Abstract
Background:
We applied various machine learning algorithms to a large national dataset to model the risk of postoperative sepsis after appendectomy to evaluate utility of such methods and identify factors associated with postoperative sepsis in these patients.
Methods:
The National Surgery Quality Improvement Program database was used to identify patients undergoing appendectomy between 2005 and 2017. Logistic regression, support vector machines, random forest decision trees, and extreme gradient boosting machines were used to model the occurrence of postoperative sepsis.
Results:
In the study, 223,214 appendectomies were identified; 2,143 (0.96%) were indicated as having postoperative sepsis. Logistic regression (area under the curve 0.70; 95% confidence interval, 0.68–0.73), random forest decision trees (area under the curve 0.70; 95% confidence interval, 0.68–0.73), and extreme gradient boosting (area under the curve 0.70; 95% confidence interval, 0.68–0.73) afforded similar performance, while support vector machines (area under the curve 0.51; 95% confidence interval, 0.50–0.52) had worse performance. Variable importance analyses identified preoperative congestive heart failure, transfusion, and acute renal failure as predictors of postoperative sepsis.
Conclusion:
Machine learning methods can be used to predict the development of sepsis after appendectomy with moderate accuracy. Such predictive modeling has potential to ultimately allow for preoperative recognition of patients at risk for developing postoperative sepsis after appendectomy thus facilitating early intervention and reducing morbidity.
Introduction
Sepsis continues to be a leading cause of preventable morbidity and mortality in surgical patients.1 Accounting for more than $20 billion dollars in annual costs to our nation’s hospital system,2 there is increasing attention of payors for providers to reduce the incidence of postoperative sepsis. Acute appendicitis is one of the most common abdominal surgical emergencies worldwide.3 Although the rate of sepsis after appendectomy is low,4 when it does occur, it confers increased morbidity and even mortality for the patient.5 The low rate of sepsis after appendectomy limits an individual surgeon’s experience with recognizing the patients at risk for this complication. In these clinical scenarios, data aggregation from electronic health records can be used for modeling and predicting these rare events using artificial intelligence (AI). There is the potential that the incorporation of AI into surgical practices would benefit patient care.
Since the Health Information Technology for Economic and Clinical Health Act passed in 2009,6 data from millions of patients nationwide is now captured in different electronic health records (EHR) across multiple institutions. The complexity, heterogeneity, size, and rate at which this data is generated, however, has out-paced the labor-intensive analytical techniques traditionally used in medicine.7 Machine learning (ML), a branch of AI, seeks to overcome the limitations of traditional statistical techniques in analyzing this type of data. Compared with conventional clinical statistical methods, which rely on a set of programmed rules, ML is a type of algorithm in which the model learns from examples such that a task can be correctly performed on the encounter of novel inputs.8 Whereby it would take a lifetime to gather robust clinical experience with treating a rare condition or to manually classify and analyze hundreds of millions of data points, ML models are able to find statistical patterns across numerous heterogeneous features relatively quickly.9
These algorithms have already been widely used in other industries for decades.10 In recent years, there has been growing interest in the application of ML in health services research, where it has begun to be perceived as an innovative tool with potential to harness the complexity of big data to provide predictive power and improve clinical decision making.11 The application of ML in the clinical setting, however, particularly within the field of surgery, has been limited for reasons including difficult interpretability. In this proof of concept study, we aim to use a widely used, nationally validated surgical dataset to evaluate the performance of different ML algorithms against traditional statistical methods in prediction of a rare postoperative sepsis after appendectomy.
Methods
Patient population
Using the American College of Surgeons National Surgical Quality Improvement Program (ACS NSQIP) database, Current Procedural Terminology codes were used to identify cases of open (44950, 44960) and laparoscopic (44970, 44979, 44950, 44960) appendectomy performed as the principle operative procedure in patients age 16 years and older between 2005 and 2017. In an effort to target only cases in which sepsis developed as a new diagnosis postoperatively, records coded for sepsis 48 hours before appendectomy were excluded from analysis. This study was approved by the Loyola University Chicago Institutional Review Board as exempt. American College of Surgeons National Surgical Quality Improvement Program and the hospitals participating in the ACS-NSQIP are the source of the data used herein; they have not verified and are not responsible for the statistical validity of the data analysis or the conclusions derived by the authors.
Definitions and variable selection
The primary outcome of interest was postoperative sepsis, defined in this study as the presence of sepsis or septic shock. Specifically, for the diagnosis of sepsis, either preoperative or postoperative, ACS-NSQIP requires the presence of documented infection or end organ ischemia in addition to at least 2 of the 5 clinical signs and symptoms of systemic inflammatory response syndrome. Septic shock was defined as the presence of sepsis in addition to documented organ or circulatory dysfunction. Independent variables included for analysis were selected a priori from those available in the NSQIP dataset as those clinical parameters felt by the practicing physicians on the research team as the ones that are likely to be associated with the development of postoperative sepsis (Table I).
Table I.
Variables used as input for prediction models
| Demographics | Age, BMI, sex, race, ethnicity |
|---|---|
| Preoperative risk factors/comorbid conditions | Smoking, insulin-dependent diabetes, noninsulin-dependent diabetes, dyspnea, ventilator dependency, COPD, ascites, CHF within 30 d preoperatively, hypertension requiring medication, acute preoperative renal failure, preoperatively on/requiring dialysis, disseminated cancer, chronic steroid use, >10% loss of body weight in last 6 mo, bleeding disorders, transfusion ≥1 unit packed pRBC within 72 hours before surgery, open wound/infection, days to operation, procedural Current Procedural Terminology code |
| Preoperative laboratory results | Serum sodium, BUN, serum creatinine, albumin, total bilirubin, aspartate transaminase, alkaline phosphatase, WBC, hematocrit, and platelet count |
BUN, blood urea nitrogen; COPD, chronic obstructive pulmonary disease, pRBC, packed red blood cells; WBC, white blood cell.
Missing data handling
Missing data patterns were imputed through multivariable imputation by chained equations to obtain a complete analytical dataset. Multivariable imputation by chained equations imputes missing values for all covariates through a chained equation process using various methods such as predictive mean matching, classification and regression trees, random forest (RF), and sample.12 We compared each of these methods on a randomly selected subset of data. In this study, we used predictive mean matching to impute continuous variables, which is also a preferred method for skewed data,13 and logistic regression for categorical variables.
Statistical methods and ML algorithms
General descriptive statistics were calculated as frequency and percentages and compared between cohorts using χ2 test for categorical variables and Student t tests for continuous variables. For the development of ML algorithms, the dataset was randomly split into 80% training14 and 20% hidden testing14 datasets across all years with measures to prevent over-fitting and ensure validation. In this setup, a hidden test set was used as a hold-out dataset where we built a model using training data and further tested its validity on the unused hidden test data.
Classification algorithms implemented on the training dataset included traditional multivariable logistic regression (LR) and 3 ML algorithms: support vector machines (SVM), random forest decision trees (RFDT), and extreme gradient boosting (XGB) machines. SVM, RFDT, and XGB machines were chosen for this analysis as these are among the most commonly used types of supervised ML methods and best used for classification and regression tasks as in the case of this study.14,15
Model performance was reported using standard metrics of epidemiology and organized by area under the receiver operating characteristic curve (AUC-ROC). Subgroup analysis was implemented using the calculated predicted probability based off the ensemble of the best model. Variable importance analysis was conducted to reveal the influence of predictors on model performance. Variable importance values were normalized (0–100) for simplicity. All data acquisition and analysis were performed in R (version 3.6.1, The R Foundation for Statistical Computing) and Python 3.5.6.
Results
Missing data
Of the preoperative variables included in the prediction models (Table I), there were 11 variables that had greater than 1% missing data (Table II). Of these 11 variables, 10 were preoperative laboratory values. The variable with the highest rate of missing data was serum albumin (31.6%) while white blood cell count had the lowest rate (3.5%). Descriptive statistics of variables before and after imputation provided similar results (data not shown).
Table II.
Characteristics of imputed variables with more than 1% missingness
| Preoperative laboratory value | No. missing | Percent missing | Mean | SD |
|---|---|---|---|---|
| Albumin | 70,527 | 31.6% | 4.18 | .493 |
| Aspartate transaminase | 57,124 | 25.6% | 24.5 | 19.8 |
| Alkaline phosphatase | 53,216 | 23.8% | 75.6 | 28.7 |
| Bilirubin | 53,207 | 23.8% | .786 | .624 |
| BMI (kg/m2) | 21,823 | 9.8% | 28.6 | 6.7 |
| Blood urea nitrogen | 18,573 | 8.3% | 13 | 5.9 |
| Sodium | 15,051 | 6.7% | 138 | 2.73 |
| Creatinine | 14,177 | 6.4% | .881 | .475 |
| Platelet | 8,637 | 3.9% | 238 | 64.5 |
| Hematocrit | 8,078 | 3.6% | 41.08 | 4.461 |
| White blood cell count | 7,752 | 3.5% | 11.707 | 4.264 |
BMI, body mass index; SD, standard deviation.
Descriptive characteristics
The NSQIP data base included 223,214 records for appendectomy, 94.64% of which were performed for a primary diagnosis of appendicitis. There were 21,839 (9.8%) cases performed using an open approach, whereas the majority (n = 201,375; 90.2%) were completed laparoscopically. Patients were 39.8 ± 16.3 years old and predominately male (50.9%) and white (70.4%). The incidence of postoperative sepsis was 0.96% (n = 2,143), of which, 7.6% developed septic shock. The 30-day mortality rate was significantly higher in patients who developed postoperative sepsis (2.6%) compared with the overall mortality rate in the study population (0.07%, P < .001). Univariate analysis (Table III) revealed that postoperative sepsis after appendectomy was more likely to occur in patients who were older (48.1 years vs 39.8 years, P < .001), black (10.8% vs 7.6%, P < .001), and had comorbid conditions, including both insulin and noninsulin dependent diabetes, congestive heart failure (CHF), hypertension, renal failure, disseminated cancer, chronic steroid use, and bleeding disorders (all P values < .001).
Table III.
Univariate analysis for rate of postoperative sepsis after appendectomy
| No postoperative sepsis | Postoperative sepsis | P value | |
|---|---|---|---|
| N | 221,073 | 2,141 | |
| Procedure type (%) | <.001 | ||
| Appendectomy (44,950) | 18,078 (8.2) | 282 (13.2) | |
| Appendectomy for ruptured appendix (44,960) | 3,259 (1.5) | 220 (10.3) | |
| Laparoscopic appendectomy (44,970) | 199,479 (90.2) | 1,631 (76.2) | |
| Unlisted laparoscopy procedure, appendix (44,979) | 257 (0.1) | 8 (0.4) | |
| Age (mean, SD) | 39.75 (16.30) | 48.09 (18.41) | <.001 |
| BMI (kg/m2; mean, SD) | 28.58 (6.69) | 29.82 (7.24) | <.001 |
| Female (%) | 108,736 (49.2) | 884 (41.3) | <.001 |
| Race | <.001 | ||
| Asian | 10,140 (5.9) | 69 (4.2) | |
| Black | 15,723 (9.1) | 218 (13.2) | |
| Other | 2,401 (1.4) | 17 (1.0) | |
| White | 144,911 (83.7) | 1,349 (81.6) | |
| Hispanic | 27,804 (15.2) | 252 (14.7) | .568 |
| Comorbidities | |||
| Smoking (%) | 40,486 (18.3) | 429 (20.0) | .043 |
| Noninsulin dependent diabetes (%) | 6,029 (2.7) | 124 (5.8) | <.001 |
| Insulin dependent diabetes (%) | 3,873 (1.8) | 106 (5.0) | <.001 |
| COPD (%) | 1,995 (0.9) | 60 (2.8) | <.001 |
| Ascites (%) | 426 (0.2) | 9 (0.4) | .033 |
| CHF (%) | 272 (0.1) | 9 (0.4) | <.001 |
| Hypertension (%) | 35,669 (16.1) | 642 (30.0) | <.001 |
| Renal failure (%) | 125 (0.1) | 7 (0.3) | <.001 |
| Dialysis (%) | 376 (0.2) | 27 (1.3) | <.001 |
| Disseminated cancer (%) | 498 (0.2) | 21 (1.0) | <.001 |
| Steroid use (%) | 2,390 (1.1) | 55 (2.6) | <.001 |
| Weight loss (%) | 405 (0.2) | 13 (0.6) | <.001 |
| Bleeding disorder (%) | 3,702 (1.7) | 117 (5.5) | <.001 |
| Transfusion ≥1 units packed red cells within 48 hours preoperatively (%) | 159 (0.1) | 3 (0.1) | .445 |
| Preoperative laboratory values | |||
| Sodium (mean, SD) | 138.34 (2.72) | 137.37 (3.16) | <.001 |
| BUN (mean, SD) | 12.78 (5.82) | 15.10 (10.42) | <.001 |
| Creatinine (mean, SD) | 0.88 (0.46) | 1.06 (1.01) | <.001 |
| Albumin (mean, SD) | 4.19 (0.49) | 3.98 (0.59) | <.001 |
| Bilirubin (mean, SD) | 0.79 (0.62) | 0.89 (0.75) | <.001 |
| SGOT (mean, SD) | 24.47 (19.87) | 25.23 (24.04) | .079 |
| Alkaline phosphatase (mean, SD) | 75.25 (28.36) | 80.57 (34.48) | <.001 |
| White blood cell count (mean, SD) | 11.68 (4.26) | 12.78 (4.86) | <.001 |
| Hematocrit (mean, SD) | 41.09 (4.46) | 40.68 (5.19) | <.001 |
| Platelet (mean, SD) | 237.91 (64.34) | 240.09 (73.73) | .12 |
| Days from admission to operation (mean, SD) | 0.40 (6.79) | 0.47 (4.48) | .604 |
| 30-d mortality (%) | 99 (0.0) | 55 (2.6) | <.001 |
| Superficial incisional surgical site infection (%) | 2195 (1.0) | 99 (4.6) | <.001 |
| Deep incisional surgical site infection (%) | 378 (0.2) | 61 (2.8) | <.001 |
| Organ space surgical site infection (%) | 2192 (1.0) | 618 (28.9) | <.001 |
| C.diff infection (%) | 166 (0.2) | 24 (2.9) | <.001 |
| Perforated (CPT = 44960) (%) | 3,259 (1.5) | 220 (10.3) | <.001 |
| Pneumonia (%) | 387 (0.2) | 114 (5.3) | <.001 |
| Urinary infection (%) | 902 (0.4) | 48 (2.2) | <.001 |
BMI, body mass index; CPT, Current Procedural Terminology; SD, standard deviation; SGOT, serum glutamic oxaloacetic transaminase.
Prediction of postoperative sepsis
Table IV summarizes the performance of each of the prediction models reported in terms of AUC-ROC. LR, RFDT, and XGB machines models provided equivocal predictive accuracy (AUC 0.70; 95% confidence interval [CI], 0.68–0.73), whereas SVM yielded significantly lower performance (AUC 0.5142; 95% CI, 0.50–0.53). The XGB machines model provided the highest accuracy of any single methodology (AUC 0.7030; 95% CI, 0.68–0.73). A final model was constructed using an ensemble of the best models including LR, RF, and XGB machines. This ensemble model yielded the highest attained predictive accuracy (AUC 0.71; 95% CI, 0.69–0.73).
Table IV.
Performance of postoperative prediction models reported as AUC (95% CI)
| LR | SVM | RFDT | XGB machines | Ensemble model (LR, RFDT, XGB) | |
|---|---|---|---|---|---|
| Fold 1 | 0.7152 (0.6844–0.7460) | 0.5996 (0.5677–0.6313) | 0.7122 (0.6813–0.7430) | 0.7165 (0.6857–0.7473) | 0.7219 (0.6912–0.7526) |
| Fold 2 | 0.7142 (0.6821–0.7463) | 0.4824 (0.4510–0.5139) | 0.7030 (0.6707–0.7353) | 0.7112 (0.6790–0.7434) | 0.7169 (0.6848–0.7489) |
| Fold 3 | 0.7133 (0.6822–0.7443) | 0.5200 (0.4887–0.5511) | 0.7152 (0.6842–0.7463) | 0.7218 (0.6909–0.7527) | 0.7266 (0.6957–0.7574) |
| Fold 4 | 0.7074 (0.6767–0.7382) | 0.4791 (0.4492–0.5090) | 0.7198 (0.6893–0.7503) | 0.7238 (0.6933–0.7542) | 0.7291 (0.6987–0.7594) |
| Fold 5 | 0.7269 (0.6962–0.7575) | 0.5122 (0.4813–0.5432) | 0.7350 (0.7045–0.7655) | 0.7295 (0.6990–0.7602) | 0.7388 (0.7084–0.7692) |
| Average 5-fold CV accuracy | 0.7150 (0.7010–0.7289) | 0.5136 (0.4997–0.5275) | 0.7170 (0.7031–0.7309) | 0.7205 (0.7067–0.7344) | 0.7266 (0.7128–0.7404) |
| 20% test data*,† | 0.6955 (0.6756–0.7304) Sensitivity: 0.6261 Specificity: 0.6509 | 0.5142 (0.5020–0.5264) Sensitivity: ‡NA Specificity: ‡NA | 0.6953 (0.6756–0.7304) Sensitivity: 0.6712 Specificity: 0.6067 | 0.7030 (0.6756–0.7304) Sensitivity: 0.6441 Specificity: 0.6667 | 0.7074 (0.6871–0.7277) Sensitivity: 0.6464 Specificity: 0.6454 |
Results for 20% test data reported as AUC with 95% CI, sensitivity and specificity.
Sensitivity and specificity reported using a cutoff value of 0.015 on predicted probabilities (0.015 was chosen as this provided the most balanced Sensitivity and Specificity on the ensemble model).
Not calculated due to poor cross-validation performance.
Subgroup analysis performed using the ensemble model revealed overall similar performance within cohorts compared with model accuracy in the study population. The ensemble model had higher predictive accuracy in females, open cases, and in white patients (Table V). Figure 1 shows the top-20 predictors for the XGB machines classifier. Variable importance analysis revealed that the factors most strongly associated with postoperative sepsis in this model were recent 30-day exacerbation or diagnosis of CHF, transfusion of 1 or more units of red blood cells within 72-hours preoperatively, and acute renal failure within 24 hours prior (variable importance scores of 100.0, 48.04, 44.81, respectively).
Table V.
Subgroup analysis performed using the ensemble model
| Variable | AUC (95% CI) |
|---|---|
| Sex | |
| Female | 0.72 (0.68–0.76) |
| Male | 0.69 (0.66–0.73) |
| Procedure (CPT code) | |
| Appendectomy (44950) | 0.64 [0.57–0.72] |
| Appendectomy for ruptured appendix (44960) | 0.61 [0.53–0.69] |
| Laparoscopic appendectomy (44970) | 0.60 (0.66–0.72) |
| Procedure type | |
| Open | 0.71 (0.66–0.77) |
| Laparoscopic | 0.68 (0.65–0.71) |
| Race | |
| White | 0.71 (0.67–0.74) |
| Black | 0.78 (0.70–0.85) |
| Asian | 0.68 (0.49–0.87) |
| Other/Unknown | 0.66 (0.60–0.71) |
CPT, Current Procedural Terminology.
Figure 1.
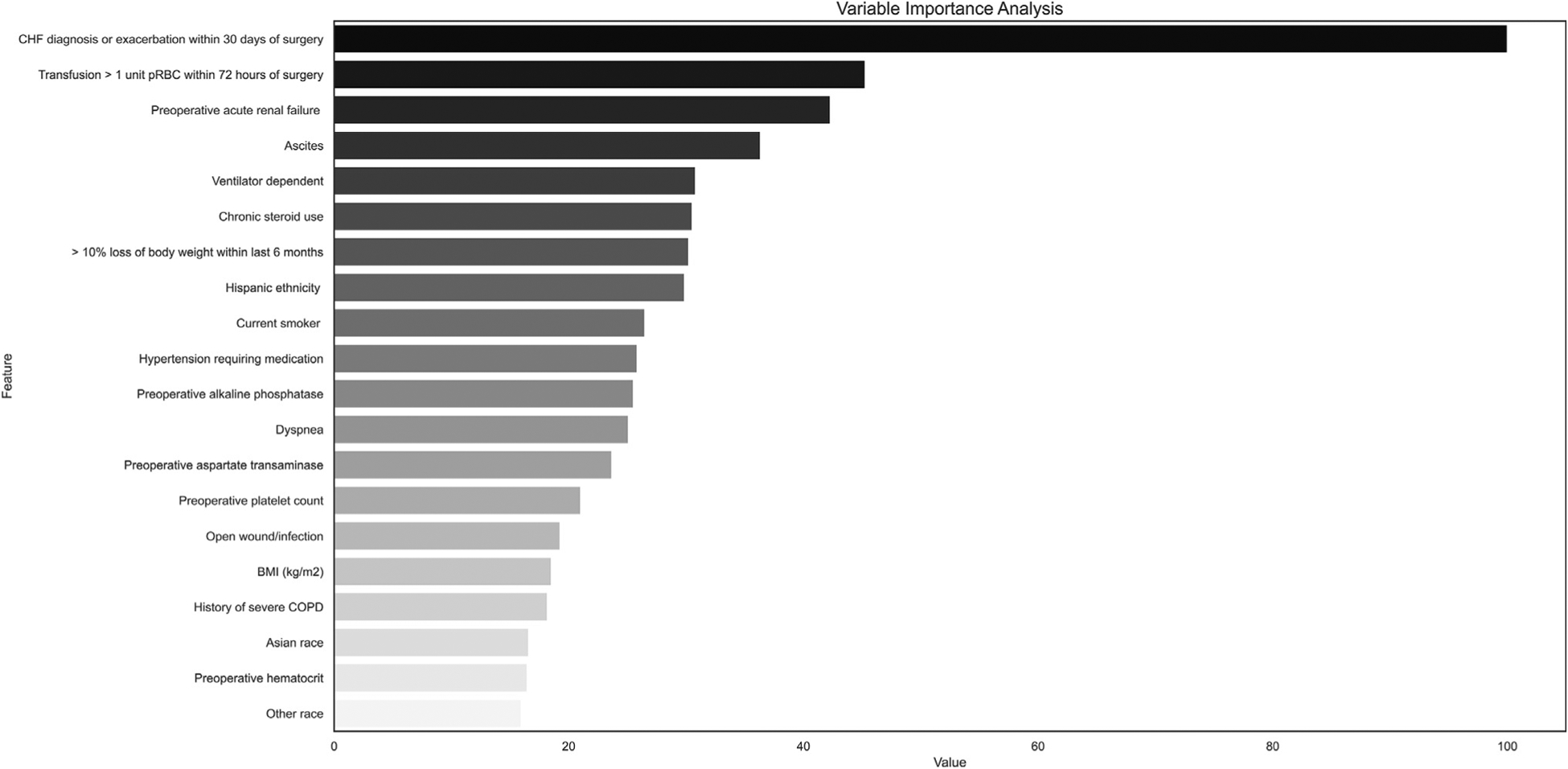
Relative influence of predictors in XGB machines classier model represented on a scale from 0 to 100% where 100% represents the predictor with the greatest influence in the model.
Association between postoperative sepsis and mortality
The rate of 30-day postoperative mortality in our cohort was <0.1%. Given the markedly low event rate, for the purposes of this study, a separate postoperative mortality prediction model was not constructed. Postoperative mortality was therefore examined using the predicted risk for postoperative sepsis as a risk for mortality from which classification accuracy statistics were calculated and summarized (Table VI). Overall, the postoperative sepsis prediction models performed well in predicting postoperative 30-day mortality, particularly the ensemble and RFDT models.
Table VI.
Prediction model performance calculated using the predicted risk for postoperative sepsis as a risk for postoperative 30-d mortality on 20% test dataset
| LR | SVM | RF | EGB | Ensemble model (LR, RF, XGB) | |
|---|---|---|---|---|---|
| Sensitivity* | 0.8214 | *NA | 0.9286 | 0.8929 | 0.8929 |
| Specificity* | 0.8723 | *NA | 0.8438 | 0.8564 | 0.8952 |
| AUC | 0.9192 (0.9016–0.9368) | 0.5 (0.4730–0.5269) | 0.963 (0.9507–0.9753) | 0.934 (0.9179–0.9501) | 0.9536 (0.9399–0.9673) |
Sensitivity and specificity reported using a cutoff value of 0.015 on predicted probabilities (0.015 was chosen as this provided the most balanced sensitivity and specificity on the ensemble model).
Discussion
In this study we hypothesized that ML algorithms, a type of AI, could be used to model prediction of postoperative sepsis in patients undergoing appendectomy using a national database. The predictive accuracy of 4 different models: LR, SVM, RFDT, XGB machines as well as an ensemble model were compared. Using only variables that could be determined prospectively prior to operative intervention, we found that overall, there was no significant improvement in ML model accuracy, measured by our median AUC-ROC of 0.70 (interquartile range, 0.70–0.71) over traditional LR (AUC 0.70; 95% CI, 0.68–0.73). Our model accuracy was highest in our ensemble classifier (AUC 0.71; 95% CI, 0.69–0.73), which used the LR, RF decision trees, and XGB machines as the inputs to develop a prediction. The results reflect the ability of an ensemble ML model to aggregate the prediction of each constituent model to achieve higher prediction accuracy than any individual model alone. In this study, the improvement in prediction accuracy afforded by the ensemble model was marginal compared with traditional LR. The similar AUC-ROC achieved between models, however, demonstrates that even in the case of predicting rare events, such as postoperative sepsis after appendectomy, there are preoperative signs of postoperative sepsis as captured even within a relatively small number of features.
Although our study was relatively limited in the number of features available to create prediction tasks, other studies have demonstrated ML to be a powerful tool in transforming large amounts of complex EHR data into prognostic models with applications in clinical decision support, resource allocation, and health care workflow design.8,16,17 Wong et al demonstrated that when applied to institutional EHR data comprising 796 variables, ML algorithms, similar to those used in our study, outperformed current clinical tools in predicting development of in-patient delirium.17 Similarly, Taylor et al found that ML outperforms LR and other traditional clinical tools such as CURB-65 for prediction of sepsis, as captured through 1,697 different International Classification of Disease, version 9, codes with EHR data from 5,278 emergency department visits.16 Rajkomar et al used a total of 46,864,534,945 raw EHR data elements of varied temporal relationships from which they were able to create prediction tools for in-patient mortality, 30-day readmission, prolonged duration of stay, and diagnosis at discharge that outperformed traditional clinical prediction tools at every time point.8
As demonstrated by the aforementioned studies, when afforded a large number of inputs, AI has the ability to make earlier and more accurate predictions. Such prediction models have the potential to facilitate early intervention, which has been shown to reduce complications, hospital duration of stay, and cost.18 In surgery, early identification afforded by AI prediction tools, such as in the case of postappendectomy sepsis, can aid surgeons in their decision making regarding the timing and type of intervention, selection and duration of perioperative antibiotics, postoperative disposition, additional monitoring, and need for follow-up surveillance imaging for detection of early abscess formation, particularly in cases that otherwise would not have raised suspicion in the majority of providers as being high risk for sepsis.
Postoperative sepsis is known to be associated with increased mortality,19 and sepsis has been found to be a predictor of postoperative 30-days mortality.20,21 Because of the low mortality rate (<0.1%) after appendectomy, we did not build a mortality prediction model in this study. Instead, we used the risk of postoperative sepsis that was predicted by the model as a risk factor for mortality. In doing so, we obtained a very high predictive accuracy (as high as AUC of 0.96) on our 20% hold-out test. This result confirms that there may be clusters of postoperative complications identified by preoperative patterns and sepsis and mortality may be placed in the same cluster.
Critiques of ML have cited the lack of interpretability and intrinsic ambiguity that is inherent to “black box” type models as limitations in their utility within clinical medicine.22 In an effort to address this concern, we performed a variable importance analysis, which allowed insight into the variables that had the greatest influence within the ML model. The fact that our results yielded factors that are known to be clinically relevant to the development of sepsis (CHF, renal failure, preoperative blood transfusion)4 validated our ML model. To draw conclusions regarding the results of the variable importance analysis, however, our model would need to be externally validated outside of the NSQIP setting (eg, demonstrating the model predictive performance in non-NSQIP data sets, such as EHR data or PCORNet’s Clinical Data Research Network data).23 If externally validated, the preoperative risk factors found to be strong predictors of postoperative sepsis may assist clinicians in optimizing modifiable risk factors in the case of elective surgery. In cases of nonmodifiable risk factors, these results may improve shared decision making with patients preoperatively to include discussion regarding predicted risk of complications, mortality, postoperative disposition and duration of stay.
There are a few important limitations to this study. First, the direct clinical implementation of our proposed model may be limited due to a high rate of false positives. Previous studies have shown, however, that integration of our model with preoperative clinical text notes may significantly improve the postoperative sepsis prediction and reduce false positive rates.21 Additionally, we chose to include all cases of appendectomy in an effort to maximize the size of the dataset. After rerunning our analysis on adults age 18 years and older, we found no significant difference in results of model performance (Appendix A). We also chose to limit our study to only those patients who were indicated as not having a diagnosis of sepsis preoperatively. We understand this as a potential point of contention, as the pathophysiology of acute appendicitis is by definition a systemic inflammatory process secondary to infection. We felt that by eliminating patients who were systemically ill enough to have a diagnosis of sepsis preoperatively, we could, in theory, isolate our prediction task to those patients who were at an early stage of the disease pathology or perforation, therefore making prediction of postoperative sepsis more difficult. In doing so, we identified CHF and acute renal failure as risk factors for postoperative sepsis which is supported by similar studies.4,5 Laparoscopy, which has been shown to potentially reduce postoperative complications,24 was not identified on variable importance analysis as being an important predictor within the models. Additionally, we were not able to include data regarding intraoperative findings, such as retrocecal or gangrenous appendix, as these may significantly contribute to the development of postoperative sepsis.
Another limitation of this study is that we chose to apply ML to a national database as opposed to EHR data. ACS-NSQIP is a very high quality and well-organized surgical database of harmonized and curated data sets, with robust methods for ensuring data fidelity on a predefined number of relevant variables. The small size and high quality of the dataset is one of the main reasons that the ML models we tested did not exceed the performance of logistic regression in this study. In more noisy and uncurated data sets, however, it is likely that ML models would significantly outperform logistic regression. Knowing this, we chose to use ML in the prediction of postoperative sepsis after appendectomy using a well-studied surgical database and relatively consistent disease process and surgical procedure. By using this platform to evaluate ML methods we hoped to reduce potential confounding biases and improve interpretability of our results without sacrificing performance of the models.25 The results of this study are hypothesis-generating and are meant to fuel deeper understanding of the application of machine learning in generating predictions of surgical complications.
In conclusion, ML methods can be used to predict the development of postoperative sepsis post-appendectomy with moderately high accuracy. In patients who initially present without sepsis, factors associated with the development of postoperative sepsis after appendectomy include recent CHF exacerbation or diagnosis, acute renal failure, and preoperative transfusion. Such predictive models may ultimately allow for recognition of patients at risk for postoperative sepsis after appendectomy prior to surgical intervention potentially facilitating early risk mitigation and improve informed decision making.
Supplementary Material
Acknowledgments
The authors thank all surgeons, registrars, physician assistants, and administrative nurses from ACS-NSQIP participating hospitals.
Funding/Support
Dr C. Bunn is supported by National Institute of Health T32 NIGMS 5T32GM008750-20. Dr S. Kulshrestha is supported by National Institute of Health T32 NIAAA 5T32AA013527-17.
Footnotes
Accepted for Presentation at the 77th Annual Meeting of the Central Surgical Association, Milwaukee, WI.
Conflicts of interest/Disclosure
The authors have no related conflicts of interest to declare.
Supplementary data
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.1016/j.surg.2020.07.045.
References
- 1.Moore LJ, Moore FA, Todd SR, Jones SL, Turner KL, Bass BL. Sepsis in general surgery: The 2005–2007 national surgical quality improvement program perspective. Arch Surg. 2010;145:695–700. [DOI] [PubMed] [Google Scholar]
- 2.Lagu T, Rothberg MB, Shieh M-S, Pekow PS, Steingrub JS, Lindenauer PK. Hospitalizations, costs, and outcomes of severe sepsis in the United States 2003 to 2007. Crit Care Med. 2012;40:754–761. [DOI] [PubMed] [Google Scholar]
- 3.Stewart B, Khanduri P, McCord C, et al. Global disease burden of conditions requiring emergency surgery. Br J Surg. 2014;101:e9–e22. [DOI] [PubMed] [Google Scholar]
- 4.Ninh A, Wood K, Bui AH, Leitman IM. Risk factors and outcomes for sepsis after appendectomy in adults. Surg Infect (Larchmt). 2019;20:601–606. [DOI] [PubMed] [Google Scholar]
- 5.Margenthaler JA, Longo WE, Virgo KS, et al. Risk factors for adverse outcomes after the surgical treatment of appendicitis in adults. Ann Surg. 2003;238:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumenthal D. Launching hitech. New Engl J Med. 2010;362:382–385. [DOI] [PubMed] [Google Scholar]
- 7.Krumholz HM. Big data and new knowledge in medicine: The thinking, training, and tools needed for a learning health system. Health Affairs. 2014;33: 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine. 2018;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obermeyer Z, Emanuel EJ. Predicting the future: Big data, machine learning, and clinical medicine. New Engl J Med. 2016;375:1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portugal I, Alencar P, Cowan D. The use of machine learning algorithms in recommender systems: A systematic review. Expert Systems with Applications. 2018;97:205–227. [Google Scholar]
- 11.Karhade AV, Thio QC, Ogink PT, et al. Development of machine learning algorithms for prediction of 30-day mortality after surgery for spinal metastasis. Neurosurgery. 2018;85:E83–E91. [DOI] [PubMed] [Google Scholar]
- 12.Azur MJ, Stuart EA, Frangakis C, Leaf PJ. Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res. 2011;20:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinke K. Multiple imputation under violated distributional assumptions: A systematic evaluation of the assumed robustness of predictive mean matching. J Educ Behav Stat. 2017;42:371–404. [Google Scholar]
- 14.Kubben P, Dumontier M, Dekker A. Fundamentals of Clinical Data Science. Cham, Switzerland: Springer Nature; 2019. [PubMed] [Google Scholar]
- 15.Steinwart I, Christmann A. Support Vector Machines. Springer Science & Business Media; 2008. [Google Scholar]
- 16.Taylor RA, Pare JR, Venkatesh AK, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: A local big dataedriven, machine learning approach. Acad Emerg Med. 2016;23:269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong A, Young AT, Liang AS, Gonzales R, Douglas VC, Hadley D. Development and validation of an electronic health record-based machine learning model to estimate delirium risk in newly hospitalized patients without known cognitive impairment. JAMA Network Open. 2018;1:e181018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugarbaker DJ, Jaklitsch MT, Bueno R, et al. Prevention, early detection, and management of complications after 328 consecutive extrapleural pneumo-nectomies. J Thorac Cardiovasc Surg. 2004;128:138–146. [DOI] [PubMed] [Google Scholar]
- 19.Fried E, Weissman C, Sprung C. Postoperative sepsis. Curr Opin Crit Care. 2011;17:396–401. [DOI] [PubMed] [Google Scholar]
- 20.Akbilgic O, Langham MR Jr, Walter AI, Jones TL, Huang EY, Davis RL. A novel risk classification system for 30-day mortality in children undergoing surgery. PLoS One. 2018;13:e0191176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akbilgic O, Homayouni R, Heinrich K, Langham MR, Davis RL. Unstructured text in EMR improves prediction of death after surgery in children. Informatics. 2019;6:4. [Google Scholar]
- 22.Cabitza F, Rasoini R, Gensini GF. Unintended consequences of machine learning in medicine. JAMA. 2017;318:517–518. [DOI] [PubMed] [Google Scholar]
- 23.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21:578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming FJ, Kim MJ, Messing S, Gunzler D, Salloum R, Monson JR. Balancing the risk of postoperative surgical infections: A multivariate analysis of factors associated with laparoscopic appendectomy from the NSQIP database. Ann Surg. 2010;252:895–900. [DOI] [PubMed] [Google Scholar]
- 25.Prosperi M, Min JS, Bian J, Modave F. Big data hurdles in precision medicine and precision public health. BMC Med Inform Decis Mak. 2018;18:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


