Abstract
The actin cytoskeleton drives cell motility and is essential for neuronal development and function. LIM and SH3 protein 1 (LASP1) is a unique actin-binding protein that is expressed in a wide range of cells including neurons, but its roles in cellular motility and neuronal development are not well understood. We report that LASP1 is expressed in rat hippocampus early in development, and this expression is maintained through adulthood. High-resolution imaging reveals that LASP1 is selectively concentrated at the leading edge of lamellipodia in migrating cells and axonal growth cones. This local enrichment of LASP1 is dynamically associated with the protrusive activity of lamellipodia, depends on the barbed ends of actin filaments, and requires both the LIM domain and the nebulin repeats of LASP1. Knockdown of LASP1 in cultured rat hippocampal neurons results in a substantial reduction in axonal outgrowth and arborization. Finally, loss of the Drosophila homologue Lasp from a subset of commissural neurons in the developing ventral nerve cord produces defasciculated axon bundles that do not reach their targets. Together, our data support a novel role for LASP1 in actin-based lamellipodial protrusion and establish LASP1 as a positive regulator of both in vitro and in vivo axon development.
INTRODUCTION
The development of complex circuits in the central nervous system requires the precise wiring of axons with their synaptic targets. Axon growth and guidance depend on the axon growth cone, a fan-shaped motile structure at the distal end of the growing axon. To reach its final destination, the growth cone must navigate vast distances in response to a multitude of biochemical and biophysical signals (Kolodkin and Tessier-Lavigne, 2011). The growth cone is highlighted by two actin-rich protrusions: filopodia, fingerlike protrusions containing long linear filamentous actin (F-actin), and lamellipodia, sheetlike thin protrusions powered by a network of branched short F-actin (Dent et al., 2011). Growth cone filopodia are believed to function in sampling the extracellular environment, whereas lamellipodia drive growth cone extension (Vitriol and Zheng, 2012; Omotade, et al., 2017). Extracellular guidance cues are translated into spatially organized intracellular signaling cascades that lead to distinct growth cone responses by targeting the structure and dynamics of the actin cytoskeleton in the growth cone (Kalil and Dent, 2005; Lowery and Van Vactor, 2009; Vitriol and Zheng, 2012). The dynamics of F-actin polymerization, depolymerization, and reorganization are tightly regulated by a large number of actin-binding proteins (Pollard and Borisy, 2003; Vitriol and Zheng, 2012; Gomez and Letourneau, 2014; Omotade et al., 2017). Actin assembly at the leading edge of the lamellipodia is a driving force behind growth cone motility (Pollard and Borisy, 2003; Dent et al., 2011; Yang et al., 2012). During growth cone protrusion, a small subset of actin-binding proteins are specifically recruited to the leading edge to facilitate the addition of actin monomers to the plus ends (also referred to as barbed ends) of actin filaments, which are concentrated at the membrane-actin cytoskeleton interface (Lowery and Van Vactor, 2009; Pollard and Cooper, 2009; Dent et al., 2011). Forward movement of the growth cone requires the action of these actin-binding proteins for new polymerization at the leading edge, together with the engagement of the “molecular clutch” to produce the traction force (Lowery and Van Vactor, 2009).
LIM and SH3 protein 1 (LASP1) is the smallest member of the nebulin family of actin-binding proteins, of which the founding member nebulin is well known for its structural role in skeletal muscles (Tomasetto et al., 1995a; Terasaki et al., 2004; Pappas et al, 2011; Orth et al., 2015). LASP1 is highly expressed in a number of tissues including the brain, and its dysregulation has been implicated in several neurological disorders, including autism spectrum disorder and schizophrenia (Stone et al., 2007; Joo et al., 2013). Therefore, LASP1 may play a role in nervous system development and function. In support of this notion, an early study identified LASP1 in the postsynaptic density fraction of brain tissues and showed the accumulation of LASP1 in axonal growth cones and dendritic spines in culture (Phillips et al., 2004). Furthermore, LASP1 has recently been shown to regulate dendritic arborization and postsynaptic spine development (Myers et al., 2020). However, no study has examined the function of LASP1 in young neurons, especially in axonal development.
LASP1 contains an N-terminal LIM domain, two internal actin-binding nebulin repeats, and a C-terminal SH3 protein-interaction domain (Tomasetto et al., 1995a; Schreiber et al., 1998). LASP1 was found to be up-regulated in numerous cancer types, including ovarian, breast, renal, colorectal, and prostate cancers (Tomasetto et al., 1995b; Grunewald et al., 2006, 2007). This increased LASP1 expression has been shown to positively correlate with cell motility and cancer metastasis (Grunewald et al., 2006, 2007). Therefore, LASP1 appears to regulate actin-based cell motility, but the mechanism by which LASP1 regulates actin-based protrusion and cellular motility is currently unclear. Moreover, if and how LASP1 functions in neurons during early development remains unknown. In this study, we investigated the expression profile, subcellular distribution, and function of LASP1 in developing neurons. We present evidence that LASP1 regulates the actin-based protrusive activities that underlie the axon growth and branching in culture, as well as plays a role in axon development in vivo.
RESULTS
LASP1 expression in neurons and its dynamic localization to the leading edge of actin-based membrane protrusions
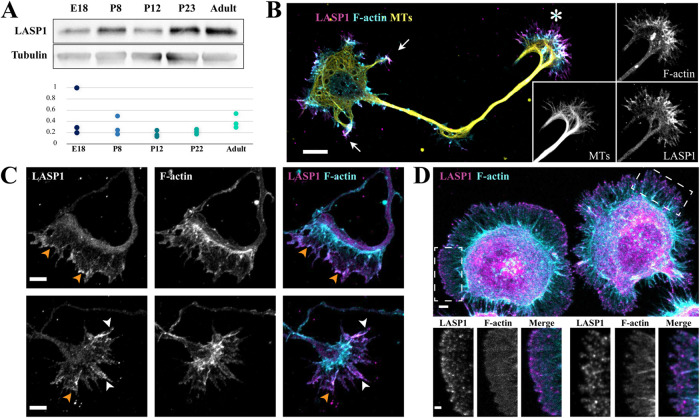
The presence of LASP1 in brain tissue has been reported previously (Orth et al., 2015), but its developmental expression profile and functions in neurons remain unknown. To address these questions, we first examined LASP1 expression in the hippocampus, a complex brain structure that has previously been found to be critical for learning and memory (Jarrard, 1993; Gonçalves et al., 2016). Immunoblotting for LASP1 was performed with hippocampal lysates harvested at embryonic day 18 (E18), postnatal day 8 (P8), P12, P23, and adulthood. These key stages capture the progression of neuronal development, from axon elongation to synapse formation and maturation (Crain et al., 1973; Harris et al., 1992; Fiala et al., 1998; Tyzio et al., 1999). We detected LASP1 expression in the E18 hippocampus, which persists through each stage to adulthood (Figure 1A). The finding that LASP1 is expressed at E18 suggests a possible function for LASP1 in early neuronal development. To investigate this possibility, we examined the subcellular distribution of LASP1 in dissociated rat hippocampal neurons after 2 d in vitro (DIV2) using immunofluorescence. By DIV2, hippocampal neurons have an established polarity with one long axon and multiple short minor processes destined to be dendrites (Dotti et al., 1988; Kaech and Banker, 2006). We found that LASP1 is highly enriched in the axonal growth cone (Figure 1B, asterisk). LASP1 signals were also detected at the tip of the minor processes (Figure 1B, arrows). The growth cone is consisted of two distinct compartments: the peripheral and central regions (P- and C-region), of which the P-region is a broad and flat area highlighted by actin-rich lamellipodia and filopodia, whereas the C-region, located behind the P-region and connected to the axonal shaft, is enriched in microtubules and cellular organelles (Vitriol and Zheng, 2012). LASP1 signals appear to be mostly concentrated in the P-region of the growth cone (Figure 1B, see the insets). A close examination supports the notion that LASP1 is largely found in the P-region, particularly at the leading edge of the growth cone lamellipodia (Figure 1C, orange arrowheads), as well as along filopodial actin bundles (Figure 1C bottom row, white arrowheads). A similar spatial pattern was found in cultured Cath.a-differentiated (CAD) cells (Qi et al., 1997), a mouse neuroblastoma cell line (Figure 1D). Within 2 h of plating on laminin-coated coverslips, CAD cells attach and spread large and broad lamella with motile lamellipodia around the cell’s periphery. Immunostaining for LASP1 in these cells shows bright, concentrated signals at the edges of the lamellipodia, as well as on some filopodia with thick F-actin bundles. Given that lamellipodia and filopodia are known to be significant drivers of actin-based membrane protrusion and cellular motility, this raises the possibility that LASP1 could regulate the actin-based projections that underlie growth cone motility and axon development.
FIGURE 1:
LASP1 is expressed in the developing brain and localizes to the axon growth cone. (A) Top, a representative Western blot showing the developmental profile of LASP1 in rat hippocampi at the indicated stages. Bottom, the plot shows normalized LASP1 levels relative to tubulin loading control from three independent blots. (B) Representative confocal images of a rat hippocampal neuron in culture at 2 d in vitro (DIV2) stained for endogenous LASP1 (magenta), F-actin (phalloidin, cyan), and α-tubulin (yellow). Individual channels of the growth cone are shown as the insets on the right. Arrows indicate the minor processes destined to be dendrites, and asterisk indicates the axonal growth cone. Scale bar: 20 µm. (C) Representative confocal images of growth cones from fixed DIV2 cultured rat hippocampal neurons, stained for endogenous LASP1 (magenta) and F-actin (phalloidin, cyan). LASP1 localizes to the leading edge of lamellipodia (orange arrowheads) and actin bundles in filopodia (white arrowheads). Scale bars: 5 µm. (D) Representative confocal images of two CAD cells labeled for LASP1 (magenta) and F-actin (phalloidin, cyan). LASP1 is seen at lamellipodia and filopodia in a similar pattern to growth cones. Scale bars: 5 µm (main panel) and 2 µm (insets).
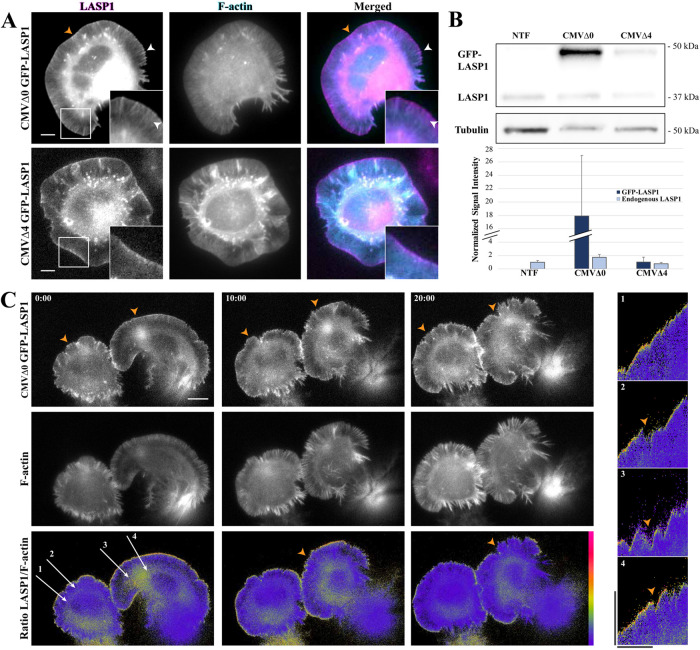
To understand the spatiotemporal dynamics behind LASP1 localization and its potential functions in actin-based motility, we took advantage of the large, motile lamellipodia seen in spreading CAD cells to perform live cell imaging. Here, GFP-tagged LASP1 is coexpressed with the F-actin marker, Lifeact-mRuby, in CAD cells (Figure 2A). To ensure that exogenous GFP-LASP1 can faithfully represent the spatiotemporal pattern of endogenous LASP1, we experimented with various levels of GFP-LASP1 expression using constructs with reduced CMV promoter strengths (Morita et al., 2012). We found that high expression under a full-strength CMV promoter (CMVΔ0 GFP-LASP1) resulted in overexpressed GFP-LASP1 that not only highlighted the leading edges of lamellipodia (Figure 2A top row, orange arrowheads) but also labeled F-actin bundles in the lamella and other structures (Figure 2A top row, white arrowheads). While this localization is similar to what has been previously reported (Nakagawa et al., 2006), it is clearly different from the endogenous LASP1 localization detected by immunofluorescence (Figure 1D). We therefore experimented with the more severely crippled CMV constructs to obtain lower levels of expression. We found that the expression of GFP-LASP1 from the CMVΔ4 construct (CMVΔ4 GFP-LASP1) most closely approximated the endogenous localization of LASP1 seen in Figure 1 (Figure 2A, bottom row). Western blotting confirmed that GFP-LASP1 expressed using CMVΔ4 is similar to the endogenous LASP1 level, whereas CMVΔ0 resulted in more than 10 times the level of GFP-LASP1 overexpression compared with the endogenous LASP1. We therefore used CMVΔ4 GFP-LASP1 for all subsequent live cell imaging.
FIGURE 2:
GFP-LASP1 localizes to protruding membranes. (A) Representative images of live CAD cells coexpressing intact (CMVΔ0 GFP-LASP1, top) or crippled (CMVΔ4 GFP-LASP1, bottom) CMV promoter-driven GFP-LASP1 (left, magenta), along with Lifeact-mRuby (F-actin, middle, cyan). GFP-LASP1 overexpressed by an intact CMV promoter localizes to the leading edge (orange arrowheads) and actin bundles (white arrowheads), whereas CMVΔ4 GFP-LASP localizes only to the leading edge, similar to endogenous LASP1. Scale bars: 5 µm. (B) Top, representative anti-LASP1 and anti-tubulin Western blots from CAD cells transfected with CMVΔ0 GFP-LASP1 or CMVΔ4 GFP-LASP1, along with nontransfected controls (NTF). Bottom, graph shows quantification of GFP-LASP1 expression, normalized to tubulin loading control, with endogenous LASP1 from the nontransfected condition set to 1. Error bars represent standard error. (C) Time-lapse images of CAD cells coexpressing CMVΔ4 GFP-LASP1 and Lifeact-mRuby. Bottom row, images show GFP to mRuby ratio, color coded with a rainbow heat map (scale to right). Images were captured every 5 s for 20 min. Scale bar: 10 µm. Right, representative kymographs (correspond to labeled arrows on left merged image). High levels of GFP-LASP1 can be found at the leading edge during cell protrusion, but GFP-LASP1 largely disappears from the edge on cell retraction (orange arrowheads). Images are representative of 10 cells across three independent culture replicates. Kymograph scale bars: vertical is 10 µm, horizontal is 10 min.
To observe the spatiotemporal dynamics of LASP1, we performed live imaging on motile CAD cells expressing GFP-LASP1 and Lifeact-mRuby (Figure 2C). To highlight the pool of LASP1 at the leading edge of the lamellipodia, we generated ratiometric images of LASP1:F-actin signals, which are color coded such that the yellow/red hot colors represent high ratio values, whereas blue/purple cool colors indicate low ratio values. It is clear that a narrow yellow band outlining the lamellipodia around the cells can be seen, suggesting high LASP1:F-actin ratio values at the leading edge of lamellipodia (Figure 2C, bottom row). It should be noted that the high LASP1:F-actin ratio values are not observed evenly along the leading edge, and there are locations that the LASP1:F-actin ratio dropped over the course of the time-lapse recording (Figure 2C, orange arrowheads). To further understand the dynamics of LASP localization to the leading edge, we produced several kymographs using the time-lapse sequence. It is clear that LASP1 becomes enriched at the leading edge during forward membrane protrusion, and disappears when the protrusion is paused or retracting (Figure 2C, right, orange arrowheads). Therefore, LASP1 localization to the leading edge of lamellipodia is tightly associated with membrane protrusion.
Mechanisms of LASP1 localization to the leading edge
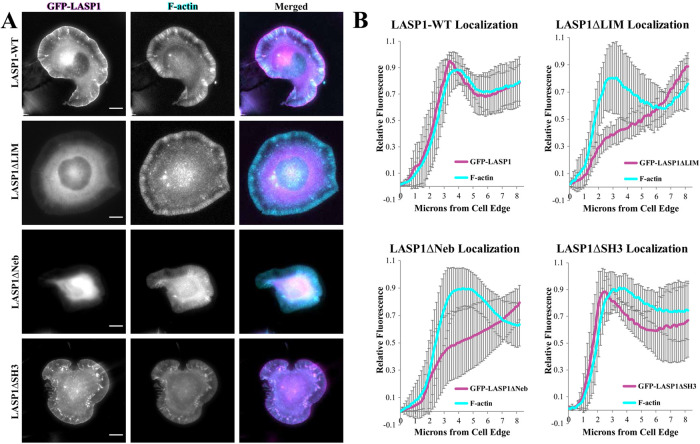
We first investigated which domains of LASP1 are required for its localization to the leading edge of protruding lamellipodia. Here, GFP-tagged LASP1 deletion mutants, in which the LIM, nebulin, or SH3 domain was removed (Stölting et al., 2012; Myers et al., 2020), were coexpressed with Lifeact-mRuby in CAD cells for live imaging. We found that removal of either the LIM domain (GFP-LASP1ΔLIM) or the nebulin repeats (GFP-LASP1ΔNeb) resulted in a loss of LASP1 localization to the leading edge (Figure 3A). However, deletion of the SH3 domain (GFP-LASP1ΔSH3) did not alter its localization to the leading edge of lamellipodia (Figure 3A). Line profiles of LASP1 fluorescence show that both the full-length GFP-LASP1 and GFP-LASP1ΔSH3 proteins exhibited a fluorescence peak at the leading edge of the lamellipodia, which was not observed for GFP-LASP1ΔLIM and GFP-LASP1ΔNeb (Figure 3B). Although the SH3 domain has been implicated in most of LASP1’s known protein interactions (Orth et al., 2015), our data indicate that the LIM domain and nebulin repeats are required for LASP1 localization to the leading edge. While the nebulin repeats are well known for F-actin binding (Pappas et al., 2011), the involvement of the LIM domain for LASP1 localization to the leading edge is of interest. Given that the leading edge of lamellipodia is the site of actin polymerization underlying membrane protrusion, it is plausible that the LIM domain and nebulin repeats work cooperatively to target LASP1 to the site of active actin polymerization and potentially regulate protrusive activity.
FIGURE 3:
The LIM domain and nebulin repeats are essential for LASP1 localization to the leading edge. (A) Live CAD cells coexpressing Lifeact-mRuby (F-actin, cyan) and wild-type GFP-LASP1, GFP-LASP1ΔLIM, GFP-LASP1ΔNebulin (GFP-LASP1ΔNeb), or GFP-LASP1ΔSH3 (magenta). Wild-type GFP-LASP1 and GFP-LASP1ΔSH3 localize to the leading edge. However, GFP-LASP1ΔLIM and GFP-LASP1ΔNeb do not localize to the leading edge and instead appear mostly soluble. Scale bars: 10 µm. (B) Line scan analysis of GFP-LASP1 localization at the leading edge (120 line scans from 30 cells across three independent culture replicates). Error bars represent standard error.
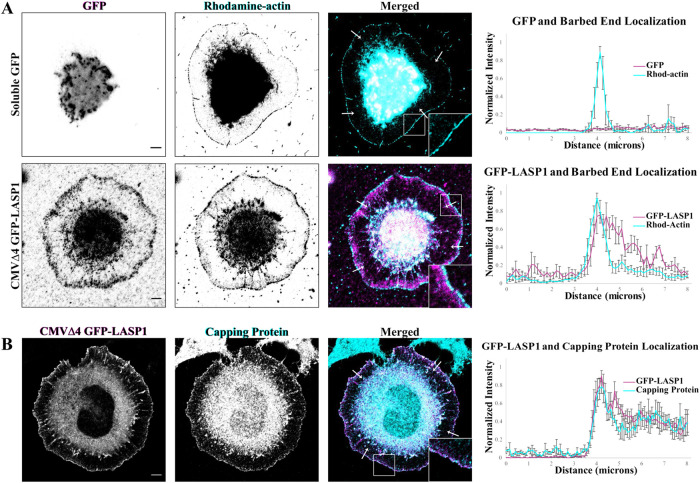
The leading edge of lamellipodia is known for its high concentration of the barbed ends of actin filaments, which are required for the rapid actin polymerization underlying membrane protrusion (Pollard and Borisy, 2003). We thus examined if LASP1 at the leading edge is colocalized with actin barbed ends. We adopted a barbed end assay (Gu et al., 2010; Marsick et al., 2010; Marsick and Letourneau, 2011) together with immunofluorescent staining of LASP1. Due to an incompatibility between the barbed end labeling and the LASP1 immunolabeling protocols, we were unable to perform immunostaining on endogenous LASP1 using our antibody after the cells have undergone barbed end labeling. To get around this issue, we expressed LASP1 at a low level using CMVΔ4 GFP-LASP1 and performed immunolabeling using an anti-GFP antibody. Consistently, the rhodamine-actin-labeled barbed ends of actin filaments were found to be concentrated in a narrow band at the leading edge of the lamellipodia. Soluble GFP signals exhibited no localization in CAD cells and were distributed throughout the cell, with the highest concentration at the cell center due to its larger volume (Figure 4A, top row). Fluorescence line profiles confirmed that there is no overlap between the normalized GFP signal and the barbed ends. Strikingly, GFP-LASP1 signals were seen to be largely overlapping with the barbed end signals, both highlighting the leading edge of the lamellipodia (Figure 4A, bottom row). Line profile quantification shows that the peak of LASP1 signal intensity overlaps with the narrow band of actin barbed ends. While LASP1 signals were seen further into the lamellipodia without barbed end signals, this may be explained by the short labeling window when only a small amount of rhodamine-actin can be incorporated into the F-actin network, or the light fixation prior to barbed end labeling that may have cross-linked LASP1 to the actin filaments. Nonetheless, the overlap between the peak of LASP1 and actin barbed ends supports the notion that LASP1 is likely targeted to sites of actin polymerization in the lamellipodia. Finally, immunostaining for GFP-LASP1 and actin capping protein (CP) also showed that these two proteins are highly colocalized in the lamellipodia (Figure 4B). Given that CP is enriched at the barbed ends of actin filaments, and it works cooperatively with the Arp2/3 family of nucleation factors for lamellipodial protrusion (Pollard et al., 2000), our data suggest that LASP1 may associate with actin barbed ends in lamellipodia to potentially regulate actin-based motility.
FIGURE 4:
LASP1 localizes to actin plus ends at the leading edges of CAD cells. (A) Representative images of CAD cells expressing soluble GFP or CMVΔ4 GFP-LASP1 (magenta), with actin plus ends labeled with rhodamine–conjugated actin (cyan). Scale bars: 5 µm. Graphs (right) depict the average of four normalized line profiles from cells on left (arrows in merged images show location of line scans). Error bars represent standard error. LASP1 localization at the leading edge overlaps with the labeled plus ends. Representative of 10 cells from three independent culture replicates. (B) Representative images showing colocalization of CMVΔ4 GFP-LASP1 (magenta) and endogenous CP in CAD cells (cyan). Scale bar: 5 µm. Graph (right) shows the average of four normalized line profiles from arrows in merged image. Error bars represent standard error.
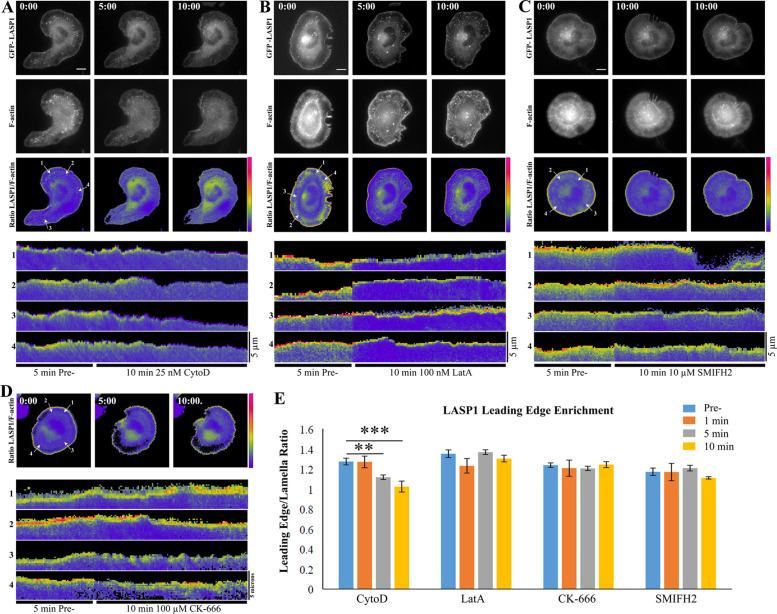
To further examine the association between LASP1 and the barbed ends of actin filaments, we applied a low concentration of Cytochalasin D (CytoD), a pharmacological inhibitor that caps the barbed ends of actin filaments to block further polymerization (Buck and Zheng, 2002). We performed live imaging on CAD cells coexpressing CMVΔ4 GFP-LASP1 and Lifeact-mRuby and applied 25 nM CytoD after a 5-min baseline period (Figure 5A). We found that within 5 min of drug application, GFP-LASP1 disappeared from the leading edge of lamellipodia in CAD cells (n = 3 cells per condition from three culture replicates; 5 min not significant, 10 min **p = 0.0029, 15 min ***p < 0.0001 by one-way ANOVA and Holm-Sidak’s multiple comparisons test) (Figure 5, A and E). It should be noted that at this low concentration, CytoD did not cause drastic changes in the actin cytoskeleton and no obvious retraction of lamellipodia was observed. Since CytoD inhibits actin polymerization, we tested if the loss of LASP1 signals at the leading edge is a result of reduced polymerization. We performed the same experimental paradigm using three different inhibitors of actin polymerization whose mechanisms of action differ significantly from CytoD: Latrunculin A (LatA), which sequesters actin monomers (100 nM LatA, Figure 5B) (Lee et al., 2013); SMIFH2, which inhibits formin-based polymerization (10 μM, Figure 5C) (Vitriol et al., 2015); and CK-666, which prevents Arp2/3-based actin nucleation (100 μM, Figure 5D) (Vitriol et al., 2015). We found that none of these drugs abolished the LASP1 localization to the leading edge of lamellipodia (n = 3 cells per condition from three culture replicates; p values not significant by one-way ANOVA and Holm-Sidak’s multiple comparisons test) (Figure 5E). These data support the notion that LASP1 is associated with barbed ends at the leading edge of protruding lamellipodia.
FIGURE 5:
LASP1 localization to the leading edge depends on barbed ends of actin filaments. (A–D) Time-lapse images of CAD cells expressing CMVΔ4 GFP-LASP1 and Lifeact-mRuby before (time 0), at 5 and 10 min after the application of 25 nM CytoD (A), 100 nM Lat A (B), 10 µM SMIFH2 (C), and 100 µM CK-666 (D). For each condition, representative images of GFP-LASP1, Lifeact-mRuby, and their ratiometric images are shown, except (D) in which only the ratiometric images are shown. For each cell, four kymographs from the lines indicated in the corresponding ratiometric image are shown in the lower portion of each panel. Each kymograph includes the time-lapse recordings of 5 min before (pre-) and 10 min after the onset of the specific drug treatment. Scale bars: 10 µm. Images are representative of three cells per condition across three independent culture replicates. (E) Quantification of LASP1 localization at the leading edge of the cell 5 min before, as well as 1, 5, and 10 min after drug application. Error bars represent standard error. **p < 0.005; ***p < 0.001. P values were calculated using a one-way ANOVA and Holm-Sidak’s multiple comparisons test.
LASP1 promotes axon elongation and branching
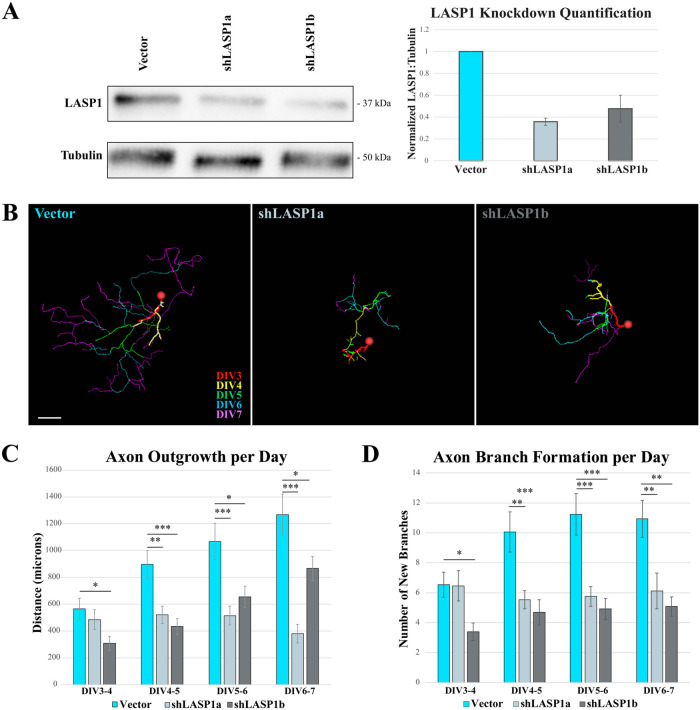
The striking dynamic pattern of LASP1 localization to the leading edge of protruding lamellipodia suggest that LASP1 may regulate actin-based motility. While CAD cells offer an advantage for high-resolution image-based analysis of LASP1 localization and dynamics, they are not the best system to reliably assess actin-based motility. Because LASP1 is localized to the leading edge of growth cone lamellipodia and filopodia (see Figure 1), we used primary hippocampal neurons in culture to examine the effects of LASP1 knockdown on growth cone motility. We developed two distinct shRNA hairpins that target nonoverlapping regions of LASP1 mRNA to knock down LASP1 in hippocampal neurons and to control for off-target effects. Here, shLASP1a and shLASP1b were designed to target the coding sequence and the 3′UTR, respectively, and were inserted into a multicistronic vector that expresses mCherry (Myers et al., 2020). We found that shLASP1a and shLASP1b can effectively knock down endogenous LASP1 in hippocampal neurons to 30.7% ± 7.0% and 44.1% ± 15.9% (mean ± SEM), respectively, after 72 h, as assessed by Western blot (Figure 6A). To measure the effect of LASP1 depletion on axon outgrowth, we plated dissociated rat hippocampal neurons in multichamber glass bottom dishes, then transfected them on DIV2 with shLASP1a, shLASP1b, or the empty vector as a control. Images of transfected neurons were captured every 24 h starting at DIV3 through DIV7. Axon outgrowth over each 24 h period was measured by tracing only the new axon growth using the Fiji Simple Neurite Tracer program (Longair et al., 2011). Representative traces were color coded for each 24 h period and collapsed across all time points (Figure 6B). While control neurons grew long, complex axonal arbors, neurons depleted of LASP1 produced shorter and more simplified axons. Quantitatively, the total axon outgrowth in LASP1 knockdown neurons was significantly lower than controls across all time points (n = approximately 30 cells per condition across three culture replicates; see Supplemental Table S1 for exact n and p values: *p < 0.05; **p < 0.005; ***p < 0.001 by one-way ANOVA Tukey HSD post-hoc test) (Figure 6C). We also counted the number of new branch points for each condition and found a significant reduction in the axonal complexity of knockdown neurons compared with controls across all time points (see Supplemental Table S2 for exact n and p values; *p < 0.05; **p < 0.005; ***p < 0.001 by one-way ANOVA Tukey HSD post-hoc test) (Figure 6D). These results suggest that LASP1 plays a significant role in promoting axon outgrowth and arborization.
FIGURE 6:
Knockdown of LASP1 truncates axon elongation and reduces branching in cultured hippocampal neurons. (A) Representative anti-LASP1 and anti-tubulin Western blots of lysates from hippocampal neurons transfected with empty plasmid (vector), shLASP1a, or shLASP1b for 72 h. Graph shows ∼60–70% reduction in LASP1 expression in cultures expressing shRNAs across three independent culture replicates. LASP1 levels are normalized to tubulin loading controls. Error bars represent standard error. (B) Axon tracings of representative rat hippocampal neurons transfected at DIV2 with empty backbone control (Vector), shLASP1a, or shLASP1b. Neurons were imaged every 24 h from DIV3 until DIV7, and each day’s growth is temporally color coded (scale lower right, Vector image). Knockdown of LASP1 reduces axon outgrowth over each 24 h period. Scale bar: 150 µm. (C) Graph depicting axon outgrowth (in µm) every 24 h. (D) Graph depicting the number of newly formed branches at each time point. Error bars represent standard error. *p < 0.05; **p < 0.005; ***p < 0.001. P values calculated using a one-way ANOVA Tukey’s HSD post-hoc test. Data collected from approximately 30 neurons from three independent cultures (see Supplemental Tables S1 and S2 for exact n and p values).
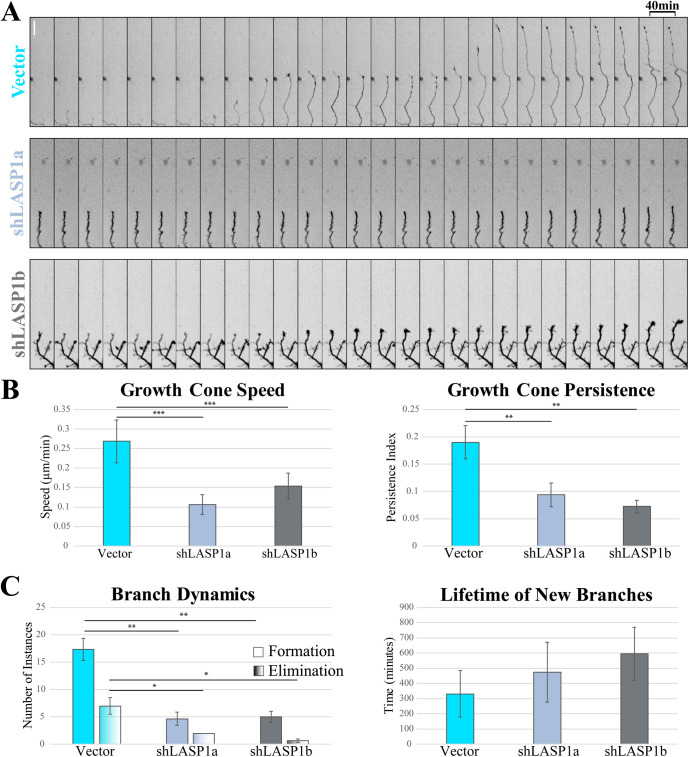
While it is clear that loss of LASP1 causes a reduction in axon length, whether this is due to increased growth cone retraction or reduced growth cone protrusion is uncertain. Furthermore, it is unclear whether LASP1 supports branch development by promoting branch formation or by preventing branch retraction. To understand the role of LASP1 in growth cone motility and branching dynamics, we plated and transfected neurons to knockdown LASP1 as described above, then captured images at DIV5 every 10 min for at least 16 h. When we examined the growth cones from control neurons, we found that they protruded in stairlike bursts over the imaging period, while growth cones from LASP1 knockdown cells were nearly static over the same timeframe (Figure 7A; see also Supplemental Video S1). To quantify this, we utilized the Imaris tracking software to semiautomatically detect and track individual growth cones (Imaris v.9.5.1, Bitplane). Analysis of growth cone motility showed that LASP1 knockdown growth cones were slower and had reduced persistence (ratio of displacement to total distance traveled) compared with controls (*p < 0.05; **p < 0.005; ***p < 0.001 as calculated using a one-way ANOVA Tukey HSD post-hoc test; see Supplemental Table S3 for exact n and p values; from three independent culture replicates) (Figure 7B). Careful examination of axon branch dynamics showed that knockdown of LASP1 significantly reduced both the formation and elimination of new axonal branches with no significant effect on their total lifetime (n = 3 neurons per condition, 3 independent culture replicates; *p < 0.05; **p < 0.005, see Supplemental Table S4 for exact p values calculated using a one-way ANOVA Tukey HSD post-hoc test) (Figure 7C). Together with the imaging data in CAD cells, our results support the notion that LASP1 functions to regulate the actin-based protrusions that underlie the growth and branching of developing axons in culture.
FIGURE 7:
Knockdown of LASP1 reduces growth cone protrusion and branch formation in cultured hippocampal neurons. (A) Montages of representative growth cones from DIV5 rat hippocampal neurons expressing Vector control, shLASP1a, or shLASP1b. Montage slices represent 40-min intervals. Scale bar: 40 µm. (B) Graph of growth cone speed (left) and persistence (right). Persistence index represents a ratio of growth cone displacement to total distance traveled. Growth cone speed and persistence are reduced in the absence of LASP1. (C) Graphs of dynamic axon branch formation/elimination (left), and lifetime (right) across all three conditions. Loss of LASP1 causes reduced branch formation and elimination, with no significant effect on the lifetime of new branches. Error bars represent standard error. *p < 0.05; **p < 0.005; ***p < 0.001 by one way ANOVA Tukey HSD post-hoc test. Experiments represent three independent culture replicates. (See Supplemental Tables 3 and 4 for exact n and p values).
Movie S1.
Time-lapse sequences of hippocampal neurons in culture showing their dynamic extension and retraction of axons and minor processes with and without knockdown of LASP1. Three sequences are shown: the control (empty vector transfection, left), knockdown using shLASP1a (middle) and shLASP1b (right). Images were taken every ten minutes for 10 hours. Control cells produced more new branches and faster growth cones than knockdown cells. Scale bar: 50 μm.
Lasp promotes axon commissure development and fasciculation in vivo
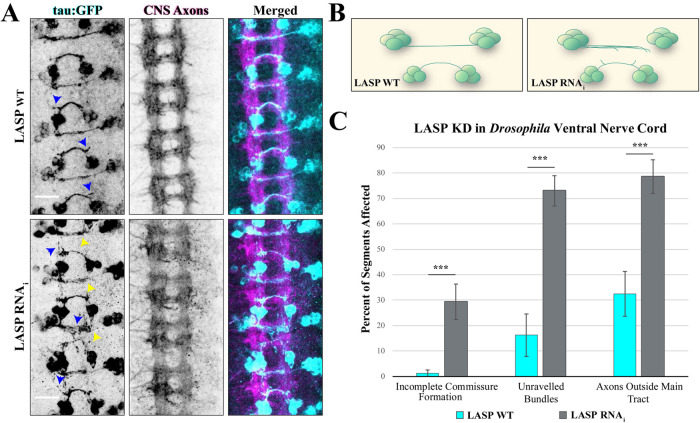
To examine the role of LASP1 in axon development in vivo, we utilized the Drosophila model system, both for its well-developed toolbox to study axon development and because it expresses a single nebulin family member called Lasp (Suyama et al., 2009; Orth et al., 2015). Drosophila Lasp is homologous to human LASP1 and shares the same domain architecture (i.e., N-terminal LIM domain, two nebulin repeats, and a C-terminal SH3 domain), making it an ideal in vivo system to study LASP1 in isolation (Lee et al., 2008; Suyama et al., 2009). Furthermore, Lasp is expressed very early in embryo development, like in our vertebrate model (Suyama et al., 2009). Within the developing Drosophila nervous system, we chose to focus on the ventral nerve cord, which at early stages of development contains stereotypic railroadlike bundles of commissural axons (Figure 8A, center images) that show clear phenotypes when axon guidance-related proteins are disrupted (Evans and Bashaw, 2010). To facilitate the identification of axonal defects, we targeted a specific subset of neurons in the ventral nerve cord using the UAS-Gal4 inducible system. For this set of experiments, Gal4 is expressed under a cell type-specific promoter, binds to the UAS element, and drives expression of the gene downstream of UAS. Here, we utilize the eagle-Gal4 element (egl-Gal4), which drives UAS-gene expression in two pairs of neuron clusters per abdominal segment of the ventral nerve cord and has been used extensively in previous studies of axon guidance receptors and signaling pathways (Garbe et al., 2007; O’Donnell and Bashaw, 2013a, b). Gal4 drives expression of GFP to visualize these neurons, Lasp RNAi to knockdown Lasp protein, and dicer to enhance RNAi efficiency. As a control, we drove expression of the GFP and dicer genes only, without the LASP RNAi. Both the Lasp-knockdown and control crosses produced viable embryos, which were fixed at stage 16 and immunostained for both GFP and central nervous system axons (BP102) as described previously (Bashaw, 2010). Z-stacks of the embryonic ventral nerve cords were analyzed for abnormalities in midline crossing. Specifically, each z-stack was blinded, then analyzed for axons and commissures that do not follow the typical trajectory described previously (O’Donnell and Bashaw, 2013a, b). The number of segments with aberrations in traditional guidance phenotypes (incomplete midline crossing, defasciculation, etc.) was measured as a percentage of the total segments in each embryo, then broken down by phenotype (Santiago et al., 2013). In the control embryos, we observed tight bundles of commissural axons that cross over to the contralateral neuron cluster, which is consistent with previous studies using this driver (Figure 8, A top and B left) (Garbe et al., 2007). However, we observed several aberrant phenotypes in the Lasp knockdown embryos (Figure 8, A bottom row, B). First, several segments displayed axons that did not reach their contralateral target. In some cases, this was due to incomplete midline crossing, while in other cases the axons crossed the midline but turned away from their destination or did not extend fully along their typical path (Figure 8A, yellow arrowheads). In total, this phenotype occurred in 30% of the measured knockdown segments on average, compared with approximately 1% of the control embryo segments (Figure 8C) (n = 10 wild-type and 18 knockdown embryos, p = 0.0008398 via Welch two-sample t test). Second, we observed two forms of defasciculation (Figure 8A, blue arrowheads). The first was an unravelling of the normally tight axon bundles, to the point that spaces were visible between the axons, which was observed in over 70% of all knockdown segments measured, indicating that Lasp promotes the tight bundling of commissural axons (p = 6.88e-06 via two-sample t test). The second form of commissure defasciculation we observed was individual axons leaving the main axon bundle, as well as the typical commissural path, and terminating far away from the synaptic target. This phenotype was documented in approximately 30% of the control embryo segments but over 75% of the knockdown segments, indicating that Lasp promotes axons continuing to grow along the path of their commissure bundles (p = 0.000298 via two-sample t test). While the incomplete commissure formation phenotype is highly consistent with our in vitro data, where axons do not extend far without LASP1, the defasciculation phenotypes indicate a possible role for LASP1 in in vivo axon guidance.
FIGURE 8:
Lasp knockdown leads to defects in axon commissure formation in Drosophila. (A) Representative images of stage 16 Drosophila embryos labeled with tau:GFP in egl-expressing ventral nerve cord axons. Top row shows dicer-only control embryo (WT-LASP); bottom row shows embryo expressing dicer plus LASP RNAi (LASP KD). Left column shows GFP-expressing egl neurons (green), middle column shows CNS axon (BP102, magenta), right column shows merged images. Arrowheads indicate defects in commissural axon development, including defasciculation (blue) and axon tracts that do not reach their targets (yellow). Scale bars: 15 µm. (B) Schematic of one segment from wild-type (left) or LASP knockdown (right) egl neurons in the Drosophila ventral nerve cord. Lasp knockdown results in numerous guidance defects, including defasciculation of axon bundles and truncated commissures. (C) Graph shows axon guidance defects from 10 control and 18 knockdown embryos collected across three independent experiments. ***p < 0.001; both defasciculation measures were analyzed using a Student’s t test. Incomplete commissure formation calculated using a Welch’s t test. Error bars represent standard error.
DISCUSSION
Actin-based cellular motility is essential for many developmental events, such as neuronal development, as well as for many pathological events, including cancer cell metastasis (Kalil and Dent, 2005; Lowery and Van Vactor, 2009). During brain development, neurons send axonal projections through the developing tissue to their respective synaptic targets. Essential for this process is the axon growth cone, an actin-rich structure at the tip of the axon that responds rapidly to extracellular cues in order to navigate toward its goal in a process known as axon guidance (Kolodkin and Tessier-Lavigne, 2011). Growth cones contain two actin-based membrane protrusions, lamellipodia and filopodia, which undergo dynamic protrusive activities as a part of the process of axon growth. The actin dynamics underlying growth cone motility is targeted by a plethora of spatially and temporally restricted signaling cascades and actin regulatory proteins to achieve directional outgrowth during axon guidance (Lowery and Van Vactor, 2009; Pollard and Cooper, 2009; Dent et al., 2011). In this study, we present evidence that LASP1, a unique member of the nebulin family of actin binding proteins, localizes to the actin polymerization zone during membrane protrusion and plays an important role in axon development. Previous studies in nonneuronal cells have implicated LASP1 in cell migration and focal adhesion, with no clear mechanism of action in actin-based cellular motility (Orth et al., 2015). LASP1 is expressed in the brain and has been implicated in a number of neurological and neurodevelopmental disorders (Phillips et al., 2004; Stone et al., 2007; Joo et al., 2013), but a role for LASP1 in early brain development has not yet been established. In this study, we present evidence that LASP1 is expressed during brain development prior to synapse formation. Further analyses show that LASP1 is dynamically enriched at the leading edge of protruding lamellipodia in motile neuronal cells. Importantly, LASP1 localization appears to depend on the free barbed ends in lamellipodial actin networks, suggesting that LASP1 plays a role in the actin polymerization underlying membrane protrusion, growth cone motility, and cellular migration. Consistently, LASP1 knockdown resulted in impaired axon growth cone protrusion speed and branch formation in cultured primary hippocampal neurons. Finally, knockdown of the Drosophila LASP1 orthologue, Lasp, resulted in a significant disruption of axon midline projections and defasciculation of commissural axon bundles in vivo. These findings support LASP1 as a novel actin regulatory protein that promotes the actin-based motility underlying axon elongation and guidance during early brain development.
Filopodia and lamellipodia are two established actin-based membrane protrusions found in motile growth cones. It is commonly believed that growth cone filopodia play a sensory role as they sample the environment to a greater extent than the growth cone itself. Filopodia contain bundles of actin filaments, and their growth depends largely on the formin family of actin nucleating factors and regulators. Lamellipodia are thin, sheetlike membrane protrusions that are believed to primarily function in growth cone movement. Lamellipodia contain a branched network of short actin filaments with their barbed (plus) ends pushing against the plasma membrane. Forward protrusion of lamellipodia is believed to driven by Arp2/3-based nucleation and polymerization at the leading edge where the addition of actin monomers to the barded ends push the membrane forward, followed by the engagement of the molecular clutch for interaction with the extracellular matrix (Suter and Forscher, 2000; Case and Waterman, 2015). Our immunofluorescence imaging showed that LASP1 is selectively enriched in the P-region of the growth cone where filopodia and lamellipodia exist. Importantly, LASP1 signals were associated with the leading edge of lamellipodia as well as some filopodia. These observations suggest that LASP1 may function in the actin-based protrusive activities underlying growth cone movement. This hypothesis is supported by our imaging work on LASP1 localization and dynamics in motile CAD cells. Here, CAD cells were used for their large and dynamic lamellipodia that enable high-resolution image-based analyses. Importantly, the spatial pattern of LASP1 in CAD cells is very similar to that of the growth cones: local enrichment at the leading edge of the lamellipodia as well as association with some filopodia. The role of LASP1 in actin-based membrane protrusive activities was supported by our live cell imaging data, as GFP-LASP1 was only enriched at the leading edges of active protrusions, but disappeared when the membrane retracted. Furthermore, this leading edge enrichment pattern was found to require both the LIM and nebulin regions of LASP1. Interestingly, Nakagawa et al. (2009) reported that the LIM domain and the first nebulin motif of LASP1’s sister protein, LASP2, work cooperatively to bind to actin filaments (Nakagawa et al., 2009). Our results are consistent with these findings and highlight the importance of both of these two domains for spatiotemporal targeting of LASP1 to the leading edge.
What is the mechanism that enables LASP1 localization to the leading edge? The observation that LASP1 accumulates at the leading edge of protruding lamellipodia suggests that LASP1 might be targeted to the region where actin polymerization is most abundant. The finding that CytoD was able to displace LASP1 from the leading edge further supports this notion. CytoD binds and caps the barbed ends of actin filaments to prevent actin polymerization (Goddette and Frieden, 1986). When applied at low concentrations, CytoD is known to be effective in capping the barbed ends of actin filaments without grossly disrupting F-actin structures (Lee et al., 2013). In our work, we found that a low concentration of CytoD (25 nM) did not disrupt the F-actin network of the lamellipodia, but effectively dislodged LASP1 from the leading edge. Importantly, none of the other reagents that inhibit actin assembly through other mechanisms, such as LatA, affected the LASP1 localization pattern. These results suggest that LASP1 is likely associated with barbed ends at the F-actin-membrane interface of lamellipodia. This notion is further supported by direct imaging of actin barbed ends, in which the band of barbed ends at the leading edge of lamellipodia overlaps with LASP1. Furthermore, our data show that LASP1 localization overlaps with CP, which is found at the leading edge of lamellipodia and has been shown to cap barbed ends (Akin and Mullins 2008). The high degree of colocalization between LASP1 and CP further supports that LASP1 localizes to the leading edge where F-actin barbed ends are in high abundance and therefore may regulate actin-based membrane protrusion. It is interesting to see that inhibition of Arp2/3 complex by CK-666 did not remove LASP1 from the leading edge. CK-666 at 100 µM was previously shown to be specific for Arp2/3 inhibition (Wu et al., 2012; Vitriol et al., 2015). Indeed, we observed an increase in the number of long parallel actin bundles in the lamellipodia after CK-666 treatment (Figure 5D), similar to previous reports (Vitriol et al., 2015; Skruber et al., 2020), confirming the effectiveness of CK-666. It is plausible that the acute nature of CK-666 application and the presence of formin-based nucleation prevented a drastic reduction in the availability of barbed ends in the lamellipodia for LASP1 localization. A similar argument could also apply to the SMIFH2 results but with one potential caveat as SMIFH2 could inhibit myosin II (Nishimura et al., 2020). Nonetheless, the lack of effect by SMIFH2 or CK-666 on LASP1 localization suggests that LASP1 is not associated with these two families of nucleation complexes for its localization to the leading edge.
Currently, the mechanism by which LASP1 localizes to the leading edge and how it regulates the actin dynamics underlying lamellipodia protrusion remains unknown. It should be noted that our colocalization data of LASP1 and barbed ends of actin filaments do not have the resolution to precisely determine if LASP1 is localized to the barbed ends or immediately adjacent. Since there is no previously published evidence for LASP1 to directly interact with the actin barbed ends, we hypothesize that LASP1 may function via these two nonexclusive mechanisms. First, LASP1 may localize immediately behind the barbed ends to stabilize the segment of newly polymerized actin filaments. Newly polymerized actin is well known to be less stable than aged filaments, due to rearrangement of the filaments’ internal structures (Hao et al., 2008). Therefore, LASP1 may be selectively targeted to the leading edge to stabilize newly generated segments of actin filaments immediately after the barbed ends. This would be a dynamic process, so when the segments of F-actin mature, LASP1 would lose its binding affinity. This would fit with our data suggesting that LASP1 binds to actin filaments at the leading edge, as well as partially supported by the fact that gross overexpression of GFP-LASP1 causes it to bind to most F-actin in the cell. However, it remains unclear how CytoD removes LASP1 from the leading edge. It is possible that CytoD capping of barbed ends, and the subsequent blockade of new assembly, lead to the elimination of unstable “newly generated segments” for LASP1 binding, although presumably this would also be true for the other polymerization inhibitors we utilized, none of which affected LASP1 localization. The second possibility is that LASP1 may function in actin uncapping or anti-capping to promote filament polymerization in collaboration with Ena/VASP. Previous studies have shown that VASP can interact with LASP1 via the LASP1 SH3 domain (Keicher et al., 2004). Given that VASP exhibits an extremely similar leading edge localization that is also membrane protrusion- and free barbed end-dependent (Bear et al., 2002), it is plausible that the function of LASP1 at the edge of lamellipodia involves VASP in some manner. However, because our data show that LASP1 leading edge enrichment does not require its SH3 domain, it is unlikely that VASP is the mechanism by which LASP1 is recruited to the leading edge. A recent study has shown that LASP1 interacts with N-WASP, which may activate Arp2/3 to stimulate actin polymerization and cell migration (Yan et al., 2020). However, the LASP1-N-WASP interaction appears to also depend on the SH3 domain of LASP1. Therefore, LASP1 localization to the leading edge of lamellipodia is unlikely to be mediated by N-WASP, though it’s possible LASP1 may promote lamellipodial protrusions through that mechanism. Future experiments employing advanced multichannel superresolution imaging, together with selective molecular manipulations, will enable us to delineate the precise mechanisms underlying LASP1 localization and functions.
Given that axonal growth and guidance depend on actin-based growth cone motility, LASP1 may play a role in actin-driven membrane protrusive activities to regulate axon development. In support of this hypothesis, LASP1 knockdown substantially impaired the axon outgrowth and branch formation of cultured hippocampal neurons over several days. Axonal outgrowth is achieved by rapid forward movement of the growth cone interspersed with pauses and retractions (Smirnov et al., 2014). Consistent with the notion that LASP1 may function in growth cone forward movement, we found that both the speed and the persistence of growth cone advance was drastically reduced in LASP1-knockdown neurons compared with control cells. Furthermore, we found that loss of LASP1 reduced the production of new branches, with no effect on their lifetime. This indicates that LASP1 is likely promoting the formation of new axon branches, as opposed to stabilizing existing branches. Together, these findings suggest that LASP1 is performing a similar function in neuronal development by promoting the protrusive growth of both new axon branches and growth cones.
To understand how our axonal outgrowth phenotype translates to an in vivo model system, we studied commissural neurons in the embryonic ventral nerve cords of Drosophila, which expresses only one LASP1 orthologue, Lasp. Knockdown of Lasp in Drosophila embryos led to several defects in the axon pathfinding of egl+ commissural axons. The high number of knockdown commissural axons that failed to reach their contralateral targets is consistent with the notion that LASP1 regulates actin-based motility. The observed defasciculation of axon bundles might be related to the role of LASPs in cell adhesion (Lin et al., 2004; Bliss et al., 2013). In vertebrates, two LASPs, LASP1 and LASP2, are expressed and have been shown to be present and function in cell adhesion complexes. It is also possible that the observed defects may be a result of Lasp functioning downstream of axon guidance signaling pathways, some of which have been shown to drive the fasciculation of growing axon bundles (Wolman et al., 2007). Future studies will be necessary to examine how Lasp is regulated by these pathways, if at all.
In summary, our study shows a novel role for LASP1 in lamellipodial protrusion and axon development through a combination of microscopic, in vitro, in vivo, and molecular approaches. Together with recent work on LASP1 in synaptic development (Myers et al., 2020), our work indicates that this unique nebulin family member plays an important role in neuronal development. Importantly, the presence of multiple phosphorylation sites in the linker region of LASP1 and its multiple protein–protein modules also highlight the potential for LASP1 to connect intricate signaling cascades to the actin dynamics underlying axon growth, guidance, and synapse formation. Future studies are needed to better understand the functions of LASP1 in neurons and the precise mechanisms underlying the regulation of actin-based motility.
MATERIALS AND METHODS
Cell culture
Primary culture of rat hippocampal neurons was performed as previously described (Omotade et al., 2018; Myers et al., 2020). Briefly, E18 rat embryos of both sexes were obtained from time-pregnant Sprague Dawley rats (Charles River Laboratories, Wilmington, MA). Brains were removed from the embryos and the hippocampi were isolated for dissociation, which consisted of treatment with trypsin for 15 min at 37°C, followed by pipette trituration in neuronal culture media (neurobasal medium containing GlutaMAX [Invitrogen], penicillin/streptomycin, and B27 supplement [Thermo Fisher Scientific]). Dissociated hippocampal neurons were plated at a density of 50,000 cells/35-mm dish on acid-washed glass coverslips (for immunohistochemistry) or 250,000 in 4-well 35-mm glass bottom dishes (Greiner) (for live cell imaging). All coverslips for hippocampal neuron cultures were coated with 0.1 mg/ml poly-D-lysine (Sigma) 24 h before dissection. Neurons were kept in a 37°C, 5% CO2 incubator thereafter in neuronal culture media, with one complete media change 24 h after plating. All animal use was carried out in compliance with National Institutes of Health (NHS) guidelines, and protocols were approved by the Emory University’s Institutional Animal Care and Use Committee.
CAD cells (Qi et al., 1997) were cultured and maintained in DMEM/F12 (Invitrogen) media supplemented with 8% fetal bovine serum (FBS; Atlanta Biologicals) and 1% Penicillin/Streptomycin (Invitrogen). For imaging experiments, CAD cells reaching 90% confluence were resuspended and plated on detergent- and HCl-washed No. 1 glass coverslips coated with 20 µg/ml laminin (CAS#: 114956-81-9, MilliporeSigma, St. Louis, MO); 10,000 cells were plated on 25-mm round coverslips and allowed to spread for 1–3 h.
Transfection and constructs
CAD cells were transfected 18–24 h prior to imaging at approximately 70% confluence using Xtreme Gene transfection reagent (MilliporeSigma) and Opti-MEM (Invitrogen) according to manufacturer instructions. The GFP-LASP1 construct has been described previously and was generously provided by Joachim Kremerskothen (Stölting et al., 2012). To reduce LASP1 expression levels, we subcloned GFP-LASP1 into crippled CMV constructs provided by Richard Kahn at Emory University, courtesy of Wesley Sundquist (University of Utah) (Morita et al., 2012; Newman et al., 2016), and GFP-LASP1 was inserted via PCR. The GFP-LASP1ΔLIM, GFP-LASP1ΔNeb, and GFP-LASP1ΔSH3 constructs have been previously described (Stölting et al., 2012; Myers et al., 2020). Lifeact-mRuby (pN1-Lifeact-mRuby) was provided by Roland Wedlich-Soldner, Max-Planck Institute of Biochemistry.
To knock down LASP1 in hippocampal neurons, we used two shRNA constructs in the pSuper-mCherry backbone, shLASP1a and shLASP1b, respectively. The following oligos were annealed and ligated into pSuper.neo+mCherry (Myers et al., 2020) digested with BglII and XhoI: shLASP1a, 5′GATCCCCAAGGTGAACTGTCTGGATAAGTTCAAGAGACTTATCCAGACAGTTCACCTTTTTTTC (Forward)/5′ TCGAGAAAAAAAGGTGAACTGTCTGGATAAGTCTCTTGAACTTATCCAGACAGTTCACCTTGGG (Reverse) (previously published) (Myers et al., 2020) and shLASP1b, 5′GATCCCCCCATTAAGGAGATCGGTTATTCAAGAGATAACCGATCTCCTTAATGGTTTTTC (Forward)/5′TCGAGAAAAACCATTAAGGAGATCGGTTATCTCTTGAATAACCGATCTCCTTAATGGGGG (Reverse). The empty pSuper vector, shLASP1a, and shLASP1b were transfected into DIV2 rat hippocampal neurons using the OZBiosciences Neuron Magnetofection kit (OZBiosciences) per manufacturer instructions.
Immunohistochemistry
Neurons were fixed on DIV2 with 4% PFA/4% sucrose in phosphate-buffered saline (PBS) without magnesium or calcium for 30 min. CAD cells were fixed 1 h after plating on coverslips using 4% PFA in PBS without magnesium or calcium for 15 min. Cells were permeablized with 0.2% Triton X-100 in PBS for 10 min, then left at room temperature in blocking solution (4% bovine serum albumin [BSA], 1% goat serum, 0.1% Triton X-100) for 1 h. The cells were then incubated with primary antibodies at 4°C overnight, followed by three washes in PBS, and labeled with fluorescent secondary antibodies at room temperature for 1 h. The following antibodies were used: LASP1 (1:500 rabbit anti-LASP1, Proteintech 10515-1-AP), α-tubulin (1:1000 mouse anti-α-tubulin DM1A clone, Sigma T6199), and/or CP (1:500 rabbit anti-CP β2, generously provided by John Hammer of the National Heart Lung and Blood Institute, NIH, Bethesda, MD) (Schafer et al., 1994). Alexa Fluor–conjugated secondary antibodies (Invitrogen) were diluted at a concentration of 1:750 in PBS containing 2% goat serum. Alexa Fluor 568-Phalloidin (Invitrogen) was applied at a 1:100 dilution in PBS for 20 min. All labeled coverslips were mounted on glass slides using Fluoromount-G (SouthernBiotech) for imaging.
Western blots
Rat hippocampi were isolated at the indicated time points and homogenized through a syringe tip in lysis buffer containing 20 mM Tris (pH 8.0), 137 mM NaCl, 1% Triton X-100, 10% glycerol, 2 mM EDTA, and cOmplete protease inhibitor cocktail (Sigma) (Omotade et al., 2018). Cultured neurons and CAD cells were lysed directly in 1× Laemmli sample buffer, then boiled for 5 min, followed by vortexing for 5 min. Equal volumes of cell lysates or equal amounts of protein from tissue homogenates, as assessed by a Bradford assay, were loaded on mini-Protean 12% Tris-glycine acrylamide gels (Bio-Rad) and then transferred to nitrocellulose. Membranes were blocked with 5% milk in PBS-Tween for 1 h, then incubated overnight at 4°C with primary antibodies. The following antibodies were used for blotting: rabbit anti-LASP1 (1:1000, Proteintech), mouse anti-α-tubulin DM1A clone (1:5000, Sigma), rabbit anti-GFP (1:5000, Invitrogen), goat anti-mouse 647 (1:10,000, Invitrogen), or goat anti rabbit HRP (1:10,000, Invitrogen). Membranes were then incubated in Alexa Fluor 647–conjugated secondary antibody (Invitrogen) for 1 h and visualized by immunofluorescence.
Live imaging and drug treatments
CAD cells were plated on laminin-coated coverslips for 1 h in Krebs-Ringers solution (150 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM HEPES, pH 7.4) supplemented with 20% FBS, then mounted in live-cell chambers. To inhibit actin polymerization, the following drugs were used at the indicated final concentrations: 25 nM CytoD (Sigma), 100 nM LatA (EMD Millipore), 10 µM SMIFH2 (EMD Millipore), and 100 μM CK-666 (EMD Millipore). Cells were imaged for 5 min prior to drug addition, then for 10 min immediately following drug addition to monitor changes in localization. Primary hippocampal neurons were cultured and imaged in phenol red-free neuronal culture medium. For multiday imaging, cells were imaged every 24 h using software xyz location markers to locate the same neurons each time. For growth cone and branch tracking, cells were imaged every 10 min for 18 h in a stage top incubator (Tokai-hit) at 37°C in 5% CO2.
Actin barbed end labeling
CAD cells transfected with either CMVΔ4 GFP-LASP1 or GFP were plated on coverslips as described above. Actin barbed ends were labeled as described previously (Marsick and Letourneau, 2011; Gu et al., 2010). Briefly, cells were lightly fixed in permeabilization buffer (138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 1% BSA, 20 mM HEPES, pH 7.5, 1 mM ATP, 0.2 mg/ml saponin) with 0.05% PFA and 0.05% glutaraldehyde for 1 min. Next, plus ends were labeled with permeabilization buffer containing 0.45 µM rhodamine-nonmuscle actin (Cytoskeleton, APHR-A) for 2 min. Coverslips were then immediately fixed with 4% PFA and 0.05% glutaraldehyde in PHEM cytoskeletal stabilization buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, 0.12M sucrose, pH 7.0) for 30 min and immunostained for GFP (Alexa-488 rabbit anti-GFP, Invitrogen A-21311).
Microscopy
CAD cells were imaged on a Nikon Ti microscope (Nikon Instruments) with a Plan Apo 60× objective (NA 1.4) and a Hamamatsu CCD camera (Hammamatsu Photonics). Hippocampal neurons were imaged using a Nikon Eclipse Ti2 microscope with a Plan Apo 20× objective (NA 0.8) and Hamamatsu CCD. Drosophila embryos were imaged as z-stacks comprised of at least 42 optical sections (0.75 μm step-size) using a Nikon C2 laser-scanning confocal system with a Nikon Eclipse Ti2 microscope and a Plan Apo 20× objective (NA 0.8); maximum intensity projections. All microscopes were equipped with Nikon Elements software.
Analysis and statistics
Images were analyzed using the Imaris Neuroscience and Filament tracking modules (growth cone and axon branch tracking), Nikon Elements (kymograph analysis), and Fiji (axon length measurements using Simple Neurite Tracer) (ImageJ, NHS). Statistics were calculated using R. Line profiles were made in Fiji and normalized to the peak maximum intensity with the exception of the soluble GFP channel in Figure 5A, which was normalized to the cell center.
To analyze changes in the leading edge enrichment of LASP1 after actin drug treatments, we made four line profiles per cell at each of the indicated times. We then generated a ratio of the mean intensity of GFP-LASP1 within 1 µm of the leading edge, to the mean intensity of GFP-LASP1 in the lamella (between 4–5 microns from the leading edge). Thus, ratio values equal to 1 represent no enrichment at the leading edge relative to the lamella, while values greater than 1 represent enrichment at the leading edge.
Axon growth cone dynamics were measured by tracking growth cones with the Imaris filament tracer option. Growth cone speed was calculated for each frame based on the position change from the previous frame. The persistence index was measured as the physical displacement of the growth cone divided by the net displacement between the starting and ending positions. New branches were tracked using the same Imaris tracking function and were classified as “eliminated” when the branch fully retracted back to its site of origin for three frames in a row.
Drosophila embryo image stacks were made into maximum intensity projections and analyzed using Nikon Elements. Data were analyzed using a one-way ANOVA with a Tukey HSD post-hoc test, unless otherwise specified. Drosophila experiments were analyzed using a Student’s t test, or a Welch’s t test when variances were unequal. All graphs represent mean ± SEM unless otherwise specified.
Drosophila
Two crosses from three fly lines were used to produce the embryos measured in this paper. They are as follows: 1) UAStau:myc:GFP/cytubGAL80, egl GAL4 (a gift from Greg Bashaw, University of Pennsylvania, Philadelphia, PA); 2) UASdicr;UAS P(TRiP) LASP RNAi (Bloomington Drosophila Stock Center, Bloomington, IN); 3) UAS dicr on 2 (a gift from Ken Moberg, Emory University, Atlanta, GA). Embryos were collected and stained for BP102 (1:100 mouse anti-BP102, Developmental Studies Hybridoma Bank) and GFP (1:500 rabbit anti-GFP, Thermofisher A-11122) using a previously published immunocytochemistry protocol (Bashaw, 2010). Briefly, embryos were collected for 4 h on apple cider vinegar agar plates, then aged for 14 h overnight. Embryos were washed briefly in deionized water, then incubated in a 50% bleach solution for 5 min to remove the chorion. The embryos were then rinsed and fixed in 3.7% formaldehyde under a layer of heptane for 15 min. The fixative layer was then removed and replaced with pure methanol. The embryos were vortexed for 1 min, then rinsed three times with methanol and transferred to PBS. Permeabilization of the embryos was achieved by adding 1% TritonX in PBS (PBSTx) and incubating for 5 min on an orbital shaker. Embryos were then blocked using 2% normal goat serum diluted in PBSTx for 10 min. Primary antibody was diluted in blocking buffer and applied overnight at 4°C on an orbital shaker. After rinsing the embryos three times in PBSTx, secondary antibodies (goat anti-rabbit 488 and goat anti-mouse 546, Invitrogen) were applied to the embryos at 1:500 in blocking buffer for 2 h at room temperature. Embryos were rinsed three times in PBSTx, then sorted for fluorescence on an epifluorescence microscope (approximately 50% of the embryos do not inherit the UAStau:myc:GFP gene). Embryos were mounted on slides in Fluoromount-G mounting media (SouthernBiotech) and sealed with nail polish prior to confocal imaging. Image stacks were collapsed into maximum intensity projections, then file names were blinded for analysis. Each segment of the ventral nerve cord was analyzed for three developmental abnormalities: 1) commissures that do not complete the journey to their contralateral target, 2) individual axons that leave the main commissural tract, and 3) commissures with axons that have spread apart, such that gaps are visible within the commissure. The number of segments with each phenotype is expressed as a percentage of the total number of segments in each individual embryo. Note that many segments had more than one phenotype and were counted as such.
Supplementary Material
Acknowledgments
This research project was supported in part by research grants from the NHS to J.Q.Z. (GM083889), a Ruth L. Kirschstein National Research Service Award (NRSA) predoctoral fellowship to S.L.P. (NS101786), and a NRSA postdoctoral fellowship to K.R.M. (NS092342), as well as by the Emory University Integrated Cellular Imaging Microscopy Core of the Emory Neuroscience National Institute of Neurological Disorders and Stroke Core Facilities grant (5P30NS055077). We thank Jenny Mai for her help with image analysis, Stephanie Zimmer for assistance with plasmid development, and Gazal Arora for her help with early LASP1 staining. Special thanks to Kenneth Moberg and his laboratory for their help with the Drosophila work. The authors declare no competing interests.
Abbreviations used:
- BSA
bovine serum albumin
- CAD
Cath.a-differentiated
- CP
capping protein
- C-region
central region
- CytoD
Cytochalasin D
- F-actin
filamentous actin
- FBS
fetal bovine serum
- LASP1
LIM and SH3 protein 1
- LatA
Latrunculin A
- PBS
phosphate-buffered saline
- P-region
peripheral region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-06-0366) on September 30, 2020.
REFERENCES
- Akin O, Mullins RD (2008). Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133, 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ (2010). Visualizing axons in the drosophila central nervous system using immunohistochemistry and immunofluorescence. Cold Spring Harb Protoc 10, https//.org/10.1101/pdb.prot5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler, FB (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509–521. [DOI] [PubMed] [Google Scholar]
- Bliss KT, Chu M, Jones-Weinert CM, Gregorio CC (2013). Investigating lasp-2 in cell adhesion: new binding partners and roles in motility. Mol Biol Cell 24, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck KB, Zheng JQ (2002). Growth cone turning induced by direct local modification of microtubule dynamics. J Neurosci 22, 9358–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM (2015). Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol 17, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain B, Cotman C, Taylor D, Lynch G (1973). A quantitative electron microscopic study of synaptogenesis in the dentate gyrus of the rat. Brain Res 63, 195–204. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB (2011). The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol 3, https//.org/10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA (1988). The establishment of polarity by hippocampal neurons in culture. J Neurosci 8, 1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TA, Bashaw GJ (2010). Axon guidance at the midline: of mice and flies. Curr Opin Neurobiol 20, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM (1998). Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci 18, 8900–8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe DS, O’Donnell M, Bashaw GJ (2007). Cytoplasmic domain requirements for Frazzled-mediated attractive axon turning at the Drosophila midline. Development 134, 4325–4334. [DOI] [PubMed] [Google Scholar]
- Goddette DW, Frieden C (1986). Actin polymerization the mechanism of action of cytochalasin D. J Biol Chem 261, 15974–15980. [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC (2014). Actin dynamics in growth cone motility and navigation. J Neurochem 129, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves JT, Schafer ST, Gage FH (2016). Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell 167, 897–914. [DOI] [PubMed] [Google Scholar]
- Grunewald TGP, Kammerer U, Schulze E, Schindler D, Honig A, Zimmer M, Butt E (2006). Silencing of LASP-1 influences zyxin localization, inhibits proliferation and reduces migration in breast cancer cells. Exp Cell Res 312, 974–982. [DOI] [PubMed] [Google Scholar]
- Grunewald TGP, Kammerer U, Winkler C, Schindler D, Sickmann A, Honig A, Butt E (2007). Overexpression of LASP-1 mediates migration and proliferation of human ovarian cancer cells and influences zyxin localisation. Br J Cancer 96, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Lee CW, Fan Y, Komolos D, Tang X, Sun C, Yu K, Hartzell HC, Cheng G, Bamburg JR, Zheng JQ (2010). ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat Neurosci 13, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YK, Brieher WM, Mitchison TJ (2008). Dynamic stabilization of actin filaments. Proc Natl Acad Sci USA 105, 16531–16536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B (1992). Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci 12, 2685–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrard LE (1993). On the role of the hippocampus in learning and memory in the rat. Behav Neural Biol 60, 9–26. [DOI] [PubMed] [Google Scholar]
- Joo J, Lee S, Nah SS, Kim YO, Kim DS, Shim SH, Hwangbo Y, Kim HK, Kwon JT, Kim JW, et al. (2013). Lasp1 is down-regulated in NMDA receptor antagonist-treated mice and implicated in human schizophrenia susceptibility. J Psychiatr Res 47, 105–112. [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G (2006). Culturing hippocampal neurons. Nat Protoc 1, 2406–2415. [DOI] [PubMed] [Google Scholar]
- Kalil K, Dent EW (2005). Touch and go: Guidance cues signal to the growth cone cytoskeleton. Curr Opin Neurobiol 15, 521–526. [DOI] [PubMed] [Google Scholar]
- Keicher C, Gambaryan S, Schulze E, Marcus K, Meyer HE, Butt E (2004). Phosphorylation of mouse LASP-1 on threonine 156 by cAMP- and cGMP-dependent protein kinase. Biochem Biophys Res Commun 324, 308–316. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Tessier-Lavigne M (2011). Mechanisms and molecules of neuronal wiring: A primer. Cold Spring Harb Perspect Biol 3, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Vitriol EA, Shim S, Wise AL, Velayutham RP, Zheng JQ (2013). Dynamic localization of g-actin during membrane protrusion in neuronal motility. Curr Biol 23, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Zhou L, Kim J, Kalbfleisch S, Schöck F (2008). Lasp anchors the Drosophila male stem cell niche and mediates spermatid individualization. Mech Dev 125, 768–776. [DOI] [PubMed] [Google Scholar]
- Lin YH, Park ZY, Lin D, Brahmbhatt AA, Rio MC, Yates JR, Klemke RL (2004). Regulation of cell migration and survival by focal adhesion targeting of Lasp-1. J Cell Biol 165, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD (2011). Simple neurite tracer: Open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D (2009). The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol 10, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsick BM, Flynn KC, Santiago-Medina M, Bamburg JR, Letourneau PC (2010). Activation of ADF/cofilin mediates attractive growth cone turning toward nerve growth factor and netrin-1. Dev Neurobiol 70, 565–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsick BM, Letourneau PC (2011). Labeling F-actin barbed ends with rhodamine-actin in permeabilized neuronal growth cones. J Vis Exp 8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Arii J, Christensen D, Votteler J, Sundquist WI (2012). Attenuated protein expression vectors for use in siRNA rescue experiments. Biotechniques 0, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KR, Yu K, Kremerskothen J, Butt E, Zheng JQ (2020) The nebulin family LIM and SH3 proteins regulate postsynaptic development and function. J Neurosci 40, 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Suzuki H, Machida S, Suzuki J, Ohashi K, Jin M, Miyamoto S, Terasaki AG (2009). Contribution of the LIM domain and nebulin-repeats to the interaction of Lasp-2 with actin filaments and focal adhesions. PLoS One 4, e7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Terasaki AG, Suzuki H, Ohashi K, Miyamoto S (2006). Short-term retention of actin filament binding proteins on lamellipodial actin bundles. FEBS Lett 580, 3223–3228. [DOI] [PubMed] [Google Scholar]
- Newman LE, Schiavon C, Kahn RA (2016). Plasmids for variable expression of proteins targeted to the mitochondrial matrix or intermembrane space. Cell Logist 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Shi S, Zhang F, Liu R, Takagi Y, Bershadsky AD, Viasnoff V, Sellers JR (2020). The formin inhibitor, SMIFH2, inhibits members of the myosin superfamily. bioRxiv 10.1101/2020.08.30.274613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell MP, Bashaw GJ (2013a) Src inhibits midline axon crossing independent of frazzled/deleted in colorectal carcinoma (DCC) receptor tyrosine phosphorylation. J Neurosci 33, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell MP, Bashaw GJ (2013b). Distinct functional domains of the Abelson tyrosine kinase control axon guidance responses to Netrin and Slit to regulate the assembly of neural circuits. Development 140, 2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotade OF, Pollitt SL, Zheng JQ (2017) Actin-based growth cone motility and guidance. Mol Cell Neurosci 84, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotade OF, Rui Y, Lei W, Yu K, Hartzell HC, Fowler VM, Zheng JQ (2018). Tropomodulin isoform-specific regulation of dendrite development and synapse formation. J Neurosci 38, 10271–10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth MF, Cazes A, Butt E, Grunewald TGP (2015). An update on the LIM and SH3 domain protein 1 (LASP1): a versatile structural, signaling, and biomarker protein. Oncotarget 6, 26–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas CT, Bliss KT, Zieseniss A, Gregorio CC (2011). The Nebulin family: An actin support group. Trends Cell Biol 21, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GR, Anderson TR, Florens L, Gudas C, Magda G, Yates JR, Colman DR (2004). Actin-binding proteins in a postsynaptic preparation: Lasp-1 is a component of central nervous system synapses and dendritic spines. J Neurosci Res 78, 38–48. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD (2000). Molecular Mechanisms Controlling Actin Filament Dynamics in Nonmuscle Cells. Annu Rev Biophys Biomol Struct 29, 545–576. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA (2009). Actin, a central player in cell shape and movement. Science 326, 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Wang JK, McMillian M, Chikaraishi DM (1997). Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci 17, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago C, Labrador J-P, Bashaw GJ (2013). The homeodomain transcription factor Hb9 controls axon guidance in Drosophila through the regulation of Robo receptors. Cell Rep 18, 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Korshunova YO, Schroer TA, Cooper JA (1994). Differential localization and sequence analysis of capping protein β- subunit isoforms of vertebrates. J Cell Biol 127, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Moog-Lutz C, Régnier CH, Chenard MP, Boeuf H, Vonesch JL, Tomasetto C, Rio MC (1998). Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol Med 4, 675–687. [PMC free article] [PubMed] [Google Scholar]
- Skruber K, Warp PV, Shklyarov R, Thomas JD, Swanson MS, Henty-Ridilla JL, Read TA, Vitriol EA (2020). Arp2/3 and Mena/VASP require Profilin 1 for actin network assembly at the leading edge. Curr Biol 30, 2651-2664.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov MS, Cabral KA, Geller HM, Urbach JS (2014). The effects of confinement on neuronal growth cone morphology and velocity. Biomaterials 35, 6750–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stölting M, Wiesner C, van Vliet V, Butt E, Pavenstädt H, Linder S, Kremerskothen J (2012). Lasp-1 regulates podosome function. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JL, Merriman B, Cantor RM, Geschwind DH, Nelson SF (2007). High density SNP association study of a major autism linkage region on chromosome 17. Hum Mol Genet 16, 704–715. [DOI] [PubMed] [Google Scholar]
- Suter DM, Forscher P (2000). Substrate-cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. J Neurobiol 44, 97–113. [PubMed] [Google Scholar]
- Suyama R, Jenny A, Curado S, Pellis-van Berkel W, Ephrussi A (2009). The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo. Development 136, 95–105. [DOI] [PubMed] [Google Scholar]
- Terasaki AG, Suzuki H, Nishioka T, Matsuzawa E, Katsuki M, Nakagawa H, Miyamoto S, Ohashi K (2004). A novel LIM and SH3 protein (lasp-2) highly expressing in chicken brain. Biochem Biophys Res Commun 313, 48–54. [DOI] [PubMed] [Google Scholar]
- Tomasetto C, Régnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R, Basset P, Rio MC (1995a). Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics 28, 367–376a. [DOI] [PubMed] [Google Scholar]
- Tomasetto C, Moog-Lutz C, Régnier CH, Schreiber V, Basset P, Rio MC (1995b) Lasp-1 (MLN 50) defines a new LIM protein subfamily characterized by the association of LIM and SH3 domains. FEBS Lett 373, 245–249b. [DOI] [PubMed] [Google Scholar]
- Tyzio R, Represa A, Jorquera I, Ben-Ari Y, Gozlan H, Aniksztejn L (1999). The establishment of GABAergic and glutamatergic synapses on CA1 pyramidal neurons is sequential and correlates with the development of the apical dendrite. J Neurosci 19, 10372–10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol EA, McMillen LM, Kapustina M, Gomez SM, Vavylonis D, Zheng JQ (2015). Two functionally distinct sources of actin monomers supply the leading edge of lamellipodia. Cell Rep 11, 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol EA, Zheng JQ (2012). Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron 73, 1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman MA, Regnery AM, Becker T, Becker CG, Halloran MC (2007). Semaphorin3D regulates axon-axon interactions by modulating levels of L1 cell adhesion molecule. J Neurosci 27, 9653–9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE (2012). Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Liu J, Zhou R, Lin C, Wu K, Yang S, Yang S, Zhou J, Xu L, Wang H, Zhao L (2020). LASP1 interacts with N-WASP to activate the Arp2/3 complex and facilitate colorectal cancer metastasis by increasing tumour budding and worsening the pattern of invasion. Oncogene 5743–5755. [DOI] [PubMed] [Google Scholar]
- Yang Q, Zhang XF, Pollard TD, Forscher P (2012). Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J Cell Biol 197, 939–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.