Abstract
The phagosome harboring the bacterial pathogen Legionella pneumophila is known to be enriched with phosphatidylinositol 4‐phosphate (PtdIns4P), which is important for anchoring a subset of its virulence factors and potentially for signaling events implicated in the biogenesis of the Legionella‐containing vacuole (LCV) that supports intracellular bacterial growth. Here we demonstrate that the effector MavQ is a phosphoinositide 3‐kinase that specifically catalyzes the conversion of phosphatidylinositol (PtdIns) into PtdIns3P. The product of MavQ is subsequently phosphorylated by the effector LepB to yield PtdIns(3,4)P2, whose 3‐phosphate is then removed by another effector SidF to generate PtdIns4P. We also show that MavQ is associated with the LCV and the ∆mavQ mutant displays phenotypes in the anchoring of a PtdIns4P‐binding effector similar to those of ∆lepB or ∆sidF mutants. Our results establish a mechanism of de novo PtdIns4P biosynthesis by L. pneumophila via a catalysis axis comprised of MavQ, LepB, and SidF on the surface of its phagosome.
Keywords: Legionella, type IV secretion, effector protein, phosphatidylinositol 4‐phosphate, kinase
Subject Categories: Membrane & Intracellular Transport; Microbiology, Virology & Host Pathogen Interaction; Signal Transduction
Legionella‐containing vacuoles are enriched with phosphatidylinositol 4‐phosphate, serving as an anchor for Dot/Icm effectors. This study describes the de novo biosynthesis of PtdIns4P on the Legionella phagosome by coordinated activities of MavQ, LepB and SidF.
Introduction
The bacterial pathogen Legionella pneumophila is the causative agent of Legionnaires’ disease that is characterized by severe pneumonia or a flu‐like sickness called Pontiac fever (Fraser et al, 1977). This bacterium can survive and replicate intracellularly in a wide spectrum of host cells ranging from free‐living protozoans to mammalian macrophages by exploiting evolutionarily conserved mechanisms (Al‐Quadan et al, 2012). Upon being internalized by host cells, the pathogen establishes a degradation‐resistant compartment termed the Legionella‐containing vacuole (LCV). The LCV does not undergo lysosomal fusion, but instead develops into a compartment that resembles the endoplasmic reticulum (ER) by intimately communicating with the secretory and, probably to a lesser extent, the endocytic and retrograde branches of the vesicle trafficking pathways of host cells (Isberg et al, 2009; Personnic et al, 2016; Barlocher et al, 2017). The formation of the LCV requires the Dot/Icm (defective in organelle trafficking/intracellular multiplication) type IV secretion system which delivers more than 330 effector proteins into host cells (Qiu & Luo, 2017). Many Dot/Icm effector proteins are involved in the subversion of host membrane trafficking pathways through targeting small GTPases of the Arf (Nagai et al, 2002), Rab (Qiu & Luo, 2017) and Ran families (Rothmeier et al, 2013), which are important regulatory knots for specific routes of vesicle trafficking. Among these, the activity of Rab1, the central regulator of vesicle trafficking between the ER and the Golgi apparatus, is regulated by multiple Dot/Icm effectors through sophisticated mechanisms. For example, SidM/DrrA is a Rab1‐specific guanine nucleotide exchange factor (GEF) (Machner & Isberg, 2006; Murata et al, 2006) which recruits Rab1 to the LCV, and LepB is a GTPase‐activating protein (GAP) (Ingmundson et al, 2007) for Rab1. By controlling the activation cycle of Rab1, SidM/DrrA facilitates the fusion of ER‐derived vesicles to the LCV (Arasaki et al, 2012). The hijacking of the activity of Rab1 and other Rab small GTPases is further regulated by several effector‐induced reversible modifications, including AMPylation by SidM/DrrA (Muller et al, 2010) and SidD (Neunuebel et al, 2011; Tan & Luo, 2011), phosphorylcholination by AnkX (Mukherjee et al, 2011; Tan et al, 2011) and Lem3 (Tan et al, 2011), phosphoribosyl‐linked serine ubiquitination by members of SidEs (Bhogaraju et al, 2016; Qiu et al, 2016; Kotewicz et al, 2017), SidJ (Bhogaraju et al, 2019; Black et al, 2019; Gan et al, 2019; Sulpizio et al, 2019), DupA and DupB (Wan et al, 2019; Shin et al, 2020). These different layers of regulation appear to occur at distinct cellular locations and times during the intracellular life cycle of L. pneumophila (Qiu & Luo, 2017).
In addition to small GTPases, phosphoinositides (PIs) are another class of critical regulatory determinants in membrane trafficking (Di Paolo & De Camilli, 2006). These lipids are minor phospholipid components (less than 10%) of the organellar membranes localizing on their cytoplasmic surfaces. The core chemical moiety of PI lipid is phosphatidylinositol (PtdIns), whose 3′, 4′ or 5′ free hydroxyl groups of the inositol ring can be phosphorylated by organelle‐specific PI kinases or dephosphorylated by distinct lipid phosphatases (Di Paolo & De Camilli, 2006). Coordinated activities of these enzymes yield seven mono‐, double‐, or triple‐phosphorylated PI derivatives (Di Paolo & De Camilli, 2006). Each of these seven PIs preferentially localizes to distinct subcellular compartments, where they exert signaling effects via the recruitment of cytosolic proteins harboring such different PI‐binding motifs as the PH (pleckstrin homology), PX (phox homology), or FYVE (Fab‐1, YGL023, Vps27, and EEA1) domains (Downes et al, 2005; Lemmon, 2008). The presence of specific PI also dictates the organelle identity and the category of the vesicle that the compartment interacts with.
Many intracellular bacterial pathogens subvert host PI metabolism by effectors that function as PI kinases or phosphatases to facilitate their survival and replication (Hilbi, 2006). L. pneumophila appears to employ several strategies to modulate PI metabolism in infected cells (Qiu & Luo, 2017; Swart & Hilbi, 2020). One branch is the biosynthesis of PtdIns4P on the surface of the LCV catalyzed by Dot/Icm effector proteins endowed with PI‐metabolizing activities (Dong et al, 2016). In this pathway, the amino terminal portion of LepB (LepB_NTD) acts as a lipid kinase that converts PtdIns3P into PtdIns(3,4)P2, which serves as the substrate of another Dot/Icm effector SidF, a PI phosphatase that specifically hydrolyzes its 3‐phosphate to yield PtdIns4P (Hsu et al, 2012). However, this model is incomplete because the source of PtdIns3P on the LCV is unknown. An earlier study by Urbanus et al (2016) reported that the L. pneumophila Dot/Icm effector MavQ (Lpg2975) (Huang et al, 2011) is a PI kinase, but the exact PI product synthesized by MavQ and the biological roles of MavQ during L. pneumophila infection has not yet been defined. Here, we demonstrate that MavQ catalyzes the formation of PtdIns3P using PtdIns as substrate. We also found that MavQ works together with LepB and SidF to sequentially synthesize PtdIns4P on the LCV surface.
Results
MavQ is a phosphatidylinositol‐specific phosphoinositide 3‐kinase (PI3K)
MavQ is a Dot/Icm substrate that is widely distributed in Legionella spp., and genes coding for this protein are present in 71 out of the 80 sequenced genomes covering 58 Legionella species and subspecies (Gomez‐Valero et al, 2019). It has been demonstrated that MavQ is a kinase potentially participating in the modulation of host PI metabolism (Urbanus et al, 2016). However, neither the PI products generated by MavQ nor the biological roles of MavQ during L. pneumophila infection have been elucidated.
It has been shown that MavQ is lethal to yeast (Nevo et al, 2014; Burstein et al, 2015; Urbanus et al, 2016). Consistent with a previous study (Urbanus et al, 2016), mutations in residues predicted to be important for catalysis or ATP binding abolished its yeast toxicity (Fig 1A). These mutants expressed similarly to the wild‐type gene in yeast (Fig 1B), indicating that the loss of toxicity is due to the lack of kinase activity. Then, we utilized the ADP‐Glo kinase assay to investigate the kinase activity of MavQ. Recombinant MavQ displayed weak but detectable ATP hydrolysis activity (Fig 2A). To probe the role of lipids in its activity, we established reactions that contained each of a panel of PIs and found that robust ATP hydrolysis occurred in reactions receiving PtdIns (Fig 2A). This activity was strictly dependent on the motifs predicted for kinase catalysis or for ATP binding, since mutations in H149, N152, or D160 abolished the ability of MavQ to hydrolyze ATP (Fig 2B). These observations are in good agreement with previous data (Urbanus et al, 2016). An extremely weak but detectable stimulation occurred toward PtdIns5P (Fig 2A). In contrast, MavQ exhibited no detectable kinase activity in reactions containing PtdIns3P, PtdIns4P, PtdIns(3,4)P2, PtdIns(3,5)P2, or PtdIns(4,5)P2 (Fig 2A). These results further support the conclusion that MavQ is a PI kinase with a high substrate specificity for PtdIns.
Figure 1. The kinase motif embedded in MavQ is critical for its activity to inhibit yeast growth.
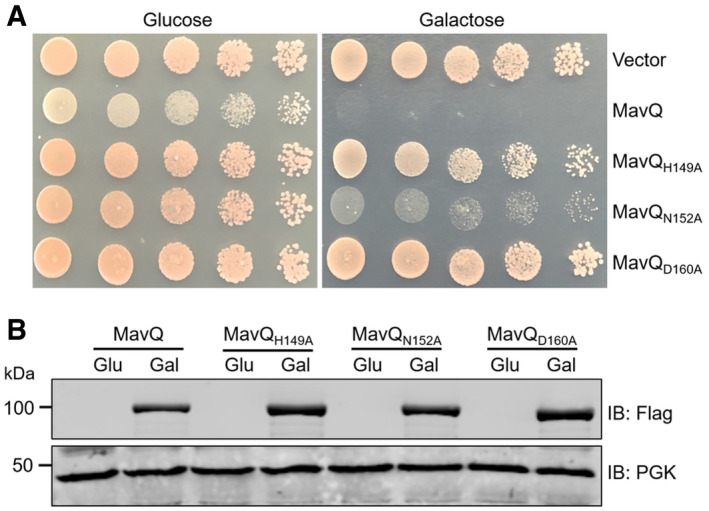
- Mutations in the kinase motif abolished the yeast toxicity of MavQ. S. cerevisiae was transformed with plasmids expressing Flag‐tagged wild‐type MavQ or the indicated MavQ mutants under the galactose‐inducible promoter. Yeast cells were spotted on synthetic medium supplemented with glucose or galactose for 3 days before image acquisition.
- The expression of MavQ in the yeast was detected by Flag antibody. The PGK (3‐phosphoglyceric phosphokinase) was used as a loading control.
Data information: Data shown are from at least three independent experiments (biological replicates, n ≥ 3).
Source data are available online for this figure.
Figure 2. MavQ is a phosphoinositide 3 Kinase that converts PtdIns into PtdIns3P.
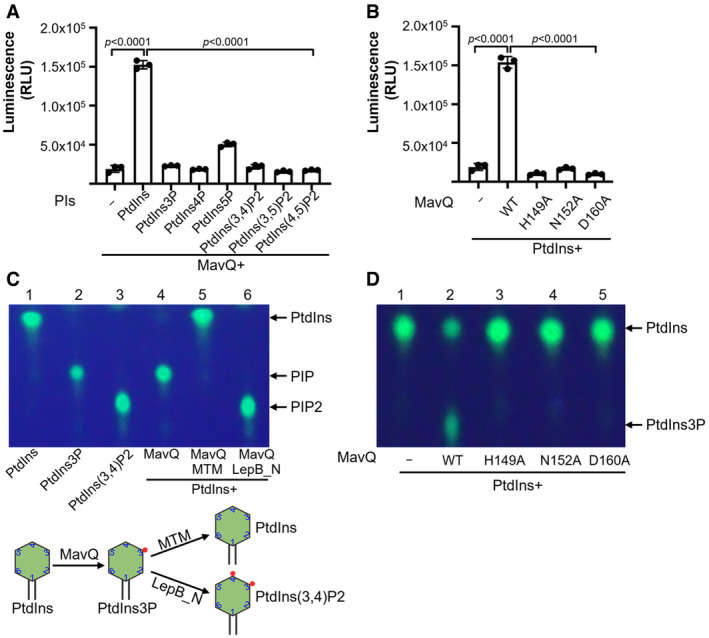
- Biochemical assays for the kinase activity of MavQ. Purified MavQ was incubated with or without specific PI substrates, and the conversion of ATP to ADP was measured by the ADP‐Glo assay. RLU, relative luminescence units.
- H149, N152, and D160 in MavQ are critical for its kinase activity. Purified wild‐type and mutant MavQ were reacted with PtdIns and ATP. The kinase activity was determined by the ADP‐Glo assay.
- The PI product produced by MavQ determined by TLC assays. di‐C8‐Bodipy‐FL‐PtdIns was used to react with MavQ (lane 4), and samples were further incubated with the 3‐phosphatase MTM which hydrolyzes both PtdIns3P and PtdIns(3,5)P2 (lane 5) or with the PI4K LepB_NTD which phosphorylates PtdIns3P into PtdIns (3,4)P2 (lane 6). A schematic illustration of the enzymatic reactions is shown in the lower panel.
- Mutations in H149, N152, and D160 abolish the ability of MavQ to convert PtdIns into PtdIns3P. Experiments were performed with the same procedure as described in panel C.
Data information: In (A and B), data are shown as mean ± SD of triplicates (technical replicates, n = 3) and are representative of three independent experiments (biological replicates, n = 3). In (C and D), data are representative of at least three independent experiments (biological replicates, n ≥ 3). Unpaired two‐tailed Student’s t‐test, p < 0.05 indicates significant difference.
Source data are available online for this figure.
Next we employed the fluorescent PI‐based thin‐layer chromatography (TLC) method (Dong et al, 2016) to determine the lipid products generated by MavQ. Incubation of PtdIns with MavQ resulted in a product with a migration pattern similar to that of PIP, indicating mono‐phosphorylation of PtdIns by MavQ (Fig 2C). This product produced by MavQ can be hydrolyzed back to PtdIns by the PtdIns3P‐specific PI phosphatase myotubularin 1 (MTM) (Taylor et al, 2000). It can also be further phosphorylated by LepB_NTD, the kinase specific for PtdIns3P (Dong et al, 2016), to yield a PIP2 product (Fig 2C). In line with results from the ADP‐Glo kinase assay, the H149, N152, and D160 mutants of MavQ each have lost the capacity to generate PIP from PtdIns (Fig 2D). Taken together, these data demonstrate that MavQ functions as a PI3K to catalyze the conversion of PtdIns into PtdIns3P.
The PI3K activity of MavQ suggests that this protein can disrupt PI, particularly PtdIns3P metabolism in eukaryotic cells. We tested this hypothesis by using the well‐described PtdIns3P fluorescence probe containing a duplicate FYVE finger from the human homologue of the hepatocyte growth factor‐regulated tyrosine kinase substrate (GFP‐2xFYVEHrs) (Gaullier et al, 1998). In mammalian cells, GFP‐2xFYVEHrs localizes to the early endosomal membrane through an interaction with PtdIns3P (Gaullier et al, 1998). In response to wortmannin, an inhibitor of eukaryotic PI3Ks (Walker et al, 2000), fluorescence signals of GFP‐2xFYVEHrs were redistributed from the early endosome network to the cytoplasm (Fig 3A and B). However, in cells transfected to express MavQ, the puncta localization pattern of GFP‐2xFYVEHrs was not affected in cells treated with wortmannin, indicating that MavQ catalyzes the production of PtdIns3P in cells. These results also suggest that MavQ is a PI3K that is insensitive to wortmannin. Indeed, preincubation of MavQ with high concentrations of wortmannin did not detectably affect its ATP hydrolysis activity, nor did it affect the production of PtdIns3P from PtdIns (Fig 3C and D). In contrast, the activity of p110α, the mammalian PI3K, which is known to be sensitive to wortmannin (Kumar & Doss, 2016), is inhibited (Fig 3C and D). This observation is similar to previously described bacterial PI3Ks (Ledvina et al, 2018), which are often wortmannin‐resistant, probably due to divergence in their structures or the mechanisms of catalysis.
Figure 3. The PI3K activity of MavQ is insensitive to wortmannin.
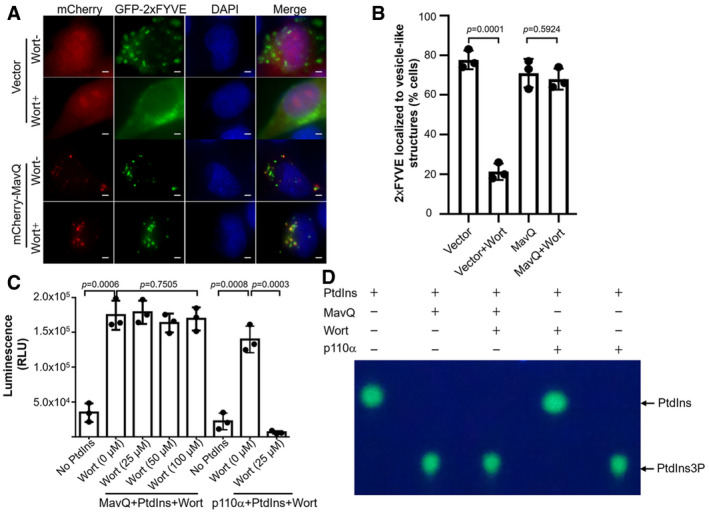
- Representative fluorescence images of HeLa cells expressing the PtdIns3P probe GFP‐2xFYVEHrs and MavQ with or without wortmannin treatment. Scale bars, 5 μm.
- Percentage of HeLa cells transfected with the indicated plasmids in which GFP‐2xFYVEHrs localizes to vesicle‐like structures. At least 100 cells (n ≥ 100) were counted for each sample. Wort, wortmannin.
- Induction of the ATP hydrolysis activity of MavQ by PtdIns is not affected by wortmannin. Recombinant MavQ was incubated with increasing amounts of wortmannin for 30 min at room temperature prior to the addition of PtdIns and ATP. The wortmannin‐sensitive PI3K p110α was used as a control. The ADP‐Glo assay was used to measure the kinase activity.
- Wortmannin does not affect the production of PtdIns3P by MavQ. Purified MavQ was pretreated with 100 μM of wortmannin for 30 min at room temperature. di‐C8‐Bodipy‐FL‐PtdIns and ATP were then added to the reaction mixtures. The TLC assay was employed to detect the generation of PtdIns3P by MavQ. The wortmannin‐sensitive PI3K p110α was included as a control.
Data information: In panel B, data show the mean ± SD of three independent experiments (biological replicates, n = 3). In panel C, data are shown as mean ± SD of triplicates (technical replicates, n = 3) and are representative of three independent experiments (biological replicates, n = 3). In panel D, data are representative of at least three independent experiments (biological replicates, n ≥ 3). Unpaired two‐tailed Student’s t‐test, p < 0.05 indicates significant difference.
Source data are available online for this figure.
MavQ is associated with the LCV and is dispensable for the intracellular growth of L. pneumophila
To detect the cellular localization of MavQ during infection, we infected bone marrow‐derived macrophages (BMDMs) with relevant L. pneumophila strains followed by immunostaining with MavQ‐specific antibodies. Staining signals corresponding to MavQ were enriched on LCVs formed by the L. pneumophila strain harboring a functional Dot/Icm transporter (Fig 4A). Approximately 60% of the LCVs harboring wild‐type bacteria stained positive for MavQ 2 h post‐infection (Fig 4B). No such signal was detected on LCVs containing the dotA mutant Lp03 or the mavQ deletion mutant (Fig 4A and B). Furthermore, expression of the MavQ from a plasmid in the ∆mavQ mutant restored such association to levels close to those of the wild‐type strain (Fig 4B). Moreover, kinetic analysis indicates that the association of MavQ with the LCV persists for more than 6 h, a duration similar to that for LepB (Ingmundson et al, 2007) and SidF (Hsu et al, 2012; Fig 4C).
Figure 4. The association of MavQ with the LCV and its role in intracellular growth of L. pneumophila in macrophages.
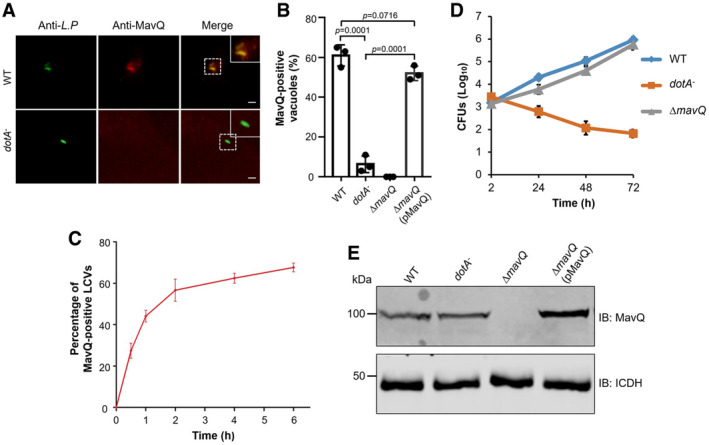
- Representative immunofluorescence images of the association of MavQ with the LCV. BMDM cells infected with wild‐type or Dot/Icm‐deficient (dotA −) L. pneumophila were sequentially stained with antibodies against L. pneumophila and MavQ. Images were taken under a fluorescence microscope. Insets represent 3× magnification of regions defined by dash lines. Scale bar, 5 μm.
- The rates of LCVs positively stained for MavQ 2 h post‐infection. At least 100 LCVs (n ≥ 100) were scored for each sample.
- Dynamics of MavQ association with LCV. BMDMs were infected with the wild‐type L. pneumophila strain at an MOI of 2 for the indicated experimental durations. At least 100 phagosomes (n ≥ 100) were counted for each sample to obtain the percentage of MavQ‐positive LCVs.
- Intracellular growth of L. pneumophila in macrophages. BMDMs were infected with indicated L. pneumophila strains at an MOI of 0.05. At the indicated time points, CFUs were determined by plating the saponin‐solubilized lysates of infected cells on CYE plates.
- Expression of mavQ in L. pneumophila strains used for infection. Total proteins of bacteria cultured to the post‐exponential phase separated by SDS–PAGE were probed with MavQ‐specific antibodies. ICDH was detected as a loading control.
Data information: In panels B and C, data are shown as mean ± SD of three independent experiments (biological replicates, n = 3). In panel D, results shown are from one representative experiment done in triplicate (technical replicates, n = 3) and are representative of three independent experiments (biological replicates, n = 3). In panel E, results show one representative of three independent experiments (biological replicates, n = 3). Unpaired two‐tailed Student’s t‐test, p < 0.05 indicates significant difference.
Source data are available online for this figure.
To assess whether mavQ plays a role in bacterial virulence, we determined intracellular growth of the ∆mavQ mutant in BMDMs. The mutant replicated similarly to the wild‐type strain during the entire experimental duration (Fig 4D and E), suggesting that deletion of mavQ did not detectably affect intracellular growth of L. pneumophila in this infection model, which is akin to mutants lacking sidF (Banga et al, 2007; Hsu et al, 2012) or lepB (Ingmundson et al, 2007; Dong et al, 2016).
MavQ works together with LepB and SidF to produce PtdIns4P on the LCV
The production of a specific PI is achieved by coordinated activities of both kinases and phosphatases. PtdIns4P is the signature PI lipid of the Golgi apparatus which is critical for organelle identity, signaling, and vesicle trafficking (Graham & Burd, 2011). It is established that LCVs are enriched with PtdIns4P and many Dot/Icm substrates are associated with the bacterial phagosome via specifically binding to this PI (Weber et al, 2006; Brombacher et al, 2009; Hsu et al, 2012; Dong et al, 2016). Two Dot/Icm effector proteins have been described to participate in the PtdIns4P production by functioning as kinase and phosphatase, respectively. LepB functions as a PI4K via a domain in its amino terminal portion, and it specifically converts PtdIns3P into PtdIns(3,4)P2 (Dong et al, 2016), which serves as the substrate for SidF, a PI 3‐phosphatase that preferentially removes the 3‐phosphate from PtdIns(3,4)P2 to generate PtdIns4P (Hsu et al, 2012).
The observation that MavQ catalyzes the conversion of PtdIns into PtdIns3P prompted us to postulate that MavQ acts upstream of LepB and SidF to form a catalysis cascade for the production of PtdIns4P on the LCV. In agreement with this notion, sequential addition of LepB_NTD and SidF in reactions containing PtdIns, MavQ, and ATP resulted in the production of PtdIns4P (Fig 5A). In addition, MavQ, LepB, and SidF are associated with the LCV during bacterial infection (Banga et al, 2007; Hsu et al, 2012; Dong et al, 2016). Thus, MavQ likely functions in concert with LepB and SidF to produce PtdIns4P on the LCV.
Figure 5. MavQ functions together with LepB and SidF to produce PtdIns4P on the LCV.
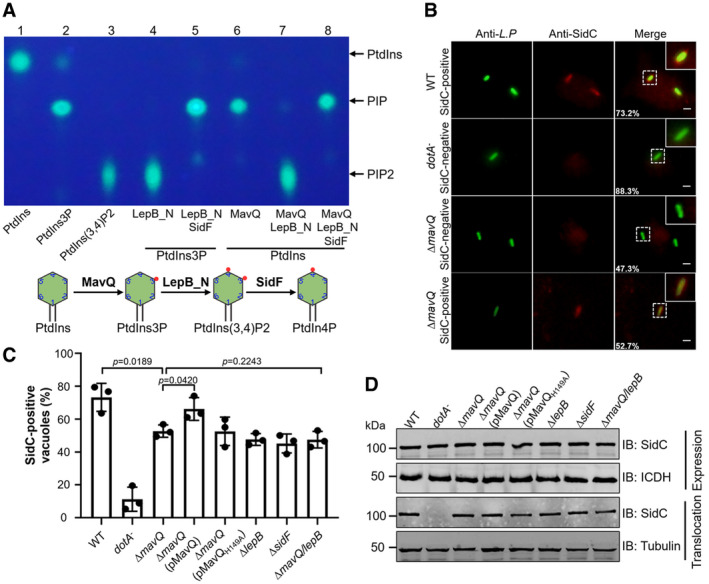
- Biochemical synthesis of PtdIns4P from PtdIns by sequential reactions catalyzed by MavQ, LepB_NTD, and SidF. MavQ was reacted with di‐C8‐Bodipy‐FL‐PtdIns, resulting in the production of PtdIns3P (lane 6). The PtdIns3P‐specific PI4K LepB‐NTD was then added into the reaction, yielding PtdIns(3,4)P2 (lane 7). The product of the sample receiving LepB_NTD was further incubated with the 3‐phosphatase SidF to yield PtdIns4P (lane 8). The lipid products were detected and visualized by TLC. A schematic illustration of the enzymatic reactions is shown in the lower panel.
- Representative images of SidC anchoring on LCVs. BMDMs infected with the indicated L. pneumophila strains at an MOI of 2 for 2 h were sequentially stained with antibodies against the bacteria and SidC. Images were acquired using a fluorescence microscope. Insets represent 3× magnification of regions defined by dash lines. Scale bar, 5 μm.
- Quantitation of SidC‐positive LCVs. At least 100 phagosomes (n ≥ 100) were scored for each sample.
- Expression and translocation of SidC. The lysates of L. pneumophila strains used for infection were probed for protein expression, and ICDH was detected as a loading control (top panel). SidC translocation was detected by probing the saponin‐soluble fractions of infected U937 cells with SidC‐specific antibodies. Tubulin was probed as a loading control (bottom panel).
Data information: In panels A and D, data show one representative of three independent experiments (biological replicates, n = 3). In panel C, results are shown as mean ± SD from three independent experiments (biological replicates, n = 3). Unpaired two‐tailed Student’s t‐test, p < 0.05 indicates significant difference.
Source data are available online for this figure.
If MavQ functions in the same axis of LepB and SidF in PtdIns4P production, a mutant lacking this gene will have phenotypes similar to the ∆lepB and ∆sidF strains in the accumulation of PtdIns4P on the LCV. One established method to determine PtdIns4P accumulation on the LCV is to detect its ability to retain lipid‐binding effectors such as SidC and SidM/DrrA (Weber et al, 2006; Brombacher et al, 2009; Hsu et al, 2012; Dong et al, 2016). Thus, we determined the association of SidC with the LCV formed by relevant L. pneumophila strains. In samples infected with the wild‐type strain, we observed that approximately 75% of the vacuoles stained positive for SidC, which was reduced to about 51% in cells infected with the ∆mavQ mutant (Fig 5B and C). The rates of SidC‐positive vacuoles for the ∆mavQ mutant are similar to that of the ∆lepB or ∆sidF mutant, which are about 47% in our experiments as well as in previous studies (Hsu et al, 2012; Dong et al, 2016; Fig 5C). Such defects in retaining SidC on the LCV displayed by the ∆mavQ mutant can be restored by expressing wild‐type MavQ but not its catalytically inactive mutant MavQH149A from a plasmid (Fig 5C). Furthermore, in samples infected by a mutant lacking both mavQ and lepB, we did not observe additional reduction in the percentage of vacuoles stained positive for SidC (Fig 5C). Consistent with this phenotype, the lack of both mavQ and lepB in the mutant did not significantly affect the intracellular replication of L. pneumophila (Fig EV1). In all cases, deletion of the PI metabolism genes did not detectably affect the expression or translocation of SidC into host cells by L. pneumophila (Fig 5D). Taken together, these results indicate that MavQ catalyzes the first of a three‐reaction cascade for the production of PtdIns4P from PtdIns on L. pneumophila phagosomes.
Figure EV1. The lack of mavQ and lepB does not affect the intracellular replication of L. pneumophila .
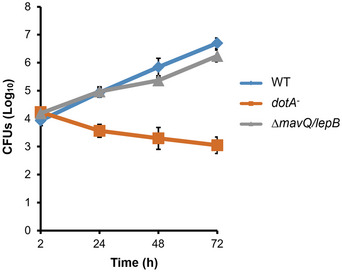
BMDMs infected with indicated L. pneumophila strains were lysed with saponin at 2, 24, 48, and 72 h post‐infection. The saponin‐solubilized lysates of infected cells were plated on CYE plates and grown at 37°C for 4 days to determine the CFUs. Results are shown as mean ± SD (technical replicates, n = 3) and are representative of three independent experiments (biological replicates, n = 3).
SidP does not affect the PI3K activity of MavQ
An earlier study (Urbanus et al, 2016) found that the PI 3‐phosphatase SidP (Toulabi et al, 2013) was capable of suppressing the toxicity of MavQ to yeast cells. Intriguingly, such inhibition is independent of its PI 3‐phosphatase activity but requires its uncharacterized carboxyl terminus domain (Urbanus et al, 2016). Furthermore, MavQ and SidP interact in yeast two‐hybrid and in affinity purification–mass spectrometry assays (Urbanus et al, 2016). Hence, SidP was considered a metaeffector of MavQ (Urbanus et al, 2016). In our yeast suppression assay, wild‐type SidP, the catalytically inactive mutant SidPR560K as well as the carboxyl fragment of SidP (SidP664–822) each indeed suppresses the toxicity of MavQ (Fig EV2). By GST pull‐down assays, we also observed interactions between these two proteins (Fig 6A). However, adding recombinant SidP into reactions containing PtdIns and MavQ did not significantly affect its kinase activity, even in reactions in which the molar ratio between SidP and MavQ was 10: 1 (Fig 6B and C). Furthermore, the generation of PtdIns3P in mammalian cells by ectopically produced MavQ was not affected by SidP. In MavQ and SidP co‐transfected cells treated with wortmannin, approximately 70% of the cells showed vesicular localization of GFP‐2xFYVEHrs, which was similar to samples transfected to express MavQ alone (Fig 6D). Thus, SidP does not interfere with the activity of MavQ, at least with regard to its PI3K enzymatic activity. Consistent with these observations, deletion or overexpression of sidP does not affect the association of SidC with the LCV (Figs 6E and EV3). Thus, it appears that SidP does not affect the PI3K activity of MavQ in biochemical assays. The biological relevance of its ability to suppress MavQ toxicity awaits further investigation.
Figure EV2. SidP suppresses the yeast toxicity of MavQ.
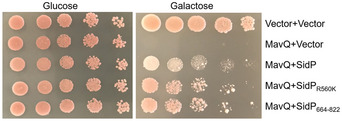
The yeast strain expressing Flag‐MavQ guided by a galactose‐inducible promoter was transformed with plasmids carrying wild‐type SidP, catalytically inactive mutant SidPR560K, or the carboxyl fragment of SidP (SidP664–822) under the glyceraldehyde‐3‐phosphate dehydrogenase (GPD) promoter. Serial diluted yeast cells were spotted onto selective media plates with 2% glucose or 2% galactose. Images were taken after 3 days of incubation at 30°C. Results shown are from one representative of three independent experiments (biological replicates, n = 3).
Figure 6. SidP does not affect the enzymatic activity of MavQ.
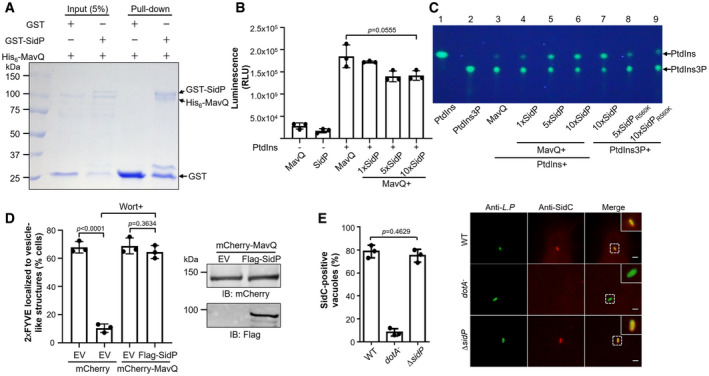
-
ADirect interactions between SidP and MavQ in the GST pull‐down assay. GST beads coated with GST‐SidP or GST were incubated with His6‐MavQ. After washing the beads with GST binding buffer, the proteins resolved by SDS–PAGE were detected by Coomassie brilliant blue staining.
-
B, CSidP does not impair the kinase activity of MavQ in vitro. MavQ was preincubated with increasing amounts of SidP, and the kinase activity of MavQ was measured by the ADP‐Glo assay (B) or the TLC assay (C).
-
DCo‐expression of SidP with MavQ in HeLa cells does not influence the production of PtdIns3P caused by MavQ. The association of the fluorescence probe GFP‐2xFYVEHrs with vesicle‐like structures was used to indicate the distribution of PtdIns3P in cells. At least 100 cells were scored in each sample (n ≥ 100) (left panel). Wort, wortmannin. Expression of MavQ and SidP in the samples was shown in the right panel. EV represents empty vector.
-
EDeletion of sidP does not impact the association of SidC with the LCV. BMDMs infected with the indicated L. pneumophila stains were stained as described in Fig 5, and the association of SidC with bacterial phagosomes was similarly determined (n ≥ 100) (left panel). Representative images of infected cells harboring bacterial phagosomes were also shown (right panel). Insets represent 3x magnification of regions defined by dash lines. Scale bar, 5 μm.
Data information: In panels A and C, data show one representative of three independent experiments (biological replicates, n = 3). In panel B, data are shown as mean ± SD of triplicates (technical replicates, n = 3) and are representative of three independent experiments (biological replicates, n = 3). In panels D and E, data are shown as mean ± SD of three independent experiments (biological replicates, n = 3). Unpaired two‐tailed Student’s t‐test, p < 0.05 indicates significant difference.
Source data are available online for this figure.
Figure EV3. Overexpression of SidP in L. pneumophila does not affect the association of SidC on the LCV.
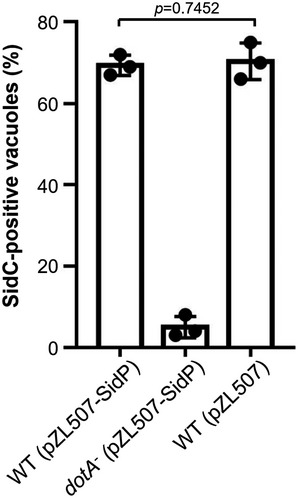
BMDMs were infected with indicated L. pneumophila strains at an MOI of 2 for 2 h. The anchoring of SidC on the LCV was detected by immunostaining with antibody specific for SidC. At least 100 phagosomes (n ≥ 100) were counted for each sample. Results shown are the mean ± SD from three independent experiments (biological replicates, n = 3). Unpaired two‐tailed Student’s t‐test, p < 0.05 indicates significant difference.
LegA5 is not required for PtdIns4P accumulation on the bacterial phagosome
It has been demonstrated that the Dot/Icm effector LegA5 (Lpg2322) is also a PI3K, which catalyzes the formation of PtdIns3P from PtdIns (Ledvina et al, 2018). To investigate whether this protein has an impact on the production of PtdIns4P on the LCV, we constructed a ∆legA5 mutant and tested its ability to retain SidC on the bacterial phagosome. We found that the rates of SidC‐positive LCVs for ∆legA5 mutant were similar to those of the wild‐type strain (Fig EV4A and B). These results suggest that LegA5, unlike MavQ, LepB, and SidF, is not involved in the production of PtdIns4P on the LCV.
Figure EV4. LegA5 is not required for the anchoring of SidC to the LCV.
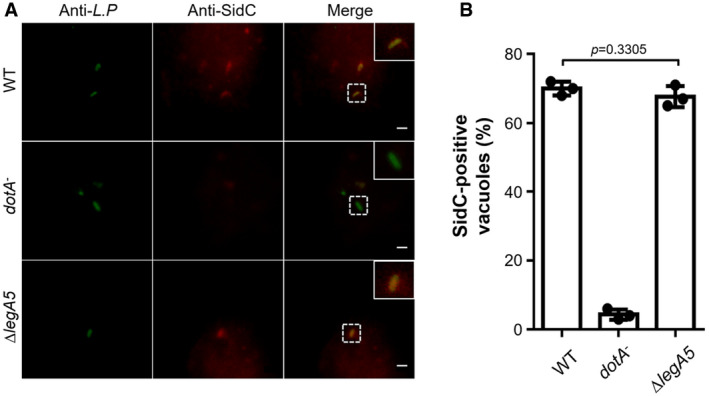
- Representative immunofluorescence images of anti‐Legionella and anti‐SidC staining of bacterial phagosomes. BMDMs were infected with the indicated L. pneumophila stains at an MOI of 2 for 2 h. SidC association on the bacterial phagosomes was detected as described in Fig 5. Insets represent 3× magnification of regions defined by dash lines. Scale bar, 5 μm.
- Percentages of SidC‐positive LCVs. At least 100 phagosomes (n ≥ 100) were scored for each sample. Results are shown as mean ± SD of three independent experiments (biological replicates, n = 3). Unpaired two‐tailed Student’s t‐test, p < 0.05 indicates significant difference.
Discussion
One hallmark of the LCV membrane is its enrichment of PtdIns4P, which appears to play important roles in the development of the compartment permissive for intracellular L. pneumophila replication. Vesicles originating from the ER have been suggested to be a major source of the membrane materials needed to support the expansion of LCVs containing multiplying bacteria (Kagan & Roy, 2002). PtdIns4P is known to serve as the anchor for a subset of Dot/Icm effectors, thus allowing their association with the LCV to promote events such as membrane remodeling and the acquisition of specific host proteins for its development (Weber et al, 2006). For example, once anchored on the LCV by binding to PtdIns4P, the multifunctional effector SidM/DrrA recruits Rab1 to facilitate the fusion of ER‐derived vesicles (Arasaki et al, 2012). Similarly, the ubiquitin E3 ligases SidC and SdcA recruit and ubiquitinate Rab10 on the LCV (Jeng et al, 2019) in a process that is regulated by the bacterial deubiquitinase Lem27 (Liu et al, 2020). Interestingly, although the mechanism is unknown, Rab10 is required for optimal bacterial replication in host cells (Hoffmann et al, 2014; Jeng et al, 2019). It is conceivable that other PtdIns4P‐binding effectors such as Lpg1101 and Lpg2603 (Hubber et al, 2014) also contribute to the development of the LCV by recruiting and co‐opting specific host proteins. In addition, the enrichment of PtdIns4P on this compartment mimics one important property of the Golgi apparatus, which may facilitate the interception of vesicles from the ER by the bacterial phagosome (Kagan & Roy, 2002).
Although PtdIns3P can be produced at other cellular locations such as the plasma membrane during signaling, this lipid is mainly found in the endosomal system (Mayinger, 2012). The PI3K activity of MavQ clearly will produce PtdIns3P on the LCV, whose membrane properties differ greatly from those of the endosome. The high level substrate specificity and physical proximity among MavQ, LepB, and SidF will maintain PtdIns3P on the LCV at low levels. Furthermore, the phosphatase SidP (Toulabi et al, 2013) may eliminate any residual PtdIns3P on the LCV. Thus, similar to eukaryotic cells (Marat & Haucke, 2016), the interplay between bacterial PI kinases and phosphatases determines the accumulation of specific PI at distinct cellular compartments in cells infected by L. pneumophila.
Two lines of evidence suggest that MavQ, LepB, and SidF function in the same pathway in the production of PtdIns4P. First, mutants lacking mavQ, lepB, or sidF exhibit similar phenotypes in the retention of the PtdIns4P‐binding effector SidC (Hsu et al, 2012; Dong et al, 2016; Fig 5B and C). Second, the mutant lacking both lepB and mavQ did not cause additional defects in the anchoring of SidC (Fig 5B and C). Thus, we propose a model for the production of PtdIns4P by these three Dot/Icm effectors on the LCV (Fig EV5). The observation that disruption of the MavQ‐LepB‐SidF cascade did not abolish the association of SidC on the LCV suggests the existence of other mechanisms involved in the production of PtdIns4P on the bacterial phagosome. Indeed, the accumulation of PtdIns4P on the LCVs also involves capturing Golgi‐derived PtdIns4P‐positive vesicles (Weber et al, 2018). During L. pneumophila infection, PtdIns3P associated with the endosomal system is unlikely the major substrate for LepB because it is destroyed by the Rab5‐activated lipase VipD (Lucas et al, 2014). Instead, additional pools of PtdIns3P may be produced by yet unidentified enzymes of host or pathogen origin. Interestingly, the Dot/Icm effector LegA5 has been shown to have PI3K activity capable of converting PtdIns into PtdIns3P (Ledvina et al, 2018). However, deletion of legA5 did not impair the association of SidC with the bacterial phagosome (Fig EV4A and B), suggesting that this effector protein does not participate in reactions involved in the enrichment of PtdIns4P on the LCV. The role of LegA5 in L. pneumophila infection awaits further investigation. PtdIns3P is also an potential anchor for effector proteins that harbor PtdIns3P‐binding domains, allowing their association with the LCV or other compartments (Jank et al, 2012; Finsel et al, 2013; Nachmias et al, 2019).
Figure EV5. A model for de novo biosynthesis of PtdIns4P from PtdIns by MavQ, LepB, and SidF on the phagosome membrane.
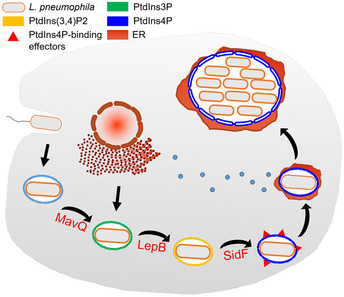
Once being internalized by phagocytosis, L. pneumophila resides in the LCV that has intimate communication with ER‐derived vesicles. The LCV develops into a PtdIns4P enriched compartment which is permissive for bacterial replication. At least three Dot/Icm effector proteins have been established to contribute to the PI metabolism on the LCV. MavQ catalyzes the conversion of PtdIns into PtdIns3P, which is then phosphorylated by the PI4K LepB to yield PtdIns(3,4)P2; the PI 3‐phosphatase SidF removes the 3‐phosphate from PtdIns(3,4)P2 to generate PtdIns4P. The accumulation of PtdIns4P is critical for the specific anchoring of Dot/Icm effectors on the LCV, such as SidM/DrrA, SidC, SdcA, Lpg1101, and Lpg2603.
Alternatively, host enzymes involved in PI metabolism may account for a portion of PtdIns4P on the LCV. It has been shown that silencing of the host PI4KIIIα leads to a cumulative reduction in PtdIns4P levels on the phagosomes harboring the ∆sidF mutant (Hubber et al, 2014), suggesting that PI4KIIIα may function in parallel to the MavQ‐LepB‐SidF axis for the biosynthesis of PtdIns4P on the LCV. Similar observations have been made for the host PI4KIIIβ (Brombacher et al, 2009). In addition, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 are also possible sources for PtdIns4P on the LCV, which can be converted into PtdIns4P and PtdIns(3,4)P2, respectively, by the PI 5‐phosphatase oculocerebrorenal syndrome of Lowe (OCRL1) (Erdmann et al, 2007). Interestingly, OCRL1 is associated with the LCV, probably by binding to LpnE, another L. pneumophila effector protein (Weber et al, 2009). Despite these observations, the exact mechanism of how these host enzymes contribute to the production of PtdIns4P on the LCV needs further investigation.
The interactions among L. pneumophila enzymes involved in PI metabolism appear complex. For example, it has been shown that SidP (Toulabi et al, 2013) suppresses the yeast toxicity of MavQ in a manner that is independent of its PI 3‐phosphatase activity (Urbanus et al, 2016). Although SidP appears to bind MavQ with high affinity, it does not affect its kinase activity in biochemical assays (Fig 6B and C). Thus, it may possess a yet unrecognized activity dedicated to the inhibition of MavQ in a way that may require a factor from host cells. Clearly, the interplay among several factors, including the cellular localization of the effector, the time that the effectors are translocated into the host cell, the physical interactions among them, and the cellular distribution of their products, will dictate the effects of PIs in the development of the LCV. Such complexity is further compounded by the presence of a cohort of PtdIns3P‐binding effectors (Jank et al, 2012; Finsel et al, 2013; Nachmias et al, 2019). Future studies aiming at elucidating the activity of these effectors and their relationships will provide further insights into the roles of PIs in the intracellular life cycle of L. pneumophila.
Materials and Methods
Bacterial strains and plasmid construction
The bacterial strains, plasmids, and primers used in this study are listed in Appendix Tables S1–S3, respectively. L. pneumophila strains used are derivatives of the Philadelphia 1 strain Lp02 and were grown on charcoal‐yeast extract (CYE) plates or in aces‐buffered yeast extract (AYE) broth. In‐frame deletion mutants of L. pneumophila were made by the plasmid pSR47S (Merriam et al, 1997). Genomic DNA of L. pneumophila was used to amplify mavQ, lepB, sidP, and sidF. The cDNA of the myotubularin 1 gene was synthesized by GenScript Biotech Corporation. Amplified DNA products were inserted into pET28a (Novagen) or pGEX‐6P‐1 (GE healthcare) for protein expression in Escherichia coli or cloned into pZL507 (Xu et al, 2010) for expression in L. pneumophila. pCMV‐4xFlag was constructed based on the backbone of pCMV‐Flag (Sigma‐Aldrich), introducing an additional 3xFlag tag before the multiple cloning site. In order to express proteins in mammalian cells, genes were ligated to pCMV‐4xFlag or pcDNA3.1‐mCherry (Kleaveland et al, 2018) (a gift from David Barte, Addgene plasmid # 128744; http://n2t.net/addgene:128744; RRID:Addgene_128744). The PI probe GFP‐2xFYVEHrs (the duplicate FYVE finger from the human homologue of the hepatocyte growth factor‐regulated tyrosine kinase substrate) (Cao et al, 2008) was a gift from Dr. Feng Shao at the National Institute of Biological Sciences, China. pSB157m was modified from the integrative yeast plasmid pSB157 (Fazzio & Tsukiyama, 2003) (courtesy of Sue Biggins, Fred Hutchinson Cancer Research Center, Seattle, WA) with the addition of a Flag‐tag and several restriction sites for inserting genes of interest. pSB157m was used to express Flag‐tagged MavQ in the Saccharomyces cerevisiae strain W303a (Thomas & Rothstein, 1989). Point mutations were introduced into genes by QuikChange II Site‐Directed Mutagenesis Kit (Agilent Technologies). The integrity of all plasmids was verified by DNA sequencing.
Protein expression and purification
Plasmids derived from pET28a or pGEX‐6P‐1 for protein production were transformed into E. coli strain BL21 (DE3). Overnight cultures were diluted at 1:50 in fresh LB medium supplemented with the appropriate antibiotics. Cultures grown to an OD600nm of 0.6–0.8 at 37°C were induced by adding isopropyl β‐D‐galactopyranoside (IPTG) to a concentration of 0.2 mM. Bacteria were further cultured for 16–18 h at 18°C in a shaker set at 250 rpm. Each step of the following operations was performed at 4°C. Cells were collected by centrifugation and lysed by a homogenizer (JN‐mini, JNBIO, Guangzhou, China). Lysed samples were centrifugated at 12,000 g for 30 min to remove cell debris and unbroken cells. The supernatants were either incubated with 1 ml prewashed Ni2+‐NTA beads (Qiagen) or Glutathione Sepharose 4 Fast Flow beads (GE healthcare) for 1 h at 4°C. For purification of His6‐tagged proteins, the Ni2+‐NTA beads were eluted by 300 mM imidazole. GST‐tagged proteins were eluted by 20 mM reduced glutathione. Purified proteins were dialyzed twice in a buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, and 1 mM dithiothreitol (DTT). Protein concentrations were measured by the Bradford Protein Assay (Bio‐Rad).
In vitro kinase and phosphatase assays
A panel of diC8 PIs purchased from Echelon Biosciences were individually dissolved in sterilized deionized water to make 1 mM stock solutions. The kinase activity was determined by the ADP‐Glo™ Kinase Assay (Promega) in 96‐well white polystyrene plates. 0.1 μg of purified enzymes were incubated with 0.5 nM PI substrates in 25 µl kinase buffer (40 mM Tris–HCl (pH 7.5), 20 mM MgCl2, 1 mM dithiothreitol (DTT), and 0.025 ng/ml of BSA) (Dong et al, 2016). Reactions were allowed to proceed in dark for 60 min at 25°C. To stop the reaction, 25 µl of ADP‐Glo reagent was added to the plates and incubated for 40 min at 25°C. Next, 50 µl of detection reagent was added and incubated for 40 min at 25°C. The plates were read on a Synergy H1 Hybrid Multi‐Mode Microplate Reader (BioTek). The phosphatase assay was conducted in a 25‐µl reaction mixture containing 1 nM of lipids and 0.1 μg of enzyme in a reaction buffer (50 mM Tris–HCl (pH 8.0) and 150 mM NaCl). After incubation at 37°C for 20 min, 100 µl of malachite green reagent (Sigma‐Aldrich) was added to the reaction. Absorbance at 620 nm was measured by a Synergy H1 Hybrid Multi‐Mode Microplate Reader (BioTek).
Thin‐layer chromatography (TLC)
The kinase reaction was performed as described above except that the substrates were replaced with Bodipy‐FL labeled diC8 PIs. For reactions containing multiple enzymes, the second enzyme was added after the completion of the reaction catalyzed by the first enzyme and incubated for another 20 min. For reactions containing phosphatases, the kinase reaction products were dried by a Speed‐Vac for 30 min at 45°C and resuspended in 20 µl of a phosphatase buffer (50 mM ammonium carbonate (pH 8.0), 2 mM DTT). 1 μg of purified phosphatase was added to the mixture and incubated at 37°C for 1 h. All reaction mixtures were dried and dissolved in 10 μl of isopropanol/methanol/acetic acid (v/v/v, 5/5/2). The TLC silica gel 60 F254 (Sigma‐Aldrich) was activated by a methanol and water (v/v, 3/2) solution containing 1% potassium oxalate for 30 min and was dried at 65°C for 1 h. Samples were spotted on the activated TLC plate and were developed in a solvent system containing chloroform/methanol/acetone/glacial acetic acid/water (v/v/v/v/v, 7/5/2/2/2) (Dong et al, 2016). Fluorescence signals were acquired by a Goodsee‐5 TLC imager.
Yeast toxicity assays
MavQ or its point mutants were inserted into pSB157m which contains a galactose‐inducible promoter and transformed into the S. cerevisiae strain W303a (Thomas & Rothstein, 1989). Transformants were selected by plating on synthetic defined (SD)‐Ura agar plates containing 2% glucose. Yeast cells cultured in SD‐Ura liquid medium containing glucose were serially diluted. 10 μl of each dilution was then spotted onto SD‐Ura agar plates containing 2% glucose or galactose. Plates were incubated at 30°C for 72 h before image acquisition. To determine the influence of SidP on the yeast toxicity of MavQ, wild‐type SidP, SidPR560K (Toulabi et al, 2013) and SidP664‐822 (Urbanus et al, 2016) were cloned into p425GPD (Mumberg et al, 1995) and transformed into W303a (pSB157m::MavQ). The spotting of the yeast cells on selection medium containing glucose or galactose was performed similarly as described above.
GST pull‐down assay
20 µg of GST‐SidP or GST was incubated with 20 µl of prewashed GST magnetic beads (Sigma‐Aldrich) in 1 ml of GST binding buffer (50 mM Tris–HCl (pH 7.5), 137 mM NaCl, 13.7 mM KCl) for 2 h at 4°C on a shaker. Unbound proteins were removed by washing the beads with GST binding buffer for 3 times. The beads were further incubated with 20 µg of His6‐MavQ in 1 ml of GST binding buffer for 2 h at 4°C on a shaker. After extensive washing, the beads were resuspended in 50 µl of 1×SDS loading buffer. Samples resolved by SDS–PAGE were stained with Coomassie brilliant blue.
Cell culture, transfection, infection, and immunostaining
HeLa cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone) supplemented with 10% FBS at 37°C in a 5% CO2 incubator. Transfection was performed using Lipofectamine 3000 (Thermo Fisher). U937 cells (ATCC) were cultured in RPMI 1640 medium supplemented with 10% FBS and were differentiated into macrophages by phorbol‐12‐myristate‐13‐acetate (PMA). Bone marrow‐derived macrophages (BMDMs) were prepared from 6‐ to 8‐week‐old female A/J mice (Model Animal Research Center of Nanjing University, Nanjing, China) using bone marrow macrophage media (RPMI1640, 20% FBS and 20% L‐cell supernatant‐conditioned medium) (Thermo Fisher). All animal procedures were approved by the Institutional Animal Care and Use Committee of Jilin University (number of permit: SY201902008).
For fluorescence imaging of GFP‐2xFYVEHrs, HeLa cells seeded on coverslips were co‐transfected with peGFP::2xFYVEHrs (Cao et al, 2008) and plasmids derived from pcDNA3.1‐mCherry for 18 h. When indicated, wortmannin was added at a final concentration of 800 nM. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X‐100 for 5 min. Nuclei were labeled by Hoechst staining.
For bacterial infection experiments, L. pneumophila strains grown in AYE broth to the post‐exponential phase (OD600nm = 3.3–3.8) were checked for motility. For intracellular growth assays, BMDMs seeded in 24‐well plates at 4 × 105/well were infected with L. pneumophila at a multiplicity of infection (MOI) of 0.05. After 2 h of incubation at 37°C, infections were synchronized by washing the samples three times with warm PBS to remove extracellular bacteria. Infection samples were further incubated at 37°C in a 5% CO2 incubator. At 2, 24, 48, and 72 h post‐infection, infected macrophages were lysed with 0.02% saponin for 30 min. The lysates were diluted with PBS and plated onto CYE plates. After incubation at 37°C for 5 days, the colony‐forming units (CFUs) were counted. To detect translocated MavQ and SidC in infected cells by immunostaining, 2 × 105 BMDMs seeded on coverslips in 24‐well plates were infected with L. pneumophila at an MOI of 2. Two hours after infection, samples washed for three times with PBS were fixed with 4% paraformaldehyde. The extracellular bacteria were stained with anti‐Legionella antibodies (Xu et al, 2010) before permeabilization. To stain the total bacteria, MavQ, and SidC, cells were permeabilized by 0.2% Triton X‐100 and were sequentially stained with antibodies specific for L. pneumophila, MavQ, and SidC. Images were acquired by an Olympus IX‐83 fluorescence microscope.
Antibodies and immunoblotting
Polyclonal antibodies against MavQ were produced by immunization of mice with recombinant His6‐MavQ (AbMax Biotechnology Co., LTD, Beijing, China). Antibodies used for immunoblotting analysis and their dilutions are anti‐mCherry (Sigma‐Aldrich, cat# AB356482, 1:2,000), anti‐Flag (Sigma‐Aldrich, Cat# F1804, 1:3,000), anti‐tubulin (DSHB, E7, 1:10,000), anti‐ICDH (1:20,000) (Liu & Luo, 2007), anti‐MavQ (1:1,000), and anti‐SidC (1:10,000) (Luo & Isberg, 2004). For immunoblotting, total proteins separated by SDS–PAGE were transferred onto nitrocellulose membranes (Pall Life Sciences). After blocking with 5% nonfat milk, proteins were detected by incubation with appropriate primary antibodies and IRDye‐conjugated secondary antibodies (Li‐Cor). The signals were detected using the Odyssey® CLx Imaging System (Li‐Cor).
Data analysis
Statistical analysis was calculated by the unpaired two‐tailed Student’s t‐tests, and p < 0.05 was considered as significant difference.
Author contributions
JZQ conceived the project. JZQ and Z‐QL designed the experiments. GL and HL performed the research. JZQ, GL, HL, and Z‐QL analyzed the data. JZQ wrote the first draft of the manuscript. JZQ and Z‐QL revised the manuscript with help from all other authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank Dr. Shao Feng at the National Institute of Biological Sciences for providing plasmids. This work was supported by the Thousand Young Talents Program of the Chinese government (JZQ) and startup fund from Jilin University and the First Hospital of Jilin University; National Natural Science Foundation of China grants (grant #: 31770149 and 31970134 to JZQ).
EMBO reports (2021) 22: e51163
Contributor Information
Zhao‐Qing Luo, Email: luoz@purdue.edu.
Jiazhang Qiu, Email: qiujz@jlu.edu.cn.
Data availability
No large primary datasets have been generated or deposited.
References
- Al‐Quadan T, Price CT, Abu Kwaik Y (2012) Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella . Trends Microbiol 20: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K, Toomre DK, Roy CR (2012) The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE‐dependent membrane fusion. Cell Host Microbe 11: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S, Gao P, Shen X, Fiscus V, Zong WX, Chen L, Luo ZQ (2007) Legionella pneumophila inhibits macrophage apoptosis by targeting pro‐death members of the Bcl2 protein family. Proc Natl Acad Sci USA 104: 5121–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlocher K, Welin A, Hilbi H (2017) Formation of the Legionella replicative compartment at the crossroads of retrograde trafficking. Front Cell Infect Microbiol 7: 482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Kalayil S, Liu Y, Bonn F, Colby T, Matic I, Dikic I (2016) Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167: 1636–1649 [DOI] [PubMed] [Google Scholar]
- Bhogaraju S, Bonn F, Mukherjee R, Adams M, Pfleiderer MM, Galej WP, Matkovic V, Lopez‐Mosqueda J, Kalayil S, Shin D et al (2019) Inhibition of bacterial ubiquitin ligases by SidJ‐calmodulin catalysed glutamylation. Nature 572: 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MH, Osinski A, Gradowski M, Servage KA, Pawlowski K, Tomchick DR, Tagliabracci VS (2019) Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE‐family ubiquitin ligases. Science 364: 787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H (2009) Rab1 guanine nucleotide exchange factor SidM Is a major phosphatidylinositol 4‐phosphate‐binding effector protein of Legionella pneumophila . J Biol Chem 284: 4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Satanower S, Simovitch M, Belnik Y, Zehavi M, Yerushalmi G, Ben‐Aroya S, Pupko T, Banin E (2015) Type III effectors in Pseudomonas aeruginosa . MBio 6: e00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao CH, Backer JM, Laporte J, Bedrick EJ, Wandinger‐Ness A (2008) Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol Biol Cell 19: 3334–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Dong N, Niu M, Hu L, Yao Q, Zhou R, Shao F (2016) Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4‐kinase activity of the Legionella LepB effector. Nat Microbiol 2: 16236 [DOI] [PubMed] [Google Scholar]
- Downes CP, Gray A, Lucocq JM (2005) Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol 15: 259–268 [DOI] [PubMed] [Google Scholar]
- Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camillil P (2007) A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell 13: 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Tsukiyama T (2003) Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol Cell 12: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Finsel I, Ragaz C, Hoffmann C, Harrison CF, Weber S, van Rahden VA, Johannes L, Hilbi H (2013) The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 14: 38–50 [DOI] [PubMed] [Google Scholar]
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, Harris J, Mallison GF, Martin SM, McDade JE et al (1977) Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med 297: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Gan NH, Zhen XK, Liu Y, Xu XL, He CL, Qiu JZ, Liu YC, Fujimoto GM, Nakayasu ES, Zhou BA et al (2019) Regulation of phosphoribosyl ubiquitination by a calmodulin‐dependent glutamylase. Nature 572: 387–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R (1998) FYVE fingers bind PtdIns(3)P. Nature 394: 432–433 [DOI] [PubMed] [Google Scholar]
- Gomez‐Valero L, Rusniok C, Carson D, Mondino S, Perez‐Cobas AE, Rolando M, Pasricha S, Reuter S, Demirtas J, Crumbach J et al (2019) More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci USA 116: 2265–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Burd CG (2011) Coordination of Golgi functions by phosphatidylinositol 4‐kinases. Trends Cell Biol 21: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbi H (2006) Modulation of phosphoinositide metabolism by pathogenic bacteria. Cell Microbiol 8: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Finsel I, Otto A, Pfaffinger G, Rothmeier E, Hecker M, Becher D, Hilbi H (2014) Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella‐containing vacuoles from macrophages. Cell Microbiol 16: 1034–1052 [DOI] [PubMed] [Google Scholar]
- Hsu F, Zhu W, Brennan L, Tao L, Luo ZQ, Mao Y (2012) Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc Natl Acad Sci USA 109: 13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O'Connor TJ, Chen C, Machner M, Montminy T, Isberg RR (2011) The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13: 227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Arasaki K, Nakatsu F, Hardiman C, Lambright D, De Camilli P, Nagai H, Roy CR (2014) The machinery at endoplasmic reticulum‐plasma membrane contact sites contributes to spatial regulation of multiple legionella effector proteins. Plos Pathog 10: e1004222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR (2007) Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450: 365–369 [DOI] [PubMed] [Google Scholar]
- Isberg RR, O'Connor TJ, Heidtman M (2009) The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jank T, Bohmer KE, Tzivelekidis T, Schwan C, Belyi Y, Aktories K (2012) Domain organization of Legionella effector SetA. Cell Microbiol 14: 852–868 [DOI] [PubMed] [Google Scholar]
- Jeng EE, Bhadkamkar V, Lbe NU, Gause H, Jiang LH, Chan J, Jian RQ, Jimenez‐Morales D, Stevenson E, Krogan NJ et al (2019) Systematic identification of host cell regulators of Legionella pneumophila pathogenesis using a genome‐wide CRISPR screen. Cell Host Microbe 26: 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Roy CR (2002) Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol 4: 945–954 [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP (2018) A Network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotewicz KM, Ramabhadran V, Sjoblom N, Vogel JP, Haenssler E, Zhang M, Behringer J, Scheck RA, Isberg RR (2017) A single Legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe 21: 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar DT, Doss CGP (2016) Investigating the inhibitory effect of wortmannin in the hotspot mutation at codon 1047 of PIK3CA kinase domain: a molecular docking and molecular dynamics approach. Adv Protein Chem Str 102: 267–297 [DOI] [PubMed] [Google Scholar]
- Ledvina HE, Kelly KA, Eshraghi A, Plemel RL, Peterson SB, Lee B, Steele S, Adler M, Kawula TH, Merz AJ et al (2018) A Phosphatidylinositol 3‐kinase effector alters phagosomal maturation to promote intracellular growth of francisella. Cell Host Microbe 24: 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA (2008) Membrane recognition by phospholipid‐binding domains. Nat Rev Mol Cell Bio 9: 99–111 [DOI] [PubMed] [Google Scholar]
- Liu YC, Luo ZQ (2007) The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 75: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Luo J, Zhen X, Qiu J, Ouyang S, Luo ZQ (2020) Interplay between bacterial deubiquitinase and ubiquitin E3 ligase regulates ubiquitin dynamics on Legionella phagosomes. Elife 9: e58114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Gaspar AH, Pallara C, Rojas AL, Fernandez‐Recio J, Machner MP, Hierro A (2014) Structural basis for the recruitment and activation of the Legionella phospholipase VipD by the host GTPase Rab5. Proc Natl Acad Sci USA 111: E3514–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR (2004) Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA 101: 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR (2006) Targeting of host rab GTPase function by the intravacuolar pathogen Legionella pneumophila . Dev Cell 11: 47–56 [DOI] [PubMed] [Google Scholar]
- Marat AL, Haucke V (2016) Phosphatidylinositol 3‐phosphates‐at the interface between cell signalling and membrane traffic. EMBO J 35: 561–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayinger P (2012) Phosphoinositides and vesicular membrane traffic. Biochim Biophys Acta 1821: 1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam JJ, Mathur R, Maxfield‐Boumil R, Isberg RR (1997) Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun 65: 2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Liu XY, Arasaki K, McDonough J, Galan JE, Roy CR (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477: 103–U122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329: 946–949 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide‐exchange factor. Nat Cell Biol 8: 971–977 [DOI] [PubMed] [Google Scholar]
- Nachmias N, Zusman T, Segal G (2019) Study of Legionella effector domains revealed novel and prevalent phosphatidylinositol 3‐phosphate binding domains. Infect Immun 87: e00153‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR (2002) A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295: 679–682 [DOI] [PubMed] [Google Scholar]
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Yergey A, Machner MP (2011) De‐AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila . Science 333: 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G (2014) Identification of Legionella pneumophila effectors regulated by the LetAS‐ RsmYZ‐ CsrA regulatory cascade, many of which modulate vesicular trafficking. J Bacteriol 196: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic N, Barlocher K, Finsel I, Hilbi H (2016) Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol 24: 450–462 [DOI] [PubMed] [Google Scholar]
- Qiu JZ, Sheedlo MJ, Yu KW, Tan YH, Nakayasu ES, Das C, Liu XY, Luo ZQ (2016) Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Luo ZQ (2017) Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 15: 591–605 [DOI] [PubMed] [Google Scholar]
- Rothmeier E, Pfaffinger G, Hoffmann C, Harrison CF, Grabmayr H, Repnik U, Hannemann M, Wolke S, Bausch A, Griffiths G et al (2013) Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. Plos Pathog 9: e1003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Mukherjee R, Liu YB, Gonzalez A, Bonn F, Liu Y, Rogov VV, Heinz M, Stolz A, Hummer G et al (2020) Regulation of phosphoribosyl‐linked serine ubiquitination by deubiquitinases DupA and DupB. Mol Cell 77: 164–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpizio A, Minelli ME, Wan M, Burrowes PD, Wu XC, Sanford EJ, Shin JH, Williams BC, Goldberg ML, Smolka MB et al (2019) Protein polyglutamylation catalyzed by the bacterial calmodulin‐dependent pseudokinase SidJ. Elife 8: e51162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart AL, Hilbi H (2020) Phosphoinositides and the Fate of Legionella in Phagocytes. Front Immunol 11: 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YH, Arnold RJ, Luo ZQ (2011) Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci USA 108: 21212–21217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YH, Luo ZQ (2011) Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475: 506–U102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE (2000) Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3‐phosphate. Proc Natl Acad Sci USA 97: 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Toulabi L, Wu X, Cheng Y, Mao Y (2013) Identification and structural characterization of a Legionella phosphoinositide phosphatase. J Biol Chem 288: 24518–24527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus ML, Quaile AT, Stogios PJ, Morar M, Rao C, Di Leo R, Evdokimova E, Lam M, Oatway C, Cuff ME et al (2016) Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol Syst Biol 12: 893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL (2000) Structural determinants of phosphoinositide 3‐kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell 6: 909–919 [DOI] [PubMed] [Google Scholar]
- Wan M, Sulpizio AG, Akturk A, Beck WHJ, Lanz M, Faca VM, Smolka MB, Vogel JP, Mao YX (2019) Deubiquitination of phosphoribosyl‐ubiquitin conjugates by phosphodiesterase‐domain‐containing Legionella effectors. Proc Natl Acad Sci USA 116: 23518–23526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H (2006) Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. Plos Pathog 2: e46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SS, Ragaz C, Hilbi H (2009) The inositol polyphosphate 5‐phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole and binds to the bacterial effector LpnE. Cell Microbiol 11: 442–460 [DOI] [PubMed] [Google Scholar]
- Weber S, Steiner B, Welin A, Hilbi H (2018) Legionella‐containing vacuoles capture PtdIns(4)P‐rich vesicles derived from the Golgi apparatus. MBio 9: e02420‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ (2010) Inhibition of host vacuolar H+‐ATPase activity by a Legionella pneumophila effector. Plos Pathog 6: e1000822 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
No large primary datasets have been generated or deposited.


