Abstract
Background
Retention of agricultural bio-mass residues without proper treatment could affect the subsequent plant growth. In the present investigation, the co-cultivation of genetically engineered T. asperellum and B. amyloliquefaciens has been employed for multiple benefits including the enrichment of lignocellulose biodegradation, plant growth, defense potential and disease resistance.
Results
The Vel1 gene predominantly regulates the secondary metabolites, sexual and asexual development as well as cellulases and polysaccharide hydrolases productions. Overexpression mutant of the Trichoderma asperellum Vel1 locus (TA OE-Vel1) enhanced the activity of FPAase, CMCase, PNPCase, PNPGase, xylanase I, and xylanase II through the regulation of transcription regulating factors and the activation of cellulase and xylanase encoding genes. Further, these genes were induced upon co-cultivation with Bacillus amyloliquefaciens (BA). The co-culture of TA OE-Vel1 + BA produced the best composition of enzymes and the highest biomass hydrolysis yield of 89.56 ± 0.61%. The co-culture of TA OE-Vel1 + BA increased the corn stover degradation by the secretion of cellulolytic enzymes and maintained the C/N ratio of the corn stover amended soil. Moreover, the TA OE-Vel1 + BA increased the maize plant growth, expression of defense gene and disease resistance against Fusarium verticillioides and Cohilohorus herostrophus.
Conclusion
The co-cultivation of genetically engineered T. asperellum and B. amyloliquefaciens could be utilized as a profound and meaningful technique for the retention of agro residues and subsequent plant growth.
Keywords: Co-cultivation, T. asperellum, B. amyloliquefaciens, Vel1, Cellulase, Lignocellulose degradation
Background
Retention of agricultural bio-mass residues after harvest is an ideal strategy to improve sustainable agriculture [1]. Lately, onsite bio-degradation of crop residue has followed to maintain the soil fertility and to decrease the argumentative effects of residual burning in the agricultural field. Nonetheless, a few investigations showed that the retention of agricultural bio-mass residues affect soil properties and crop yields [2]. For instance, inadequate biomass degradation influences planting and seedling development, which augment the plant pests and pathogens [3, 4]. A promising solution for this issue is to inoculate the lignocellulolytic biomass degrading microbes into the soil. However, only a few studies have been focused on the onsite degradation of lignocellulolytic biomass using the microbes. Hence, it is the proper time to develop a technology for onsite biomass degradation, plant growth promotion and disease control.
Trichoderma asperellum has been considered as a beneficial fungus for plant growth, disease control, and the production of lignocellulolytic degrading enzymes [5]. Genetic engineering of the genes required for the regulation of lignocellulolytic enzyme synthesis of Trichoderma could provide an opportunity to improve both biomass degradation and plant growth. The expression of genes involved in lignocellulose degradation has been regulated by the co-ordination of numerous transcription factors [6]. Among them, Vel1 positively regulates the cellulase production [7]. Karimi Aghcheh et al. [8] studied that the knockout of Vel1 entirely declines the production and expression of cellulases related genes. In addition, the Vel1 gene also regulate the morphogenesis, secondary metabolites and mycoparasitism of the Trichoderma [9].
Besides, co-cultivation technology is an advantage to stimulate the synergistic expression of metabolic pathways of two microbes [10]. Through co-cultivation, microbes develop different mechanisms to use substrates either by symbiotic or antagonistic interactions. The co-cultivation activates the silent genes and thereby induce the enzyme production. Substantial improvements have been made on the co-cultivation technology by co-cultivating the genetically engineered microbes to increase the production [11]. This methodology widens the prospects for the biosynthesis of complex proteins to utilize the natural substrates such as lignocellulolytic biomass. In our previous study, we proved that the co-cultivation of B. amyloliquefaciens 1841 and T. asperellum GDFS1009 activated several genes and induced the production of secondary metabolites and enzymes, including cellulase [12, 13]. B. amyloliquefaciens used in the co-cultivation is a plant growth promoting rhizobacteria [12–14]. In our previous study, we observed that the role of T. atroviride Vel1 was enhanced by the B. amyloliquefaciens in the co-culture [9]. In light of the above findings, it has been anticipated that the application of the co-culture containing Vel1 overexpression Trichoderma asperellum and B. amyloliquefaciens into the soil could enhance the genetic regulation on the cellulase and hemicellulase production and improve the bio-degradation of lignocellulolytic biomass in soil and subsequently improve plant growth and disease resistance of the plants grown in the same soil.
To prove our hypothesis, we developed the co-cultivation of Vel1 over expressed mutant T. asperellum GDFS1009 and B. amyloliquefaciens 1841 (TA OE-Vel1 + BA) to improve the cellulase production using the combination of genetic engineering and co-cultivation technology. We showed that the crude enzyme produced by TA OE-Vel1 + BA enhanced the hydrolysis of corn stover biomass than the axenic culture. We further demonstrate that the co-culture of T. asperellum OE Vel1 mutant and B. amyloliquefaciens enhanced the in-vivo lignocellulolytic degradation, plant growth, defense potential and disease control.
Results
Increased cellulase production by genetically engineering of T. asperellum Vel1 gene
The strong promoter TrpC was used to improve the expression of Vel1. The over expression cassette containing TrpC promoter, Vel1 ORF and TrpC terminator was cloned into pCAMBIA1300 and transferred to T. asperellum using A. tumefaciens-mediated transformation (ATMT). The recombinant vector pCAMBIA1300 Vel1 OE and transformants are shown in Fig. 1. In total, 126 T. asperellum recombinant strains were obtained through the ATMT transformation. Among them, 5 recombinant strains showed higher filter paper activity (FPAse) than the wild type strain. The growth of T. asperellum recombinants on the cellulose containing medium was displayed in Additional file 1: Table S1. The fastest growing T. asperellum recombinants were selected among 5 transformants with the maximum cellulase activities.
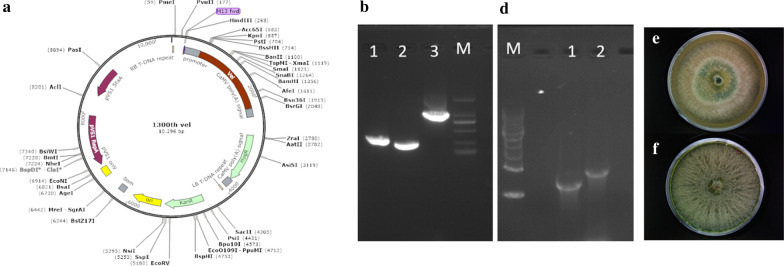
Fig. 1.
Overexpression of Vel1 gene in T. asperellum GDFS1009. a Schematic diagram of the constructed plasmid pCAMBIA1300 -Vel1. b PCR amplification of TrpC promoter, TrpC terminator and Vel1 ORF (1: TrpC promoter; 2: TrpC terminator; 3: Vel1 ORF; M DS 2000 marker) c PCR results for transformant identification (M: 1 KB marker; 1: Vel1 ORF; 2: over expression cassette containing TrpC promoter, Vel1 ORF and TrpC terminator). d The phenotypes of Wild type (TA) and e phenotype of Vel1 gene overexpress transformants grown on PDA plates
Influence of cellulase production by different samples
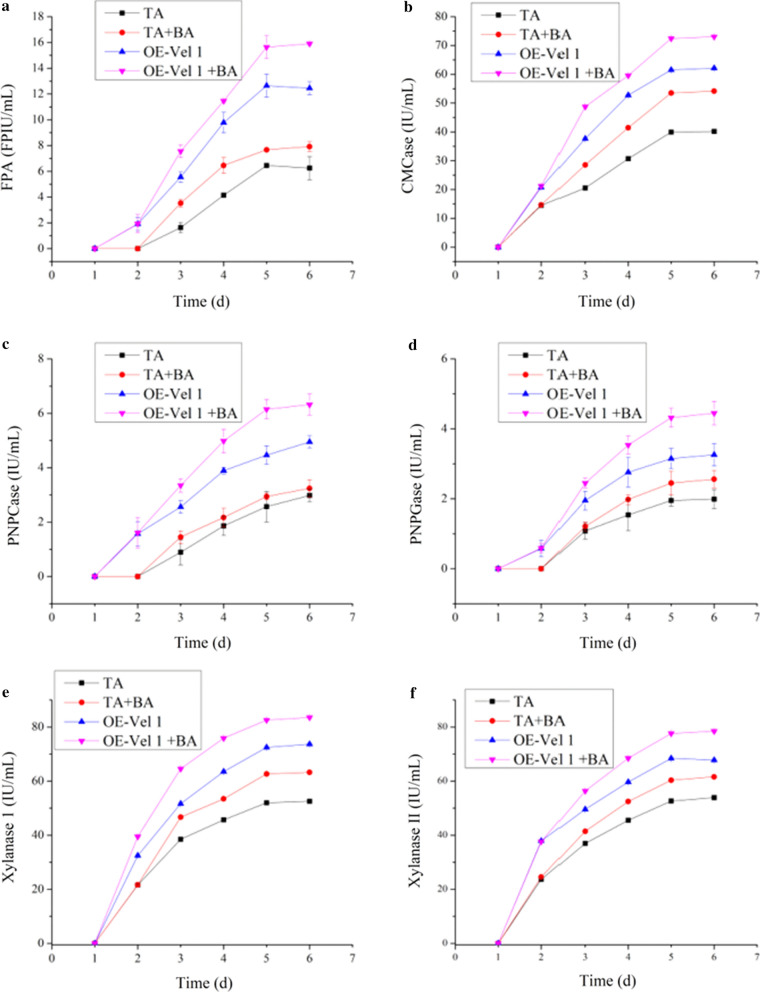
The four samples (Additional file 1: Fig. S1) such as the axenic culture of T. asperellum, co- culture of T. asperellum and B. amyloliquefaciens, the axenic culture of the TA OE-Vel1, and the co-culture of TA OE-Vel1 and B. amyloliquefaciens were analyzed to know the best sample to produce lignocellulolytic enzyme for the degradation of lignocellulolytic biomass. The genetically engineered T. asperellum (TA OE-Vel1) showed higher activities of filter paper activity (FPAase), carboxymethyl cellulase (CMCase), 4-nitrophenylcellobiosidase (PNPCase), p-nitrophenyl-β-D-glucopyranoside (PNPGase), xylanase I and xylanase II (Fig. 2) compare to the T. asperellum, yielding values of 12.45 ± 0.05 FPIU, 62.14 ± 0.34, 4.95 ± 0.23, 3.26 ± 0.32, 73.67 ± 0.37, and 67.8 ± 0.36 IU/mL after 6 days of fermentation, respectively. On the other hand, the axenic culture of B. amyloliquefaciens failed to produce all of these activities. This revealed that the Vel1 gene improved the activity of enzymes related to cellulose and hemicellulose hydrolysis. In-addition, B. amyloliquefaciens also induced the expression of lignocellulolytic enzymes [12]. The enzyme activities including FPAase, CMCase, PNPCase, PNPGase, xylanase I and xylanase II were enriched by the TA + BA compared to the TA (Fig. 2). The enzyme production might be owing to the different substrates. After 6 days of fermentation, the FPAase, CMCase, PNPCase, PNPGase, xylanase I and xylanase II activity of the co-culture of TA OE-Vel1 and B. amyloliquefaciens were 7.92 ± 0.04 FPIU, 54.16 ± 0.46, 3.24 ± 0.32, 2.56 ± 0.25, 63.23 ± 0.37, and 61.57 ± 0.43 IU/mL respectively. It was identified that the co-cultivation of T. asperellum and B. amyloliquefaciens was a fantastic combination to obtain the higher activity of FPAase, CMCase, PNPCase, PNPGase, xylanase I and xylanase II activity. For the first time, in this investigation, the co-culture of the genetically engineered T. asperellum and B. amyloliquefaciens was attempted to synthesize the highest enzyme production by linking the recombination technology and co-cultivation. As shown in Fig. 2, the FPAase, CMCase, PNPCase, PNPGase, xylanase I and xylanase II activity of the co-culture of TA OE-Vel1 and B. amyloliquefaciens were 15.91 ± 0.14 FPIU, 73.04 ± 0.16, 6.32 ± 0.39, 4.45 ± 0.32, 83.56 ± 0.43, and 78.45 ± 0.38 IU/mL respectively. These enzyme activities were considerably increased 1.1–1.3 fold than the TA OE-Vel1. Also, the enzyme activities were increased than the co-culture of T. asperellum and B. amyloliquefaciens. The results showed that this sample upgraded the synthesis of cellulase. Further, the results recommend that this kind of modified co-cultivation is more valuable than that of recombination technology and co-cultivation.
Fig. 2.
Comparison of cellulase production by different approaches.a FPase, b CMCase, c pNPcase, d pNPGase, e xylanase I, and f xylanase II. Values are the average of biological triplicates. Error bars represent the standard error
Influence of the transcription regulating genes, cellulase and xylanase encoding gene expression by different samples
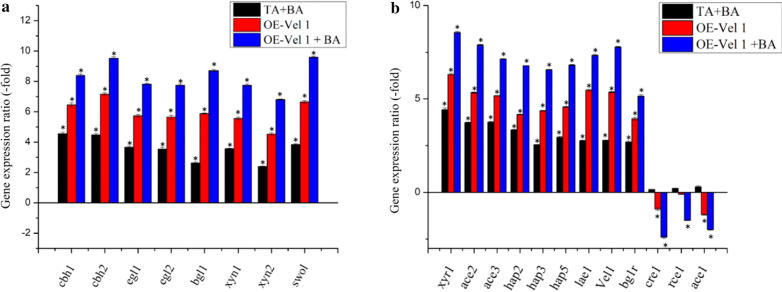
The expression pattern of cbh1, cbh2, egl1, egl2, bgl1, xyn1 and xyn2 were compared to know the regulatory level of cellobiohydrolases, endoglucanases, β-glucosidase and xylanase under different approaches. The expressions of these genes were strongly upregulated in the order of TA OE-Vel1 + BA > TA OE-Vel1 > TA + BA relative to the axenic culture of T. asperellum (Fig. 3). The expression level of the major cellulase gene, including cellobiohydrolases (cbh1 and cbh2) endoglucanases (egl1 and egl2), and β-glucosidase (bgl1) were similar to production of cellulolytic enzymes (Fig. 3a). The cellulase and xylanase encoding genes were coordinated by the group of transcription factors (TFs), including both inducer and inhibitors. The expression of cellulase regulatory genes by the Vel1 has been explored by studying the expression of nine positive regulators and three repressor genes. The stimulation of cellulase was initially verified by the transcription analysis of xyr1, ace II, and ace III, which are the most important inducers of cellulase and xylanase production [15–17]. As shown in Fig. 3b, the relative quantification of the xyr1, ace II, and ace III gene were upregulated by the over-expression of the Vel1 gene. The expression of xyr1, ace II and ace III were increased to 8.56, 7.98 and 7.14 fold, respectively in the TA OE-Vel1 + BA then the axenic culture of T. asperellum. Meanwhile, the relative transcription folds of these genes were only 4.4, 3.7 and 3.7 in TA + BA. In addition, the transcription factors BglR and Hap2/3/5 complex also positively regulated the cellulase and xylanase. The transcription level of the BglR and the Hap2/3/5 complex was also upregulated in co-ordination with other genes. Among the negatively transcription regulating factors, cre-1 is the carbon catabolite repressor, which completely inhibits the expression of the cellulase and xylanase genes [18]. Relatively, ace1 inhibits the C2H2 zinc finger and negatively regulate the genes encoding cellulase and xylanase. Also, the rce1 is the negative regulator by provoking Xyr1 [19]. To detect the influence of ace I, rce 1, and cre 1, the expression level of these genes was quantified. The results showed that these genes were downregulated with the overexpression of the Vel1 gene and TA OE-Vel1 + BA. The downregulation of ace I, rce 1, and cre 1 might be involved in the upregulation of cbh1, cbh2, egl1, egl2, and bgl1 through the overexpression of Vel1 gene [20].
Fig. 3.
Transcriptional changes of the genes coding cellobiohydrolases, endoglucanases, β-glucosidase, xylanase, accessory proteins and transcription factors of different approaches. a expression of cbh1, cbh2, egl1, egl2, bgl1, xyn1 xyn2 and swo1, b expression of transcription factors including XYR1, ACE II, ACE III, BglR, Hap2/3/5, ACEI, RCE1, and CRE1. Fold changes in TA + BA, TA OE-Vel1 and TA OE-Vel1 + BA were relatively compared to the wild type axenic culture (TA) at 72 h. Values are the average of biological triplicates. Error bars represent the standard error. Asterisks refer significant differences from monoculture of wild type strain (TA) (*p < 0.05, Student’s t test)
Hydrolysis of cellulosic biomass by the differently sourced cellulases
The pretreated corn stover was hydrolyzed using crude enzymes of different samples (Fig. 4). The enzymes obtained from the TA OE-Vel1 + BA showed maximum hydrolysis. This may be because of the high production of hydrolytic enzymes by the co-culture TA OE-Vel1 + BA. The over-expression of the Vel1 gene in T. asperellum enriched the cellulase production. At 72 h, TA OE-Vel1 + BA produced the hydrolysis yield of 89.56 ± 0.61%, which was greater than the co-culture of T. asperellum and B. amyloliquefaciens and the axenic culture of genetically engineered T. asperellum. The hydrolysis yield generated by the TA OE-Vel1 + BA and TA + BA was higher than the axenic culture of T. asperellum. However, TA OE-Vel1 + BA showed a better hydrolysis yield than TA + BA. This might be due to the reason of the activation of transcription factors and enzyme coding genes by the over expression of the Vel1 gene and by the co-cultivation with B. amyloliquefaciens as an inducer. Consequently, the enzyme production and hydrolysis yield were higher in the TA OE-Vel1 + BA sample.
Fig. 4.
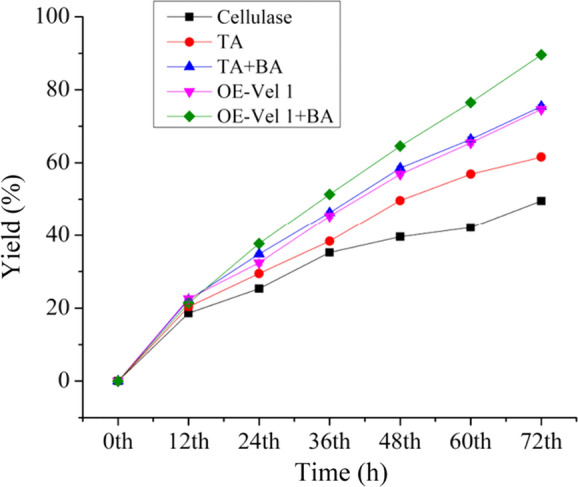
Hydrolysis of pretreated corn Stover by the crude enzyme of different approaches. Values are the average of biological triplicates. Error bars represent the standard error
Synergistic effect of corn stover amendments and microbial inoculation on lignocellulose degradation, plant growth and defense response
Throughout pot experiment, maize plants were grown healthy without any toxic symptoms. The axenic and co-culture were used to enrich plant growth in soil with and without the amendments of corn stover (Additional file 1: Fig. S2). The growth parameters of plants grown in soil samples amended with corn stover differed significantly (P ≤ 0.05) from the plants grown in non-amended soil as assessed by Duncan’s new multiple range test. TA OE-Vel1 + BA and TA + BA co-culture exhibited a remarkable effect on both plant growth and lignocellulolytic degradation. However, the plant growth in the untreated corn stover amended soil was reduced (Table 1). Overall, biodegradation of corn stover amended soil with TA OE-Vel1 + BA co-culture (T13) increased the shoot height and root height of the maize plants when compared to non-amended soil and other treatments (Table 1). Shoot height and root height of maize plants grown in corn stover amended soil treated with co-culture (T12) were 1.68 and 1.31 fold higher, respectively than control (T1). likewise, the shoot height and root height of maize plants grown in non-amended soil treated with co-culture (T5) and TA OE-Vel1 + BA co-culture (T6) were also higher than control (T1). Likewise, the fresh and dry biomass of shoot and root was also influenced by corn stover amendments treated with TA OE-Vel1 + BA (T13) and TA + BA (T12). The influence of TA OE-Vel1 + BA and TA + BA on the corn stover amendments improved the plant height and biomass of maize than all other treatment. The influence of TA OE-Vel1 + BA and TA + BA co-culture on disease index against Fusarium verticillioides and Cohilohorus herostrophus was also observed in both amended and non-amended soil compared to the control (T7 and T14). On the other hand, the disease index of T12 and T13 were 7 times higher than that of control (T14).
Table 1.
Effect of axenic, co-culture and modular co-culture of T. asperellum and B. amyloliquefaciens on the plant growth and biological control against Fusarium verticillioides and Cohilohorus herostrophus under both corn stover amended and non-amended soil in green house conditions
| Treatments | Length (cm) | Wet weight (gm) | Dry weight (gm) | Disease index (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Shoot | Root | Shoot | Root | Shoot | Root | Root rot | Leaf spot | |
| T1 | 41.22.41i | 22.54 ± 0.36f | 1.36 ± 0.04f | 0.03 ± 0.07f | 0.1 ± 0.02g | 0.01 ± 0.021e | ND | ND |
| T2 | 54.26.3f | 19.55 ± 0.6h | 1.57 ± 0.09f | 0.12 ± 0.08d | 0.17 ± 0.01f | 0.04 ± 0.06b | 30c | 30c |
| T3 | 51.98 ± 0.41g | 31.17 ± 0.41c | 2.51 ± 0.07e | 0.14 ± 0.01d | 0.33 ± 0.01d | 0.0344 ± 0.01c | 40d | 40d |
| T4 | 55.46 ± 0.45f | 25.8 ± 0.48c | 3.26 ± 0.05d | 0.12 ± 0.05d | 0.33 ± 0.09d | 0.04 ± 0.08c | 30c | 30c |
| T5 | 59.87 ± 0.5d | 25.51 ± 0.2e | 3.42 ± 0.07d | 0.15 ± 0.01d | 0.45 ± 0.07c | 0.05 ± 0.01b | 30c | 30c |
| T6 | 65.55 ± 0.4c | 28.99 ± 0.5d | 4.4 ± 0.09c | 0.15 ± 0.01d | 0.46 ± 0.02c | 0.05 ± 0.01b | 20b | 20b |
| T7 | 34.74 ± 0.20j | 17.7 ± 0.29 l | 2.5 ± 0.11e | 0.04 ± 0.07ef | 0.25 ± 0.01e | 0.024 ± 0.05d | 60e | 60e |
| T8 | 43.94 ± 0.56h | 21.02 ± 0.50g | 2.72 ± 0.13e | 0.08 ± 0.02e | 0.28 ± 0.01e | 0.02 ± 0.01d | ND | ND |
| T9 | 55.37 ± 0.37f | 22.62 ± 0.47f | 3.51 ± 0.15d | 0.14 ± 0.015d | 0.44 ± 0.01c | 0.05 ± 0.01eb | 30d | 30d |
| T10 | 55.89 ± 0.37e | 24.64 ± 0.51e | 3.53 ± 0.14d | 0.14 ± 0.06d | 0.33 ± 0.17d | 0.05 ± 0.01b | 40b | 40b |
| T11 | 68.88 ± 0.3b | 35.55 ± 0.46b | 5.68 ± 0.08c | 0.22 ± 0.02c | 0.44 ± 0.01c | 0.05 ± 0.03b | 20a | 20a |
| T12 | 69.53 ± 0.36b | 29.62 ± 0.51d | 5.56 ± 0.15b | 0.34 ± 0.15b | 0.59 ± 0.02b | 0.07 ± 0.016a | 10a | 10a |
| T13 | 74.98 ± 0.57a | 44.94 ± 0.28a | 6.54 ± 0.14a | 0.45 ± 0.017a | 0.64 ± 0.011a | 0.07 ± 0.0017a | 10a | 10a |
| T14 | 42.2 ± 0.3j | 16.3 ± 0.42J | 2.618 ± 0.1e | 0.08 ± 0.004e | 0.25 ± 0.001e | 0.03 ± 0.001c | 70d | 70d |
Results are average of five replicates for each treatment; the values given are the standard error of the mean. Different superscripts in the same column are significantly different (P < 0.05) based on the ANOVA
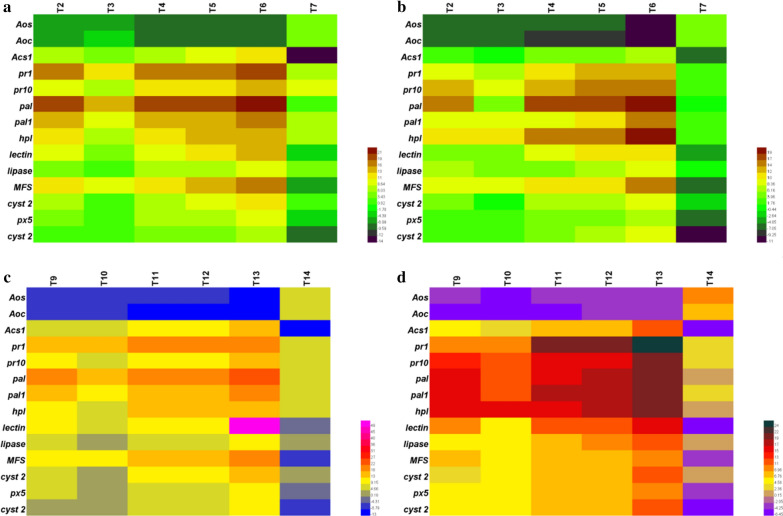
To further understand the plant response to the corn stover amended soil inoculated with co-culture, we studied the induction of defense-related gene expression using semi-quantitative reverse transcriptase (RT)-PCR (Fig. 5). The actin gene has been used as an internal control. Fourteen genes related to different plant defense pathways were selected: allene oxide synthase (AOS), allene oxide cyclase (AOC) (jasmonic acid), 1-aminocyclopropane-1-carboxylic acid synthase (ACS) (ethylene), pathogenesis-related protein 1 (PR1) and pathogenesis-related protein 10 (PR10) (systemic acquired resistance), phenylalanine ammonia-lyase (PAL) and (PAL1) (salicylic acid), hydroperoxide lyase (HPL), lectin, lipase, multiflux efflux synthase (MFS), cystatin ii proteinase inhibitor (Cyst2), peroxidase (PX5), cystatin proteinase inhibitor (Cyst) and thiolase (other defense-related genes). The regulation of these genes by axenic, TA OE-Vel1 + BA and TA + BA co-culture were examined locally at the root and systematically at leaves. The defense gene expression against Fusarium verticillioides and Cohilohorus herostrophus on maize roots and leaves are shown in Fig. 5. The AOS and AOC gene was upregulated in both roots and leaves of plants infected with Fusarium verticillioides and Cohilohorus herostrophus, respectively, but it was gradually reduced in the plants treated with TA OE-Vel1 + BA (T6 and T13), TA + BA (T5 and T12) co-culture, and axenic culture (T2, T3, T4, T9, T10 and T11) in both amended and non-amended soil. The upregulation of AOS and AOC revealed that the plants were highly infected by the Fusarium verticillioides and Cohilohorus herostrophus in T7 and T14. Based on the expression profiles, the ACS genes was highly induced by TA OE-Vel1 + BA application in the root of T6 and T13 (Fig. 5a and c). Followed by the co-culture of TA + BA induced the ACS genes of maize plants. Interestingly, the expression of these genes was downregulated in T7 and T14. TA OE-Vel1 + BA and TA + BA inoculated maize plants expressed the defense genes locally on the plant root and systematically in leaves as a response of Cohilohorus herostrophus (Fig. 5b and d). The expression of systemic acquired resistance pathway-related genes such as PR1 and PR10 in roots and leaves of maize plants inoculated with TA OE-Vel1 + BA and TA + BA co-culture were upregulated than other treatments of both amended and non-amended soil. The PAL and PAL1 were upregulated in the following order T13 > T12 > T11 > T9 > T10 and T6 > T5 > T4 > T2 > T3 in both corn stover amended and non-amended soil, respectively. The upregulation of the genes such as HPL, lectin, lipase, MFS, Cyst2, PX5, Cyst, and thiolase was also enhanced by the TA OE-Vel1 + BA and TA + BA co-culture compared to the control.
Fig. 5.
Induction of defense gene expression in maize roots and shoots by the axenic, co-culture and modular co-culture of T. asperellum and B. amyloliquefaciens against Fusarium verticillioides and Cohilohorus herostrophus. Heat map profile of defense gene expression in a root and b leaves of non-amended soil were showed in terms of fold changes compared to the control (T1). Heat map profile of defense gene expression in c root and d leaves of corn stover amended soil were showed in terms of fold changes compared to the control (T8). Results are average of five replicates for each treatment; the values given are the standard error of the mean
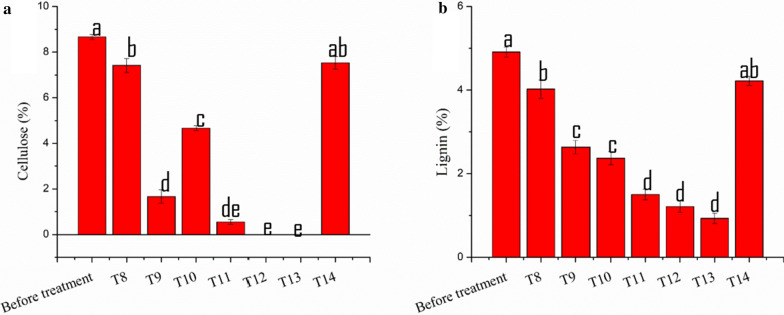
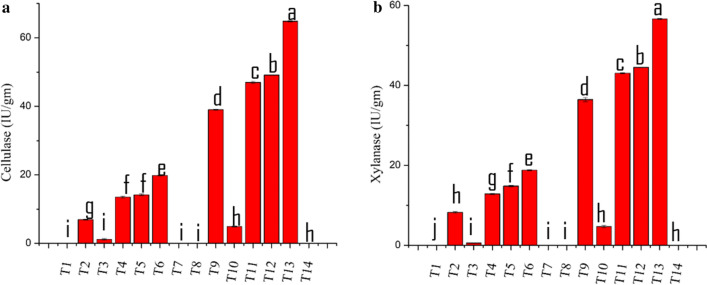
SOM, TOC, TN and C/N content in the soil of each treatment are shown in Table 2. There were no notable changes in the content of soil SOM, TOC, TN and C/N in T1 to T7, which was not amended with the corn stover. Amendment of corn stover increased the soil SOM and TOC in the treatment T8, T14, and T10, which has not been treated with the Trichoderma. Biodegradation of corn stover amendment treated with TA OE-Vel1 + BA and TA + BA co-culture reduced the SOM and TOC content of the soil. At the end of the experiments, the C/N ratio of the TA OE-Vel1 + BA and TA + BA treated corn stover amendment soil was rapidly decreased and it was closer to the standard value compared soil treated with axenic culture and control. In comparison with all other treatments, T13 and T12 treatment showed better degradation. In connection to the improvement of the C/N ratio, the cellulose content of the T13 and T12 was completely reduced by the TA OE-Vel1 + BA and TA + BA co-culture (Fig. 6a). Similarly, the lignin content was also reduced in T13 and T12 compared to other treatments and before treatment (Fig. 6b). The cellulase and xylanase content of the T13 and T12 was increased compared to other treatments (Fig. 7a and b).
Table 2.
Effect of axenic, co-culture and modular co-culture of T. asperellum and B. amyloliquefaciens on the soil chemical properties such as SOM, TOC, TN and C/N ratio on both corn stover amended and non-amended soil
| Treatments | SOM | TOC | TN | C/N |
|---|---|---|---|---|
| T1 | 358.11 ± 034d | 716.23 ± 0.68d | 31.25 ± 0.05ab | 22.91 ± 0.02e |
| T2 | 348.65 ± 0.72f | 697.3 ± 0.45f | 31.24 ± 0.06ab | 22.32 ± 0.02f |
| T3 | 353.03 ± 0.36e | 706.07 ± 0.72e | 31.16 ± 0.09ab | 22.65 ± 0.04ef |
| T4 | 347.49 ± 0.19f | 694.98 ± 0.38f | 31.26 ± 0.03ab | 22.22 ± 0.07f |
| T5 | 353.12 ± 0.26e | 706.24 ± 0.52e | 31.42 ± 0.04a | 22.47 ± 0.02ef |
| T6 | 346.71 ± 0.26f | 693.42 ± 0.52f | 31.68 ± 0.15a | 22.88 ± 0.09fg |
| T7 | 348.43 ± 0.32f | 696.86 ± 0.65f | 31.26 ± 0.04ab | 22.28 ± 0.05f |
| T8 | 428.7 ± 0.21a | 857.4 ± 0.43a | 27.85 ± 0.4f | 30.79 ± 0.4a |
| T9 | 347.66 ± 0.30f | 695.33 ± 0.60f | 29.26 ± 0.14de | 23.76 ± 0.1d |
| T10 | 377.8 ± 0.35c | 755.6 ± 0.7c | 26.07 ± 0.19 g | 28.98 ± 0.2b |
| T11 | 342.5 ± 0.6g | 685 ± 0.30g | 30.75 ± 0.28bc | 22.27 ± 0.09f |
| T12 | 307.73 ± 0.25i | 615.46 ± 0.51i | 28.85 ± 0.28e | 21.33 ± 0.2g |
| T13 | 326.78 ± 0.30h | 653.56 ± 0.60h | 30.32 ± 0.11c | 21.55 ± 0.05g |
| T14 | 419.15 ± 0.31b | 838.3 ± 0.62b | 29.57 ± 0.29d | 28.35 ± 0.2c |
Results are average of five replicates for each treatment; the values given are the standard error of the mean. Different superscripts in the same column are significantly different (P < 0.05) based on the ANOVA
Fig. 6.
Effect of axenic, co-culture and modular co-culture of T. asperellum and B. amyloliquefaciens on the hydrolysis of cellulose (a) and lignin (b) content of corn stover amended soil. Results are average of five replicates for each treatment; the values given are the standard error of the mean. Different letters on the parentheses are significantly different (P ≤ 0.05)
Fig. 7.
Effect of axenic, co-culture and modular co-culture of T. asperellum and B. amyloliquefaciens on the production of cellulase and xylanase of corn stover amended and non-amended soil. Results are average of five replicates for each treatment; the values given are the standard error of the mean. Different letters on the parentheses are significantly different (P ≤ 0.05)
Discussion
The establishment of genetic engineering technology has allowed the production of an extensive array of bio-products by exploiting several microbes as biocatalysts [21]. Even though considerable application of bacterial or fungus cultures has been well studied, genetic engineering technology has satisfied the growing need for complex biosynthesis enzymes. On the other hand, co-cultivation of microorganism has been effectively used to convert the complex substrates into the simple biomolecules for the industrial applications [22–25]. In recent times, the genetically engineered microbial strains are used in co-cultivation to enhance the specific metabolites through metabolic pathways [11]. In connection with the point previously mentioned, integration of co-cultivation and genetic engineering compromises the biosynthesis of enzymes required for the hydrolysis of agricultural biomass onsite to improve sustainable agriculture. This new approach provides a space to develop new engineered biosynthetic pathways in co-cultivation environment [26].
In the present study, we investigated the role of T. asperellum Vel1 gene on production of the carbohydrate active enzymes, including FPAase, CMCase, PNPCase, PNPGase, xylanase I and xylanase II. The Carbohydrate Active Enzyme (CAZYmes) refers to the enzymes, which are required to hydrolyze the polysaccharides. Vel1 is a comprehensive regulator of the numerous fungi, especially Trichoderma. Interestingly, Vel1 is involved in the regulation of asexual sporulation, secondary metabolism, and mycoparasitism of Trichoderma [29]. Karimi Aghcheh et al., [8] confirmed that the expression of cellulase requires Vel1 gene. Similarly, we observed that the overexpression of the Vel1 gene increased the production of cellulase. Our results further showed that the cellulases and xylanases are co-regulated by the Vel1. The enzyme activity of the different approaches revealed that it coincides with the expression of genes such as cellulase and xylanase encoding genes and transcription regulating factors. The expression pattern of cbh1, cbh2, egl1, egl2, bgl1, xyn1 and xyn2 was analyzed to know the regulatory level of cellobiohydrolases, endoglucanases, β-glucosidase and xylanase. The results assumed that overexpression of the Vel1 gene positively influenced the expression of these genes to increase the activity of cellobiohydrolases, endoglucanases, β-glucosidase, and xylanase.
The regulation of cellulase and hemicellulase gene expression of the Trichoderma is extremely synchronized by the transcription regulatory factors [20]. In Trichoderma, ten transcription factors were identified as important for regulation of cellulase gene expression [20]. Among them, XYR1, ACE II and ACE III are significant transcriptional regulators. [15–17, 27]. Also, the HAP2/3/5 complex stimulates the open chromatin structure required for the transcription stimulation [20]. BglR stimulates the β-glycosidases genes. Interestingly, in our study, these genes were upregulated in co-ordination with the Vel1 gene. ACE1 and RCE1 are the transcriptional repressor of cellulase gene expression [19, 28]. The carbon catabolic repression has been regulated by the negative regulation of CRE1 gene [18]. Transcript data of ACEI, RCE1, and CRE1 of the present study were downregulated in the TA OE-Vel1 strain (Fig. 3). Similarly, the co-cultivation (TA + BA and TA OE-Vel1 + BA) downregulated the expression of ACEI, RCE1, and CRE1. The decreased transcription of ACEI, RCE1, and CRE1 by co-cultivation could positively upregulated expression of cbh1, cbh2, egl1, egl2, and bgl1. Overall, the present investigation demonstrated that the regulatory action of the Vel1 on the production of CAZymes. The additional role of the Vel1 gene concerning the synthesis of cellulolytic enzymes is fascinating due to the interaction of several transcription regulatory genes.
However, interaction between the Vel1 gene, CAZymes and transcription regulating factors is not acquainted so far. Conversely, the interactions between sporulation and cellulase production have been proven in T. reesei [8]. In this connection, it is exciting that several cellulase related genes and its transcription regulatory genes are clustered in the genome of T. asperellum. It had proved that these genes were positively regulating cellulase and xylanase production. Further, the results proved that the Vel1 gene is a superior regulatory gene to synchronize the expression of cellulases and other related transcription factors [9, 29]. Moreover, we observed that the co-cultivation is also positively influencing this synchronization.
The co-cultivation with B. amyloliquefaciens offered differentiated cellular environs to induce the genes involved in metabolic pathways [12, 13]. The metabolic pathway might comprise several enzymes, and these properties could vary depending on the circumstance. Axenic cultivation offers an identical environment and it might not be appropriate to express all genes. Similar to our study, the co-cultivation of an engineered Escherichia coli and Saccharomyces cerevisiae also enhanced cellulase production [30]. In addition to that, modular co-culture engineering decreases the intrusion of biosynthesis of other metabolites and induce the specific genes. In the present study, B. amyloliquefaciens induced the gene expression of cellulase and xylanase encoding genes.
The enzymes synthesized by the different samples generated more glucose than the commercial cellulase. It showed that on-site enzyme production has several advantages including cost-effective, CAZymes composition and concentration [31]. Further, it improved the hydrolysis efficiency of pre-treated corn stover. Consequently, the samples applied for cellulase synthesis and hydrolysis of pre-treated corn stover were compared and shown in Additional file 1: Table S2. The enzymes produced by TA OE-Vel1 + BA co-culture (sample 4) were confirmed to be efficient and cost-effective to produce glucose from the corn stover. The co-cultivation of genetically engineered T. asperellum and B. amyloliquefaciens is a promising method to hydrolyse the lignocellulolytic biomass for agricultural purposes. Finally, the co-culture of TA OE-Vel1 + BA increased the consumption of complex substrates and enhanced the hydrolysis rate of pretreated corn stover. Furthermore, the co-culture of the genetically engineered T. asperellum and B. amyloliquefaciens have a better impact on the colonization of agro residues due to the multiple functions along with the maximum production of hydrolysis enzymes. Hence this has been used to recycle the crop residues in pot based experiments for the betterment of soil fertility, plant growth and disease resistance.
Retention of biomass residue after harvest is an important module of sustainable agriculture practice. Presently, in China the practice of maize retention has been followed to improve the soil properties and yield of the subsequent crop [32]. The accumulation of crop residues lacking appropriate soil management could lead to problems including temporary loss of nitrogen and moisture. Hence, new technology for the retention of crop residue is required to improve the growth of a subsequent crop. The application of microbes into the soil after retaining the maize residues showed the benefits on the soil quality and subsequent plant growth. The application of Streptomyces microflavus and Aspergillus niger enriched the degradation of lignocellulose biomass and stimulates soil nutrient availability for the subsequent plant growth [32, 33]. Hence, 25% of maize residue was incorporated into the soil and treated with the axenic, TA OE-Vel1 + BA and TA + BA co-culture to enrich the biomass degradation for subsequent plant growth, defense potential and disease control. TA OE-Vel1 + BA and TA + BA co-culture increased the residue decomposition through the production of lignocellulolytic enzymes and thereby it increased the nutrients content of the soil.
In general, the incorporation of maize residues increases the soil C/N ratio and reduce the availability of nitrogen to plants [34]. Hence, some researchers suggested to incorporate the inorganic nitrogen into the soil to maintain the optimal C/N for plant growth [34]. In the present study the TA OE-Vel1 + BA and TA + BA co-culture reduced the C/N ratio closer to the standard value compared to the axenic culture without any supplementation of inorganic nitrogen. Besides, few studies showed that the retention of maize residues in the field increased the wheat growth, grain filling and yield compared to the non-retention field [32, 35]. Similarly, the plant growth parameters are higher in the corn stover amended soil compared to the non-amended soils. The improvement in enzyme production by the TA OE-Vel1 + BA and TA + BA co-culture significantly correlated with the soil quality and plant growth (Additional file 1: Table S3). The Vel1 overexpression gene upregulated the expression of plant defense gene. Mukherjee and Kenerley (2010) observed that the knockout of Vel1 gene did not mycoparasite the pathogens R. solani and P. ultimum due to the downregulation of secondary metabolite producing genes. The present investigations revealed that the overexpression of the Vel1 gene might induce the secondary metabolite producing genes of Trichoderma and mycoparasitism to upregulate the defense potential of plants. In addition, the B. amyloliquefaciens present in the co- culture is an another plant growth promoting and biocontrol agent. This B. amyloliquefaciens not only induced the production of Trichoderma enzymes and corn stover degradation, but it was also involved in the plant growth and biocontrol efficiency when it was applied in the soil. Previously, Karuppiah et al. [12] evidenced that the co-culture stimulated the defense potential of plants against pathogens. Likewise, we observed that the TA OE-Vel1 + BA and TA + BA improved the plant defense gene expression than the axenic cultures. Overall, the in-vivo study revealed that the amendment of corn stover with the TA OE-Vel1 + BA and TA + BA co-culture of T. asperellum and B. amyloliquefaciens into soil increased the production of soil enzymes and cornstover degradation. The increment of soil organic carbon and maintenance of the C/N ratio increased the soil fertility and maize growth than the non-amended soil. The application of these plant growth and biocontrol microbes in the form of co-culture not only increased the plant growth but also enhancing the expression of defense genes against Fusarium verticillioides and Cohilohorus herostrophus pathogens and reduced the disease index compared to other treatments. This residue management technology could be useful to retain the agricultural biomass after harvesting to preserve the soil structure, and productivity and it can be an option of eco-friendly technology for the effective utilization of plant residue.
Conclusions
The Vel1 gene expression is increased with the help of the TrpC promotor. The enzyme activity such as FPAase, CMCase, PNPCase, PNPGase, xylanase I and xylanase II were increased by the over expression of Vel1. It also increased the hydrolysis of pretreated corn stover (1.8 fold). The present study confirmed that the regulation of cellulase and xylanase was coordinated by the regulation of several transcription factors (Fig. 3). Further, it was also confirmed that Vel1 gene co-ordinate the regulation of the transcription factor to induce the cellulase and xylanase encoding genes for the maximum production of enzymes. The co-cultivation of genetically modified T. asperellum and B. amyloliquefaciens increased (2.1 fold) the production of cellulase and xylanase to hydrolyse the cellulolytic biomass. Our results revealed that the treatment of TA OE-Vel1 + BA on the corn stover amended soil increased the soil lignocellulolytic enzyme activity and corn stover degradation. The TA OE-Vel1 + BA positively influenced the growth of maize plants and disease resistance against Fusarium verticillioides and Cohilohorus herostrophus. The co-cultivation of genetically engineered T. asperellum and B. amyloliquefaciens could be used as a novel and advanced technique to return the crop residue into the field to improve the soil fertility along with the plant growth and disease resistance. This technique could be an eco-friendly technology for efficient consumption of crop residue and plant growth in the field for the next level of sustainable agriculture.
Methods
Microbial strains
The T. asperellum GDFS1009 (CGMCC NO. 9512) and B. amyloliquefaciens 1841 (CGMCC NO. 15465) were acquired from our culture collection facility and Sichuan University, respectively and stored in the CGMCC, China. T. asperellum GDFS1009 and B. amyloliquefaciens were cultured on potato dextrose (PD) and luria bertani (LB) agar, respectively. Agrobacterium tumefaciens AGL-1 has been used for the agrobacterium mediated transformation of pCAMBIA1300 Vel1 OE vector into T. asperellum. Fusarium verticillioides and Cohilohorus heterostrophus were sourced from our laboratory microbial collection center and used as a target pathogen to induce the root rot and leaf spot diseases in maize plants.
Engineering of Vel1 gene overexpression
A 2.17 kb of DNA portion comprising the TrpC promoter, and TrpC terminator from the pCAMBIA1300 vector and Vel1 ORF (pCAMBIA1300) of T. asperellum GDFS1009 was over-lapped using PCR. This gene cassette was introduced into pCAMBIA1300 to generate the pCAMBIA1300 Vel1 OE via Hieff CloneTM One Step Cloning Kit. The T. asperellum Vel1OE strains were attained by transforming the pCAMBIA1300 Vel1OE into T. asperellum GDFS1009. The mutants were confirmed by PCR. Primers used for the engineering of Vel1 gene are shown in Additional file 1: Table S4.
Cellulase production by the mono and co-culture
For the mono-culture, T. asperellum (TA) (106/mL spores) or the recombinant T. asperellum (TA OE-Vel1) (106/mL spores) were cultured in the minimal broth containing 2% avicel as described by [36]. For co-cultivation, 0.1% of the B. amyloliquefaciens 1841 (BA) was added into the 48th hour T. asperellum and recombinant T. asperellum pre-culture medium and named as TA + BA and TA OE-Vel1 + BA respectively. Further, it was cultured in the incubator shaker at 30◦C until 72h.
Enzyme activity
Filter paper activity (FPAase) was analyzed based on the standard method explained by Ghose [37]. The endoglucanase (CMCase) activity was tested according to Bailey and Nevalainen [38]. Cellobiohydrolase (pNPCase), and β-glucosidase (pNPGase) were assessed by the methodology of Zhang et al. [36]. Xylanase I and II were assessed at pH 3.7 and 5.0 respectively according to the method of Sticker et al. [39]. All testing was executed in biological triplicates.
Gene expression analysis
RNA isolation and cDNA synthesis were carried out as described by Karuppiah et al. [11]. The expression of cellulose and hemicellulose hydrolysis related genes and transcription regulators were estimated as described by Karuppiah et al. [11]. The real-time data were normalized with the 18S rRNA gene by means of the 2−ΔΔCt method. All tests were carried out in three independent experiments in triplicate. The list of primers used has given appears in Additional file 1: Table S5.
Pretreatment and hydrolysis of corn stover
The corn stover was pretreated by the method of Tsegaye et al. [40]. The crude enzymes of four different samples were used to hydrolyze the lignocellulose biomassas suggested by Zhang et al. [36]. The glucose content of the hydrolysate was estimated using HPLC (Waters 410, Waters, MA) and eluted with 0.004 M H2SO4 at a flow rate of 0.6 mL/min.
Synergistic effect of corn stover amendments and microbial inoculation on lignocellulose degradation, plant growth and defense response
Experimental setup
The pot trial was conducted in a greenhouse in 15 cm diameter pots comprising of horticulture soil amended with or without 25 mg corn stover g−1 soil (25%). The corn stover was air-dried, finely powdered, and uniformly mixed into the soil. The pot trial was comprised of 14 different experiments with triplicates. The treatments were: T1- control, T2-TA, T3-BA, T4- TA OE-Vel1, T5-co-culture of TA and BA, T6- co-culture of TA OE-Vel1 and BA, T7- pathogen, T8- 25% corn stover amendment (control), T9- 25% corn stover amendment + TA, T10- 25% corn stover amendment + BA, T11- 25% corn stover amendment + TA OE-Vel1, T12- 25% corn stover amendment + co-culture of TA and BA, T13- 25% corn stover amendment + co-culture of TA OE-Vel1and BA, T14- 25% corn stover amendment + pathogen. The axenic and co-cultures were evenly applied in 25% corn stover amended and non-amended soil. All experimental pots were watered everyday untill 15 days. After that, maize seeds were seeded and grow until 30 days, 1 × 106 conidia ml−1 of Fusarium verticillioides and Cohilohorus heterostrophus was applied into the pot soil and leaves, respectively on T2—T7 and T9—T14. The plant growth and disease index were evaluated according to the method of Karuppiah et al. [11] and Wang et al. [41]. The disease index was accessed based on the leaf spot and root rot disease of each treatment using the grading technique from grade 0 to grade 5. 0: no disease; 1: no more than 10%; 2: 11–30%; 3: 31–50%; 4: 51–70%; and 5: > 70%. the disease index was calculated as follows: DI = Σ (sum of plants in each disease stage × grade value)/sum of all plants × uppermost grade × 100) [41].
Expression of defense genes by the axenic and co-culture in both corn stover amended and non-amended soils against the pathogens were analyzed using qPCR. Roots and leaves of each treated maize plants were individually collected and the RNA was extracted with the Vazyme fastpure plant total RNA isolation kit. The cDNA was synthesized using Vazyme HiScript III 1st Strand cDNA Synthesis Kit (+ gDNA wiper). The amplification was performed as described previously by [13] using Roche light cycler 96. The Actin gene was used to normalize the gene expression. Data are expressed using the 2−ΔΔCT method.
Determination of soil organic matter, total organic carbon, lignin, cellulose, and C/N ratio.
Soil organic matter (SOM) was estimated gravimetrically by the loss‐on‐ignition technique and expressed as mg −g dry weight as suggested by Danise et al. [42]. Total organic carbon (TOC) was calculated using the results of SOM with 2 as a conversion factor from SOC to TOC [42]. Total nitrogen (TN) was estimated using the Kjeldahl method [43]. C:N ratio was calculated using the results TOC and TN. Lignin and cellulose content of the soil was estimated using the Updegra and acetyl‐bromide spectrophotometric technique, respectively as suggested by Danise et al. [42].
Determination of cellulase and xylanase activity in soil
Humic materials of the soil samples were removed using active carbon and PolychIal AT according to the method of Kanazawa and Miyashita [44]. Soil cellulase activity was determined with the modification of Kanazawa and Miyashita [44] method. Briefly, 10 gm of soil and 200 mg of avicel were dissolved with phosphate buffer in a conical flask and incubated for 24 h at 28 °C. After incubation, the samples were centrifuged and reducing sugar content of the 0.5 m1 of the supernatants was estimated using 3 ml of DNS reagent. To determine the soil xylanse activity, 10 gm of soil and 200 mg of xylan were dissolved with phosphate buffer in a conical flask and incubated for 24 h at 28 °C. After incubation, the samples were centrifuged, and reducing sugar content was measured as described above [45]. One unit of cellulase and xylanse activity is explained as the quantity of enzyme requisite to liberate 1 µmol reducing sugars per gram of soil.
Statistical analysis
The graphs were plotted using origin 6.0. Results displayed were mean of triplicate values through standard error. The Student’s T-test was conducted to differentiate the gene expression among the control and test samples. Two-way ANOVA, post hoc LSD, and Duncan test were used to determine the statistical significance between each samples through SPSS 2.0. Pearson’s correlation was performed between the variables of all treatments used in pot experiments using SPSS 2.0.
Statistical significances were determined using one-way analysis of variance (ANOVA) and post hoc Duncan multiple range tests (DMRTs).
Supplementary Information
Additional file 1: Table S1. The growth and cellulase production of Trichoderma asperellum recombinants on CBH screening medium. Table S2. Comprehensive information about the enzyme activity and corn stover hydrolysis of different samples. Table S3. Correlation plot of Pearson correlation coefficient for all measured variables in pot experiments. ** Correlation is significant at the 0.01 level (2-tailed). *Correlation is significant at the 0.05 level (2-tailed). Table S4. Primers used in construction of over-expression strains. Table S5. Sequences of the primers used for the real-time PCR. Figure S1. Work flow of the present study. Figure S2. Table 1 Effect of axenic, co-culture and modular co-culture of T. asperellum and B. amyloliquefaciens on the plant growth and biological control against Fusarium verticillioides and Cohilohorus herostrophus under both corn stover amended and non-amended soil in green house conditions. (T8-T14) cornstover amended soil; (T1-T7) cornstover non amended soil.
Acknowledgements
Not applicable
Abbreviations
- ACS
1-Aminocyclopropane-1-carboxylic acid synthase
- AOC
Allene oxide cyclase
- AOS
Allene oxide synthase
- BA
Bacillus amyloliquefaciens
- bgl1
β-Glucosidase
- C/N
Cabon/Nitrogen
- cbh1
Cellobiohydrolases 1
- cbh2
Cellobiohydrolases 2
- CMCase
Endoglucanase
- Cyst
Cystatin proteinase inhibitor
- Cyst2
Cystatin ii proteinase inhibitor
- egl1
Endoglucanases 1
- egl2
Endoglucanases 2
- FPAase
Filter paper activity
- HPL
Hydroperoxide lyase
- MFS
Multiflux efflux synthase
- PAL1
Phenylalanine ammonia-lyase (PAL)
- PNPCase
Cellobiohydrolase
- PNPGase
β-Glucosidase
- PR1
Pathogenesis-related protein 1
- PR10
Pathogenesis-related protein 10
- PX5
Peroxidase
- SOM
Soil organic matter
- TA
Trichoderma asperellum
- TA OE-Vel1
Overexpression mutant of Trichoderma asperellum Vel1 locus
- TA OE-Vel1 + BA
Co-culture of TA OE-Vel1 and BA
- TA + BA
Co-culture of Trichoderma asperellum and Bacillus amyloliquefaciens
- TN
Total nitrogen
- TOC
Total organic carbon
- Vel1
Velvet 1
- xyn1
Xylanase 1
- xyn2
Xylanase 2
Authors’ Contributions
VK and JC conceived and designed the experiments. VK carried out the main work, analyzed the data and drafted the manuscript. LZ, HL, and MV participated in the research. JC supervised the work. JC and MV revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0200403; 2017YFD0201108), National Natural Science Foundation of China (31672072; 31872015); The Key Project of Science and Technology of Shanghai (18391902400); Earmarked Fund for China Agriculture Research System (CARS-02).
Availability of data and materials
All data generated during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-021-01540-3.
References
- 1.Sharma S, Thind HS, Yadvinder S, Sidhu HS, Jat ML, Parihar CM. Effects of crop residue retention on soil carbon pools after 6 years of rice–wheat cropping system. Environ Earth Sci. 2019;78:296. doi: 10.1007/s12665-019-8305-1. [DOI] [Google Scholar]
- 2.Guo L, Zheng S, Cao C, Li C. Tillage practices and straw-returning methods affect topsoil bacterial community and organic C under a rice-wheat cropping system in central China. Sci Rep. 2016;6:33155. doi: 10.1038/srep33155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichel R, Wei J, Islam MS, Schmid C, Wissel H, Schröder P, Schloter M, Brüggemann N. Potential of wheat straw, spruce sawdust, and lignin as high organic carbon soil amendments to improve agricultural nitrogen retention capacity: an incubation study. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu X. A meta-analysis of the effects of crop residue return on crop yields and water use efficiency. PLoS ONE. 2020;15:e0231740. doi: 10.1371/journal.pone.0231740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian Y, Zhong L, Sun Y, Sun N, Zhang L, Liu W, Qu Y, Zhong Y. Enhancement of cellulase production in trichoderma reesei via disruption of multiple protease genes identified by comparative secretomics. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.02784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Druzhinina I, Kubicek C. Genetic engineering of Trichoderma reesei cellulases and their production. Microb Biotechnol. 2017;10(6):1485–1499. doi: 10.1111/1751-7915.12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu K, Dong Y, Wang F, Jiang B, Wang M, Fang X. Regulation of cellulase expression, sporulation, and morphogenesis by velvet family proteins in Trichoderma reesei. Appl Microbiol Biotechnol. 2016;100:769–779. doi: 10.1007/s00253-015-7059-2. [DOI] [PubMed] [Google Scholar]
- 8.Karimi Aghcheh R, Németh Z, Atanasova L, Fekete E, Paholcsek M, Sándor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP. The VELVET A ortholaogue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS ONE. 2014;9:e112799. doi: 10.1371/journal.pone.0112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karuppiah V, Li Y, Sun J, Vallikkannu M, Chen J. Vel1 regulates the growth of Trichoderma atroviride during co-cultivation with Bacillus amyloliquefaciens and is essential for wheat root rot control. Biol Control. 2020;151:104374. doi: 10.1016/j.biocontrol.2020.104374. [DOI] [Google Scholar]
- 10.Jawed K, Yazdani SS, Koffas MAG. Advances in the development and application of microbial consortia for metabolic engineering. Metab Eng Commun. 2019;9:e00095. doi: 10.1016/j.mec.2019.e00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben Said S, Tecon R, Borer B, Or D. The engineering of spatially linked microbial consortia—potential and perspectives. Curr Opin Biotechnol. 2020;62:137–145. doi: 10.1016/j.copbio.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karuppiah V, Sun J, Li T, Vallikkannu M, Chen J. Co-cultivation of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841 causes differential gene expression and improvement in the wheat growth and biocontrol activity. Front Microbiol. 2019;10:1068–1068. doi: 10.3389/fmicb.2019.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karuppiah V, Vallikkannu M, Li T, Chen J. Simultaneous and sequential based co-fermentations of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841: a strategy to enhance the gene expression and metabolites to improve the bio-control and plant growth promoting activity. Microb Cell Fact. 2019;18:185. doi: 10.1186/s12934-019-1233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhury SP, Hartmann A, Gao X, Borriss R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—a review. Front Microbiol. 2015;6:780. doi: 10.3389/fmicb.2015.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dos Santos CL, de Paula RG, Antoniêto ACC, Persinoti GF, Silva-Rocha R, Silva RN. Understanding the role of the master regulator XYR1 in Trichoderma reesei by global transcriptional analysis. Front Microbiol. 2016;7:175–175. doi: 10.3389/fmicb.2016.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aro N, Saloheimo A, Ilmen M, Penttila M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J Biol Chem. 2001;276:24309–24314. doi: 10.1074/jbc.M003624200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Chen Y, Wu C, Liu P, Wang W, Wei D. The transcription factor ACE3 controls cellulase activities and lactose metabolism via two additional regulators in the fungus Trichoderma reesei. J Biol Chem. 2019;294:18435–18450. doi: 10.1074/jbc.RA119.008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoniêto ACC, de Paula RG, Castro LDS, Silva-Rocha R, Persinoti GF, Silva RN. Trichoderma reesei CRE1-mediated carbon catabolite repression in re-sponse to sophorose through RNA sequencing analysis. Curr Genomics. 2016;17:119–131. doi: 10.2174/1389202917666151116212901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y, Zheng F, Wang L, Zhao G, Chen G, Zhang W, Liu W. Rce1, a novel transcriptional repressor, regulates cellulase gene expression by antagonizing the transactivator Xyr1 in Trichoderma reesei. Mol Microbiol. 2017;105(1):65–83. doi: 10.1111/mmi.13685. [DOI] [PubMed] [Google Scholar]
- 20.de Paula RG, Antonieto ACC, Ribeiro LFC, Carraro CB, Nogueira KMV, Lopes DCB, Silva AC, Zerbini MT, Pedersoli WR, Costa MDN, Silva RN. New genomic approaches to enhance biomass degradation by the industrial fungus Trichoderma reesei. Int J Genomics. 2018;2018:1974151. doi: 10.1155/2018/1974151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adrio JL, Demain AL. Genetic improvement of processes yielding microbial products. FEMS Microbiol Rev. 2006;30:187–214. doi: 10.1111/j.1574-6976.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- 22.Kaushal R, Sharma N, Tandon D. Cellulase and xylanase production by co-culture of Aspergillus niger and Fusarium oxysporum utilizing forest waste. Türk Biyokimya Dergisi/Turkish J Biochem. 2012;37:35–41. [Google Scholar]
- 23.Hero JS, Pisa JH, Perotti NI, Romero CM, Martínez MA. Endoglucanase and xylanase production by Bacillus sp AR03 in co-culture. Prep Biochem Biotechnol. 2017;47:589–596. doi: 10.1080/10826068.2017.1280826. [DOI] [PubMed] [Google Scholar]
- 24.Hu HL, van den Brink J, Gruben BS, Wösten HAB, Gu JD, de Vries RP. Improved enzyme production by co-cultivation of Aspergillus niger and Aspergillus oryzae and with other fungi. Int Biodeterior Biodegrad. 2011;65:248–252. doi: 10.1016/j.ibiod.2010.11.008. [DOI] [Google Scholar]
- 25.Jiang Y, Wu R, Zhou J, He A, Xu J, Xin F, Zhang W, Ma J, Jiang M, Dong W. Recent advances of biofuels and biochemicals production from sustainable resources using co-cultivation systems. Biotechnol Biofuels. 2019;12:155. doi: 10.1186/s13068-019-1495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Wang X. Modular co-culture engineering, a new approach for metabolic engineering. Metab Eng. 2016;37:114–121. doi: 10.1016/j.ymben.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Silva-Rocha R, Castro LDS, Antoniêto ACC, Guazzaroni M-E, Persinoti GF, Silva RN. Deciphering the Cis-regulatory elements for XYR1 and CRE1 regulators in Trichoderma reesei. PLoS ONE. 2014;9:e99366. doi: 10.1371/journal.pone.0099366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saloheimo A, Aro N, Ilmén M, Penttilä M. ACEI of Trichoderma reesei Is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee PK, Kenerley CM. Regulation of morphogenesis and biocontrol properties in Trichoderma virens by a VELVET Protein, Vel1. Appl Environ Microbiol. 2010;76:2345–2352. doi: 10.1128/AEM.02391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, York SW, Ingram LO, Shanmugam KT. Simultaneous fermentation of biomass-derived sugars to ethanol by a co-culture of an engineered Escherichia coli and Saccharomyces cerevisiae. Biores Technol. 2019;273:269–276. doi: 10.1016/j.biortech.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Zoglowek M, Ahring B, Lübeck PS, Lübeck M. Co-cultivation of Trichoderma reesei RutC30 with three black Aspergillus strains facilitates efficient hydrolysis of pretreated wheat straw and shows promises for on-site enzyme production. Biores Technol. 2014;169:143–148. doi: 10.1016/j.biortech.2014.06.082. [DOI] [PubMed] [Google Scholar]
- 32.Kong L. Maize residues, soil quality, and wheat growth in China A review. Agron Sustain Dev. 2014;34:405–416. doi: 10.1007/s13593-013-0182-5. [DOI] [Google Scholar]
- 33.Huang W, Bai Z, Hoefel D, Hu Q, Lv X, Zhuang G, Xu S, Qi H, Zhang H. Effects of cotton straw amendment on soil fertility and microbial communities. Front Environ Sci Eng. 2012;6:336–349. doi: 10.1007/s11783-011-0337-z. [DOI] [Google Scholar]
- 34.Alijani K, Bahrani MJ, Kazemeini SA. Short-term responses of soil and wheat yield to tillage, corn residue management and nitrogen fertilization. Soil Tillage Res. 2012;124:78–82. doi: 10.1016/j.still.2012.05.005. [DOI] [Google Scholar]
- 35.Zhang X, Li H, He J, Wang Q, Golabi MH. Influence of conservation tillage practices on soil properties and crop yields for maize and wheat cultivation in Beijing, China. Soil Res. 2009;47:362–371. doi: 10.1071/SR08110. [DOI] [Google Scholar]
- 36.Zhang J, Zhang G, Wang W, Wang W, Wei D. Enhanced cellulase production in Trichoderma reesei RUT C30 via constitution of minimal transcriptional activators. Microb Cell Fact. 2018;17:75. doi: 10.1186/s12934-018-0926-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghose TK. Measurement of cellulase activities. Pure Appl Chem. 1987;59(2):257–268. doi: 10.1351/pac198759020257. [DOI] [Google Scholar]
- 38.Bailey MJ, Nevalainen KMH. Induction, isolation and testing of stable Trichoderma reesei mutants with improved production of solubilizing cellulase. Enzyme Microb Technol. 1981;3:153–157. doi: 10.1016/0141-0229(81)90076-4. [DOI] [Google Scholar]
- 39.Stricker AR, Trefflinger P, Aro N, Penttilä M, Mach RL. Role of Ace2 (Activator of Cellulases 2) within the xyn2 transcriptosome of Hypocrea jecorina. Fungal Genet Biol. 2008;45:436–445. doi: 10.1016/j.fgb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Tsegaye B, Balomajumder C, Roy P. Alkali delignification and Bacillus sp BMP01 hydrolysis of rice straw for enhancing biofuel yields. Bull Natl Res Cent. 2019;43:136. doi: 10.1186/s42269-019-0175-x. [DOI] [Google Scholar]
- 41.Wang Y-J, Wei X-Y, Jing X-Q, Chang Y-L, Hu C-H, Wang X, Chen K-M. The fundamental role of NOX family proteins in plant immunity and their regulation. Int J Mol Sci. 2016;17(6):805. doi: 10.3390/ijms17060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danise T, Fioretto A, Innangi M. Spectrophotometric methods for lignin and cellulose in forest soils as predictors for humic substances. Eur J Soil Sci. 2018;69:856–867. doi: 10.1111/ejss.12678. [DOI] [Google Scholar]
- 43.Bremner JM, Tabatabai MA. Use of an ammonia electrode for determination of ammonium in Kjeldahl analysis of soils. Commun Soil Sci Plant Anal. 1972;3:159–165. doi: 10.1080/00103627209366361. [DOI] [Google Scholar]
- 44.Kanazawa S, Miyashita K. A modified method for determination of cellulase activity in forest soil. Soil Sci Plant Nutr. 1986;32:71–79. doi: 10.1080/00380768.1986.10557482. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The growth and cellulase production of Trichoderma asperellum recombinants on CBH screening medium. Table S2. Comprehensive information about the enzyme activity and corn stover hydrolysis of different samples. Table S3. Correlation plot of Pearson correlation coefficient for all measured variables in pot experiments. ** Correlation is significant at the 0.01 level (2-tailed). *Correlation is significant at the 0.05 level (2-tailed). Table S4. Primers used in construction of over-expression strains. Table S5. Sequences of the primers used for the real-time PCR. Figure S1. Work flow of the present study. Figure S2. Table 1 Effect of axenic, co-culture and modular co-culture of T. asperellum and B. amyloliquefaciens on the plant growth and biological control against Fusarium verticillioides and Cohilohorus herostrophus under both corn stover amended and non-amended soil in green house conditions. (T8-T14) cornstover amended soil; (T1-T7) cornstover non amended soil.
Data Availability Statement
All data generated during this study are included in this published article.