Abstract
Patients in whom mismatch repair (MMR)-deficient cancer develops in the absence of pathogenic variants of germline MMR genes or somatic hypermethylation of the MLH1 gene promoter are classified as having suspected Lynch syndrome (SLS). Germline whole-genome sequencing (WGS) and targeted and genome-wide tumor sequencing were applied to identify the underlying cause of tumor MMR deficiency in SLS. Germline WGS was performed on samples from 14 cancer-affected patients with SLS, including two sets of first-degree relatives. MMR genes were assessed for germline pathogenic variants, including complex structural rearrangements and noncoding variants. Tumor tissue was assessed for somatic MMR gene mutations using targeted, whole-exome sequencing or WGS. Germline WGS identified pathogenic MMR variants in 3 of the 14 cases (21.4%), including a 9.5-megabase inversion disrupting MSH2 in a mother and daughter. Excluding these 3 MMR carriers, tumor sequencing identified at least two somatic MMR gene mutations in 8 of 11 tumors tested (72.7%). In a second mother–daughter pair, a somatic cause of tumor MMR deficiency was supported by the presence of double somatic MSH2 mutations in their respective tumors. More than 70% of SLS cases had double somatic MMR mutations in the absence of germline pathogenic variants in the MMR or other DNA repair–related genes on WGS, and, therefore, were confidently assigned a noninherited cause of tumor MMR deficiency.
Lynch syndrome (LS) is an autosomal-dominantly inherited disorder caused by germline variants affecting the DNA mismatch–repair (MMR) genes MLH1, MSH2, MSH6, and PMS2.1 Carriers of pathogenic variants in MMR genes have an increased risk for multiple cancer types, predominantly colorectal cancer (CRC) and endometrial cancer.2 A key characteristic of LS-related tumors is DNA-MMR deficiency, evidenced by microsatellite instability (MSI),3 and a loss of MMR protein expression. However, MMR deficiency is not unique to LS and can occur when both alleles of an MMR gene are deactivated by somatic changes [eg, MLH1 promoter methylation, loss of heterozygosity (LOH), or mutation]. It is important to distinguish MMR-deficient cancers that arise from LS and those that arise sporadically, because patients with LS and their relatives can benefit from intensive clinical management and surveillance.
Individuals with MMR-deficient tumors with no evidence of MLH1 promoter methylation and in whom no MMR germline pathogenic variants have been identified are defined as having suspected LS (SLS). This group may represent over 50% of the MMR-deficient CRCs and endometrial cancers in population-based studies4 and poses significant clinical challenges due to the intensive screening of family members, that is, not based on carrier status. Potential explanations of SLS include: i) artefactual loss of MMR protein expression or incorrect interpretation of immunohistochemistry (IHC) staining,5,6 ii) the presence of MMR germline pathogenic variants that have not been identified or that are outside of the current testing scope,7,8 iii) germline pathogenic variants in non-MMR genes, which include MUTYH9 and POLE/POLD1,10 and iv) unidentified pathologic processes leading to MMR deficiency.7 Studies have shown that somatic inactivation of both MMR gene alleles (double or biallelic somatic mutation) is observed in up to 70% of SLS-related cancers.11, 12, 13, 14 Therefore, diagnostic approaches that can resolve the underlying etiology of SLS are needed for optimal risk assessment and management.
The utility of germline whole-genome sequencing (WGS) and targeted tumor sequencing in the determination of germline and somatic causes of MMR deficiency was assessed in 14 SLS cases from the Australasian Colorectal Cancer Family Registry15,16 and the Australian National Endometrial Cancer Study.17 Additionally, WGS of tumor samples from two mother–daughter pairs with SLS was used to estimate tumor-related features such as MSI, tumor mutation burden (TMB), and somatic mutational signatures, contributing to the diagnosis of an inherited or sporadic etiology in their families.
Materials and Methods
Study Participants
For this study, SLS was defined as meeting the following criteria: i) the presence of one or more MMR-deficient tumors, determined by loss of expression of one or more MMR proteins on IHC [eg, patterns of loss of MLH1/PMS2 or MSH2/MSH6 or MSH6 only), ii) absence of evidence of MLH1 promoter methylation in tumors that showed loss of expression of the MLH1 and PMS2 proteins, and iii) absence of pathogenic variants detected on germline Sanger sequencing or multiplex ligation-dependent probe amplification of the MMR gene(s) indicated as defective by the pattern of protein loss on IHC.
To enrich this WGS study for potential LS cases that might have been missed on conventional germline screening approaches, 13 participants were selected from 18 identified as having SLS from a clinic-based recruitment arm of the Australasian Colorectal Cancer Family Registry15,16 study and 1 SLS participant from the Australian National Endometrial Cancer Study17 based on having blood- and tumor-derived DNA material available for testing and who demonstrated one or more features associated with LS, namely: i) having a diagnosis of early-onset (age <50 years) cancer, ii) development of multiple tumors, iii) having a first-degree relative with an LS-spectrum tumor, and/or iv) meeting the Amsterdam I or II or Revised Bethesda clinical criteria. A family history of cancer meeting the Amsterdam I and II18 clinical criteria or Revised Bethesda guidelines19 was derived as previously described.
Two LS individuals with previously identified pathogenic MMR gene variants underwent WGS as positive controls: control sample 1 carried a pathogenic intronic MSH2 variant (c.212-478T>G) within a highly repetitive region that has been shown to disrupt splicing8; control sample 2 carried a 1928-bp deletion of MSH2 exon 6 (chr2:47640695-47642623) (Supplemental Figure S1A). Written informed consent for blood and tumor sample collection for research purposes was obtained from all participants.
MMR Molecular Testing
All tumors from the study participants were MMR deficient as determined on IHC staining for the MLH1, MSH2, MSH6, and PMS2 proteins as previously described.20,21 A subset of tumors were tested for MSI using a 10-marker panel as previously described.21 Tumor methylation of the MLH1 gene promoter was performed using MethyLight assays as previously described.17,20,22 Testing for the BRAF p.V600E somatic mutation was performed on samples of CRCs and adenomas using fluorescence allele-specific amplification with products being resolved via capillary separation.23 Germline testing for MLH1, MSH2, MSH6, and PMS2 gene variants was performed using Sanger sequencing and multiplex ligation-dependent probe amplification, including testing for 3′ deletions in EPCAM as previously described.17,20
Germline and Tumor Sequencing
Germline WGS was performed on peripheral blood–derived DNA from the 14 individuals with SLS and 2 MSH2 pathogenic variant–positive controls in this study using the TruSeq DNA library-preparation guide (Illumina, San Diego, CA), yielding a mean insert size of 300 to 400 bp. The libraries were sequenced by Macrogen (Seoul, South Korea) on a HiSeq 2000 sequencer (Illumina) to a mean coverage of 30×.
To confirm the germline MSH2 exon 1 to 7 inversion, primers were designed for an inversion-specific PCR that spanned the specific break points, amplifying a 238-bp product if the inversion was present. A control amplicon was designed for exon 3 of the TDG gene such that a 344-bp product was amplified irrespective of whether the MSH2 inversion was present. Table 1 contains the primers that were used. The inversion PCR amplified a product with the following sequence:
Table 1.
Primers Used in Inversion-Specific PCR
| Primer type | Primer sequence |
|---|---|
| Inversion primers | |
| Forward | 5′-GGGGCCATAATCCAGTCCTT-3′ |
| Reverse | 5′-ATGTGTGCCTGCATATGTGT-3′ |
| TDG primers | |
| Forward | 5′-CAAAACAACCAGTGGAACCCA-3′ |
| Reverse | 5′-ACCTCATGAAGCTGACACCA-3′ |
TDG, thymine DNA glycosylase.
5′-TGTGGAGCTGGGGCCATAATCCAGTCCTTATGTGATTACTGTGAAGTTATCCTTTTCCCCCAACATCTACTTATGAATAAAGAGTTTATTAAGTGGTTAACTGCAAGGCAAGATTGCTCACAGTACTCTCAATGACACTCCAGGCTCAATGGCCTAG[break point]AGGACatatatgtgtatatatacaCAtatatacgtatatatatacacacacacatatatatacacatatgcaggcacacat-3′ (uppercase, sequence 9.5-megabase (Mb) upstream of MSH2 intron 7; lowercase, MSH2 intron 7 sequence; bold, inserted sequences that were not present in the reference sequence; underline, primers used to detect inversion).
The PCR-amplified bands from SLS case 12 (SLS12), who carried the MSH2 exons 1 to 7 inversion, were sequenced along with those from a positive control (which came from the Cardiff Molecular Genetics Laboratory in the United Kingdom) using the primers designed by Rhees et al.24 The sequencing trace in Supplemental Figure S2 confirmed that the inversion carried by SLS12 was the same as that in the positive control.
All SLS-related tumors were screened for somatic mutations in the MLH1, MSH2, MSH6, PMS2, MLH3, MSH3, PMS1, EXO1, EPCAM, and SETD2 genes using a custom-designed AmpliSeq (Thermo Fisher Scientific, Waltham, MA) targeted resequencing assay as follows. Formalin-fixed, paraffin-embedded (FFPE) tissue was macrodissected to enrich for tumor cells. DNA extraction was performed using QIAamp DNA FFPE tissue kit and standard protocols (Qiagen, Hilden, Germany). FFPE-derived DNA was determined to have degradation sufficient for proceeding with library preparation without further fragmentation. End repair and adapter ligation were performed with an NEBNext Ultra II kit (New England BioLabs, Ipswich, MA) according to the manufacturer's protocols. Individually barcoded libraries were pooled in equimolar amounts, and 1.1 nmol/L of combined libraries were sequenced using a NovaSeq 6000 S4-flowcell at 2 × 150-bp reads with a mean depth of 40× in the capture target regions (Illumina, San Diego, CA). The customized AmpliSeq panel was designed to screen for MMR genes in the FFPE tumor DNA samples through the generation of a PCR-based library (125 to 175 bp) and sequencing on the Ion Torrent system (Thermo Fisher Scientific, Waltham, MA). A customized panel was employed and consisted of 443 amplicons across two pools including the 4 core MMR genes (MLH1, MSH2, MSH6, and PMS2) plus 6 additional genes involved in the mismatch-repair pathway (MSH3, MLH3, EPCAM, PMS1, EXO1, and SETD2), comprising a total capture size of approximately 35.07 Kb (targeting exonic and splice sites only) and achieving 94.5% mean coverage of the 10 genes [ranging from 96.9% (MLH1) to 80.5% (PMS2)]. DNA samples were obtained from archival FFPE tumor material, with 10 ng of double-stranded DNA per sample per pool used. Technical success of somatic-mutation detection was high, with 14 of 15 FFPE tumor DNA samples in this study passing quality control for library construction and sequencing and 84% of reads being on target. The mean read depth per sample was 2800. Replicate testing provided identical results. MMR gene mutations were validated using Sanger sequencing as previously described.15,17,25, 26, 27 Somatic mutations with a variant allele frequency of ≥0.07 were confidently validated on Sanger sequencing. Amplicon-specific primers are provided in Supplemental Table S1.
For additional somatic resolution, a service provider (GenomeScan, Leiden, the Netherlands) was used for WGS (n = 4) or whole-exome sequencing (WES) (n = 1) on samples from a subset of tumors based on the identification of a single somatic MMR gene mutation from the targeted panel analysis (SLS5 and SLS13) or from a mother–daughter pair (SLS9 and SLS10). Capture of the whole exome was performed on a single FFPE CRC tissue DNA sample and matched peripheral blood–derived DNA (SLS5) using SureSelect Clinical Research Exome software version 2 (Agilent, Santa Clara, CA), with sequencing performed on the NovaSeq 6000 system comprising 150-bp paired-end reads. Mean on-target depths of coverage were 99 in FFPE tumor DNA samples and 110 in blood-derived DNA samples. Whole genomes were sequenced to a depth of 30× from a 500- to 700-bp library generated using a NEBNext Ultra II chemistry kit (New England BioLabs). Clustering and sequencing of 1.1 nmol/L of sample-specific libraries were performed on the NovaSeq 6000 system.
Bioinformatic Analysis of Sequencing Data
For both germline and tumor DNA samples, raw FastQ sequence quality control was confirmed using FastQC version 0.11.7,28 and paired-end FastQ files were aligned to the human reference genome (hg19)29 using Burrows-Wheeler Aligner software version 0.7.12.30 Germline single-nucleotide variants (SNVs) and short insertions and deletions (indels) were called using the GATK Best Practices Pipeline software version 3.4-46.31 Somatic SNVs and short indels were called using Strelka software version 2.9.2,32 Platypus software version 0.8.1,33 and Mutect software version 2,34 using default settings. Each somatic SNV or somatic indel variant was annotated by the consensus of the three callers, and high-confidence calls were determined as those reported by at least two of three callers to reduce false-positive results. Germline and somatic variants were annotated using Variant Effect Predictor software version 77 (Ensembl35) and SnpEff software version 4.1,36 including in silico variant effect predictions from REVEL (rare exome variant ensemble learner)37 and CADD (combined annotation dependent depletion),38 and population frequencies from gnomAD (Genome Aggregation Database).39 Pathogenicity classifications from InSiGHT40 (https://insight-database.org, last accessed September 18, 2018) for the MMR genes and ClinVar41 were associated with variants where available. Germline structural variants were detected with DELLY software version 0.7.1,42 LUMPY software version 0.2.11,43 and GRIDSS software version 0.11.5,44 and somatic structural variants were detected with GRIDSS software version 2.2.144 and Manta software version 1.5.0.45 In all cases, high-confidence structural variant calls were selected using the application of the quality filters recommended in the documentation of each tool. The concordance between the calls made by each tool was calculated, allowing a window of uncertainty in break end coordinates of ±50 bp. Germline and somatic copy number variants were detected using HMMCopy software version 1.22.0.46
SNVs were assessed for predicted effect on splicing using HumanSplicingFinder software version 3.147 and MaxEntScan version 1.048 within the gene(s) indicated to be deficient on IHC. MSI was assessed computationally using MSIsensor software version 0.549 using recommended thresholds: >3.5, high level; 1 to 3.5, low level; and <1, microsatellite stable. Somatic mutational signatures from the set of 30 standard profiles50 published on the COSMIC website (https://cancer.sanger.ac.uk/cosmic/signatures_v2, last accessed August 2, 2019) were calculated using the DeconstructSigs method51 for each tumor sample using their identified somatic SNVs. TMB was calculated as the rate of somatic SNVs per megabase in the coding region of the genome, with a TMB of >10 considered hypermutated and with a TMB of >100 considered ultra-hypermutated.52 Tumor LOH was assessed on LOH software version 0.3,53 a tool that compares the allele fraction of germline variants within tumor samples.
The selected thresholds for variant population frequency in gnomAD version 2.1 and predicted pathogenicity scores from CADD and REVEL were derived from the analysis of missense variants classified as pathogenic (class 5) or likely pathogenic (class 4) from the InSiGHT database and published recommendations for the in silico prediction tools,37,38 respectively. All likely pathogenic and pathogenic missense variants in the four main MMR genes in the InSiGHT database were annotated with their population frequencies from gnomAD and their predicted pathogenicity from CADD and REVEL to estimate suitable thresholds for variant filtering. A mean population frequency of 2.9 × 10−5 (SD 4.6 × 10−5) was observed, with an extreme outlier maximum value of 1.7 × 10−4, a mean CADD score of 29.3 (minimum, 17.6; SD 3.8), and a mean REVEL score of 0.9 (minimum, 0.3; SD 0.1). All variants retained after filtering were manually inspected in the Integrative Genomics Viewer version 2.4.1348 to assess the quality of read alignments and supporting evidence.
Candidate Gene Prioritization
Germline and somatic variants were annotated and prioritized by their intersection with three tiers of genes (Supplemental Table S2), with 2-Kb flanking regions added around the transcription start and stop sites. The highest-priority genes (Tier 1, n = 5) contained the four MMR genes plus EPCAM. Variants intersecting promoter and enhancer regions of the MMR gene indicated to be defective in an individual's tumor were annotated using promoter and enhancer regions taken from the GeneHancer database.54 Tier 2 comprised genes assessed by the ClinGen Hereditary Colorectal Cancer and Polyposis Susceptibility Gene Curation Panel to have definitive, strong, or moderate evidence supporting an association with hereditary CRC and/or polyposis (APC, ATM, AXIN2, BLM, BMPR1A, GREM1, MLH3, MUTYH, POLD1, POLE, PTEN, SMAD4, STK11) or other syndromes with rare manifestation of CRC and/or polyposis (FLCN, TP53, CDH1)55 or recently reported recessive polyposis genes (MSH356, NTHL157,58) (n = 18). Tier 3 contained a curated list of DNA-repair genes reported in the literature59,60 (n = 265). In SLS cases showing MLH1/PMS2 loss, all pseudogenes associated with PMS2 were searched for high-impact germline and somatic variants to account for the possibility of misattributed variants arising from errors in sequencing alignment.
Variant Filtering and Prioritization
Germline SNVs and indels identified in Tier 1 or 2 genes were prioritized if they were predicted to have an impact on function (truncating, frameshift, and splice site) or nonsynonymous variants resulting in a missense substitution with a CADD score of ≥15 or a REVEL score of ≥0.6. Tier 3 germline SNVs and indels were prioritized if they were loss-of-function or high-impact variants only. Filtering for population frequency encompassed prioritizing variants with a gnomAD allele frequency of ≤2 × 10−4 [except for known pathogenic variants in MUTYH NM_001128425.1: c.536A>G p.Tyr179Cys (allele frequency, 1.5 × 10−3) and NM_001128425.1: c.1187G>A. p.Gly396Asp (allele frequency, 3 × 10−3)] and in NTHL1 NM_002528.5: c.268C>T p.Gln90Ter (allele frequency, 1.4 × 10−3) (transcripts available from https://genome.ucsc.edu/cgi-bin/hgTables, last accessed August 2, 2019). All germline SNVs and indels were annotated with ClinVar clinical classifications, and those with a review status of at least two-stars (criteria provided, multiple submitters, no conflicts) classified as benign or likely benign were excluded. Prioritized germline variants were validated on Sanger sequencing and tested in relatives with a DNA sample available.
High-confidence somatic SNVs and indels called by at least two of three callers with a variant allele frequency of ≥0.10 were retained for further analysis. In targeted AmpliSeq tumor sequencing, SNVs and indels were called with a variant allele frequency of ≥0.07 (variant allele frequency fraction that could be validated on Sanger sequencing). For Tier 1 genes, coding SNVs and indels with an InSiGHT classification of ≥3 were prioritized. Intronic variants predicted to disrupt splicing or intersecting a promoter or enhancer region of the MMR gene indicated as defective on IHC were also prioritized. For Tier 2 and 3 genes, only loss-of-function variants were considered unless genes were known to harbor somatic hotspots (eg, within the exonuclease domains of POLE and POLD1).
Results
The characteristics of the 14 study participants with SLS are described in Table 2. Age at diagnosis of MMR-deficient tumor ranged from 35 to 78 years (median, 52 years) and 10 were female (71%). A family history of cancer meeting Amsterdam I or II clinical criteria was observed in 3 cases (21.4%). Two sets of mother–daughter pairs (SLS9 and SLS10, SLS11 and SLS12) were included in the cohort (Table 2 and Figure 1, A and B). Seven tumors showed loss of MLH1/PMS2 expression and were negative for the BRAFV600E somatic mutation in addition to being negative for tumor MLH1 gene promoter hypermethylation.
Table 2.
Characteristics of the Participants and Their Tumors That Were Included in This Study
| SLS ID | Sex | Age at diagnosis, years | Tumor ID | Tumor type | MMR IHC loss | MSI status | Family cancer history (MMR IHC result) | Clinical criteria | Final classification∗ | Evidence supporting classification |
|---|---|---|---|---|---|---|---|---|---|---|
| C1 | F | 19 | CRC | MSH2/MSH6 | MSI-H | Lynch | MSH2: c.212-478T>G (germline) | |||
| C2 | M | 37 | CRC | MSH2/MSH6 | MSI-H | Lynch | MSH2 deletion of exon 6 (germline) | |||
| SLS1 | F | 52 | T1 | CRC: adenocarcinoma of transverse colon | MLH1/PMS2 | NT | Sister CRC (normal) at 42 years, breast at 53 years | None | Double MLH1 somatic |
MLH1: c.879C>G p.Tyr293Ter (somatic) MLH1: c.1975C>T p.Arg659Ter (somatic) |
| SLS2 | M | 51 | T2A | Kidney | MLH1/PMS2 | NT | None | None | NC | |
| 51 | T2B | Colonic tubular adenoma | Normal | NT | ||||||
| SLS3 | M | 45 | T3 | CRC: adenocarcinoma of transverse colon | MLH1/PMS2 | MSI-H | Sister CRC at 40 years; brother CRC (normal) at 45 years, pancreas at 60 years; brother kidney (normal) at 57 years, prostate at 63 years; brother brain at 59 years | Revised Bethesda | Single MLH1 somatic | MLH1: c.332C>T p.Ala111Val (somatic) |
| SLS4 | F | 68 | T4A | Breast | Normal | MSS | Brother CRC at unknown age; sister CRC at 72 and 77 years; sister CRC (normal) at 67 and CRC (MLH1/PMS2 loss†) at 78 years | None | Double MLH1 somatic | Multiple somatic MLH1 variants including truncating mutation MLH1: c.1036C>T p.Gln346Ter, MMRd not present across all tumors |
| 78 | T4B | Colonic tubulovillous adenoma | MLH1/PMS2 | NT | ||||||
| SLS5 | F | 42 | T5 | CRC: adenocarcinoma of transverse colon | MLH1/PMS2 | MSI-H | Mother CRC at 45 years; father CRC at 54 years | Amsterdam II | Double MLH1 somatic |
MLH1: c.1989+1G>A (somatic) MLH1: LOH (somatic) |
| SLS6 | F | 41 | T6A | CRC | NT | NT | None | Revised Bethesda | Lynch | MLH1: c.1958T>G p.Leu653Arg (germline) |
| 53 | T6B | CRC: adenocarcinoma of transverse colon | MLH1/PMS2 | NT | ||||||
| 53 | T6C | CRC | MLH1/PMS2 | NT | ||||||
| 76 | T6D | Sebaceous carcinoma | MLH1/PMS2 | NT | ||||||
| SLS7 | M | 66 | T7A | CRC: adenocarcinoma of rectum | MSH2/MSH6 | MSI-H | Daughter CRC at 45 years, mesothelioma (normal) at 48 years; sister endometrial at 37 years; brother bladder at 81 years; sister lung at 58 years | Amsterdam II | Double MSH2 somatic | MSH2: c.1351C>T p.Gln451Ter (somatic) |
| 79 | T7B | Colonic tubular adenoma | Normal | NT | MSH2: c.1204C>T p.Gln402Ter (somatic) | |||||
| SLS8 | M | 62 | T8A | Tubulovillous adenoma of rectum | MSH2/MSH6 | MSI-H | Mother CRC at 93 years | None | Double MSH2 somatic | MSH2: c.859G>T p.Gly287Ter (somatic) |
| 62 | T8B | CRC | Normal | NT | MSH2: c.2465_2466delinsAG p.Cys822Ter (somatic) | |||||
| 75 | T8C | Prostate | NT | NT | ||||||
| SLS9‡ | F | 54 | T9A | Duodenum | MSH6 | NT | Daughter ovarian at 40 years and endometrial at 40 years | None | Double MSH2 somatic | Multiple MSH2 somatic mutations, not shared with first-degree relative, MMRd not present across all tumors |
| 58 | T9B | CRC: mucinous adenocarcinoma of ascending colon | MSH2/MSH6 | NT | ||||||
| 67 | T9C | Brain | Normal | NT | ||||||
| SLS10‡ | F | 40 | T10A | Ovary | MSH2/MSH6 | MSI-H | Double MSH2 somatic | Multiple MSH2 somatic mutations, not shared with first-degree relative, mutational signatures 14 and 20 and POLD1 somatic mutation supporting somatic etiology | ||
| 40 | T10B | Endometrium | MSH2/MSH6 | NT | ||||||
| SLS11§ | F | 40 | T11A | CRC: mucinous adenocarcinoma of caecum | MSH2/MSH6 | NT | Daughter CRC at 35 years; brother CRC at 29 years and 46 years; brother CRC at 59 years | Amsterdam I | Lynch | MSH2: 9.5 Mb Inversion ex1-7 (germline) |
| 51 | T11B | Breast | NT | NT | ||||||
| SLS12§ | F | 35 | T12 | CRC: adenocarcinoma of sigmoid colon | MSH2/MSH6 | MSI-H | Lynch | MSH2: 9.5 Mb Inversion ex1-7 (germline) | ||
| SLS13 | F | 41 | T13 | CRC: adenocarcinoma of transverse colon | MSH2/MSH6 | NT | None | Revised Bethesda | Single MSH2 somatic | MSH2: c.2006-3T>G (somatic) |
| SLS14 | F | 66 | T14 | Endometrium | MSH2/MSH6 | NT | Brother CRC at 63 years | none | Double MSH2 somatic |
MSH2: c.1276+2delT (somatic) MSH2: c.2131C>T p.Arg711Ter (somatic) |
Family Cancer History, cancer in first degree relatives only shown; MMR IHC results shown in parentheses.
F, female; M, male; CRC, colorectal cancer; MMR, mismatch repair; MSI, microsatellite instability; MSI-H, microsatellite instability-high; MSS, microsatellite stable; NT, not able to be tested; SLS, suspected Lynch syndrome.
Positive controls. Control 1: intronic mutation disrupting splicing. Control 2: deletion of exon 6 in MSH2.
Final Classification—classification for each SLS case after germline whole-genome sequencing and tumor sequencing analysis detailed in this study. Double and single somatic mutation classification considered to be non-Lynch syndrome and of sporadic etiology. NC, tumor failed testing and therefore no classification could be achieved.
Sister developed two CRCs: one at age 67 years with normal MMR protein expression and one at age 78 years with loss of MLH1/PMS2 protein expression resulting from MLH1 promoter hypermethylation in the tumor.
Relative pair: SLS10 is the daughter of SLS9.
Relative pair: SLS12 is the daughter of SLS11.
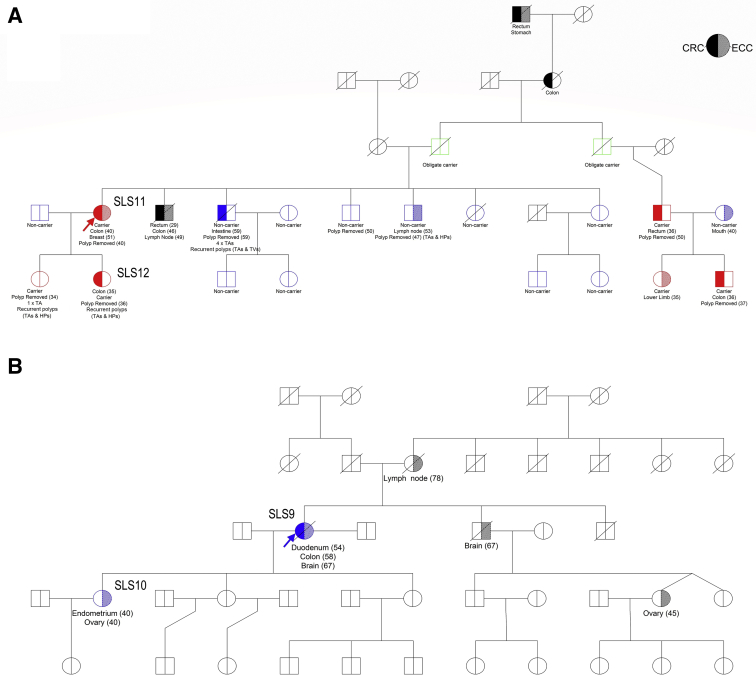
Figure 1.
Family pedigrees of two sets of mother–daughter pairs. A: SLS11 and SLS12. B: SLS9 and SLS10. Probands SLS11 (Family A) and SLS9 (Family B) are indicated with arrows. In Family A, an inversion-specific PCR test was used for genotyping 14 additional relatives, identifying four additional carriers (two CRC affected, and one in whom adenomas developed at age 34 years) and two additional obligate carriers (both unaffected). HP, hyperplastic polyp; TA, tubular adenoma; TV, tubulovillous adenoma
Germline Whole-Genome Sequencing Analysis
Germline variants retained after filtering and prioritization are provided in Supplemental Table S3. Of all 14 SLS individuals with samples screened on WGS, 3 carried predicted MMR pathogenic germline variants, including a MLH1 missense variant c.1958T>G, p.Leu653Arg in SLS6, recently reclassified as class 4 (likely pathogenic) by InSiGHT, and a 9.5-Mb inversion encompassing exons 1 to 7 of MSH2 (chr2:38121107–chr2:47669532) in mother–daughter pair (SLS11 and SLS12) (Supplemental Figure S1, B–D). These two germline pathogenic variants were not detected previously due to the MLH1 missense variant being incorrectly annotated and classified as benign and because the 9.5-Mb inversion was not detectable with earlier versions of the multiplex ligation-dependent probe amplification detection kit. An inversion-specific PCR test was used to genotype 14 additional relatives, identifying 4 additional carriers (2 CRC affected, and 1 who developed adenomas at age 34 years) and 2 additional obligate carriers (both unaffected) (Figure 1A). Sanger sequencing of the inversion-specific PCR product confirmed that this was the same inversion as previously reported24 (Supplemental Figure S2). In SLS cases showing MLH1/PMS2 loss, no high-impact variants affecting PMS2 or its pseudogenes were identified.
Somatic MMR Gene Analysis
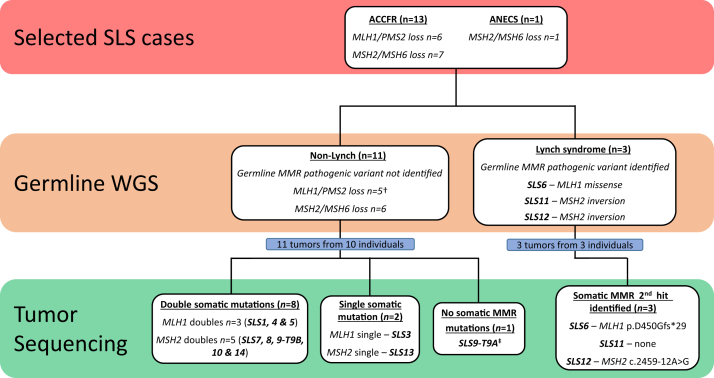
Targeted tumor sequencing of the MMR genes was completed in 14 tumors from 13 SLS cases [tumor (T) 2A failed testing] (Table 2); 11 of the tested tumors occurred in the colon or rectum. To further explore somatic mutations and better resolve LOH across the MMR genes, a subset of 5 tumors was selected for additional WGS (n = 4) or WES (n = 1), comprising the mother–daughter pair (SLS9 and SLS10) in whom no pathogenic germline variants were identified, and from 2 (of the 3) tumors that showed only a single somatic MMR gene mutation after targeted sequencing analysis suggesting that these tumors remained unresolved under a two-hit hypothesis for the MMR genes. In the MMR genes indicated to be defective on IHC, at least two somatic mutations were identified in 9 of the 14 tumors (64.3%), 1 somatic mutation was identified in 3 (21.4%), and no somatic mutations were identified in 2 (14.3%) (Supplemental Table S3 and Figure 2). No somatic structural variants or copy number variants intersecting with the four MMR genes were detected.
Figure 2.
A summary of testing approach and outcomes and the final classification of each SLS case from the germline whole-genome sequencing (WGS) and tumor-sequencing analyses in this study. †SLS2: kidney tumor (T2A) failed tumor sequencing. ‡Additional tumor WGS information: no MMR deficiency–related mutational signatures, microsatellite stable by microsatellite instability sensor, tumor mutation burden not considered hypermutated; therefore, tumor not considered to have MMR deficiency and likely false-positive loss of MSH6 expression on IHC. ACCFR, Australasian Colorectal Cancer Family Registry; ANECS, Australian National Endometrial Cancer Study.
Two tumors from SLS9 were tested (T9A-duodenal and T9B-CRC) by targeted sequencing and WGS. In T9A, IHC indicated loss of MSH6; however, no MMR gene variants were found that might have disrupted MSH6 in that tumor (Supplemental Table S3). In T9B, which demonstrated a loss of MSH2/MSH6, multiple somatic MSH2 mutations were observed, including somatic variants predicted to disrupt splicing through the activation of both cryptic splicing donor and acceptor sites, plausibly related to the somatic inactivation of both alleles of that gene (Supplemental Table S3). Similarly, ovarian cancer tumor (T10A) from SLS10 showed a loss of MSH2/MSH6 expression where the targeted tumor sequencing identified multiple somatic MSH2 variants that were confirmed on WGS (Supplemental Table S3).
WES was performed on the CRC tumor T5. The first 50 Mb of the p-arm of chromosome 3, containing MLH1, showed clear evidence of LOH (Supplemental Figure S3A). Additionally, a MLH1:c.1989+1G>A variant with high variant allele frequency was identified on both the targeted (79%) and WES (58%) analyses of the tumor. The combination of this somatic variant and LOH suggests biallelic MLH1 inactivation in the tumor and is concordant with the IHC results (Supplemental Table S3). None of the other four tumors with WGS showed evidence of LOH (Supplemental Figure S3).
Tumor Whole-Genome and Exome Sequencing Analysis
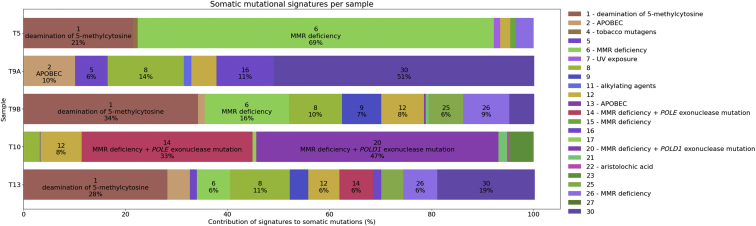
High-impact somatic mutations in genes from Tiers 2 and 3 were identified in each of the five tumors with WGS/WES (Supplemental Table S3). Notably, the three CRCs tested (T5, T9B, and T13) had stop-gain mutations in APC (Supplemental Table S3). The ovarian tumor (T10A) had a missense mutation in the DNA polymerase gene POLD1:p.Glu318Lys, which is situated at the exonuclease active site within the encoded protein. In addition, features associated with tumor MMR-deficiency were investigated, namely somatic mutational signatures (signatures 6, 14, 15, 20, and 26, as defined on the COSMIC website, https://cancer.sanger.ac.uk/cosmic/signatures_v2, last accessed August 2, 2019), TMB and MSI, estimated from the sequencing data. The somatic mutational signatures are shown in Figure 3, where signatures 6, 20, and 26 were observed in four (T5, T9B, T10A, and T13) of the five tumors, ranging in contribution from 4% to 69% and supporting tumor MMR-deficiency status determined on MMR IHC (Supplemental Table S3). Only the ovarian tumor (T10A) showed significant contributions from signatures 20 and 14, both of which have been associated with MMR deficiency and defective polymerase proofreading (COSMIC version 3.1). The duodenal tumor (T9A) did not exhibit any MMR deficiency–associated signatures despite a loss of MSH6 protein expression on initial MMR IHC testing. The MSI status derived from WGS/WES by MSIsensor predicted four of the five tumors tested to be MSI-high, whereas the duodenal tumor T9A was predicted to be microsatellite stable (Supplemental Table S3). Four tumors (T5, T9B, T10A, and T13) exhibited high TMBs consistent with a hypermutator phenotype (or ultra-hypermutator in the case of the ovarian tumor T10A), whereas the duodenal tumor T9A was not considered to be hypermutated (Supplemental Table S3). A summary of testing approach and outcomes, and the final classification of each SLS case from the germline WGS and tumor-sequencing analyses in this study, are shown in Figure 2 and Table 2, respectively.
Figure 3.
The somatic mutational signature components for tumor samples T5, T9A, T9B, T10, and T13 indicated by the signature number, suggested etiology, and the percentage contribution to the overall mutational composition for each sample. APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like.
Discussion
In this study, germline WGS was applied to tumor samples from 14 patients classified as having SLS and who were selected for having clinical features suggestive of LS (eg, diagnosis at <50 years of age, family history meeting Amsterdam clinical criteria), to determine whether an investigation of the DNA-MMR genes beyond current conventional testing approaches could improve the detection of MMR germline pathogenic variants. WGS analysis identified MMR germline pathogenic variants in 3 of the 14 SLS cases (21.4%), although these specific variants would have been detected with current targeted MMR gene-testing approaches. In addition, at least 1 MMR-deficient tumor from 13 SLS cases (1 tumor failed) was analyzed for somatic mutations within the MMR genes using targeted sequencing and further tested on WGS or WES in 5 of these tumors. At least two somatic mutations in MMR genes indicated as defective on IHC were identified in 8 of the 11 tumors tested (72.7%), excluding the three identified carriers of germline pathogenic variants. These collective findings demonstrate that germline WGS and tumor MMR gene analysis can be used to confidently assign a noninherited cause of MMR deficiency related to double somatic MMR mutations, even in SLS cases with clinical features suggestive of LS, and add to the growing number of studies that have shown that double somatic MMR mutations are a common cause of MMR deficiency in SLS.11, 12, 13,61, 62, 63, 64 Furthermore, the application of tumor WGS/WES to the subset of 5 SLS tumors enabled the determination of additional features associated with defective DNA MMR including somatic-mutation signatures 6, 20, and 26; MSI-high status; and high TMB, providing further evidence of an MMR-deficient tumor phenotype and allaying concerns over a false-positive MMR IHC result.
Germline WGS analysis of the mother–daughter pair SLS11 and SLS12, each with a MSH2/MSH6-deficient CRC, identified an inversion encompassing exons 1 to 7 of MSH2. This inversion was first reported in the literature by Wagner et al65 in 2002 and later by Rhees et al24 in 2014 and by Mork et al66 in 2017 using various methods, although none employing WGS. Although the evolution of clinical genetic testing has ensured that this inversion can now be detected by more recent versions of the multiplex ligation-dependent probe amplification assay, this study demonstrates that WGS is also a valid approach with the advantage of nonspecific structural variant detection.
In a second mother–daughter pair (SLS9 and SLS10), germline and tumor WGS (Figure 1B) identified no germline coding variants affecting MSH2 or MSH6. In addition to exonic variants, WGS enabled the investigation of intronic and promoter/enhancer variants in the MMR genes. A germline MSH6:NG_007111.1:g.3634G>A variant that resided 20 bp upstream from the MSH6 promoter/enhancer element GH02J047781 (the highest-scoring element in the GeneHancer database for MSH6) was identified in both individuals. This variant was not observed in gnomAD, highlighting its rarity. Further segregation of this variant in relatives was not possible. WGS was performed on three tumor samples (T9A, T9B, and T10A) from these two patients to further resolve tumor etiology. T9B-CRC and T10A–ovarian cancer both acquired somatic frameshift mutations within a coding microsatellite of MSH6. Although these somatic MSH6 mutations may represent the second hit subsequent to the germline variant (MSH6:NG_007111.1:g.3634G>A), it is also possible that this mutation may be a consequence of MMR deficiency, rather than a cause. Somatic expansion of this repeat in MSH6 exon 5 has been previously reported.67 Two of the three tumors tested in these patients showed a loss of MSH2/MSH6 expression, suggesting that the defect lies in MSH2. A rare germline MSH2:c.366+761A>G variant predicted by HumanSplicingFinder and MaxEntScan to activate a cryptic splice donor site was identified in SLS10 but not in the mother (SLS9). T9B-CRC and T10A–ovarian cancer both harbored multiple somatic MSH2 mutations that could potentially inactivate both alleles of MSH2, supporting a sporadic etiology of both of these tumors. The T9A-duodenal tumor in the mother showed solitary loss of MSH6 expression, although no somatic MSH2 or MSH6 mutations were identified.
Four of the five tumors tested by WGS/WES (T5, T9B, T10A, and T13) demonstrated high proportions of mutational signatures 6, 20, and 26 (Figure 3), associated with defective DNA MMR.50 Additionally, the TMB for these four tumors indicated hypermutation and high levels of MSI were predicted by MSIsensor. These findings provide mechanistic evidence of defective MMR in these tumors, consistent with the IHC results. For the duodenal tumor T9A, no MMR deficiency–related somatic mutational signatures were observed, the tumor was predicted to be microsatellite stable, and the TMB was <10 mutations/Mb. These results suggest that T9A did not have defective DNA MMR despite the loss of MSH6 indicated on IHC, representing a possible false-positive IHC result. Therefore, SLS9 was classified as having one MMR-deficient tumor with functional evidence of defective MMR (T9B-CRC) and one MMR-deficient tumor without functional evidence of defective MMR (T9A-duodenal); the latter was not suggestive of an LS etiology.
The ovarian cancer T10A demonstrated strong somatic mutational signatures 20 (47%) and 14 (33%) and was ultra-hypermutated (117.7 mutations/Mb). The findings from a recent study demonstrated that tumors exhibiting both somatic mutational signatures 20 and 14 and an ultra-hypermutation phenotype are associated with defective MMR and (clonal) exonuclease domain mutations in the DNA polymerase gene POLD1, where the POLD1 somatic mutation precedes the loss of MMR.68 Concordant with the somatic mutational signature profile, somatic mutation in the exonuclease domain of POLD1 was observed, together with multiple predicted somatic mutations in MSH2. These findings suggest a somatic etiology underlying the MMR deficiency in T10A. The combined germline and somatic WGS findings in the mother–daughter pair SLS9 and SLS10 argue in favor of somatic MSH2 loss in these individuals.
Several studies have shown that double somatic mutations (including LOH), resulting in biallelic MMR gene inactivation, are a common cause of MMR deficiency in SLS.11, 12, 13,61, 62, 63, 64 Excluding both mother–daughter relative pairs and the MLH1 pathogenic variant carrier, six of the eight remaining tumors/SLS cases (SLS1, SLS4, SLS5, SLS7, SLS8, and SLS14) were classified as double somatics, suggesting a somatic cause of their tumor MMR deficiency and supported by an absence of predicted MMR germline pathogenic variants in five of these six individuals (SLS1, SLS4, SLS5, SLS8, and SLS14). Additionally, in the double somatic cases SLS4, SLS7, and SLS8, second or third primary tumors developed in which IHC showed normal retained expression of the MMR proteins. This finding is unlike those in LS, in which synchronous or metachronous tumors develop concordant loss of MMR protein expression. Of interest, five of the seven double somatic SLS cases had a first-degree relative who developed an LS-related cancer. In a recent study by Pearlman et al,14 patients with LS in whom CRC developed were 15 times more likely to have a family history of cancer meeting Amsterdam II criteria, and 6 times more likely to develop multiple LS-associated cancers, compared with patients in whom MMR-deficient CRC developed as a result of double somatic MMR mutations. In the present study, two SLS cases with double somatic CRCs (SLS5 and SLS7) had a family history fulfilling the Amsterdam II criteria, and multiple tumors developed in three (SLS4, SLS7, and SLS8). These findings suggest that a family cancer history and the presence of multiple synchronous or metachronous tumors, while good predictors of LS, do not rule out a somatic etiology for tumor MMR deficiency.
Germline pathogenic variants within the exonuclease domain of the POLE or POLD1 gene or biallelic mutations within the MUTYH gene may indirectly lead to tumor MMR deficiency.9,69,70 A single SLS carrier (SLS8) was identified as heterozygous for the MUTYH:p.Arg426Cys predicted pathogenic missense variant. No evidence of a second germline MUTYH mutation was found; however, the corresponding tumor (T8A) had two somatic loss-of-function mutations in MSH2. A previous analysis of the role of germline MUTYH mutations in SLS showed a significant association for biallelic MUTYH carriers but not for monoallelic carriers,69 further supporting a somatic etiology for this MMR-deficient tumor phenotype. Recent studies of SLS have identified germline bioinformatically predicted pathogenic variants in MUTYH,61,63 EXO1,71 and POLD171 as well as BUB1,61 SETD2,61 FAN1,61 RFC1,71 RPA1,71 MLH3,71 MSH3,63 AXIN1,63 and AXIN2.63 Of these candidate SLS genes, predicted pathogenic variants were found only in EXO1 (exonuclease 1), a gene that plays an important role in MMR via strand excision,72 identified in SLS7, SLS13, and SLS14. Of interest, the carrier of EXO1:c.-125G>A variant (SLS13), predicted by Variant Effect Predictor to be in a potential splice region, also had a somatic mutation in EXO1:p.Arg668Ter. Two bioinformatically predicted pathogenic missense germline variants in the Tier 2 gene ATM were identified in two individuals, although one of these occurred in SLS6, who carried the germline MLH1:p.Leu653Arg pathogenic variant.
This study had several strengths. The germline WGS approach enabled the detection of complex structural variants, identifying the inversion of MSH2 exons 1 to 7, and the detection of MMR variants in the promoter/enhancer and intronic regions. Current targeted next-generation sequencing (ie, multigene panel testing) approaches may miss these and other types of pathogenic variants due to incomplete genome coverage (eg, intronic and promoter regions), difficulties with low DNA complexity (eg, recognized difficulty in detecting the recurring MSH2:c.942+3 A>T pathogenic variant73), and challenges in identifying complex mutations74,75 with commonly used sequencing technologies. Germline WGS was able to detect a previously identified deep intronic pathogenic variant in control sample 1 within a highly repetitive intronic region of the MSH2 gene.8 This variant was predicted to potentially disrupt splicing by both HumanSplicingFinder and MaxEntScan, validating the approach employed in this study. Rare intronic variants that are predicted to disrupt splicing in the likely-defective MMR gene were also detected in three germline samples, and a rare germline MSH6 promoter region variant shared by a mother and daughter was detected. Validation of the effect of these bioinformatically prioritized variants is challenging and is an area where further research is needed.
The application of targeted MMR gene sequencing to all but one of the MMR-deficient tumors from the SLS cases enabled the detection of double somatic MMR mutations. The concordance for identifying coding MMR gene somatic mutations was high between targeted MMR gene sequencing and WGS/WES; however, the additional information gained from WGS/WES in the five tumors tested provided clinically useful insights into the tumor etiology and confirmation of MMR deficiency. There still remain significant challenges in classifying somatic variants,76,77 particularly in noncoding regions of the genome, yet the impact of noncoding somatic alterations on tumorigenesis is increasingly recognized.78 More widespread somatic MMR gene testing and international data sharing will facilitate improvements in both somatic and germline MMR gene variant classification.40,76 A limitation of the study was the possibility that there are genes outside of those included in the three tiers that influence MMR gene and/or protein expression. For example, overexpression of miR-155 has been shown to significantly down-regulate the MMR proteins, inducing a mutator phenotype and MSI.79 Furthermore, potential somatic mosaicism was not explored, which, although rare, has been previously reported in individuals with LS.11,80,81
Conclusion
Germline WGS identified three carriers of MMR pathogenic variants, including a 9.5-Mb inversion in multiple family members, although these specific pathogenic variants would have been detected with current targeted MMR gene-testing approaches. The potential advantage of WGS in identifying novel complex variants should be considered in conjunction with the limitations of novel and noncoding variant interpretation, as discussed in the previous paragraph, before WGS should be recommended as routine clinical testing. This study further highlights the diagnostic benefit of tumor sequencing of the MMR genes in SLS cases. Double somatic MMR gene mutations, and therefore a likely sporadic etiology, were identified as the cause of tumor MMR deficiency in >70% of the SLS cases in this study (excluding the three identified LS carriers) in whom germline WGS confirmed the absence of other novel germline complex MMR gene variants. Additionally, WGS or WES of the SLS tumors provided additional interrogation for LOH and enabled the determination of tumor MMR deficiency–related features of MSI, TMB, and somatic mutational signatures. The combined analysis of germline and tumor WGS in a SLS mother–daughter pair provided evidence against LS as the cause of tumor MMR deficiency in these two relatives. As costs of high-throughput DNA sequencing continue to fall, the application of a tumor-sequencing approach has the potential to replace the current LS screening methodology based on tumor IHC, PCR-based MSI analysis, and germline multigene panel sequencing and has been supported recently.82 Testing for double somatic MMR mutations is currently not a part of routine clinical practice, and is not a part of the recommendations of the American College of Medical Genetics and Genomics guidelines for genetic testing for LS.
Acknowledgments
We thank the members of the Colorectal Oncogenomics Group; the participants and staff from the Colon-CFR, in particular, Maggie Angelakos, Samantha Fox, and Allyson Templeton for their support of the manuscript; and the Melbourne Bioinformatics on its Peak Computing Facility for computation and bioinformatics support.
Footnotes
Supported by National Health and Medical Research Council of Australia (NHMRC) project grant GNT1125269 (D.D.B.); an NHMRC R.D. Wright Career Development Fellowship (D.D.B.); the University of Melbourne Research at Melbourne Accelerator Program (D.D.B.); a State of Victoria Department of Health and Human Services Victorian Health and Medical Research Fellowship (B.J.P.); a Government of Australia Research Training Program scholarship (P.G.); NHMRC Senior Research Fellowship ID1061779 (A.B.S.); NIH National Cancer Institute award UM1CA167551 (M.A.J.); and cooperative agreements with the following Colon Cancer Family Registry centers (project C-AU-1014-01): Australasian Colorectal Cancer Family Registry NCI/NIH grants U01 CA074778 (M.A.J.) and U01/U24 CA097735 (M.A.J.); and the Victorian Cancer Registry.
Disclosures: D.D.B. received fees in 2017 and 2018 as a consultant and member of the Tumor Agnostic (dMMR) Advisory Board at Merck Sharp and Dohme for pembrolizumab.
The content of this article does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Colon Cancer Family Registry (Colon-CFR). Any mention of trade names, commercial products, or organizations do not imply endorsement by the US Government or the Colon-CFR.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.12.003.
Author Contributions
D.D.B., M.C., and C.R. conceived and designed the study; M.C., J.E.J., R.W., and J.C. curated samples and performed laboratory testing; S.P. B.J.P., K.M., and P.G. implemented the bioinformatics analysis pipeline software; B.J.P., K.M., M.C., and D.D.B. prepared the manuscript; R.A.H., S.J., A.K.W., A.B.S., F.A.M., I.M.W., J.L.H., and M.A.J. acquired the study data; all authors revised the manuscript for important intellectual content, and read and approved the final manuscript.
Supplemental Data
A: DNA sequencing read alignments of the 1928-bp deletion of MSH2 exon 6 (chr2:47640695-47642623) in control sample 2. B and C: DNA sequencing read alignments for the two break ends of the 9.5-Mb inversion encompassing exons 1 to 7 of MSH2 (chr2:38121107-chr2:47669532) in the mother–daughter pair of SLS11 and SLS12. Red bars indicate discordant read pairs and illustrate that these large structural variants are readily detected in this type of sequencing data. D: Diagram of the position of the 9.5-Mb inversion encompassing exons 1 to 7 of MSH2 on chromosome 2.
Sequencing trace confirming that the inversion carried by SLS12 is the same as that in the positive control. Break points are marked with lines. The sequence between the lines is a novel inserted sequence.
A–E: Plots depicting the presence or the absence of loss of heterozygosity (LOH) in each tumor across the first 90 million bases (MB) of the p-arm of chromosome 3 (A), which encompasses the MLH1 gene; and the p-arm of chromosome 2 (B–E), which encompasses the EPCAM, MSH2, and MSH6 genes. Green bars indicate regions where the variant allele fraction of germline variants within tumor samples deviates from the germline state and thus provides strong evidence of LOH. Tumor sample T5 shows loss of MLH1 via IHC (Table 2) and demonstrates significant regions of LOH surrounding the MLH1 gene. Tumor sample T9A shows loss of MSH6 via IHC (Table 2), and tumor samples T9B, T10, and T13 show loss of MSH2 via IHC (Table 2); however, none of these tumors demonstrates evidence of LOH in the regions surrounding these genes.
References
- 1.Ligtenberg M.J., Kuiper R.P., Chan T.L., Goossens M., Hebeda K.M., Voorendt M., Lee T.Y., Bodmer D., Hoenselaar E., Hendriks-Cornelissen S.J., Tsui W.Y., Kong C.K., Brunner H.G., van Kessel A.G., Yuen S.T., van Krieken J.H., Leung S.Y., Hoogerbrugge N. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 2.Win A.K., Young J.P., Lindor N.M., Tucker K.M., Ahnen D.J., Young G.P., Buchanan D.D., Clendenning M., Giles G.G., Winship I., Macrae F.A., Goldblatt J., Southey M.C., Arnold J., Thibodeau S.N., Gunawardena S.R., Bapat B., Baron J.A., Casey G., Gallinger S., Le Marchand L., Newcomb P.A., Haile R.W., Hopper J.L., Jenkins M.A. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol. 2012;30:958–964. doi: 10.1200/JCO.2011.39.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thibodeau S.N., Bren G., Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan D.D., Rosty C., Clendenning M., Spurdle A.B., Win A.K. Clinical problems of colorectal cancer and endometrial cancer cases with unknown cause of tumor mismatch repair deficiency (suspected Lynch syndrome) Appl Clin Genet. 2014;7:183–193. doi: 10.2147/TACG.S48625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klarskov L., Ladelund S., Holck S., Roenlund K., Lindebjerg J., Elebro J., Halvarsson B., von Salome J., Bernstein I., Nilbert M. Interobserver variability in the evaluation of mismatch repair protein immunostaining. Hum Pathol. 2010;41:1387–1396. doi: 10.1016/j.humpath.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Markow M., Chen W., Frankel W.L. Immunohistochemical pitfalls: common mistakes in the evaluation of Lynch syndrome. Surg Pathol Clin. 2017;10:977–1007. doi: 10.1016/j.path.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Boland C.R. The mystery of mismatch repair deficiency: lynch or lynch-like? Gastroenterology. 2013;144:868–870. doi: 10.1053/j.gastro.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clendenning M., Buchanan D.D., Walsh M.D., Nagler B., Rosty C., Thompson B., Spurdle A.B., Hopper J.L., Jenkins M.A., Young J.P. Mutation deep within an intron of MSH2 causes Lynch syndrome. Fam Cancer. 2011;10:297–301. doi: 10.1007/s10689-011-9427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morak M., Heidenreich B., Keller G., Hampel H., Laner A., de la Chapelle A., Holinski-Feder E. Biallelic MUTYH mutations can mimic Lynch syndrome. Eur J Hum Genet. 2014;22:1334–1337. doi: 10.1038/ejhg.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsayed F.A., Kets C.M., Ruano D., van den Akker B., Mensenkamp A.R., Schrumpf M., Nielsen M., Wijnen J.T., Tops C.M., Ligtenberg M.J., Vasen H.F., Hes F.J., Morreau H., van Wezel T. Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur J Hum Genet. 2015;23:1080–1084. doi: 10.1038/ejhg.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sourrouille I., Coulet F., Lefevre J.H., Colas C., Eyries M., Svrcek M., Bardier-Dupas A., Parc Y., Soubrier F. Somatic mosaicism and double somatic hits can lead to MSI colorectal tumors. Fam Cancer. 2013;12:27–33. doi: 10.1007/s10689-012-9568-9. [DOI] [PubMed] [Google Scholar]
- 12.Mensenkamp A.R., Vogelaar I.P., van Zelst-Stams W.A., Goossens M., Ouchene H., Hendriks-Cornelissen S.J., Kwint M.P., Hoogerbrugge N., Nagtegaal I.D., Ligtenberg M.J. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014;146:643–646.e648. doi: 10.1053/j.gastro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Haraldsdottir S., Hampel H., Tomsic J., Frankel W.L., Pearlman R., de la Chapelle A., Pritchard C.C. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014;147:1308–1316.e1301. doi: 10.1053/j.gastro.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearlman R., Haraldsdottir S., de la Chapelle A., Jonasson J.G., Liyanarachchi S., Frankel W.L., Rafnar T., Stefansson K., Pritchard C.C., Hampel H. Clinical characteristics of patients with colorectal cancer with double somatic mismatch repair mutations compared with Lynch syndrome. J Med Genet. 2019;56:462–470. doi: 10.1136/jmedgenet-2018-105698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcomb P.A., Baron J., Cotterchio M., Gallinger S., Grove J., Haile R., Hall D., Hopper J.L., Jass J., Le Marchand L., Limburg P., Lindor N., Potter J.D., Templeton A.S., Thibodeau S., Seminara D. Colon cancer family registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins M.A., Win A.K., Templeton A.S., Angelakos M.S., Buchanan D.D., Cotterchio M., Figueiredo J.C., Thibodeau S.N., Baron J.A., Potter J.D., Hopper J.L., Casey G., Gallinger S., Le Marchand L., Lindor N.M., Newcomb P.A., Haile R.W., Colon Cancer Family Registry Cohort Investigators Cohort profile: the colon cancer family registry cohort (CCFRC) Int J Epidemiol. 2018;47:387–388i. doi: 10.1093/ije/dyy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchanan D.D., Tan Y.Y., Walsh M.D., Clendenning M., Metcalf A.M., Ferguson K., Arnold S.T., Thompson B.A., Lose F.A., Parsons M.T., Walters R.J., Pearson S.A., Cummings M., Oehler M.K., Blomfield P.B., Quinn M.A., Kirk J.A., Stewart C.J., Obermair A., Young J.P., Webb P.M., Spurdle A.B. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32:90–100. doi: 10.1200/JCO.2013.51.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasen H.F., Watson P., Mecklin J.P., Lynch H.T. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 19.Umar A., Boland C.R., Terdiman J.P., Syngal S., de la Chapelle A., Ruschoff J., Fishel R., Lindor N.M., Burgart L.J., Hamelin R., Hamilton S.R., Hiatt R.A., Jass J., Lindblom A., Lynch H.T., Peltomaki P., Ramsey S.D., Rodriguez-Bigas M.A., Vasen H.F., Hawk E.T., Barrett J.C., Freedman A.N., Srivastava S. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchanan D.D., Clendenning M., Rosty C., Eriksen S.V., Walsh M.D., Walters R.J., Thibodeau S.N., Stewart J., Preston S., Win A.K., Flander L., Ouakrim D.A., Macrae F.A., Boussioutas A., Winship I.M., Giles G.G., Hopper J.L., Southey M.C., English D., Jenkins M.A. Tumor testing to identify lynch syndrome in two Australian colorectal cancer cohorts. J Gastroenterol Hepatol. 2017;32:427–438. doi: 10.1111/jgh.13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindor N.M., Burgart L.J., Leontovich O., Goldberg R.M., Cunningham J.M., Sargent D.J., Walsh-Vockley C., Petersen G.M., Walsh M.D., Leggett B.A., Young J.P., Barker M.A., Jass J.R., Hopper J., Gallinger S., Bapat B., Redston M., Thibodeau S.N. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 22.Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Koh H., Simms L., Barker M., Leggett B., Levine J., Kim M., French A.J., Thibodeau S.N., Jass J., Haile R., Laird P.W. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 23.Buchanan D.D., Sweet K., Drini M., Jenkins M.A., Win A.K., English D.R. Risk factors for colorectal cancer in patients with multiple serrated polyps: a cross-sectional case series from genetics clinics. PLoS One. 2010;5:e11636. doi: 10.1371/journal.pone.0011636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhees J., Arnold M., Boland C.R. Inversion of exons 1-7 of the MSH2 gene is a frequent cause of unexplained Lynch syndrome in one local population. Fam Cancer. 2013;13:219–225. doi: 10.1007/s10689-013-9688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poynter J.N., Siegmund K.D., Weisenberger D.J., Long T.I., Thibodeau S.N., Lindor N., Young J., Jenkins M.A., Hopper J.L., Baron J.A., Buchanan D., Casey G., Levine A.J., Le Marchand L., Gallinger S., Bapat B., Potter J.D., Newcomb P.A., Haile R.W., Laird P.W. Colon cancer family registry I: molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh M.D., Buchanan D.D., Cummings M.C., Pearson S.A., Arnold S.T., Clendenning M., Walters R., McKeone D.M., Spurdle A.B., Hopper J.L., Jenkins M.A., Phillips K.D., Suthers G.K., George J., Goldblatt J., Muir A., Tucker K., Pelzer E., Gattas M.R., Woodall S., Parry S., Macrae F.A., Haile R.W., Baron J.A., Potter J.D., Le Marchand L., Bapat B., Thibodeau S.N., Lindor N.M., McGuckin M.A., Young J.P. Lynch syndrome-associated breast cancers: clinicopathologic characteristics of a case series from the colon cancer family registry. Clin Cancer Res. 2010;16:2214–2224. doi: 10.1158/1078-0432.CCR-09-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clendenning M., Walsh M.D., Gelpi J.B., Thibodeau S.N., Lindor N., Potter J.D., Newcomb P., LeMarchand L., Haile R., Gallinger S., Colorectal Cancer Family R., Hopper J.L., Jenkins M.A., Rosty C., Young J.P., Buchanan D.D. Detection of large scale 3' deletions in the PMS2 gene amongst colon-CFR participants: have we been missing anything? Fam Cancer. 2013;12:563–566. doi: 10.1007/s10689-012-9597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews S. 1st ed. 2012. FastQC: A Quality Control Tool for High Throughput Sequence Data. 0.10. [Google Scholar]
- 29.Genome Reference Consortium . 2009. hg19, GRCh37 Genome Reference Consortium Human Reference 37(GCA_000001405.1) [Google Scholar]
- 30.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11 10 11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saunders C.T., Wong W.S., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 33.Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S.R.F., Consortium W.G.S., Wilkie A.O.M., McVean G., Lunter G. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson B.A., Spurdle A.B., Plazzer J.P., Greenblatt M.S., Akagi K., Al-Mulla F. InSiGht: application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., Karapetyan K., Katz K., Liu C., Maddipatla Z., Malheiro A., McDaniel K., Ovetsky M., Riley G., Zhou G., Holmes J.B., Kattman B.L., Maglott D.R. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rausch T., Zichner T., Schlattl A., Stutz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cameron D.L., Schroder J., Penington J.S., Do H., Molania R., Dobrovic A., Speed T.P., Papenfuss A.T. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017;27:2050–2060. doi: 10.1101/gr.222109.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Kallberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 46.Ha G., Roth A., Lai D., Bashashati A., Ding J., Goya R., Giuliany R., Rosner J., Oloumi A., Shumansky K., Chin S.F., Turashvili G., Hirst M., Caldas C., Marra M.A., Aparicio S., Shah S.P. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012;22:1995–2007. doi: 10.1101/gr.137570.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desmet F.O., Hamroun D., Lalande M., Collod-Beroud G., Claustres M., Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 49.Niu B., Ye K., Zhang Q., Lu C., Xie M., McLellan M.D., Wendl M.C., Ding L. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Australian Pancreatic Cancer Genome Initiative, ICGC Breast Cancer Consortium, ICGC MMML-Seq Consortium, ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal R., McGranahan N., Herrero J., Taylor B.S., Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell B.B., Light N., Fabrizio D., Zatzman M., Fuligni F., de Borja R. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–1056.e1010. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georgeson P., Walsh M.D., Clendenning M., Daneshvar S., Pope B.J., Mahmood K., Joo J.E., Jayasekara H., Jenkins M.A., Winship I.M., Buchanan D.D. Tumour mutational signature in sebaceous skin lesions from individuals with Lynch syndrome. Mol Genet Genomic Med. 2019;7:e00781. doi: 10.1002/mgg3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M., Lancet D., Cohen D. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:bax028. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seifert B.A., McGlaughon J.L., Jackson S.A., Ritter D.I., Roberts M.E., Schmidt R.J., Thompson B.A., Jimenez S., Trapp M., Lee K., Plon S.E., Offit K., Stadler Z.K., Zhang L., Greenblatt M.S., Ferber M.J. Determining the clinical validity of hereditary colorectal cancer and polyposis susceptibility genes using the clinical genome resource clinical validity framework. Genet Med. 2019;21:1507–1516. doi: 10.1038/s41436-018-0373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adam R., Spier I., Zhao B., Kloth M., Marquez J., Hinrichsen I., Kirfel J., Tafazzoli A., Horpaopan S., Uhlhaas S., Stienen D., Friedrichs N., Altmuller J., Laner A., Holzapfel S., Peters S., Kayser K., Thiele H., Holinski-Feder E., Marra G., Kristiansen G., Nothen M.M., Buttner R., Moslein G., Betz R.C., Brieger A., Lifton R.P., Aretz S. Exome sequencing identifies biallelic MSH3 germline mutations as a recessive subtype of colorectal adenomatous polyposis. Am J Hum Genet. 2016;99:337–351. doi: 10.1016/j.ajhg.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weren R.D., Ligtenberg M.J., Kets C.M., de Voer R.M., Verwiel E.T., Spruijt L., van Zelst-Stams W.A., Jongmans M.C., Gilissen C., Hehir-Kwa J.Y., Hoischen A., Shendure J., Boyle E.A., Kamping E.J., Nagtegaal I.D., Tops B.B., Nagengast F.M., Geurts van Kessel A., van Krieken J.H., Kuiper R.P., Hoogerbrugge N. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 58.Grolleman J.E., de Voer R.M., Elsayed F.A., Nielsen M., Weren R.D.A., Palles C. Mutational signature analysis reveals NTHL1 deficiency to cause a multi-tumor phenotype. Cancer Cell. 2019;35:256–266.e255. doi: 10.1016/j.ccell.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 59.Knijnenburg T.A., Wang L., Zimmermann M.T., Chambwe N., Gao G.F., Cherniack A.D. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018;23:239–254 e236. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood R.D., Mitchell M., Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Vargas-Parra G.M., Gonzalez-Acosta M., Thompson B.A., Gomez C., Fernandez A., Damaso E., Pons T., Morak M., Del Valle J., Iglesias S., Velasco A., Solanes A., Sanjuan X., Padilla N., de la Cruz X., Valencia A., Holinski-Feder E., Brunet J., Feliubadalo L., Lazaro C., Navarro M., Pineda M., Capella G. Elucidating the molecular basis of MSH2-deficient tumors by combined germline and somatic analysis. Int J Cancer. 2017;141:1365–1380. doi: 10.1002/ijc.30820. [DOI] [PubMed] [Google Scholar]
- 62.Salvador M.U., Truelson M.R.F., Mason C., Souders B., LaDuca H., Dougall B., Black M.H., Fulk K., Profato J., Gutierrez S., Jasperson K., Tippin-Davis B., Lu H.M., Gray P., Shah S., Chao E.C., Ghahramani N., Landsverk M., Gau C.L., Chen D., Pronold M. Comprehensive paired tumor/germline testing for Lynch syndrome: bringing resolution to the diagnostic process. J Clin Oncol. 2019;37:647–657. doi: 10.1200/JCO.18.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jansen A.M., Geilenkirchen M.A., van Wezel T., Jagmohan-Changur S.C., Ruano D., van der Klift H.M., van den Akker B.E., Laros J.F., van Galen M., Wagner A., Letteboer T.G., Gomez-Garcia E.B., Tops C.M., Vasen H.F., Devilee P., Hes F.J., Morreau H., Wijnen J.T. Whole gene capture analysis of 15 CRC susceptibility genes in suspected Lynch syndrome patients. PLoS One. 2016;11:e0157381. doi: 10.1371/journal.pone.0157381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geurts-Giele W.R., Leenen C.H., Dubbink H.J., Meijssen I.C., Post E., Sleddens H.F., Kuipers E.J., Goverde A., van den Ouweland A.M., van Lier M.G., Steyerberg E.W., van Leerdam M.E., Wagner A., Dinjens W.N. Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J Pathol. 2014;234:548–559. doi: 10.1002/path.4419. [DOI] [PubMed] [Google Scholar]
- 65.Wagner A., van der Klift H., Franken P., Wijnen J., Breukel C., Bezrookove V., Smits R., Kinarsky Y., Barrows A., Franklin B., Lynch J., Lynch H., Fodde R. A 10-Mb paracentric inversion of chromosome arm 2p inactivates MSH2 and is responsible for hereditary nonpolyposis colorectal cancer in a North-American kindred. Genes Chromosomes Cancer. 2002;35:49–57. doi: 10.1002/gcc.10094. [DOI] [PubMed] [Google Scholar]
- 66.Mork M.E., Rodriguez A., Taggart M.W., Rodriguez-Bigas M.A., Lynch P.M., Bannon S.A., You Y.N., Vilar E. Identification of MSH2 inversion of exons 1-7 in clinical evaluation of families with suspected Lynch syndrome. Fam Cancer. 2017;16:357–361. doi: 10.1007/s10689-016-9960-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shia J., Zhang L., Shike M., Guo M., Stadler Z., Xiong X., Tang L.H., Vakiani E., Katabi N., Wang H., Bacares R., Ruggeri J., Boland C.R., Ladanyi M., Klimstra D.S. Secondary mutation in a coding mononucleotide tract in MSH6 causes loss of immunoexpression of MSH6 in colorectal carcinomas with MLH1/PMS2 deficiency. Mod Pathol. 2013;26:131–138. doi: 10.1038/modpathol.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haradhvala N.J., Kim J., Maruvka Y.E., Polak P., Rosebrock D., Livitz D., Hess J.M., Leshchiner I., Kamburov A., Mouw K.W., Lawrence M.S., Getz G. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun. 2018;9:1746. doi: 10.1038/s41467-018-04002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castillejo A., Vargas G., Castillejo M.I., Navarro M., Barbera V.M., Gonzalez S., Hernandez-Illan E., Brunet J., Ramon y C.T., Balmana J., Oltra S., Iglesias S., Velasco A., Solanes A., Campos O., Sanchez Heras A.B., Gallego J., Carrasco E., Gonzalez Juan D., Segura A., Chirivella I., Juan M.J., Tena I., Lazaro C., Blanco I., Pineda M., Capella G., Soto J.L. Prevalence of germline MUTYH mutations among Lynch-like syndrome patients. Eur J Cancer. 2014;50:2241–2250. doi: 10.1016/j.ejca.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 70.Jansen A.M., van Wezel T., van den Akker B.E., Ventayol Garcia M., Ruano D., Tops C.M., Wagner A., Letteboer T.G., Gomez-Garcia E.B., Devilee P., Wijnen J.T., Hes F.J., Morreau H. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet. 2016;24:1089–1092. doi: 10.1038/ejhg.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xavier A., Olsen M.F., Lavik L.A., Johansen J., Singh A.K., Sjursen W., Scott R.J., Talseth-Palmer B.A. Comprehensive mismatch repair gene panel identifies variants in patients with Lynch-like syndrome. Mol Genet Genomic Med. 2019;7:e850. doi: 10.1002/mgg3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keijzers G., Liu D., Rasmussen L.J. Exonuclease 1 and its versatile roles in DNA repair. Crit Rev Biochem Mol Biol. 2016;51:440–451. doi: 10.1080/10409238.2016.1215407. [DOI] [PubMed] [Google Scholar]
- 73.Mu W., Lu H.M., Chen J., Li S., Elliott A.M. Sanger confirmation is required to achieve optimal sensitivity and specificity in next-generation sequencing panel testing. J Mol Diagn. 2016;18:923–932. doi: 10.1016/j.jmoldx.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 74.Liu Q., Hesson L.B., Nunez A.C., Packham D., Williams R., Ward R.L., Sloane M.A. A cryptic paracentric inversion of MSH2 exons 2-6 causes Lynch syndrome. Carcinogenesis. 2016;37:10–17. doi: 10.1093/carcin/bgv154. [DOI] [PubMed] [Google Scholar]
- 75.Morak M., Koehler U., Schackert H.K., Steinke V., Royer-Pokora B., Schulmann K., Kloor M., Hochter W., Weingart J., Keiling C., Massdorf T., Holinski-Feder E., German HNPCC consortium Biallelic MLH1 SNP cDNA expression or constitutional promoter methylation can hide genomic rearrangements causing Lynch syndrome. J Med Genet. 2011;48:513–519. doi: 10.1136/jmedgenet-2011-100050. [DOI] [PubMed] [Google Scholar]
- 76.Shirts B.H., Konnick E.Q., Upham S., Walsh T., Ranola J.M.O., Jacobson A.L., King M.C., Pearlman R., Hampel H., Pritchard C.C. Using somatic mutations from tumors to classify variants in mismatch repair genes. Am J Hum Genet. 2018;103:19–29. doi: 10.1016/j.ajhg.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., Nikiforova M.N. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X., Meyerson M. Illuminating the noncoding genome in cancer. Nat Cancer. 2020;1:864–872. doi: 10.1038/s43018-020-00114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valeri N., Gasparini P., Fabbri M., Braconi C., Veronese A., Lovat F., Adair B., Vannini I., Fanini F., Bottoni A., Costinean S., Sandhu S.K., Nuovo G.J., Alder H., Gafa R., Calore F., Ferracin M., Lanza G., Volinia S., Negrini M., McIlhatton M.A., Amadori D., Fishel R., Croce C.M. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci U S A. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pastrello C., Fornasarig M., Pin E., Berto E., Pivetta B., Viel A. Somatic mosaicism in a patient with Lynch syndrome. Am J Med Genet A. 2009;149A:212–215. doi: 10.1002/ajmg.a.32620. [DOI] [PubMed] [Google Scholar]
- 81.Geurts-Giele W.R., Rosenberg E.H., Rens A.V., Leerdam M.E.V., Dinjens W.N., Bleeker F.E. Somatic mosaicism by a de novo MLH1 mutation as a cause of Lynch syndrome. Mol Genet Genomic Med. 2019;7:e00699. doi: 10.1002/mgg3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hampel H., Pearlman R., Beightol M., Zhao W., Jones D., Frankel W.L., Goodfellow P.J., Yilmaz A., Miller K., Bacher J., Jacobson A., Paskett E., Shields P.G., Goldberg R.M., de la Chapelle A., Shirts B.H., Pritchard C.C. Assessment of tumor sequencing as a replacement for Lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol. 2018;4:806–813. doi: 10.1001/jamaoncol.2018.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: DNA sequencing read alignments of the 1928-bp deletion of MSH2 exon 6 (chr2:47640695-47642623) in control sample 2. B and C: DNA sequencing read alignments for the two break ends of the 9.5-Mb inversion encompassing exons 1 to 7 of MSH2 (chr2:38121107-chr2:47669532) in the mother–daughter pair of SLS11 and SLS12. Red bars indicate discordant read pairs and illustrate that these large structural variants are readily detected in this type of sequencing data. D: Diagram of the position of the 9.5-Mb inversion encompassing exons 1 to 7 of MSH2 on chromosome 2.
Sequencing trace confirming that the inversion carried by SLS12 is the same as that in the positive control. Break points are marked with lines. The sequence between the lines is a novel inserted sequence.
A–E: Plots depicting the presence or the absence of loss of heterozygosity (LOH) in each tumor across the first 90 million bases (MB) of the p-arm of chromosome 3 (A), which encompasses the MLH1 gene; and the p-arm of chromosome 2 (B–E), which encompasses the EPCAM, MSH2, and MSH6 genes. Green bars indicate regions where the variant allele fraction of germline variants within tumor samples deviates from the germline state and thus provides strong evidence of LOH. Tumor sample T5 shows loss of MLH1 via IHC (Table 2) and demonstrates significant regions of LOH surrounding the MLH1 gene. Tumor sample T9A shows loss of MSH6 via IHC (Table 2), and tumor samples T9B, T10, and T13 show loss of MSH2 via IHC (Table 2); however, none of these tumors demonstrates evidence of LOH in the regions surrounding these genes.