Abstract
8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), a major product of DNA oxidation, is a pre-mutagenic lesion which is prone to mispair, if left unrepaired, with 2′-deoxyadenosine during DNA replication. While unrepaired or incompletely repaired 8-oxodG has classically been associated with genome instability and cancer, it has recently been reported to have a role in the epigenetic regulation of gene expression.
Despite the growing collection of genome-wide 8-oxodG mapping studies that have been used to provide new insight on the functional nature of 8-oxodG within the genome, a comprehensive view that brings together the epigenetic and the mutagenic nature of the 8-oxodG is still lacking.
To help address this gap, this review aims to provide (i) a description of the state-of-the-art knowledge on both the mutagenic and epigenetic roles of 8-oxodG; (ii) putative molecular models through which the 8-oxodG can cause genome instability; (iii) a possible molecular model on how 8-oxodG, acting as an epigenetic signal, could cause the translocations and deletions which are associated with cancer.
Keywords: 8-oxodG, Base excision repair, TOP1-DPC, Single-Strand break, Double-strand break, Genome instability, Cancer
1. Introduction
DNA is a dynamic molecule that is continuously subjected to changes, many of which may involve the alteration of both the DNA backbone and the normal nucleobases [1]. Some of these alterations reflect normal modifications of DNA, but others represent DNA damage that has been implicated to have a role in pathological processes such as neurodegeneration [2], ageing [3], and cancer [4]. It has been estimated that, under physiological conditions, every cell, every day has to address the formation of approximately 70,000 DNA nucleobase lesions that, if unrepaired, represent a serious threat to genome integrity [1,5,6].
Among the various agents that damage DNA, reactive oxygen species (ROS) deserve particular attention due to the almost omnipresence, and their ability to both compromise the structure/function of DNA, and alter the associated physiological processes [7,8]. Some ROS, such as the hydroxyl radical (•OH), are also free radicals, and contain an atom or a molecule with an unpaired electron that makes them highly reactive and capable of oxidizing molecules upon contact. Sources of ROS can be exogenous, such as ultraviolet (UV) light, ionizing radiation (IR), toxins, chemicals, and pollutants, or endogenous, such as cellular metabolism in the mitochondria and peroxisomes [9–11] (Fig. 1).
Fig. 1.
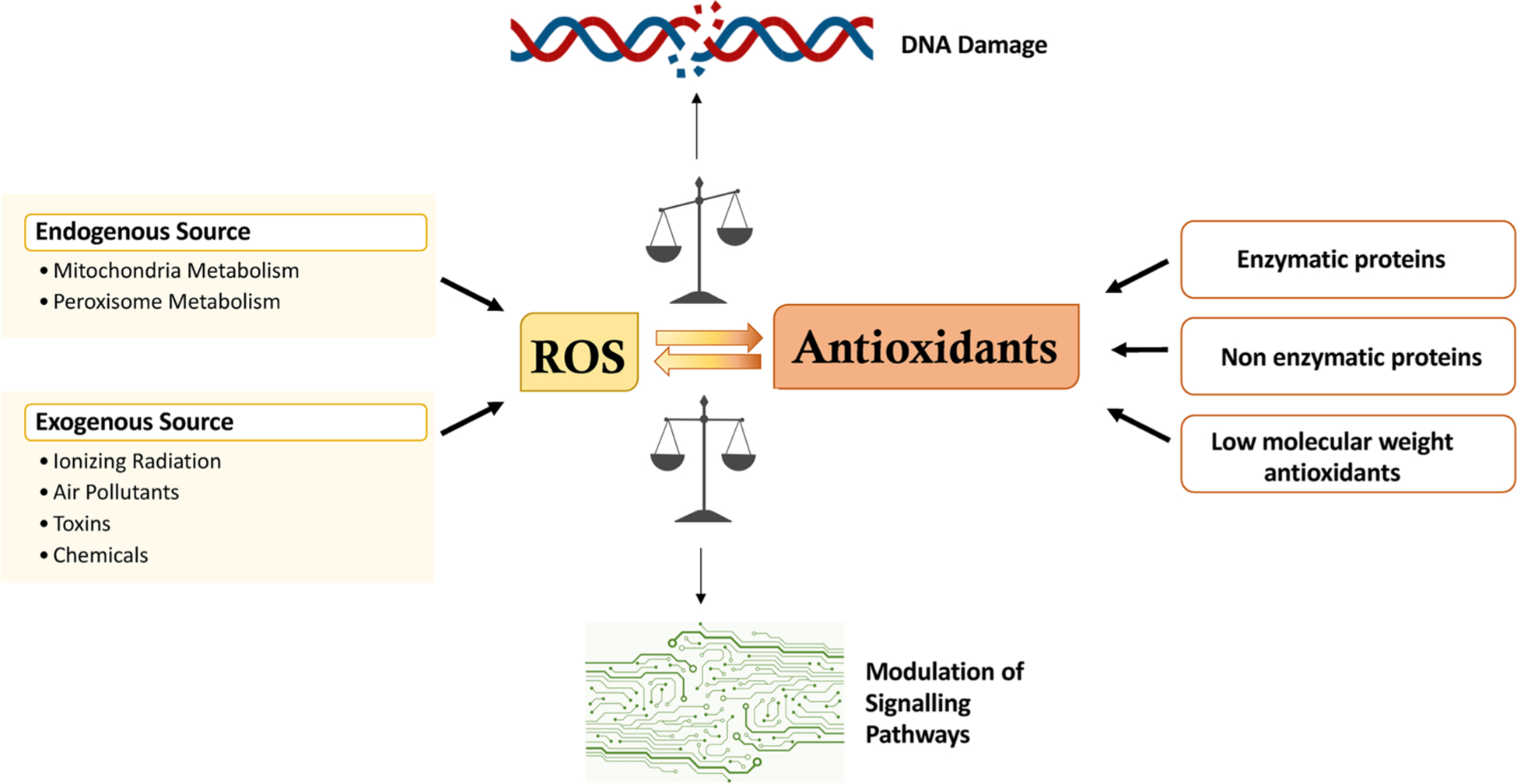
Schematic representation of balance between ROS production and antioxidant defense. ROS can be produced by endo- or exogenous sources. The oxidative damage is prevented by enzymatic or non-enzymatic proteins and low molecular weight antioxidants. Under a physiological cellular state, the level of ROS is stable in a dynamic equilibrium balanced by oxidants and antioxidants and the physiological levels of ROS modulate the signalling pathways. When the levels of oxidants and antioxidants are imbalanced, the ROS increase and produce different types of DNA damage ranging from base modifications and transversions to genomic instability.
To counter the production of ROS, cells possess enzymatic proteins (such as the superoxide dismutases, SOD, the glutathione and ascorbate peroxidases and peroxisomal catalase) that, acting in concert with non-enzymatic proteins (e.g. peroxiredoxins, thioredoxins, glutaredoxin and metallothionein) [12–16], and together with low molecular weight antioxidants (e.g. glutathione, ascorbate, carotenoids and melatonin) [16, 17], are able to prevent the formation of oxidatively generated damage to important macromolecules such as lipids, proteins and nucleic acids [18] (Fig. 1).
While ROS play a physiological role in cell function such as an intracellular signalling molecule [19–22], an overproduction of ROS, if not counteracted by antioxidants, represents a serious problem for cell health [23]. To maintain a healthy cell status, oxidants and antioxidants should be in a state of equilibrium. Imbalance of this equilibrium in favour of the oxidants causes oxidative stress (OS) that, with an increase of ROS levels, induces improper signalling and increased oxidation of macromolecules [23–25]. In particular, ROS can have a dramatic impact on DNA generating a spectrum of types of damage ranging from nucleobase and sugar modifications, to modification and breakage of the phosphate backbone [26–29] (Fig. 1). To mitigate these consequences, cells possess a number of pathways, such as DNA repair, cell cycle arrest, autophagy and apoptosis, which are all involved in the protective DNA damage response (DDR) [23,30,31].
2. Endogenous sources of ROS
ROS are produced continuously upon exposure to external agents such as radiation and pollutants, along with endogenous metabolism [9–11]. Specifically, ROS are mainly generated from oxidative metabolism within the mitochondria, where the majority of oxygen is converted to water and only a small percent (0.2–2 %) to superoxide anions () [32] (Fig. 2). Superoxide anions can be converted to hydrogen peroxide (H2O2) in the cytosol, either spontaneously or through catalysis by SOD [33]. Next, H2O2 can be reduced to H2O, or partially reduced to OH•, through the metal catalysed Haber-Weiss reaction in the presence of reduced transition metals [e.g. Fe2+ (Fenton reaction) and Cu+] [34]. •OH molecules are also formed from the decay of the reactive nitrogen species such as the peroxynitrite (ONOO-). •OH is the main strong cellular oxidant and, even though it has been well documented in vivo to cause DNA nucleobase modifications, and single-strand breaks (SSBs) [35], it remains unclear how cytosol-produced •OH can travel into the nucleus. Indeed, such is the reactivity of the hydroxyl radical that it reacts immediately with any molecule close to its site of formation, making it difficult to explain how it can be produced in the cytosol and then diffuse to the nucleus [36]. To address this, it has been proposed that H2O2, being a molecule much more able to able to diffuse [37], and less reactive, than •OH, can diffuse from the site of production into the nucleus, or alternatively can be also produced directly in the nucleus where, through the Fenton, or other metal-dependent reactions, can give rise to local •OH, and hence DNA damage.
Fig. 2.
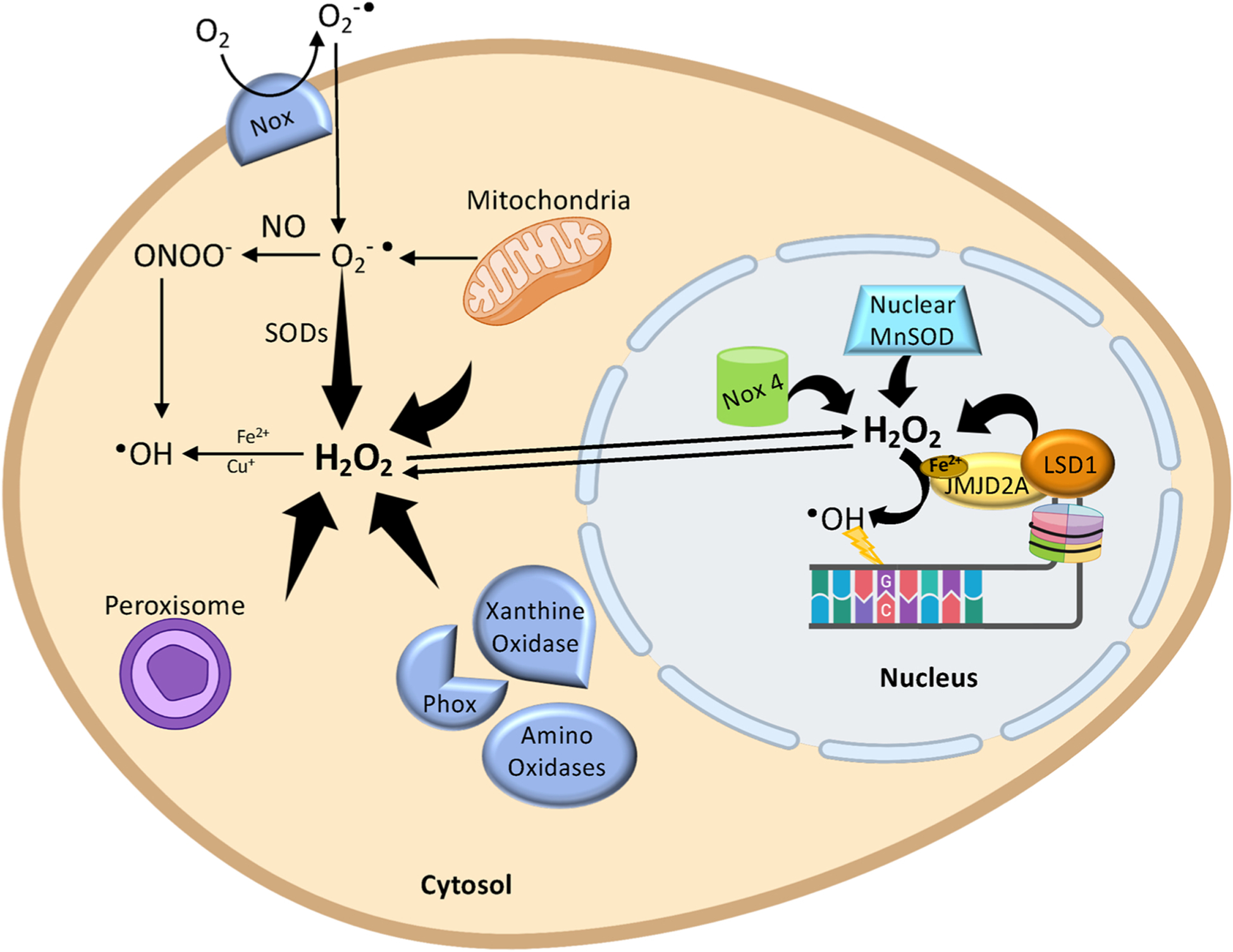
The formation of reactive oxygen species (ROS) in the cell. First, the transmembrane NADPH oxidases (NOX) and mitochondria generate superoxide anion (O2) which diffuses in the cytoplasm. Then, O2can be converted to peroxynitrite (ONOO-) in the presence of NO or can be converted to H2O2 by superoxide dismutase (SOD). The cytosolic H2O2 may also be generated from different oxidases (Amino oxidases, Phox and Xanthine oxidase) or from peroxisome or from mitochondria. Next, H2O2 can be partially reduced to •OH, through the metal-catalysed Haber-Weiss reaction in the presence of reduced transition metals [e.g. Fe2+ (Fenton reaction) and Cu+]. •OH molecules are also formed from the ONOO-. The H2O2 may diffuse from the cytosol to the nucleus or can be produced directly into the nucleus by LSD1 demethylation reaction or by other nuclear oxidases. Finally, through the Fenton reaction, the H2O2 is converted into OH•, which in turn oxidizes 2′-deoxyguanosine (created with biorender.com).
Sources of nuclear H2O2 have been identified and, besides nuclear oxidases (e.g. NOX4 and MnSOD) [38,39], Lysine-Specific histone Demethylases 1 and 2 (LSD1 and LSD2) have also been recently associated with the production of H2O2 in the nucleus [40,41](Fig. 2). LSD1 and 2 are able to demethylate the mono- and di-methylated Lys4 and Lys9 of histone H3 through a flavin adenine dinucleotide (FAD)-dependent oxidative reaction [40–42]. During the demethylation, the LSD1 and 2 enzymes use as cofactor the FAD that is reduced to FADH2 and then reoxidized to FAD by oxygen with the generation of formaldehyde (the methylating agent) and H2O2 (the oxidizing agent). Intriguingly, the LSD1-associated production of H2O2 promotes the formation of 8-oxodG [43–50]. This suggests that in the nucleus, even if levels of free iron are lowered by the pool of nuclear ferritin [51], compared to other intracellular compartments, H2O2 may still be converted to •OH through the Fenton reaction [34], probably involving iron-containing complexes associated with the DNA structure [52,53] (Fig. 2). Supportive of this suggestion is a report that members of the Jumonji-type demethylases, a superfamily of oxygenases containing a Fe2+ ion in their catalytic domain, can form complexes with LSD1 and H2O2 produced by LSD1 activity reduced in the hydroxide ion by the oxidation of the Jumonji-contained Fe2+ ion [49]. In close proximity of DNA molecules, this event may be responsible for the oxidation of nucleobases. Indeed, depletion of iron-containing JMJD2A, a Jumonji demethylase that interacts with LSD1 to activate TGF-β1-induced genes, inhibits the formation of nuclear 8-oxodG [49]. Together, these data support the hypothesis that •OH can be generated from H2O2 produced at sites close enough to react with DNA.
3. 2′-Deoxyguanosine is a major target for DNA oxidation
All DNA nucleobases are susceptible to damage by ROS [9,54,55], either at the level of the free nucleotide (in the dNTP pool) [56], or as the 2′-deoxynucleoside in the context of the DNA molecule. Deoxyguanosine (dG) is particularly vulnerable to oxidation due to its low oxidation potential [57,58]. The primary products of dG oxidation are 8-oxo-7, 8-dihydro-2′-deoxyguanosine (8-oxodG) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapydG) (Fig. 3). Moreover, 8-oxodG can be oxidized further producing guanidinohydantoin and spiroiminodihydantoin. The main products of dA oxidation are 8-oxo-7, 8-dihydro-2′-deoxyadenosine (8-oxodA), 4,6-diamino-5-formamidopyrimidine (FapydA) and 2-hydroxy-2′-deoxyadenosine (2-oxodA). Pyrimidines can also be oxidized, and this leads to the formation of products such as 5-hydroxy-2′-deoxycytosine (5–OHdC), 5, 6-dihydro-2′-deoxythymidine (DHdT) and 5-hydroxymethyl-2′-deoxydeoxyuridine (5hmdU) [59] (Fig. 3).
Fig. 3.
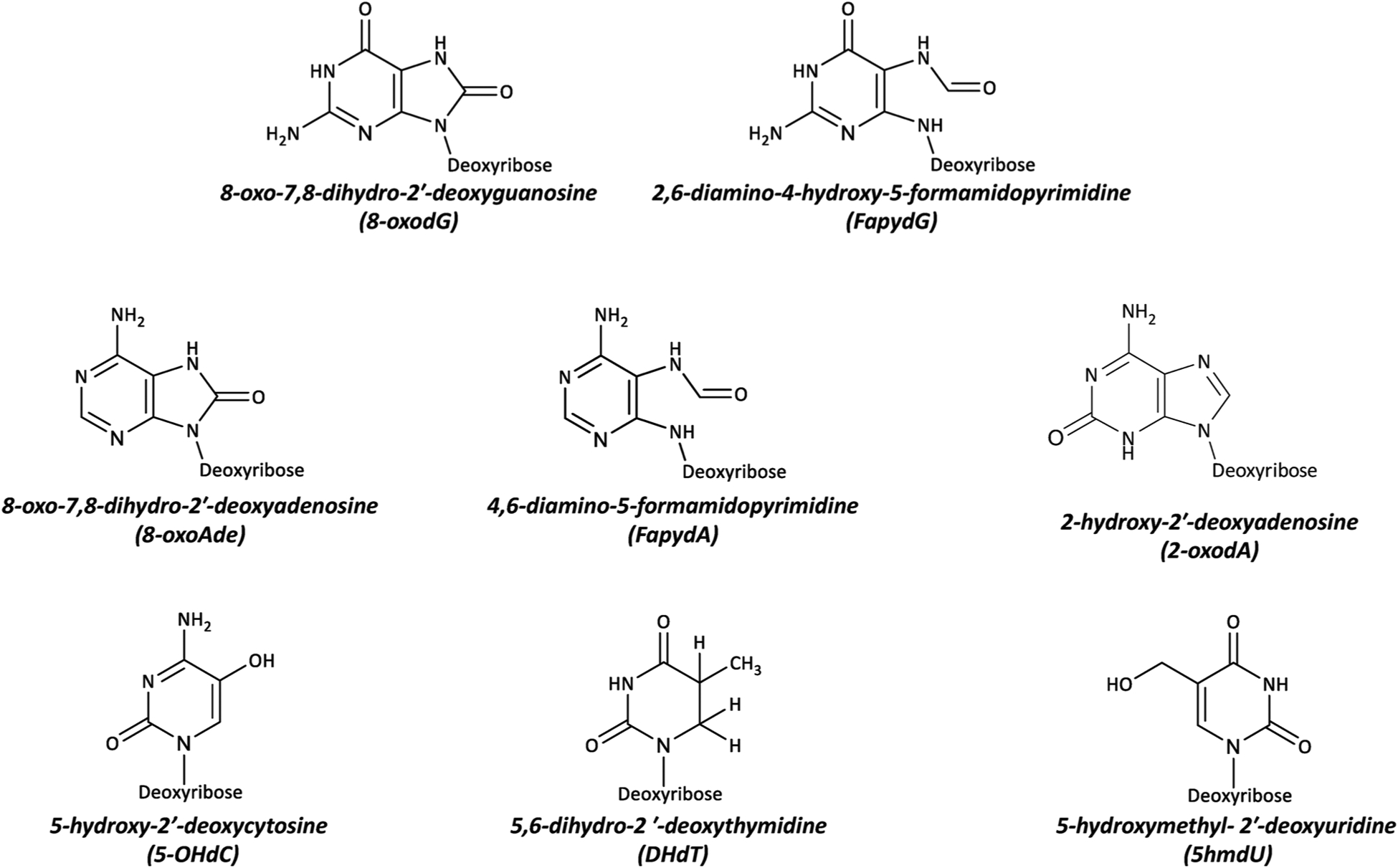
The most common DNA lesions generated by interaction of ROS with the deoxynucleosides.
8-oxodG is understood to be the most abundant [5,59], oxidatively modified DNA lesion and is generated by the introduction of an oxo group on the C8 position, and addition of a hydrogen atom on the N7 of the imidazole ring of dG (Fig. 4A).
Fig. 4.
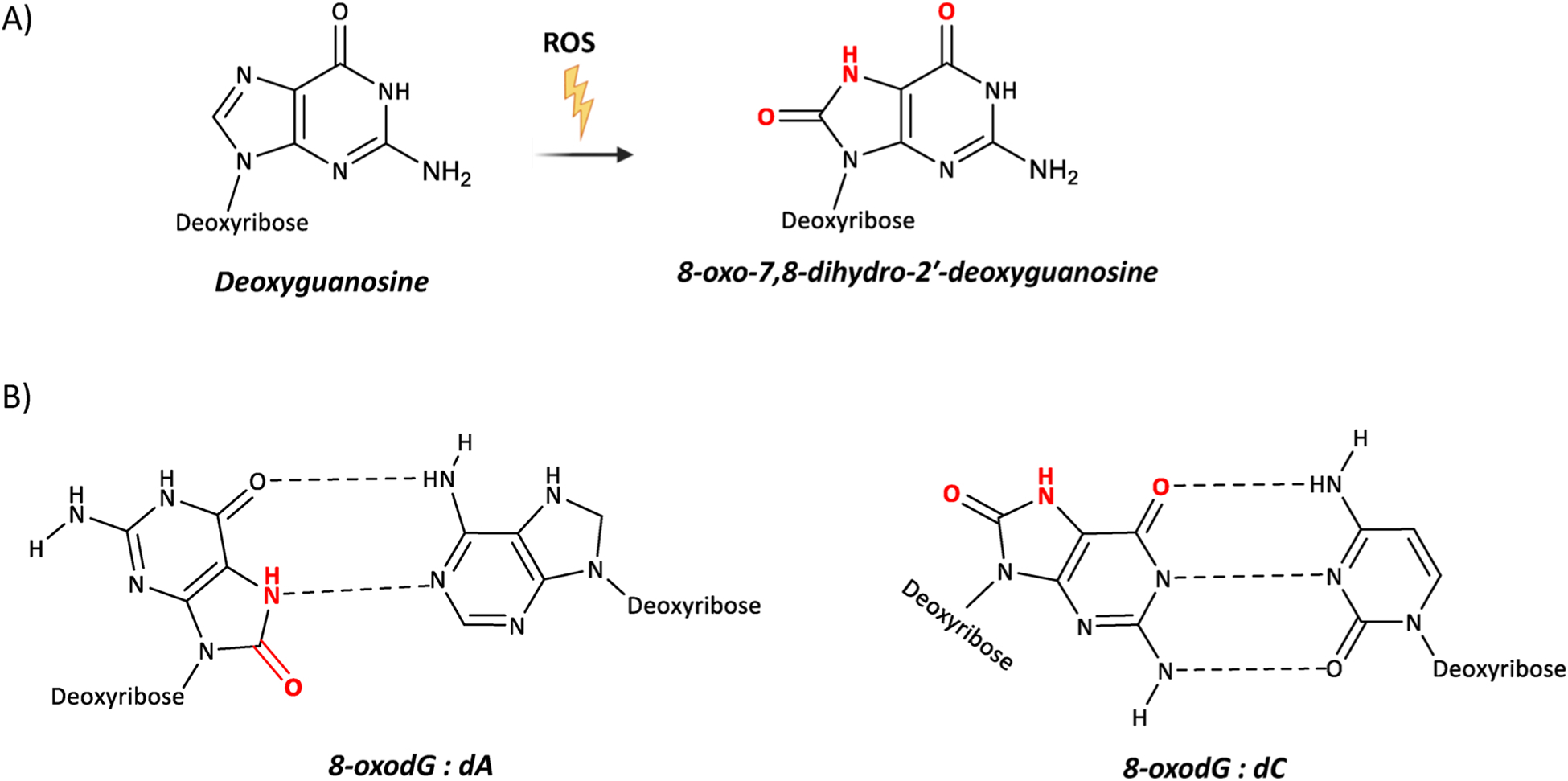
Chemical structures of 8-oxodG and its altered base-pairs. (A) ROS oxidize 2′-deoxyguanosine and generate 8-oxo-7,8-dihydro-deoxyguanosine (8-oxodG). (B) 8-oxodG in the syn conformation forms Hoogsteen base pair with 2′-deoxyadenosine, whereas 8-oxodG in the anti conformation forms Watson-Crick base pair with 2′-deoxycytidine.
8-oxodG is described as a premutagenic DNA lesion because when present in DNA during replication it leads to a dC:dG to dA:dT transversion [60–64] (Fig. 4B). As for all the other nucleobases, 8-oxodG is able to assume the anti or syn conformation, depending on the angle around the glycosidic bond. When oxygen is present on the C8 position, the syn conformation is more energetically favourable. What nucleobase base-pairs with 8-oxodG depends upon whether the syn and anti conformation is adopted (Fig. 4B). Indeed, 8-oxodG in the syn conformation is able to structurally mimic deoxythymidine (dT), and this, during DNA replication, accounts for a pro-mutagenic of 8-oxodG (syn): dA (anti) Hoogsteen base mispairing [60–64]. The 8-oxodG:dA mispair often evades DNA repair (e.g. proofreading activity of replicative polymerases) because it structurally mimics the dT:dA base pair, and does not result in any distortion of the DNA helix structure [65]. Conversely, the 8-oxodG:dC Watson-Crick base pair, induces a modest distortion of the DNA helix structure, and this is more easily recognized by the DNA repair proteins, leading to the removal of 8-oxodG.
4. Base excision repair of 8-oxodG
Base excision repair (BER) is the main mechanism of pre-replicative removal of 8-oxodG [5,66]. The role of BER is to specifically recognize and repair (largely non-bulky) DNA nucleobase alterations and SSBs [67,68] which, if left unrepaired, could be converted into DSBs during DNA replication [69].
BER can proceed via two different pathways – so-called short-patch and long-patch repair, which involve subsets of repair proteins that operate independently [68,70,71]. The majority of repair events involve the classic short-patch mechanism (characterised by the replacement of a single damaged nucleotide) [70,71] (Fig. 5). Long patch BER, involves the processing of 2–12 nucleotides, including the damaged one. The choice of which of these two subpathways is followed depends upon the DNA glycosylase which initiates BER [71,72]. BER DNA glycosylases can be either monofunctional, or bifunctional, and with either β-lyase activity or both β and δ lyase activity [9,71,73].
Fig. 5.
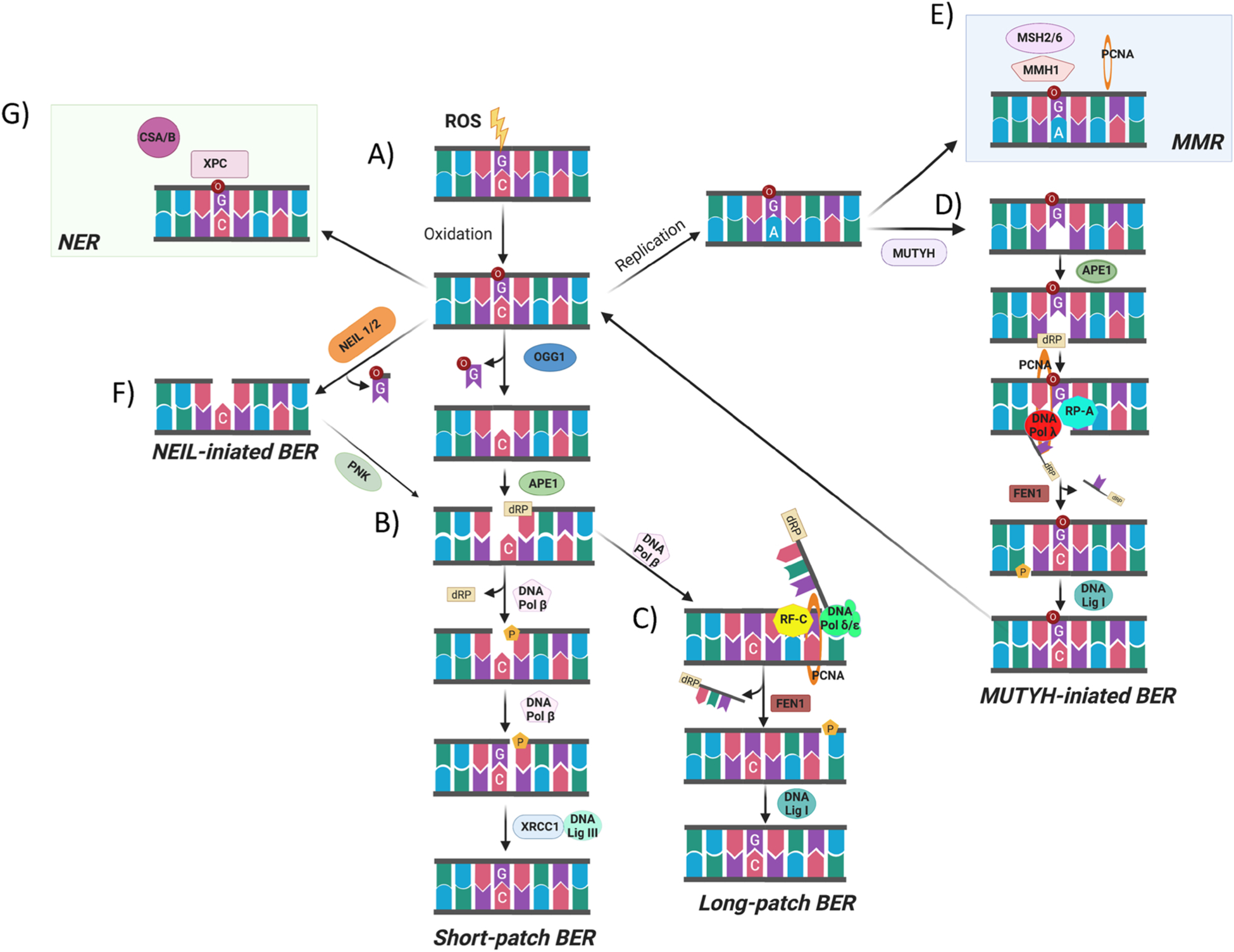
Scheme of 8-oxodG repair pathways. (A) When ROS attack DNA, this leads to the oxidation of 2′-deoxyguanosine and the formation of 8-oxodG which can pair with deoxycytidine. (B) The majority of 8-oxodG is recognized and removed by the bifunctional DNA glycosylase OGG1 through the short patch BER. Upon the removal of the oxidized nucleobase, the DNA polymerase β repairs the single-nucleotide gap by inserting a 2′-deoxyguanosine and then the DNA ligase III/XRCC1 complex mediates the final ligation step. (C) If the 5-deoxyribose phosphate (5′-dRP) terminus cannot be processed by SSB-end-processing enzymes, the DNA pol β incorporates the first nucleotide into the nick and the DNA pol δ/ε performs strand displacement synthesis creating a 5′-flap structure which is, in a PCNA-dependent manner, recognized and excised by the endonuclease activity of FEN-1. Finally, the DNA ligase I coordinates the final ligation step in long-patch BER. (D) When the 8-oxodG is not recognized by OGG1 before the S-phase, the replicative polymerase could incorporate 2′-deoxyadenosine and the resulting 8-oxodG:dA mispair may be processed by MMR components such as MSH2/6,MMH1and PCNA, to remove the mis-paired native 2′-deoxynucleoside. (E) Alternatively, the 2′-deoxyadenosine introduced during the replication could be recognized and removed by MYH mediated BER. (F) NEIL1 or NEIL2 could also initiate the 8-oxodG repair process. (G) the CSA and CSB together with XPC, as components of the TC-NER pathway, have been proposed also to repair 8-oxodG (created with biorender.com).
Monofunctional DNA glycosylases perform excision of the nucleobase only, releasing the modified nucleobase and creating an apurinic/apyrimidinic (AP) site. In contrast, the bifunctional glycosylases excise the modified nucleobase and subsequently hydrolyze, via the associated AP lyase activity, the DNA backbone. This occurs through a β-elimination step forming a 3′-α,β-unsaturated aldehyde adjacent to a 5′-phosphate. Some bifunctional DNA glycosylases can also perform a δ-elimination step, in which convert the 3′-aldehyde to a 3′-phosphate [9,68,70,71,73].
The predominant BER mechanism for the removal of 8-oxodG is initiated through the specific bifunctional glycosylase 8-oxodG DNA glycosylase 1 (OGG1), which recognizes and then excises the 8-oxodG from the sugar-phosphate backbone. OGG1 does have a weak AP lyase activity, and cleaves the DNA at the abasic site via a β-elimination mechanism generating a 3′-phospho-α,β-unsaturated aldehyde terminus (3′-dRP) and 5′-phosphate (AP) site. Apurinic/apyrimidinic endonuclease 1 (APE1) then cleaves the DNA phosphate backbone creating an SSB forming a polymerase-ready 3′-OH residue. At this point, short patch BER proceeds with the repair of the arising SSBs; specifically, DNA polymerase β removes the downstream 5′-sugar-phosphate, using its dRP-lyase activity, and repairs the single-nucleotide gap inserting one nucleotide. Then, the DNA ligase III/X-ray repair cross-complementing protein 1 (XRCC1) complex ligates the 3′-OH group of the newly inserted nucleotide with the downstream 5′-phosphate finishing the short-patch BER process [9,70,71].
If the 5′-termini cannot be processed by SSB end-processing enzymes, the long-patch pathway leads to the repair of the SSB by replacing a stretch of 2–12 nucleotides (Fig. 5). In long-patch BER, DNA pol β incorporates the first nucleotide into the nick, while the replicative DNA pol δ is required in the elongation step to perform strand displacement synthesis. In this scenario, replication factor C (RF—C) loads the sliding clamp for DNA polymerases the Proliferating Cell Nuclear Antigen (PCNA) and then Flap endonuclease 1 (FEN1) excises the displaced oligonucleotide. Finally, DNA ligase I coordinates the final ligation step in long-patch BER [9,71,72].
Frequently during DNA replication, the replicative DNA polymerase, such as DNA pol δ/ε, misincorporates dAMP opposite 8-oxodG instead of the correct dCMP [73]. The resulting 8-oxodG:dA mismatch is repaired by MutY glycosylase Homologue (MYH)-initiated long-patch BER [61, 74]. The monofunctional MYH excises, unconventionally, the undamaged Ade before another round of replication, giving the cells a second opportunity to avoid the fixation of the mutation caused from dC:dG to dA:dT transversion, by forming a substrate for OGG1. Following the removal of the unmodified nucleobase by MYH, the DNA ends are subsequently processed by APE1, as in canonical BER, resulting in a nick with 3′-OH and 5′-dRP moieties. The DNA polymerase λ in complex with the cofactors PCNA and replication protein A (RP-A) promote the incorporation of correct dC opposite 8-oxodG and additional one nucleotide during the elongation reaction. After the lesion bypass, RP-A and DNA pol λ dissociate, instead FEN1 through the interaction with PCNA cleaves the 5′ flap. Subsequently, the DNA ligase I, interacting with PCNA, binds the created nicked intermediate and ligates the 5′-P ends. Overall, the MYH-initiated BER is an example of the inter-relationship between long-patch BER and short patch BER [73,75].
In addition to MYH and OGG1, two bifunctional DNA glycosylases NEIL (Nei-like)-1 and -2, with an associated β,δ-elimination activity, can also repair 8-oxodG via BER [61,76,77](Fig. 5). Both NEIL1 and NEIL2 interact with the BER proteins and initiate an APE-independent repair mechanism by excising 8-oxodG and catalyzing the β,δ-elimination of the abasic site leaving a 3′-phosphate at the resulting strand break. The phosphatase polynucleotide kinase (PNK), present in mammalian cells but not in Escherichia coli (E. coli), removes the 3′-phosphate in the gap creating the substrate for DNA synthesis required by DNA polβ. The final step is DNA ligation-mediated by XRCC1/LigIII [61,76,77].
Besides BER, an intricate network of other repair pathways exist for the repair of oxidatively generated DNA lesions. This includes Mis-Match Repair (MMR) [78] and Transcription-Coupled Nucleotide Excision Repair (TC-NER) [61,79] (Fig. 5). Mounting evidence suggests that the MMR machinery (a collection of repair-associated proteins) plays a role in the post-replicative removal of 8-oxodG opposite dA, competing with MYH for the binding to, and processing of, the mispairing. Furthermore, it has been shown that the dA:8-oxodG mispair is recognized and bound by the human MMR factors MutS homologues 2 (hMSH2) and 6 (hMSH6), and that hMSH6 interacts with MYH [80]. It has also been reported that both MYH and MMR proteins interact with PCNA, which acts as a coordinator of 8-oxodG:dA repair [81,82]. Another MMR protein, MutL homologue 1 (hMLH1), has also been reported to play a role in the repair of 8-oxodG. Evidence for this comes from MMR-defective cells, in which the hMLH1 gene is silenced, show a four-fold higher level of 8-oxodG than MMR-proficient cells [83].
Additionally, a number of components of the TC-NER pathway have been proposed to have a role in the repair of 8-oxodG, specifically both the Cockayne syndrome B (CSB) and A (CSA) proteins, together with Xeroderma pigmentosum complementation group C (XPC) protein [84]. In particular, CSB could catalyse the removal of 8-oxodG and that it could interact with BER proteins, such as APE1 [85]. Similarly to CSB, CSA is also involved in the repair of 8-oxodG but its exact role is still to be determined [86]. Finally, the XPC protein has also been shown to enhance the activity of OGG1 [84]. Thus, XPC might also be involved in the BER of 8-oxodG, probably through the active displacement of OGG1 DNA glycosylase after it has produced an AP site.
5. Genome-wide mapping and the significance of the location for 8-oxodG
It is seemingly well established that 8-oxodG is not distributed uniformly across the genome, not least due to the dynamic equilibrium between local rate of generation and local repair efficiency. On this basis, the observed distribution of 8-oxodG provides a snapshot of such a state of equilibrium. The first genomic view of the 8-oxodG distribution was provided by Nakabeppu and coworkers using immunofluorescence detection of 8-oxodG with a monoclonal antibody on human metaphase chromosomes from human peripheral lymphocytes [87]. The authors demonstrated that 8-oxodG is not uniformly distributed in normal human cells. Indeed, they cytogenetically mapped the position of the 8-oxodG signal, at megabase resolution, to the boundary regions of R and/or G chromosomal bands that are known as transition zones of DNA replication timing. Moreover, they also found that chromosomal regions with a high density of 8-oxodG are located within regions with a high frequency of recombination and single nucleotide polymorphisms (SNPs), thus suggesting that 8-oxodG could contribute to the genomic diversity in humans [87].
Subsequently, Toyokuni’s group provided a higher resolution map of 8-oxodG in the genome of normal rat kidney cells, by combining immunoprecipitation of 8-oxodG-containing DNA with microarray hybridization [88]. They showed that 8-oxodG is preferentially located at gene deserts and not associated with the transcription activity of genes. Moreover, they suggested that the spatial location of genomic DNA in the nucleus determines its susceptibility to oxidation as a strong correlation between 8-oxodG levels with lamina-associated domains (LADs) was found [88].
Recently, genome-wide strategies have been reported by different laboratories to map steady-state levels of 8-oxodG in the yeast, mouse and human genomes [89–91]. Burrow’s laboratory developed OG-Seq to identify the 8-oxodG sites in the mouse genome [89]. OG-Seq is based on the chemical labelling of 8-oxodG with biotin for affinity purification. This approach allowed the mapping of 8-oxodG, with a 150 bp genomic resolution, in both wild-type and OGG1−/− MEFs. This study provided the first genome-wide evidence that certain, specific gene loci (including promoters, 5′-UTRs, 3′-UTRs, exons and introns) are enriched for 8-oxodG when compared with the intergenic regions to a random distribution of the 8-oxodG peaks throughout the genome. In addition, the authors also demonstrated that 8-oxodG-containing peaks harboured more G-quadruplex (G4) and 5′-GG-3′ reactive sequences than expected by chance [89].
The single-nucleotide-resolution mapping of 8-oxodG has been reported for Saccharomyces cerevisiae genome by Sturla’s lab [90]. The authors developed the Click-code-seq technique to insert a biocompatible locator code, readable by high-throughput sequencing, by coupling the specificity of DNA repair enzymes with the efficiency of a click DNA ligation reaction. In this study, it was reported that 8-oxodG accumulates at sites of high nucleosome occupancy when compared to nucleosome-free linker regions. Moreover, local sequence context analysis performed at the flanking regions of the 8-oxodG revealed that the first dG in a 5′-GG-3′ dinucleotide is most easily oxidized [90].
Poetsch et al. developed AP-seq for genome-wide mapping of apurinic sites and 8-oxodG in human cells at a resolution of approximately 300 base pairs [91]. The authors used an aldehyde reactive probe (ARP) that, under recommended conditions, specifically reacts with the aldehyde group of the AP-site, arose from excision of 8-oxodG, and introduces a covalent biotin tag into the DNA at the damage site. Then, the biotin-tagged DNA fragments are pulled down using streptavidin-labelled magnetic beads and sequenced using high-throughput technology. Genomic features, as well as functional elements, have a role in shaping the local distribution of oxidatively generated damage. Among genomic features, the GC content has a major role. Indeed, 8-oxodG formation tends to increase as local GC content rises up around 47 % and decreases when GC content goes beyond this value, suggesting that nucleobase composition alone cannot explain the accumulation of 8-oxodG in regions with higher GC content. Moreover, 8-oxodG is reported to be enriched at genic regions within introns and in functional elements, such as transposable and repetitive elements and G4 structures [91].
We recently developed OxiDIP-seq to isolate and map 8-oxodG-enriched DNA fragments in human and mouse cells with a resolution of about 200–300 bp [92]. This is a pull-down-based approach using an 8-oxodG-specific antibody to enrich 8-oxodG-containing DNA fragments, followed by high-throughput sequencing. We found that 42 % of the identified 8-oxodG peaks are localized at gene loci, with both promoter regions and gene body regions enriched for 8-oxodG. In particular, the accumulation of 8-oxodG at gene loci is associated with activation of the DNA damage response (DDR) and the occurrence of DSBs [92]. Moreover, we found G4-enrichment at 8-oxodG-containing regions and a complex association between 8-oxodG and GC content. Promoter regions with high (> 47 %) GC content display low levels of 8-oxodG [93]. This suggested to us that other mechanisms, such as the epigenetics involved in the regulation of transcription and replication regulation, may be involved in the accumulation of 8-oxodG. Intriguingly, we demonstrated that DNA replication and transcription have a role in shaping local distribution 8-oxodG [92,93].
Balasubramaniam’s group recently published the “snAP-seq” method to map AP sites in the human genome at single-nucleotide resolution [94]. As AP sites are intermediates in the repair of 8-oxodG, this approach may be used partly as a proxy for the distribution of 8-oxodG. The approach uses Click chemistry to attach biotin to AP sites using the Hydrazino-iso-Pictet-Spengler (HIPS) reaction. Despite the ability of the technique to identify AP sites at single-nucleotide resolution, snAP-seq was not able to detect a consensus position between the mapped AP sites, at least when applied to HeLa cells [94]. This suggests that AP sites do not accumulate site-specifically at single-nucleotide level in the population of in vitro cultured cells. In this case, usage of a peak calling bioinformatic tool allowed the identification of DNA stretches where AP sites accumulate in control and APE1 knocked-down cells [94]. According to previous reports [89,91–93], AP sites are enriched at genomic locations associated with open chromatin, suggesting that these regions are more prone to the formation of AP sites, compared to other forms of DNA damage [94].
Fang and Zou established the “enTRAP-seq” protocol to identify 8-oxodG in mouse embryonic fibroblasts with a resolution of 250bp [95]. This approach is based on the ability of an OGG1 mutant (K249Q) to trap, as a stable complex, the 8-oxodG-containing DNA fragments for subsequent enrichment via affinity purification. Notably, enTRAP-seq revealed enrichment of 8-oxodG in regulatory elements such as promoters, 5′UTR, CpG islands and G4, thus supporting the findings reported by other previous studies [95].
Recently, two studies reported the genome-wide mapping of the oxidation-associated AP sites [96,97]. The first work mapped AP sites, along with the binding of OGG1 and APE1 proteins, and G4 structures and revealed that 8-oxodG-derived AP sites occur predominantly at G4 sequences [96]. Furthermore, activation of the BER pathway at 8-oxodG-enriched G4 sequences triggers the formation of G4 structures through the binding of APE1 to G4 sequences. Moreover, APE1 promotes both the folding and stabilization of G4 structures at promoter regions as well as the loading of transcription factors to mediate gene expression [96]. The second work provides a highly sensitive and quantitative approach (named “Nick-seq”) to map, at single-nucleotide resolution, oxidation-induced AP sites in DNA from E. coli treated with a sub-lethal dose of hydrogen peroxide [97]. This study showed that the oxidation-induced AP sites are non-randomly distributed and preferentially associated with DNA regions undergoing replication or transcription during H2O2 stress, suggesting that transcriptionally active and single-strand DNA regions are vulnerable to oxidatively-induced DNA damage [97].
All the above described experimental genome-wide methodology used to map oxidatively generated DNA damage possess benefits and weaknesses. Differences in methods, with their advantages and disadvantages, have been recently reviewed in [98].
6. 8-oxodG as an epigenetic mark
8-oxodG and its repair intermediates play a role in the regulation of gene expression and as such, they may have epigenetic-like features. Several molecular mechanisms have been described for the gene regulatory function of 8-oxodG and have been described elsewhere [53, 98–100].
Multiple studies showed that the formation of 8-oxodG within G-quadruplex structures is important for the epigenetic gene regulation. Indeed, the oxidatively generated modification of dG in potential G-quadruplex forming sequences (PQSs) leads to an increase in the transcription levels of the associated region. Specifically, when the OGG1 recognizes and excises 8-oxodG from the duplex, it creates an AP site which leads to the formation of a G-quadruplex structure in the PQS [26, 101]. The G-quadruplex structure extends the binding of the catalytically inactive APE1 on the AP site and this favours the recruitment of other transcription factors (TF) for gene activation [26,101]. This mechanism has been proposed for the control of expression of various genes, such as VEGF, PCNA, NTHL, HIF1-α and NEIL [99,102–105]. Conversely, other studies have demonstrated that the oxidation of dG in PQSs can also downregulate gene expression [106–108]. Indeed, recently it has been shown that the oxidation of dG in PQSs modulates both magnitude and direction of gene expression change (activation or repression) through a mechanism that could depend on the distance of the PQS from the transcription start site and its strand of occupancy (coding versus non-coding strand).
It has also been proposed that the binding of OGG1 to 8-oxodG in promoter regions of NF-κB target genes induces gene expression [109, 110]. The binding of enzymatically inactive OGG1 to 8-oxodG-containing promoter regions induces a bend in the DNA helix that facilitates the recruitment of specific TFs (NF-κB/RelA and Sp1) and the assembly of the transcriptional machinery [109,110]. Furthermore, increased levels of 8-oxodG have been found in binding regions of other TFs in association with the recruitment of the co-transcription factor LSD1 [43–47, 49,50,111]. In this regard, another mechanism has been described to explain the epigenetic function of the 8-oxodG. Specifically, the TF mediates the recruitment to its binding regions of the LSD1 enzyme whose activity, as already described above, promotes transcription activation via 8-oxodG formation. Indeed, it has been proposed that the DNA nicks generated upon the removal of 8-oxodG by the BER machinery facilitate the entrance of the endonuclease Topoisomerase IIβ (TOPIIβ), and induce a permissive chromatin architecture (i.e. relaxation) for transcription initiation. This process of requiring LSD1-mediated DNA oxidation has been proven necessary for the transcription of the target genes of the estrogen receptor (ER), Myc, androgen receptor and TGF-β1 respectively [43–47,49,50,111].
Finally, 8-oxodG could also perform its epigenetic function in concert with the DNA methylation. Indeed, even if the mechanisms are not yet well defined, the repair of 8-oxodG is linked to DNA methylation. Taken together, these findings reveal that 8-oxodG clearly has potential roles in gene regulation.
7. 8-oxodG as a source of genomic instability
8-oxodG is a useful biomarker of oxidative stress [112] and its accumulation in the genome has been associated with cancer initiation and progression and has been proposed as a prognostic factor in breast cancer [113]. Genetic knock-out mouse models have been particularly useful to identify which proteins of the BER pathway play a crucial role in genome maintenance. Indeed, OGG1+/− and −/− mice have been generated [114–118]. These mice are viable and fertile and despite the lack of a pathological phenotype, they show an increase in nuclear and mitochondrial 8-oxodG levels, with an elevated dC:dG to dA:dT transversion rates at 18 months after birth. In addition, OGG1+/− and −/− mice show a slightly elevated predisposition for lung cancer and, when exposed to the genotoxic agents; they also show a multiorgan enhanced susceptibility for cancer development [114–118]. Similarly, MYH+/− mice are viable and fertile and show only a slight predisposition to develop intestinal cancer [118,119]. Interestingly, OGG1−/− and MYH−/− mice display a strong susceptibility for lung and ovarian cancers and lymphomas [118]. These data suggest that the secondary 8-oxodG repair mechanisms are capable of compensating for the loss of either OGG1 or MYH individually, under physiological conditions. However, they are insufficient under conditions of an exacerbated threat to genetic integrity, e.g. via genotoxin exposure, or when more than one BER pathway, or backup repair mechanism, is compromised. The data also suggests that OGG1 and MYH are key to maintaining genomic instability and prevention of certain cancers. Finally, lacking other proteins involved in the repair of 8-oxodG, such as APE1, Pol β, XRCC1, DNA ligase I and III display embryonic lethality suggesting that the proteins at the core of the repair of 8-oxodG (and indeed other adducts) have a crucial role in the preservation of the correct programs of transcription, replication and in the maintenance of genomic stability [64,120–125].
8-oxodG contributes to ROS-induced genome instability via several mechanisms many of which are associated with the effects of unrepaired 8-oxodG, as well as the accumulation of SSBs unrepaired intermediates deriving from the incompleted repair of 8-oxodG repair.
7.1. Unrepaired 8-oxodG as a source of genome instability
In the syn conformation, 8-oxodG has the ability to mimic dT and, if not repaired, during DNA replication, it represents the main source of dC:dG to dA:dT transversion mutations [61,126,127] (Fig. 6). This can be formally demonstrated by using a unique system named Tracing DNA Adducts by TArgeted Mutagenesis (TATAM system) [128]. Indeed, using the TATAM system, 8-oxodG was stably introduced in the genome of a human lymphoblastoid cell line and the genetic mutation generated by the adduct integration was traced. A single 8-oxodG was able to generate a spectrum of mutations, but predominantly dC:dG to dA:dT trans versions (Fig. 6) and single-base deletions. To further link 8-oxodG to genome instability, the dC:dG to dA:dT transversion has been identified to be the predominant somatic mutation, in a study that analysed the coding regions of 518 cancer-related genes in 210 different human cancers. Independent studies [129] also demonstrated that the dC:dG to dA:dT transversion represents one-third of the 22,910 somatic substitutions identified in a lung cancer cell line [130] and the second most predominant mutational signature in melanoma cells [131].
Fig. 6.
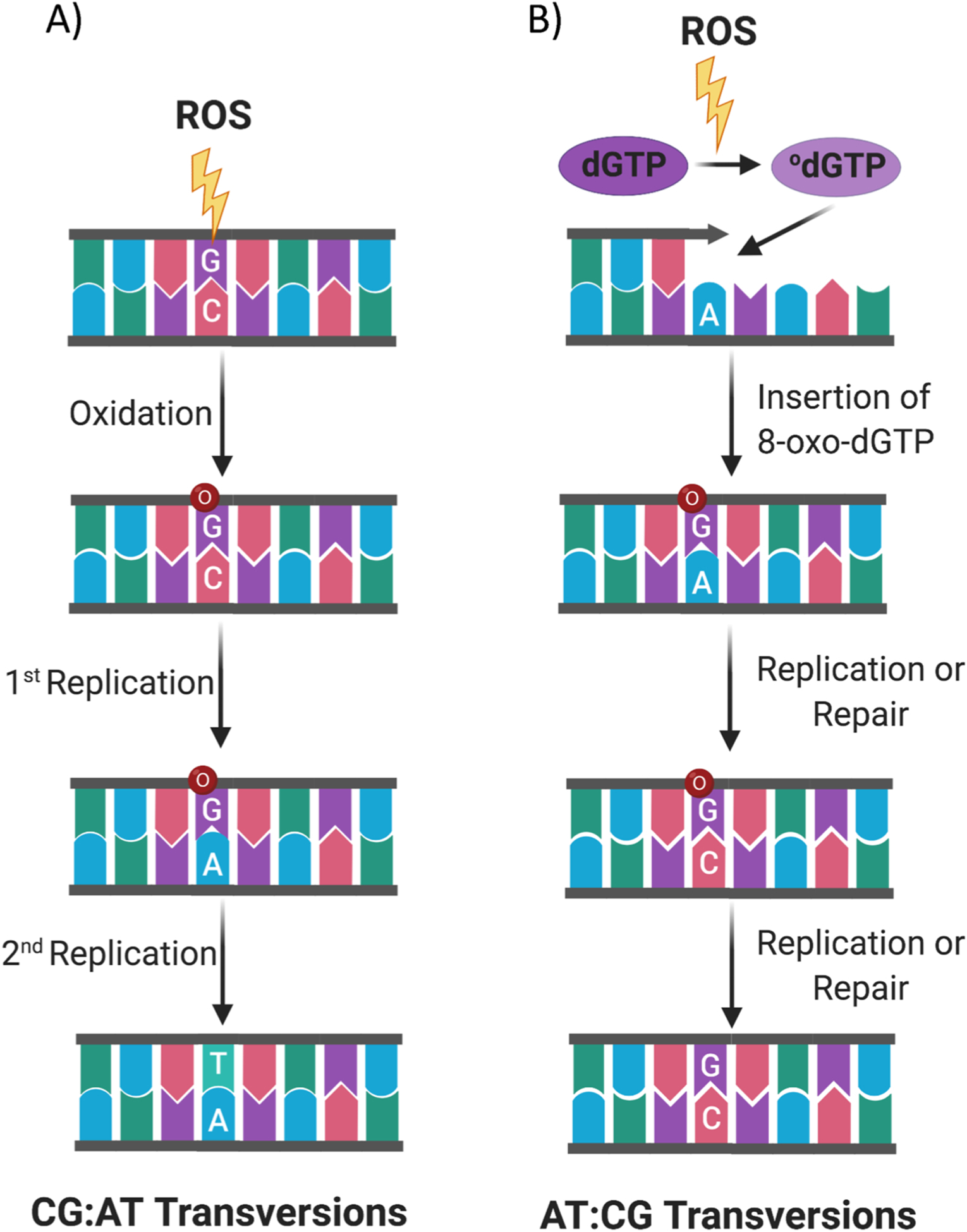
Two mechanisms of 8-oxodG induced transversions. (A) ROS can directly attack DNA, oxidize 2′-deoxyguanosine and form the dC:8-oxodG base pair. Subsequently, when replication occurs across the DNA region containing 8-oxodG, often the 8-oxodG is erroneously bypassed and this results in the insertion of 2′-deoxyadenosine insertion, forming a dA:8-oxodG base pair. If the dA:8-oxodG mispair is left unrepaired during a second round of replication it will produce a dA:dT base pair leading a dC:dG to dA:dT transversion mutation. (B) ROS can also oxidize dGTP, in the dNTP pool, to 8-oxodGTP, which is then erroneously incorporated during DNA replication, giving rise to the 8-oxodG:dA mismatch. If replication or inappropriate MYH excision of 2′-deoxyadenosine occurs, the 8-oxodG pairs with cytidine. 8-oxodG:dC mispairs are a substrate for OGG1-initiated repair which, in this context, produces the dA: dT to dC:dG mutations (created with biorender.com).
8-oxodG can lead also to dA:dT to dC:dG mutations (Fig. 6), and can occur when 8-oxodGTP, from the deoxyribonucleotide pool, is erroneously incorporated during DNA replication and the mismatch is not recognized by polymerase proofreading and MYH. Subsequently, unrepaired 8-oxodG pairs with deoxycytidine in the next round of replication, and causes dA:dT to dC:dG mutations [132–136]. Such dA:dT to dC:dG mutations are found in oesophageal adenocarcinoma and inflammatory Barrett’s oesophagus [137,138].
Intriguingly, 8-oxodG has been reported to impact on, and affect the catalytic activity of, human topoisomerase 1 (TOP1) and in particular when the 8-oxodG lesion occurs just downstream (at the +1 position in the scissile DNA strand) to the TOP1 cleavage site [139]. TOP1 is an essential enzyme, which is able to address DNA topological problems (e.g. supercoils, catenates and knots) by cleaving one DNA strand through the formation of transient enzyme-DNA cleavage complexes (TOPcc), with phosphotyrosine linkages between the catalytic tyrosyl residue of the enzymes and the DNA ends [140,141]. TOP1 plays an important role in the transcription and replication processes. It resolves DNA overwinding ahead of RNA and DNA polymerases, enabling DNA translocation [142,143], and overwinding resulting from the convergence of replication forks and transcription bubbles [144,145]. TOP1 enzymes also prevent replication fork stalling promoting the advancement of replication forks [146]. Notably, the crystal structure of human TOP1 in a non-covalent complex with an 8-oxodG-containing DNA oligonucleotide shows that the O8 atom of the 8-oxodG, clashes sterically with Thr-718 residue in the TOP1 cleavage site, which reorganizes the active site of TOP1 into an inactive conformation [139]. If the 8-oxodG is left unrepaired it traps TOPcc in a topoisomerase DNA-protein crosslinks (TOP-DPC) complex that threatens genome integrity. The TOP-DPC complex is genotoxic, as it favours the accumulation of spontaneous SSB, but may be resolved by the Tyrosyl-DNA phosphodiesterase 1 (TDP1) [140,141,147]. Recently, Sordet and coworkers identified a mechanism for the formation of transcription-associated DSBs in non-replicating cells. In particular, the authors showed that a transcription-induced DSB is formed by two SSB on opposing DNA strands. One SSB arises from the cleavage of an R-loop by XPF/XPG endonucleases and the other from the repair of a TOP-DPC by the TDP1 pathway [145]. Since 8-oxodG accumulates at R-loop-containing regions in non-replicating cells, and also induces the TOP-DPC complex formation, we speculate that 8-oxodG may also contribute to the formation of the SSBs, and then to DSBs, in the above-proposed mechanism [145]. Alternatively, within a cluster of Oxidatively-generated Clustered DNA Lesions (OCDL), a DSB could be formed from two close SSBs where the first one could derive from the BER of the 8-oxodG and the second from an 8-oxodG-mediated TOP1-DPC (Fig. 7A).
Fig. 7.
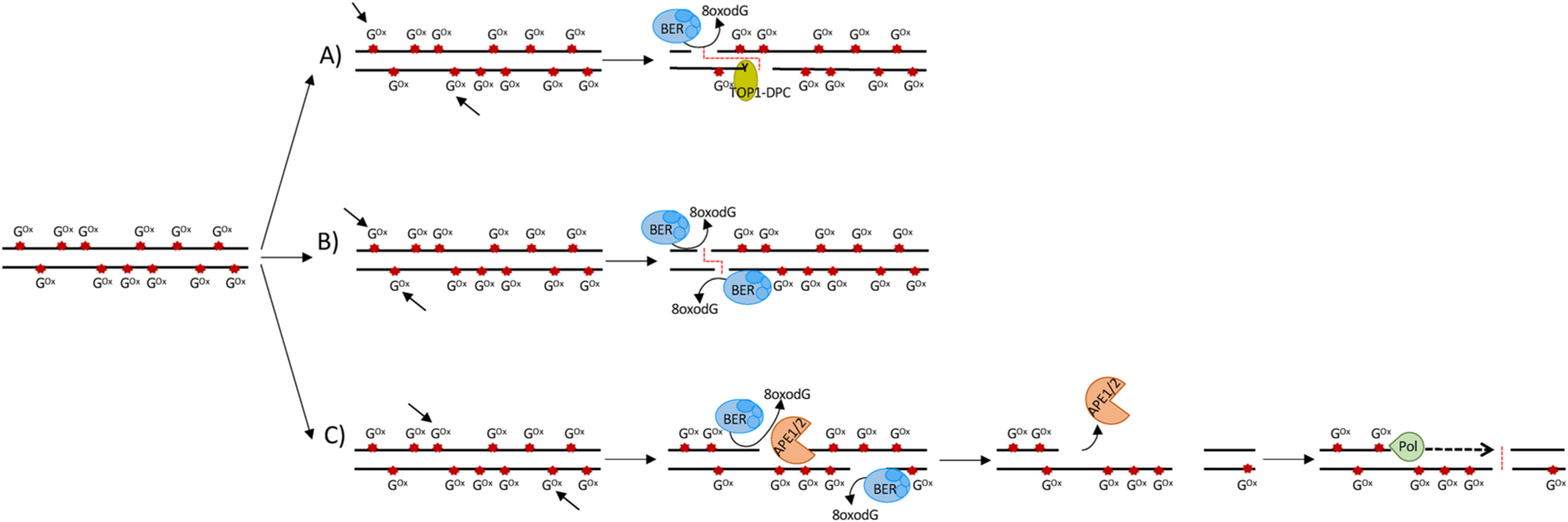
Proposed alternative molecular models for 8-oxodG-mediated DSB formation at ODCL. A DSB could be formed from two SSBs localized on opposite DNA strands. These two SSBs could be generated by either: (A) the BER of a 8-oxodG on one strand and 8-oxodG-mediated TOP-DPC formation on the other strand; (B) BER of two proximal 8-oxodG localized on opposite strands; (C) BER of two distant 8-oxodG localized on different strands, followed by DNA resection and fill-in processes, mediated by the 3′–5′exonuclease activity of APE1/APE2 and by a DNA polymerase, respectively. Arrows indicate the 8-oxodG involved in the generation of SSBs. The red dashed lines indicate DSBs.
Future studies are required to demonstrate whether this form of 8-oxodG-associated genome instability is mechanistically linked to TOP-DPC formation and TDP1 pathway activation.
7.2. Incomplete repair of 8-oxodG as a source of genome instability
The correct balance of the activity of the BER proteins is essential to the cell to carry out efficient and error-free DNA repair [73]. In fact, an imbalanced, or uncoordinated, BER activity can result in the incomplete repair of 8-oxodG, with a consequent accumulation of SSBs, various intermediate products of repair, and DSBs resulting in genome instability.
It has been demonstrated that a low BER activity can cause an accumulation of SSBs intermediates, genome instability and lead to neoplasia [50,68,69,148,149]. High BER activity, on the other hand, can also be detrimental for the cell, as it has been shown to interfere with DNA replication and transcription processes [149–153].
Genetic alterations in genes coding for BER proteins, and damage to repair proteins themselves [154], can result in an unbalancing of the BER pathway via impairment of one or more enzyme activities, contributing to genome instability, neurodegeneration, ageing and cancer development. Indeed, in relation to the cancer predisposition, even if hereditary deficiencies have not yet reported for the OGG1 activity, the polymorphisms and somatic mutations affecting the OGG1 gene (such as R46Q, R131Q) [155], and its function, have been associated with several types of cancer [156]. In addition, germline mutations in MYH have been found and associated with colon, breast and pancreatic cancers and also with some pediatric cancers, such as glioma and astrocytoma [74]. Decreased levels of MYH, OGG1 and Nudix (Nucleoside Diphosphate Linked Moiety X)-Type Motif 1 (e.g. MTH1) have been reported in adenocarcinoma, hepatocarcinoma and prostate carcinoma [157–159]. In addition to the protective role against mutations and tumour formation, MYH is also able to initiate apoptosis in oxidatively stressed cells [160]. Mechanistically, p53 mediates the transcriptional upregulation of MYH. This leads to MYH hyperactivation which in turn causes an accumulation of SSBs in the nuclear genome and PARP1-activation of the apoptotic process [161].
Among the BER proteins, defects in APE1 activity have been extensively demonstrated to be implicated in cancer [162]. Overexpression of APE1 leads to SSB accumulation and genomic instability in XRCC1-deficient cells [163]. Interestingly, p53 is able to coordinate the BER activity by downregulating the transcription of APE1 via Sp1 activation [164]. Conversely, impairment of p53 function, a characteristic of many cancers, leads to upregulation of APE1 and increased genomic instability [165]. Very recently, using Xenopus egg extract and an in vitro reconstitution system, it has been reported that APE1 can ‘sense’ the presence of SSBs and initiate, with APE2, a two-step 3′–5′ SSB end resection. After this, the SSB is eventually repaired by activation of the ATR– Chk1 DDR pathway [166].
As for other BER proteins, mutations have been documented in the DNA polymerases (Pol) β and λ, which affect their activities, and lead to tumour formation [73]. While few polymorphisms of Pol β are associated with various cancers [167,168], the T221 P and R438W polymorphisms in Pol λ have been demonstrated to decrease its nucleobase substitution fidelity and increase mutation frequency in breast cancer [169]. Interestingly, the stability of both Pol β and λ is dependent on the ARF/p53 pathway [170–172]. At steady-state levels, Pol β and λ protein are ubiquitinated by the E3 ubiquitin ligase Mule (ARF-BP1/HectH9) and degraded via the proteasome (Fig. 8). ARF is capable of counteracting such a process by interacting with, and inhibiting, the E3 ubiquitin ligase Mule activity and thus impairing the proteasomal degradation of Pol β and λ with a consequent upregulation of the flow of BER enzymes into the nucleus from newly synthesized BER enzymes. ARF, therefore, represents an important player in the regulation of BER, for while it supports efficient BER by increasing the levels of Pol β and λ and, at the same time, it can decrease BER activity through the inhibition of the E3 ubiquitin ligase MDM2 and consequent upregulation of P53. p53 activation, in turn, downregulates APE1 (Fig. 8).
Fig. 8.
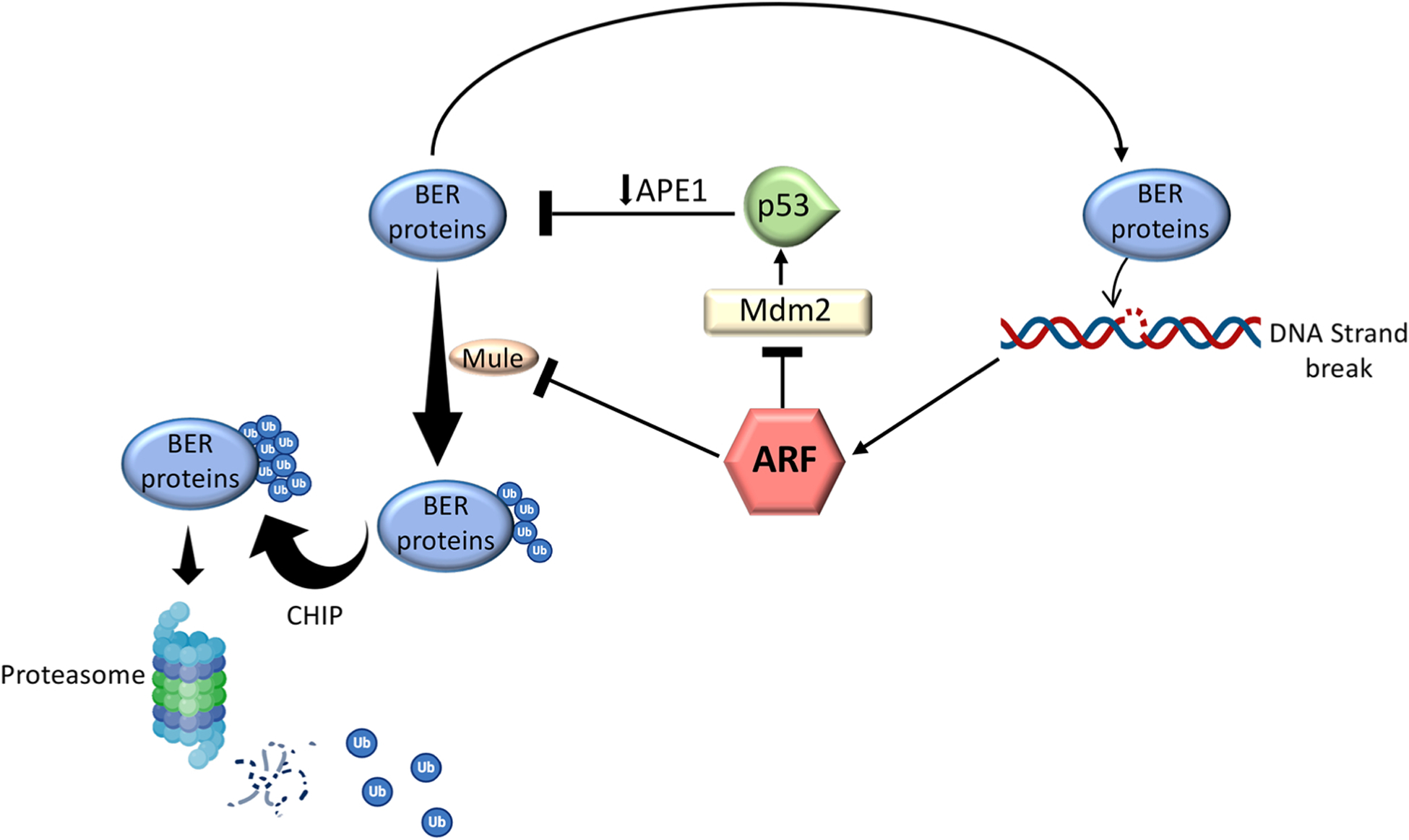
ARF regulation of the levels of BER proteins. In the absence of DNA damage detection, the concomitant intracellular level of ARF is decreased and the synthesized BER proteins are ubiquitinated by Mule and subsequently polyubiquitinated by CHIP (Carboxy-terminus of Hsc70 Interacting Protein) for proteasomal degradation. Following DNA damage detection, ARF accumulates in the cells and, by inhibiting Mule activity, increases the nuclear levels of the BER enzymes. Accumulation of ARF after DNA damage detection can also decrease the levels of BER proteins via a negative feedback loop where ARF inhibits the MDM2 protein and stabilizes the p53 protein which in turn downregulates the transcription of the APE1 gene.
Among the BER proteins, dysfunction of XRCC1 has been extensively studied in carcinogenesis [173,174]. Knock-down of the XRCC1 gene, as well as the R280H germline variant [175], have been shown to cause: (i) deficiencies in BER; (ii) the accumulation of unrepaired SSBs; (iii) genome instability and (iv) cell transformation. In the literature, there are a plethora of data regarding XRCC1 expression levels in many different types of cancer and this is reviewed in detail in [73]. Briefly, the data appear to be contradictory as both high, as well as a low, expression levels of XRCC1 have been detected in tumour samples, and have been associated with both a poor and a good overall survival. Further studies are required to better clarify how the activity of XRCC1 correlates with cancer progression.
Mechanistically, the model of genome instability mediated by unbalanced BER of 8-oxodG requires the accumulation of persistent SSBs. Indeed, closely opposed SSBs, derived from closely opposed 8-oxodG within OCDLs, are able to produce DSBs [176] (Fig. 7B). Alternatively, two distant oxidatively-generated SSBs could produce a DSB when DNA resection takes place, mediated by the 3′–5′exonuclease activity of APE1/APE2, to generate ssDNA. The subsequent infilling of the ssDNA, mediated by a DNA polymerase, could give rise to a DSB (Fig. 7C). Notably, DSBs associated with the 8-oxodG repair are predominantly repaired by the error-prone NHEJ pathway [176]. In accordance with this view, we recently mapped the accumulation of endogenous 8-oxodG in human fragile promoters showing characteristic features of genomic instability, such as GC skewness, G4 structures, R-loops, bidirectional transcription [93]. These oxidized promoters showed an increased occurrence of SSBs, DSBs, together with DDR activation, and recruitment of the NHEJ protein, XRCC4. We also noted translocation breakpoints, of the kind commonly identified in cancer. Moreover, we found that transcription and replication have a role in shaping the spatial distribution of local oxidatively damaged DNA [92,93]. Indeed, our data showed that: (i) the accumulation of 8-oxodG in genes containing 8-oxodG at promoter regions correlated with the occupancy of the processive isoform of RNAPII and with levels of active transcription; (ii) a subset of promoters maintain this accumulation of 8-oxodG, even in the absence of DNA replication; (iii) more highly transcribed genes do not show 8-oxodG enrichment; (vi) the accumulation of 8-oxodG also occurs at some other fragile regions (e.g. DNA replication origins located within the gene body of long genes, as well as at a subset of fragile promoters) where transcription and replication conflicts have been frequently observed. We, therefore, hypothesize that transcription/replication-generated fragile structures (e.g. R-loop-s-containing regions) could contribute to genome instability as they favour the formation of ODCLs where the BER of two proximal 8-oxodG can generate SSBs and, consequently, DSBs (Fig. 7).
8. Conclusions and perspectives
8-oxodG is widely described as a type of DNA damage, but it is increasingly clear that there is more to it than that. Indeed, while unrepaired 8-oxodG causes transversions [60–64,126], it can also lead to the formation of DSBs [69,93,176,177], and thought this genomic instability [50,68,69,148,149]. Furthermore, there is growing evidence that 8-oxodG has a role in the epigenetic regulation of transcription [26, 43,44,47,49,50,109,110].This epigenetic role could be intrinsically associated with its nature as DNA damage. Mechanistically, the repair of 8-oxodG, as deliberate DNA damage generated locally by LSD1 activity, is capable of leading to SSB formation which, in gene promoter regions, relaxes the local chromatin structure and/or determines chromatin looping and/or G4-formation. This chromatin reorganization, in turn, facilitates the crosstalk between several protein complexes involved in transcription regulation.
Interestingly, in the context of a subset of promoter regions of transcribed genes, the occurrence of 8-oxodG is associated with RNAPII occupancy, R-loops and G4 structures [93]. R-loops have been found to be a peculiarity of 8-oxodG-enriched DNA regions and this is probably because of the presence of ssDNA that is more easily oxidized than dsDNA, and more prone to form G4 structures. Oxidized promoters are also associated with the accumulation of ɣ-H2AX, SSBs, DSBs, NHEJ proteins and genetic translocations/deletions [93]. These data suggest a model in which 8-oxodG serves as a driver of the transcription process, but under deregulated conditions, or within genomic intrinsic fragile structures, 8-oxodG could cause the formation of DSBs and chromosomal translocations/deletions.
This model, along with the described genetic and epigenetics features associated with 8-oxodG-enriched promoters (namely the “8-oxodG model” - Fig. 9, left panel), is analogous to the molecular model established for Class Switch Recombination [178] (here named as the “dU model” - Fig. 9, right panel), where the Activation-Induced cytidine Deaminase (AID) activity causes accumulation of deoxyuridine (dU) and recruitment of a specific BER enzyme, the uracil-DNA glycosylase UNG, which generates an AP site that is first processed to an SSB and then to a DSB by the APE1/APE2/MMR protein activities.
Fig. 9.
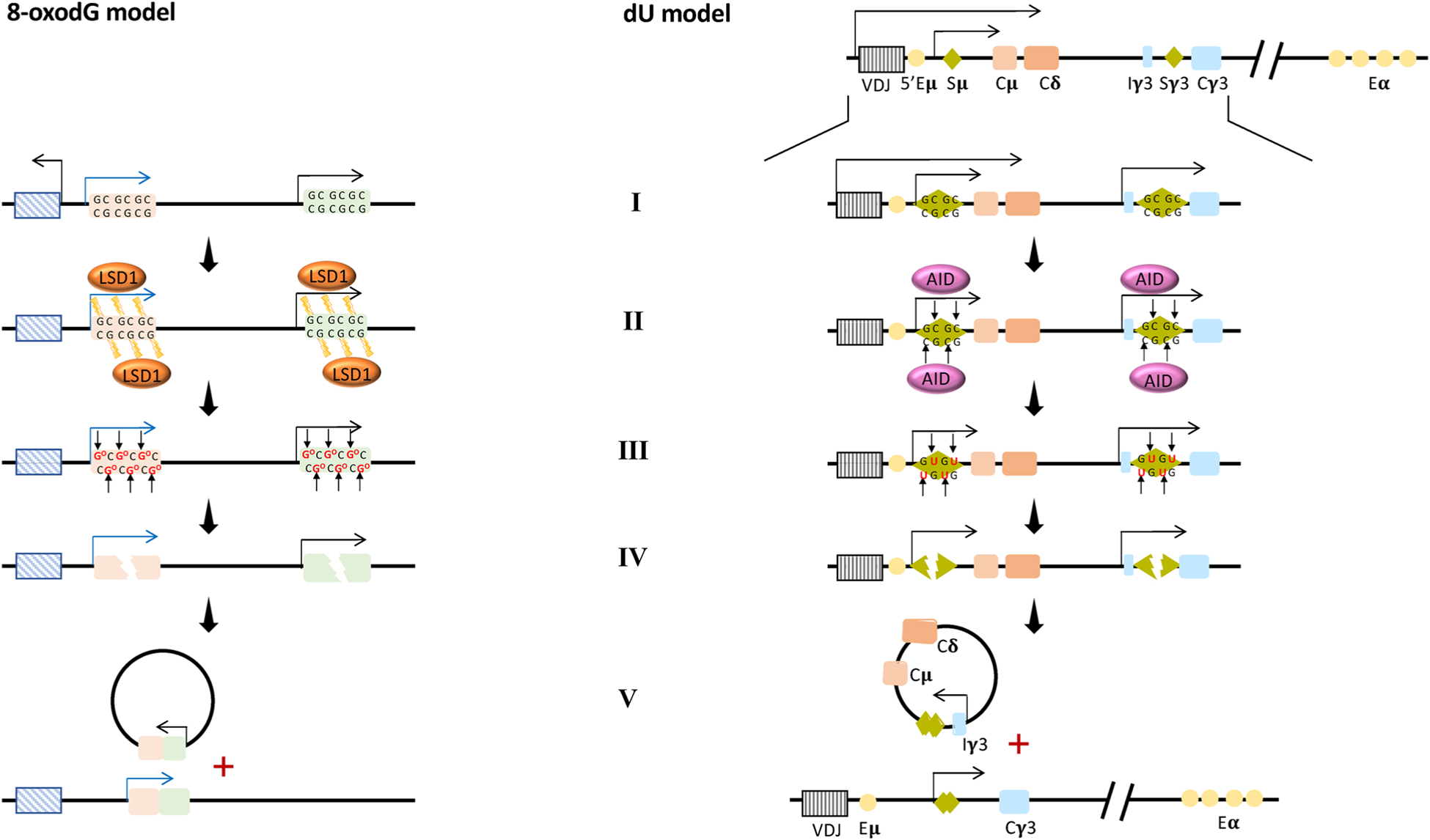
Proposed model of 8-oxodG-mediated genome instability (8-oxodG model) analagous to the AID/dU-mediated model of Class Switch Recombination (dU model).
8-oxodG model: Schematic representation of a genetic locus showing two genes subject to 8-oxodG-mediated transcription initiation. AID/dU model: Organization of the IgH locus in mice [including the antigen recognition V(D)J gene segment, the switch (S) regions, the constant (C) region exon segments, and the enhancers (E)] and associated CSR process.
(I) Specific stimuli initiate the transcription process that determines R-loops formation and RNAPII accumulation at respectively the S regions of the dU model and at oxidized promoters of the 8-oxodG model. (II–III) The recruitment of Activation-induced Cytidine Deaminase (AID) at the S regions in the dU model and the recruitment of LSD1 at the promoters of the genes loci in the 8-oxodG model (II) determine the accumulation of deoxyuridine respectively at the S regions in the dU model and of 8-oxodG at the promoters in the 8-oxodG model (III). (IV) The recruitment of specific BER enzymes: the uracil-DNA glycosylase UNG in the dU model and OGG1 in 8-oxodG model. These remove dU and 8-oxodG, respectively, generating multiple AP sites. These AP sites are processed to SSB and then to DSB by the APE1/APE2/MMR protein activities in both models. (V) Finally, two regions (each one containing one DSB) are brought together as a result of chromatin looping and ligated by the NHEJ proteins, causing a genetic translocation and deletion in both the models.
Based on the many similarities shared by the above two processes, one associated with 8-oxodG and the other with dU, it is conceivable that the molecular rules of the AID-UNG-APE1 action could be applied to the LSD1-OGG1-APE1 action. Moreover, as these two models act on a 2′-deoxynucleoside associated with the dC:dG base pair (with 8-oxodG model targeting 8-oxodG and dU model addressing the dC) and both could cause SSBs, it is conceivable that, when they operate simultaneously on the dC:dG base pair, and can be a source of DSBs.
It is hoped that this provocative comparison is a source of inspiration for developing new insight into 8-oxodG for which its epigenetic function could be the other (positive) side of its well-known role in genome instability. However, it is still far from clear how and if the emerging epigenetic role of 8-oxodG is linked with its ability to induce or influence genome instability. A future challenge will be to find answers to the significant number of open questions on the full, biological role of 8-oxodG for moving forward the pharmacological targeting of the oxidatively damaged DNA, and its repair in therapies for cancer and other diseases in which oxidative stress is implicated.
Acknowledgements
This work is supported by POR Campania FESR 2014-2020 “SATIN” to S.A., AIRC IG 23066 to B.M. M.S.C. is supported, in part, by the National Institute of Environmental Health Sciences of the National Institutes of Health under award number R15ES027196. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We wish to thank Dr. Luigi Lania for helpful discussions and critical review of the manuscript.
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- [1].Lindahl T, Instability and decay of the primary structure of DNA, Nature (1993), 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- [2].Kim GH, Kim JE, Rhie SJ, Yoon S, The role of oxidative stress in neurodegenerative diseases, Exp. Neurobiol (2015), 10.5607/en.2015.24.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Beckman KB, Ames BN, The free radical theory of aging matures, Physiol. Rev (1998), 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- [4].Klaunig JE, Kamendulis LM, The role of oxidative stress in carcinogenesis, Annu. Rev. Pharmacol. Toxicol (2004), 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- [5].Lindahl T, Barnes DE, Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol, 2000, 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- [6].Tubbs A, Nussenzweig A, Endogenous DNA damage as a source of genomic instability in Cancer, Cell (2017), 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hoeijmakers JHJ, DNA damage, aging, and cancer, N. Engl. J. Med (2009), 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- [8].Di Meo S, Reed TT, Venditti P, Victor VM, Role of ROS and RNS sources in physiological and pathological conditions, Oxid. Med. Cell. Longev (2016), 10.1155/2016/1245049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].van Loon B, Markkanen E, Hübscher U, Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine, DNA Repair (Amst). 9 (2010) 604–616, 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- [10].Fry RC, Begley TJ, Samson LD, Genome-wide responses to DNA-damaging agents, Annu. Rev. Microbiol (2005), 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- [11].Mena S, Ortega A, Estrela JM, Oxidative stress in environmental-induced carcinogenesis, Mutat. Res. - Genet. Toxicol. Environ. Mutagen (2009), 10.1016/j.mrgentox.2008.09.017. [DOI] [PubMed] [Google Scholar]
- [12].Nicolussi A, D’Inzeo S, Capalbo C, Giannini G, Coppa A, The role of peroxiredoxins in cancer, Mol. Clin. Oncol (2017), 10.3892/mco.2017.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sabharwal SS, Waypa GB, Marks JD, Schumacker PT, Peroxiredoxin-5 targeted to the mitochondrial intermembrane space attenuates hypoxia-induced reactive oxygen species signalling, Biochem. J (2013), 10.1042/BJ20130740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Matsuzawa A, Thioredoxin and redox signaling: roles of the thioredoxin system in control of cell fate, Arch. Biochem. Biophys (2017), 10.1016/j.abb.2016.09.011. [DOI] [PubMed] [Google Scholar]
- [15].Rouhier N, Lemaire SD, Jacquot JP, The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation, Annu. Rev. Plant Biol (2008), 10.1146/annurev.arplant.59.032607.092811. [DOI] [PubMed] [Google Scholar]
- [16].Vašák M, Meloni G, Chemistry and biology of mammalian metallothioneins, J. Biol. Inorg. Chem (2011), 10.1007/s00775-011-0799-2. [DOI] [PubMed] [Google Scholar]
- [17].Loren P, Sánchez R, Arias ME, Felmer R, Risopatrón J, Cheuquemán C, Melatonin scavenger properties against oxidative and nitrosative stress: impact on gamete handling and in vitro embryo production in humans and other mammals, Int. J. Mol. Sci (2017), 10.3390/ijms18061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Winterbourn CC, Reconciling the chemistry and biology of reactive oxygen species, Nat. Chem. Biol (2008), 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- [19].Waypa GB, Smith KA, Schumacker PT, O2 sensing, mitochondria and ROS signaling: the fog is lifting, Mol. Aspects Med. (2016), 10.1016/j.mam.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Winterbourn CC, Hampton MB, Thiol chemistry and specificity in redox signaling, Free Radic. Biol. Med (2008), 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- [21].Davalli P, Mitic T, Caporali A, Lauriola A, D’Arca D, ROS, Cell Senescence, and Novel Molecular Mechanisms in Aging and Age-Related Diseases, Oxid. Med. Cell. Longev (2016), 10.1155/2016/3565127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging, Cell (2013), 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davalli P, Marverti G, Lauriola A, D’Arca D, Targeting oxidatively induced DNA damage response in cancer: opportunities for novel cancer therapies, Oxid. Med. Cell. Longev 2018 (2018), 10.1155/2018/2389523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schieber M, Chandel NS, ROS function in redox signaling and oxidative stress, Curr. Biol (2014), 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sies H, Berndt C, Jones DP, Oxidative stress, Annu. Rev. Biochem (2017), 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- [26].Fleming AM, Ding Y, Burrows CJ, Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair, Proc. Natl. Acad. Sci. U. S. A (2017), 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cooke MS, Evans MD, Dizdaroglu M, Lunec J, Oxidative DNA damage: mechanisms, mutation, and disease, FASEB J. 17 (2003) 1195–1214, 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- [28].Evans MD, Cooke MS, Factors contributing to the outcome of oxidative damage to nucleic acids, BioEssays. 26 (2004) 533–542, 10.1002/bies.20027. [DOI] [PubMed] [Google Scholar]
- [29].Cooke MS, Evans MD, 8-Oxo-deoxyguanosine: Reduce, reuse, recycle? Proc. Natl. Acad. Sci. U. S. A (2007) 10.1073/pnas.0706878104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD, ROS and the DNA damage response in cancer, Redox Biol. (2019), 10.1016/j.redox.2018.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jackson SP, Bartek J, The DNA-damage response in human biology and disease, Nature (2009), 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hansford RG, Hogue BA, Mildaziene V, Dependence of H2O2 formation by rat heart mitochondria on substrate availability and donor age, J. Bioenerg. Biomembr (1997), 10.1023/A:1022420007908. [DOI] [PubMed] [Google Scholar]
- [33].Fukai T, Ushio-Fukai M, Superoxide dismutases: role in redox signaling, vascular function, and diseases, Antioxidants Redox Signal. (2011), 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fenton HJH, LXXIII. - Oxidation of tartaric acid in presence of iron, J. Chem. Soc. Perkin Trans. I (1894), 10.1039/CT8946500899. [DOI] [Google Scholar]
- [35].Curtin NJ, DNA repair dysregulation from cancer driver to therapeutic target, Nat. Rev. Cancer (2012), 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- [36].Aust AE, Eveleigh JF, Mechanisms of DNA oxidation. Proc. Soc. Exp. Biol. Med, 1999, 10.1046/j.1525-1373.1999.d01-141.x. [DOI] [PubMed] [Google Scholar]
- [37].Giorgio M, Trinei M, Migliaccio E, Pelicci PG, Hydrogen peroxide: A metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol (2007) 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- [38].Kuroda J, Nakagawa K, Yamasaki T, Nakamura KI, Takeya R, Kuribayashi F, Imajoh-Ohmi S, Igarashi K, Shibata Y, Sueishi K, Sumimoto H, The superoxide-producing NAD(P)H oxidase Nox4 in the nucleus of human vascular endothelial cells, Genes Cells (2005), 10.1111/j.1365-2443.2005.00907.x. [DOI] [PubMed] [Google Scholar]
- [39].Kwa. Tsang C, Liu Y, Thomas J, Zhang Y, Zheng XFS, Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance, Nat. Commun (2014), 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y, Histone demethylation mediated by the nuclear amine oxidase homolog LSD1, Cell (2004), 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- [41].Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, Shi YG, Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 Methylation, Mol. Cell (2010), 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Forneris F, Binda C, Battaglioli E, Mattevi A, LSD1: oxidative chemistry for multifaceted functions in chromatin regulation, Trends Biochem. Sci (2008), 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- [43].Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV, DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression, Science 319 (80) (2008) 202–206, 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- [44].Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B, LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription, Oncogene (2010), 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- [45].Amente S, Lania L, Avvedimento EV, Majello B, DNA oxidation drives Myc mediated transcription, Cell Cycle 9 (2010) 3002–3004, 10.4161/cc.9.15.12499. [DOI] [PubMed] [Google Scholar]
- [46].Amente S, Lania L, Majello B, The histone LSD1 demethylase in stemness and cancer transcription programs, Biochim. Biophys. Acta - Gene Regul. Mech (2013), 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- [47].Mo W, Zhang J, Li X, Meng D, Gao Y, Yang S, Wan X, Zhou C, Guo F, Huang Y, Amente S, Avvedimento EV, Xie Y, Li Y, Identification of novel AR-Targeted MicroRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate Cancer, PLoS One (2013), 10.1371/journal.pone.0056592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Majello B, Gorini F, Saccà CD, Amente S, Expanding the role of the histone lysine-specific demethylase lsd1 in cancer, Cancers (Basel). (2019), 10.3390/cancers11030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pezone A, Taddei ML, Tramontano A, Dolcini J, Boffo FL, De Rosa M, Parri M, Stinziani S, Comito G, Porcellini A, Raugei G, Gackowski D, Zarakowska E, Olinski R, Gabrielli A, Chiarugi P, Avvedimento EV, Targeted DNA oxidation by LSD1-SMAD2/3 primes TGF-β1/ EMT genes for activation or repression, Nucleic Acids Res. 48 (2020) 8943–8958, 10.1093/nar/gkaa599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sengupta S, Wang H, Yang C, Szczesny B, Hegde ML, Mitra S, Ligand-induced gene activation is associated with oxidative genome damage whose repair is required for transcription, Proc. Natl. Acad. Sci. U. S. A 117 (2020) 22183–22192, 10.1073/pnas.1919445117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Thompson KJ, Fried MG, Ye Z, Boyer P, Connor JR, Regulation, mechanisms and proposed function of ferritin translocation ot cell nuclei, J. Cell. Sci (2002). [DOI] [PubMed] [Google Scholar]
- [52].Wu Y, Brosh RM, DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster, Nucleic Acids Res. (2012), 10.1093/nar/gks039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Giorgio M, Dellino IG, Gambino V, Roda N, Pelicci PG, On the epigenetic role of guanosine oxidation, Redox Biol. (2020), 10.1016/j.redox.2019.101398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cooke MS, Evans MD, Dizdaroglu M, Lunec J, Oxidative DNA damage: mechanisms, mutation, and disease, FASEB J. (2003), 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- [55].Evans MD, Dizdaroglu M, Cooke MS, Oxidative DNA damage and disease: induction, repair and significance, Mutat. Res. - Rev. Mutat. Res (2004), 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- [56].Burak MJ, Guja KE, Garcia-Diaz M, Nucleotide binding interactions modulate dNTP selectivity and facilitate 8-oxo-dGTP incorporation by DNA polymerase lambda, Nucleic Acids Res. (2015), 10.1093/nar/gkv760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Steenken S, Jovanovic SV, How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution, J. Am. Chem. Soc (1997), 10.1021/ja962255b. [DOI] [Google Scholar]
- [58].Baik MH, Silverman JS, Yang IV, Ropp PA, Szalai VA, Yang W, Thorp HH, Using density functional theory to design DNA base analogues with low oxidation potentials, J. Phys. Chem. B (2001), 10.1021/jp010643g. [DOI] [Google Scholar]
- [59].Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T, DNA Repair and Mutagenesis, 2nd ed., ASM Press, 2006. [Google Scholar]
- [60].Koga Y, Taniguchi Y, Sasaki S, Synthesis of the oligoribonucleotides incorporating 8-oxo-guanosine and evaluation of their base pairing properties, Nucleosides, Nucleotides and Nucleic Acids. (2013), 10.1080/15257770.2013.767461. [DOI] [PubMed] [Google Scholar]
- [61].Boiteux S, Coste F, Castaing B, Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: properties and biological roles of the Fpg and OGG1 DNA N-glycosylases, Free Radic. Biol. Med (2017), 10.1016/j.freeradbiomed.2016.11.042. [DOI] [PubMed] [Google Scholar]
- [62].Batra VK, Beard WA, Hou EW, Pedersen LC, Prasad R, Wilson SH, Mutagenic conformation of 8-oxo-7,8-dihydro-2′-dGTP in the confines of a DNA polymerase active site, Nat. Struct. Mol. Biol (2010), 10.1038/nsmb.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shibutani S, Takeshita M, Grollman AP, Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG, Nature (1991), 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- [64].Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hübscher U, 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins, Nature (2007), 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- [65].Hsu GW, Ober M, Carell T, Beese LS, Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase, Nature (2004), 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- [66].Lindahl T, Repair of intrinsic DNA lesions, Mutat. Res. Genet. Toxicol. Environ. Mutagen (1990), 10.1016/0165-1110(90)90022-4. [DOI] [PubMed] [Google Scholar]
- [67].Caldecott KW, Mammalian DNA single-strand break repair: an X-ra(y)ted affair, BioEssays (2001), 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- [68].Fortini P, Dogliotti E, Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways, DNA Repair (Amst). (2007), 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- [69].Tubbs A, Nussenzweig A, Endogenous DNA damage as a source of genomic instability in Cancer, Cell 168 (2017) 644–656, 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E, Two pathways for base excision repair in mammalian cells, J. Biol. Chem (1996), 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- [71].Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E, The type of DNA glycosylase determines the base excision repair pathway in mammalian cells, J. Biol. Chem (1999), 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- [72].Klungland A, Lindahl T, Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1), EMBO J. (1997), 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Markkanen E, Not breathing is not an option: how to deal with oxidative DNA damage, DNA Repair (Amst). 59 (2017) 82–105, 10.1016/j.dnarep.2017.09.007. [DOI] [PubMed] [Google Scholar]
- [74].Markkanen E, Dorn J, Hübscher U, MUTYH DNA glycosylase: the rationale for removing undamaged bases from the DNA, Front. Genet (2013), 10.3389/fgene.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Svilar D, Goellner EM, Almeida KH, Sobol RW, Base excision repair and lesion-dependent subpathways, Antioxid. Redox Signal 14 (2011) 2491–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK, NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells, DNA Repair (Amst). (2006), 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Dou H, Mitra S, Hazra TK, Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2, J. Biol. Chem (2003), 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- [78].Yang H, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, Miller JH, Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch, Nucleic Acids Res. (2001), 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Fousteri M, Mullenders LHF, Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects, Cell Res. (2008), 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- [80].Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL, Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6, J. Biol. Chem (2002), 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- [81].Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL, Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair, J. Biol. Chem (2001), 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- [82].Li GM, Mechanisms and functions of DNA mismatch repair, Cell Res. (2008), 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- [83].Colussi C, Parlanti E, Degan P, Aquilina G, Barnes D, Macpherson P, Karran P, Crescenzi M, Dogliotti E, Bignami M, The Mammalian Mismatch Repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool, Curr. Biol (2002), 10.1016/S0960-9822(02)00863-1. [DOI] [PubMed] [Google Scholar]
- [84].D’Errico M, Parlanti E, Teson M, De Jesus BMB, Degan P, Calcagnile A, Jaruga P, Bjørås M, Crescenzi M, Pedrini AM, Egly JM, Zambruno G, Stefanini M, Dizdaroglu M, Dogliotti E, New functions of XPC in the protection of human skin cells from oxidative damage, EMBO J. (2006), 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM, Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates, Nucleic Acids Res. (2007), 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM, Orioli D, Frosina G, Zambruno G, Dizdaroglu M, Stefanini M, Dogliotti E, The role of CSA in the response to oxidative DNA damage in human cells, Oncogene (2007), 10.1038/sj.onc.1210232. [DOI] [PubMed] [Google Scholar]
- [87].Ohno M, Miura T, Furuichi M, Tominaga Y, Tsuchimoto D, Sakumi K, Nakabeppu Y, A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome, Genome Res. (2006), 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Yoshihara M, Jiang L, Akatsuka S, Suyama M, Toyokuni S, Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus, DNA Res. (2014), 10.1093/dnares/dsu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ding Y, Fleming AM, Burrows CJ, Sequencing the Mouse Genome for the Oxidatively Modified Base 8-Oxo-7,8-dihydroguanine by OG-Seq, J. Am. Chem. Soc 139 (2017) 2569–2572, 10.1021/jacs.6b12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wu J, McKeague M, Sturla SJ, Nucleotide-resolution genome-wide mapping of oxidative DNA damage by click-code-Seq, J. Am. Chem. Soc (2018), 10.1021/jacs.8b03715. [DOI] [PubMed] [Google Scholar]
- [91].Poetsch AR, Boulton SJ, Luscombe NM, Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis 06 Biological Sciences 0604 Genetics, Genome Biol. (2018), 10.1186/s13059-018-1582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Amente S, Di Palo G, Scala G, Castrignanò T, Gorini F, Cocozza S, Moresano A, Pucci P, Ma B, Stepanov I, Lania L, Pelicci PG, Dellino GI, Majello B, Genome-wide mapping of 8-oxo-7,8-dihydro-2′-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells, Nucleic Acids Res. (2019), 10.1093/nar/gky1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Gorini F, Scala G, Di Palo G, Dellino GI, Cocozza S, Pelicci PG, Lania L, Majello B, Amente S, The genomic landscape of 8-oxodG reveals enrichment at specific inherently fragile promoters, Nucleic Acids Res. 48 (2020) 4309–4324, 10.1093/nar/gkaa175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Liu ZJ, Martínez Cuesta S, Van Delft P, Balasubramanian S, Sequencing abasic sites in DNA at single-nucleotide resolution, Nat. Chem 11 (2019) 629–637, 10.1038/s41557-019-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Fang Y, Zou P, Genome-wide mapping of oxidative DNA damage via engineering of 8-Oxoguanine DNA glycosylase, Biochemistry 59 (2020) 85–89, 10.1021/acs.biochem.9b00782. [DOI] [PubMed] [Google Scholar]
- [96].Roychoudhury S, Pramanik S, Harris HL, Tarpley M, Sarkar A, Spagnol G, Sorgen PL, Chowdhury D, Band V, Klinkebiel D, Bhakat KK, Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome, Proc. Natl. Acad. Sci. U. S. A (2020), 10.1073/pnas.1912355117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Cao B, Wu X, Zhou J, Wu H, Liu L, Zhang Q, DeMott MS, Gu C, Wang L, You D, Dedon PC, Nick-seq for single-nucleotide resolution genomic maps of DNA modifications and damage, Nucleic Acids Res. 48 (2020) 6715–6725, 10.1093/nar/gkaa473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Poetsch AR, The genomics of oxidative DNA damage, repair, and resulting mutagenesis, Comput. Struct. Biotechnol. J (2020), 10.1016/j.csbj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Fleming AM, Burrows CJ, 8-Oxo-7,8-dihydroguanine, friend and foe: epigenetic-like regulator versus initiator of mutagenesis, DNA Repair (Amst). (2017), 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang R, Hao W, Pan L, Boldogh I, Ba X, The roles of base excision repair enzyme OGG1 in gene expression, Cell. Mol. Life Sci (2018), 10.1007/s00018-018-2887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fleming AM, Zhu J, Ding Y, Burrows CJ, 8-oxo-7,8-dihydroguanine in the context of a gene promoter G-Quadruplex is an on-off switch for transcription, ACS Chem. Biol (2017), 10.1021/acschembio.7b00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Fleming AM, Zhu J, Howpay Manage SA, Burrows CJ, Human NEIL3 gene expression regulated by epigenetic-like oxidative DNA modification, J. Am. Chem. Soc (2019), 10.1021/jacs.9b01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Pastukh V, Ruchko M, Gorodnya O, Wilson GL, Gillespie MN, Sequence-specific oxidative base modifications in hypoxia-inducible genes, Free Radic. Biol. Med (2007), 10.1016/j.freeradbiomed.2007.08.027. [DOI] [PubMed] [Google Scholar]
- [104].Redstone SCJ, Fleming AM, Burrows CJ, Oxidative Modification of the Potential G-Quadruplex Sequence in the PCNA Gene Promoter Can Turn on Transcription, Chem. Res. Toxicol (2019), 10.1021/acs.chemrestox.8b00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhu J, Fleming AM, Burrows CJ, The RAD17 promoter sequence contains a potential tail-dependent G-Quadruplex that downregulates gene expression upon oxidative modification, ACS Chem. Biol (2018), 10.1021/acschembio.8b00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Membrino A, Cogoi S, Pedersen EB, Xodo LE, G4-DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy, PLoS One (2011), 10.1371/journal.pone.0024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Cogoi S, Xodo LE, G4 DNA in ras genes and its potential in cancer therapy, Biochim. Biophys. Acta - Gene Regul. Mech (2016), 10.1016/j.bbagrm.2016.02.002. [DOI] [PubMed] [Google Scholar]
- [108].Cogoi S, Ferino A, Miglietta G, Pedersen EB, Xodo LE, The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription, Nucleic Acids Res. (2018), 10.1093/nar/gkx1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, Boldogh I, Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor κB-driven gene expression, J. Biol. Chem (2016), 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pan L, Hao W, Zheng X, Zeng X, Ahmed Abbasi A, Boldogh I, Ba X, OGG1-DNA interactions facilitate NF-k B binding to DNA targets, Sci. Rep (2017), 10.1038/srep43297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zuchegna C, Aceto F, Bertoni A, Romano A, Perillo B, Laccetti P, Gottesman ME, Avvedimento EV, Porcellini A, Mechanism of retinoic acid-induced transcription: histone code, DNA oxidation and formation of chromatin loops, Nucleic Acids Res. (2014), 10.1093/nar/gku823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Sova H, Jukkola-Vuorinen A, Puistola U, Kauppila S, Karihtala P, 8-Hydroxydeoxyguanosine: a new potential independent prognostic factor in breast cancer, Br. J. Cancer 102 (2010) 1018–1023, 10.1038/sj.bjc.6605565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Valavanidis A, Vlachogianni T, Fiotakis C, 8-Hydroxy-2′ -deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis, J. Environ. Sci. Heal. - Part C Environ. Carcinog. Ecotoxicol. Rev (2009), 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- [114].Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE, Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage, Proc. Natl. Acad. Sci. U. S. A (1999), 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, Masumura KI, Nohmi T, Nishimura S, Noda T, Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice, Proc. Natl. Acad. Sci. U. S. A (2000), 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, Sekiguchi M, Nakabeppu Y, Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption, Cancer Res. (2003). [PubMed] [Google Scholar]
- [117].Kakehashi A, Ishii N, Okuno T, Fujioka M, Gi M, Wanibuchi H, Enhanced susceptibility of Ogg1 mutant mice to multiorgan carcinogenesis, Int. J. Mol. Sci (2017), 10.3390/ijms18081801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, Miller JH, Deficiencies in mouse myh and Ogg1 result in tumor predisposition and g to t mutations in codon 12 of the K-Ras oncogene in lung tumors, Cancer Res. (2004), 10.1158/0008-5472.CAN-03-3834. [DOI] [PubMed] [Google Scholar]
- [119].Sakamoto K, Tominaga Y, Yamauchi K, Nakatsu Y, Sakumi K, Yoshiyama K, Egashira A, Kura S, Yao T, Tsuneyoshi M, Maki H, Nakabeppu Y, Tsuzuki T, MUTYH-null mice are susceptible to spontaneous and oxidative stress-induced intestinal tumorigenesis, Cancer Res. (2007), 10.1158/0008-5472.CAN-06-4802. [DOI] [PubMed] [Google Scholar]
- [120].Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T, The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice, Proc. Natl. Acad. Sci. U. S. A (1996), 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, Meneses J, Pedersen RA, Chen DJ, A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity, Mutat. Res. - DNA Repair (1998), 10.1016/S0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- [122].Sobol RW, Horton JK, Kühn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH, Requirement of mammalian DNA polymerase-β in base-excision repair, Nature (1996), 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- [123].Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA, Requirement for the Xrcc1 DNA base excision repair gene during early mouse development, Dev. Biol (1999), 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- [124].Bentley DJ, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW, DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability, Nat. Genet (1996), 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- [125].Puebla-Osorio N, Lacey DB, Alt FW, Zhu C, Early embryonic lethality due to targeted inactivation of DNA ligase III, Mol. Cell. Biol (2006), 10.1128/mcb.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Bruner SD, Norman DP, Verdine GL, Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA, Nature 403 (2000) 859–866, 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- [127].Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G, Bass AJ, Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity, Nat. Genet (2013), 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yasui M, Kanemaru Y, Kamoshita N, Suzuki T, Arakawa T, Honma M, Tracing the fates of site-specifically introduced DNA adducts in the human genome, DNA Repair (Amst). 15 (2014) 11–20, 10.1016/j.dnarep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- [129].Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR, Patterns of somatic mutation in human cancer genomes, Nature (2007), 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, Varela I, Nik-Zainal S, Davies HR, Ordõez GR, Mudie LJ, Latimer C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J, Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF, Peckham HE, Tsung EF, Costa GL, Lee CC, Minna JD, Gazdar A, Birney E, Rhodes MD, McKernan KJ, Stratton MR, Futreal PA, Campbell PJ, A small-cell lung cancer genome with complex signatures of tobacco exposure, Nature (2010), 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordõez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR, A comprehensive catalogue of somatic mutations from a human cancer genome, Nature (2010), 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Maki H, Sekiguchi M, MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis, Nature (1992), 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- [133].Inoue M, Kamiya H, Fujikawa K, Ootsuyama Y, Murata-Kamiya N, Osaki T, Yasumoto K, Kasai H, Induction of chromosomal gene mutations in Escherichia coli by direct incorporation of oxidatively damaged nucleotides: new evaluation method for mutagenesis by damaged dna precursors in vivo, J. Biol. Chem (1998), 10.1074/jbc.273.18.11069. [DOI] [PubMed] [Google Scholar]
- [134].Satou K, Kawai K, Kasai H, Harashima H, Kamiya H, Mutagenic effects of 8-hydroxy-dGTP in live mammalian cells, Free Radic. Biol. Med (2007), 10.1016/j.freeradbiomed.2007.02.024. [DOI] [PubMed] [Google Scholar]
- [135].Satou K, Hori M, Kawai K, Kasai H, Harashima H, Kamiya H, Involvement of specialized DNA polymerases in mutagenesis by 8-hydroxy-dGTP in human cells, DNA Repair (Amst). (2009), 10.1016/j.dnarep.2008.12.009. [DOI] [PubMed] [Google Scholar]
- [136].Suzuki T, Kamiya H, Mutations induced by 8-hydroxyguanine (8-oxo-7,8-dihydroguanine), a representative oxidized base, in mammalian cells, Genes Environ. (2017), 10.1186/s41021-016-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Murugaesu N, Wilson GA, Birkbak NJ, Watkins TBK, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, Rowan A, Phillimore B, Biggs J, Begum S, Matthews N, Hochhauser D, Hanna GB, Swanton C, Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy, Cancer Discov. (2015), 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O’Donovan M, Malhotra S, Di Pietro M, Ivakhno S, He M, Weaver JMJ, Lynch AG, Kingsbury Z, Ross M, Humphray S, Bentley D, Fitzgerald RC, Hayes SJ, Ang Y, Welch I, Preston S, Oakes S, Save V, Skipworth R, Tucker O, Davies J, Crichton C, Schusterreiter C, Underwood T, Noble F, Stacey B, Kelly J, Byrne J, Haydon A, Sharland D, Owsley J, Barr H, Lagergren J, Gossage J, Davies A, Mason R, Chang F, Zylstra J, Sanders G, Wheatley T, Berrisford R, Bracey T, Harden C, Bunting D, Roques T, Nobes J, Loo S, Lewis M, Cheong E, Priest O, Parsons SL, Soomro I, Kaye P, Saunders J, Pang V, Welch N, Catton JA, Duffy JP, Ragunath K, Lovat L, Haidry R, Miah H, Kerr S, Eneh V, Butawan R, Lewis M, Cheong E, Kumar B, Igali L, Walton S, Dann A, Safranek P, Hindmarsh A, Sudjendran V, Scott M, Cluroe A, Miremadi A, Mahler-Araujo B, Nutzinger B, Peters C, Abdullahi Z, Crawte J, MacRae S, Noorani A, Elliott RF, Bower L, Edwards P, Tavare S, Eldridge M, Bornschein J, Secrier M, Yang TP, O’Neill JR, Adamczuk K, Lao-Sirieix P, Grehan N, Smith L, Lishman S, Beardsmore D, Dawson S, Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma, Nat. Genet (2015), 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Pourquier P, Ueng LM, Fertala J, Wang D, Park HJ, Essigmann JM, Bjornsti MA, Pommier Y, Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages: 7,8-dihydro-8-oxoguanine and 5-hydroxycytosine, J. Biol. Chem 274 (1999) 8516–8523, 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]
- [140].Sun Y, Saha S, Wang W, Saha LK, Huang SYN, Pommier Y, Excision repair of topoisomerase DNA-protein crosslinks (TOP-DPC), DNA Repair (Amst). 89 (2020) 102837, 10.1016/j.dnarep.2020.102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Sun Y, Saha LK, Saha S, Jo U, Pommier Y, Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways, DNA Repair (Amst). 94 (2020) 102926, 10.1016/j.dnarep.2020.102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Garg LC, DiAngelo S, Jacob ST, Role of DNA topoisomerase I in the transcription of supercoiled rRNA gene, Proc. Natl. Acad. Sci. U. S. A (1987), 10.1073/pnas.84.10.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Collins I, Weber A, Levens D, Transcriptional consequences of topoisomerase inhibition, Mol. Cell. Biol (2001), 10.1128/mcb.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Manzo SG, Hartono SR, Sanz LA, Marinello J, De Biasi S, Cossarizza A, Capranico G, Chedin F, DNA Topoisomerase I differentially modulates R-loops across the human genome, Genome Biol. (2018), 10.1186/s13059-018-1478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Cristini A, Ricci G, Britton S, Salimbeni S, yin S, Huang N, Marinello J, Calsou P, Pommier Y, Favre G, Capranico G, Gromak N, Sordet O, Dual processing of R-Loops and topoisomerase I induces transcription-dependent DNA double-strand breaks, Cell Rep. (2019), 10.1016/j.celrep.2019.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Chen SH, Chan NL, Hsieh TS, New mechanistic and functional insights into DNA topoisomerases, Annu. Rev. Biochem (2013), 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- [147].Yang SW, Burgin AB, Huizenga BN, Robertson CA, Yao KC, Nash HA, A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases, Proc. Natl. Acad. Sci. U. S. A (1996), 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH, XRCC1 and DNA polymerase B in cellular protection against cytotoxic DNA single-strand breaks, Cell Res. (2008), 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Nemec AA, Wallace SS, Sweasy JB, Variant base excision repair proteins: Contributors to genomic instability, Semin. Cancer Biol (2010), 10.1016/j.semcancer.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Yamtich J, Nemec AA, Keh A, Sweasy JB, A germline polymorphism of DNA polymerase Beta Induces genomic instability and cellular transformation, PLoS Genet. (2012), 10.1371/journal.pgen.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Servant L, Bieth A, Hayakawa H, Cazaux C, Hoffmann JS, Involvement of DNA polymerase β in DNA replication and mutagenic consequences, J. Mol. Biol (2002), 10.1006/jmbi.2001.5307. [DOI] [PubMed] [Google Scholar]
- [152].Fréchet M, Canitrot Y, Bieth A, Dogliotti E, Cazaux C, Hoffmann JS, Deregulated DNA polymerase β strengthens ionizing radiation-induced nucleotidic and chromosomal instabilities, Oncogene (2002), 10.1038/sj.onc.1205295. [DOI] [PubMed] [Google Scholar]
- [153].Albertella MR, Lau A, O’Connor MJ, The overexpression of specialized DNA polymerases in cancer, DNA Repair (Amst). (2005), 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- [154].Karran P, Brem R, Protein oxidation, UVA and human DNA repair, DNA Repair (Amst). (2016), 10.1016/j.dnarep.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Anderson PC, Daggett V, The R46Q, R131Q and R154H polymorphs of human DNA glycosylase/β-lyase hOgg1 severely distort the active site and DNA recognition site but do not cause unfolding, J. Am. Chem. Soc (2009), 10.1021/ja809726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Karahalil B, Bohr VA, Wilson DM, Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk, Hum. Exp. Toxicol (2012), 10.1177/0960327112444476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Kohno Y, Yamamoto H, Hirahashi M, Kumagae Y, Nakamura M, Oki E, Oda Y, Reduced MUTYH, MTH1, and OGG1 expression and TP53 mutation in diffuse-type adenocarcinoma of gastric cardia, Hum. Pathol (2016), 10.1016/j.humpath.2016.01.006. [DOI] [PubMed] [Google Scholar]
- [158].Krupa R, Czarny P, Wigner P, Wozny J, Jablkowski M, Kordek R, Szemraj J, Sliwinski T, The relationship between single-nucleotide polymorphisms, the expression of DNA damage response genes, and hepatocellular carcinoma in a polish population, DNA Cell Biol. (2017), 10.1089/dna.2017.3664. [DOI] [PubMed] [Google Scholar]
- [159].Shinmura K, Kato H, Kawanishi Y, Yoshimura K, Igarashi H, Goto M, Tao H, Inoue Y, Sugiyama T, Furuse H, Ozono S, Sugimura H, Reduced expression of the DNA glycosylase gene MUTYH is associated with an increased number of somatic mutations via a reduction in the DNA repair capacity in prostate adenocarcinoma, Mol. Carcinog (2017), 10.1002/mc.22509. [DOI] [PubMed] [Google Scholar]
- [160].Oka S, Nakabeppu Y, DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis, Cancer Sci. (2011), 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- [161].Oka S, Leon J, Tsuchimoto D, Sakumi K, Nakabeppu Y, MUTYH, an adenine DNA glycosylase, mediates p53 tumor suppression via PARP-dependent cell death, Oncogenesis. (2014), 10.1038/oncsis.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Shah F, Logsdon D, Messmann RA, Fehrenbacher JC, Fishel ML, Kelley MR, Exploiting the Ref-1-APE1 node in cancer signaling and other diseases: from bench to clinic, Npj Precis. Oncol (2017), 10.1038/s41698-017-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Sossou M, Flohr-Beckhaus C, Schulz I, Daboussi F, Epe B, Radicella JP, APE1 overexpression in XRCC1-deficient cells complements the defective repair of oxidative single strand breaks but increases genomic instability, Nucleic Acids Res. 33 (2005) 298–306, 10.1093/nar/gki173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Zaky A, Busso C, Izumi T, Chattopadhyay R, Bassiouny A, Mitra S, Bhakat KK, Regulation of the human AP-endonuclease (APE1/Ref-1) expression by the tumor suppressor p53 in response to DNA damage, Nucleic Acids Res. (2008), 10.1093/nar/gkm1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Poletto M, Legrand AJ, Fletcher SC, Dianov GL, P53 coordinates base excision repair to prevent genomic instability, Nucleic Acids Res. 44 (2016) 3165–3175, 10.1093/nar/gkw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Lin Y, Raj J, Li J, Ha A, Hossain MA, Richardson C, Mukherjee P, Yan S, APE1 senses DNA single-strand breaks for repair and signaling, Nucleic Acids Res. 48 (2020) 1925–1940, 10.1093/nar/gkz1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Donigan KA, Sun KW, Nemec AA, Murphy DL, Cong X, Northrup V, Zelterman D, Sweasy JB, Human POLB gene is mutated in high percentage of colorectal tumors, J. Biol. Chem (2012), 10.1074/jbc.M111.324947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Starcevic D, Dalal S, Sweasy JB, Is there a link between DNA polymerase β and cancer? Cell Cycle (2004). [PubMed] [Google Scholar]
- [169].Swett RJ, Elias A, Miller JA, Dyson GE, Andrés Cisneros G, Hypothesis driven single nucleotide polymorphism search (HyDn-SNP-S), DNA Repair (Amst). (2013), 10.1016/j.dnarep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL, CHIP-Mediated Degradation and DNA Damage-Dependent Stabilization Regulate Base Excision Repair Proteins, Mol. Cell (2008), 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- [171].Parsons JL, Tait PS, Finch D, Dianova II, Edelmann MJ, Khoronenkova SV, Kessler BM, Sharma RA, McKenna WG, Dianov GL, Ubiquitin ligase ARF-BP1/Mule modulates base excision repair, EMBO J. (2009), 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Khan S, Guevara C, Fujii G, Parry D, p14ARF is a component of the p53 response following ionizing irradiation of normal human fibroblasts, Oncogene (2004), 10.1038/sj.onc.1207824. [DOI] [PubMed] [Google Scholar]
- [173].Markkanen E, Fischer R, Ledentcova M, Kessler BM, Dianov GL, Cells deficient in base-excision repair reveal cancer hallmarks originating from adjustments to genetic instability, Nucleic Acids Res. (2015), 10.1093/nar/gkv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Nassour J, Martien S, Martin N, Deruy E, Tomellini E, Malaquin N, Bouali F, Sabatier L, Wernert N, Pinte S, Gilson E, Pourtier A, Pluquet O, Abbadie C, Defective DNA single-strand break repair is responsible for senescence and neoplastic escape of epithelial cells, Nat. Commun (2016), 10.1038/ncomms10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Sizova DV, Keh A, Taylor BF, Sweasy JB, The R280H X-ray cross-complementing 1 germline variant induces genomic instability and cellular transformation, DNA Repair (Amst). (2015), 10.1016/j.dnarep.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Sharma V, Collins LB, Chen T, Herr N, Sun W, Swenberg JA, Nakamura J, Oxidative stress at low levels can induce clustered DNA lesions leading to NHEJ mediated mutations, Oncotarget 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Amente S, Di Palo G, Scala G, Castrignanò T, Gorini F, Cocozza S, Moresano A, Pucci P, Ma B, Stepanov I, Lania L, Pelicci PG, Dellino GI, Majello B, Genome-wide mapping of 8-oxo-7,8-dihydro-2′-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells, Nucleic Acids Res. 47 (2019) 221–236, 10.1093/nar/gky1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Daniel JA, Nussenzweig A, The AID-induced DNA damage response in chromatin, Mol. Cell 50 (2013) 309–321, 10.1016/j.molcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


