Abstract
Curcumin-loaded polymeric micelles composed of poly(ethylene glycol)-b-poly(N-2-benzoyloxypropyl methacrylamide) (mPEG-b-p(HPMA-Bz)) were prepared to solubilize and improve the pharmacokinetics of curcumin. Curcumin-loaded micelles were prepared by a nanoprecipitation method using mPEG5kDa-b-p(HPMA-Bz) copolymers with varying molecular weight of the hydrophobic block (5.2, 10.0, and 17.1 kDa). At equal curcumin loading, micelles composed of mPEG5kDa-b-p(HPMA-Bz)17.1kDa showed better curcumin retention in both phosphate-buffered saline (PBS) and plasma at 37 °C than micelles based on block copolymers with smaller hydrophobic blocks. No change in micelle size was observed during 24 h incubation in plasma using asymmetrical flow field-flow fractionation (AF4), attesting to particle stability. However, 22–49% of the curcumin loading was released from the micelles during 24 h from formulations with the highest to the lowest molecular weight p(HPMA-Bz), respectively, in plasma. AF4 analysis further showed that the released curcumin was subsequently solubilized by albumin. In vitro analyses revealed that the curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles were internalized by different types of cancer cells, resulting in curcumin-induced cell death. Intravenously administered curcumin-loaded, Cy7-labeled mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles in mice at 50 mg curcumin/kg showed a long circulation half-life for the micelles (t1/2 = 42 h), in line with the AF4 results. In contrast, the circulation time of curcumin was considerably shorter than that of the micelles (t1/2α = 0.11, t1/2β = 2.5 h) but ∼5 times longer than has been reported for free curcumin (t1/2α = 0.02 h). The faster clearance of curcumin in vivo compared to in vitro studies can be attributed to the interaction of curcumin with blood cells. Despite the excellent solubilizing effect of these micelles, no cytostatic effect was achieved in neuroblastoma-bearing mice, possibly because of the low sensitivity of the Neuro2A cells to curcumin.
Keywords: nanomedicine, pHPMA, in vitro uptake and localization, human neuroblastoma xenograft model, pharmacokinetics parameters, pharmacodynamics
1. Introduction
Curcumin is a polyphenolic compound that is mainly isolated from the rhizome of Curcuma longa.1 A plethora of studies have shown that curcumin exerts a wide variety of pharmacological effects against several pathologies, including cancer.2 Curcumin interacts with numerous vital pathways in cancer cells, resulting in antimutagenic, cytostatic, cytotoxic, and antimetastatic effects.3 Also, clinical studies have consistently demonstrated that curcumin is safe and well tolerated at high doses, exhibiting no dose-limiting toxicity.4 However, curcumin has poor aqueous solubility (log P = 2.5),5 degrades under neutral-to-alkaline conditions,6,7 and is heavily biotransformed and rapidly eliminated, altogether accounting for poor pharmacokinetics that translate to insufficient curcumin accumulation in tumors to instill significant therapeutic responses.8 Consequently, much research has been devoted to improve curcumin uptake and prolong circulation time.
Polymer-based nanocarriers have been investigated to improve the solubility and therapeutic efficacy of hydrophobic anticancer drugs.9,10 In particular, polymeric micelles have attracted substantial attention for solubilization of hydrophobic drugs.11,12 The nanometer-size range of polymeric micelles facilitates the enhanced permeability and retention (EPR) effect, commonly serving as the pathophysiological basis for intratumoral accumulation of nanoparticulate drug-delivery systems.13 Encapsulation of curcumin in polymeric micelles has also been employed to tackle its poor solubility and stability issues.14,15 To this end, a variety of amphiphilic polymers such as block copolymers of poly(2-oxazoline)s, poly(ethylene glycol)-b-poly(ε-caprolactone), and poly(ethylene glycol)-b-poly(lactic-co-glycolic acid) have been used to prepare curcumin-loaded micelles, leading to promising results in vitro and in vivo.16−18
Numerous curcumin delivery systems have been designed and evaluated in vivo with respect to pharmacokinetics. The reported 1.3- to 5-fold increase in curcumin area under the curve is at best classified as a marginal increase compared to the improvement rates of other nano-encapsulated drugs versus their respective free form controls (manuscript in preparation). It is therefore warranted to develop more stable nanoformulations to fully exploit curcumin’s pharmacodynamic potency. One frequently employed strategy to improve the stability of polymeric micelles is centered on physical interactions between the hydrophobic core and the payload, such as π–π stacking interactions between aromatic groups.19 Gong et al.20 successfully applied this approach to improve the pharmacokinetics of curcumin using N-(tert-butoxycarbonyl)-l-phenylalanine end-capped mPEG-PCL (mPEG-PCL-Phe(Boc)).
Also, the stability of drug-delivery systems needs to be investigated to gain insight into in vitro–in vivo relationships. Drug release studies in relevant media can provide valuable information regarding the performance of a formulation in retaining the encapsulated drug in vitro and in vivo.21 However, studying the stability of drug-delivery systems loaded with hydrophobic drugs in biological fluids such as plasma is challenging because of the difficulties in recovering and separating nanoparticles from other components of the release medium, such as (lipo)proteins. Most often, dialysis or repeated high-speed centrifugation is required for complete separation.22,23 One possible solution to overcome this challenge is to apply asymmetrical flow field-flow fractionation (AF4). This technique separates nanoparticles mainly based on their hydrodynamic size.24 AF4 columns do not have a stationary phase, which reduces possible interactions and enables studying highly sensitive samples in biological media. Additionally, this technique can separate self-assembled particles such as micelles from other components without disrupting their structure.25
In previous studies, mPEG-b-p(HPMA-Bz) block copolymers were successfully employed as micellar drug-delivery systems and prepared on a relatively large scale.26−28 A comprehensive study of formulation and processing parameters of polymeric micelles based on these block copolymers allowed proper control of micelle size.29 Moreover, mPEG-b-p(HPMA-Bz) micelles are associated with excellent particle stability and good drug retention due to π–π stacking interactions.26,27,30,31 The promising results achieved with paclitaxel-loaded mPEG-b-p(HPMA-Bz) micelles in terms of pharmaceutical aspects (loading and stability) and in vivo therapeutic efficacy motivated us to assess this platform for its suitability as a delivery system for curcumin.
Accordingly, curcumin-loaded micelles with different (block copolymer) sizes were prepared and evaluated for stability in buffer and plasma. Particle stability due to π–π stacking interactions in the micelle core was studied by solid-state NMR. Also, the uptake of curcumin-containing mPEG-b-p(HPMA-Bz) micelles was analyzed in various human cancer cell lines. Subsequently, the circulation kinetics and biodistribution of mPEG-b-p(HPMA-Bz) micelles and encapsulated curcumin were assessed in mice. Finally, the therapeutic efficacy of curcumin-loaded micelles was evaluated in a mouse model of human neuroblastoma.
2. Materials and Methods
2.1. Materials and Animals
The (mPEG5kDa)2-ABCPA macroinitiator and HPMA-Bz were synthesized and characterized according to previously described protocols.28,29N-(2-aminoethyl)methacrylamide hydrochloride (AEMA), triethylamine, HEPES, lithium chloride (LiCl), sulforhodamine B (SRB) sodium salt, trichloroacetic acid, curcuminoid mixture, human serum albumin (HSA), and Roswell Park Memorial Institute (RPMI) 1640 culture medium were purchased from Sigma-Aldrich (St. Louis, MO). Phosphate-buffered saline (PBS) 10× solution was acquired from Fisher Bioreagents (Pittsburgh, PA). Tris(hydroxymethyl)aminomethane was obtained from Serva (Heidelberg, Germany). Stellate Cell Growth Supplements (SteCGS) were purchased from ScienCell (Carlsbad, CA). Endothelial Basal Medium (EBM-2) supplemented with growth factors (Growth Medium 2 SupplementMix) were obtained from PromoCell (Heidelberg Germany). l-Glutamine, penicillin, and streptomycin were acquired from Lonza (Basel, Switzerland). Accutase was purchased from Global Cell Solutions (Charlottesville, VA). Trypsin-EDTA, PBS, and gentamicin/amphotericin B were acquired from Gibco (Waltham, MA). PEG standards for gel permeation chromatography (GPC) calibration were obtained from Agilent Technologies (Santa Clara, CA). Cyanine 7 (Cy7) NHS ester and cyanine 5 (Cy5) NHS ester were obtained from Lumiprobe (Hannover, Germany). Radioimmunoprecipitation assay (RIPA) lysis buffer (10×, 0.5 M Tris-HCl, pH = 7.4, 1.5 M NaCl, 2.5% deoxycholic acid, 10% NP-40, 10 mM EDTA) was purchased from Merck KGaA (Darmstadt, Germany). Six- and 24-well plates were acquired from Greiner Bio-One (Cellstar; Kremsmünster, Austria). Fetal bovine serum (FBS) was obtained from Bodinco (Alkmaar, The Netherlands). All solvents were purchased from Biosolve (Valkenswaard, The Netherlands) and used as received. Syringe filters of regenerated cellulose were ordered from Phenomenex (Torrance, CA). Dialysis bags with 8 kDa cutoff were purchased from Spectrum Chemical (SpectraPor; New Brunswick, NJ). Female BALB/c mice (18–22 g) and female A/J mice (18–22 g) were purchased from Charles River (Den Bosch, The Netherlands) and Envigo (Horst, The Netherlands), respectively.
2.2. Cell Culture
Murine neuroblastoma (Neuro2A) cells were purchased from ATCC (Manassas, VA). Human extrahepatic cholangiocarcinoma (TFK-1) was ordered from DMSZ (Brunswick, Germany). Human extrahepatic cholangiocarcinoma (Sk-ChA-1) and gallbladder adenocarcinoma (Mz-ChA-1) cells were licensed to Michal Heger by the University Hospital Zurich, Switzerland. Human biliary adenocarcinoma (EGI-1) cells were provided by the Tytgat Institute for Liver and Intestinal Research to Michal Heger (Amsterdam UMC, location AMC). Primary human umbilical vein endothelial cells (HUVECs) were obtained from Lonza (Verviers, Belgium). Primary human pancreatic stellate cells (hPSC) were obtained from ScienCell Research Laboratories (Carlsbad, CA). TFK-1, Mz-ChA-1, Sk-ChA-1, EGI-1 and Neuro2A cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 culture medium supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine. hPSCs were cultured in complete stellate cell medium supplemented with 2% FBS, 1% penicillin/streptomycin, and 1% SteCGS. HUVECs were cultured in EBM-2 medium supplemented with growth factors (Growth Medium 2 SupplementMix) and antibiotics/fungicidals (gentamicin/amphotericin B) up to passage number 6. Cells were maintained at 37 °C in a 5% CO2 humidified atmosphere (standard culture conditions). EGI-1, Mz-ChA-1, SK-ChA-1, and TFK-1 cells were cultured in 75 cm2 cell culture flasks and passaged once per week at a ratio of 1:10, 1:2, 1:8, and 1:6, respectively.
2.3. Synthesis of mPEG-b-p(HPMA-Bz) Polymers
mPEG-b-p(HPMA-Bz) block copolymers were synthesized via free-radical polymerization and characterized by 1H-NMR and GPC as described previously.28−30 Three different mPEG-b-p(HPMA-Bz) block copolymers with fixed mPEG5kDa and different molecular weights of the hydrophobic block were synthesized by varying the molar feed ratios of macroinitiator/monomer (1:200, 1:100, 1:50) using acetonitrile (ACN) as a solvent. Polymerization was performed under nitrogen at 70 °C for 24 h. The polymers were collected by precipitation in ice-cold diethyl ether and dried under vacuum. The polymer characteristics were published previously30 and reported in the Supporting Information (SI) Table S1.
2.4. Synthesis and Characterization of Fluorescently Labeled mPEG-b-p(HPMA-Bz)
mPEG-b-p(HPMAm-Bz98%-co-AEMAm2%) with a macroinitiator/monomer molar ratio of 1:200 was synthesized as described previously.26 The primary amine groups in the hydrophobic block were reacted with the Cy7-NHS ester or Cy5-NHS ester. In short, the polymer (100 mg) was transferred into a glass vial and dissolved in 1.4 mL of dimethyl sulfoxide (DMSO). Next, 0.58 mL of dye stock solution (10 mg/mL) and 3.2 μL of dry triethylamine were added. The reaction was conducted in the dark at room temperature overnight. The fluorescently labeled polymers were dialyzed against a tetrahydrofuran (THF)/water mixture (1:1 v/v) for 72 h. The medium was refreshed five times to remove uncoupled dye using an 8 kDa dialysis membrane. The final product was obtained as dark green and blue powders after lyophilization. GPC was performed to confirm the dye conjugation to the polymer, as described,26 using refractive index (RI) and UV detectors (detection wavelength, 700 and 650 nm for Cy7 and Cy5, respectively). The results are reported in SI Figure S3.
2.5. Preparation and Characterization of Empty/Curcumin-Loaded Micelles
Empty and curcumin-loaded mPEG-b-p(HPMA-Bz) micelles were prepared by a nanoprecipitation procedure.29 In short, mPEG-b-p(HPMA-Bz) (30 mg/mL) alone or together with curcumin (varying concentrations depending on the target load) were dissolved in THF and added dropwise at a 1:1 volume ratio to HEPES-buffered saline (HBS; containing 20 mM HEPES and 150 mM NaCl, pH = 7.4) while stirring. Curcumin micelles were prepared in dim light to avoid photodegradation. Subsequently, THF was removed by evaporation overnight in a fume hood. The micelle dispersions were filtered using 0.2 μm regenerated cellulose membranes to remove unencapsulated curcumin and any polymer aggregates. The formulations used for cell culture and animal studies were additionally dialyzed for 24 h using an 8 kDa dialysis membrane to completely remove the organic solvent. Previously, it was shown that the residual THF content can be reduced to acceptable levels by evaporation and subsequent dialysis to achieve an intravenous formulation with THF content below the threshold concentration (720 ppm) as stipulated by International Council of Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use.29 The curcumin-loaded polymeric micelles were characterized for size, size distribution, curcumin content, and polymer concentration. Micelle size was determined by dynamic light scattering (DLS) using a Zetasizer (model ZS90, Malvern Instruments, Malvern, U.K.). Curcumin was quantified as reported earlier.6 The encapsulation efficacy (EE) and loading capacity (LC) were calculated as follows
GPC analysis was conducted to measure the polymer concentration using two serial PLgel 5 μm MIXED-D GPC columns (Polymer Laboratories, Agilent Technologies) at 65 °C. Dimethylformamide (DMF) containing 10 mM LiCl was used as eluent and results were obtained with a refractive index detector (RI) (Waters 2414, Waters Corporation, Milford, MA) and UV detection (280 nm) (waters 2489, Waters Corporation). Aqueous micelle dispersions were diluted 10-fold in DMF and the absorbance was measured at 280 nm. Standard curves were obtained with the same polymer in DMF in a 0.1–10 mg/mL concentration range.
2.6. Analysis of π–π Stacking in Curcumin-Loaded Micelles by Solid-State NMR Spectroscopy
1H-NMR spectra of mPEG-b-p(HPMA-Bz) copolymer and curcumin dissolved in DMSO-d6 as the solvent were recorded using a Bruker 600 MHz spectrometer (Billerica, MA). The DMSO peak at 2.52 ppm was used for calibration. The following chemical shifts for mPEG-b-p(HPMA-Bz) copolymer were obtained (SI Figure S1): 8.0 (b, 2H, aromatic CH), 7.55 (b, 1H, aromatic CH), 7.65 (b, 2H, aromatic CH), 7.35 (b, CO-NH-CH2), 5.0 (b, NH-CH2-CH(CH3)-O-(Bz)), 3.40–3.60 (b, mPEG5000 methylene protons, O-CH2-CH2), 3.1 (b, NH-CH2-CH) and 0.6–2.2 (b, the rest of the protons are from the methyl and backbone CH2 protons). The chemical shifts of curcumin (SI Figure S2) were determined as follows: 9.66 (s, 2H, aromatic OH), 7.58 (d, 2H, CO-CH-CH), 7.35 (s, 2H, C-CH-C-OCH3), 7.16 (d, 2H, aromatic CH), 6.84 (d, 2H, aromatic CH), 6.78 (d, 2H, CO-CH-CH), 6.08 (s, 1H, OH-C-CH-CO), and 3.84 (s, 6H, aromatic OCH3).
Empty mPEG-b-p(HPMA-Bz) micelles (30 mg/mL) and 9% (w/w) curcumin-loaded micelles (30 mg/mL) were prepared in D2O according to Section 2.5. Solid-state NMR experiments were performed using a Bruker Avance III spectrometer operating at a 1H Larmor frequency of 500 MHz equipped with a 4 mm double-resonance probe head at MAS rates varying from 340 to 1120 Hz to distinguish isotropic signals from spinning side bands. Spectral referencing was done using adamantane. Complementary static NMR experiments were performed using a Bruker Avance III spectrometer operating at a 1H Larmor frequency of 600 MHz equipped with a 5 mm triple resonance probe head.
2.7. Curcumin Retention in mPEG-b-p(HPMA-Bz) Micelles Dispersed in Phosphate-Buffered Saline
The stability of the curcumin micelles in PBS (11.9 mM phosphates, 137 mM sodium chloride, and 2.5 mM potassium chloride, pH = 7.4), measured by the retention of curcumin in the micelles, was determined as described previously.32 In short, mPEG-b-p(HPMA-Bz) micelles containing 2.0, 4.8, and 9% curcumin (w/w) were diluted 5-fold in PBS and incubated at 37 °C with constant shaking. The polymer concentration was kept at ∼6 mg/mL for the different formulations while curcumin concentration was around 0.6, 0.3, and 0.12 mg/mL from the highest to the lowest curcumin loading, respectively. The samples were protected from light to avoid photodegradation. At predefined time points, 100 μL aliquots were removed and centrifuged at 5000g for 10 min to spin down precipitated curcumin. Next, the supernatant was diluted at least 10× with methanol and vortexed to disrupt the micelles and solubilize the loaded curcumin. The curcumin concentration was determined by high-performance liquid chromatography (HPLC).6 HPLC analysis was performed on a Waters instrument equipped with a C18 column (SunFire, 5 μm, 150 mm × 4.6 mm; Waters Corporation, Milford, MA). A gradient system was applied using 5:95 (v/v) ACN/water and 100% ACN as eluent A and B, respectively. The pH of the eluents was adjusted by addition of 0.25% (v/v) acetic acid. The gradient ran from 90% A to 70% B in 15 min at a flow rate of 1.2 mL/min. The injection volume was 20 μL. Curcumin was detected at 425 and 254 nm.
2.8. Stability of Curcumin-Loaded Micelles in Plasma
Curcumin-loaded micelles were prepared as described in Section 2.5 using 3.0 mg of curcumin and 30 mg of the different polymers. The curcumin-loaded micelles were diluted 10× in human plasma (total volume of 400 μL) and incubated at 37 °C in a block thermomixer (Eppendorf, Hamburg, Germany) at 450 rpm. The samples were protected from light to avoid photodegradation. At predefined time points, 20 μL aliquots were collected and analyzed using AF4. Separation was performed using an AF2000 separation system (Postnova Analytics, Landsberg, Germany) that consisted of PN1130 isocratic pumps, a degasser, two UV detectors PN3211 (280, 425 nm), and an online-coupled DLS (Zetasizer Nano ZS, Malvern Instruments). The separation channel included a spacer with 350 μm thickness, 27 cm channel length, deltoid shaped channel profile, and a 10 kDa cutoff regenerated cellulose membrane (Postnova Analytics). PBS was used as the mobile phase. To separate the micelles from plasma components, a cross-flow program using a time delay exponential decay was set up. After applying an injection flow of 0.20 mL/min, the sample was focused for 4 min at a focus flow rate of 2.3 mL/min and cross-flow of 2 mL/min. At the end of the focusing step and a transition time of 1 min, the cross-flow was kept constant at 2 mL/min for 7 min. Next, the cross-flow was decreased at an exponential decay of 0.5 to 0.1 mL/min over 20 min. Finally, the cross-flow was kept constant at 0.1 mL/min for 15 min. During the entire run (both focus and elution step), the detector flow rate was 0.5 mL/min. The stability is reported as a percentage of release and calculated as
2.9. Association and Uptake Analysis of Free Curcumin, Curcumin-Loaded Micelles, and Cy5-Labeled Micelles by Flow Cytometry and Confocal Microscopy
Ten percent (w/w) Cy5-labeled mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles (3.0 mg of Cy5-labeled polymer and 27 mg of unlabeled polymer) and curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles (3.0 mg of curcumin and 30 mg of polymer) were prepared in HBS as described in Section 2.5. Cells were seeded at a density of 5 × 105 cells/well in six-well plates and incubated for 18 h. The cell culture medium was subsequently replaced by fresh medium containing 0.3 mg/mL of 10% (w/w) Cy5-labeled micelles, free curcumin (dissolved in DMSO; the final concentration of DMSO was 0.4%), or curcumin-loaded micelles (20 μM) dispersed in HBS. Before flow cytometry, the cells were rinsed twice with PBS, trypsinized using 0.05% trypsin-EDTA, and collected in culture medium. The Cy5 and curcumin fluorescence intensity of approximately 5000 single cells per condition was quantified on a BD FACSCanto II (BD Biosciences, Franklin Lakes, NJ) equipped with dedicated filter sets. Different detector voltages were used for the free curcumin and micellar curcumin groups. Data were processed in FlowJo (BD Biosciences) and plotted using GraphPad Prism (GraphPad Software, La Jolla, CA).
Cells were seeded on glass coverslips in six-well plates at a density of 5 × 105 cells/well and incubated for 18 h. The culture medium was subsequently supplemented with the indicated formulations at 0.3 mg/mL (Cy5-labeled micelles) or 20 μM (free curcumin or curcumin-loaded micelles). Cells were then incubated for 0.5, 1, and 4 h and imaged after washing in PBS with a 40× oil immersion objective using a Leica TCS SP8 SMD scanning unit (Leica Microsystems, Wetzlar, Germany) mounted on a Leica DMI6000 inverted microscope enclosed in an incubator equilibrated at 37 °C. Curcumin and Cy5 fluorescence were excited at 405 and 633 nm, respectively.
2.10. Curcumin and Micelle In Vitro Cytotoxicity
The cytotoxicity of free curcumin, empty mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles, and curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles was investigated in vitro in different cancer cell lines (Neuro2A, EGI-1, TFK-1, SK-ChA-1, and Mz-ChA-1) and noncancerous cells. Cell viability was assessed with the SRB total protein assay.33 EGI-1, TFK-1, SK-ChA-1, Neuro2A, and HUVECs were seeded in 24-well plates at a density of 5 × 104 cells/well, and Mz-ChA-1 cells were seeded at 25 × 104 cells/well due to their slow growth rate. The cells were cultured for 24 h under standard culture conditions. Next, empty and curcumin-loaded micelles dispersed in HBS and curcumin dissolved in DMSO were added at various concentrations (curcumin and polymer concentrations ranged 0–200 μM and 0–800 μg/mL, respectively). The final concentration of DMSO used to solubilize curcumin was 0.4% (v/v) in cell culture medium. Cells incubated with HBS were used as control. After 24, 48, and 72 h of incubation, the wells were washed with PBS and total protein content was determined with the SRB assay as described by Vichai et al.33 Absorbance at 564 nm was recorded using a BioTek Synergy HT microplate reader and the relative cell viability was calculated as follows
The 50% inhibitory concentration (IC50) was derived from a nonlinear regression model (curve fit) based on a sigmoidal inhibitor versus normalized response curve (variable slope) and calculated using GraphPad Prism.
2.11. Acute Toxicity of Empty Micelles in Healthy BALB/c Mice
All animal studies were conducted in compliance with guidelines provided by national regulations and approved by Utrecht University’s institutional review board for animal experiments. Empty polymeric micelles (50 mg/mL) were prepared using mPEG5kDa-b-p(HPMA-Bz)17.1kDa in HBS as described in Section 2.5. The empty micelles were injected intravenously into the BALB/c mice via the tail vein at escalating doses of 100, 300, and 500 mg/kg (100–200 μL, n = 3 per dose). Control mice received 200 μL HBS intravenously. Body weight and general health conditions were monitored before, directly after, and 24 h after administration. Also, blood cell counts were measured after 24 h. At the end of the experiment, the animals were sacrificed via deep isoflurane anesthesia followed by cervical dislocation.
2.12. Circulation Kinetics and Biodistribution of Cy7-Labeled Curcumin Micelles in Mice
Cy7-labeled micelles loaded with curcumin were prepared by dissolving 300 mg of unlabeled mPEG5kDa-b-p(HPMA-Bz)17.1kDa, 4.5 mg of Cy7-labeled polymer (1.5% w/w), and 30.9 mg of curcumin in 4 mL of THF. One milliliter of the resulting solution was subsequently added to 1 mL of HBS. THF was removed by evaporation overnight in a fume hood followed by dialysis against HBS for 1 day to completely remove the organic solvent. Finally, the micellar dispersions were filtered using 0.2 μm regenerated cellulose membranes to remove unencapsulated curcumin and any polymer aggregates. Higher starting polymer and curcumin concentrations were used to compensate for curcumin and polymer loss during dialysis and to achieve the intended injection dose (maximum volume of injection was 200 μL per mouse). The characteristics of the micellar dispersions are reported in (SI Table S2).
For the pharmacokinetics (PK) and biodistribution studies, the formulation was intravenously administered into female BALB/c mice (100–200 μL injection volume), corresponding to a final concentration of 50 mg curcumin/kg. Blood samples were collected into EDTA containing tubes after 1 min (set as 100% injected dose), 10 min, and 1 h via submandibular puncture (50–80 μL), and after 4 and 24 h via cardiac puncture following animal sacrifice as described in Section 2.11. Plasma samples were prepared by centrifugation at 1500g for 10 min. The liver, spleen, heart, kidneys, lungs, and brain were dissected after 4 and 24 h and fluorescence of the excised organs was imaged using a Pearl Impulse small animal imaging system (LI-COR Biosciences, Bad Homburg, Germany). The tissues were rinsed with PBS and snap-frozen in liquid nitrogen and stored at −80 °C until further analysis. The plasma samples (1 volume) were mixed with 4 volumes of ACN and vortexed for 2 min, followed by centrifugation (15 000g, 10 min). Curcumin content in the supernatant was determined by reversed-phase HPLC (RP–HPLC) on a Waters system equipped with a C18 column (SunFire, 5 μm, 150 mm × 4.6 mm). The eluent consisted of 50% ACN and 50% water containing 0.25% acetic acid as a pH modifier. The injection volume and flow rate were 20 μL and 1.2 mL/min, respectively. Curcumin was detected at 425 and 254 nm. The data were analyzed with Empower software (Waters Corporation). Calibration was done using curcumin in 80/20% ACN/water with concentrations ranging from 0.1 to 100 μg/mL.
The concentration of Cy7-labeled micelles in the plasma samples was measured at 800 nm on an Odyssey imaging system (LI-COR Biosciences) using a calibration curve of Cy7-labeled micelles in PBS in a concentration range of 0.2–20 μg/mL. Blood samples were diluted 10–40 times in PBS and the intensity of Cy7 was extrapolated from the linear fit function and corrected for the dilution factor.
To determine the curcumin and Cy7-polymer content in different organs, the excised tissue samples were treated as follows. RIPA buffer (200 μL) was added to ∼100 mg of tissue and the mixture was homogenized at a speed of 6000/s for 60 s using a tissue homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France). The homogenized tissue was treated with ACN (1:4 volume ratio) and vortexed for 2 min, followed by centrifugation at 15 000g for 10 min. The curcumin concentration in the supernatant was analyzed by HPLC as described in Section 2.7. The concentration of Cy7-polymer in different tissues was analyzed as described above by measuring the fluorescence of diluted tissue homogenates in RIPA buffer. Cy7-polymer dissolved in DMSO and subsequently diluted in tissue homogenate suspension obtained from untreated mice was used for calibration (0.2–40 μg/mL).
Pharmacokinetics parameters were calculated using the PkSolver 2.0 add-in template in Microsoft Excel. Calculations were based on noncompartmental and two-compartmental analysis for the micelles and curcumin, respectively.34
2.13. Therapeutic Efficacy of Curcumin Micelles in Tumor-Bearing Mice
The anticancer properties of the curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles (9% w/w, prepared in HBS) were studied in a syngeneic murine subcutaneous neuroblastoma (Neuro2A) model. Tumors were grown from cells subcutaneously injected (3 × 106 cells/100 μL PBS, pH = 7.4) into the right flank of female A/J mice. Nine days after inoculation, the mice received an intravenous bolus of curcumin-loaded micelles at 50 mg curcumin/kg, empty micelles (500 mg polymer/kg), or HBS solution (maximum 200 μL/injection) for 10 consecutive days (n = 5 per group). The infusion of solutions was performed slowly. Body weight and tumor size were monitored daily for 1 month. Tumor volume was calculated using V = (A × 0.52) × B2, where A and B are the largest and the smallest superficial diameters, respectively.35 The mice were sacrificed as described in Section 2.11 upon reaching one of the following human end points: weight loss of >25%, tumor volume reaching 2000 mm3, tumor breaking out of the skin, or when the animal was deemed moribund.
2.14. Statistical Analysis
Statistical analysis was performed in Prism (GraphPad Software, San Diego, CA). A Kruskal–Wallis test with Dunn’s post-hoc analysis was used to compare the differences in IC50 values between different incubation times. A Mann–Whitney U test was used to compare the size of micelles before and after curcumin loading. Statistical significance is designated as *P ≤ 0.05, **P value ≤ 0.01.
3. Results and Discussion
3.1. Characterization of Curcumin-Loaded mPEG-b-p(HPMA-Bz) Micelles
The mPEG-b-p(HPMA-Bz) polymers contained a hydrophilic mPEG5kDa block and different hydrophobic blocks (molecular weights of 5.2, 10.0, and 17.1 kDa). The polymers were synthesized by free-radical polymerization,30 and their characteristics are reported in Table S1.
Curcumin was loaded into the micelles by a nanoprecipitation technique with an encapsulation efficiency of >90% (Table 1). Previously, mPEG-b-p(HPMA-Bz) micelles based on 21 kDa copolymer had a maximal loading capacity of 19% without compromising the entrapment efficiency (87–97%).27 In this study, a lower loading capacity of 9% was chosen to avoid the risk of compromising the solubilization capacity of the hydrophobic core. Also, the size of curcumin-loaded micelles was between 40 and 60 nm for the lowest to the highest-molecular-weight polymers, respectively, and comparable to that of previously studied paclitaxel-loaded micelles.30 A PDI of ≤ 0.14 indicates a narrow size distribution of the particles.36,37
Table 1. Physicochemical Characteristics of Empty and Curcumin-Loaded Micelles (9% w/w Loading) Prepared Using Copolymers with a Different Molecular Weight of the Hydrophobic Blocka,b,c.
| empty
micelles |
curcumin-loaded
micelles |
|||||
|---|---|---|---|---|---|---|
| polymer | Z-ave (nm) | PDI | Z-ave (nm) | PDI | EE (%) | LC (%) |
| mPEG5kDa-b-p(HPMA-Bz)17.1kDa | 52 ± 1 | 0.09 ± 0.02 | 59 ± 3 | 0.09 ± 0.02 | 91 ± 5 | 9.0 ± 0.6 |
| mPEG5kDa-b-p(HPMA-Bz)10.0kDa | 46 ± 1 | 0.05 ± 0.01 | 48 ± 2 | 0.03 ± 0.01 | 94 ± 6 | 8.8 ± 0.5 |
| mPEG5kDa-b-p(HPMA-Bz)5.2kDa | 40 ± 1 | 0.08 ± 0.05 | 38 ± 2 | 0.14 ± 0.10 | 90 ± 12 | 9.0 ± 1.4 |
Z-ave, Z-average hydrodynamic diameter; PDI, polydispersity index.
Data are presented as mean ± SD (n ≥ 3).
Differences between Z-ave were not significant (P > 0.05, Mann–Whitney U-test).
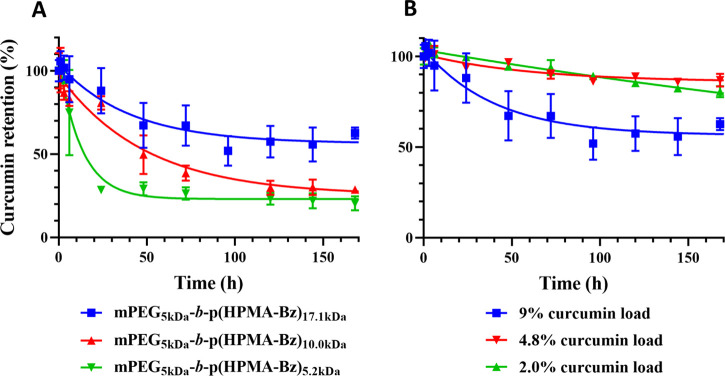
Curcumin retention in the mPEG5kDa-b-p(HPMA-Bz) micelles was studied in PBS at 37 °C by determining micellar curcumin content as a function of incubation time. The results are plotted as both percentage and curcumin concentration in Figure 1 and SI Figure S5, respectively. Figure 1A depicts the stability of micelles containing a 9% (w/w) curcumin load that were formulated from polymers with different molecular weights of the hydrophobic block. The curcumin micelles composed of mPEG5kDa-b-p(HPMA-Bz)17.1kDa showed the highest curcumin retention, retaining approximately 60% of curcumin after 168 h. Micelles comprised of mPEG5kDa-b-p(HPMA-Bz)10.0kDa and mPEG5kDa-b-p(HPMA-Bz)5.2kDa retained 25–30% of the loaded curcumin after 168 h incubation. The rate of curcumin release from the micelles was inversely proportional to the size of the hydrophobic blocks. Most of the curcumin in micelles with the smallest hydrophobic block (5.2 kDa; 70%) was released during the first 2 days. In line with studies using different polymers38,39 and our previous study on paclitaxel-loaded mPEG-b-p(HPMA-Bz),30 micelles based on block copolymers with a larger hydrophobic block retained the loaded drug more effectively. Particle stability as measured by curcumin retention seems to be correlated with the ratio of aromatic rings present in the larger polymers relative to curcumin (Table 2). It can therefore be hypothesized that strong hydrophobic and π–π interactions contribute to the stability of the curcumin-loaded micelles. The presence of π–π interactions in the core of micelles was studied by solid-state 1H NMR (Figures 5 and 6, vide infra).
Figure 1.
(A) Stability of curcumin-loaded mPEG5kDa-b-p(HPMA-Bz) micelles composed of polymers with varying molecular weight of the hydrophobic block (17.1, 10.0, and 5.2 kDa). Micelles contained 9% (w/w) curcumin and were dispersed in PBS, pH = 7.4, at 37 °C during 168 h. (B) Stability of mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles as a function of curcumin load (9, 4.8, and 2.0% w/w) during 168 h incubation in PBS, pH = 7.4, at 37 °C. Data are presented as mean ± SD (n = 3).
Table 2. HPMA-Bz Monomers/Curcumin Ratio in Different Curcumin-Loaded Micelle Formulations at a Feed of 3.0 mg/mL Curcumin and 30 mg/mL Polymera.
| polymer | curcumin (mM) | HPMA-Bz (mM) | ratio HPMA-Bz/curcumin (mol/mol) |
|---|---|---|---|
| mPEG5kDa-b-p(HPMA-Bz)17.1kDa | 7.4 | 94 | 12.7 |
| mPEG5kDa-b-p(HPMA-Bz)10.0kDa | 7.7 | 81 | 10.6 |
| mPEG5kDa-b-p(HPMA-Bz)5.2kDa | 7.3 | 62 | 8.4 |
An exemplary calculation to obtain the ratio of HPMA-Bz/curcumin is provided for mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles in the Supporting Information.
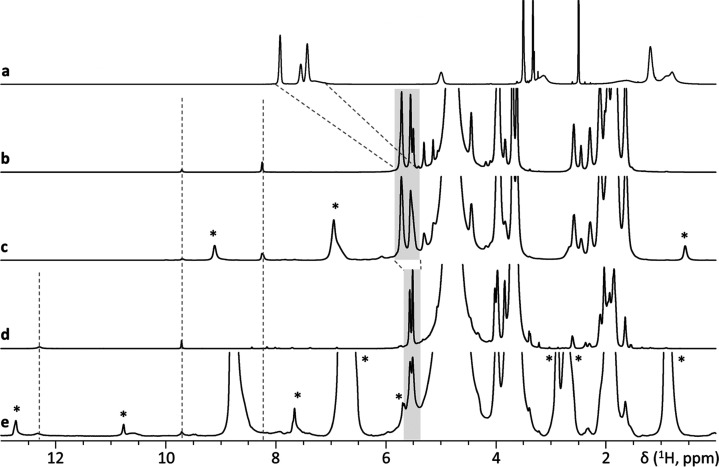
Figure 5.
One-dimensional solid-state 1H NMR spectra obtained using different MAS rates compared to solution-state NMR data. (a) 1H NMR spectrum of mPEG5kDa-b-p(HPMA-Bz)17.1kDa copolymer in solution using DMSO as the solvent. (b) Empty micelles dispersed in D2O (solution state). (c) Empty micelles dispersed in D2O (MAS frequency: 1070 Hz). (d) Curcumin-loaded micelles dispersed in D2O (solution state). (e) Curcumin-loaded micelles dispersed in D2O (MAS frequency 1110 Hz). Asterisks (*) indicate spinning side bands.
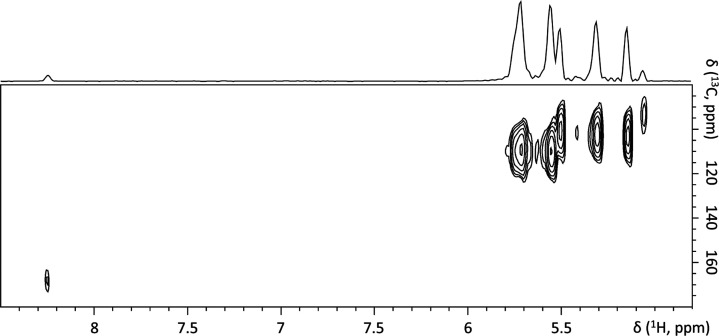
Figure 6.
Spectral cutout of a 2D 1H-13C heteronuclear single quantum correlation (HSQC) spectrum of empty micelles dispersed in D2O (static conditions).
Based on the retention properties in PBS, studies were continued with the curcumin-mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles that were deemed most stable. Next, the effect of curcumin load on retention rate was investigated in PBS. The characteristics of the micelles with 2.0 and 4.8% curcumin load are reported in SI Table S5. Figure 1B reveals that lower curcumin loads lead to improved micelle stability. Compared to a 9% (w/w) load, mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles containing 2.0 and 4.8% curcumin exhibited around 20% curcumin release during 168 h incubation at 37 °C, while 40% of curcumin was released from mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles containing 9% (w/w) curcumin. Similarly, Lübtow et al.40 evaluated the stability of micelles based on ABA triblock copolymers comprised of hydrophilic poly(2-methyl-2-oxazoline) shell A and poly(2-oxazoline) and poly(2-oxazine)-based hydrophobic blocks B. Although the stability study was different (e.g., higher drug loading and longer monitoring time frame) than the present study, the long-term stability of many of their micelle formulations was higher at lower initial drug feed. Another study reported that the release rate of hydrophobic drugs from hydrotropic polymeric micelles based on N,N-diethylnicotinamide increased with higher drug loading.41
As reported previously by Sheybanifard et al.,30 using DLS and TEM analysis, and as confirmed by AF4 in this study (Figure 3), mPEG-b-p(HPMA-Bz) micelles retained their structural stability upon release of the payload from the micelles. Also, it was demonstrated previously by Naksuriya et al.6 that the hydrophobic core of mPEG-b-p(HPMA-Bz) micelles significantly reduced the oxidative degradation of curcumin, compared to solubilized curcumin in basic aqueous medium. Protection from degradation is also apparent for curcumin encapsulated in mPEG5kDa-b-p(HPMA-Bz) micelles. Only a small additional peak with a retention time of around 5 min in the HPLC chromatograms at 254 nm was detected in samples collected at later time points (SI Figure S6), which may be ascribed to the formation of dioxygenated bicyclopentadione (the main degradation product of curcumin) as demonstrated by Naksuriya et al.6 It is arguable that oxidative modification and alkaline degradation of curcumin mainly occurred after curcumin has dissociated from the micelles, once it became deprotonated.
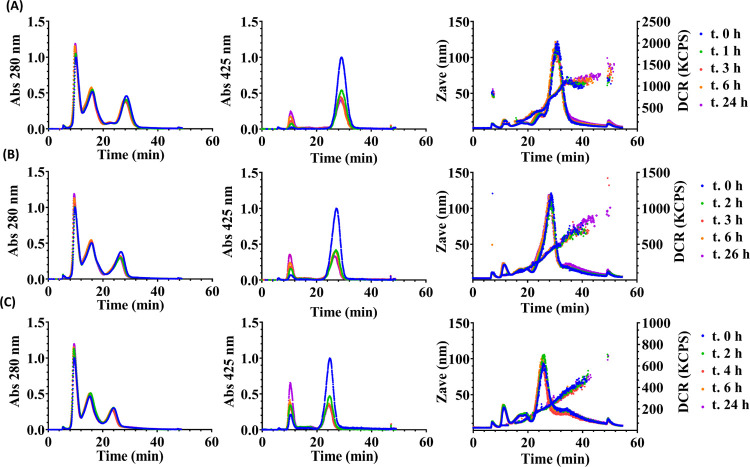
Figure 3.
AF4 fractograms of mPEG5kDa-b-p(HPMA-Bz) micelles with different molecular weights of the hydrophobic block loaded with 9% (w/w) curcumin. (A) Curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles, (B) curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)10.0kDa micelles, and (C) curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)5.2kDa micelles incubated in plasma at 37 °C. Fractograms were recorded at 280 nm (left panel), 425 nm (middle panel), and by the DLS detector (Z-average size and derived count rate (DCR), right panel). Absorbance was normalized to the signal at time zero (the left and middle panels). Abbreviations: Abs, absorbance; KCPS, kilocounts per second.
To mimic quasi-physiological conditions, the stability of curcumin-loaded micelles was studied in human plasma using the AF4 technique. To validate this method, samples consisting of HSA (green line, Figure 2A), plasma spiked with curcumin/DMSO (red lines, Figure 2A), and curcumin-loaded micelles composed of mPEG-b-p(HPMA-Bz) with different molecular weights of the hydrophobic block were fractionated and analyzed by measuring absorbance at 280 nm (Figure 2A; to detect both micelles and protein) and at 425 nm (Figure 2B; to detect curcumin only). Figure 2A confirms that AF4 is a suitable method to separate micelles of different sizes from (lipo)proteins such as albumin, the most abundant plasma protein. The first peak in the plasma sample is albumin, as evidenced by overlaying the fractograms of HSA and plasma (green and red lines, Figure 2A). Moreover, the fractogram of curcumin-spiked plasma at 425 nm (red line, Figure 2B) demonstrates that curcumin binds to and co-elutes with albumin. The observation is in line with previous reports showing that albumin has binding sites for curcumin.42,43
Figure 2.
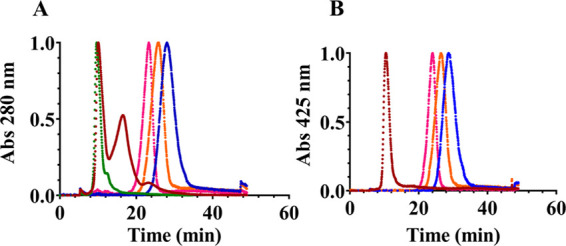
AF4 fractograms of (red) curcumin-spiked plasma, (green) human serum albumin, (blue) curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles, (orange) curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)10.0kDa micelles, and (pink) curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)5.2kDa micelles. Fractograms were recorded at 280 nm (A) and 425 nm (B). The absorbance (Abs) at each wavelength was normalized to maximum intensity.
Curcumin-loaded micelles were mixed with plasma at a final curcumin concentration of 0.3 mg/mL, which is in the sink condition range (0.22–0.44 mg/mL) considering one to two binding sites in albumin (molecular weight 66.5 kDa) at a physiological concentration of around 40 mg/mL. Figure 3 (right panels) shows that the size and size distribution of different micelles did not change up to 24 h incubation in plasma. Also, UV absorbance at 280 nm corresponding to plasma (lipo)proteins and micelles showed a negligible reduction over time (left panels). On the other hand, the absorbance specific for curcumin (425 nm, middle panels) decreased in the micellar fraction, particularly during the first hour. At the same time, curcumin gradually appeared in the albumin fraction, indicating curcumin relocalization from particle to protein.
The rate at which the curcumin area under the curve (AUC) in the albumin fraction increased at the expense of the micellar fraction is reported as percentage of release (Figure 4). At the first measuring point, i.e., after the addition of curcumin-loaded micelles to plasma and before incubation at 37 °C (taken as time zero), 1.2–10.7% curcumin release was observed for the largest to the smallest micelles, respectively. This may be ascribed to curcumin release from the micelles within approximately 10 min after addition to plasma until complete separation by AF4. The release was slower for micelles prepared from mPEG5kDa-b-p(HPMA-Bz)17.1kDa with the longest hydrophobic block (22% in 24 h) versus mPEG5kDa-b-p(HPMA-Bz)10.0kDa (34% release) and mPEG5kDa-b-p(HPMA-Bz)5.2kDa (49% release). It is remarked that the decrease in AUC of curcumin in the micellar fraction over time is not fully compensated by an increase in curcumins’ AUC in the albumin fraction. Partial precipitation and interaction of curcumin with the AF4 membrane may coincide with albumin binding. Disposition of curcumin on the membrane was very evident as the membrane turned yellow over time. In correspondence with the stability data (Figure 1), the AF4 results confirm that curcumin has better retention in micelles prepared from the mPEG5kDa-b-p(HPMA-Bz) with a higher molecular weight of the hydrophobic block in the presence of plasma. Correspondingly, Moquin et al.44 evaluated the stability of AB3 miktoarm star micelles loaded with curcumin in different media such as Dulbecco’s modified Eagle medium (DMEM) enriched with FBS using AF4, and observed that curcumin binds to serum proteins. However, the authors did not clearly explain the role of albumin.
Figure 4.
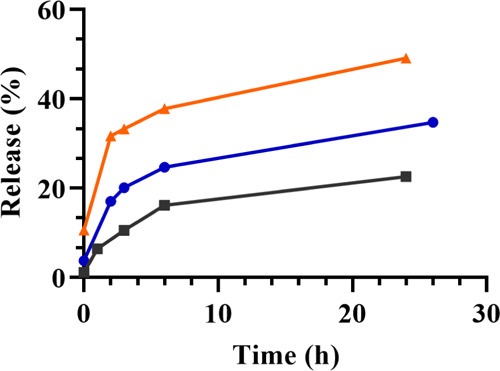
Release of curcumin from mPEG5kDa-b-p(HPMA-Bz) micelles in plasma at 37 °C. Release (%) was calculated as (area under the curve of curcumin in albumin fraction at 425 nm)/(area under the curve of curcumin in the micelle and albumin fractions at 425 nm) × 100%, based on the areas under the curve presented in Figure 3. Curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa, mPEG5kDa-b-p(HPMA-Bz)10.0kDa, and mPEG5kDa-b-p(HPMA-Bz)5.2kDa micelles are depicted as black, blue, and orange, respectively.
3.2. Detection of π–π Stacking in Curcumin-Loaded Micelles
Previously, the presence of π–π stacking interactions among aromatic rings in the core of empty micelles based on a thermosensitive polymer was demonstrated by one- (1D) and two-dimensional (2D) solid-state 1H-NMR.32 In this study, the changes in the solid-state 1H NMR spectrum of mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles upon loading of curcumin were investigated using a similar approach by applying slow MAS in the order of 1 kHz to prevent micelles destruction (Figure 5).
Figure 5a shows the spectrum of copolymer dissolved in DMSO. In line with our previous study, upon formation of micelles in D2O using nanoprecipitation as described in Section 2.5, our solid-state 1H-NMR data showed that aromatic peaks significantly broadened and shifted toward higher field (from 7.4–7.9 to 5.5–5.7 ppm) as expected for π–π interactions.45,46 In addition, weak and broad peaks appeared at 8.25 and 9.31 ppm (Figure 5b,c). Figure 6 demonstrates that signals resonating at 5.5–5.7 ppm in the spectrum of micelles correlate with signals at 100–120 ppm in 13C dimension. There are no other correlations at higher ppm values except for a thus far unassigned 13C signal at ∼170 ppm. In the spectra of curcumin-loaded micelles (Figure 5d,e), peaks of the aromatic protons shifted further to high field. In addition, the signal at 8.21 ppm either disappeared or further broadened beyond detection and a new very low-field and broad signal appeared at 12.30 ppm. The observed spectral changes may indicate the presence of π–π stacking interactions in the hydrophobic core of the micelles which further strengthen upon addition of curcumin. However, to unambiguously prove these interactions, additional 2D NMR experiments, possibly at a higher magnetic field, will be needed.
3.3. Cellular Uptake of mPEG5kDa-b-p(HPMA-Bz)17.1kDa Micelles In Vitro
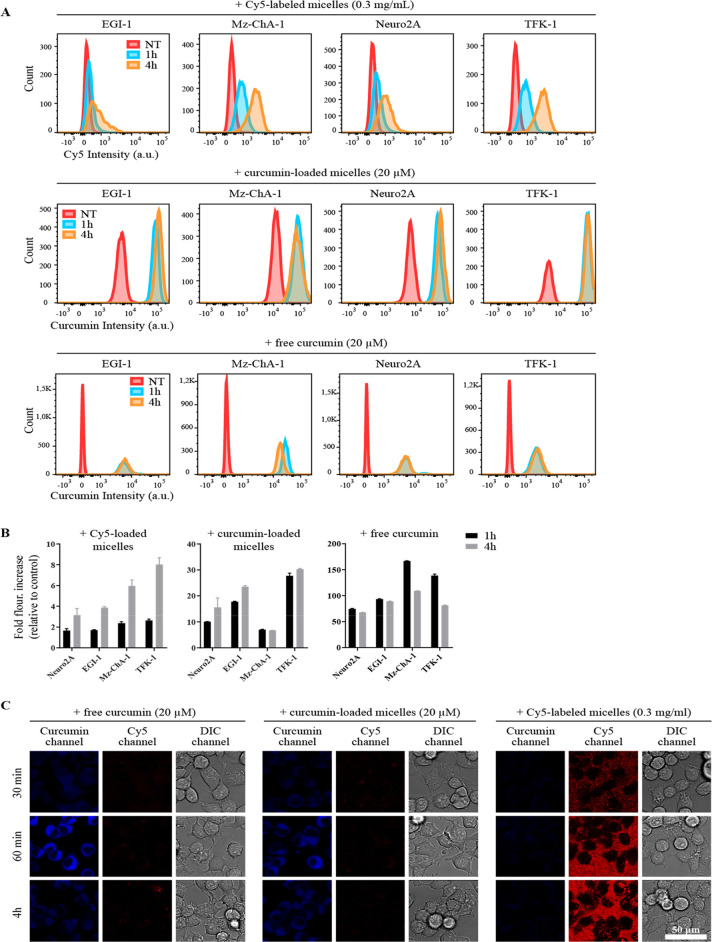
In light of the stability data, further studies focused solely on the mPEG5kDa-b-p(HPMA-Bz)17.1kDa formulation. Cell association and internalization were studied by flow cytometry and confocal imaging, respectively. Figure 7 demonstrates that the micelles were internalized by different types of cancer cells. The internalization of the micelles occurred in a time-dependent manner (Figure 7A,B). Confocal microscopy revealed that free curcumin, empty micelles, and curcumin-loaded micelles were intracellularly detected in Neuro2A cells during a 4 h time span (Figure 7C). At a log P of 2.5, free curcumin can permeate cell membranes passively, which is in line with the chemical behavior of compounds with a log P > 1.6.47 Quantification of curcumin by flow cytometry showed different association kinetics for curcumin in its free form and loaded in micelles. Free curcumin was detected intracellularly within 1 h of incubation. After 4 h incubation, the intracellular curcumin level remained constant in Neuro2A and EGI-1 cells, suggesting equilibrium between curcumin in the culture medium and the cytosol. In TFK-1 and M-ChA-1 cells, curcumin fluorescence intensity had decreased, possibly due to degradation. In line with this study, Sun et al.48 reported similar rapid cell uptake of free curcumin followed by a decline in curcumin fluorescence intensity over time due to degradation of unprotected curcumin. Figure 7B shows that the intracellular curcumin fluorescence intensity was substantially lower (by 3- to 24-fold) for curcumin-loaded micelles incubated at the same curcumin concentration with the cells compared to the free curcumin. This lower fluorescence from intracellular curcumin is possibly attributable to either lower uptake of the micelles or fluorescence quenching of micellar curcumin (SI Figure S8), which suggests that the kinetics of curcumin uptake are governed by micelle uptake. It should be noted that curcumin is strongly solvatochromic, so differential interaction of curcumin with its chemical environment should not be discounted as a (partial) basis for the differences in fluorescence intensity. Importantly, the intracellular fluorescence intensity increased with time upon incubation of the cells with curcumin-loaded micelles, pointing to a relatively slow internalization of the loaded micelles. Taken together, the results demonstrate that the intracellular presence of curcumin is at least partly related to the uptake of intact curcumin-loaded micelles. However, some release of curcumin from the micelles in the cell culture medium and/or in the cytosol cannot be excluded.
Figure 7.
Time-based uptake of free curcumin, empty mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles, and curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles. (A) Representative flow cytograms of cancer cells that had been exposed to free curcumin, curcumin-loaded micelles, and Cy5-labeled micelles. Cells were incubated with nonsupplemented medium containing curcumin in free form or loaded in micelles at 20 μM or Cy5-loaded micelles (0.3 mg/mL) for 1 or 4 h. (B) Fluorescence quantification of results from (A). The bars represent the mean ± SD fold-increase in fluorescence intensity relative to nontreated samples (n = 3 per group). (C) Confocal imaging of uptake of free curcumin, 9% (w/w) curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles, and Cy5-labeled mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles. Neuro2A cells seeded on glass coverslips were incubated in medium supplemented with the indicated formulations for 0.5, 1, and 4 h and imaged by confocal microscopy. Note the binding of the Cy5-labeled micelles to the surface of the coverslips despite five washing steps, visible as granular red staining in-between cells. Nevertheless, this nonspecific binding does not mask the increasing uptake of micelles at later time points, discernible as dotted staining of cell membranes. Galleries constitute representative images. Scale bar: 50 μm. Abbreviations: NT, not treated; DIC, differential interference contrast.
3.4. In Vitro Cytotoxicity of Curcumin-Loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa Micelles
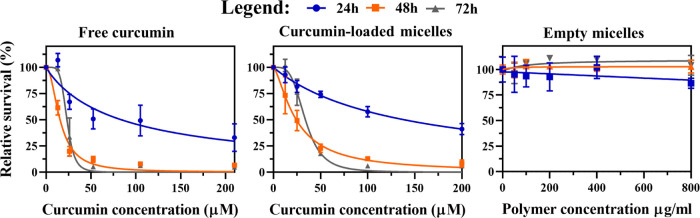
Cytotoxicity of empty and curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles was evaluated in human neuroblastoma (Neuro2A) cells and compared to the cytotoxicity of free curcumin. The data are summarized in Figure 8.
Figure 8.
Cytotoxicity of free curcumin, curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles, and empty mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles in Neuro2A cells incubated for 24, 48, and 72 h. Cell viability was measured with the SRB total protein assay. Data were normalized to the average value of the control (untreated) cells at the respective incubation time. The highest curcumin concentration in mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles (200 μM) corresponds to a polymer concentration of 800 μg/mL. Data were fitted using a nonlinear regression model (curve fit) based on a sigmoidal inhibitor versus normalized response curve (variable slope) and are presented as mean ± SD (n = 4 per incubation time).
Empty micelles did not notably affect Neuro2A cell viability up to a polymer concentration of 800 μg/mL, which is in agreement with previous studies.26,27 The cytotoxicity of free curcumin and curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles was dose- and time-dependent, whereby the most profound cell death occurred during the first 48 h. Curcumin interferes with several vital pathways and, almost without exception, induces apoptosis in tumor cells3 that includes Neuro2A cells. Sidhar et al.49 showed that curcumin can induce reexpression of brain expressed X-linked (Bex) genes and thereby activate p53 and cause apoptosis in Neuro2A cells. Also, the induction of apoptosis in Neuro2A cells via mitochondrial pathways was reported by Jana et al.50
The IC50 values decreased over time and plateaued around 20 μM after 48 and 72 h (Table 3), corresponding to similar IC50 values reported in the literature for other cancer cell lines.3 The higher IC50 value observed at 72 h incubation can be explained by several phenomena. First, curcumin was shown to exit the micellar complex (Figures 1 and 4), which also seems to occur following particle internalization (Figure 7). Free curcumin in the intracellular milieu becomes deprotected and may be degraded and/or metabolized to molecular entities that are not cytotoxic,3 a process that culminates in a sublethal intracellular curcumin concentration within 48 h. Second and in parallel, the remaining viable cells recover and/or activate cell survival pathways,51 which leads to cell proliferation and higher total protein content at 72 h relative to 48 h, and hence a higher IC50 value.
Table 3. IC50 Values (μM) of Free Curcumin and Curcumin-Loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa Micelles in Neuro2A, EGI-1, Mz-CHA-1, Sk-ChA-1, and TFK-1 Cells at Different Curcumin Exposure Timesa,b,c.
| free
curcumin |
curcumin-loaded
micelles |
|||||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| Neuro2A | 83.8 ± 13.5 | 15.6 ± 0.8* | 23.3 ± 1.2 | 138.7 ± 9.0 | 24.3 ± 1.8** | 33.9 ± 1.2 |
| EGI-1 | 21.7 ± 0.5 | 18.2 ± 0.3 | 19.1 ± 0.3 | 38.4 ± 4.4 | 20.6 ± 0.5 | 21.8 ± 0.5 |
| Mz-ChA-1 | 47.6 ± 1.6 | 38.4 ± 1.4 | 37.8 ± 1.1 | 132.8 ± 6.2 | 80.1 ± 2.6 | 69.2 ± 2.6** |
| Sk-ChA-1 | 33.9 ± 1.3 | 21.5 ± 0.4 | 21.3 ± 0.4 | 46.3 ± 1.6 | 36.3 ± 1.1 | 33.4 ± 1.1* |
| TFK-1 | 27.7 ± 0.9 | 20.1 ± 0.9 | 17.5 ± 0.5 | 76.7 ± 4.8 | 36.9 ± 1.7 | 30.8 ± 0.7** |
Data are presented as mean ± SD (free curcumin (n = 3), free curcumin in Neuro2A (n = 4), and curcumin-loaded micelles (n = 4) per time point).
Statistical analysis versus 24 h incubation; *P value ≤ 0.05 and **P value ≤ 0.01 (Kruskal–Wallis test with Dunn’s post-hoc correction).
Statistical analysis between 48 and 72 h incubation time was not significant (Kruskal–Wallis test with Dunn’s post-hoc correction).
The cytotoxicity study was repeated in cell lines derived from human cholangiocarcinoma (EGI-1, Sk-ChA-1, and TFK-1) and gallbladder adenocarcinoma (Mz-ChA-1) to validate the Neuro2A results. Mz-ChA-1 did not show cell death up to a polymer concentration of 800 μg/mL, while TFK-1, EGI-1, and Sk-ChA-1 exhibited a moderate decrease in relative survival at the highest polymer concentration (SI Figure S7). In line with the Neuro2A toxicity data, the IC50 values for free and micellar curcumin in the biliary cancer cell lines decreased and plateaued after 48 h (Table 3). The IC50 values of free curcumin in Neuro2A, EGI-1, Sk-ChA-1, and TFK-1 after 48 and 72 h were comparable and gyrated around 20 μM.
Neuro2A and Mz-ChA-1 cells showed higher IC50 values (84 and 48 μM, respectively) compared to the other cell lines, particularly during the first 24 h of exposure (Table 3). This might be due to the slower growth rate and thus slower metabolic rate of Neuro2A and Mz-ChA-1 cells (doubling time of 3–4 days)52 compared to the other cell lines (around 2 days).52,53 Metabolically hyperactive cancer cells are generally more susceptible to pleiotropic agents such as curcumin because these agents can attack multiple biochemical hubs that culminate in metabolic catastrophe and corollary cell death.54 Generally, the IC50 values of free curcumin for these cell lines are within the range (1–100 μM) of those reported for different types of cancer cell lines in the literature, with a mean ± SD of 21 ± 17 μM.3 Among the cell lines, Neuro2A and Mz-ChA-1 with the lowest growth rates showed the highest IC50 values for curcumin-loaded micelles (above 130 μM after 24 h exposure), affirming this inverse metabolic rate-chemical susceptibility relationship. It should be noted that the IC50 substantially decreased at longer exposure times, particularly for Neuro2A cells, reaching around 24–33 μM (Table 3). In support of these findings, relatively high resistance to both free and curcumin-loaded γ-cyclodextrin liposomal nanoparticles was also reported in a primary cell line established from an untreated patient osteosarcoma biopsy. Chemotherapeutic resistance was attributed to the very slow growth rate and low uptake capacity of the cells with inherently low metabolic activity.55
The higher IC50 value for curcumin-loaded mPEG5kDA-b-p(HPMA-Bz) micelles versus free curcumin is echoed by previous observations by Naksuriya et al.,27 and is likely due to the slow release of curcumin from the micelles either in the medium or intracellularly after internalization of the micelles (Figures 7 and 8).
3.5. Pharmacokinetics and Biodistribution of Curcumin-Loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa Micelles
To detect any possible acute toxicity, mice received an escalating polymer dose of the empty mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles. The mice did not exhibit signs of discomfort, changes in blood cell count, or weight loss 24 h after intravenous administration (SI Figures S11 and S12) for any of the tested doses (100, 300, 500 mg/kg). Also, the cytocompatibility of the micelles was confirmed in vitro in noncancer cells such as fibroblasts (up to 800 μg/mL) and HUVECs even at polymer concentrations up to 3 mg/mL (Figures S9 and S10). Therefore, the in vitro and in vivo results suggest that the micelles have good cytocompatibility and do not induce short-term toxicity.
Next, the circulation kinetics and biodistribution of Cy7-labeled micelles loaded with 9% curcumin were studied in BALB/c mice at a polymer dose of 500 mg/kg, which is equivalent to 50 mg/kg of curcumin. Figure 9 depicts curcumin and Cy7-labeled micelle concentration in plasma at different time points after intravenous injection. More than 50% of the injected dose of micelles was still in the circulation after 24 h, underscoring the in vivo stability of the micelles. This is in agreement with the in vitro stability results obtained by AF4 (Figure 3) and previous studies on mPEG-b-p(HPMA-Bz) micelles by Varela-Moreira et al. and Shi et al.26,56 However, 90% of the loaded curcumin was rapidly eliminated during the first hour. Accordingly, AF4 experiments demonstrated that curcumin-loaded micelles incubated in plasma at 37 °C were stable in terms of size and polydispersity for at least 24 h, but that a significant decrease in curcumin content in the micellar fraction occurred within 1 h (Figure 3). The release of curcumin from the micelles during the first hour after injection was substantially faster than the release rate anticipated by the stability studies in plasma. After intravenous administration, curcumin-loaded micelles are in contact with (lipo)proteins and different blood cells. It has been demonstrated by Bolger et al.57 that curcumin distributes across blood cells and is even metabolized, particularly in erythrocytes, which can explain the observed discrepancy between the AF4 and PK data of the curcumin-loaded micelles.
Figure 9.
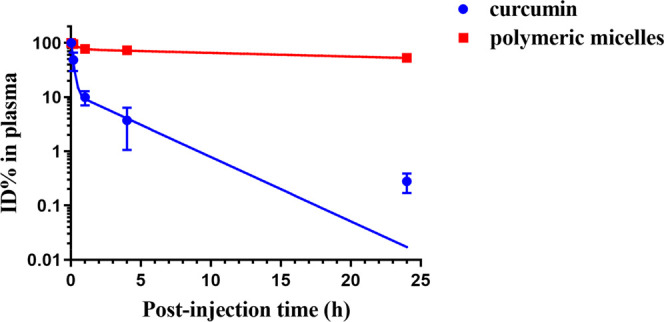
Circulation kinetics of 9% (w/w) curcumin-loaded Cy7-labeled mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles (500 mg/kg, equivalent to 50 mg curcumin/kg) in BALB/c mice. Plasma samples collected at different time points were used to quantify the percentage of the initial dose (ID%) present in the systemic circulation. Data are presented as mean ± SD (n = 3 per time point). Symbols: experimental data of Cy7-labeled polymeric micelles (red squares) and curcumin (blue dots); red and blue lines: fitted curves of compartmental analysis of plasma concentrations.
Noncompartmental and two-compartmental analyses were employed to determine the PK parameters of the micelles and curcumin, respectively (Table 4). The PK profile of curcumin in plasma demonstrated that the initial (t1/2α) and terminal elimination phase (t1/2β) were 6.7 min and 2.5 h, respectively. The α and β phases are primarily attributed to the drug distribution from the central compartment (circulation) and elimination by metabolism and excretion, respectively.58 Based on the curcumin plasma concentration (ID%; Figure 9) and PK (Table 4), the crucial part of the circulation kinetics of curcumin encompasses the first 4 h, where most of the curcumin was eliminated from the circulation and during which time the initial half-life is defined. In contrast, mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles exhibited a prolonged circulation time with a t1/2 of 42 h (Table 4). These results are comparable to the circulation kinetics of intravenously administered PEGylated liposomes.59
Table 4. Pharmacokinetic Parameters of mPEG5kDa-b-p(HPMA-Bz)17.1kDa Micelles and Curcumin after Intravenous Administration of 9% (w/w) Curcumin-Loaded Micelles Containing 1.5% (w/w) Cy7-Labeled Polymer, Analyzed by Noncompartmental and Two-Compartmental Models, Respectivelya,b.
|
t1/2 (h) |
||||||||
|---|---|---|---|---|---|---|---|---|
| component | α-phase | β-phase | t1/2 (h) | AUC0–∞ (μg·h/mL) | V1 (mL/kg) | V2 (mL/kg) | V (mL/kg) | CL (mL/h/kg) |
| curcumin | 0.11 | 2.5 | 319 | 86 | 344 | 157 | ||
| micelles | 42 | 6713 | 70 | 1.2 | ||||
The formulation was injected at 50 mg curcumin/kg body weight, corresponding to 7.8 mg/kg Cy7-labeled polymer.
t1/2, half-life (initial half-life t1/2α and terminal half-life t1/2β); V, volume of distribution; CL, clearance; AUC0–∞, extrapolated area under the curve (AUC) from time zero to infinity.
The results were compared to the pharmacokinetics of free curcumin (compartmental and noncompartmental analysis) reported in the literature. A t1/2α of 0.023 ± 0.001 h (average value ± standard deviation of three independent studies) has been reported for free curcumin in healthy mice,18,60,61 which is around 5 times shorter than the t1/2α of curcumin after administration of the curcumin-loaded mPEG-b-p(HPMA-Bz) micelles (Table 4). In addition, the AUC values reported in the literature were normalized to the injected dose (AUC/mg/kg), which ranged between 0.036 and 4.08 (μg·h/mL)/(mg/kg) for free curcumin in the mice and had a mean ± SEM AUC of 1.1 ± 0.5 μg·h/mL based on nine studies, indicating a broad range in reported AUC values.16,18,20,60−65 Among these studies, Gao et al. reported an AUC value of 60.3 μg·h/mL for free curcumin at a similar curcumin administration dose (50 mg/kg) in mice using noncompartmental analysis.63 The normalized AUCs of curcumin nanoformulations in two studies were 6.0 and 5.9 (μg·h/mL)/(mg/kg) compared to normalized AUCs of 1.2 for free curcumin (μg·h/mL)/(mg/kg) in both studies, respectively.16,63 These nanoformulations increased the AUC by a factor 5. In this study, the normalized AUC of curcumin-loaded mPEG-b-p(HPMA-Bz) micelles was 6.4 (μg·h/mL)/(mg/kg). Therefore, it can be concluded that curcumin-loaded mPEG-b-p(HPMA-Bz) micelles improved the circulation kinetics of curcumin to the same extent as other nanoformulations.
The PK values of the particles were not reported in relevant publications.16,18,20,60−65 It is therefore not possible to relate the curcumin retention with the circulation time of the particles. However, since the curcumin concentration declined rapidly in the circulation and the AUC ratio of nanoformulated curcumin to free curcumin is around 5,16,63 these nanoformulations (including mPEG-b-p(HPMA-Bz) micelles) thus mainly acted as an efficient solubilizer. Similar to the mPEG-b-p(HPMA-Bz) micelles of the present study, Gong et al.20 utilized an intermolecular π–π stacking strategy using mPEG-PCL-Phe(Boc)-based micelles to improve the PK of curcumin. These micelles prolonged the circulation kinetics of curcumin compared to both mPEG-PCL micelles and the free form by ∼3-fold. The normalized AUC for the mPEG-PCL-based curcumin formulation was 0.23 (μg·h/mL)/(mg/kg), which is lower than the AUC reported for mPEG-b-p(HPMA-Bz) micelles (6.4 (μg·h/mL)/(mg/kg)). Accordingly, mPEG-b-p(HPMA-Bz) micelles can result in higher curcumin exposure per injected dose. The difference might be ascribed to the number of aromatic rings in the polymers since only the end of the mPEG-PCL polymers was modified.
The volume of distribution of mPEG-b-p(HPMA-Bz) micelles was 70 mL/kg (Table 4), indicating that the micelles were retained in the circulation considering that an average mouse has a blood volume of 85–96 mL/kg66 or blood volume of around 1.5–2.5 mL (6–8% of the body weight).67 In contrast, the larger volume of distribution (334 mL/kg) for curcumin shows that it leaves the vascular space extensively.68
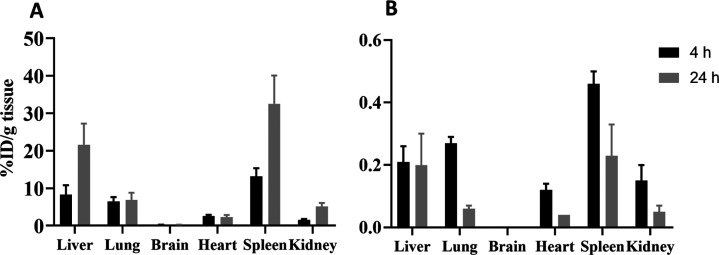
Figures 10A and 11 show that 20 and 30% of the injected dose of Cy7-labeled micelles accumulated in organs that extensively harbor cells of the mononuclear phagocyte system (MPS), namely, the liver and spleen, respectively, 24 h post-administration. Approximately 5% of Cy7-labeled polymer was detected in the kidneys, suggesting disintegration of micelles into polymer unimers that have a molecular size below the size threshold for glomerular filtration.69 The accumulation of Cy7-labeled mPEG-b-p(HPMA-Bz) polymers in the liver, spleen, and kidneys increased over time due to clearance by the MPS and liberation of unimers from the micelles. It is important to note that part of the polymeric micelle disposition in hypervascularized tissue (e.g., liver, lungs, and kidneys) can be ascribed to the micelles that are still in the blood circulation since these micelles had a long circulation time (Table 4).
Figure 10.
Accumulation of curcumin-loaded Cy7-labeled mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles in tissues of BALB/c mice at 4 and 24 h after intravenous injection. (A) Percentage of injected dose (ID%)/g tissue of Cy7-labeled polymer and (B) ID%/g tissue of curcumin. Data represent mean ± SD (n = 3 per time interval).
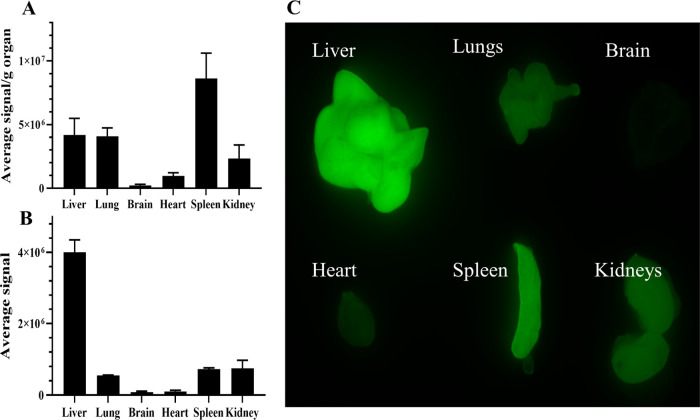
Figure 11.
Ex vivo fluorescence reflectance imaging (FRI) analysis of the accumulation of the Cy7-labeled mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles in organs 24 h after intravenous administration. Images were obtained at λex = 785 nm with λem = 820 nm (n = 3), i.e., at wavelengths at which there is no tissue autofluorescence (data not shown). The extent of Cy7-labeled micelle accumulation is plotted per gram of tissue (A) and per organ (B).
Figure 10B shows that curcumin accumulated in the spleen, liver, and lungs (i.e., MPS-enriched tissues) after 4 h circulation. The curcumin concentration amounted to 0.2–3 μg/g tissue in the different organs at 4 and 24 h post-administration (SI Figure S13). In line with other curcumin biodistribution studies, curcumin cleared from the tissues over time (Figure 10B).16,18,70 The difference between curcumin and micelles biodistribution patterns may be attributable to their dissociation and different circulation kinetics. Unlike the stable micelles, curcumin cleared faster from plasma.
3.6. Therapeutic Efficacy of Curcumin-Loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa Micelles
The potential therapeutic benefit of the curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles was evaluated in Neuro2A tumor-bearing mice. A syngeneic tumor model was selected to avoid xenograft models without a fully functional immune system. Varela-Moreira et al.56 demonstrated that compromising the status of the immune system can significantly change the circulation kinetics of mPEG-b-p(HPMA-Bz) micelles. Also, Neuro2A (neuroblastoma) tumors are reasonably vascularized and can present therapeutic advantages in the form of an EPR effect.71,72
Nine days after inoculation, the mice received 10 consecutive daily injections of curcumin-loaded micelles. Tumor volume and body weight were monitored over time. The group that received curcumin-loaded micelles did not show tumor growth suppression or growth delay compared to the control groups receiving empty micelles and HBS (SI Figure S14). Since the size of tumors on the day of starting the treatment was relatively large (526 ± 180 mm3), it was hypothesized that the curcumin dosage was insufficient to exert a therapeutic response, as a result of which the mice reached humane end points. Therefore, in a follow-up experiment, treatment was started at a time when the tumors were palpable (7 days after the inoculation of the tumor cells). Although the overall number of mice that developed tumors was lower (6 out of 27 mice) due to the inoculation with fewer tumor cells, no anticancer effect was observed under otherwise similar therapeutic conditions (SI Figure S15). Contrary to curcumin-loaded nanosized delivery systems that exhibited antitumor effects in different human tumor murine models, the curcumin-loaded mPEG5kDa-b-p(HPMA-Bz) micelles did not impart tumor inhibition despite similar PK profiles as other tested nanoformulations. The absence of therapeutic efficacy may stem from the possibility that curcumin is extracted too rapidly from the micelles and degraded and/or cleared from the circulation before ample accumulation could have occurred in the tumor to instill notable tumoricidal effects. Another confounding variable may be the relatively low sensitivity of the Neuro2A cells to curcumin-loaded micelles, as was shown in vitro (Table 3), which translates to higher curcumin levels needing to be achieved in the Neuro2A tumors than in other types of subcutaneous tumor xenografts for a therapeutic effect.
Taken together, mPEG-b-p(HPMA-Bz) micelles are excellent solubilizers for curcumin with a PK profile that is comparable to other curcumin nanoformulations. To date, the majority of the developed curcumin nanoformulations acts as solubilizers since they only improved the AUC by a factor of 1.3- to 5-fold compared to curcumin in its free form, as pointed out in the introduction. It should be emphasized that this increase was sufficient to confer antitumor activity in non-Neuro2A tumor-bearing mouse models,16,20,62,63,70 similar to the well-known paclitaxel nanomedicines Genexol and Abraxane. These formulations hardly improve the PK of loaded paclitaxel but do have valuable therapeutic benefits.73−76 It is therefore possible that curcumin-loaded mPEG-b-p(HPMA-Bz) micelles are effective in other human tumor models that were proven susceptible to curcumin and other nanomedicines.
4. Conclusions
Curcumin-loaded mPEG5KDa-b-p(HPMA-Bz) micelles with different block copolymer sizes were prepared, characterized, and subjected to stability analysis. The micelles were stable in plasma, but curcumin was rapidly transferred from the polymeric micelles to albumin. The polymeric micelles with the highest-molecular-weight hydrophobic block were found to be the most stable formulation, exhibiting comparable pharmacokinetics in mice as other curcumin nanoformulations reported in the literature. Nevertheless, the curcumin-loaded micelles did not stall tumor growth or reduce tumor size in a murine human neuroblastoma model, despite exhibiting in vitro cytotoxicity. It might be possible to achieve therapeutic efficacy in this tumor model using combination therapy with other chemotherapeutic agents.77,78 In the final analysis, curcumin-loaded mPEG5KDa-b-p(HPMA-Bz)17.1kDa micelles can either be repurposed to target more amenable cancers such as hematological malignancies or cancers of the lymphatic and immune system3 or injected directly into the tumor for local uptake and gradual release of the chemotherapeutic. Alternatively, the formulation can be subjected to more research focused on developing more stable curcumin-loaded micelles.
Acknowledgments
This work was funded by the European Union’s Horizon 2020 research and innovation program Marie Sklodowska-Curie Innovative Training Networks (ITN) under grant no. 676137. Michal Heger is currently supported by grants from the Dutch Cancer Foundation (KWF project # 10666), National Natural Science Foundation of China (#81872220), a Zhejiang Provincial Foreign Expert Program Grant, Zhejiang Provincial Key Natural Science Foundation of China (#Z20H160031), and a grant for the establishment of the Jiaxing Key Laboratory for Photonanomedicine and Experimental Therapeutics. The SK-ChA-1, Mz-ChA-1 cell lines were kindly received under a license agreement to Michal Heger from Alexander Knuth and Claudia Matter (University Hospital Zurich, Switzerland). The AF4 data used in this research were generated through access to the Nanobiotechnology Laboratory under the Framework of access to the Joint Research Centre Physical Research Infrastructures of the European Commission (Stability of polymeric micelles in serum, MicStab, Research Infrastructure Access Agreement RIAA 35050/7).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.molpharmaceut.0c01114.
Characteristics of the mPEG5kDa-b-p(HPMA-Bz)x block copolymers as determined by 1H-NMR and GPC; 1H-NMR of mPEG5kDa-b-p(HPMA-Bz)17.1kDa and curcumin in DMSO; GPC chromatograms of Cy7- and Cy5-labeled mPEG-b-p(HPMA-Bz); characterization of Cy7-labeled micelles loaded with curcumin; effect of dialysis on curcumin-loaded micelles; validation of HPLC method; physicochemical characteristics of mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles loaded with 4.8 and 2.0% w/w curcumin; curcumin concentration over time loaded in mPEG5kDa-b-p(HPMA-Bz) micelles composed of polymers with varying molecular weight of the hydrophobic block and curcumin loading; representative HPLC chromatograms of curcumin samples obtained during stability study in PBS at 37 °C; representative calculation for the ratio of HPMA-Bz/curcumin; in vitro cytotoxicity of free curcumin, curcumin-loaded micelles, and empty micelles on EGI-1, Mz-ChA-1, Sk-ChA-1, and TFK-1 cell lines; fluorescence spectra of curcumin-loaded micelles with different loadings; cytotoxicity of empty micelles in hPSC and HUVECs; possible acute toxicity of empty micelles (mice body weight before and 24 h after intravenous administration of empty micelles with doses of 100, 300, and 500 mg/kg); blood cell count in mice after 24 h treatment with doses of 100, 300, and 500 mg/kg; curcumin concentration in excised tissues after intravenous injection of curcumin-loaded mPEG5kDa-b-p(HPMA-Bz)17.1kDa micelles; therapeutic efficacy of curcumin-loaded micelles in fast-growing Neuro2A tumor model (3 × 106 cells/100 μL PBS, pH = 7.4); and therapeutic efficacy of curcumin-loaded micelles in slow-growing Neuro2A tumor model (1 × 106 cells/100 μL PBS, pH = 7.4) (PDF)
The authors declare the following competing financial interest(s): Part of this research has been done in collaboration with the European Commission in Italy. The work was conducted by Dr. Robin B. Capomaccio while working at the European Commission with no connection to AstraZeneca. Dr. Robin B. Capomaccio is currently an employee at AstraZeneca. No further work on the publication has been conducted using AstraZeneca resources. Michal Heger is co-founder of Nurish.Me and has equity in that company (whose business activities are unrelated to the present work).
Supplementary Material
References
- Patil S. S.; Bhasarkar S.; Rathod V. K. Extraction of curcuminoids from Curcuma longa: comparative study between batch extraction and novel three phase partitioning. Prep. Biochem. Biotechnol. 2019, 49, 407–418. 10.1080/10826068.2019.1575859. [DOI] [PubMed] [Google Scholar]
- Shanmugam M. K.; Rane G.; Kanchi M. M.; Arfuso F.; Chinnathambi A.; Zayed M. E.; Alharbi S. A.; Tan B. K.; Kumar A. P.; Sethi G. The multifaceted role of curcumin in cancer prevention and treatment. Molecules 2015, 20, 2728–2769. 10.3390/molecules20022728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger M.; van Golen R. F.; Broekgaarden M.; Michel M. C. The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharmacol. Rev. 2014, 66, 222–307. 10.1124/pr.110.004044. [DOI] [PubMed] [Google Scholar]
- Vareed S. K.; Kakarala M.; Ruffin M. T.; Crowell J. A.; Normolle D. P.; Djuric Z.; Brenner D. E. Pharmacokinetics of curcumin conjugate metabolites in healthy human subjects. Cancer Epidemiol., Biomarkers Prev. 2008, 17, 1411–1417. 10.1158/1055-9965.EPI-07-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa S.; Atsumi T.; Ishihara M.; Kadoma Y. Cytotoxicity, ROS-generation activity and radical-scavenging activity of curcumin and related compounds. Anticancer Res. 2004, 24, 563–570. [PubMed] [Google Scholar]
- Naksuriya O.; van Steenbergen M. J.; Torano J. S.; Okonogi S.; Hennink W. E. A kinetic degradation study of curcumin in its free form and loaded in polymeric micelles. AAPS J. 2016, 18, 777–787. 10.1208/s12248-015-9863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.; Gordon O. N.; Edwards R. L.; Luis P. B. Degradation of curcumin: from mechanism to biological implications. J. Agric. Food Chem. 2015, 63, 7606–7614. 10.1021/acs.jafc.5b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P.; Kunnumakkara A. B.; Newman R. A.; Aggarwal B. B. Bioavailability of curcumin: problems and promises. Mol. Pharmaceutics 2007, 4, 807–818. 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- Wicki A.; Witzigmann D.; Balasubramanian V.; Huwyler J. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J. Controlled Release 2015, 200, 138–157. 10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- Bobo D.; Robinson K. J.; Islam J.; Thurecht K. J.; Corrie S. R. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm. Res. 2016, 33, 2373–2387. 10.1007/s11095-016-1958-5. [DOI] [PubMed] [Google Scholar]
- Houdaihed L.; Evans J. C.; Allen C. Overcoming the road blocks: advancement of block copolymer micelles for cancer therapy in the clinic. Mol. Pharmaceutics 2017, 14, 2503–2517. 10.1021/acs.molpharmaceut.7b00188. [DOI] [PubMed] [Google Scholar]
- Cabral H.; Miyata K.; Osada K.; Kataoka K. Block copolymer micelles in nanomedicine applications. Chem. Rev. 2018, 118, 6844–6892. 10.1021/acs.chemrev.8b00199. [DOI] [PubMed] [Google Scholar]
- Fang J.; Nakamura H.; Maeda H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Delivery Rev. 2011, 63, 136–151. 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Naksuriya O.; Okonogi S.; Schiffelers R. M.; Hennink W. E. Curcumin nanoformulations: A review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment. Biomaterials 2014, 35, 3365–3383. 10.1016/j.biomaterials.2013.12.090. [DOI] [PubMed] [Google Scholar]
- Mehanny M.; Hathout R. M.; Geneidi A. S.; Mansour S. Exploring the use of nanocarrier systems to deliver the magical molecule; Curcumin and its derivatives. J. Controlled Release 2016, 225, 1–30. 10.1016/j.jconrel.2016.01.018. [DOI] [PubMed] [Google Scholar]
- Gong C.; Deng S.; Wu Q.; Xiang M.; Wei X.; Li L.; Gao X.; Wang B.; Sun L.; Chen Y.; Li Y.; Liu L.; Qian Z.; Wei Y. Improving antiangiogenesis and anti-tumor activity of curcumin by biodegradable polymeric micelles. Biomaterials 2013, 34, 1413–1432. 10.1016/j.biomaterials.2012.10.068. [DOI] [PubMed] [Google Scholar]
- Lübtow M. M.; Nelke L. C.; Seifert J.; Kühnemundt J.; Sahay G.; Dandekar G.; Nietzer S. L.; Luxenhofer R. Drug induced micellization into ultra-high capacity and stable curcumin nanoformulations: Physico-chemical characterization and evaluation in 2D and 3D in vitro models. J. Controlled Release 2019, 303, 162–180. 10.1016/j.jconrel.2019.04.014. [DOI] [PubMed] [Google Scholar]
- Song Z.; Feng R.; Sun M.; Guo C.; Gao Y.; Li L.; Zhai G. Curcumin-loaded PLGA-PEG-PLGA triblock copolymeric micelles: Preparation, pharmacokinetics and distribution in vivo. J. Colloid Interface Sci. 2011, 354, 116–123. 10.1016/j.jcis.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Shi Y.; Lammers T.; Storm G.; Hennink W. E. Physico-chemical strategies to enhance stability and drug retention of polymeric micelles for tumor-targeted drug delivery. Macromol. Biosci. 2017, 17, 1600160 10.1002/mabi.201600160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F.; Chen D.; Teng X.; Ge J.; Ning X.; Shen Y. L.; Li J.; Wang S. Curcumin-loaded blood-stable polymeric micelles for enhancing therapeutic effect on erythroleukemia. Mol. Pharmaceutics 2017, 14, 2585–2594. 10.1021/acs.molpharmaceut.6b01171. [DOI] [PubMed] [Google Scholar]
- D’Souza S. A review of in vitro drug release test methods for nano-sized dosage forms. Adv. Pharm. 2014, 2014, 304757 10.1155/2014/304757. [DOI] [Google Scholar]
- Abouelmagd S. A.; Sun B.; Chang A. C.; Ku Y. J.; Yeo Y. Release kinetics study of poorly water-soluble drugs from nanoparticles: Are we doing it right?. Mol. Pharmaceutics 2015, 12, 997–1003. 10.1021/mp500817h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil D.; Frank-Kamenetskii A.; Barry J.; Reukov V.; Xiang Y.; Das A.; Varma A. K.; Kindy M. S.; Banik N. L.; Vertegel A. Albumin-assisted method allows assessment of release of hydrophobic drugs from nanocarriers. Biotechnol. J. 2018, 13, 1700337 10.1002/biot.201700337. [DOI] [PubMed] [Google Scholar]
- Contado C. Field flow fractionation techniques to explore the “nano-world”. Anal. Bioanal. Chem. 2017, 409, 2501–2518. 10.1007/s00216-017-0180-6. [DOI] [PubMed] [Google Scholar]
- Wagner M.; Holzschuh S.; Traeger A.; Fahr A.; Schubert U. S. Asymmetric flow field-flow fractionation in the field of nanomedicine. Anal. Chem. 2014, 86, 5201–5210. 10.1021/ac501664t. [DOI] [PubMed] [Google Scholar]
- Shi Y.; van der Meel R.; Theek B.; Oude Blenke E.; Pieters E. H. E.; Fens M. H. A. M.; Ehling J.; Schiffelers R. M.; Storm G.; van Nostrum C. F.; Lammers T.; Hennink W. E. Complete regression of xenograft tumors upon targeted delivery of paclitaxel via Π–Π stacking stabilized polymeric micelles. ACS Nano 2015, 9, 3740–3752. 10.1021/acsnano.5b00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naksuriya O.; Shi Y.; van Nostrum C. F.; Anuchapreeda S.; Hennink W. E.; Okonogi S. HPMA-based polymeric micelles for curcumin solubilization and inhibition of cancer cell growth. Eur. J. Pharm. Biopharm. 2015, 94, 501–512. 10.1016/j.ejpb.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Bresseleers J.; Bagheri M.; Storm G.; Metselaar J. M.; Hennink W. E.; Meeuwissen S. A.; van Hest J. C. M. Scale-up of the manufacturing process to produce docetaxel-loaded mPEG-b-p(HPMA-Bz) block copolymer micelles for pharmaceutical applications. Org. Process Res. Dev. 2019, 23, 2707–2715. 10.1021/acs.oprd.9b00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri M.; Bresseleers J.; Varela-Moreira A.; Sandre O.; Meeuwissen S. A.; Schiffelers R. M.; Metselaar J. M.; van Nostrum C. F.; van Hest J. C. M.; Hennink W. E. Effect of formulation and processing parameters on the size of mPEG-b-p(HPMA-Bz) polymeric micelles. Langmuir 2018, 34, 15495–15506. 10.1021/acs.langmuir.8b03576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheybanifard M.; Beztsinna N.; Bagheri M.; Miriam Buhl E.; Bresseleers J.; Varela-Moreira A.; Shi Y.; van Nostrum C. F.; van der Pluijm G.; Storm G.; Hennink W. E.; Lammers T.; Metselaar J. M. Systematic evaluation of design features enables efficient selection of Π electron-stabilized polymeric micelles. Int. J. Pharm. 2020, 119409 10.1016/j.ijpharm.2020.119409. [DOI] [PubMed] [Google Scholar]
- Zhuang W.-R.; Wang Y.; Cui P.-F.; Xing L.; Lee J.; Kim D.; Jiang H.-L.; Oh Y.-K. Applications of π-π stacking interactions in the design of drug-delivery systems. J. Controlled Release 2019, 294, 311–326. 10.1016/j.jconrel.2018.12.014. [DOI] [PubMed] [Google Scholar]
- Shi Y.; van Steenbergen M. J.; Teunissen E. A.; Novo L.; Gradmann S.; Baldus M.; van Nostrum C. F.; Hennink W. E. Pi-pi stacking increases the stability and loading capacity of thermosensitive polymeric micelles for chemotherapeutic drugs. Biomacromolecules 2013, 14, 1826–1837. 10.1021/bm400234c. [DOI] [PubMed] [Google Scholar]
- Vichai V.; Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Huo M.; Zhou J.; Xie S. PKSolver: An add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Programs Biomed. 2010, 99, 306–314. 10.1016/j.cmpb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Fens M. H.; Hill K. J.; Issa J.; Ashton S. E.; Westwood F. R.; Blakey D. C.; Storm G.; Ryan A. J.; Schiffelers R. M. Liposomal encapsulation enhances the antitumour efficacy of the vascular disrupting agent ZD6126 in murine B16.F10 melanoma. Br. J. Cancer 2008, 99, 1256–1264. 10.1038/sj.bjc.6604675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J.; McKenna S. A.; Patel T. R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S. DLS and zeta potential – What they are and what they are not?. J. Controlled Release 2016, 235, 337–351. 10.1016/j.jconrel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Lin W.-J.; Juang L.-W.; Lin C.-C. Stability and release performance of a series of pegylated copolymeric micelles. Pharm. Res. 2003, 20, 668–673. 10.1023/A:1023215320026. [DOI] [PubMed] [Google Scholar]
- Hussein Y. H. A.; Youssry M. Polymeric micelles of biodegradable diblock copolymers: Enhanced encapsulation of hydrophobic drugs. Materials 2018, 11, 688. 10.3390/ma11050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübtow M. M.; Haider M. S.; Kirsch M.; Klisch S.; Luxenhofer R. Like dissolves like? A comprehensive evaluation of partial solubility parameters to predict polymer-drug compatibility in ultrahigh drug-loaded polymer micelles. Biomacromolecules 2019, 20, 3041–3056. 10.1021/acs.biomac.9b00618. [DOI] [PubMed] [Google Scholar]
- Huh K. M.; Lee S. C.; Cho Y. W.; Lee J.; Jeong J. H.; Park K. Hydrotropic polymer micelle system for delivery of paclitaxel. J. Controlled Release 2005, 101, 59–68. 10.1016/j.jconrel.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Barik A.; Mishra B.; Kunwar A.; Indira Priyadarsini K. Interaction of curcumin with human serum albumin: Thermodynamic properties, fluorescence energy transfer and denaturation effects. Chem. Phys. Lett. 2007, 436, 239–243. 10.1016/j.cplett.2007.01.006. [DOI] [Google Scholar]
- Basu A.; Suresh Kumar G. Elucidating the energetics of the interaction of non-toxic dietary pigment curcumin with human serum albumin: A calorimetric study. J. Chem. Thermodyn. 2014, 70, 176–181. 10.1016/j.jct.2013.10.037. [DOI] [Google Scholar]
- Moquin A.; Sharma A.; Cui Y.; Lau A.; Maysinger D.; Kakkar A. Asymmetric AB3 miktoarm star polymers: Synthesis, self-Assembly, and study of micelle stability using AF4 for efficient drug delivery. Macromol. Biosci. 2015, 15, 1744–1754. 10.1002/mabi.201500186. [DOI] [PubMed] [Google Scholar]
- Mafra L.; Santos S. M.; Siegel R.; Alves I.; Almeida Paz F. A.; Dudenko D.; Spiess H. W. Packing interactions in hydrated and anhydrous forms of the antibiotic ciprofloxacin: a solid-state NMR, X-ray diffraction, and computer simulation study. J. Am. Chem. Soc. 2012, 134, 71–74. 10.1021/ja208647n. [DOI] [PubMed] [Google Scholar]
- Carignani E.; Borsacchi S.; Bradley J. P.; Brown S. P.; Geppi M. Strong intermolecular ring current influence on 1H chemical shifts in two crystalline forms of naproxen: a combined solid-state NMR and DFT study. J. Phys. Chem. C 2013, 117, 17731–17740. 10.1021/jp4044946. [DOI] [Google Scholar]
- Bennion B. J.; Be N. A.; McNerney M. W.; Lao V.; Carlson E. M.; Valdez C. A.; Malfatti M. A.; Enright H. A.; Nguyen T. H.; Lightstone F. C.; Carpenter T. S. Predicting a drug’s membrane permeability: A computational model validated with in vitro permeability assay data. J. Phys. Chem. B 2017, 121, 5228–5237. 10.1021/acs.jpcb.7b02914. [DOI] [PubMed] [Google Scholar]
- Sun J.; Bi C.; Chan H. M.; Sun S.; Zhang Q.; Zheng Y. Curcumin-loaded solid lipid nanoparticles have prolonged in vitro antitumour activity, cellular uptake and improved in vivo bioavailability. Colloids Surf., B 2013, 111, 367–375. 10.1016/j.colsurfb.2013.06.032. [DOI] [PubMed] [Google Scholar]
- Sidhar H.; Giri R. K. Induction of Bex genes by curcumin is associated with apoptosis and activation of p53 in N2a neuroblastoma cells. Sci. Rep. 2017, 7, 41420 10.1038/srep41420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana N. R.; Dikshit P.; Goswami A.; Nukina N. Inhibition of proteasomal function by curcumin induces apoptosis through mitochondrial pathway. J. Biol. Chem. 2004, 279, 11680–11685. 10.1074/jbc.M310369200. [DOI] [PubMed] [Google Scholar]
- Broekgaarden M.; Weijer R.; van Gulik T. M.; Hamblin M. R.; Heger M. Tumor cell survival pathways activated by photodynamic therapy: a molecular basis for pharmacological inhibition strategies. Cancer Metastasis Rev. 2015, 34, 643–690. 10.1007/s10555-015-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuth A.; Gabbert H.; Dippold W.; Klein O.; Sachsse W.; Bitter-Suermann D.; Prellwitz W.; Meyer zum Büschenfelde K. H. Biliary adenocarcinoma. Characterisation of three new human tumor cell lines. J. Hepatol. 1985, 1, 579–596. 10.1016/S0168-8278(85)80002-7. [DOI] [PubMed] [Google Scholar]
- Saijyo S.; Kudo T.; Suzuki M.; Katayose Y.; Shinoda M.; Muto T.; Fukuhara K.; Suzuki T.; Matsuno S. Establishment of a new extrahepatic bile duct carcinoma cell line, TFK-1. Tohoku J. Exp. Med. 1995, 177, 61–71. 10.1620/tjem.177.61. [DOI] [PubMed] [Google Scholar]
- Lee K. W.; Bode A. M.; Dong Z. Molecular targets of phytochemicals for cancer prevention. Nat. Rev. Cancer 2011, 11, 211–218. 10.1038/nrc3017. [DOI] [PubMed] [Google Scholar]
- Dhule S. S.; Penfornis P.; Frazier T.; Walker R.; Feldman J.; Tan G.; He J.; Alb A.; John V.; Pochampally R. Curcumin-loaded γ-cyclodextrin liposomal nanoparticles as delivery vehicles for osteosarcoma. Nanomedicine 2012, 8, 440–451. 10.1016/j.nano.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Moreira A.; van Straten D.; van Leur H. F.; Ruiter R. W. J.; Deshantri A. K.; Hennink W. E.; Fens M. H. A. M.; Groen R. W. J.; Schiffelers R. M. Polymeric micelles loaded with carfilzomib increase tolerability in a humanized bone marrow-like scaffold mouse model. Int. J. Pharm. 2020, 100049 10.1016/j.ijpx.2020.100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger G. T.; Licollari A.; Tan A.; Greil R.; Vcelar B.; Majeed M.; Helson L. Distribution and metabolism of Lipocurc (liposomal curcumin) in dog and human blood cells: species selectivity and pharmacokinetic relevance. Anticancer Res. 2017, 37, 3483–3492. [DOI] [PubMed] [Google Scholar]
- Brown D.; Tomlin M.. Pharmacokinetic Principles. In Pharmacology & Pharmacokinetics: A Basic Reader; Tomlin M., Ed.; Springer-Verlag: London, 2010; pp 13–51. [Google Scholar]
- Gabizon A. A.; Pappo O.; Goren D.; Chemla M.; Tzemach D.; Horowitz A. T. Preclinical studies with doxorubicin encapsulated in polyethyleneglycol-coated liposomes. J. Liposome Res. 1993, 3, 517–528. 10.3109/08982109309150734. [DOI] [Google Scholar]
- Sun M.; Gao Y.; Guo C.; Cao F.; Song Z.; Xi Y.; Yu A.; Li A.; Zhai G. Enhancement of transport of curcumin to brain in mice by poly(n-butylcyanoacrylate) nanoparticle. J. Nanopart. Res. 2010, 12, 3111–3122. 10.1007/s11051-010-9907-4. [DOI] [Google Scholar]
- Zhai S.; Ma Y.; Chen Y.; Li D.; Cao J.; Liu Y.; Cai M.; Xie X.; Chen Y.; Luo X. Synthesis of an amphiphilic block copolymer containing zwitterionic sulfobetaine as a novel pH-sensitive drug carrier. Polym. Chem. 2014, 5, 1285–1297. 10.1039/C3PY01325A. [DOI] [Google Scholar]
- Duan J.; Zhang Y.; Han S.; Chen Y.; Li B.; Liao M.; Chen W.; Deng X.; Zhao J.; Huang B. Synthesis and in vitro/in vivo anti-cancer evaluation of curcumin-loaded chitosan/poly(butyl cyanoacrylate) nanoparticles. Int. J. Pharm. 2010, 400, 211–220. 10.1016/j.ijpharm.2010.08.033. [DOI] [PubMed] [Google Scholar]
- Gao X.; Zheng F.; Guo G.; Liu X.; Fan R.; Qian Z.-y.; Huang N.; Wei Y.-q. Improving the anti-colon cancer activity of curcumin with biodegradable nano-micelles. J. Mater. Chem. B 2013, 1, 5778–5790. 10.1039/c3tb21091j. [DOI] [PubMed] [Google Scholar]
- Sun D.; Zhou J.-K.; Zhao L.; Zheng Z.-Y.; Li J.; Pu W.; Liu S.; Liu X.-S.; Liu S.-J.; Zheng Y.; Zhao Y.; Peng Y. Novel curcumin liposome modified with hyaluronan targeting CD44 plays an anti-leukemic role in acute myeloid leukemia in vitro and in vivo. ACS Appl. Mater. Interfaces 2017, 9, 16857–16868. 10.1021/acsami.7b02863. [DOI] [PubMed] [Google Scholar]
- Chen S.; Li Q.; Li H.; Yang L.; Yi J.-Z.; Xie M.; Zhang L.-M. Long-circulating zein-polysulfobetaine conjugate-based nanocarriers for enhancing the stability and pharmacokinetics of curcumin. Mater. Sci. Eng., C 2020, 109, 110636 10.1016/j.msec.2020.110636. [DOI] [PubMed] [Google Scholar]
- Riches A. C.; Sharp J. G.; Thomas D. B.; Smith S. V. Blood volume determination in the mouse. J. Physiol. 1973, 228, 279–284. 10.1113/jphysiol.1973.sp010086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- accessed July 24
- Paul A.Drug Distribution. In Introduction to Basics of Pharmacology and Toxicology; Raj G. M.; Raveendran R., Eds.; Springer: Singapore, 2019; pp 89–98. [Google Scholar]
- Seymour L. W.; Duncan R.; Strohalm J.; Kopeček J. Effect of molecular weight (Mw) of N-(2-hydroxypropyl)methacrylamide copolymers on body distribution and rate of excretion after subcutaneous, intraperitoneal, and intravenous administration to rats. J. Biomed. Mater. Res. 1987, 21, 1341–1358. 10.1002/jbm.820211106. [DOI] [PubMed] [Google Scholar]
- Hong J. Y.; Liu Y. Y.; Xiao Y.; Yang X. F.; Su W. J.; Zhang M. Z.; Liao Y. H.; Kuang H. X.; Wang X. T. High drug payload curcumin nanosuspensions stabilized by mPEG-DSPE and SPC: in vitro and in vivo evaluation. Drug Delivery 2017, 24, 109–120. 10.1080/10717544.2016.1233589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balza E.; Carnemolla B.; Mortara L.; Castellani P.; Soncini D.; Accolla R. S.; Borsi L. Therapy-induced antitumor vaccination in neuroblastomas by the combined targeting of IL-2 and TNFalpha. Int. J. Cancer 2010, 127, 101–110. 10.1002/ijc.25018. [DOI] [PubMed] [Google Scholar]
- Vader P.; van der Meel R.; Symons M. H.; Fens M. H.; Pieters E.; Wilschut K. J.; Storm G.; Jarzabek M.; Gallagher W. M.; Schiffelers R. M.; Byrne A. T. Examining the role of Rac1 in tumor angiogenesis and growth: a clinically relevant RNAi-mediated approach. Angiogenesis 2011, 14, 457–466. 10.1007/s10456-011-9229-x. [DOI] [PubMed] [Google Scholar]
- Sparreboom A.; Scripture C. D.; Trieu V.; Williams P. J.; De T.; Yang A.; Beals B.; Figg W. D.; Hawkins M.; Desai N. Comparative preclinical and clinical pharmacokinetics of a cremophor-free, nanoparticle albumin-bound paclitaxel (ABI-007) and paclitaxel formulated in Cremophor (Taxol). Clin. Cancer Res. 2005, 11, 4136–43. 10.1158/1078-0432.CCR-04-2291. [DOI] [PubMed] [Google Scholar]
- Kim S. C.; Kim D. W.; Shim Y. H.; Bang J. S.; Oh H. S.; Kim S. W.; Seo M. H. In vivo evaluation of polymeric micellar paclitaxel formulation: toxicity and efficacy. J. Controlled Release 2001, 72, 191–202. 10.1016/S0168-3659(01)00275-9. [DOI] [PubMed] [Google Scholar]
- Varela-Moreira A.; Shi Y.; Fens M. H. A. M.; Lammers T.; Hennink W. E.; Schiffelers R. M. Clinical application of polymeric micelles for the treatment of cancer. Mater. Chem. Front. 2017, 1, 1485–1501. 10.1039/C6QM00289G. [DOI] [Google Scholar]
- Youn Y. S.; Bae Y. H. Perspectives on the past, present, and future of cancer nanomedicine. Adv. Drug Delivery Rev. 2018, 130, 3–11. 10.1016/j.addr.2018.05.008. [DOI] [PubMed] [Google Scholar]
- Tan B. L.; Norhaizan M. E. Curcumin combination chemotherapy: The implication and efficacy in cancer. Molecules 2019, 24, 2527 10.3390/molecules24142527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai K.; Brockmüller A.; Kubatka P.; Shakibaei M.; Büsselberg D. Curcumin’s beneficial effects on neuroblastoma: Mechanisms, challenges, and potential solutions. Biomolecules 2020, 10, 1469 10.3390/biom10111469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.