Abstract
The Ziziphus species are naturally tolerant to a range of abiotic stresses. Therefore, it is expected that they are an enriched source of genes conferring stress tolerance. Heat shock proteins (Hsps) play a significant role in plants in imparting tolerance against abiotic stress conditions. To get an insight into potential Hsp function in Ziziphus, we performed a genome-wide analysis and expression study of Hsp70 and Hsp100 gene families in Ziziphus jujuba. We identified 21 and 6 genes of the ZjHsp70 and ZjHsp100 families, respectively. Physiochemical properties, chromosomal location, gene structure, motifs, and protein domain organization were analysed for structural and functional characterization. We identified the contribution of tandem and segmental gene duplications in expansions of ZjHsp70s and ZjHsp100s in Z. jujuba. Promoter analysis suggested that ZjHsp70s and ZjHsp100s perform diverse functions related to abiotic stress. Furthermore, expression analyses revealed that most of the Z. jujuba Hsp genes are differentially expressed in response to heat, drought, and salinity stress. Our analyses suggested ZjHsp70-3, ZjHsp70-5, ZjHsp70-6, ZjHsp70-16, ZjHsp70-17, ZjHsp70-20, ZjHsp100-1, ZjHsp100-2, and ZjHsp100-3 are potential candidates for further functional analysis and with regard to breeding new more resilient strains. The present analysis laid the foundation for understanding the molecular mechanism of Hsps70 and Hsp100 gene families regulating abiotic stress tolerance in Z. jujuba.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12192-020-01179-w.
Keywords: Genome-wide analysis, Heat shock protein (Hsp), Gene family, Expression analysis, qRT-PCR, Z. jujuba
Introduction
In recent decades, abiotic stress has caused severe economic losses of crop production globally. Abiotic stress hampers the biochemical and physiological processes of crop plants, which negatively affects their growth and development, thus cumulatively affecting crop yield as well as food quality (Wahid 2007; Hasanuzzaman et al. 2013). The global climate model has proposed that temperatures will rise by 1.4 to 5.8 °C by the end of the twenty-first century due to the increase in atmospheric greenhouse gases (Planton et al. 2016). So far, the progressively changing climate has affected food security (Coumou and Rahmsdorf 2012), mainly in South Asia and sub-Saharan Africa, but as well on the global level (Battisti and Naylor 2009). As the sessile nature of crop plants means that they cannot relocate to avoid stress, they have therefore evolved molecular tolerance mechanisms to stressful conditions (Khan et al. 2019). Heat shock proteins (Hsps) are produced in response to stress as a molecular defence mechanism that is very important for the survival and growth of plants (Hasanuzzaman et al. 2013). Members of the Hsp gene family play significant roles in developmental processes in addition to abiotic stress conditions such as drought, heat, salinity, cold, and heavy metal toxicity (Khan et al. 2019). The Hsps have been associated with responses to the infection by pathogens such as nematodes, playing a role in programmed cell death (PCD) during the remodelling of leaves (Maimbo et al. 2007; Rowarth et al. 2020). The Hsps act as essential molecular chaperones in the transport and assembly of nascent proteins, refolding, and exclusion of denatured proteins in a cell under abiotic stress conditions and enhance tolerance (Hasanuzzaman et al. 2013). They are classified into five major families: Hsp100, Hsp90, Hsp70, Hsp60, and small Hsp (10–40 kDa) based on their molecular weight (Khan et al. 2019; Hasanuzzaman et al. 2013).
The Hsp70 protein is a central part of the cellular network of folding catalysts and chaperones (Mayer and Bukau 2005). It contains two important functional domains, NBD (N-terminal ATPase domain) of ~ 44-kDa, and a ~ 18-kDa SBD (substrate-binding domain) (Dragovic et al. 2006). The various roles of the Hsp70 gene family have been characterised in several plants. For instance, in Arabidopsis thaliana, the cpHsc70-1 mutant showed defective phenotypes when germinated seeds were subjected to heat stress (Su and Li 2008). The M. sativa MsHsp70 was induced by heat stress and transgenic lines of A. thaliana overexpressing MsHsp70 have enhanced tolerance to drought stress (Li et al. 2017). Hsp70s have also been demonstrated to play significant functions under stress condition in wheat (Duan et al. 2011), cucumber (Li et al. 2014), pepper (Guo et al. 2014), and rubber trees, etc. (Zhang et al. 2009). Similarly, the Hsp100/Clp (Caseinolytic Protease) proteins belong to the AAA+ ATPase superfamily and these chaperones are also crucial in modulating plant thermotolerance traits (Mishra and Grover 2016). Hsp100 proteins contain several domains (amino (N)–terminal, nucleotide-binding domain 1 (NBD1), middle domain, nucleotide-binding domain 2 (NBD2), and a carboxyl (C)–terminal domain). This gene family is divided into ClpATPases class I (ClpA, ClpB, ClpC, ClpD, ClpE) with two ATP binding domains and ClpATPases class II (ClpM, ClpN, ClpX, ClpY) with a single ATP binding domain (Mishra and Grover 2016). The A. thaliana genome contains 7 class-1 Clp/Hsp100 protein-encoding genes including 4 ClpB proteins 2 ClpC, 1 ClpD protein, but no members of the ClpA or the ClpE subclass (Lee et al. 2007). Mutants of A. thaliana, rice, and maize plants with defective Hsp100 expression showed severe sensitivity to heat stress (Hong and Vierling 2000; Nieto-Sotelo et al. 2002; Lin et al. 2014). Additionally, overexpression of Hsp100 in A. thaliana and rice increased plant thermotolerance (Queitsch et al. 2000; Katiyar-Agarwal et al. 2003).
Ziziphus is native to Asia and mainly found in central and Southwest Asia, central India, and China (Outlaw et al. 2002; Pandey et al. 2010; Sabir et al. 2020). The Ziziphus species including Z. jujuba (Chinese jujuba) and Z. nummularia are well adapted to the arid and semiarid region and tolerant to a range of abiotic stresses including heat, drought, salinity, chilling, and metal toxicity (Pandey et al. 2010; Sabir et al. 2020). Therefore, they are expected to house a repository of genes responsible for abiotic stress tolerance (Pandey et al. 2010; Padaria et al. 2016; Panzade et al. 2020). For example, transcriptomic profiling in Z. nummularia identified several drought-responsive genes (Yadav et al. 2018a; Yadav et al. 2018b). Furthermore, the heat stress-inducible isoform ZnJClpB1-C of Hsp100 and the abscisic acid-stress-ripening gene (ASR) from Z. nummularia have been shown to enhance heat stress tolerance in N. tabacum and drought stress tolerance in E.coli cells (Padaria et al. 2016; Panzade et al. 2020). Similarly, SbHsp70-1 from the stress-tolerant cereal crop Sorghum bicolor also confers thermal tolerance in E. coli (Mulaudzi-Masuku et al. 2015).
In this study, we surveyed the newly available genome sequence of Z. jujuba for Hsp70 and Hsp100 gene family members and conducted stress challenge experiments as the first step in identifying the role of these Hsp families in stress tolerance.
Materials and methods
Genome-wide identification of Hsp70 and Hsp100 gene family members in Z. jujuba
The Hsp70 and Hsp100 gene family members in Z. jujuba were identified by performing a BLASTP search against Z. jujuba genome database (https://www.genome.jp/kegg-bin/show_organism?org=zju) by using orthologous amino acid sequences of Arabidopsis thaliana, Rhamnella rubrinervis, and Cannabis sativa, as queries (e-value of 1 × 10−5 and identity of > 60% as the threshold). The Hidden Markov model (HMM) of the Hsp70 and Hsp100 protein family also used to search the candidate amino acid sequences. All of the retrieved amino acid sequences were analysed for the presence of the Hsp70 and Hsp100 conserved protein domains using SMART (http://smart.embl-heidelberg.de/) tool (Letunic et al. 2012). Protein sequences with redundancy and truncated domains were removed. Then, the coding sequences (CDS), genomic sequences, amino acid sequences, chromosomal location data, and nucleotide sequences of 2000-bp upstream from the translation initiation codon were retrieved from the Z. jujuba genome database (Table S1).
Predicted analysis of physico-chemical properties and sub-cellular localization
The full-length protein coding sequences of Hsp70 and Hsp100 gene family members were analysed in silico for various physico-chemical parameters (amino acid composition, theoretical isoelectric point (pI), molecular weight, grand average of hydropathicity (GRAVY), aliphatic index, etc.) using the Expasy ProtParam tool (http://us.expasy.org/tools/protp aram.html) (Garg et al. 2016). Sub-cellular localization was predicted using CELLO v.2.5 (http://cello.life.nctu.edu.tw/cello2go/alignment.php) (Yu et al. 2014).
Analyses of gene structure, motifs, and domain architecture
The intron/exon structure of the Z. jujuba Hsp70 and Hsp100 gene family was analysed based on the alignment of genomic DNA sequences with corresponding CDS sequences, by using the Genes Structure Display Server tool (GSDS, http://gsds.cbi.pku.edu.cn/index.php) (Hu et al. 2015). The conserved motifs of Hsp70 and Hsp100 protein families were identified using amino acid sequences in the MEME tool (Multiple Expectation Maximization for Motif Elicitation, http://meme-suite.org/tools /meme). The parameters were used as follows: Optimum motifs: 6 to 50 amino acid residues, the maximum number of motifs: 20, a number of repetitions: any (Bailey and Elkan 1995). Domains of Hsps protein were analysed by using the SMART tool (http://smart.embl-heidelberg.de/) (Letunic et al. 2012).
Microsatellite mining and identification
The SSR Locator (http://microsatellite.org/ssr.php?info) software was used to extract microsatellites (Da Maia et al. 2008) with mono-, di-, tri-, tetra-, penta, hepta-, octa-, nova-, or decanucleotide motifs. SSRs with a total length of ≥ 20 bp with the parameters described by Gao et al. (2011) were used to identify potential SSRs across the ZjHsp70 and ZjHsp100 genes.
Multiple sequence alignment and phylogenetic analysis
In the present study, two phylogenetic trees were constructed to classify and assess the phylogenetic relationships of the Z. jujuba Hsp70 and Hsp100 gene families. The first tree was limited solely to Z. jujuba Hsp70 and Hsp100 amino acid sequences and the second contained Hsp70 and Hsp100 amino acid sequences from Ziziphus jujuba, Arabidopsis thaliana, Cannabis sativa, and Rhamnella rubrinervis (retrieved from NCBI and TAIR database (Table S2)). Multiple sequence alignments of all Hsp70 and Hsp100 amino acid sequences were carried out using ClustalW with default parameters (Larkin et al. 2007). The phylogenetic trees were constructed using the Neighbour-Joining (NJ) method implemented in MEGA 7.0.26 software, with a bootstrap value 1000 (Tamura et al. 2013).
Chromosomal locations and duplication of Hsp70 and Hsp100 in Z. jujuba
The chromosomal localization of Hsp70 and Hsp100 genes family was analysed using MapChart software (Voorrips 2002), according to the chromosomal position data available at the Ziziphus database. Coding protein pairs with ≥ 50% identity and covering ≥ 90% protein length were considered as duplicated genes (Wang et al. 2017). If genes from the same species were placed in the same clade of the tree, these were further analysed to identify whether the potential duplication was of the tandem or segmental type. If the paralogous pair was separated by five or fewer genes within a 100-kb region, they were designated as tandemly duplicated genes. If the paralogs were separated by > 5 genes or they were mapped onto duplicated chromosomal blocks, they were designated as segmental duplications (Zhang and Li 2018; Liu et al. 2018; Zhang et al. 2015a, b).
Promoter analysis of Hsp70 and Hsp100 gene families
Potential cis-elements of the promoter regions were analysed in the 2000-bp sequences upstream of the TSS (transcriptional start site) of each of the Hsp70 and Hsp100 gene using the PLACE database (http://www.dna.affrc.go.jp/PLACE/).
Plant material, growth conditions, and stress treatments
Seeds of Indian jujuba variety “Maharwali” were sterilised for 10 min using a 10% hypochlorous acid and rinsed with distilled water three times. Upon scarification, seeds were grown in a growth chamber (at National Phytotron Facility, Indian Agricultural Research Institute, New Delhi, India) maintained at 22 °C, 16-h light/8-h dark photoperiod, RH 60–70%. One-month-old seedlings were exposed to various stresses. The heat stress at 42 ± 1 °C was produced by raising the temperature at 1 °C per 10 min until the temperature reaches 42 °C and the same temperature was continued for 2 h (Vishwakarma et al. 2018). The drought and salinity stress treatments were produced by seedlings being transferred to 400 mM mannitol or 300 mMNaCl for 5 h. The seedlings, which were grown under normal condition and kept in water for the same duration at 25 ± 2 °C, were used as controls (Singh et al. 2016; Liu et al. 2018). Total RNA isolation was carried out from leaves using the Spectrum™ Plant Total RNA Kit and RNase-free DNase I (Sigma-Aldrich, USA) was used to get eradicate genomic DNA as per the manufacturer’s protocol. First-strand synthesis of cDNA was performed using the SuperScript III First-Strand Synthesis System (Sigma-Aldrich, USA). Specific oligonucleotide primers were designed using the IDT primer quest tool (Table S3). Real-time expression analysis was carried out using three technical replicates of each biological replicate using the Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific, USA) according to the manufacturer’s recommendations. Quantifications were normalised using the actin housekeeping gene of Z. nummularia as an internal control (Sun et al. 2009). The steps used for amplification were 95 °C for 30s followed by 32 cycles at 95 °C for 3 s and 62 °C for 30s. Ct values were estimated using the 2–ΔΔCT method (Livak and Schmittgen 2001).
Statistical analysis
Student’s t test was used to evaluate the data at a significance level of p < 0.05.
Results
Genome wide identification of the Hsp70 and Hsp100 gene family members in Z. jujuba
A total of 21 Hsp70 and 6 Hsp100 unique full-length genes were identified in Z. jujuba genome, which were designated as ZjHsp70-1 to ZjHsp70-21 and ZjHsp100-1 to ZjHsp100-6. The CDS sequences of ZjHsp70 and ZjHsp100 genes ranged from 1448 to 2754 base pairs and 2427 to 2901 base pairs, respectively, with predicted protein lengths ranging from 481 to 917 and 808 to 966 amino acids with varying sub-cellular locations predicted (Table 1 and Table S1). The majority of ZjHsp70s were predicted to be localised to the cytoplasm. The subcellular locations of ZjHsp100s were more varied with three localised to the cytoplasm, two in the chloroplasts, and one to the mitochondrion.
Table 1.
Features of Hsp70 and Hsp100 in Z. jujuba
| Name | Locus ID | Chr | CDS (bp) | Exon | No. of aa | MW (kDa) | Subcellular localization |
|---|---|---|---|---|---|---|---|
| Hsp70-1 | zju:107432045 | 1 | 2310 | 11 | 769 | 85.92 | Nuclear |
| Hsp70-2 | zju:107405039 | 1 | 2571 | 10 | 856 | 94.34 | Cytoplasmic |
| Hsp70-3 | zju:107406481 | 1 | 1968 | 1 | 655 | 71.85 | Cytoplasmic |
| Hsp70-4 | zju:107405906 | 1 | 1956 | 1 | 651 | 71.21 | Cytoplasmic |
| Hsp70-5 | zju:107426626 | 1 | 1965 | 1 | 654 | 71.74 | Chloroplast |
| Hsp70-6 | zju:107412565 | 2 | 2139 | 8 | 712 | 75.88 | Cytoplasmic |
| Hsp70-7 | zju:107411081 | 2 | 2133 | 3 | 710 | 78.27 | Cytoplasmic |
| Hsp70-8 | zju:107411863 | 2 | 1965 | 6 | 654 | 72.53 | ER |
| Hsp70-9 | zju:107411934 | 2 | 1977 | 6 | 658 | 73.41 | ER |
| Hsp70-10 | zju:107411076 | 2 | 1953 | 2 | 650 | 71.18 | Nuclear |
| Hsp70-11 | zju:107413527 | 3 | 1947 | 2 | 648 | 70.80 | Cytoplasmic |
| Hsp70-12 | zju:107413482 | 3 | 2487 | 3 | 828 | 92.32 | ER |
| Hsp70-13 | zju:107413529 | 3 | 1965 | 2 | 654 | 71.51 | Cytoplasmic |
| Hsp70-14 | zju:107416864 | 4 | 2754 | 15 | 917 | 101.49 | Cytoplasmic |
| Hsp70-15 | zju:107423197 | 7 | 1719 | 3 | 572 | 61.85 | Cytoplasmic |
| Hsp70-16 | zju:107426507 | 9 | 1659 | 3 | 552 | 62.94 | Cytoplasmic |
| Hsp70-17 | zju:107426748 | 9 | 2049 | 6 | 682 | 73.38 | Mitochondrial |
| Hsp70-18 | zju:107429814 | 11 | 2001 | 9 | 666 | 73.36 | ER |
| Hsp70-19 | zju:107431116 | 11 | 1941 | 2 | 646 | 70.82 | Cytoplasmic |
| Hsp70-20 | zju:107407469 | u | 1770 | 6 | 589 | 65.05 | Cytoplasmic |
| Hsp70-21 | zju:107408891 | u | 1448 | 3 | 481 | 53.61 | ER |
| Hsp100-1 | zju:107422833 | 1 | 2793 | 10 | 930 | 104.81 | Cytoplasmic |
| Hsp100-2 | zju:107415872 | 4 | 2637 | 9 | 878 | 99.08 | Cytoplasmic |
| Hsp100-3 | zju:107419106 | 5 | 2733 | 7 | 910 | 101.39 | Cytoplasmic |
| Hsp100-4 | zju:107420312 | 6 | 2466 | 9 | 822 | 91.40 | Mitochondrial |
| Hsp100-5 | zju:107423408 | 7 | 2901 | 12 | 966 | 105.97 | Chloroplast |
| Hsp100-6 | zju:107403384 | u | 2427 | 8 | 808 | 89.49 | Chloroplast |
Physio-biochemical properties
Results indicated that there were significant variations among ZjHsp70s and ZjHsp100s proteins in physical and chemical properties (Table S1). As per the instability index, 4 out of 21 ZjHsp70s and 3 out of 6 ZjHSsp100s were considered as unstable proteins (ZjHsp70-1, ZjHsp70-2, ZjHsp70-14, ZjHsp70-17, ZjHsp100-3, ZjHsp100-5, and ZjHsp100-6), (Instability index > 40.4). The aliphatic index is a feature of thermostability, and ZjHsp70-15 and ZjHsp100-3 had the highest aliphatic index of 102.6 and 96.24, respectively.
Analyses of phylogenetic relationship, gene structure, motifs, and domain architecture
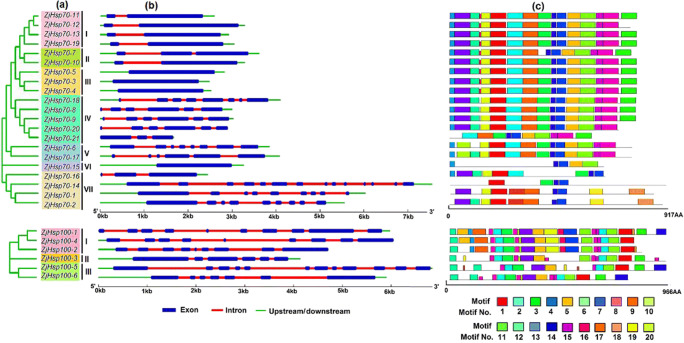
Based on sequence divergence the Z. jujuba, Hsp70 and ZjHsp100 gene family members were grouped into seven and three sub-groups respectively (Fig. 1a). Group I and VII were clustered in the Hsp110/SSE subfamily, with the group I members located in the ER, and cytoplasm and group VII members located in cytoplasm and nucleus. Group II and VI were clustered in the DnaK subfamily and located in both the cytoplasm and nucleus. Members of group III were located in cytoplasm. Group IV members were also located in the cytoplasm and ER. To get detailed information on structural variations among the Hsp70s and Hsp100s genes in Z. jujiba, we analysed the intron/exon structure. The length and number of introns in all ZjHsp70 and ZjHsp100 genes were observed to differ from 0 to 13 and 5 to 11, respectively (Fig. 1b). The conserved motifs of ZjHsp70 and ZjHsp100 genes varied considerably between family members (Fig. 1c, Table S4). The type, order, and number of motifs were found to be similar in the proteins within a single sub-family and different in the proteins between sub-families..
Fig. 1.
Phylogenetic relationships, gene structure and conserved motifs analysis in the ZjHsp70 and ZjHsp100 family protein in Z. jujuba. a The unrooted phylogenetic tree shows sub-families of ZjHsp70s (I–VII) and ZjHsp100s (I–III) are highlighted with different coloured backgrounds. b Gene structure analysis of ZjHsp70 and ZjHsp100 gene family members. The sizes of the exons and introns can be estimated using the scale given at the bottom. c Conserved motif analysis of the ZjHsp70 and ZjHsp100 family protein in Z. jujuba. Each coloured box represents a motif in each of the ZjHsp70 and ZjHsp100 proteins
The result of in silico domain identification revealed that all the ZjHsp70 proteins possess the conserved Hsp70 (PF00012.17) and MreB_Mbl (MreB/Mbl protein) domains. However, the MreB_Mbl domain was absent in ZjHsp70-14 and ZjHsp70-15 proteins, and interestingly ZjHsp70-16 has two Hsp70 and MreB_Mbl domains (Table S5). Similarly, all ZjHsp100 class proteins possessed the conserved domains belonging to Clp_N (Clp amino-terminal domain, pathogenicity island component), AAA (ATPase family associated with various cellular activities), AAA_2 (Cdc48 subfamily), and ClpB_D2-small (C-terminal, D2-small domain, of ClpB protein) domain. However, ZjHsp100-2 and ZjHsp100-4 lack the ClpB_D2-small domain.
Identification and occurrence patterns of different SSRs
A total of 13 and 3 perfect SSR loci (ranging in size from 20 to 30 bp) were identified within ZjHsp70 and ZjHsp100 genes (Table 2) using SSR locator software. In case of ZjHsp70, SSRs were divided into five types composed of mono-, di-, tri-, tetra-, and hexa-motifs. However, in ZjHsp100, only mono- and di- types of SSR motifs were found. The highest number of SSR loci was located in introns (56%) followed by 5′ UTR (38%), with only one SSR loci identified in the 3′ UTR.
Table 2.
SSRs loci identified in Hsp70 and Hsp100 genes in Z. jujuba
| S no. | Name | SSR type | SSR start | SSR end | SSR length (bp) | SSR position |
|---|---|---|---|---|---|---|
| 1 | ZjHsp70-1 | (T)22 | 354 | 376 | 22 | 5′ UTR |
| 2 | ZjHsp70-1 | (T)22 | 2368 | 2390 | 22 | Intron |
| 3 | ZjHsp70-2 | (T)20 | 3893 | 3913 | 20 | Intron |
| 4 | ZjHsp70-6 | (A)20 | 161 | 181 | 20 | 5′ UTR |
| 5 | ZjHsp70-8 | (T)20 | 201 | 221 | 20 | Intron |
| 6 | ZjHsp70-14 | (T)26 | 405 | 431 | 26 | 5′ UTR |
| 7 | ZjHsp70-14 | (T)20 | 460 | 480 | 20 | 5′ UTR |
| 8 | ZjHsp70-15 | (ATTTTT)4 | 3194 | 3218 | 24 | 3′ UTR |
| 9 | ZjHsp70-17 | (TCT)8 | 569 | 593 | 24 | Intron |
| 10 | ZjHsp70-17 | (TTTA)5 | 694 | 714 | 20 | Intron |
| 11 | ZjHsp70-18 | (T)20 | 3845 | 3865 | 20 | 5′ UTR |
| 12 | ZjHsp70-20 | (TA)14 | 239 | 267 | 28 | Intron |
| 13 | ZjHsp70-20 | (AT)12 | 1054 | 1078 | 24 | Intron |
| 14 | ZjHsp100-2 | (TG)15 | 612 | 642 | 30 | Intron |
| 15 | ZjHsp100-4 | (A)22 | 24 | 46 | 22 | 5′ UTR |
| 16 | ZjHsp100-6 | (TC)13 | 1712 | 1738 | 26 | Intron |
Comparative analysis of the ZjHsp70s and ZjHsp100s in Z.jujuba, A. thaliana, C. sativa, and R. rubrinervis
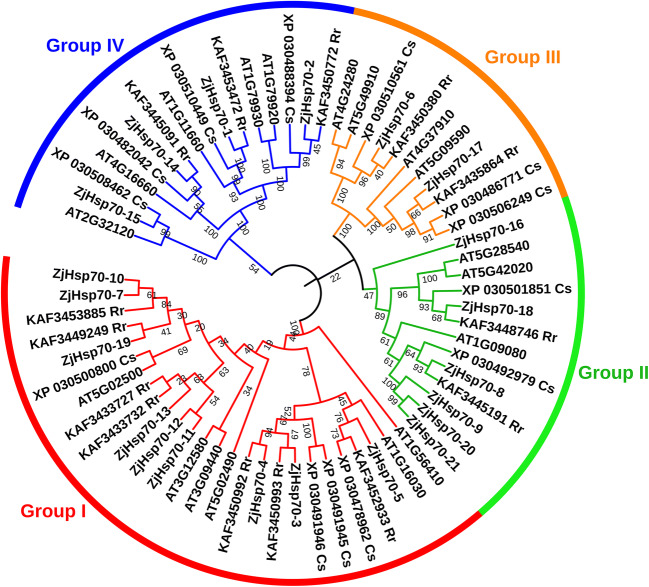
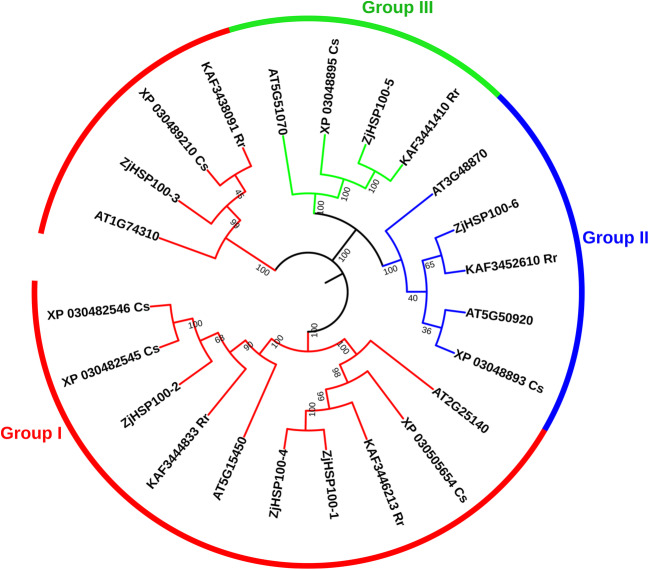
Comparative genomics allows the analysis of the same protein families between diverse plant species. In the present study, a phylogenetic relationship of ZjHsp70s and ZjHsp100s was studied with the orthologous protein sequences from A. thaliana, C. sativa, and R. rubrinervis (Figs. 2 and 3). All the Hsp70s were divided into four groups (I, II, III, IV), with group I members in the majority with 26 genes. Groups II, III, and IV contained 13, 11, and 16 members respectively. The Hsp100s were divided into 3 groups. Group I was the largest, containing 14 genes. Group II included 5 members and group III consisted of 4 members (Table S6). The combined phylogenetic tree further identified the paralogous and orthologous relationships between the Hsp70 and Hsp100 gene family members. Mainly the paralogous pairs of each species contained duplicated genes, indicating the events of species-specific Hsp70 and Hsp100 gene duplication. Ten pairs of paralogous genes Hsp70 were identified. A total of 13 pairs of orthologous genes derived from a common ancestral gene have been identified in different plant species, with the highest numbers (10) identified in Z. jujuba and R. rubrinervis. In the case of ZjHsp100, one pair of paralogous genes and a total of 5 pairs of orthologous genes were identified.
Fig. 2.
Phylogenetic relationship analysis of ZjHsp70 with its orthologs. Phylogenetic tree constructed by using amino acid sequences of Z. jujuba, A. thaliana, R. rubrinervis, and C. sativa. The red, green, orange, and blue colour of each node represents group I, group III, group III, and group IV respectively
Fig. 3.
Phylogenetic analysis of ZjHsp100 with its orthologs. Phylogenetic tree constructed by using amino acid sequences of Z. jujuba, A. thaliana, R. rubrinervis, and C. sativa. The red, blue, and green colour of each node represents group I, group III, and group III respectively
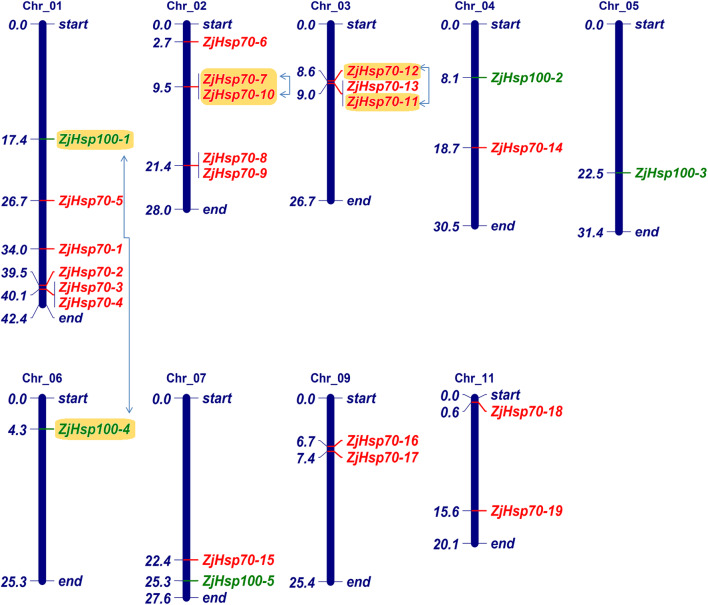
Chromosomal locations and gene duplication
Among the 12 chromosomes of Z. jujuba genome, the Hsp70 and Hsp100 genes were unevenly dispersed across a total of 7 and 5 chromosomes respectively (Fig. 4). The uneven distribution of ZjHsp70 and ZjHsp100 genes indicates a genetic divergence in the process of evolution. The results illustrated that there were three and one paralogous pairs observed in the ZjHsp70 and ZjHsp100 gene family respectively (Figs. 2 and 3). One of the three ZjHsp70 paralog gene pairs, ZjHsp70-7:ZjHsp70-10, was identified as tandem duplication found on chromosome 2 in a head-to-tail orientation in 24.36 kb and no other genes were present in between them. In addition, a segmental duplication was observed in the paralogous pair ZjHsp70-11:ZjHsp70-12 as they were separated by 381.19 kb and present in tail-to-head orientation on chromosome 3. Similarly, the second segmental duplication in the paralogs ZjHsp100-1:ZjHsp100-4 was also separated by a large distance of 13,138 kb, with ZjHsp100-1 on chromosome 1 segmentally linked to ZjHsp100-4 on chromosome 6. These results revealed that segmental duplication has played an important role in the expansion of the Hsp70 and Hsp100 gene family in Z. jujuba. Due to the lack of chromosomal location data of the paralogous pair ZjHsp70-20:ZjHsp70-21, the exact type of duplication could not be determined.
Fig. 4.
Chromosomal locations and duplications of ZjHsp70 and ZjHsp100 gene family members on Z jujuba chromosomes. The red and green colouring indicates ZjHsp70 and ZjHsp100 genes, respectively. Segmentally duplicated genes are indicated by blue arrow; Tandemly duplicated ZjHsp genes are indicated by yellow boxes
Promoter analysis
In order to identify the upstream cis-regulatory elements of Hsp70 and Hsp100 genes in Z. jujuba, the upstream regulatory regions (URRs) were analysed using the Plant Place tool (Table S7). The analysis showed the presence of 35 and 33 types of different cis-regulatory elements was situated in Hsp70 and Hsp100 genes of Z. jujuba respectively. Among them, many cis-regulatory elements were gene-specific. The identified cis-regulatory elements were grouped into three categories based on their different roles, as putative abiotic stress-responsive, hormone-responsive, and light-responsive elements.
In the URRs of the ZjHsp70 gene family, abiotic stress-responsive cis-regulatory elements including heat stress-responsive, dehydration-responsive, salt stress-responsive elements, plant growth and development, and stress response were prominent. The gibberellin responsive elements, cytokinin response regulator (RR) binding motif, abscisic acid-responsive element, salicylic acid (SA)-induced elements, were the major hormonal responsive cis-regulatory elements observed. The URRs of Hsp70-1, Hsp70-3, Hsp70-5, Hsp70-6, Hsp70-11, Hsp70-13, Hsp70-15, Hsp70-16, and Hsp70-18 genes contained the highest numbers of nearly all identified cis-regulatory elements. The URRs of the Hsp100 gene family of Z. jujuba contained prominent abiotic stress-responsive, light-responsive, and hormonal responsive cis-regulatory elements as found in the Hsp70 gene family. The URRs of ZjHsp100-1, ZjHsp100-5, and ZjHsp100-6 genes contained the highest numbers of nearly all identified cis-regulatory elements. This in silico analysis of cis-regulatory elements indicated that these genes are likely involved in diverse transcriptional regulatory functions.
Expression analysis of ZjHsp70 and ZjHsp100 genes under abiotic stress conditions
In the present analysis, to get insight into which ZjHsp70 and ZjHsp100 genes are potentially involved in the response to abiotic stress conditions, the expression profiles of all ZjHsp70 and ZjHsp100 genes were analysed using acute stress experiments, as previously used in Liu et al. 2018, Singh et al. 2016, Mulaudzi-Masuku et al. 2015, to provide a comparative analysis. Expression levels of ZjHsp70-3, ZjHsp70-5, ZjHsp70-6, ZjHsp70-16, ZjHsp70-17, and ZjHsp70-20 were upregulated in response to three of the abiotic stresses exposed (Fig. 5), with ZjHsp70-1, ZjHsp70-8, and ZjHsp70-12 upregulated in response to the salinity and drought stress conditions. jHsp70-4, ZjHsp70-10, and ZjHsp70-14 gene were observed to be upregulated in response to the drought stress conditions, whilst ZjHsp70-19 was observed to be upregulated in heat and salinity and downregulated in drought stress. Among the ZjHsp100 genes, ZjHsp100-1, ZjHsp100-3, ZjHsp100-4, and ZjHsp100-6 were upregulated in response to heat stress (Fig. 5) and ZjHsp100-2 and ZjHsp100-3 upregulated in response to drought stress. Interestingly, ZjHsp100-1 and ZjHsp100-4 genes were upregulated but ZjHsp100-2, ZjHsp100-3, ZjHsp100-5, and ZjHsp100-6 genes were downregulated in response to salinity stress condition. These results demonstrated that ZjHsp70 and Hsp100 may potentially play collective roles in abiotic stress tolerance.
Fig. 5.
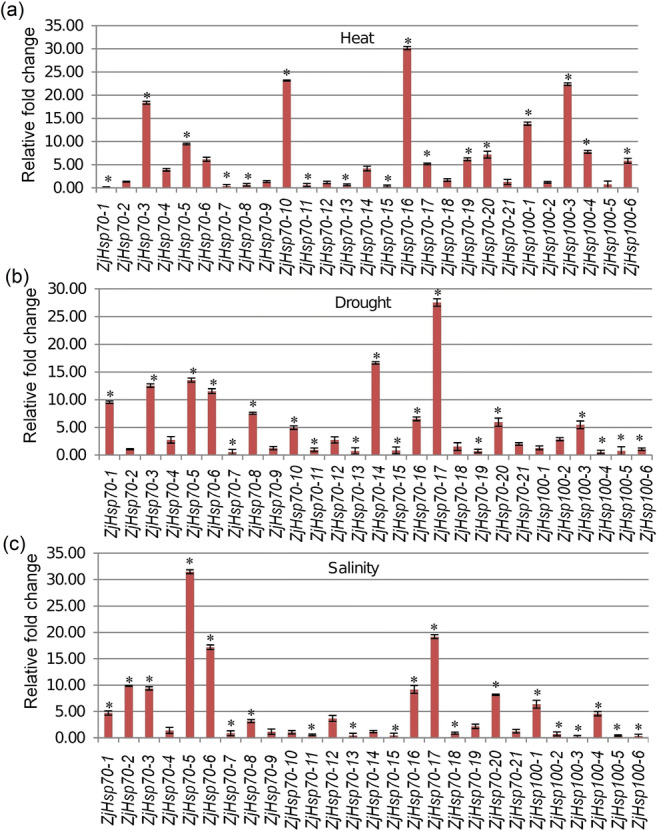
Expression analyses of ZjHsp70 and ZjHsp100 gene family members under heat (a), drought (b), and salinity (c) stress conditions. Statistical significance within genes was indicated with an asterisk. Statistical significance was indicated with an asterisk (P < 0.05). Error bars represents ± SD from three technical replicates
Discussion
Ziziphus species have significant abiotic stress-tolerant abilities; suggesting they may be potential repositories of genes responsible for abiotic stress tolerance. Therefore, we performed in silico analysis to characterise Hsp70 and Hsp100 gene families in Z. jujuba and also analysed gene expression of candidate Hsps to identify potential stress-responsive genes, which could assist the bio-prospecting of genes via expression study and further functional analysis.
We identified 21 and 6 members of the Hsp70 and Hsp100 gene family in Z. jujuba respectively. The number of identified genes in Z. jujuba is comparatively less than rice (32 Hsp70, 9 Hsp100), cotton (30 Hsp70), and A. thaliana (18 Hsp70, 6 Hsp90) (Lin et al. 2001; Jung et al. 2013; Zhang et al. 2014). This might be because all of the genomes of these other species contain more predicted genes (i.e. cotton 41,330, 2287 Mb (Yang et al. 2020), M. domestica 15,345, 690 Mb (Scott et al. 2014), rice 49,066, 430 Mb (Mustafiz et al. 2016) as compared to Z. jujuba (32,808, 437.65 Mb) (Liu et al. 2014). The ZjHsp70 and ZjHsp100 family proteins have diverse biophysical properties. The Hsp70 and Hsp100 in Z. jujuba comprised a multi-genic family and different members were localised in different cellular organelles, as has been identified previously in other species such as S. tuberosum, N. tobacum, and S. italica (Liu et al. 2018; Song et al. 2019; Singh et al. 2016; Lee et al. 2007). This indicated the different patterns of organelle localization may be associated with particular molecular mechanisms in relation to abiotic stress tolerance.
The instability index indicates the in silico stability of protein, and 17 out of 21 Z. jujuba proteins were found to be stable proteins (Gonzalez-Diaz et al. 2007). The single ZjHsp70-6 protein localised in chloroplasts was the most stable among the 17 stable proteins, which suggested their potential role in maintaining homeostasis of photosystem II under stress condition (Su and Li 2010). The role of members situated in the chloroplast mostly includes contributions to the formation of thylakoid membrane, maintaining the normal process of photosystem II under heat stress, and assisting in protein translocation (Liu et al. 2007; Zhong et al. 2013). A similar result was observed in N. tobacum where members localised in the chloroplast (NtHsp70-46 to NtHsp70-49) were stable proteins (Song et al. 2019). The ZjHsp70 genes were unevenly situated on chromosomes, generally near to both ends of the chromosomes (Fig. 4), similar to Hsp70 genes mapped in N. tabacum and A. thaliana (Song et al. 2019). The ZjHsp100 genes were randomly present associated across 5 different chromosomes across the genome. Gene duplication events of entire genomes, chromosomal segments, and individual genes play a role in the expansion of gene family members (Cannon et al. 2004; Maere et al. 2005). These events also contribute to the evolution of novel gene functions and potentially play an important role enhancing plant environmental adaptability (Huang et al. 2015). We identified 3 and 1 pairs of ZjHsp70 and ZjHsp100 respectively that have undergone tandem and segmental gene duplication, indicating the importance of these events in the amplification of the ZjHsp gene family members.
Furthermore, we analysed the gene structures of Hsp70 and Hsp100 gene families. Their intron-exon organization was highly similar within the sub-families, as defined by the phylogenetic analyses (Fig. 1a). For example, in sub-family I, all of the ZjHsp70 members had a single intron, whilst in sub-family III, no introns were observed. These observations indicate that some introns were lost and gained during the structural evolution between the ZjHsp70 genes, as previously identified in S. tuberosum (Liu et al. 2018; Zhang et al. 2015a, b). This variation in intron/exon structures between the different gene family groups provides the possibility for alternative splicing events, which could potentially increase the diversity of gene function (Barbazuk et al. 2008). In the motif study, the number, type, and order of motifs in the ZjHsp70 and ZjHsp100 proteins between the sub-families were also similar, but differed between the proteins from the other sub-families.
Large numbers of SSRs are distributed in eukaryotic genomes and may play an important role in gene regulation (Gao et al. 2011; Xue et al. 2018). SSR markers have been used in marker-assisted selection to enhance the efficiency of heat stress tolerance plant breeding in various crops including barley, potato, and rice (Jha et al. 2014; Shamsudin et al. 2016). In this study, a total of 16 perfect potential SSRs were mined from both ZjHsp70 and ZjHsp100 genes. Most SSRs were predicted in the intergenic region (5′ UTRs and 3′ UTRs). Similar observations were reported in the previous studies in Brassica rapa (Hong et al. 2007), Rosaceae species (Moe et al. 2011), rice, and A. thaliana (Fujimori et al. 2003; Lawson and Zhang 2006).
A comparative phylogenetic tree was constructed to obtain insight into the evolutionary relationships of Hsps and to identify paralogs within species and orthologs between species. We used the amino acid sequences of Hsp70 and Hsp100 genes from C. sativa and R. rubrinervis as a close relatives and sequences from A. thaliana as an out group. Based on the phylogenetic tree, ZjHsp70s and ZjHsp100 could be divided into four and three groups respectively based on their sequence relatedness (Liu et al. 2018; Chaudhary et al. 2019; Singh et al. 2016). A total of 10 pairs of paralogs in the Hsp70s were identified which included 3 pairs from Z. jujuba, 4 pairs from A. thaliana, 2 pairs from C. sativa, and 1 pair from R. rubrinervis. In the case of ZjHsp100s, 1 pair of paralogous genes from Z. jujuba and 1 pair from C. sativa were identified. This implies that the expansion of these gene families in each species was species-specific approach (Liu et al. 2018). The cis-regulatory elements in promoter regions play an important role in the functional divergence of the Hsp genes. Analysis of the cis-regulatory element showed three types of cis-regulatory elements, including hormonal responsive elements, stress-responsive elements, and light-responsive elements (Song et al. 2019). Among the majority of Hsp70 and Hsp100 promoter sequences, cis-regulatory elements related to abiotic stress were identified (Song et al. 2019). In the recent past, Song et al. (2019) also reported these three types of cis-regulatory elements in N. tabacum. For instance, the CCAATBOX1, LTRE1HVBLT49, and LTRECOREATCOR15 contribute to metabolic regulation of heat stress (Deng et al. 2018). The MYCCONSENSUSAT and MYBCORE contribute to metabolic regulation of drought stress and GT1GMSCAM4 contributes to metabolic regulation of salinity stress (Senavirathne et al. 2017). However, the DOFCOREZM motif is the most overrepresented cis-element in both Hsp70 and Hsp100 gene family involved in plant metabolism and drought condition (Corrales et al. 2014). Similarly the overrepresented DOFCOREZM cis-element was observed in the Hsp70 of P. patens a high dehydration tolerant plant, which can be recover even after dehydration to 92% loss of fresh weight (Tang et al. 2016). Due to the presence of diverse abiotic stress-responsive cis-elements, ZjHsp70 and ZjHsp100 could play significant roles in response to diverse abiotic stresses. Hence, the present investigation offers a theoretical foundation for further studies of the actual mechanism of action (Song et al. 2019). Nonetheless, no correlation between the cis-regulatory elements of the promoter sequences of ZjHsp and the expression pattern of genes was observed. This may be due to several reasons. Firstly, it may be due to the spatiotemporal pattern of gene expression during the growth and development of plants. Secondly, there may be unidentified cis-regulatory elements which contribute to the diversity of the expression patterns observed, or finally, it may be due to the particular nature (level and duration) of the stress challenge (Song et al. 2019).
Previous analysis has also reported that Hsp70 and Hsp100 genes play important roles under abiotic stress conditions including heat, drought, and salinity in other plants (Chaudhary et al. 2019; Liu et al. 2018; Singh et al. 2016; Mulaudzi-Masuku et al. 2015). In this study, the expression profiles of the ZjHsp70 and ZjHsp100 genes in response to heat, salinity, and drought stress were analysed (Fig. 5). The results showed that most of the ZjHsp70 and ZjHsp100 genes were induced by abiotic stresses. Not all Hsp70s have the ability to increase the stress tolerance of plants, some are intimately involved in the development of plants (Noel et al. 2007), which would not be detected in this study.
Conclusion
In the present study, a total of 21 and 6 Hsp70 and Hsp100 gene family members were identified in Z. jujuba using an in silico approach. Analysis of their gene distributions, genomic organization, gene structures, and protein structures suggested a complex evolutionary history for these two Hsp families. Segmental and tandem duplications were identified suggesting their role in the expansion of ZjHsp70 and ZjHsp100 gene families. Based on promoter analysis, the results indicated that ZjHsp70s and ZjHsp100s were involved in abiotic stress tolerance and development, which was validated in certain genes by expression analysis in response to heat, drought, and salinity. Thus, the present investigation suggested that ZjHsp70-3, ZjHsp70-5, ZjHsp70-6, ZjHsp70-16, ZjHsp70-17, ZjHsp70-20, ZjHsp100-1, ZjHsp100-2, and ZjHsp100-3 are potential candidate genes for further functional analysis in crop improvement research.
Supplementary information
Summary of all Hsp70 and Hsp100 gene sequences, functional domains and biochemical characteristics in Z. jujuba (XLSX 105 kb)
Orthologous protein sequences of Hsp70 and Hsp100 gene family used for phylogenetic relationship analysis (XLSX 30 kb)
List of primers used for qRT-PCR analysis of ZjHsp70 and ZjHsp100 gene family (XLSX 12 kb)
Identified conserved motif sequences of ZjHsp70 and ZjHsp100 protein family in Z. jujuba (XLSX 11 kb)
Summary of domains present in ZjHsp100 and ZjHsp70 proteins (XLSX 11 kb)
Identified paralogous and orthologous pairs of ZjHsp70s and ZjHsp100s gene sequences of Z. jujuba, P. mume and A. thaliana (XLSX 11 kb)
Details of cis-regulatory elements observed in ZjHsp70 and ZjHsp100 gene family (XLSX 20 kb)
Acknowledgements
We gratefully acknowledge the National Phytotron Facility, IARI, for providing space for growing plants.
Authors’ contributions
KPP conceived, designed experiments, performed the most of analysis, experiments, wrote and revised the manuscript. SSK conducted the analysis and help in manuscript writing. NRC and BH also help in manuscript writing.
Funding
KP gratefully acknowledges the Indian Agricultural Research Institute (IARI) for the IARI-Junior Research Fellowship grant and the Department of Biotechnology (DBT), Government of India, for DBT-Senior research fellowship grant.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
N/A
Consent to participate
N/A
Consent for publication
N/A
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kishor Prabhakar Panzade, Email: kishor.panzade@gmail.com.
Sonam S. Kale, Email: sonamkale09@gmail.com
Narendra R. Chavan, Email: agribiotech@themgmgroup.com
Bhupal Hatzade, Email: bhupal.hatzade@ajeetseed.co.in, Email: iarilab10@gmail.com.
References
- Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Bio l3:21-29 [PubMed]
- Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- Battisti DS, Naylor RL. Historical warnings of future food insecurity with unprecedented seasonal heat. Science. 2009;323:240–244. doi: 10.1126/science.1164363. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Baranwal VK, Kumar R, Sircar D, Chauhan H. Genome-wide identification and expression analysis of Hsp70, Hsp90, and Hsp100 heat shock protein genes in barley under stress conditions and reproductive development. Funct Integr Genomics. 2019;19:1007–1022. doi: 10.1007/s10142-019-00695-y. [DOI] [PubMed] [Google Scholar]
- Corrales AR, Nebauer SG, Carrillo L, Fernández-Nohales P, Marqués J, Renau-Morata B, Granell A, Pollmann S, Vicente-Carbajosa J, Molina RV, Medina J. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J Exp Bot. 2014;65(4):995–1012. doi: 10.1093/jxb/ert451. [DOI] [PubMed] [Google Scholar]
- Coumou D, Rahmstorf S (2012) A decade of weather extremes. Nature Climate Change 2:491-496
- Da Maia LC, Palmieri DA, de Souza VQ, Kopp MM, de Carvalho FIF, Costa de Oliveira A (2008) SSR Locator: tool for simple sequence repeat discovery integrated with primer design and PCR simulation. Int J Plant Genomics 412696 [DOI] [PMC free article] [PubMed]
- Deng Z, Chen J, Wei Y, Liu H, Yang H, Dai L, Li D. Two translationally controlled tumor protein (TCTP) genes from Hevea brasiliensis play overlapping and different roles in development and stress response. Ind Crop Prod. 2018;114:137–145. [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan YH, Guo J, Ding K, Wang SJ, Zhang H, Dai XW, Chen YY, Govers F, Huang LL, Kang ZS. Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Mol Biol Rep. 2011;38(1):301–307. doi: 10.1007/s11033-010-0108-0. [DOI] [PubMed] [Google Scholar]
- Fujimori S, Washio T, Higo K, Ohtomo Y, Murakami K, Matsubara K, Kawai J, Carninci P, Hayashizaki Y, Kikuchi S, Tomita M. A novel feature of microsatellites in plants: a distribution gradient along the direction of transcription. FebsLett. 2003;554:17–22. doi: 10.1016/s0014-5793(03)01041-x. [DOI] [PubMed] [Google Scholar]
- Gao C, Tang Z, Yin J, An Z, Fu D, Li J. Characterization and comparison of gene-based simple sequence repeats across Brassica species. Mol Gen Genomics. 2011;286:161–170. doi: 10.1007/s00438-011-0636-x. [DOI] [PubMed] [Google Scholar]
- Garg VK, Avashthi H, Tiwari A, Jain PA, Ramkete PW, Kayastha AM, Singh VK. MFPPI–multi FASTA ProtParam interface. Bioinformation. 2016;12:74–77. doi: 10.6026/97320630012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Diaz H, Perez-castillo Y, Podda G, Uriarte E. Computational chemistry comparison of stable/nonstable protein mutants classification models based on 3D and topological indices. J Comput Chem. 2007;28:1990–1995. doi: 10.1002/jcc.20700. [DOI] [PubMed] [Google Scholar]
- Guo M, Zhai YF, Lu JP, Chai L, Chai WG, Gong ZH, Lu MH. Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int J Mol Sci. 2014;15:19741–19759. doi: 10.3390/ijms151119741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Nahar K, Alam M, Roychowdhury R, Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci. 2000;97:4392–4397. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CP, Piao ZY, Kang TW, Batley J, Yang TJ, Hur YK, Bhak J, Park BS, Edwards D, Lim YP. Genomic distribution of simple sequence repeats in Brassica rapa. Mol Cell. 2007;23:349–356. [PubMed] [Google Scholar]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li MY, Wang F, Xu ZS, Huang W, Wang GL, Ma J, Xiong AS. Heat shock factors in carrot: genome-wide identification, classification, and expression profiles response to abiotic stress. Mol Biol Rep. 2015;42:893–905. doi: 10.1007/s11033-014-3826-x. [DOI] [PubMed] [Google Scholar]
- Jha UC, Bohra A, Singh NP. Heat stress in crop plants: its nature, impacts and integrated breeding strategies to improve heat tolerance. Plant Breed. 2014;133:679–701. [Google Scholar]
- Jung KH, Gho HJ, Nguyen MX, Kim SR, An G. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct Integr Genomics. 2013;13:391–402. doi: 10.1007/s10142-013-0331-6. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Grover A. Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol. 2003;51:677–686. doi: 10.1023/a:1022561926676. [DOI] [PubMed] [Google Scholar]
- Khan SU, Waqas H, Shehzad SA, Imran M. Theoretical analysis of tangent hyperbolic nanoparticles with combined electrical MHD, activation energy and Wu’s slip features: a mathematical model. Phys Scr. 2019;94:125211. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lawson MJ, Zhang L. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol. 2006;7:R14. doi: 10.1186/gb-2006-7-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Rioflorido I, Hong SW, Larkindale J, Waters ER, Vierling E. The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J. 2007;49:115–127. doi: 10.1111/j.1365-313X.2006.02940.x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu SS, Yi CY, Wang F, Zhou J, Xia XJ, et al. Hydrogen peroxide mediates abscisic acid-induced HSP 70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ. 2014;37:2768–2780. doi: 10.1111/pce.12360. [DOI] [PubMed] [Google Scholar]
- Li Z, Long R, Zhang T, Wang Z, Zhang F, Yang Q, et al. Molecular cloning and functional analysis of the drought tolerance gene MsHSP70 from alfalfa (Medicago sativa L.) J Plant Res. 2017;130:387–396. doi: 10.1007/s10265-017-0905-9. [DOI] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:201–208. doi: 10.1379/1466-1268(2001)006<0201:gaoths>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MY, Chai KH, Ko SS, Kuang LY, Lur HS, Charng YY. A positive feedback loop between heat shock protein101 and heat stress-associated 32-kd protein modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol. 2014;164:2045–2053. doi: 10.1104/pp.113.229609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Willmund F, Golecki JR, Cacace S, He B, Markert C, Schroda M. The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 2007;50:265–277. doi: 10.1111/j.1365-313X.2007.03047.x. [DOI] [PubMed] [Google Scholar]
- Liu MJ, Zhao J, Cai QL, Liu GC, Wang JR, Zhao ZH, et al. The complex jujube genome provides insights into fruit tree biology. Nat Commun. 2014;5:1–12. doi: 10.1038/ncomms6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pang X, Cheng Y, Yin Y, Zhang Q, Su W, et al. The Hsp70 gene family in Solanum tuberosum: genome-wide identification, phylogeny, and expression patterns. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-34878-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25: 402-408 [DOI] [PubMed]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 2007;145:1588–1599. doi: 10.1104/pp.107.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RC, Grover A. ClpB/Hsp100 proteins and heat stress tolerance in plants. Crit Rev Biotechnol. 2016;36:862–874. doi: 10.3109/07388551.2015.1051942. [DOI] [PubMed] [Google Scholar]
- Moe KT, Chung JW, Cho YI, Moon JK, Ku JH, Jung JK, Lee J, Park YJ. Sequence information on simple sequence repeats and single nucleotide polymorphisms through transcriptome analysis of mungbean. J Integr Plant Biol. 2011;53:63–73. doi: 10.1111/j.1744-7909.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- Mulaudzi-Masuku T, Mutepe RD, Mukhoro OC, Faro A, Ndimba B. Identification and characterization of a heat-inducible Hsp70 gene from Sorghum bicolor which confers tolerance to thermal stress. Cell Stress Chaperones. 2015;20:793–804. doi: 10.1007/s12192-015-0591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafiz A, Kumari S, Karan R. Ascribing functions to genes: journey towards genetic improvement of rice via functional genomics. Curr Genomics. 2016;17:155–176. doi: 10.2174/1389202917666160202215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Martinez LM, Ponce G, Cassab GI, Alagon A, Meeley RB, et al. Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. Plant Cell. 2002;14:1621–1633. doi: 10.1105/tpc.010487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, Witte CP, Bhat R, Pochon N, Colby T, Parker JE. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell. 2007;19:4061–4076. doi: 10.1105/tpc.107.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outlaw WH, Zhang S, Riddle KA, Womble AK, Anderson LC, Outlaw WM, et al. The jujube (Ziziphusjujuba Mill.), a multipurpose plant. Econ Bot. 2002;56:198–200. [Google Scholar]
- Padaria JC, Yadav R, Tarafdar A, Lone SA, Kumar K, Sivalingam PN. Molecular cloning and characterization of drought stress responsive abscisic acid-stress-ripening (Asr1) gene from wild jujube, Ziziphus nummularia (Burm. f.) Wight & Arn. Mol Biol Rep. 2016;43:849–859. doi: 10.1007/s11033-016-4013-z. [DOI] [PubMed] [Google Scholar]
- Pandey A, Singh R, Radhamani J, Bhandari DC. Exploring the potential of Ziziphus nummularia (Burm. f.) Wight et Arn. from drier regions of India. Genet Resour Crop Evol. 2010;57:929–936. [Google Scholar]
- Panzade KP, Vishwakarma H, Padaria JC (2020) Heat stress inducible cytoplasmic isoform of ClpB 1 from Z. nummularia exhibits enhanced thermotolerance in transgenic tobacco. Mol Biol Rep 1–11 [DOI] [PubMed]
- Planton S. Driouech F, Rhaz KE, Lionello P (2016) The climate of the Mediterranean regions in the future climate projections. The Mediterranean region under climate change. A scientific update is report on climate change, 83
- Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowarth NM, Dauphinee AN, Denbigh GL, Gunawardena AH. Hsp70 plays a role in programmed cell death during the remodelling of leaves of the lace plant (Aponogeton madagascariensis) J Exp Bot. 2020;71:907–918. doi: 10.1093/jxb/erz447. [DOI] [PubMed] [Google Scholar]
- Sabir MA, Rasheed F, Zafar Z, Khan I, Nawaz MF, Haq I, Bilal M (2020) A consistent CO2 assimilation rate and an enhanced root development drives the tolerance mechanism in Ziziphus jujuba under soil water deficit. Arid Land Res Manag 1-13
- Scott JG, Warren WC, Beukeboom LW, Bopp D, Clark AG, Giers SD, Hediger M, Jones AK, Kasai S, Leichter CA, Li M, Meisel RP, Minx P, Murphy TD, Nelson DR, Reid WR, Rinkevich FD, Robertson HM, Sackton TB, Sattelle DB, Thibaud-Nissen F, Tomlinson C, van de Zande L, Walden KKO, Wilson RK, Liu N. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 2014;15:466. doi: 10.1186/s13059-014-0466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senavirathne WMA, Jayatilake DV, Herath V, Wickramasinghe HAM. Evaluation of genetic diversity of cis-acting elements of Abscisic acid responsive element binding protein (ABRE-BP) in selected Sri Lankan rice varieties. Trop Agric Res. 2017;28:120–132. [Google Scholar]
- Shamsudin NAA, Swamy BM, Ratnam W, Cruz MTS, Raman A, Kumar A. Marker assisted pyramiding of drought yield QTLs into a popular Malaysian rice cultivar, MR219. BMC Genet. 2016;17:30. doi: 10.1186/s12863-016-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Jaishankar J, Muthamilarasan M, Shweta S, Dangi A, Prasad M. Genome-wide analysis of heat shock proteins in C4 model, foxtail millet identifies potential candidates for crop improvement under abiotic stress. Sci Rep. 2016;6:1–14. doi: 10.1038/srep32641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Pan F, Lou X, Wang D, Yang C, Zhang B, Zhang H. Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum. Mol Biol Rep. 2019;46:1941–1954. doi: 10.1007/s11033-019-04644-7. [DOI] [PubMed] [Google Scholar]
- Su PH, Li HM. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 2008;146:1231–1241. doi: 10.1104/pp.107.114496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell. 2010;22:1516–1531. doi: 10.1105/tpc.109.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HF, Meng YP, Cui GM, Cao QF, Li J, Liang AH. Selection of housekeeping genes for gene expression studies on the development of fruit bearing shoots in Chinese jujube (Ziziphus jujube Mill.) Mol Biol Rep. 2009;36:2183–2190. doi: 10.1007/s11033-008-9433-y. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T, Yu A, Li P, Yang H, Liu G, Liu L. Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses. Sci Rep. 2016;6:33650. doi: 10.1038/srep33650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma H, Junaid A, Manjhi J, Singh GP, Gaikwad K, Padaria JC. Heat stress transcripts, differential expression, and profiling of heat stress tolerant gene TaHsp90 in Indian wheat (Triticuma estivum L.) cv C306. PLoS One. 2018;13:e0198293. doi: 10.1371/journal.pone.0198293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wahid A. Physiological implications of metabolite biosynthesis for net assimilation and heat-stress tolerance of sugarcane (Saccharum officinarum) sprouts. J Plant Res. 2007;120:219–228. doi: 10.1007/s10265-006-0040-5. [DOI] [PubMed] [Google Scholar]
- Wang P, Song H, Li C, Li P, Li A, Guan H, et al. Genome-wide dissection of the heat shock transcription factor family genes in Arachis. Front Plant Sci. 2017;8:106. doi: 10.3389/fpls.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Chen B, Wang R, Win AN, Li J, Chai Y. Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and its parental species. Appl Biochem Biotechnol. 2018;184:582–598. doi: 10.1007/s12010-017-2563-8. [DOI] [PubMed] [Google Scholar]
- Yadav R, Lone SA, Gaikwad K, Singh NK, Padaria JC. Transcriptome sequence analysis and mining of SSRs in Jhar Ber (Ziziphus nummularia (Burm. f.) Wight & Arn) under drought stress. Sci Rep. 2018;8:1–10. doi: 10.1038/s41598-018-20548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Verma OP, Padaria JC. Transcript profiling and gene expression analysis under drought stress in Ziziphus nummularia (Burm. f.) Wright & Arn. Mol Biol Rep. 2018;45:163–174. doi: 10.1007/s11033-018-4149-0. [DOI] [PubMed] [Google Scholar]
- Yang Z, Qanmber G, Wang Z, Yang Z, Li F. Gossypium genomics: trends, scope, and utilization for cotton improvement. Trends Plant Sci. 2020;25:488–500. doi: 10.1016/j.tplants.2019.12.011. [DOI] [PubMed] [Google Scholar]
- Yu CS, Cheng CW, Su WC, Chang KCC, Huang SW, Hwang JK, Lu CH, Gajendra P. Raghava S (2014) CELLO2GO: A Web Server for Protein subCELlular LOcalization Prediction with Functional Gene Ontology Annotation. PLoS ONE 9:e99368 [DOI] [PMC free article] [PubMed]
- Zhang Z, Li X. Genome-wide identification of AP2/ERF superfamily genes and their expression during fruit ripening of Chinese jujube. Sci Rep. 2018;8:1–16. doi: 10.1038/s41598-018-33744-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Zhu JH, Zhang QQ, Cai YB. Molecular characterization of an ethephon-induced Hsp70 involved in high and low-temperature responses in Hevea Brasiliensis. Plant Physiol Biochem. 2009;47:954–959. doi: 10.1016/j.plaphy.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang M, Chen J, Rong J, Ding M. Genome-wide analysis of HSP70 superfamily in Gossypium raimondii and the expression of orthologs in Gossypium hirsutum. Yi chuan Hereditas. 2014;36:921–933. doi: 10.3724/SP.J.1005.2014.0921. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li J, Zhang L, Wang Y, Zheng H, et al. Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics. 2015;16:1–19. doi: 10.1186/s12864-015-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao HK, Dong QL, Zhang YY, Wang YM, Li HY. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.) Front Plant Sci. 2015;6:773. doi: 10.3389/fpls.2015.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell. 2013;25:2925–2943. doi: 10.1105/tpc.113.111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of all Hsp70 and Hsp100 gene sequences, functional domains and biochemical characteristics in Z. jujuba (XLSX 105 kb)
Orthologous protein sequences of Hsp70 and Hsp100 gene family used for phylogenetic relationship analysis (XLSX 30 kb)
List of primers used for qRT-PCR analysis of ZjHsp70 and ZjHsp100 gene family (XLSX 12 kb)
Identified conserved motif sequences of ZjHsp70 and ZjHsp100 protein family in Z. jujuba (XLSX 11 kb)
Summary of domains present in ZjHsp100 and ZjHsp70 proteins (XLSX 11 kb)
Identified paralogous and orthologous pairs of ZjHsp70s and ZjHsp100s gene sequences of Z. jujuba, P. mume and A. thaliana (XLSX 11 kb)
Details of cis-regulatory elements observed in ZjHsp70 and ZjHsp100 gene family (XLSX 20 kb)