Abstract
Human papillomavirus (HPV) has been the leading cause of cervical cancer for over 25 years. Approximately 5.5–11% of all cervical cancers are reported to be HPV-negative, which can be attributed to truly negative and false-negative results. The truly HPV-negative cervical cancers are almost all cervical adenocarcinomas with unclear etiology. False HPV negativity can arise from histological misclassification, latent HPV infection, disruption of the targeting fragment, non-high risk HPV infection, and HPV testing methods. HPV-negative cervical cancers are often diagnosed at an advanced FIGO stage and have a poor prognosis; thus, the management of these cases requires greater attention.
Keywords: human papillomavirus, human papillomavirus (HPV) testing, false-negative, human papillomavirus (HPV)-negative, cervical cancer
Introduction
Cervical cancer is the fourth most common malignancy among women worldwide, accounting for approximately 7% of all cancer cases in women (1, 2). Persistent infection with human papillomavirus (HPV), particularly high-risk genotypes of HPV, is considered the major cause of cervical cancer. HPV DNA replicates from free DNA in the basal cells of the cervix during the initial period of HPV infection, and then integrates into the host genome as the infection progresses, with subsequent upregulation of E6 and E7 oncogene expression (3). HPV can be found in almost all cervical squamous cell carcinomas and precancerous lesions, including high grade squamous intraepithelial lesions (HSILs) or grade 2–3 cervical intraepithelial neoplasia (CIN). Although the sensitivity of HPV testing has improved significantly in recent years, a small fraction of cervical cancers are continued to be reported as HPV-negative. HPV-negative cervical cancer is often diagnosed at an advanced FIGO stage and associated with poor prognosis. Insights into the etiology, therapy, and prognosis of HPV-negative cervical cancer may help develop appropriate strategies for its management in patients.
Currently, there is no clear definition of HPV-negative cervical cancer to describe cases diagnosed by pathological features in the absence of HPV-infection via HPV testing. The existence of cervical adenocarcinoma independent of HPV infection has been recognized by the majority of researchers (4–6). It is estimated that approximately 5.5–11% of cervical cancers worldwide are HPV-negative (7–10). A review of 243 studies and 30,848 women with invasive cervical cancer that were reported between 1990 and 2010 revealed a gradual decrease in the number of HPV-negative cases (11). In this meta-analysis, the incidence of HPV-positivity in 1990–1999, 2000–2005, and 2006–2010 was 85.9%, 87.9%, and 92.9%, respectively. The downward trend in HPV-negativity could be related to improvements in HPV testing and non-cervical cancer classification. A recent study using next-generation sequencing (NGS) to characterize primary cervical cancer revealed that HPV-negative cervical cancer accounted for approximately 5% of all cervical cancer cases (12). However, only a few rare pathological types of cervical cancer are truly HPV-negative (13–15). In studies involving HPV testing, the true incidence of HPV-negative invasive cervical cancer might be overestimated (16).
This review provides a comprehensive overview of the attributable reasons, clinical characteristics, treatment, and prognostic measures for HPV-negative cervical cancer, with the aim to assist in the development of effective therapeutic strategies to improve clinical outcomes.
Human Papillomavirus-Negative Cervical Cancer: Attributable Reasons
For HPV-negative cervical cancers, clinicians should consider whether the cervical cancer is HPV-independent, a misclassification of non-cervical cancer, or an HPV false-negative case.
Human Papillomavirus-Independent Cervical Cancer
HPV-independent cervical cancer, considered to be “truly” HPV-negative, is not associated with HPV infection. Cervical squamous cell carcinoma is rarely HPV-negative (17), and a confirmed HPV-independent cervical squamous cell carcinoma has not yet been reported. For cervical adenocarcinoma, the HPV negativity rate is approximately 15–38% (10, 18, 19). The HPV positivity rate in carcinoma in situ varies according to different histological features (17). Although the exact mechanism underlying HPV-independent cervical cancer is unclear, most researchers consider it to be caused by mutations in tumor-associated genes such as TP53, PIK3CA, and CDKN2A (20).
Misclassification of Non-Cervical Cancer
Cervical cancers include the direct extension of endometrial carcinoma or those arising from distant metastasis of other primary HPV-negative tumors. Research has shown that almost 68% of HPV-negative cervical cancers were misdiagnosed as primary cervical cancer (7). A study examining HPV-negative cervical adenocarcinoma indicated that more than 50% of cases could not be distinguished from endometrial carcinoma based on histological features alone (21). Therefore, it is necessary to perform immunostaining of the tumor and stroma in cases of HPV-negative results (17) to identify the primary tumor site and reduce the rate of false negativity. A combination of estrogen receptor (ER), progesterone receptor (PR), vimentin, and CD10 negativity along with carcinoembryonic antigen (CEA), diffuse p16, CD34, and HPV positivity suggests cervical adenocarcinoma, while a combination of ER, PR, vimentin, diffuse p16, and CD10 positivity along with CEA, CD34, and HPV negativity suggests uterine adenocarcinoma (17, 22). Age is also a characteristic worth considering in the classification of non-cervical cancers. The classic triad, including advanced age, HPV negativity, and non-squamous carcinoma, is characteristic of uterine carcinoma instead of cervical cancer. Compared with other sites, such as the gynecologic tract, it is rare for the uterine cervix to be a metastatic site considering its anatomy (23); however, 3.7% of female genital metastatic tumors reportedly involve the uterine cervix (24).
Human Papillomavirus False-Negative Cervical Cancer
Latent Human Papillomavirus Infection
Natural infection of HPV has a latency period, in which viral replication is restricted by the immune system and HPV gene expression is in a silent state. However, the natural history of HPV from infection to cervical cancer remains unclear (25). A 5-year follow-up study involving sensitive HPV DNA testing revealed that most HPV infections disappeared within two years, except those with precancerous lesions or worse (26). Latent infections often have a low incidence of tumorigenesis and a higher chance of false-negativity as the viral load is too low to be detected using HPV testing. However, nearly 0.05% of HPV-negative cases reportedly progressed to grade 3 CIN (CIN3) or cervical cancer in the subsequent 3–5 years (26).
Loss of Human Papillomavirus Fragments During Integration
The HPV L1 fragment is highly conserved in different HPV genotypes; thus, it is targeted by consensus or genotype-specific primers in many HPV detection tests. Integration of the HPV genome into the host genome involves disruption of E1, E2, L2, or L1 fragments (27, 28). HPV testing targeting L1 may be less reliable than that targeting E6/E7 oncogenes, as L1 expression can be lost while E6/E7 expression is always present. The positivity rate of HPV16 and HPV18 detected targeting the HPV L1 region was 91.7% and 72.1%, respectively, suggesting that HPV L1 testing missed 8.3% of HPV16 and 27.9% of HPV18 infections compared with HPV testing targeting E6/E7 (29). However, other researchers believe that HPV L1 testing would rarely result in a false-negative outcome because HPV integration with disruption of L1 coexists with other forms of HPV insertion into the host genome involving disruption of E1/E2 in the same sample.
Studies investigating the mechanisms of HPV infection have demonstrated that a few tumors no longer express the HPV E6/E7 oncogene (HPV-inactive) during cancer development (30). Although the HPV-inactive status is oncogenic, it results in an HPV false-negative outcome using HPV E6/E7 mRNA testing. A study by Banister et al. revealed overall DNA methylation to be decreased, while WNT/β-catenin and Sonic Hedgehog signaling was upregulated in HPV-inactive cervical cancers (30). The somatic mutation profiles differ considerably between HPV-active and HPV-inactive tumors, with more somatic mutations present in HPV-inactive tumors (especially in the TP53, ARID, WNT, and PI3K pathways) (30). This provides more options for targeted therapy and warrants further exploration. Targeting WNT, PI3K, or TP53 mutations may effectively treat HPV-inactive tumors, leading to improved survival outcomes of these patients.
Cervical Cancer Caused By Non-High Risk Human Papillomavirus
Several studies have reported the association between cervical cancer and infection with low-risk HPV types 6, 11, 42, 44, and 70 (31–33). Whether low-risk HPV causes cervical cancer or acquired by accident is unknown. Petry et al. estimated that 1%–2% of primary cervical cancers were associated with non-high risk HPV (non-hr-HPV) infection (7), a far higher percentage than the one estimated in the large international collection of invasive cervical cancer (32, 33). Currently, most HPV tests target hr-HPV subtypes and are unable to detect non-hr HPV infection, resulting in partial HPV false-negative results.
False-Negative Human Papillomavirus Testing
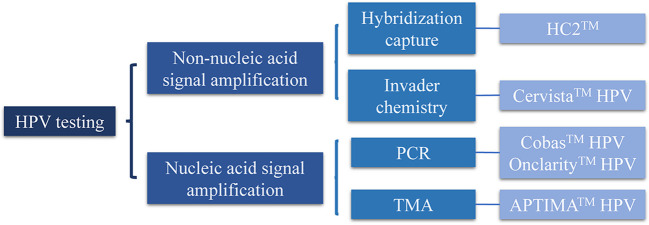
HPV testing can be divided into two categories, namely, nucleic and non-nucleic acid signal amplification ( Figure 1 ). Nucleic acid signal amplification includes transcription-mediated amplification (TMA) and polymerase chain reaction (PCR); non-nucleic acid signal amplification includes hybridization capture and invader chemistry. Currently, the US Food and Drug Administration (FDA) has approved five HPV tests for cervical cancer screening, namely, Hybrid Capture 2™ (HC2™), Cervista™ HPV HR, Cervista™ HPV16/18, Cobas™ HPV, APTIMA™ HPV (28), and BD’s Onclarity™ HPV ( Figure 1 , Table 1 ) (34–50).
Figure 1.
Classification of human papillomavirus (HPV) testing. PCR, polymerase chain reaction; TMA, transcription-mediated amplification.
Table 1.
Comparison of FDA-approved human papillomavirus (HPV) testing.
| HPV testing(Reference) | Methodology | HPV genotypes | Internal control | Sensitivity CIN2+,% | Specificity CIN2+,% | Analytical Sensitivity |
|---|---|---|---|---|---|---|
| HC2™ (34–39) | Hybrid capture | 13 hr-HPV | No | 87–98 | 20–85 | 1,000–5,000 copies/reaction |
| Cervista™ HPV HR and HPV16/18 (38, 40–42) | Invader chemistry | 14 hr-HPV | Histone 2 gene | 90–92.8 | 44.2–47 | 625–7,500 copies/reaction for different hr-HPV types |
| Cobas™ HPV (36, 39, 43–45) | PCR | 12 hr-HPV and HPV16/18 | β-globin | >88.2 | 59.3–70.5 | 150–1,200 copies/ml for different hr-HPV types |
| APTIMA™ HPV (34, 37, 44, 46) | TMA | 14 hr-HPV | Exogenous RNA | 87.8–100 | >85 | 17–488 copies/reaction |
| BD’s Onclarity™ HPV (47–50) | PCR | 14 hr-HPV | β-globin | 84.6–96.1 | 46.2–89.1 | 692–2,990 copies/ml for different HPV types |
CIN2+, grade 2 of cervical intraepithelial neoplasia or even worse; HC2, Hybrid Capture 2; hr-HPV, high-risk HPV; PCR, polymerase chain reaction; TMA, transcription-mediated amplification.
Non-nucleic acid signal amplification methods, including HC2™, Cervista™ HPV HR, and Cervista™ HPV16/18, have lower sensitivity but higher cut-off values than nucleic acid signal amplification methods, resulting in false-negative HPV test outcomes, especially when samples have low viral loads. The lack of an internal control in HC2™ also increases the false-negativity rate probably due to DNA contamination or degradation (28, 51). HC2™ covers E1, E2, E4, E5, E6, E7, L1, L2, and LCR fragments of hr-HPV. However, the other three methods targeting L1 alone are prone to false-negative results because of the disruption of L1 fragment during HPV genome integration. APTIMA™ HPV, which differs from HPV DNA testing, detects E6/E7 mRNA of 14 hr-HPV types. Since the expression of E6/E7 mRNA increases after HPV genome integration into the host genome, positive APTIMA™ HPV testing results always indicate cervical cancer or adverse outcomes. It is, therefore, not advisable to use APTIMA™ HPV in primary screening, because E6/E7 mRNA is expressed mainly after HPV integration into the host genome, resulting in a window period between HPV infection and detection. This window phase may increase the rate of HPV false-negative outcomes. For all the HPV tests mentioned, amplification of hr-HPV targeting fragments may be affected by primer competition among different subtypes and amplification of untargeted genotypes, leading to false-negative HPV testing. In conclusion, false-negative HPV testing is associated with lower sensitivity, HPV targeting fragments, hr-HPV genotypes detected, and detection of HPV DNA or RNA.
In addition to the method used, HPV false-negative results are also related to sampling errors. Poor cell viability from necrotic and/or inflammatory sites often results in HPV false-negative outcomes. Faulty sample collection methods, including samples mixed with blood or lubricant, as well as fixation procedure can result in false negativity. Therefore, the accuracy of HPV testing in published studies should be questioned discreetly. Some studies employed HPV testing to investigate previously stored cervical cancer specimens, but it is unknown whether such specimens precisely reflect HPV infection in the patients. A retrospective study demonstrated that samples from elderly patients or those stored for a longer duration had lower HPV-positivity rates (13). The effect of storage time on HPV positivity was more distinct in adenocarcinoma than in squamous cell carcinoma (13). Other factors that affect HPV positivity include the time between excision to fixation and fixator type (13). In a retrospective study, the use of unbuffered formalin fixation was an important factor influencing HPV-negative results (13).
In summary, the most important reason for false-negative HPV testing results is the significant difference among HPV detection methods, which is not realized by all clinicians (29). Testing procedures and sample quality, as in DNA/RNA degradation of formalin-fixed and paraffin-embedded samples, can also lead to HPV false-negative results.
Clinical Features of Human Papillomavirus-Negative Cervical Cancer
Age
A global study involving 760 cases of cervical adenocarcinoma revealed that older patient age at initial diagnosis was associated with a lower positivity rate of HPV DNA testing (13). A similar trend was identified in squamous cell carcinoma without any clear reason. One possible explanation is that viral vitality is gradually lost during tumor progression, especially in older patients with more time to develop cancer. Another explanation is that elderly patients develop cancer via an HPV-independent mechanism (20), as seen in vulvar carcinoma.
Molecular and Pathological Features
Pathological type influences the results of HPV detection, as demonstrated by differences in HPV infection rates between cervical squamous cell carcinoma and adenocarcinoma. Globally, 12.7% of squamous cell carcinoma and 15%–38% of cervical adenocarcinoma are HPV-negative (10, 19). The parakeratosis or hyperkeratosis status of squamous cell carcinoma can lead to false-negative HPV testing results (52). Additionally, HPV-negative cervical cancer whose histopathology is almost all adenocarcinoma is possibly missed by HPV testing (8, 18, 53). The positivity rate of some HPV genotypes in cervical adenocarcinoma was reportedly low (17). Moreover, the HPV DNA load in cervical adenocarcinoma was lower than that in squamous cell carcinoma, challenging HPV detection in adenocarcinoma (17). The glandular epithelium is not susceptible to persistent HPV infection; accumulation of free HPV DNA as well as copy numbers of integrated HPV were low even in HPV-infected glandular epithelium (13). In contrast, HPV-infected squamous cell carcinoma tended to have higher copy numbers of HPV DNA and integrated virus.
The common pathological types of HPV-positive adenocarcinoma are intestinal, villoglandular, signet-ring cell, and endometrioid adenocarcinoma, which originates from the cervical squamous columnar junction zone, accounting for nearly 90% of all cervical adenocarcinomas ( Table 2 ) (13–15, 17, 18, 54). The pathological types of HPV-negative adenocarcinoma are gastric, clear cell, serous, and mesonephric adenocarcinomas ( Table 2 ). These types are quite rare and their occurrence might not be HPV-related (13).
Table 2.
Pathological types of cervical adenocarcinoma and its human papillomavirus (HPV)-positive rate.
| Pathological types (Reference) | Percentage of cervical adenocarcinoma, % | HPV-positive rate, % (17) |
|---|---|---|
| Endocervical (usual) type (15, 18) | 73–79 | 80–100 |
| Intestinal (15) | 3–8 | 83–100 |
| Villoglandular (15) | 0.8–6 | 100 |
| Signet-ring cell (15) | 0.3 | 100 |
| Endometrioid (13, 15) | 1.1–1.6 | 27.3 |
| -From squamous columnar junction zone | — | 100 |
| -From upper endocervix and lower uterine segment | — | 0 |
| Gastric (15, 18) | 1.5–10 | 0 |
| Clear cell (13, 18) | 4.4–6.3 | 20–27.6 |
| Serous (13, 15) | 0.5–3.5 | 25–30.4 |
| Mesonephric (15) | 0.3 | 0 |
The pathogenesis of these HPV-independent pathological types is correlated with specific mutations of the genome. The PI3K-AKT pathway may be involved in the development of clear cell adenocarcinoma; immunostaining results for p-AKT and p-mTOR were positive in 50% of cases (17, 55). In elderly patients with this adenocarcinoma subtype, PTEN expression was lost in 50% of patients, while EGFR and HER2 expression increased in 75% and 50% of patients, respectively (17, 55). Gastric adenocarcinoma has been associated with somatic and germline mutations of STK11 and TP53 (Peutz-Jeghers Syndrome) (14). PIK3CA, PTEN, and CTNNB1 mutations have been frequently reported for endometrioid adenocarcinoma (20). In mesonephric adenocarcinoma, 81% of patients harbored KRAS or NRAS mutations, while 62% carried mutations of ARID1A, ARID1B, or SMARCA4, but none of PIK3CA or PTEN (17, 56). The characteristic mutation of mesonephric adenocarcinoma differed from the common mutations of cervical adenocarcinoma, with 7% of cases harboring KRAS/NRAS mutations (56). Therefore, RAS/MAPK pathway inhibitors may provide potential treatment options for mesonephric adenocarcinoma.
FIGO Stage
HPV-negative patients are prone to develop advanced FIGO stage and lymphatic space invasion prior to diagnosis, resulting in poor prognosis ( Table 3 ) (57, 58). A multicenter study revealed that 62.5% of HPV-negative adenocarcinomas were stage II or higher, while 83.7% of usual type cases were stage I at diagnosis, which concurred with previous studies (60). Further, HPV-negative cases in this study exhibited a larger tumor size than HPV-positive cases.
Table 3.
Studies of FIGO stage and prognosis of human papillomavirus (HPV)-negative cervical cancers.
| Study (Reference) | Cases (HPV negative/overall) | HPV testing | Advanced FIGO stage (HPV negative vs. HPV positive) | Lymphatic metastasis (HPV negative vs. HPV positive) | DFS (HPV negative vs. HPV positive) | OS (HPV negative vs. HPV positive) |
|---|---|---|---|---|---|---|
| Nicolas et al. (57) | 21/214 | PCR | 91% vs. 57%, p<0.01 | 67% vs. 36%, p<0.01 | 59.8 m (95%CI 32.0–87.6 m) vs. 132.2 m (95%CI 118.6–145.8 m), p<0.01 | 77.0 m (95%CI 47.2–106.8 m) vs. 153.8 m (95%CI 142.0–165.6 m), p=0.01 |
| Van der Marel et al. (58) | 8/136 | HC2™, PCR L1&E7 | 87.5% vs. 52.3%, p=0.053 | 37.5% vs. 17.2%, p=0.150 | 51.9 m (95%CI 12.2–91.7 m) vs. 109.9 m (95%CI 98.2–121.5 m), p=0.010 | 67.7 m (95%CI 20.0–106.9 m) vs. 108.9 m (95%CI 97.7–120.0 m), p=0.225 |
| Feng et al. (59) | 43/122 | PCR (HPV 16/18) | — | — | — | 5 year: HR=1.250 (95%CI 0.562–2.784), p=0.584 8 year: HR=1.530 (95%CI 0.697–3.362), p=0.289 |
PCR, polymerase chain reaction; HC2, Hybrid Capture 2; DFS, disease-free survival; OS, overall survival; m, months; HR, hazard ratio; 95%CI, 95% confidence interval.
Treatment of Human Papillomavirus-Negative Cervical Cancer
Currently, HPV-negative cervical cancer has no specific therapy and thus consults with HPV-positive cervical cancer treatment strategies. Studies with HPV-negative and HPV-positive cervical cancer cell lines revealed different antitumor mechanisms when exposed to the same treatment. For example, a histone deacetylase (HDAC) inhibitor repressed E6 activity to promote apoptosis in HPV-positive cervical cancer cells but caused G2 phase arrest in HPV-negative cervical cancer cells, while dehydroepiandrosterone caused apoptosis in HPV-positive cervical cancer cells and necrosis in HPV-negative cervical cancer cells (61, 62). Based on the etiology of HPV-related cervical cancer, gene expression in 74 cell lines demonstrated significantly higher p16 expression while that of phosphorylated retinoblastoma protein (pRb) was lower in HPV-positive cell lines, compared with HPV-negative cell lines (63). Abemaciclib, a CDK4/6 inhibitor, suppressed CDK4/6-Rb-E2F and mTOR pathways, resulting in superior treatment in HPV-negative cancer (64). The overall survival (OS) rate of patients who received surgery combined with other oncologic treatment differed significantly between HPV-positive and HPV-negative cases, while that of patients who received surgery alone did not (60), suggesting that adjuvant chemoradiotherapy may benefit HPV-negative cases. True HPV-negative cervical cancers are associated with specific pathological types, therefore understanding their tumorigenesis will contribute to the selection of suitable therapies for cervical cancer. Much attention has focused on exploring TP53, ARID, WNT, and PI3K pathways, which mutate frequently in cervical adenocarcinoma, to develop effective targeted therapies. Recently, lncRNA has emerged as a research hotspot for HPV-negative cervical cancer treatment (65, 66).
Prognosis of Human Papillomavirus-Negative Cervical Cancer
As early as 1990, a study of 106 early-stage invasive cervical cancer cases using PCR revealed that the risk of overall relapse did not differ among different HPV genotypes in HPV-positive patients, but was 2.6 times higher in HPV-negative patients and their risk of distant metastasis was 4.5 times higher than HPV-positive patients. The 24-month relapse-free survival rate of HPV-positive patients was higher than that of HPV-negative patients (77% vs. 40%) (67). A meta-analysis of 2838 cervical cancer cases from 17 studies revealed that HPV positivity correlated with better prognosis (OS: HR=0.610, p=0.001; disease-free survival: HR=0.362, p<0.001) ( 68). To date, three other studies have reached the same conclusion that HPV-negative cervical cancer is associated with poor prognosis ( Table 3 ) (57–59). Nevertheless, a 10-year follow-up study of 204 patients with cervical cancer revealed that the 5-year OS rate of HPV-negative and HPV-positive patients was 82% and 58% (p=0.003), respectively, indicating that HPV infection was significantly correlated with poor OS of patients (69). Further investigation is warranted to elucidate the effects of negative HPV testing on prognosis.
Discussion
HPV-negative cervical cancers are divided into truly negative and false-negative categories. Truly negative cervical cancers have an HPV-independent pathogenesis with specific pathological types, of which HPV vaccination and testing probably have little effect on their prevention. The diagnosis of truly negative cervical cancer mainly depends on cytological screening and observation of histological features, combined with cytological multiple staining. Further investigation of the pathways and biomarkers of the different pathological types is required to develop a basis for precise therapy. For false-negative cervical cancer, retesting should be considered using other HPV testing methods according to their characteristics after analyzing the reason for false-negative HPV results. HPV-positive results after retesting may be due to initial testing failures for hr-HPV or the inability of the standard HPV test to detect other HPV genotypes (7). Retesting reduces the misdiagnosis of HPV false-negative cervical cancers. Most HPV-negative samples from reported studies were formalin-fixed and paraffin-embedded, which affects HPV DNA quality and causes false-negative results. Improving HPV detection strategies by developing standardized and high-quality HPV tests is crucial to reduce false negativity. Different procedures for sample collection, storage, and testing affect HPV test outcomes, therefore operating procedures for HPV testing need to be standardized. It is beneficial to choose more sensitive HPV testing verified by universal standards and to consider the cut-off values of HPV testing, especially in persistent HPV infections with low viral activity (70). Further, the laboratory conducting HPV testing should be authorized by institutions and meet international standards (71).
Cervical adenocarcinoma is the major pathological type of HPV-negative cervical cancer, most likely caused by mutations of PI3K-AKT or other pathways. Preclinical studies have demonstrated that tumorigenesis differs between HPV-positive and HPV-negative cervical cancers, which presents the possibility of developing targeted therapies for HPV-negative patients. This may provide a basis for cervical cancer treatment research in the future. Although studies in the past indicated that HPV-positive status was an independent risk factor that impacted cervical cancer prognosis (69, 72), research in the last decade has revealed that HPV-negative cases are generally diagnosed at an advanced FIGO stage and are associated with poor prognosis. Large-scale multicenter studies need to be conducted to further elucidate the relationship between HPV negativity and cervical cancer.
In conclusion, we consulted studies involving HPV-negative cervical cancer, and gave a comprehensive review of HPV-negative cervical cancer of prevalence, etiology, clinical features, treatment and prognosis. Although HPV-negative cervical cancer reveals different characteristics from HPV-positive one, most studies ignore the HPV status of cervical cancer, which restricts a profound insights of HPV-negative cervical cancer. Clarifying the different categories of HPV-negative cervical cancers is crucial to the development of suitable treatments and to guide studies investigating HPV-negative cervical cancers. The presence of HPV-independent cervical cancers should not affect the promotion of HPV testing and vaccination. Clinicians should classify and treat HPV-negative cases cautiously, and consider the correlation between advanced stage and poor prognosis of cervical cancer to provide women with negative HPV testing better management and effective treatment.
Author Contributions
YZ and GW provided the concept and designed the study. BX and JG wrote the draft of the manuscript. YZ and YS gave critical revision of the manuscript. YZ and JG provided economic support to the study. The authors meet all the criteria set out in the journal’s authorship criteria. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Nos. 81974463 and 81672573).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (2020) 70(1):7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3. Woodman CBJ, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer (2007) 7(1):11–22. 10.1038/nrc2050 [DOI] [PubMed] [Google Scholar]
- 4. Rodríguez-Carunchio L, Soveral I, Steenbergen RDM, Torné A, Martinez S, Fusté P, et al. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. BJOG (2015) 122(1):119–27. 10.1111/1471-0528.13071 [DOI] [PubMed] [Google Scholar]
- 5. Pirog EC. Diagnosis of HPV-negative, gastric-type adenocarcinoma of the endocervix. Methods Mol Biol (2015) 1249:213–9. 10.1007/978-1-4939-2013-6_16 [DOI] [PubMed] [Google Scholar]
- 6. Molijn A, Jenkins D, Chen W, Zhang X, Pirog E, Enqi W, et al. The complex relationship between human papillomavirus and cervical adenocarcinoma. Int J Cancer (2016) 138(2):409–16. 10.1002/ijc.29722 [DOI] [PubMed] [Google Scholar]
- 7. Petry KU, Liebrich C, Luyten A, Zander M, Iftner T. Surgical staging identified false HPV-negative cases in a large series of invasive cervical cancers. Papillomavirus Res (2017) 4:85–9. 10.1016/j.pvr.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blatt AJ, Kennedy R, Luff RD, Austin RM, Rabin DS. Comparison of cervical cancer screening results among 256,648 women in multiple clinical practices. Cancer Cytopathol (2015) 123(5):282–8. 10.1002/cncy.21544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guan P, Howell-Jones R, Li N, Bruni L, de Sanjosé S, Franceschi S, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer (2012) 131(10):2349–59. 10.1002/ijc.27485 [DOI] [PubMed] [Google Scholar]
- 10. Clifford GM, Smith JS, Plummer M, Muñoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer (2003) 88(1):63–73. 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li N, Franceschi S, Howell-Jones R, Snijders PJF, Clifford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer (2011) 128(4):927–35. 10.1002/ijc.25396 [DOI] [PubMed] [Google Scholar]
- 12. Cancer Genome Atlas Research N. Albert Einstein College of M. Analytical Biological S. Barretos Cancer H. Baylor College of M. Beckman Research Institute of City of H et al. Integrated genomic and molecular characterization of cervical cancer. Nature (2017) 543(7645):378–84. 10.1038/nature21386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pirog EC, Lloveras B, Molijn A, Tous S, Guimerà N, Alejo M, et al. HPV prevalence and genotypes in different histological subtypes of cervical adenocarcinoma, a worldwide analysis of 760 cases. Modern Pathol (2014) 27(12):1559–67. 10.1038/modpathol.2014.55 [DOI] [PubMed] [Google Scholar]
- 14. Pirog EC, Park KJ, Kiyokawa T, Zhang X, Chen W, Jenkins D, et al. Gastric-type Adenocarcinoma of the Cervix: Tumor With Wide Range of Histologic Appearances. Adv Anat Pathol (2019) 26(1):1–12. 10.1097/PAP.0000000000000216 [DOI] [PubMed] [Google Scholar]
- 15. Stolnicu S, Barsan I, Hoang L, Patel P, Terinte C, Pesci A, et al. International Endocervical Adenocarcinoma Criteria and Classification (IECC): A New Pathogenetic Classification for Invasive Adenocarcinomas of the Endocervix. Am J Surg Pathol (2018) 42(2):214–26. 10.1097/pas.0000000000000986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rossi PG, Ronco G, Dillner J, Elfström KM, Snijders PJF, Arbyn M, et al. Why follow-back studies should be interpreted cautiously: The case of an HPV-negative cervical lesion. Cancer Cytopathol (2016) 124(1):66–7. 10.1002/cncy.21622 [DOI] [PubMed] [Google Scholar]
- 17. Pirog EC. Cervical Adenocarcinoma: Diagnosis of Human Papillomavirus-Positive and Human Papillomavirus-Negative Tumors. Arch Pathol Lab Med (2017) 141(12):1653–67. 10.5858/arpa.2016-0356-RA [DOI] [PubMed] [Google Scholar]
- 18. Holl K, Nowakowski AM, Powell N, McCluggage WG, Pirog EC, Collas De Souza S, et al. Human papillomavirus prevalence and type-distribution in cervical glandular neoplasias: Results from a European multinational epidemiological study. Int J Cancer (2015) 137(12):2858–68. 10.1002/ijc.29651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol (2010) 11(11):1048–56. 10.1016/s1470-2045(10)70230-8 [DOI] [PubMed] [Google Scholar]
- 20. Jenkins D, Molijn A, Kazem S, Pirog EC, Alemany L, de Sanjosé S, et al. Molecular and pathological basis of HPV-negative cervical adenocarcinoma seen in a global study. Int J Cancer (2020) 147(9):2526–36. 10.1002/ijc.33124 [DOI] [PubMed] [Google Scholar]
- 21. Hopenhayn C, Christian A, Christian WJ, Watson M, Unger ER, Lynch CF, et al. Prevalence of human papillomavirus types in invasive cervical cancers from 7 US cancer registries before vaccine introduction. J Low Genit Tract Dis (2014) 18(2):182–9. 10.1097/LGT.0b013e3182a577c7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tjalma W. HPV negative cervical cancers and primary HPV screening. Facts Views Vis Obgyn 10(2):107–13. 10.5858/arpa.2016-0356-RA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karpathiou G, Chauleur C, Hathroubi S, Peoc’h M. Secondary Tumors of the Gynecologic Tract: A Clinicopathologic Analysis. Int J Gynecol Pathol (2019) 38(4):363–70. 10.1097/PGP.0000000000000516 [DOI] [PubMed] [Google Scholar]
- 24. Mazur MT, Hsueh S, Gersell DJ. Metastases to the female genital tract. Analysis of 325 cases. Cancer (1984) 53(9):1978–84. [DOI] [PubMed] [Google Scholar]
- 25. Gravitt PE, Winer RL. Natural History of HPV Infection across the Lifespan: Role of Viral Latency. Viruses (2017) 9(10):267. 10.3390/v9100267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katki HA, Kinney WK, Fetterman B, Lorey T, Poitras NE, Cheung L, et al. Cervical cancer risk for women undergoing concurrent testing for human papillomavirus and cervical cytology: a population-based study in routine clinical practice. Lancet Oncol (2011) 12(7):663–72. 10.1016/S1470-2045(11)70145-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res (2014) 24(2):185–99. 10.1101/gr.164806.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsakogiannis D, Gartzonika C, Levidiotou-Stefanou S, Markoulatos P. Molecular approaches for HPV genotyping and HPV-DNA physical status. Expert Rev Mol Med (2017) 19:e1. 10.1017/erm.2017.2 [DOI] [PubMed] [Google Scholar]
- 29. Tjalma WAA, Depuydt CE. Cervical cancer screening: which HPV test should be used–L1 or E6/E7? Eur J Obstet Gynecol Reprod Biol (2013) 170(1):45–6. 10.1016/j.ejogrb.2013.06.027 [DOI] [PubMed] [Google Scholar]
- 30. Banister CE, Liu C, Pirisi L, Creek KE, Buckhaults PJ. Identification and characterization of HPV-independent cervical cancers. Oncotarget (2017) 8(8):13375–86. 10.18632/oncotarget.14533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. González-Bosquet E, Muñoz A, Suñol M, Lailla JM. Cervical cancer and low-risk HPV; a case report. Eur J Gynaecol Oncol (2006) 27(2):193–4. [PubMed] [Google Scholar]
- 32. Guimerà N, Lloveras B, Alemany L, Iljazovic E, Shin HR, Jung-Il S, et al. Laser capture microdissection shows HPV11 as both a causal and a coincidental infection in cervical cancer specimens with multiple HPV types. Histopathology (2013) 63(2):287–92. 10.1111/his.12137 [DOI] [PubMed] [Google Scholar]
- 33. Guimerà N, Lloveras B, Lindeman J, Alemany L, van de Sandt M, Alejo M, et al. The occasional role of low-risk human papillomaviruses 6, 11, 42, 44, and 70 in anogenital carcinoma defined by laser capture microdissection/PCR methodology: results from a global study. Am J Surg Pathol (2013) 37(9):1299–310. 10.1097/PAS.0b013e31828b6be4 [DOI] [PubMed] [Google Scholar]
- 34. Ratnam S, Coutlee F, Fontaine D, Bentley J, Escott N, Ghatage P, et al. Aptima HPV E6/E7 mRNA test is as sensitive as Hybrid Capture 2 Assay but more specific at detecting cervical precancer and cancer. J Clin Microbiol (2011) 49(2):557–64. 10.1128/JCM.02147-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luu HN, Dahlstrom KR, Mullen PD, VonVille HM, Scheurer ME. Comparison of the accuracy of Hybrid Capture II and polymerase chain reaction in detecting clinically important cervical dysplasia: a systematic review and meta-analysis. Cancer Med (2013) 2(3):367–90. 10.1002/cam4.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cui M, Chan N, Liu M, Thai K, Malaczynska J, Singh I, et al. Clinical performance of Roche Cobas 4800 HPV Test. J Clin Microbiol (2014) 52(6):2210–1. 10.1128/JCM.00883-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haedicke J, Iftner T. A review of the clinical performance of the Aptima HPV assay. J Clin Virol (2016) 76 Suppl 1:S40–S8. 10.1016/j.jcv.2015.10.027 [DOI] [PubMed] [Google Scholar]
- 38. Tao K, Yang J, Yang H, Guo Z-H, Hu Y-M, Tan Z-Y, et al. Comparative study of the cervista and hybrid capture 2 methods in detecting high-risk human papillomavirus in cervical lesions. Diagn Cytopathol (2014) 42(3):213–7. 10.1002/dc.23025 [DOI] [PubMed] [Google Scholar]
- 39. Heideman DAM, Hesselink AT, Berkhof J, van Kemenade F, Melchers WJG, Daalmeijer NF, et al. Clinical validation of the cobas 4800 HPV test for cervical screening purposes. J Clin Microbiol (2011) 49(11):3983–5. 10.1128/JCM.05552-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Day SP, Hudson A, Mast A, Sander T, Curtis M, Olson S, et al. Analytical performance of the Investigational Use Only Cervista HPV HR test as determined by a multi-center study. J Clin Virol (2009) 45 Suppl 1:S63–72. 10.1016/S1386-6532(09)70010-1 [DOI] [PubMed] [Google Scholar]
- 41. Einstein MH, Martens MG, Garcia FAR, Ferris DG, Mitchell AL, Day SP, et al. Clinical validation of the Cervista HPV HR and 16/18 genotyping tests for use in women with ASC-US cytology. Gynecol Oncol (2010) 118(2):116–22. 10.1016/j.ygyno.2010.04.013 [DOI] [PubMed] [Google Scholar]
- 42. Boehmer G, Wang L, Iftner A, Holz B, Haedicke J, von Wasielewski R, et al. A population-based observational study comparing Cervista and Hybrid Capture 2 methods: improved relative specificity of the Cervista assay by increasing its cut-off. BMC Infect Dis (2014) 14:674. 10.1186/s12879-014-0674-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rao A, Young S, Erlich H, Boyle S, Krevolin M, Sun R, et al. Development and characterization of the cobas human papillomavirus test. J Clin Microbiol (2013) 51(5):1478–84. 10.1128/JCM.03386-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castle PE, Eaton B, Reid J, Getman D, Dockter J. Comparison of human papillomavirus detection by Aptima HPV and cobas HPV tests in a population of women referred for colposcopy following detection of atypical squamous cells of undetermined significance by Pap cytology. J Clin Microbiol (2015) 53(4):1277–81. 10.1128/JCM.03558-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Castle PE, Stoler MH, Wright TC, Sharma A, Wright TL, Behrens CM. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol (2011) 12(9):880–90. 10.1016/S1470-2045(11)70188-7 [DOI] [PubMed] [Google Scholar]
- 46. Castle PE, Reid J, Dockter J, Getman D. The reliability of high-risk human papillomavirus detection by Aptima HPV assay in women with ASC-US cytology. J Clin Virol (2015) 69:52–5. 10.1016/j.jcv.2015.05.006 [DOI] [PubMed] [Google Scholar]
- 47. Wright TC, Jr., Stoler MH, Agreda PM, Beitman GH, Gutierrez EC, Harris JM, et al. Clinical performance of the BD Onclarity HPV assay using an adjudicated cohort of BD SurePath liquid-based cytology specimens. Am J Clin Pathol (2014) 142(1):43–50. 10.1309/ajcp53kmhnrdicbl [DOI] [PubMed] [Google Scholar]
- 48. Cuschieri K, Geraets DT, Moore C, Quint W, Duvall E, Arbyn M. Clinical and Analytical Performance of the Onclarity HPV Assay Using the VALGENT Framework. J Clin Microbiol (2015) 53(10):3272–9. 10.1128/jcm.01366-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakamura M, Nakade K, Orisaka S, Iwadare J, Mizumoto Y, Fujiwara H. Comparison Study of BD Onclarity HPV With digene HC2 High-Risk HPV DNA Test and Roche Cobas 4800 HPV for Detecting High-Risk Human Papillomavirus in Japan. Am J Clin Pathol (2019) 151(3):263–9. 10.1093/ajcp/aqy124 [DOI] [PubMed] [Google Scholar]
- 50. BD . Onclarity™ HPV Assay. (2019). https://legacy.bd.com/resource.aspx?IDX=30114.
- 51. Poljak M, Kocjan BJ, Oštrbenk A, Seme K. Commercially available molecular tests for human papillomaviruses (HPV): 2015 update. J Clin Virol (2016) 76 Suppl 1S3–S13. 10.1016/j.jcv.2015.10.023 [DOI] [PubMed] [Google Scholar]
- 52. Xiao G-Q, Emanuel POM. Cervical parakeratosis/hyperkeratosis as an important cause for false negative results of Pap smear and human papillomavirus test. Aust N Z J Obstet Gynaecol (2009) 49(3):302–6. 10.1111/j.1479-828X.2009.00998.x [DOI] [PubMed] [Google Scholar]
- 53. Tao X, Zheng B, Yin F, Zeng Z, Li Z, Griffith CC, et al. Polymerase Chain Reaction Human Papillomavirus (HPV) Detection and HPV Genotyping in Invasive Cervical Cancers With Prior Negative HC2 Test Results. Am J Clin Pathol (2017) 147(5):477–83. 10.1093/ajcp/aqx027 [DOI] [PubMed] [Google Scholar]
- 54. Sal V, Kahramanoglu I, Turan H, Tokgozoglu N, Bese T, Aydin O, et al. Primary signet ring cell carcinoma of the cervix: A case report and review of the literature. Int J Surg Case Rep (2016) 21:1–5. 10.1016/j.ijscr.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ueno S, Sudo T, Oka N, Wakahashi S, Yamaguchi S, Fujiwara K, et al. Absence of human papillomavirus infection and activation of PI3K-AKT pathway in cervical clear cell carcinoma. Int J Gynecol Cancer (2013) 23(6):1084–91. 10.1097/IGC.0b013e3182981bdc [DOI] [PubMed] [Google Scholar]
- 56. Mirkovic J, Sholl LM, Garcia E, Lindeman N, MacConaill L, Hirsch M, et al. Targeted genomic profiling reveals recurrent KRAS mutations and gain of chromosome 1q in mesonephric carcinomas of the female genital tract. Modern Pathol (2015) 28(11):1504–14. 10.1038/modpathol.2015.103 [DOI] [PubMed] [Google Scholar]
- 57. Nicolas I, Marimon L, Barnadas E, Saco A, Rodriguez-Carunchio L, Fuste P, et al. HPV-negative tumors of the uterine cervix. Mod Pathol (2019) 32(8):1189–96. 10.1038/s41379-019-0249-1 [DOI] [PubMed] [Google Scholar]
- 58. van der Marel J, van Baars R, Quint WGV, Berkhof J, del Pino M, Torné A, et al. The impact of human papillomavirus genotype on colposcopic appearance: a cross-sectional analysis. BJOG (2014) 121(9):1117–26. 10.1111/1471-0528.12668 [DOI] [PubMed] [Google Scholar]
- 59. Feng D, Xu H, Li X, Wei Y, Jiang H, Xu H, et al. An association analysis between mitochondrial DNA content, G10398A polymorphism, HPV infection, and the prognosis of cervical cancer in the Chinese Han population. Tumour Biol (2016) 37(4):5599–607. 10.1007/s13277-015-4429-4 [DOI] [PubMed] [Google Scholar]
- 60. Stolnicu S, Hoang L, Chiu D, Hanko-Bauer O, Terinte C, Pesci A, et al. Clinical Outcomes of HPV-associated and Unassociated Endocervical Adenocarcinomas Categorized by the International Endocervical Adenocarcinoma Criteria and Classification (IECC). Am J Surg Pathol (2019) 43(4):466–74. 10.1097/pas.0000000000001224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feng S, Yang Y, Lv J, Sun L, Liu M. Valproic acid exhibits different cell growth arrest effect in three HPV-positive/negative cervical cancer cells and possibly via inducing Notch1 cleavage and E6 downregulation. Int J Oncol (2016) 49(1):422–30. 10.3892/ijo.2016.3508 [DOI] [PubMed] [Google Scholar]
- 62. Girón RA, Montaño LF, Escobar ML, López-Marure R. Dehydroepiandrosterone inhibits the proliferation and induces the death of HPV-positive and HPV-negative cervical cancer cells through an androgen- and estrogen-receptor independent mechanism. FEBS J (2009) 276(19):5598–609. 10.1111/j.1742-4658.2009.07253.x [DOI] [PubMed] [Google Scholar]
- 63. Kalu NN, Mazumdar T, Peng S, Shen L, Sambandam V, Rao X, et al. Genomic characterization of human papillomavirus-positive and -negative human squamous cell cancer cell lines. Oncotarget (2017) 8(49):86369–83. 10.18632/oncotarget.21174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu Y, Zhao R, Fang S, Li Q, Jin Y, Liu B. Abemaciclib sensitizes HPV-negative cervical cancer to chemotherapy via specifically suppressing CDK4/6-Rb-E2F and mTOR pathways. Fundam Clin Pharmacol (2020). 10.1111/fcp.12574 [DOI] [PubMed] [Google Scholar]
- 65. Ta W, Zhang Y, Zhang S, Sun P. LncRNA ANCR downregulates hypoxia−inducible factor 1α and inhibits the growth of HPV−negative cervical squamous cell carcinoma under hypoxic conditions. Mol Med Rep (2020) 21(1):413–9. 10.3892/mmr.2019.10792 [DOI] [PubMed] [Google Scholar]
- 66. Ju W, Luo X, Zhang N. LncRNA NEF inhibits migration and invasion of HPV-negative cervical squamous cell carcinoma by inhibiting TGF-β pathway. Biosci Rep (2019) 39(4)BSR20180878. 10.1042/bsr20180878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Riou G, Favre M, Jeannel D, Bourhis J, Le Doussal V, Orth G. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA. Lancet (1990) 335(8699):1171–4. 10.1016/0140-6736(90)92693-c [DOI] [PubMed] [Google Scholar]
- 68. Li P, Tan Y, Zhu L-X, Zhou L-N, Zeng P, Liu Q, et al. Prognostic value of HPV DNA status in cervical cancer before treatment: a systematic review and meta-analysis. Oncotarget (2017) 8(39):66352–9. 10.18632/oncotarget.18558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pilch H, Günzel S, Schäffer U, Tanner B, Brockerhoff P, Maeurer M, et al. The presence of HPV DNA in cervical cancer: correlation with clinico-pathologic parameters and prognostic significance: 10 years experience at the Department of Obstetrics and Gynecology of the Mainz University. Int J Gynecol Cancer (2001) 11(1):39–48. 10.1046/j.1525-1438.2001.011001039.x [DOI] [PubMed] [Google Scholar]
- 70. Weaver B, Shew M, Qadadri B, Tu W, Tong Y, Denski C, et al. Low-level persistence of human papillomavirus 16 DNA in a cohort of closely followed adolescent women. J Med Virol (2011) 83(8):1362–9. 10.1002/jmv.22116 [DOI] [PubMed] [Google Scholar]
- 71. Vassilakos P, Tran PL, Sahli R, Low N, Petignat P. HPV-negative CIN3 and cervical cancer in Switzerland: any evidence of impact on screening policies? Swiss Med Wkly (2017) 147:w14559. 10.4414/smw.2017.14559 [DOI] [PubMed] [Google Scholar]
- 72. Pilch H, Günzel S, Schäffer U, Tanner B, Brockerhoff P, Maeurer M, et al. Human papillomavirus (HPV) DNA in primary cervical cancer and in cancer free pelvic lymph nodes–correlation with clinico-pathological parameters and prognostic significance. Zentralbl Gynakol (2001) 123(2):91–101. 10.1055/s-2001-12411 [DOI] [PubMed] [Google Scholar]