Abstract
SLC45A2 encodes a putative transporter expressed primarily in pigment cells. SLC45A2 mutations cause oculocutaneous albinism type 4 (OCA4) and polymorphisms are associated with pigmentation variation, but the localization, function, and regulation of SLC45A2 and its variants remain unknown. We show that SLC45A2 localizes to a cohort of mature melanosomes that only partially overlaps with the cohort expressing the chloride channel OCA2. SLC45A2 expressed ectopically in HeLa cells localizes to lysosomes and raises lysosomal pH, suggesting that in melanocytes SLC45A2 expression, like OCA2 expression, results in the deacidification of maturing melanosomes to support melanin synthesis. Interestingly, OCA2 overexpression compensates for loss of SLC45A2 expression in pigmentation. Analyses of SLC45A2- and OCA2-deficient mouse melanocytes show that SLC45A2 likely functions later during melanosome maturation than OCA2. Moreover, the light skin-associated SLC45A2 allelic F374 variant restores only moderate pigmentation to SLC45A2-deficient melanocytes due to rapid proteasome-dependent degradation resulting in lower protein expression levels in melanosomes than the dark skin-associated allelic L374 variant. Our data suggest that SLC45A2 maintains melanosome neutralization that is initially orchestrated by transient OCA2 activity to support melanization at late stages of melanosome maturation, and that a common allelic variant imparts reduced activity due to protein instability.
INTRODUCTION
Melanins are the main source of pigmentation in the skin, hair, and eyes of mammals and other vertebrates. In humans, melanins serve as a barrier to the harmful effects of ultraviolet radiation and play an important role in the development and functioning of the retina (d’Ischia et al., 2015). Melanin synthesis takes place in skin and eye melanocytes and in ocular pigment epithelia within specialized organelles called melanosomes (Seiji et al., 1963; Marks and Seabra, 2001; Hearing, 2005). Heritable defects in melanin synthesis underlie the various forms of oculocutaneous albinism (OCA), characterized by impaired vision and increased susceptibility to skin and ocular cancers (Montoliu et al., 2014). To date, nonsyndromic OCA has been linked to inactivating mutations in eight different genes (Montoliu et al., 2014; Pennamen et al., 2020). Sequence variation at the loci for some of these genes has been linked to variability in skin, hair, and eye color among humans (Lamason et al., 2005; Stokowski et al., 2007; Branicki et al., 2008; Han et al., 2008; Liu et al., 2015; Crawford et al., 2017; Martin et al., 2017; Adhikari et al., 2019). Nevertheless, the molecular function of the majority of the OCA genes has not yet been fully characterized.
OCA type 4 (OMIM #606574) represents 3–12% of total OCA patients in population studies (Gronskov et al., 2009; Wei et al., 2010, 2015; Mauri et al., 2017; Lasseaux et al., 2018) and is due to mutations in the SLC45A2 gene encoding the putative transmembrane transporter SLC45A2 (aka membrane-associated transporter protein, MATP, or antigen isolated from immuno-selected melanoma-1, AIM1) (Newton et al., 2001). Mutations in the homologous gene underlie pigment dilution in a number of vertebrate species, including gorilla, several breeds of dog, tigers, horses, mice, shrew, chickens, pigeons, quail, frogs, fish, and perhaps cattle (Fukamachi et al., 2001; Newton et al., 2001; Mariat et al., 2003; Gunnarsson et al., 2007; Minvielle et al., 2009; Tsuboi et al., 2009; Tsetskhladze et al., 2012; Dooley et al., 2013; Prado-Martinez et al., 2013; Xu et al., 2013; Domyan et al., 2014; Winkler et al., 2014; Wijesena and Schmutz, 2015; Caduff et al., 2017; Rothammer et al., 2017; DeLay et al., 2018), and polymorphisms at the SLC45A2 locus are associated with skin tone differences and skin aging in several human population studies (Yuasa et al., 2006; Soejima and Koda, 2007; Stokowski et al., 2007; Branicki et al., 2008; Han et al., 2008; Cerqueira et al., 2014; Lopez et al., 2014; Liu et al., 2015; Jonnalagadda et al., 2016; Fracasso et al., 2017; Law et al., 2017; Adhikari et al., 2019). OCA4 patients have very low levels of pigmentation and phenotypically resemble OCA2 patients who lack the melanosomal chloride channel, OCA2 (Bellono et al., 2014), suggesting that SLC45A2 plays an important role in melanogenesis (Montoliu et al., 2014). Moreover, primary melanocytes from mice carrying the inactivating underwhite (uw) mutation of Slc45a2 (Newton et al., 2001; Du and Fisher, 2002) are severely hypopigmented (Costin et al., 2003), indicating a melanocyte-intrinsic defect; this would be consistent with the known restriction of SLC45A2 expression to pigment cells and a few other cell types (Harada et al., 2001; Baxter and Pavan, 2002; Loftus et al., 2002; Bin et al., 2015). However, while the 12-transmembrane domain SLC45A2 protein bears weak homology to sucrose transporters in plants and Drosophila (Lemoine, 2000; Newton et al., 2001; Meyer et al., 2011), its function in melanocytes is not understood.
When expressed in yeast, mouse SLC45A2 functions at the plasma membrane as an acid-dependent importer of sugars (sucrose, glucose, or fructose) into the cytosol (Bartölke et al., 2014), suggesting that if SLC45A2 localized to acidic organelles it might facilitate export of a sugar and protons from the lumen to the cytosol. Neutralization of acidic early stage melanosomes is a critical process for melanogenesis (Raposo et al., 2001; Bellono et al., 2014), as the key enzyme in melanogenesis, Tyrosinase (TYR), is minimally active at pH < 6 (Ancans et al., 2001; Halaban et al., 2002). Consistent with a function in proton export and neutralization of acidic organelles, pigmentation of a zebrafish Slc45a2 mutant was rescued upon inhibition of endolysosomal and melanosomal acidification by treatment with bafilomycin A1 (BafA1) or by knockdown of the atp6v1 subunit of the vacuolar ATPase (vATPase; Dooley et al., 2013), and knockdown of SLC45A2 in a pigmented melanoma cell line was reported to result in increased acidification of early stage melanosomes (Bin et al., 2015). However, despite limited evidence for SLC45A2 on melanosomes (Bin et al., 2015), the localization of SLC45A2 within melanocytes has not yet been firmly established and it is not clear whether the effects of SLC45A2 mutants reflect a direct effect of SLC45A2 function on the lumenal environment of melanosomes or an indirect effect due to impaired function of other organelles during melanosome maturation, as appears to be the case for the endolysosomal transporter MFSD12 (Crawford et al., 2017). Moreover, if SLC45A2 indeed functions to neutralize melanosome pH, its function must be coordinated with that of OCA2, a major regulator of melanosomal pH (Bellono et al., 2014). While mice lacking expression of either SLC45A2 or OCA2 each have dramatic coat color dilution (Dickie, 1964; Sweet et al., 1998), mice with hypomorphic mutations in both Slc45a2 and Oca2 are more severely hypopigmented than either mutant alone (Lehman et al., 2000), suggesting that the encoded proteins have distinct functions.
Differences in skin and hair pigmentation among European, Chinese, South American, and South Asian human populations have been ascribed to a single genetic variant in SLC45A2 associated with the SNP rs16891982 (c.1122G>C). This results in a missense mutation p.Leu374Phe (ClinVar ID: 194990) and is correlated with lighter skin, hair, and eyes (Yuasa et al., 2006; Soejima and Koda, 2007; Stokowski et al., 2007; Branicki et al., 2008; Han et al., 2008; Cerqueira et al., 2014; Lopez et al., 2014; Liu et al., 2015; Jonnalagadda et al., 2016; Adhikari et al., 2019). The SLC45A2-F374 variant is nearly fixed in light-skinned human populations (Soejima and Koda, 2007). Residue 374 lies within the eighth predicted transmembrane domain of SLC45A2 (Newton et al., 2001). When introduced into a homologous plant sucrose transporter, the amino acid corresponding to SLC45A2-F374 resulted in a 90% decrease in transporter activity without influencing the affinity for substrate (Reinders and Ward, 2015), and slc45a2 with an introduced F374 mutation was unable to rescue pigmentation in a slc45a2 mutant zebrafish (Tsetskhladze et al., 2012). However, the mechanism underlying the decreased activity of the SLC45A2-F374 variant is not understood.
Analyses of SLC45A2 localization and function in pigment cells have been hindered by the lack of suitable specific antibodies. Here we assess the localization and function of SLC45A2-L374 and -F374 variants in melanosome biogenesis by analyzing epitope-tagged human SLC45A2 expressed in wild-type (WT) melanocytes, Slc45a2-deficient melanocytes from uw mice, and HeLa cells, and by comparing the phenotypes of uw and OCA2-deficient pink-eyed dilute melanocytes. We show that expression of SLC45A2, like that of OCA2, increases the pH of lysosomes and/or late endosomes when expressed ectopically in HeLa cells and that in melanocytes SLC45A2 localizes to melanosomes, where it likely neutralizes the melanosome lumen. We also show that OCA2 and SLC45A2 localize to distinct subsets of melanosomes and that the F374 variant accelerates SLC45A2 degradation but does not alter its localization. Our data indicate that melanosome neutralization is a critical process for the maintenance of eumelanin pigmentation that is regulated at more than one step during melanosome maturation.
RESULTS
Functionality of HA-SLC45A2
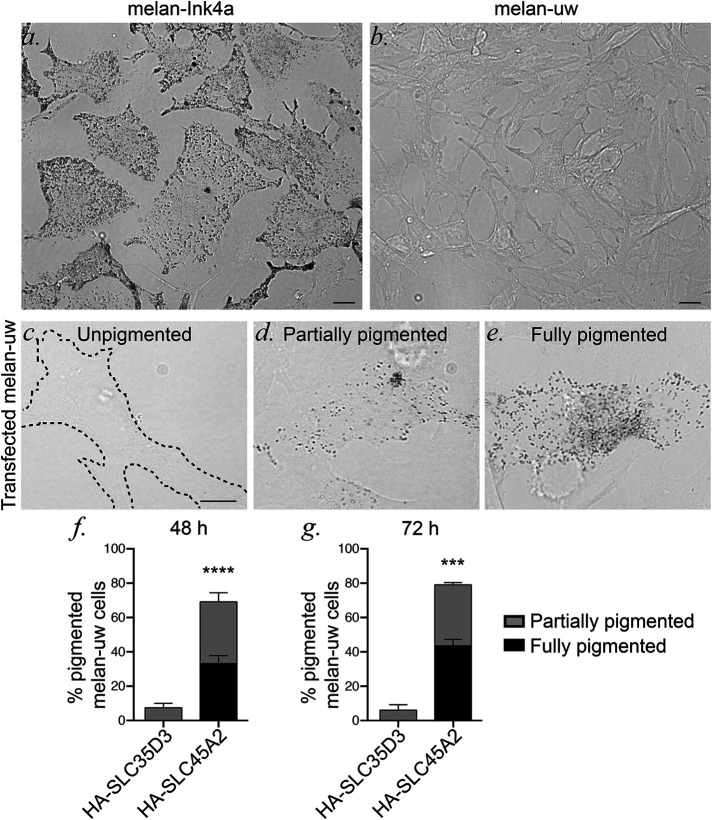
Available antibodies to SLC45A2 were not suitable in our hands for immunolocalization analyses. Therefore, we used epitope-tagging to investigate SLC45A2 localization in melanocytes. Human SLC45A2 was tagged at the N- or C-terminus with either the HA11 epitope tag or EGFP and cloned into a retroviral vector. To determine whether the tagged transgenes were functional, they were expressed by recombinant retroviral infection in Slc45a2-deficient melan-uw cells from uw mice (Dickie, 1964). The Slc45a2uw allele encodes a nonfunctional truncated SLC45A2 protein and an undetectable transcript (Newton et al., 2001; Du and Fisher, 2002). Consequently, uw mice (Dickie, 1964; Sweet et al., 1998; Lehman et al., 2000) and primary melanocytes derived from them (Costin et al., 2003) are severely hypopigmented. Likewise, melan-uw cells are very pale compared with WT immortalized melanocytes (melan-Ink4a) from C57BL/6-Ink4a–/– mice (Sviderskaya et al, 2002) (Figure 1, a and b). While expression of the EGFP-tagged proteins failed to consistently restore pigmentation in melan-uw cells (data not shown), expression of the N-terminally HA-tagged human SLC45A2 (HA-SLC45A2) restored partial or full pigmentation in a substantial fraction of cells by 48–72 h postinfection, whereas pigmentation was not restored by expression of a similarly tagged unrelated polytopic protein—the putative sugar-nucleotide transporter, SLC35D3, which plays no role in pigmentation (Chintala et al., 2007) (Figure 1, c–g). Additionally, quantitative analysis of melanin content in melan-uw cells stably expressing HA-SLC45A2 showed pigmentation at levels comparable to those of melan-Ink4a cells (see Figure 5f). These data indicate that HA-SLC45A2 is fully functional in melanocytes and therefore validate its use in defining the localization and biosynthesis of SLC45A2.
FIGURE 1:
HA-SLC45A2 functionally restores pigmentation in Slc45a2-deficient uw melanocytes. Widefield microscopy with bright field illumination of melan-Ink4a (a) and melan-uw (b) cells to visualize pigmentation. Scale bar, 10 μm. Mouse melan-uw cells were transiently transfected with plasmids encoding human SLC45A2 with an N-terminal HA epitope (HA-SLC45A2) or with a comparably HA-tagged CTRL polytopic protein, SLC35D3 (HA-SLC35D3). At 48–72 h after transfection, HA-expressing cells were scored for their level of pigmentation. (c–e) Examples of unpigmented (c, like untransfected or CTRL-transfected cells), partially pigmented (d) and fully pigmented (e) cells. Scale bar, 10 μm. (f, g) Quantification of the percentage of HA-expressing cells with partial (gray) or full (black) pigmentation at 48 (f) or 72 (g) h after transfection. Note that all cells except melan-Ink4a were treated with 200 pM cholera toxin (see Materials and Methods). Data represent mean ± SEM from three independent experiments and the following total sample sizes: 234 (HA-SLC45A2) and 109 (HA-SLC35D3) at 48 h; 173 (HA-SLC45A2) and 108 (HA-SLC35D3) at 72 h. ***p < 0.001; ****p < 0.0001 by unpaired two-tailed t test.
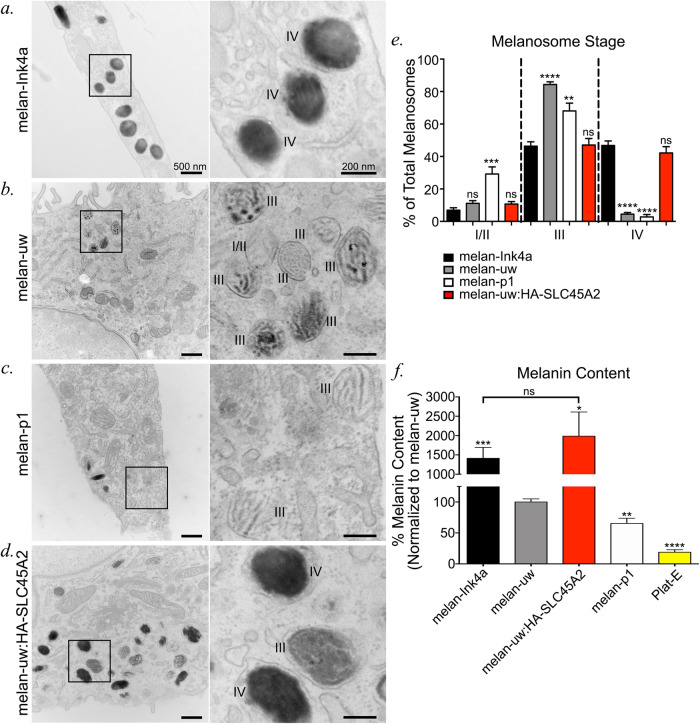
FIGURE 5:
SLC45A2 is required for melanosomes to progress from stage III to stage IV. (a–d) Melan-Ink4a (a), melan-uw (b), melan-p1 (c), and melan-uw cells stably expressing HA-SLC45A2 (melan-uw:HA-SLC45A2; d) were fixed and analyzed by conventional transmission electron microscopy. Insets of boxed regions on the left were magnified 4.5× on the right. I/II, stage I/II melanosomes; III, stage III melanosomes; IV, stage IV melanosomes. Scale bars, 500 nm (left) and 200 nm (right). (e) The number of stage I/II, stage III, and stage IV melanosomes per cell was quantified for each of the cell lines and is shown as a percentage of total counted melanosomes per cell. For each stage, data represent the mean ± SEM from the following number of independent experiments, cells, and melanosomes: melan-Ink4a (2, 54, 717), melan-uw (3, 49, 1292), melan-p1 (2, 27, 626), and melan-uw:HA-SLC45A2 (3, 76, 1927). Statistical analyses were performed by Welch’s ANOVA with multiple comparisons to melan-Ink4a for each stage and Dunnett T3 test for multiple comparisons. (f) Melanin content in cell lysates was determined by spectrometry relative to total protein content. Plat-E cells are included as a nonpigmented cell CTRL. The data are presented as a percentage normalized to melanin content in melan-uw samples ± SEM and represent at least three independent experiments performed in duplicate or triplicate. Statistical significance was determined by Welch’s ANOVA with Dunnett T3 test for multiple comparisons. Except where indicated, statistical comparisons were made relative to melan-uw. Note that all cells except melan-Ink4a and Plat-E were treated with 200 pM cholera toxin. **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, no significant difference.
HA-SLC45A2 localizes to the limiting membrane of pigmented melanosomes and is partially enriched in a membrane subdomain
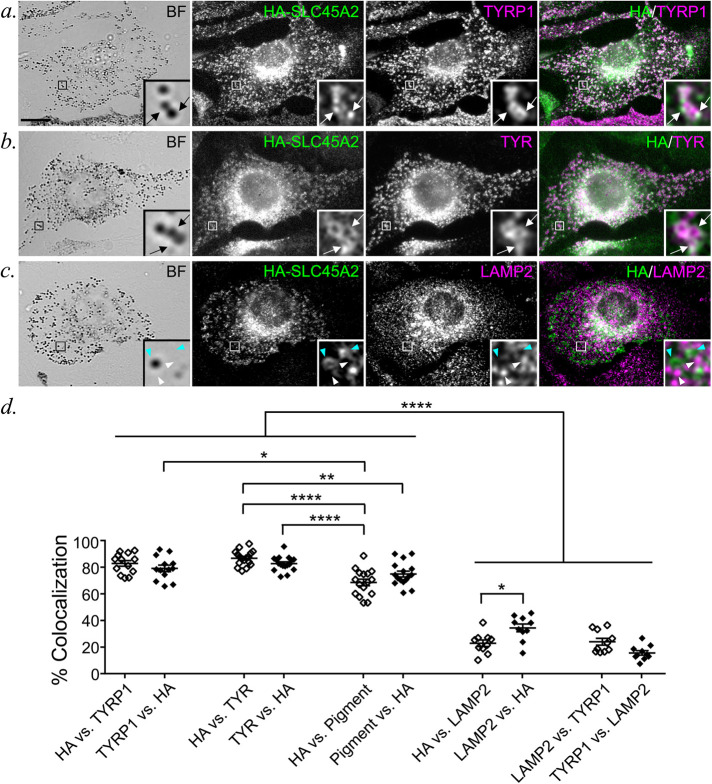
To define SLC45A2 localization in melanocytes, HA-SLC45A2 was expressed either stably from recombinant retroviruses in melan-uw cells or by transient transfection in WT melan-Ink4a melanocytes. Cells were then fixed and processed for immunofluorescence microscopy (IFM) with image deconvolution (dIFM) using antibodies to the HA tag and to endogenous markers. In Slc45a2-deficient melan-uw cells, HA-SLC45A2 localized primarily to pigment granules (Figure 2, a, b, and d) often in a ring pattern around the granule (Figure 2b), in two to three puncta associated with the granule exterior (Figure 2a), or both (Figure 2c). The distinct patterns did not correlate with SLC45A2 expression levels. The melanosomes labeled by HA-SLC45A2 overlapped with those harboring the melanosomal enzymes TYR and TYR-related protein-1 (TYRP1) (Figure 2, a, b, and d; overlap was 82.7 ± 1.3% with TYR and 79.2 ± 2.5% with TYRP1). By contrast, HA-SLC45A2-labeled structures overlapped poorly with LAMP2, a membrane protein of lysosomes but not melanosomes, which are distinct organelles in eumelanin-generating melanocytes (Raposo et al., 2001) (Figure 2, c and d; overlap was 34.4% ± 2.9% with LAMP2). Essentially identical results were observed for HA-SLC45A2 transiently expressed in melan-Ink4a cells (Supplemental Figure EV1), which express endogenous SLC45A2. The degree of HA-SLC45A2 labeling on punctate subdomains versus the melanosome limiting membrane was similar in transiently transduced melan-Ink4a and stably transduced melan-uw cells. Together, these data indicate that SLC45A2 localizes to mature melanosomes, where it is often enriched in subdomains associated with the limiting membrane.
FIGURE 2:
HA-SLC45A2 localizes to melanosomes when expressed in SLC45A2-deficient melan-uw melanocytes. (a–c) Melan-uw cells stably expressing HA-tagged SLC45A2 were fixed; labeled for HA (green) and for TYRP1 (a), TYR (b), or LAMP2 (c) (magenta); and analyzed by dIFM and BF to visualize melanin. Insets of boxed regions are magnified 7.5×. Scale bar, 10 μm. SLC45A2 that localized to TYRP1- or TYR-labeled compartments (white arrows) or SLC45A2 (cyan arrowheads) and LAMP2 (white arrowheads) on separate compartments are indicated. (d) The percentage of compartments with both markers in a–c was quantified by manual counting of at least 13 cells each from three separate experiments. Colocalization is represented as mean ± SEM of label 1 vs. label 2, in which the number of compartments containing both label 1 and label 2 is presented as a percentage of the total number of compartments containing label 2. The percentage of colocalization between TYRP1 and LAMP2 was quantified as a negative CTRL. Data are from three independent experiments with the following total sample sizes: 13 (TYRP1 vs. HA; HA vs. TYRP1; LAMP2 vs. HA; HA vs. TYRP1; TYRP1 vs. LAMP2; LAMP2 vs. TYRP1) and 17 (TYR vs. HA; HA vs. TYR; HA vs. Pigment; Pigment vs. HA). Statistical significance was determined using one-way ANOVA with Tukey post-hoc test for multiple comparisons; only significant differences are indicated. *p < 0.05; **p < 0.01; ****p < 0.0001.
SLC45A2 localizes to lysosomes and regulates lysosomal pH when ectopically expressed in HeLa cells
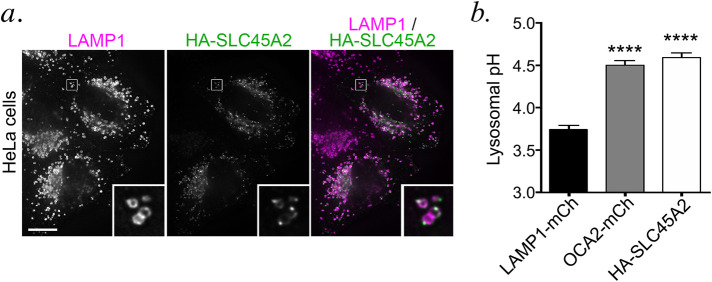
When expressed in non-melanocytic cells, many melanosomal proteins localize to late endosomes and lysosomes (Bouchard et al., 1989; Vijayasaradhi et al., 1995; Calvo et al., 1999; Simmen et al., 1999; Berson et al., 2001; Piccirillo et al., 2006; Sitaram et al., 2009; Ambrosio et al., 2016; Bellono et al., 2016). Indeed, when expressed in HeLa cells and analyzed by dIFM, HA-SLC45A2 was detected on LAMP1-containing lysosomes in distinct punctate microdomains associated with the limiting membrane (Figure 3a), similar to the distribution in some melanosomes in melanocytes. This indicates that SLC45A2 localizes to lysosomes when expressed ectopically in HeLa cells and retains its ability to accumulate in a membrane subdomain.
FIGURE 3:
HA-SLC45A2 expressed in HeLa cells localizes to lysosomes and neutralizes lysosomal pH. (a). HeLa cells transiently transfected with HA-SLC45A2 were fixed, labeled for HA (green) and the lysosomal protein LAMP1 (magenta), and analyzed by dIFM. Merged image is at right. Insets of boxes are magnified fivefold. Scale bar, 10 μm. (b) HeLa cells transiently transfected with LAMP1-mCherry alone, mCherry-OCA2, or HA-SLC45A2 and LAMP1-mCherry were incubated with LysoSensor DND-160 and analyzed by fluorescence microscopy with excitation wavelength 405. The W1/W2 emission ratio for mCherry-labeled endolysosomes was calculated and compared with that from a standard curve to define pH as described in Materials and Methods. Data represent mean ± SEM from three independent experiments with the following total sample sizes: 319 LAMP1-mCherry+ endolysosomes, 391 OCA2-mCherry+ endolysosomes, and 299 HA-SLC45A2/LAMP1-mCherry+ endolysosomes. Statistical significance was determined using Welch’s ANOVA with Games-Howell test for multiple comparisons; only significant differences are indicated. ****p < 0.0001.
SLC45A2 was predicted to be a transmembrane transporter that influences melanosome pH (Newton et al., 2001; Bin et al., 2015). We have been unable to directly assess melanosomal pH in melanocytes using common cellular probes for lysosomal pH such as Lysotracker and LysoSensor because in our hands melanocytes fail to accumulate these probes intracellularly. However, we previously showed that the OCA2 chloride channel expressed in HeLa cells localizes to lysosomes and neutralizes their lumenal pH, paralleling its role in neutralizing the lumenal pH of melanosomes to enhance TYR activity and melanogenesis in melanocytes (Bellono et al., 2014). We therefore tested whether SLC45A2 expression affects lysosomal acidification in HeLa cells using the pH-sensitive ratiometric dye LysoSensor DND-160. HeLa cells were transfected with LAMP1-mCherry alone as a negative control (CTRL), mCherry-OCA2 as a positive CTRL, or LAMP1-mCherry and HA-SLC45A2 together to identify HA-SLC45A2-expressing cells by live imaging (cotransfection efficiency measured by immunostaining was >90%). Cells preincubated with LysoSensor DND-160 were imaged at two emission wavelengths, and an average emission ratio was calculated for individual mCherry-LAMP1-positive compartments (see Materials and Methods). The corresponding pH for the average emission ratio was determined using a pH curve generated in each experiment by incubation in buffers with different pH values. Our results show that the average pH of lysosomes containing HA-SLC45A2 was nearly one pH unit higher than the pH of CTRL cells and similar to the pH of mCherry-OCA2-containing lysosomes (Figure 3b). These data indicate that, like OCA2, SLC45A2 can function to raise organellar pH and suggest that in melanocytes SLC45A2 functions physiologically to raise melanosomal pH.
SLC45A2 and OCA2 function at distinct stages of melanosome maturation
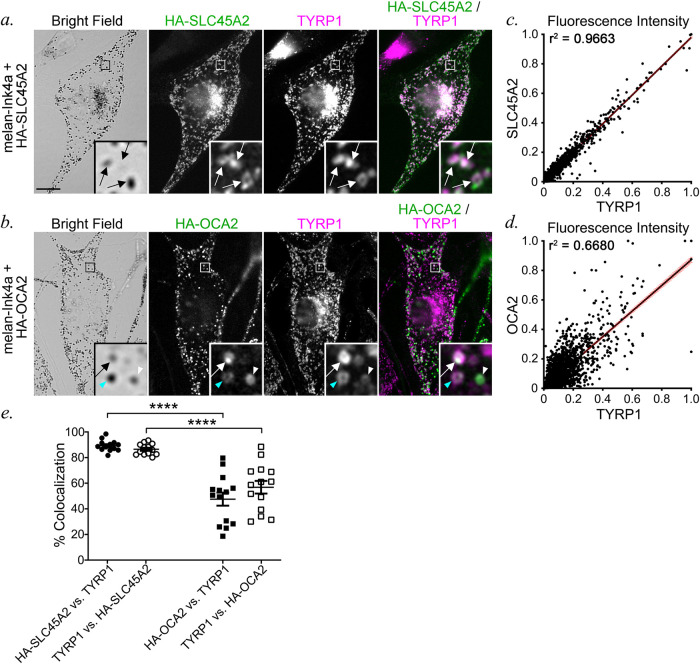
The similar function of SLC45A2 and OCA2 in neutralizing organellar pH led us to ask whether these transporters function at the same or distinct stages of melanosome maturation. We first used dIFM to compare the distribution of HA-SLC45A2 and HA-OCA2 to melanosomes labeled by the mature melanosome marker TYRP1 in transiently transfected melan-Ink4a cells. HA-SLC45A2 and TYRP1 were largely present in the same compartments (86.58 ± 1.25% of HA-SLC45A2 in TYRP1-containing structures and 89.17 ± 1.32% of TYRP1 in HA-SLC45A2-containing structures; Figure 4, a and e), and the fluorescence signal intensity within positive compartments was highly correlated (Figure 4c). By contrast, although TYRP1 and OCA2 each primarily localize to subsets of pigment granules (Vijayasaradhi et al., 1991, 1995; Raposo et al., 2001; Sitaram et al., 2009, 2012), they label only partially overlapping populations (58.57 ± 4.87% of HA-OCA2 in TYRP1-containing compartments and 48.66 ± 5.32% of TYRP1 in OCA2-containing compartments; Figure 4, b and e). Moreover, within those overlapping compartments, the fluorescence signal intensities of HA-OCA2 and TYRP1 over background correlated substantially less well than those of HA-SLC45A2 and TYRP1 (Figure 4d). These data suggest that OCA2 and SLC45A2 are enriched in melanosomes of distinct maturation stages, with SLC45A2 present in largely the same subset of mature melanosomes as TYRP1, while OCA2 occupies a partially different subset of melanosomes.
FIGURE 4:
SLC45A2 and OCA2 occupy partially distinct melanosome subsets. (a, b) Melan-Ink4a cells transiently expressing either HA-SLC45A2 (a) or HA-OCA2 (b) were fixed, labeled for HA (green) and TYRP1 (magenta), and analyzed by dIFM and BF. Insets of boxed regions are magnified 7.5×. Scale bar, 10 μm. White arrows, colocalization of HA-SLC45A2 or HA-OCA2 to TYRP1-containing compartments at comparable relative intensities; white arrowhead, compartments with high HA-OCA2 and low TYRP1; cyan arrowhead, compartments with low HA-OCA2 and high TYRP1. (c, d) Quantification of the object-based fluorescence intensity of HA-SLC45A2 vs. TYRP1 (c) or HA-OCA2 vs. TYRP1 (d) in 10 cells each from three independent experiments. Correlation coefficients (r2) of fluorescent intensities for each pair of markers are indicated. (e) Manual quantification (mean ± SEM) of the percentage of colocalization between TYRP1 and either HA-SLC45A2 or HA-OCA2 and TYRP1, shown as a percentage of total TYRP1-, SLC45A2-, or OCA2-containing compartments. Values are presented as label 1 vs. label 2, in which compartments containing both label 1 and label 2 are indicated as a percentage of total compartments containing label 2. Data are quantified from three independent experiments with the following total sample sizes: 13 (HA-SLC45A2) and 14 (HA-OCA2). Statistical significance was determined using Welch’s ANOVA with Dunnett T3 test for multiple comparisons; only significant differences are indicated. ****p < 0.0001.
The distinct localization patterns for OCA2 and SLC45A2 suggest that they might regulate the lumenal pH and pigmentation of melanosomes at different maturation stages. We therefore tested for potential differences in melanosome pigmentation in melan-uw cells and immortalized melan-p1 melanocytes from OCA2-deficient pink-eyed dilute mice. Electron microscopy can be used to define four stages of melanosome maturation based on morphology and content of melanin pigments (Seiji et al., 1963). Stages I and II lack pigment, but are distinguished by the presence of disorganized intralumenal fibers (stage I) or well-organized fibrils in parallel intralumenal sheets (stage II). Stage III is defined by the appearance of pigment on the fibrils and stage IV by pigment throughout the organelle. Electron microscopy analyses showed that relative to either WT melan-Ink4a or rescued melan-uw cells stably expressing HA-SLC45A2 (melan-uw:HA-SLC45A2), both melan-uw and melan-p1 cells harbored melanosomes of similar size and number, but with more stage III melanosomes and fewer stage IV melanosomes (Figure 5, a–e). However, only melan-p1 cells contained significantly more stage I/II melanosomes than the CTRLs (Figure 5e). Moreover, although we could not quantify it, the amount of pigment on the fibrils in stage III melanosomes in melan-p1 cells consistently appeared to be lower than in melan-uw cells (Figure 5, b vs. c). Accordingly, quantitative melanin content assays showed that while both melan-uw and melan-p1 had dramatically lower melanin content than melan-Ink4a or melan-uw:HA-SLC45A2 cells, the melanin content of melan-p1 cells was significantly lower than that of melan-uw (Figure 5f). These data indicate that loss of SLC45A2 results in a milder phenotype than loss of OCA2. Taken together, with the data showing that SLC45A2 and OCA2 are found on different subsets of melanosomes (Figure 4), the most likely scenario is that OCA2 is required at an earlier stage of melanization than SLC45A2 during melanosome maturation.
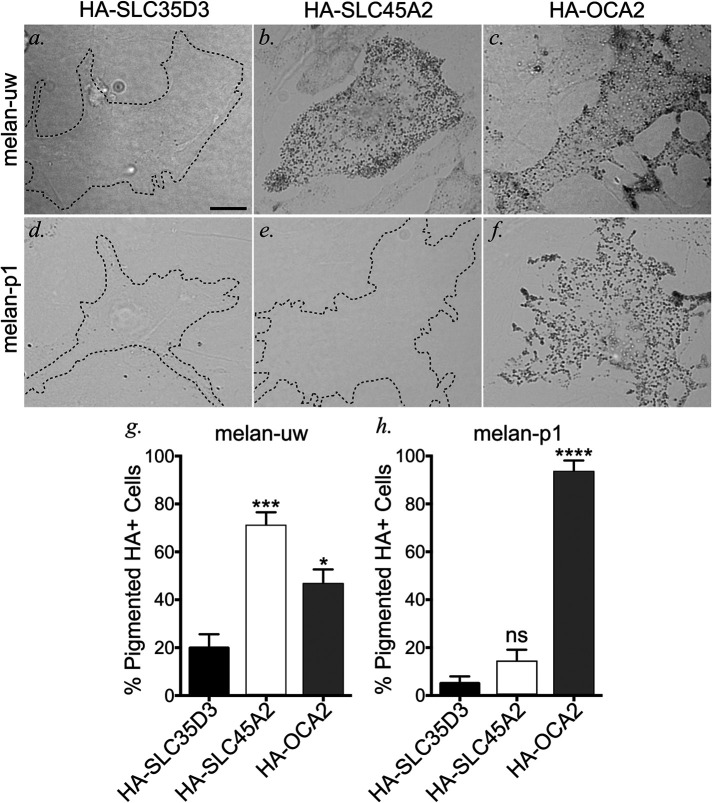
OCA2 overexpression partially compensates for loss of SLC45A2 expression
Given that both OCA2 and SLC45A2 are capable of raising lysosomal pH when expressed in HeLa cells, we tested whether overexpression of either protein could compensate for the loss of the other. HA-SLC45A2, HA-OCA2, or HA-SLC35D3 as a negative CTRL (see Figure 1) were transiently expressed by transfection in either Slc45a2-deficient melan-uw melanocytes or Oca2-deficient melan-p1 melanocytes, and the fraction of HA-positive cells that were pigmented after 3 d was quantified. As expected, very few melan-uw cells expressing HA-SLC35D3 were pigmented above background (19.88 ± 5.76%), whereas most HA-SLC45A2 expressing cells were pigmented (70.7 ± 5.85%; Figure 6, a, b, and g). Surprisingly, a large fraction of HA-OCA2-expressing melan-uw cells were also pigmented (46.37 ± 6.29%; Figure 6, c and g), suggesting that OCA2 overexpression can compensate for the loss of SLC45A2. By contrast, whereas HA-OCA2 expression restored pigmentation to melan-p1 cells (93.17 ± 4.97%), HA-SLC45A2 expression was as ineffective at restoring pigmentation in melan-p1 cells as the negative CTRL HA-SLC35D3 (Figure 6, d–f, h). Thus, overexpression of SLC45A2 cannot compensate for the loss of OCA2. These data are consistent with a model in which sequential function of OCA2 and then SLC45A2 is required to modulate and maintain a nearly neutral pH in melanosomes.
FIGURE 6:
OCA2 overexpression compensates for loss of SLC45A2-dependent melanosome neutralization in melanocyte pigmentation. SLC45A2-deficient melan-uw cells or OCA2-deficient melan-p1 cells were transiently transfected with negative CTRL HA-SLC35D3 (a, d), HA-SLC45A2 (b, e), or HA-OCA2 (c, f), fixed and labeled for HA (not shown). BF was used to visualize pigmented melanosomes within HA-positive cells. Scale, 10 μm. (g, h) The number of HA-containing pigmented cells was quantified and is depicted as a mean percentage of total HA-containing (HA+) cells ± SEM. Quantification is from the following independent experiments and total sample sizes: melan-uw + HA-SLC35D3 (3, 67), melan-uw + HA-SLC45A2 (9, 320), melan-uw + HA-OCA2 (9, 308), melan-p1 + HA-SLC35D3 (3, 62), melan-p1 + HA-SLC45A2 (4, 137), and melan-p1 + HA-OCA2 (4, 90). Statistical significance was determined using one-way ANOVA with Holm-Sidak’s test for multiple comparisons. *p < 0.05; ***p < 0.001; ****p < 0.0001; ns, not significant.
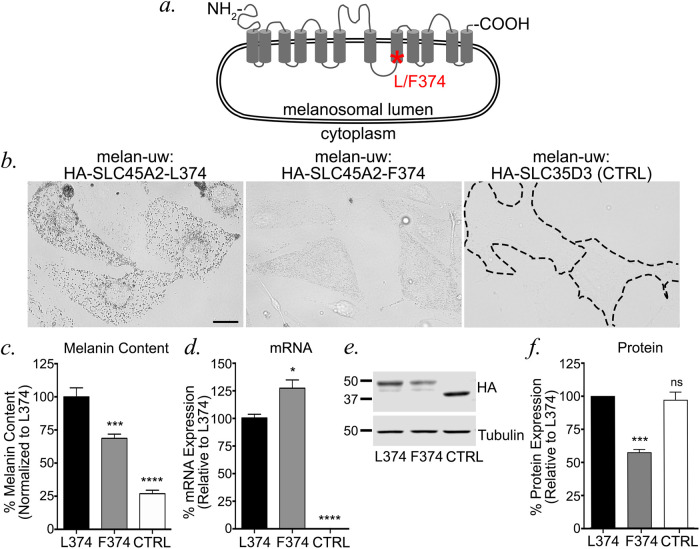
The light skin-associated SLC45A2-F374 variant protein is expressed at lower levels than the dark skin-associated SLC45A2-L374 variant
The major human SLC45A2 allele associated with light skin tone encodes a phenylalanine (F) instead of a leucine (L) at amino acid position 374, within the eighth transmembrane domain of the predicted 12-transmembrane domain-containing protein (Figure 7a). How does this single amino acid substitution impact pigmentation? We reasoned that the F374 variant could affect protein folding/stability, localization to melanosomes, or transporter function within melanosomes. To begin to distinguish between these possibilities, we generated stable melan-uw cells expressing the HA-tagged dark skin-associated L374 variant (as used in Figures 1 – 6), the HA-tagged F374 variant, or HA-SLC35D3 as a CTRL. Bright field microscopy (BF) analysis revealed that cells expressing HA-SLC45A2-F374 were darker than CTRL cells but lighter than cells expressing HA-SLC45A2-L374 (Figure 7b), consistent with published results (Cook et al., 2009). The observed phenotype was confirmed by quantitative melanin content assay, in which cells expressing HA-SLC45A2-F374 harbored significantly more pigment than CTRL cells but less than cells expressing HA-SLC45A2-L374 (Figure 7c). The lower level of pigmentation in cells expressing HA-SLC45A2-F374 did not reflect reduced SLC45A2 mRNA expression, as quantitative RT-PCR analyses showed that melan-uw cells expressing HA-SLC45A2-F374 had 1.25-fold more SLC45A2 mRNA than the WT HA-SLC45A2-L374-expressing cells (Figure 7d). However, immunoblotting analyses of cell lysates for the HA-tagged proteins revealed that the HA-SLC45A2-L374 stable cell line expressed twofold more HA-SLC45A2 than cells stably expressing the HA-SLC45A2-F374 variant (Figure 7, e and f). These data indicate that the reduced pigmentation conferred by the F374 variant is reflected by a posttranslational reduction in SLC45A2 protein content.
FIGURE 7:
The human light skin-associated SLC45A2-F374 variant is expressed in melanosomes at lower levels than the dark variant. (a) Schematic diagram of human SLC45A2 primary structure, with transmembrane (gray cylinders) and luminal and cytosolic domains (gray lines) indicated. An asterisk indicates the position of L/F374. (b) Stably transfected melan-uw cells expressing HA-SLC45A2-L374 (left), HA-SLC45A2-F374 (center), or HA-SLC35D3 as a negative CTRL (right) were imaged by BF to emphasize pigmentation levels. Bar, 10 μm. (c–f) Stably transfected melan-uw cells expressing HA-SLC45A2-L374, HA-SLC45A2-F374, or HA-SLC35D3 (CTRL) were analyzed for (c) melanin content by quantitative spectroscopy assay, (d) SLC45A2 mRNA expression by quantitative RT-PCR, or (e, f) HA protein expression by immunoblotting relative to tubulin as a loading CTRL. A representative immunoblot is shown in e; quantification of HA expression normalized to tubulin levels over three experiments is shown in f. All values are normalized to 100% for cells expressing HA-SLC45A2-L374. Each experiment was repeated at least three times. Statistical analyses were determined by Welch’s ANOVA with Dunnett T3 test for multiple comparisons (c, d) or one-way ANOVA with Dunnett’s test for multiple comparisons (f).*p < 0.05; ***p < 0.001; ****p < 0.0001; ns, no significant difference.
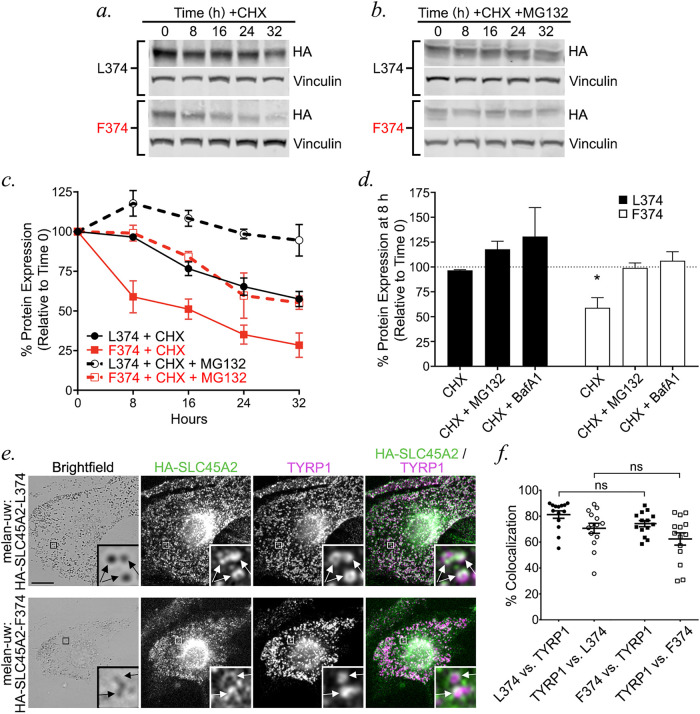
To test whether the reduced protein content of the F374 variant reflected increased protein degradation, we assessed the protein stability of each variant over time in stably transduced melan-uw cells. Cells were treated with cycloheximide (CHX) to block new protein synthesis, and the amount of HA-SLC45A2 remaining in cell lysates at different time points was assessed by immunoblotting. While the levels of both variants were substantially reduced by 32 h following CHX treatment, HA-SLC45A2-F374 was degraded at a higher rate than HA-SLC45A2-L374 (L374 half-life, 53.2 h; F374 half-life, 8.5 h) and was nearly completely eliminated by 32 h (Figure 8, a and c). To determine whether the rapid decline in the HA-SLC45A2-F374 variant protein levels were dependent on proteasomal degradation, cells were treated with the proteasome inhibitor MG132 during the CHX chase. MG132 treatment nearly completely blocked the gradual disappearance of HA-SLC45A2-L374 and also blocked the rapid degradation of HA-SLC45A2-F374 at early chase times (Figure 8, b and c). MG132 treatment did not, however, block the slow degradation of HA-SLC45A2-F374 at later time points (Figure 8, b and c). These data indicate that normal turnover of L374 and of the labile fraction of F374 requires proteasome activity, but that slow, long-term degradation of F374 occurs by a proteasome-independent mechanism. To test for the role of endolysosomal degradation, cells were treated with the vATPase inhibitor BafA1 during the CHX chase. BafA1 treatment caused significant cell death by 24 h, but within the first 16 h blocked degradation of both HA-SLC45A2-L374 and -F374, suggesting that degradation was largely mediated in endolysosomes (Supplemental Figure EV2). Importantly, dIFM analysis showed that HA-SLC45A2-F374 colocalized with TYRP1 to a similar extent as HA-SLC45A2-L374 (Figure 8, d and e), indicating that a smaller cohort of nondegraded HA-SLC45A2-F374 variant localizes properly to melanosomes where it functions in supporting elevated pH.
FIGURE 8:
The F374 variant is less stable than the L374 variant and is degraded by proteasome-dependent endolysosomal degradation. (a, b) Melan-uw cells stably expressing either HA-SLC45A2-L374 (L374: a, b, top) or -F374 (F374: a, b, bottom) were cultured with CHX) (25 μg/ml) alone (a) or combined with proteasome inhibitor MG132 (25 μM; b) for the indicated times (in hours). At the indicated times, cells were harvested and whole cell lysates were fractionated by SDS–PAGE and analyzed by immunoblotting for HA and for vinculin as a loading CTRL. Relevant regions of the gels are shown. (c) Band intensities for HA-SLC45A2 were quantified over three independent experiments, normalized to vinculin band intensities, and presented as a percentage of the signal observed at time 0 ± SEM. (d) Band intensities for HA-SLC45A2 after treatment for 8 h with CHX alone or with MG132 or 25 nM BafA1 (blots are shown in Supplemental Figure EV2) were quantified over three independent experiments, normalized to vinculin band intensities, and presented as a percentage of the signal for each isoform at time 0 ± SEM. Statistical significance of values relative to those at time 0 was determined using ordinary one-way ANOVA with Dunnett post-hoc test for multiple comparisons. Only statistically significant differences are noted. (e, f) Melan-uw cells stably expressing either HA-SLC45A2-L374 (e, top) or HA-SLC45A2-F374 (e, bottom) were fixed, labeled for HA (green) and TYRP1 (magenta), and analyzed by dIFM and by BF. Merged HA/TYRP1 image is at right; insets of boxed regions are magnified 7.5×. HA-SLC45A2 and TYRP1 localizing to the same compartments are indicated with arrows. Scale, 10 μm. (f) The degree of TYRP1 and SLC45A2 localization to the same compartments was quantified manually from 14 cells over two independent experiments and is presented as label 1 vs. label 2, in which the colocalization between label 1 and label 2 is a percentage of total label 2. Quantification is from two independent experiments with 14 cells counted per sample. Statistical significance was determined using Kruskal–Wallis test with Dunn’s post-hoc test for multiple comparisons. ns, no significant difference; *p < 0.05.
Taken together, these data suggest that the light skin-associated SLC45A2-F374 variant is less stable than the dark skin-associated SLC45A2-L374 variant due to endolysosomal proteolysis that is proteasome-dependent, resulting in lower SLC45A2 activity in melanosomes.
DISCUSSION
SLC45A2 is a critical determinant of skin and eye pigmentation, but until now its localization and functional role in melanogenesis have not been clearly delineated. Using a functional epitope-tagged form of SLC45A2 expressed in immortalized SLC45A2-deficient epidermal mouse melanocytes, we show here that SLC45A2 localizes to a subset of mature melanosomes marked by TYR and TYRP1 expression, where it partially accumulates in subdomains of the organelle outer membrane. When expressed ectopically in HeLa cells, SLC45A2 localizes to subdomains of the lysosomal membrane and functions to increase lysosomal pH, supporting a previously proposed role in neutralizing melanosomes (Bin et al., 2015). Although the melanosomal chloride channel OCA2 also functions to neutralize melanosome pH (Bellono et al., 2014), SLC45A2 localizes to a partially distinct cohort of melanosomes, and SLC45A2-deficient melanocytes harbor melanosomes that are more pigmented than OCA2-deficient cells—together suggesting that SLC45A2 functions at a later melanosome maturation stage than OCA2. Accordingly, overexpression of OCA2 in SLC45A2-deficient melanocytes compensated for SLC45A2 deficiency in pigment production, whereas SLC45A2 overexpression could not compensate for OCA2 deficiency. Although it is possible that these data reflect the distinct abilities of the OCA2 channel and the SLC45A2 transporter to modulate pH on the same melanosome subsets, the localization of these two proteins to partially distinct subsets leads us to conclude that SLC45A2 functions to neutralize lumenal pH at a later stage of melanosome maturation than OCA2. We also identified the mechanism underlying the pigmentation phenotype of the light skin-associated F374 variant of SLC45A2. Although SLC45A2-F374 localizes to melanosomes like the dark skin-associated L374 variant, SLC45A2-F374 is less stable and more rapidly degraded than -L374, resulting in reduced SLC45A2-F374 protein levels at steady state. Together, our data 1) indicate that SLC45A2 regulates melanogenesis by controlling melanosome pH in a manner that is nonredundant with OCA2 and 2) uncover the mechanism by which a common genetic variant imparts light skin color.
Our data indicate that SLC45A2 localizes to the melanosomal membrane, where we propose that it directly functions to increase lumenal pH. This is analogous to other proteins on the melanosome membrane, such as the chloride channel OCA2 and the cation channel TPC2, which also directly regulate melanosome pH through ion transport (Sitaram et al., 2009; Bellono et al., 2014, 2016; Ambrosio et al., 2016), and contrasts with ion transporters/channels that may indirectly regulate melanosome pH through effects on other organelles. For example, the putative solute transporter MFSD12 negatively regulates pigmentation from lysosomes (Crawford et al., 2017) and NCKX5 encoded by SLC24A5, the gene deficient in OCA6, appears to positively regulate pigmentation from the trans Golgi network (Ginger et al., 2008; Rogasevskaia et al., 2019). Together, these observations suggest that melanosome pH is regulated both directly and indirectly via a complex network of putative channels/transporters. Further work is necessary to identify other regulators of melanosome pH and to understand how these proteins cooperate to fine-tune the lumenal pH of melanosomes, thereby controlling melanin synthesis.
When expressed at the plasma membrane in yeast, SLC45A2 functions as a sucrose/proton symporter that transports sucrose from the extracellular space into the cytosol in a manner that requires a proton gradient and is maximal at a slightly acidic pH (Bartölke et al., 2014). Interestingly, SLC45A2 and other SLC45 family members can transport not only the disaccharide sucrose but also the monosaccharides glucose, fructose, and mannose (Bartölke et al., 2014). If it were to function similarly in melanocytes, SLC45A2 would expel protons and sugar molecules from the melanosome lumen into the cytosol, hence increasing melanosome pH. Whether sugars are substrates for SLC45A2 under physiological conditions in melanosomes remains to be determined, but given that lysosomal enzymes are present within maturing melanosomes (Diment et al., 1995; Raposo et al., 2001), it is possible that monosaccharides released from glycoproteins by lysosomal glycosidases in early stage melanosomes could serve as substrates for sugar/proton symporter activity. Our data show that expression of SLC45A2 in lysosomes of HeLa cells is sufficient to increase pH, indicating that its proton-dependent symporter activity is sufficient to raise lumenal pH by counteracting the activity of the vATPase, which pumps protons into the lumen of both melanosomes and lysosomes in an ATP-dependent manner (Bhatnagar and Ramalah, 1998; Tabata et al., 2008; Mindell, 2012). Sugar cotransport from the melanosome into the cytosol would also decrease the osmotic concentration inside the melanosome, requiring counter-ion transport to balance solute concentrations. Further investigation into the melanosomal channels, transporters, and exchangers is required to understand how the melanosome regulates not only its pH but also its osmolarity.
Unlike melanosomal proteins such as TYRP1 that are detected relatively uniformly on the melanosome membrane by IFM, SLC45A2 is sometimes detected in a punctate pattern, corresponding to specific subdomains of the melanosome membrane. The nature of these subdomains and the mechanism by which SLC45A2 assembles into them remain unclear. The punctate localization is conserved on lysosomes on ectopic expression of SLC45A2 in nonpigmented HeLa cells, suggesting that the assembly of SLC45A2 into subdomains does not require interactions with melanocyte-specific proteins. Some cells expressing HA-SLC45A2 displayed a mixture of punctate and ringlike SLC45A2 patterns on melanosomes. This might either reflect an artifact of overexpression or the consequence of a regulated process of aggregate assembly that may be influenced by melanosome contents or other features of melanosome maturation. The composition, regulation, and function of these SLC45A2-containing structures warrant further investigation.
The high colocalization between SLC45A2 and TYRP1 suggests that SLC45A2 is codelivered with TYRP1 to maturing melanosomes, likely via BLOC-1-dependent membrane transport (Setty et al., 2007, 2008). In contrast, despite similar requirements for BLOC-1 for delivery to melanosomes (Sitaram et al., 2012), there is significantly less overlap between OCA2 and TYRP1, and in structures in which they overlap their abundance is inversely related. Moreover, the relative melanin content of SLC45A2- and OCA2-deficient melanocytes is different, such that SLC45A2-deficient melanocytes harbor more mature melanosomes and slightly higher pigmentation than OCA2-deficient melanocytes. While these latter data alone could be interpreted to reflect distinct activities of OCA2 and SLC45A2 in facilitating increased melanosomal pH, together with the localization data they suggest that OCA2 functions at an earlier melanosomal maturation stage compared with SLC45A2. This would imply either that the melanosome delivery of OCA2 and TYRP1/SLC45A2 is staggered or that OCA2 is cleared soon after delivery. The latter is supported by the short half-life of OCA2 (Sitaram et al., 2009), the dependence of this half-life on melanization (Donatien and Orlow, 1995), and the accumulation of OCA2 on internal membranes in mature melanosomes (Sitaram et al., 2012). Based on these observations, we propose a model in which OCA2, SLC45A2, and TYRP1 are delivered together to maturing melanosomes—which harbor active vATPase to acidify their lumen (Bhatnagar and Ramalah, 1998; Tabata et al., 2008). OCA2 would facilitate a rapid efflux of chloride ions, immediately reducing membrane potential and thereby slowing vATPase activity. As the melanosomes mature, OCA2 clearance from the limiting membrane coupled with continued SLC45A2 arrival would support a slow but continued melanosome neutralization mediated by SLC45A2 proton export, counteracting residual vATPase activity. If this model were true, then OCA2 overexpression, by prolonging exposure on the melanosomal membrane, would antagonize vATPase activity in a more sustained way, perhaps resulting in slow proton efflux from the melanosome through another means such as membrane recycling (Dennis et al., 2016). This model would thus explain our observation that OCA2 overexpression can compensate for SLC45A2 loss but that SLC45A2 overexpression—with its limited capacity for proton export—cannot compensate for OCA2 loss.
The SLC45A2-F374 variant is the predominant allele in northern Europe and is strongly associated with light skin and hair and blue eyes (Soejima et al., 2006; Yuasa et al., 2006; Lao et al., 2007; Sabeti et al., 2007; Soejima and Koda, 2007; Branicki et al., 2008; Cook et al., 2009; Lucotte et al., 2010; Lucotte and Yuasa, 2013; Mukherjee et al., 2013; Lopez et al., 2014). However, the mechanism responsible for the reduced pigmentation associated with SLC45A2-F374 was previously unclear. We show here that compared with SLC45A2-L374, expression of SLC45A2-F374 in SLC45A2-deficient melanocytes results in a more meager increase in pigmentation, despite higher levels of SLC45A2 mRNA. Our results demonstrate that this is due to instability of the SLC45A2-F374 protein, leading to a higher rate of degradation relative to SLC45A2-L374 through a proteasome- and vATPase-dependent mechanism. Nevertheless, the remaining cohort of SLC45A2-F374 localizes appropriately to melanosomes. Thus, we propose that the reduced pigmentation associated with the F374 allele is due to insufficient levels of SLC45A2-F374 on the melanosomal membrane to maintain optimal neutral pH for maximal TYR activity, thus resulting in decreased melanin synthesis. Whether SLC45A2-F374 is rapidly degraded following melanosomal delivery or is missorted for endolysosomal degradation remains to be determined.
In conclusion, our data significantly advance our understanding of SLC45A2 and its function on melanosomes and begin to unravel the stepwise contributions of SLC45A2 and OCA2 to lumenal pH regulation and proper melanosome maturation. We also reveal the molecular basis explaining why the SLC45A2-F374 variant is a hypomorphic allele and propose a mechanism by which this allele contributes to lighter pigmentation in humans. The regulation of melanosome pH through the coordinated function of transporters, channels, and other proteins requires further investigation.
MATERIALS AND METHODS
Reagents
Unless otherwise specified, chemicals were obtained from Sigma-Aldrich (St. Louis, MO) and tissue culture reagents from Life Technologies/Thermo Fisher Scientific (Waltham, MA). Protease inhibitors were purchased from Roche Diagnostics (Rotkreuz, Switzerland), gene amplification primers from Integrated DNA Technologies (Coralville, IA), GoTaq DNA polymerase from Promega (Madison, WI), and restriction enzymes and T4 DNA ligase from New England Biolabs (Ipswich, MA).
Cell culture
Immortalized melanocyte cell lines melan-p1 (OCA2 deficient) from C3H-Oca2cp/25H (pink-eyed dilute) mice (Sviderskaya et al., 1997) and WT melan-Ink4a-Arf1 (formerly called melan-Ink4a-1; referred to here as melan-Ink4a or WT) from C57BL/6J-Ink4a-Arf−/− (Cdkn2 null) mice (Sviderskaya et al., 2002) have been described and were derived from the skins of neonatal mice. Three immortal SLC45A2-deficient melanocyte lines, melan-uw-1, -2, and -3, were similarly derived from the skins of neonatal C57BL/6J-Ink4a-Arf–/– Slc45a2uw/uw (uw) mice. Mice were housed and interbred by Lynn Lamoreux (Texas A&M University, College Station, TX), and skins were shipped on ice to London. Only the melan-uw-2 line (referred to here as melan-uw) was used here because they were more uniform in pigmentation both before and after stable expression of HA-SLC45A2. All cells were cultured at 37°C and 10% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 200 nM 12-O-tetradecanoylphorbol-13-acetate (TPA). Melan-uw cells grow poorly in the absence of adenylate cyclase activators, which also stimulate the transcription of melanogenic genes. To properly compare them, melan-uw cells, melan-p1 cells, and all stable cell lines derived from these cells were additionally supplemented with 200 pM cholera toxin. Melan-Ink4a cell medium was not supplemented with cholera toxin, but we expect that this might contribute to underestimating the differences between Melan-Ink4a and the other cell lines due to the increased expression of melanogenic genes in the latter. Cell lines were authenticated by restoration of pigmentation on expression of the WT form of their defective genes and verified to be negative for mycoplasma every 2–3 mo using the MycoAlert Mycoplasma Detection Kit (Lonza).
Retrovirus production from transiently transfected Plat-E cells (Morita et al., 2000) and retroviral transduction of melanocyte cell lines were carried out as described previously (Setty et al., 2007; Meng et al., 2012). Briefly, Plat-E cells were transfected with retroviral DNA constructs using Lipofectamine 2000 (Thermo Fisher), and the medium was replaced the next day. Retrovirus-containing supernatants were collected 48 h later, filtered, and added to melan-Ink4a cells or melan-uw cells in a 1:1 ratio with fresh medium. The medium was replaced the next day, and pools of stable transductants were selected 24 h later by adding 300 μg/ml hygromycin B to the medium. Stable transfectants were occasionally treated with 200 µg/ml hygromycin B for 2–3 d to maintain selective pressure for the transgene. For transient transfections, cells on glass coverslips were transfected with DNA constructs using Lipofectamine 3000 or Lipofectamine 2000 (Thermo Fisher), and the medium was replaced the next day. Cells were fixed using 4% formaldehyde/ phosphate-buffered saline (PBS) 48 or 72 h after transfection and stored at 4°C for immunolabeling and analysis.
DNA constructs
We first generated a GFP-SLC45A2 (L374) fusion protein by inserting human SLC45A2 cDNA, generated by RT-PCR of RNA isolated from darkly pigmented human epidermal melanocytes, into the BamHI/Xhol sites of pcDNA4/TO (Invitrogen/Life Technologies) in frame with GFP and a Gly-Ala-Gly-Ala linker previously inserted in the AflII/HindIII sites (sequence available on request). To generate HA-tagged SLC45A2, the insert from pcDNA4/TO-GFP-SLC45A2 was amplified by PCR adding XhoI and NotI restriction sites at the 5′ and 3′ ends, respectively, and subcloned into the respective sites of pCI (Promega). An N-terminal Kozak consensus start site, HA tag, and Gly-Ser linker were subsequently added by PCR using a distinct forward primer (5′-gcatatctcgagatgTACCCATACGATGTTCCAGATTACGCTggctcaggatctgggatgggtagcaacagtgggc-3′) and the same reverse primer. The HA-SLC45A2 insert was subsequently subcloned as a XhoI-NotI fragment into pBMN-(XN)-IRES-hygro to generate the retroviral vector encoding HA-SLC45A2. The F374 variant was made by site-directed mutagenesis of the pBMN-(XN)-IRES-hygro-HA-SLC45A2 (L374) construct using the Clontech site-directed mutagenesis kit. All recombinant plasmids were verified by sequencing by the University of Pennsylvania Cell Center, the Nucleic Acid/Protein Research Core Facility at the Children’s Hospital of Pennsylvania, or Macrogen.
The plasmid-based expression vector pCR3-OCA2-WT-HA-UTR2 and corresponding retroviral vector pBMN-OCA2-WT-HA-UTR2 encoding human OCA2 with an exofacial HA epitope were described in Sitaram et al. (2009), pCDM8.1-HA-SLC35D3 encoding human SLC35D3 with an N-terminal HA epitope tag was described in Meng et al. (2012), and human LAMP1-mCherry in pCDNA3.1 was a generous gift from Sergio Grinstein (University of Toronto and Hospital for Sick Children, Toronto, ON, Canada).
Melanin content quantification
Melanin quantification by spectroscopy was done essentially as described (Delevoye et al., 2009). Briefly, melanocytes seeded on 6-cm dishes were trypsinized, pelleted, and sonicated in melanin buffer (50 mM Tris, 2 mM EDTA, and 150 mM NaCl, pH 7.4) supplemented with protease inhibitor cocktail (Roche). Insoluble material was pelleted for 15 min at 16,000 × g (4°C), rinsed in ethanol/diethyl ether (1:1), and dissolved in 2 M NaOH/20% dimethyl sulfoxide at 60°C. The optical density at 492 nm was measured to estimate melanin content and normalized to protein concentration as determined by BCA protein determination kit (Thermo Fisher). Plat-E cells, an unpigmented cell line, were used as a negative CTRL. Statistical significance from at least three independent experiments was determined by Welch’s ANOVA with Dunnett T3 test for multiple comparisons.
Antibodies
Primary antibodies used and their sources (listed in parentheses) include: mouse monoclonal antibody TA99/Mel5 to TYRP1 (American Type Culture Collection; Rockville, MD); rat monoclonal antibody 3F10 to the HA11 epitope (Sigma); mouse monoclonal antibody 16B12 to the HA11 epitope (BioLegend); mouse monoclonal antibody H4A3 to human LAMP1 (Developmental Studies Hybridoma Bank, Iowa City, IA); rabbit anti-LAMP2 (Abcam; for mouse melanocytes); rabbit anti-TYR (Pep7h, to the C-terminal 17 amino acids of human TYR [ Calvo et al., 1999]); mouse monoclonal antibody to γ-Tubulin (Sigma); and rabbit antibody to vinculin (E1E9V; Cell Signaling). Species- and/or mouse isotype–specific secondary antibodies from donkey or goat and conjugated to Alexa Fluor 488 or Alexa Fluor 594 used in IFM or conjugated to Alexa Fluor 680 or Alexa Fluor 790 for immunoblots were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA).
BF, IFM, and colocalization analyses
IFM analyses of fixed cells were done essentially as described (Dennis et al., 2016). Briefly, cells were plated on Matrigel (BD)-coated coverslips, fixed with 4% formaldehyde (VWR Scientific, Radnor, PA) in PBS, labeled with primary and secondary antibodies diluted in PBS/0.02% saponin/0.01% bovine serum albumin, mounted onto slides using Prolong Gold (Thermo Fisher), and analyzed on a Leica DMI-6000 microscope equipped with a 40× or 63× objective lens (Leica; 1.4 NA), a Hamamatsu Photonics ORCA-Flash4.0 sCMOS C11440-22CU digital camera, and Leica Application Suite X (LAS X) software. Images in sequential z-planes (0.2 μm step size) were deconvolved with Microvolution software and further analyzed using ImageJ (http://fiji.sc/Fiji; National Institutes of Health [NIH]).
Because SLC45A2 localized partially to subdomains on the melanosome membrane and thus did not fully overlap other melanosomal markers, previously used automated methods to quantify colocalization did not yield meaningful values; similarly, OCA2 localized largely to the interior of melanosomes (Sitaram et al., 2009, 2012) and was thus partially distinct from the distribution of TYRP1 or TYR cytoplasmic domain on the melanosome membrane. Therefore, quantification of the degree of overlap between two channels on individual organelles was performed manually on deconvolved, Z-projected images of cells in ImageJ. In brief, single-channel images and a merged image were synchronized using “Analyze > Tools > Synchronize Windows;” signals that were localized to the same organelle were counted using the “Multi-Point” tool; remaining signals in single channels were counted using the Multi-Point tool; and the total colocalized structures was calculated as a percentage of total structures for each channel. Statistical significance was determined using one-way ANOVA with Tukey’s multiple comparisons test, Welch’s ANOVA with Dunnett T3 post-hoc test, or Kruskal–Wallis test with Dunn’s multiple comparisons test where appropriate.
Scoring of pigmentation in transfected melan-uw and melan-p1 cells was done blinded. Fixed, stained coverslips were randomly assigned numbers and samples were revealed only after images were taken and scored. Statistical significance was determined using one-way ANOVA with Holm-Sidak’s test for multiple comparisons.
Fluorescence intensity analyses
Object-based fluorescence intensity was measured using ImageJ as described in https://www.unige.ch/medecine/bioimaging/files/1914/1208/6000/Quantification.pdf. Briefly, 8-bit, single-channel images were opened in ImageJ and brightness and contrast adjusted for each image. The image was duplicated and a threshold applied using “Image > Adjust > Auto Local Threshold.” To multiply binary images of the two channels of interest, we used “Process > Image Calculator > Multiply.” To redirect intensity measurements from binary image to fluorescent image, we used “Analyze > Set measurements” and pulled down the “Redirect to:” menu to choose the appropriate fluorescent image, making sure “Area” and “Integrated density” were also checked. With “binary image” clicked, we used “Analyze > Analyze Particles” to measure object intensity. This was repeated for each channel, and for each sample values were normalized to the highest integrated density value in each channel. At least 10 cells from each of two independent experiments were analyzed.
Lysosomal pH measurements
HeLa cells were transfected one day prior to imaging experiments with LAMP1-mCherry alone, mCherry-OCA2 alone, or HA-SLC45A2 and LAMP1-mCherry to identify endolysosomes, and incubated with 1 μM LysoSensor-ND160 (Thermo Fisher) for 5 min. LysoSensor was excited at 405 nm and its emission detected at 417–483 nm (W1) and 490–540 nm (W2). The ratio of LysoSensor W1/W2 emission in endolysosomes expressing LAMP1-mCherry was assigned a pH value based on a calibration curve generated prior to each experiment using solutions containing 125 mM KCl, 25 mM NaCl, 24 μM Monensin, and varying concentrations of MES to adjust the pH to 3.5, 4.5, 5, 5.5, 6.5, 7, or 7.5. The fluorescence ratio was linear from pH 5–7.0. Statistical significance was determined by Welch’s ANOVA with Games-Howell post-hoc test for multiple comparisons.
Electron microscopy
Melanocytes were cultured in 10-cm dishes and fixed in situ with Karnovsky’s fixative (4% paraformaldehyde [VWR Scientific, Radnor, PA], 4 mM calcium chloride, 72 mM sodium cacodylate, pH 7.4) containing 0.5% glutaraldehyde (Polysciences, Warrington, PA) for 1–2 h at room temperature. This solution was then removed and replaced with Karnovsky’s fixative containing 2% glutaraldehyde, and the cells were fixed overnight at room temperature. Cells were collected by scraping using a cell scraper and centrifugation at 27 × g for 5 min, resuspended in Karnovsky’s fixative containing 0.5% glutaraldehyde, and stored at 4°C until processing. After subsequent buffer washes, the samples were postfixed in 2.0% osmium tetroxide with 1.5% K3Fe(CN)6 for 1 h at room temperature and then rinsed in deionized water prior to en bloc staining with 2% uranyl acetate. After dehydration through a graded ethanol series, the tissue was infiltrated and embedded in EMbed-812 (Electron Microscopy Sciences, Fort Washington, PA) at the Electron Microscopy Resource Laboratory (University of Pennsylvania). Thin sections were stained with uranyl acetate and SATO lead and examined with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera (Hamamatsu, Bridgewater, NJ) and AMT Advantage NanoSprint500 software (Advanced Microscopy Techniques, Woburn, MA).
For melanosome staging, a combined total of at least 20 cells from at least two independent experiments and over 600 melanosomes was analyzed for each cell type, and the analysis was done blinded. The number of stage I/II, stage III, and stage IV melanosomes was quantified, and the percentage of melanosomes at each stage was calculated for each cell. These data were analyzed and plotted using Prism 6 (GraphPad, La Jolla, CA) and statistical significance was determined by Welch’s ANOVA with multiple comparisons (samples compared with melan-Ink4a) and Dunnett T3 test for multiple comparisons.
Immunoblotting
Melan-uw cells stably expressing HA-SLC45A2-L374, HA-SLC45A2-F374, or HA-SLC35D3 cultured in 60-mm Petri dishes were treated with 25 μg/ml CHX ± 25 µM MG-132 or ± 25 nM BafA1 and collected for immunoblot every 8 h for 32 h. For each time point the cells were rinsed in cold PBS before pelleting by centrifugation. After collection of all time points, cell pellets were resuspended in 1× Laemmli buffer (Laemmli, 1970), lysed by sonication, and heated at 37°C for 5 min. Protein concentration for each sample was measured using the BCA assay (Pierce) according to the manufacturer’s protocol; 20 μg protein from each sample was fractionated by SDS–PAGE on 10% polyacrylamide gels and transferred to PVDF membranes (Immobilon-FL, Millipore). Membranes were blocked with 5% milk in Tris-buffered saline (TBS), incubated overnight with antibodies to HA or vinculin diluted in TBS/0.5% Tween, washed with TBS/0.5% Tween, and analyzed using Alexa Fluor 680– or 790–conjugated secondary antibodies and Odyssey imaging system (LI-COR, Lincoln, NE). For immunoblotting without drug treatments, the method was the same except that CHX, MG-132, and BafA1 were not added and antibodies to tubulin were used for the loading CTRL. Statistical significance was determined using one-way ANOVA with Dunnett’s test for multiple comparisons. Protein half-lives were calculated using one phase decay.
Quantitative RT-PCR
Total RNA was extracted from melan-uw cells stably expressing HA-SLC45A2, HA-SLC45A2-F374, or HA-SLC35D3 as a CTRL using the RNeasy Plus Kit (Qiagen), and 3 µg was reverse transcribed (RT) using the SuperScript III kit (Life Technologies). The resulting cDNA was used for qPCR. Primers were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) to span an exon–exon junction to avoid amplification of any contaminating genomic DNA and were as follows: SLC45A2 (NM_016180.3) - F: CCCTGTACACTGTGCCCTTT and R: CTTCCCTCTCACGCTGTTGT. Reactions were prepared according to the manufacturer’s protocol using SYBR Select Master Mix (Invitrogen/Thermo Fisher Scientific) and cycled on a Real-Time PCR System (Bio-Rad). β-actin was used as an internal CTRL and all reactions were run in triplicate. mRNA levels were quantified by calculating average 2-∆∆Ct values, where Ct is the cycle number for the CTRL and target transcript at the chosen threshold. The ∆Ct = Cttarget–Ctβ-actin was calculated by subtracting the average Ct of β-actin from the average Ct of the target transcript. The ∆Ctmean was calculated for the reference sample (L374) and ∆∆Ct = ∆Ct–∆Ctmean was calculated by subtracting the ∆Ctmean from the ∆Ct. The relative mRNA expression of samples compared with L374 samples was calculated by 2-∆∆Ct. Statistical significance was determined using Welch’s ANOVA with Dunnett T3 test for multiple comparisons.
Statistical analyses
Except where noted, we performed each experiment three times and sample sizes are as indicated in each figure legend. Statistical data are presented as mean ± SEM. Statistics were calculated in GraphPad Prism using ordinary unpaired Student’s t test, ordinary one-way ANOVA, Welsh’s ANOVA, or Kruskal–Wallis test with post-hoc correction as specified. Welsh’s ANOVA or Kruskal–Wallis test was used instead of ordinary one-way ANOVA when the data displayed heteroscedasticity or did not display a normal distribution, respectively, which was determined using Graphpad Prism. Significant differences between CTRL or experimental samples are indicated (****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05). Only p < 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
The authors are grateful to Lynn Lamereux for generating Slc45a2 uw/uw Ink4a-Arf –/– mice, Donald Koroma for consultation on experimental details, and Sergio Grinstein for the gift of the LAMP1-mCherry expression construct. We acknowledge funding from NIH grants R01 AR048155 (M.S.M.), R01 AR071382 (E.O. and M.S.M.), and F32 AR062476 (M.K.D.) from the National Institute of Arthritis, Skin and Musculoskeletal Diseases, and T32 GM007229 Training Program in Cell and Molecular Biology from the National Institute of General Medical Sciences (L.L. and A.J.L.), and from Wellcome Trust grant 108429/Z/15/Z (E.V.S. and D.C.B.).
Abbreviations used:
- AIM1
antigen isolated from immuno-selected melanoma-1
- BafA1
bafilomycin A1
- BF
bright field microscopy
- CHX
cycloheximide
- CTRL
control
- dIFM
immunofluorescence microscopy with image deconvolution
- IFM
immunofluorescence microscopy
- MATP
membrane-associated transporter protein
- OCA
oculocutaneous albinism
- PBS
phosphate-buffered saline
- TBS
Tris-buffered saline
- TYR
tyrosinase
- TYRP1
tyrosinase-related protein-1
- uw
underwhite
- vATPase
vacuolar adenosine triphosphatase
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-03-0200) on September 23, 2020.
REFERENCES
- Adhikari K, Mendoza-Revilla J, Sohail A, Fuentes-Guajardo M, Lampert J, Chacon-Duque JC, Hurtado M, Villegas V, Granja V, Acuna-Alonzo V, et al. (2019). A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nat Commun 10, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio AL, Boyle JA, Aradi AE, Christian KA, Di Pietro SM (2016). TPC2 controls pigmentation by regulating melanosome pH and size. Proc Natl Acad Sci USA 113, 5622–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancans J, Tobin DJ, Hoogduijn MJ, Smit NP, Wakamatsu K, Thody AJ (2001). Melanosomal pH controls rate of melanogenesis, eumelanin/phaeomelanin ratio and melanosome maturation in melanocytes and melanoma cells. Exp Cell Res 268, 26–35. [DOI] [PubMed] [Google Scholar]
- Bartölke R, Heinisch JJ, Wieczorek H, Vitavska O (2014). Proton-associated sucrose transport of mammalian solute carrier family 45: an analysis in Saccharomyces cerevisiae. Biochem J 464, 193–201. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Pavan WJ (2002). The oculocutaneous albinism type IV gene Matp is a new marker of pigment cell precursors during mouse embryonic development. Mech Dev 116, 209–212. [DOI] [PubMed] [Google Scholar]
- Bellono NW, Escobar IE, Lefkovith AJ, Marks MS, Oancea E (2014). An intracellular anion channel critical for pigmentation. eLife 3, e04543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellono NW, Escobar IE, Oancea E (2016). A melanosomal two-pore sodium channel regulates pigmentation. Sci Rep 6, 26570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Harper D, Tenza D, Raposo G, Marks MS (2001). Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol Biol Cell 12, 3451–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar V, Ramalah A (1998). Characterization of Mg2+-ATPase activity in isolated B16 murine melanoma melanosomes. Mol Cell Biochem 189, 99–106. [DOI] [PubMed] [Google Scholar]
- Bin BH, Bhin J, Yang SH, Shin M, Nam YJ, Choi DH, Shin DW, Lee AY, Hwang D, Cho EG, et al. (2015). Membrane-associated transporter protein (MATP) regulates melanosomal pH and influences tyrosinase activity. PLoS One 10, e0129273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard B, Fuller BB, Vijayasaradhi S, Houghton AN (1989). Induction of pigmentation in mouse fibroblasts by expression of human tyrosinase cDNA. J Exp Med 169, 2029–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branicki W, Brudnik U, Draus-Barini J, Kupiec T, Wojas-Pelc A (2008). Association of the SLC45A2 gene with physiological human hair colour variation. J Hum Genet 53, 966–971. [DOI] [PubMed] [Google Scholar]
- Caduff M, Bauer A, Jagannathan V, Leeb T (2017). A single base deletion in the SLC45A2 gene in a Bullmastiff with oculocutaneous albinism. Anim Genet 48, 619–621. [DOI] [PubMed] [Google Scholar]
- Calvo PA, Frank DW, Bieler BM, Berson JF, Marks MS (1999). A cytoplasmic sequence in human tyrosinase defines a second class of di-leucine-based sorting signals for late endosomal and lysosomal delivery. J Biol Chem 274, 12780–12789. [DOI] [PubMed] [Google Scholar]
- Cerqueira CC, Hunemeier T, Gomez-Valdes J, Ramallo V, Volasko-Krause CD, Barbosa AA, Vargas-Pinilla P, Dornelles RC, Longo D, Rothhammer F, et al. (2014). Implications of the admixture process in skin color molecular assessment. PLoS One 9, e96886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, et al. (2007). The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood 109, 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, Brown DL, Duffy DL, Pastorino L, Bianchi-Scarra G, et al. (2009). Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol 129, 392–405. [DOI] [PubMed] [Google Scholar]
- Costin G-E, Valencia JC, Vieira WD, Lamoreux ML, Hearing VJ (2003). Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J Cell Sci 116, 3203–3212. [DOI] [PubMed] [Google Scholar]
- Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A, Thompson S, Lo Y, et al. (2017). Loci associated with skin pigmentation identified in African populations. Science 358, eaan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLay BD, Corkins ME, Hanania HL, Salanga M, Deng JM, Sudou N, Taira M, Horb ME, Miller RK (2018). Tissue-Specific Gene Inactivation in Xenopus laevis: Knockout of lhx1 in the Kidney with CRISPR/Cas9. Genetics 208, 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C, Hurbain I, Tenza D, Sibarita J-B, Uzan-Gafsou S, Ohno H, Geerts WJC, Verkleij AJ, Salamero J, Marks MS, et al. (2009). AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J Cell Biol 187, 247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Ischia M, Wakamatsu K, Cicoira F, Di ME, Garcia-Borron JC, Commo S, Galván I, Ghanem G, Kenzo K, Meredith P, et al. (2015). Melanins and melanogenesis: from pigment cells to human health and technological applications. Pigment Cell Melanoma Res 28, 520–544. [DOI] [PubMed] [Google Scholar]
- Dennis MK, Delevoye C, Acosta-Ruiz A, Hurbain I, Romao M, Hesketh GG, Goff PS, Sviderskaya EV, Bennett DC, Luzio JP, et al. (2016). BLOC-1 and BLOC-3 regulate VAMP7 cycling to and from melanosomes via distinct tubular transport carriers. J Cell Biol 214, 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie MM (1964). Underwhite. Mouse News Lett 30, 30. [Google Scholar]
- Diment S, Eidelman M, Rodriguez GM, Orlow SJ (1995). Lysosomal hydrolases are present in melanosomes and are elevated in melanizing cells. J Biol Chem 270, 4213–4215. [DOI] [PubMed] [Google Scholar]
- Domyan ET, Guernsey MW, Kronenberg Z, Krishnan S, Boissy RE, Vickrey AI, Rodgers C, Cassidy P, Leachman SA, Fondon JW 3rd, et al. (2014). Epistatic and combinatorial effects of pigmentary gene mutations in the domestic pigeon. Curr Biol 24, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donatien PD, Orlow SJ (1995). Interaction of melanosomal proteins with melanin. Eur J Biochem 232, 159–164. [DOI] [PubMed] [Google Scholar]
- Dooley CM, Schwarz H, Mueller KP, Mongera A, Konantz M, Neuhauss SC, Nüsslein-Volhard C, Geisler R (2013). Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigment Cell Melanoma Res 26, 205–217. [DOI] [PubMed] [Google Scholar]
- Du J, Fisher DE (2002). Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J Biol Chem 277, 402–406. [DOI] [PubMed] [Google Scholar]
- Fracasso NCA, de Andrade ES, Wiezel CEV, Andrade CCF, Zanao LR, da Silva MS, Marano LA, Donadi EA, Castelli EC, Simoes AL, et al. (2017). Haplotypes from the SLC45A2 gene are associated with the presence of freckles and eye, hair and skin pigmentation in Brazil. Leg Med (Tokyo) 25, 43–51. [DOI] [PubMed] [Google Scholar]
- Fukamachi S, Shimada A, Shima A (2001). Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nature Genet 28, 381–385. [DOI] [PubMed] [Google Scholar]
- Ginger RS, Askew SE, Ogborne RM, Wilson S, Ferdinando D, Dadd T, Smith AM, Kazi S, Szerencsei RT, Winkfein RJ, et al. (2008). SLC24A5 encodes a trans-Golgi network protein with potassium-dependent sodium-calcium exchange activity that regulates human epidermal melanogenesis. J Biol Chem 283, 5486–5495. [DOI] [PubMed] [Google Scholar]
- Gronskov K, Ek J, Sand A, Scheller R, Bygum A, Brixen K, Brondum-Nielsen K, Rosenberg T (2009). Birth prevalence and mutation spectrum in danish patients with autosomal recessive albinism. Invest Ophthalmol Vis Sci 50, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Gunnarsson U, Hellström AR, Tixier-Boichard M, Minvielle F, Bed’hom B, Ito S, Jensen P, Rattink A, Vereijken A, Andersson L (2007). Mutations in Slc45a2 cause plumage color variation in chicken and Japanese quail. Genetics 175, 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R, Patton RS, Cheng E, Svedine S, Trombetta ES, Wahl ML, Ariyan S, Hebert DN (2002). Abnormal acidification of melanoma cells induces tyrosinase retention in the early secretory pathway. J Biol Chem 277, 14821–14828. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, et al. (2008). A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet 4, e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Li YF, El-Gamil M, Rosenberg SA, Robbins PF (2001). Use of an in vitro immunoselected tumor line to identify shared melanoma antigens recognized by HLA-A*0201-restricted T cells. Cancer Res 61, 1089–1094. [PubMed] [Google Scholar]
- Hearing VJ (2005). Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci 37, 3–14. [DOI] [PubMed] [Google Scholar]
- Jonnalagadda M, Norton H, Ozarkar S, Kulkarni S, Ashma R (2016). Association of genetic variants with skin pigmentation phenotype among populations of west Maharashtra, India. Am J Hum Biol 28, 610–618. [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lamason RL, Mohideen M-APK, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. (2005). SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786. [DOI] [PubMed] [Google Scholar]
- Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M (2007). Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet 71, 354–369. [DOI] [PubMed] [Google Scholar]
- Lasseaux E, Plaisant C, Michaud V, Pennamen P, Trimouille A, Gaston L, Monferme S, Lacombe D, Rooryck C, Morice-Picard F, et al. (2018). Molecular characterization of a series of 990 index patients with albinism. Pigment Cell Melanoma Res 31, 466–474. [DOI] [PubMed] [Google Scholar]
- Law MH, Medland SE, Zhu G, Yazar S, Vinuela A, Wallace L, Shekar SN, Duffy DL, Bataille V, Glass D, et al. (2017). Genome-wide association shows that pigmentation genes play a role in skin aging. J Invest Dermatol 137, 1887–1894. [DOI] [PubMed] [Google Scholar]
- Lehman AL, Silvers WK, Puri N, Wakamatsu K, Ito S, Brilliant MH (2000). The underwhite (uw) locus acts autonomously and reduces the production of melanin via a unique pathway. J Invest Dermatol 115, 601–606. [DOI] [PubMed] [Google Scholar]
- Lemoine R (2000). Sucrose transporters in plants: update on function and structure. Biochim Biophys Acta 1465, 246–262. [DOI] [PubMed] [Google Scholar]
- Liu F, Visser M, Duffy DL, Hysi PG, Jacobs LC, Lao O, Zhong K, Walsh S, Chaitanya L, Wollstein A, et al. (2015). Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum Genet 134, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen YA, Wu XS, Jiang Y, Bittner M, Hammer JA, III, Pavan WJ (2002). Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci USA 99, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez S, Garcia O, Yurrebaso I, Flores C, Acosta-Herrera M, Chen H, Gardeazabal J, Careaga JM, Boyano MD, Sanchez A, et al. (2014). The interplay between natural selection and susceptibility to melanoma on allele 374F of SLC45A2 gene in a South European population. PLoS One 9, e104367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucotte G, Mercier G, Dieterlen F, Yuasa I (2010). A decreasing gradient of 374F allele frequencies in the skin pigmentation gene SLC45A2, from the north of West Europe to North Africa. Biochem Genet 48, 26–33. [DOI] [PubMed] [Google Scholar]
- Lucotte G, Yuasa I (2013). Near fixation of 374l allele frequencies of the skin pigmentation gene SLC45A2 in Africa. Biochem Genet 51, 655–665. [DOI] [PubMed] [Google Scholar]
- Mariat D, Taourit S, Guérin G (2003). A mutation in the MATP gene causes the cream coat colour in the horse. Genet Sel Evol 35, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MS, Seabra MC (2001). The melanosome: membrane dynamics in black and white. Nature Rev Mol Cell Biol 2, 738–748. [DOI] [PubMed] [Google Scholar]
- Martin AR, Lin M, Granka JM, Myrick JW, Liu X, Sockell A, Atkinson EG, Werely CJ, Moller M, Sandhu MS, et al. (2017). An unexpectedly complex architecture for skin pigmentation in Africans. Cell 171, 1340–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri L, Manfredini E, Del Longo A, Veniani E, Scarcello M, Terrana R, Radaelli AE, Calo D, Mingoia G, Rossetti A, et al. (2017). Clinical evaluation and molecular screening of a large consecutive series of albino patients. J Hum Genet 62, 277–290. [DOI] [PubMed] [Google Scholar]
- Meng R, Wang Y, Yao Y, Zhang Z, Harper DC, Heijnen HFG, Sitaram A, Li W, Raposo G, Weiss MJ, et al. (2012). SLC35D3 delivery from megakaryocyte early endosomes is required for platelet dense granule biogenesis and differentially defective in Hermansky-Pudlak syndrome models. Blood 120, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Vitavska O, Wieczorek H (2011). Identification of an animal sucrose transporter. J Cell Sci 124, 1984–1991. [DOI] [PubMed] [Google Scholar]
- Mindell JA (2012). Lysosomal acidification mechanisms. Annu Rev Physiol 74, 69–86. [DOI] [PubMed] [Google Scholar]
- Minvielle F, Cecchi T, Passamonti P, Gourichon D, Renieri C (2009). Plumage colour mutations and melanins in the feathers of the Japanese quail: a first comparison. Anim Genet 40, 971–974. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Grønskov K, Wei AH, Martínez-García M, Fernández A, Arveiler B, Morice-Picard F, Riazuddin S, Suzuki T, Ahmed ZM, et al. (2014). Increasing the complexity: new genes and new types of albinism. Pigment Cell Melanoma Res 27, 11–18. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T (2000). Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7, 1063–1066. [DOI] [PubMed] [Google Scholar]
- Mukherjee M, Mukerjee S, Sarkar-Roy N, Ghosh T, Kalpana D, Sharma AK (2013). Polymorphisms of four pigmentation genes (SLC45A2, SLC24A5, MC1R and TYRP1) among eleven endogamous populations of India. J Genet 92, 135–139. [DOI] [PubMed] [Google Scholar]
- Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH (2001). Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet 69, 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennamen P, Tingaud-Sequeira A, Gazova I, Keighren M, McKie L, Marlin S, Halem SG, Kaplan J, Delevoye C, Lacombe D, et al. (2020). Dopachrome tautomerase variants in patients with oculocutenous albinism. Genes in Medicine (inpress). [DOI] [PubMed] [Google Scholar]
- Piccirillo R, Palmisano I, Innamorati G, Bagnato P, Altimare D, Schiaffino MV (2006). An unconventional dileucine-based motif and a novel cytosolic motif are required for the lysosomal and melanosomal targeting of OA1. J Cell Sci 119, 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J, Hernando-Herraez I, Lorente-Galdos B, Dabad M, Ramirez O, Baeza-Delgado C, Morcillo-Suarez C, Alkan C, Hormozdiari F, Raineri E, et al. (2013). The genome sequencing of an albino Western lowland gorilla reveals inbreeding in the wild. BMC Genomics 14, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS (2001). Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J Cell Biol 152, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Ward JM (2015). Investigating polymorphisms in membrane-associated transporter protein SLC45A2, using sucrose transporters as a model. Mol Med Rep 12, 1393–1398. [DOI] [PubMed] [Google Scholar]
- Rogasevskaia TP, Szerencsei RT, Jalloul AH, Visser F, Winkfein RJ, Schnetkamp PPM (2019). Cellular localization of the K(+) -dependent Na(+) -Ca(2+) exchanger NCKX5 and the role of the cytoplasmic loop in its distribution in pigmented cells. Pigment Cell Melanoma Res 32, 55–67. [DOI] [PubMed] [Google Scholar]
- Rothammer S, Kunz E, Seichter D, Krebs S, Wassertheurer M, Fries R, Brem G, Medugorac I (2017). Detection of two non-synonymous SNPs in SLC45A2 on BTA20 as candidate causal mutations for oculocutaneous albinism in Braunvieh cattle. Genet Sel Evol 49, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. (2007). Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiji M, Fitzpatrick TM, Simpson RT, Birbeck MSC (1963). Chemical composition and terminology of specialized organelles (melanosomes and melanin granules) in mammalian melanocytes. Nature 197, 1082–1084. [DOI] [PubMed] [Google Scholar]
- Setty SRG, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS (2008). Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature 454, 1142–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SRG, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC, et al. (2007). BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell 18, 768–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen T, Schmidt A, Hunziker W, Beermann F (1999). The tyrosinase tail mediates sorting to the lysosomal compartment in MDCK cells via a di-leucine and a tyrosine-based signal. J Cell Sci 112, 45–53. [DOI] [PubMed] [Google Scholar]
- Sitaram A, Dennis MK, Chaudhuri R, De Jesus-Rojas W, Tenza D, Setty SRG, Wood CS, Sviderskaya EV, Bennett DC, Raposo G, et al. (2012). Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol Biol Cell 23, 3178–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A, Piccirillo R, Palmisano I, Harper DC, Dell’Angelica EC, Schiaffino MV, Marks MS (2009). Localization to mature melanosomes by virtue of cytoplasmic dileucine motifs is required for human OCA2 function. Mol Biol Cell 20, 1464–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima M, Koda Y (2007). Population differences of two coding SNPs in pigmentation-related genes SLC24A5 and SLC45A2. Int J Legal Med 121, 36–39. [DOI] [PubMed] [Google Scholar]
- Soejima M, Tachida H, Ishida T, Sano A, Koda Y (2006). Evidence for recent positive selection at the human AIM1 locus in a European population. Mol Biol Evol 23, 179–188. [DOI] [PubMed] [Google Scholar]
- Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, Filsell W, Ginger RS, Green MR, van der Ouderaa FJ, et al. (2007). A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet 81, 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sviderskaya EV, Bennett DC, Ho L, Bailin T, Lee ST, Spritz RA (1997). Complementation of hypopigmentation in p-mutant (pink-eyed dilution) mouse melanocytes by normal human P cDNA, and defective complementation by OCA2 mutant sequences. J Invest Dermatol 108, 30–34. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, Easty DJ, Cheong SC, Beach D, DePinho RA, Bennett DC (2002). p16Ink4a in melanocyte senescence and differentiation. J Natl Cancer Inst 94, 446–454. [DOI] [PubMed] [Google Scholar]
- Sweet HO, Brilliant MH, Cook SA, Johnson KR, Davisson MT (1998). A new allelic series for the underwhite gene on mouse chromosome15. J Heredity 89, 546–551. [DOI] [PubMed] [Google Scholar]
- Tabata H, Kawamura N, Sun-Wada G-H, Wada Y (2008). Vacuolar-type H+-ATPase with the a3 isoform is the proton pump on premature melanosomes. Cell Tissue Res 332, 447–460. [DOI] [PubMed] [Google Scholar]
- Tsetskhladze ZR, Canfield VA, Ang KC, Wentzel SM, Reid KP, Berg AS, Johnson SL, Kawakami K, Cheng KC (2012). Functional assessment of human coding mutations affecting skin pigmentation using zebrafish. PLoS One 7, e47398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi K, Hayashi Y, Jogahara T, Ogura G, Murata Y, Oda S (2009). Oculocutaneous albinism in Suncus murinus: establishment of a strain and identification of its responsible gene. Exp Anim 58, 31–40. [DOI] [PubMed] [Google Scholar]
- Vijayasaradhi S, Doskoch PM, Houghton AN (1991). Biosynthesis and intracellular movement of the melanosomal membrane glycoprotein gp75, the human b (brown) locus product. Exp Cell Res 196, 233–240. [DOI] [PubMed] [Google Scholar]
- Vijayasaradhi S, Xu YQ, Bouchard B, Houghton AN (1995). Intracellular sorting and targeting of melanosomal membrane proteins: identification of signals for sorting of the human brown locus protein, gp75. J Cell Biol 130, 807–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei A, Wang Y, Long Y, Wang Y, Guo X, Zhou Z, Zhu W, Liu J, Bian X, Lian S, et al. (2010). A comprehensive analysis reveals mutational spectra and common alleles in Chinese patients with oculocutaneous albinism. J Invest Dermatol 130, 716–724. [DOI] [PubMed] [Google Scholar]
- Wei AH, Zang DJ, Zhang Z, Yang XM, Li W (2015). Prenatal genotyping of four common oculocutaneous albinism genes in 51 Chinese families. J Genet Genomics 42, 279–286. [DOI] [PubMed] [Google Scholar]
- Wijesena HR, Schmutz SM (2015). A missense mutation in SLC45A2 is associated with albinism in several small long haired dog breeds. J Hered 106, 285–288. [DOI] [PubMed] [Google Scholar]
- Winkler PA, Gornik KR, Ramsey DT, Dubielzig RR, Venta PJ, Petersen-Jones SM, Bartoe JT (2014). A partial gene deletion of SLC45A2 causes oculocutaneous albinism in Doberman pinscher dogs. PLoS One 9, e92127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Dong GX, Hu XS, Miao L, Zhang XL, Zhang DL, Yang HD, Zhang TY, Zou ZT, Zhang TT, et al. (2013). The genetic basis of white tigers. Curr Biol 23, 1031–1035. [DOI] [PubMed] [Google Scholar]
- Yuasa I, Umetsu K, Harihara S, Kido A, Miyoshi A, Saitou N, Dashnyam B, Jin F, Lucotte G, Chattopadhyay PK, et al. (2006). Distribution of the F374 allele of the SLC45A2 (MATP) gene and founder-haplotype analysis. Ann Hum Genet 70, 802–811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.