Abstract
This study aimed to evaluate the feasibility of diagnosing periodontitis via the identification of 18 bacterial species in mouth-rinse samples. Patients (n = 110) who underwent dental examinations in the Department of Periodontology at the Veterans Health Service Medical Center between 2018 and 2019 were included. They were divided into healthy and periodontitis groups. The overall number of bacteria, and those of 18 specific bacteria, were determined via real-time polymerase chain reaction in 92 mouth-rinse samples. Differences between groups were evaluated through logistic regression after adjusting for sex, age, and smoking history. There was a significant difference in the prevalence (healthy vs. periodontitis group) of Aggregatibacter actinomycetemcomitans (2.9% vs. 13.5%), Treponema denticola (42.9% vs. 69.2%), and Prevotella nigrescens (80% vs. 2.7%). Levels of Treponema denticola, Prevotella nigrescens, and Streptococcus mitis were significantly associated with severe periodontitis. We demonstrated the feasibility of detecting periopathogenic bacteria in mouth-rinse samples obtained from patients with periodontitis. As we did not comprehensively assess all periopathogenic bacteria, further studies are required to assess the potential of oral-rinsing solutions to indicate oral infection risk and the need to improve oral hygiene, and to serve as a complementary method for periodontal disease diagnosis.
Keywords: polymerase chain reaction, periodontitis, bacteria
1. Introduction
Subgingival plaque bacteria are the main etiology of periodontitis. Complex interactions between certain pathogens are key in the development of periodontal disease [1]. Microbial complexes in the subgingival biofilm are classified into five groups: red, green, orange, yellow, and purple. In particular, the red group, which is composed of Tannerella forsythia, Treponema denticola, and Porphyromonas gingivalis, has been determined as one of the main causes of periodontal disease [1].
Several studies have shown that the presence and number of these bacteria are related to disease prediction criteria such as probing depth, bone loss, attachment loss, and bleeding on probing [2,3]. Various bacterial species besides those of the red complex have been found to be key in the development and progression of periodontitis; among these species, P. gingivalis, Prevotella intermedia, and Aggregatibacter actinomycetemcomitans have been shown to have the strongest association with periodontal disease [4]. A previous study showed that P. gingivalis, T. denticola, and A. actinomycetemcomitans, when present in saliva, contributed to pocket deepening [5]. In order to detect the bacteria associated with periodontal disease, plaque is usually collected from a specific tooth and analyzed [4,6]. Most studies have analyzed bacterial groups using plaque samples [7].
Multiplex real time-polymerase chain reaction (RT-PCR) allows RT measurement of amplified deoxyribonucleic acid (DNA) using a fluorescent substance. In general PCR, the final product is observed via agarose gel electrophoresis; therefore, accurate bacterial quantification is impossible. However, multiplex RT-PCR can be used to quantitatively analyze the product amplified per PCR cycle.
Mouth-rinsing solutions have been used in various sialochemistry studies [8,9]. Recently, some studies assessing the prevalence and levels of specific bacterial species have been conducted using PCR analysis of mouth-rinsing solutions [10]. However, very few studies have investigated the link between the diagnosis of periodontitis and the oral bacteria present in a mouth-rinsing solution. Additionally, the phosphate-buffered saline solution used in previous studies has been reported to cause discomfort.
The purpose of this study was to examine the correlation between periodontal disease and 18 different bacteria by conducting a RT-PCR analysis of mouth-rinsing solutions and to evaluate the usefulness of this diagnostic method.
2. Materials and Methods
2.1. Patient Selection
Patients who visited the Department of Periodontology at the Veterans Health Service Medical Center between 2018 and 2019 for various reasons underwent routine examination. Due to the lack of prior studies conducted with rinsing solutions, we decided to use this method to compare bacterial species prevalence and levels in healthy patients and patients with severe periodontal disease. After examination, 110 patients were selected to participate in the study.
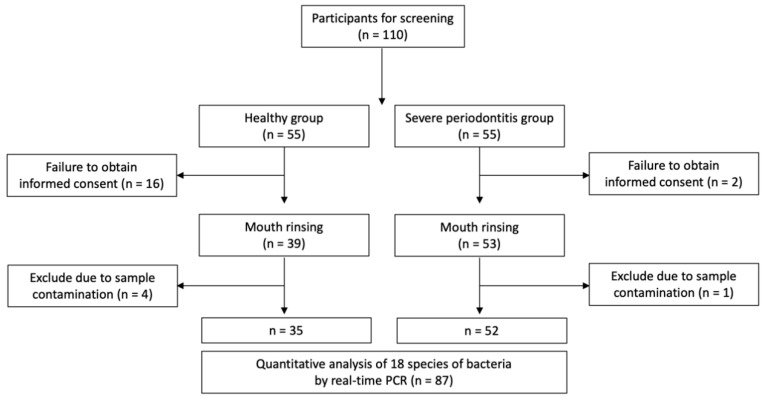
However, 18 patients refused to participate in the mouth-rinsing test. Hence, 92 patients were finally included in this study. Additionally, five subjects were excluded from the study because their mouth-rinsing solutions were contaminated in the process of transferring the collected samples (Figure 1). The study protocol was approved by the institutional review board of Veterans Health Service Medical Center (BOHUN No. 2018-03-002). All participants provided written informed consent. This study was conducted according to the Helsinki Declaration of 1975 and its later revisions.
Figure 1.
Study flow chart.
2.2. Sample Size Determination
The sample size was calculated using G*Power 3.1 software [11]. Comparisons between the two groups were conducted at a two-sided alpha level of 5% and a power of 90%. It was determined that a sample size of 42 participants per group would provide a power of 90% for the detection of between-group differences. However, considering a drop-out rate of 25%, a sample size of 55 patients per group was finalized.
2.3. Periodontal Examination
Each patient underwent an assessment of the probing depth and gingival recession at six sites per tooth using a periodontal probe (PCP-12, Hu-Friedy, Rotterdam, The Netherlands) by one examiner. Attachment loss was also measured.
After the dental examination, the presence and severity of periodontal disease were determined according to the Centers for Disease Control and Prevention/American Academy of Periodontology definitions [12]. We performed an additional examination using a mouth-rinse solution in both healthy patients and those with severe periodontal disease. Severe periodontitis was defined as two or more interproximal sites with a clinical attachment loss ≥ 6 mm, which are not the same area, and one or more interproximal sites with a probing depth ≥ 5 mm.
2.4. Sample Collection and DNA Extraction
Mouth-rinse samples were collected in the morning after regular brushing. Each subject rinsed their mouth with 10 mL of Easygen gargle (YD Global Life Science, Seongnam, Korea) for 60 s, after which the gargling liquid containing the patient’s saliva was collected as previously described [13]. DNA was extracted from the gargle sample using a Qiagen column (DNA Mini Kit, Qiagen, Hilden, Germany), according to the manufacturer’s instructions.
2.5. Multiplex Quantitative RT-PCR (qPCR)
The qPCR was performed with the EasyPerio molecular kit (YD Global Lifescience, Seongnam, Korea), according to the manufacturer’s instructions. The kit consisted of 8 different oligo mixes and 2 × master mixes. This was designed according to the typical multiplex qPCR method [14]. The CFX96 Touch™ RT-PCR Detection System (Bio-Rad, Hercules, CA, USA) was used for qPCR. The sequential steps in the PCR procedure were as follows: pre-denaturation for 30 s at 95 °C; 40 cycles of 5 s denaturation at 95 °C; and 30 s extension and annealing at 62 °C. Fluorescence scanning was performed after the extension and annealing step. Information on the primers and probes is displayed in Table 1. In this way, DNA of 18 species of bacteria was extracted and analyzed by RT-qPCR. The 18 species of bacteria were the following: A. actinomycetemcomitans, P. gingivalis, T. forsythia, T. denticola, Fusobacterium nucleatum, P. intermedia, Parvimonas micra, Campylobacter rectus, Eubacterium nodatum, Eikenella corrodens, Streptococcus mitis, Streptococcus mutans, Lactobacillus casei, Staphylococcus aureus, Enterococcus faecalis, Actinomyces viscosus, Prevotella nigrescens, and Streptococcus sobrinus.
Table 1.
Primers and probes of the 18 species of bacteria analyzed.
| Bacteria | Target Gene |
Primer/ Probe |
Sequence (5′-3′) | Ref. | Bacteria | Target Gene |
Primer/ Probe |
Sequence (5′-3′) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
|
Aggregatibacter
actinomycetemcomitans |
leukotoxin | Forward | CG**********GA | [15] |
Eubacterium
nodatum |
hypothetical protein | Forward | TG**********GA | [16] |
| Reverse | AT**********CA | Reverse | AA**********AT | ||||||
| Probe | [FAM]GG**********CC[BHQ1] | Probe | [TR]TT**********GG[BHQ2] | ||||||
|
Porphyromonas
gingivalis |
hemagglutinin | Forward | AC**********GC | [17] |
Eikenella
corrodens |
prolineiminopeptidase | Forward | GC**********TG | [16] |
| Reverse | GC**********CT | Reverse | GC**********TT | ||||||
| Probe | [HEX]CG**********GA[BHQ1] | Probe | [Cy5]AC**********AT[BHQ2] | ||||||
|
Tannerella
forsythia |
karilysin protease | Forward | TG**********CC | [18] |
Streptococcus
mitis |
16S ribosomal RNA | Forward | GT**********CG | [19] |
| Reverse | TT**********CA | Reverse | TA**********AT | ||||||
| Probe | [TR]CC**********GG[BHQ2] | Probe | [FAM]TA**********CC[BHQ1] | ||||||
|
Treponema
denticola |
OpdB | Forward | AG**********AG | [20] |
Streptococcus
mutans |
PTS EII | Forward | CA**********CA | [21] |
| Reverse | GC**********AT | Reverse | TG**********CC | ||||||
| Probe | [Cy5]CG**********TC[BHQ2] | Probe | [HEX]TG**********GG[BHQ1] | ||||||
|
Fusobacterium
nucleatum |
16S ribosomal RNA | Forward | GG**********TC | [22] |
Streptococcus
sobrinus |
Ftsk | Forward | GG**********CC | [23] |
| Reverse | CT**********GC | Reverse | AC**********GG | ||||||
| Probe | [FAM]AA**********CG[BHQ1] | Probe | [TR]AG**********GC[BHQ2] | ||||||
|
Prevotella
intermedia |
hemagglutinin | Forward | CA**********AC | [15] |
Lactobacillus
casei |
att | Forward | CA**********GT | [24] |
| Reverse | CA**********TC | Reverse | AC**********CC | ||||||
| Probe | [HEX]CC**********AC[BHQ1] | Probe | [Cy5]TG**********GT[BHQ2] | ||||||
|
Prevotella
nigrescens |
gyrase subunit B |
Forward | AG**********CT | [16] |
Staphylococcus
aureus |
clumping factor A |
Forward | GC**********AA | [25] |
| Reverse | GC**********CT | Reverse | GA**********TT | ||||||
| Probe | [TR]GC**********AA[BHQ2] | Probe | [FAM]TG**********CA[BHQ1] | ||||||
|
Parvimonas
micra |
16S ribosomal RNA | Forward | GA**********AG | [15] |
Enterococcus
faecalis |
gelE-sprE operon | Forward | GA**********TT | [26] |
| Reverse | GG**********CC | Reverse | CG**********AC | ||||||
| Probe | [FAM]GG**********CA[BHQ1] | Probe | [HEX]GC**********GA[BHQ1] | ||||||
|
Campylobacter
rectus |
GroEL | Forward | AA**********GG | [16] |
Actinomyces
viscosus |
nanH | Forward | GC**********CG | [21] |
| Reverse | TC**********GA | Reverse | GA**********CA | ||||||
| Probe | [HEX]GG**********GT[BHQ1] | Probe | [TR]GA**********AA[BHQ2] |
2.6. Bacterial Quantification
Standard curves were generated using the 18 plasmids at five different concentrations. The plasmids’ DNA contained specific sequences of each microorganism. Each bacterial gene used for plasmid construction is listed in Table 1. The copy numbers of each oral-bacterial DNA were calculated by substituting the cycle threshold values obtained from the qPCR into the quantitative formula obtained through the standard curve.
2.7. Statistical Analysis
This study evaluated whether there was a significant difference in the prevalence and levels of bacterial species between healthy individuals and those with periodontitis. Sex and smoking history were expressed as frequencies and percentages, and age, as means and standard deviations. The total number of bacteria was reported as median and interquartile range, and the number of each bacterial species was reported after normalization (dividing by the total number of bacteria in each sample). Differences in prevalence between groups were evaluated through logistic regression. Spearman’s rank correlation was used to examine the association between the levels of the different target species. Only two species that had at least five complete observations were estimated with the correlation coefficient. Logistic regression models were applied with disease status (healthy or with periodontal disease) as the dependent variable and the bacterial category as the independent variable. The bacterial category comprised three levels. Level 0 represented PCR-negative subjects, while levels 1 and 2 were categorized according to the median of the number of bacterial cells in PCR-positive subjects; levels 1 and 2 were assigned to values less than or greater than the median, respectively.
The Firth’s penalized maximum-likelihood bias-reduction method was used to estimate the odds ratio when there was a complete separation [27,28]. All regression analyses were adjusted for known confounders of periodontitis, including age, sex, and smoking history.
Statistical analyses were performed using R 3.5.1 (R Development Core Team; R Foundation for Statistical Computing, Vienna, Austria). p values < 0.05 were considered statistically significant.
3. Results
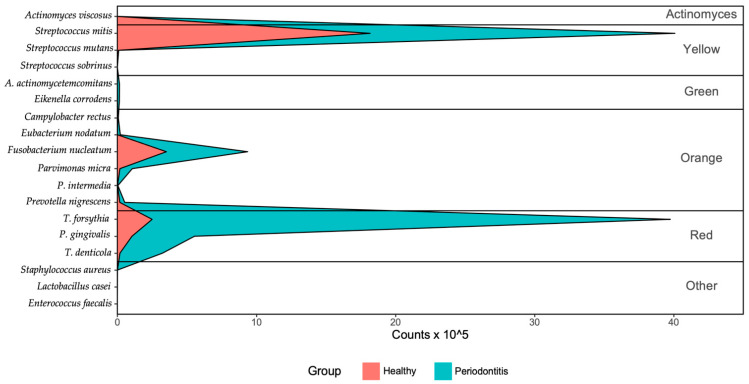
There were 35 individuals in the healthy group and 52 in the severe periodontitis group (Table 2). Figure 2 shows the mean counts of bacteria in the two groups. The number of bacteria of the red, yellow, and orange groups was higher in patients with periodontal disease than in the healthy group. The results of the quantitative analysis of the 18 species of bacteria are shown in Table 3. S. mitis, P. micra, and F. nucleatum were found in all subjects in the healthy group. P. nigrescens and C. rectus were found in 80% of the subjects in the healthy group. Among bacteria in the red complex group, P. gingivalis was found in 45.7%, T. forsythia in 74.3%, and T. denticola in 42.9% of the subjects in the healthy group. E. faecalis and A. viscosus were not detected in any of the healthy subjects. Similar to the healthy group, S. mitis, P. micra, and F. nucleatum were found in all subjects in the severe periodontitis group. P. gingivalis, P. nigrescens, T. forsythia, and T. denticola were detected in 90.4%, 82.7%, 73.1%, and 69.2% of individuals with severe periodontitis, respectively. After adjusting for age, sex, and smoking history, there were differences in the prevalence of A. actinomycetemcomitans, T. denticola, and P. nigrescens between the healthy group and severe periodontitis group. Among the red complex group bacteria, only T. denticola prevalence was significantly different between groups (Table 3).
Table 2.
Participant demographics.
| Characteristic | Healthy Group (n = 35) |
Severe Periodontitis Group (n = 52) | |
|---|---|---|---|
| Age (Years, mean ± SD) | 39.0 ± 17.9 | 56.2 ± 15.2 | |
| Sex | Male | 29 (83%) | 44 (85%) |
| Female | 6 (17%) | 8 (15%) | |
| Smoking | Non-smokers | 31 (89%) | 46 (88%) |
| Current smokers | 4 (11%) | 6 (12%) | |
Abbreviations: SD, Standard deviation.
Figure 2.
Mean bacterial cells in the healthy group and periodontal disease group.
Table 3.
Prevalence of target species and their quantities in polymerase chain reaction-positive subjects.
| Bacteria | Healthy Group (n = 35) | Severe Periodontitis Group (n = 52) |
|---|---|---|
| Aggregatibacter actinomycetemcomitans | ||
| Prevalence, n (%) a | 1 (2.9) | 7 (13.5) |
| Median bacterial cells proportion (%) (IQR) | 0.46 (0.46–0.46) | 0.75 (0.46–1.07) |
| Porphyromonas gingivalis | ||
| Prevalence, n (%) | 16 (45.7) | 47 (90.4) |
| Median bacterial cells proportion (%) (IQR) | 3.43 (1.74–5.86) | 3.83 (2.27–8.28) |
| Tannerella forsythia | ||
| Prevalence, n (%) | 26 (74.3) | 38 (73.1) |
| Median bacterial cells proportion (%) (IQR) | 3.07 (0.65–7.74) | 26.07 (4.49–50.82) |
| Treponema denticola | ||
| Prevalence, n (%) a | 15 (42.9) | 36 (69.2) |
| Median bacterial cells proportion (%) (IQR) | 0.59 (0.18–2.34) | 2.91 (1.36–5.39) |
| Fusobacterium nucleatum | ||
| Prevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells proportion (%) (IQR) | 18.73 (13.31–23.15) | 12.89 (7.23–20.02) |
| Prevotella intermedia | ||
| Prevalence, n (%) | 8 (22.9) | 15 (28.8) |
| Median bacterial cells proportion (%) (IQR) | 0.22 (0.05–0.46) | 0.11 (0.05–0.19) |
| Prevotella nigrescens | ||
| Prevalence, n (%) a | 28 (80.0) | 43 (82.7) |
| Median bacterial cells proportion (%) (IQR) | 0.73 (0.4–1.94) | 0.47 (0.14–1.23) |
| Parvimonas micra | ||
| Prevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells proportion (%) (IQR) | 0.5 (0.29–0.82) | 0.99 (0.45–1.81) |
| Campylobacter rectus | ||
| Prevalence, n (%) | 28 (80.0) | 45 (86.5) |
| Median bacterial cells proportion (%) (IQR) | 0.11 (0.06–0.18) | 0.08 (0.04–0.13) |
| Eubacterium nodatum | ||
| Prevalence, n (%) | 3 (8.6) | 14 (26.9) |
| Median bacterial cells proportion (%) (IQR) | 0.21 (0.12–0.27) | 0.71 (0.3–1.34) |
| Eikenella corrodens | ||
| Prevalence, n (%) | 4 (11.4) | 15 (28.8) |
| Median bacterial cells proportion (%) (IQR) | 0.07 (0.04–0.45) | 0.28 (0.15–0.71) |
| Streptococcus mitis | ||
| Prevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells proportion (%) (IQR) | 73.72 (63.61–79.49) | 59.13 (37.87–70.34) |
| Streptococcus mutans | ||
| Prevalence, n (%) | 23 (65.7) | 35 (67.3) |
| Median bacterial cells proportion (%) (IQR) | 0.03 (0.02–0.1) | 0.03 (0.01–0.15) |
| Streptococcus sobrinus | ||
| Prevalence, n (%) | 1 (2.9) | 5 (9.6) |
| Median bacterial cells proportion (%) (IQR) | 0.06 (0.06–0.06) | 0 (0–0.01) |
| Lactobacillus casei | ||
| Prevalence, n (%) | 6 (17.1) | 18 (34.6) |
| Median bacterial cells proportion (%) (IQR) | 0.01 (0–0.04) | 0 (0–0.01) |
| Staphylococcus aureus | ||
| Prevalence, n (%) | 15 (42.9) | 4 (7.7) |
| Median bacterial cells proportion (%) (IQR) | 0.02 (0.01–0.14) | 0.03 (0–0.07) |
| Enterococcus faecalis | ||
| Prevalence, n (%) | 0 (0.0) | 0 (0.0) |
| Median bacterial cells proportion (%) (IQR) | NA (NA–NA) | NA (NA–NA) |
| Actinomyces viscosus | ||
| Prevalence, n (%) | 0 (0.0) | 0 (0.0) |
| Median bacterial cells proportion (%) (IQR) | NA (NA–NA) | NA (NA–NA) |
| Total number of cellsPrevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells (IQR) | 36,126,518 (16,199,034–92,716,204) | 108524910 (69,243,624.5–177,393,988.25) |
Abbreviations: IQR, interquartile range; NA, not available. a Significant difference between groups at p < 0.05, analyzed using the logistic regression analysis.
Table 4 shows the correlations between the different bacterial species in all participants. Correlation coefficients ranged from −1 to 1, with numbers greater than 0 indicating positive correlations and numbers lower than 0 indicating negative correlations. A. actinomycetemcomitans and P. gingivalis showed a correlation of 0.96 and a p value lower than 0.05, indicating a significant positive correlation. P. gingivalis had a positive correlation with E. nodatum and a negative correlation with S. mitis. T. forsythia was negatively correlated with F. nucleatum, P. nigrescens, and S. mitis. F. nucleatum was positively correlated with P. nigrescens, S. mitis, and L. casei. P. intermedia was positively correlated with P. nigrescens and C. rectus.
Table 4.
Interspecies correlations in all subjects.
| Aa | Pg | Tf | Td | Fn | Pi | Pn | Pm | Cr | En | Ec | Sm | Smu | Ss | Lc | Sa | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa | 0.96 * | −0.49 | 0.3 | 0.38 | 0.98 * | 0.91 * | 0.57 | 0.07 | −0.26 | ||||||||
| Pg | −0.04 | 0.25 | −0.01 | −0.06 | 0.16 | 0.23 | −0.02 | 0.57 * | −0.3 | −0.34 * | 0.16 | 0.5 | −0.05 | −0.27 | −0.06 | ||
| Tf | 0.19 | −0.56 * | −0.21 | −0.28 * | 0.03 | −0.02 | 0.1 | −0.37 | −0.87 * | −0.22 | −0.54 | −0.2 | −0.21 | 0.14 | |||
| Td | −0.17 | 0.53 * | 0.05 | 0.61 * | −0.08 | 0.8 * | −0.24 | −0.42 * | −0.02 | −0.17 | 0.34 | −0.4 | −0.07 | ||||
| Fn | 0.21 | 0.25 * | −0.15 | 0.18 | −0.17 | 0.07 | 0.25 * | 0.02 | 0.9 * | 0.52 * | 0.02 | −0.13 | |||||
| Pi | 0.53 * | 0.14 | 0.51 * | 0.51 | 0.09 | −0.09 | 0.43 | 0.32 | |||||||||
| Pn | 0.15 | 0.23 | −0.16 | −0.13 | 0.07 | −0.18 | 0.75 | −0.03 | −0.32 | −0.21 | |||||||
| Pm | 0.18 | 0.13 | −0.27 | −0.23 * | 0 | 0.17 | −0.02 | −0.23 | −0.02 | ||||||||
| Cr | −0.31 | −0.24 | 0.03 | −0.15 | 0.88 * | 0.06 | −0.24 | −0.16 | |||||||||
| En | −0.13 | 0.35 | 0.3 | 0.42 | |||||||||||||
| Ec | 0.24 | 0.19 | −0.17 | −0.16 | |||||||||||||
| Sm | 0.12 | 0.33 | 0.04 | 0.23 | −0.03 | ||||||||||||
| Smu | −0.12 | −0.13 | −0.15 | −0.09 | |||||||||||||
| Ss | −0.62 | ||||||||||||||||
| Lc | −0.16 | ||||||||||||||||
| Sa | −0.21 | ||||||||||||||||
| Total |
Abbreviations: Aa, Aggregatibacter actinomycetemcomitans; Pg, Porphyromonas gingivalis; Tf, Tannerella forsythia; Td, Treponema denticola; Fn, Fusobacterium nucleatum; Pi, Prevotella intermedia; Pm, Parvimonas micra; Cr, Campylobacter rectus; En, Eubacterium nodatum; Ec, Eikenella corrodens; Sm, Streptococcus mitis; Smu, Streptococcus mutans; Lc, Lactobacillus casei; Sa, Staphylococcus aureus; Pn, Prevotella nigrescens; Ss, Streptococcus sobrinus; and * p < 0.05.
Table 5 shows the correlations between bacterial species in the healthy group. T. forsythia was positively correlated with C. rectus and negatively correlated with S. mitis. T. denticola was negatively correlated with S. mitis.
Table 5.
Interspecies correlations in healthy subjects.
| Aa | Pg | Tf | Td | Fn | Pi | Pn | Pm | Cr | En | Ec | Sm | Smu | Ss | Lc | Sa | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa | 0.96 * | −0.81 | 0.24 | 0.55 | 0.98 * | 0.92 * | 0.56 | 0.18 | −0.34 | ||||||||
| Pg | −0.03 | 0.22 | 0.02 | −0.04 | 0.12 | 0.22 | 0 | 0.57 * | −0.32 | −0.3 * | 0.14 | 0.71 | −0.05 | −0.07 | |||
| Tf | 0.11 | −0.67 * | −0.13 | −0.34 | −0.12 | −0.13 | 0.05 | −0.49 | −0.88 * | −0.29 | 0.11 | −0.31 | 0.05 | ||||
| Td | −0.12 | 0.55 | 0.07 | 0.62 * | −0.09 | 0.78 * | −0.34 | −0.33 * | −0.08 | −0.17 | 0.4 | −0.16 | |||||
| Fn | 0.26 | 0.33 * | −0.1 | 0.24 | −0.12 | 0.28 | 0.43 * | 0.38 * | 0.33 | 0.19 | 0.01 | ||||||
| Pi | 0.75 * | 0.21 | 0.56 * | 0.58 | 0.15 | −0.16 | 0.36 | ||||||||||
| Pn | 0.23 | 0.4 * | −0.22 | −0.14 | 0.15 | −0.15 | −0.26 | −0.17 | −0.23 | ||||||||
| Pm | 0.22 | −0.09 | −0.32 | −0.11 | −0.08 | −0.37 | 0.11 | −0.12 | |||||||||
| Cr | −0.32 | −0.22 | 0.12 | −0.18 | 0.95 * | 0.22 | −0.14 | ||||||||||
| En | −0.13 | 0.31 | 0.3 | 0.45 | |||||||||||||
| Ec | 0.2 | 0.18 | −0.23 | −0.24 | |||||||||||||
| Sm | 0.12 | −0.2 | 0.24 | 0.04 | |||||||||||||
| Smu | 0.01 | −0.23 | |||||||||||||||
| Ss | −0.49 | ||||||||||||||||
| Lc | −0.12 | ||||||||||||||||
| Sa | |||||||||||||||||
| Total |
Abbreviations: Aa, Aggregatibacter actinomycetemcomitans; Pg, Porphyromonas gingivalis; Tf, Tannerella forsythia; Td, Treponema denticola; Fn, Fusobacterium nucleatum; Pi, Prevotella intermedia; Pm, Parvimonas micra; Cr, C. Campylobacter rectus; En, Eubacterium nodatum; Ec, Eikenella corrodens; Sm, S. Streptococcus mitis; Smu, Streptococcus mutans; Lc, Lactobacillus casei; Sa, Staphylococcus aureus; Pn, Prevotella nigrescens; Ss, Streptococcus sobrinus; and * p < 0.05.
Table 6 shows the correlations between bacterial species in the periodontal disease group. P. gingivalis had a significant positive correlation with A. actinomycetemcomitans. F. nucleatum was negatively correlated with T. forsythia.
Table 6.
Interspecies correlations in subjects with severe periodontitis.
| Aa | Pg | Tf | Td | Fn | Pi | Pn | Pm | Cr | En | Ec | Sm | Smu | Ss | Lc | Sa | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa | 0.96 * | −0.81 | 0.24 | 0.55 | 0.98 * | 0.92 * | 0.56 | 0.18 | −0.34 | ||||||||
| Pg | −0.03 | 0.22 | 0.02 | −0.04 | 0.12 | 0.22 | 0 | 0.57 * | −0.32 | −0.3 * | 0.14 | 0.71 | −0.05 | −0.07 | |||
| Tf | 0.11 | −0.67 * | −0.13 | −0.34 | −0.12 | −0.13 | 0.05 | −0.49 | −0.88 * | −0.29 | 0.11 | −0.31 | 0.05 | ||||
| Td | −0.12 | 0.55 | 0.07 | 0.62 * | −0.09 | 0.78 * | −0.34 | −0.33 * | −0.08 | −0.17 | 0.4 | −0.16 | |||||
| Fn | 0.26 | 0.33 * | −0.1 | 0.24 | −0.12 | 0.28 | 0.43 * | 0.38 * | 0.33 | 0.19 | 0.01 | ||||||
| Pi | 0.75 * | 0.21 | 0.56 * | 0.58 | 0.15 | −0.16 | 0.36 | ||||||||||
| Pn | 0.23 | 0.4 * | −0.22 | −0.14 | 0.15 | −0.15 | −0.26 | −0.17 | −0.23 | ||||||||
| Pm | 0.22 | −0.09 | −0.32 | −0.11 | −0.08 | −0.37 | 0.11 | −0.12 | |||||||||
| Cr | −0.32 | −0.22 | 0.12 | −0.18 | 0.95 * | 0.22 | −0.14 | ||||||||||
| En | −0.13 | 0.31 | 0.3 | 0.45 | |||||||||||||
| Ec | 0.2 | 0.18 | −0.23 | −0.24 | |||||||||||||
| Sm | 0.12 | −0.2 | 0.24 | 0.04 | |||||||||||||
| Smu | 0.01 | −0.23 | |||||||||||||||
| Ss | −0.49 | ||||||||||||||||
| Lc | −0.12 | ||||||||||||||||
| Sa | |||||||||||||||||
| Total |
Abbreviations: Aa, Aggregatibacter actinomycetemcomitans; Pg, Porphyromonas gingivalis; Tf, Tannerella forsythia; Td, Treponema denticola; Fn, Fusobacterium nucleatum; Pi, Prevotella intermedia; Pm, Parvimonas micra; Cr, Campylobacter rectus; En, Eubacterium nodatum; Ec, Eikenella corrodens; Sm, S. Streptococcus mitis; Smu, Streptococcus mutans; Lc, Lactobacillus casei; Sa, Staphylococcus aureus; Pn, Prevotella nigrescens; Ss, Streptococcus sobrinus; and * p < 0.05.
Table 7 shows the categorization of the number of bacteria into three levels. P. gingivalis, T. denticola, P. micra, S. mitis, L. casei, S. aureus, E. nodatum, and total bacteria were significantly associated with severe periodontitis at certain levels. However, after adjusting for factors such as sex, age, and smoking, only T. denticola, P. nigrescens, and S. mitis were significant. T. denticola significance was only noted at level 2, in which the risk of periodontal disease was 7.3 times higher compared to level 0. P. nigrescens was significantly associated with severe periodontitis at levels 1 and 2; the risk of periodontal disease at level 2 was 22.5 times higher than that at level 0. S. mitis significance was only observed at level 2.
Table 7.
Association of severe periodontitis according to levels of target species.
| Levels | No. of Subjects | No. (%) with Severe Periodontitis | Crude OR (95% CI) | p Value | Adjusted OR (95% CI) | p Value | |
|---|---|---|---|---|---|---|---|
| Aa | |||||||
| 0 | 79 | 45 (57.0) | 1 | 1 | |||
| 1 | 4 | 3 (75.0) | 1.8 (0.3–18.9) | 0.557 | 13.7 (0.9–389.2) | 0.059 | |
| 2 | 4 | 4 (100.0) | 6.8 (0.7–915.8) | 0.111 | 1.6 (0.1–232.1) | 0.750 | |
| Pg | |||||||
| 0 | 24 | 5 (20.8) | 1 | - | 1 | - | |
| 1 | 31 | 23 (74.2) | 10.9 (3.1–39.0) | <0.001 | 3.3 (0.4–26.6) | 0.271 | |
| 2 | 32 | 24 (75.0) | 11.4 (3.2–40.6) | <0.001 | 1.1 (0.1–8.7) | 0.942 | |
| Tf | |||||||
| 0 | 23 | 14 (60.9) | 1 | - | 1 | - | |
| 1 | 32 | 13 (40.6) | 0.4 (0.1–1.3) | 0.141 | 0.1 (0.0–1.1) | 0.059 | |
| 2 | 32 | 25 (78.1) | 2.3 (0.7–7.5) | 0.169 | 3.7 (0.3–48.1) | 0.319 | |
| Td | |||||||
| 0 | 36 | 16 (44.4) | 1 | - | 1 | - | |
| 1 | 25 | 14 (56.0) | 1.6 (0.6–4.4) | 0.375 | 5.3 (0.6–44.8) | 0.129 | |
| 2 | 26 | 22 (84.6) | 6.9 (2.0–24.0) | 0.002 | 7.3 (1.1–47.4) | 0.035 | |
| Fn | |||||||
| 1 | 43 | 30 (69.8) | 1 | - | 1 | - | |
| 2 | 44 | 22 (50.0) | 0.4 (0.2–1.0) | 0.062 | 1.0 (0.2–4.3) | 0.969 | |
| Pi | |||||||
| 0 | 64 | 37 (57.8) | 1 | - | 1 | - | |
| 1 | 11 | 7 (63.6) | 1.3 (0.3–4.8) | 0.717 | 0.7 (0.1–7.6) | 0.762 | |
| 2 | 12 | 8 (66.7) | 1.5 (0.4–5.3) | 0.568 | 0.5 (0.1–3.4) | 0.504 | |
| Pn | |||||||
| 0 | 16 | 9 (56.2) | 1 | - | 1 | - | |
| 1 | 35 | 25 (71.4) | 1.9 (0.6–6.7) | 0.289 | 120.4 (5.3–2725.4) | 0.002 | |
| 2 | 36 | 18 (50.0) | 0.8 (0.2–2.5) | 0.677 | 22.5 (2.0–260.6) | 0.012 | |
| Pm | |||||||
| 1 | 43 | 19 (44.2) | 1 | - | 1 | - | |
| 2 | 44 | 33 (75.0) | 3.8 (1.5–9.4) | 0.004 | 4.4 (1.0–20.1) | 0.057 | |
| Cr | |||||||
| 0 | 14 | 7 (50.0) | 1 | - | 1 | - | |
| 1 | 36 | 25 (69.4) | 2.3 (0.6–8.1) | 0.203 | 3.5 (0.5–27.3) | 0.226 | |
| 2 | 37 | 20 (54.1) | 1.2 (0.3–4.0) | 0.795 | 1.6 (0.2–11.0) | 0.628 | |
| En | |||||||
| 0 | 70 | 38 (54.3) | 1 | - | 1 | - | |
| 1 | 8 | 5 (62.5) | 1.3 (0.3–6.1) | 0.694 | 0.3 (0.1–2.0) | 0.227 | |
| 2 | 9 | 9 (100.0) | 16.0 (1.9–2097.0) | 0.006 | 4.3 (0.4–609.5) | 0.280 | |
| Ec | |||||||
| 0 | 68 | 37 (54.4) | 1 | - | 1 | - | |
| 1 | 9 | 6 (66.7) | 1.7 (0.4–7.3) | 0.490 | 2.6 (0.2–30.9) | 0.441 | |
| 2 | 10 | 9 (90.0) | 7.5 (0.9–62.8) | 0.061 | 13.6 (0.5–380.9) | 0.124 | |
| Sm | |||||||
| 1 | 43 | 33 (76.7) | 1 | - | 1 | - | |
| 2 | 44 | 19 (43.2) | 0.2 (0.1–0.6) | 0.001 | 0.1 (0.0–0.8) | 0.024 | |
| Smu | |||||||
| 0 | 29 | 17 (58.6) | 1 | - | 1 | - | |
| 1 | 29 | 18 (62.1) | 1.2 (0.4–3.3) | 0.788 | 2.5 (0.4–18.3) | 0.354 | |
| 2 | 29 | 17 (58.6) | 1.0 (0.4–2.8) | 1 | 0.4 (0.1–2.5) | 0.316 | |
| Ss | |||||||
| 0 | 81 | 47 (58.0) | 1 | 1 | |||
| 1 | 3 | 3 (100.0) | 5.1 (0.5–692.1) | 0.205 | 1.6 (0.1–244.0) | 0.755 | |
| 2 | 3 | 2 (66.7) | 1.2 (0.2–13.7) | 0.856 | 1.4 (0.1–193.5) | 0.873 | |
| Lc | |||||||
| 0 | 63 | 34 (54.0) | 1 | - | 1 | - | |
| 1 | 12 | 11 (91.7) | 9.4 (1.1–77.0) | 0.037 | 1.4 (0.1–14.1) | 0.795 | |
| 2 | 12 | 7 (58.3) | 1.2 (0.3– 4.2) | 0.780 | 0.3 (0.0–1.9) | 0.192 | |
| Sa | |||||||
| 0 | 68 | 48 (70.6) | 1 | - | 1 | - | |
| 1 | 9 | 2 (22.2) | 0.1 (0.0–0.6) | 0.011 | 0.2 (0.0–2.0) | 0.162 | |
| 2 | 10 | 2 (20.0) | 0.1 (0.0–0.5) | 0.006 | 0.4 (0.0–7.4) | 0.525 | |
| Total | |||||||
| 1 | 43 | 18 (41.9) | 1 | - | 1 | - | |
| 2 | 44 | 34 (77.3) | 4.7 (1.9–12.0) | 0.001 | 1.4 (0.3–5.8) | 0.673 |
Level 0 indicates polymerase chain reaction (PCR)-negative subjects. Level 1 indicates that the number of bacterial cells is less than the median number in PCR-positive subjects. Level 2 indicates that the number of bacterial cells is equal to or greater than the median number in PCR-positive subjects. Logistic regression analysis was performed after adjusting for known confounders: sex, age, and smoking history. Abbreviations: OR, odds ratio; CI, confidence interval; Aa, Aggregatibacter actinomycetemcomitans; Pg, Porphyromonas gingivalis; Tf, Tannerella forsythia; Td, Treponema denticola; Fn, Fusobacterium nucleatum; Pi, Prevotella intermedia; Pm, Parvimonas micra; Cr, Campylobacter rectus; En, Eubacterium nodatum; Ec, Eikenella corrodens; Sm, Streptococcus mitis; Smu, Streptococcus mutans; Lc, Lactobacillus casei; Sa, Staphylococcus aureus; Pn, Prevotella nigrescens; and Ss, Streptococcus sobrinus.
4. Discussion
To the best of our knowledge, this preliminary study is the first to quantify bacteria with PCR in a mouth-rinsing solution, as opposed to a subgingival plaque or saliva sample. Newer diagnostic methods have been developed with more detailed stages and grades corresponding to the related treatment protocol [29]. While periodontal probing is the traditional method used for diagnosing periodontal disease, the detection of periopathogenic bacteria with PCR may potentially serve as an adjunct assessment. Nevertheless, to date, no standardized methods have been proposed for the diagnosis of periodontal disease based on gargled solutions [30].
Several studies on periodontal pathogens have been conducted using RT-PCR analysis. P. gingivalis, T. forsythia, T. denticola, and P. intermedia have been reported to be mainly prevalent in Asian populations [31,32]. However, A. actinomycetemcomitans prevalence varies widely. In this study, a low A. actinomycetemcomitans prevalence was observed. Previous studies have reported even lower levels in this and other previous studies compared to other pathogens [2,33]. In line with the results of previous studies, we found significant differences between the groups of bacteria known to be related to periodontal disease. The prevalence of A. actinomycetemcomitans, T. denticola, P. nigrescens, and S. mitis were significantly different between the healthy and periodontal disease groups.
A. actinomycetemcomitans is a common pathogen in aggressive periodontitis, and it is known to have mutually inhibitory effects on Streptococcus sanguis, Streptococcus uberis, and A. viscosus [34]. A. actinomycetemcomitans is involved in the pathogenesis of aggressive periodontitis in younger patients [35]. T. denticola and P. nigrescens are both known to be related to periodontitis. A previous study showed clear evidence of increased immune responses to T. denticola, P. nigrescens, and F. nucleatum in 89 patients with chronic periodontitis [36]. F. nucleatum is frequently detected in the subgingival plaque of patients with chronic periodontitis and is often found associated with periodontal pockets. A. actinomycetemcomitans, T. forsythia, T. denticola, and P. gingivalis are strongly associated with periodontal disease, disease progression, and treatment failure. P. intermedia, P. micra, C. rectus, E. nodatum, P. nigrescens, and F. nucleatum can also act as pathogens if their concentrations exceed certain thresholds [37].
Periodontal disease is a result of complex interactions between the periodontal pathogens and normal flora [38]. This fact rationalizes the use of mouth-rinsing solution for bacterial analysis, as it provides mixed bacterial samples. Nevertheless, the presence of periodontal pathogens in the gingival crevices by itself does not cause or initiate periodontal inflammation. The bacterial load in an area with periodontal disease is higher than that in a healthy area; these bacteria are called periodontopathic [39]. P. gingivalis and T. forsythia are some of the main pathogens of periodontitis, but no significant difference was found between the healthy and periodontal disease groups in this study. The distribution, as well as the number of bacterial species varies in diseased and healthy periodontal tissues. In this study, S. mitis, a Gram-positive strain present in healthy tissues, had a 100% prevalence in both normal and severe periodontitis groups. F. nucleatum, which belongs to the red complex group and is strongly associated with periodontal disease, also had a 100% prevalence in both groups. Therefore, although these bacterial species may be proportionally less dominant, they are present in the oral cavity as a constituent of the normal flora [40]. Our findings revealed a significant positive correlation between A. actinomycetemcomitans and P. gingivalis. This indicates that both bacterial species affect each other’s growth [38]. In addition, P. gingivalis and E. nodatum also showed a positive correlation, indicating that the higher the number of P. gingivalis, the higher the number of E. nodatum. Conversely, T. forsythia was negatively correlated with F. nucleatum, P. nigrescens, and S. mitis. Hence, these bacteria may inhibit each other’s growth.
After dividing bacterial levels according to whether they were above or below the median, and adjusting for confounding factors (e.g., sex, age, and smoking habit), T. denticola, P. nigrescens, and S. mitis were significantly associated with periodontitis. These results indicate that the risk of periodontal disease is increased if the levels of T. denticola and P. nigrescens are high. It can also be inferred that the higher the level of S. mitis, the lower the risk of developing periodontal disease.
This study has some limitations because we could not verify the reproducibility of our results. Moreover, in order for the mouth-rinsing solution analysis to be of diagnostic value, a certain number of bacteria must be detected to indicate disease. Implementation of the new classification system described above was not possible when recruiting participants in this study. We could only divide participants into two groups: healthy and severe periodontitis. Due to the lack of previous studies on diagnostic methods using mouth-rinsing solutions, we tried to evaluate differences between the two groups using the existing classification method. This should be complemented in the next study. There were limitations in adjusting for age, sex, and smoking history, because of the small sample size. Among the correction variables, age is an important variable related to periodontal disease, but in this study, the sample size was not large enough to consider the correction variable, even though it had already been adjusted.
As mentioned earlier, periodontitis is a disease with various factors caused by subgingival bacterial colonies such as A. actinomycetemcomitans, T. forsythia, and P. gingivalis. [41]. However, P. gingivalis was not significantly associated with periodontitis after adjustment for confounding factors in this study. P. gingivalis has been shown to have a higher prevalence in deep pockets [1,42,43]. Therefore, the results of our study may reflect the low ability of mouth rinsing to sample P. gingivalis in these regions. Because the number of bacteria needed to cause periodontal disease may vary depending on the host’s immune system, additional research methods are needed, such as comparing with crevicular fluid and gingival biopsy to show reliable results [44].
This study suggests that the analysis of mouth-rinsing solution might be a promising diagnostic method, and further studies with greater sensitivity should be conducted with larger samples to determine its perceived usefulness. Diagnosing the severity of periodontitis by analyzing gargled mouth-rinse solutions is less invasive than collecting plaque samples. We hope that the analysis of mouth-rinsing solutions will become an accepted diagnostic method for periodontal disease. A limitation of this study is that only three periopathogenic bacteria, among a total of 18 species, exhibited a significant difference between the healthy and periodontal disease groups; nevertheless, the advantages of the detection method are obvious.
In summary, the findings of this study are as follows: (1) similar to previous studies, bacteria known to cause periodontal disease were detected with mouth-rinsing solutions in patients with severe periodontal disease; (2) significant differences were found in the prevalence (healthy vs. periodontal disease group) of A. actinomycetemcomitans (2.9% vs. 13.5%), T. denticola (42.9% vs. 69.2%), and P. nigrescens (80% vs. 82.7%); and (3) T. denticola, P. nigrescens, and S. mitis levels were significantly different between groups in the quantitative analysis.
We did not comprehensively assess all periopathogenic bacteria in this study; therefore, additional research is required to assess the potential of oral-rinsing solutions to reflect oral-infection risk and the need to improve oral hygiene, as well as to serve as a complementary method for periodontal disease diagnosis. Similar to the results of plaque analysis, which has been conducted in many studies, the results obtained by detecting bacteria in mouth-rinsing solutions show that there is a relationship between specific bacteria and severe periodontal disease. While mouth-rinsing solutions are non-invasive, simple, and capable of detecting a wide range of bacterial species, they are limited by the lack of clear diagnostic criteria. Therefore, in order for this diagnostic method to be effective, research aimed at establishing the criteria for the type and number of bacteria should be conducted. Recently, the concept of the diagnosis of periodontitis has been improved to complement the treatment stage. If this simple diagnostic kit is quantified and developed, it is expected to be helpful in future treatment planning.
Author Contributions
Conceptualization, D.-W.L.; methodology, D.-W.L. and J.-H.Y.; software, Y.L.; validation, J.-W.O. and J.-H.K.; formal analysis, Y.L.; investigation, D.-W.L.; resources, D.-W.L.; data curation, J.-H.K. and J.-W.O.; writing—original draft preparation, J.-H.K. and J.-W.O.; writing—review and editing, D.-W.L.; visualization, J.-H.Y.; supervision, S.-H.C.; project administration, D.-W.L.; funding acquisition, D.-W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by VHS Medical Center Research Grant, Republic of Korea, grant number VHSMC18015.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Veterans Health Service Medical Center (BOHUN No. 2018-03-002).
Informed Consent Statement
Informed consent was obtained from all participants involved in the study.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Socransky S.S., Haffajee A.D., Cugini M.A., Smith C., Kent R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 2.Papapanou P.N., Teanpaisan R., Obiechina N.S., Pithpornchaiyakul W., Pongpaisal S., Pisuithanakan S., Baelum V., Fejerskov O., Dahlen G. Periodontal microbiota and clinical periodontal status in a rural sample in southern Thailand. Eur. J. Oral Sci. 2002;110:345–352. doi: 10.1034/j.1600-0722.2002.21361.x. [DOI] [PubMed] [Google Scholar]
- 3.Pradhan-Palikhe P., Mäntylä P., Paju S., Buhlin K., Persson G.R., Nieminen M.S., Sinisalo J., Pussinen P.J. Subgingival Bacterial Burden in Relation to Clinical and Radiographic Periodontal Parameters. J. Periodontol. 2013;84:1809–1817. doi: 10.1902/jop.2013.120537. [DOI] [PubMed] [Google Scholar]
- 4.Hyvärinen K., Laitinen S., Paju S., Hakala A., Suominen-Taipale L., Skurnik M., Könönen E., Pussinen P.J. Detection and quantification of five major periodontal pathogens by single copy gene-based real-time PCR. Innate Immun. 2009;15:195–204. doi: 10.1177/1753425908101920. [DOI] [PubMed] [Google Scholar]
- 5.Paju S., Pussinen P.J., Suominen-Taipale L., Hyvönen M., Knuuttila M., Könönen E. Detection of Multiple Pathogenic Species in Saliva is Associated with Periodontal Infection in Adults. J. Clin. Microbiol. 2008;47:235–238. doi: 10.1128/JCM.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mineoka T., Awano S., Rikimaru T., Kurata H., Yoshida A., Ansai T., Takehara T. Site-Specific Development of Periodontal Disease is Associated with Increased Levels of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythiain Subgingival Plaque. J. Periodontol. 2008;79:670–676. doi: 10.1902/jop.2008.070398. [DOI] [PubMed] [Google Scholar]
- 7.Torrungruang K., Jitpakdeebordin S., Charatkulangkun O., Gleebbua Y. Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Treponema denticola/Prevotella intermedia Co-Infection are Associated with Severe Periodontitis in a Thai Population. PLoS ONE. 2015;10:e0136646. doi: 10.1371/journal.pone.0136646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshizawa J.M., Schafer C.A., Schafer J.J., Farrell J.J., Paster B.J., Wong D.T.W. Salivary Biomarkers: Toward Future Clinical and Diagnostic Utilities. Clin. Microbiol. Rev. 2013;26:781–791. doi: 10.1128/CMR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguirre A., Testa-Weintraub L., Banderas J., Haraszthy G., Reddy M., Levine M. Sialochemistry: A Diagnostic Tool? Crit. Rev. Oral Biol. Med. 1993;4:343–350. doi: 10.1177/10454411930040031201. [DOI] [PubMed] [Google Scholar]
- 10.Liguori G., Lucariello A., Colella G., de Luca A., Marinelli P. Rapid identification of Candida species in oral rinse solutions by PCR. J. Clin. Pathol. 2006;60:1035–1039. doi: 10.1136/jcp.2006.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faul F., Erdfelder E., Buchner A., Lang A.-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 12.Eke P.I., Page R.C., Wei L., Thornton-Evans G., Genco R.J. Update of the Case Definitions for Population-Based Surveillance of Periodontitis. J. Periodontol. 2012;83:1449–1454. doi: 10.1902/jop.2012.110664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E.-H., Joo J.-Y., Lee Y.J., Koh J.-K., Choi J.-H., Shin Y., Cho J., Park E., Kang J., Lee K., et al. Grading system for periodontitis by analyzing levels of periodontal pathogens in saliva. PLoS ONE. 2018;13:e0200900. doi: 10.1371/journal.pone.0200900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez A., Rodríguez M., Córdoba J.J., Andrade M.J. PCR Primer Design. Volume 1275. Humana Press; New York, NY, USA: 2015. Design of primers and probes for quantitative real-time PCR methods; pp. 31–56. [DOI] [PubMed] [Google Scholar]
- 15.Boutaga K., van Winkelhoff A.J., Vandenbroucke-Grauls C.M.J.E., Savelkoul P.H.M. Periodontal pathogens: A quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol. Med. Microbiol. 2005;45:191–199. doi: 10.1016/j.femsim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Elkaïm R., Dahan M., Kocgozlu L., Werner S., Kanter D., Kretz J.G., Tenenbaum H. Prevalence of periodontal pathogens in subgingival lesions, atherosclerotic plaques and healthy blood vessels: A preliminary study. J. Periodontal Res. 2007;43:224–231. doi: 10.1111/j.1600-0765.2007.01018.x. [DOI] [PubMed] [Google Scholar]
- 17.Boutaga K., van Winkelhoff A.J., Vandenbroucke-Grauls C.M.J.E., Savelkoul P.H.M. Comparison of real-time PCR and culture for detection of Porphyromonas gingivalis in subgingival plaque samples. J. Clin. Microbiol. 2003;41:4950–4954. doi: 10.1128/JCM.41.11.4950-4954.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki N., Yoshida A., Saito T., Kawada M., Nakano Y. Quantitative Microbiological Study of Subgingival Plaque by Real-Time PCR Shows Correlation between Levels of Tannerella forsythensis and Fusobacterium spp. J. Clin. Microbiol. 2004;42:2255–2257. doi: 10.1128/JCM.42.5.2255-2257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki N., Nakano Y., Yoshida A., Yamashita Y., Kiyoura Y. Real-Time TaqMan PCR for Quantifying Oral Bacteria during Biofilm Formation. J. Clin. Microbiol. 2004;42:3827–3830. doi: 10.1128/JCM.42.8.3827-3830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asai Y., Jinno T., Igarashi H., Ohyama Y., Ogawa T. Detection and Quantification of Oral Treponemes in Subgingival Plaque by Real-Time PCR. J. Clin. Microbiol. 2002;40:3334–3340. doi: 10.1128/JCM.40.9.3334-3340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawashdeh R.Y., Malkawi H.I., Al-Hiyasat A.S., Hammad M.M. A fast and sensitive molecular detection of Streptococcus mutans and Actinomyces viscosus from dental plaques. Jordan J. Biol. Sci. 2008;1:135–139. [Google Scholar]
- 22.Yun J.-H., Park J.-E., Kim D.-I., Lee S.-I., Choi S.-H., Cho K.-S., Lee D.-S. Identification of putative periodontal pathogens in Korean chronic periodontitis patients. J. Korean Acad. Periodontol. 2008;38:143–152. doi: 10.5051/jkape.2008.38.2.143. [DOI] [Google Scholar]
- 23.Yoshida A., Suzuki N., Nakano Y., Kawada M., Oho T., Koga T. Development of a 5′ Nuclease-Based Real-Time PCR Assay for Quantitative Detection of Cariogenic Dental Pathogens Streptococcus mutans and Streptococcus sobrinus. J. Clin. Microbiol. 2003;41:4438–4441. doi: 10.1128/JCM.41.9.4438-4441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haarman M., Knol J. Quantitative Real-Time PCR Analysis of Fecal Lactobacillus Species in Infants Receiving a Prebiotic Infant Formula. Appl. Environ. Microbiol. 2006;72:2359–2365. doi: 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai H., Procop G.W., Kobayashi N., Togawa D., Wilson D.A., Borden L., Krebs V., Bauer T.W. Simultaneous Detection of Staphylococcus aureus and Coagulase-Negative Staphylococci in Positive Blood Cultures by Real-Time PCR with Two Fluorescence Resonance Energy Transfer Probe Sets. J. Clin. Microbiol. 2004;42:5739–5744. doi: 10.1128/JCM.42.12.5739-5744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granger K., Rundell M.S., Pingle M.R., Shatsky R., Larone D.H., Golightly L.M., Barany F., Spitzer E.D. Multiplex PCR-Ligation Detection Reaction Assay for Simultaneous Detection of Drug Resistance and Toxin Genes from Staphylococcus aureus, Enterococcus faecalis, and Enterococcus faecium. J. Clin. Microbiol. 2009;48:277–280. doi: 10.1128/JCM.01411-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Firth D. Bias Reduction of Maximum Likelihood Estimates. Biometrika. 1993;80:27. doi: 10.1093/biomet/80.1.27. [DOI] [Google Scholar]
- 28.Heinze G., Schemper M. A solution to the problem of separation in logistic regression. Stat. Med. 2002;21:2409–2419. doi: 10.1002/sim.1047. [DOI] [PubMed] [Google Scholar]
- 29.Tonetti M.S., Greenwell H., Kornman K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018;89:S159–S172. doi: 10.1002/JPER.18-0006. [DOI] [PubMed] [Google Scholar]
- 30.Park N.J., Zhou X., Yu T., Brinkman B.M., Zimmermann B.G., Palanisamy V., Wong D.T. Characterization of salivary RNA by cDNA library analysis. Arch. Oral Biol. 2007;52:30–35. doi: 10.1016/j.archoralbio.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papapanou P.N., Baelum V., Luan W.-M., Madianos P.N., Chen X., Fejerskov O., Dahlén G. Subgingival Microbiota in Adult Chinese: Prevalence and Relation to Periodontal Disease Progression. J. Periodontol. 1997;68:651–666. doi: 10.1902/jop.1997.68.7.651. [DOI] [PubMed] [Google Scholar]
- 32.Kuboniwa M., Amano A., Kimura K.R., Sekine S., Kato S., Yamamoto Y., Okahashi N., Iida T., Shizukuishi S. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol. Immunol. 2004;19:168–176. doi: 10.1111/j.0902-0055.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 33.He J., Huang W., Pan Z., Cui H., Qi G., Zhou X., Chen H. Quantitative analysis of microbiota in saliva, supragingival, and subgingival plaque of Chinese adults with chronic periodontitis. Clin. Oral Investig. 2011;16:1579–1588. doi: 10.1007/s00784-011-0654-4. [DOI] [PubMed] [Google Scholar]
- 34.Slots J. Subgingival microflora and periodontal disease. J. Clin. Periodontol. 1979;6:351–382. doi: 10.1111/j.1600-051X.1979.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 35.Haubek D., Ennibi O.-K., Poulsen K., Vaeth M., Poulsen S., Kilian M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet. 2008;371:237–242. doi: 10.1016/S0140-6736(08)60135-X. [DOI] [PubMed] [Google Scholar]
- 36.Papapanou P.N., Neiderud A.M., Disick E., Lalla E., Miller G.C., Dahlén G. Longitudinal stability of serum immunoglobulin G responses to periodontal bacteria. J. Clin. Periodontol. 2004;31:985–990. doi: 10.1111/j.1600-051X.2004.00599.x. [DOI] [PubMed] [Google Scholar]
- 37.Van Winkelhoff A.J., Loos B.G., van der Reijden W.A., van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J. Clin. Periodontol. 2002;29:1023–1028. doi: 10.1034/j.1600-051X.2002.291107.x. [DOI] [PubMed] [Google Scholar]
- 38.Kolenbrander P.E., Palmer R.J., Periasamy S., Jakubovics N.S. Oral multispecies biofilm development and the key role of cell–cell distance. Nat. Rev. Microbiol. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 39.Socransky S.S., Haffajee A.D. The Bacterial Etiology of Destructive Periodontal Disease: Current Concepts. J. Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 40.Aas J.A., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genco R., Kornman K., Williams R., Offenbacher S., Zambon J.J., Ishikawa I., Listgarten M., Michalowicz B., Page R., Schenkein H., et al. Periodontal Diseases: Pathogenesis and Microbial Factors. J. Am. Dent. Assoc. 1998;129:58S–62S. doi: 10.1016/S0002-8177(15)30088-X. [DOI] [PubMed] [Google Scholar]
- 42.Kawada M., Yoshida A., Suzuki N., Nakano Y., Saito T., Oho T., Koga T. Prevalence of Porphyromonas gingivalis in relation to periodontal status assessed by real-time PCR. Oral Microbiol. Immunol. 2004;19:289–292. doi: 10.1111/j.1399-302X.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- 43.Klein M.I., Gonçalves R.B. Detection ofTannerella forsythensis(Bacteroides forsythus) and Porphyromonas gingivalisby Polymerase Chain Reaction in Subjects with Different Periodontal Status. J. Periodontol. 2003;74:798–802. doi: 10.1902/jop.2003.74.6.798. [DOI] [PubMed] [Google Scholar]
- 44.Lee N.-H., Lee E., Kim Y.-S., Kim W.-K., Lee Y.-K., Kim S.-H. Differential expression of microRNAs in the saliva of patients with aggressive periodontitis: A pilot study of potential biomarkers for aggressive periodontitis. J. Periodontal Implant. Sci. 2020;50:281–290. doi: 10.5051/jpis.2000120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.