Abstract
Epimorphic regeneration commonly relies on the activation of reserved stem cells to drive new cell production. The planarian Schmidtea mediterranea is among the best regenerators in nature, thanks to its large population of adult stem cells, called neoblasts. While neoblasts have long been known to drive regeneration, whether a subset of neoblasts is reserved for this purpose is unknown. Here, we revisit the idea of reserved neoblasts by approaching neoblast heterogeneity from a regulatory perspective. By implementing a new fluorescence‐activated cell sorting strategy in planarians, we identify a population of neoblasts defined by low transcriptional activity. These RNAlow neoblasts are relatively slow‐cycling at homeostasis and undergo a morphological regeneration response characterized by cell growth at 48 h post‐amputation. At this time, RNAlow neoblasts proliferate in a TOR‐dependent manner. Additionally, knockdown of the tumour suppressor Lrig‐1, which is enriched in RNAlow neoblasts, results in RNAlow neoblast growth and hyperproliferation at homeostasis, and ultimately delays regeneration. We propose that slow‐cycling RNAlow neoblasts represent a regeneration‐reserved neoblast population.
Keywords: planarians, quiescence, regeneration, stem cells, TORC1
Subject Categories: Regenerative Medicine
A population of slow‐cycling RNAlow neoblasts in the planarian Schmidtea mediterranea grows and enters the cell cycle following amputation in a TOR‐dependent manner. These findings suggest that planarians use a slow‐cycling adult stem cell strategy in their regenerative biology.
Introduction
Regeneration is a widespread phenomenon across the animal kingdom and often relies on a proliferative response, termed “epimorphosis”, to replace lost cells (Morgan, 1898; Iismaa et al, 2018). This epimorphic response can be sourced in a variety of ways. These include cell cycle re‐entry of mature cells through dedifferentiation, altered cell cycle dynamics within proliferative stem cell populations, or the activation of reserved stem cells (Clevers & Watt, 2018). Elucidating the contribution from each is critical for understanding the mechanisms behind regeneration in different injury contexts.
By approaching epimorphosis from an evolutionary perspective, we can take advantage of highly regenerative organisms to discover conserved factors that facilitate successful regeneration. The flatworm planarian, Schmidtea mediterranea, has the remarkable ability to regenerate all tissues after an injury (Newmark & Sánchez Alvarado, 2002). This regenerative response is facilitated by a large population of adult stem cells, called neoblasts. Neoblasts are believed to be the only cycling cells in planarians and to be heterogeneous in terms of lineage potential (van Wolfswinkel et al, 2014; Scimone et al, 2014; Molinaro & Pearson, 2016, 2018; Zhu & Pearson, 2016). Research in the last decade has definitively identified pluripotent adult stem cells in the planarian, called clonogenic neoblasts (cNeoblasts), which can individually rescue lethally irradiated animals and restore epimorphic regeneration (Wagner et al, 2011). This discovery was followed by the identification of TSPAN‐1+ pluripotent cells, which demonstrated a 25% rescue efficiency when individually transplanted into irradiated hosts (Zeng et al, 2018). While the topic of neoblast heterogeneity has been a primary area of study in recent years, the majority of this research has focused on questions of lineage potential. Whether the neoblast population is heterogenous in terms of proliferation control, and how this might influence regeneration, are important questions to consider for understanding the epimorphic strategies that result in successful regeneration.
After decades of debates throughout the 20th century, the possibility that a reserved, injury‐induced subset of neoblasts exists in planarians has been given little consideration in the molecular era. In a pioneering study from 2000, Newmark and Sánchez‐Alvarado found that continuous BrdU administration resulted in labelling of 99% of neoblasts after 3 days, leading to the conclusion that essentially all neoblasts are constantly cycling during homeostasis (Newmark & Sánchez Alvarado, 2000). However, in the decades since this seminal study the planarian field has seen many important molecular and technological advancements, including the identification of molecular neoblast markers (such as piwi‐1 (Reddien et al, 2005)) and the use of fluorescence‐activated cell sorting (FACS) for isolating neoblasts (Hayashi et al, 2006). These tools have facilitated a growing body of research on neoblast heterogeneity, yet studies on neoblast proliferation control are limited. Thus, we looked to re‐evaluate this question of reserved neoblasts by approaching neoblast heterogeneity from a regulatory perspective.
Neoblasts drive epimorphosis in planarians. Following amputation, the neoblast population exhibits a robust proliferative response characterized by two peaks in mitosis at 6‐ and 48‐h post‐amputation (hpa) (Wenemoser & Reddien, 2010). This mitotic response is dependent on TOR (target of rapamycin) signalling through TOR complex 1 (TORC1), which is required for regeneration (Wenemoser & Reddien, 2010; Peiris et al, 2012; Tu et al, 2012). Although TOR is known to be involved in controlling the rate of proliferation of neoblasts during regeneration, the mechanism of this control is still unknown.
Recent work in other systems has uncovered a conserved role for TOR signalling in facilitating wound healing by promoting the activation of quiescent stem cells. Quiescence is defined as a state of low proliferative and transcriptional activity, which serves to limit mutations and delay stem cell ageing by minimizing the accumulation of molecular damage caused by cellular activities (Derényi & Szöllsi, 2017; Clevers & Watt, 2018; Tümpel & Rudolph, 2019). During quiescence, cells rest outside of the active cell cycle in the G0 phase. In vertebrates many tissue hierarchies begin with quiescent multipotent stem cells. These cells are maintained in G0 during homeostasis in adults, often through the function of highly conserved tumour suppressors. Following injury, the increased tissue needs dictate the activation of these cells: quiescent stem cells undergo cell growth, increase metabolic activity and enter the cell cycle to aid in tissue regeneration (Tian et al, 2011; Nagashima et al, 2013; Rodgers et al, 2014; Lee et al, 2018; Lo Sicco et al, 2018). In many contexts, this activation is mediated by TOR signalling through association with its accessory protein, Raptor, in the TORC1 complex (Wullschleger et al, 2006; Gan & DePinho, 2009; Rodgers et al, 2014; Lee et al, 2018). Given the TOR‐dependency and incredibly rapid nature of the planarian regeneration response, we hypothesize that TOR signalling promotes regeneration through the activation of a subset of reserved neoblasts.
In this study we report the presence of a population of neoblasts defined by low transcriptional activity, which we refer to as RNAlow neoblasts. Compared to other neoblasts, RNAlow neoblasts are relatively small and divide less frequently during homeostasis. Upon injury, RNAlow neoblasts undergo robust cell growth by 48 h post‐amputation and enter the cell cycle in a TOR‐dependent manner. The conserved tumour suppressor, Lrig‐1, is required to prevent RNAlow neoblast growth at homeostasis, and knockdown of Lrig‐1 results in homeostatic hyperproliferation and delayed regeneration. Taken together, our data highlight novel heterogeneity among planarian neoblasts and suggest that planarians use a slow‐cycling adult stem cell strategy in their regenerative biology.
Results
Planarian neoblasts have variable rates of division
To begin assaying for proliferation heterogeneity, we tested the hypothesis that neoblast division rates are uniform by performing a DNA label‐retention assay using the thymidine analog BrdU to assess division rates within the neoblast population. BrdU was administered through feeding to avoid potential confounding effects of the neoblast injury response. In line with previous findings, we found that 62 ± 9% of piwi‐1 + cells incorporated BrdU by 1 day post‐pulse (dpp). Maximal double labelling was observed at 2 dpp, then piwi‐1+ cells rapidly diluted the BrdU, dropping to 9.1 ± 2% BrdU+ by 14 dpp. Surprisingly, we observed that 1.3 ± 0.3% of piwi‐1 + cells retained the label for at least 35 dpp (Fig 1A and B). To ensure that label retention was not a consequence of BrdU‐induced cell cycle arrest, we used a double‐pulse experiment where the same animals were sequentially pulsed with two thymidine analogs, BrdU and EdU, 14 days apart. We observed BrdU+EdU+ cells in the parenchyma, demonstrating that BrdU+ neoblasts can still proliferate at least 14 days from the initial labelling (Fig 1C). Thus, given the gradual decline in BrdU label retention over the span of 5 weeks, we conclude that the rate of division within the neoblast population is not uniform, but is rather more heterogeneous than previously thought.
Figure 1. Planarian neoblasts have variable rates of division.
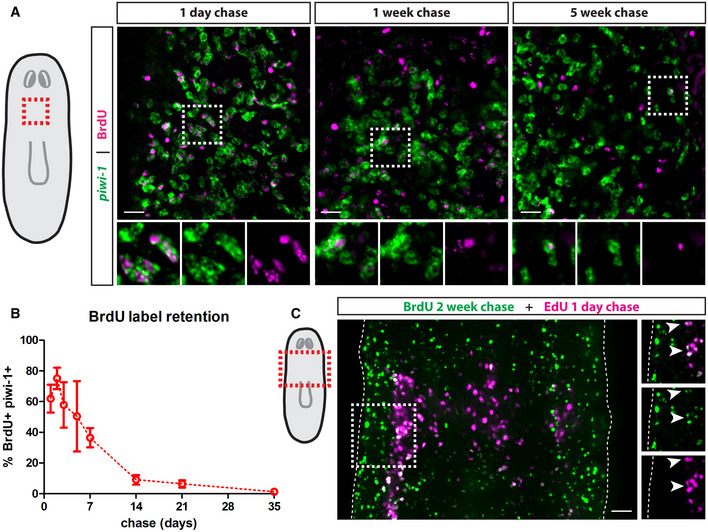
- Whole‐mount FISH for piwi‐1 and immunostaining for BrdU at 1‐day, 1‐week and 5‐weeks post‐BrdU administration. White boxes denote the magnified regions shown in the bottom panels. Scale bars, 30 µm.
- Quantification of the percentage of BrdU label‐retaining neoblasts. Data are presented as mean ± s.d. (n = 4–6 animals per time point).
- Double‐pulse experiment showing colocalization of BrdU after a 2‐week chase period with EdU after a 1‐day chase period. White boxes denote the magnified region shown on the left. Dashed lines mark the edges of the animal and white arrows mark colocalization. Scale bar, 50 µm.
Data information: All images are single confocal planes taken from the prepharyngeal region as indicated in the diagrams.
Source data are available online for this figure.
Planarians have RNAlow neoblasts
The identification of relatively long‐term label‐retaining neoblasts prompted us to investigate neoblast cell cycle status in greater detail. Flow cytometry is a widely used technique for assaying cell cycle dynamics, and the classic Hoechst staining protocol used in the planarian field robustly resolves proliferating neoblasts in the S/G2/M phases of the cell cycle based on their > 2C DNA content (Hayashi et al, 2006). In order to further resolve G0/G1 neoblasts, we adopted a FACS strategy widely used in other systems that considers RNA levels based on fluorescence of the RNA dye pyronin Y, on the basis that slow‐cycling stem cells (often in the G0 phase) typically have lower transcriptional output (Darzynkiewicz et al, 1980; Kim & Sederstrom, 2015; Eddaoudi et al, 2018). Intriguingly, using these dyes, we defined a highly reproducible RNAlow FACS gate in planarians (Fig 2A). The cells in the RNAlow gate registered as having ≤ 2C DNA content by Hoechst fluorescence and fell within the X2 Hoechst side population (SP) when backgated (Fig EV1A). To determine whether this gate contained neoblasts, we lethally irradiated animals (60 Gy) and found that the RNAlow gate was completely depleted by 1 day post‐irradiation (dpi), a time point by which neoblasts are lost (Fig 2A, see Discussion). Additionally, by single cell RNA‐sequencing (scRNAseq) of 96 cells from the RNAlow gate, we found that 59.4% expressed piwi‐1 and 89.6% expressed at least one of piwi‐1, piwi‐2 or piwi‐3, all of which are known to be expressed in neoblasts (Reddien et al, 2005; Palakodeti et al, 2008).
Figure 2. Sorting neoblasts by DNA/RNA content reveals a relatively slow‐cycling population.
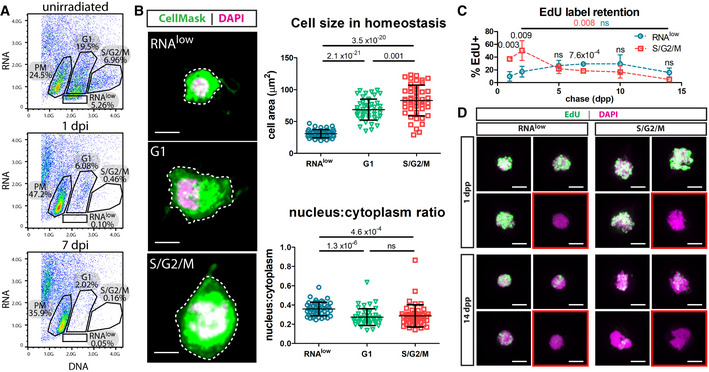
- Dissociated cells from unirradiated, 1 dpi or 7 dpi animals (60 Gy of irradiation) were stained with Hoechst (DNA) and Pyronin Y (RNA) and sorted by FACS to reveal an RNAlow population. Percentages indicate the proportion of events within each gate.
- Cell size and nucleus:cytoplasm ratio measured by CellMask (cytoplasm) and DAPI (nucleus) staining of FACS sorted cells at homeostasis (n ≥ 47 per sample). Representative images are shown, with dashed lines to indicate the area measured. Scale bars, 5 µm.
- EdU pulse‐chase time course of label retention in sorted RNAlow and S/G2/M neoblasts (n ≥ 3 individual experiments per time point). Red and blue P values indicate the statistical significance of EdU dilution from 2 to 14 dpp in S/G2/M or RNAlow neoblasts, respectively. Black P values indicate the statistical significance of EdU retention in RNAlow neoblasts compared to S/G2/M neoblasts.
- Maximum projection confocal images showing examples of cells from the beginning and end of the EdU label‐retention time course in (C). Red boxes denote examples of EdU− cells. Scale bars, 5 µm.
Data information: Data in (B and C) are presented as mean ± s.d. Statistical significance was assessed using Welch's t‐tests (P values are indicated; ns, not significant [P > 0.05]). Data are representative of ≥ 3 independent experiments.
Figure EV1. Backgating and RNA‐sequencing of DNA/RNA FACS gates.
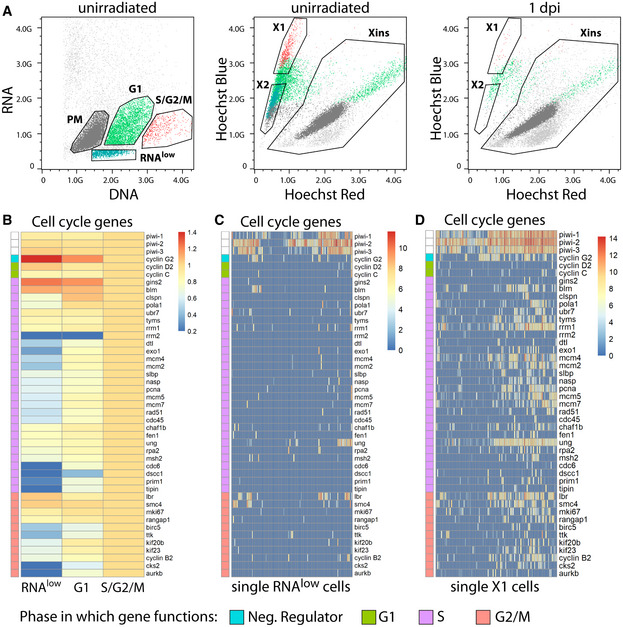
-
ABackgating of the RNAlow, G1, S/G2/M, and PM populations from Hoechst/pyronin Y stained cells (left) onto a classic Hoechst side population plot (middle, unirradiated; right, 1 day post 60 Gy irradiation). Plots are representative of n > 3 independent experiments.
-
BExpression of cell cycle genes in bulk RNAseq data of RNAlow, G1 and S/G2/M cells. Heatmap scale, fold change of log2 normalized expression compared to S/G2/M.
-
C, DExpression of cell cycle genes in scRNAseq of 96 RNAlow neoblasts (C) or 96 previously sequenced X1 neoblasts (D, data ref: Molinaro & Pearson, 2016). Heatmap scale, log2 normalized expression level.
Data information: In (B–D), the cell cycle phase in which each gene primarily functions is indicated by the colour bar along the left side of each heatmap.
We defined three other gates using this FACS strategy. Above the RNAlow gate was a population of cells with higher RNA and 2C DNA content. This gate was partially depleted by 1 dpi and largely ablated by 7 dpi (Fig 2A), leading us to hypothesize that it may contain a mix of G1 neoblasts and differentiating post‐mitotic progeny. These cells populated the region connecting the X1 (S/G2/M) and X2 (Hoechst SP) gates, with spill‐over into both gates, and were also found in the X‐insensitive (Xins) gate (Fig EV1A). As expected, cells with >2C DNA content by Hoechst fluorescence (the S/G2/M gate, classically the X1 gate, Fig EV1A), were depleted by 1 dpi (Fig 2A). Lastly, an irradiation‐insensitive population of cells was also resolved using our staining strategy, which we refer to as the post‐mitotic (PM) gate (Fig 2A). These cells largely populated the Xins gate (Fig EV1A). Notably, due to the relatively small size of neoblasts, separation of the cell cycle gates by pyronin Y fluorescence required high laser power on the FACS instrument; thus, many other cell types (with higher RNA content) may not be represented on our FACS plots.
To ensure that the RNAlow cells were not simply immediate neoblast progeny, which are also depleted soon after irradiation, we blocked differentiation via RNAi knockdown of mex3‐1, a gene known to be required for differentiation of all neoblast progeny (Zhu et al, 2015). The RNAlow gate was unaffected (Appendix Fig S1A); however, we observed a ~ 1.7‐fold decrease in the number of cells in the G1 gate, suggesting contribution from differentiating cells in this gate. Additionally, bulk RNAseq data revealed enriched expression of the prog gene family in the G1 gate (Appendix Fig S1B). Prog genes mark early post‐mitotic progenitors of the epithelial lineage (van Wolfswinkel et al, 2014; Zhu et al, 2015), supporting our hypothesis that this gate contains both G1 neoblasts and early neoblast progeny.
RNAlow neoblasts are relatively small and slow‐cycling
We proceeded to morphologically characterize cells from these newly defined neoblast FACS gates. By sorting cells from the RNAlow, G1 and S/G2/M gates and staining them with CellMask (cytoplasmic dye) and DAPI (nuclear dye), we found that RNAlow cells were significantly smaller and had a significantly higher nucleus:cytoplasm ratio than G1 or S/G2/M cells (Fig 2B). This was an intriguing finding, as these physical characteristics are reminiscent of previously described planarian pluripotent cNeoblasts (Wagner et al, 2011) and are common of quiescent stem cells in other systems (Rumman et al, 2015). In light of this, we next sought to determine whether the RNAlow population was relatively slow‐cycling compared to other neoblasts. We performed an EdU label‐retention assay, in which we collected cells from the RNAlow and S/G2/M gates after various chase periods and determined the percentage of cells in each gate that retained detectable EdU levels. Due to the contamination with post‐mitotic progeny cells, the G1 gate was excluded from this analysis. We observed that significantly more cells from the S/G2/M gate initially incorporated EdU than cells from the RNAlow gate (Fig 2C and D). As expected, actively cycling S/G2/M cells rapidly diluted the label, decreasing from 50 ± 15% EdU+ at 2 dpp to 4.9 ± 3.4% EdU+ by 14 dpp (Fig 2C). We did not observe significant label dilution over this same time period in RNAlow cells (17 ± 8.4% EdU+ at 2 dpp, 15 ± 7.0% EdU+ at 14 dpp, Fig 2C), indicating that RNAlow neoblasts divide less frequently than the S/G2/M population.
As a second approach for assaying proliferation status we examined the expression of cell cycle genes by RNA‐sequencing (RNAseq). Bulk RNAseq of the RNAlow, G1 and S/G2/M gates revealed that RNAlow cells generally had the lowest overall expression of cell cycle genes, with the notable exception of ccng2, which is a negative regulator of cell cycle progression (Horne et al, 1997) (Fig EV1B). As expected, expression of most cell cycle genes was higher in the G1 gate and highest in the S/G2/M gate. Consistent with this, the expression of cell cycle genes was very low in scRNAseq data of RNAlow cells (Fig EV1C), but readily detectable in previously published scRNAseq data of X1 cells (Fig EV1D, X1 Data ref: Molinaro & Pearson, 2016). Interestingly, we found that cells from the RNAlow gate exhibited enriched expression of several genes with previously described roles in maintaining stem cell quiescence (Cheung & Rando, 2013), especially the tumour suppressor Lrig‐1, which is involved in maintaining quiescence in a variety of systems, including murine intestinal stem cells and human epidermal stem cells (Table 1) (Jensen & Watt, 2006; Powell et al, 2012). Taken together, from these data we have identified a novel neoblast population with a set of definable characteristics, including low RNA content, small size, and relatively slow cell cycle dynamics.
Table 1.
Relative expression levels of previously described genes with roles in maintaining quiescent stem cells.
| Function | Gene | FC over G1 | FC over S/G2/M | FC over PM |
|---|---|---|---|---|
| Chromatin regulation | smarca2 | 0.81 | 1.55 | 0.87 |
| Regulation of transcription | foxo3 | 0.94 | 2.01 | 1.37 |
| ezh1 | 1.20 | 1.51 | 2.20 | |
| ctdsp1 | 1.14 | 1.90 | 0.77 | |
| thra | 1.75 | 5.28 | 2.90 | |
| tef | 0.62 | 2.35 | 0.83 | |
| RNA processing | dicer | 0.97 | 1.35 | 1.94 |
| Metabolism | lkb1 | 0.93 | 1.58 | 1.36 |
| atg7 | 0.96 | 2.48 | 1.32 | |
| Tumour suppression | p53 | 0.32 | 0.39 | 0.17 |
| rb | 1.15 | 1.35 | 1.83 | |
| Lrig1 | 1.79 | 8.98 | 2.46 |
Expression is reported as the fold change (FC) gene expression from bulk RNAseq data of cells from the RNAlow gate compared to the G1, S/G2/M or PM gates. Bold numbers, FC > 1.5.
RNAlow neoblasts undergo injury‐induced growth
We proceeded to characterize the response of these RNAlow neoblasts to injury. In other systems, slow‐cycling stem cells become activated following injury, and this is commonly characterized by cell growth, which acts to trigger cell cycle entry (Conlon & Raff, 1999; Gan & DePinho, 2009). To determine whether RNAlow neoblasts undergo a morphological injury response, we amputated animals and measured cell size at 6 hpa and 48 hpa, which correspond to the injury‐induced peaks of mitosis (Wenemoser & Reddien, 2010). Compared to intact (uninjured) controls, RNAlow neoblasts exhibited a 12% increase in cell area at 6 hpa, and a 22% increase by 48 hpa by CellMask staining (Fig 3A). Automated cell size measurements using a Coulter Counter demonstrated accompanying increases in cell diameter (3.5%) and volume (12%) (Appendix Fig S2). Interestingly, this growth response was unique to the RNAlow population, as G1 and S/G2/M cells did not grow following injury (Fig 3A). Furthermore, we found that RNAlow neoblast growth was transient at the population level, as we observed a return to homeostatic levels by 96 hpa (Fig 3B).
Figure 3. RNAlow neoblasts grow during regeneration.
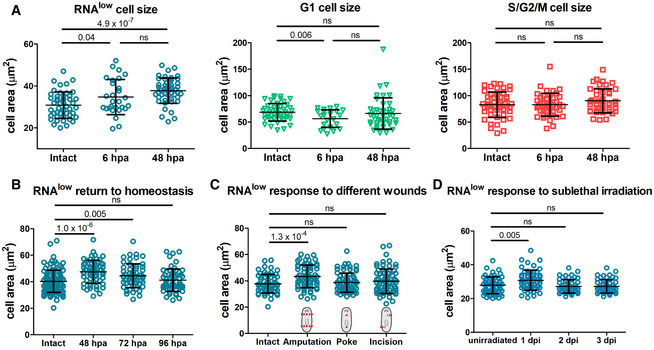
- Area measurements of cells from the RNAlow, G1 and S/G2/M gates in intact animals and at 6 and 48 hpa (n ≥ 28 for each sample in the RNAlow plot, n ≥ 24 in the G1 plot, n ≥ 42 in the S/G2/M plot).
- RNAlow neoblast size in intact animals and at 48, 72 and 96 hpa (n ≥ 54).
- RNAlow neoblast size in intact animals and at 48 h following amputation, poke or incision injuries (n ≥ 61).
- RNAlow neoblast size in unirradiated animals and at 1, 2 and 3 dpi (12.5 Gy. of irradiation, n ≥ 52).
Data information: Data are presented as mean ± s.d. Statistical significance was assessed using Welch's t‐tests (P values are indicated; ns, not significant [P > 0.05]). Data are representative of three (A) or two (B–D) independent experiments.
We next asked whether injury‐induced cell growth was specific to regeneration contexts by assaying cell size following non‐missing tissue injuries (which do not elicit a mitotic peak at 48 h post injury [hpi]). We did not observe an increase in the size of RNAlow neoblasts at 48 hpi following poke or incision wounds compared to uninjured controls (Fig 3C), suggesting missing tissue may be involved in triggering cell growth.
Because the injury response by RNAlow neoblasts was greatest at the time of the second mitotic peak, we considered whether the status of the neoblast compartment factored into the decision to respond. We used sublethal irradiation (12.5 Gy) to deplete the majority of the neoblast compartment and observed a transient increase in the average size of RNAlow neoblasts by 1 dpi followed by a return to homeostatic levels by 2 dpi (Fig 3D). Thus, the physical growth injury response by RNAlow neoblasts is expedited following specific depletion of the neoblast compartment. We also noticed a trend in which the RNAlow gate tended to deplete at a slower rate than the S/G2/M gate, while the G1 gate exhibited the least depletion; however, only the difference between the G1 and S/G2/M gates at 3 dpi was statistically significant (one‐way ANOVA: F(2,6) = 6.94, P = 0.027; post‐hoc Turkey HSD: P = 0.027; Fig EV2A and B).
Figure EV2. Gate depletion kinetics following sublethal irradiation.
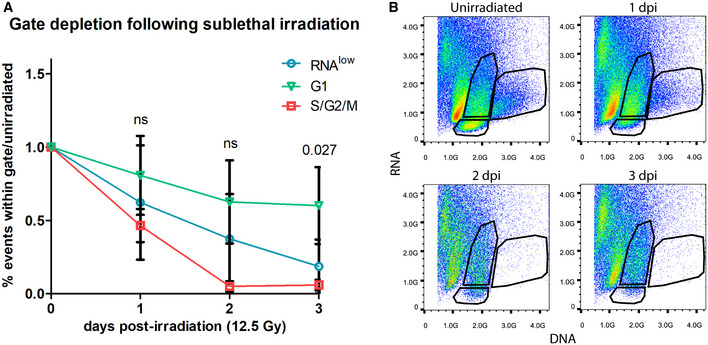
- Quantification of the proportion of events within the RNAlow, G1 or S/G2/M FACS gates in unirradiated controls and at 1–3 dpi (12.5 Gy of irradiation, n = 3 biological replicates). Data are normalized to unirradiated controls and presented as mean ± s.d. A one‐way ANOVA was conducted to compare the depletion of each gate at a given time point. Gate depletion was not significant at 1 dpi (F(2,6) = 1.12, P = 0.38) or 2 dpi (F(2,6) = 4.31, P = 0.069). Gate depletion was significant at 3 dpi (F(2,6) = 6.94, P = 0.027). A post‐hoc Turkey HSD test identified a significant difference between the G1 and S/G2/M gates at 3 dpi (P = 0.027).
- Representative FACS plots of the time course in (A). Plots are representative of n = 3 independent experiments.
RNAlow neoblasts extend projections in response to injury
While assaying cell size, we observed from CellMask staining that ~ 50% of cells from all three neoblast‐containing gates, at homeostasis, displayed projections, which are known physical features of cNeoblasts (Fig 4A) (Wagner et al, 2011). Interestingly, at 48 hpa, we observed a dramatic increase in the proportion of RNAlow neoblasts displaying projections (75.6 ± 1.6%) compared to intact controls (48.0 ± 1.7%) (Fig 4A). This corresponded with a significant increase in the average projection length from 4.6 ± 2.5 to 9.3 ± 7.0 µm (Fig 4B). Furthermore, the average number of projections and the sum total projection length per RNAlow neoblast were significantly increased by 48 hpa, but not at 6 hpa (Fig 4C). These phenotypes were not observed for G1 or S/G2/M cells (Fig 4B, D and E). Thus, taken together, the morphological response of RNAlow neoblasts to amputation involves cell growth and the extension of projections.
Figure 4. RNAlow neoblasts extend projections following amputation.
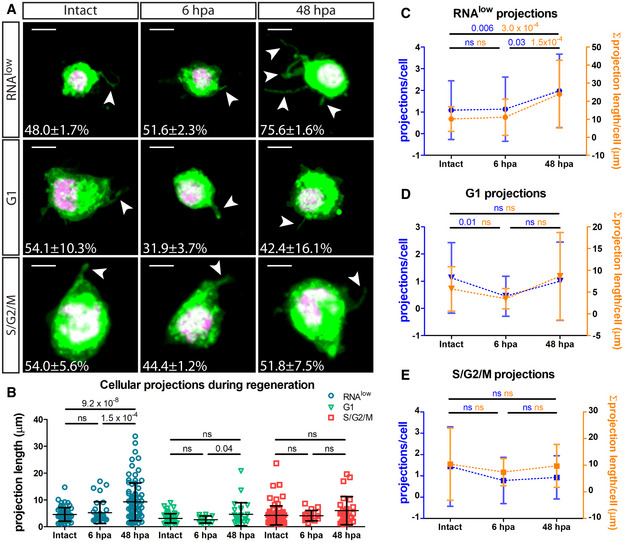
-
ARepresentative images of RNAlow, G1 and S/G2/M cells stained with CellMask and DAPI in intact animals and at 6 and 48 hpa. All images shown are from samples processed in the same experiment. Intact images are duplicated from Fig 2B. White arrows mark projections. Percentage of cells displaying projections in each condition is noted (n > 100 cells counted). Scale bars, 5 µm.
-
BAverage projection length in RNAlow, G1 and S/G2/M cells from intact animals and at 6 and 48 hpa (n ≥ 13).
-
C–EQuantification of the number of projections per cell (blue data) and the sum total projection length per cell (orange data) in RNAlow (C), G1 (D) or S/G2/M (E) cells isolated from intact animals and at 6 and 48 hpa (n ≥ 23 for projections/cell, n ≥ 10 for Σ projection length/cell).
Data information: Data are presented as mean ± s.d. Statistical significance was assessed using Welch's t‐tests (P values are indicated; ns, not significant [P > 0.05]). Data are representative of two independent experiments.
RNAlow neoblasts express markers of pluripotency and multiple cell lineages
We next examined RNAlow neoblast lineage heterogeneity using our scRNAseq dataset. We examined expression levels of different early lineage markers and observed expression of epithelial, gut, and neural progenitor markers (van Wolfswinkel et al, 2014; Molinaro & Pearson, 2016) within RNAlow neoblasts; however, clustering the cells by these genes did not yield clear lineage‐defined clusters (Fig EV3A). This contrasted with a previously published dataset of 96 single X1 cells, in which epithelial and neural clusters could be resolved (Fig EV3B, Data ref: Molinaro & Pearson, 2016). Indeed, expression of diverging lineage markers was often detected within individual RNAlow neoblasts, suggesting that some of these cells may not be specified to any one lineage.
Figure EV3. The RNAlow gate likely contains a mix of lineage‐committed and pluripotent neoblasts.
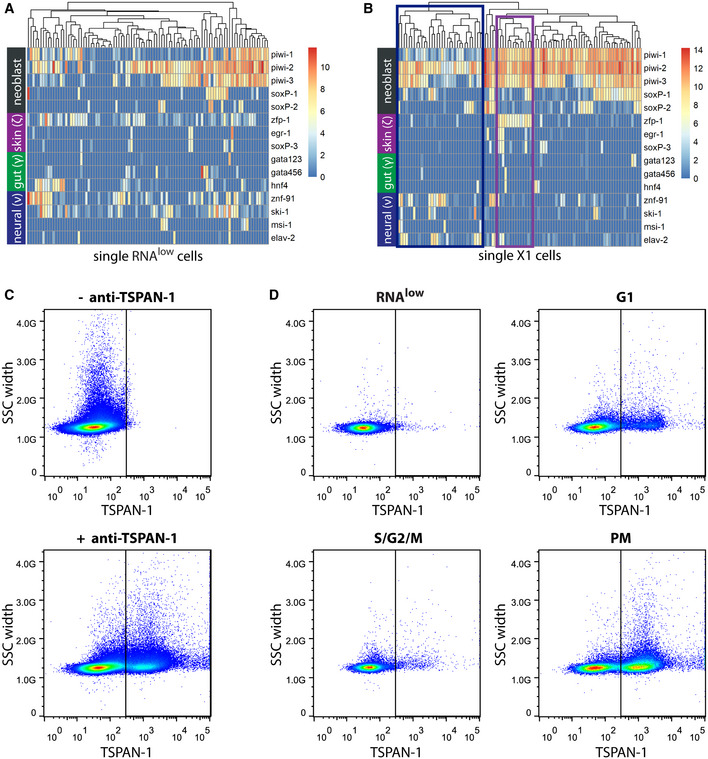
-
A, BExpression levels of general and lineage‐committed neoblast markers in scRNAseq data of cells from the RNAlow gate (A) or previously sequenced X1 neoblasts (B, data ref: Molinaro & Pearson, 2016).Heatmap scale, log2 normalized expression level. Blue box, putative neural progenitor cluster; purple box, putative epithelial progenitor cluster.
-
CFACS plots showing TSPAN‐1 immunostaining of whole dissociated animals without (top) or with (bottom) anti‐TSPAN‐1 antibody.
-
DFACS plots showing TSPAN‐1 immunostaining of cells from the RNAlow, G1, S/G2/M, or PM gates.
Data information: In (C and D), plots are representative of n = 4 independent experiments. SSC, side scatter.
We next hypothesized that the RNAlow neoblast population might include pluripotent neoblasts. In a recent study on neoblast heterogeneity by Zeng et al (2018), scRNAseq of X1 neoblasts was used to discover a putative marker for pluripotent neoblasts, tspan‐1. Using an antibody against TSPAN‐1, the authors showed that transplantation of a single TSPAN‐1+ cell could rescue lethally irradiated animals 25% of the time, demonstrating their pluripotency. We performed immunostaining using an anti‐TSPAN‐1 antibody (a kind gift from Dr. Alejandro Sánchez‐Alvarado) on dissociated cells and subsequently stained them with Hoechst and pyronin Y. We observed clear TSPAN‐1− and TSPAN‐1+ populations (21.4 ± 2.7% TSPAN‐1+, Fig EV3C). When we considered the neoblast gates independently, we found that a subset of cells in each gate expressed TSPAN‐1: 1.4 ± 0.7% of RNAlow, 9.3 ± 1.8% of G1 and 6.1 ± 2.6% of S/G2/M cells were TSPAN‐1+ (Fig EV3D). Interestingly, we observed high expression of TSPAN‐1 in the PM gate (35.0 ± 10.6% TSPAN‐1+). This is consistent with tspan‐1 expression in a previously published whole planarian scRNAseq cell atlas (Appendix Fig S3, Data ref: Fincher et al, 2018) and may, in part, account for the reported 75% of TSPAN‐1+ cells that were unable to rescue lethally irradiated animals following transplantation.
RNAlow neoblasts enter mitosis in a TOR‐dependent manner
We next sought to characterize a functional output of this morphological injury response. Although RNAlow neoblasts largely do not express proliferation genes at homeostasis, their rapid growth response following amputation prompted us to hypothesize that they may proliferate in this context. Despite many attempts, the cytotoxicity of pyronin Y precluded transplantation of RNAlow cells; therefore, we assayed cell cycle entry by inducing mitotic arrest with the M‐phase blocker nocodazole (NDZ) (van Wolfswinkel et al, 2014). From this experiment, we hypothesized that following injury RNAlow neoblasts would enter the cell cycle and become arrested in mitosis (i.e. with > 2C DNA content), resulting in depletion of the RNAlow gate and a concurrent increase in the S/G2/M gate (Fig 5A). By immunostaining, we observed a robust increase in the number of mitotic (H3P+) cells following 1 day of NDZ treatment, as expected (Appendix Fig S4A). Extended NDZ exposure resulted in increased apoptosis as measured by TUNEL staining, which we predicted reflects neoblast death following prolonged cell cycle arrest (Appendix Fig S4B); therefore, NDZ treatments were limited to 1 day for all subsequent experiments. When assayed by flow cytometry of NDZ‐treated intact and regenerating animals, we consistently observed an approximately 50% reduction in the RNAlow gate at 48 hpa compared to uninjured controls (Fig 5B). This was concurrent with a 2.8‐fold increase in the proportion of S/G2/M cells, supporting our hypothesis that RNAlow neoblasts enter the cell cycle during regeneration.
Figure 5. RNAlow neoblasts exhibit TOR‐dependent cell cycle entry during regeneration.
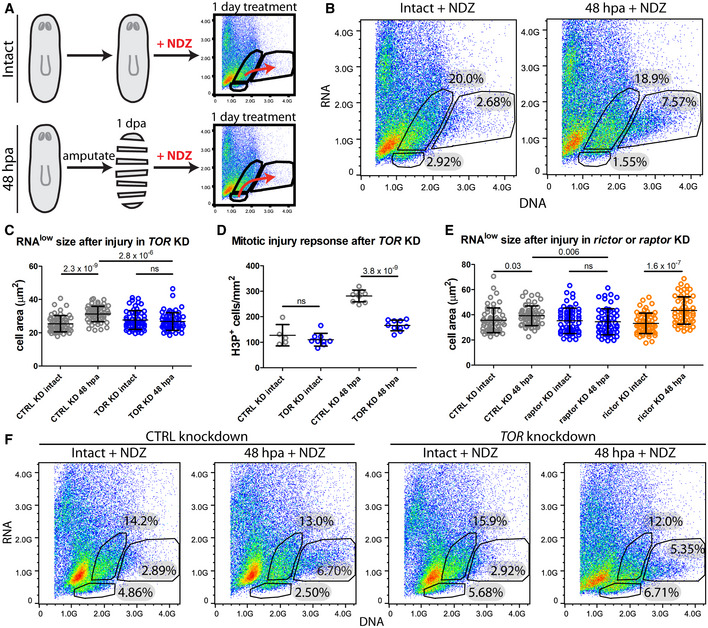
- Schematic of the experimental design.
- FACS plots of Hoechst and Pyronin Y stained cells isolated from intact and 48 hpa regenerating animals following 1 day of NDZ treatment.
- RNAlow neoblast size in intact animals and at 48 hpa in control or TOR KD animals (n ≥ 55).
- Quantification of H3P immunostaining in intact animals and at 48 hpa in control or TOR KD animals (n = 5–10).
- RNAlow neoblast size in intact animals and at 48 hpa in control, raptor or rictor KD animals (n ≥ 54).
- FACS plots of Hoechst and Pyronin Y stained cells isolated from intact and regenerating animals following 1 day of nocodazole treatment in control or TOR KD animals. Percentages indicate the proportion of events within each gate.
Data information: Data in (C–E) are presented as mean ± s.d. Statistical significance was assessed using Welch's t‐tests (P values are indicated; ns, not significant [P > 0.05]). In (C and D), animals were amputated at 5fd3. In (E), Animals were amputated at 7fd3. Data are representative of three (B) or two (C–F) independent experiments.
We next sought to identify molecular factors regulating the injury‐induced cell cycle entry of RNAlow neoblasts. Because robust cell growth occurs by 48 hpa, we tested whether genes required for the second mitotic peak, such as follistatin (fst) (Tewari et al, 2018) or epidermal growth factor receptor 3 (egfr‐3) (Lei et al, 2016), might function in this response. However, RNAi knockdown of either gene failed to impair the growth response of RNAlow neoblasts at 48 hpa (Fig EV4A and B).
Figure EV4. fst and egfr‐3 are not required for the RNAlow neoblast growth response to injury.
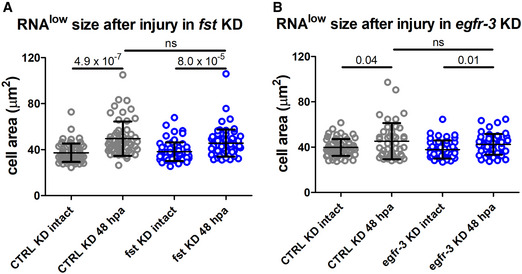
- RNAlow neoblast size in intact animals and at 48‐hpa in control or fst KD animals (n ≥ 56).
- RNAlow neoblast size in intact animals and at 48‐hpa in control or egfr‐3 KD animals (n ≥ 43).
Data information: Data presented as mean ± s.d. Statistical significance was assessed using Welch's t‐tests (P values are indicated; ns, not significant [P > 0.05]). In (A), animals were amputated at 4fd3. In (B), animals were amputated at 8fd3. The data are representative of 3 (A) or 2 (B) independent experiments.
Another gene implicated in the planarian regenerative response is the homolog of the mammalian target of rapamycin (TOR). TOR is a highly conserved regulator of cell growth (Laplante & Sabatini, 2009) and has been implicated in the activation of reserved stem cells in a variety of contexts, including in mouse muscle stem cells (MuSCs) and hematopoietic stem cells (HSCs) (Gan & DePinho, 2009; Rodgers et al, 2014). Previous work in planarians demonstrated that RNAi knockdown of TOR abolished the proliferative response following amputation, resulting in a complete failure to regenerate, and slowed neoblast ageing (by measure of telomere length) in homeostatic animals (Wenemoser & Reddien, 2010; González‐Estévez et al, 2012; Peiris et al, 2012; Tu et al, 2012; Iglesias et al, 2019). We asked whether TOR might exert its function at the level of RNAlow neoblasts. We amputated TOR(RNAi) animals and assayed cell growth at 48 hpa. Whereas regenerating control(RNAi) animals exhibited significant cell growth compared to uninjured controls, the size of RNAlow neoblasts remained unchanged in regenerating TOR(RNAi) animals (Fig 5C). Consistent with previous reports, this corresponded with a significantly reduced mitotic response as measured by H3P immunostaining (Fig 5D), hinting that a TOR‐dependent RNAlow neoblast response may be at least partially responsible for injury‐induced hyperproliferation.
Mechanistically, TOR exerts its functions as the catalytic subunit of two distinct complexes, TORC1 and TORC2, via interaction with the Raptor and Rictor accessory proteins, respectively (Wullschleger et al, 2006). Previous work in planarians has shown that the role of TOR in regeneration is carried out specifically through TORC1 (Tu et al, 2012; Peiris et al, 2012). Indeed, we observed significant growth of RNAlow neoblasts in both control(RNAi) and rictor(RNAi) animals at 48 hpa, but not in raptor(RNAi) animals (Fig 5E). Therefore, we conclude that TORC1, but not TORC2, is required for the RNAlow neoblast injury‐induced cell growth response following amputation.
To determine whether TOR‐mediated cell growth was required for cell cycle entry, we treated intact and 48‐hpa control(RNAi) and TOR(RNAi) animals with NDZ. In contrast to control(RNAi) animals, we observed that the RNAlow gate from TOR(RNAi) regenerating animals was not reduced following NDZ treatment, indicating that RNAlow neoblasts failed to enter the cell cycle and leave the gate by 48 hpa (Fig 5F). Importantly, previous work has demonstrated that while TOR signalling is implicated in apoptosis during homeostasis and remodelling in planarians, TOR(RNAi) animals exhibit normal cell death during the early stages of regeneration (Tu et al, 2012). Thus, the lack of RNAlow gate depletion in regenerating TOR(RNAi) animals also offers strong support that RNAlow gate depletion is not a result of NDZ‐induced apoptosis. Notably, we still observed an increase in the proportion of cells in the S/G2/M gate in regenerating TOR(RNAi) animals, which likely reflects the continued contribution from G1 neoblasts as this gate decreases in all NDZ conditions by 48 hpa. Taken together, these data support our hypothesis that TOR signalling through TORC1 is a conserved requirement for the injury response of cells in the RNAlow state, promoting the proliferation of RNAlow neoblasts during planarian regeneration.
Lrig‐1 is required to restrict RNAlow neoblast growth at homeostasis
As mentioned above, both ccng2 and Lrig‐1 exhibited enriched expression in cells from the RNAlow gate by RNAseq. Whole‐mount in situ hybridization (WISH) revealed broad expression patterns for both genes, including expression in the parenchyma and central nervous system (Fig 6A and Appendix Fig S5A). From a published scRNAseq atlas of all planarian tissues, approximately 43% of Lrig‐1 + cells and 56% of ccng2 + cells also expressed piwi‐1, indicating expression within neoblasts (Fincher et al, 2018).
Figure 6. Lrig‐1 is required for sustained regeneration.
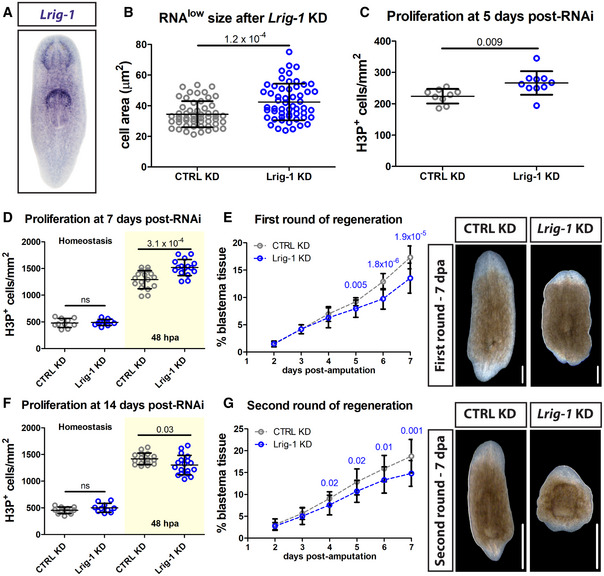
- Representative WISH images for Lrig‐1.
- RNAlow neoblast size in intact animals following control or Lrig‐1 knockdown (n ≥ 53 technical replicates).
- Quantification of H3P immunostaining in intact animals following control or Lrig‐1 knockdown (n = 9–10 technical replicates).
- Quantification of H3P immunostaining at homeostasis (fixed at 7 days post‐RNAi, white region) and at 48 hpa (amputated at 5 days post‐RNAi and fixed at 7 days post‐RNAi, yellow region) during the first round of regeneration in control or Lrig‐1 KD animals (n = 10–17).
- Quantification of unpigmented blastema area (as % total tissue area) in regenerating control or Lrig‐1 KD animals from 2 to 7 dpa during the first round of regeneration (n = 10 animals per condition per time point). The same animals were used for all time points. Representative images from 7 dpa are shown. Scale bars, 500 µm.
- Quantification of H3P immunostaining at homeostasis (fixed at 14 days post‐RNAi, white region) and at 48 hpa (amputated at 12 days post‐RNAi and fixed at 14 days post‐RNAi, yellow region) during a second round of regeneration in control or Lrig‐1 KD animals (n = 10–18).
- Quantification of unpigmented blastema area (as % total tissue area) in regenerating control or Lrig‐1 KD animals from 2 to 7 dpa during the second round of regeneration (n = 9–10 animals per condition per time point). The same animals were used for all time points. Representative images from 7 dpa are shown. Scale bars, 500 µm.
Data information: Data in (B–G) are presented as mean ± s.d. Statistical significance was assessed using Welch's t‐tests (P values are indicated; ns, not significant [P > 0.05]). In (B and C), data were obtained at 5fd5. In (D–G), animals were initially amputated at 5fd5. Second amputations were performed at 7 dpa following the first amputation. Data are representative of three (B and C) or two (D‐G) independent experiments (with the exception of the homeostatic data in (D and F), for which a single experiment was performed). In (D–G), combined data from both independent experiments is shown.
We assayed RNAlow cell size as a proxy for cell cycle entry. At 5 days following the final RNAi feed (5 days post‐RNAi), RNAlow cell size was indistinguishable from controls in ccng2(RNAi) animals (Appendix Fig S5B); however, we observed a significant increase in RNAlow cell size following knockdown of Lrig‐1 at homeostasis (Fig 6B). Similarly, proliferation levels were normal in ccng2(RNAi) animals (Appendix Fig S5C), whereas hyperproliferation was observed in Lrig‐1(RNAi) animals at this time (Fig 6C). We did not observe an overt differentiation phenotype in either Lrig‐1(RNAi) or ccng2(RNAi) animals, as assessed by quantifying chat + neurons, piwi‐1+ neoblasts, and epithelial density via DAPI staining (Fig EV5A–C).
Figure EV5. No effect of Lrig‐1 knockdown on differentiation at homeostasis.
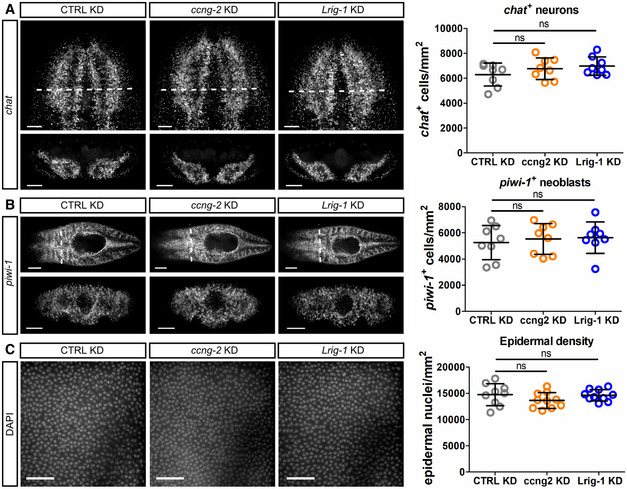
-
A–CRepresentative confocal images and quantifications of chat + neurons (A), piwi‐1 + neoblasts (B) and cell nuclei in the epidermis marked by DAPI (C) following control, ccng2, or Lrig‐1 knockdown at homeostasis (n = 8 technical replicates for all). Scale bars, 100 µm in (A); 250 µm in (B) (top panels) and 100 µm in (B) (bottom panels); 50 µm in (C).
Data information: Data presented as mean ± s.d. Statistical significance was assessed using Welch's t‐tests (ns, not significant [P > 0.05]). Animals were fixed at 5fd5. In (A and B), images are maximum projections and bottom panels are cross‐sections taken from the region indicated by the dashed white lines in the corresponding top panel. In (C), images are single confocal planes at the level of the epidermis. The data are representative of one independent experiment.
To assess the role of Lrig‐1 in regeneration, we amputated animals at 5 days post‐RNAi knockdown and assayed the mitotic injury response by H3P immunostaining at 48 hpa (i.e. 7 days post‐RNAi). Lrig‐1(RNAi) regenerating fragments exhibited significantly higher proliferation levels at 48 hpa compared to control animals (Fig 6D). Notably, although we observed homeostatic hyperproliferation at 5 days post‐RNAi, by 7 days homeostatic proliferation levels were normal in Lrig‐1(RNAi) animals, suggesting that the increased hyperproliferation compared to controls at 48 hpa was injury‐induced. This led us to hypothesize that Lrig‐1(RNAi) animals might exhibit enhanced epimorphic regeneration at early time points following injury. To test this, we quantified the proportion of unpigmented blastema tissue relative to the total size of the regenerating fragment for 7 days following head and tail amputation. Contrary to our hypothesis, Lrig‐1(RNAi) animals initially exhibited normal blastema production until 4 dpa, and subsequently produced significantly less blastema tissue (Fig 6E).
Because we observed reduced blastema formation only after 4 days of regeneration, we further challenged the same knockdown animals by amputating again at 7 days post the first amputation (12 days post‐RNAi). Interestingly, in contrast to the first round of regeneration, in this second round we observed a significantly diminished proliferative injury response in Lrig‐1(RNAi) animals compared to controls at 48 hpa (Fig 6F). Again, this phenotype was injury‐induced, as homeostatic animals at the corresponding time point (14 days post‐RNAi, no amputations) did not exhibit a hypoproliferation phenotype. During this second round of regeneration, Lrig‐1(RNAi) animals began exhibiting reduced blastema formation 1 day earlier, beginning at 3 dpa (Fig 6G). Thus, Lrig‐1 is required for maintaining small RNAlow neoblast size at homeostasis and for sustaining regeneration in planarians.
Discussion
In this study, we approached neoblast heterogeneity from a regulatory perspective to gain insight into the mechanisms of proliferation control that influence epimorphic regeneration. Upon discovering that a subset of neoblasts retain BrdU labelling for at least 5 weeks, we took a flow cytometry approach to study the cell cycle status of the neoblast population in greater detail. Using Hoechst and pyronin Y staining, we identified an RNAlow neoblast population with relatively low proliferative activity in homeostasis. This FACS strategy is commonly used in other systems to identify quiescent stem cells in the G0 phase of the cell cycle, which have characteristically low transcriptional output (Darzynkiewicz et al, 1980; Kim & Sederstrom, 2015; Eddaoudi et al, 2018). Thus, we were intrigued that these small RNAlow neoblasts exhibited many characteristics reminiscent of quiescent stem cells, such as a relatively slow division rate, similarities in morphology and gene expression profiles, and most notably a TORC1‐dependent proliferation response to injury. Additionally, we found that the homeostatic state of these cells is maintained at least in part by the conserved tumour suppressor Lrig‐1, which has known roles in maintaining quiescence in other systems (Jensen & Watt, 2006; Jensen et al, 2009; Powell et al, 2012; Simion et al, 2014; Nam & Capecchi, 2020). Because of these similarities, we suggest that these RNAlow neoblasts are in a “G0‐like” state, and we hypothesize that this state functions to maintain a reserved stem cell population in planarians (Fig 7). However, it is important to note that we cannot formally exclude the possibility that the RNAlow gate represents a subset of small G1 neoblasts.
Figure 7. Proposed model of RNAlow neoblast regulation.
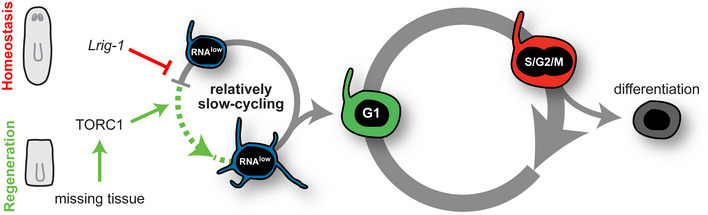
At homeostasis, RNAlow neoblasts are relatively slow‐cycling and their size is restricted by Lrig‐1. During regeneration, RNAlow neoblasts grow, extend projections, and enter the cell cycle by 48 hpa in a TORC1‐dependent manner.
Whereas many quiescent stem cell populations (such as LT‐HSCs) are highly radio‐resistant, we observed that RNAlow neoblasts are sensitive to high doses of irradiation. By using a sublethal dose of irradiation we observed that RNAlow neoblasts elicit a cell growth response very rapidly following this injury (within 1 day). Given the correlation between cell growth and cell cycle entry that we observed during regeneration, it is difficult to confidently assess the radio‐sensitivity of this population, as both cell death and cell cycle entry are possible explanations for the observed gate depletion. Depletion of the RNAlow gate tended to occur more slowly than depletion of the S/G2/M gate following sublethal irradiation; however, analysis of earlier time points will likely be required to identify any significant differences in depletion rates and to elucidate the underlying cause of the depletion.
In addition to cell growth, we noticed that RNAlow neoblasts extended cellular projections at 48 hpa. To our knowledge, this is not a common morphological phenotype associated with the activation of reserved stem cells, and at this stage the function of these projections is unknown. Interestingly, we also found that a small subset of RNAlow neoblasts expressed the putative pluripotent neoblast marker, TSPAN‐1, which has known roles in cell migration (Zeng et al, 2018). From the work of Zeng et al, transplanted TSPAN‐1+ cells reportedly did not become mitotically active until 2 days post transplantation, which aligns with the time at which we observed the increase in cellular projections. This is also consistent with the previously described localization of the 48 hpa mitotic peak to the wound site (Wenemoser & Reddien, 2010). Additionally, cellular projections extending from planarian neoblasts have been readily observed following partial irradiation experiments, in which neoblasts from a shielded region of the animal migrate into adjacent irradiated regions to repopulate the stem cell compartment and contribute to tissue maintenance (Abnave et al, 2017). Thus, we hypothesize that RNAlow neoblasts extend projections at 48 hpa to facilitate cell migration.
In this report we identified two regulators of RNAlow neoblasts. The first, TOR signalling, is required for the RNAlow neoblast injury response, as knockdown of TOR was sufficient to prevent cell growth and cell cycle entry at 48 hpa. In many vertebrate contexts activation from quiescence is mediated by TOR signalling through TORC1 (Gan & DePinho, 2009; Rodgers et al, 2014; Lee et al, 2018). In mouse muscle stem cells, fibro‐adipogenic progenitors, skeletal stem cells and hematopoietic stem cells, TOR signalling directs the “alerting” of these quiescent populations to injury, resulting in their growth and facilitating proliferation of those cells in the vicinity of the wound (Lee et al, 2018). Thus, in addition to its known regulatory functions on the neoblast compartment as a whole, we propose that TORC1 plays a similar role in planarians and promotes regeneration in part by facilitating the “activation” of RNAlow neoblasts. In a similar vein, we speculate that the delayed regeneration following knockdown of Lrig‐1 may result from a failure to reserve RNAlow neoblast activity for regeneration. At present, the homeostatic Lrig‐1 knockdown phenotype (RNAlow neoblast growth and hyperproliferation) and enriched expression in RNAlow neoblasts lend support to this hypothesis. Furthermore, we speculate that the augmented proliferative response observed in Lrig‐1(RNAi) animals upon an initial amputation may result from RNAlow neoblasts already being in an “alert” state, resulting in an increased contribution by the RNAlow neoblast population to the injury response. We also speculate that the diminished proliferative response observed in Lrig‐1(RNAi) animals following a second amputation challenge may be a consequence of a depleted reserved neoblast pool; however, future work will be required to directly test the contribution of RNAlow neoblasts to regeneration. Likewise, a detailed interrogation of cell cycle dynamics will be required to elucidate the precise division rates of different neoblast populations and to demonstrate whether and the degree to which RNAlow neoblasts are reserved for regeneration. Given the similarities with reserved stem cells observed so far, continuing to study the mechanisms underlying the regenerative response of RNAlow neoblasts offers an exciting new avenue for discovering conserved pathways involved in epimorphic regeneration.
Materials and Methods
In situ hybridization
Whole‐mount in situ hybridization was performed as previously described (Pearson et al, 2009; Currie et al, 2016). Briefly, animals were killed with 5% NAC solution followed by fixation in 4% formaldehyde solution for 20 min. Animals were then rinsed with PBST followed by a 2 min reduction step (50 mM DTT, 1% NP‐40, 0.5% SDS in PBS). Following PBST rinses and dehydration, animals were bleached overnight in 6% H2O2 in methanol. Bleached animals were hybridized with riboprobe overnight at 56°C, washed with 2×, 0.2× and 0.05× SSC (2 20 min washes each), blocked in 10% horse serum for 2 h and incubated with anti‐dig‐AP (Roche 16646820, 1:4,000) and/or anti‐FITC‐POD (Roche 28847220, 1:300) antibody overnight at 4°C. Probes were detected by FastBlue (Fast Blue B Salt [Sigma D9805] with naphthol AS‐MX phosphate [Sigma 855]) or tyramide amplification developments and subsequently rinsed in PBST and stored in antifade solution. Colorimetric stains (Fig 6A) were developed using 4‐nitro blue tetrazolium chloride (NBT, Roche 11383213001) with 5‐bromo‐4‐chloro‐3‐indolyl‐phosphate (BCIP, Roche 11383221001), post‐fixed for 10 min, washed for 5 min in ethanol, and stored in glycerol. All quantifications were done using the ImageJ software cell counter tool (http://rsb.info.nih.gov/ij/) with the exception of piwi‐1 cell counts in Fig 1 and the quantifications in Fig EV5, which were done with Imaris software (Bitplane, South Windsor, CT, USA). All image processing for figures was done in Adobe Photoshop.
FACS isolation of RNAlow neoblasts, TSPAN‐1 staining and irradiation
Approximately 30 animals per sample were rinsed with cold CMF + 1% BSA (CFMB). Animals were then thoroughly dounced with a sterile plastic pestle and strained through a 40 µm strainer in 3 ml of CMFB. Cells were stained with Hoechst 342 (25 µg/ml) for 20 min, then Pyronin Y (Sigma 213519) was added directly to the cell suspension to a final concentration of 50 µM and cells were left to incubate with the dyes for an additional 10 min. Cells were then centrifuged (300 g for 5 min, low brake) and resuspended in 500 µl of cold CMFB. Cells were sorted on a Beckman‐Coulter MoFlo XDP sorter. For all experiments, the RNAlow gate was set using samples from uninjured animals. Generally, the upper boundary of the RNAlow gate corresponded with the lower boundary of the S/G2/M gate. Additionally, lethally irradiated controls were used to confirm the boundary between the RNAlow gate (depleted by 1 dpi) and the G1 gate (largely still present at 1 dpi). Due to technical variability in sample preparation (including staining conditions and gate setting), in all cases control and experimental samples were prepared at the same time, and representative results from a single FACS run are shown. For TSPAN‐1 staining, cells were incubated for 1 h with anti‐TSPAN‐1, rinsed with cold CMFB and incubated with anti‐rabbit‐APC (Invitrogen A10931, 1:500) for 30 min. Hoechst & pyronin Y were added as above beginning in the last 30 min of secondary antibody incubation. For irradiation experiments, planarians were exposed to 60 Gy (lethal) or 12.5 Gy (sublethal) of γ‐irradiation from a 137Cs source. FACS plots were created and analysed using FlowJo software (https://www.flowjo.com/).
CellMask staining
Sorted cells were transferred in 200 µl CMFB to a 96 well Cellview microplate (Greiner Bio‐one, 655891) and allowed to settle. Cells were then fixed in 4% formaldehyde in PBS for 15 min, followed by a rinse in PBS. Cells were incubated in CellMask staining solution (1:500 in PBS) for 20 min, followed by 20 min in DAPI staining solution (1:1,000 in PBS). Cells were then kept indefinitely at 4°C in glycerol.
Label‐retention assays
Animals were gradually adapted to high salt medium (5 g/l Instant Ocean Aquarium Salt) for 2 days prior to BrdU or EdU administration. BrdU (Sigma B5002‐5G, 50 mg/ml in DMSO) was fed in liver paste at a concentration of 10 mg/ml and stained as previously described (Zhu & Pearson, 2018). Briefly, after the desired chase period animals were killed with 5% NAC solution followed by fixation in 4% formaldehyde solution for 20 min. Animals were then rinsed with PBST followed by a 2 min reduction step. Following PBST rinses and dehydration, animals were bleached overnight in 6% H2O2 in methanol. For samples undergoing FISH, probe incubation, washes and developments were performed at this point. Samples were then incubated in acid for 45 min (2N HCl, 0.5% Triton‐X), neutralized with 0.1 M sodium borate and blocked (0.5% Roche Western Blocking Reagent, 0.6% BSA, 5 mM thymidine, in PBST) overnight at 4°C. BrdU was detected with mouse anti‐BrdU (Millipore MAB3222, 1:300), followed by anti‐mouse HRP (Jackson Immunoresearch 115‐036‐006, 1:500) and tyramide amplification. Only worms with robust staining throughout the body were used for quantification.
EdU (Sigma T511293, 50 mg/ml in DMSO) was dissolved in water to a final concentration of 5 mg/ml and injected into the gut of animals. Injected animals were maintained in high salt water for the desired chase period, then fixed or processed for FACS sorting as described above. For whole‐mount staining, fixation was followed by a 10 min Proteinase K treatment (Invitrogen 25530‐049, 1:1,000), 10 min post‐fix, PBS rinse, 30 min click reaction (1 mM CuSO4, 0.01 mM azide‐fluor 488 (Click Chemistry Tools 1276‐1), 10 mM ascorbic acid), PBS rinse and 1 h incubation in DAPI staining solution. Samples were then kept indefinitely at 4°C in glycerol. EdU detection was performed first, followed by BrdU immunostaining as described above. For EdU detection on sorted cells, fixation was followed by a PBS rinse, 30 min click reaction, PBS rinse and 20 min incubation in DAPI staining solution. Samples were then kept indefinitely at 4°C in glycerol.
H3P immunolabelling and TUNEL staining
For H3P immunostaining, animals were killed with 5% NAC and fixed in Carnoy's solution for 2 h at 4°C. Animals were then bleached overnight in 6% H2O2 in methanol. Bleached animals were rehydrated and blocked for 2 h in 10% horse serum, then incubated with the rabbit monoclonal antibody to H3ser10p from Millipore (05‐817R‐1) at 1:1,000 overnight at 4°C. Animals were then washed for 2 h with PBST and incubated overnight with anti‐rabbit‐HRP antibody (Jackson Immunoresearch 111‐036‐047, 1:500). Samples were washed again for 2 h with PBST followed by a 10 min wash in borate buffer and a tyramide amplification development. TUNEL was performed as previously described (Pellettieri et al, 2010; Stubenhaus & Pellettieri, 2018). H3P and TUNEL stains were imaged on a Leica M165 fluorescent dissecting microscope and quantified using the ImageJ software cell counter tool (http://rsb.info.nih.gov/ij/).
Cell size measurements
For cell area measurements, cells stained with CellMask and DAPI were imaged on a Leica DMIRE2 inverted fluorescence microscope with a Hamamatsu Back‐Thinned EM‐CCD camera and spinning disc confocal scan head with a 60× objective using Volocity software. Area and projection measurements were taken on full projections using ImageJ software (http://rsb.info.nih.gov/ij/). Cell projections were excluded from area measurements. All image processing for figures was done in Adobe Photoshop. Automated cell diameter and volume measurements were obtained using the Multisizer 4 Coulter Counter (Beckman‐Coulter, Mississauga, ON).
RNA‐sequencing and analysis
Cells were stained and sorted as described above. Over 200k cells were collected for each bulk RNA‐sequencing sample. RNA extraction was performed using Trizol (Invitrogen 15596026) and library preparation was performed as previously described using Illumina TruSeq kits (Zhu & Pearson, 2018). Samples were sequenced to a depth of approximately 60 M single‐end 100‐base reads on an Illumina HiSeq2500. For single cell RNA‐sequencing, 96 cells from the RNAlow FACS gate were collected in strip tubes and single cell cDNA libraries were prepared using the Smartseq2 protocol as previously described (Molinaro & Pearson, 2016). Single‐cell samples were sequenced to a depth of > 3 M single‐end 75‐base reads on an Illumina NextSeq500. All samples were aligned to the transcriptome under NCBI BioProject PRJNA215411 using Bowtie2 with 15‐bp 3′ sequence trimming (Langmead & Salzberg, 2012). Aligned counts for bulk RNAseq and scRNAseq data are provided in Datasets EV1 and EV2. Bulk RNAseq data were converted to RPM for gene expression analyses.
RNA‐interference
HT115 bacteria transformed with the gene or control vector were grown in 2xYT at 37°C to an OD of 0.7–0.9 and then induced with 1 mM IPTG for 2 h and pelleted. Pelleted bacteria were mixed with calf liver paste and fed to starved planarians every 3 days for the indicated number of feeds, denoted as XfdY, where X is the total number of feeds and Y is the number of days after the last feed. For regeneration experiments, animals were amputated 3 days after the last feed. For FACS experiments, cells were sorted at least 3 days after the last RNAi feed.
Nocodazole treatment
Animals were soaked in nocodazole solution (1:2,500 nocodazole (Cell Signaling Technology 2190S, stock = 1 mg/ml in DMSO) + 1% DMSO in planarian water) for 20–24 h prior to FACS or staining.
Statistical analysis
All data are plotted as mean ± standard deviation (s.d.). Significance was calculated using a Welch's t‐test unless otherwise stated.
Animal husbandry
Asexual individuals of S. mediterranea CIW4 strain were reared as previously described (Newmark & Sánchez Alvarado, 2000).
Author contributions
AMM designed and performed most experiments and data analysis and wrote the manuscript. NLM contributed to experimental design, performed TUNEL experiments and EdU injections, and performed all revision experiments. NLM and BJP revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Review Process File
Source Data for Figure 1
Acknowledgements
AMM was supported by Canadian Institute for Health Research (CIHR) Frederick Banting and Charles Best Canada Graduate Scholarship (#GSD‐152379) as well as Natural Sciences and Engineering Research Council of Canada (NSERC) grant #RGPIN‐2016‐06354. NLM was supported by a student Restracomp award from the Hospital for Sick Children, as well as CIHR grant #PJT‐159611. BJP was supported by Ontario Institute for Cancer Research (OICR) grant #IA‐026. We would like to thank Dr. Alejandro Sánchez‐Alvarado for the gift of the anti‐TSPAN‐1 antibody. We would like to thank Drs. Ricardo Zayas, Labib Rouhana, and Jason Pellettieri for helpful comments on the manuscript. We would like to thank the Flow Cytometry core facility at the Hospital for Sick Children (https://lab.research.sickkids.ca/fcf/) for technical assistance to optimize the new FACS strategies in this manuscript.
EMBO reports (2021) 22: e50292
Data availability
Transcript IDs for all genes referenced in this manuscript are provided in Appendix Table S1.
The datasets produced in this study are available in the following databases:
-
‐
RNA‐seq data: Gene Expression Omnibus GSE157860 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157860)
References
- Abnave P, Aboukhatwa E, Kosaka N, Thompson J, Hill MA, Aboobaker AA (2017) Epithelial‐mesenchymal transition transcription factors control pluripotent adult stem cell migration in vivo in planarians. Development 144: 3440–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Rando TA (2013) Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Watt FM (2018) Defining adult stem cells by function, not by phenotype. Annu Rev Biochem 87: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Conlon I, Raff M (1999) Size control in animal development. Cell 96: 235–244 [DOI] [PubMed] [Google Scholar]
- Currie KW, Brown DDR, Zhu S, Xu C, Voisin V, Bader GD, Pearson BJ (2016) HOX gene complement and expression in the planarian Schmidtea mediterranea . Evodevo 7: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Traganos F, Melamed MR (1980) New cell cycle compartments identified by multiparameter flow cytometry. Cytometry 1: 98–108 [DOI] [PubMed] [Google Scholar]
- Derényi I, Szöllsi GJ (2017) Hierarchical tissue organization as a general mechanism to limit the accumulation of somatic mutations. Nat Commun 8: 14545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddaoudi A, Canning SL, Kato I (2018) Flow cytometric detection of g0 in live cells by Hoechst 33342 and Pyronin Y staining. In Cellular Quiescence, Lacorazza HD (ed.), pp 49–57. New York, NY: Humana Press; [DOI] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW (2018) Cell type transcriptome atlas for the planarian Schmidtea mediterranea . Science 360: 1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW (2018) Gene Expression OmnibusGene Expression Omnibus GSE111764 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111764) [DATASET]
- Gan B, DePinho RA (2009) mTORC1 signaling governs hematopoietic stem cell quiescence. Cell Cycle 8: 1003–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Estévez C, Felix DA, Smith MD, Paps J, Morley SJ, James V, Sharp TV, Aboobaker AA (2012) SMG‐1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet 8: e1002619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K (2006) Isolation of planarian X‐ray‐sensitive stem cells by fluorescence‐activated cell sorting. Dev Growth Differ 48: 371–380 [DOI] [PubMed] [Google Scholar]
- Horne MC, Donaldson KL, Goolsby GL, Tran D, Mulheisen M, Hell JW, Wahl AF (1997) Cyclin G2 is up‐regulated during growth inhibition and B cell antigen receptor‐mediated cell cycle arrest. J Biol Chem 272: 12650–12661 [DOI] [PubMed] [Google Scholar]
- Iglesias M, Felix DA, Gutiérrez‐Gutiérrez Ó, De Miguel‐Bonet M, Sahu S, Fernández‐Varas B, Perona R, Aboobaker AA, Flores I, González‐Estévez C (2019) Downregulation of mTOR signaling increases stem cell population telomere length during starvation of immortal planarians. Stem Cell Rep 13: 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iismaa SE, Kaidonis X, Nicks AM, Bogush N, Kikuchi K, Naqvi N, Harvey RP, Husain A, Graham RM (2018) Comparative regenerative mechanisms across different mammalian tissues. NPJ Regen Med 3: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM (2009) Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Watt FM (2006) Single‐cell expression profiling of human epidermal stem and transit‐amplifying cells: Lrig1 is a regular of stem cell quiescence. Proc Natl Acad Sci USA 103: 11958–11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Sederstrom JM (2015) Assaying cell cycle status using flow cytometry. Curr Protoc Mol Biol 2015: 28.6.1–28.6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped‐read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM (2009) mTOR signaling at a glance. J Cell Sci 122: 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Santo AIE, Zwingenberger S, Cai L, Vogl T, Feldmann M, Horwood NJ, Chan JK, Nanchahal J (2018) Fully reduced HMGB1 accelerates the regeneration of multiple tissues by transitioning stem cells to GAlert. Proc Natl Acad Sci USA 115: E4463–E4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Thi‐Kim VuH, Mohan RD, McKinney SA, Seidel CW, Alexander R, Gotting K, Workman JL, Sánchez Alvarado A (2016) Egf signaling directs neoblast repopulation by regulating asymmetric cell division in planarians. Dev Cell 38: 413–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Sicco C, Reverberi D, Villa F, Pfeffer U, Quarto R, Cancedda R, Tasso R (2018) Circulating healing (CH) cells expressing BST2 are functionally activated by the injury‐regulated systemic factor HGFA. Stem Cell Res Ther 9: 300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro AM, Pearson BJ (2016) In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol 17: 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro AM, Pearson BJ (2016) Gene Expression Omnibus GSE79866, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79866). [DATASET]
- Molinaro AM, Pearson BJ (2018) Myths vs. FACS: what do we know about planarian stem cell lineages? Int J Dev Biol 62: 527–535 [DOI] [PubMed] [Google Scholar]
- Morgan TH (1898) Experimental studies of the regeneration of Planaria maculata . Roux Arch Dev Biol 7: 364–397 [Google Scholar]
- Nagashima M, Barthel LK, Raymond PA (2013) A self‐renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N‐cadherin to regenerate retinal neurons. Development 140: 4510–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HS, Capecchi MR (2020) Lrig1 expression prospectively identifies stem cells in the ventricular‐subventricular zone that are neurogenic throughout adult life. Neural Dev 15: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A (2000) Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol 220: 142–153 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A (2002) Not your father’s planarian: a classic model enters the era of functional genomics. Nat Rev Genet 3: 210–219 [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR (2008) The PIWI proteins SMEDWI‐2 and SMEDWI‐3 are required for stem cell function and piRNA expression in planarians. RNA 14: 1174–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A (2009) Formaldehyde‐based whole‐mount in situ hybridization method for planarians. Dev Dyn 238: 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris TH, Weckerle F, Ozamoto E, Ramirez D, Davidian D, García‐Ojeda ME, Oviedo NJ (2012) TOR signaling regulates planarian stem cells and controls localized and organismal growth. J Cell Sci 125: 1657–1665 [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A (2010) Cell death and tissue remodeling in planarian regeneration. Dev Biol 338: 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE et al (2012) The pan‐ErbB negative regulator lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A (2005) SMEDWI‐2 is a PIWI‐like protein that regulates planarian stem cells. Science 310: 1327–1330 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR et al (2014) MTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature 510: 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumman M, Dhawan J, Kassem M (2015) Concise review: Quiescence in adult stem cells: biological significance and relevance to tissue regeneration. Stem Cells 33: 2903–2912 [DOI] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, Reddien PW (2014) Neoblast specialization in regeneration of the planarian Schmidtea mediterranea . Stem Cell Rep 3: 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simion C, Cedano‐Prieto ME, Sweeney C (2014) The LRIG family: enigmatic regulators of growth factor receptor signaling. Endocr Relat Cancer 21: R431–R443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenhaus B, Pellettieri J (2018) Detection of apoptotic cells in planarians by whole‐mount TUNEL. In Methods in Molecular Biology, Rink JC (ed.), pp 435–444. New York, NY: Humana Press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari AG, Stern SR, Oderberg IM, Reddien PW (2018) Cellular and molecular responses unique to major injury are dispensable for planarian regeneration. Cell Rep 25: 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ (2011) A reserve stem cell population in small intestine renders Lgr5‐positive cells dispensable. Nature 478: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Pearson BJ, Sánchez Alvarado A (2012) TORC1 is required to balance cell proliferation and cell death in planarians. Dev Biol 365: 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tümpel S, Rudolph KL (2019) Quiescence: good and bad of stem cell aging. Trends Cell Biol 29: 672–685 [DOI] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW (2011) Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332: 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW (2010) Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol 344: 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW (2014) Single‐cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15: 326–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Zeng A, Li H, Guo L, Gao X, McKinney S, Wang Y, Yu Z, Park J, Semerad C, Ross E et al (2018) Prospectively isolated tetraspanin+ neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173: 1593–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Hallows SE, Currie KW, Xu C, Pearson BJ (2015) A mex3 homolog is required for differentiation during planarian stem cell lineage development. Elife 4: e07025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Pearson BJ (2016) (Neo)blast from the past: new insights into planarian stem cell lineages. Curr Opin Genet Dev 40: 74–80 [DOI] [PubMed] [Google Scholar]
- Zhu SJ, Pearson BJ (2018) Smed‐myb‐1 specifies early temporal identity during planarian epidermal differentiation. Cell Rep 25: 38–46 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW (2018) Gene Expression OmnibusGene Expression Omnibus GSE111764 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111764) [DATASET]
- Molinaro AM, Pearson BJ (2016) Gene Expression Omnibus GSE79866, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79866). [DATASET]
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Review Process File
Source Data for Figure 1
Data Availability Statement
Transcript IDs for all genes referenced in this manuscript are provided in Appendix Table S1.
The datasets produced in this study are available in the following databases:
-
‐
RNA‐seq data: Gene Expression Omnibus GSE157860 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157860)


