Abstract
Multiple sclerosis (MS) is a neuroinflammatory disorder characterized by autoimmune-mediated inflammatory lesions in CNS leading to myelin damage and axonal loss. MS is a heterogenous disease with variable and unpredictable disease course. Due to its complex nature, MS is difficult to diagnose and responses to specific treatments may vary between individuals. Therefore, there is an indisputable need for biomarkers for early diagnosis, prediction of disease exacerbations, monitoring the progression of disease and for measuring responses to therapy. Genomic and proteomic studies have sought to understand the molecular basis of MS and find biomarker candidates. Advances in next-generation sequencing and mass-spectrometry techniques have yielded an unprecedented amount of genomic and proteomic data; yet, translation of the results into the clinic has been underwhelming. This has prompted the development of novel data science techniques for these large datasets to identify biologically relevant relationships and ultimately point towards useful biomarkers. Herein we discuss optimization of omics study designs, advances in the generation of omics data, and systems biology approaches aimed at improving biomarker discovery and translation to the clinic for MS.
Keywords: Genomic, Proteomic, Systems Biology, Biomarkers, Multiple Sclerosis, Networks
1. Introduction
Multiple sclerosis (MS) is an autoimmune neuroinflammatory disorder that affects nearly one million Americans and 2.5 million individuals worldwide [1, 2]. MS is characterized by the occurrence of histopathological lesions in the central nervous system (CNS) detected by MRI [3]. Histopathologically, these lesions are composed of inflammatory cells, including infiltrating myelin reactive T cells, B cells/plasma cells, macrophages, and dendritic cells (DC) that release inflammatory cytokines, chemokines and antibodies [4]. Accordingly, there is a consensus in the field that MS is driven primarily by T cells with important contributions of B cells and innate immune cells [5, 6]. Furthermore, CNS resident cells, including astrocytes and microglia also contribute to disease pathology by sustaining a proinflammatory milieu. The focal areas of inflammation in the CNS cause myelin degradation and axonal loss that manifests itself clinically in individuals by an array of symptoms, including numbness, fatigue, visual and emotional disturbance, and paralysis [7].
While MS etiology is still not resolved, several genetic and environmental factors are known to influence susceptibility. Over 200 genes have been associated with MS susceptibility, the most strongly implicated being HLA types with HLA-DR2 (DRB1*15:01) showing the strongest correlation with occurrence of disease [8, 9]. Environmental factors such as geographic location (i.e. distance from the equator), smoking, salt intake, and infections have also been linked to MS incidence, which implies that the disease risk of an individual is most likely the result of several genetic risk factors and environmental influences [10, 11].
MS is a heterogenous clinical condition whose diagnosis is broadly classified into three types based on disease presentation: relapsing remitting MS (RRMS), primary progressive MS (PPMS) and secondary progressive MS (SPMS) [12]. The diagnosis of MS typically begins with an initial finding of clinically isolated syndrome (CIS) or radiologically isolated syndrome (RIS), and definitive diagnosis of MS requires evidence of dissemination of lesions in the CNS in space and time as well as exclusion of other potential disorders [13]. The foremost guideline for diagnosis, the McDonald criteria, has undergone several revisions [14–16]. These iterations reflect the challenge of diagnosing MS without a definitive laboratory test. Once the diagnosis of MS is established, the task of monitoring the disease is equally extensive and includes MRI and disability measures such as the Expanded Disability Status Scale (EDSS) [17–19]. However, MRI and clinical measures of disease state and progression are limited in terms of defining the underlying pathology nor are they predictive for recurrence or progression. Therefore, disease prognosis is currently limited by a lack of definitive molecular biomarkers to monitor disease activity and progression.
Disease-modifying therapies for MS have been effective in managing acute exacerbations and slowing the progression of the disease [20]. Unfortunately, responses to treatments are variable between patients and there are few biomarkers to measure the efficacy of therapeutics in individuals [21]. Importantly, while disease-modifying therapies may slow progression of RRMS to SPMS, there are currently no therapies that can completely prevent it, which is also hampered by a lack of understanding of the mechanisms that drive progressive disease and a lack of biomarkers to monitor progression [22, 23]. Furthermore, MS lacks definitive markers to measure drug responses precisely and quickly in patients.
Taken together, there is an urgent need to develop biomarkers for MS at key stages in the disease process: MS diagnosis, prediction of relapses in RRMS, predicting the progression of RRMS to SPMS, monitoring the progression of PPMS and SPMS as well as for measuring drug efficacy. We posit that the large genomic and proteomic data sets generated in the search of trying to better understand the pathogenesis of MS provide an invaluable resource that can be explored for the discovery of novel biomarkers. However, as of now, the potential of these large datasets to yield novel biomarkers for diagnosis or prognosis of MS has yet to be fully realized in clinical practice.
The biomarker pipeline entails an initial discovery phase consisting of large omics studies that can be mined for promising candidate molecules that are then selected for preclinical validation, and ultimately for clinical validation and assay development [24–26]. Major bottlenecks in this process are present at each stage, beginning during preclinical validation. Along this line, there is less market incentive for the development of commercial biomarkers than there is for drug development which necessitates streamlined and efficient methods of biomarker discovery and commercialization [27]. Nonetheless, identifying surrogate endpoint biomarkers for drug efficacy may allow to significantly shorten the time and money spent on clinical trials and thus allow faster transition to drug production [27].
Herein, we will discuss different approaches and pipelines for generation and analysis of large omics data sets to assist with biomarker discovery and with translating these markers to the clinic, as summarized in Figure 1. We posit that improved discovery and subsequent analysis methods using modern data science tools as well as animal models can facilitate overcoming these bottlenecks and help deliver more robust biomarker candidates for clinical validation.
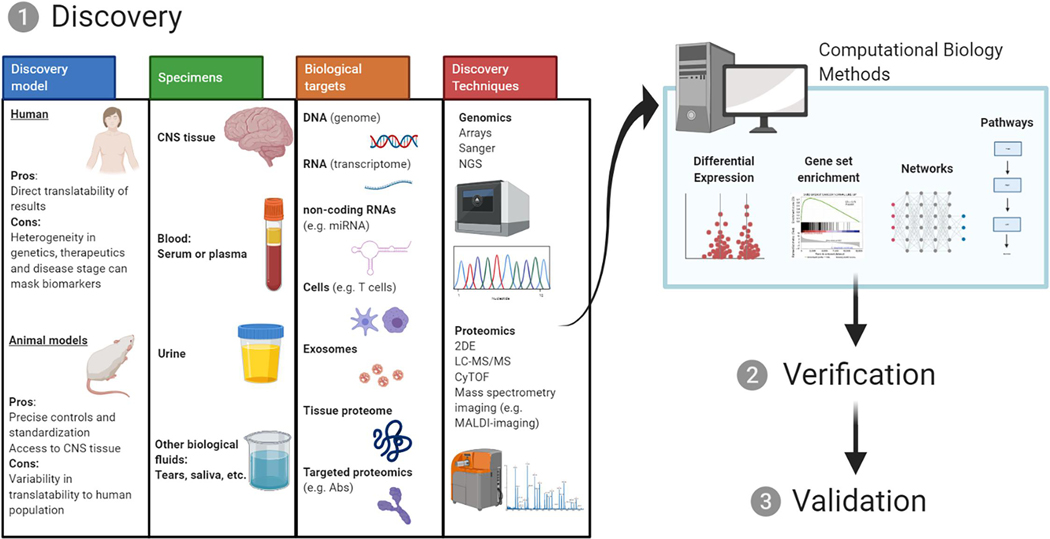
Fig. 1.
Study design and analysis methods for biomarker discovery. Discovery is the first stage in the pipeline of biomarker development, followed by verification and validation. The discovery stage requires careful consideration for the selection of discovery model, specimens, biological targets and discovery techniques. After the generation of large omics datasets, improved computational biology analysis methods can yield potential candidate molecules for the verification and validation stages and clinical testing.
2. Biological samples for candidate biomarker discovery
2.1. Post-mortem tissue
Investigations of MS genomes and proteomes for the purpose of biomarker discovery have varied widely in use of source materials, such as MS patient post-mortem tissues, cerebrospinal fluid (CSF), patient serum, and tissue samples generated in mouse models such as EAE [28, 29]. Several studies have utilized MS post-mortem tissue to investigate the genome and proteome of lesions, identifying molecular signatures of lesions and pointing to potential therapeutic targets [30–32]. Though CNS tissue sampling is generally not feasible in MS patients, discovery using CNS-derived or associated materials (e.g. CSF) can offer important insights into disease mechanisms as pathogenesis occurs in the CNS and thus may eventually lead to biomarkers detectable in the blood by virtue of the increased permeability of the blood-brain barrier (BBB) in MS [33, 34]. Utilizing approaches such imaging mass spectrometry of CNS tissue may permit brain regional discrimination of proteins [35]. Similarly, slide sequencing allows for high resolution spatial RNA-Sequencing (RNA-Seq) of brain regions and may be useful for investigation of CNS regional transcriptional changes, though this has yet to be realized in MS biomarker research [32].
2.2. Cerebrospinal fluid
Many biomarker discovery studies have focused on CSF because it is a direct reflection of the disease processes in the CNS [36]. The most prominent MS molecular biomarker historically is the presence of oligoclonal bands (OCB) in the CSF as seen by isoelectric focusing [37, 38]. OCB consist of immunoglobulins, predominantly IgG secreted by plasma cells in the CNS, and their presence is indicative of CNS inflammation and is predictive of conversion of CIS to MS [39–41]. One of the recent and clinically more advanced potential MS biomarkers, neurofilament light (NfL), a marker of neuronal damage and disease activity, is also detected in the CSF [42]. In addition, genomic studies of the CSF have sequenced the B cell receptor (BCR) and T cell receptor (TCR) repertoires in the CSF of MS patients [43, 44]. Although the CSF has proven to be a prolific source for biomarker discovery, CSF sampling is invasive and for clinical purposes other body fluids are preferable.
2.3. Blood
Blood is a reflection of all tissues and cell types and it is an ideal source for routine monitoring of biomarkers in patients because obtaining it is minimally invasive [45]. Blood consists of a cellular component, such as leukocytes and red blood cells, and the liquid plasma component comprised of proteins, salts, lipids, amino acids, vitamins and carbohydrates [46]. Blood from patients is readily accessible and several research groups have searched for biomarkers and disease protein fingerprints in the serum [47, 48]. The BBB is more permeable in MS patients than healthy controls and its permeability correlates with relapses [34]. Leakage proteins from the CNS can be detected in the blood potentially because of altered glial-lymphatic clearance in MS patients [49, 50]. For instance, NfL can be detected in blood, and its blood levels correlate with CNS levels thus providing a more feasible alternative to obtaining CSF for sampling [51, 52].
Genomic studies in the blood have focused on whole blood profiling as well as profiling individual cells to identify differentially expressed transcripts in MS and unique T cell/B cell gene expression signatures in MS patients [53, 54]. Furthermore, single-cell RNA-Seq (scRNA-Seq) analysis of immune cells in blood and CSF of MS patients identified changes in gene signatures and pathways in T cells of MS patients [55]. Moreover, MS blood was reported to contain exosomes which originated in the CNS, in which their content correlates to disease activity [56]. The utility of these genes and pathways as biomarkers for disease will need further investigation, but nonetheless the blood might be a useful and important tissue to investigate genomic biomarkers.
The presence of highly abundant proteins (e.g. albumin, globulins, and apolipoprotein B) in the blood presents a challenge for mass spectrometry proteomic investigations. For example, albumin and immunoglobulins account for about 75% of the total protein mass, and together with about 20 additional proteins, accounts for approximately 99% of the total blood proteome. Thus, low-abundant proteins such as tissue-specific and leakage proteins correspond to only 1% of all blood proteins [57]. This phenomenon leads to what is termed as “masking” of lower abundance proteins in proteomic methods such as in LC-MS/MS. This challenge prompted the development of depletion methods to remove the top abundant proteins from blood; however, masking remains a difficult challenge even after utilizing depletion methods [58]. With advances in quantitation and separation techniques as well as higher sensitivity mass spectrometry, low abundance proteins can be detected without fractionation, but whether these technologies can yield clinically relevant disease-specific biomarkers has yet to be conclusively determined [59].
2.4. Alternative body fluids
Though less frequently represented as source materials in the literature, urine, tears and saliva can be obtained with minimally invasive procedures and are relatively easy to store [28, 29, 60]. Oligoclonal bands are present in the tears of MS patients, indicative of cross talk between blood plasma, from which lacrimal glands produce tears, and the CNS [61, 62]. Vesicles in tears were shown to be derived from microglia and neurons as further evidence of the information exchange between the CNS and tears as well as pointing towards the potential use of tears in diagnostic tests [63]. Similarly, MBP-like molecules as identified by cross reactivity with MBP peptides are detectable in urine and correlate with increased CNS lesion load [64]. Proteomic profiling showed that the saliva proteome in MS patients is distinct from healthy controls [65]. The omics field has not prioritized discovery of these fluids as of yet, but they are a potential source for discovery and validation of biomarkers.
2.5. Considerations for the use of human samples in biomarker discovery
Beyond the selection of tissues for biomarker discovery, the patient populations from which patient samples are obtained for biomarker discovery is an area that requires careful consideration. The selection of patient samples for MS omic studies is often driven by availability, and standardization and comparison of samples within studies and between studies can be challenging. Many groups apply their own criteria and methods of inclusion/exclusion of patient samples from studies, but they vary widely between studies and are less standardized for biomarker studies than for clinical trials [66]. One acceptable method employed is the use of longitudinal, prospective studies following patients over time, but they often are limited by high demand for precious patient samples, small sample size, short collection periods, high rate of drop outs and variability in therapies of the patients [67, 68]. Longitudinal studies of molecular MS biomarkers have generally been implemented with limited scope, such as monitoring specific markers such as NfL as opposed to periodic sampling for the generation of longitudinal omics data for biomarker discovery [69]. We advocate for more standardized sampling protocols and better study design in order to make discoveries reproducible and allow to tease out biologically relevant findings in these large datasets. These standards should take into consideration time since diagnosis as well as therapies applied, and patients should best be stratified by synchronizing starting or end points such as specific treatments.
Another consideration in study design concerns the demographics of the population used for sampling. The majority of MS studies are conducted in Caucasian populations, whereas MS can occur in ethnically diverse populations and there is evidence suggesting that within the preponderance of female diagnoses, Hispanic/Latino and African-American females are diagnosed with CIS and MS at higher rates than Caucasian females [70–72]. Here, genomic and transcriptomic analysis, and conceivably in combination with proteomic studies, may allow stratification of large and seemingly diverse data sets using key traits, such as HLA haplotypes, to provide novel insights.
In consideration of optimized subject selection for biomarker discovery, monitoring of first-degree relatives for the purpose of MS onset biomarkers may hold significant potential. First degree relatives of MS patients have a 20–40 times greater risk of developing MS than the background population [73, 74]. A longitudinal study has been undertaken to monitor the development of MS in first-degree relatives of patients with MS, with its initial finding two years into the study showing that first-degree relatives in this cohort exhibit a 30 times greater chance of developing MS [75]. Thus, genomic/proteomic analysis of materials derived from similarly designed and sufficiently powered studies could be very revealing in terms of potential biomarkers for disease onset.
2.6. Animal models
As an alternative to the use of the above-mentioned human tissues and fluids, several studies have used animal models, such as experimental autoimmune encephalomyelitis (EAE) in rodents [76–82]. The use of animal models allows for investigation of the CNS and other tissues during disease and circumvents the difficulties of obtaining human tissues. Furthermore, the use of animal models of MS can help discovery as well as verification of biomarker candidates by allowing investigation of the omics landscape in the CNS, confirming expression in the blood, and permitting correlation of the putative biomarkers with immune phenotype, disease pathology, and clinical signs in a controlled environment [82–84]. As to whether the EAE omics studies are a faithful reflection of processes in MS it is worth noting that most of the advancements in understanding the pathology of MS and development of therapeutics have resulted from animal models, and studies in EAE could therefore conceivably also yield data relevant to human disease [85]. In addition, the relative genetic homogeneity of syngeneic animal models and synchronicity of timed animal studies could potentially allow for the teasing out of relevant disease biomarkers that might not be seen in tissue samples garnered from genetically diverse and heterogenous human MS populations at highly variable disease states and time points.
It is a valid concern that studies in syngeneic mice may miss the mark in terms of the genetic diversity observed in humans. Thus, non-human primate (NHP) models of EAE may be informative under certain circumstances. Of note, the EAE model was initially characterized in NHPs and transitioned to studies primarily in rodents [85, 86]. Moreover, the phylogenetic similarity to humans is reflected in their similar immune responses and tissue pathology [87]. EAE has been documented in several NHPs and marmosets are of particular interest in research studies because of strong clinical and histopathological resemblance to MS [88, 89]. However, studies in NHPs are cost prohibitive, fall under extensive regulatory guidelines, and raise additional ethical concerns. Nevertheless, biomarker studies in NHPs may represent a valid approach under the appropriate circumstances.
Rodent models of EAE, for example using mice, have proven of enormous value for unraveling the pathophysiological mechanisms of neuroinflammatory disease. Nonetheless, rodent models could benefit from further optimization to more closely reflect the genetic diversity seen in humans and thus better translate to the human populations where MS occurs [90]. Outbred mouse strains are commercially available and there is evidence to suggest that studies using outbred strains of mice may correlate better to human disease as they are commonly used in pharmacological studies [90–92]. Alternatively, “humanized” mice expressing MS-associated molecules, such as the HLA-DR2 allele and human T cell receptors (TCRs) have been used to better approximate human MS in the mouse and they also hold potential for use in biomarker studies [93–95]. Another way in which to align mouse studies more closely to human disease is to select EAE models that have clinical phenotypes that recapitulate MS phenotypes. SJL mice are commonly used in studies of RRMS and NOD mice have been used to study EAE progression [96–98]. Overall, there is still room in the field for biomarker studies using novel mouse models that better reflect certain key aspects of human MS, such as SPMS.
3. Genomic Approaches For Biomarker Discovery
For as long as tools for DNA analysis have existed, researchers have used them to investigate the role that genes play in health and human diseases. The identification of different HLA alleles in the 1960s using serological techniques such as microlymphocytotoxicity assays quickly ushered in the discovery that the development of MS is strongly correlated with certain HLA haplotypes [99, 100]. HLA alleles remain the most important risk factor for MS confirmed over the following decades by single nucleotide polymorphism (SNP) genome-wide association studies (GWAS) using SNP microarrays and more recently with next-generation sequencing (NGS) [8, 101]. GWAS has provided additional insights into susceptibility loci, disease pathogenesis, potential therapeutic targets and putative biomarkers [102, 103].
Genomics employs tools to gather genetic data and bioinformatics methodologies to interpret these frequently enormous data sets [104]. In MS, genomics has provided fundamental insights into disease susceptibility, immune and neurodegenerative mechanisms that drive the disease, therapeutic targets and potential biomarkers. These discoveries and their basis have been extensively reviewed elsewhere [29, 105–107]. Additionally, each methodology requires specific bioinformatic approaches and computation analysis pipelines to mitigate intrinsic technical and biological caveats, and these have been discussed in detail elsewhere (for both genomic and proteomic methods) [108–112]. Figure 2 outlines general pipelines in quantitative genomic and proteomic practices. More detailed pipelines for each genomic and proteomic techniques are available through web-based tolls including SequencEnG for genomic [113] and ProteoSuite for proteomic [114]. Table 1 summarizes the key genomic methods used for biomarker discovery and their relative advantages and caveats for biomarker discovery. Here, we will discuss recent advances in genomic methods and key genetics targets applicable to MS research and biomarker discovery.
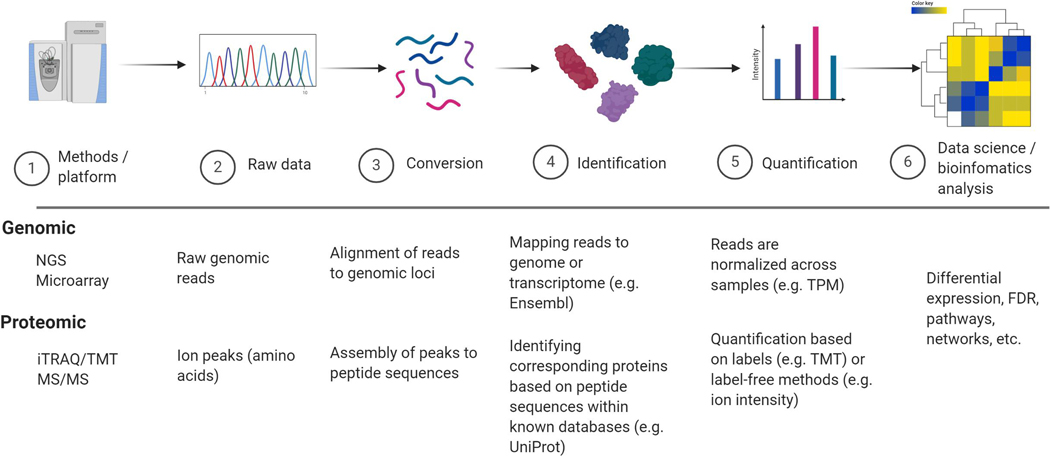
Fig. 2.
Pipelines in quantitative genomic and proteomic. Depending on instrumentation and platform (1) raw data is generated (2). Raw data is converted (or “translated”) to genomic loci or peptide sequences (3) followed by identification using specific genome or proteome databases (4), normalization, and quantification (5). Last, data can be investigated using computational and statistical tools (6).
Table 1:
Advantages and disadvantages of genomic and proteomic methodologies
| Advantages | Disadvantages | Previous applications | |
|---|---|---|---|
| Genomic | |||
| DNA SNP microarray [102, 129, 229] | Precise, quantitative, and cost effective | Not entirely discovery-driven, needs specific targets | Used in GWAS to identify HLA and other gene variants associated with MS |
| NGS Whole Genome Sequencing [230] | Unbiased full coverage of the genome | Large amount of information, lack of standardization for biomarker discovery | Used to identify genetic variants associated with PPMS |
| NGS Whole Exosome sequencing [56, 231] | Finds differences in functional genes, cost effective alternative to whole genome sequencing | Misses important genetic variations that occur outside of protein-coding regions | Used to identify genetic biomarkers indicative of disease course |
| Single cell NGS [55, 123, 232, 233] | High resolution of individual cell heterogeneity | High cost and lack of standardized computational pipelines | Used to identify transcriptional changes in blood cells in MS |
| Single nucleus NGS [124, 125, 234] | Data from CNS cell types that are difficult to isolate for single cell analysis | Misses cytosolic information | Used to identify lineage-specific alterations in transcription during MS |
| miRNA sequencing [144, 146, 147] | Offer insight into disease-related modification of gene expression. miRNA levels are putative biomarkers | Difficulty in reaching statistical significance in studies | Used to identify several miRNAs as markers of MS disease state and severity |
| Slide sequencing [32] | Regional discrimination of gene expression | No standardized pipeline for using resulting data for biomarker discovery | Used to identify spatial differences in gene expression in the brain |
| Epigenetic techniques [150, 235] | Insight into links between environmental exposures and MS occurrence and trajectory | Lack of longitudinal studies that definitively link environmentally induced epigenetic changes to MS disease course | Used to identify modifications associated with MS occurrence and severity |
| Proteomic | |||
| 2DE and 2-DIGE [157, 158, 236, 237] | Can be used for separation prior to mass spectrometry | Displays approximately ~200–2000 spots, does not detect many regulatory molecules | Used for the early characterization of IgG in MS patient CSF |
| Shotgun proteomics [30] | Unbiased, high-resolution data | Qualitative data only | Used to characterize proteins in MS lesions |
| Quantitative shotgun proteomics [81, 82, 164, 167] | Unbiased, high-resolution data that allows for statistics and bioinformatics approaches |
No standardize pipeline for analyzing the resulting data | Used to identify differentially expressed protein in the brain, CSF, and serum during MS/EAE |
| CyTOF [170, 171] | Quantitative data on the single cell level | Limited by availability of labelled antibodies | Used to identify unique cell protein signatures in neuroinflammation and MS |
| Imaging mass spectrometry [35] | Regional discrimination of protein expression | Not quantitative | Used to map proteins in brain lesions |
| Post-translational modification methods [176] | Adds to diversity of proteomic data by identifying differences not related to amino acid sequence | Large diversity of modifications that can be hard to target and quantify | Identification of modified epitopes potentially contributing to autoimmunity |
3.1. Advanced genomic techniques and methodologies
Next generation sequencing encompasses high-throughput sequencing methods that deliver DNA and RNA sequences at the whole genome scale [115]. This is in contrast to the foundational sequencing methods, Maxam-Gilbert sequencing and the widely used Sanger dideoxy termination sequencing method that reads about 1000bp per experiment [104]. Sanger sequencing is the means by which the human genome was sequenced over the course of a decade and, at that time, at great expense [116]. Modern NGS platforms can sequence an entire human genome in less than a day and at a fraction of the cost [104]. Several NGS technologies are available such as sequencing by synthesis (Illumina), Ion semiconductor (Ion Torrent), single-nucleotide addition (pyrosequencing), and single-molecule real-time sequencing (Pacific Biosciences) [117]. Each one of these platforms is tailored and more suitable for different applications with different types of analysis [118, 119]. Illumina platforms contribute to most of the NGS data worldwide due to greater availability of bioinformatics tools to analyze these data, base-call quality, high yield and low cost [120, 121]. The use of NGS has not only confirmed HLA allele susceptibility previously determined by other technologies, but it has opened the opportunity for better understanding disease pathology and the discovery of biomarkers. Researchers who previously used Sanger sequencing to identify an antibody gene signature in B cells predictive of the development of MS were able to confirm the same signature using large-scale NGS pyrosequencing [44]. They confirmed the fidelity of NGS compared to Sanger and pointed towards a more clinically relevant testing technology. Researchers have also been able to use NGS to answer very specific questions, for instance, the identification of Vitamin D gene variants and changes in Vitamin D signaling pathway in MS patients [122].
The sensitivity of NGS also allows for the characterization of the genome on a single cell basis [123]. Single nucleus RNA-Seq (snRNA-Seq) of human post-mortem tissue identified differences in oligodendrocyte profiles in MS patients compared with healthy controls, with oligodendrocytes showing higher expression of myelin genes and oligodendrocyte transcription factor 1 (OLIG1), and snRNA-Seq of different locations of CNS lesions of MS patients revealing lineage-specific signatures of stressed oligodendrocytes, reactive astrocytes and activated microglia associated with region-specific changes [124, 125]. Single-cell RNA-Seq (scRNA-Seq) of leukocytes in the CNS and blood of MS patients revealed increased transcriptional diversity of leukocytes and increased cell diversity in the CNS [55]. Taken together, these works highlight the potential of different NGS platforms to reveal sensitive and disease-related tissue specific changes in MS to uncover novel disease processes, and the data sets generated may aid in MS biomarker discovery.
3.2. Investigations of disease etiology and susceptibility
The association between HLA haplotypes and MS has been described in the 1970s using serological methods [99, 126]. Single nucleotide polymorphism (SNP) genotyping corroborated these and other studies showing that certain HLA haplotypes associate with MS, with HLA-DR2 (DRB1*1501) being most strongly correlated with disease incidence [8, 127, 128]. HLAs were the only known genetic determinants of MS until GWAS studies using data from SNP arrays identified over 200 additional genetic risk loci, including polymorphisms in immune-related genes such as CXCR5, TNFRSF1A, IL2RA and IL7RA [102, 129]. However, the full spectrum of gene variation cannot be discovered using only DNA arrays and advances in NGS techniques have the potential to reveal the entire spectrum of sequence variations between MS and healthy patients as well as between different MS disease states [130]. Compared with serological haplotyping, NGS has allowed for high-resolution genotyping of HLA alleles associated with MS and enabled the comparison of both coding and non-coding regions of the genes confirming previously known MS-risk alleles, permitting the identification of protective alleles, and implicating roles for other MHC alleles in MS, including MHC class I alleles such as HLA-B*39:01 and 15:01 [101, 131]. Higher-resolution measurements have also allowed for the discrimination of alleles that were previously indistinguishable, thus resolving controversies as to whether certain alleles are harmful or protective, such as in the case of HLA-DRB1*04 alleles where the literature was inconclusive [101]. NGS of haplotypes allowed for four-field allele typing of DRB1*04:01:01:01SG and revealed that this allele is associated with susceptibility or protection depending on the haplotype with which it is associated and that this allele alone confers neither protection or susceptibility [101].
Cancer researchers have made great strides in using NGS technology to characterize tumor-specific immune landscapes, and we posit MS research will benefit using these approaches. Cancer research groups have used NGS RNA-Seq data to characterize the immune landscape through sensitive HLA typing and characterization of BCR and TCR repertoires in cancer specimens [132–134]. Through this a strong correlation was identified between T cell diversity and tumor load and expression of novel tumor antigens [133]. Furthermore, this approach revealed novel tumor evasion mechanisms and immunoglobulin heavy chain sequences in tumor-infiltrating B cells that could lead to novel immunotherapies [134]. We anticipate similar advances using these techniques in the field of MS research.
3.3. Exosomal sequencing and microRNA
Exosomes are membrane-bound vesicles secreted by different cell types that are used for cell-to-cell communication and are known to play roles in immune modulation [135]. Exosomes contain proteins, lipids and RNA from the cell of origin that not only allow for tracing back to their origin, but can also provide insights into physiological and disease processes [136–140]. Furthermore, microRNA (miRNA) which are normally located in the cytoplasm of cells, can be packaged and exported in exosomes [141]. miRNAs are small endogenous RNA that regulate gene expression post-transcriptionally by modulating mRNA stability and decay [142]. Blood exosome sequencing has become increasingly prominent in genetics studies and biomarker discovery, particularly for tissues with limited access such as the CNS [140]. For instance, in MS serum exosomes that diffuse out of the CNS contain myelin oligodendrocyte protein (MOG) and these exosomes correlate to disease severity [143]. miRNA specifically identified in exosomes in the CNS and serum of patients have significant potential as biomarkers [140, 144, 145]. NGS platforms can be used to characterize the expression of miRNA [140]. Moreover, NGS platforms have been used to characterize miRNA contents in the CSF and serum of MS patients, and miRNA expression has been correlated with disease status, pointing towards their potential as prognostic biomarkers [140, 144–146]. Along these lines, specific miRNAs such as miR-155 have been implicated as indicators of disease and also as putative therapeutic targets [144, 147].
3.4. Epigenetics
Only a subset of individuals with MS-related genes develop MS and the field of epigenetics may therefore hold the potential for adding to our understanding of the mechanisms by which environmental factors drive the development of disease. DNA methylation and histone modifications are epigenetic factors that do not influence the sequence of DNA, but still exert an effect on phenotype [148, 149]. The prominent risk gene, HLA DRB1*15:01 is hypomethylated in MS patients, and hypermethylation resulting in an overall decreased expression of the gene is believed to be protective [150]. In addition, risk factors associated with the occurrence of MS such as vitamin D deficiency and Epstein-Barr virus infection are known to promote epigenetic changes [151, 152]. It is also known that aberrant DNA methylation can exacerbate inflammation in MS [153]. Potentially, new methods for the analysis of epigenetic changes on the background of genetic susceptibility genes may provide inroads to development of biomarkers for disease progression and serve to develop new drugs and determine their efficacy.
In summary, genomic studies have been mostly geared towards identifying susceptibility alleles and pathogenic mechanisms in MS. However, these datasets may also be useful to identify prognostic and diagnostic markers of MS. For instance, it was reported that SNPs in HLA and non-HLA genes are predictive of MS occurrence and of clinical outcomes [154]. Similarly, sequencing of miRNA provided proof-of-concept that transcripts can serve as biomarkers for disease activity [144, 145]. We posit that a better understanding of mechanisms of disease as elucidated by genomics will point to potential biomarkers of disease, particularly when combined with proteomic data and analysis using modern data science techniques.
4. Proteomic Approaches For Biomarker Discovery
Although genomic tools have been extensively used in research due to their high sensitivity and breadth/depth of information, proteomic methods hold great promise for biomarker discovery supported by an increasing number of tools for protein identification and quantification [24]. A culmination of advancements in instrumentation and data acquisition have allowed for methods with increased sensitivity and specificity, thereby leading to large proteomic data sets which are becoming more similar in scope to genomic data [155, 156]. Shown in Table 1 are key proteomic methods and their benefits and weaknesses for biomarker discovery.
4.1. Technology and Instrumentation
Early attempts at understanding the changes in the proteome during MS used 2-dimensional gel electrophoresis (2DE) separation for the identification and comparison of specific targets within samples [157, 158]. As technology progressed, 2DE separation was used in tandem with mass spectrometry yielding more comprehensive and reproducible insights into proteome changes occurring in MS [159–161]. Driven by further advancements in technology such as the development of the Orbitrap and improvement in time-of-flight (TOF) mass spectrometers, researchers have favored more high throughput shotgun proteomic approaches [162, 163]. Shotgun proteomics is the evaluation of complex mixtures of proteins digested into peptides using a combination of high performance liquid chromatography combined with mass spectrometry [162]. In the search for differentially expressed proteins shotgun proteomics has been combined with various quantitation techniques.
4.2. Quantitation
Quantitative proteomics has allowed for novel statistical and bioinformatics approaches for the identification of clinically relevant proteins and disease pathways [164]. Tandem mass tags (TMT) and isobaric tags for relative and absolute quantification (iTRAQ) are now widely used in MS studies [165, 166]. Using iTRAQ labels, researchers identified differentially expressed proteins in rat EAE spinal cords that are most likely upregulated due to inflammatory infiltration and microglial activation [77]. iTRAQ labelling of human serum identified a panel of proteins enriched in patients with aggressive MS as well as a panel of proteins enriched in asymptomatic MS [48]. TMT labeling of mouse EAE brains allowed for the identification of a panel of CNS proteins predictive of the onset of clinical EAE symptoms that was subsequently validated by their serum concentrations [82]. TMT labeling of five separate EAE brain regions and spinal cord revealed that the spinal cord showed the highest level of differentially expressed protein expression compared to naïve CNS with the altered proteins representing predominantly inflammatory processes [81].
Label-free absolute quantification has also gained momentum, though not as widely implemented, in investigations of MS and in other diseases [167]. Datasets generated by these quantitative approaches have revealed many potential biomarkers; yet, molecular biomarkers have been slow to reach the clinic and establish their utility. The enormous size of the generated datasets, both by proteomics and genomics, have presented a veritable challenge in developing analysis techniques that yield candidate markers that are biologically and clinically relevant.
4.3. Single-cell mass cytometry
Another field that is emerging in MS research is single-cell cytometry by time-of-flight mass spectrometry (CyTOF). In CyTOF, cells are labelled with antibody (Ab)-conjugated isotope reporters. In contrast to fluorophores their detection is not limited by spectral overlap, allowing for the detection of dozens of cellular features by each individual cell and specifically allowing for high-dimensional immunophenotyping of individual cells [168, 169]. For example, a comparison of myeloid cells in CNS of EAE mice identified differences in signaling molecules and cytokine production within myeloid populations as compared with naïve animals [170]. Moreover, CyTOF profiling of peripheral blood mononuclear cells and CSF cells in MS patients identified a T helper cell subset characterized by high expression of granulocyte-macrophage colony-stimulating Factor (GM-CSF) and C-X-C motif chemokine receptor 4 (CXCR4), and this population was reduced following disease-modifying therapy, thus pointing to these cells as markers of drug response [171]. Though CyTOF is considered a high-throughput method, the markers which can be measured are finite and depend on the availability of Ab-conjugates. Until the time that single cell proteomics is firmly established, CyTOF can provide semi high-throughput insights into protein expression on cells that can identify mechanisms, therapeutic targets and biomarkers of disease [172].
4.4. Post-translational modifications
Adding to the diversity of proteomic data, an important field of inquiry is the role of post-translational modifications (PTMs) in MS pathogenesis and investigation of modified proteins as potential biomarkers. PTMs encompass numerous covalent modifications of proteins after biosynthesis, including glycosylation, methylation, phosphorylation and citrullination, the presence of which can be detected through Western blotting, modification-specific ELISAs and mass spectrometry by virtue of their effect on amino acid residue and protein mass [173, 174]. Investigations in EAE have implicated PTMs of self-antigens in the etiopathogenesis of MS and other autoimmune diseases, as they change recognition of T cell epitopes when they occur on residues that contact the TCR and could contribute to the breakdown of peripheral tolerance [175–177].
PTMs of myelin basic protein (MBP) have been studied extensively, revealing MBP PTMs that correlate with MS disease severity including methylated Arg107, and overall increase in deamination and reduction in phosphorylation compared with healthy controls [178]. Citrullination of Arginine in MBP also has implications in MS pathogenesis, with the ratio of citrullinated MBP to MBP much higher in MS patients, thus affecting the overall structure and function of MBP [179]. Citrullination of Arginine has been implicated in several autoimmune diseases and modification of citrullination has been proposed as a potential therapeutic target [180, 181].
Beyond modifications of MBP, very few PTMs of other CNS targets have been investigated. In EAE, PTMs in proteins regulating signal transduction and axonal integrity have been observed, with citrullination of Arg27 on glial fibrillary acidic protein (GFAP) contributing to pathophysiology of astrocytes [182]. It has also been observed that phosphorylation of the protein collapsin response mediator protein-2 (CRMP-2) is abundant in degenerating spinal cords in EAE animals, and limiting its phosphorylation is protective [183]. In summary, investigation of PTMs in MS is a field that merits more research and the use of quantitation techniques and high-resolution proteomics has the potential to yield unique insights into the heterogeneity of PTMs in MS and their role in pathogenesis.
In concluding this section we would like to point out that while we have highlighted the advantages of genomic and proteomic techniques individually, novel bioinformatic approaches are in development to apply these methods in concert to extract biologically relevant information in a field known as proteogenomics [184, 185]. Proteogenomics is used to integrate genomic and proteomic data using data science methods, and we posit that this approach will be useful to discover novel biomarkers [184, 185]. Thus, we suggest the integration of multi-omic techniques in parallel to utilizing computation biology methods to unravel the complexities of large datasets to aid in biomarker discovery as highlighted next.
5. Improved Analysis Approaches For Biomarker Discovery
Altogether, genomic and proteomic investigations have yielded large quantitative datasets encompassing numerous molecules with the potential for elucidating MS susceptibility and pathogenesis. These datasets can also be explored to identify novel biomarkers for the disease when utilizing data science and system-biology approaches. Currently there are no established guidelines on how to approach omics data set analyses in the field of MS biomarker discovery. Therefore, herein we highlight some of the major data science analysis approaches which may allow the discovery of novel biomarker candidates from large omics data. These include single-molecule differential regulation and the utility of systems biology approaches.
A key step in biomarker discovery is narrowing down the number of putative targets from a large-omics dataset by identifying differentially regulated molecules [24]. Several different statistical approaches can be used in this process, which include both traditional statistics (e.g. ANOVA) and systems analysis tool (e.g. network analysis). These approaches are mostly complementary to each other and can be used in synergy for a more rigorous analysis of biomarker discovery. The role of different statistical methods in approaching biological questions has been reviewed elsewhere [186]. Methods include ANOVA with correction for false-discovery rate, and different types of regression models, as well as some less-common statistical models and novel methods which are exponentially growing [187–189].
5.1. Initial approach: Single molecule(s) with statistically significant expression changes
The most straight forward approach in identifying biomarker candidates from large datasets is by investigating expression fold-changes accompanied by high statistical significance. This approach together with effect size statistics has been extensively used for MS biomarker discovery [29, 82, 190–192]. Along these lines, our group has previously utilized this approach to identify potential MS biomarker candidates by investigating the CNS proteome during EAE [193, 194]. The concept underlying this approach is that molecules with statistically significant expression changes are likely to be disease-related (or treatment-related) and thus may yield biomarker candidates with high sensitivity. Furthermore, this approach allows identification of molecules which have not been previously associated with disease mechanisms from omics datasets. Building off this starting point, systems biology analyses allows for rigorous analysis to identify disease related mechanisms and processes which can further aid in prioritizing biomarker candidates, as well as in identifying additional ones, as summarized next.
5.2. Refinement: Utilizing systems biology and data science
Systems biology approaches can aid in biomarker discovery by revealing key molecules in an omics database which contribute to different disease-related processes [195]. Additionally, these approaches can identify key regulators (e.g. network hubs) independent of specific molecule fold-changes or expression change p-values, thus unveiling molecules which may drive several disease processes in which their own (the molecule’s) p-value may not be significantly altered in the specific dataset. Moreover, this approach may allow identification of additional biomarker candidates by revealing an important disease process, for example identifying a metabolic pathway that is altered during disease by exploring a genomic dataset. However, rather than using the genes in these pathways as biomarkers, one can explore metabolites and products of these pathways as potential biomarkers. This approach might be particularly useful in proteomic datasets which lack the ability to identify and quantify low-abundance proteins [196, 197]. Furthermore, identifying novel related pathways associated with clinical features of MS can aid in better understanding of pathological processes of the disease and in personalized biomarker discovery for disease prognosis or therapeutic responses. We will next highlight and discuss several systems analysis methods which are commonly used in biomarker discovery including gene set, pathway and network analysis approaches, though other approaches are also available [195, 198–201].
5.3. Gene set enrichment analysis (GSEA)
GSEA is an commonly used method which compiles all genes involved in a particular process independent of their interactions [202, 203]. Gene sets can be comprised of transcriptional-regulation, pathways, ontologies, diseases, cell types, etc. of curated lists which share a common biological entity [203]. These gene sets are curated from databases with broad scope and are based on multiple types of evidence. There are several databases available for GSEA including Molecular Signatures Database (MSigDB) which contains curated gene sets of many leading databases, such as Reactome and Kyoto Encyclopedia of Genes and Genomes (KEGG) [204–207]. Following GSEA each gene set receives an enrichment score and a p-value, which together determine the likelihood of that gene set being altered (or significantly enriched) in the dataset. Network analysis and GSEA are conceptually related and can sometimes be used interchangeably. Gene sets can be converted to networks, and networks can be converted to gene sets [208]. However, unlike networks, gene sets can be positively or negatively enriched in a dataset. GSEA has been used for an array of purposes such as the discovery of the MS susceptibility risk loci CD6, IRF8 and TNFRSF1A [209]. Other applications included the identification of profiles distinguishing myelin-reactive T cells in MS and healthy controls and identifying transcriptional profiles predictive of fingolimod therapy responsiveness [54, 210].
These analysis methodologies represent largely untapped resources for better understanding genetic and proteomic alterations in MS. Tools for such analysis are rapidly evolving and include the ability to integrate regulatory pathways into differential network analysis of gene expression data [211, 212]. Importantly, MS studies can utilize these tools to integrate both genomic and proteomic datasets to elucidate biologically relevant alterations that might reveal biomarkers that could be useful in the diagnosis and care of MS patients.
5.4. Pathway analysis
Pathway analysis applies lists of altered genes to biological pathways and attempts to integrate multiple gene/protein alterations in concert to yield lists of altered pathway activities [213]. Pathways can be analyzed both as networks and as gene sets. Other types of analyses are also available to determine pathway enrichment [214]. Along these lines, in MS, pathway analysis has been used on SNP array data to look broadly at the landscape of pathways in MS, which yielded for example information on the upregulation of the JAK-STAT and TCR signaling pathways [215]. Pathway analysis has also been used to investigate specific processes, such as the transcriptional changes in oligodendrocytes during remyelination in the cuprizone model, revealing that cholesterol-synthesis pathways are upregulated [216]. Others have identified the IFN-γ signaling pathway to be most significantly dysregulated in transcriptomic datasets of healthy controls versus RRMS, SPMS or PPMS [217].
5.5. Network analysis
Network analysis groups proteins that share similar functionality and indicate dependency between molecules, though not necessarily in a linear pathway [218]. For instance, a network can be a group of proteins which are up- or down-regulated in a cell or tissue upon exposure to treatment. A network can also be a group of molecules which share a less defined relationship, for example a “neurological disorder” network which encompasses many different genes and proteins which were shown to be differently regulated in such diseases. Some of the most common types of networks include protein-protein interactions, metabolic networks, and transcriptional regulatory networks [208, 213]. Thus, it is important to carefully select the networks that are applied to the dataset for biomarker discovery, for instance, using a protein interaction network in a genomic dataset may not yield robust and genuinely relevant candidates. There are different statistical approaches to determine if a network is affected in a dataset, which has been comprehensively reviewed elsewhere [208]. Generally, the higher the coverage of a network, the more likely the network is affected. Network analysis has been used to gauge the general genetic landscape of MS using SNP array data and has also revealed heterogeneity of MS-risk networks between cell types within the same individual [219, 220]. Network comparisons have also revealed shared gene expression networks between MS and ischemic stroke [221].
In summary, systems biology analysis and integration of large multi-layer omics datasets may facilitate discovery of conserved networks and pathways across studies.
5.6. Opportunities for Future Advancements in Methodologies
Integrating pathway and network analysis as well as other computational methods provides a number of advantages for biomarker discovery. For instance, integration of different analysis approaches, such as pathways, allows focusing on fewer hits in large datasets during the discovery and validation phases (e.g. a single pathway which is altered rather than 10s or 100s of molecules of that pathway) [222]. Moreover, pathway and network analysis results are non-redundant and can minimize variation when analyzing datasets from large patient cohorts, for instance a single molecule can vary largely across individuals; however a signaling pathway will remain constantly altered, and therefore will yield more robust biomarker candidates. This is also true, when using single cell data analysis, for example a specific subset of cells might be altered in many patients (e.g. inflammatory monocytes); however, on the gene level there might be significant variations across a population. Furthermore, pathway analysis can lead to faster biological hypothesis generation and point to novel biomarkers which may not have been obvious in the raw database due to technical challenges, such as ‘protein masking’ or low read depth. Furthermore, the integration of equitable and unbiased bioinformatic and computational biology approaches to explore proteomics and genomic datasets will allow researchers to better understand MS disease mechanisms, and consequently assist development of novel therapeutics and sensitive biomarkers. Along these lines, mutations (genetic changes which may impact protein function such as missense or frame-shift mutations; not polymorphisms) have been mostly ignored as biomarkers for MS; however improved computational algorithms and genomic techniques (such as whole-exome sequencing (WGS)) may yield information on mutations associated with MS [223]. This could potentially lead to identification of novel genetic links associated with MS pathogenesis and progression, as well as provide novel targets and biomarkers for drug responses. Along these lines, a recent study that performed WGS analysis across multi-incident MS families identified several rare mutations associated with MS susceptibility in these families [224]. Using pathway analysis, this group identified several immune-related pathways which were “enriched” in mutated genes including in Wnt signaling, complement, and inflammasomes [224]. However, these mutations need to be further corroborated in larger cohorts and in different tissues of MS patients. Additionally, the functional effects of these mutations need to be evaluated. It would be interesting to investigate de novo mutations within specific CNS resident cells (e.g. astrocytes) or immune cells (e.g. T cells) which may exist in MS patients. Moreover, using advanced data science tools and proteogenomic can further aid in identifying those genetic alterations which directly affect the proteomic landscape of patients [225].
In addition to using pathway and network analysis to improve biomarker discovery, new computational techniques allow to overcome many challenges in biomarker discovery using large datasets. For example, the utility of animal models in the discovery phase may not be translatable to human datasets, however development and implantation of new computational methods, including pathway analysis, fostered genome-wide annotation of functional DNA elements and therefore enabled extensive comparison between human and mouse genomes, allowing to extrapolate animal model studies into human biomarker discovery (e.g. some genes/proteins which exists in mice do not exist in human but the entire pathway is altered) [226]. Furthermore, the development and implementation of new computational methodologies, including machine learning techniques may allow for more rigorous selection of biomarkers and biomarker candidates by applying novel features (i.e. biomarker) selection tools which may outperform traditional statistics [188, 227, 228].
Together, these methods of prioritization may ultimately lead to the identification of both novel and authentic biomarkers. We postulate that implementation of these approaches holds enormous potential for MS biomarker discovery.
6. Conclusions
The development of omics techniques has offered the potential to better understand the factors that drive susceptibility to MS as well as provide insights into mechanisms that drive the pathogenesis of disease. Importantly, within these omics datasets there is the potential for discovering specific and sensitive biomarkers to aid diagnosis and treatment of patients. Researchers have generated vast MS-related datasets, but little of this large body of work has translated into use in the clinic as diagnostic tools and biomarkers. The ability to analyze these large datasets has lagged behind the ability to collect the data. As no single gene is causative of MS, it is likely that a systems biology approach that shifts from focusing on individual genes and proteins of interest to gene sets, pathways and networks will yield greater progress in facilitating clinically relevant findings. Additionally, improved data science techniques can allow for standardization in the analysis of disparate datasets generated across different platforms and preclinical models. We anticipate that the greater utilization of bioinformatics tools for pathway and network analysis will propel forward the discovery of clinically useful biomarkers in MS.
There is an urgent need to develop biomarkers for key aspects of MS diagnosis, prognosis, and treatment.
Large datasets hold the potential for discovering biomarkers to aid diagnosis and treatment.
The ability to analyze large datasets has lagged behind the ability to generate them.
Data science tools will propel forward the discovery of clinically useful biomarkers.
Acknowledgments:
We thank Dr. Yufeng Wang (UT San Antonio) and Dr. William E. Haskins (Gryphon Bio) for critically reviewing this manuscript and helpful suggestions. Figure was created in BioRender.com. This work was supported by grants NS084201 and AI144731 from the National Institutes of Health and grants RG5501 and RG1602 from the National Multiple Sclerosis Society (T.G.F.). C.C.H. was supported by RISE R25GM060655.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wallin MT, et al. , The prevalence of MS in the United States. A population-based estimate using health claims data, 2019. 92(10): p. e1029–e1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallin MT, et al. , Global, regional, and national burden of multiple sclerosis 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet Neurology, 2019. 18(3): p. 269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Absinta M, Sati P, and Reich DS, Advanced MRI and staging of multiple sclerosis lesions. Nat Rev Neurol, 2016. 12(6): p. 358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruck W, The pathology of multiple sclerosis is the result of focal inflammatory demyelination with axonal damage. J Neurol, 2005. 252 Suppl 5: p. v3–9. [DOI] [PubMed] [Google Scholar]
- 5.Raphael I, et al. , T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine, 2015. 74(1): p. 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salou M, et al. , Expanded CD8 T-cell sharing between periphery and CNS in multiple sclerosis. Ann Clin Transl Neurol, 2015. 2(6): p. 609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakshi R, et al. , Fatigue in multiple sclerosis and its relationship to depression and neurologic disability. Multiple Sclerosis Journal, 2000. 6(3): p. 181–185. [DOI] [PubMed] [Google Scholar]
- 8.Haines J, Linkage of the MHC to familial multiple sclerosis suggests genetic heterogeneity. The Multiple Sclerosis Genetics Group. Human Molecular Genetics, 1998. 7(8): p. 1229–1234. [DOI] [PubMed] [Google Scholar]
- 9.Barcellos LF, et al. , HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. American journal of human genetics, 2003. 72(3): p. 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinshenker BG, et al. , THE NATURAL HISTORY OF MULTIPLE SCLEROSIS: A GEOGRAPHICALLY BASED STUDY: I. CLINICAL COURSE AND DISABILITY. Brain, 1989. 112(1): p. 133–146. [DOI] [PubMed] [Google Scholar]
- 11.Steelman AJ, Infection as an Environmental Trigger of Multiple Sclerosis Disease Exacerbation.Frontiers in immunology, 2015. 6: p. 520–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lublin FD, et al. , Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology, 2014. 83(3): p. 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald WI, et al. , Recommended diagnostic criteria for multiple sclerosis: Guidelines from the international panel on the diagnosis of multiple sclerosis. Annals of Neurology, 2001. 50(1): p. 121–127. [DOI] [PubMed] [Google Scholar]
- 14.Polman CH, et al. , Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Annals of neurology, 2011. 69(2): p. 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polman CH, et al. , Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Annals of Neurology, 2005. 58(6): p. 840–846. [DOI] [PubMed] [Google Scholar]
- 16.Thompson AJ, et al. , Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. The Lancet Neurology, 2018. 17(2): p. 162–173. [DOI] [PubMed] [Google Scholar]
- 17.Miller DH, et al. , Magnetic resonance imaging in monitoring the treatment of multiple sclerosis: concerted action guidelines. Journal of Neurology, Neurosurgery & Psychiatry, 1991. 54(8): p. 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtzke JF, On the origin of EDSS. Multiple Sclerosis and Related Disorders, 2015. 4(2): p. 95–103. [DOI] [PubMed] [Google Scholar]
- 19.Kurtzke JF, Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology, 1983. 33(11): p. 1444–52. [DOI] [PubMed] [Google Scholar]
- 20.Gholamzad M, et al. , A comprehensive review on the treatment approaches of multiple sclerosis: currently and in the future. Inflammation Research, 2019. 68(1): p. 25–38. [DOI] [PubMed] [Google Scholar]
- 21.Harris VK and Sadiq SA, Biomarkers of therapeutic response in multiple sclerosis: current status. Mol Diagn Ther, 2014. 18(6): p. 605–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Angelis F, John NA, and Brownlee WJ, Disease-modifying therapies for multiple sclerosis. BMJ, 2018. 363: p. k4674. [DOI] [PubMed] [Google Scholar]
- 23.Dobson R. and Giovannoni G, Multiple sclerosis - a review. Eur J Neurol, 2019. 26(1): p. 27–40. [DOI] [PubMed] [Google Scholar]
- 24.Rifai N, Gillette MA, and Carr SA, Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechnology, 2006. 24(8): p. 971–983. [DOI] [PubMed] [Google Scholar]
- 25.Goossens N, et al. , Cancer biomarker discovery and validation. Translational cancer research, 2015. 4(3): p. 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robeson RH, Siegel AM, and Dunckley T, Genomic and Proteomic Biomarker Discovery in Neurological Disease. Biomark Insights, 2008. 3: p. 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stern AD, Alexander BM, and Chandra A, Innovation Incentives and Biomarkers. Clin Pharmacol Ther, 2018. 103(1): p. 34–36. [DOI] [PubMed] [Google Scholar]
- 28.Comabella M. and Montalban X, Body fluid biomarkers in multiple sclerosis. The Lancet Neurology, 2014. 13(1): p. 113–126. [DOI] [PubMed] [Google Scholar]
- 29.Raphael I, et al. , Body fluid biomarkers in multiple sclerosis: how far we have come and how they could affect the clinic now and in the future. Expert Review of Clinical Immunology, 2015. 11(1): p. 69–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han MH, et al. , Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature, 2008. 451(7182): p. 1076–1081. [DOI] [PubMed] [Google Scholar]
- 31.Broadwater L, et al. , Analysis of the mitochondrial proteome in multiple sclerosis cortex.Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 2011. 1812(5): p. 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elkjaer ML, et al. , Molecular signature of different lesion types in the brain white matter of patients with progressive multiple sclerosis. Acta neuropathologica communications, 2019. 7(1): p. 205–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lassmann H, Brück W, and Lucchinetti CF, The Immunopathology of Multiple Sclerosis: An Overview. Brain Pathology, 2007. 17(2): p. 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cramer SP, et al. , Abnormal blood–brain barrier permeability in normal appearing white matter in multiple sclerosis investigated by MRI. NeuroImage: Clinical, 2014. 4: p. 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maccarrone G, et al. , MALDI imaging mass spectrometry analysis—A new approach for protein mapping in multiple sclerosis brain lesions. Journal of Chromatography B, 2017. 1047: p. 131–140. [DOI] [PubMed] [Google Scholar]
- 36.Deisenhammer F, et al. , The Cerebrospinal Fluid in Multiple Sclerosis. Frontiers in immunology, 2019. 10: p. 726–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul A, Comabella M, and Gandhi R, Biomarkers in Multiple Sclerosis. Cold Spring Harb Perspect Med, 2019. 9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziemssen T, Akgün K, and Brück W, Molecular biomarkers in multiple sclerosis. Journal of Neuroinflammation, 2019. 16(1): p. 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Correale J. and Molinas M. de los Milagros Bassani, Oligoclonal bands and antibody responses in Multiple Sclerosis. Journal of Neurology, 2002. 249(4): p. 375–389. [DOI] [PubMed] [Google Scholar]
- 40.Ziemssen T. and Ziemssen F, The role of the humoral immune system in multiple sclerosis (MS) and its animal model experimental autoimmune encephalomyelitis (EAE). Autoimmunity Reviews, 2005. 4(7): p. 460–467. [DOI] [PubMed] [Google Scholar]
- 41.Amato MP and Ponziani G, A prospective study on the prognosis of multiple sclerosis. Neurological Sciences, 2000. 21(2): p. S831–S838. [DOI] [PubMed] [Google Scholar]
- 42.Khalil M, et al. , Neurofilaments as biomarkers in neurological disorders. Nature Reviews Neurology, 2018. 14(10): p. 577–589. [DOI] [PubMed] [Google Scholar]
- 43.Lossius A, et al. , High-throughput sequencing of TCR repertoires in multiple sclerosis reveals intrathecal enrichment of EBV-reactive CD8+ T cells. Eur J Immunol, 2014. 44(11): p. 3439–52. [DOI] [PubMed] [Google Scholar]
- 44.Rounds WH, et al. , The antibody genetics of multiple sclerosis: comparing next-generation sequencing to sanger sequencing. Frontiers in neurology, 2014. 5: p. 166–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geyer PE, et al. , Revisiting biomarker discovery by plasma proteomics. Molecular systems biology, 2017. 13(9): p. 942–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thadikkaran L, et al. , Recent advances in blood-related proteomics. PROTEOMICS, 2005. 5(12): p. 3019–3034. [DOI] [PubMed] [Google Scholar]
- 47.Avasarala JR, Wall MR, and Wolfe GM, A distinctive molecular signature of multiple sclerosis derived from MALDI-TOF/MS and serum proteomic pattern analysis. Journal of Molecular Neuroscience, 2005. 25(1): p. 119–125. [DOI] [PubMed] [Google Scholar]
- 48.Tremlett H, et al. , Serum proteomics in multiple sclerosis disease progression. Journal of Proteomics, 2015. 118: p. 2–11. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez JI, Cayrol R, and Prat A, Disruption of central nervous system barriers in multiple sclerosis. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 2011. 1812(2): p. 252–264. [DOI] [PubMed] [Google Scholar]
- 50.Rasmussen MK, Mestre H, and Nedergaard M, The glymphatic pathway in neurological disorders. Lancet Neurol, 2018. 17(11): p. 1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuhle J, et al. , Blood neurofilament light chain as a biomarker of MS disease activity and treatment response. Neurology, 2019. 92(10): p. e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varhaug KN, et al. , Neurofilament Light Chain as a Biomarker in Multiple Sclerosis. Frontiers in Neurology, 2019. 10(338). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nickles D, et al. , Blood RNA profiling in a large cohort of multiple sclerosis patients and healthy controls. Human molecular genetics, 2013. 22(20): p. 4194–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao Y, et al. , Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Science Translational Medicine, 2015. 7(287): p. 287ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schafflick D, et al. , Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nature Communications, 2020. 11(1): p. 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pusic AD, Pusic KM, and Kraig RP, What are exosomes and how can they be used in multiple sclerosis therapy? Expert Rev Neurother, 2014. 14(4): p. 353–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaros JA, et al. , Affinity depletion of plasma and serum for mass spectrometry-based proteome analysis. Methods Mol Biol, 2013. 1002: p. 1–11. [DOI] [PubMed] [Google Scholar]
- 58.Tu C, et al. , Depletion of abundant plasma proteins and limitations of plasma proteomics. Journal of proteome research, 2010. 9(10): p. 4982–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dey KK, et al. , Deep undepleted human serum proteome profiling toward biomarker discovery for Alzheimer’s disease. Clinical proteomics, 2019. 16: p. 16–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gebregiworgis T, et al. , A Urinary Metabolic Signature for Multiple Sclerosis and Neuromyelitis Optica. Journal of Proteome Research, 2016. 15(2): p. 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devos D, et al. , Silver stained isoelectrophoresis of tears and cerebrospinal fluid in multiple sclerosis. J Neurol, 2001. 248(8): p. 672–5. [DOI] [PubMed] [Google Scholar]
- 62.Calais G, et al. , Tear analysis in clinically isolated syndrome as new multiple sclerosis criterion. Mult Scler, 2010. 16(1): p. 87–92. [DOI] [PubMed] [Google Scholar]
- 63.Pieragostino D, et al. , Proteomics characterization of extracellular vesicles sorted by flow cytometry reveals a disease-specific molecular cross-talk from cerebrospinal fluid and tears in multiple sclerosis. J Proteomics, 2019. 204: p. 103403. [DOI] [PubMed] [Google Scholar]
- 64.Whitaker JN, et al. , Urinary myelin basic protein-like material as a correlate of the progression of multiple sclerosis. Ann Neurol, 1995. 38(4): p. 625–32. [DOI] [PubMed] [Google Scholar]
- 65.Manconi B, et al. , Top-down proteomic profiling of human saliva in multiple sclerosis patients. J Proteomics, 2018. 187: p. 212–222. [DOI] [PubMed] [Google Scholar]
- 66.Dubuisson N, et al. , Inclusion criteria used in trials of people with progressive multiple sclerosis. Multiple Sclerosis Journal, 2018. 26(3): p. 279–283. [DOI] [PubMed] [Google Scholar]
- 67.Sawcer S, The complex genetics of multiple sclerosis: pitfalls and prospects. Brain, 2008. 131(12): p. 3118–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amato MP, Zipoli V, and Portaccio E, Multiple sclerosis-related cognitive changes: a review of cross-sectional and longitudinal studies. J Neurol Sci, 2006. 245(1–2): p. 41–6. [DOI] [PubMed] [Google Scholar]
- 69.Hyun JW, et al. , Longitudinal analysis of serum neurofilament light chain: A potential therapeutic monitoring biomarker for multiple sclerosis. Mult Scler, 2019: p. 1352458519840757. [DOI] [PubMed] [Google Scholar]
- 70.Langer-Gould A, et al. , The incidence of clinically isolated syndrome in a multi-ethnic cohort. J Neurol, 2014. 261(7): p. 1349–55. [DOI] [PubMed] [Google Scholar]
- 71.Langer-Gould A, et al. , Incidence of multiple sclerosis in multiple racial and ethnic groups. Neurology, 2013. 80(19): p. 1734–9. [DOI] [PubMed] [Google Scholar]
- 72.Amezcua L, Oksenberg JR, and McCauley JL, MS in self-identified Hispanic/Latino individuals living in the US. Multiple sclerosis journal - experimental, translational and clinical, 2017. 3(3): p. 2055217317725103–2055217317725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sadovnick AD, et al. , Evidence for genetic basis of multiple sclerosis. The Lancet, 1996. 347(9017): p. 1728–1730. [DOI] [PubMed] [Google Scholar]
- 74.Sadovnick AD, Baird PA, and Ward RH, Multiple sclerosis: updated risks for relatives. Am J Med Genet, 1988. 29(3): p. 533–41. [DOI] [PubMed] [Google Scholar]
- 75.Xia Z, et al. , Genes and Environment in Multiple Sclerosis project: A platform to investigate multiple sclerosis risk. Annals of Neurology, 2016. 79(2): p. 178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mahesula S, et al. , Immunoenrichment microwave and magnetic proteomics for quantifying CD47 in the experimental autoimmune encephalomyelitis model of multiple sclerosis. Electrophoresis, 2012. 33(24): p. 3820–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu T, et al. , Identification of differentially expressed proteins in experimental autoimmune encephalomyelitis (EAE) by proteomic analysis of the spinal cord. Journal of proteome research, 2007. 6(7): p. 2565–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hueber W. and Robinson WH, Genomics and proteomics: Applications in autoimmune diseases. Pharmacogenomics and personalized medicine, 2009. 2: p. 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fazeli AS, et al. , Proteome analysis of brain in murine experimental autoimmune encephalomyelitis. PROTEOMICS, 2010. 10(15): p. 2822–2832. [DOI] [PubMed] [Google Scholar]
- 80.Rosenling T, et al. , The experimental autoimmune encephalomyelitis model for proteomic biomarker studies: from rat to human. Clin Chim Acta, 2011. 412(11–12): p. 812–22. [DOI] [PubMed] [Google Scholar]
- 81.Hasan M, et al. , Quantitative Proteome Analysis of Brain Subregions and Spinal Cord from Experimental Autoimmune Encephalomyelitis Mice by TMT-Based Mass Spectrometry. PROTEOMICS, 2019. 19(5): p. 1800355. [DOI] [PubMed] [Google Scholar]
- 82.Raphael I, et al. , Serum Neuroinflammatory Disease-Induced Central Nervous System Proteins Predict Clinical Onset of Experimental Autoimmune Encephalomyelitis. Frontiers in immunology, 2017. 8: p. 812–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson AP, et al. , The experimental autoimmune encephalomyelitis (EAE) model of MS: utility for understanding disease pathophysiology and treatment. Handbook of clinical neurology, 2014. 122: p. 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Terry RL, Ifergan I, and Miller SD, Experimental Autoimmune Encephalomyelitis in Mice. Methods in molecular biology (Clifton, N.J.), 2016. 1304: p. 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Constantinescu CS, et al. , Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). British journal of pharmacology, 2011. 164(4): p. 1079–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rivers TM, Sprunt DH, and Berry GP, OBSERVATIONS ON ATTEMPTS TO PRODUCE ACUTE DISSEMINATED ENCEPHALOMYELITIS IN MONKEYS. J Exp Med, 1933. 58(1): p. 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brok HP, et al. , Non-human primate models of multiple sclerosis. Immunol Rev, 2001. 183: p. 173–85. [DOI] [PubMed] [Google Scholar]
- 88.Raine CS, et al. , Demyelination in primate autoimmune encephalomyelitis and acute multiple sclerosis lesions: A case for antigen-specific antibody mediation. Annals of Neurology, 1999. 46(2): p. 144–160. [DOI] [PubMed] [Google Scholar]
- 89.t Hart BA, et al. , Non-invasive measurement of brain damage in a primate model of multiple sclerosis. Trends in Molecular Medicine, 2004. 10(2): p. 85–91. [DOI] [PubMed] [Google Scholar]
- 90.Tuttle AH, et al. , Comparing phenotypic variation between inbred and outbred mice. Nature Methods, 2018. 15(12): p. 994–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chia R, et al. , The origins and uses of mouse outbred stocks. Nature Genetics, 2005. 37(11): p. 1181–1186. [DOI] [PubMed] [Google Scholar]
- 92.Yalcin B, et al. , Commercially available outbred mice for genome-wide association studies. PLoS genetics, 2010. 6(9): p. e1001085–e1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gregersen JW, et al. , Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature, 2006. 443(7111): p. 574–577. [DOI] [PubMed] [Google Scholar]
- 94.Madsen LS, et al. , A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat Genet, 1999. 23(3): p. 343–7. [DOI] [PubMed] [Google Scholar]
- 95.Ji N, et al. , Small Molecule Inhibitor of Antigen Binding and Presentation by HLA-DR2b as a Therapeutic Strategy for the Treatment of Multiple Sclerosis. The Journal of Immunology, 2013. 191(10): p. 5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ignatius Arokia Doss PM, et al. , The Non-Obese Diabetic Mouse Strain as a Model to Study CD8+ T Cell Function in Relapsing and Progressive Multiple Sclerosis. Frontiers in Immunology, 2015. 6(541). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levy H, Assaf Y, and Frenkel D, Characterization of brain lesions in a mouse model of progressive multiple sclerosis. Experimental Neurology, 2010. 226(1): p. 148–158. [DOI] [PubMed] [Google Scholar]
- 98.McCarthy DP, Richards MH, and Miller SD, Mouse models of multiple sclerosis: experimental autoimmune encephalomyelitis and Theiler’s virus-induced demyelinating disease. Methods in molecular biology (Clifton, N.J.), 2012. 900: p. 381–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sachs JA, HLA antigens in multiple sclerosis. Proceedings of the Royal Society of Medicine, 1977. 70(12): p. 869–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singal DP, et al. , SEROTYPING FOR HOMOTRANSPLANTATION XVII. PRELIMINARY STUDIES OF HL-A SUBUNITS AND ALLELES. Transplantation, 1968. 6(8): p. 904–912. [DOI] [PubMed] [Google Scholar]
- 101.Creary LE, et al. , Deconstruction of HLA-DRB1*04:01:01 and HLA-DRB1*15:01:01 class II haplotypes using next-generation sequencing in European-Americans with multiple sclerosis. Multiple sclerosis (Houndmills, Basingstoke, England), 2019. 25(6): p. 772–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cotsapas C. and Mitrovic M, Genome-wide association studies of multiple sclerosis. Clinical & translational immunology, 2018. 7(6): p. e1018–e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science, 2019. 365(6460): p. eaav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bohlander SK, ABCs of genomics. Hematology, 2013. 2013(1): p. 316–323. [DOI] [PubMed] [Google Scholar]
- 105.Hollenbach JA and Oksenberg JR, The immunogenetics of multiple sclerosis: A comprehensive review. Journal of autoimmunity, 2015. 64: p. 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Habek M, Borovecki F, and Brinar VV, Genomics in multiple sclerosis. Clin Neurol Neurosurg, 2010. 112(7): p. 621–4. [DOI] [PubMed] [Google Scholar]
- 107.Axisa PP and Hafler DA, Multiple sclerosis: genetics, biomarkers, treatments. Curr Opin Neurol, 2016. 29(3): p. 345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lähnemann D, et al. , Eleven grand challenges in single-cell data science. Genome Biology, 2020. 21(1): p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Svensson V, Vento-Tormo R, and Teichmann SA, Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc, 2018. 13(4): p. 599–604. [DOI] [PubMed] [Google Scholar]
- 110.Stegle O, Teichmann SA, and Marioni JC, Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet, 2015. 16(3): p. 133–45. [DOI] [PubMed] [Google Scholar]
- 111.Yang IS and Kim S, Analysis of Whole Transcriptome Sequencing Data: Workflow and Software. Genomics Inform, 2015. 13(4): p. 119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deutsch EW, et al. , A guided tour of the Trans-Proteomic Pipeline. Proteomics, 2010. 10(6): p. 1150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, et al. , SequencEnG: an interactive knowledge base of sequencing techniques. Bioinformatics, 2019. 35(8): p. 1438–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gonzalez-Galarza FF, et al. , A critical appraisal of techniques, software packages, and standards for quantitative proteomic analysis. OMICS, 2012. 16(9): p. 431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Levy SE and Myers RM, Advancements in Next-Generation Sequencing. Annu Rev Genomics Hum Genet, 2016. 17: p. 95–115. [DOI] [PubMed] [Google Scholar]
- 116.International Human Genome Sequencing, C., Finishing the euchromatic sequence of the human genome. Nature, 2004. 431(7011): p. 931–945. [DOI] [PubMed] [Google Scholar]
- 117.Metzker ML, Sequencing technologies - the next generation. Nat Rev Genet, 2010. 11(1): p. 31–46. [DOI] [PubMed] [Google Scholar]
- 118.Metzker ML, Sequencing technologies — the next generation. Nature Reviews Genetics, 2010. 11(1): p. 31–46. [DOI] [PubMed] [Google Scholar]
- 119.Lahens NF, et al. , A comparison of Illumina and Ion Torrent sequencing platforms in the context of differential gene expression. BMC Genomics, 2017. 18(1): p. 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Erguner B, Ustek D, and Sagiroglu MS. Performance comparison of Next Generation sequencing platforms. IEEE. [DOI] [PubMed] [Google Scholar]
- 121.Pereira R, Oliveira J, and Sousa M, Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics. J Clin Med, 2020. 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pytel V, et al. , Exonic variants of genes related to the vitamin D signaling pathway in the families of familial multiple sclerosis using whole-exome next generation sequencing. Brain and Behavior, 2019. 9(4): p. e01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Papalexi E. and Satija R, Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol, 2018. 18(1): p. 35–45. [DOI] [PubMed] [Google Scholar]
- 124.Jäkel S, et al. , Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature, 2019. 566(7745): p. 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schirmer L, et al. , Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature, 2019. 573(7772): p. 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bertrams HJ and Kuwert EK, Association of Histocompatibility Haplotype HLA-A3-B7 with Multiple Sclerosis. The Journal of Immunology, 1976. 117(5 Part 2): p. 1906. [PubMed] [Google Scholar]
- 127.Olerup O. and Hillert J, HLA class II-associated genetic susceptibility in multiple sclerosis: A critical evaluation. Tissue Antigens, 1991. 38(2): p. 1–15. [DOI] [PubMed] [Google Scholar]
- 128.Sawcer S, et al. , Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature, 2011. 476(7359): p. 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Risk Alleles for Multiple Sclerosis Identified by a Genomewide Study. New England Journal of Medicine, 2007. 357(9): p. 851–862. [DOI] [PubMed] [Google Scholar]
- 130.Luo L, Boerwinkle E, and Xiong M, Association studies for next-generation sequencing. Genome Res, 2011. 21(7): p. 1099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ogawa K, et al. , Next-generation sequencing identifies contribution of both class I and II HLA genes on susceptibility of multiple sclerosis in Japanese. J Neuroinflammation, 2019. 16(1): p. 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Boegel S, et al. , HLA typing from RNA-Seq sequence reads. Genome Medicine, 2012. 4(12): p. 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li B, et al. , Landscape of tumor-infiltrating T cell repertoire of human cancers. Nature Genetics, 2016. 48(7): p. 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hu X, et al. , Landscape of B cell immunity and related immune evasion in human cancers. Nature Genetics, 2019. 51(3): p. 560–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tan L, et al. , Recent advances of exosomes in immune modulation and autoimmune diseases. Autoimmunity, 2016. 49(6): p. 357–365. [DOI] [PubMed] [Google Scholar]
- 136.Li X, et al. , Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL bioengineering, 2019. 3(1): p. 011503–011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Doyle LM and Wang MZ, Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells, 2019. 8(7): p. 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Larssen P, et al. , Tracing Cellular Origin of Human Exosomes Using Multiplex Proximity Extension Assays. Molecular & cellular proteomics : MCP, 2017. 16(3): p. 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang J, et al. , A next generation sequencing based approach to identify extracellular vesicle mediated mRNA transfers between cells. BMC genomics, 2017. 18(1): p. 987–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Selmaj I, et al. , Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Annals of Neurology, 2017. 81(5): p. 703–717. [DOI] [PubMed] [Google Scholar]
- 141.Leung AKL, The Whereabouts of microRNA Actions: Cytoplasm and Beyond. Trends in cell biology, 2015. 25(10): p. 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu TX and Rothenberg ME, MicroRNA. J Allergy Clin Immunol, 2018. 141(4): p. 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Galazka G, et al. , Multiple sclerosis: Serum-derived exosomes express myelin proteins. Mult Scler, 2018. 24(4): p. 449–458. [DOI] [PubMed] [Google Scholar]
- 144.Gandhi R, et al. , Circulating MicroRNAs as biomarkers for disease staging in multiple sclerosis. Annals of Neurology, 2013. 73(6): p. 729–740. [DOI] [PubMed] [Google Scholar]
- 145.Ebrahimkhani S, et al. , Exosomal microRNA signatures in multiple sclerosis reflect disease status. Scientific Reports, 2017. 7(1): p. 14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liguori M, et al. , Association between miRNAs expression and cognitive performances of Pediatric Multiple Sclerosis patients: A pilot study. Brain and behavior, 2019. 9(2): p. e01199–e01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Murugaiyan G, et al. , Silencing MicroRNA-155 Ameliorates Experimental Autoimmune Encephalomyelitis. The Journal of Immunology, 2011. 187(5): p. 2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chuang JC and Jones PA, Epigenetics and MicroRNAs. Pediatric Research, 2007. 61(7): p. 24–29. [DOI] [PubMed] [Google Scholar]
- 149.Urdinguio RG, Sanchez-Mut JV, and Esteller M, Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. The Lancet Neurology, 2009. 8(11): p. 1056–1072. [DOI] [PubMed] [Google Scholar]
- 150.Kular L, et al. , DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nature Communications, 2018. 9(1): p. 2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhu H, et al. , A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J Pediatr, 2013. 162(5): p. 1004–9.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Buschle A. and Hammerschmidt W, Epigenetic lifestyle of Epstein-Barr virus. Seminars in Immunopathology, 2020. 42(2): p. 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Celarain N. and Tomas-Roig J, Aberrant DNA methylation profile exacerbates inflammation and neurodegeneration in multiple sclerosis patients. Journal of Neuroinflammation, 2020. 17(1): p. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pan G, et al. , Role of genetic susceptibility variants in predicting clinical course in multiple sclerosis: a cohort study. Journal of Neurology, Neurosurgery & Psychiatry, 2016. 87(11): p. 1204–1211. [DOI] [PubMed] [Google Scholar]
- 155.Aslam B, et al. , Proteomics: Technologies and Their Applications. Journal of Chromatographic Science, 2017. 55(2): p. 182–196. [DOI] [PubMed] [Google Scholar]
- 156.Hasin Y, Seldin M, and Lusis A, Multi-omics approaches to disease. Genome Biol, 2017. 18(1): p. 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Walsh MJ and Tourtellotte WW, Temporal invariance and clonal uniformity of brain and cerebrospinal IgG, IgA, and IgM in multiple sclerosis. J Exp Med, 1986. 163(1): p. 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tourtellotte WW and Shapshak P, Application of 2 dimensional electrophoresis (2DE) in the search for the multiple sclerosis (MS) antigen. Journal of Neuroimmunology, 1988. 17(3): p. 263. [Google Scholar]
- 159.Chiasserini D, et al. , CSF proteome analysis in multiple sclerosis patients by two-dimensional electrophoresis. European Journal of Neurology, 2008. 15(9): p. 998–1001. [DOI] [PubMed] [Google Scholar]
- 160.Hammack BN, et al. , Proteomic analysis of multiple sclerosis cerebrospinal fluid. Mult Scler, 2004. 10(3): p. 245–60. [DOI] [PubMed] [Google Scholar]
- 161.Almeras L, et al. , New antigenic candidates in multiple sclerosis: Identification by serological proteome analysis. PROTEOMICS, 2004. 4(7): p. 2184–2194. [DOI] [PubMed] [Google Scholar]
- 162.Zhang Y, et al. , Protein analysis by shotgun/bottom-up proteomics. Chemical reviews, 2013. 113(4): p. 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jain MR, et al. , Altered proteolytic events in experimental autoimmune encephalomyelitis discovered by iTRAQ shotgun proteomics analysis of spinal cord. Proteome Science, 2009. 7(1): p. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schubert OT, et al. , Quantitative proteomics: challenges and opportunities in basic and applied research. Nature Protocols, 2017. 12(7): p. 1289–1294. [DOI] [PubMed] [Google Scholar]
- 165.Kroksveen AC, et al. , Proteomics of human cerebrospinal fluid: discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J Proteomics, 2011. 74(4): p. 371–88. [DOI] [PubMed] [Google Scholar]
- 166.Singh V, et al. , Proteomics technologies for biomarker discovery in multiple sclerosis. Journal of Neuroimmunology, 2012. 248(1): p. 40–47. [DOI] [PubMed] [Google Scholar]
- 167.Opsahl JA, et al. , Label-free analysis of human cerebrospinal fluid addressing various normalization strategies and revealing protein groups affected by multiple sclerosis. PROTEOMICS, 2016. 16(7): p. 1154–1165. [DOI] [PubMed] [Google Scholar]
- 168.Spitzer MH and Nolan GP, Mass Cytometry: Single Cells, Many Features. Cell, 2016. 165(4): p. 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Böttcher C, et al. , Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nature Neuroscience, 2019. 22(1): p. 78–90. [DOI] [PubMed] [Google Scholar]
- 170.Ajami B, et al. , Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nature Neuroscience, 2018. 21(4): p. 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Galli E, et al. , GM-CSF and CXCR4 define a T helper cell signature in multiple sclerosis. Nature Medicine, 2019. 25(8): p. 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marx V, A dream of single-cell proteomics. Nature Methods, 2019. 16(9): p. 809–812. [DOI] [PubMed] [Google Scholar]
- 173.Olsen JV and Mann M, Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics, 2013. 12(12): p. 3444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Witze ES, et al. , Mapping protein post-translational modifications with mass spectrometry. Nature Methods, 2007. 4(10): p. 798–806. [DOI] [PubMed] [Google Scholar]
- 175.van Stipdonk MJ, et al. , T cells discriminate between differentially phosphorylated forms of alphaB-crystallin, a major central nervous system myelin antigen. International Immunology, 1998. 10(7): p. 943–950. [DOI] [PubMed] [Google Scholar]
- 176.Anderton SM, Post-translational modifications of self antigens: implications for autoimmunity. Curr Opin Immunol, 2004. 16(6): p. 753–8. [DOI] [PubMed] [Google Scholar]
- 177.Dharmasaroja P, Specificity of autoantibodies to epitopes of myelin proteins in multiple sclerosis. Journal of the Neurological Sciences, 2003. 206(1): p. 7–16. [DOI] [PubMed] [Google Scholar]
- 178.Kim JK, et al. , Multiple Sclerosis. Molecular & Cellular Proteomics, 2003. 2(7): p. 453. [DOI] [PubMed] [Google Scholar]
- 179.Moscarello MA, et al. , Myelin in multiple sclerosis is developmentally immature. The Journal of Clinical Investigation, 1994. 94(1): p. 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.György B, et al. , Citrullination: A posttranslational modification in health and disease. The International Journal of Biochemistry & Cell Biology, 2006. 38(10): p. 1662–1677. [DOI] [PubMed] [Google Scholar]
- 181.Yang L, Tan D, and Piao H, Myelin Basic Protein Citrullination in Multiple Sclerosis: A Potential Therapeutic Target for the Pathology. Neurochemical Research, 2016. 41(8): p. 1845–1856. [DOI] [PubMed] [Google Scholar]
- 182.Grant JE, et al. , Post-translational modifications in the rat lumbar spinal cord in experimental autoimmune encephalomyelitis. J Proteome Res, 2007. 6(7): p. 2786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Petratos S, et al. , Limiting multiple sclerosis related axonopathy by blocking Nogo receptor and CRMP-2 phosphorylation. Brain, 2012. 135(6): p. 1794–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vogel C. and Marcotte EM, Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews Genetics, 2012. 13(4): p. 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kumar D, et al. , Integrating transcriptome and proteome profiling: Strategies and applications. PROTEOMICS, 2016. 16(19): p. 2533–2544. [DOI] [PubMed] [Google Scholar]
- 186.Vaux DL, Basic Statistics in Cell Biology. Annual Review of Cell and Developmental Biology, 2014. 30(1): p. 23–37. [DOI] [PubMed] [Google Scholar]
- 187.Leclercq M, et al. , Large-Scale Automatic Feature Selection for Biomarker Discovery in High-Dimensional OMICs Data. Front Genet, 2019. 10: p. 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Salekin S, et al. , Early response index: a statistic to discover potential early stage disease biomarkers. BMC Bioinformatics, 2017. 18(1): p. 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang K, Geng W, and Zhang S, Network-based logistic regression integration method for biomarker identification. BMC Systems Biology, 2018. 12(9): p. 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ridolfi E, et al. , Expression and Genetic Analysis of MicroRNAs Involved in Multiple Sclerosis. Int J Mol Sci, 2013. 14(3): p. 4375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Romme Christensen J, et al. , Cellular sources of dysregulated cytokines in relapsing-remitting multiple sclerosis. J Neuroinflammation, 2012. 9: p. 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hagman S, et al. , Disease-associated inflammatory biomarker profiles in blood in different subtypes of multiple sclerosis: prospective clinical and MRI follow-up study. J Neuroimmunol, 2011. 234(1–2): p. 141–7. [DOI] [PubMed] [Google Scholar]
- 193.Raphael I, et al. , Microwave & Magnetic (M2) Proteomics Reveals CNS-Specific Protein Expression Waves that Precede Clinical Symptoms of Experimental Autoimmune Encephalomyelitis. Scientific Reports, 2014. 4(1): p. 6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Raphael I, et al. , Microwave and magnetic (M2) proteomics of the experimental autoimmune encephalomyelitis animal model of multiple sclerosis. ELECTROPHORESIS, 2012. 33(24): p. 3810–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Villoslada P. and Baranzini S, Data integration and systems biology approaches for biomarker discovery: challenges and opportunities for multiple sclerosis. J Neuroimmunol, 2012. 248(1–2): p. 58–65. [DOI] [PubMed] [Google Scholar]
- 196.Chandramouli K. and Qian PY, Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics, 2009. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Biron DG, et al. , The pitfalls of proteomics experiments without the correct use of bioinformatics tools. Proteomics, 2006. 6(20): p. 5577–96. [DOI] [PubMed] [Google Scholar]
- 198.Creixell P, et al. , Pathway and network analysis of cancer genomes. Nature methods, 2015. 12(7): p. 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lempiäinen H, et al. , Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Scientific Reports, 2018. 8(1): p. 3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cirincione AG, Clark KL, and Kann MG, Pathway networks generated from human disease phenome. BMC Medical Genomics, 2018. 11(3): p. 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Caberlotto L. and Lauria M, Systems biology meets -omic technologies: novel approaches to biomarker discovery and companion diagnostic development. Expert Rev Mol Diagn, 2015. 15(2): p. 255–65. [DOI] [PubMed] [Google Scholar]
- 202.Mootha VK, et al. , PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics, 2003. 34(3): p. 267–273. [DOI] [PubMed] [Google Scholar]
- 203.Subramanian A, et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, 2005. 102(43): p. 15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liberzon A, et al. , Molecular signatures database (MSigDB) 3.0. Bioinformatics, 2011. 27(12): p. 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liberzon A, et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems, 2015. 1(6): p. 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kanehisa M. and Goto S, KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res, 2000. 28(1): p. 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jassal B, et al. , The reactome pathway knowledgebase. Nucleic Acids Res, 2020. 48(D1): p. D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ma’ayan A, Introduction to network analysis in systems biology. Science signaling, 2011. 4(190): p. tr5–tr5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.De Jager PL, et al. , Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nature Genetics, 2009. 41(7): p. 776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Moreno-Torres I, et al. , Immunophenotype and Transcriptome Profile of Patients With Multiple Sclerosis Treated With Fingolimod: Setting Up a Model for Prediction of Response in a 2-Year Translational Study. Frontiers in immunology, 2018. 9: p. 1693–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Grimes T, Potter SS, and Datta S, Integrating gene regulatory pathways into differential network analysis of gene expression data. Scientific Reports, 2019. 9(1): p. 5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lichtblau Y, et al. , Comparative assessment of differential network analysis methods. Brief Bioinform, 2017. 18(5): p. 837–850. [DOI] [PubMed] [Google Scholar]
- 213.Yu C, et al. , A strategy for evaluating pathway analysis methods. BMC Bioinformatics, 2017. 18(1): p. 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cirillo E, Parnell LD, and Evelo CT, A Review of Pathway-Based Analysis Tools That Visualize Genetic Variants. Front Genet, 2017. 8: p. 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Baranzini Sergio E., et al. , Network-Based Multiple Sclerosis Pathway Analysis with GWAS Data from 15,000 Cases and 30,000 Controls. The American Journal of Human Genetics, 2013. 92(6): p. 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Voskuhl RR, et al. , Gene expression in oligodendrocytes during remyelination reveals cholesterol homeostasis as a therapeutic target in multiple sclerosis. Proceedings of the National Academy of Sciences, 2019. 116(20): p. 10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cervantes-Gracia K. and Husi H, Integrative analysis of Multiple Sclerosis using a systems biology approach. Scientific Reports, 2018. 8(1): p. 5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yan J, et al. , Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Briefings in bioinformatics, 2018. 19(6): p. 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Baranzini SE, et al. , Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature, 2010. 464(7293): p. 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.International Multiple Sclerosis Genetics, C., A systems biology approach uncovers cell-specific gene regulatory effects of genetic associations in multiple sclerosis. Nature communications, 2019. 10(1): p. 2236–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Li H, et al. , Shared Gene Expression Between Multiple Sclerosis and Ischemic Stroke. Front Genet, 2018. 9: p. 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Khatri P, Sirota M, and Butte AJ, Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol, 2012. 8(2): p. e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wee Y, et al. , The bioinformatics tools for the genome assembly and analysis based on third-generation sequencing. Brief Funct Genomics, 2019. 18(1): p. 1–12. [DOI] [PubMed] [Google Scholar]
- 224.Vilarino-Guell C, et al. , Exome sequencing in multiple sclerosis families identifies 12 candidate genes and nominates biological pathways for the genesis of disease. PLoS Genet, 2019. 15(6): p. e1008180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mertins P, et al. , Proteogenomics connects somatic mutations to signalling in breast cancer. Nature, 2016. 534(7605): p. 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Breschi A, Gingeras TR, and Guigo R, Comparative transcriptomics in human and mouse. Nat Rev Genet, 2017. 18(7): p. 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hayashida M. and Akutsu T, Complex network-based approaches to biomarker discovery. Biomark Med, 2016. 10(6): p. 621–32. [DOI] [PubMed] [Google Scholar]
- 228.Latterich M. and Schnitzer JE, Streamlining biomarker discovery. Nat Biotechnol, 2011. 29(7): p. 600–2. [DOI] [PubMed] [Google Scholar]
- 229.LaFramboise T, Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic acids research, 2009. 37(13): p. 4181–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jia X, et al. , Genome sequencing uncovers phenocopies in primary progressive multiple sclerosis. Annals of neurology, 2018. 84(1): p. 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gil-Varea E, et al. , Exome sequencing study in patients with multiple sclerosis reveals variants associated with disease course. Journal of neuroinflammation, 2018. 15(1): p. 265–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Goldman SL, et al. , The Impact of Heterogeneity on Single-Cell Sequencing. Frontiers in Genetics, 2019. 10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hwang B, Lee JH, and Bang D, Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental & Molecular Medicine, 2018. 50(8): p. 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bakken TE, et al. , Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLOS ONE, 2018. 13(12): p. e0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Huynh JL and Casaccia P, Epigenetic mechanisms in multiple sclerosis: implications for pathogenesis and treatment. The Lancet Neurology, 2013. 12(2): p. 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Elkabes S. and Li H, Proteomic strategies in multiple sclerosis and its animal models. Proteomics. Clinical applications, 2007. 1(11): p. 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Auer M, et al. , Utility of Two-Dimensional Difference Gel Electrophoresis in Diagnosis of Multiple Sclerosis. Diagnostics (Basel, Switzerland), 2018. 8(3): p. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]