Abstract
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor β (TGFβ) superfamily and have crucial roles during development; including mesodermal patterning and specification of renal, hepatic, and skeletal tissues. In vitro developmental models currently rely upon costly and unreliable recombinant BMP proteins that do not enable dynamic or precise activation of the BMP signaling pathway. Here, we report the development of an optogenetic BMP signaling system (optoBMP) that enables rapid induction of the canonical BMP signaling pathway driven by illumination with blue light. We demonstrate the utility of the optoBMP system in multiple human cell lines to initiate signal transduction through phosphorylation and nuclear translocation of SMAD1/5, leading to upregulation of BMP target genes including Inhibitors of DNA binding ID2 and ID4. Furthermore, we demonstrate how the optoBMP system can be used to fine-tune activation of the BMP signaling pathway through variable light stimulation. Optogenetic control of BMP signaling will enable dynamic and high-throughput intervention across a variety of applications in cellular and developmental systems.
Keywords: BMP signaling, cell signaling, chondrocytes, light-oxygen-voltage (LOV)-sensing domain, optogenetics
Optogenetic technologies, which enable control of cell signaling and physiology through light, are rapidly expanding in scope beyond initial restrictions to light sensitive channelrhodopsins. Optogenetic approaches enable truly dynamic cellular perturbation through spatiotemporally precise optical stimulation techniques that can be finely tuned through modulation of light wavelength, intensity, and frequency. Here, we report the development and initial application of an optogenetic receptor system responsive to blue light, enabling activation of the bone morphogenetic protein (BMP) signaling pathway in the absence of a ligand in many cell types.
BMPs form part of the transforming growth factor β (TGFβ) superfamily of signaling molecules: a group of structurally related proteins that perform key cellular regulatory functions throughout development and during tissue homeostasis.1 TGFβ molecules are dimeric when active and initiate signal transduction through binding to a Type I or II receptor. Ligand binding leads to the formation of a heterotetrameric signaling complex consisting of two Type I and two Type II receptors that assemble depending upon ligand specificity.2 In canonical signaling, the TGFβ superfamily receptors recruit and activate receptor (R)-SMAD proteins, defined by the presence of a short SSXS activation motif which enables their interaction with Type I receptors.3
Despite close structural homology, TGFβ superfamily receptors are defined as TGFβ-like or BMP-like depending upon their interaction with one of two R-SMAD groups; SMAD2/3 in TGFβ-like and SMAD1/5/8 in BMP-like signaling. Phosphorylated R-SMAD proteins form a complex with SMAD4 facilitating their translocation to and accumulation within the nucleus. R-SMAD–SMAD4 complexes bind directly to DNA where they, in conjunction with other regulatory transcription factors, control the expression of an extensive variety of genes despite the relatively small number of upstream pathway components.3 In addition to SMAD-related canonical signaling, both TGFβ and BMP receptor complexes recruit and activate a variety of other intracellular signaling mediators. Noncanonical signaling, primarily through various mitogen-activated protein kinases (MAPKs), contributes to the induction of specific TGFβ/BMP cellular responses such as epithelial to mesenchymal transition, proliferation, and migration.4
The ability to elicit a diverse range of cellular responses allows TGFβ and BMP signals to have multiple roles in directing cell fate during development, including the specification of germ layers and subsequent specialization of a wide variety of cell types. Indeed, many established directed differentiation protocols of human pluripotent stem cells (hPSCs) require the activation or inhibition of TGFβ/BMP signals throughout multiple stages to drive differentiation.5−9 However, control of the BMP signaling pathway is primarily limited to the use of inconsistent and costly recombinant growth factors; with synthetic small molecules not yet considered potent enough for tissue engineering approaches.10
In vitro investigative and developmental models primarily rely upon activation or inhibition of signaling pathways to elicit a downstream cellular response; predominantly through the use of soluble recombinant growth factors or synthetic chemical compounds that mimic their activity. However, the use of stimulatory molecules to investigate complex biological systems is limited by cellular receptor expression and molecule instability.11 Furthermore, the exogenous addition of stimulatory molecules, including growth factors or small molecules, does not provide the fidelity or temporal precision required for complex systems such as developmental models. Biological tools that enable dynamic perturbation of cellular stimuli can more precisely examine the interplay of signaling factors that occurs within complex molecular networks. For example, technological advances in microfluidic- and bioactive biomaterial-based approaches have the capacity to provide increased control of signal inputs but are still limited by their complexity and low throughput.12,13
Optogenetic approaches enable the use of light as a stimulus, which confers key advantages over traditional pharmacological manipulation, including the flexibility of light illumination and dosage precision in space and time. Although initial optogenetic approaches were limited by the utility of light-sensitive channelrhodopsins, the discovery and application of photoreceptor sensory domains with simple activation mechanics have enabled the generation of more malleable optogenetic tools.14,15 Commonly employed photoreceptors include flavoprotein blue light response sensors and red light sensitive phytochromes.16,17 The light-oxygen-voltage (LOV) flavoprotein family of photoreceptors is particularly attractive in photoreceptor engineering due to the small size and the versatility of optogenetic manipulations.18 Optogenetic tools that utilize the LOV sensing domain have been used to enable optical control of cellular signaling pathways, gene expression regulation, and protein localization.19−21
Precise and fine control of TGFβ-superfamily molecules has focused upon TGFβ-like rather than BMP-like signals through various means; including chemical-mediated dimerization of chimeric receptors, sequestering and release of TGFβ ligands by biomaterials and magnetic induction of TGFβ signaling.22−24 In addition, several recent studies have described optogenetic induction of TGFβ-like signaling,25,26 although induction of BMP-like signaling has not yet been reported. The combination of factors described above triggered us to develop an optogenetic BMP-like signaling system as a cost-effective and high-throughput tool to dynamically perturb the BMP signaling pathway.
Results and Discussion
Development of an Inducible Optogenetic BMP Signaling System
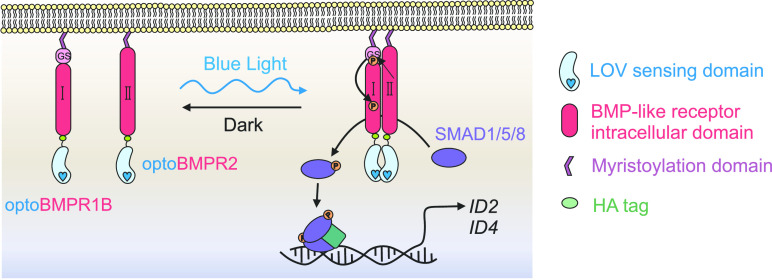
Activation of the canonical BMP signaling cascade is controlled by the tetramerization of Type I and II receptors in the presence of an appropriate ligand. We hypothesized that BMP-like Type I and II receptor heterodimerization would be sufficient to initiate signaling, as previous work has indicated that TGFβ-like Type I and II receptor dimerization can activate the canonical TGFβ-like SMAD2/3 signaling pathway.23,26 Therefore, we constructed two optogenetic BMP-like receptors; optoBMPR1B and optoBMPR2 (Figure 1A,B). We fused the aureochrome1 light oxygen voltage (LOV) sensing domain derived from Vaucheria frigida(27) with the intracellular regions of both receptors at the C-terminal end and anchored both to the plasma membrane with a myristoylation motif. The V. frigida LOV sensing domain dimerizes upon blue light stimulation; thus forcing the Type I and II receptors into close proximity to initiate signal transduction (Figure 1A). Such a strategy has previously proven successful in the light-induced dimerization of TGFβ-like nodal receptors and several receptor tyrosine kinases.21,26 We then incorporated both receptors into lentiviral doxycycline-inducible vectors to enable transgenic genomic integration combined with temporal control over receptor expression (Figure S1A,B).
Figure 1.
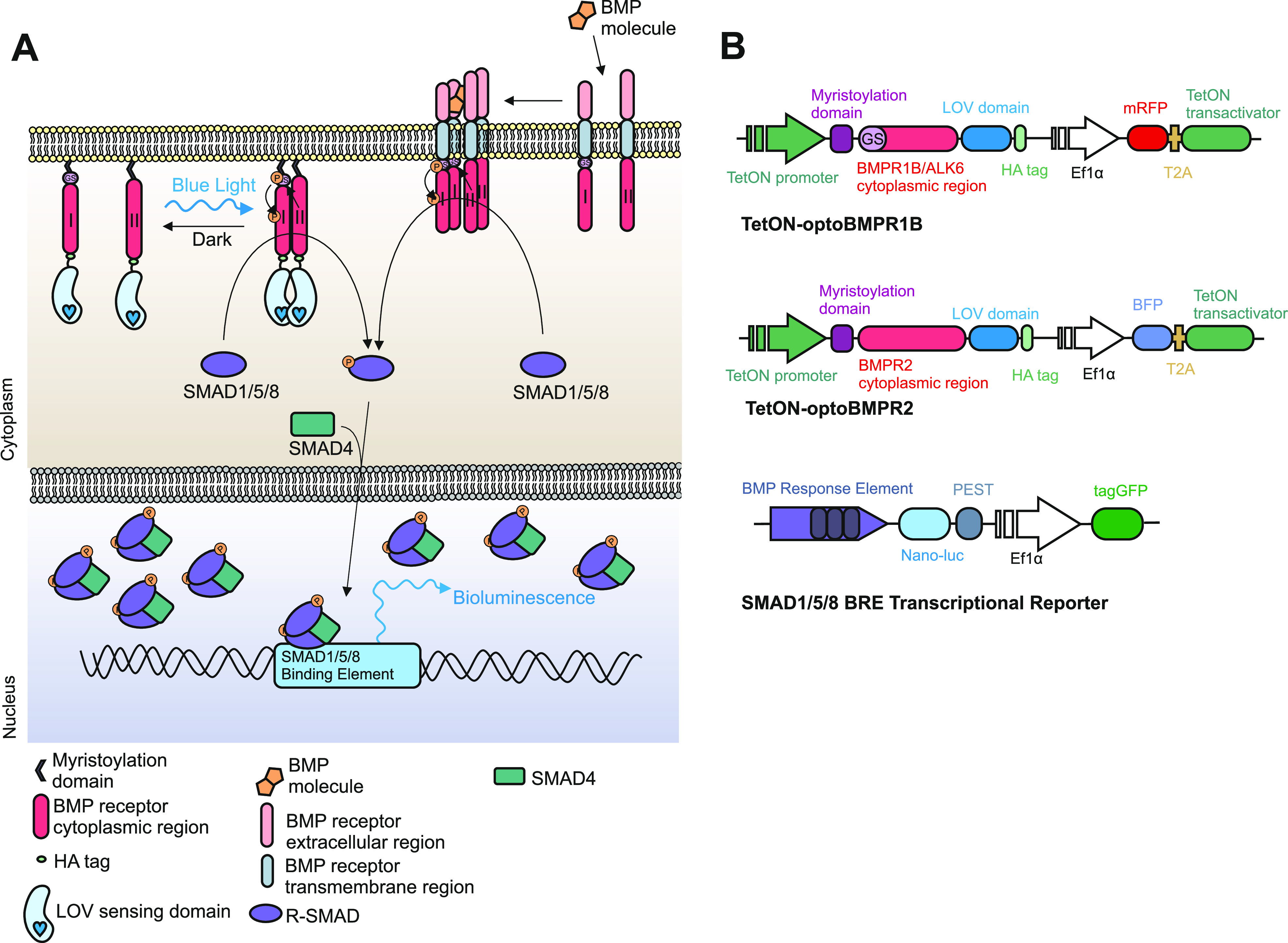
Development of opto-BMP system. (A) Schematic representation of the optogenetic BMP (optoBMP) system. (B) Vector illustrations of the (top) optogenetic BMP receptors and (bottom) SMAD1/5/8 response element reporter (BRE). Optogenetic receptors were designed as follows: the cytoplasmic region of either BMPR1B/ALK6 or BMPR2 was inserted between a myristoylation signal peptide and the light oxygen voltage (LOV) domain. A hemeagglutinin (HA) tag is located at the C-terminus. Optogenetic receptors were then inserted into a doxycycline inducible second generation lentiviral backbone vector with distinct fluorescent protein markers.
OptoBMP Can Be Used to Modulate SMAD1/5/8 Transcriptional Kinetics
We initially characterized activation of the optoBMP system in HEK293T cells through analysis of real-time SMAD1/5/8 transcriptional kinetics using a BMP-like response element reporter (BRE) (Figure S1C). After 24 h doxycycline treatment to induce expression of the optoBMP system, optoBMP-HEK293T-BRE cells were illuminated with blue light (470 nm, 0.25 mW/cm2) for 15 min. After 4 h, a significant, over 2-fold induction of the BRE above prestimulation was observed in illuminated cells while cells that were kept in the dark did not appear to respond (Figure 2A). Optical BRE induction was not significantly different from that obtained with BMP2. Furthermore, we were able to correlate BRE response amplitude with variable light irradiance, demonstrating the ability to fine-tune the optoBMP response with blue light (Figure 2B). Analysis of real-time SMAD1/5/8 transcriptional kinetics demonstrated a unique light-driven response profile in comparison to BMP2-stimulated controls (Figure 2C). BMP2 or light illumination resulted in a response peak after ∼4 h, but light illumination resulted in a significantly more rapid induction of the BRE, as illustrated at 30 min after initial stimulation (Figure 2D).
Figure 2.
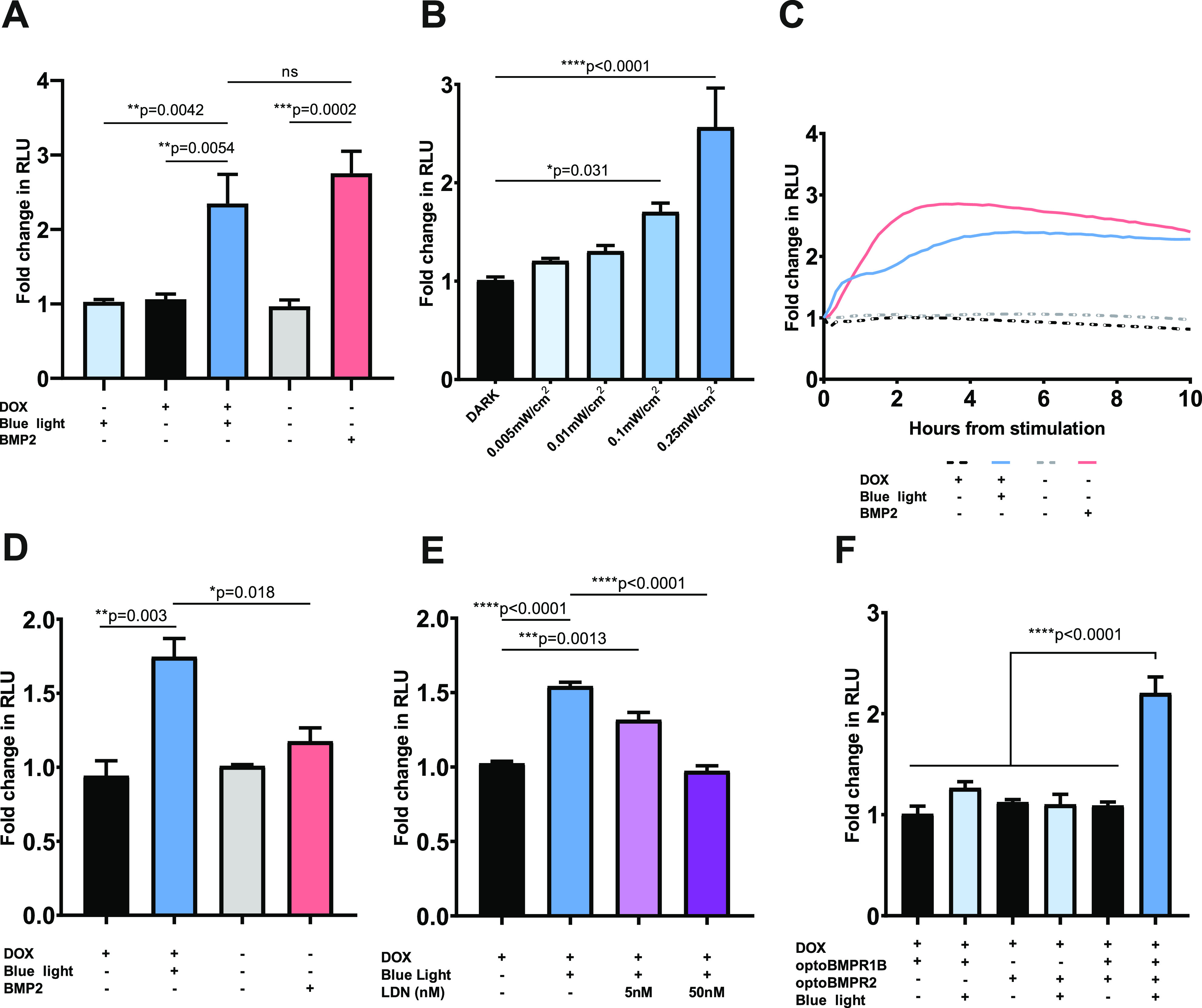
Characterization of opto-BMP system in optoBMP-HEK293T-BRE cells. (A) Analysis of BRE induction 4 h after stimulation with blue light or 50 ng/mL BMP2. N = 3 wells per condition across five independent experiments. (B) Manipulation of BRE induction through stimulation with variable blue light irradiance. N = 3 wells per condition across four independent experiments. (C) Analysis of BRE kinetics over 10 h after stimulation with blue light or 50 ng/mL BMP2. Response after 30 min shown in panel D. N = three wells per condition across three independent experiments. (D) Analysis of BRE induction 30 min after stimulation with blue light or 50 ng/mL BMP2. N = 3 wells per condition across three independent experiments. (E) Analysis of BRE induction 30 min after stimulation with blue light and addition of LDN193189. N = 2 wells per condition across three independent experiments. (F) Analysis of BRE induction in cell lines with different optogenetic receptor expressed 4 h after stimulation with blue light. N = 3 wells per condition across three independent experiments. Data information: In panels A–F data are presented as mean fold change in NanoLuc Luciferase activity (RLU) from prestimulation across independent experiments. In panels A, B, D, E, and F bars represent mean values + standard error of the mean (SEM). P values were generated using an ordinary one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001, ns = not significant).
To ensure the optoBMP system functioned through BMP-like receptor activation, we treated optoBMP-HEK293T-BRE cells with LDN193189; which directly inhibits the kinase activity of BMP-like Type I receptors.28 Addition of LDN193189 at a low concentration (5 nM) is sufficient to inhibit BMPR1A/ALK3 kinase activity but not that of BMPR1B/ALK6, which we utilized in the design of the optoBMP system.29 We observed significant BRE induction in cells treated with 5 nM LDN193189 30 min after light illumination in comparison to cells that remained in the dark (Figure 2E). Upon application of 50 nM LDN193189, BRE induction was abolished, indicating that the optoBMP system initiates canonical BMP-like signal transduction through light-induced opto-BMPR1B kinase activity. Optogenetic activation of opto-BMPR1B additionally requires the presence of opto-BMPR2, with neither receptor functional when expressed individually (Figure 2F). Thus, although light-induced LOV dimerization may result in the formation of optogenetic receptor homodimers or oligomers, the optoBMP system likely functions through transactivation of Type I kinase activity by Type II receptors.
Through targeting the most upstream pathway components for optical control, the optoBMP system almost entirely utilizes native cellular machinery. As the binding of SMAD1/5/8 to transcriptional targets are influenced and directed by cell-dependent expressed cofactors, the design of the optoBMP system does not interfere with this process. Currently available small molecule compounds that target the BMP pathway subvert receptor activation and thus may disrupt or improperly engage in additional recruitment of signaling effectors that bind to active SMAD1/5/8.10,30 Therefore, the optoBMP system has greater potential to investigate the roles of BMP signals in any natural cellular context. As expression and activation of the optoBMP system can be induced at any time upon doxycycline and light illumination, respectively, the system subverts native cellular regulatory systems driving the expression of specific receptors or the internalization and degradation of ligand–receptor complexes.31 In addition, BMP ligands are promiscuous in their receptor activation, and thus light illumination provides precise dosage and downstream receptor specificity.
Blue Light Stimulation of optoBMP Drives a BMP-like Response in a Chondrogenic Cell Line
To investigate the potential of the optoBMP system in a relevant context, we transferred the system into the immortalized chondrocyte cell line TC28a2 to test the response in a skeletal model.32 BMP signals are critical in chondrocyte and osteocyte development and maintenance.33 Optogenetic receptor expression (detected through the tagged hemeagglutinin (HA) signal) was only seen in doxycycline treated cells (Figure 3A), confirming the doxycycline-dependent induction of optogenetic receptor expression. We then investigated if activation of the optoBMP system induced native SMAD1/5/8 signal transduction. Nuclear accumulation of P-SMAD1/5 was analyzed 2 h after 15 min of blue light illumination (Figure 3B). Single cell quantification of the mean nuclear intensity of P-SMAD1/5 indicated a significant increase in comparison to controls that remained in dark conditions (Figure S2A). P-SMAD1/5 nuclear intensity was observed to be higher in BMP2-treated controls that were stimulated continuously for 2 h, but the percentage of positive cells in both light and BMP2-stimulated conditions after 2 h was not significantly different (Figure 3C).
Figure 3.
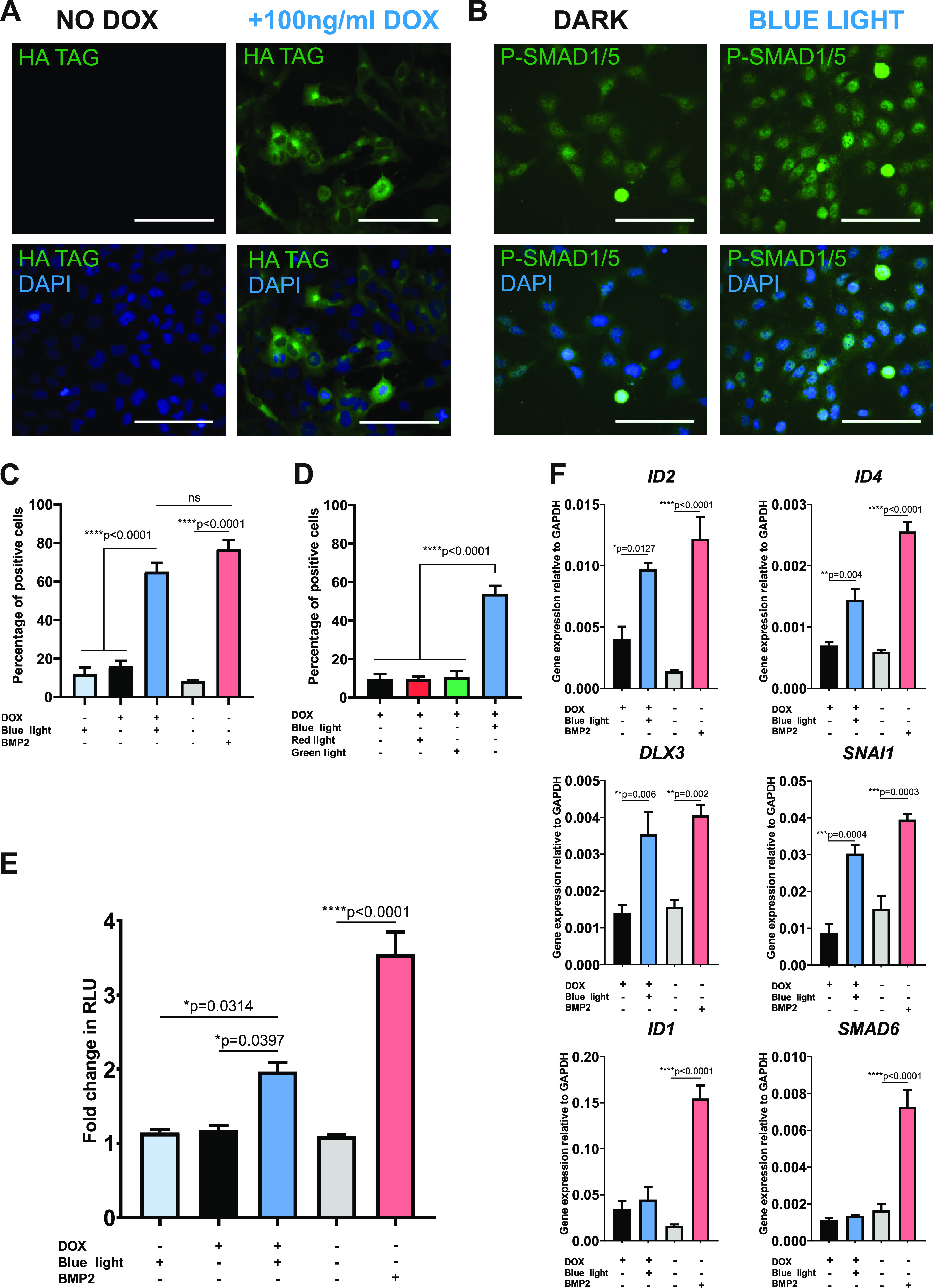
Induction of a BMP-like response in chondrogenic optoBMP-TC28a2 cells. (A) Representative immunofluorescence images of cells stained for HA tag with or without addition of 100 ng/mL doxycycline for 24 h. (B) Representative immunofluorescence images of cells stained for P-SMAD1/5 2 h after 15 min blue light illumination (right) or having been kept in the dark (left). (C) Percentage of P-SMAD1/5 positive cells calculated through single-cell quantification of mean nuclear P-SMAD1/5 fluorescence intensity. Cells were stimulated with blue light illumination or 50 ng/mL BMP2. Threshold for positivity was calculated through analysis of unstimulated controls. N = 3 different fields of view per condition across three independent experiments. (D) Percentage of P-SMAD1/5 positive cells when stimulated with different light wavelengths. Cells were stimulated with either red, blue, or green light (0.25 mW/cm2) or kept in the dark. N = 3 different fields of view per condition across three independent experiments. (E) Analysis of BRE induction in optoBMP-TC28a2-BRE cells 4 h after stimulation with blue light or 50 ng/mL BMP2. Bars represent mean fold change in RLU value 4 h from prestimulation + SEM. N = 3 wells per condition across three independent experiments. (F) Gene expression analyses of BMP-target genes. Cells were stimulated with blue light illumination or 50 ng/mL BMP2 and analyzed after 4 h. Gene expression was normalized to GAPDH. N = 4 independent experiments. Data information: Scale bars in panels A and B represent 100 μm. Data presented in panels C–F represent mean values + SEM. P values were generated using an ordinary one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.0005, ****p < 0.0001, ns = not significant).
We observed a small increase in basal SMAD1/5/8 signaling upon doxycycline addition in the absence of light, indicating a low level of spontaneous optoBMP activation, most likely as a result of the spatial proximity of receptors both expressed at the cell membrane. Alternatively, or in combination, continuous doxycycline-driven receptor expression may result in receptor excess. However, blue light illumination resulted in significant enhancement of SMAD1/5 phosphorylation and induction of transcriptional activity in comparison to cells kept in the dark. Although not an issue in this study, spontaneous optoBMP activation without light may be eliminated through a similar approach to that used by Li et al.25 in which one of the receptors is localized to the cytoplasm, hence allowing signaling solely thorough light-activated membrane localization. Supplementation of doxycycline, and therefore expression of the optoBMP system, was required to enable blue-light induction of P-SMAD1/5 nuclear accumulation. The blue light specificity of the optoBMP system was then demonstrated by subjecting optoBMP-TC28a2 cells to variable light wavelength stimulations (Figure S2B). There was no significant difference in mean percentage of cells positive for nuclear P-SMAD1/5 between cells left in the dark and cells illuminated with red (670 nm) or green (560 nm) light (Figure 3D). In addition, activation of the optoBMP system with blue light did not induce TGFβ-like SMAD2 nuclear translocation, demonstrating specificity in targeting the BMP signaling arm of the TGFβ superfamily (Figure S3).
To illustrate the ability of the optoBMP system to elicit a BMP-like transcriptional response in optoBMP-TC28a2 cells, we initially measured the SMAD1/5/8 response through BRE induction. As with optoBMP-HEK293T cells, we found that 15 min of blue light illumination resulted in significant induction of a SMAD1/5/8 response after 4 h (Figure 3E). We then investigated whether BRE induction correlated with upregulation of direct BMP-pathway target genes 4 h after blue light illumination. Blue light induction of optoBMP resulted in significant upregulation of Inhibitor of Differentiation 2 (ID2), ID4, Distal-Less Homeobox 3 (DLX3) and Snail Family Transcriptional Repressor 1 (SNAI1) which are all known to be downstream targets of SMAD1/5/8 (Figure 3F).34−36
We then investigated whether activation of the optoBMP system initiated involvement of noncanonical signal transduction through MAPK. Phosphorylation of extracellular signal-related kinase (ERK1/2) did not appear to be different between nonstimulated cells and those stimulated with light or BMP2 (Figure S4A). However, light illumination and BMP2 stimulation resulted in significant induction of Cyclooxygenase-2 (PTGS2); which is facilitated through BMP-activation of noncanonical p38 MAPK37 (Figure S4B). Additionally, both light illumination and BMP2 stimulation induced a small but not significant increase in H6 Family Homeobox 3 (HMX3) expression, which has additionally been demonstrated to be controlled through noncanonical BMP signals.38JUNB expression, which can also be BMP-responsive through noncanonical mediators,37 was not significantly altered. Therefore, the optoBMP system has the capacity to stimulate activation of both canonical and some noncanonical downstream BMP targets in a comparable manner to that of BMP2 stimulation.
Blue light illumination did not appear to result in upregulation of known BMP response genes ID1 or inhibitory SMAD6, which acts as part of a negative feedback loop, both of which are upregulated following BMP2 stimulation.39 The apparent differences between optoBMP- and BMP2 ligand-driven responses may be a result of unique signaling receptor complexes. The lack of native BMPR1B/ALK6 expression in TC28a2 cells indicates that a BMP signaling response is driven through a BMPR1A/ALK3-BMPR2 receptor complex unlike the optoBMP system (Figure S5). Although BMPR1B/ALK6 activation has been demonstrated to result in ID1 induction in several cell lines, cellular context is known to be important in directing downstream BMP signaling and therefore may not totally align with optogenetic receptor complex activation in TC28a2 cells.40,41 Moreover, BMP2 stimulation used here involved 4 h of sustained signaling in comparison to a short 15 min light-induced activation of the BMP pathway. In future work we will aim to mimic BMP-ligand induced activation through the use of longer durations of light illumination.
Application of optoBMP in a Human Development Model
As human embryonic stem cells (hESCs) have proved a useful model system for studying development to many different lineages, they are a suitable system for testing optogenetic regulation of development. To demonstrate the potential of the optoBMP system in this context we transduced hESC line MAN1342 with the optoBMP system. We initially performed verification of pluripotency through immunofluorescent staining of established pluripotent markers OCT4 and NANOG (Figure 4A). Then, to determine functionality of the optoBMP system in hESCs, we analyzed nuclear accumulation of P-SMAD1/5 after blue light illumination (Figure 4B). Single cell quantification of nuclear P-SMAD1/5 indicated significant induction of the canonical BMP pathway when compared to cells kept in the dark or stimulated with light in the absence of doxycycline (Figure 4C). To further illustrate optogenetic activation in hESCs we analyzed BMP target gene expression. Again we observed significant upregulation of direct BMP target genes including ID2, ID3, and DLX3 after optogenetic stimulation (Figure 4D). Interestingly, several genes appeared upregulated by light illumination and not by BMP2, including differentiation genes SNAI1 and SOX9. As ID, SNAIL, and SOX genes have crucial roles during developmental processes, these preliminary data suggest that the optoBMP system could be utilized in hPSC development models as a substitute for recombinant growth factor supplementation.34,43
Figure 4.
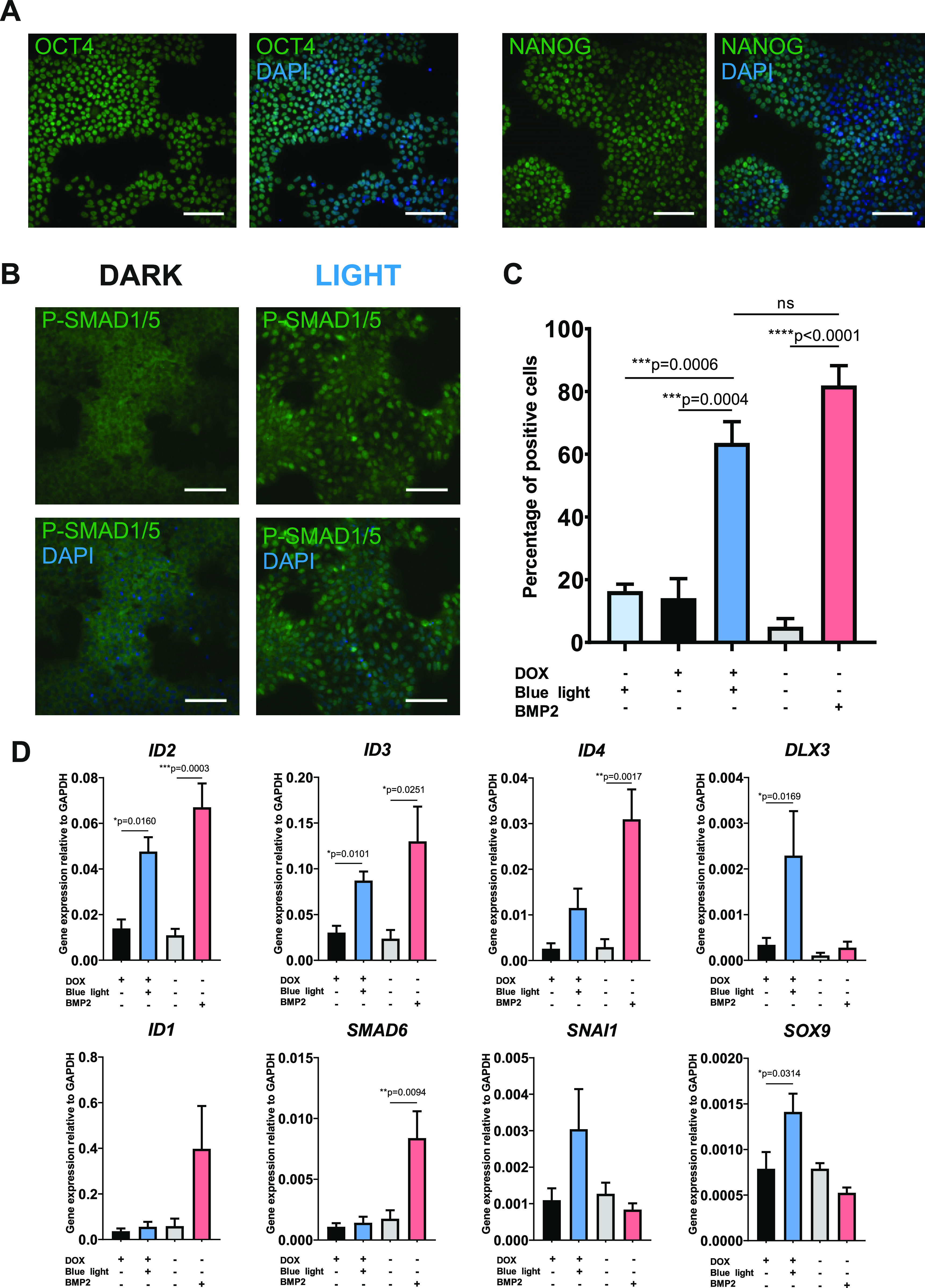
Optogenetic activation of canonical BMP signaling in pluripotent optoBMP-hESCs. (A) Verification of pluripotency. Representative immunofluorescence images of optoBMP-hESCs stained for pluripotent markers OCT4 and NANOG. (B) Representative immunofluorescence images of cells stained for P-SMAD1/5 2 h post-15 min blue light illumination. (C) Percentage of P-SMAD1/5 positive cells calculated through single-cell quantification of mean nuclear P-SMAD1/5 intensity. Threshold for positivity calculated through analysis of unstimulated controls. N = 3 different fields of view per condition across three independent experiments. (D) Gene expression analyses of BMP-target genes. Cells were stimulated with 15 min of blue light illumination or 50 ng/mL BMP2. Gene expression was normalized to GAPDH. N = 4 independent experiments. Data information: Scale bars in panels A and B represent 100 μm. Data presented in panel C represent mean values + SEM. P values were generated using an using an ordinary one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.0005, ****p < 0.0001, ns = not significant) in panel C and unpaired two-tailed students t-tests in panel D (*p < 0.05, **p < 0.01, ***p < 0.0005).
In this study we demonstrate that short light pulses are sufficient to alter BMP signaling using the optoBMP system. In the future it will be interesting to investigate the effect of complex light stimulation patterns and longer illumination periods, as used in several other optogenetic studies.25,44 Complex cellular responses, such as directing cell fate decisions and differentiation, often require sustained and specifically timed stimulation, as illustrated by the recent in vitro application of an optogenetic Wnt tool in hPSCs.45 In addition, the ability to precisely manipulate the amplitude and spatial location of cell signaling pathways can be used to investigate developmental signaling networks in vivo. For example, recent studies in Drosophila melanogaster and zebrafish have demonstrated the utility of optogenetic tools in the investigation of embryonic signaling dynamics.26,46−48 Furthermore, the high-throughput nature and pathway specificity of optogenetic approaches additionally lend themselves to live cell screens in drug discovery pipelines and further dissection of signaling pathways.49−52 Given the involvement of BMP signals in both development and disease, the optoBMP system can provide a powerful investigative tool in a number of settings.
The optoBMP system in its current form carries some limitations. The two engineered optogenetic receptors are currently incorporated into individual expression vectors, thus it is difficult to simultaneously express both in difficult-to-transfect cell lines. Random genomic integration via lentiviral transduction may also result in early quiescence or other growth abnormalities; particularly in clonal cell lines. Modification of the optoBMP expression system through CRISPR/Cas9-mediated incorporation into genomic safe harbor locus AAVS1 would be useful in the future to enable uniform and ubiquitous expression without lentiviral transgene silencing; an issue which has been commonly reported in the differentiation of pluripotent stem cells.53−55
In summary, we report the development of a novel optogenetic signaling system that enables selective dose and time dependent control of BMP-like signals through canonical SMAD1/5/8 and some noncanonical MAPK signal transduction with blue light-sensitive BMP receptors. We demonstrate that the optoBMP system can reliably recapitulate initiation of the canonical BMP signaling cascade in several cell lines through phosphorylation and nuclear translocation of SMAD1/5/8 leading to BMP-like transcriptional activation, including upregulation of direct BMP gene targets. Extension of the optogenetic toolkit to target the BMP pathway will provide the means to dynamically probe BMP signals in a wide variety of systems.
Methods
DNA Vector Assembly
All vectors were constructed through NEB HiFi Assembly or restriction enzyme cloning. Both optogenetic receptors were assembled through an initial generation of a vector backbone through PCR amplification of opto-mFGFR (a gift from Harald Janovjak, Addgene plasmid #58745) to omit the mFGFR coding region. The intracellular coding regions of human BMPR1B/ALK6 (fully sequenced cDNA clone from Source Bioscience; vector #IRATp970H1175D) and BMPR2 (Source Bioscience, #IRATp970B1178D) were then PCR amplified and inserted into the vector backbone through NEB HiFi Assembly (New England Biolabs, #E2621). Each optogenetic receptor was then subsequently inserted into a second-generation lentiviral shuttle doxycycline inducible vector through NEB HiFi Assembly. Briefly, vector inserts were generated through PCR amplification of each optogenetic receptor with 30bp overhangs. Vector backbones, pCDH-TRE3G-MCS-EF1a-tagRFP-T2A-TetON3G or pCDH-TRE3G-MCS-EF1a-tagBFP-T2A-TetON3G for Type I or II receptor, respectively, were then digested with EcoRI and vector inserts were annealed in through NEB HiFi Assembly. Final lentiviral shuttle vectors were designated TetOn-optoBMPR1B and TetOn-optoBMPR2.
A modified version of a BMP reporter vector previously described56 was constructed. The BMP response element (BRE) consisted of multiple SMAD binding elements arranged in tandem in forward and reverse orientations, placed upstream of the AAV minimal late promoter.57 The BRE was cloned upstream of a destabilized form of NanoLuc Luciferase (Promega) containing a C-terminal protein degradation sequence (PEST sequence, NLucP). The completed BRE-NLucP was cloned into a modified version of the lentiviral expression vector pCDH-EF1-T2A-copGFP, resulting in the construct pCDH-BRE-nLUCP-EF1a-copGFP. Vector maps were generated using Snapgene software (Figure S1).
Cell Culture
HEK293T (ATCC, #CRL-11268), TC28a232 and SW1353 (ATCC, #HTB-94) cell lines were cultured in 75 cm2 cell culture flasks (Corning) with DMEM (Gibco, #11960044) containing 10% w/v fetal bovine serum (Merck, #12103C), 1% w/v l-glutamine (Gibco, #25030081) and 1% w/v penicillin/streptomycin (Gibco, #15140122). For routine maintenance, cells were subcultured into flasks containing fresh warmed medium at a passage ratio of 1:10. Briefly, cells were washed with phosphate buffered saline (PBS; Merck, #D8537) before cell dissociation with 3 mL of TryPLE Express solution (Gibco, #12604021). Cells were then centrifuged at 700g for 3 min before pellet resuspension and continued passage.
MAN13 human embryonic stem cells (hESCs)42 were cultured in 6-well plates (Corning) coated with 5 μg/mL human recombinant vitronectin (Life Technologies, #A14700) in mTesR1 (StemCell Technologies, #5850) with medium replaced every 2 days. For routine maintenance culture, cells were washed with PBS before dissociation using 0.5 mM EDTA solution (Invitrogen, #15575-038). Cells were centrifuged at 700g for 3 min before being subcultured into fresh medium containing 1X RevitaCell Supplement (Life Technologies, #A2644501).
Lentiviral Particle Production and Cell Line Generation
All lentiviral particles were generated through transfection of HEK293T cells using calcium chloride precipitation. Briefly, HEK293T cells were seeded in 15 cm2 dishes at 8 × 106 cells/dish 24 h before transfection with 9 μg of psPAX2, 6 μg of pMD2.G (VSV), 12 μg of shuttle plasmid (TetOn-optoBMPR1B, TetOn-optoBMPR2, or pCDH-BRE-nLUCP-EF1a-copGFP) per dish. DNA was added to 2.5 M CaCl2 before mixing with HEPES-buffered saline (containing sodium chloride, HEPES acid, and disodium phosphate). The calcium phosphate–DNA coprecipitate was then added dropwise to cells. Medium containing lentiviral particles was collected over 48 h and concentrated at 6000g overnight at 4 °C. The lentiviral pellet was then resuspended in 10 mL of ice-cold PBS and was centrifuged in a high-speed SW40-Ti rotor (Beckman Coulter) at 50 000g for 90 min at 4 °C. Finally, the concentrated pellet was resuspended in 200 μL of ice-cold PBS and stored at −80 °C. Viral titers were calculated through serial dilution of viral preparations before transduction of HEK293T cells and flow cytometry analysis of appropriate fluorescent markers. HEK293T and TC28a2 cells were transduced with viral particles at a multiplicity of infection (MOI) that did not exceed 20 IU/cell, while MAN13 cells were transduced at an MOI that did not exceed 10 IU/cell before being sorted through FACS (BD FACS Aria Fusion).
Optogenetic and Chemical Stimulation
Prior to optical stimulation experiments, all cell lines were seeded in black-walled 96 well plates at a density of 1 × 104 cells/well or in black-walled 24 well plates at 5 × 104 cells/well and were maintained in the dark after induction of optogenetic receptor expression through 24 h incubation in serum-free medium containing 100 ng/mL doxycycline hyclate (Merck, #D9891). Optical stimulation was performed at room temperature using either a custom-built Arduino-controlled LED array device using 470 nm LEDs58 or CoolLED pE4000 at 460 nm. Light irradiance was controlled through a piece of custom software (LED array) or directly from the equipment control panel (CoolLED). Light illumination was performed at 0.25 mW/cm2 for 15 min unless otherwise highlighted. Irradiance was measured using a spectroradiometer (SpectroCAL MKII; Cambridge Research Systems). Alongside optogenetic stimulation where noted, cells were stimulated with 50 ng/mL BMP2 (R&D Systems, #355-GMP) or 10 ng/mL TGFβ3 (Peprotech, 100-36E). Inhibition of the BMP signaling pathway was achieved through addition of LDN193187 dihydrochloride (Tocris, #6053) in combination with optical stimulation at concentrations noted. Alongside LDN193187, cells were treated with DMSO vehicle control.
Nano-Glo Luciferase Assay System
Optogenetic HEK293T and TC28a2 cells were seeded in black-walled 96-well plates before 24 h serum starvation and induction of optogenetic receptor expression. Extended NanoLuc luciferase substrate Vivazine (Promega, #N2580) was added to live cells (dilution to 1X in serum-free DMEM) 2 h prior to stimulations. After optogenetic or chemical stimulations were performed, expression of NanoLuc luciferase was detected in an Alligator Luminescence System (Cairn Research) using an appropriate exposure time (10–30 min). Bioluminescent micrographs taken over a period of time were stacked using ImageJ software. A region of interest (ROI) was then drawn around each well and relative luminescent units (RLU) were calculated for each micrograph through Z-stack analysis. Luminescence was normalized to background and to the luminescence value prior to the start of stimulation.
Immunocytochemistry and Imaging
Cells were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature and subsequently washed in PBS three times. Fixed cells were then permeabilized and blocked in 0.1% Triton-X (Merck, #9002-93-1), 10% donkey serum (DS) (Merck, #D9663) in PBS for 30 min at room temperature, before a further three PBS washes. Cells were incubated with primary antibodies (Cell Signaling Technologies; #3724 HA tag 1:700 dilution, #9516 P-SMAD1/5 1:200 dilution, #5339 SMAD2 1:200 dilution, #2890 OCT4-A 1:200 dilution, #4903 NANOG 1:200 dilution) in 1% DS in PBS at 4 °C overnight before being subsequently washed three times in PBS. Cells were then treated with AlexaFluor-488- or Alexa-Fluor-594-labeled secondary antibodies (Life Technologies #A32790/#A32754, 1.200 dilution in PBS 1% DS) for 45 min at room temperature before a final three PBS washes. Nuclei were visualized through staining with 10 μg/mL 4,6-diamindino-2-phenylindole (DAPI - Life Technologies, #D1306) in PBS for 5 min at room temperature followed by three washes in PBS. Fluorescent micrographs were taken using a Zeiss Axioimager D2 upright microscope and captured using a Coolsnap HQ2 camera (Photometrics) through micromanager software v1.4.23.
Image Analysis
Image processing and quantification was performed using ImageJ software. Nuclear quantification of P-SMAD1/5 was performed through an initial identification of nuclear outlines through the DAPI channel (358 nm). This was achieved through the “Analyze Particles” function of ImageJ after converting the image to a binary output. Each nuclear outline identified by ImageJ was then applied to the P-SMAD1/5 channel (488 nm) before using the ROI manager to measure the mean gray value within each nuclear outline. Values that fell 2×SD from the mean were omitted from analysis to eliminate false positives or negatives. A threshold mean intensity value to determine a positive P-SMAD1/5 response was calculated from analysis of nonstimulated control images.
RNA Extraction, Reverse Transcription, and qPCR Analysis
RNA extraction and purification was performed using a Qiagen RNeasy mini-kit (Qiagen, #74104) according to the manufacturer’s instructions. Briefly, cells were dissociated and the cell pellet was resuspended in 350 μL of RLT lysis buffer. Cell lysate was then subjected to a series of on-column washes, after the addition of 350 μL of 70% ethanol, before elution in nuclease-free water (Invitrogen, #4387937). After quantification of RNA concentration, 2 μg of RNA was converted to cDNA using a high capacity cDNA reverse transcription kit (Thermo Fisher Scientific, #4368813). qPCR reaction was then prepared using PowerUp SYBR green master mix (ThermoFisher Scientific, #A25742) with 10 ng of cDNA per reaction and 400 nM final concentration forward and reverse primers. qPCR reaction was run using a BioRad C1000Touch Thermal Cycler using the following cycling conditions: denaturation at 95 °C for 10 min, 39 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 35 s, final extension at 72 °C for 10 min, and melt curve analysis at 65 °C for 5 s and 95 °C for 30 s. Raw qPCR data were normalized to GAPDH using the 2–ΔCT method. Primer sequences are listed in Table 1.
Table 1. List of Primer Sequences Used for qPCR Reactions. oBMPR1B and oBMPR2 Indicate Synthetic Optogenetic Transgenes.
| gene | forward primer | reverse primer |
|---|---|---|
| GAPDH | ATGGGGAAGGTGAAGGTCG | TAAAAGCAGCCCTGGTGACC |
| ID1 | TTACTCACGCCTCAAGGAGC | TCAGCGACACAAGATGCGAT |
| ID2 | CTGCAGCACGTCATCGACTA | CCACACAGTGCTTTGCTGTC |
| ID3 | ACTCAGCTTAGCCAGGTGGA | AAGCTCCTTTTGTCGTTGGA |
| ID4 | CCTGCAGCACGTTATCGACT | GTCGCCCTGCTTGTTCAC |
| DLX3 | CCGCGTACGATCTACTCCAG | CACCTCCCCGTTCTTGTAGA |
| SNAI1 | CCCAATCGGAAGCCTAACTA | GGACAGAGTCCCAGATGAGC |
| SMAD6 | GGGCCCGAATCTCCGC | AGAATTCACCCGGAGCAGTG |
| PTGS2 | ATCCTGAATGGGGTGATGAG | GCCACTCAAGTGTTGCACAT |
| HMX3 | GTGGTACCCCTACACCCTGA | GGAGAGTCGCGGTCTGTG |
| JUNB | CGGCAGCTACTTTTCTGGTC | GTGTAGGCGTCGTCGTGAT |
| BMPR1A | ATGCTTCATGGCACTGGGAT | TGTGGTTTCTCCCTGGTCATC |
| BMPR1B | GAGGATGACTCTGGGTTGCC | AGGCAGTGTAGGGTGTAGGT |
| BMPR2 | TGGGACAATATTATGCTCGAAA | CTGAATTGAGGGAGGAGTGG |
| oBMPR1B | AAGTTACGCCCCTCATTCCC | CAGAGCCTTCACGAGACTGT |
| oBMPR2 | TACCTTGCAGAAGGAGGCAC | ACTGGCGTAGACGATAGGGT |
Western Blotting
Optogenetic TC28a2 cells were seeded in black-walled 24 well plates at 5 × 104 cells/well before 24 h serum starvation and induction of optogenetic receptor expression. Cells were then subjected to optogenetic or BMP2 stimulation as described previously. Protein was harvested 2 h poststimulation through briefly washing in PBS and lysing cells in 1X cell lysis buffer with 6 μM Phenylmethanesulfonyl Fluoride (Cell Signaling Technologies, #9803 and #8553). Briefly, cells were incubated on ice in lysis buffer for 5 min before lysate was scraped into 1.5 mL tubes and centrifuged at 14 000g at 4 °C for 10 min. The supernatant was stored at −20 °C.
Protein concentration was measured using a Pierce BCA assay kit (Thermo Fisher Scientific, #23225) before boiling 2 μg of protein at 95 °C for 10 min in lane marker reducing buffer (Thermo Fisher Scientific, #39000). Protein samples were then subjected to gel electrophoresis at 100 V on a 10% Bis-Tris gel (Thermo Fisher Scientific, #NW00100BOX) alongside broad range markers (11–245 kDa, NEB P7712S). Protein was transferred to a nitrocellulose membrane using an iBlot-2 Gel Transfer and iBlot-2 Transfer stacks (Thermo Fisher Scientific, #IB21001 and #IB23001) before blocking in 1X Tris buffered saline (TBS, Merck, #T5912) containing 5% bovine serum albumin (BSA) and 0.1% Tween (Merck, #P1379) for 1 h at room temperature. Membranes were then incubated at 4 °C overnight with primary antibodies (Cell Signaling Technologies, #9102 ERK1/2, #4370 P-ERK1/2) in TBS containing 0.1% Tween (TBS-0.1%Tween), before washing three times in TBS-0.1%Tween. Membranes were then treated with secondary antibodies (1:15,000 IRDye 680RD Donkey anti-Rabbit; LI-COR 925–68071) in TBS-0.1%Tween containing 5% BSA for 45 min at room temperature before a final three washes in TBS-0.1%Tween and detection of protein using an Odyssey CLx imaging system (LI-COR).
Statistical Analyses and Figure Generation
Figures were generated using CorelDraw Graphics Suite or GraphPad Prism 8 software. Statistical analyses were performed using GraphPad Prism 8. Comparison between two groups were performed using an unpaired two-tailed student’s t-test and more than two groups with an ordinary one-way analysis of variance (ANOVA) and subsequent Tukey’s multiple comparisons test. A p value of <0.05 was indicative of significance.
Acknowledgments
The authors acknowledge financial support from the Engineering and Physical Sciences Research Council (EPSRC) and Medical Research Council (MRC) Centre for Doctoral Training (CDT) in Regenerative Medicine (EP/L014904/1) studentship to P.H. with additional support from Arthritis Research UK (Grant R20786) and Medical Research Council (Grant MR/S002553/1). TC28a2 cells were a gift from Dr L. Reynard (Newcastle University). The authors are grateful to Franck Martial for technical support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.0c00315.
Vector maps of viral shuttle plasmids; single cell quantification of nuclear P-SMAD1/5 fluorescence in optoBMP-TC28a2 cells; analysis of SMAD2 nuclear translocation in optoBMP-TC28a2 cells; activation of noncanonical signaling after blue light stimulation in optoBMP-TC28a2 cells; gene expression analyses of BMP-like receptors in optoBMP-TC28a2 cells (PDF)
Author Contributions
The project was conceived by S.J.K. and R.L; experimental design was created by S.J.K. and P.H. P.H. performed all experiments and data analysis. S.C. generated lentiviral TetON and BRE reporter plasmids. S.W. generated optoBMP-hESC line. N.B. performed Western blot analysis. Data were interpreted by P.H., S.W., C.A.S., R.L., and S.J.K. The manuscript was written by P.H., S.W., C.A.S., and S.J.K. All figures were generated by P.H.
The authors declare no competing financial interest.
Supplementary Material
References
- Massagué J. (2012) TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630. 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-G.; Hata A.; Lo R.; Wotton D.; Shi Y.; Pavletich N.; Massague J. (1998) Determinants of specificity in TGF-β signal transduction. Genes Dev. 12, 2144–2152. 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. (1998) TGFβ Signal Transduction. Annu. Rev. Biochem. 67, 753–791. 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Zhang Y. E. (2017) Non-Smad signaling pathways of the TGF-β family. Cold Spring Harbor Perspect. Biol. 9, a022129. 10.1101/cshperspect.a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldershaw R. A.; Baxter M. A.; Lowe E. T.; Bates N.; Grady L. M.; Soncin F.; Brison D. R.; Hardingham T. E.; Kimber S. J. (2010) Directed differentiation of human embryonic stem cells toward chondrocytes. Nat. Biotechnol. 28, 1187–1194. 10.1038/nbt.1683. [DOI] [PubMed] [Google Scholar]
- Hay D. C.; Fletcher J.; Payne C.; Terrace J. D.; Gallagher R. C. J.; Snoeys J.; Black J. R.; Wojtacha D.; Samuel K.; Hannoun Z.; Pryde A.; Filippi C.; Currie I. S.; Forbes S. J.; Ross J. A.; Newsome P. N.; Iredale J. P. (2008) Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc. Natl. Acad. Sci. U. S. A. 105, 12301–12306. 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K. M. M.; Chen A.; Koh P. W. W.; Deng T. Z. Z.; Sinha R.; Tsai J. M. M.; Barkal A. A. A.; Shen K. Y. Y.; Jain R.; Morganti R. M. M.; Shyh-Chang N.; Fernhoff N. B. B.; George B. M. M.; Wernig G.; Salomon R. E. E. A.; Chen Z.; Vogel H.; Epstein J. A. A.; Kundaje A.; Talbot W. S. S.; Beachy P. A. A.; Ang L. T. T.; Weissman I. L. L. (2016) Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 166, 451–467. 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Nimkingratana P.; Smith C. A.; Cheng A.; Hardingham T. E.; Kimber S. J. (2019) Enhanced chondrogenesis from human embryonic stem cells. Stem Cell Res. 39, 101497. 10.1016/j.scr.2019.101497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher J.; Dickinson A.; Cain S.; Hu Y.; Bates N.; Harvey A.; Ren J.; Zhang W.; Moreton F. C.; Muir K. W.; Ward C.; Touyz R. M.; Sharma P.; Xu Q.; Kimber S. J.; Wang T. (2019) Patient-Specific iPSC Model of a Genetic Vascular Dementia Syndrome Reveals Failure of Mural Cells to Stabilize Capillary Structures. Stem Cell Rep. 13, 817–831. 10.1016/j.stemcr.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genthe J. R.; Min J.; Farmer D. M.; Shelat A. A.; Grenet J. A.; Lin W.; Finkelstein D.; Vrijens K.; Chen T.; Guy R. K.; Clements W. K.; Roussel M. F. (2017) Ventromorphins: A New Class of Small Molecule Activators of the Canonical BMP Signaling Pathway. ACS Chem. Biol. 12, 2436–2447. 10.1021/acschembio.7b00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A.; Briquez P.; Hubbell J.; Cochran J. (2016) Engineering growth factors for regenerative medicine applications. Acta Biomater. 30, 1–12. 10.1016/j.actbio.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata Y.; Lutolf M. P. (2017) Multiscale microenvironmental perturbation of pluripotent stem cell fate and self-organization. Sci. Rep. 7, 1–11. 10.1038/srep44711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFail A. J.; Chu C. R.; Izzo N.; Marra K. G. (2006) Controlled release of bioactive TGF-β1 from microspheres embedded within biodegradable hydrogels. Biomaterials 27, 1579–1585. 10.1016/j.biomaterials.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Ziegler T.; Möglich A. (2015) Photoreceptor engineering. Front. Mol. Biosci. 2, 1–25. 10.3389/fmolb.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy A. M.; Gerrard E. J.; Legrand J. M. D.; Hobbs R. M.; Janovjak H. (2019) Engineering Strategy and Vector Library for the Rapid Generation of Modular Light-Controlled Protein–Protein Interactions. J. Mol. Biol. 431, 3046–3055. 10.1016/j.jmb.2019.05.033. [DOI] [PubMed] [Google Scholar]
- Conrad K. S.; Manahan C. C.; Crane B. R. (2014) Photochemistry of flavoprotein light sensors. Nat. Chem. Biol. 10, 801–809. 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A.; Weiner O. D.; Lim W. A.; Voigt C. A. (2009) Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudasaini A.; El-Arab K. K.; Zoltowski B. D. (2015) LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Front. Mol. Biosci. 2, 1–15. 10.3389/fmolb.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Chen X.; Yang Y. (2012) Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9, 266–269. 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- Guntas G.; Hallett R. A.; Zimmerman S. P.; Williams T.; Yumerefendi H.; Bear J. E.; Kuhlman B. (2015) Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U. S. A. 112, 112–117. 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusch M.; Schelch K.; Riedler R.; Reichhart E.; Differ C.; Berger W.; Inglés-Prieto Á.; Janovjak H. (2014) Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 33, 1713–1726. 10.15252/embj.201387695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves A. I.; Rotherham M.; Markides H.; Rodrigues M. T.; Reis R. L.; Gomes M. E.; El Haj A. J. (2018) Triggering the activation of Activin A type II receptor in human adipose stem cells towards tenogenic commitment using mechanomagnetic stimulation. Nanomedicine 14, 1149–1159. 10.1016/j.nano.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Luo K.; Lodish H. (1996) Signalling by chimeric erythropoietin-TGFB receptors: homodimerization of the cytoplasmic domain of the type I TGF-B receptor and heterodimerization with the type II receptor are both required for intracellular signal transduction. EMBO J. 15, 4485–4496. 10.1002/j.1460-2075.1996.tb00826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Lin B.; Kim S.; Choi B.; Evseenko D.; Lee M. (2015) TGF-β1 conjugated chitosan collagen hydrogels induce chondrogenic differentiation of human synovium-derived stem cells. J. Biol. Eng. 9, 1–11. 10.1186/1754-1611-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Lee M.; Kim N.; Wu G.; Deng D.; Kim J. M.; Liu X.; Heo W. Do; Zi Z. (2018) Spatiotemporal Control of TGF-β Signaling with Light. ACS Synth. Biol. 7, 443–451. 10.1021/acssynbio.7b00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako K.; Pradhan S. J.; Barone V.; Ingles-Prieto A.; Muller P.; Ruprecht V.; Čapek D.; Galande S.; Janovjak H.; Heisenberg C.-P. (2016) Optogenetic Control of Nodal Signaling Reveals a Temporal Pattern of Nodal Signaling Regulating Cell Fate Specification during Gastrulation. Cell Rep. 16, 866–877. 10.1016/j.celrep.2016.06.036. [DOI] [PubMed] [Google Scholar]
- Takahashi F.; Yamagata D.; Ishikawa M.; Fukamatsu Y.; Ogura Y.; Kasahara M.; Kiyosue T.; Kikuyama M.; Wada M.; Kataoka H. (2007) AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl. Acad. Sci. U. S. A. 104, 19625–19630. 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny G. D.; Yu P. B.; Laha J. K.; Xing X.; Liu J. F.; Lai C. S.; Deng D. Y.; Sachidanandan C.; Bloch K. D.; Peterson R. T. (2008) Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg. Med. Chem. Lett. 18, 4388–4392. 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanvitale C. E.; Kerr G.; Chaikuad A.; Ramel M. C.; Mohedas A. H.; Reichert S.; Wang Y.; Triffitt J. T.; Cuny G. D.; Yu P. B.; Hill C. S.; Bullock A. N. (2013) A New Class of Small Molecule Inhibitor of BMP Signaling. PLoS One 8, e62721. 10.1371/journal.pone.0062721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford S. T. J.; Ranghini E. J.; Grimley E.; Lee P. H.; Dressler G. R. (2019) High-throughput screens for agonists of bone morphogenetic protein (BMP) signaling identify potent benzoxazole compounds. J. Biol. Chem. 294, 3125–3136. 10.1074/jbc.RA118.006817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.; Chen Y. G. (2012) Regulation of TGF-β receptor activity. Cell Biosci. 2, 9. 10.1186/2045-3701-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring M. B.; Birkhead J. R.; Suen L. F.; Yamin R.; Mizuno S.; Glowacki J.; Arbiser J. L.; Apperley J. F. (1994) Interleukin-1 beta-modulated gene expression in immortalized human chondrocytes. J. Clin. Invest. 94, 2307–2316. 10.1172/JCI117595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar V. S.; Gamer L. W.; Rosen V. (2016) BMP signalling in skeletal development, disease and repair. Nat. Rev. Endocrinol. 12, 203–221. 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- Hollnagel A.; Oehlmann V.; Heymer J.; Rüther U.; Nordheim A. (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem. 274, 19838–19845. 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- Yang G.; Yuan G.; Li X.; Liu P.; Chen Z.; Fan M. (2014) BMP-2 induction of Dlx3 expression is mediated by p38/Smad5 signaling pathway in osteoblastic MC3T3-E1 cells. J. Cell. Physiol. 229, 943–954. 10.1002/jcp.24525. [DOI] [PubMed] [Google Scholar]
- Savary K.; Caglayan D.; Caja L.; Tzavlaki K.; Bin Nayeem S.; Bergström T.; Jiang Y.; Uhrbom L.; Forsberg-Nilsson K.; Westermark B.; Heldin C. H.; Ferletta M.; Moustakas A. (2013) Snail depletes the tumorigenic potential of glioblastoma. Oncogene 32, 5409–5420. 10.1038/onc.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susperregui A. R. G.; Gamell C.; Rodríguez-Carballo E.; Ortuño M. J.; Bartrons R.; Rosa J. L.; Ventura F. (2011) Noncanonical BMP signaling regulates cyclooxygenase-2 transcription. Mol. Endocrinol. 25, 1006–1017. 10.1210/me.2010-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S.; Wang B.; Mansour S. L.; Schoenwolf G. C. (2016) BMP regulates regional gene expression in the dorsal otocyst through canonical and non-canonical intracellular pathways. Development 143, 2228–2237. 10.1242/dev.137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A.; Lagna G.; Massagué J.; Hemmati-Brivanlou A. (1998) Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 12, 186–197. 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.; Kirkbride K.; Sheu R.; Blobe G. (2009) The Transforming Growth Factor-β Type III Receptor Mediates Distinct Subcellular Trafficking and Downstream Signaling of Activin-like Kinase (ALK)3 and ALK6 Receptors. Mol. Biol. Cell 20, 4362–4370. 10.1091/mbc.e09-07-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M.; Otsuka F.; Nakamura K.; Inagaki K.; Suzuki J.; Miura D.; Fujio H.; Matsubara H.; Date H.; Ohe T.; Makino H. (2004) Characterization of the Bone Morphogenetic Protein (BMP) system in human pulmonary arterial smooth muscle cells isolated from a sporadic case of primary pulmonary hypertension: Roles of BMP type IB receptor (activin receptor-like kinase-6) in the mitotic action. Endocrinology 145, 4344–4354. 10.1210/en.2004-0234. [DOI] [PubMed] [Google Scholar]
- Ye J.; Bates N.; Soteriou D.; Grady L.; Edmond C.; Ross A.; Kerby A.; Lewis P. A.; Adeniyi T.; Wright R.; Poulton K. V.; Lowe M.; Kimber S. J.; Brison D. R. (2017) High quality clinical grade human embryonic stem cell lines derived from fresh discarded embryos. Stem Cell Res. Ther. 8, 1–13. 10.1186/s13287-017-0561-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseenko D.; Zhu Y.; Schenke-Layland K.; Kuo J.; Latour B.; Ge S.; Scholes J.; Dravid G.; Li X.; MacLellan W. R.; Crooks G. M. (2010) Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 107, 13742–13747. 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt K. P.; Olson E. J.; Castillo-Hair S. M.; Hartsough L. A.; Landry B. P.; Ekness F.; Yokoo R.; Gomez E. J.; Ramakrishnan P.; Suh J.; Savage D. F.; Tabor J. J. (2016) An open-hardware platform for optogenetics and photobiology. Sci. Rep. 6, 1–13. 10.1038/srep35363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repina N. A.; Bao X.; Zimmermann J. A.; Joy D. A.; Kane R. S.; Schaffer D. V. (2019) Optogenetic control of Wnt signaling for modeling early embryogenic patterning with human pluripotent stem cells. bioRxiv 665695 10.1101/665695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. E.; Toettcher J. E. (2019) Signaling Dynamics Control Cell Fate in the Early Drosophila Embryo. Dev. Cell 48, 361. 10.1016/j.devcel.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan R.; Necakov A.; Trylinski M.; Harish R. K.; Krueger D.; Esposito E.; Schweisguth F.; Neveu P.; De Renzis S. (2019) Optogenetic inhibition of Delta reveals digital Notch signalling output during tissue differentiation. EMBO Rep. 20, e47999. 10.15252/embr.201947999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. E.; Goyal Y.; Pannucci N. L.; Schüpbach T.; Shvartsman S. Y.; Toettcher J. E. (2017) The Spatiotemporal Limits of Developmental Erk Signaling. Dev. Cell 40, 185–192. 10.1016/j.devcel.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglés-Prieto Á.; Reichhart E.; Muellner M. K.; Nowak M.; Nijman S. M. B.; Grusch M.; Janovjak H. (2015) Light-assisted small-molecule screening against protein kinases. Nat. Chem. Biol. 11, 952–954. 10.1038/nchembio.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo D.; Seo Y.; Jung H.; Kim S.; Kim N.; Park S. M.; Lee H.; Lee S.; Cho K. H.; Heo W. Do (2019) Locally Activating TrkB Receptor Generates Actin Waves and Specifies Axonal Fate. Cell Chem. Biol. 26, 1652–1663. 10.1016/j.chembiol.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Siuda E. R.; McCall J. G.; Al-Hasani R.; Shin G.; Park S.; Schmidt M. J.; Anderson S. L.; Planer W. J.; Rogers J. A.; Bruchas M. R. (2015) Optodynamic simulation of β-adrenergic receptor signalling. Nat. Commun. 6 (8480), 9480 10.1038/ncomms9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimas A.; Ambrosi C. M.; Yu J.; Williams J. C.; Bien H.; Entcheva E. (2016) OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat. Commun. 7, 11542. 10.1038/ncomms11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdellah K.; Gutierrez-Guerrero A.; Cobo M.; Muñoz P.; Martín F. (2014) A chimeric HS4-SAR insulator (IS2) that prevents silencing and enhances expression of lentiviral vectors in pluripotent stem cells. PLoS One 9, e84268. 10.1371/journal.pone.0084268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff N.; Lachmann N.; Ackermann M.; Kohlscheen S.; Brendel C.; Maetzig T.; Niemann H.; Antoniou M. N.; Grez M.; Schambach A.; Cantz T.; Moritz T. (2013) A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells 31, 488–499. 10.1002/stem.1316. [DOI] [PubMed] [Google Scholar]
- Herbst F.; Ball C. R.; Tuorto F.; Nowrouzi A.; Wang W.; Zavidij O.; Dieter S. M.; Fessler S.; Van Der Hoeven F.; Kloz U.; Lyko F.; Schmidt M.; Von Kalle C.; Glimm H. (2012) Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol. Ther. 20, 1014–1021. 10.1038/mt.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart-Cairns M. P.; Lim K. T. W.; Zuk A.; Godwin A. R. F.; Cain S. A.; Sengle G.; Baldock C. (2019) Internal cleavage and synergy with twisted gastrulation enhance BMP inhibition by BMPER. Matrix Biol. 77, 73–86. 10.1016/j.matbio.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O.; Ten Dijke P. (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277, 4883–4891. 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- Ballister E. R.; Rodgers J.; Martial F.; Lucas R. J. (2018) A live cell assay of GPCR coupling allows identification of optogenetic tools for controlling Go and Gi signaling. BMC Biol. 16, 1–16. 10.1186/s12915-017-0475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.