Abstract
Cells adapt to drastic changes in genome quantity during evolution and cell division by adjusting the nuclear size to exert genomic functions. However, the mechanism by which DNA content within the nucleus contributes to controlling the nuclear size remains unclear. Here, we experimentally evaluated the effects of DNA content by utilizing cell-free Xenopus egg extracts and imaging of in vivo embryos. Upon manipulation of DNA content while maintaining cytoplasmic effects constant, both plateau size and expansion speed of the nucleus correlated highly with DNA content. We also found that nuclear expansion dynamics was altered when chromatin interaction with the nuclear envelope or chromatin condensation was manipulated while maintaining DNA content constant. Furthermore, excess membrane accumulated on the nuclear surface when the DNA content was low. These results clearly demonstrate that nuclear expansion is determined not only by cytoplasmic membrane supply but also by the physical properties of chromatin, including DNA quantity and chromatin structure within the nucleus, rather than the coding sequences themselves. In controlling the dynamics of nuclear expansion, we propose that chromatin interaction with the nuclear envelope plays a role in transmitting chromatin repulsion forces to the nuclear membrane.
INTRODUCTION
Genome size varies across species and generally correlates with various body composition parameters at wider scales including whole body, organ, tissue, cell, and nuclear sizes. It is likely that even within the same species, size parameters in polyploid organisms are generally higher than those in normal diploid organisms (Fankhauser, 1945; Levy and Heald, 2015). Additionally, within some tissues such as the sepal in plants, cells composing the tissue display a variation of ploidy, and the sizes of these cells correlate with nuclear size and DNA content (Robinson et al., 2018). These size parameters from genome to body are reciprocally scaled and appear to exhibit a hierarchical scaling relationship (Uppaluri et al., 2016; Robinson et al., 2018); however, there are many exceptions showing less correlation. For example, the body size of the axolotl with an approximately 30 Gb genome is much smaller than that of an elephant possessing an approximately 4 Gb genome. The diversity in genome size is believed to result from multiple events of genomic editing such as whole genome duplication during evolution, cell fusion, and endoreduplication at the cellular levels, although the mechanisms underlying this hierarchical scaling relationship from genome to body size remain unknown. Regardless, cells must tolerate these evolutionary and environmental alterations in DNA content at the cellular level, and the control of nuclear size to allow containment of genetic material must be initially influenced by alterations in DNA content. Indeed, if confined to the same nuclear volume, the genome density between budding yeast (∼12 Mb) or axolotl would differ by approximately 3000-fold. Therefore, it is assumed that cells maintain a moderate space within the nucleus to allow for DNA functions, such as transcription and replication, by adjusting the nuclear size.
The control of nuclear size is fundamental for cellular organization and development (Edens et al., 2013). As most chromatin domains are in close contact with the nuclear inner membrane (van Steensel and Belmont, 2017), alterations in nuclear size are expected to exert a large impact on chromatin conformation within the nucleus and on DNA functions. Indeed, manipulation of the nuclear size induced experimentally by ectopic expression of effective proteins for nuclear expansion influences cell cycle timing and transcriptional activity during early development in Xenopus laevis embryos (Jevtic´ and Levy, 2015, 2017). Additionally, abnormalities in nuclear size and morphology coincide with diseases such as cancer and aging (Zink et al., 2004; Scaffidi and Misteli, 2006). In cancer cells particularly, the nucleus is often enlarged concomitantly with large amplifications and duplications of the genome (Landry, Pyl, et al., 2013). Although the alteration in coding sequences of nuclear-lamina–associated genes contributes significantly to the hypertrophy of the nucleus, the increase in DNA content is thought to exert a non-negligible effect on this process.
The nuclear size generally correlates well with the nuclear DNA content. When comparing the nuclear size in the same cell type across species within certain vertebrate classes, the nuclear size correlates positively with the DNA content (Cavalier-Smith, 1982; Gregory, 2001). This correlation had led some researchers to propose a concept originally called the “nucleoskeletal theory,” in which the nuclear size is controlled by the DNA content (Cavalier-Smith, 1978). Additionally, another concept, termed here the “cytoplasmic theory,” asserts that nuclear size is regulated by cytoplasmic factors, including nuclear constituents, and is independent of nuclear DNA content. This theory is supported by correlations between nuclear size and the size of the entire cell (Jorgensen et al., 2007; Neumann and Nurse, 2007). For example, during early-stage embryo development in multicellular organisms, the nuclear size decreases as cell size decreases during embryogenesis despite constant DNA content (Levy and Heald, 2010; Jevtic´ and Levy, 2015; Tsichlaki and FitzHarris, 2016; Mukherjee et al., 2020). Additionally, in polyploid fission yeast, the cell size and nuclear size are altered simultaneously, and the nuclear-to-cytoplasmic volume ratio (N/C ratio) remains constant (Neumann and Nurse, 2007). Given these observations, the cytoplasmic theory has been considered to be more effective in determining nuclear size than the nucleoskeletal theory has been. Furthermore, certain determinants supporting this theory have been identified within the cytoplasm. In Xenopus, the nuclear size is controlled by the quantity of lamins, which are the main constituents of the nuclear lamina and are imported from the cytoplasm through nuclear pore complexes (NPCs) (Levy and Heald, 2010; Jevtic´ et al., 2015). Another known factor is the quantity of lipid membranes that compose the lipid bilayer of the nuclear membrane and are supplied from the endoplasmic reticulum in a process regulated by dynein motors and microtubules (MTs) (Hara and Merten, 2015; Kume et al., 2019; Mukherjee et al., 2020).
In the context of the nucleoskeletal theory, DNA and chromatin have both been considered as putative determinants of nuclear size (Cavalier-Smith, 2005). Following chromosome segregation during mitosis, constituents of the nuclear envelope seal rapidly around condensed sister chromatids via spatiotemporally regulated molecular interactions with chromatin. Indeed, chromatin is known to bind directly to inner nuclear membrane proteins and lipid membranes (Gruenbaum et al., 2005; Ulbert et al., 2006; Anderson and Hetzer, 2007; Zierhut et al., 2014). Considering chromatin interaction with the nuclear envelope and the correlation between nuclear size and DNA content among species and cells exhibiting varying ploidy within individual species, DNA content should exert a non-negligible role on nuclear size determination in vivo. Mechanisms by which the DNA content contributes to nuclear size determination, however, remain poorly understood. To gain insights into these mechanisms, we here evaluated the effect of DNA content on nuclear size by experimentally manipulating the DNA content of a cell-free reconstruction system derived from X. laevis egg extracts and time-lapse imaging of in vivo developing embryos.
RESULTS
Manipulation of DNA replication alters nuclear expansion dynamics
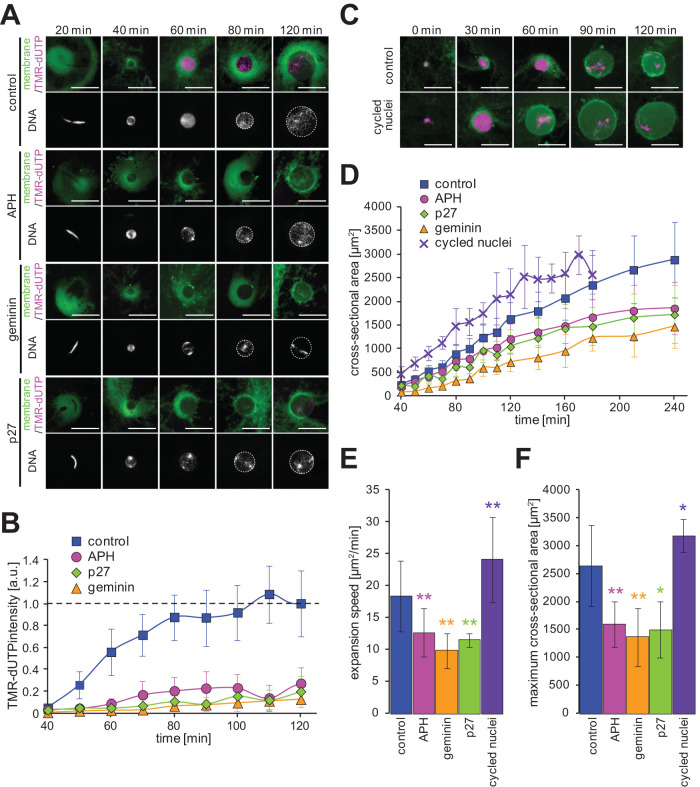
We reconstructed nuclei in the conventional cell-free system of X. laevis egg extracts. In this system, demembranated sperm chromatin of X. laevis was mixed with interphase cytoplasmic extracts from X. laevis unfertilized eggs to provide a source of nuclear DNA. Within 40 min of initial incubation, small spherical nuclei were assembled, and the nuclei continued to expand when cycloheximide was added to block spontaneous mitotic transition (Figure 1A). Upon addition of fluorescently tagged deoxyuridine triphosphates (dUTPs) to the extract, incorporation of dUTPs into the newly synthesized DNA was detectable after 40–50 min of incubation, and signal intensity within the nucleus increased until saturation at 70–80 min (Figure 1B). Nuclear expansion is expected to continue until the limited quantity of nuclear membrane constituents within the extract is exhausted (Goehring and Hyman, 2012; Hara and Merten, 2015). When the limited amount of membrane constituents within the cytoplasm is utilized, the plateau size is achieved. Corroboratively, expansion of the nuclear cross-sectional area slowed gradually after 120 min of incubation, and the value reached a plateau after a much longer incubation time (Supplemental Figure S1A). Since the estimated plateau values correlated well with the maximum value during 240 min of incubation (Supplemental Figure S1B), we used this maximum value for considering the plateau value in this study.
FIGURE 1:
Nuclear expansion dynamics after manipulation of DNA replication. (A) DNA, membrane, and incorporated TMR-dUTPs were visualized in the reconstructed nuclei in the presence of DNA replication inhibitors (APH, geminin, and p27) after the indicated incubation time. Merged images (top) of membrane (green) with TMR-dUTPs (magenta) and DNA (bottom) are shown. Broken circles represent the position of the nuclear membrane. Bars, 50 µm. (B) Dynamics of the intensities of the incorporated TMR-dUTPs in whole nuclei. The intensity was calculated by multiplying the measured mean TMR-dUTP intensity (/µm2) by the measured nuclear cross-sectional area. Each calculated value was divided by the mean value of the individual extract preparation after 120 min of incubation with X. laevis sperm chromatin. Control: n = 11; APH: n = 8; geminin: n = 3; p27: n = 4. (C) DNA (magenta) and membrane (green) were visualized in the nuclei from replicated chromatin in the second interphase of the cycling extract (cycled nuclei) and from sperm chromatin in the first interphase (control) after the indicated incubation time. Bars, 50 µm. (D) Dynamics of the measured mean cross-sectional area in the nuclei with X. laevis sperm chromatin in the presence of each DNA replication inhibitor (control: n = 34; APH: n = 20; geminin: n = 5; p27: n = 3) or with replicated chromatin in the cycling extract (cycled nuclei: n = 8). Average values are connected by a line in each data set. (E) Calculated expansion speeds and (F) measured maximum values of the nuclear cross-sectional area during incubation. Averages of values from each extract preparation are plotted. Error bars, SD. Asterisks, P value from Wilcoxon test compared with samples in the control. * and **, statistically significant difference, P < 0.05 and P < 0.001, respectively.
To address the effects of the nuclear DNA content on the regulation of nuclear size, we inhibited DNA replication in the cell-free extracts. In the presence of aphidicolin (APH), an inhibitor of DNA polymerase α, geminin recombinant proteins, which inhibit formation of the prereplication complex via binding to Cdt1, or p27 recombinant proteins, a cyclin-dependent kinase inhibitor, the fluorescently tagged dUTPs were poorly incorporated into the chromatin within the reconstructed nuclei (Figure 1, A and B). Under each condition, the nuclei were assembled in a manner identical to the control condition, and the nuclei appeared spherical with slight alterations in chromatin distribution. For the analysis of nuclear expansion dynamics using the measured cross-sectional area of nuclei at each incubation time, we calculated the speed of nuclear expansion. To rule out the effects of the doubling DNA content at the DNA replication phase and the decrease in the nuclear expansion speed by reaching plateau values, we calculated the slope of mean nuclear cross-sectional area over the time from 60 to 120 min of incubation for considering expansion speed. As a result, in the presence of either inhibitor, the calculated speed and maximum value decreased significantly (Figure 1, D–F). Additionally, in this experiment the number of reconstructed nuclei per volume of cytoplasmic extract did not differ among the samples in the absence and presence of either inhibitor, suggesting that the speed and the maximum value are reduced despite the same availability of constituents within the cytoplasm. It should be noted that the observed nuclear cross-sectional area varied among individual preparations of cytoplasmic extracts. Therefore, we set criteria to allow us to use the measured data for further analysis (see Materials and Methods), and we also confirmed the reduction in the expansion speed and maximum value in the presence of each inhibitor using the normalized values of the nuclear cross-sectional area for each extract preparation (Supplemental Figure S1C). Furthermore, the measured nuclear cross-sectional area at the initial measurable time points (40 min of incubation) revealed fewer differences between the control and replication-inhibited samples (Figure 1D), possibly due to the same DNA content between them before the initiation of DNA replication (Figure 1B). This result suggested that the detected nuclear expansion speed and maximum value are independent of the initial nuclear size immediately after sealing nuclear membranes around chromatids like at telophase.
Next, to analyze the nuclear expansion by increasing DNA content, we used a cycling extract. Generally, this extract supplemented with sperm chromatin can progress spontaneously through the M−S−M cell cycle several times. During the incubation of this extract, sister chromatids can be segregated by mitotic spindle elongation; however, the segregated sister chromatids often are not separated enough due to the lack of cytokinesis in the cell-free system, resulting in the assembly of one nucleus from two sister chromatids during the second interphase (Supplemental Video 1). Thus, the majority of the nuclei at the second interphase initially possess diploid genomes and duplicate these genomes by DNA replication during incubation. To observe the expansion dynamics of the nuclei exhibiting increased DNA content, we blocked the progression from this second interphase to the next mitosis by adding cycloheximide (Figure 1C; hereafter reconstructed nuclei in this condition are referred to as “cycled nuclei”). As a result, the nuclear expansion speed and maximum value in the cycled nuclei increased significantly compared with that observed in the nuclei of X. laevis sperm chromatin in the conventional interphase extract (Figure 1, D–F) and in the same cycling extract of the first interphase (Supplemental Figure S1D). Overall, these results suggested that the two parameters, expansion speed and plateau value, are dependent on DNA content within the nucleus.
Movie S1.
Time-lapse imaging of the cycling extract with X. laevis sperm chromatin by phase contrast with Hoechst33342 staining (magenta) and GFP-NLS (green). The round green signals could be recognized as an interphase nucleus and subsequent magenta signals without green signal could be recognized as mitotic phase after nuclear envelope breakdown. The extract containing chromatin were incubated under the imaging chamber (Hara and Merten, 2015)
Use of different genomes alters nuclear expansion dynamics
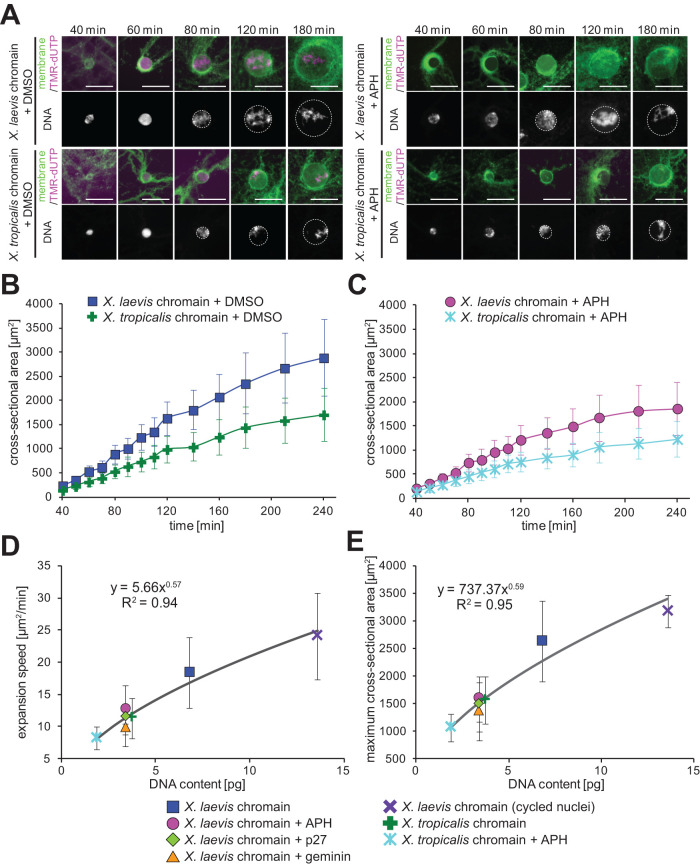
The alteration in the DNA content described above was achieved by manipulating the DNA replication process. To rule out potential effects of the DNA replication itself, we manipulated the DNA content by using demembranated sperm chromatin derived from the sister species Xenopus tropicalis. The genome size of X. tropicalis sperm is ∼1.7 Gb, corresponding to ∼1.9 pg of double-strand DNA and almost half that of X. laevis (∼3.1 Gb and ∼3.4 pg, respectively). Previous studies have utilized X. tropicalis sperm chromatin to recapitulate interphase nuclei within X. laevis cytoplasmic extracts (Levy and Heald, 2010). When the X. tropicalis sperm chromatin was used in our experimental system, spherical nuclei were reconstructed with normal DNA replication and expanded as incubation progressed, similar to what was observed using X. laevis sperm (Figure 2A). Quantification of the expansion dynamics revealed that the calculated expansion speed and maximum value were both significantly reduced when using X. tropicalis sperm chromatin rather than X. laevis chromatin (Figure 2, B, D, and E). The measured nuclear cross-sectional area observed from the use of X. tropicalis chromatin was nearly 60% that of those obtained using X. laevis sperm at every incubation time, implying that the difference in absolute values between these data may be indistinguishable or very small at early incubation times. This tendency is consistent with the small reduction in nuclear surface area detected after 90 min of incubation in a previous study (Levy and Heald, 2010). In the presence of the DNA replication inhibitor, APH, both parameters were also more reduced in the context of X. tropicalis sperm chromatin than were observed using X. laevis chromatin (Figure 2, C–E). It should be noted that the same DNA content dependency of nuclear expansion dynamics was detected when normalizing the values by the use of X. laevis sperm chromatin in each extract preparation (Supplemental Figure S2, A and B). Furthermore, in our observations, the reconstructed nuclei were compressed between two glass slides to some degree (see Materials and Methods), suggesting that the observed DNA content dependency of nuclear expansion dynamics is achieved by a difference in the spreading of the nucleus without alterations in the nuclear size itself. To evaluate this possibility, the reconstructed nuclei were observed under noncompressed conditions (see Materials and Methods). As a result, the DNA content dependency of nuclear expansion was still confirmed (Supplemental Figure S2C), suggesting that the observed DNA content dependency of nuclear expansion dynamics was not derived from nuclear spreading under our observation with nuclear compression. We next plotted all mean values of each calculated expansion speed and the maximum value in different conditions against the DNA content within the nucleus (Figure 2, D and E). From these plots, both parameters correlated significantly with the DNA content, suggesting that nuclear expansion is dependent on DNA content, and not on species-specific genome sequences or the DNA replication process. Although the fits using power law regression is preliminary due to the insufficient short range of DNA content (2–15 pg) used in this study, this hypoallometric scaling relationship, indicating that the exponent of DNA content in the regression equation is lower than 1, suggests that the contribution on nuclear expansion per DNA content is not linear and is concomitantly reduced with increasing DNA content.
FIGURE 2:
Nuclear expansion dynamics using different sperm chromatins. (A) DNA, membrane, and incorporated TMR-dUTPs were visualized in the reconstructed nuclei from X. laevis or X. tropicalis sperm chromatin in the presence of DMSO (buffer control) or APH after the indicated incubation time. Merged images (top) of membrane (green) and TMR-dUTP (magenta) and DNA (bottom) are shown. Broken circles represent the position of the nuclear membrane. Bars, 50 µm. (B) Dynamics of the measured mean cross-sectional area in the nuclei from X. laevis (blue, n = 34) or X. tropicalis (green, n = 12) sperm chromatin. (C) Dynamics of the measured mean cross-sectional area in the nuclei from X. laevis (pink, n = 20) or X. tropicalis (light blue, n = 12) sperm chromatin in the presence of APH. Average values are connected by a line in each data set. Error bars, SD. (D) Calculated expansion speeds and (E) measured maximum values of the nuclear cross-sectional area during incubation were plotted against the DNA content within the nucleus. Averages of values in each DNA content are fitted to the power law regression. Error bars, SD.
DNA-content–dependent nuclear expansion in X. laevis embryos
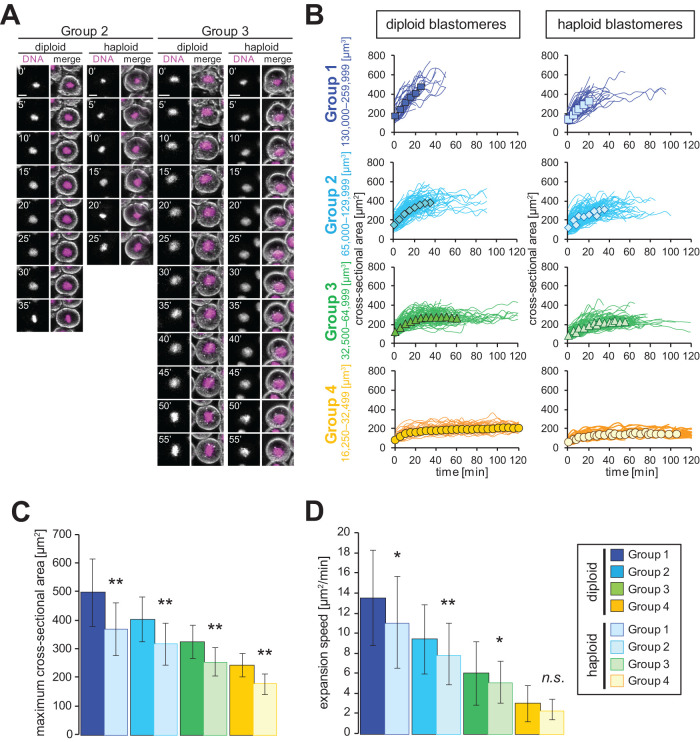
Next, we examined whether DNA-content–dependent nuclear expansion is conserved in vivo. It is, however, difficult to visualize the nucleus within the blastomeres of X. laevis developing embryos containing opaque yolk platelets and pigments. To overcome this, we utilized translucent yolk-less blastomeres for time-lapse imaging (Iwao et al., 2005). We prepared the translucent blastomeres from diploid X. laevis embryos by normal insemination with X. laevis sperm and from haploid X. laevis embryos by insemination with UV-irradiated X. laevis sperm (Figure 3A). Since the size of the obtained translucent blastomeres was not constant due to the instability of cleavage furrow position just after centrifugation of embryos during the preparation of translucent blastomeres (see Materials and Methods), the nuclear expansion dynamics was compared between diploid and haploid blastomeres in each group categorized by cell volume at defined ranges (Figure 3B). From the measured values of nuclear cross-sectional area, we obtained a value for maximum size or, alternatively, we used a value at the final observation time point before nuclear envelope breakdown (NEBD) in cases where nuclear expansion did not reach a plateau. For each group, the maximum value was larger in diploid than in haploid blastomeres (Figure 3C), which is consistent with previous observations using fixed blastomeres (Jevtic´ and Levy, 2017). Additionally, when calculating the nuclear expansion speed prior to reaching a plateau, the speed of nuclear expansion in diploid blastomeres was higher than that of haploid blastomeres in all groups with larger cell volumes (Figure 3D). It should be noted that decreases in the expansion speed and maximum size in haploid blastomeres were still observed in the ungrouped data (Supplemental Figure S3, B and C). Furthermore, the initial sectional area of the nucleus, a value that corresponds to the size of chromatids at telophase, was different between haploid and diploid blastomeres (Figure 3B). Nonetheless, the increases in the nuclear cross-sectional area from the initial values were also higher and more rapid in diploid blastomeres than those in haploid blastomeres (Supplemental Figure S3A). These results suggest that both the plateau value and the expansion speed in in vivo X. laevis developing embryos are altered in a DNA-content–dependent manner.
FIGURE 3:
DNA-content–dependent nuclear expansion in in vivo X. laevis embryos. (A) Snapshots of representative time-lapse images in translucent blastomeres from diploid or haploid X. laevis embryos in each group of the cell volume. DNA visualized by Hoechest 33342 (left) and merged images with phase contrast image (right) are shown. Images were taken at 5 min intervals. Bars, 20 µm. (B) Dynamics of individual nuclear cross-sectional area (line) and average cross-sectional area (symbols) in each group of the cell volume from diploid (filled symbols) or haploid (open symbols) blastomeres after initiation of interphase. The cell volume was estimated from the measured cross-sectional area by considering the cell as a sphere, and the analyzing groups were categorized as Group 1 (blue: n = 48 [diploid], 67 [haploid]; 130,000−259,999 µm3), Group 2 (light blue: n = 116 [diploid], 131 [haploid]; 65,000−129,999 µm3), Group 3 (green: n = 95 [diploid], 108 [haploid]; 32,500−64,999 µm3), and Group 4 (orange: n = 46 [diploid], 52 [haploid]; 16,250−32,499 µm3). The values are connected by a line for each individual observation. (C) Average measured maximum values (plateau values or values at the final measurement prior to NEBD) or (D) calculated expansion speed of the nuclear cross-sectional area in diploid or haploid blastomeres from each group. Error bars, SD. Asterisks, P value from Wilcoxon test compared with the diploid data in each group. * and **, statistically significant difference, P < 0.05 and P < 0.001, respectively. n.s., no significant difference.
Membrane accumulation on the nuclear surface in response to reduced DNA content
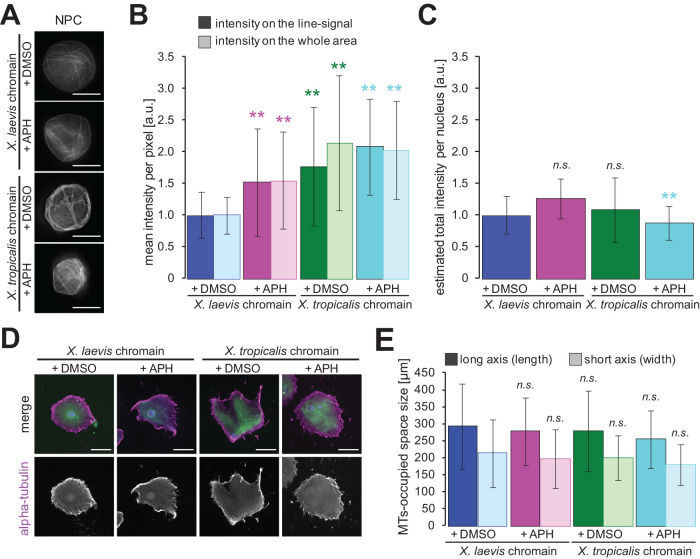
To analyze the differences in nuclei with different DNA contents, we next visualized the nuclear membrane by immunostaining for NPCs (Figure 4A). In the images of the reconstructed nuclei after spinning down on an observation slide, we detected multiple lines of continuous NPC-positive signals as well as the dot-like pattern on relatively flat regions. The signal intensity on the line pattern was higher than that of the dot-like pattern, suggesting that overlapping and accumulating nuclear membranes result in the observed line patterns. Although the line pattern may be generated artificially by the spinning down during sample preparation, we also confirmed membrane accumulation on the nuclear surface in the fixed X. laevis in vivo embryo without spinning down (Supplemental Figure S4A), as previously observed (Jevtic´ and Levy, 2015). When comparing these membrane signals among the nuclei with different DNA contents, the signal intensity on the line pattern appears to increase mainly in samples with reduced DNA content (Figure 4A). Furthermore, we also confirmed substantial enhancement of membrane intensity stained by a membrane-specific dye, 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3)], on the rim of the reconstructed nuclei with lower DNA content under noncompressed conditions (Supplemental Figure S2C) and on an observation slide after spinning down (Supplemental Figure S4B). To evaluate the DNA-content–dependent tendency, we quantified the mean intensities per pixel on the line pattern and throughout the whole nuclear area (Figure 4B). Both intensities increased when the DNA content was reduced, indicating that the amount of membranes on the nuclear surface accumulated more in cases of lower DNA content. Interestingly, when estimating the total membrane amount of a whole observed nuclear area, for which the obtained mean intensity was multiplied by the measured nuclear sectional area, we found that the calculated values were almost constant among nuclei with different DNA content (Figure 4C). These data provoked the assumption that the total amount of membrane constituents, which are supplied from the cytoplasm, is almost the same regardless of the DNA content. Furthermore, quantifying the membrane intensity of DiOC6(3) in the reconstructed nuclei revealed the same tendencies of mean intensities and the calculated total intensity against DNA content (Supplemental Figure S4, C and D), supporting that a surplus amount of nuclear membrane constituents, including lipid membranes and NPCs, is accumulated on the nuclear surface, particularly in cases of lower DNA content.
FIGURE 4:
Accumulation of membranes on the surface of nuclei with less DNA content. (A) NPCs on the reconstructed nuclei from X. laevis or X. tropicalis sperm chromatin in the presence of DMSO (buffer control) or APH were visualized after 120 min of incubation. Bars, 20 µm. (B) Average intensities of NPC-positive membrane signals per pixel on the continuous line pattern (filled bars) and throughout the whole nuclear section (dotted bars) of samples with different DNA contents. X. laevis chromatin with DMSO: n = 85 (line), n = 82 (area); X. laevis chromatin with APH: n = 92 (line), n = 90 (area); X. tropicalis chromatin with DMSO: n = 93 (line), n = 93 (area); X. tropicalis chromatin with APH: n = 80 (line), n = 67 (area). The measured intensity value from an individual image is normalized to the mean value of the control condition (X. laevis sperm chromatin without APH) in each extract preparation. (C) Estimated total intensities of NPC-positive membrane signals throughout the nuclear section among samples. The total intensity was estimated by multiplying the mean intensity by the measured nuclear cross-sectional area in each image, and the calculated value was normalized to the mean value of the control condition. (D) α-Tubulin (magenta), membranes (green), and DNA (blue) were visualized around the reconstructed nuclei with different DNA contents after 120 min of incubation. Merged images (top) and α-tubulin (bottom) are shown. Bars, 100 µm. (E) Average length (along the long axis) and width (along the short axis) of the MT-occupied space. Averaged values in each sample with different DNA contents were calculated. X. laevis chromatin with DMSO: n = 54 (length), n = 53 (width); X. laevis chromatin with APH: n = 132 (length), n = 134 (width); X. tropicalis chromatin with DMSO: n = 95 (length), n = 91 (width); X. tropicalis chromatin with APH: n = 122 (length), n = 121 (width). Error bars, SD. Asterisks, P value from Wilcoxon test compared with that in the control condition with X. laevis chromatin. **, statistically significant difference, P < 0.001. n.s., no significant difference.
A previous study suggested that the supply of lipid membranes, modulated by MTs and dynein, regulates nuclear expansion speed and correlates with the size of MT-occupied space around the nucleus (Hara and Merten, 2015; Mukherjee et al., 2020). To confirm the ability of the membrane supply, we next visualized MTs around the reconstructed nuclei with different DNA contents (Figure 4D). The measured size of the MT-occupied space was not significantly different among the four different conditions of DNA content (Figure 4E), supporting our assumption that the amount of nuclear membrane constituents is not determined by the DNA content. Overall, in cases of lower DNA content, the nucleus did not expand sufficiently and the accumulation of surplus membranes on the nuclear surface may be induced.
Physical properties of chromatin within the nucleus modulate nuclear expansion dynamics
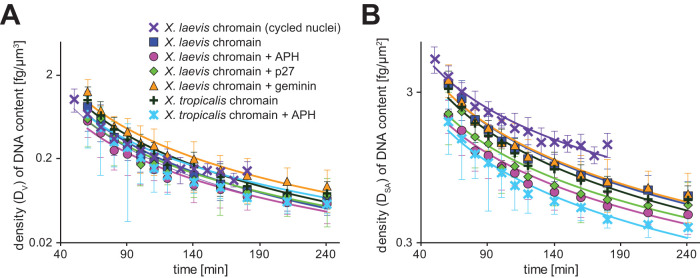
To further consider how DNA within the nucleus can affect nuclear expansion, we calculated the density (Dv), which is the ratio of the DNA content (C) to the estimated nuclear volume (V), in each DNA content condition during nuclear expansion after completion of DNA replication (Figure 5A). When plotting these data against incubation time (T) in a semilog plot, the data set from the X. laevis sperm chromatin without any inhibitors (Figure 1) was plotted as a regression line where D = ∼1130 × T∼1.83. Interestingly, all other data sets obtained from various conditions of DNA content in the same cytoplasmic X. laevis extracts (Figures 1 and 2) were plotted surrounding the regression line from the X. laevis chromatin. Conversely, when calculating the density of the DNA content per calculated nuclear surface area (DSA), each data set was not plotted in proximity and DNA-content–dependent differences were still present (Figure 5B). These data revealed that although the effect of DNA content on nuclear expansion kinetics is masked when DNA content is normalized to the nuclear volume, it is maintained when it is normalized to the nuclear surface area. These tendencies suggest the possibility that chromatin interaction with the nuclear envelope contributes in determining nuclear expansion dynamics.
FIGURE 5:
Dynamics of the ratio of DNA content to nuclear size parameters during nuclear expansion. (A) Dynamics of density DV of DNA content per estimated nuclear volume in samples with different DNA content in X. laevis cytoplasmic extract. (B) Dynamics of density DSA of DNA content per estimated nuclear surface area in the samples with different DNA content in X. laevis cytoplasmic extract. Average values from each extract preparation are plotted. Each data set is fitted to the power regression line. Error bars, SD.
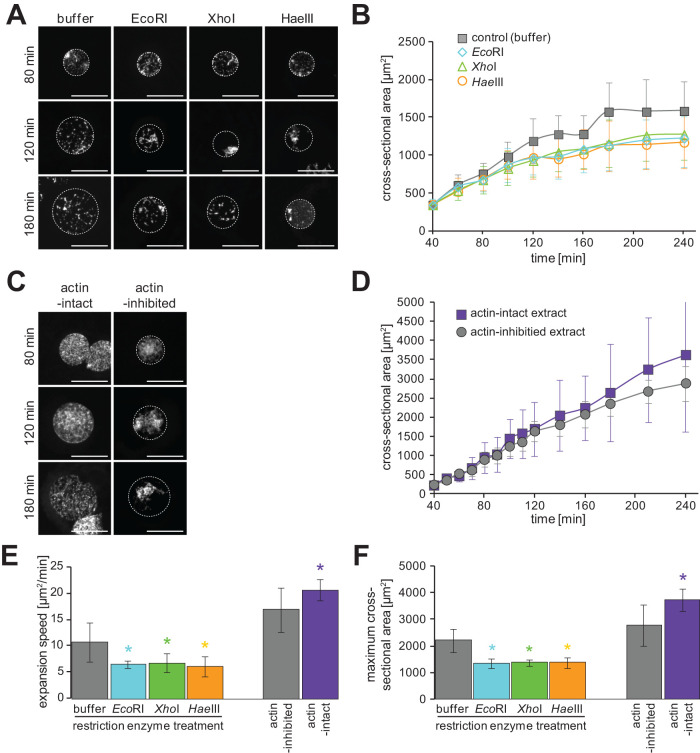
To experimentally assess the contribution of chromatin interaction with the nuclear envelope, we first digested linker DNA by adding restriction enzymes after preassembly of small spherical nuclei according to the previous reports using restriction enzymes in the X. laevis cell-free extract system (Kobayashi et al., 2002). In the presence of the restriction enzymes, a portion of the chromatin was dispersed within the nucleoplasm, particularly after the use of the four-cutter enzyme HaeIII. Furthermore, chromatin was condensed to form multiple chromatin clusters within the nucleus, where some clusters are located near the nuclear membrane (Figure 6A). These changes in chromatin structure reduced the frequency and strength of chromatin interactions with the nuclear envelope. Compared with the control condition, after treatment with an appropriate buffer for each enzyme, the expansion speed and maximum value of the nuclear cross-sectional area decreased significantly in the presence of either enzyme (Figure 6, B, E, and F). Nonetheless, the treatment with the restriction enzyme reduced the intensity of incorporated fluorescently labeled dUTPs into the chromatin as previously analyzed (Supplemental Figure S5A; Kobayashi et al., 2002). Subsequently, the small DNA fragments generated by treatment with restriction enzymes may leak out from the nucleus, suggesting that the observed decreases in nuclear expansion dynamics are caused by the reduction of DNA quantity through immature DNA replication and/or leaking DNA. To evaluate the contribution of immature DNA replication, we also supplemented the restriction enzymes with the sample in the presence of APH (Supplemental Figure S5B). Although the difference in nuclear size appeared to be less prominent than that observed without APH, the decreases in nuclear expansion speed and maximum values were also detected (Supplemental Figure S5, C and D). This result suggests that the observed reduction in nuclear expansion parameters is caused by the changes in chromatin structure via DNA fragmentation treated with restriction enzymes independently of the immature DNA replication. To further analyze the effects of chromatin interaction with the nuclear envelope, we enhanced chromatin tethering to the nuclear envelope by using distinct cytoplasmic extracts. A previous study demonstrated that chromatin tethering is regulated by filamentous actin localized to the nuclear periphery in X. laevis early embryos and in cell-free egg extracts, termed “actin-intact extracts,” with accumulating filamentous actin underneath the nuclear envelope (Oda et al., 2017). After incubating the actin-intact extracts with X. laevis sperm chromatin, much chromatin was tethered to the nuclear envelope and was distributed evenly throughout the nucleoplasm compared with the large detachment of highly condensed chromatin from the nuclear envelope in the conventional “actin-inhibited extract” (Figure 6C). The reconstructed nucleus with an enhanced interaction of chromatin with the nuclear envelope expanded more rapidly, and its maximum nuclear cross-sectional area was larger, although the difference in the nuclear cross-sectional area was not apparent at shorter incubation times compared with the actin-inhibited extracts as previously observed (Oda et al., 2017; Figure 6, D–F). It should be noted that when reconstructing the nucleus with different DNA content using the actin-intact extracts, DNA content dependency of nuclear expansion dynamics was still observed (Supplemental Figure S5E). Although the accumulation of filamentous actin underneath the nuclear envelope may possibly modulate nuclear expansion dynamics through changing the rigidity of the nuclear envelope (Oda et al., 2017), the status of chromatin interaction with the nuclear envelope correlates with nuclear expansion dynamics among the samples treated with restriction enzymes or using the actin-intact extracts.
FIGURE 6:
Nuclear expansion dynamics after modification of chromatin interaction with the nuclear envelope. (A) Representative images of reconstructed nuclei from X. laevis sperm chromatin in the presence of restriction enzymes (EcoRI, XhoI, or HaeIII) with the appropriate buffer after incubation for the indicated time. (B) Dynamics of the measured mean cross-sectional area of reconstructed nuclei in the presence of the indicated restriction enzymes, indicated by different colors. (C) Representative images of reconstructed nuclei from X. laevis sperm chromatin in the actin-intact extract after the indicated incubation time. DNA was stained with Hoechst 33342. Broken circles represent the position of the nuclear membrane. Bars, 50 µm. (D) Dynamics of the measured mean cross-sectional area of reconstructed nuclei in the actin-intact extracts (purple) or conventional actin-inhibited extracts (gray). Average values from each extract preparation are connected by a line in each data set. (E) Calculated expansion speeds and (F) measured maximum values of nuclear cross-sectional area of samples treated with buffer control (gray, n = 6), EcoRI (light blue, n = 6), XhoI (green, n = 5), HaeIII (orange, n = 5) and using actin-inhibited extract (gray, n = 37) or actin-inhibited extract (purple, n = 6). Average values from each extract preparation are shown. Error bars, SD. Asterisk, P value from Wilcoxon test compared with that in each control condition. *, statistically significant difference, P < 0.05.
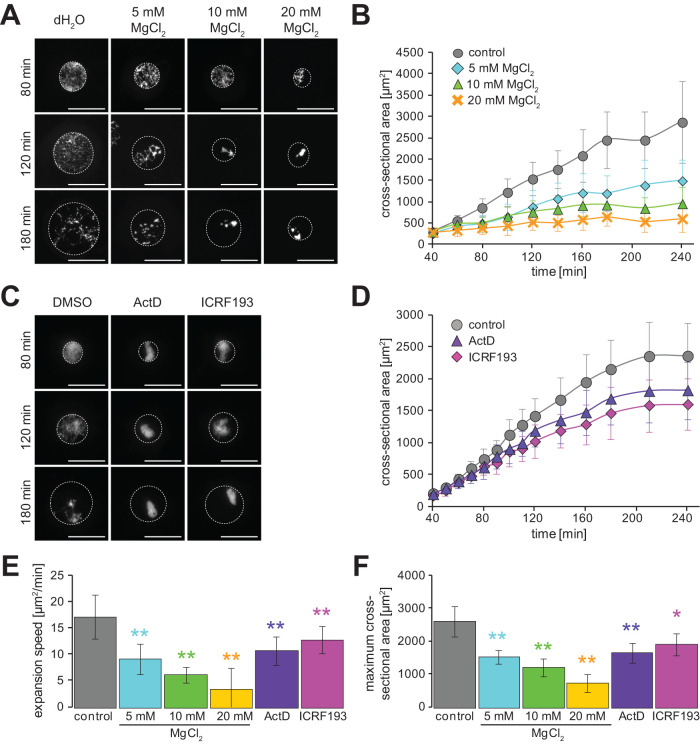
Supplementation with restriction enzymes also induced a slight condensation in chromatin as well as DNA fragmentation (Figure 6A). This excess condensation might be caused by a high concentration of bivalent cations such as Mg2+, which are present in the buffer for restriction enzymes, resulting in a reduction in electron repulsion forces of chromatin. This feature implies that not only the chromatin interaction with the nuclear envelope, but also chromatin condensation within the nucleus, possibly contribute to controlling nuclear expansion dynamics. To evaluate the effect of chromatin condensation, we next manipulated the chromatin condensation status without fragmentation of DNA. First, we changed the concentration of Mg2+ ions without adding restriction enzymes. In the presence of excess Mg2+ ions in extracts containing preassembled small nuclei, the chromatin appeared to be more highly condensed in a dose-dependent manner (Figure 7A). Under these conditions, nuclear expansion speed and maximum values revealed a greater reduction with higher Mg2+ ion concentrations (Figure 7, B, E, and F), which is consistent with previous observations of the reduction in size of isolated nuclei from culture cells upon addition of Mg2+ ions (Chan et al., 2017; Shimamoto et al., 2017). It should be noted that when the excess Mg2+ ions are added to the extract immediately after mixing sperm chromatin, the sperm chromatin could not be decondensed and the nucleus was rarely expanded (Supplemental Figure S6A). Furthermore, although the addition of Mg2+ ions also partially reduced the efficiency of DNA replication even after preassembly of the nucleus (Supplemental Figure S6B), both nuclear expansion parameters were still reduced in a dose-dependent manner based on Mg2+ ion concentration even under inhibiting DNA replication (Supplemental Figure S6C). To further support the effect of chromatin condensation, we next induced excess condensation of chromatin without changing the status of cations by supplementation with actinomycin D, a DNA intercalator, or ICRF-193, a known inhibitor of topoisomerase II. With supplementation with either inhibitor immediately after mixing sperm chromatin with the X. laevis cytoplasmic extracts, the sperm chromatin was not decondensed, and the nuclear membrane was rarely sealed around the chromatin (Supplemental Figure S6A). However, when we added either inhibitor after reconstructing the round-shaped nuclei by swelling the DNA from highly compacted sperm chromatin, the compaction of chromatin was induced within the nucleus (Figure 7C). Under these conditions, the nuclear expansion speed and maximum value were reduced compared with those of the control condition (Figure 7, D–F). It should be noted that DNA replication was completed with a slight delay, even in the presence of actinomycin D (Supplemental Figure S6D), which is known to inhibit DNA replication (Guy and Taylor, 1978). Although the above induced hypercondensation of chromatin by addition of either excess cation or condensation inhibitors was accompanied by a large detachment of chromatin from the nuclear envelope, chromatin detachment was also observed apparently in the reconstructed nucleus using the actin-inhibited extracts under the control condition for the inhibitors (Figure 7, A and C). Therefore, these data suggest that the physical status of chromatin structure, including not only chromatin interaction with the nuclear envelope but also chromatin condensation within the nucleus, is involved in the control of nuclear expansion dynamics.
FIGURE 7:
Nuclear expansion dynamics after manipulation of chromatin condensation. (A) Representative images of the reconstructed nuclei from X. laevis sperm chromatin in the presence of MgCl2 at the indicated concentrations or dH2O for buffer control after the indicated incubation time. (B) Dynamics of the measured mean cross-sectional area of reconstructed nuclei in the presence of extra Mg2+ ions. Control: n = 4; 5 mM: n = 4; 10 mM: n = 4; 20 mM MgCl2: n = 4. (C) Representative images of the reconstructed nuclei of X. laevis sperm chromatin in the presence of actinomycin D (ActD), ICRF-193, or DMSO for buffer control after the indicated incubation time. DNA was stained with Hoechst 33342. Broken circles represent the position of the nuclear membrane. Bars, 50 µm. (D) Dynamics of the measured mean nuclear cross-sectional area with X. laevis sperm chromatin in the presence of ActD (purple, n = 7), ICRF-193 (pink, n = 7), or DMSO (gray, n = 7) for buffer control. Average values are connected by a line in each data set. (E) Calculated expansion speeds and (F) measured maximum values of nuclear cross-sectional area with X. laevis sperm chromatin in the presence of buffer control, restriction enzymes, or inhibitors of chromatin structures. Average values from each extract preparation are shown. Error bars, SD. Asterisks, P value from Wilcoxon test compared with that in the control condition. * and **, statistically significant difference, P < 0.05 and P < 0.001, respectively.
Nuclear import ability correlates with DNA content and chromatin structure
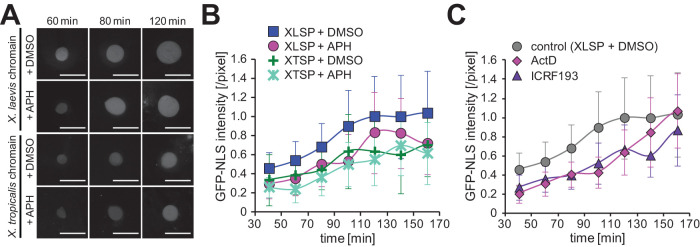
To finally evaluate the possibility that the DNA content and chromatin structures contribute to nuclear expansion via changing nuclear import ability, we next quantified general nuclear import of nuclear localization signal (NLS) proteins. After mixing green fluorescent protein (GFP)-NLS recombinant proteins with the extract, these proteins were imported into the nuclei in all four conditions of DNA content (Figure 8A). The mean intensity of the imported GFP-NLS per nuclear cross-sectional area increased over incubation time in all conditions of DNA content (Figure 8B). Although the intensities of GFP-NLS in individual reconstructed nuclei were fluctuating even within the same incubation time (Supplemental Figure S4E), the mean intensity increased more rapidly under conditions with more DNA content at corresponding incubation times (Figure 8B). This correlated change in the nuclear import activity with DNA content is consistent with that in nuclear expansion dynamics. Furthermore, quantification of the mean intensity of GFP-NLS under the manipulation of chromatin condensation by supplementation with actinomycin D or ICRF-193 revealed a reduction in nuclear import activity compared with the control (Figure 8C). Therefore, these results indicate that changes in nuclear expansion dynamics by modulating DNA content and chromatin condensation status correlate with changes in nuclear import activity.
FIGURE 8:
Dynamics of nuclear import ability after modulating DNA content or chromatin condensation status. (A) Representative images of GFP-NLS proteins in the reconstructed nuclei for the four different conditions of DNA content after the indicated incubation time. Bars, 50 µm. (B) Dynamics of the mean GFP-NLS intensity per nuclear sectional area among four different conditions of DNA content (X. laevis sperm chromatin with DMSO [XLSP + DMSO, n = 11] and with aphidicolin [XLSP + APH, n = 6], X. tropicalis sperm chromatin with DMSO [XTSP + DMSO, n = 5] and with aphidicolin [XTSP + APH, n = 6]) and (C) samples with inducing chromosome condensation (ActD: n = 3: ICRF-193: n = 3; DMSO control: n = 11). The measured intensity value was divided by that of the control (XLSP + DMSO) condition after 120 min of incubation for each preparation. Average values from each extract preparation are connected by a line in each data set. Error bars, SD.
DISCUSSION
For many years, a correlation between nuclear size and DNA content has been observed among individual animals with different ploidy and among different species. In this study, we utilized a cell-free system with manipulations of DNA content to characterize mechanisms by which DNA content controls nuclear expansion speed and plateau size (Figures 1 and 2). Additionally, although even cytoplasmic volume per chromatin was maintained, differences in DNA content could alter the dynamics of nuclear expansion. In in vivo X. laevis embryos, the manipulation of DNA content could alter the expansion speed and maximum nuclear size (Figure 3), clearly indicating that the DNA content can influence the N/C ratio in our experimental setup. Despite this, the N/C ratio in diploid and haploid in vivo cells remains constant in fission yeast (Neumann and Nurse, 2007) and in plant sepals (Robinson et al., 2018). This discrepancy in N/C ratio might be caused by the changes in cytoplasmic volume, depending on DNA content. Generally, the ploidy is coupled to the control of cell size, although the underlying mechanisms have remained unknown. Our experimental setup exhibited a manipulation of DNA content while maintaining the same cytoplasmic volume in developing embryos and in the cell-free system, suggesting that a change in the N/C ratio is detectable only in these conditions. Therefore, in previously reported in vivo situations, simultaneous changes in DNA ploidy and cytoplasmic volume might mask the contribution of DNA content to nuclear size control.
On the basis of the observation of the accumulation of excess membrane in the presence of low DNA content (Figure 4) and experimental evidence of the contribution of chromatin to nuclear expansion in this study (Figures 6 and 7), we proposed that the physical properties of chromatin can modulate the magnitude of forces in expanding the nucleus from the inside. Induced excess condensation by adding cations or inhibitors decreased the expansion speed and plateau size of the reconstructed nucleus (Figure 7). Additionally, a more condensed chromatin status in the DNA-replication–blocked nucleus by supplementation with geminin recombinant proteins (Supplemental Figure S2D) is a plausible explanation for the slightly slower expansion speed and smaller maximum size compared with dispersed chromatin in the nucleus supplemented with APH or p27 recombinant proteins (Figure 1, D and E). Therefore, we assume that the repulsion forces among the negatively charged chromatin help to expand the nuclear membrane outward from inside the nucleus. In this assumption, both DNA quantity and condensation status could modulate the magnitude of the repulsion forces. This assumption is supported by previous experimental evidence regarding changes in nuclear deformation upon modification of chromatin condensation (Stephens et al., 2019). Induced hyper- or hypocondensation of global chromatin via histone tail modifications, perturbation of heterochromatin proteins, or manipulation of cation concentration in the isolated nuclei from cells and nuclei in cells in vivo alters nuclear rigidity (Furusawa et al., 2015; Chan et al., 2017; Shimamoto et al., 2017; Stephens et al., 2017, 2018). In addition to the direct effect from chromatin condensation status, a change in nuclear import ability accompanied by a change in chromatin status is another possibility for the control of nuclear expansion dynamics. The chromatin within the nucleus is known to serve as a scaffold to bind Ran guanine nucleotide exchange factor, RCC1, which is known to exchange RanGDP (Ran guanosine diphosphate) to RanGTP (Ran guanosine triphosphate) (Bischoff and Ponstingl, 1991; Nemergut et al., 2001). A high concentration of RanGTP within the nucleus promotes the dissociation of the importin–NLS–cargo complex and export of the importins to the cytoplasm (Görlich et al., 1996). If the RCC1 quantity in the nucleus is elevated in cases with higher DNA content or hypocondensation of chromatin, the expected RCC1 quantity could account for the large amount of imported NLS proteins observed within the nucleus (Figure 8), coupled with rapid nuclear expansion. In addition to the contribution of chromatin condensation, another important aspect regarding the mechanisms that control chromatin-mediated nuclear expansion dynamic is how to transmit the expansion forces from the chromatin to the nuclear envelope. One plausible idea is that chromatin interaction with the nuclear envelope could work as a force transmitter. In this assumption, an increase in chromatin interaction can transmit more expansion forces from the chromatin to the nuclear envelope, implying an increase in nuclear expansion, nuclear rigidity, and nuclear membrane tightening. Actually, the observed nuclear expansion parameters correlated positively with the strength of chromatin interaction with nuclear envelope among samples with reduced chromatin interaction by digesting DNA or with enhanced chromatin tethering in the actin-intact extracts (Figure 6). Supporting this idea, previous studies indicated that a lack of or disturbance in chromatin tethering to the nuclear envelope induces excess nuclear deformation and a less stiff nucleus in cells in vivo (Schreiner, Koo, et al., 2015; Stephens et al., 2017, 2018). Some inner nuclear membrane proteins, including lamin, LEM domain (LAP2, emerin, MAN1 domain) family proteins, and filamentous actin, have been identified to bind to the chromatin (Gruenbaum et al., 2005; Zierhut et al., 2014; Oda et al., 2017). Thus, a lesser physical interaction of chromatin with the nuclear envelope could reduce the amount of expansion forces transmitted from the chromatin to the nuclear envelope and induce an excess accumulation of nuclear membrane without tightening. Further studies in direct force measurements against the nucleus during the nuclear expansion phase and under perturbations with chromatin structure would be informative for understanding how chromatin can impact nuclear expansion dynamics.
Regardless, the physical properties of chromatin alone, including chromatin condensation and chromatin interaction with the nuclear envelope, are not able to determine nuclear expansion dynamics. A supply of membrane constituents, such as inner nuclear membrane proteins and lipid membranes, from the cytoplasm per se is definitely a prerequisite to expand and form the nucleus (Levy and Heald, 2010; Hara and Merten, 2015). Therefore, we assume that the contribution of the physical properties of chromatin balances with the supply of nuclear membrane constituents to determine nuclear expansion dynamics. When an imbalance occurs due to a reduction in DNA quantity, the nuclear membrane could not expand rapidly, failing to maintain tension with an excess accumulation of membrane constituents (Figure 4). Furthermore, when an imbalance occurs due to an excess supply of membrane constituents, lamins or lipid membranes, without changing the DNA quantity, a similar surplus membrane accumulation could be observed (Jevtic´ et al., 2015; Walters, Amoateng, et al., 2019). Although the mechanisms by which cells comprehensively regulate nuclear size remain unclear, it is clear that nuclear size determination in vivo is likely coordinated by multiple factors, including the supply of membrane constituents, the contribution of chromatin, and others, rather than being regulated by a single factor (Kume et al., 2017; Cantwell and Nurse, 2019; Walters, Amoateng, et al., 2019).
Our major finding in this study, which is the contribution of physical properties of DNA and chromatin to nuclear size control, agrees with the original nucleoskeletal theory proposed by Cavalier-Smith (2005) that includes the contributions of DNA content and chromatin structure based on interspecies comparisons between genome size and nuclear size. DNA structure itself does not differ across species, and therefore the contributions of physical properties of DNA to nuclear size control should be conserved among eukaryotes. From an evolutionary viewpoint, abrupt changes in the DNA content such as deletion and duplication can rapidly modulate nuclear size regardless of the exact changes in the DNA sequences and their transcripts. The alteration in nuclear size is assumed to modulate the status of chromatin structures such as formation of heterochromatin, euchromatin, and topological domains due to the limited area for contact of chromatin to the nuclear envelope, resulting in accompanied modulation of DNA function as a long-term secondary effect. Thus, organisms might adapt the DNA content in addition to altering encoding genes to modulate the intranuclear functions in the context of evolutionary processes.
MATERIALS AND METHODS
Reconstruction of nuclei using cell-free Xenopus egg extracts
Crude cytostatic factor (CSF) metaphase-arrested extracts from unfertilized X. laevis eggs were prepared as described in a previous study (Iwabuchi, Ohsumi, et al., 2000). To reconstruct the interphase nuclei, crude CSF extracts were supplemented with 1 µM tetramethyl-rhodamine-dUTP (TMR-dUTP; Roche; 11534378910; to visualize DNA replication), 100 µg/ml cycloheximide (Sigma; C7698; to inhibit spontaneous transition to mitosis), 2 µM GFP-NLS recombinant proteins (to visualize nuclear import), and 0.6 mM CaCl2 (to release the extract into interphase). Subsequently, demembranated sperm (150–200/µl) were mixed with the extract and the samples were incubated for the indicated time period at 22°C. Extracts showing the formation of nuclei with aberrant morphology were excluded from further experiments. Mitotic cycling extracts from activated X. laevis eggs were prepared as previously described (Ohsumi et al., 2006). To recapitulate the cell cycle, the cycling extracts were supplemented with demembranated sperm (150–200/µl) and incubated for the indicated time period at 22°C. Normally the cell cycle (M–S–M transition) is repeated at least three times at ∼60 min intervals. In the case of arrest at the first interphase just after monitoring the M phase behavior (like condensed chromosomes), 100 µg/ml cycloheximide was added and incubated for the indicated time period at 22°C. In the case of arrest at the second interphase after monitoring the second mitotic behavior, cycloheximide was added. To prepare the actin-intact extract, the unfertilized eggs of X. laevis were crushed without cytochalasin B (Sigma; C6762) as previously described (Oda et al., 2017). Demembranated sperm chromatin from X. laevis was prepared as described (Ohsumi et al., 2006). Demembanated sperm chromatin from X. tropicalis (obtained from the X. tropicalis National Bio-Resource Project, Japan) was prepared using the same protocol as for X. laevis.
Observation of the reconstructed nuclei
The reconstructed nuclei were fixed and stained by mixing the same volume of 4% formaldehyde and 15% glycerol in the extraction buffer (EB; 100 mM KCl, 5 mM MgCl2, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH, pH 7.5) containing 5 µg/ml Hoechst 33342 (Invitrogen; H1399) and 10 µg/ml DiOC6(3) (Sigma; 318426). For the measurement of nuclear size or imported GFP-NLS proteins, ∼4 µl of the mixture was put on a glass slide and covered with a coverslip (without staining with DiOC6(3), in the case of GFP-NLS observation). These observation slides were subjected to fluorescence imaging using an Eclipse Ti-E (Nikon) wide-field microscope equipped with a 40× objective lens and sCMOS camera Zyla4.2 (Andor) motorized by Micromanager. For the measurement of nuclear size under noncompressive conditions (Supplemental Figure S2C), ∼10 µl of the fixed mixture containing the reconstructed nuclei stained with Hoechst 33342 and DiOC6(3) was put on a glass slide between two lines of spacer tape (commercial 100-µm-thick, double-sided tape) and covered with a coverslip, sealing the open parts with VALAP. For immunostaining of tubulin, extracts containing the reconstructed nuclei were fixed and stained as described previously (Hara and Merten, 2015). Briefly, 10 µl of egg extract containing nuclei was gently mixed by inversion with 1 ml of 30% glycerol and 4% formaldehyde in BRB-80 (80 mM piperazinediethanesulphonic acid [PIPES], 1 mM ethylene glycol bis(2-aminoethyl ether)-N,N,N’,N’-tetraacetic acid [EGTA], 1 mM MgC12, pH 6.8). After incubation for 15 min at room temperature, the fixed nuclei fractions were sedimented on ethanol-coated coverslips through a 3 ml glycerol cushion (40% glycerol in BRB-80 buffer) by centrifugation at 2000 × g and 16°C for 15 min. The coverslips were subsequently stained with an anti–α-tubulin antibody (DM1A clone; Thermo Fisher Scientific; MS581P0; 1000× dilution), Alexa 633 anti-mouse immunoglobulin G antibody (Thermo Fisher Scientific; A11003; 1000× dilution), 10 µg/ml Hoechst 33342, and 10 µg/ml DiOC6(3). The samples were subjected to z-sectional imaging using the above-mentioned wide-field microscope equipped with a 20× or 40× objective lens and processed deconvolution using the Microvolution ImageJ plug-in (Microvolution, USA). For visualizing the nuclear membrane using DiOC6(3), extracts containing reconstructed nuclei were fixed with 5% formaldehyde in EB and sedimented on poly-L-lysine–coated coverslips through a 3 ml sucrose cushion (30% sucrose in EB) by centrifugation at 200–1000 × g at 16°C for 15 min. Coverslips were stained with 10 µg/ml Hoechst 33342 and 1 µg/ml DiOC6(3). For visualizing nuclei with staining NPCs, the above-mentioned coverslips with sedimented nuclei were stained using α-NPC antibody (mAb414; BioLegend; 902901). Imaging of DiOC6(3) signals and NPCs on the nuclei was performed using the above-mentioned wide-field microscopy setup with a 100× objective lens and processed for deconvolution as described above.
Observation of nuclear expansion of in vivo translucent blastomeres
The translucent blastomeres with fewer yolks and pigment granules were prepared and cultured as previously described (Iwao et al., 2005) with slight modifications. Briefly, the fertilized and dejellied embryos of X. laevis at the four-cell stage just before the third cleavage furrow formation were floated on 32% Ficoll (wt/vol) in Steinberg’s solution (58 mM NaCl, 0.67 mM KCl, 0.34 mM CaCl2, 0.85 mM MgSO4, and 4.6 mM Tris-HCl, pH 7.4) and centrifuged at 700 × g for 10 min at 20°C. The centrifuged embryos were incubated in Steinberg’s solution for 1 h and transferred into Ca-Mg–free medium (88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, and 7.5 mM Tris-HCl, pH 8.0). After incubating for 1 h, fertilization membranes were manually removed with fine forceps and translucent spherical blastomeres of the animal hemispheres were isolated with a glass rod in the Ca-Mg–free medium. The isolated translucent blastomeres were observed on the fibronectin (0.1 mg/ml) and laminin (0.1 mg/ml)-coated glass bottom dish within a small drop of the Ca-Mg–free medium with 0.5−1 µg/ml Hoechst 33342. Time-lapse imaging was carried out with the above-mentioned wide-field fluorescence microscopy with a 20× objective lens. To obtain haploid embryos, the UV-irradiated sperm was fertilized on the laid eggs as previously described (Kawahara, 1978). Briefly, the sperm suspension was irradiated with 81 mJ/cm2 UV radiation and used for insemination. A portion of the inseminated eggs was incubated until the four-cell stage just before cleavage furrow formation and were subject to the above-mentioned translucence procedures. The remaining part of the treated embryos was kept incubating until swimming tadpoles were formed. Only if more than half of the embryos survived and most of the survived embryos displayed a typical haploid syndrome (Hamilton, 1963), the data of translucent blastomers were used for further analyses as a haploid sample. For sampling the data from the time-lapse images, we manually excluded the blastomeres displaying chromosome mis-segregation and large numbers of condensed chromosomes (from diploid embryos) in mitosis since the resulting translucent blastomeres are expected to be prepared from the mixtures of haploid with diploid embryos.
Reagents
For inhibiting DNA replication, 50–100 µM APH (Wako; 011-09811) and 5–15 µM p27 recombinant protein were simultaneously added to the extracts with demembranated sperm chromatin. Geminin recombinant protein (0.4–1.2 µM) was added into the CSF extract and preincubated for 15 min at 22°C, in advance of adding CaCl2, cycloheximide, and demembranated sperm chromatin. Both recombinant proteins were kindly gifted by Atsuya Nishiyama (University of Tokyo). For manipulating the chromatin structure, 5−20 mM MgCl2, 10 µg/ml actinomycin D (Wako; 018-21264), or 141 µM ICRF-193 (Santa Cruz; sc-200889) was added to the extract containing preassembled nuclei after 30–40 min of incubation. For digesting DNA inside the nucleus, 0.2 U/µl EcoRI, 0.2 U/µl XhoI, or 0.1 U/µl HaeIII (NEB; R3101, R3101, R0146) was added with the equipped buffer (NEB; 1× CutSmart buffer) into the extract containing preassembled nuclei.
Quantification of nuclear size and cellular parameters
The cross-sectional area of the images of the reconstructed nucleus in a fixed sample on a glass slide was measured using an ellipsoid function in ImageJ software (National Institutes of Health) throughout this study. The ellipsoid region of interest (ROI) was set on the rim of DiOC6(3)-positive nuclear membranes or along the inner side of the membrane accumulation around the DNA signal if the rim signal was not clear. When measuring the nuclear cross-sectional area on the fixed slide, the detected area was slightly larger than that in the noncompressive conditions (Supplemental Figure S2C), suggesting that the nuclei were compressed to some degree in the fixed slide in this study. For quantification of the nuclear expansion dynamics, we calculated the mean value of the cross-sectional area at each incubation time point and utilized the data set of the mean values in each extract preparation. The values were measured in at least 20 nuclei for each experimental condition, such as different time points and supplements. Each experimental condition was repeated using at least three individual extracts. To reduce the effects of variations in extract preparation, we used only data showing a more than 35-µm diameter in the reconstructed nuclei with X. laevis sperm chromatin without any inhibitor after 120-min incubation for further analyses as described previously (Hara and Merten, 2015). Moreover, for normalization, the calculated nuclear parameters at a particular time point and condition (e.g., extract treated with APH or X. tropicalis sperm chromatin for 80-min incubation) were divided by the mean nuclear cross-sectional area obtained after 120 min of incubation in the control condition using exactly the same extract preparation (e.g., nuclei with the buffer supplementation (dimethyl sulfoxide [DMSO]) using X. laevis sperm chromatin for 120-min incubation). Mean values of the mean nuclear sectional area as well as of the mean SDs from every individual experiment are illustrated in the graphs. For calculating the speed of nuclear expansion using the data set of mean nuclear sectional area, we calculated a slope of the regression line in the nuclear sectional area against time from 60 to 120 min of incubation in each extract preparation. For calculating the maximum values, we choose the maximum mean value from the data sets during 240 min of incubation.
As for in vivo blastomere imaging, the cross-sectional area of the Hoechst 33342–positive nucleus and the cross-sectional area of the cell detected using the phase contrast image were measured. Using a time-series data set of the measured nuclear cross-sectional area in individual blastomeres, the slope of the regression line in the nuclear cross-sectional area against time was calculated and the plateau value was chosen. For calculating the increase in the nuclear cross-sectional area after completion of the nuclear formation, we subtracted the values recognized as initial time series of interphase (for in vivo imaging) from values after the indicated time. The cell volume was estimated by considering the cell as a sphere.
For quantification of MT-occupied space, the size was quantified on the basis of the Gaussian fitting for intensity histogram as described previously (Hara and Merten, 2015). For quantification of imported GFP-NLS proteins, the images were acquired with the same exposure conditions from the samples that were compared. From the images, the mean intensity per pixel inside the nuclear region was measured by ImageJ with an ellipsoid function and subtracted the background intensity. Furthermore, the whole intensity inside the nucleus was calculated by multiplying the mean intensity by the measured cross-sectional area. To normalize variations among the extract preparations, the mean value of each intensity was divided by the mean value in the sample containing nuclei with X. laevis sperm chromatin without any inhibitor after 120 min of incubation for each extract preparation. For quantification of the intensity of DiOC6(3)- or NPC-positive signals, images were acquired with z-sections, including the surface of the nuclei adhering to the coverslip, and with the same exposure conditions among samples. For the images after performing deconvolution, only a single plane of well-focused nuclear surface was subjected to further quantification. The mean intensities of the signal on line-like patterns and throughout the nuclear region within the rim of the nucleus were measured using ImageJ software with the Straight line and Ellipsoid ROIs, respectively. After subtracting the background intensity, these values were used. The whole intensity of these signals throughout the nucleus was estimated by multiplying the calculated intensity throughout the nuclear region by the measured nuclear cross-sectional area.
For calculating the density of DNA inside the nucleus, the calculated weight of DNA (1 base pair of double helix = 660 pg/mol) inside the nucleus was divided by the estimated nuclear volume or nuclear surface area, using only the data after 60 min of incubation (50 min of incubation in the case using the cycling extract), recognizing the completion of DNA replication. The nuclear surface area and volume were estimated by considering the nucleus as a sphere.
Statistical analysis
For evaluation of correlation, the coefficient of determination, R2, was calculated by Excel software. Significant differences among samples were determined by nonparametric Wilcoxon tests using R software.
Supplementary Material
Acknowledgments
We are grateful to Atsuya Nishiyama for providing recombinant proteins and other members of our lab for fruitful discussion. Xenopus tropicalis was provided by the Amphibian Research Center (Hiroshima University) through the National Bio-Resource Project of MEXT, Japan. This study was supported by the Home for Innovative Researchers and Academic Knowledge Users (HIRAKU) consortium, a research grant from the Astellas Foundation for Research on Metabolic Disorders, a Sasakawa Scientific Research Grant from the Japan Science Society, JSPS KAKENHI Grant Numbers JP16K14727, JP20H03253, and research grants from the Inamori Foundation and the Yamaguchi University Foundation (Y.H.).
Abbreviations used:
- APH
aphidicolin
- CSF
crude cytostatic factor
- DiOC 6(3)
3,3′-dihexyloxacarbocyanine iodide
- DMSO
dimethyl sulfoxide
- dUTPs
deoxyuridine triphosphates
- GDP
guanosine diphosphate
- GFP
green fluorescent protein
- GTP
guanosine triphosphate
- LEM domain
LAP2, emerin, MAN1 domain
- MT
microtubule
- NLS
nuclear localization signal
- TMR
tetramethyl-rhodamine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-02-0113) on September 30, 2020.
REFERENCES
Boldface names denote co–first authors.
- Anderson DJ, Hetzer MW (2007). Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol 9, 1160–1166. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82. [DOI] [PubMed] [Google Scholar]
- Cantwell H, Nurse P (2019). A systematic genetic screen identifies essential factors involved in nuclear size control. PLoS Genet 15, e1007929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T (1978). Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci 34, 247–278. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T (1982). Skeletal DNA and the evolution of genome size. Annu Rev Biophys Bioeng 11, 273–302. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T (2005). Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot 95, 147–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CJ, Li W, Cojoc G, Guck J (2017). Volume transitions of isolated cell nuclei induced by rapid temperature increase. Biophys J 112, 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens LJ, White KH, Jevtic P, Li X, Levy DL (2013). Nuclear size regulation: from single cells to development and disease. Trends Cell Biol 23, 151–159. [DOI] [PubMed] [Google Scholar]
- Fankhauser G (1945). Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment of cell number and cell shape. J Exp Zool 100, 445–455. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M (2015). Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun 6, 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Hyman AA (2012). Organelle growth control through limiting pools of cytoplasmic components. Curr Biol 22, 330–339. [DOI] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR (1996). Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Gregory TR (2001). The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells Mol Dis 27, 830–843. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL (2005). The nuclear lamina comes of age. Nat Rev Mol Cell Biol 6, 21–31. [DOI] [PubMed] [Google Scholar]
- Guy AL, Taylor JH (1978). Actinomycin D inhibits initiation of DNA replication in mammalian cells. Proc Natl Acad Sci USA 75, 6088–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton L (1963). An experimental analysis of the development of the haploid syndrome in embryos of Xenopus laevis . J Embryol Exp Morphol 11, 267–278. [PubMed] [Google Scholar]
- Hara Y, Merten CA (2015). Dynein-based accumulation of membranes regulates nuclear expansion in Xenopus laevis egg extracts. Dev Cell 33, 562–575. [DOI] [PubMed] [Google Scholar]
- Iwabuchi M, Ohsumi K, Yamamoto TM, Sawada W, Kishimoto T (2000). Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M-M transition in Xenopus oocyte extracts. EMBO J 19, 4513–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwao Y, Uchida Y, Ueno S, Yoshizaki N, Masui Y (2005). Midblastula transition (MBT) of the cell cycles in the yolk and pigment granule-free translucent blastomeres obtained from centrifuged Xenopus embryos. Dev Growth Differ 47, 283–294. [DOI] [PubMed] [Google Scholar]
- Jevtic´ P, Edens LJ, Li X, Nguyen T, Chen P, Levy DL (2015). Concentration-dependent effects of nuclear lamins on nuclear size in Xenopus and mammalian cells. J Biol Chem 290, 27557–27571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic´ P, Levy DL (2015). Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr Biol 25, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic´ P, Levy DL (2017). Both nuclear size and DNA amount contribute to midblastula transition timing in Xenopus laevis . Sci Rep 7, 7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B (2007). The size of the nucleus increases as yeast cells grow. Mol Biol Cell 18, 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H (1978). Production of triploid and gynogenetic diploid Xenopus by cold treatment. Dev Growth Differ 20, 227–236. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Tada S, Tsuyama T, Murofushi H, Seki M, Enomoto T (2002). Focus-formation of replication protein A, activation of checkpoint system and DNA repair synthesis induced by DNA double-strand breaks in Xenopus egg extract. J Cell Sci 115(Pt 15), 3159–3169. [DOI] [PubMed] [Google Scholar]
- Kume K, Cantwell H, Burrell A, Nurse P (2019). Nuclear membrane protein Lem2 regulates nuclear size through membrane flow. Nat Commun 10, 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Cantwell H, Neumann FR, Jones AW, Snijders AP, Nurse P (2017). A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control. PLoS Genet 13, 1006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al. (2013). The genomic and transcriptomic landscape of a HeLa cell line. G3 3, 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R (2010). Nuclear size is regulated by importing α and Ntf2 in Xenopus . Cell 143, 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R (2015). Biological scaling problems and solutions in amphibians. Cold Spring Harb Perspect Biol 8, 019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee RN, Sallé J, Dmitrieff S, Nelson KM, Oakey J, Minc N, Levy DL (2020). The perinuclear ER scales nuclear size independently of cell size in early embryos. Dev Cell 54, 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG (2001). Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540–1543. [DOI] [PubMed] [Google Scholar]
- Neumann FR, Nurse P (2007). Nuclear size control in fission yeast. J Cell Biol 179, 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Shirai N, Ura N, Ohsumi K, Iwabuchi M (2017). Chromatin tethering to the nuclear envelope by nuclear actin filaments: a novel role of the actin cytoskeleton in the Xenopus blastula. Genes Cells 22, 376–391. [DOI] [PubMed] [Google Scholar]
- Ohsumi K, Yamamoto TM, Iwabuchi M (2006). Oocyte extracts for the study of meiotic M-M transition. Methods Mol Biol 322, 445–458. [DOI] [PubMed] [Google Scholar]
- Robinson DO, Coate JE, Singh A, Hong L, Bush M, Doyle JJ, Roeder AHK (2018). Ploidy and size at multiple scales in the Arabidopsis sepal. Plant Cell 30, 2308–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T (2006). Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SGJ, King MC (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun 6, 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Tamura S, Masumoto H, Maeshima K (2017). Nucleosome– nucleosome interactions via histone tails and linker DNA regulate nuclear rigidity. Mol Biol Cell 28, 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell 28, 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF (2019). Chromatin’s physical properties shape the nucleus and its functions. Curr Opin Cell Biol 58, 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liua PZ, Banigan EJ, Almassalhad LM, Backmand V, Adame SA, Goldmane RD, Marko JF (2018). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell 29, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlaki E, FitzHarris G (2016). Nucleus downscaling in mouse embryos is regulated by cooperative developmental and geometric programs. Sci Rep 6, 28040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S, Platani M, Boue S, Mattaj IW (2006). Direct membrane protein– DNA interactions required early in nuclear envelope assembly. J Cell Biol 173, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppaluri S, Weber SC, Brangwynne CP (2016). Hierarchical size scaling during multicellular growth and development. Cell Rep 17, 345–352. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS (2017). Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AD, Amoateng K, Wang R, Chen JH, McDermott G, Larabell CA, Gadal O, Cohen-Fix O (2019). Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume. Mol Biol Cell 30, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut, C., Jenness, C., Kimura, H., Funabiki, H. (2014). Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat Struct Mol Biol 21 , 617–625. [Google Scholar]
- Zink D, Fischer AH, Nickerson JA (2004). Nuclear structure in cancer cells. Nat Rev Cancer 4, 77–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.