Abstract
The ongoing outbreak of the coronavirus infection has killed more than 2 million people. Herein, we demonstrate Rhodamine 6G (Rh-6G) dye conjugated DNA aptamer attached gold nanostar (GNS) based distance dependent nanoparticle surface energy transfer (NSET) spectroscopy has capability for rapid diagnosis of specific SARS-CoV-2 spike recombinant antigen or SARS-CoV-2 spike protein pseudotyped baculovirus within 10 minutes time. Since Rh-6G attached single stand DNA aptamer wrapped GNS, 99% dye fluorescence was quenched due to the NSET process. In the presence of spike antigen or virus, fluorescence signal persists due to the aptamer-spike protein binding. Specifically, the limit of detection for NSET assay has been determined to be 130 femto gram/mL for antigen and 8 particles/mL for virus. Finally, we have demonstrated that DNA aptamer attached GNS can stop virus infection by blocking the angiotensin-converting enzyme 2 (ACE2) receptor binding capability and destroying the lipid membrane of the virus.
Keywords: COVID-19 antigen detection, SARS-CoV-2 identification and inactivation, Spike protein specific aptamer attached gold nanostar, Nanoparticle surface energy transfer
Graphical Abstract
The respiratory syndrome coronavirus-2 (SARS-CoV-2) epidemic has spread worldwide very rapidly, and it threatens the world economy, health and social life1-10. As per the world health organization (WHO), more than 106 million people around the world have suffered by coronavirus disease of 2019 (COVID-19) and around 2.3 million have died worldwide1-5. For any disease, the immediate requirement is the fast and effective diagnostic of the virus, which is the key to prevent infection for our society5-15. In the current letter, we report that spike protein specific aptamer attached gold nanostar (GNS) can be used for rapid diagnosis of specific SARS-CoV-2 spike recombinant antigen or virus itself via distance dependent nanoparticle surface energy transfer (NSET) process 16-22.
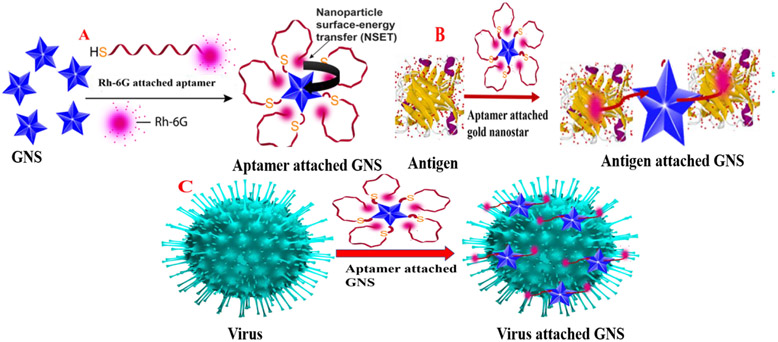
Since gold nanoparticle exhibit 9–10 orders of magnitude higher quenching efficiency than typical small molecule dye–quencher pairs16-24, nanoscopic surface energy transfer (NSET) spectroscopy using GNS has capability to be used as biophysical tools beyond Förster resonance energy transfer (FRET) 16-22. It is now well documented that since gold nanostars exhibit very high extinction coefficient, single GNS based optical nano-probe are comparable with the optical organic probe made with more than 106 dye molecules together16-27. Recent published data indicate that the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein (S protein) is responsible for virus entry and disease pathogenesis3-15. Therefore, we have used spike protein specific aptamer attached GNS for specific diagnosis and effective inhibition of the virus. As shown in the Figure 1A, in the absence of spike protein, Rh-6G attached single stand (ss) DNA aptamer wrapped gold nanoparticle. Due to the above fact, fluorescence signal from Rh-6G dye is quenched by GNS via NSET process. In contrast, as shown in Figures 1B and 1C, when SARS-CoV-2 antigen or virus particles are added, due to the aptamer-spike protein binding, the distance between GNS and dye increases and as a result, fluorescence signal persists. We have used the observed NSET signal change in the absence or presence of antigen or virus for the detection purpose. Reported data show that spike protein specific aptamer attached GNS based NSET can be used for the diagnosis of COVID-19 spike antigen at 130 femto gram (fg)/mL concentration level and virus at 8 particles/mL level.
Figure 1:
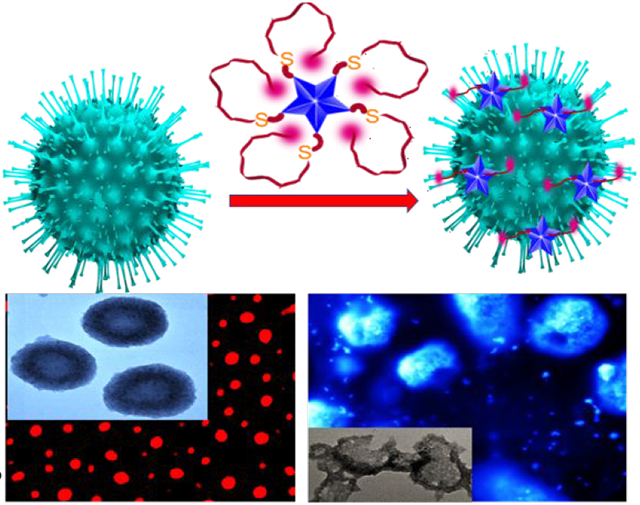
A) Schematic representation of the design criteria for Rh-6G conjugated DNA aptamer attached gold nano star (GNS) based NSET. Due to the wrapping of ss-DNA on GNS, the NSET occurs between GNA and Rh-6G. B) Scheme shows that spike protein specific ss DNA aptamer binds with COVID-19 antigen, which allows DNA to be unwrapped from GNS. C) Scheme shows that spike protein specific DNA binds with SARS-CoV-2 spike protein pseudotyped baculovirus, which allows ss DNA to be unwrapped from GNS.
For the detection and inactivation, SARS-CoV-2 spike protein pseudotyped baculovirus has been used27-28. Since the S protein of virus and host angiotensin-converting enzyme 2 (ACE2) binding is the key for the infection spread, we have used aptamer attached GNS as an inhibitor for blocking virus spread8-15,27-30. In the current letter, we demonstrate that the spike protein specific DNA aptamer attached GNS (without Rh-6G) can be used to block the viral entry into cells. For this purpose, we have used ACE2 expressing HEK293T cell line 27-28. Reported scanning and transmission electron microscope and fluorescence microscopy data show that spike protein specific DNA aptamer attached GNS can be used to stop the spread of the virus.
For the design of Rh-6G conjugated ss-DNA aptamer attached GNS, which is specific for binding with spike protein, we have used 5′-ATCCAGAGTGACGCAGCATTTCATCGGGTCCAAAAGGGGCTGCTCGGGATTGCGGATATGGACACGT -3′, DNA aptamer which has been reported recently to be specific for the SARS-CoV-2 spike glycoprotein 12. As shown in the Figure 1A, ss-DNA (HS–(CH2)6–3′-oligo-5′-Rh-6G) was attached to GNS via thiol-gold chemistry, as we and others have reported before 16-22. For this purpose, initially we have synthesized GNS using silver assisted seedless growth method in the presence of HEPES buffer 23-25. We have reported the synthesis details in the supporting information. After that, GNSs were characterized by UV-Vis spectroscopy and TEM as reported in Figures S1A-B in the supporting information.
TEM data show that for GNS particle the spherical core diameter is around 15 ± 3 and branch length is around 12 ± 4 nm. The extinction spectra show that freshly prepared GNS exhibit strong plasm on band with λmax ~560 nm 23. In the next step, ssDNA (HS–(CH2)6–Oligo-Rh-6G) aptamer was attached to GNS via thiol-gold chemistry. We have reported the synthesis details in the supporting information. Next, to determine how many aptamers are bound to GNS, we have separated aptamer from GNS by dissolving nanostar in 10 μM potassium cyanide. In the next step, from fluorescence recovery intensity measurement, we have found out that around 60-80 aptamers are attached to each GNS.
As reported in Figure S1C in the supporting information and Figure 2A, the fluorescence signal from Rh-6G attached aptamer is totally quenched when it has been attached to GNS. As shown in Figure 1A, when a Rh-6G conjugated ss DNA aptamer is attached to GNS surface via -SH linkage, 5’-Rh-6G ends of -ss DNA loop back onto the GNS surface16-22. In this condition, strong NSET 16-22 occurs between dye donor and GNS acceptor, which provided 99% of quenching efficiency, as reported in Figure 2A. As we and others have reported before, when Rh-6G is placed at a short distance on GNS, which possesses a strong plasmon field, the electrons of Rh-6G participating in the excitation/emission process will interact with the field16-22. As a result, we have observed strong NSET and it is because each GNS has capability to be used as an excellent quencher, which is equivalent to several million dye molecules16-22. To understand whether in the Rh-6G conjugated ss (single strand) DNA aptamer GNS surface, 5’-Rh-6G ends of -ss DNA can loopback onto GNS, we have added complimentary DNA which will form double helix structure.
Figure 2:
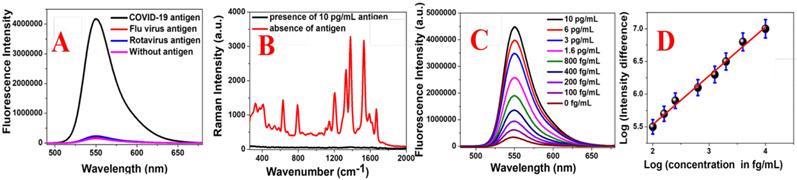
A) Fluorescence spectra from Rh-6G conjugated spike protein specific DNA aptamer attached GNS in the absence of antigen and presence of SARS-CoV-2 spike recombinant antigen (10 pg/mL), flu virus antigen (100 pg/mL) and rotavirus antigen (100 pg/mL). The ratio of DNA aptamer and COVID-19 antigen was kept as 1: 1. B) Figure shows how SERS intensity from Rh-6G conjugated DNA aptamer attached GNS varies with the addition of antigen. The ratio of DNA aptamer and COVID-19 antigen was kept as 1: 1. C) Fluorescence spectra from Rh-6G conjugated spike protein specific DNA aptamer attached GNS in the presence of SARS-CoV-2 spike recombinant antigen (10 pg/mL), flu virus antigen (100 pg/mL) and rotavirus antigen (100 pg/mL). C) Plot indicates the variation of log of (NSET intensity change due to the addition of spike recombinant antigen) with the log of (amount of spike recombinant antigen).
Since -ds (double stand) DNA is not conformationally flexible for bending, 5’-Rh-6G ends of -ds DNA cannot loop back onto GNS. Due to the above fact, the distance between GNS and Rh-6G dye increases. As a result, we have observed high fluorescence after the formation of -ds DNA, as reported in Figure S1E in the supporting information. The above experimental data is a clear evidence that in -ss DNA aptamer, the rhodamine 6G at 3’ end can loop back onto the GNS surface.
To understand better, whether 5’-Rh-6G ends of -ss DNA aptamers are adsorbed onto the GNS surface, surface enhanced Raman intensity (SERS) enhancement experiment has been explored23-25. As reported in Figure 2B, excellent SERS intensity enhancement has been observed. The observed Raman mode from Rh-6G attached -ss DNA aptamers is N–C–C bending mode of the ethylamine group at 376 cm−1, as we and others reported before23-25. Similarly, we have also observed Raman modes due to the bending of C–C–C ring at 615 cm−1 and C–H out-of-plane at 778 cm−1, as we and others reported before23-25. From the observed Raman modes, we have estimated that the Raman enhancement factor is ~3.68 × 107, which clearly indicates that 5’-Rh-6G ends of -ss DNA aptamers are adsorbed onto the GNS surface.
As shown in Figure 1B, due to the antigen-aptamer binding, the distance between GNS and Rh-6G dye increases when coronavirus antigens are added to spike protein specific -ss DNA conjugated GNS. As a result, fluorescence signal persists as reported in Figure S1D in the supporting information and as Figure 2C. The observed NSET intensity change in the presence of specific antigen has been used for the detection purpose. It is now well documented that aptamers are capable of binding with antigen protein via non-covalent interaction such as hydrogen bonding, van der Waals forces and other interactions 11-12. Recently reported molecular dynamics simulations with experimental observation study indicates that that our aptamers bind to several amino acid residues of receptor-binding domain of the of the SARS-CoV-2 S protein12.
Reported simulation study indicates that cytosine base at 53 position, guanine base at 54 position, and adenine base at 66 position of the aptamer form a network of hydrogen bonds with glutamine, lysine, and tyrosine amino acids from receptor-binding domain of the of the SARS-CoV-2 S protein12. Similarly, the thymine base at 41 position of the aptamer forms a network of hydrogen bonds with threonine from receptor-binding domain of the of the SARS-CoV-2 S protein12. Due these antigen-aptamer interactions, the aptamer became straight and the distance between the GNS and the Rh-6G dye increased.
To understand better, whether 5’-Rh-6G ends of DNA aptamers are far from the GNS surface when specific antigen has been added, we have also performed SERS experiment23-25. As reported in Figure 2B, in the presence of spike antigen, we have not observed any Raman signal from Rh-6G conjugated -ss DNA aptamer attached GNS. The observed SERS data clearly indicate that Rh-6G in -ss DNA aptamer is not adsorbed onto GNS surface after aptamer binds with spike antigen and the distance between Rh-6G and GNS is several nanometers. As a result, we have not observed any SERS signal.
In last few months, FDA has authorized antigen tests for COVID-19 infection 2. Our experimental data demonstrated that distance dependent NSET can be used to detect the same virus antigens within 10 minutes of experimental time. For finding the limit of detection (LOD) of the reported distance dependent NSET assay, we have determined the NSET intensity change in the presence of different amounts of spike antigen. As shown in Figure 2C, distance dependent NSET assay can recognize COVID-19 specific antigen even at the concentration of 100 fg/mL. As reported in Figure 2D, the log of NSET intensity difference (Intensity after and before COVID-19 spike antigen) varies linearly with the log of the concentration of COVID-19 spike antigen. We have used the Equation 1 for finding the LOD for the distance dependent NSET assay 16-25.
| (1) |
In our experiment the standard deviation of the blank () has been measured in the absence of antigen. The slope of the calibration curve (S) has been calculated from linear curve reported in Figure 2D. LOD for the distance dependent NSET assay has been determined to be ~ 130 fg/mL for spike antigen.
Selectivity of the 5’-Rh-6G conjugated DNA attached GNS based distance dependent NSET assay has been demonstrated using flu virus antigen and rotavirus antigen separately. As shown in Figure 2A, we have not observed any distance dependent NSET intensity variation for flu or rotavirus antigen, although we have used 100 pg/mL antigen. On the other hand, excellent distance dependent NSET intensity variation can be noted when only 10 pg/mL spike antigen has been added. The above experimental data clearly indicate that NSET assay can be used for specific recognition of SARS-CoV-2 spike recombinant antigen.
As shown in Figure 1C, spike protein specific DNA conjugated GNS based NSET diagnosis of virus is based on binding affinity of virus spike protein and aptamer. As reported in Figures 3B-3D and S2C in the supporting information, due to the above interaction, the distance dependent NSET intensity enhances abruptly. Reported distance dependence NSET intensity variation has been used for the detection of the virus. Since the size of virus (120-160 nm) is much higher than GNS, electron microscopy images reported in Figures 3B and S2A-2B in the supporting information show that several aptamers attached GNS are bound on the virus. To understand better, whether 5’-Rh-6G ends of -ss DNA aptamers are far from the GNS surface in the presence of virus, we have measured Raman signal with or without virus addition. As reported in Figure S2D in the supporting information, no Raman signal has been observed from Rh-6G conjugated -ss DNA aptamer attached GNS, when viruses are present. The observed SERS data clearly indicate that Rh-6G in -ss DNA aptamers are not adsorbed on GNS surface after aptamer binds with virus.
Figure 3:
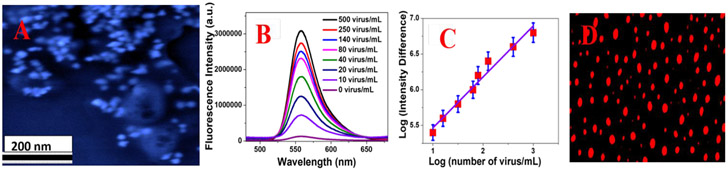
A) SEM image of pseudo baculovirus attached GNS. B) How the fluorescence spectra from Rh-6G conjugated DNA aptamer attached GNS varies when different amounts of virus are added. C) Plot shows how the log of (fluorescence intensity difference in the presence and absence of baculovirus) varies with the log of (concentration of baculovirus). D) Fluorescence image of pseudo baculovirus when they are attached with Rh-6G conjugated DNA aptamer bound GNS.
Our experimental data indicate that distance dependence NSET based virus diagnosis can be performed within 10 minutes using spike. To understand whether 5’-Rh-6G conjugated DNA attached GNS are bound onto virus surface, we have also performed fluorescence imaging experiment.
As reported in Figure 3D, we can clearly see the red color image from the virus attached nanosystem, which indicates that Rh-6G conjugated -ss DNA attached GNS bound virus and Rh-6G are far from GNS surface.
Sensitivity of the distance dependent NSET assay for specific virus detection has been determined by varying the virus amount from 10 virus/mL to 500 virus/mL. As shown in Figure 3B, distance dependent NSET assay has capability to identify coronavirus even at 10 virus/mL concentration level. As reported in Figure 3C, the log of NSET intensity difference (intensity before and after virus addition) varies linearly with the log of the concentration of the virus. Using the linear curve reported in Figure 3F and Equation 1, the LOD for distance dependent NSET assay has been determined to be ~ 8 virus/mL.
To understand whether Rh-6G conjugated spike protein specific DNA aptamer attached GNS based NSET assay can be used for the detection of COVID-19 specific antigen or virus, we have performed experiment using antigen or virus infected artificial nasal mucus fluid sample.
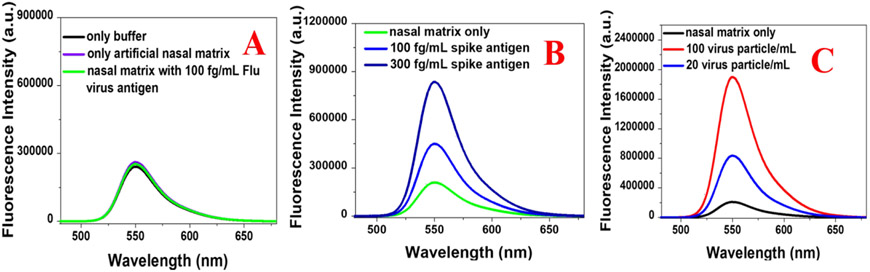
Artificial nasal mucus fluid was purchased from Biochemazone, Canada and the nasal matrix was infected by adding different concentrations of COVID-19 specific antigen or virus respectively. As shown in Figure 4A, we have not observed NSET intensity change in the presence of only nasal matrix or nasal matrix infected with flu virus antigen. On the other hand, as reported in Figure 4B, Rh-6G conjugated spike protein specific DNA aptamer attached GNS based NSET assay has capability to detect COVID-19 specific antigen even in the concentration of 100 fg/mL. Similarly, as reported in Figure 4C, Rh-6G conjugated spike protein specific DNA aptamer attached GNS based NSET assay has capability to detect virus even in the concentration of 20 virus/mL.
Figure 4:
A) Fluorescence spectra from Rh-6G conjugated spike protein specific DNA aptamer attached GNS in the presence of only buffer, only artificial nasal matrix and artificial nasal matrix infected with flu virus antigen (100 fg/mL). B) Fluorescence spectra from Rh-6G conjugated spike protein specific DNA aptamer attached GNS in the presence of artificial nasal matrix infected with SARS-CoV-2 spike recombinant antigen (100 fg/mL and 300 fg/mL). C) Fluorescence spectra from Rh-6G conjugated spike protein specific DNA aptamer attached GNS in the presence of artificial nasal matrix infected with pseudo baculovirus (20 virus particle/mL and 100 virus particle/mL)
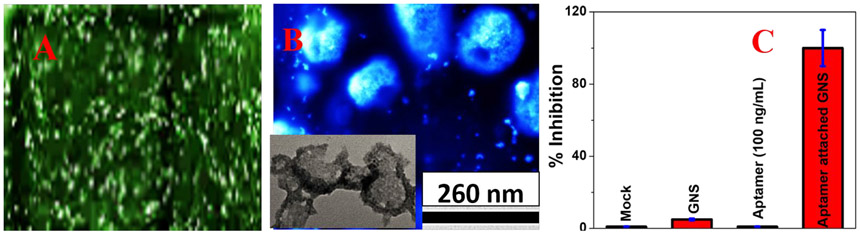
Next, we have determined whether spike protein specific-ss DNA aptamer attached GNS can be used to inhibit viral replication. For this purpose, ACE2 expressing HEK293T cell line has been used27-28. In this experiment, we have used spike protein specific -ss DNA aptamer attached GNS without Rh-6G. We have discussed experimental details in the supporting information. Reported virus inactivation data in Figures 5A-5C and Figure S3 in the supporting information indicate that ss DNA aptamer attached GNS can block viral replication. 100% inhibition efficiency was observed for ss DNA aptamer attached GNS at the concentration of 100 ng/mL. On the other hand, less than 1% inhibition efficiency was achieved for only GNS or aptamer at the same concentration level. The observed excellent inhibition efficiency for aptamer attached GNS can be because of the binding of the aptamer attached GNS to virus resulting in inability of virus to bind with ACE2 27-30. Experimental data shown in Figure 5B indicates that aptamer attached GNS can destroy the lipid membrane of pseudo baculovirus, so that the virus particle collapses27-30 and as a result, it stops spreading.
Figure 5:
A) Fluorescence image shows strong green fluorescent protein expression from target cells when spike protein specific ss DNA aptamer attached GNS is absent. B) SEM image indicates damage of virus lipid membrane of pseudo baculovirus, when virus and bio-conjugated nanoparticles are incubated for an hour. Inserted TEM image also indicates the same. C) Inhibition efficiency variation when only buffer (Mock), only GNS, 100 ng/mL spike protein specific ss DNA aptamer only and 100 ng/mL spike protein specific ss DNA aptamer attached GNS have been used. For this experiment, each of them were separately incubated with virus for an hour.
In conclusion, in this work we report that spike protein specific aptamer attached GNS can be used for detection and inactivation of corona virus. Our finding indicates that rhodamine 6G (Rh-6G) dye conjugated DNA aptamer attached GNS based distance dependent NSET has capability to detect spike antigen or virus itself very rapidly and results can be obtained in less than 10 minutes. Furthermore, NSET assay is highly sensitive where LOD as low as 130 fg/mL can be achieved for SARS-CoV-2 spike recombinant antigen and in case of virus the LOD is only 8 particles/mL. Finally, we demonstrate that 100% virus inhibition efficiency for aptamer attached GtgNS occurs via blocking cell attachment process and damage of lipid membrane. Although reported experimental data indicate that the aptamer conjugated GNS have the capability for diagnosis and inhibition of corona virus, till now we are in infancy for this research. In the next phase, we plan to move beyond the phase of demonstrating in the laboratory and find a way to move for clinical applications.
Supplementary Material
ACKNOWLEDGEMENTS
Dr. Ray is supported by NSF-RAPID grant # DMR-2030439 and NSF-PREM grant # DMR-1826886. Dr. Ray thanks NIH-NIMHD grant # 1U54MD015929-01 for cell culture, bioimaging and biological assay facilities. Dr. Tandon is supported by NASA award (80NSSC19K1603) and COVID-19 funds from his institute.
Footnotes
SUPORTING INFORMATIONS: Synthetic details for aptamer conjugated GNS formation and their characterization as well as other experiments are available as supporting information. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Jackson LA; Anderson EJ; Rouphael NG; Roberts PC; Makhene M; Coler RN; McCullough MP; Chappell JD; Denison MR; Stevens LJ; et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med 2020, 383, 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrosillo N; Viceconte G; Ergonul O; Ippolito G; Petersen E COVID-19, SARS and MERS: Are They Closely Related?. Clin. Microbiol. Infect 2020, 26, 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C; Horby PW; Hayden FG; Gao GF A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F; Zhao S; Yu B; Chen Y-M; Wang W; Song Z-G; Hu Y; Tao Z-W; Tian J-H; Pei Y-Y; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrapp D; Wang N; Corbett KS; Goldsmith JA; Hsieh C-L; Abiona O; Graham BS; McLellan JS Cryo-EM Structure of the 2019-Ncov Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L; Lin D; Sun X; Curth U; Drosten C; Sauerhering L; Becker S; Rox K; Hilgenfeld R Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science 2020, 368, 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandhi M; Yokoe DS; Havlir DV Asymptomatic Transmission, the Achilles’ Heel of Current Strategies to Control Covid-19. N. Engl. J. Med 2020, 382, 2158–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter LJ; Garner LV; Smoot JW; Li Y; Zhou Q; Saveson CJ; Sasso JM; Gregg AC; Soares DJ; Beskid TR; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci 2020, 6, 591–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C; Li W; Drabek D; Okba NMA; van Haperen R; Osterhaus ADME; van Kuppeveld FJM; Haagmans BL; Grosveld F; Bosch B-J A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun 2020, 11 (1), 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehranfar A, Izadya M; Theoretical Design of Functionalized Gold Nanoparticles as Antiviral Agents against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), J. Phys. Chem. Lett 2020, 11, 10284–10289 [DOI] [PubMed] [Google Scholar]
- 11.Liu R; He L; Hu Y; Luo Z; Zhang J; A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chemical Science 2020, 11, 12157–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Y; Song J; Wei X; Huang M; Sun M; Zhu L; Lin B; Shen H; Zhu Z; Yang C Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem 2020, 92, 9895–9900 [DOI] [PubMed] [Google Scholar]
- 13.Moitra P; Alafeef M; Dighe K; Frieman MB; Pan D Selective Naked-Eye Diagnosis of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo G; Lee G; Kim MJ; Baek SH; Choi M; Ku KB; Lee CS; Jun S; Park D; Kim SJ; Lee JO; et al. Rapid Diagnosis of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142 [DOI] [PubMed] [Google Scholar]
- 15.Alafeef M; Dighe K; Moitra P; Pan D; Rapid, Ultrasensitive, and Quantitative Diagnosis of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, Article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ray PC; Fan Z; Crouch RA; Sinha SS; Pramanik A Nanoscopic Optical Rulers Beyond the FRET Distance Limit: Fundamentals and Applications, Chem. Soc. Rev 2014, 43, 6370–6404 [DOI] [PubMed] [Google Scholar]
- 17.Riskowski RA; Armstrong RE; Greenbaum NL; and Strouse GF; Triangulating Nucleic Acid Conformations Using Multicolor Surface Energy Transfer, ACS Nano, 2016, 10, 2, 1926–1938 [DOI] [PubMed] [Google Scholar]
- 18.Grifin J; Singh AK; Senapati D; Rhodes P ; Mitchell K; Robinson B; Yu E; Ray PC Size- and Distance-Dependent Nanoparticle Surface-Energy Transfer (NSET) Method for Selective Sensing of Hepatitis C Virus RNA Chem. - Eur. J 2009, 15, 342–351 [DOI] [PubMed] [Google Scholar]
- 19.Vangara A; Pramanik A,; Gao Y; Gates K; Begum S; Ray PC; Fluorescence Resonance Energy Transfer Based Highly Efficient Theranostic Nanoplatform for Two-Photon Bioimaging and Two-Photon Excited Photodynamic Therapy of Multiple Drug Resistance Bacteria, ACS Appl. Bio Mater, 2018, 1, 298–309 [DOI] [PubMed] [Google Scholar]
- 20.Carnevale KJF; Riskowski RA; Strouse GF; A Gold Nanoparticle Bio-Optical Transponder to Dynamically Monitor Intracellular pH. ACS Nano 2018, 12, 5956–5968 [DOI] [PubMed] [Google Scholar]
- 21.Chavva SS; Nellore BPV; Pramanick A; Sinha SS; Jones S; Ray PC; Designing a multicolor long range nanoscopic ruler for the imaging of heterogeneous tumor cells, Nanoscale, 2016, 8, 13769–13780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kundu S: Patra, S.; Nanoscale Strategies for Light Harvesting, Chem. Rev 2017, 117, 2, 712–757 [DOI] [PubMed] [Google Scholar]
- 23.Pramanik A; Mayer J; Patibandla S; Gates K; Gao Ye, Davis D; Seshadri R; Ray PC; Mixed-Dimensional Heterostructure Material-Based SERS for Trace Level Identification of Breast Cancer-Derived Exosomes, ACS Omega 2020, 5, 27, 16602–16611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha SS; Jones S; Pramanik A; Ray PC; Nanoarchitecture Based SERS for Biomolecular Fingerprinting and Label-Free Disease Markers Diagnosis, Acc. Chem. Res, 2016, 49,2725–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandra K; Culver KSB; Werner SE; Lee RC; Odom TW Manipulating the Anisotropic Structure of Gold Nanostars using Good’s Buffers. Chem. Mater 2016, 28, 6763–6769. [Google Scholar]
- 26.DeRussy MB; Aylward MA; Fan Z; Ray PC; Tandon R; Inhibition of cytomegalovirus infection and photothermolysis of infected cells using bioconjugated gold nanoparticles, Scientific Reports 2014, 4, Article number: 5550, doi: 10.1038/srep05550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tandon R, Mitra D, Sharma P, McCandless MG, Stray SJ, Bates JT, Marshall GD. Effective screening of SARS-CoV-2 neutralizing antibodies in patient serum using lentivirus particles pseudotyped with SARS-CoV-2 spike glycoprotein. Sci Rep. 2020. 5, 10(1), 19076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pramanik A; Gao Ye; Patibandla S; Mitra D; McCandless MG; Fassero LA; Gates K; Tandon R; and Ray PC: Rapid Diagnosis and Effective Inhibition of Corona Virus Using Spike Antibody Attached Gold Nanoparticle, Nanoscale Advances, 2021. (ASAP; Article) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du T; Zhang J; Li C; Song T; Li P; Liu J; Du X; Wang S; Gold/Silver Hybrid Nanoparticles with Enduring Inhibition of Coronavirus Multiplication through Multisite Mechanisms, Bioconjugate Chem. 2020, 31, 11, 2553–2563 [DOI] [PubMed] [Google Scholar]
- 30.Medhi R; Srionoi P; Ngo N; Tan H; Lee TR; Nanoparticle-Based Strategies to Combat COVID-19, ACS Appl. Nano Mater 2020, 3, 9, 8557–8580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.