
| MT | 80.9 [19.31] | N/a |
aIntra- and extracellular metabolites extracted from dry fungal biomass (419 mg) and fermentation media (100 ml Potato dextrose broth), respectively. EA—ethyl acetate, CH—chloroform, HX–hexane, and MT—methanol. The % (w/w) and % (w/v) yield, for the Intra- and Extracellular metabolites, respectively, are shown in brackets
N/a not applicable
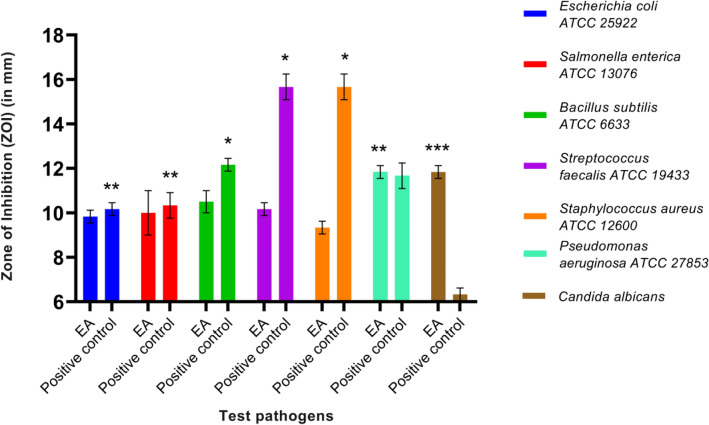
There was a significant difference in the ZOI between the EA fraction and the DMSO (control) for all the seven pathogens tested (t-test, P < 0.05, Fig. 5). Furthermore, comparing the EA fraction with the antibiotic gentamicin (positive control), there was no significant difference in ZOI against P. aeruginosa ATCC 27853 (10 µg gentamicin vs. 500 µg crude extract). This would suggest that the metabolite present in the EA fraction was as good as the standard antibiotic gentamicin used against Pseudomonas aeruginosa ATCC 27853 (t-test, P > 0.05, Fig. 6). Moreover, the EA fraction showed a significant ZOI against the yeast pathogen, C. albicans, in comparison to fluconazole (positive control) or DMSO (negative control) (Figs. 6 and 7, t-test, P < 0.05). These results showed that the extracellular crude extract from the fungal endophyte, isolate MUS1, contains antimicrobial metabolites.
Fig. 5.
Zone of inhibition (ZOI) produced by the ethyl acetate (EA) fraction (500 µg of crude extract/well) from the fungal isolate MUS1. The crude extract was extracted from fermented broth using Potato dextrose broth (PDB) as a culture medium using ethyl acetate (EA). The dried crude extract was dissolved in dimethyl sulfoxide (DMSO). Blank DMSO was taken as a Negative control. The EA fraction produced a bigger ZOI for the seven human pathogens tested, compared to the negative control. Data are presented as Mean ± SD (n = 3). A ZOI value of 6 (size of an agar-well ~ 6 mm) was assigned for the diameter of the agar-well for when no inhibition was seen. *The significant difference in the ZOI between EA extract and Negative control on the test pathogens at P < 0.05, meaning effective control of pathogen in comparison to the negative control
Fig. 6.
Zone of inhibition (ZOI) produced by the ethyl acetate (EA) fraction (500 µg of crude extract /well) isolated from the fungal isolate MUS1. The crude extract was extracted from fermented broth using Potato dextrose broth (PDB) as a culture medium using ethyl acetate (EA). The crude extract was dissolved in dimethyl sulfoxide (DMSO). Gentamicin (10 µg/disc) and fluconazole (25 µg/disc) were used as Positive controls against bacteria and yeast pathogens, respectively. Blank DMSO was taken as a Negative control. Except for Streptococcus and Staphylococcus spp., the EA fraction performed as well or better than the controls. Data are presented as Mean ± SD (n = 3). A ZOI value of 6 (size of an agar-well ~ 6 mm) was assigned for the diameter of the agar-well when no inhibition was seen. *The significant difference in the ZOI between Positive control (Gentamicin disc, 10 µg) and EA extract (500 µg) on the bacterial pathogens at P < 0.05 (has antibacterial activity but less potent as compared to the antibiotic used as positive control). **No significant difference in the ZOI between Positive control (Gentamicin disc, 10 µg) and EA extract (500 µg) on the bacterial pathogens at P > 0.05 (has antibacterial activity having similar potency to the antibiotic used as positive control). ***A significant difference in the ZOI between EA extract and Positive control (Fluconazole disc, 25 µg) on the yeast pathogen at P < 0.05 (has antifungal activity and more potent as compared to the antifungal used as positive control)
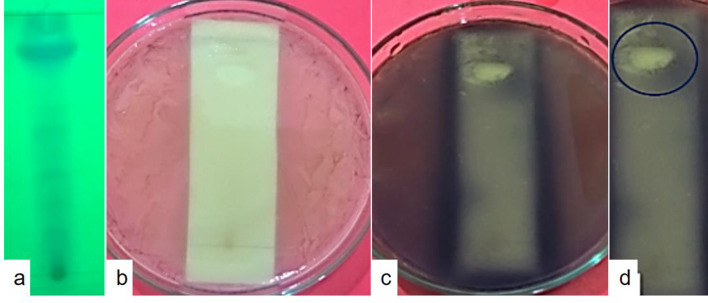
Fig. 7.
TLC-bioautography assay against Pseudomonas aeruginosa ATCC 27853. The active component in the crude extract responsible for antimicrobial activity was identified by the TLC-bioautography agar overlay method, which is a combination of contact and direct bioautography. a TLC profile of metabolites under long UV light. The crude extract was extracted from fermented broth using Potato dextrose broth (PDB) as a culture medium using ethyl acetate (EA). The EA crude extract ~ 10 µl was spotted onto TLC plates (Silica gel 60 F254, Merck) with at least 1 cm between the spots and placed in a solvent mixture of Chloroform: Methanol (9:1, v/v). Once the mobile phase reached the top, the TLC plate was removed, the solvent evaporated and observed under short and long UV. b Bioautography assay. The TLC plate was placed in a 90 mm Petri-plate and covered with molten Mueller–Hinton agar medium to a thickness of 2–3 mm. Once solid, the agar was overlaid with 106 CFU/ml of P. aeruginosa ATCC 27853. The plates were incubated at 37 ± 2 °C for 18–24 h. c Sprayed with MTT solution, (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide, 0.5 mg ml−1) and d Colorless inhibition zone (marked with a circle) against a purple background, showing the active component in the crude extract responsible for antimicrobial activity. Metabolically-active microorganisms convert the tetrazolium salt (MTT) into the corresponding intensely colored formazan (dehydrogenase activity of the microorganisms). As expected, dead bacteria cannot reduce the MTT, hence unable to produce the characteristic color change from yellowish to purple
The EA fraction that showed a potent antimicrobial activity during the AWD assay, was further analyzed for MIC against the human-pathogens, P. aeruginosa ATCC 27853, and C. albicans. The EA fraction of isolate MUS1, showed a MIC of 250 and 125 µg/ml against P. aeruginosa ATCC 27853 and C. albicans, respectively. The MIC for gentamicin against P. aeruginosa ATCC 27853 was 3.125 µg/ml. No standard MIC was analyzed for C. albicans as it was resistant to the antifungal agent, fluconazole.
The inhibitory activity of the EA fraction from isolate MUS1, as shown in the TLC bioautography assay, was attributed to one major constituent, spot 8 (Rf = 0.82), which gave a clear zone of inhibition seen against a purple background (Table 2 and Fig. 7).
Table 2.
TLC-bioautography assay against Pseudomonas aeruginosa ATCC 27853
| Spot number | Rf value | ZOI |
|---|---|---|
| 1 | 0.25 | |
| 2 | 0.34 | |
| 3 | 0.4 | |
| 4 | 0.45 | |
| 5 | 0.51 | |
| 6 | 0.55 | |
| 7 | 0.6 | |
| 8 | 0.82a | Positive |
| 9 | 0.98 |
a Inhibitory activity, after TLC separation of the ethyl acetate (EA) fraction from isolate MUS1, was attributed to one major constituent, spot 8 (Rf = 0.82), which gave a clear zone of inhibition (ZOI) seen against a purple background
Rf retardation factor
Antioxidant activity
The antioxidant activity of the EA fraction of isolate MUS1 was evaluated by the DPPH-radical scavenging assay. As shown in the graph (Fig. 8), an increase in the concentration of the EA fraction, increased the percentage of DPPH-radical inhibition. For example, at 100 µg/ml concentration, the % DPPH-radical scavenging activity was 83.15 ± 0.39, 81.62 ± 0.06, and 62.36 ± 0.29 for ascorbic acid, BHT, and the EA fraction from isolate MUS1, respectively (Fig. 8). Ascorbic acid and BHT are used as controls. Similarly, the concentration required for 50% inhibition of DPPH free radical (IC50) was 56.15, 62.87, and 81.52 µg/ml for ascorbic acid, BHT, and the EA fraction of isolate MUS1, respectively (Fig. 9).
Fig. 8.
DPPH-radical scavenging assay. The % scavenging ability of the ethyl acetate (EA) fraction from isolate MUS1, compared to the standard compounds, Ascorbic acid, and Butylated hydroxytoluene (BHT), is shown. The crude extract was extracted from fermented broth using Potato dextrose broth (PDB) as a culture medium using EA. Data are presented as Mean ± SD
Fig. 9.
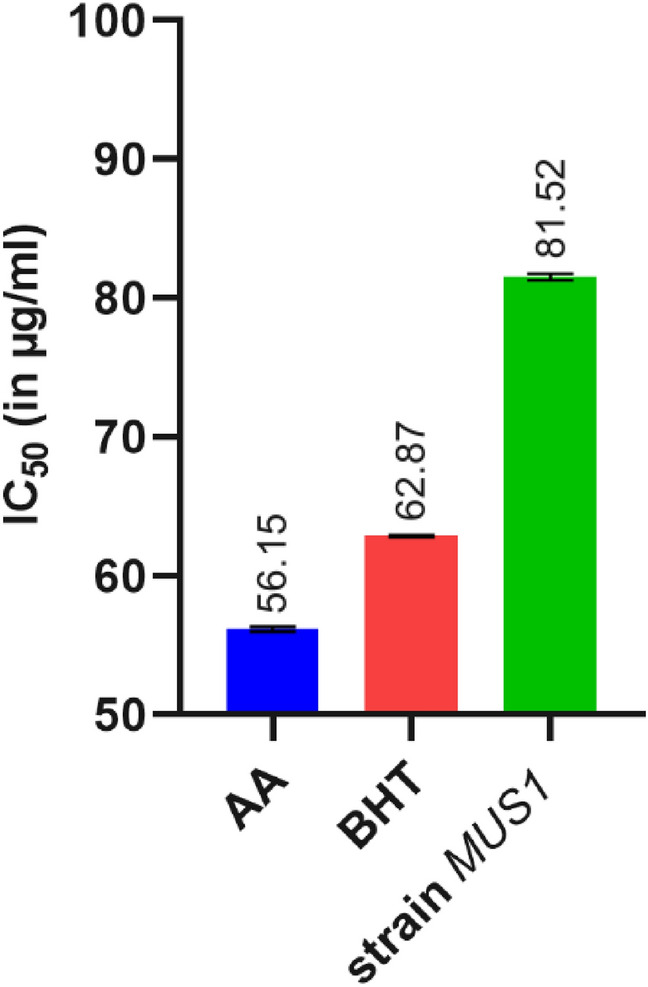
Graphical representation of the measure of half-maximal inhibitory concentration (IC50) for 50% DPPH scavenging activity (Mean ± SD), for Ascorbic acid (AA), Butylated hydroxytoluene (BHT) and isolate MUS1 under investigation
Total phenol and flavonoid content
The TPC and TFC were analyzed for extracts obtained from three different solvents and expressed in gallic acid equivalents (GAE) and rutin equivalents (RE) in µg per mg of dry fungal crude extract, respectively (Table 3). Both, the TPC and TFC, were higher in the EA extract compared to the CH and HX extracts. Moreover, taking into account the DPPH-radical scavenging activity of the EA extract (Fig. 8), it would suggest that the antioxidant activity of the EA extract could be due to phenolic and flavonoid metabolites present in the extract.
Table 3.
Total phenolic and flavonoid content for isolate MUS1
| Solvent fraction | Total phenolics (µg GAE/mg dry extract) | Total flavonoids (µg RE/mg dry extract) |
|---|---|---|
| Ethyl Acetate | 16.90 ± 0.075 | 11.59 ± 0.148 |
| Chloroform | 10.06 ± 0.121 | 10.57 ± 0.074 |
| Hexane | 0.93 ± 0.068 | 1.97 ± 0.074 |
Values are means of three biological replicates (n = 3)
Taxol analysis
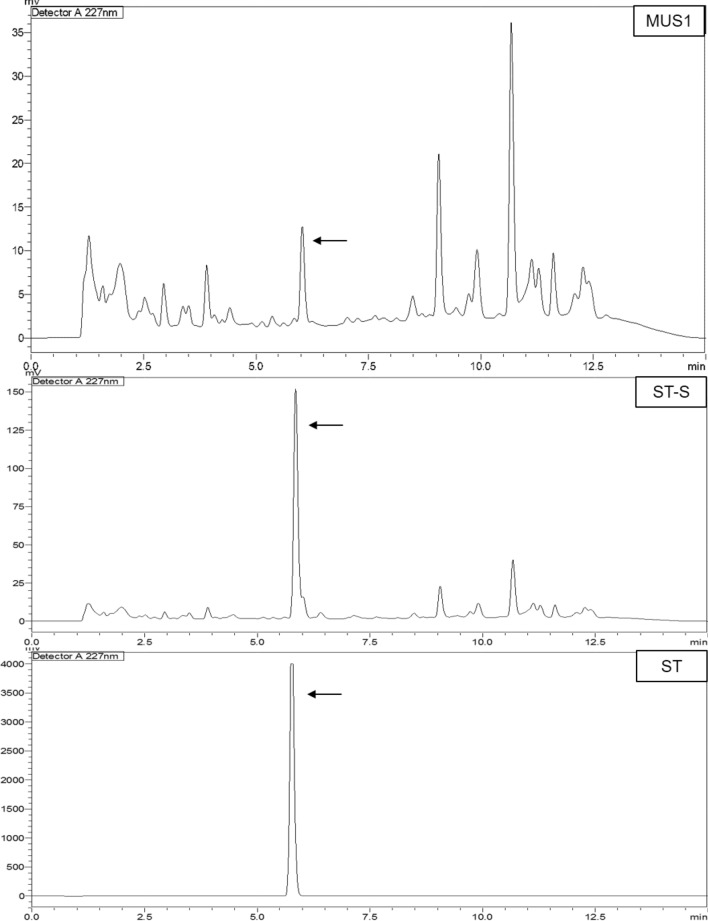
TLC analysis of the EA fraction of isolate MUS1, showed two putative fungal-Taxol spots (Rf = 0.75 and 0.76) sharing the same Rf value as the Taxol standard (Rf = 0.75) (Table 4). All three spots, which were scrapped and purified from the TLC plate, were further analyzed by RP-HPLC equipped with a detector system (UV 227 nm). Both, the authentic Taxol standard as well as the sample from isolate MUS1, had a retention time of 5.85 min (Fig. 10). The concentration of Taxol obtained was calculated to be 282.05 µg/l of fermentation broth.
Table 4.
TLC analysis of the ethyl acetate fraction for Taxol production by isolate MUS1
| Spot number | Rf values | |
|---|---|---|
| MUS1 | Taxol standard (ST-S) | |
| 1 | 0.18 | |
| 2 | 0.31 | |
| 3 | 0.75a | 0.75a |
| 4 | 0.76a | |
aPutative fungal-Taxol spots (0.75 and 0.76) along with the Taxol standard, were scraped from the TLC plate and further subjected to reverse phase-HPLC analysis
Rf retardation factor
Fig. 10.
HPLC chromatograms showing the same retention time (5.85 min, shown with arrow) for the ST, ST-S, and the MUS1 spot obtained in the TLC assay. The crude extract was extracted from fermented broth using Potato dextrose broth (PDB) as a culture medium using ethyl acetate (EA). The EA-extracted crude extract was run on TLC plates against standard Taxol (ST—directly prepared by dissolving standard Taxol powder in methanol). Both spots ST-S (an ST co-run with MUS1, scrapped from TLC and membrane filtered later) and MUS1 were scrapped from the silica, membrane filtered, and analyzed using reverse-phase HPLC (RP-HPLC). ST and ST-S are the authentic Taxol standard while MUS1 corresponds to the putative Taxol compound obtained from the EA fraction of isolate MUS1 (also scrapped from TLC and membrane filtered). A similar chromatogram before and after the Taxol peak in MUS1 and ST-S represents the fingerprints of scrapped silica
Discussion
Forest trees and Medicinal plants confer the largest niche for endophytic fungal diversity (Schueffler and Anke 2011). The endophytic fungi belong to ascomycetes, basidiomycetes, zygomycetes, or oomycetes. It is known that endophytic fungi isolated from plants show antimicrobial and antioxidant activities (Gupta et al. 2020; Pan et al. 2017; Sharma et al. 2016; Yashavantha Rao et al. 2015), including other bioactivities such as enzyme activity (Devi et al. 2012), volatile biocontrol agents (Strobel et al. 2001) and volatile metabolites having fuel potentials (Singh et al. 2011; Strobel et al. 2008; Tomsheck et al. 2010). Moreover, endophytic fungi can produce similar or unique bioactive secondary metabolites compared to host plant metabolites including Taxol (Schueffler and Anke 2011; Shrestha et al. 2001; Strobel and Daisy 2003; Venieraki et al. 2017). The increasing demand for secondary metabolites in international markets poses a severe threat to many plant species (Gupta et al. 2020). However, having the ability to produce plant secondary metabolites or similar analogs by endophytic fungi they can be considered as a unique bioresource for the production of secondary metabolites alternative to plant metabolites.
The Himalayan yew, Taxus wallichiana Zucc. is a medicinal plant, a natural source of the anticancer drug Taxol and its related analogs (Wani et al. 1971). It also harbors several fungal endophytes, some of which have been reported to produce Taxol in the artificial medium (Flores-Bustamante et al. 2010; Zhou et al. 2010). Chemodiversity, ethnobotanical use of the plant, and region from where the plant is obtained are the important criteria for the isolation of endophytes to explore them for novel compounds with potential bioactivity (Dhayanithy et al. 2019). Despite the relatively large number of studies that have been done on endophytic fungi isolated from T. wallichiana, the focus has been on Taxol and, therefore, little information is available on other bioactive metabolites including antimicrobial and antioxidant properties of metabolites produced by these fungal isolates. Therefore, in the current study, we have studied the endophytic fungal population from the T. wallichiana plant for its bioactive properties and potential for Taxol production.
From 1993 to 2019, several fungal endophytes are reported. Alternaria, Aspergillus, Chaetomium, Colletotrichum, Fusarium, Rhizoctonia, Penicillium, Pestalotiopsis, Phoma, Phomopsis, Rhizopus, Talaromyces, Trichoderma, Xylaria, etc. species represent the major isolated endophytic fungi from several plant species (Gupta et al. 2020). In our current study, in addition to Annulohypoxylon sp. strain MUS1, other fungal genera isolated and identified included Alternaria sp., Aspergillus sp., Fusarium sp., Mucor sp., Penicillium sp., and Trichoderma sp., which is consistent with the published literature on the diversity of endophytic fungi isolated from Taxus wallichiana (Flores-Bustamante et al. 2010; Gupta et al. 2020; Kusari et al. 2014). Some of the isolates in the current study remained unidentified as many of the endophytic fungal isolates are not able to sporulate in artificial media and are thus categorized as ‘sterile fungi’ or ‘sterile mycelia’, and such isolates are difficult to identify at genus or species level. Hence, fungal barcoding (ITS sequencing) is more reliable compared to phenotypic identification, as morphological characters are unstable and may be affected by environmental factors (Gupta et al. 2020).
Recently, it was reported that crude extracts obtained with ethyl acetate from a fermented broth of Fusarium sp. isolated from T. baccata showed significant antimicrobial activity against all pathogens tested (i.e., Bacillus subtilis, Staphylococcus epidermis, Staphylococcus aureus, Klebsiella pneumoniae, Shigella flexneri, Escherichia coli, Candida albicans, and Candida tropicalis) (Tayung and Jha 2010). Similarly, among the 16 endophytic fungi isolated during our study, Annulohypoxylon sp. strain MUS1 showed a potential for the production of antimicrobial and antioxidant metabolites. The AWD assay used in our study alluded to the presence of a metabolite or group of metabolites in the EA fraction that had antimicrobial activity against seven different human pathogens, which included Streptococcus faecalis, Staphylococcus aureus, Bacillus subtilis, Escherichia coli, Salmonella enterica, Pseudomonas aeruginosa, and the yeast Candida albicans. Natural products have served as powerful therapeutics against pathogenic bacteria since the golden age of antibiotics of the mid-twentieth century. However, the increasing frequency of antibiotic-resistant infections demonstrates the urgency of new antibiotics are critical for modern medicine (Rossiter et al. 2017). In such scenarios, natural metabolites from endophytic fungi, such as isolate MUS1 metabolites, can be a boon to fight antibiotic resistance in the coming days. Given the importance of natural antimicrobial metabolite, agar overlay bioautography was carried out to isolate the bioactive component responsible for antimicrobial activity from the mixture of metabolites in EA extract. Zones of microbial growth inhibition are visualized with the aid of a dehydrogenase activity-detecting reagent (tetrazolium salts). Metabolically active microorganisms convert the tetrazolium salt (MTT) into the corresponding intensely colored formazan attributed to the dehydrogenase activity of the microorganisms. Hence, colorless zone/s of bioactive component/s are visualized against the colored background (Dewanjee et al. 2015). One can isolate, purify, and characterize the bioactive component from the mixture of secondary metabolites from the combined use of bioautography and TLC (Fig. 7, Table 2).
Along with antimicrobial activity, fungal endophytes also show good promise as sources for metabolites having antioxidant capacities. For example, it was recently reported that fungal endophytes isolated from the bulbs of Fritillaria unibracteata var. wabuensis, have a new potential as a natural resource for antioxidant metabolites (Pan et al. 2017). Likewise, in our study the radical scavenging assay using DPPH showed that the antioxidant activity of the EA fraction from isolate MUS1, is comparable to standard antioxidants, viz., ascorbic acid, and BHT. The ability to scavenge DPPH free radicals indicates the potential use of the secondary metabolites as protective agents against cellular damage, as free radicals damage cell structures through reaction with nucleic acids, lipids, and proteins (Marcelo et al. 2018). The free radical scavengers from endophyte MUS1 thus play an important role in maintaining the integrity and functions of biomolecules of cells. The EA fraction of isolate MUS1 also showed the greatest amount for phenolics and flavonoids content (Table 3). Naturally occurring phenolic and flavonoid metabolites are of great interest to the pharmaceutical industries as well as the food or cosmetic industries, as these metabolites exhibit many biological functions including anti-inflammatory, antiallergic, hepatoprotective, antithrombotic, antiviral, anticarcinogenic, and vasodilatory actions (Baba and Malik 2015; Kowalska et al. 2017). Although there is a report for a Hypoxylon sp., isolated from Persea indica, producing volatile organic metabolites that have potential as fuels or fuel additives (Tomsheck et al. 2010), to our knowledge, this is the first report on the isolation and related bioactivities of an Annulohypoxylon sp. from Taxus spp.
Since the first report in 1993 for endophytic-Taxol production, many studies have been published showing that many of the endophytic fungi isolated from Taxus spp. are also capable of producing Taxol (Flores-Bustamante et al. 2010; Gupta et al. 2020; Zhou et al. 2010). Therefore, the 16 endophytic fungi isolated during our study were also evaluated for the production of Taxol using TLC against standard Taxol. Only one fungus, isolate MUS1, showed the ability for the production of Taxol, and this was confirmed using RP-HPLC analysis. The concentration of Taxol obtained was calculated to be 282.05 µg/l, which agrees with the published values for other endophytic fungi able to produce Taxol (Flores-Bustamante et al. 2010; Zhou et al. 2010).
Production of secondary metabolites is influenced by fermentation parameters i.e., culture medium, available nutrients, pH, temperature, salinity, and glucose concentrations (Marcelo et al. 2018). Given that metabolite production can vary according to culture conditions, the fungal isolate MUS1 was not discarded yet and will continue to be evaluated for the production and characterization of bioactive metabolites.
Conclusion
Among the 16 endophytic fungal isolates, the Annulohypoxylon sp. strain MUS1 not only showed the ability to produce anticancer compound Taxol, but also the production of antimicrobial and antioxidant metabolites. The EA crude extract of the isolate MUS1, significantly inhibited the fluconazole-resistant pathogenic yeast, Candida albicans, as well as has an inhibitory effect on a range of Gram-positive and Gram-negative bacterial pathogens and has potent radical scavenging activity. Once these bioactive metabolites have been identified, they could be further developed to be used by the pharmaceutical and food industries. Though further analysis is required to elucidate the molecular structure of the metabolites present in isolate MUS1, our results underscore the importance of preserving the Himalayan yew in Nepal, as they harbor microorganisms that could be the source of potent bioactive metabolites in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
We would like to acknowledge the Department of Biotechnology, Kathmandu University (KU), Nepal for the facilitation of the research works. We would also like to acknowledge Mr. Basant Awasthi, Teaching Assistant, Department of Geomatics Engineering, KU for his contribution to making the location map of Mustang. Further, we want to thank Prof. Dr. Janardan Lamichhane, Department of Biotechnology, KU, and Associate Professor Dr. Rajani Shakya, Department of Pharmacy, KU for the HPLC analysis.
Abbreviations
- ATCC
American Type Culture Collection
- BLAST
Basic Local Alignment Search Tool
- BLASTn
Nucleotide BLAST
- HIV
Human immunodeficiency virus
- MEGAX
Molecular Evolutionary Genetics Analysis Version X
- NCBI
National Center for Biotechnology Information
- UV–Vis
Ultraviolet–Visible
Author contributions
DPG designed the study, carried out the majority of research activities, wrote the Manuscript. HV and AA equally contributed to conducting research activities and manuscript editing. JO, KL, ME are involved in molecular analysis of isolated and purified endophytes and editing the manuscript. MRGG participated in designing the research, guiding DPG to carry out the research, and provided financial support. All authors read and approved the final manuscript.
Funding
We are grateful to the Swedish Research Council for their grant (Ref/348-2012-6138/Agreement/C0613801) for this project.
Compliance with ethical standards
Ethics approval and consent to participate
Not applicable.
Conflict of interest
The authors declare that they have no conflicts of interest in the publication.
Accession Numbers
The sequence obtained in this study was deposited in the NCBI GenBank database (Accession number: MN699475).
References
- Baba SA, Malik SA. Determination of total phenolic and flavonoid content, antimicrobial and antioxidant activity of a root extract of Arisaema jacquemontii Blume. J Taibah Univ Sci. 2015;9(4):449–454. doi: 10.1016/j.jtusci.2014.11.001. [DOI] [Google Scholar]
- Barnett HL, Hunter BB. Illustrated genera of imperfect fungi. St. Paul: American Phytopathological Society (APS Press); 1998. [Google Scholar]
- Bascom-Slack CA, Ma C, Moore E, Babbs B, Fenn K, Greene JS, et al. Multiple, novel biologically active endophytic actinomycetes isolated from upper Amazonian rainforests. Microb Ecol. 2009;58(2):374–383. doi: 10.1007/s00248-009-9494-z. [DOI] [PubMed] [Google Scholar]
- Devi NN, Prabakaran JJ, Wahab F. Phytochemical analysis and enzyme analysis of endophytic fungi from Centella asiatica. Asian Pac J Trop Biomed. 2012;2(3, Supplement):S1280–S1284. doi: 10.1016/S2221-1691(12)60400-6. [DOI] [Google Scholar]
- Dewanjee S, Gangopadhyay M, Bhattacharya N, Khanra R, Dua TK. Bioautography and its scope in the field of natural product chemistry. J Pharm Anal. 2015;5(2):75–84. doi: 10.1016/j.jpha.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayanithy G, Subban K, Chelliah J. Diversity and biological activities of endophytic fungi associated with Catharanthus roseus. BMC Microbiol. 2019;19(1):22. doi: 10.1186/s12866-019-1386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Bustamante ZR, Rivera-Orduña FN, Martínez-Cárdenas A, Flores-Cotera LB. Microbial paclitaxel: advances and perspectives. J Antibiot. 2010;63(8):460–467. doi: 10.1038/ja.2010.83. [DOI] [PubMed] [Google Scholar]
- Garyali S, Kumar A, Reddy MS. Taxol production by an endophytic fungus, Fusarium redolens, isolated from Himalayan yew. J Microbiol Biotechnol. 2013;23(10):1372–1380. doi: 10.4014/jmb.1305.05070. [DOI] [PubMed] [Google Scholar]
- Głowniak K, Mroczek T. Investigations on preparative thin-layer chromatographic separation of taxoids from Taxus baccata L. J Liq Chromatogr Relat Technol. 1999;22(16):2483–2502. doi: 10.1081/JLC-100101816. [DOI] [Google Scholar]
- Gupta S, Chaturvedi P, Kulkarni MG, Van Staden J. A critical review on exploiting the pharmaceutical potential of plant endophytic fungi. Biotechnol Adv. 2020;39:107462. doi: 10.1016/j.biotechadv.2019.107462. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wang J, Li G, Zheng Z, Su W. Antitumor and antifungal activities in endophytic fungi isolated from pharmaceutical plants Taxus mairei, Cephalataxus fortunei and Torreya grandis. FEMS Immunol Med Microbiol. 2001;31(2):163–167. doi: 10.1111/j.1574-695X.2001.tb00513.x. [DOI] [PubMed] [Google Scholar]
- Kowalska H, Czajkowska K, Cichowska J, Lenart A. What's new in biopotential of fruit and vegetable by-products applied in the food processing industry. Trends Food Sci Technol. 2017;67:150–159. doi: 10.1016/j.tifs.2017.06.016. [DOI] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari S, Singh S, Jayabaskaran C. Rethinking production of Taxol(R) (paclitaxel) using endophyte biotechnology. Trends Biotechnol. 2014;32(6):304–311. doi: 10.1016/j.tibtech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Li D-W. Microscopic methods for analytical studies of fungi. In: Gupta VK, Tuohy MG, Ayyachamy M, Turner KM, O’Donovan A, editors. Laboratory protocols in fungal biology: current methods in fungal biology. New York: Springer; 2013. pp. 113–131. [Google Scholar]
- Li D, Fu D, Zhang Y, Ma X, Gao L, Wang X, et al. Isolation, purification, and identification of taxol and related taxanes from taxol-producing fungus Aspergillus niger subsp. taxi. J Microbiol Biotechnol. 2017;27(8):1379–1385. doi: 10.4014/jmb.1701.01018. [DOI] [PubMed] [Google Scholar]
- Liu K, Ding X, Deng B, Chen W. Isolation and characterization of endophytic taxol-producing fungi from Taxus chinensis. J Ind Microbiol Biotechnol. 2009;36(9):1171–1177. doi: 10.1007/s10295-009-0598-8. [DOI] [PubMed] [Google Scholar]
- Marcelo A, Geronimo RM, Vicente CJB, Callanta RBP, Bennett RM, Ysrael MC, Dedeles GR. TLC screening profile of secondary metabolites and biological activities of Salisapilia tartarea S1YP1 isolated from philippine mangroves. J Oleo Sci. 2018;67(12):1585–1595. doi: 10.5650/jos.ess18129. [DOI] [PubMed] [Google Scholar]
- Pan F, Su TJ, Cai SM, Wu W. Fungal endophyte-derived Fritillaria unibracteata var. wabuensis: diversity, antioxidant capacities in vitro and relations to phenolic, flavonoid or saponin compounds. Sci Rep. 2017;7:42008. doi: 10.1038/srep42008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja HA, Miller AN, Pearce CJ, Oberlies NH. Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod. 2017;80(3):756–770. doi: 10.1021/acs.jnatprod.6b01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossiter SE, Fletcher MH, Wuest WM. Natural products as platforms to overcome antibiotic resistance. Chem Rev. 2017;117(19):12415–12474. doi: 10.1021/acs.chemrev.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueffler A, Anke T. Antimicrobial Compounds from Tree Endophytes. In: Pirttilä AM, Frank AC, editors. Endophytes of forest trees: biology and applications. Dordrecht: Springer; 2011. pp. 265–294. [Google Scholar]
- Schwalbe R, Steele-Moore L, Goodwin AC. Antimicrobial susceptibility testing protocols. 1. Boca Raton: CRC Press; 2007. [Google Scholar]
- Sharma D, Pramanik A, Agrawal PK. Evaluation of bioactive secondary metabolites from endophytic fungus Pestalotiopsis neglecta BAB-5510 isolated from leaves of Cupressus torulosa D.Don. 3 Biotech. 2016;6(2):210. doi: 10.1007/s13205-016-0518-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha K, Strobel GA, Shrivastava SP, Gewali MB. Evidence for paclitaxel from three new endophytic fungi of Himalayan yew of Nepal. Planta Med. 2001;67(4):374–376. doi: 10.1055/s-2001-14307. [DOI] [PubMed] [Google Scholar]
- Singh SK, Strobel GA, Knighton B, Geary B, Sears J, Ezra D. An endophytic Phomopsis sp. possessing bioactivity and fuel potential with its volatile organic compounds. Microb Ecol. 2011;61(4):729–739. doi: 10.1007/s00248-011-9818-7. [DOI] [PubMed] [Google Scholar]
- Stierle A, Strobel G, Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science. 1993;260(5105):214–216. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 2003;67(4):491–502. doi: 10.1128/mmbr.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G, Yang X, Sears J, Kramer R, Sidhu RS, Hess WM. Taxol from Pestalotiopsis microspora, an endophytic fungus of Taxus wallachiana. Microbiology (Reading) 1996;142(Pt 2):435–440. doi: 10.1099/13500872-142-2-435. [DOI] [PubMed] [Google Scholar]
- Strobel GA, Dirkse E, Sears J, Markworth C. Volatile antimicrobials from Muscodor albus, a novel endophytic fungus. Microbiology (Reading) 2001;147(Pt 11):2943–2950. doi: 10.1099/00221287-147-11-2943. [DOI] [PubMed] [Google Scholar]
- Strobel GA, Knighton B, Kluck K, Ren Y, Livinghouse T, Griffin M, et al. The production of myco-diesel hydrocarbons and their derivatives by the endophytic fungus Gliocladium roseum (NRRL 50072) Microbiology. 2008;154(11):3319–3328. doi: 10.1099/mic.0.2008/022186-0. [DOI] [PubMed] [Google Scholar]
- Tayung K, Jha DK. Antimicrobial endophytic fungal assemblages inhabiting bark of Taxus baccata L. of Indo-Burma mega biodiversity hotspot. Indian J Microbiol. 2010;50(Suppl 1):74–81. doi: 10.1007/s12088-010-0056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsheck AR, Strobel GA, Booth E, Geary B, Spakowicz D, Knighton B, et al. Hypoxylon sp., an endophyte of Persea indica, producing 1,8-cineole and other bioactive volatiles with fuel potential. Microb Ecol. 2010;60(4):903–914. doi: 10.1007/s00248-010-9759-6. [DOI] [PubMed] [Google Scholar]
- Venieraki A, Dimou M, Katinakis P. Endophytic fungi residing in medicinal plants have the ability to produce the same or similar pharmacologically active secondary metabolites as their hosts. Hellenic Plant Protect J. 2017 doi: 10.1515/hppj-2017-0006. [DOI] [Google Scholar]
- Wani MC, Taylor HL, Wall ME, Coggon P, McPhail AT. Plant antitumor agents. VI. Isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J Am Chem Soc. 1971;93(9):2325–2327. doi: 10.1021/ja00738a045. [DOI] [PubMed] [Google Scholar]
- Watanabe T. Pictorial atlas of soil and seed fungi: morphologies of cultured fungi and key to species. 3. Boca Raton: CRC Press; 2010. [Google Scholar]
- Wijeratne EM, Bashyal BP, Gunatilaka MK, Arnold AE, Gunatilaka AA. Maximizing chemical diversity of fungal metabolites: biogenetically related Heptaketides of the endolichenic fungus Corynespora sp. (1) J Nat Prod. 2010;73(6):1156–1159. doi: 10.1021/np900684v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashavantha Rao HC, Santosh P, Rakshith D, Satish S. Molecular characterization of an endophytic Phomopsisliquidambaris CBR-15 from Cryptolepis buchanani Roem. and impact of culture media on biosynthesis of antimicrobial metabolites. 3 Biotech. 2015;5(2):165–173. doi: 10.1007/s13205-014-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhu H, Liu L, Lin J, Tang K. A review: recent advances and future prospects of taxol-producing endophytic fungi. Appl Microbiol Biotechnol. 2010;86(6):1707–1717. doi: 10.1007/s00253-010-2546-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.