Abstract
An individual’s susceptibility to atherosclerotic cardiovascular disease (ASCVD) is influenced by numerous clinical and lifestyle factors, motivating the multifaceted approaches that are currently endorsed for primary and secondary ASCVD prevention. With growing knowledge of the genetic basis of ASCVD – in particular, coronary artery disease (CAD) – and its contribution to disease risk, there is increased interest in understanding the potential clinical utility of a genetic predictor that might further refine the assessment and management of ASCVD risk. Rapid scientific and technological advances have enabled widespread genotyping efforts and dynamic research in the field of CAD genetic risk prediction. In this review, we describe how genomic analyses of CAD have been leveraged to create polygenic risk scores (PRS). We then discuss evaluations of the clinical utility of these scores, pertinent mechanistic insights gleaned, and practical considerations relevant to PRS implementation in the health care setting.
Keywords: heritability, genome wide association study, coronary artery disease, polygenicity, atherosclerosis
Subject Terms: Coronary Artery Disease, Atherosclerosis
FOUNDATIONAL THEORIES – THE GENETIC ARCHITECTURE OF CORONARY ARTERY DISEASE
Heritability of coronary atherosclerosis
The heritability of atherosclerotic cardiovascular disease (ASCVD) has long been postulated given the observed aggregation of coronary artery disease (CAD) and its sequelae of myocardial infarction (MI) in families, particularly when disease onset occurs early in life.1–4 In the Framingham Heart Study Offspring Cohort, a parental history of premature CAD was associated with roughly two-fold odds of incident cardiovascular disease after adjustment for traditional clinical risk factors, suggesting an independent heritable basis for cardiovascular disease susceptibility.5 While familial aggregation may imply enrichment of shared deleterious DNA sequence variants or non-genetic factors (e.g. health-related behaviors, food access, parental income, and neighborhood), studies of high-risk families and twin populations have since approximated the heritability of CAD – defined specifically as the total proportion of phenotypic variation explained by genetic factors – at 40% to 60%.6 At the individual level, the relative contribution of inherited over acquired risk factors is likely greatest among individuals in whom CAD arises prematurely.6, 7
Monogenic (Mendelian) determinants of CAD
Investigations into the genetic determinants of CAD have uncovered distinct models of inheritance that speak to a complex genetic architecture. For some individuals and families, genetic risk follows an apparent, classical Mendelian inheritance pattern, with disease manifesting at a relatively young age and with a smaller contribution from environmental risk factors.8 In such cases, risk may be mediated by a rare (minor allele frequency [MAF] < 0.5%), high-impact mutation in a single gene (“monogenic”) that results most commonly in familial hypercholesterolemia (FH) – a clinical syndrome characterized by marked elevations in plasma concentrations of low-density lipoprotein cholesterol (LDL-C) and occasionally associated physical stigmata such as corneal arcus and tendon xanthomas. FH follows an autosomal incomplete dominant pattern of inheritance, where the number of abnormal alleles inherited (0, 1, or 2) correlates directly with the severity of the FH phenotype.9 Candidate gene studies and linkage analyses of patients with FH have localized the causative defects to pathogenic mutations in genes encoding the LDL receptor (LDLR), apolipoprotein B (APOB) and the proprotein convertase subtilisin/kexin type 9 (PCSK9), all of which directly or indirectly interfere with LDL receptor-mediated uptake of cellular LDL-C particles from the bloodstream.10–14 However, the prevalence of heterozygous FH is approximately 1 in 300–500 individuals. And despite an abundance of clinical criteria for FH, most individuals do not have pathogenic mutations in the aforementioned FH genes. 15, 16 The residual etiologies are likely a combination of: (1) novel undetected genes responsible for FH, (2) alternative heritable mechanisms, which will be discussed below, and (3) phenocopy, or non-genetic mechanisms (e.g., maladaptive health-related behaviors). FH may often be suspected in the setting of severe hypercholesterolemia and retrospective analyses imply a strongly favorable clinical effect of early statins.17 As such, current guidelines recommend early statin preventive therapy regardless of other risk factors when FH is apparent.18, 19
Autosomal recessive Mendelian hypercholesterolemia syndromes are much rarer and are typically assessed in most clinical FH genetic panel tests. Homozygous genotypes of pathogenic mutations in LDLRAP1 may lead to an FH phenotype. Furthermore, sitosterolemia is a condition that leads to plant sterol accumulation in the blood and tissues (due to homozygous genotypes of pathogenic mutations in ABCG5 or ABCG8) and may also lead to a similar phenotype. The aggregate prevalence of these genotypes is 1 in 5,000,000.9
Polygenic determinants of CAD and genome-wide association studies
While monogenic CAD risk has provided key mechanistic insights for CAD, it does not explain the more common familial aggregation of CAD in the population. For most of the population, inherited CAD risk is the product of many common (MAF > 5%) genetic variants of small effect sizes (“polygenic”) working in aggregate and alongside environmental and lifestyle factors.20 CAD-associated common genetic variants have been identified through increasingly large, population-based genome-wide association studies (GWAS), enabled by marked scientific and technological advancements over the past two decades. Specifically, the completion of the Human Genome Project and the systematic classification of millions of single nucleotide polymorphisms (SNPs; i.e. through the International HapMap project) have prompted corresponding developments in high-throughput, low-cost DNA microarrays to assay pre-specified genotypes, as well as imputation strategies to statistically infer unknown genotypes from known ones.21–23 Parallel computational innovations have facilitated pipelines to interrogate the large amounts of human genetic data generated, permitting efficient, population genetic analyses of CAD and many other traits and diseases.
Fundamentally, a disease-specific GWAS compares the DNA profiles of disease cases and disease-free control participants to detect statistically significant enrichment or depletion of assayed alleles among cases versus controls. Due to the simultaneous assessment of a million or more independent SNPs for association with a disease or trait, correction for multiple-testing yields a stringent P-value threshold of less than 5×10−8 to achieve “genome-wide” statistical significance.24 To minimize detecting spurious associations driven by the ascertainment of cases and controls, genetic ancestry differences (i.e., population stratification) is accounted for in analyses and independent consistent replication is typically pursued. Therefore, GWAS of CAD and other diseases have relied upon the formation of global, disease-specific consortia to recruit sufficient individuals with and without the disease of interest to both maximize power and limit false discoveries.
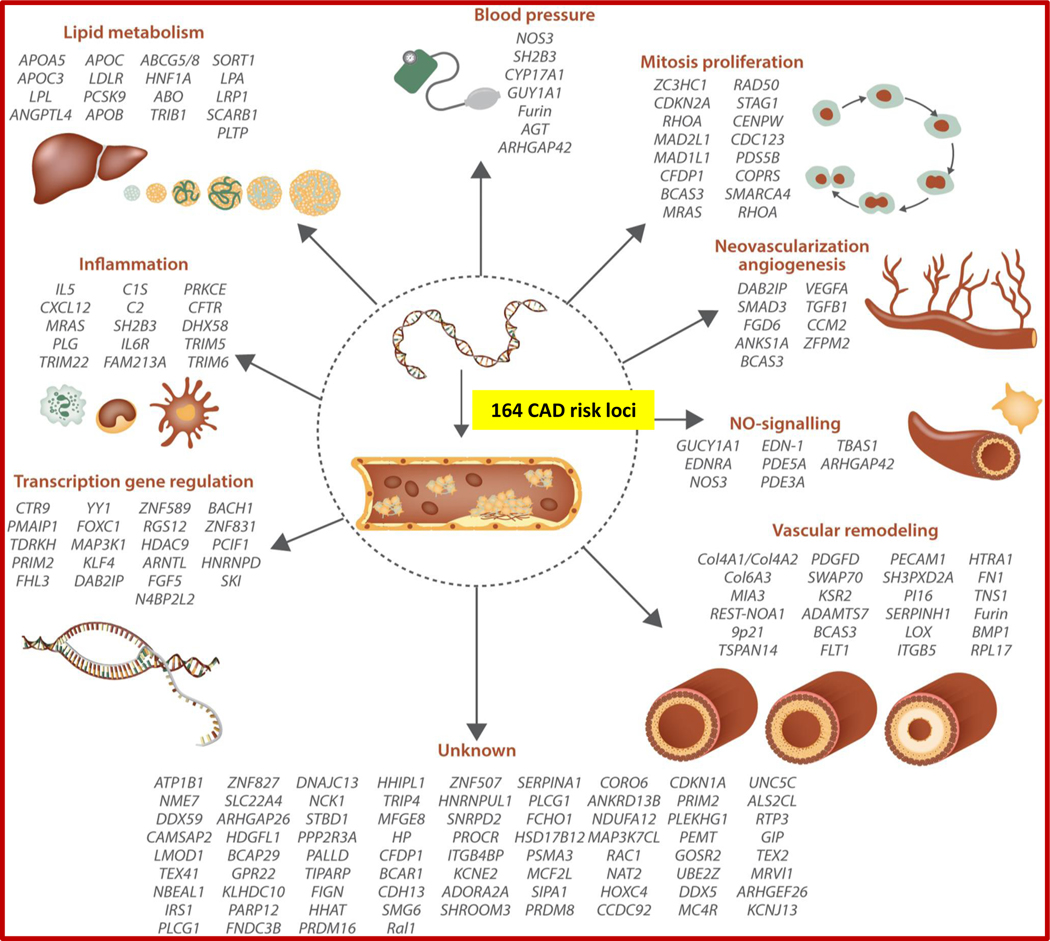
The first common genetic variants associated with CAD at a level of genome-wide significance were identified by three independent groups in 2007 within a 58-kb interval in chromosome 9p21.25–27 Subsequent GWAS and meta-analyses have involved international collaboratives such as the Myocardial Infarction Genetics Consortium (MIGen), the Coronary Artery Disease Genome-Wide Replication and Meta-Analysis (CARDIoGRAM) consortium, and the Coronary Artery Disease Genetics Consortium (C4D). The merging of the CARDIoGRAM and C4D collaboratives in 2013 yielded CARDIoGRAMplusC4D, which has since served as the leading international consortium for CAD discovery genetics.28–31 These collaborative efforts have largely been predicated on the amalgamation of distinct case-control studies to augment power for genetic discovery and evaluate for consistent associations across cohorts. The advent of the United Kingdom (UK) Biobank – a population-based biobank of 500,000 genotyped individuals with linked health registry data – provided another rich source of CAD cases and controls and ensuing genetic association analyses incorporating data from the UK Biobank have further expanded our understanding of the polygenic basis of CAD.32,33–36 Collectively to-date, discovery genetic analyses over the past 13 years have identified 164 common genetic loci associated with CAD at levels of genome-wide significance (Figure 1).37
Figure 1 –
Genome-wide association studies have identified 164 genetic risk loci associated with coronary artery disease, with many classified into distinct mechanistic pathways. From Erdmann et al. Cardiovascular Research 2018.37
The successful identification of many genetic risk loci for CAD has also lent credence to the “common disease-common variant” hypothesis for CAD heritability.38–42 For example, a GWAS led by the CARDIoGRAMplusC4D consortia in 2015 leveraged data from the 1000 Genomes Project, permitting the analysis of lower frequency (MAF = 0.5 – 5%) genetic variants in addition to common genetic variants. While the study uncovered both known and novel susceptibility loci for CAD, nearly all association signals reaching genome-wide significance were among the common genetic variants tested, further suggesting that low-frequency variants do not contribute substantially to the population-level heritability of CAD.43 Nevertheless, associated low-frequency variants, typically with larger effects than observed for associated common variants and more often in coding sequences, have yielded important mechanistic insights into the pathogenesis of CAD.44, 45
POLYGENIC RISK SCORES – BASIC PRINCIPLES and METHODS
The liability threshold model for binary traits
The results of GWAS have therefore affirmed both statistical and evolutionary theories surrounding the genetics of complex traits – namely, that the genetic architecture of complex diseases (like CAD) comprises a preponderance of common, low-impact genetic variants, with more modest contribution from rare variants of large effect.41, 42 In aggregate, the associated risk conferred by these (largely common) genetic polymorphisms aligns with the well-established “liability threshold model” of disease, which proposes a continuous and normal risk distribution for binary outcomes arising from numerous genetic and non-genetic factors, and the existence of a theoretical risk threshold above which a given disease or phenotype typically manifests.46, 47
Genetic factors contributing to an individuals’ predilection for a complex trait can be represented by a polygenic score, a single, normally-distributed quantitative factor that captures the aggregate genetic influences of many common genetic variants. When the genetic predisposition captured by a polygenic score applies to a binary disease outcome, the term “polygenic risk score” (PRS) may be used, reflecting the net susceptibility to disease (“risk”) conferred by the numerous common genetic variants accounted for. In this review, we focus on polygenic scoring for binary disease outcomes – in particular, PRS for CAD. While the predictive accuracy of any PRS is bounded by the underlying heritability of a disease, several factors significantly influence PRS performance – i.e. the precision of common variant association estimates from GWAS; the specific populations in which the PRS is developed and applied; as well as various methodologic considerations when assembling a PRS.
Methods for PRS construction
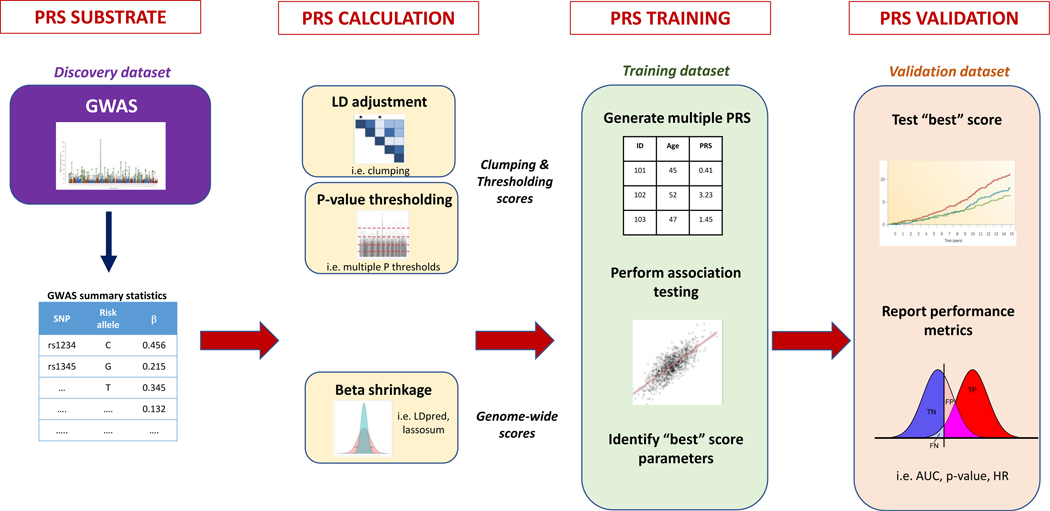
In their current form, PRS are primarily derived from GWAS estimates of individual SNP-trait associations. More specifically, the output of a GWAS is a set of “summary statistics” for the association between each of the (often millions of) genotyped or imputed SNPs assessed and a given trait or disease (Figure 2). For each genetic variant, summary statistics typically include: a variant identifier, its position in the genome, classification of risk (“effect”) versus protective (“non-effect”) alleles, the effect size of the per-effect allele association with the trait, a measure of confidence in the effect size estimate (i.e. standard error), and the statistical p-value. Given differences in allelic references across genotyping platforms, effect allele frequencies are often used to harmonize discovery genotyping platforms with the testing genotyping platforms. Summary statistics of GWAS have been made publicly available by many GWAS consortia, including the CARDIoGRAMplusC4D consortium for CAD, enabling PRS method development, calculation, and evaluation by the broad scientific community.43, 48
Figure 2 –
Development of polygenic risk scores. Modified from Choi et al. bioRxiv 2018.52
At the most basic level, an “unweighted” PRS sums the total number of risk alleles within an individual’s genome across a set of SNPs known to be significantly associated with the disease of interest. For example, an unweighted PRS comprising 10 common SNPs that are highly associated with disease yields a distribution of total scores in the general population that can range anywhere from 0 (no risk alleles) to 20 (two copies of the risk allele at each SNP). An unweighted PRS – variably termed an “unweighted allele score” – therefore assumes that all incorporated genetic variants have equivalent effect sizes.49 However, as the genetic architectures of most complex traits include unequal variant effects, this somewhat simplistic assumption limits the performance of unweighted PRS models.50, 51
More commonly, PRS incorporate the distinct effect sizes (or “weights”) of common variant associations with the disease of interest. Mathematically, a “weighted” PRS for each study participant j, is represented by the number of risk alleles at each variant i, multiplied by the respective, per-allele effect size (derived from GWAS summary statistics) and summed across M variants: . The resultant raw scores of a weighted, common variant PRS also tend to approximate a normal distribution at the population level. As expected, weighted PRS generally exhibit improved predictive accuracy as compared to their unweighted counterparts.50
Several other methodologic factors must be considered when calculating a PRS, including: (1) the selection of an optimal p-value threshold to guide variant inclusion; (2) the appropriate handling of linkage disequilibrium (LD), the correlation between SNPs located in the same region of the genome that may be inherited together due to lower rates of recombination; and (3) the prior probability of a SNP being causal for the outcome (as opposed to being correlated with the causal SNP(s)).52, 53
A selected p-value threshold determines the total number of SNPs incorporated into a PRS model. For example, a threshold of genome-wide significance denotes that only SNPs with GWAS association p-value < 5×10−8 are included in PRS calculation, and all remaining SNPs are excluded. A stringent p-value cutoff (such as genome-wide significance) yields a higher proportion of causal variants at a lower false positive rate, whereas a more lenient threshold yields a greater total number of genetic variants, albeit with less trait specificity. Achieving an optimal p-value threshold (often used as a “tuning parameter”) in a training dataset is therefore meant to balance the signal-to-noise ratio associated with using either a smaller number of highly-significant SNPs with more precise effect estimates, or a larger set of SNPs that more comprehensively account for trait heritability, but where the average, per-SNP effect estimates are less precise.
For disease prediction, the central goal is less about the discovery of associated variants and more about explaining phenotypic variation. Accordingly, recent evidence suggests that including more genetic variants (through the use of liberal significance cutoffs) leads to improved PRS performance for most complex diseases.54–56 Nonetheless, there is no single p-value threshold that maximizes PRS performance in all circumstances, as the optimal set of parameters is highly dependent on the genetic architecture of the disease, the specific attributes of the GWAS summary data from which the input variants are drawn (i.e. sample size, density of genotyping and imputation) and any differences between the discovery cohort and the target population. Therefore, it is now customary as part of PRS development to calculate and test many scores of varying sizes employing a range of p-value thresholds – from genome-wide significant (p < 5×10−8) to all independent SNPs (p < 1) – within a training dataset to arrive at the optimal parameters for a specific PRS.52, 57
A related, and equally important consideration, is accounting for the correlation between SNPs that are in close proximity (linkage disequilibrium). Failure to do so may result in the overrepresentation of genetic variants within regions of high LD, and marked reductions in PRS performance. There are two fundamental approaches to handling LD in the context of PRS generation. The first involves removing one SNP from a pair in high LD, either at random (“pruning”) or by preferentially preserving the SNP more highly associated with the outcome (“clumping”).52 This process retains a set of SNPs that are largely independent of one another, although it requires the somewhat arbitrary selection of a correlation threshold to nominate SNPs as being in LD.58 The second approach is to permit the inclusion of correlated SNPs, but to adjust (or “shrink”) effect estimates based on their correlation structure – for example, two correlated SNPs would both be preserved in a PRS model, but their respective effect estimates would be downsized commensurate with the degree of correlation. The former approach is typically employed alongside p-value thresholding, where SNPs are input into a PRS assuming they meet specified thresholds for both genetic correlation (and are therefore conditionally independent) and statistical significance – so-called “clumping and thresholding” scores.59 The latter approach is pertinent to expanded – i.e. “genome-wide” – PRS with more relaxed or no p-value thresholds, permitting incorporation of a larger number of genetic variants that may require adjustments for LD. The development of novel computational tools to better account for LD, including LDpred and Lassosum, have enabled more widespread generation and testing of expanded PRS (Table).60, 61
Table –
Overview and examples of CAD PRS types
| CAD PRS score types | Brief explanation | Examples from literature |
|---|---|---|
| Unweighted PRS (“unweighted allele score”) | Sum total of the number of common variant risk alleles within an individual’s genome known to be significantly associated with the disease of interest (or a linked trait), without incorporation of the distinct regression effect estimates for each common variant association. |
Kathiresan et al. NEJM 2008
67 • First evaluation of polygenic risk prediction for CAD • PRS of 9 unweighted LDL/HDL-associated SNPs associated with incident CAD in the Malmo Diet and Cancer Study (MDCS) Thanassoulis et al. Circ Cardiovasc Genet 2012 71 • PRS comprising 13 unweighted CAD SNPs associated with higher CAC in Framingham Heart Study |
| Weighted, limited clumping & thresholding PRS | Common variant risk alleles are multiplied (“weighted”) by their corresponding regression effect sizes for the disease of interest and summed to yield a single score. “Limited” scores typically impose stricter p-value thresholds and comprise only those SNPs associated with disease at levels of genome-wide significance. |
Ripatti et al. Lancet 2010
68
• PRS comprising 13 weighted genome-wide significant CAD SNPs associated with incident CAD in MDCS and FINRISK Khera et al. NEJM 2016 89 • PRS of 50 weighted genome-wide significant CAD SNPs associated with clinical and subclinical CAD across four cohorts • Marked attenuation of polygenic risk observed with adherence to healthy lifestyle behaviors Mega et al. Lancet 2015 100 • PRS of 27 weighted genome-wide significant SNPs associated with incident primary and secondary CV events in JUPITER, ASCOT, CARE, and PROVE-IT TIMI 22 clinical trials • Benefit of primary and secondary prevention statin therapy most pronounced in high polygenic risk group Natarajan et al. Circulation 2017 90 •PRS of 57 weighted genome-wide significant SNPs associated with CV outcomes in WOSCOPS trial •Risk reductions with statin therapy most prominent among high PRS group |
| Weighted, expanded clumping & thresholding PRS | As above, a weighted sum of risk alleles using corresponding, allele-specific regression effect estimates. However, “expanded” scores typically include SNPs below the threshold of genome-wide significance, by imposing more liberal thresholds for statistical significance (p-value) and/or SNP correlation (R2) |
de Vries et al. Int J Epidemiol 2015
77
• 152 CAD SNPs (49 genome-wide significant and 103 uncorrelated/ suggestive) • Associated with incident CAD in the Rotterdam study Abraham et al. Eur Heart J 2016 78 • PRS comprising 49,310 independent CAD SNPs (R2 < 0.7) • Improved prediction for incident CAD over prior, limited CAD PRS, and beyond Framingham Risk Score |
| Weighted, genome-wide PRS | Weighted sum of risk alleles using corresponding, allele-specific regression effect estimates. Significance threshold is relaxed or absent to permit incorporation of a greater number of SNPs, and effect estimates are adjusted (i.e. via “shrinkage”) based on extent of correlation with the other SNPs included. |
Khera et al. Nat Gen 2018
60
• Genome-wide PRS comprising 6.6 million genetic variants using LDpred algorithm • Captured extremes of polygenic risk for CAD in UK Biobank – levels of risk equivalent to monogenic FH mutations Inouye et al. JACC 2018 80 • Genome-wide PRS comprising 1.7 million genetic variants derived from 3 separate CAD discovery efforts (“metaGRS”) • Predicted incident CAD in UK Biobank better than any single clinical risk factor Damask et al. Circulation 2020 102 • Genome-wide PRS of ~6.6 million genetic variants applied to ODYSSEY-OUTCOMES trial of PCSK-9 inhibition • Patients at high polygenic risk enriched for recurrent CV events and greater treatment benefit from PCSK-9 inhibitor Aragam et al. JACC 2019 87 • Genome-wide PRS of ~6.6 million genetic variants applied to three health care systems (Partners, Penn, Mt. Sinai) • Patients at high polygenic risk not fully captured by current guideline-based prevention algorithms |
Evaluation of PRS performance
The product of PRS calculation and training is a defined set of genetic variants and associated effect sizes that can be applied to an independent validation dataset – one not included in the input GWAS or the training of PRS parameters – to evaluate score performance. Standard epidemiologic measures are employed to assess the association between PRS and outcomes. For binary diseases, it is conventional to report odds ratios (OR) or hazards ratios (HR) per standard deviation (SD) change in the PRS, the proportion of phenotypic variation explained (R2 or pseudo-R2), area under the receiver operating characteristic curve (AUC) or C-statistic, and the association P-value. Assuming a statistically significant P-value, the AUC and C-statistic typically convey the ability of the PRS to discriminate between diseased and non-diseased individuals, as bounded by the heritability of the disease. However, while AUC, C-statistic and related metrics – i.e. net reclassification index and integrated discrimination index – have been historically emphasized and may provide adequate population-level assessments of model discrimination, they do not inform tangibly on the prognosis of an individual or subgroup. Prognosis is perhaps most relevant for guiding clinical management, and may be better captured by abovementioned effect estimates of disease risk (such as the odds or hazards ratios).62–65 Moreover, to translate PRS clinically requires concurrent, disease-specific consideration of traditional risk factors, a rubric to define “high genetic risk” with corresponding strategies for risk modification, and an appropriate framework for integrating PRS-based risk stratification into routine clinical management.66
UTILITY OF POLYGENIC RISK SCORES FOR THE PREDICTION AND PREVENTION OF ASCVD
Prediction of clinical ASCVD by initial CAD PRS
The original assessment of CAD polygenic risk prediction predated most early genetic discovery efforts for CAD, and involved an unweighted PRS comprising 9 SNPs associated with LDL or HDL cholesterol at levels of genome-wide significance. This unweighted, “lipid allele score” associated with incident cardiovascular events – myocardial infarction, stroke, and cardiovascular death – in the Malmo Diet and Cancer Study (MDCS) after adjustment for traditional risk factors such as baseline lipid levels, but did not improve risk discrimination when added to these clinical predictors.67 Following the identification of CAD risk loci by initial GWAS, an analysis of a weighted PRS comprising 13 genome-wide significant CAD-associated SNPs in the prospective MDCS and FINRISK cohorts demonstrated a strong association with incident CAD – a hazard ratio of 1.66 comparing those in the highest versus lowest quintile of the PRS after adjusting for clinical covariates; although, the CAD PRS did not improve risk discrimination beyond these traditional risk factors and family history.68
Ensuing analyses of unweighted and weighted CAD PRS utilizing anywhere between 11 and 50 distinct, genome-wide significant CAD-associated SNPs yielded varying levels of association with prevalent and incident CAD.69–75 Notably, a study by Tada et al. in MDCS showed that the association between a CAD PRS of 50 genome-wide significant SNPs and incident CAD was robust to adjustments for self-reported family history, and that addition of this PRS to family history and other clinical risk factors improved model discrimination and reclassification, albeit modestly. Importantly, this study also demonstrated the predictive advantage of a 50-variant PRS over a 27-variant PRS (including only genome-wide significant SNPs in both cases) as evidenced by a steeper gradient of risk when comparing the highest and lowest quintiles of the PRS (HR = 1.92 and 1.70, respectively, with 50-SNP and 27-SNP PRS).76
Studies have since shown the predictive benefit of expanding CAD PRS to include SNPs that fall below the threshold of genome-wide significance, likely due to better capture of CAD heritability. For example, an assessment of CAD risk prediction in the Rotterdam Study showed a slight discriminative advantage when using a more permissive 152-SNP PRS (49 genome-wide significant CAD SNPs and an additional 103 uncorrelated, suggestive CAD SNPs with false discovery rate < 5%) over a PRS limited to 49 genome-wide significant CAD SNPs.77 Abraham et al. subsequently performed iterative training of PRS models based on differing correlation (R2) thresholds for LD and arrived at an optimal PRS comprising 49,310 independent SNPs. This expanded PRS was tested in multiple prospective cohorts from FINRISK and the Framingham Heart Study, and showed marked predictive and discriminative improvement over CAD PRS models limited to genome-wide significant or suggestive SNPs, and beyond established clinical risk scores (i.e. the Framingham Risk Score).78
Prediction of clinical ASCVD by genome-wide CAD PRS
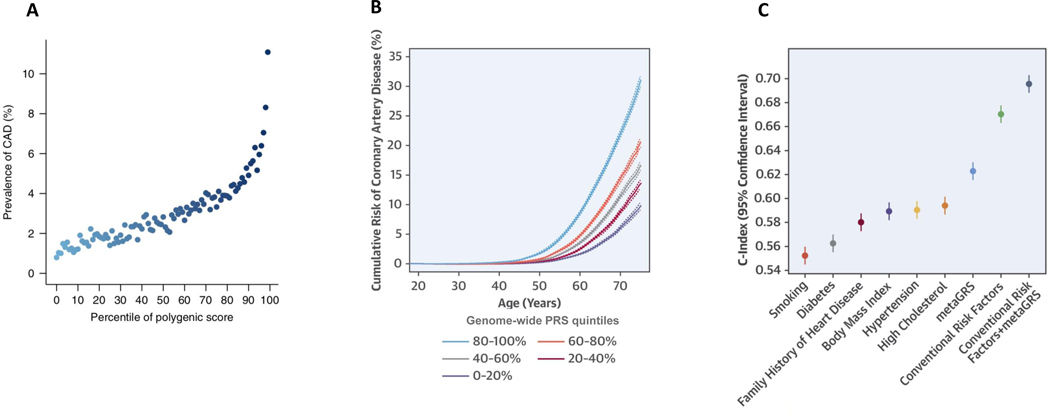
As described above, methodologic advances have now enabled the generation of genome-wide PRS incorporating millions of common genetic variants. In particular, two studies focusing on the UK Biobank (N ~ 500,000) have demonstrated the potential of genome-wide PRS to enhance CAD risk prediction. Khera et al. leveraged the LDpred algorithm – a Bayesian approach to PRS calculation that accounts for patterns of correlation between SNPs using a LD reference panel – to generate a CAD PRS comprising 6.6 million common genetic variants.60 This genome-wide PRS compared favorably to previously reported CAD PRS – i.e. the abovementioned 50-SNP and 49,310-SNP scores – as evidenced by more robust p-values and higher, per-SD effect estimates for the association with combined prevalent and incident CAD. Notably, this expanded score better identified individuals at the extremes of disease risk. Specifically, individuals in the top 1% of the 6.6 million variant PRS were at nearly 5-fold odds of developing CAD when compared to the remaining 99% of the UK Biobank population; by comparison, membership in the top 1% of the 49,310-SNP score by Abraham et al. conferred just under 3-fold odds of CAD (Figure 3).79
Figure 3 –
Genome-wide polygenic risk scores: (A) identify a marked, inherited predisposition to CAD; (B) provide lifetime estimates of risk; and (C) add to the discriminative ability of clinical risk factors. Panel (A) from Khera et al. Nature Genetics 2018.78 Panels (B) and (C) from Inouye et al. JACC 2018.79
Subsequently, Inouye et al. constructed a PRS comprising 1.7 million common genetic variants reflecting the weighted average of genetic information from three orthogonal CAD discovery efforts. Compared to prior scores, this “metaGRS” captured a greater proportion of CAD heritability (~27%), and demonstrated marked improvements in risk prediction and discrimination, while confirming clinical risk factor-independent associations with incident CAD. As tested in the UK Biobank, the discriminative capacity (C-statistic) of the metaGRS alone was greater than that of any other, individual clinical risk factor (i.e. hypertension, hyperlipidemia, diabetes, body mass index, family history of heart disease, or current smoking), and added to the net discrimination of all six conventional risk factors combined. In addition, this study rigorously characterized the marked differences in lifetime trajectories of CAD risk between strata of the metaGRS (Figure 3). Such age-independent risk trajectories would be ascertainable early in life and before the manifestation of other clinical risk factors, suggesting that genetic assessments may permit the early identification of patients at high lifetime risk of CAD, and the implementation of corresponding prevention strategies to mitigate risk.80, 81
Additional testing of genome-wide PRS in separate cohorts has yielded mixed results, although the specific methodologies and outcome measures utilized, and the sample sizes assessed have likely influenced the conclusions drawn from each study. An analysis of the above two CAD PRS generated by LDpred and metaGRS in a French-Canadian population (N ~ 11,000) demonstrated robust associations with prevalent CAD (AUC = 0.72 – 0.89), but more modest associations with incident and recurrent events (AUC 0.56 – 0.60). The latter observation was thought to be the result of a focus on prevalent CAD in the discovery CAD GWAS studies from which each score was derived, as well as a high rate of preventive therapies (i.e. 76% statin prescription rate) in the study population, which may have influenced incident analyses of primary events.82
In an analysis of the Atherosclerosis Risk in Communities (ARIC) and Multiethnic Study of Atherosclerosis (MESA) cohorts (Total N = 7,237), the 6.6 million variant LDpred CAD PRS associated strongly with prevalent CAD, and more modestly with incident CAD, but did not add significantly to conventional clinical factors including the ACC/AHA Pooled Cohort Equations (PCE), as assessed by metrics of model discrimination, calibration and risk reclassification. Of note, for analyses in ARIC, the authors required that participants complete their fourth study visit without developing CAD in order to be included in incident disease analyses, which may have reduced the power of the study to detect meaningful changes in the C-statistic. Nonetheless, when evaluated separately, the PRS (plus age and sex) achieved a comparable C-statistic to the PCE (which includes age and sex) in both ARIC (PRS: 0.669 versus PCE: 0.701) and MESA (PRS: 0.672 versus PCE: 0.660) despite marked temporal differences in when each risk score may be ascertained – i.e. the PRS is available at the time of birth, before the onset of clinical risk factors required to compute the PCE (which has been validated for individuals with a minimum age of 40).83
A similar analysis in the UK Biobank (N = 352,660) examined a genome-wide CAD PRS in the context of two clinical risk scores – the PCE and the analogous, UK-recommended QRISK3 score. The authors constructed genome-wide PRS using the aforementioned lassosum method, which employs a penalized regression model and an external reference panel to account for LD.61 The best performing CAD PRS in the independent training set comprised 1,037,385 SNPs. Incident event analyses with the best-performing score demonstrated equivalent C-statistics between the age/sex-adjusted lassosum CAD PRS (0.76) and the PCE (0.76), and a statistically significant but modest increase in the C-statistic when the two were combined (0.78).84
A third, and separate study, analyzed a genome-wide CAD PRS (6.6 million variant LDpred PRS) and the PCE in the UK Biobank and also in MDCS (N ~ 28,000). As above, this analysis showed comparable C-statistics between age/sex-adjusted CAD PRS and the PCE, and a small increase in the C-statistic when the two were combined. However, within each of the four standard PCE risk categories routinely used in the clinical setting (“Low,” “Borderline,” “Intermediate,” and “High” risk), incident event rates were markedly different based on PRS strata. Specifically, within each PCE subgroup, there was a 2–4-fold higher rate of incident CAD among those at high (top 20% of PRS) versus low polygenic risk (bottom 20% of PRS) over the 10-year follow-up period, suggesting the ability of the CAD PRS to substantially stratify disease risk trajectories within each PCE risk category.85
The latter findings re-demonstrate the known challenges of relying on metrics of discrimination when assessing the incremental prognostic benefit of an added predictor.63 However, despite somewhat differing study conclusions, these analyses uniformly demonstrate the robust predictive power of genome-wide PRS – on par with the widely-used PCE across multiple populations – and affirm their role as the “first-risk factor” for CAD, with the ability to prognosticate risk from a young age and over an extended time horizon (Figure 4).81 Accordingly, the potential utility of CAD PRS among younger individuals is typically acknowledged and requires further, prospective testing.
Figure 4 –
Polygenic risk score as the “first risk factor” for coronary artery disease.
However, the conflicting study conclusions have sparked controversy around the clinical utility of CAD PRS in middle-aged adults.86 Given robust evidence around traditional risk calculators and their readily-available constituent clinical factors in middle-aged adults, these clinical factors should continue to serve as the foundation of CAD risk assessments for this age demographic. PRS are also poised to be readily available, and may provide complementary information to support clinical decision-making, ideally within a guideline-supported framework. The aforementioned study in UK Biobank and MDCS is pertinent in this regard as it demonstrated a marked gradient in longitudinal risk when considering the CAD PRS within guideline-supported categories of clinical risk as defined by the PCE.85 In particular, a two- to four-fold risk difference was noted between PRS strata among participants at borderline to intermediate clinical risk, a group for whom updated ACC/AHA prevention guidelines already endorse the use of CAD “risk-enhancing factors” (i.e. C-reactive protein, Lipoprotein(a), family history of premature MI) to help guide statin initiation.19 Indeed, the extensive literature reviewed above may support the incorporation of PRS as yet another risk-enhancer to up-classify risk in scenarios of clinical equipoise. In fact, accruing evidence now suggests that a CAD PRS measures risk not otherwise captured by well-established prevention algorithms.87 Therefore, the data speak to combining complementary genetic and clinical information within guideline-supported frameworks to better capture different trajectories of disease risk, and to facilitate earlier prevention strategies for higher risk groups. Such individuals may be identified genetically earlier in life prior to the onset of traditional risk factors, or in mid-life alongside established clinical risk models.
Association of CAD PRS with subclinical ASCVD
The genetic determinants of CAD align with those of subclinical coronary atherosclerosis. A discovery genetic effort of coronary artery calcification (CAC) and carotid intima-media thickness (CIMT) in over 77,000 individuals from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium demonstrated genome-wide significant signals at known CAD loci such as at CDKN2B (the 9p21 locus), PHACTR1, APOB, and APOE.88 Perhaps unsurprisingly, then, polygenic predictors of CAD have been shown to associate with subclinical atherosclerosis in multiple vascular beds. In a study of two generations of the Framingham Heart Study, a gradient of risk for high CAC was observed across tertiles of an unweighted 13-SNP PRS.71 Separately, a CAD PRS (of 50 weighted, genome-wide significant SNPs) was found to associate strongly with CAC as measured in the BioImage study.89 In an assessment of CAC from the Coronary Artery Risk Development in Young Adults (CARDIA) observational cohort and CIMT from BioImage, a 1-SD increase in a CAD PRS (of 57 weighted, genome-wide significant SNPs) associated with significantly increased odds of having CAC (OR = 1.32; p = 0.02), and a 9.7% increase in carotid plaque burden (p = 0.01); notably, a more restricted 27-SNP PRS did not associate with carotid plaque burden in BioImage.90
A relevant clinical application of these genetic associations may be in the use of a PRS to guide preventive screening for subclinical coronary or carotid atherosclerosis. In a recent study of over 6,000 participants from the MESA observational cohort, a CAD PRS (of 157 genome-wide significant SNPs) strongly predicted non-zero CAC thereby improving the yield of screening cardiovascular computed tomography (CT).91 Accordingly, a CAD PRS may facilitate earlier and more targeted, imaging-based assessments of individuals at high polygenic risk for CAD, for whom the presence of CAC may inform considerably the timing of statin initiation.
Of note, a PRS may also prognosticate risk in the absence of demonstrable, subclinical coronary atherosclerosis. In a study of patients undergoing coronary angiography from the Penn Medicine Biobank, an expanded, genome-wide CAD PRS comprising over 130,000 SNPs was associated with the burden and severity of angiographically-confirmed coronary stenoses. In addition, high polygenic risk was strongly associated with all-cause mortality, and this persisted even among the subset of patients without angiographic CAD.92 These findings support that an elevated CAD PRS portends a poorer prognosis, and that this risk may, in part, be complementary to that gleaned from anatomical assessments of coronary stenoses.
Monogenic versus polygenic risk for CAD
Current national guidelines for the primary prevention of CAD endorse preventive statin therapy for individuals harboring a rare monogenic FH mutation, present in 1 in 250 individuals in the general population and conferring a 3-fold risk of CAD.19, 93 Devising an analogous, polygenic risk framework can be challenging due to the continuous spectrum of polygenic susceptibility as compared to the dichotomous risk classification of a monogenic mutation. One approach involves determining a polygenic risk threshold that confers a “monogenic equivalent” level of disease risk. As described above, Khera et al. uncovered individuals at the extremes of polygenic risk for CAD in the UK Biobank by applying a genome-wide PRS comprising 6.6 million genetic variants as generated by the LDpred algorithm. This PRS performed better than prior CAD PRS that comprised more limited sets of genetic variants. Notably, individuals in the top 8% of the PRS distribution (~1 in 12 individuals) were at 3-fold odds of CAD as compared to the rest of the population – a level of risk roughly equivalent to harboring a pathogenic FH mutation.93, 94 In addition to being more prevalent, individuals at high polygenic risk did not have appreciably different clinical risk profiles from the general population, unlike individuals with FH who are often identified by severe hypercholesterolemia.79
In a follow-up analysis, the authors pursued whole genome sequencing of the VIRGO (Variation in Recovery: Role of Gender on Outcomes of Young AMI Patients) and MESA studies to compare the relative prevalence and clinical significance of monogenic and polygenic risk with regards to early-onset MI. The analysis included 2,081 hospitalized, premature MI patients from VIRGO and 3,761 population-based controls from MESA, all of whom had FH mutation status ascertained and an LDpred, genome-wide CAD PRS calculated. A pathogenic FH mutation was present in 1.7% of all patients with premature MI, and conferred 3.8-fold increased odds of disease; by comparison, high polygenic risk (top 5% of the PRS) was present in 17.3% of these patients and conferred 3.7-fold increased odds of disease. Again, patients with a FH mutation demonstrated markedly elevated serum LDL-C levels (mean 206 mg/dL) while those at high polygenic risk did not (mean 132 mg/dL; population mean 122 mg/dL).95 These and other analyses now suggest that an elevated CAD PRS may confer risk on par with a pathogenic FH mutation, but that the former is at least 10-times more common in the general population and accounts for a larger portion of early-onset MI cases.96
However, for eventual clinical implementation, it will be important to consider the combined influences of monogenic and polygenic risk on lifelong susceptibility to CAD. As these two pathways of risk appear to be independent of one another, it is conceivable that individuals inheriting both a pathogenic FH mutation and a high polygenic risk profile for CAD (i.e. a large burden of common variant risk alleles) are at substantially elevated risk compared to the general population. Conversely, some individuals may harbor a pathogenic mutation for FH, but a low-risk polygenic profile, which might offset some of the disease susceptibility conferred by the inherited monogenic mutation.
Initial observations have supported these theories through simultaneous interrogation of monogenic and polygenic influences on CAD risk and/or intermediate risk factors. For example, in the abovementioned whole genome sequencing analysis of early-onset MI, patients with both a FH mutation and a high CAD PRS had greater LDL-C levels than those with either high monogenic or polygenic risk only.95 Similarly, two studies have shown that CAD PRS strongly predict incident cardiovascular disease among patients with FH, suggesting that a polygenic background may significantly modulate the penetrance of a pathogenic FH mutation.78, 97
A more recent analysis utilizing newly-available exome sequencing data from the UK Biobank investigated the interaction between rare, monogenic FH variants and polygenic CAD risk as assessed by a genome-wide PRS. While carriers of a monogenic FH mutation had a collective 3-fold odds of developing CAD compared to non-carriers, there was a marked gradient of risk among mutation carriers that depended on their respective polygenic backgrounds. Specifically, as compared to non-carriers, the odds of CAD among FH mutation carriers ranged from 1.30-fold for those in the lowest quintile of the PRS distribution to 12.59-fold for those in the highest PRS quintile. Models to estimate the odds of developing disease by age 75 yielded probabilities ranging from 4.8% (FH mutation non-carriers in the lowest PRS strata) to 77.7% (FH mutation carriers in the highest PRS strata).98 These observations provide robust evidence for the interplay between monogenic and polygenic determinants of CAD risk, and emphasize the importance of jointly considering these two genetic risk profiles when prognosticating disease risk in the clinical setting.
Mitigation of polygenic CAD risk through favorable lifestyle and medications
The clinical utility of any risk prognostication tool is dependent both on its predictive accuracy and its potential actionability. Indeed, recent studies have attested to the actionability of polygenic risk for CAD, namely the ability to modify risk through both non-pharmacologic and pharmacologic strategies.
In an observational study across four independent cohorts (N = 55,685), a CAD PRS (of 50 genome-wide significant SNPs) strongly associated with clinical and subclinical CAD, but genetic risk was uniformly attenuated by adherence to a healthy lifestyle, defined as engaging in at least three out of four healthy lifestyle behaviors (no smoking, no obesity, healthy diet, and regular physical activity). Specifically, individuals in the highest quintile of polygenic risk who were adherent to a healthy lifestyle achieved a consistent ~50% reduction in disease risk as well as significant reductions in CAC as compared to individuals at high genetic risk who did not commit to a favorable lifestyle. Notably, the high PRS/favorable lifestyle subgroup achieved a level of disease risk on par with (or below) those at low genetic risk but with unfavorable lifestyle habits.89 A similar analysis in the UK Biobank (N= 339,003) yielded concordant findings including the observed log-additive effect of genetic risk and healthy behaviors on the odds of developing incident cardiovascular diseases, affirming the value of lifestyle modification for the population at-large, while also emphasizing its particular importance for those with a marked genetic predisposition for cardiovascular disease.99
In their aforementioned assessment of the UK Biobank, Inouye et al. demonstrated attenuation of polygenic CAD risk among individuals who self-reported statin therapy.80 This observation is well supported by post-hoc genetic analyses of clinical trial cohorts that have demonstrated the marked benefit of lipid-lowering therapies to reduce incident cardiovascular events in those at high polygenic risk. In an analysis of the JUPITER (Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin) and ASCOT (Anglo-Scandinavian Cardiac Outcomes Trial) primary prevention statin trials, Mega et al. observed a strong association between CAD PRS (27 weighted genome-wide significant SNPs) and incident cardiovascular events independent of traditional risk factors. Furthermore, a graded increase in statin benefit was noted moving from low (bottom 20% of PRS), to intermediate (middle 60% of PRS) and high (top 20% of PRS) polygenic risk strata, as evidenced by greater absolute and relative risk reductions among those at high polygenic risk, and a corresponding decrease in the number needed to treat.100 A similar phenomenon was observed in a genetic analysis of the WOSCOPS (West of Scotland Coronary Prevention Study) primary prevention statin trial, where a CAD PRS (57 weighted genome-wide significant SNPs) was associated with cardiovascular outcomes, and risk reductions with statin therapy were most pronounced among those at high polygenic risk for CAD (top 20% of PRS). A study-level meta-analysis of the three primary prevention trials – JUPITER, ASCOT, and WOSCOPS – indicated an absolute risk reduction in those at high polygenic risk of 3.6% versus 1.3% in all others, and a corresponding relative risk reduction of 46% in the high polygenic risk subgroup versus 26% in all others.90 These findings suggest the utility of CAD PRS to identify patients most likely to respond to primary prevention statin therapy, thereby reducing the number needed to treat to achieve population-wide treatment benefit.
Polygenic prediction and prevention of secondary cardiovascular events
Prior studies of CAD PRS have shown variable performance with regards to the prediction of recurrent cardiovascular events.35, 69, 82 Secondary prevention clinical trials facilitate rigorous assessments of recurrent event prediction and response to therapy through the interrogation of well-phenotyped study populations with meticulous event adjudication. In the above analysis by Mega et al., the 27-SNP CAD PRS was also assessed in the context of two secondary prevention trials – CARE and PROVE-IT TIMI 22 – investigating the clinical benefit of statin therapy in patients who had suffered an acute coronary syndrome. Compared to the low genetic risk subgroup, the multivariable-adjusted hazards for recurrent coronary artery disease were 1.65 (p = 0.0030) for those at intermediate polygenic risk, and 1.81 (p = 0.0029) for those at high polygenic risk. Absolute and relative risk reductions were again graded across genetic risk strata, with those at high polygenic risk deriving the largest benefit from secondary prevention statin therapy.100
More recently, a genetic substudy of the FOURIER trial (Further Cardiovascular Outcomes Research With PCSK9 Inhibition in Subjects With Elevated Risk) assessed the same 27-SNP CAD PRS in a population with established ASCVD (prior history of MI, nonhemorrhagic stroke, or symptomatic peripheral artery disease) randomized to PCSK-9 inhibitor therapy or placebo on top of baseline statin therapy. The CAD PRS associated with incident major vascular and coronary events in the placebo arm of this secondary prevention population after adjustment for clinical risk factors, with those at intermediate (middle 60% of PRS) and high (top 20% of PRS) polygenic risk demonstrating progressively greater hazards of disease as compared to the low genetic risk subgroup. Importantly, the CAD PRS successfully stratified the population by treatment effect, with those at high polygenic risk deriving 2-fold greater benefit from PCSK-9 inhibitor therapy as compared to the overall trial population. Notably, PCSK-9 inhibitor therapy appeared to offset the risk conferred by a polygenic susceptibility to CAD – in contrast to the placebo arm, cardiovascular event rates in the treatment arm were comparable between those at high and low polygenic risk.101 Although follow-up studies are required, these initial investigations of secondary prevention clinical trial populations attest to a potential role for polygenic risk assessments to identify high risk patients likely to benefit from tailored secondary prevention strategies.
BIOLOGICAL INSIGHTS GLEANED FROM STUDIES OF POLYGENIC ARCHITECTURE
Determining mechanistic links between individual common genetic loci and disease
GWAS have identified 164 common genetic loci associated with CAD, furthering our understanding of the genetic architecture of CAD, and motivating studies to probe the biological pathways linking specific genetic loci to distinct mechanisms of disease. Post-GWAS analyses seeking to translate GWAS findings to biological function typically begin with a range of bioinformatic analyses of lead SNP associations to help choose the genetic signals that most warrant detailed experimental follow-up. For example, novel SNPs arising from GWAS are routinely interrogated in available tissue and disease-specific transcriptome datasets.102–105 The finding of an association between a risk allele and differences in gene expression may suggest a functional effect warranting further experimental investigation.
Both computational and functional approaches have been used to categorize many of the uncovered common CAD loci into distinct mechanistic pathways. In one study featuring both approaches, Klarin et al. performed a GWAS and meta-analysis of the UK Biobank and CARDIoGRAMplusC4D and identified 15 novel loci associated with CAD at a level of genome-wide significance. A phenome-wide association study (PheWAS) was then performed, where each lead SNP was tested for association with a range of phenotypes to better understand the potential spectrum of phenotypic consequences for a given genetic variant.106 PheWAS revealed associations between the novel locus CCDC92 and traits of insulin resistance and lipodystrophy within adipose tissue, serving as a putative mechanistic link to the development of CAD. For the novel locus ARGHEF26, which lacked significant PheWAS associations to CAD risk factors or other traits, experimental knock down by small interfering RNA led to decreased transendothelial migration and adhesion of leukocytes, suggesting a plausible biological pathway by which this genetic locus might mediate coronary atherosclerosis.33 Notably, PheWAS have been pursued in other studies of novel CAD loci and demonstrated that many loci are associated with one or more known CAD risk factors, but for a large portion, the underlying mechanisms remain unknown (Figure 1).37, 107
Assessments of shared genetic architecture
Beyond assessments of individual loci to understand particular genetic mechanisms contributing to disease biology, the aggregate assessment of many disease-associated common genetic variants may inform more broadly on shared genetic architectures. PRS – i.e. genome-wide scores and other scores comprising many genetic variants – enable such assessments as they are more appropriate than individual risk alleles for assessing genetic overlap between a disease and related traits.
In one such analysis, Ntalla et al. studied the association between a 300-SNP CAD PRS and a range of cardiovascular and non-cardiovascular conditions in the UK Biobank. In particular, the authors found strong associations with peripheral artery disease (PAD), stroke, and abdominal aortic aneurysm even after the removal of CAD cases suggesting a common genetic predisposition among these distinct disease entities.108 Indeed, the noted association in prior studies between CAD PRS and subclinical atherosclerosis in multiple vascular beds supports the theory that shared processes – with shared genetic determinants – mediate the formation of atherosclerotic plaques.71, 88, 90
However, more comprehensive assessments of shared heritability may be best achieved through studies of genetic correlation – the genetic relationship between two traits typically determined by comparing sets of trait-specific GWAS data.109 The development of computational tools such as linkage disequilibrium score regression (LDSC) now permits efficient comparisons across many traits through use of readily-available GWAS summary statistics.110, 111 LD Hub is a publicly-available server that hosts LDSC calculations of genetic correlations for a broad range of published GWAS summary statistics.112
The rate-limiting step is therefore the conduct of a GWAS for a trait or disease of interest to permit downstream comparisons with summary statistics of other traits. For certain vascular conditions outside the coronary circulation, where genetic discovery has been slowed by modest case counts, significant progress has been enabled by the Million Veteran Program (MVP), a population-based, mega-biobank with genotype data linked to health registry data from the United States Department of Veterans Affairs Health System.113 A recent, landmark GWAS of PAD combined data from MVP and the UK Biobank (Total N ~ 600,000 with ~36,000 PAD cases) to identify 19 PAD-associated genetic loci, including 18 loci not previously reported.114 Eleven of the 19 reported loci were also associated with disease in other vascular beds (coronary and cerebral), including genes related to LDL-C, lipoprotein(a), and lipoprotein lipase (LDLR, LPA, LPL). Furthermore, strong genetic correlations were observed when comparing summary-level genetic data from this PAD analysis with prior GWAS results for CAD (from the CARDIoGRAMplusC4D consortium) and large-artery stroke (from the MEGASTROKE consortium) further implicating shared pathways that likely mediate arterial atherosclerosis across all three ASCVD phenotypes.43, 115
An ensuing genetic analysis of venous thromboembolism (VTE) in MVP and UK Biobank has provided additional insights into the potential mechanistic overlap between differing vascular pathologies. This discovery analysis of ~26,000 cases and ~620,000 controls uncovered 22 novel genetic loci associated with VTE. Notably, LDSC analyses revealed genetic correlations between VTE and all three of the above ASCVD phenotype (CAD, PAD, and large artery stroke). Furthermore, genetic analyses suggested a causal association between LDL-C – but not HDL-C or triglycerides – and VTE.116 These assessments of the genetic architectures of distinct cardiovascular phenotypes using robust GWAS data suggest potential shared biologic pathways between atherosclerotic and venous thrombotic diseases that warrant future investigations.
PRACTICAL CONSIDERATIONS AND POTENTIAL CHALLENGES WITH PRS IMPLEMENTATION
Implementation studies of polygenic CAD risk disclosure
While additional validation of PRS is required – including prospective outcome studies that integrate both clinical and genetic tools for risk prognostication – the evidence to date suggests a future role for assessments of polygenic CAD risk to identify and target high-risk individuals earlier in life for more aggressive primary and secondary preventive strategies. But as these preventive strategies – i.e. favorable lifestyle practices and statin medications – rely on sustained behavioral changes to yield meaningful benefit, an important part of effective genomic and precision medicine will involve the ability of genetic risk disclosure to promote healthy behaviors in the general population.
Initial studies aimed at motivating behavior change through PRS disclosure have yielded mixed results. One study showed no adverse effects of PRS disclosure on shared decision making and patient satisfaction, while another demonstrated the feasibility of risk disclosure in the outpatient setting but found no improvement in patient adherence to preventive regimens.117, 118 In the Myocardial Infarction Genes (MI-GENES) trial, 203 participants at intermediate risk for CAD were randomized to the disclosure of clinical risk alone or in combination with polygenic risk. At 6 months, behavioral patterns (i.e. diet and exercise) did not differ between the two groups, but participants informed of their genetic risk were more likely to be initiated on statin therapy by their providers and achieved lower levels of LDL-C.119
Preliminary data from the GeneRISK study of over 7,300 individuals in Finland suggest that combined clinical and genomic risk disclosure may, in fact, motivate behavioral modification (i.e. weight loss and smoking cessation), particularly among those at high polygenic risk for CAD. However, a key aspect of this study was the communication of risk estimates through a web-based interactive tool (KardioKompassi) that allowed participants to compare personal risk profiles against the average risk of the total study population, and to model changes in personal risk with the adoption of specific lifestyle changes.120 It should be noted that difficulties motivating patient behavior after disclosure of high CAD risk is not a new phenomenon, and has been observed previously in studies of imaging-based risk assessments, i.e. coronary CT and carotid ultrasonography.121–124 Thus, the use of digital tools to more effectively interface with patients and communicate risk may prove critical to future efforts seeking to harness genomic and clinical risk disclosure to encourage healthy lifestyle habits.
PRS standardization and normalization across settings
Standardization of any risk stratification tool is essential for consistent implementation across settings. Different versions of the CAD PRS have been utilized in the various studies described in this review. As discussed, there are many steps involved in PRS construction that may produce differences in scores – i.e. the number of variants included, the per-SNP effect estimates based on the particular GWAS summary statistics utilized (which, in turn, are influenced by differences in GWAS characteristics), the specific computational method used for PRS generation and handling of LD, and the training dataset used to select optimal PRS parameters. The use of a standard PRS for a given condition may facilitate clinical translation, although differences in PRS performance across genetic ancestries (as described below) may challenge the notion of a single PRS that is effective for all individuals, barring the development of methods to offset these ancestry-specific variations in score performance.125
Furthermore, even with a standardized PRS meant to be employed universally, the application of a PRS to different populations presents various logistical considerations. For example, depending on the genotyping array and imputation panel used in a particular setting, differing numbers of SNPs may overlap with the PRS resulting in differences in raw scores. Additional thought must then be given to normalization of score distributions across individuals of different genetic ancestries, and across different settings (i.e. in different health systems) to designate comparable thresholds of risk, and whether/how an external reference PRS distribution might be utilized against which all others are plotted.
Ancestry-specific differences in PRS performance
To date, the majority of GWAS and imputation reference panels have been in individuals of European genetic ancestry, with a resultant bias in developed PRS towards such persons of European descent.126–129 This has significantly limited the transferability of PRS derived from Eurocentric genetic discovery efforts, which tend to perform less well in individuals of non-European ancestries.130–132 Indeed, for CAD, a genome-wide PRS has been demonstrated to associate with disease across diverse ancestry groups, although the highest predictive accuracy was seen among study participants of European genetic background.95 Promoting genetic discovery in non-European populations will be a prime component of future prediction efforts, as PRS derived from and for specific non-European ancestries have been shown to outperform those based on largely European genetic data.133 This lack of transethnic transferability is thought to be the result of ancestry-specific differences in LD structure as opposed to true, ancestry-specific differences in the genetic architecture of disease.129, 134 Accordingly, the generation of many ancestry-specific PRS (requiring alignment of an individual to a specific genetic ancestry) and a single pan-ancestry PRS have both been proposed as potential paths forward.135–137 Regardless, it will be imperative to improve trans-ancestry PRS prediction through a combination of non-European GWAS and novel computational methods to permit the clinical implementation of polygenic risk prediction in an equitable manner.
CONCLUSIONS AND FUTURE DIRECTIONS
Rapid progress over the past two decades in the understanding of complex trait biology and CAD genetics has enabled an improved understanding of disease mechanisms and the development of robust polygenic predictors capable of charting lifetime trajectories of CAD risk. Coupled with growing evidence on strategies to mitigate this inherited susceptibility, and more accessible and affordable array-based genotyping, we are now poised to leverage genomic data at a population level to facilitate the early prediction and prevention of CAD. However, future studies will be required to further refine PRS methodologies, better integrate both genomic and clinical predictors of CAD, and to address other pending implementation challenges – including ancestry-specific differences in PRS performance – to fully realize the promise of precision cardiovascular care guided by genomic risk stratification.
Acknowledgments
SOURCES OF FUNDING
K.G.A. is supported by an award from the American Heart Association Institute for Precision Cardiovascular Medicine (17IFUNP33840012). P.N. is supported by grants from the National Heart, Lung, and Blood Institute (R01HL142711, R01HL148565, R01HL148050), Fondation Leducq (TNE-18CVD04), and Hassenfeld Scholar Award from the Massachusetts General Hospital.
DISCLOSURES
P.N. reports grant support from Amgen, Apple, and Boston Scientific, and is a scientific advisor to Apple.
NON-STANDARD ABBREVIATIONS AND ACRONYMS
- ASCVD
atherosclerotic cardiovascular disease
- CAC
coronary artery calcification
- CAD
coronary artery disease
- CIMT
carotid intima-media thickness
- FH
familial hypercholesterolemia
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- MAF
minor allele frequency
- PAD
peripheral artery disease
- PCE
Pooled Cohort Equations
- PCSK-9
proprotein covertase subtilisin/kexin type 9
- PRS
polygenic risk score
- SNP
single nucleotide polymorphism
- VTE
venous thromboembolism
REFERENCES
- 1.Herapath CE and Perry CB. The Coronary Arteries in a Case of Familial Liability to Sudden Death. Br Med J. 1930;1:685–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gertler MM, Garn SM and White PD. Young candidates for coronary heart disease. J Am Med Assoc. 1951;147:621–5. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CB and Cohen BH. The familial occurrence of hypertension and coronary artery disease, with observations concerning obesity and diabetes. Ann Intern Med. 1955;42:90–127. [DOI] [PubMed] [Google Scholar]
- 4.White PD. Genes, the heart and destiny. N Engl J Med. 1957;256:965–9. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd-Jones DM, Nam BH, D’Agostino RB, Sr., Levy D, Murabito JM, Wang TJ, Wilson PW and O’Donnell CJ. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–11. [DOI] [PubMed] [Google Scholar]
- 6.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI and De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–54. [DOI] [PubMed] [Google Scholar]
- 7.Nora JJ, Lortscher RH, Spangler RD, Nora AH and Kimberling WJ. Genetic--epidemiologic study of early-onset ischemic heart disease. Circulation. 1980;61:503–8. [DOI] [PubMed] [Google Scholar]
- 8.Badano JL and Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–89. [DOI] [PubMed] [Google Scholar]
- 9.Rader DJ, Cohen J and Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL and Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown MS and Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. [DOI] [PubMed] [Google Scholar]
- 12.Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM and McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci U S A. 1989;86:587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG and Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–6. [DOI] [PubMed] [Google Scholar]
- 14.Cohen JC, Boerwinkle E, Mosley TH, Jr. and Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. [DOI] [PubMed] [Google Scholar]
- 15.Khera AV, Won HH, Peloso GM, Lawson KS, Bartz TM, Deng X, van Leeuwen EM, Natarajan P, Emdin CA, Bick AG, Morrison AC, Brody JA, Gupta N, Nomura A, Kessler T, Duga S, Bis JC, van Duijn CM, Cupples LA, Psaty B, Rader DJ, Danesh J, Schunkert H, McPherson R, Farrall M, Watkins H, Lander E, Wilson JG, Correa A, Boerwinkle E, Merlini PA, Ardissino D, Saleheen D, Gabriel S and Kathiresan S. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benn M, Watts GF, Tybjaerg-Hansen A and Nordestgaard BG. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur Heart J. 2016;37:1384–94. [DOI] [PubMed] [Google Scholar]
- 17.Besseling J, Hovingh GK, Huijgen R, Kastelein JJP and Hutten BA. Statins in Familial Hypercholesterolemia: Consequences for Coronary Artery Disease and All-Cause Mortality. J Am Coll Cardiol. 2016;68:252–260. [DOI] [PubMed] [Google Scholar]
- 18.Gidding SS, Champagne MA, de Ferranti SD, Defesche J, Ito MK, Knowles JW, McCrindle B, Raal F, Rader D, Santos RD, Lopes-Virella M, Watts GF, Wierzbicki AS, American Heart Association Atherosclerosis H, Obesity in Young Committee of Council on Cardiovascular Disease in Young CoC, Stroke Nursing CoFG, Translational B, Council on L and Cardiometabolic H. The Agenda for Familial Hypercholesterolemia: A Scientific Statement From the American Heart Association. Circulation. 2015;132:2167–92. [DOI] [PubMed] [Google Scholar]
- 19.Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, Braun LT, de Ferranti S, Faiella-Tommasino J, Forman DE, Goldberg R, Heidenreich PA, Hlatky MA, Jones DW, Lloyd-Jones D, Lopez-Pajares N, Ndumele CE, Orringer CE, Peralta CA, Saseen JJ, Smith SC Jr., Sperling L, Virani SS and Yeboah J. 2018. AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol. 2018. [Google Scholar]
- 20.Altshuler D, Daly MJ and Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J and International Human Genome Sequencing C. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 22.International HapMap C, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R and Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howie B, Fuchsberger C, Stephens M, Marchini J and Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudbridge F and Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, Wtccc C. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH and Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A and Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3. [DOI] [PubMed] [Google Scholar]
- 28.Myocardial Infarction Genetics C, Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O’Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, MacRae CA, O’Donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall A, Wellcome Trust Case Control C, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown P, Patterson CC, Schunkert H, Erdmann E, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O’Donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM and Altshuler D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Cardiogenics, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O’Donnell CJ, McPherson R, Erdmann J, Consortium CA and Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coronary Artery Disease Genetics C. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–44. [DOI] [PubMed] [Google Scholar]
- 31.Consortium CAD, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Consortium D, Consortium C, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, Mu TC, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wellcome Trust Case Control C, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O’Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H and Samani NJ. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T and Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, Segre AV, Ardlie KG, Khera AV, Kaushik VK, Natarajan P, Consortium CAD and Kathiresan S. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, Jiang T, Hamby SE, Di Angelantonio E, Assimes TL, Bottinger EP, Chambers JC, Clarke R, Palmer CNA, Cubbon RM, Ellinor P, Ermel R, Evangelou E, Franks PW, Grace C, Gu D, Hingorani AD, Howson JMM, Ingelsson E, Kastrati A, Kessler T, Kyriakou T, Lehtimaki T, Lu X, Lu Y, Marz W, McPherson R, Metspalu A, Pujades-Rodriguez M, Ruusalepp A, Schadt EE, Schmidt AF, Sweeting MJ, Zalloua PA, AlGhalayini K, Keavney BD, Kooner JS, Loos RJF, Patel RS, Rutter MK, Tomaszewski M, Tzoulaki I, Zeggini E, Erdmann J, Dedoussis G, Bjorkegren JLM, Consortium E-C, CardioGramplusC4D, group UKBCCCw, Schunkert H, Farrall M, Danesh J, Samani NJ, Watkins H and Deloukas P. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. [DOI] [PubMed] [Google Scholar]
- 35.Verweij N, Eppinga RN, Hagemeijer Y and van der Harst P. Identification of 15 novel risk loci for coronary artery disease and genetic risk of recurrent events, atrial fibrillation and heart failure. Sci Rep. 2017;7:2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Harst P and Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erdmann J, Kessler T, Munoz Venegas L and Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res. 2018;114:1241–1257. [DOI] [PubMed] [Google Scholar]
- 38.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–9. [DOI] [PubMed] [Google Scholar]
- 39.Risch N and Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. [DOI] [PubMed] [Google Scholar]
- 40.Chakravarti A. Population genetics--making sense out of sequence. Nat Genet. 1999;21:56–60. [DOI] [PubMed] [Google Scholar]
- 41.Reich DE and Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–10. [DOI] [PubMed] [Google Scholar]
- 42.Schork NJ, Murray SS, Frazer KA and Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJF, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O’Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ and Farrall M. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA and Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myocardial Infarction G, Investigators CAEC, Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, Konig IR, Weeke PE, Webb TR, Auer PL, Schick UM, Lu Y, Zhang H, Dube MP, Goel A, Farrall M, Peloso GM, Won HH, Do R, van Iperen E, Kanoni S, Kruppa J, Mahajan A, Scott RA, Willenberg C, Braund PS, van Capelleveen JC, Doney AS, Donnelly LA, Asselta R, Merlini PA, Duga S, Marziliano N, Denny JC, Shaffer CM, El-Mokhtari NE, Franke A, Gottesman O, Heilmann S, Hengstenberg C, Hoffman P, Holmen OL, Hveem K, Jansson JH, Jockel KH, Kessler T, Kriebel J, Laugwitz KL, Marouli E, Martinelli N, McCarthy MI, Van Zuydam NR, Meisinger C, Esko T, Mihailov E, Escher SA, Alver M, Moebus S, Morris AD, Muller-Nurasyid M, Nikpay M, Olivieri O, Lemieux Perreault LP, AlQarawi A, Robertson NR, Akinsanya KO, Reilly DF, Vogt TF, Yin W, Asselbergs FW, Kooperberg C, Jackson RD, Stahl E, Strauch K, Varga TV, Waldenberger M, Zeng L, Kraja AT, Liu C, Ehret GB, Newton-Cheh C, Chasman DI, Chowdhury R, Ferrario M, Ford I, Jukema JW, Kee F, Kuulasmaa K, Nordestgaard BG, Perola M, Saleheen D, Sattar N, Surendran P, Tregouet D, Young R, Howson JM, Butterworth AS, Danesh J, Ardissino D, Bottinger EP, Erbel R, Franks PW, Girelli D, Hall AS, Hovingh GK, Kastrati A, Lieb W, Meitinger T, Kraus WE, Shah SH, McPherson R, Orho-Melander M, Melander O, Metspalu A, Palmer CN, Peters A, Rader D, Reilly MP, Loos RJ, Reiner AP, Roden DM, Tardif JC, Thompson JR, Wareham NJ, Watkins H, Willer CJ, Kathiresan S, Deloukas P, Samani NJ and Schunkert H. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet. 1965;29:51–76. [Google Scholar]
- 47.Falconer DS. The inheritance of liability to diseases with variable age of onset, with particular reference to diabetes mellitus. Ann Hum Genet. 1967;31:1–20. [DOI] [PubMed] [Google Scholar]
- 48.CARDIoGRAMplusC4D Consortium.
- 49.Burgess S and Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42:1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho DSW, Schierding W, Wake M, Saffery R and O’Sullivan J. Machine Learning SNP Based Prediction for Precision Medicine. Front Genet. 2019;10:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hettige NC, Cole CB, Khalid S and De Luca V. Polygenic risk score prediction of antipsychotic dosage in schizophrenia. Schizophr Res. 2016;170:265–70. [DOI] [PubMed] [Google Scholar]
- 52.Choi SW, Shin T, Mak H and Reilly PFO. A guide to performing Polygenic Risk Score analyses. bioRxiv, 101101/416545. 2018. [Google Scholar]
- 53.Chatterjee N, Shi J and Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jostins L and Barrett JC. Genetic risk prediction in complex disease. Hum Mol Genet. 2011;20:R182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, Gerad/Perades, consortia I, Morgan K, Passmore P, Holmes C, Powell J, Brayne C, Gill M, Mead S, Goate A, Cruchaga C, Lambert JC, van Duijn C, Maier W, Ramirez A, Holmans P, Jones L, Hardy J, Seshadri S, Schellenberg GD, Amouyel P and Williams J. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.So HC and Sham PC. Exploring the predictive power of polygenic scores derived from genome-wide association studies: a study of 10 complex traits. Bioinformatics. 2017;33:886–892. [DOI] [PubMed] [Google Scholar]
- 57.Martin AR, Daly MJ, Robinson EB, Hyman SE and Neale BM. Predicting Polygenic Risk of Psychiatric Disorders. Biol Psychiatry. 2019;86:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME and Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prive F, Vilhjalmsson BJ, Aschard H and Blum MGB. Making the Most of Clumping and Thresholding for Polygenic Scores. Am J Hum Genet. 2019;105:1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, Genovese G, Loh PR, Bhatia G, Do R, Hayeck T, Won HH, Schizophrenia Working Group of the Psychiatric Genomics Consortium DB, Risk of Inherited Variants in Breast Cancer s, Kathiresan S, Pato M, Pato C, Tamimi R, Stahl E, Zaitlen N, Pasaniuc B, Belbin G, Kenny EE, Schierup MH, De Jager P, Patsopoulos NA, McCarroll S, Daly M, Purcell S, Chasman D, Neale B, Goddard M, Visscher PM, Kraft P, Patterson N and Price AL. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mak TSH, Porsch RM, Choi SW, Zhou X and Sham PC. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 2017;41:469–480. [DOI] [PubMed] [Google Scholar]
- 62.Wray NR, Yang J, Goddard ME and Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–35. [DOI] [PubMed] [Google Scholar]
- 64.Pepe MS, Fan J, Feng Z, Gerds T and Hilden J. The Net Reclassification Index (NRI): a Misleading Measure of Prediction Improvement Even with Independent Test Data Sets. Stat Biosci. 2015;7:282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kerr KF, McClelland RL, Brown ER and Lumley T. Evaluating the incremental value of new biomarkers with integrated discrimination improvement. Am J Epidemiol. 2011;174:364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torkamani A, Wineinger NE and Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. [DOI] [PubMed] [Google Scholar]
- 67.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C and Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–9. [DOI] [PubMed] [Google Scholar]
- 68.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki ML, Nieminen MS, Melander O, Salomaa V, Peltonen L and Kathiresan S. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patel RS, Sun YV, Hartiala J, Veledar E, Su S, Sher S, Liu YX, Rahman A, Patel R, Rab ST, Vaccarino V, Zafari AM, Samady H, Tang WH, Allayee H, Hazen SL and Quyyumi AA. Association of a genetic risk score with prevalent and incident myocardial infarction in subjects undergoing coronary angiography. Circ Cardiovasc Genet. 2012;5:441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tikkanen E, Havulinna AS, Palotie A, Salomaa V and Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:2261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D’Agostino RB, Hwang SJ and O’Donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5:113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ganna A, Magnusson PK, Pedersen NL, de Faire U, Reilly M, Arnlov J, Sundstrom J, Hamsten A and Ingelsson E. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol. 2013;33:2267–72. [DOI] [PubMed] [Google Scholar]
- 73.Hughes MF, Saarela O, Stritzke J, Kee F, Silander K, Klopp N, Kontto J, Karvanen J, Willenborg C, Salomaa V, Virtamo J, Amouyel P, Arveiler D, Ferrieres J, Wiklund PG, Baumert J, Thorand B, Diemert P, Tregouet DA, Hengstenberg C, Peters A, Evans A, Koenig W, Erdmann J, Samani NJ, Kuulasmaa K and Schunkert H. Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One. 2012;7:e40922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krarup NT, Borglykke A, Allin KH, Sandholt CH, Justesen JM, Andersson EA, Grarup N, Jorgensen T, Pedersen O and Hansen T. A genetic risk score of 45 coronary artery disease risk variants associates with increased risk of myocardial infarction in 6041 Danish individuals. Atherosclerosis. 2015;240:305–10. [DOI] [PubMed] [Google Scholar]
- 75.Weijmans M, de Bakker PI, van der Graaf Y, Asselbergs FW, Algra A, Jan de Borst G, Spiering W, Visseren FL and Group SS. Incremental value of a genetic risk score for the prediction of new vascular events in patients with clinically manifest vascular disease. Atherosclerosis. 2015;239:451–8. [DOI] [PubMed] [Google Scholar]
- 76.Tada H, Melander O, Louie JZ, Catanese JJ, Rowland CM, Devlin JJ, Kathiresan S and Shiffman D. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur Heart J. 2016;37:561–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Vries PS, Kavousi M, Ligthart S, Uitterlinden AG, Hofman A, Franco OH and Dehghan A. Incremental predictive value of 152 single nucleotide polymorphisms in the 10-year risk prediction of incident coronary heart disease: the Rotterdam Study. Int J Epidemiol. 2015;44:682–8. [DOI] [PubMed] [Google Scholar]
- 78.Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, Tikkanen E, Perola M, Schunkert H, Sijbrands EJ, Palotie A, Samani NJ, Salomaa V, Ripatti S and Inouye M. Genomic prediction of coronary heart disease. Eur Heart J. 2016;37:3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT and Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, Ye S, Webb TR, Rutter MK, Tzoulaki I, Patel RS, Loos RJF, Keavney B, Hemingway H, Thompson J, Watkins H, Deloukas P, Di Angelantonio E, Butterworth AS, Danesh J, Samani NJ and Group UKBCCCW. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J Am Coll Cardiol. 2018;72:1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Natarajan P. Polygenic Risk Scoring for Coronary Heart Disease: The First Risk Factor. J Am Coll Cardiol. 2018;72:1894–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wunnemann F, Sin Lo K, Langford-Avelar A, Busseuil D, Dube MP, Tardif JC and Lettre G. Validation of Genome-Wide Polygenic Risk Scores for Coronary Artery Disease in French Canadians. Circ Genom Precis Med. 2019;12:e002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mosley JD, Gupta DK, Tan J, Yao J, Wells QS, Shaffer CM, Kundu S, Robinson-Cohen C, Psaty BM, Rich SS, Post WS, Guo X, Rotter JI, Roden DM, Gerszten RE and Wang TJ. Predictive Accuracy of a Polygenic Risk Score Compared With a Clinical Risk Score for Incident Coronary Heart Disease. JAMA. 2020;323:627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, Dehghan A, Muller DC, Elliott P and Tzoulaki I. Predictive Accuracy of a Polygenic Risk Score-Enhanced Prediction Model vs a Clinical Risk Score for Coronary Artery Disease. JAMA. 2020;323:636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hindy G, Aragam K, Chaffin M, Orho-Melander M, Melander O, Khera AV and Kathiresan S. Integration Of A Genome-wide Polygenic Score With ACC/AHA Pooled Cohorts Equation In Prediction Of Coronary Artery Disease Events In >285,000 Participants. Circulation. 2019;140: A16565. [Google Scholar]
- 86.Khan SS, Cooper R and Greenland P. Do Polygenic Risk Scores Improve Patient Selection for Prevention of Coronary Artery Disease? JAMA. 2020;323:614–615. [DOI] [PubMed] [Google Scholar]
- 87.Aragam KG, Chaffin M, Hindy G, Cagan A, Weng LC, Lubitz S, Smoller J, Karlson E, Khera AV, Kathiresan S and Natarajan P. Clinical Characteristics and Treatment Patterns of Patients at High Polygenic Risk for Coronary Artery Disease. J Am Coll Cardiol. 2019;73: 9 Supplement 1. [Google Scholar]
- 88.Natarajan P, Bis JC, Bielak LF, Cox AJ, Dorr M, Feitosa MF, Franceschini N, Guo X, Hwang SJ, Isaacs A, Jhun MA, Kavousi M, Li-Gao R, Lyytikainen LP, Marioni RE, Schminke U, Stitziel NO, Tada H, van Setten J, Smith AV, Vojinovic D, Yanek LR, Yao J, Yerges-Armstrong LM, Amin N, Baber U, Borecki IB, Carr JJ, Chen YI, Cupples LA, de Jong PA, de Koning H, de Vos BD, Demirkan A, Fuster V, Franco OH, Goodarzi MO, Harris TB, Heckbert SR, Heiss G, Hoffmann U, Hofman A, Isgum I, Jukema JW, Kahonen M, Kardia SL, Kral BG, Launer LJ, Massaro J, Mehran R, Mitchell BD, Mosley TH Jr., de Mutsert R, Newman AB, Nguyen KD, North KE, O’Connell JR, Oudkerk M, Pankow JS, Peloso GM, Post W, Province MA, Raffield LM, Raitakari OT, Reilly DF, Rivadeneira F, Rosendaal F, Sartori S, Taylor KD, Teumer A, Trompet S, Turner ST, Uitterlinden AG, Vaidya D, van der Lugt A, Volker U, Wardlaw JM, Wassel CL, Weiss S, Wojczynski MK, Becker DM, Becker LC, Boerwinkle E, Bowden DW, Deary IJ, Dehghan A, Felix SB, Gudnason V, Lehtimaki T, Mathias R, Mook-Kanamori DO, Psaty BM, Rader DJ, Rotter JI, Wilson JG, van Duijn CM, Volzke H, Kathiresan S, Peyser PA, O’Donnell CJ and Consortium C. Multiethnic Exome-Wide Association Study of Subclinical Atherosclerosis. Circ Cardiovasc Genet. 2016;9:511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ, Fuster V, Boerwinkle E, Melander O, Orho-Melander M, Ridker PM and Kathiresan S. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N Engl J Med. 2016;375:2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, Sartori S, Fuster V, Reilly DF, Butterworth A, Rader DJ, Ford I, Sattar N and Kathiresan S. Polygenic Risk Score Identifies Subgroup With Higher Burden of Atherosclerosis and Greater Relative Benefit From Statin Therapy in the Primary Prevention Setting. Circulation. 2017;135:2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Severance LM, Contijoch FJ, Carter H, Fan CC, Seibert TM, Dale AM and McVeigh ER. Using a genetic risk score to calculate the optimal age for an individual to undergo coronary artery calcium screening. J Cardiovasc Comput Tomogr. 2019;13:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levin MG, Kember RL, Judy R, Birtwell D, Williams H, Arany Z, Giri J, Guerraty M, Cappola T, Regeneron Genetics C, Chen J, Rader DJ and Damrauer SM. Genomic Risk Stratification Predicts All-Cause Mortality After Cardiac Catheterization. Circ Genom Precis Med. 2018;11:e002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abul-Husn NS, Manickam K, Jones LK, Wright EA, Hartzel DN, Gonzaga-Jauregui C, O’Dushlaine C, Leader JB, Lester Kirchner H, Lindbuchler DM, Barr ML, Giovanni MA, Ritchie MD, Overton JD, Reid JG, Metpally RP, Wardeh AH, Borecki IB, Yancopoulos GD, Baras A, Shuldiner AR, Gottesman O, Ledbetter DH, Carey DJ, Dewey FE and Murray MF. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354. [DOI] [PubMed] [Google Scholar]
- 94.Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, Wiklund O, Hegele RA, Raal FJ, Defesche JC, Wiegman A, Santos RD, Watts GF, Parhofer KG, Hovingh GK, Kovanen PT, Boileau C, Averna M, Boren J, Bruckert E, Catapano AL, Kuivenhoven JA, Pajukanta P, Ray K, Stalenhoef AF, Stroes E, Taskinen MR, Tybjaerg-Hansen A and European Atherosclerosis Society Consensus P. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013;34:3478-90a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, Lichtman JH, D’Onofrio G, Mattera JA, Dreyer RP, Spertus JA, Taylor KD, Psaty BM, Rich SS, Post WS, Gupta N, Gabriel S, Lander E, Chen YI, Talkowski ME, Rotter JI, Krumholz HM and Kathiresan S. Whole Genome Sequencing to Characterize Monogenic and Polygenic Contributions in Patients Hospitalized with Early-Onset Myocardial Infarction. Circulation. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Theriault S, Lali R, Chong M, Velianou JL, Natarajan MK and Pare G. Polygenic Contribution in Individuals With Early-Onset Coronary Artery Disease. Circ Genom Precis Med. 2018;11:e001849. [DOI] [PubMed] [Google Scholar]
- 97.Paquette M, Chong M, Theriault S, Dufour R, Pare G and Baass A. Polygenic risk score predicts prevalence of cardiovascular disease in patients with familial hypercholesterolemia. J Clin Lipidol. 2017;11:725–732 e5. [DOI] [PubMed] [Google Scholar]
- 98.Fahed AC, Wang M, Homburger JR, Patel AP, Bick AG, Neben CL, Lai C, Brockman D, Philippakis A, Ellinor PT, Cassa CA, Lebo M, Ng K, Lander ES, Zhou AY, Kathiresan S and Khera AV. Polygenic background modifies penetrance of monogenic variants conferring risk for coronary artery disease, breast cancer, or colorectal cancer. medRxiv 19013086; doi: https://doiorg/101101/19013086. 2019. [Google Scholar]
- 99.Said MA, Verweij N and van der Harst P. Associations of Combined Genetic and Lifestyle Risks With Incident Cardiovascular Disease and Diabetes in the UK Biobank Study. JAMA Cardiol. 2018;3:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield M, Devlin JJ, Nordio F, Hyde C, Cannon CP, Sacks F, Poulter N, Sever P, Ridker PM, Braunwald E, Melander O, Kathiresan S and Sabatine MS. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marston NA, Kamanu FK, Nordio F, Gurmu Y, Roselli C, Sever PS, Pedersen TR, Keech AC, Wang H, Lira Pineda A, Giugliano RP, Lubitz SA, Ellinor PT, Sabatine MS and Ruff CT. Predicting Benefit From Evolocumab Therapy in Patients With Atherosclerotic Disease Using a Genetic Risk Score: Results From the FOURIER Trial. Circulation. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T and Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Franzen O, Ermel R, Cohain A, Akers NK, Di Narzo A, Talukdar HA, Foroughi-Asl H, Giambartolomei C, Fullard JF, Sukhavasi K, Koks S, Gan LM, Giannarelli C, Kovacic JC, Betsholtz C, Losic B, Michoel T, Hao K, Roussos P, Skogsberg J, Ruusalepp A, Schadt EE and Bjorkegren JL. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM and Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Webb TR, Erdmann J, Stirrups KE, Stitziel NO, Masca NG, Jansen H, Kanoni S, Nelson CP, Ferrario PG, Konig IR, Eicher JD, Johnson AD, Hamby SE, Betsholtz C, Ruusalepp A, Franzen O, Schadt EE, Bjorkegren JL, Weeke PE, Auer PL, Schick UM, Lu Y, Zhang H, Dube MP, Goel A, Farrall M, Peloso GM, Won HH, Do R, van Iperen E, Kruppa J, Mahajan A, Scott RA, Willenborg C, Braund PS, van Capelleveen JC, Doney AS, Donnelly LA, Asselta R, Merlini PA, Duga S, Marziliano N, Denny JC, Shaffer C, El-Mokhtari NE, Franke A, Heilmann S, Hengstenberg C, Hoffmann P, Holmen OL, Hveem K, Jansson JH, Jockel KH, Kessler T, Kriebel J, Laugwitz KL, Marouli E, Martinelli N, McCarthy MI, Van Zuydam NR, Meisinger C, Esko T, Mihailov E, Escher SA, Alver M, Moebus S, Morris AD, Virtamo J, Nikpay M, Olivieri O, Provost S, AlQarawi A, Robertson NR, Akinsansya KO, Reilly DF, Vogt TF, Yin W, Asselbergs FW, Kooperberg C, Jackson RD, Stahl E, Muller-Nurasyid M, Strauch K, Varga TV, Waldenberger M, Wellcome Trust Case Control C, Zeng L, Chowdhury R, Salomaa V, Ford I, Jukema JW, Amouyel P, Kontto J, Investigators M, Nordestgaard BG, Ferrieres J, Saleheen D, Sattar N, Surendran P, Wagner A, Young R, Howson JM, Butterworth AS, Danesh J, Ardissino D, Bottinger EP, Erbel R, Franks PW, Girelli D, Hall AS, Hovingh GK, Kastrati A, Lieb W, Meitinger T, Kraus WE, Shah SH, McPherson R, Orho-Melander M, Melander O, Metspalu A, Palmer CN, Peters A, Rader DJ, Reilly MP, Loos RJ, Reiner AP, Roden DM, Tardif JC, Thompson JR, Wareham NJ, Watkins H, Willer CJ, Samani NJ, Schunkert H, Deloukas P, Kathiresan S, Myocardial Infarction G and Investigators CAEC. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated With Coronary Artery Disease. J Am Coll Cardiol. 2017;69:823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ntalla I, Kanoni S, Zeng L, Giannakopoulou O, Danesh J, Watkins H, Samani NJ, Deloukas P, Schunkert H and Group UKBCCCW. Genetic Risk Score for Coronary Disease Identifies Predispositions to Cardiovascular and Noncardiovascular Diseases. J Am Coll Cardiol. 2019;73:2932–2942. [DOI] [PubMed] [Google Scholar]
- 109.van Rheenen W, Peyrot WJ, Schork AJ, Lee SH and Wray NR. Genetic correlations of polygenic disease traits: from theory to practice. Nat Rev Genet. 2019;20:567–581. [DOI] [PubMed] [Google Scholar]
- 110.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, Patterson N, Daly MJ, Price AL and Neale BM. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen C, Psychiatric Genomics C, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control C, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL and Neale BM. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Early G, Lifecourse Epidemiology Eczema C, Pourcain BS, Warrington NM, Finucane HK, Price AL, Bulik-Sullivan BK, Anttila V, Paternoster L, Gaunt TR, Evans DM and Neale BM. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, Whitbourne S, Deen J, Shannon C, Humphries D, Guarino P, Aslan M, Anderson D, LaFleur R, Hammond T, Schaa K, Moser J, Huang G, Muralidhar S, Przygodzki R and O’Leary TJ. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–23. [DOI] [PubMed] [Google Scholar]
- 114.Klarin D, Lynch J, Aragam K, Chaffin M, Assimes TL, Huang J, Lee KM, Shao Q, Huffman JE, Natarajan P, Arya S, Small A, Sun YV, Vujkovic M, Freiberg MS, Wang L, Chen J, Saleheen D, Lee JS, Miller DR, Reaven P, Alba PR, Patterson OV, DuVall SL, Boden WE, Beckman JA, Gaziano JM, Concato J, Rader DJ, Cho K, Chang KM, Wilson PWF, O’Donnell CJ, Kathiresan S, Program VAMV, Tsao PS and Damrauer SM. Genome-wide association study of peripheral artery disease in the Million Veteran Program. Nat Med. 2019;25:1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD Jr., Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen WM, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu FC, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jimenez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee JM, Lemmens R, Leys D, Lewis CM, Lin WY, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O’Donnell MJ, Psaty BM, Pulit SL, Rannikmae K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Consortium AF, Cohorts for H, Aging Research in Genomic Epidemiology C, International Genomics of Blood Pressure C, Consortium I, Starnet Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Bjorkegren JLM, Codoni V, Civelek M, Smith NL, Tregouet DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD, BioBank Japan Cooperative Hospital G, Consortium C, Consortium E-C, Consortium EP-I, International Stroke Genetics C, Consortium M, Neurology Working Group of the CC, Network NSG, Study UKYLD, Consortium M, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT Jr., Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S and Dichgans M. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klarin D, Busenkell E, Judy R, Lynch J, Levin M, Haessler J, Aragam K, Chaffin M, Haas M, Lindstrom S, Assimes TL, Huang J, Min Lee K, Shao Q, Huffman JE, Kabrhel C, Huang Y, Sun YV, Vujkovic M, Saleheen D, Miller DR, Reaven P, DuVall S, Boden WE, Pyarajan S, Reiner AP, Tregouet DA, Henke P, Kooperberg C, Gaziano JM, Concato J, Rader DJ, Cho K, Chang KM, Wilson PWF, Smith NL, O’Donnell CJ, Tsao PS, Kathiresan S, Obi A, Damrauer SM, Natarajan P, Consortium I and Veterans Affairs’ Million Veteran P. Genome-wide association analysis of venous thromboembolism identifies new risk loci and genetic overlap with arterial vascular disease. Nat Genet. 2019;51:1574–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jouni H, Haddad RA, Marroush TS, Brown SA, Kruisselbrink TM, Austin EE, Shameer K, Behnken EM, Chaudhry R, Montori VM and Kullo IJ. Shared decision-making following disclosure of coronary heart disease genetic risk: results from a randomized clinical trial. J Investig Med. 2017;65:681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Knowles JW, Assimes TL, Kiernan M, Pavlovic A, Goldstein BA, Yank V, McConnell MV, Absher D, Bustamante C, Ashley EA and Ioannidis JP. Randomized trial of personal genomics for preventive cardiology: design and challenges. Circ Cardiovasc Genet. 2012;5:368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kullo IJ, Jouni H, Austin EE, Brown SA, Kruisselbrink TM, Isseh IN, Haddad RA, Marroush TS, Shameer K, Olson JE, Broeckel U, Green RC, Schaid DJ, Montori VM and Bailey KR. Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect on Low-Density Lipoprotein Cholesterol Levels (the MI-GENES Clinical Trial). Circulation. 2016;133:1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Widen E, Surakka I, Mars N, Pollanen P, Hotakainen K, Partanen J, Aro J and Ripatti S. Returning cardiovascular disease risk prediction back to individuals motivates beneficial lifestyle changes: Preliminary results from the GeneRISK-study. Abstract C012 European Society of Human Genetics Conference; June 2018; Milan, Italy. [Google Scholar]
- 121.Knowles JW and Ashley EA. Cardiovascular disease: The rise of the genetic risk score. PLoS Med. 2018;15:e1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.O’Malley PG, Feuerstein IM and Taylor AJ. Impact of electron beam tomography, with or without case management, on motivation, behavioral change, and cardiovascular risk profile: a randomized controlled trial. JAMA. 2003;289:2215–23. [DOI] [PubMed] [Google Scholar]
- 123.McEvoy JW, Blaha MJ, Nasir K, Yoon YE, Choi EK, Cho IS, Chun EJ, Choi SI, Rivera JJ, Blumenthal RS and Chang HJ. Impact of coronary computed tomographic angiography results on patient and physician behavior in a low-risk population. Arch Intern Med. 2011;171:1260–8. [DOI] [PubMed] [Google Scholar]
- 124.Johnson HM, Einerson J, Korcarz CE, Aeschlimann SE and Stein JH. Long-term effects of carotid screening on patient outcomes and behaviors. Arch Intern Med. 2011;171:589–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lambert SA, Abraham G and Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet. 2019;28:R133–R142. [DOI] [PubMed] [Google Scholar]
- 126.Bustamante CD, Burchard EG and De la Vega FM. Genomics for the world. Nature. 2011;475:163–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hindorff LA, Bonham VL, Brody LC, Ginoza MEC, Hutter CM, Manolio TA and Green ED. Prioritizing diversity in human genomics research. Nat Rev Genet. 2018;19:175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morales J, Welter D, Bowler EH, Cerezo M, Harris LW, McMahon AC, Hall P, Junkins HA, Milano A, Hastings E, Malangone C, Buniello A, Burdett T, Flicek P, Parkinson H, Cunningham F, Hindorff LA and MacArthur JAL. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol. 2018;19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM and Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reisberg S, Iljasenko T, Lall K, Fischer K and Vilo J. Comparing distributions of polygenic risk scores of type 2 diabetes and coronary heart disease within different populations. PLoS One. 2017;12:e0179238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim MS, Patel KP, Teng AK, Berens AJ and Lachance J. Genetic disease risks can be misestimated across global populations. Genome Biol. 2018;19:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Curtis D. Polygenic risk score for schizophrenia is more strongly associated with ancestry than with schizophrenia. Psychiatr Genet. 2018;28:85–89. [DOI] [PubMed] [Google Scholar]
- 133.Chang X, Salim A, Dorajoo R, Han Y, Khor CC, van Dam RM, Yuan JM, Koh WP, Liu J, Goh DY, Wang X, Teo YY, Friedlander Y and Heng CK. Utility of genetic and non-genetic risk factors in predicting coronary heart disease in Singaporean Chinese. Eur J Prev Cardiol. 2017;24:153–160. [DOI] [PubMed] [Google Scholar]
- 134.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD and Kenny EE. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet. 2017;100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Onengut-Gumuscu S, Chen WM, Robertson CC, Bonnie JK, Farber E, Zhu Z, Oksenberg JR, Brant SR, Bridges SL Jr., Edberg JC, Kimberly RP, Gregersen PK, Rewers MJ, Steck AK, Black MH, Dabelea D, Pihoker C, Atkinson MA, Wagenknecht LE, Divers J, Bell RA, Youth SfDi, Type 1 Diabetes Genetics C, Erlich HA, Concannon P and Rich SS. Type 1 Diabetes Risk in African-Ancestry Participants and Utility of an Ancestry-Specific Genetic Risk Score. Diabetes Care. 2019;42:406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Perry DJ, Wasserfall CH, Oram RA, Williams MD, Posgai A, Muir AB, Haller MJ, Schatz DA, Wallet MA, Mathews CE, Atkinson MA and Brusko TM. Application of a Genetic Risk Score to Racially Diverse Type 1 Diabetes Populations Demonstrates the Need for Diversity in Risk-Modeling. Sci Rep. 2018;8:4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Starlard-Davenport A, Allman R, Dite GS, Hopper JL, Spaeth Tuff E, Macleod S, Kadlubar S, Preston M and Henry-Tillman R. Validation of a genetic risk score for Arkansas women of color. PLoS One. 2018;13:e0204834. [DOI] [PMC free article] [PubMed] [Google Scholar]