Abstract
UNC-104 is the Caenorhabditis elegans homolog of kinesin-3 KIF1A known for its fast shuffling of synaptic vesicle protein transport vesicles in axons. SYD-2 is the homolog of liprin-α in C. elegans known to activate UNC-104; however, signals that trigger SYD-2 binding to the motor remain unknown. Because SYD-2 is a substrate of PTP-3/LAR PTPR, we speculate a role of this phosphatase in SYD–2-mediated motor activation. Indeed, coimmunoprecipitation assays revealed increased interaction between UNC-104 and SYD-2 in ptp-3 knockout worms. Intramolecular FRET analysis in living nematodes demonstrates that SYD-2 largely exists in an open conformation state in ptp-3 mutants. These assays also revealed that nonphosphorylatable SYD-2 (Y741F) exists predominately in folded conformations, while phosphomimicking SYD-2 (Y741E) primarily exists in open conformations. Increased UNC-104 motor clustering was observed along axons likely as a result of elevated SYD-2 scaffolding function in ptp-3 mutants. Also, both motor velocities as well as cargo transport speeds were visibly increased in neurons of ptp-3 mutants. Lastly, epistatic analysis revealed that PTP-3 is upstream of SYD-2 to regulate its intramolecular folding.
INTRODUCTION
The transport of vital neuronal factors such as nucleic acids, lipids, enzymes, and organelles (synaptic vesicles, mitochondria, etc.) is essential for proper neuronal survival and functions. UNC-104 is the homolog of KIF1A in Caenorhabditis elegans known to be a fast-anterograde transporter of STVs (synaptic vesicle protein transport vesicles) regulated by active zone protein SYD-2 (liprin-α) (Hall and Hedgecock, 1991; Wagner et al., 2009; Wu et al., 2016). Heritable genetic defects in motor proteins and their adaptors are linked to various neurological diseases such as adult-onset neurodegenerative disease, Parkinson’s disease (PD), Alzheimer’s disease (AD), Charcot-Marie-Tooth disease, tauopathies, and amyotrophic lateral sclerosis (Goldstein, 2001; Hirokawa and Takemura, 2003; Franker and Hoogenraad, 2013; Brady and Morfini, 2017; Lucanus and Yip, 2018; Sheng et al., 2018). Understanding how motor proteins are regulated will provide insights on how these diseases develop. The following mechanisms reflect a few examples of how kinesins are regulated: 1) Intramolecular folding deactivates kinesins (Siddiqui and Straube, 2017). 2) Cargo binding activates kinesins (Niwa et al., 2016). 3) Physical pulling via opposing motor dynein activates kinesins (Chen et al., 2019). 4) Binding of small adaptor proteins such as SYD-2 and LIN-2 activates kinesin (Wagner et al., 2009; Wu et al., 2016). 5) Phosphorylation regulates kinesins (Gibbs et al., 2015). 6) Posttranslational modifications of microtubules (MT) as well as MT-associated proteins (MAPs) regulate kinesins (Tien et al., 2011; Shanmugam et al., 2018). 7) Differential organization of MT in subcellular regions regulates kinesin directionality (Yogev et al., 2016). In this study, we will focus on how SYD-2 regulates UNC-104 via intramolecular folding coordinated by PTP-3 phosphatase. A broad set of kinases such as GSK3β, MAPKs, ERK1/2, p38 MAPKs, JNKs, Cdk5, Cdk1, Akt, PKA, CK2, PKC, and PINK1 regulates axonal transport either directly (affecting motors) or indirectly (via adaptor proteins) (Blangy et al., 1995; Gibbs et al., 2015; Goldstein et al., 2017, 2019). On the other hand, how kinesins are regulated by phosphatases remains largely unknown. One example is that kinesin-like proteins Eg5 and MKlp2 are substrates of PTEN and PP2A phosphatases that function in cell division (McIlvain et al., 1994; He et al., 2016; Fung et al., 2017; Liu et al., 2017).
PTP-3 is an ortholog of human LAR PTPRD (leukocyte common antigen-related protein tyrosine phosphatase transmembrane receptor type D) and LAR PTPRF (leukocyte common antigen-related protein tyrosine phosphatase transmembrane receptor type F). It is composed of three N-terminal Ig domains, eight FN3 domains, a transmembrane domain, and two C-terminal PTP domains D1 and D2 (with D2 possessing very little or null phosphatase catalytic activity) (Nam et al., 1999). Northern blot analysis identified two isoforms of PTP-3 that express broadly in various tissues at early worm stages, but is restricted to the nervous system in the adult worm. Worms carrying a ptp-3 mutation have the posterior body bulged at the larval stage L1 based on defects in epidermal morphogenesis (Harrington et al., 2002). It has also been demonstrated that the PTP-3A isoform is localized to synaptic regions along with other active zone proteins such as UNC-10; however, the PTP-3B isoform is localized to extrasynaptic regions. Null mutations of ptp-3 lead to synaptic defects as well as defects in axon guidance and neuroblast migration, while overexpression of the PTP-3B isoform can rescue axon guidance and neuroblast migration defects, but not synaptic defects (Ackley et al., 2005). LIP.1 (LAR-interacting protein 1, later renamed to liprin-α) functionally interacts with the D2 domain of LAR and is often localized at focal adhesion. Functional interactions between liprins and LAR have been known for more than two decades (Serra-Pages et al., 1995, 1998); however, only recently a proteomic study had revealed that a tyrosine at position 741 in SYD-2 may act as a potential target for PTP-3 (Mitchell et al., 2016).
In C. elegans, a syd-2 mutation ju37 was found to cause diffused localization of synaptic markers such as SNB-1, syntaxin, and RAB-3. Other phenotypes caused by syd-2 mutations (such as ju37 or ok217) include reduced locomotion, defective egg laying, as well as egg retention and hatch-bag phenotypes (Jin, 1999; Patel et al., 2006). Previous studies from our lab and collaborators revealed that the physiological function of SYD-2 is regulated by an intramolecular folding via an interaction between the N-terminal CC (coiled coil) domains and C-terminal SAM (sterile alpha motif) domains (Wagner et al., 2009; Chia et al., 2013). In C. elegans, SYD-2 acts primarily as a scaffolding protein to regulate synapse formation, stability, and organization along with other active zone proteins such as ELKS-1, RYS-1, UNC-10, and sentryn (Yeh et al., 2005; Dai et al., 2006; Patel and Shen, 2009; Stigloher et al., 2011; Taru and Jin, 2011; Kittelmann et al., 2013; Edwards et al., 2015, 2018). Apart from this function, we have previously demonstrated that SYD-2 acts as an adaptor protein to regulate UNC-104′s motility by interacting with the motor’s FHA domain through its SAM domains. This interaction likely stabilizes the motor’s dimeric state thus enhancing motor motility (Wagner et al., 2009; Wu et al., 2016). Based on this knowledge, we hypothesize that extrasynaptical PTP-3B may be a crucial factor to regulate UNC-104 motor activity via modulating the intramolecular folding of SYD-2 and likely by dephosphorylating tyrosine 741.
RESULTS
Functional and genetic interactions among UNC-104, SYD-2, and PTP-3
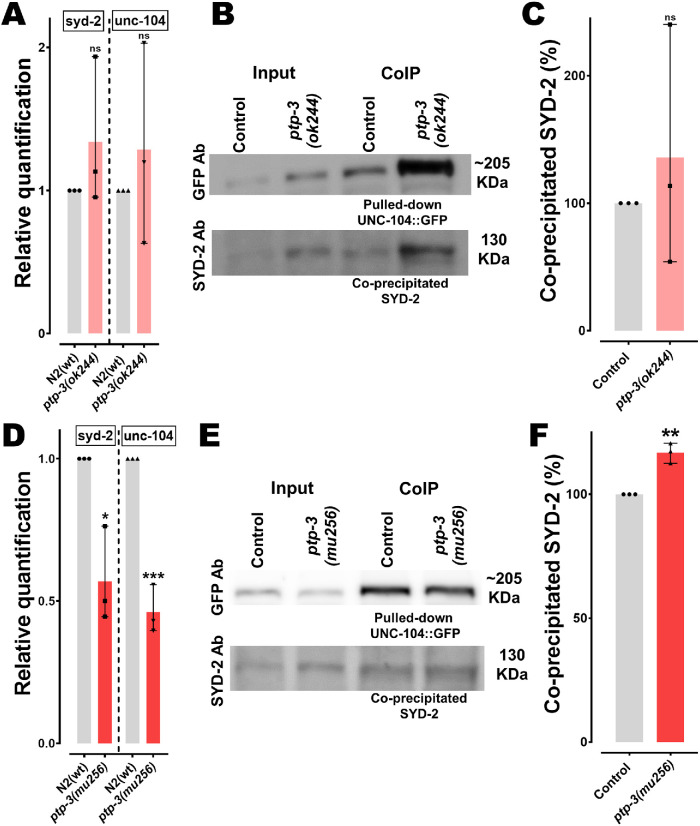
To understand if genetic interactions among the proposed PTP-3/SYD-2/UNC-104 pathway exist, we measured mRNA levels of both syd-2 and unc-104 genes in two different ptp-3 mutant strains, ptp-3(ok244) and ptp-3(mu256). As mentioned in Materials and Methods, the ok244 allele encodes for a frameshift and premature termination in the ptp-3 gene resulting in a lack of the PTP-3A isoform, while the PTP-3B isoform still remains expressed (Supplemental Figure S1). On the other hand, the mu256 allele has been characterized to be a loss-of-function mutation and worms do not express both isoforms (Ackley et al., 2005). The same study revealed that PTP-3A primarily locates at synapses while PTP-3B localizes at extrasynaptic sites. From Figure 1A it is evident that worms carrying the ok244 allele do not reveal significant differences in syd-2 and unc-104 mRNA expression, while in mu256 mutants, both syd-2 and unc-104 genes are significantly down-regulated (Figure 1D). These results may point to a genetic relation not only between ptp-3 and syd-2 (as expected) but also, unexpectedly, between ptp-3 and unc-104. To understand whether PTP-3 plays a role in reported functional interactions between UNC-104 and SYD-2 (Wagner et al., 2009; Hsu et al., 2011), we performed coimmunoprecipitation (IP) assays. Interestingly, UNC-104/SYD-2 interactions are pronounced in ptp-3(mu256) mutants (Figure 1, E and F; with pulled-down UNC-104::GFP used for normalization). Since the amount of coprecipitated SYD-2 in ptp-3(ok244) mutants varied greatly (due to unknown reasons), we cannot conclude whether this mutant affects UNC-104/SYD-2 interactions (Figure 1, B and C).
FIGURE 1:
Changes in syd-2/unc-104 gene expression levels and SYD-2/UNC-104 protein interactions in ptp-3 mutants. (A, D) Relative quantification of mRNA expression levels of syd-2 and unc-104 from lysates of young adult worms (N2 control, ptp-3(ok244) and ptp-3(mu256) allelic mutants). Quantitative PCR was performed thrice and each trial was performed in triplicates. (B, E) Upper lane: Western blot of pulled-down UNC-104::GFP using anti-GFP antibody. Bottom lane: Blot of co-IP SYD-2 detected using anti-SYD-2 polyclonal antibody from lysates of ok244 and mu256 mutants. (C, F) Quantification of coprecipitated SYD-2 (normalized with UNC-104::GFP) from different genetic backgrounds. Error bars: ± max. and min. range. Unpaired Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001; t, df = 1.126, 4 (A, syd-2); 0.7039, 4 (A, unc-104); 4.389, 4 (D, syd-2); 11, 4 (D, unc-104); 0.6547, 4 (C) and 7.182, 4 (F).
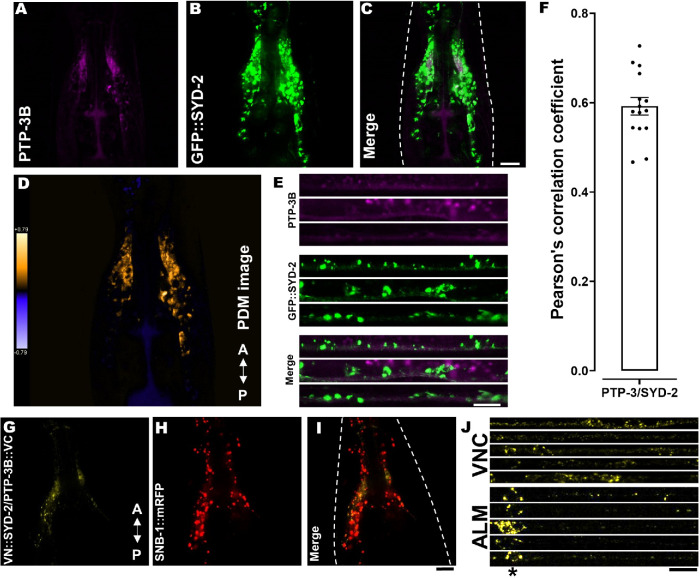
To examine whether extrasynaptic PTP-3B isoform would function to regulate SYD-2, and thereby affect UNC-104 motor motility, we performed both colocalization as well as BiFC (bimolecular fluorescent complementation) assays (Hsu et al., 2011). Indeed, we revealed positive Pearson’s correlation coefficient values when quantifying PTP-3B and SYD-2 colocalization in the C. elegans nervous system (Figure 2, A–F and Supplemental Figure S3, A and B; nerve ring and ventral nerve cord, VNC) as well as positive BiFC signals (Figure 2, G–J and Supplemental Figure S3C; nerve ring, VNC and ALM neuron). Critically, the PTP-3B/SYD-2 BiFC pair also colocalizes with a synaptic vesicle marker SNB-1 in the nerve ring (Figure 2, H and I). These data reveal that PTP-3 and SYD-2 possess the potential for physical and functional interactions in the nervous system of C. elegans not only at synapses (as reported by others for PTP-3A/SYD-2; Ackley et al., 2005) but also within axons, supporting our hypothesis for a potential role of PTP-3B/SYD-2 in UNC–104-mediated axonal trafficking.
FIGURE 2:
SYD-2/PTP-3B colocalization and BiFC analysis. (A–E) PTP-3B::CFP (pseudo-colored in magenta) and GFP::SYD-2 colocalization in nerve rings (A–D) and VNCs (E). (D) Pseudo-color PDM image (see Materials and Methods) indicating quantifiable colocalization between SYD-2 and PTP-3B in the nerve ring. (F) Pearson’s correlation coefficient quantification of data shown in C. (G) Positive PTP-3B/SYD-2 BiFC signal in the nerve ring of living worms indicating that SYD-2 and PTP-3B are at least 7–10 nm close to each other (see Materials and Methods). (H) Worm expressing SNB-1::mRFP under a pan-neuronal promoter. (I) Merged image from G and H. (J) Stacks of straightened VNCs and ALM neurons with positive PTP-3B/SYD-2 BiFC signals. *Location of somata in ALM neurons. Dotted, white line marks the contour of the worm head. Scale bars: 10 µm. N = 10 worms in F. Error bar: ± max. and min. range. A indicates the anterior direction and P the posterior direction. For BiFC-positive and -negative controls, refer to Supplemental Figure S3, C and D.
Lack of PTP-3B triggers increased unfolded states of SYD-2
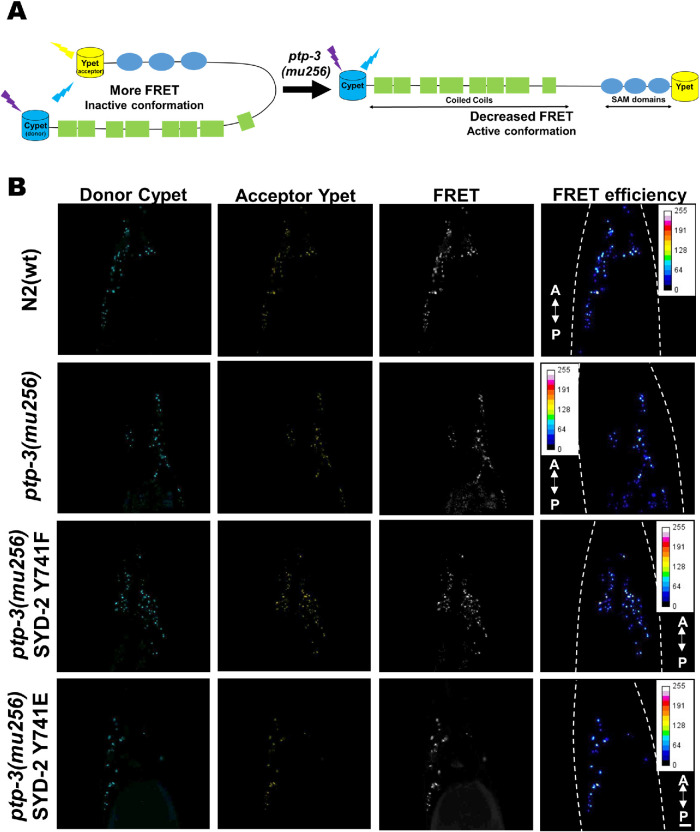
Because SYD-2 has been proposed to exist in functional (active) unfolded and (inactive) folded states (Chia et al., 2013), we propose that intramolecular folding of SYD-2 affects its binding propensities to UNC-104. As mentioned above, we assume that in ptp-3 mutants diminished dephosphorylation of SYD-2 at Y741 would lead to increased unfolded states that would enhance its interactions with UNC-104. To approach this question, we employed intramolecular Förster/Fluorescence resonance energy transfer (FRET) assays (Kalab et al., 2006) as depicted in Figure 3A. From Figure 3, B and C, it is evident that the FRET ratio (IFRET/ID) of the Cypet::SYD-2::Ypet fusion is significantly reduced in ptp-3(mu256) mutants, indicating that unfolded states of SYD-2 are more pronounced in the absence of PTP-3. Critically, also knocking down the ptp-3 gene using RNA interference (RNAi) led to comparable observations (Figure 3C; Supplemental Figure S4, C and D). As a control, we designed a nonphosphorylatable SYD-2 mutant Y741F, and its FRET ratios are comparable to that of Cypet::SYD-2::Ypet expressed in N2 wild type animals. Moreover, a phosphomimicking SYD-2 Y741E construct led to FRET ratios comparable to that of the unmodified Cypet::SYD-2::Ypet construct expressed in ptp-3(mu256) mutants (Figure 3, B and C). Also, a Cypet::Ypet FRET sensor without the syd-2 gene insert results in nonsignificant FRET ratios when expressed in either N2 wild type or ptp-3(mu256) mutants (Supplemental Figure S4, A and B). These findings demonstrate that SYD-2 appears in a more pronounced open conformation in the absence of PTP-3 and that this mechanism is likely a function of the phosphatase activity of PTP-3 on tyrosine 741 of SYD-2.
FIGURE 3:
SYD-2 exists predominantly in an open conformation in ptp-3(mu256) mutants. (A) Pictorial representation of the designed intramolecular FRET assay for this study. (B) Images of C. elegans nerve rings from the respective fluorescence channel. (C) Quantification of FRET efficiencies of Cypet::SYD-2::Ypet in different genetic backgrounds, where ptp-3 RNAi, ptp-3(mu256) and phosphomimicking SYD-2 Y741E all lead to a significant decrease in FRET ratios while the nonphosphorylatable SYD-2 Y741F reveals wild type signals. Scale bar: 10 µm. N = 26 images (C). Error bars: ± max. and min. range. One-way ANOVA with Fisher’s LSD test. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. F (DFn, DFd) = 15.83 (7, 200). A indicates anterior direction and P the posterior direction. Dotted, white line marks the contour of the worm head.
Abnormal sorting of STVs and UNC-104 motor distribution in ptp-3(mu256) mutants
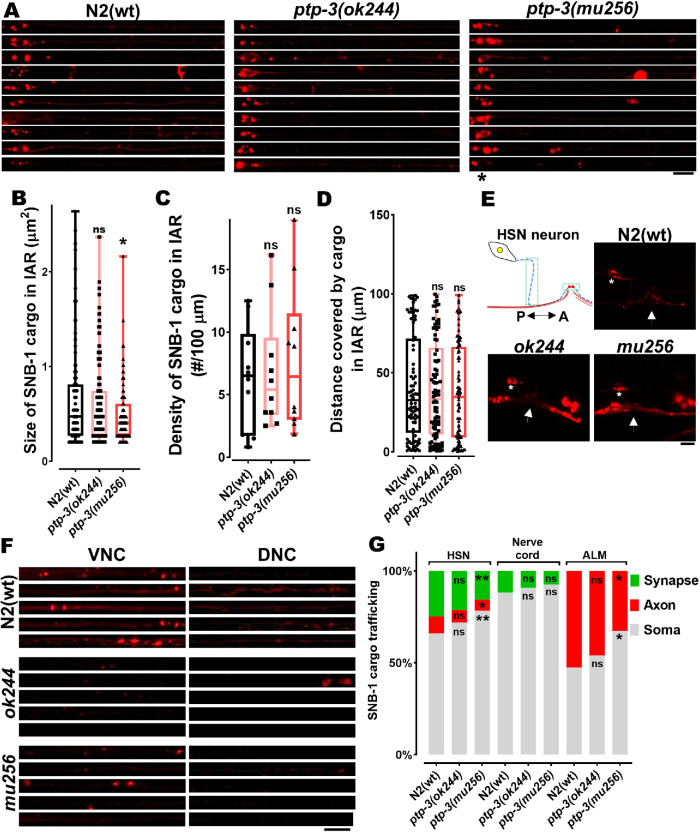
To understand whether this mechanism attributes to physiological significances, we analyzed STV distributions (with SNB-1 as a marker) in neurons in the presence or absence of PTP-3B (Figure 4A). SNB-1 particle (cluster) sizes (Figure 4B) are significantly reduced in the IAR (initial axonal region) of mu256 mutant axons, while SNB-1 cargo density (Figure 4C), as well as travel distances (Figure 4D), remain unchanged in these mutants. Interestingly, we observed a SNB-1 particle retention phenotype in somata of HSN and ALM neurons (Figure 4, E–G). While cargo obviously accumulates in the soma of HSN neurons, it is at the same time reduced at synapses in these neurons (Figure 4, E and G; note that synapses of ALM neurons are not easily accessible for quantification). This cargo retention phenotype can be partially explained by the reduced expression of unc-104 gene in mu256 mutants (Figure 1D).
FIGURE 4:
STVs accumulate in somata of ptp-3 mutants. (A) Straightened ALM neurons taken from different worms within a population and stacked on each other. (B–D) Quantification of size (B), densities (C), and travel distances (D) of SNB-1 containing synaptic vesicles from images displayed in A. (E) Cartoon of HSN neuron with its presynaptic area and representative images of SNB-1 cargo distribution in wild type and mutants. *Soma of neuron. White arrow indicates the location of the vulva. Green boxes indicate the region used for quantification of fluorescence intensity of axons (blue dashed line) and synapses (red circles). P indicates the posterior direction and A the anterior direction. (F) Representative images of VNC and DNC from different study groups, straightened and stacked. (G) Quantification (fluorescence intensity) of the amount of cargo in the cell body and the IAR of ALM, nerve cord (VNC somata and DNC synapses), and HSN neurons (soma, axon, and synapse). Scale bars: 10 µm. Box plot with median and error bars: ± max. and min. range. One-way ANOVA with Fisher’s LSD multiple comparison test. *p < 0.05 and **p < 0.01. F (DFn, DFd) = 3.426 (2, 320) (B); 3.862 (2, 27) (G, ALM); 4.327 (2, 43) (G, HSN soma); 2.455 (2, 43) (G, HSN axon) and 3.909 (2, 43) (G, HSN synapse).
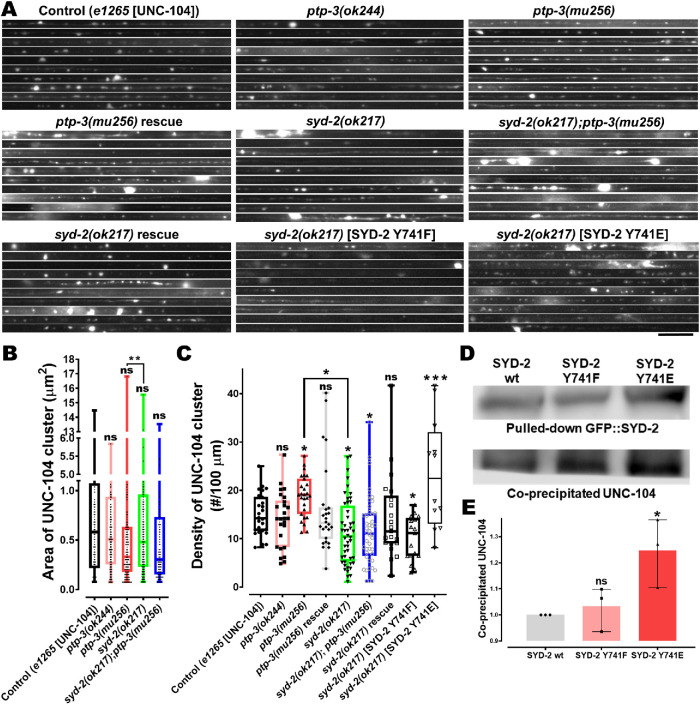
We then analyzed the distribution patterns of UNC-104 motor in the long axons of sublateral neurons in ptp-3 and syd-2 mutants as well as in ptp-3;syd-2 double mutants. We found that the densities of UNC-104 motor cluster increased significantly in ptp-3(mu256) mutants compared with wild types, and this phenotype can be rescued when overexpressing PTP-3B in mu256 mutants (Figure 5, A and C). Critically, UNC-104 motor clustering was reduced in syd-2 knockout animals (Figure 5C) consistent with previous findings (Wagner et al., 2009). As expected, overexpression of non-phosphorylatable SYD-2 Y741F was unable to rescue the ok217 effect. On the other hand, the phosphomimicking version of SYD-2 led to significantly increased UNC-104 clustering (Figure 5C), likely due to increased UNC-104 scaffolding along axons via SYD-2 (Wagner et al., 2009) induced by increased interaction between UNC-104 and SYD-2 Y741E (Figure 5, D and E). Epistasis analysis employing syd-2;ptp-3 double mutants revealed that lack of PTP-3 does not affect the syd-2 phenotype (Figure 5, A and C); thus, PTP-3 is likely located upstream of SYD-2.
FIGURE 5:
UNC-104 clustering is increased in ptp-3 mutants and decreased in syd-2 mutants. (A) UNC-104::mRFP accumulations and distribution patterns in straightened segments of sublateral neurons taken from different genetic backgrounds stacked on each other with: control = unc-104(e1265) allelic mutants rescued with UNC-104::mRFP; ptp-3(ok244) = ok244 allelic mutants expressing UNC-104::mRFP; ptp-3(mu256) = mu256 allelic mutants expressing UNC-104::mRFP; ptp-3(mu256) rescue = mu256 allelic mutants expressing both PTP-3B::CFP and UNC-104::mRFP; syd-2(ok217) = ok217 allelic mutants expressing UNC-104::mRFP; syd-2(ok217);ptp-3(mu256) = allelic double mutants expressing UNC-104::mRFP; syd-2(ok217) rescue = ok217 allelic mutants expressing both GFP::SYD-2 and UNC-104::mRFP; syd-2(ok217)[SYD-2 Y741F] = ok217 allelic mutant expressing both nonphosphorylatable GFP::SYD-2 Y741F and UNC-104::mRFP; syd-2(ok217)[SYD-2 Y741E] = ok217 allelic mutant expressing both phosphomimicking GFP::SYD-2 Y741E and UNC-104::mRFP. (B) UNC-104 cluster sizes in sublateral neurons. (C) UNC-104 densities along sublateral neurons. (D) Representative immunoblots from co-IP of UNC-104 from phosphodeficient (Y741F) and phosphomimicking (Y741E) SYD-2. (E) Quantification of coprecipitated UNC-104 either with phosphodeficient or phosphomimicking SYD-2. Scale bar: 10 µm. Box plot with median and error bars: ± max. and min. range. One-way ANOVA with Fisher’s LSD multiple comparison test. *p < 0.05, **p < 0.005, ***p < 0.001, and ****p < 0.0001. F (DFn, DFd) = 7.073 (8, 238) (C); 6.543 (2,6) (E).
PTP-3 knockout results in increased anterograde motor and cargo velocities
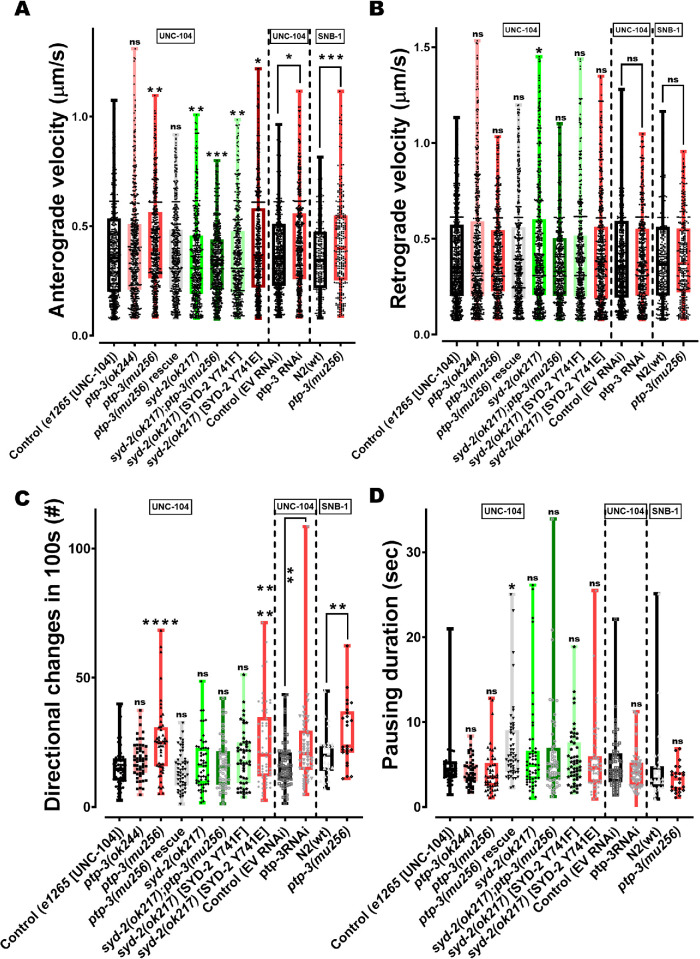
Based on the idea that PTP-3 suppresses the function of SYD-2 (that, in turn, would deactivate UNC-104), it is crucial to analyze UNC-104′s motility behavior in ptp-3 mutants. Indeed, kymograph analysis revealed that UNC-104 velocities in ptp-3(mu256) mutants significantly increased in anterograde directions (Figure 6A; see also Supplemental Figures S6 and S8). Because UNC-104 is a plus-end directed motor, it is not surprising that retrograde directions barely changed (Figure 6B). However, it needs to be noted that UNC-104 may be directly engaged in a tug-of-war with dynein (Chen et al., 2019) and/or that multiple motors of different directionalities may bind to the same cargo vesicle resulting in saltatory movements of that vesicle. Therefore, we can also not exclude that our data would comprise multiple motors events. Interestingly, bidirectional movements of UNC-104 in ptp-3(mu256) mutants increased (Figure 6C), pointing to increased tug-of-war events. It is critical to note that changes in UNC-104 motility in ptp-3(mu256) mutants are comparable to those in syd-2(ok217) worms rescued with phosphomimicking SYD-2 Y741E, and that these changes can be partially reverted by overexpressing full-length PTP-3B in mu256 mutants (Figure 6, A–D). As expected, UNC-104 moving properties did not change in ptp-3(ok244) mutants as a result of the unaffected isoform PTP-3B in these mutants (Supplemental Figure S1).
FIGURE 6:
Both motor and cargo speeds are increased in ptp-3(mu256) mutants. (A) UNC-104 and SNB-1 anterograde velocities in various genetic backgrounds as well as after ptp-3 RNAi. (B) UNC-104 and SNB-1 retrograde velocities in various genetic backgrounds as well as after ptp-3 RNAi. (C) Quantification of motor and cargo reversals (number of directional changes within 100s). (D) Quantification of motor and cargo pausing duration. (E, F) Quantification of total anterograde (E) and (F) retrograde run length. (G) Quantification of net run length. Analyzed UNC-104 particles: > 1500 events, SNB-1 particles: > 750 events. See Supplemental Figure S5 for definition of motility parameters. Box plots with median and error bars: ± max. and min. range. One-way ANOVA with Fisher’s LSD multiple comparison test for UNC-104 in mutant strains and t test with Welch’s correction for RNAi and SNB-1. *p < 0.05, **p < 0.005, ***p < 0.001 and ****p < 0.0001. F (DFn, DFd) = 12.60 (7, 4346) (A); 3.175 (7, 4544) (B); 7.156 (7, 416) (C); 3.496 (7, 419) (D); 3.119 (7, 85) (E); 3.923 (7, 85) (F) and 3.035 (7, 97) (G).
As reported earlier, UNC-104 anterograde velocities are significantly reduced in syd-2(ok217) knockout animals (Wagner et al., 2009), which is comparable to ok217 worms rescued with nonphosphorylatable SYD-2 Y741F (Figure 6A). Moreover, this effect cannot be rescued by the ptp-3 mutation, once more confirming that PTP-3 is likely located upstream of SYD-2 in the same pathway. Importantly, moving characteristics of UNC-104′s major cargo SNB-1 are comparable with that observed for the motor alone: both anterograde velocities as well as tug-of-war (reflected by particle directional changes) of SNB-1 particles are increased in mu256 mutants (Figure 6, A and C; Supplemental Figure S8, C and D). These observations emphasize the consistency of these measurements and underline the conclusion that the presence of PTP-3 would act to reduce axonal trafficking via the inhibition of SYD-2′s activating function on UNC-104. Besides investigating these effects in knockout worms, RNAi knockdown of the ptp-3 gene in N2 worms leads to comparable results (Figure 6, A–D). Note that we have shown in various publications that RNAi works efficiently in neuronal tissues (e.g., Wu et al., 2016), contrasting the common belief of worm neurons being refractive to RNAi.
Besides evaluating parameters such as motor velocities and reversals, we determined increased anterograde (total) run length of UNC-104 in PTP-3 knockout animals (Figure 6E); however, no differences could be detected for retrograde (total) run lengths (Figure 6F) as well as for net run lengths (Figure 6G). Indeed, these results are consistent with the unaffected travel distances of STVs in ptp-3 mutants (Figure 4D).
Note that for motility experiments, we utilized unc-104(e1265) worms rescued with UNC-104::mRFP as control whose motility parameter does not differ from N2 worms expressing UNC-104::mRFP (Supplemental Figure S7).
DISCUSSION
Identification of a novel pathway that regulates anterograde axonal transport
The role of SYD-2 as an UNC-104 activator, its potential to undergo intramolecular folding, as well as its amino acid Y741 as a potential target for PTP-3 dephosphorylation have been reported previously (Wagner et al., 2009; Chia et al., 2013; Mitchell et al., 2016). From these studies, we hypothesize that PTP-3 may act as an upstream regulator of SYD-2 to control binding to UNC-104 and above we provide an extensive set of evidence supporting this hypothesis. Using SILAC-based quantitative phosphoproteomics, Mitchell et al. (2016) demonstrated that SYD-2 is a substrate of PTP-3, and that in ptp-3 mutants, SYD-2′s amino acid Y741 (located near the central stalk domain; Supplemental Figure S1) is 40% hyperphosphorylated (Mitchell et al., 2016). Although, it is a standard practice to employ alanine and glutamic acid to design nonphosphorylatable or phosphomimicking mutants, one study demonstrated that phenylalanine is a more appropriate substitution to create nonphosphorylatable proteins. The same study implied that glutamic acid substitution for tyrosine is insufficient to reproducibly gain the desired phosphomimicking effect (Anthis et al., 2009). However, we have clearly demonstrated that Y/E phosphomimicking is largely comparable to wild type hyperphosphorylated SYD-2 in ptp-3(mu256) mutant backgrounds (Figure 3C). Combined with the co-IP data (Figures 1 and 5), it can be inferred that the open conformation of SYD-2 leads to exposed SAM domains (necessary for UNC-104 binding; Wagner et al., 2009) resulting in elevated interactions with UNC-104 leading to increased motor activation.
Also noteworthy is that other regulators such as RSY-2 may regulate SYD-2, and it is known that this protein acts to suppress SYD-2-dependent synapse assembly, specifically because of unexposed N- and C-terminal domains of SYD-2 incapable of scaffolding with other active zone proteins (Patel and Shen, 2009; Chia et al., 2013). Also, Chia et al. (2013) concluded that although only CC domains of SYD-2 are sufficient to assemble functional synapses, a L1086F mutation in the SAM domain of full-length SYD-2 results in loss of synapses, probably due to increased folding of SYD-2 (Chia et al., 2013). Several proteins are known to interact with either only SYD-2′s CC domains (ELKS-1, UNC-10 and GIT-1) or only SYD-2′s SAM domains (PTP-3 and GRIP) (Wyszynski et al., 2002; Ko et al., 2003; Ackley et al., 2005; Dai et al., 2006). Intramolecular folding of SYD-2 likely masks such interaction sites preventing complex interactions with such proteins. Critically, proteins such as UNC-104 and LIN-2 have been shown to directly interact with both SYD-2′s CC and SAM domains at functionally different degrees (Wagner et al., 2009; Wu et al., 2016).
Liprin-α has been shown to interact with ATP-agarose beads, leading to the assumption that it may possess the ability of autophosphorylation (Serra-Pages et al., 2005); however, there is no known kinase-domain in liprins needed for such autoregulation. More compelling evidence for another SYD-2 regulator is CASK/LIN-2 that has been shown to cooperatively act (together with SYD-2) to regulate UNC-104 motility, specifically on the motor’s superprocessivity (Wu et al., 2016). Also, it has been shown that a calcium/calmodulin-dependent protein kinase II colocalizes with liprin-α to regulate the degradation of liprin-α via phosphorylation (Hoogenraad et al., 2007). Lastly, a recent study shows that UNC-16, a MAP–kinase-like protein regulates the CDK-5/SAD-1/SYD-2 triplet protein system to inhibit organelle trafficking in neurons (Edwards et al., 2015). Further studies are required to unravel the type of kinase that phosphorylates amino acid Y741 in SYD-2.
Physiological effects of the PTP-3/SYD-2/UNC-104 pathway
Because mRNA levels of syd-2 and unc-104 were significantly reduced in mu256 mutants (Figure 1D), it is possible that STV trafficking is compromised in those animals, consistent with other studies (Hall and Hedgecock, 1991; Nonet, 1999; Kumar et al., 2010; Shen et al., 2014; Edwards et al., 2015). Indeed, mild accumulation of STVs in somas of ALM and HSN neurons, as well as reduced STVs in axons or synapses, can be seen in PTP-3 knockout worms (Figure 4G). Based on this finding it is obvious that fewer motors are available for axonal transport, yet these motors may experience a higher degree of activation (moving at faster speeds; Figure 6A). This increased motor activation phenotype is likely based on an elevated number of activated SYD-2 molecules due to increased open states in mu256 mutants (Figure 3). Critically, one study in Drosophila Dlar mutants revealed that synaptic boutons are reduced with concomitant decreased excitatory postsynaptic potential (Kaufmann et al., 2002). Another study in rat neurons demonstrated that both LAR RNAi and dominant-negative disruption of LAR results in a reduced number of excitatory synapses as well as dendritic spines. The same study uncovered that LAR/liprin-α interaction is essential for dendritic targeting of AMPA receptors at postsynaptic terminals (Dunah et al., 2005). These findings may explain the observed decreased sizes of STVs (or of STV cluster) in the axon of ALM neurons (Figure 4B), and it is noteworthy that one study relates the size of STVs to the osmotic pressure (regulated by neurotransmitter concentration) within the STVs (Qu et al., 2009). Indeed, in Drosophila, the amount of neurotransmitters in vesicles increased along with STV size upon overexpression of vesicular glutamate transporter (Daniels et al., 2004, 2006). These findings may lead to the assumption that reduced sizes of STV cluster (Figure 4B) may be due to reduced sizes of single STVs (within these clusters) as a result of diminished amounts of neurotransmitters inside the vesicles.
Abnormal STV sorting (Figure 4G) may trigger reduced expression of syd-2 and unc-104 (Figure 1D), and a recent study in Drosophila uncovered activation of Wnd/DLK MAP kinase signaling after disruption of Unc-104′s function (as well as upon overexpression of presynaptic proteins), leading to restrained expression of active zone components (Li et al., 2017). Impeded synaptic formation may also explain the mild egg retention phenotype in ptp-3(mu256) mutants (Supplemental Figure S2). Egg laying in C. elegans is regulated via HSN neurons that innervate vulval muscles. Proper formation of synapses via the interplay of SYG-1, SYD-1, and SYD-2 is essential for egg-laying, and in syd-1 as well as syd-2 mutants observed egg-laying defects often result in hatch-bag phenotypes (Supplemental Figure S2) (Jin, 1999; Patel et al., 2006).
SYD-2 has been described to inherit two major roles in UNC-104: one is the motor scaffolding function and the second is motor activation. In N2 wild type animals, UNC-104 appears clustered along axons which is evidently not related to overexpression effects, and furthermore, axonal patterns of these cluster are different from the regular (string of pearls) appearance of en passant synapses (Wagner et al., 2009; Tien et al., 2011). Based on this knowledge, one can deduce that open SYD-2 states in ptp-3 mutants (Figure 3) may trigger increased UNC-104 clustering along axons. Indeed, motor accumulation analysis exposed not only that UNC-104 clustering along axons is reduced in syd-2 mutants (Figure 5, A and C) but also that PTP-3 may act upstream of SYD-2 (based on double mutant analysis) (Figure 5C). This finding is in accordance with investigations from Drosophila demonstrating that Dliprin-α is essential for Dlar-dependent synapse assembly (Kaufmann et al., 2002).
Because axonal distribution patterns of SNB-1 and UNC-104 in axons (Figures 4 and 5) merely reflect a static snapshot of cargo displacements, dynamic motility analysis of cargo and motor is hence vital as well. Critically, motility data as shown in Figure 6 go well along with the findings from cluster pattern analysis (Figures 4 and 5), and both clearly reveal the suppressing role of PTP-3 on SYD-2 and its function in axonal transport (summarized in Figure 7). Moreover, motility features of the motor alone clearly mirror the motility features of the cargo alone (Figure 6) underlining the validity of these observations.
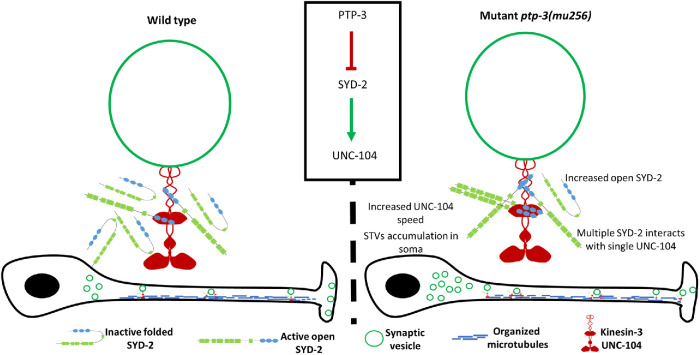
FIGURE 7:
PTP-3 regulates binding of SYD-2 to kinesin-3 UNC-104 to modulate fast axonal transport. From this study, it is evident that PTP-3 is upstream of SYD-2 to regulate UNC-104 activity. PTP-3 dephosphorylates SYD-2 at position Y741 resulting in intramolecular folding of SYD-2 concomitantly inactivating UNC-104. Lack of PTP-3 results in more open configurations of SYD-2 resulting in increased interactions with UNC-104 promoting increased UNC-104 scaffolding along axons and boosting speeds of present UNC-104 motors as well as STVs. Mild accumulation of STVs in the soma is likely the result of reduced gene expression of syd-2 and unc-104 in ptp-3 mutants.
Also worth discussing are the elevated motor reversals in ptp-3 mutants (Figure 6C) pointing to increased tug-of-war events. Future studies need to investigate the role of dynein in these observations such as that motor reveals may remain unaltered in dynein mutants. Nevertheless, based on the “paradox of codependence” and “mechanical activation” theories (Hancock, 2014), increased directional changes may be based on hyperactivated UNC-104 and thus mechanically activating dynein (if both bound to one vesicle). Though this might be valid for short-term runs, the determined considerable increase in total anterograde run lengths in ptp-3 mutants (Figure 6E) could rather be based on the motor coordination model for longer runs. These ideas go well along with current views that tug-of-war events are typically observed for short runs (Maday et al., 2014).
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Plasmid constructs and C. elegans strains
Worm strains were maintained and analyzed at 20°C on NGM (nematode growth medium) agar plates seeded with uracil auxotroph Escherichia coli OP50 strain serving as the nematode food source (Brenner, 1974). All analyses in this study were performed on young adult worms unless otherwise stated. Worm strains N2, RB633 ptp-3(ok244), CZ3761 ptp-3(mu256), and ZM607 syd-2(ok217) were obtained from CGC (Caenorhabditis Genetic Center, MN), and unc-104(e1265);Ex[Punc-104::unc-104::mrfp], unc-104(e1265);Ex[Punc-104::unc-104::gfp], and syd-2(ok217);Ex[Punc-104::unc-104::mrfp] were previously described (Wagner et al., 2009; Tien et al., 2011; Wu et al., 2016). Worm strains OIW 57 ptp-3(ok244);nthEx57[Punc-104::unc-104::mrfp], OIW 58 ptp-3(mu256);nthEx58[Punc-104::unc-104::mrfp], OIW 59 ptp-3(ok244);nthEx59[Punc-104::unc-104::gfp], and OIW 60 ptp-3(mu256);nthEx60[Punc-104::unc-104::gfp] were generated by crossing mutant strains with existing strain unc-104(e1265);nthEx[Punc-104::unc-104::mrfp] or unc-104(e1265);nthEx[Punc-104::unc-104::gfp]; double mutant OIW 61 syd-2(ok217);ptp-3(mu256);nthEx61[Punc-104::unc-104::mrfp] was generated by crossing syd-2(ok217) mutant with males of OIW 58 ptp-3(mu256);nthEx58[Punc-104::unc-104::mrfp]. Transgenic line OIW 62 ptp-3(mu256);nthEx62[Punc-104::unc-104::mrfp; Punc-104::ptp-3b::cfp] was generated by microinjection of Punc-104::unc-104::mrfp (Wagner et al., 2009) and Punc-104::ptp-3b::cfp (100 ng/µl each). Note that for the pan-neuronal Punc-104 promotor to fully operate, relatively high dosages of plasmids are necessary (Klopfenstein et al., 2002). PTP-3B containing first six introns and exons followed by cDNA was cloned into already existing Punc-104::cfp plasmid (generated by replacing gfp in pPD95.77 with cfp and subsequently cloned with Punc-104) by homologous recombination (In-Fusion HD cloning, Takara Bio USA) using CTCTAGAGGATCCCCATGGGAACGCCGGCT (forward) and CCTTTGGCCAATCCCTTCGAGAAATTGTCATAGGCGGC (reverse) primers. Genomic DNA of snb-1 gene was amplified and cloned into Punc-104::mrfp using homolog recombination with CTCTAGAGGATCCCCATGGACGCTCAAGGAGATGCC (forward) and CCTTTGGCCAATCCCTTTTTTCCTCCAGCCCATAAAACGATGAT (reverse) primers to generate Punc-104::snb-1::mrfp plasmid and microinjected at 80 ng/µl to generate OIW 63 N2(wt);nthEx63[Punc-104::snb-1::mrfp], OIW 64 ptp-3(ok244);nthEx64[Punc-104::snb-1::mrfp], and OIW 65 ptp-3(mu256);nthEx65[Punc-104::snb-1::mrfp] transgenic lines. For colocalization studies, OIW 66 ptp-3(mu256);nthEx66[Punc-104::gfp::syd-2;Punc-104::ptp-3b::cfp] worm was generated by injection of Punc-104::gfp::syd-2 (Wu et al., 2016) and Punc-104::ptp-3b::cfp (both at 100 ng/µl) plasmids. Plasmid constructs Punc-104::unc-104::gfp, Punc-104::unc-104∆PH::gfp, and Punc-86::snb-1::mrfp, as well as strains CB1265 unc-104(e1265);Ex[Punc-104::unc-104::gfp;Punc-86::snb-1::mrfp] and CB1265 unc-104(e1265);Ex[Punc-104::unc-104∆PH::gfp;Punc-86::snb-1::mrfp] were previously described (Wagner et al., 2009). Plasmid ptp-3b::vc155 was constructed by subcloning the ptp-3b gene from Punc-104::ptp-3b::cfp into Punc-104::vc155 and microinjected along with Punc-104::vn173::syd-2 (Hsu et al., 2011) and Punc-104::snb-1::mrfp to generate OIW 67 ptp-3(mu256);nthEx67[Punc-104::vn173::syd-2;Punc-104::ptp-3b::vc155;Punc-104::snb-1::mrfp] (75, 75, and 50 ng/µl, respectively). Plasmid constructs Punc-104::unc-104::vc155, Punc-104::vc155 and Pmyo-2::mrfp were previously described (Tien et al., 2011; Wu et al., 2016; Shanmugam et al., 2018). BiFC-positive control worm OIW 68 N2(wt);nthEx68[Punc-104::snb-1::mrfp;Punc-104::unc-104::vc155;Punc-104::vn173::syd-2] was developed by microinjection (50, 75, and 75 ng/µl, respectively) and BiFC-negative control OIW 69 N2(wt);nthEx69[Pmyo-2::mrfp;Punc-104::vn173::syd-2;Punc-103::vc155] was generated by microinjection (15, 50, and 50 ng/µl, respectively). For FRET experiments, the syd-2 gene from an already existing plasmid construct Punc-104::gfp::syd-2 (Wu et al., 2016) was subcloned into Punc-104::cypet::ypet using homologous recombination with GATCCCCGGGATTGGAAATGAGCTACAGCAATGGAAAC (forward) and CTTTGGGTCCTTTGGTGGTATATAAATGAAACTCGTAGGATTTTGC (reverse) primer set. The created plasmid was microinjected at a concentration of 100 ng/µl with snb-1::mrfp as a coinjection marker to generate OIW 70 N2(wt);nthEx70[Punc-104::cypet::syd-2::ypet;Punc-104::snb-1::mrfp] strain. SYD-2 Y741F mutation was generated from existing Punc-104::gfp::syd-2 (Wu et al., 2016) using 5′ phosphorylated TCGGCAATCCGCAGTTTGTTG (forward) and 5′ phosphorylated AAATATCATACCGTCGGTCACCGG (reverse) primers resulting in Punc-104::gfp::syd-2 Y741F. From this construct, SYD-2 Y741F was subcloned into Punc-104::cypet::ypet to generate Punc-104::cypet::syd-2 Y741F::ypet. SYD-2 Y741E mutation was generated by overlap extension PCR using ATGAGCTACAGCAATGGAAACATAAATTG (forward) and GCGGATTGCCTTCAATATCATACCGTCGGTCACCG (reverse) for 5′ segment of syd-2 and CGGTATGATATTGAAGGCAATCCGCAGTTTG (forward) and CTAGGTATATAAATGAAACTCGTAGGATTTTGCT (reverse) for 3′ segment of syd-2 and subcloned to generate Punc-104::cypet::syd-2 Y741E::ypet and Punc-104::gfp::syd-2 Y741E. Other FRET mutant strains OIW 71 ptp-3(mu256);nthEx71[Punc-104::cypet::syd-2::ypet;Punc-104::snb-1::mrfp], OIW 72 ptp-3(mu256);nthEx72[Punc-104::cypet::syd-2Y741F::ypet;Punc-104::snb-1::mrfp], OIW 73 N2(wt);nthEx73Punc-104::cypet::syd-2Y741F::ypet;Punc-104::snb-1::mrfp], OIW 74 ptp-3(mu256);nthEx74[Punc-104::cypet::syd-2Y741E::ypet;Punc-104::snb-1::mrfp], and OIW 75 N2(wt);nthEx75[Punc-104::cypet::syd-2Y741E::ypet;Punc-104::snb-1::mrfp] were generated by microinjection of the aforementioned FRET constructs at 120 ng/µl along with snb-1::mrfp coinjection marker. Rescue of syd-2(ok217) was performed with microinjection of 70 ng/µl of the respective plasmid into already existing syd-2(ok217);nthEx[Punc-104::unc-104::mrfp] to generate OIW 76 syd-2(ok217);nthEx76[Punc-104::unc-104::mrfp;Punc-104::gfp::syd-2], OIW 77 syd-2(ok217);nthEx77[Punc-104::unc-104::mrfp;Punc-104::gfp::syd-2 Y741F], and OIW 78 syd-2(ok217);nthEx78[Punc-104::unc-104::mrfp;Punc-104::gfp::syd-2 Y741E]. The plasmid Punc-104::Cypet::Ypet (80 ng/µl) was microinjected along with Pmyo-2::mrfp (10 ng/µl) into N2 worms to generate OIW 79 N2(wt);nthEx79[Punc-104::Cypet::Ypet;Pmyo-2::mrfp] and ptp-3(mu256) to generate OIW 80 ptp-3(mu256);nthEx80[Punc-104::Cypet::Ypet;Pmyo-2::mrfp]. Also, Punc-104::Cypet::syd-2 was microinjected along with Punc-104::snb-1::mrfp to generate OIW 81 N2(wt);nthEx81[Punc-104::Cypet::syd-2;Punc104::snb-1::mrfp], and Punc-104::syd-2::ypet was microinjected with snb-1::mrfp to obtain OIW 82 N2(wt);nthEx82[Punc-104::syd-2::ypet;Punc-104::snb-1::mrfp]. OIW 83 N2(wt);nthEx83[Punc-104::unc-104::mrfp] was generated by microinjection of 75 ng/µl of Punc-104::unc-104::mrfp.
Mutant strain ZM607 syd-2(ok217) carries a large N-terminal deletion in the syd-2 gene leading to a null mutation (Wagner et al., 2009). This deletion was confirmed via PCR using the following primer set: CGCGGGAATTATGCCTATTA (forward) and TTGCATCTGCAAAAGAAACG (reverse). Mutant strain RB633 ptp-3(ok244) carries a deletion of three FN3 domains leading to a frameshift and premature termination of the PTP-3A isoform. The deletion was confirmed via PCR using the following primer set: ACCCAAACGTTACCGAACAG (forward) and CCAGGGACTGCAGGAAAATA (reverse). Mutant strain CZ3761 ptp-3(mu256) is characterized by an insertion of a single nucleotide in exon 25 of the ptp-3 gene resulting in a frameshift leading to a premature stop at AA1777 (Supplemental Figure S1) (Ackley et al., 2005). This single nucleotide insertion was confirmed by sequencing and in addition, a G to T (resulting in coding the same amino acid—leucine) mutation four nucleotides upstream of the aforementioned insertion was found. Nevertheless, the single nucleotide insertion should still result in a premature stop leading to a null mutation as described in Ackley et al. (2005). Note that ptp-3(mu256) mutant worms revealed mild egg retention phenotypes (Supplemental Figure S2).
Microinjection of C. elegans plasmids
Microinjection was carried out as previously described (Evans, 2006) with the modification that either L4-staged worms or predominantly young adult worms were microinjected. Briefly, immobilized worms (immersed in a small drop of mineral oil [Sigma, M5904] on 2.5% agarose-coated coverslips) were microinjected using the DIC function of an Olympus IX81 microscope fitted with a needle micromanipulator controlled by an Eppendorf FemtoJet Express. Worms were recovered using recovery buffer or S buffer and then placed onto NGM plates. F1 generations were screened for the respective transgenic genes and maintained separately to clone out a stable line at 20°C.
Real-time PCR
Messenger RNA was isolated from lysates of young adult worms using TRIzol reagent (Invitrogen) following reverse transcription using the SuperScript First-Strand Synthesis system (Invitrogen). The primer set used for qPCR of the unc-104 gene was GAAGGAAATAAAGCGAGAAC (forward) and CTGCCAAATCAACCAAAG (reverse), and the primer set for the syd-2 gene was CGGAACAATACTCGACTTC (forward) and GCCACACGCTCCATT (reverse). Internal control for qPCR was the cdc-42 gene as previously described (Hoogewijs et al., 2008) and primer set was CTGCTGGACAGGAAGATTACG (forward) and TCGGACATTCTCGAATGAAG (reverse). ABI Power SYBR Green PCR Master Mix (ABI, USA) was used in conjunction with a PCR machine model ABI StepOne Plus Real-time PCR (Applied Biosystems). The experiment was repeated three times, and unpaired Student’s t test was used to compare the study groups.
Co-IP assays
For co-IP assays (Figure 1), 2 µg of anti-GFP (GFP antibody GT859, GeneTex) antibody was incubated with protein G-coated magnetic beads (PureProteome protein G magnetic beads, LSKMAGG10, EMD Millipore) for 1–3 h in IP buffer at 16°C. After washes, 500 µg or 1 mg of total protein extract was used to pull down the target proteins overnight. After subsequent washes, beads were boiled in the presence of SDS–PAGE loading solution and separated on 6% SDS–PAGE. Separated proteins were then transferred to a PVDF membrane (IPVH00010, Millipore) overnight at 4°C in a wet transfer system (Hoefer) at 35 mA. The blot was blocked with 5% bovine serum albumin and probed with primary and secondary antibodies before visualization of the band using CyECL Western blotting substrate H (ECL003-H, Cyrus Bioscience) in ImageQuant LAS4000mini. Anti-GFP antibody (B-2, SC-9996, mouse monoclonal, Santa Cruz Biotechnology) was diluted at 1:10,000 and anti-SYD-2 antibody (cL-19, SC-15655, goat polyclonal, Santa Cruz Biotechnology, USA) at 1:500. Secondary HRP antibody was used at a dilution of 1:10,000 (anti-mouse IgG HRP secondary, GTX213111-01, GeneTex) and 1:2500 (anti-goat IgG HRP secondary, GTX228416-01, GeneTex), respectively. Co-IP experiments were repeated 3 times and a single representative cropped blot is presented (Figure 1). Coprecipitated SYD-2 protein was normalized to the pulled down UNC-104::GFP (Figure 1). For relative quantification of coprecipitated protein levels, the normalized signals from the mutant group were once again normalized to the control group and represented as percentages (Figure 1). For co-IP in Figure 5D, we used anti-GFP antibody (GFP antibody GT859, GeneTex) for IP, while blots were probed with a commissioned (rabbit, polyclonal) anti-UNC-104 antibody from GeneTex (Taiwan). Co-IP UNC-104 proteins were normalized to IP (pulled-down) GFP::SYD-2.
Colocalization analysis and BiFC assays
For colocalization analysis (Figure 2), images were collected from live worms using an inverted Zeiss LSM 780 laser scanning confocal microscope. Pearson’s correlation coefficient values were then calculated from a region of interest (ROI) using ImageJ plugin Intensity Correlation Analysis after background subtraction using the plugin ROI. Pearson’s values range from –1 to +1, with values toward +1 representing two fluorophores with coexisting patterns (increasing correlation) and values close to –1 representing fluorophores with segregated patterns of localization, while 0 stands for random localization. In addition to Pearson’s values, we present PDM images with: Product of the Differences from the Mean = (cyan intensity – mean cyan intensity) * (green intensity – mean green intensity). Positive controls for colocalization (worms expressing UNC-104::GFP/SNB-1::mRFP) reveal Pearson’s values comparable to PTP-3B/SYD-2, while negative controls (worms expressing UNC-104∆PH::GFP/SNB-1::mRFP) exhibit significant reduction in Pearson’s value (Supplemental Figure S3, A and B).
The BiFC assay is a powerful tool to understand if two proteins are in close proximity for assumed functional interactions in cells. In brief, a YFP protein (Venus) is split into two halves (N-terminal Venus VN and C-terminal Venus VC) and tagged to the two test proteins. Complementation of Venus will lead to a fluorescent signal if the two test proteins are at least 7–10 nm close to each other (Hu and Kerppola, 2003; Hsu et al., 2011). In this study, we designed fusions of PTP-3B::VC155 and VN173::SYD-2 (Figure 2). The BiFC-positive control (Supplemental Figure 3C) is based on well-described interactions between UNC-104 and SYD-2 (see Introduction) and the BiFC-negative control is based on the expression of a SYD-2/empty vector BiFC pair (Supplemental Figure S3D). Note that to avoid bleed-through artifacts in negative control, we refrained from overexpressing any (red) fluorescent neuronal marker. Instead, we used a pharyngeal marker (Pmyo-2::mrfp) to locate the position of the nerve ring (yellow, dotted line in Supplemental Figure S3D) and screened through all confocal planes for potential BiFC signals at that location.
RNAi knockdown
RNAi was performed by feeding worms with bacterial strains expressing dsRNA for a particular gene of interest (RNAi feeding method; Kamath et al., 2000). RNAi feeding clones were obtained from the Julie Ahringer’s C. elegans RNAi feeding library (Source BioScience LifeScience, USA; provided by the C. elegans Core Facility, Taiwan). Feeding strains were grown overnight at 37°C on NGM plates containing ampicillin (100 µg/ml) and 1 mM IPTG. F0 worms were transferred to the respective RNAi plate and incubated at 20°C. Worms were then transferred to new plates every day and final data analysis was carried out using young adults from the cloned F1 progeny.
Intramolecular FRET analysis
For intramolecular FRET experiments, modified versions of CFP (donor) and YFP (acceptor), such as Cypet and Ypet, were used and tagged to the N- and C-terminus of SYD-2 (FRET sensor), respectively. Cypet and Ypet have been reported to exhibit very high ratiometric signals during FRET when compared to their traditional counterparts (Nguyen and Daugherty, 2005). Plasmids encoding Cypet (pCypet-Rac1(WT), #22782) and Ypet (pYpet-PBD, #22781) sequences were taken from the Addgene repository (Addgene, USA) and subcloned into worm expression plasmids as mentioned above. For intramolecular FRET, images were obtained from live worms using an inverted Zeiss LSM 780 laser scanning confocal microscope with 63×/1.40 Oil DIC M27 objective lens (shared microscope facility at Biomedical Science and Engineering Center, NTHU, Taiwan). Images were acquired at the emission wavelength of 483 nm (donor channel; 459–508 nm filter set) and 574 nm (FRET channel; 518–630 nm filter set) on excitation at 405 nm for Cypet, as well as at 574 nm (acceptor channel; 518–630 nm filter set) on excitation at 514 nm for Ypet, respectively. Ratiometric analysis was used to understand the intramolecular folding of SYD-2 protein as described previously (Wouters et al., 2001; Kalab et al., 2006; Spiering et al., 2013), which does not require bleed-through subtraction (Supplemental Figure S4E) since the ratio of donor/acceptor is 1 in intramolecular FRET (as opposed to intermolecular FRET with varying ratios and thus requiring bleed-through subtraction). Thus, fluorescence intensity from FRET channel (IFRET) was divided by the fluorescence intensity from donor channel (ID) to generate a ratio (IFRET/ID) for relative comparisons between different genetic backgrounds. Differences created by presumed slight changes in expression levels between individual worms (due to the extrachromosomal array nature of the FRET sensor) were normalized in the ratiometric analysis (IFRET/ID). In intramolecular FRET, lower ratio value represents open conformation of FRET sensor and higher ratio value indicates intramolecularly folded conformation of FRET sensor. For negative controls, we expressed Punc-104::Cypet::Ypet without the syd-2 gene in either N2(wt) or ptp-3(mu256) genetic background (that did not produce any significant difference in FRET ratios; Supplemental Figure S4, A and B). FRET pseudo-color images were generated by the “Fret analyzer” plugin from ImageJ and are accompanied by a 16 color LUT calibration scale. FRET ratios from different study groups were assessed with one-way ANOVA using Fisher’s LSD comparison test.
Fluorescence intensity measurements of STVs in neurons
Quantification of SNB-1::mRFP fluorescence in somas, axons, synapses, VNC, and DNC (dorsal nerve cord) (Figure 4) was carried out using ImageJ based on a published protocol (McCloy et al., 2014). Parameters such as area, integrated density, minimum/maximum gray value, and mean gray value were chosen and then the appropriate ROI, as well as the background, were selected and measured. The following formula was used to correct for background fluorescence: integrated density – (area of selected region * mean gray value from the background). The total fluorescence intensity measured within a neuronal region was considered 100% for normalization of variations in the expression of extrachromosomal arrays between individual worms; thus, the percentage of cargo measured in subcellular regions equals to: (Fluorescence intensity in subcellular region/Total fluorescent intensity in the neuron) * 100. One-way ANOVA with Fisher’s LSD multiple comparison test was used to compare the study groups.
Axonal motor and cargo cluster analysis
For quantification of UNC-104 motor clusters (Figure 5) along sublateral neurons, images were obtained using an Olympus IX81 microscope connected to an Andor iXon DV897 EMCCD camera and analyzed using ImageJ. Note that particles of sizes less than 3 pixels were excluded from analysis to reject random noise. For density measurements, the number of particles along axons was manually counted and the length of the axon was noted. The following formula was used for density calculation: (Total number of particles in the neurite segment/Length of that neurite segment) * 100. Analyzed particles from different groups were subjected to one-way ANOVA with multiple comparisons using Fisher’s LSD test. For the quantification of SNB-1 particles (Figure 4), we used an inverted Zeiss LSM 780 laser scanning confocal microscope to collect the data and ImageJ’s “particle analysis tool” for data quantification. The area of SNB-1 particles was analyzed as mentioned above. For analysis of distances traveled by SNB-1 cargo, a line from the axon hillock to the detected (distal) SNB-1 fluorescent particle was drawn using ImageJ’s line tool and then the measurement was executed. Travel distances were normalized by the total length of the axon seen in the field of observation and converted into percentage of distances traveled by the particles (Figure 4D). For fluorescent particle density measurements (UNC-104 and SNB-1), the following formula was used: (Number of particles in the axon/Total length of axon) * 100. One-way ANOVA with multiple comparisons using Fisher’s LSD test was used to compare the study groups.
Motility analysis
For quantification of motility parameters of fluorescently-tagged proteins in axons of ALM neurons, worms were immobilized between cover glasses coated with 2% agarose (without using any anesthetics such as levamisole). Time-lapse imaging was carried out employing an Olympus IX81 microscope fitted with a DSU Nipkow spinning disk connected to an Andor iXon DV897 EMCCD camera. Image acquisition rate was 3–4 frames per second. Obtained images were then analyzed by the kymograph method using ImageJ software. Here, the “straighten” plugin was used to straighten curved axons followed by the execution of the “reslice stack” function to generate a kymograph. A particle pause is regarded if the velocity of a particle is less than 0.07 µm/s. Vertical lines in a kymograph indicate stationary particles, whereas horizontally angled lines correspond to moving particles. An “event” is defined as an occurrence of a movement of a fluorescent particle after a reversal or a pause. Anterograde or retrograde run length is the distance covered by the fluorescent particle in the acquired kymograph. Directional changes (either from anterograde to retrograde or from retrograde to anterograde) are defined as changes in directions of a single event. The term motility is defined as a movement occurrence of any event during the time of imaging (refer to Supplemental Figure S5 for details). Analyzed particles and obtained values from different groups were tested with one-way ANOVA with multiple comparisons using Fisher’s LSD test. Histograms (Supplemental Figure S8) were plotted to identify outliers.
Supplementary Material
Acknowledgments
We thank Eric Hwang (National Chiao Tung University, Hsinchu, Taiwan) for his guidance in intramolecular FRET experiments. We thank the C. elegans Core Facility (CECF) Taiwan (funded by the Ministry of Science and Technology, MOST) for providing microinjection setups and worm observation systems. We acknowledge MOST grants NSC 100-2311-B-007-004 and MOST 103-2311-B-007-004-MY3 to O.I.W.
Abbreviations used:
- BiFC
bimolecular fluorescence complementation
- CC
coiled coil
- DNC
dorsal nerve cord
- IAR
initial axonal region
- IP
immunoprecipitation
- LAR PTPRD
leukocyte common antigen-related protein tyrosine phosphatase transmembrane receptor type D
- LAR PTPRF
leukocyte common antigen-related protein tyrosine phosphatase transmembrane receptor type F
- MAP
microtubule-associated protein
- MT
microtubule
- NGM
nematode growth medium
- PD
Parkinson’s disease
- PDM
product of the differences from the mean
- RNAi
RNA interference
- ROI
region of interest
- SAM
sterile alpha motif
- STV
synaptic vesicle protein transport vesicle
- VNC
ventral nerve cord.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-10-0591) on November 4, 2020.
REFERENCES
- Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD, Jin Y (2005). The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J Neurosci 25, 7517–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthis NJ, Haling JR, Oxley CL, Memo M, Wegener KL, Lim CJ, Ginsberg MH, Campbell ID (2009). Beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation. J Biol Chem 284, 36700–36710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Brady ST, Morfini GA (2017). Regulation of motor proteins, axonal transport deficits and adult-onset neurodegenerative diseases. Neurobiol Dis 105, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CW, Peng YF, Yen YC, Bhan P, Shanmugam MM, Klopfenstein DR, Wagner OI (2019). Insights on UNC-104-dynein/dynactin interactions and their implications on axonal transport in Caenorhabditis elegans. J Neurosci Res 97, 185–201. [DOI] [PubMed] [Google Scholar]
- Chia PH, Patel MR, Wagner OI, Klopfenstein DR, Shen K (2013). Intramolecular regulation of presynaptic scaffold protein SYD-2/liprin-alpha. Mol Cell Neurosci 56, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Taru H, Deken SL, Grill B, Ackley B, Nonet ML, Jin Y (2006). SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat Neurosci 9, 1479–1487. [DOI] [PubMed] [Google Scholar]
- Daniels RW, Collins CA, Chen K, Gelfand MV, Featherstone DE, DiAntonio A (2006). A single vesicular glutamate transporter is sufficient to fill a synaptic vesicle. Neuron 49, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Collins CA, Gelfand MV, Dant J, Brooks ES, Krantz DE, DiAntonio A (2004). Increased expression of the Drosophila vesicular glutamate transporter leads to excess glutamate release and a compensatory decrease in quantal content. J Neurosci 24, 10466–10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunah AW, Hueske E, Wyszynski M, Hoogenraad CC, Jaworski J, Pak DT, Simonetta A, Liu G, Sheng M (2005). LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nat Neurosci 8, 458–467. [DOI] [PubMed] [Google Scholar]
- Edwards SL, Morrison LM, Manning L, Stec N, Richmond JE, Miller KG (2018). Sentryn acts with a subset of active zone proteins to optimize the localization of synaptic vesicles in Caenorhabditis elegans. Genetics 210, 947–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Yorks RM, Morrison LM, Hoover CM, Miller KG (2015). Synapse-assembly proteins maintain synaptic vesicle cluster stability and regulate synaptic vesicle transport in Caenorhabditis elegans. Genetics 201, 91–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TC (2006). Transformation and microinjection. In: The C. elegans Research Community, ed. Book Worm, 10.1895/wormbook.1.7.1. [DOI] [Google Scholar]
- Franker MA, Hoogenraad CC (2013). Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J Cell Sci 126, 2319–2329. [DOI] [PubMed] [Google Scholar]
- Fung SYS, Kitagawa M, Liao PJ, Wong J, Lee SH (2017). Opposing activities of Aurora B kinase and B56-PP2A phosphatase on MKlp2 determine abscission timing. Curr Biol 27, 78–86. [DOI] [PubMed] [Google Scholar]
- Gibbs KL, Greensmith L, Schiavo G (2015). Regulation of axonal transport by protein kinases. Trends Biochem Sci 40, 597–610. [DOI] [PubMed] [Google Scholar]
- Goldstein LS (2001). Kinesin molecular motors: transport pathways, receptors, and human disease. Proc Natl Acad Sci USA 98, 6999–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Goldman D, Valk E, Loog M, Holt LJ, Gheber L (2019). Synthetic-evolution reveals narrow paths to regulation of the saccharomyces cerevisiae mitotic kinesin-5 Cin8. Int J Biol Sci 15, 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Siegler N, Goldman D, Judah H, Valk E, Koivomagi M, Loog M, Gheber L (2017). Three Cdk1 sites in the kinesin-5 Cin8 catalytic domain coordinate motor localization and activity during anaphase. Cell Mol Life Sci 74, 3395–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM (1991). Kinesin-related gene unc-104 is require for axonal transport of synaptic vesicles in C. elegans. Cell 65, 837–847. [DOI] [PubMed] [Google Scholar]
- Hancock WO (2014). Bidirectional cargo transport: moving beyond tug of war. Nature reviews. Mol Cell Biol 15, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington RJ, Gutch MJ, Hengartner MO, Tonks NK, Chisholm AD (2002). The C. elegans LAR-like receptor tyrosine phosphatase PTP-3 and the VAB-1 Eph receptor tyrosine kinase have partly redundant functions in morphogenesis. Development 129, 2141–2153. [DOI] [PubMed] [Google Scholar]
- He J, Zhang Z, Ouyang M, Yang F, Hao H, Lamb KL, Yang J, Yin Y, Shen WH (2016). PTEN regulates EG5 to control spindle architecture and chromosome congression during mitosis. Nat Commun 7, 12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R (2003). Biochemical and molecular characterization of diseases linked to motor proteins. Trends Biochem Sci 28, 558–565. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Feliu-Mojer MI, Spangler SA, Milstein AD, Dunah AW, Hung AY, Sheng M (2007). Liprinalpha1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev Cell 12, 587–602. [DOI] [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR (2008). Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CC, Moncaleano JD, Wagner OI (2011). Subcellular distribution of UNC-104(KIF1A) on binding to adaptors as UNC-16(JIP3), DNC-1(DCTN1/Glued) and SYD-2(LIPRIN-alpha) in C. elegans neurons. Neuroscience 176, 39–52. [DOI] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M (1999). The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans . Nature 401. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Frasser AG, Ahringer J. (2000). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2, research0002.0001-0002.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann N, DeProto J, Ranjan R, Wan H, Vactor DV (2002). Drosophilla liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis. Neuron 34, 27–38. [DOI] [PubMed] [Google Scholar]
- Kittelmann M, Hegermann J, Goncharov A, Taru H, Ellisman MH, Richmond JE, Jin Y, Eimer S (2013). Liprin-alpha/SYD-2 determines the size of dense projections in presynaptic active zones in C. elegans. J Cell Biol 203, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Tomishige M, Stuurman N, Vale RD (2002). Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell 109, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Kim S, Valtschanoff JG, Shin H, Lee J-R, Sheng M, Premont RT, Weinberg RJ, Kim E (2003). Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J Neurosci 23, 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Choudhary BC, Metpally R, Zheng Q, Nonet ML, Ramanathan S, Klopfenstein DR, Koushika SP (2010). The Caenorhabditis elegans Kinesin-3 motor UNC-104/KIF1A is degraded on loss of specific binding to cargo. PLoS Genet 6, e1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang YV, Adib A, Stanchev E, Xiong DT, Klinedinst X, Soppina S, Jahn P, Hume TR, Rasse RI, et al. (2017). Restraint of presynaptic protein levels by Wnd/DLK signaling mediates synaptic defects associated with the kinesin-3 motor Unc-104. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang Z, Liang H, Zhao X, Liang L, Wang G, Yang J, Jin Y, McNutt MA, Yin Y (2017). Protein phosphatase 2A (PP2A) regulates EG5 to control mitotic progression. Sci Rep 7, 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanus AJ, Yip GW (2018). Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene 37, 833–838. [DOI] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL (2014). Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84, 292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A (2014). Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13, 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlvain JM Jr, Burkhardt JK, Hamm-Alvarez S, Argon Y, Sheetz MP (1994). Regulation of kinesin activity by phosphorylation of kinesin-associated proteins. J Biol Chem 269, 19176–19182. [PubMed] [Google Scholar]
- Mitchell CJ, Kim MS, Zhong J, Nirujogi RS, Bose AK, Pandey A (2016). Unbiased identification of substrates of protein tyrosine phosphatase ptp-3 in C. elegans. Mol Oncol 10, 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H-J, Poy F, Krueger NX, Saito H, Frederick CA (1999). Crystal structure of the tandem phosphatase domains of RPTP LAR. Cell 97, 449–457. [DOI] [PubMed] [Google Scholar]
- Nguyen AW, Daugherty PS (2005). Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol 23, 355–360. [DOI] [PubMed] [Google Scholar]
- Niwa S, Lipton DM, Morikawa M, Zhao C, Hirokawa N, Lu H, Shen K (2016). Autoinhibition of a neuronal kinesin UNC-104/KIF1A regulates the size and density of synapses. Cell Rep 16, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML (1999). Visualization of synaptic specialization in live C. elegans with synaptic vesicle protein-GFP fusions. J Neurosci Methods 89, 33–40. [DOI] [PubMed] [Google Scholar]
- Patel MR, Lehrman EK, Poon VY, Crump JG, Zhen M, Bargmann CI, Shen K (2006). Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nat Neurosci 9, 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Shen K (2009). RSY-1 is a local inhibitor of presynaptic assembly in C. elegans . Science 323, 1500–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Akbergenova Y, Hu Y, Schikorski T (2009). Synapse-to-synapse variation in mean synaptic vesicle size and its relationship with synaptic morphology and function. J Comp Neurol 514, 343–352. [DOI] [PubMed] [Google Scholar]
- Serra-Pages C, Kedersha NL, Fazikas L, Medley QG, Debant A, Streuli M (1995). The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J 14, 2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Pages C, Medley QG, Tang M, Hart A, Streuli M (1998). Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem 273, 15611–15620. [DOI] [PubMed] [Google Scholar]
- Serra-Pages C, Streuli M, Medley QG (2005). Liprin phosphorylation regulates binding to LAR: Evidence of liprin autophosphorylation. Biochemistry 44, 15715–15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam MM, Bhan P, Huang H-Y, Hsieh J, T-en H, Wu G-H, Punjabi H, Aplicano VDL, Chen C-W, Wagner OI (2018). Cilium lenght and intraflagellar transport regulation by kinases PKG-1 and GCK-2 in Caenorhabditis elegans sensory neurons. Mol Cell Biol 38, e00612–e00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Zhang X, Chai Y, Zhu Z, Yi P, Feng G, Li W, Ou G (2014). Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev Cell 30, 625–636. [DOI] [PubMed] [Google Scholar]
- Sheng L, Hao SL, Yang WX, Sun Y (2018). The multiple functions of kinesin-4 family motor protein KIF4 and its clinical potential. Gene 678, 90–99. [DOI] [PubMed] [Google Scholar]
- Siddiqui N, Straube A (2017). Intracellular cargo transport by kinesin-3 motors. Biochemistry (Mosc) 82, 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiering D, Bravo-Cordero JJ, Moshfegh Y, Miskolci V, Hodgson L (2013). Quantitative ratiometric imaging of FRET-biosensors in living cells. Methods Cell Biol 114, 593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigloher C, Zhan H, Zhen M, Richmond J, Bessereau JL (2011). The presynaptic dense projection of the Caenorhabditis elegans cholinergic neuromuscular junction localizes synaptic vesicles at the active zone through SYD-2/liprin and UNC-10/RIM-dependent interactions. J Neurosci 31, 4388–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taru H, Jin Y (2011). The Liprin homology domain is essential for the homomeric interaction of SYD-2/Liprin-alpha protein in presynaptic assembly. J Neurosci 31, 16261–16268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien NW, Wu GH, Hsu CC, Chang CY, Wagner OI (2011). Tau/PTL-1 associates with kinesin-3 KIF1A/UNC-104 and affects the motor’s motility characteristics in C. elegans neurons. Neurobiol Dis 43, 495–506. [DOI] [PubMed] [Google Scholar]
- Wagner OI, Esposito A, Kohler B, Chen CW, Shen CP, Wu GH, Butkevich E, Mandalapu S, Wenzel D, Wouters FS, Klopfenstein DR (2009). Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans. Proc Natl Acad Sci USA 106, 19605–19610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters FS, Verveer PJ, Bastiaens PIH (2001). Imaging biochemistry inside cells. Trends Cell Biol 11, 203–211. [DOI] [PubMed] [Google Scholar]
- Wu GH, Shanmugam MM, Bhan P, Huang YH, Wagner OI (2016). Identification and characterization of LIN-2(CASK) as a regulator of Kinesin-3 UNC-104(KIF1A) motility and clustering in neurons. Traffic 17, 891–907. [DOI] [PubMed] [Google Scholar]
- Wyszynski M, Kim E, Dunah AW, Passafaro M, Valtschanoff JG, Serra-Pages C, Streuli M, Weinberg RJ, Sheng M (2002). Interaction between GRIP and liprin-alpha/SYD2 is required for AMPA receptor targeting. Neuron 34. [DOI] [PubMed] [Google Scholar]
- Yeh E, Kawano T, Weimer RM, Bessereau JL, Zhen M (2005). Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J Neurosci 25, 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev S, Cooper R, Fetter R, Horowitz M, Shen K (2016). MT organization determines axonal transport dynamics. Neuron 92, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.