Abstract
The novel coronavirus pandemic has rapidly spread around the world since December 2019. Various techniques have been applied in identification of SARS-CoV-2 or COVID-19 infection including computed tomography imaging, whole genome sequencing, and molecular methods such as reverse transcription polymerase chain reaction (RT-PCR). This review article discusses the diagnostic methods currently being deployed for the SARS-CoV-2 identification including optical biosensors and point-of-care diagnostics that are on the horizon. These innovative technologies may provide a more accurate, sensitive and rapid diagnosis of SARS-CoV-2 to manage the present novel coronavirus outbreak, and could be beneficial in preventing any future epidemics. Furthermore, the use of green synthesized nanomaterials in the optical biosensor devices could leads to sustainable and environmentally-friendly approaches for addressing this crisis.
Keywords: SARS-CoV-2, COVID-19, Optical biosensor, Point-of-care, Early diagnosis
Graphical abstract
Abbreviation
- 4-aminothiophenol
(4-ATP)
- Acute respiratory syndrome coronavirus
(SARS-CoV)
- Alpha-fetoprotein
(AFP)
- Angiotensin-converting enzyme 2
(ACE2)
- Clustered regularly interspaced short palindromic repeats
(CRISPR)
- DNA endonuclease-targeted CRISPR trans-reporter
(DETECTR)
- Double stranded DNA
(dsDNA)
- emergency use authorization
(EUA)
- Enzyme-linked immunosorbent assay
(ELISA)
- Food and drug administration
(FDA)
- Gold NPs
(AuNPs)
- Gold superparticle
(GSP)
- Hemagglutinin
(HA)
- Heptad repeats
(HR)
- Horseradish peroxidase
(HRP)
- Lateral flow immunoassay
(LFIA)
- Limit of detection
(LOD)
- Localized surface plasmon resonance
(LSPR)
- Loop-mediated isothermal amplification
(LAMP)
- Lung computed tomography
(CT)
- Middle East Respiratory Syndrome
(MERS-CoV)
- Multicomponent reactions
(MCRs)
- Nanoparticles
(NPs)
- Neuraminidase
(NA)
- Open reading frames
(ORFs)
- Plasmonic photothermal therapy
(PPT)
- Point-of-care
(POC)
- Poly-dimethyl sulfoxide
(PDMS)
- Polyethylene glycol
(PEG)
- Poly-methyl-methacrylate
(PMMA)
- Quantitative real-time PCR
(RT-qPCR)
- Quantum dots
(QDs)
- Receptor-binding domain
(RBD)
- Recombinant polymerase amplification
(RPA)
- Reverse transcription polymerase chain reaction
(RT-PCR)
- Specific High-sensitivity Enzymatic Reporter Unlocking
(SHERLOCK)
- Surface enhanced Raman spectroscopy
(SERS)
- Surface plasmon resonance
(SPR)
- Transmission electron microscopy
(TEM)
- Volatile organic compounds
(VOCs)
- World Health Organization
(WHO)
- Zirconium quantum dots
(ZrQDs)
1. Introduction
The sudden appearance of the novel coronavirus (SARS-CoV-2) in Wuhan, China, in December 2019, was the third type of coronavirus to cause serious infections in humans. It has had unprecedented adverse effects, not only on the healthcare services, but also on the world economy (Udugama et al., 2020). Earlier, acute respiratory syndrome coronavirus (SARS-CoV) was identified in Asia in 2003, and an outbreak of Middle East Respiratory Syndrome (MERS-CoV) in Saudi Arabia in 2012 (Wang and Anderson, 2019; Ksiazek et al., 2003). Until now, despite the decline in the newly diagnosed patients in many countries, the World Health Organization (WHO) has warned that the spread of the SARS-COV-2 still has “growth potential”. As of 1st Sep.2020, more than 60 million people have been infected with COVID-19 worldwide with more than more than 1·4 million deaths as of November 2020 (Bhaskaran et al., 2021). Therefore, it is necessary to expeditiously develop new accurate diagnostic methods with high sensitivity to reduce the mortality rate (Cheng et al., 2020). Lung computed tomography (CT) is used as an initial diagnostic procedure in individuals with suspected SARS-CoV-2, however, this technique can solely make possible to assess the patient's lungs, but cannot easily be applied to monitor the results of therapy (Ai et al., 2020; Çinkooğlu et al., 2020). Molecular techniques such as reverse transcription polymerase chain reaction (RT-PCR) are employed to detect the genetic material of the virus in patients with symptoms of active infection. PCR testing can confirm the diagnosis of coronavirus when two specific SARS-CoV-2 genes have been identified. If only one of these genes is detected, the diagnosis is uncertain (Li et al., 2020a; Corman et al., 2020). PCR tests may produce false-negative results that fail to identify the presence of SARS-COV-2 (Wikramaratna et al., 2020), which may be due to human error or problems with the test procedure itself.
In order to protect vulnerable individuals, it is essential to rapidly establish the infection status of patients with SARS-COV-2, as well as for the hospital and emergency personnel. The current RT-PCR test requires several steps and needs several hours for sampling and the evaluation (Wikramaratna et al., 2020). On the other hand, the need for patients to attend hospitals and clinics to provide samples, itself increases the risk of contracting the SARS-COV-2. Therefore, more research is needed to improve diagnostic methods to reduce the mortality by providing more accurate and faster point-of-care (POC) diagnosis, and which could be produced on a commercial scale. So far, several rapid POC tests have been commercialized or developed to diagnose SARS-CoV-2 (Vashist, 2020; Lassaunière et al., 2020). Evaluation of an antibody IgG/IgM-based POC rapid test showed only low sensitivity (36.4%) (Döhla et al., 2020). Therefore, due to the need for higher sensitivity and a rapid diagnosis, antigen-based systems or optical nanosensors could be deployed as a fast and safe method for detection of SARS-CoV-2.
Optical biosensors can be categorized into different types, such as surface plasmon resonance (SPR), surface enhanced Raman spectroscopy (SERS), or chemiluminescence (Pirzada and Altintas, 2020). LSPR and SERS-based optical biosensors are suggested for POC detection systems due to their high diagnostic sensitivity and ability to be commercialized (Sun et al., 2020; Granger et al., 2016).
Metallic Nanoparticles (NPs) are widely applied in biomedical field because their large surface lets the guest molecules such as drugs to get absorbed, entraped, or bind within the surface of the particles. Metallic NPs, such as Au, Ti, Fe, Se, and Metal Oxide NPs, namely zinc oxide (ZnO), iron oxide (Fe2O3/Fe3O4), titanium oxide (TiO2) etc. have attracted great interests as potential in drug/gene delivery and biosensors due to importance applications in the domain of clinical and therapy. Green synthesis of metallic and metal oxide NPs using different greener reducing agents can provide an eco-friendly conditions for biomedical applications especially in the biosensors. Recently, several studies have been performed on the greener synthesis of metallic and metal oxide NPs with different limit of detection and abilities to transfer the drug/gene to the targeted cell that showed excellent biocompatibility without any toxicity in vitro tests.
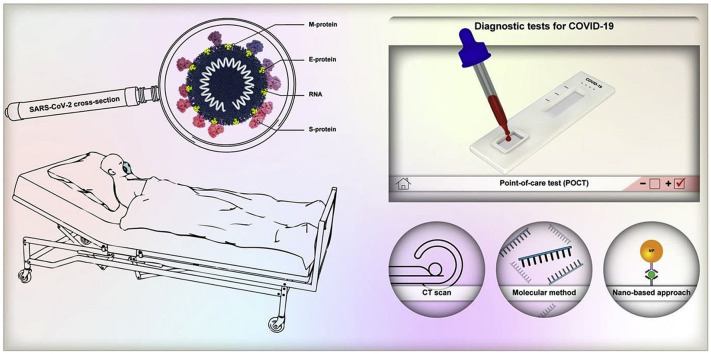
Herein, the current serological and molecular techniques for the detection of SARS-CoV-2 are discussed, and the emerging diagnostic systems based on optical biosensors and POC testing are summarized; the techniques discussed being shown in Fig. 1 . Furthermore, in this review, a novel approach based on the use of green synthesized nanomaterials for the early diagnosis and detection of SARS-CoV-2 has been discussed in-depth.
Fig. 1.
Schematics for the current Lung computed tomography (CT), molecular assays for detection of COVID-19, and emerging diagnostics assays based on optical –based biosensor and POC testing.
2. Characteristics of the SARS-CoV-2 genome and antigenic proteins
Coronavirus are enveloped viruses, belonging to the Coronaviridae family, which are divided into four genera, α, β, γ, and δ; SARS-CoV-2 belongs to the β genus (Yang and Wang, 2020). Transmission electron microscopy (TEM) of human airway epithelial cells isolated from a coronavirus patient in Wuhan, China, showed that the virus particles were 60 to 140 nm in diameter and had distinctive spikes of 9 to 12 nm (Zhu et al., 2020). SARS-CoV-2 is a positive sense RNA strand of ~30,000 nucleotides, and contains about 6–11 open reading frames (ORFs) (Guo et al., 2020).
ORF 2–10 encode four essential structural proteins, including the nucleocapsid (N), spike (S) glycoprotein, matrix protein, and envelope (E) protein (Ujike and Taguchi, 2015). The N protein is responsible for packaging the RNA genome, while the E protein enables assembly of the virus, and is important for the pathogenesis. The S protein, for the interaction of the virus with cellular membrane, binds to the angiotensin-converting enzyme 2 (ACE2) receptor on the surface of human cells. Glycoprotein S consists of two subunits, S1 and S2; S1 regulates the tropism of the virus to host cells with its receptor-binding domain (RBD), while S2 facilitates virus-cell membrane fusion via the heptad repeats 1 (HR1) and HR2 domain (Guo et al., 2020).
The analysis of the spike protein showed the presence of NH2…..S-Cys-G-S-Cys-Cys-K-COOH sequence, with disulfide bonds between cysteine amino-acids (C323, C366, C467, etc.) (Li et al., 2005). Targeting these disulfide bonds could be an option for designing optical sensors for the detection of SARS-CoV-2.
The subunits of SARS-CoV-2 and SARS-CoV showed 88% similarity in sequence, and their binding to the ACE2 receptor is largely similar (Walls et al., 2020). However, the RBD domain in the S protein is the most variable part of the viral genome. Some amino-acids in human SARS-CoV-2 (such as, F486, L455, Q493, Y505, etc.) are known to play an important role in binding to ACE2 receptors, as well as in determining the host range of the viruses (Ou et al., 2020; Andersen et al., 2020). These amino-acids are different between SARS-CoV-2 and SARS-CoV, and also in different virus species that infect specific hosts (Fig. 2 ) (Andersen et al., 2020). Since cross-species diagnosis of the virus could play a significant role in reducing its spread, this feature could be taken into account in the design of specific biosensors.
Fig. 2.
Schematic illustration of four structures of antigenic proteins of coronavirus, nucleic acid and protein sequence of SARS-CoV-2 spike protein (red area), and main residues in the spike protein that bonding to the ACE2 receptor (blue area) (Andersen et al., 2020). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Current routine diagnostic methods for detection of SARS-CoV-2: molecular and immunoassay based-techniques
There is a consensus that it may take between 1- 2 weeks from the time the person becomes infected to the onset of symptoms. During this time an individual has the opportunity to infect other people (Qin et al., 2020). We also need to consider the time required to perform the test and the individual to receive the results. In all countries of the world, testing is important for the early diagnosis of COVID-19 and the immediate isolation of patients diagnosed with the virus, and efforts are being made to develop diagnostic kits and methods with high-sensitivity and specificity. So far, several molecular and serological methods and diagnostic kits have been reported for the detection of COVID-19.
3.1. RT-PCR-based techniques
After the RNA sequence was determined by whole genome sequencing carried out in China in December 2019, the first diagnostic SARS-CoV-2 kit was developed in Berlin based on RT-PCR which can detect the coronavirus RNA in the first week of infection. This test is based on specific probes and is used only for the detection of RNA of the SARS-CoV-2 virus based on specific genes such as S, E, N and RdRP genes (Corman et al., 2020). Since viral samples are small and can contain a viral load of only a few particles, false-negative test results and low sensitivity are distinct possibility. The sensitivity of chest CT (98%) was found to be higher than the RT-PCR (71%) in patients at Taizhou Enze Medical Center, China (Fang et al., 2020).
Following the epidemic of SARS-CoV-2, many companies have developed quantitative real-time PCR (RT-qPCR) detection kits for the clinical diagnosis of virus around the globe. These experiments were performed based on specific primers for the ORF1ab/RdRp, S, N, and E sequences (Li et al., 2020a). Corman et al. developed RT-qPCR assays based on TaqMan fluorescence signal for the detection of the ORF1ab and N regions of the virus with a sensitivity of below 10 copies/reaction in 1 h and 15 min, respectively (Corman et al., 2020). Up to now, different RT-PCR SARS-CoV-2 testing kits are accessible commercially of which 29 kits have been approved by United States Food and Drug Administration (FDA) (Garg et al., 2020). Kasteren et al. compared seven commercially available RT-PCR testing kits from varius companies and showed ≥96% efficiency for the detection of low concentrations of SARS-CoV-2 RNA (Limit of detection (LOD) of 3.8–23 copies/mL) in clinical samples with 100% specificity. They described that all seven commercial RT-PCR kits could be used for the routine detection of COVID-19 through diagnostic laboratories (Van Kasteren et al., 2020). The sensitivity and LOD of six approved RT-PCR SARS-CoV-2 testing kits was investigated by Wang et al. High sensitivity and LoDs of 484 copies/mL was shown for four kits such as Liferiver Biotech, BGI Genomics, SanSure and DAAN and the LoD of BioGerm kit was 968 copies/mL. However, they showed the LOD of 7744 copies/mL for GeneoDx kit that displayed lowly sensitivity, which can be attributed to the technical deficiencies in the product's design (Wang et al., 2020). An overview of the some available RT-PCR SARS-CoV-2 testing kits and their properties is presented in Table 1 .
Table 1.
Overview of RT-PCR –based kits in order to detection of SARS-COV-2.
| Manufacturer/Kit | Genes | Sensitivity | Specitivity | Detection Time | Limit of Detection (LOD) |
|---|---|---|---|---|---|
| CerTest Biotec/VIASURE SARS-CoV-2 Real Time PCR Detection Kit | ORF1ab, N | up to 97.5% | 100% | 120 min | 2000 copies/mL |
| Seegene/The Allplex 2019-nCoV | RdRp, N, E | Up to 90.0% | 100% | Results within 2 h after extraction | 100 copies/mL |
| KH Medical/RADI COVID-19 Kit | RdRp, S | 100% | 100% | 80 min | 0.66 copies/μL |
| Liferiver/2019-nCoV realtime multiplex RT-PCR | ORF1ab, N, E | High | High | Short time | 1 × 103 copies/mL |
| BGI/Real-time Fluorescent RT-PCR Kit | ORF1ab | 94.3% | 100% | 3 h | 100 copies/mL |
| Altona Diagnostics/RealStar® SARS-CoV-2 RT-PCR Kit | E, S | Up to 97% | 100% | – | 3.8 copies/mL |
RT-qPCR techniques generally display high specificity and sensitivity in the detection of SARS-CoV, but these approaches are not without limitations. In five patients, whose RT-qPCR test results were negative for SARS-CoV-2, chest CT scans showed positive results and eventually these 5 patients were found to be infected with the COVID-19 (Xie et al., 2020). Other disadvantages of this technique is its time-consuming nature, needs for special instruments, highly technical operation and a possible long waiting time for results (Li et al., 2020a).
3.2. LAMP-based techniques
The loop-mediated isothermal amplification (LAMP) technique is a unique nucleic acid amplification method that amplifies reverse-transcribed DNA in a short time, with high specificity under isothermal conditions. In addition, this technique also can be visual, there is no need special instruments, and a constant temperature metal bath is enough. In this method, a DNA polymerase and set of four-six specific primers, amplify target genes up to 109 copies (Nagamine et al., 2002; Nguyen et al., 2020). The LAMP method is more sensitive than conventional PCR technique, and although it is less sensitive compared to qPCR it is faster than the other assays evaluated. In spite of the low sensitivity, the LAMP method showed increased specificity compared to other techniques such as qPCR as a one-step detection method (Khan et al., 2018; F Zhang et al., 2020a). Yan et al. described a reverse transcription LAMP (RT-LAMP) method tested in 130 patients, that was simple, rapid, with specificity and sensitivity of 2 × 101 and 2 × 102 copies/reaction using orf1ab-4 and S-123 primers. The mean time for this method was 26.28 ± 4.48 min, and it could detect the virus in suspicious and high-risk individuals. This method was much faster and simpler than RT-PCR and did not need any complex equipment (Yan et al., 2020). Since time and simplicity is crucial in the diagnosis of epidemic diseases, the LAMP technique could be a good option for the POC applications.
POC tests are used to rapidly and accurately diagnose patients without any need to send clinical samples to medical laboratories, and precludes the need for advanced laboratory equipment (Kozel and Burnham-Marusich, 2017). Nucleic acid extraction, amplification and detection can be combined into one integrated cartridge in POC devices, which will provide rapid, simple, and safe results. LAMP and lateral flow immunoassay (LFIA) could be useful in POC devices to detect COVID-19. The high sensitivity and rapid performance of the LAMP technique makes it a good choice for POC devices (Kozel and Burnham-Marusich, 2017; Vashist, 2020).
In February 2020, Professor Jinzhao Song and colleagues at the University of Pennsylvania described a simple POC closed-tube molecular test for COVID-19 that could be performed easily and rapidly in a clinic without any need for expensive equipment. In fact, this device combined paper and LAMP measurement technology and could be used for the sensitive and rapid detection of COVID-19. This experiment was based on closed-tube Penn-RAMP, a two-stage isothermal double stranded DNA (dsDNA) amplification technology that used recombinant polymerase amplification (RPA) in a single tube. In this study, first complete genome sequences of various COVID-19 isolates were aligned, and the conserved sequences with the highest homology to the target sequence were selected to design the primers. Due to the lack of access to authentic COVID-19 samples, inactivated HIV virus particles and other pathogens from the nasal mucosa were used to establish the test. The samples were added to a tube for amplification, and LAMP was performed at 63 °C, and then the Penn-RAMP process was carried out at 38 °C and 63 °C. Penn-RAMP alone is highly sensitive compared to LAMP and RT-PCR alone, as well as 100 times more sensitive than a single LAMP test. The Penn-RAMP method has other advantages; in addition to being easy to perform, it does not have a high cost and can be performed in both clinical and home settings. However, when the LAMP technique is used alone, it requires complex equipment and a constant temperature (Mohamed et al., 2020).
However, molecular techniques are not without their problems, and require a complicated setup with multiple primers, which might be difficult in some settings, and false positive results can occur due to the use of non-specific dyes (Vijay J. Gadkar et al., 2018).
3.3. Immunoassay-based techniques
Several immunoassay methods have been developed to detect COVID-19 virus proteins or to detect serum antibodies. Most commercial immunoassay kits are based on detecting IgM or IgG antibodies produced a few days to two weeks after the onset of viral infection (Laing et al., 2016; Amanat et al., 2020). Amanat et al. developed an enzyme-linked immunosorbent assay (ELISA) test for specific and sensitive detection of SARS-COV-2 at 3 days post-symptom onset which was performed using recombinant antigenic proteins of SARS-CoV-2 and detected seroconversion in human serum and plasma (Amanat et al., 2020). There are many commercial companies in the field of COVID immunoassays and rapid IgM/IgG tests for the rapid detection of SARS-CoV-2, including Jiangsu Medomics Medical Technology, Biomerica, Orient Gen, Sugentech Biotech, Nanjing, China (Li et al., 2020b; Amanat et al., 2020; Vashist, 2020). Because the production of IgM and IgG occurs two weeks after the onset of infection, depending on the age and sex of the individual, and the severity of the disease, other biomarkers and diagnostic methods are required for the rapid diagnosis of SARS-CoV-2. Antibody-based diagnostic tests can also distinguish between other causes of infection, including different coronaviruses (Zhao et al., 2020a).
LFIA is a user-friendly assay format that does not require technical expertise, and can provide a low-cost POC approach for SARS-CoV-2 detection. The lateral flow assay uses a paper or nitrocellulose membrane strip, which is coated with metallic NPs (AuNP)-conjugated Antibodies to capture viral proteins in the serum. After adding the patient sample (blood, plasma, or serum) to the membrane, viral proteins are drawn across the strip. As the virus antigens react with the colloidal gold NPs (AuNPs) -Antibodies in the first line, the complex formed is captured on the strip by IgG and IgM antibody within seconds, and the color of Au NPs change from red to blue because of the plasmon band coupling after aggregation. The results of the test can be read in 10–15 min. Although many companies around the world are developing rapid serological tests, one of the major problems is the cross-reaction between antibodies, which could have a false positive result to other coronaviruses types, and the loss of antibody. As a result, the accuracy of these tests should be determined by further testing (Tomás et al., 2019; Zhao et al., 2020a).
Researchers are currently studying other diagnostic techniques, including chip-based testing, microfluidics, and nanotechnology-based biosensors for the simple and rapid detection of SARS-CoV-2, as well as developing fully automated in vitro POC diagnosis methods that can perform both, the molecular and immunological tests on a single analytical platform.
Table 2 lists the currently deployed diagnostic techniques with the times, advantages and disadvantages for the detection of SARS-CoV-2.
Table 2.
Advantages and disadvantages of current techniques in detection of SARS-CoV-2.
| Tests | Analysis of Time | Advantages | Disadvantages | Ref |
|---|---|---|---|---|
| High-throughput genome sequencing | Around 1–2 weeks | Sequencing of fully genome | Need an expensive equipment | Soon et al. (2013) |
| RT-PCR | About of 1–1.5 h | Good specificity | False-negative test results, lower sensitivity than CT scan in some patients, need a complex equipment | (Corman et al., 2020; Wikramaratna et al., 2020) |
| RT-qPCR | About of 1–1.5 h | Highly sensitive technique for the detection and quantification of specific nucleic acids | Time-consuming, False-negative test results | Li et al. (2020a) |
| LAMP | About 30min | LAMP is quick and does not need costly reagents or appliance, cost-effective and high specificity. high specificity, efficiency, and rapidity, diagnostic sensitivity >95%. | Primer design is complex, unable to perform multiplex amplification, need a non-specific dyes and false positive results | (Khan et al., 2018; Vijay J. Gadkar et al., 2018) |
| ELISA | About 2 h with kit | High sensitivity (20–80%) | Antibody instability, high cost to synthesis antibody, IgG and IgM production takes place two weeks after the onset of infection | (Zhao et al., 2020a; Sakamoto et al., 2018) |
| Computed tomography (CT) | Rapid analysis | Rapid detection assay with 97% sensitivity | Low specific | Ai et al. (2020) |
3.4. CRISPR technology
The discovery of clustered regularly interspaced short palindromic repeats (CRISPR) was a important step forward in gene editing and diagnostic applications. CRISPR is a type of adaptive immunity applied through bacteria to protect themselves against infection with bacteriophages (Chertow, 2018). Among different diagnostic techniques, the CRISPR/Cas system is being studied for specific and rapid diagnosis of SARS-COV-2.
In recent years, researchers have developed a high specific and sensitive detection technology named Specific High-sensitivity Enzymatic Reporter Unlocking (SHERLOCK), which uses the activity of the crRNA/Cas13a protein complex to detection DNA and RNA molecules through amplification of the nucleic acid by an LAMP reaction, then detection of the products using CRISPR mediated reporter unlocking (Joung et al., 2020). The SHERLOCK system based on CRISPR/Cas13 has been applied to POC detection of SARS-CoV-2 by Zhang et al. who applied two crRNAs that diagnose two gene of COVID-19 such as ORF1ab and S gene in a range 10–100 copies/μL within 1 h (Zhang et al., 2020b). Recently, researcher showed that the DNA endonuclease-targeted CRISPR trans-reporter (DETECTR), as an inexpensive, visual and fast test could detect SARS-CoV-2 in <40 min through applying a lateral flow strip depend on a CRISPR system. This assay is based on E, N, and RNase P genes and includes three steps such as LAMP reaction at 62 °C for 30 min, Cas12 detection reaction at 37 °C, which takes 10 min, and RT lateral flow which takes approximately 2 min. The LOD of this assay being 10 copies/μL (Broughton et al., 2020). Another study also indicated that a CRISPR/Cas13-based assay could be applied to destroy SARS-CoV-2 sequences in human lung epithelial cells, killing more than 90% of coronavirus (Abbott et al., 2020).
3.5. Commercial POC-based molecular and immunoassay techniques for the diagnosis of SARS-CoV-2
At present many companies are trying to develop and commercialize POC in vitro diagnostic systems for the early diagnosis and control of COVID-19. The FDA has issued emergency use authorization (EUA) for 20 different SARS-CoV-2 tests, including those from Chembio, Abbott, BioMedomics, etc. A list of some POC devices for COVID-19 that have been, or are in the process of being commercialized, is provided in Table 3 .
Table 3.
The list of current COVID-19 POC test producing by different companies.
| Companies | Product | Time | Method | Approved Status |
|---|---|---|---|---|
| Abbott ID | ID now TM COVID-19 | 5 min | Molecular POC test | FDA, EUA |
| BioMedomics | COVID-19 IgM/IgG Rapid Test | 10–15 min | Lateral flow immunoassays (LFA) | FDA |
| Affinity Health Partners | COVID-19 test | 10 min | Immunoassay | FDA |
| Chembio (Medford, NY, USA) | DPP COVID-19 serological POC test | 15 min | Immunoassay | FDA |
| Mesa Biotech Inc. | Accula SARS-CoV-2 test | 30 min | PCR technology | FDA |
| Cepheid (Sunnyvale, CA, USA) | GeneXpert® Systems | 45 min | Real-time RT-PCR | FDA |
| MiCo BioMed | – | 1 h | LabChip-based real-time PCR microfluidic lab-on-a-chip technology | Seeking FDA approval |
| Fluxergy LLC | Fluxergy Analyzer system | 1 h | Polymerase chain reaction and microfluidics technology | Seeking FDA approval |
The first FDA-approved POC system was developed by Cepheid (Cepheid GeneXpert® Systems) for the rapid molecular detection of SARS-CoV-2 which could provide results within 45 min and is based on real-time RT-PCR. It is used for qualitative detection of SARS-CoV-2 nucleic acids in a nasopharyngeal swab or nasal wash from individuals suspected of being infected with COVID-19. The system employs disposable cartridges containing the RT-PCR reagents and acting as a platform for the RT-PCR process. The risk of contamination between samples is reduced due to its use of self-contained cartridges. The test costs between $10-$20 per sample (Vashist, 2020; Soon et al., 2013).
The COVID-19 IgM/IgG Rapid Test system was one of the first rapid POC LIFA systems for the diagnosis of COVID-19 developed by BioMedomics Co., USA, NC. This system is capable of rapid results in 10–15 min and is based on IgG and IgM antibodies present in plasma serum and blood samples. Requiring only a very small sample (20 μL of finger blood or 10 μg of serum/plasma), this test may be used anywhere Although the sensitivity of this diagnostic test is 88.66%, tests like LAMP have a higher sensitivity, consequently, this system could be combined with techniques such as LAMP to develop POC systems with greater sensitivity for COVID-19 detection (Sakamoto et al., 2018; Carter et al., 2020).
The Abbott ID NOW ™ test is one of the newest POC tests for COVID-19 detection that can give results within 5 min (positive results in 5 min and negative ones in 13 min). This test is based on the portable NOW platform using isothermal nucleic acid amplification technology and targeting the RdRp Gene. Given its small size, this system may be used anywhere, such as clinics and doctor's offices, with nasal, pharyngeal, or oral samples (Vashist, 2020; Rao et al., 2017). It is hoped that the development of more rapid commercial POC systems will help identify the virus and control its spread in many countries.
4. Optical-based nanosensors for rapid and sensitive detection of SARS-CoV-2
Although RT-PCR and the chest-CT scan provide highly specific and sensitive detection of COVID-19, they are not suitable for POC devices, due to the need for the expensive equipment, the requirement for travel to the testing centers, and for hospitals to transfer virus contaminated samples.
Nanotechnology is being used in the development of highly-sensitive, accurate, and efficient sensors to eliminate the need for trained staff in identifying viruses and other nanoscale pathogens (20–900 nm). Due to the unique properties of NPs, such as large surface area to volume ratio, and the presence of many reactive sites, nanomaterials can be efficient, more sensitive, and accurate for virus detection. The integration of nanosensors with other diagnostic methods provides more versatile diagnostic systems for the detection of high-risk pathogens both in and out of the laboratory (e.g., POC detection) (Quesada-González and Merkoçi, 2018; Hamdy et al., 2018; Ghasemi et al., 2018). Consequently, nanosystems have several advantages in the diagnosis of infectious disease. Because early and highly sensitive diagnosis is important in disease prevention and timely treatment, research advances in nanotechnology have been applied to virus detection. Optical biosensors can be used as a simple, safe and rapid method for early detection of epidemic viruses such as COVID-19, and can be deployed using various types of spectrometric readout, such as fluorescence, absorption, or SERS. Among all of these sensors, localized surface plasmon resonance (LSPR) and SERS display real potential as a sensitive optical sensor for virus detection, and have the most promise as POC detection systems, because they are label-free, operate in real time and have high sensitivity (Lee et al., 2017; Tokel et al., 2014).
4.1. SERS–based nanosensors
In recent years, many groups have investigated the application of biosensors for the accurate and highly-sensitive detection of various biomarkers, and also viruses such as the coronavirus family and influenza virus (Kaya et al., 2020; Nikaeen et al., 2020). In the following section, some nanosensors that are based on optical signal detection strategies, and their role in the rapid and sensitive detection of the novel coronavirus (COVID-19), are discussed. One point of view suggests that the optimum detection and destruction of nanoscale viruses requires the use of nanoscale materials such as NPs.
SERS has gained considerable interest as a potential strategy for the diagnosis at a molecular level due to its non-invasive nature, combined with high sensitivity and specificity. In the SERS technique, after the analyte molecule is absorbed onto a SERS substrate, the Raman signal of the molecule is dramatically increased due to the plasmon resonance at the surface of a gold nanostructure. Unlike fluorescence, which shows broad bands, the spectral band-width of Raman signals are narrow. This high resolution of the peaks in SERS spectra allows simultaneous detection of multiple molecules in the same sample. Other benefits of SERS are its rapid analysis rate, simplicity of sample management, and the ability to produce commercial devices. However, SERS has some limitations, namely the degradation of the substrate over time leading to a reduction in the signal intensity (Mosier-Boss, 2017; Laing et al., 2016).
For nucleic acid-based techniques such as RT-PCR the first step involves the extraction of high quality DNA or RNA which reduces false negative results. The use of poly(aminoester) carboxyl-(PC)-coated magnetic NPs (pcMNPs) for RNA extraction, provides an automated, simple extraction method taking only 20 min, and reducing the amount of contamination during extraction, as well as combining the lysis/binding processes into a single step. The MNP-RNA complex can be used directly in the RT-PCR technique without any elution step, making it faster and easier for COVID-19 diagnosis. In addition, the use of this complex in SERS-based RNA detection could be useful in high-sensitivity home diagnosis (Zhao et al., 2020b).
Metal organic frameworks (MOF) represent a class of polymers consisting of metal ions or clusters coordinated to organic ligands to produce one, two, or three-dimensional structures. MOF have been investigated for SERS due to their unique structure and chemical and thermal stability. MOF can serve as solid-phase extraction materials with the ability for rapid concentration and size-selectivity (Lai et al., 2020). Hundreds to thousands of different MOFs could be included in a library of porous materials for different biological applications (Maurin et al., 2017). The effects of morphology, size and optical properties of metal NPs such as gold and silver have been explored with the goal of optimizing SERS substrates. AuNPs (1–200 nm) have been widely applied in the field of virus detection due to their unique optical/electrical properties. Surface plasmon oscillations in AuNPs can increase Raman scattering upon optical excitation (Ou et al., 2019; Qiao et al., 2018). The examination of gas and liquid phase biomarkers in exhaled breath and coughs could detect respiratory viruses, such as influenza and coronavirus as a non-invasive strategy for rapid diagnosis using the SERS method (Leung et al., 2020). As shown in Fig. 3 A, the self-assembled AuNPs formed a “gold superparticle” (GSP) structure with approximately 170 nm diameter. In order to increase the adsorption of gaseous or liquid molecules, a ZIF-8 metal-organic-framework layer was coated onto GSPs. These MOFs lowered the electromagnetic field around the GSP surface, and with a reduced gas flow rate, they provided sufficient time for the adsorption of volatile organic compounds (VOCs) that could serve as lung cancer biomarkers onto the SERS substrate. Reactions that formed a Schiff base between the amine groups of Raman-active 4-aminothiophenol (4-ATP) molecules absorbed onto GSPs and the aldehyde groups present in analytes, allowed the detection of aldehyde-containing VOCs at the 10 ppb level (Fig. 3B and C). This SERS sensor showed the ability to detect cancer biomarker at early stages, and also had the ability to serve as a simple biosensor for early and highly sensitive detection of COVID-19 based on the structure of viral antigens and their interaction with the GSP/ZIF-8 substrate (Qiao et al., 2018). There is a great interest in using green synthesized NPs for this purpose, and in this case, by using the sustainable and green synthesized nanomaterials/NPs, the limit of detection of the biosensor could increase substantially.
Fig. 3.
Schematic illustration of Au NPs assembled into GSPs, (A) GSP@ZIF-8 core-shell structure formation; (B) Detection of VOCs by SERS spectroscopy; (C) Interaction between gaseous aldehyde and covalent linkage with amines groups of Raman active 4-ATP molecule absorbed in GSPs substrate (Qiao et al., 2018).
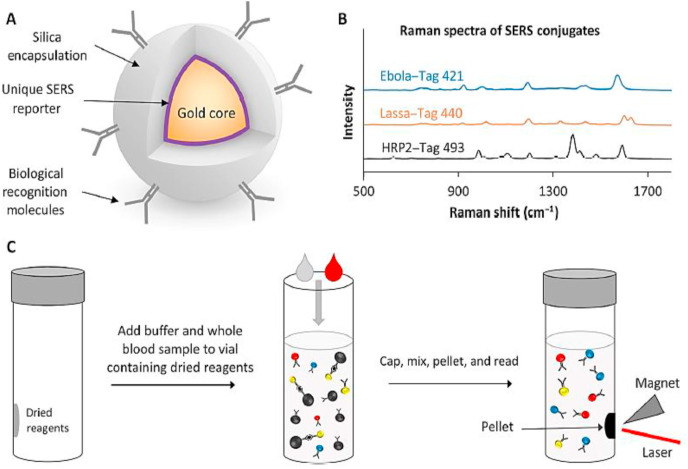
In order to overcome some limitations of immunoassay techniques, the combination of an immunoassay with SERS nanotags could provide a stable POC system for the simultaneous detection of multiple viral antigens in a blood sample within 30 min. SERS nanotags could be useful for detecting possible infection with multiple pathogens, particularly in regions of the world, which are highly susceptible to outbreaks of infectious disease. SERS nanotags are based on a core of 60-nm AuNPs functioning as a reporter, surrounded by a silica shell. In fact, each reporter can serve as a unique fingerprint and provides identifiable SERS spectra with different antigens (Fig. 4 A, B). SERS nanotags are stable under varying environmental conditions and do not require a cold-chain for storage and transfer, while simplifying the assay and reducing analysis time by eliminating the sample preparation and washing steps (Sebba et al., 2018).
Fig. 4.
Schematic illustration of SERS nano tags (A); and different SERS spectra in presence of Ebola, Lassa, and malaria antigens (B); sandwich immunoassay, binding Antibodies - SERS nanotag to Antibodies - magnetic microparticle in presence of antigen (C) (Sebba et al., 2018).
4.2. LSPR-based nanosensors
SPR and LSPR systems are promising for detecting various types of clinical analytes (Masson, 2017). In SPR, electrons are excited at the interface of a thin metal film and a dielectric medium. LSPR is an indirect approach to enhance SPR sensitivity and can be applied to label-free sensing (Malekzad et al., 2018). Colorimetric detection using AuNPs was developed due to the effect of LSPR on the optical properties. AuNPs show a red color when they are dispersed, but when they become aggregated the color changes to blue with maximal absorbance at longer wavelength (Ahmadi et al., 2018).
Colorimetric techniques based on AuNPs have been established for the rapid and simple detection of coronavirus family members in 10 min, and can entirely eliminate the need for instrumentation or skilled personnel. This nanosensor assay could accomplish the detection of MERS-CoV nucleic acids with a sensitivity limit of detection as low as 1 pmol/μL. Therefore, it could be very useful for the early diagnosis of coronavirus, which is critical for such a contagious epidemic (Kim et al., 2019). As shown in Fig. 5 A, in absence of a target, disulfide bond formation occurs between the thiolate-labeled probes. Thereafter, with the addition of salt, aggregation of Au NPs and a color change (from red to blue) can be observed along with LSPR peak changes. In the presence of a target, the thiolate probes react with the target to form a dsDNA structure, which binds to the surface of AuNPs through disulfide bonds, thus protecting the AuNPs from salt-induced aggregation and no color changes occurs (Fig. 5B). In fact, the very high sensitivity of AuNP-based colorimetric techniques for nucleic acid detection have stimulated the use of AuNPs for the sensitive detection of SARS-CoV-2, or in combination with other properties of AuNPs.
Fig. 5.
Schematic illustration of colorimetric detection of MERS-CoV nucleic acids; (a) aggregation of Au NPs in the absence of targets by addition of salt; (b) preventing Au NPs aggregation by disulfide induced self-assembly in the presence of targets (Kim et al., 2019).
Cysteine and methionine binds to AuNPs through their sulfhydryl (-SH) groups and can induce the aggregation of NPs via hydrogen bond formation or zwitterionic interactions. This allows the observation of color changes (from red to blue) with the naked eyes (Ahmadi et al., 2018). Since, the amino acid cysteine is present in the sequence of the antigenic proteins of SARS CoV-2, when the virus is present in the gas phase (exhaled breath, sneeze, cough) or in the solid phase (adhering to clothes, mask, or surfaces), it can be rapidly detected by using the optical properties of Au NPs. However, this method is not completely specific since cysteine amino acids may exist in other pathogen proteins, it could be useful in emergency detection leading to further RT-PCR testing.
The likelihood that viruses (such as influenza and coronavirus) can be transmitted from animals to humans is an important issue. Because the transmission of viruses between different animal species and thence to humans is possible and even probable, the ability to differentiate between strains of human viruses and those of other animal species is of paramount importance in the present pandemic (Parrish et al., 2008; Bolles et al., 2011). Various strains of influenzas viruses can be detected using thiolate polyethylene glycol (PEG)-gold NPs (16 nm) functionalized with sialic acids containing carbohydrates (glyco-NPs). The influenza virus contains two different types of surface glycoproteins, including neuraminidase (NA) and hemagglutinin (HA). Glyco-NPs that were functionalized with trivalent α2,6-thio-linked sialic acid and mixed with human influenza virus X31 (H3N2), generated a color change that could be observed in 30 min. HA binds to the residues of α-2,6 and α-2,3 sialic acids in human and bird cells, respectively. The HA expressed by avian influenza virus binds specifically to α-2,3-sialic acid, which is preferentially expressed in the intestinal tract, while HA proteins from human-influenza viruses bind to α-2,6 sialic acid in the respiratory tract epithelial cells. As a result, based on differences in the sialic acid residues, this biosensor could distinguish between avian and human influenza viruses between 0 to 3.0 μg/mL (Marín et al., 2013). The attachment of polyethylene glycol (PEG) chains can improve the stability of larger size AuNPs and increase the performance of nanosensors, because they prevent the nonspecific adsorption of peptides and proteins and increase the water solubility. In fact, glyco-AuNPs are a versatile hybrid nanosystem for diagnostic applications. In addition, the use of green synthesized AuNPs can effectively help to reduce the cost of the synthesis procedure, which significantly can help the early detection and treatment steps (Schulz et al., 2013; Compostella et al., 2017).
The presence of multivalent functional groups on the surface of PEGylated NPs can play a key role in the highly selective binding to different strains of the virus. Nevertheless, heterogeneous mixtures of functional groups could increase the discriminatory ability of the biosensor (Otten et al., 2016). For example, trivalent α2, 6-thio-linked sialic acid specifically bound to H3N2 influenza virus and could distinguish between diverse strains of influenza. Thiol-linked sialic acids in heterogeneous mixtures could also differentiate between virus strains from different animal species.
Otten et al. used heterogeneous mixtures of glycans on the surface of glyco-AuNPs to distinguish between members of a small panel of legume lectins in a green media (Otten et al., 2016). This approach may also be used in the specific detection and differentiation between SARS-CoV and SARS-CoV-2 viruses, and different types of coronavirus from animal species. The use of PEGylated-AuNPs as a biosensor to differentiate between the two amino acids cysteine and aspartic acid was described by Chen et al. Aspartic acid was added to a solution containing NPs bound to cysteine (by SH bonds) and bound via its amine and carboxyl groups. Moreover, this study showed that increasing the size of the NPs from 13 to 45 nm could lower the detection limit (from 18 μM to 1.8 nM) (Chen et al., 2018).
SARS-CoV-2 virus binds to ACE2 receptors through specific amino acids in the S1 antigen protein (L455, F486, Q493, S494, N501, and Y505). Specific amino acids in human ACE2 (K31, E35, D38, M82, and K353) are able to form hydrogen bonds with the S1 protein (Luan et al., 2020; Walls et al., 2020). Furthermore, additional binding may involve the carboxyl groups of the ACE2 receptor interacting with the carboxyl and amine groups of the S1 antigen. Based on these studies, it may be possible to specifically detect COVID-19 by designing different optical biosensors, including PEG-Au NPs, essentially based on specific binding between the amino acids (for instance D38 and S494). In addition, the use of heterogeneous mixtures of amino acids may allow better differentiation between viruses from different animal species.
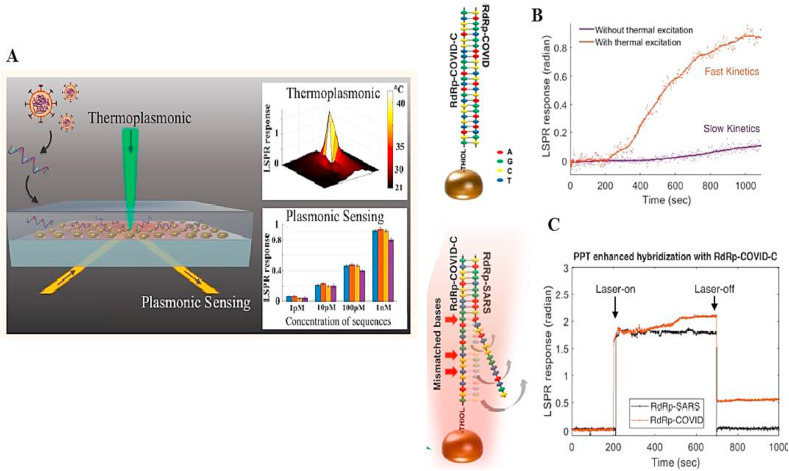
The tunability of the optical-thermal properties of plasmonic AuNPs, enables plasmonic photothermal therapy (PPT), where the wavelength of the laser can be matched with the enhanced absorption in the visible and NIR spectral regions due to the SPR oscillations (Huang et al., 2007; Qiu et al., 2020). An optical nanosensor combining the LSPR and photothermal effect was developed to discriminate between two similar SARS-CoV-2 gene sequences with high sensitivity (detection limit of 0.22 pM). In this study, an Au film was coated with thiol-cDNA ligands and self-assembled into a nanostructured chip. In this sensor, in addition to plasmonic sensing, laser irradiation at a wavelength corresponding to the plasmonic resonance frequency led to substantial heat being generated on the AuNI chip. This PPT heat effect raised the hybridization temperature and allowed the precise discrimination between two similar sequences. Fig. 6 B shows the hybridization of a RdRp gene of SARS-CoV-2 with a complementary cDNA sequence. The photothermal effect could increase the hybridization kinetics between the two sequences, and the response-slope of LSPR with photothermal excitation was steeper than that without the photothermal excitation. Furthermore, this nanosensor was able to discriminate between two similar and non-complementary sequences by the photothermal effect (Fig. 6C) (Qiu et al., 2020).
Fig. 6.
(A) Schematic illustration of dual functional plasmonic photothermal sensor for detection of SARS-CoV-2; (B) hybridization of RdRp gene sequence with complementary cDNA sequence with or without the PPT effect; (C) Discrimination of two similar sequences using PPT effect (Qiu et al., 2020).
4.3. Fluorescence-based nanosensors
Fluorescent biosensors have high sensitivity and selectivity, and a shorter response time usage in fields such as medical diagnostics. Fluorescence biosensors use fluorescent molecules as the signal output component. The specific detection is observed through changes in the fluorescence intensity or else by creation of fluorescence polarization (Wang et al., 2011; Jeong et al., 2018).
Quantum dots (QDs) are colloidal NPs of semiconductor origin, which have become highly popular in the nanosensing and bio-imaging fields, due to their stable optical properties, broad range excitation wavelength and tunable narrow fluorescence emission (Zhu et al., 2013). In comparison with organic fluorophores, QDs have size-tunable optical features (e.g. emission spectrum), and by adjusting their size, coating and attached ligands, can be tailored to detect various targets with high sensitivity for different samples (Han et al., 2001).
Ahmed et al. (2018) developed a fluorescence-based optical diagnostic nanosensor using chiral zirconium quantum dots (ZrQDs) to detect coronavirus in samples from patients with bronchitis infections. These NPs could form a strong and stable bonding with antiviral antibodies and allowed the highly sensitive detection of coronaviruses. The antibodies were conjugated with ZrQDs and magneto-plasmonic MNPs, and then after the addition of antibody-conjugated and the target virusMNPs, led to the formation of nanostructured magnetoplasmonic-fluorescent structures under the influence of an applied external magnetic field. This allowed the magnetic separation of the virus, and the photoluminescence intensity of the nanohybrids was used to measure the concentration. The low toxicity of these NPs, as well as their good binding with antiviral antibodies suggested that these NPs could be a promising nanosensor for the detection of new coronavirus species. With a diagnostic sensitivity that was 10 times higher than the ELISA technique (100 EID/50 μL for NPs versus 1000 EID/50μL for ELISA), these NPs could be useful in diagnosis of pandemic viruses (Ahmed et al., 2018).
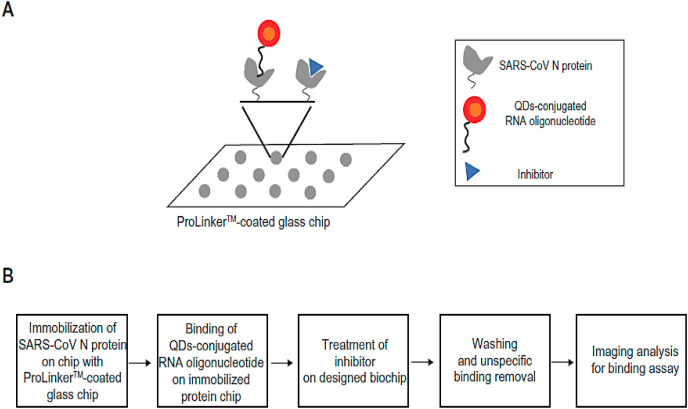
Although antibodies are often applied in sensing assays, the sensitivity can be limited by nonspecific interactions between the antibody and the target. Antibodies are also sensitive to heat and require special reaction conditions that can limit their use (Dobson et al., 2016; Akazawa-Ogawa et al., 2018). Aptamers are single-strand synthetic RNA, DNA, or peptide sequences that can be designed to specifically recognize targets by repeated selective enrichment procedures. Their advantages include sustainability, efficiency, and high flexibility in design, so they can be used in a wide variety of biological sensors (Ahmadi et al., 2019; Maghsoudi et al., 2019). QDs could be coated with oligonucleotide aptamers (linking the aptamer amine group to the QD carboxyl group) that recognized the N protein of SARS-CoV virus. In this study, a Prolinker™ coated protein chip was used to immobilize a sample containing SARS-CoV N protein by its carboxyl groups. When the SARS N protein was present in the sample the binding of the QD-aptamer could be quantified by the fluorescent emission from the chip. The LOD was 0.1 pg/mL for this biosensor, and thus it could also be tested for detection of SARS-CoV-2 (Roh and Jo, 2011).
Some polyphenolic compounds, such as catechin gallate and gallocatechin gallate, have shown some inhibitory potential against SARS-CoV as shown in Fig. 7 A and B. At a concentration of 0.05 μg/mL, gallocatechin gallate and catechin gallate showed more than 40% inhibition activity as measured by a biochip system (Roh, 2012). However, the mechanism of these compounds binding to the SARS-CoV N protein is still unclear. In this study, SARS-CoV spike proteins could be used instead of N protein, because the spike proteins are longer in length, and could provide a stronger bond by reducing the distance between the moieties (Song et al., 2019).
Fig. 7.
Schematic illustration of inhibitor screening of SARS-CoV N protein using QDs-RNA on prolinker™–coated biochip (Roh, 2012).
The LSPR signal from plasmonic metal NPs such as AuNPs and AgNPs has been used to amplify the fluorescence intensity signal of QDs, leading to lower detection limits (Adegoke et al., 2020). This feature of AuNPs was used to increase the sensitivity of detection of the NS1 protein of Zika virus using a QD-labeled antibody. The AuNPs acted as a signal amplifier and the QDs functioned as the fluorophore signal. When the target NS1 protein was present, the LSPR phenomenon of the AuNPs increased the intensity of the QD emission because the two particles were sufficiently close together (few nm), thus increasing the sensitivity. The results of this study showed high sensitivity and rapidity for the detection of Zika virus over a wide range of concentrations (10 fg/mL to 1 ng/mL) and a LOD of 1.28 fg/mL with 10–10(7) RNA copies/mL (Takemura et al., 2019a). The simultaneous use of a mixture of AuNPs and AgNPs could be even more effective and reduce the LOD to 1.7 copies/mL, Nevertheless, the cost of developing a POC diagnostic platform using LSPR in low or middle-income countries may be a challenge (Adegoke et al., 2017).
Microfluidic technology is an effective methodology in POC devices, wherein small volumes of liquid sample can circulate in micron-sized channels, allowing for the use of smaller quantities of samples and reagents. This miniaturization results in much faster detection systems than conventional techniques (Nasseri et al., 2018; Jung et al., 2015). Microfluidic PCR chips have been developed to detect pathogens and viruses, and if properly designed, can replace conventional techniques such as RT-PCR and reduce the costs associated with diagnostics (Ahrberg et al., 2016). Because the world has been so deeply impacted by the coronavirus, and developing countries have fewer resources, the cost of diagnosis is a significant factor to consider. When the rate of virus spread is high, the lack of cost-effective diagnostic tools can rapidly escalate the costs for health care systems.
Microfluidic chips can be constructed from different materials, such as silicon/glass, polymers such as poly-dimethyl sulfoxide (PDMS) or poly-methyl-methacrylate (PMMA), and even from paper (Kaneda et al., 2012; Nge et al., 2013; Soum et al., 2019). Xia et al. (2019) described a PDMS-ZnO nanorod microfluidics chip that was based on a smartphone for POC, rapid, low-cost and sensitive colorimetric detection of viruses in samples within 1.5 h (Higashi et al., 2019). The channels of these nanostructures were made up of PDMS, and the ZnO nanorods were embedded in the channel walls, which enhanced the capture of the virus and improved the detection sensitivity. Viral specific antibodies were attached to the microchannel surface, and after the virus had bound to the antibody, a second anti-virus antibody labeled with AuNPs attached to the virus. In this system, the binding of gold served as a signal amplifier to improve the detection limit. This system allowed rapid detection of the virus by the naked eye with a sensitivity of 2.7 × 104 EID 50/μL. This nanostructured microfluidic chip with its smartphone-enabled plasmonic detector system could be low cost, user-friendly and highly sensitive for the detection of SARS-CoV-2 virus antigenic proteins.
4.4. Chemiluminescence -based nanosensors
Chemiluminescence refers to light emission from the excited state product of a chemical reaction, when it returns to the ground state. In fact, when an electron drops to a lower energy level from a excited or higher energy level, the energy is lost in the form of light, termed a chemiluminescent reaction (Christodouleas et al., 2011). In recent years, it has been found that chemiluminescence reactions can be catalyzed by metal NPs in immunoassays for antigen determination in assistance of using green media. This approach is not only sensitive and selective, but also simple, cost-effective and rapid (Zhou et al., 2016; Li et al., 2013). Li et al. developed a rapid, specific and sensitive detection technique for H1N1 influenza virus based on a chemiluminescence metalloimmunoassay using AgNPs as a label, with a LOD of 0.1 pg/mL. In this system, the binding of monoclonal antibodies onto polystyrene was initially carried out by physical adsorption. When HNO3 was added to the solution, the silver salts were released into the solution and the chemiluminescence signal was produced by the Ag+–Na2S2O8 Mn2+–H3PO4 luminol system (Li et al., 2013).
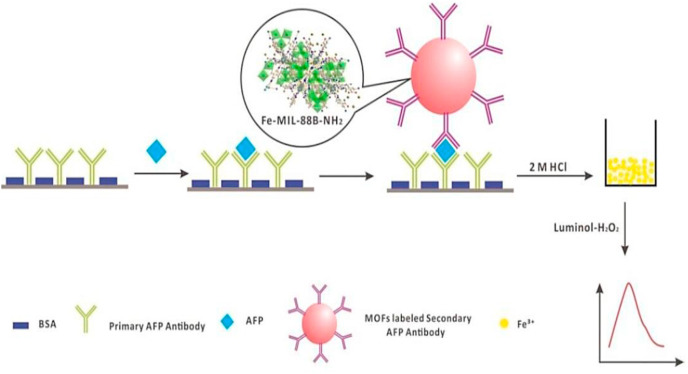
MOFs can serve as labeling agents, due to their features such as good porosity, low toxicity, feasibility of modification on the outer surface, and pore modification through organic linkers (e.g. –COOH, –NH2, etc.). MOFs have a network structure with controlled chemical functionality, and the metallic centers of these particles allow their sensitive quantification by chemiluminescence approaches (Zhou et al., 2016; Miller et al., 2016). These properties make them ideally suited for detecting larger biomolecules, such as proteins, DNA, or RNA from pathogenic viruses. Zhou et al. (2016) reported a sensitive chemiluminescence metalloimmunoassay for the detection of alpha-fetoprotein (AFP), that used a metalloimmunoassay labelling agent such as MOF-NPs. MOF-NPs (Fe-MIL-88B–NH2) which was synthesized by a microwave-assisted procedure, with a size of 50 ± 5 nm in length and 30 ± 5 nm in width. The particles were bound to the secondary antibody through the carboxyl group of NH2-BDC and the amino group of the secondary AFP antibody. After the complete dissolution of the assay plate in hydrochloric acid and the release of Fe+3, the signal was detected by a flow-injection luminol-H2O2 chemiluminescence system (Zhou et al., 2016) (Fig. 8 ). This system showed high sensitivity, low cost, good reproducibility, and was stable for long-term storage, even at room temperature. It was superior compared to systems that used other labels such as AgNPs or Fe3O4@Au (Riccò et al., 2018). This MOF-based chemiluminescence biosensor could be used for highly sensitive and low-cost detection of COVID-19.
Fig. 8.
Schematic illustration of chemiluminescence metalloimmunoassay of AFP using MOFs NPs (Fe-MIL-88B–NH2 NPs) as labelling agent (Zhou et al., 2016).
Table 4 lists some different optical nanosensors for the detection of various viral targets with high sensitivity.
Table 4.
Selected examples of optical biosensor for viral diagnosis.
| Optical Biosensor Types of | Types of sensors | Virus/Sample | Diagbostic Components | Detection limit | Ref |
|---|---|---|---|---|---|
| SERS –based nano sensor | Gold shell-isolated NPs (Au-SHINs) | Zika virus | anti-Zika NS1 antigens | 10 ng/mL | Camacho et al. (2018) |
| Gold NPs | Hepatitis B | DNA sequence | 0.1 fM | Zengin et al. (2017) | |
| SERS nanotags | Ebola, Lassa, and malaria | Virus antigenes | 5 × 105 PFU/mL | Sebba et al. (2018) | |
| SPR/LSPR-based nano sensor | SPR chip | Hepatitis B | Surface antigen | 0.00781 mg/L | Tam et al. (2016) |
| Gold NPs | SARS-CoV-2 | RdRp gene sequence | 0.22 pM | Qiu et al. (2020) | |
| Aptasensor | Avian influenza virus H5N1 | Aptamer specific against H5N1 | 0.128 HAU | Bai et al. (2012) | |
| CdSe–ZnS-based quantum dots | Norovirus | Virus particles | 0.01 ng/mL | Ashiba et al. (2017) | |
| Au/Ag sensing layers | Human enterovirus 71 (EV71) | capsid protein VP1 | 67 vp/mL | Prabowo et al. (2017) | |
| Gold NPs | MERS-CoV | Nucleic acid sequence | 1 pmol/μL | Kim et al. (2019) | |
| Fluorescent-based nano sensor | Ag@SiO2 NPs | H5N1 influenza virus | Anti-recombinant hemagglutinin protein of H5N1 aptamer | 3.5 ng/mL | Pang et al. (2014) |
| Au/Fe3O4 and CdSeS QDs | Norovirus | Particles virus | 0.48 pg/m | Takemura et al. (2019b) | |
| QDs | SARS-CoV | SARS-CoV N protein | 0.1 pg/mL | Rao et al. (2017) | |
| Chemiluminescence -based nano sensor | Ag NPs | H1N1 influenza | Virus antigenes | 0.1 pg/mL | Li et al. (2013) |
5. Routes for improvement
5.1. Multicomponent reactions
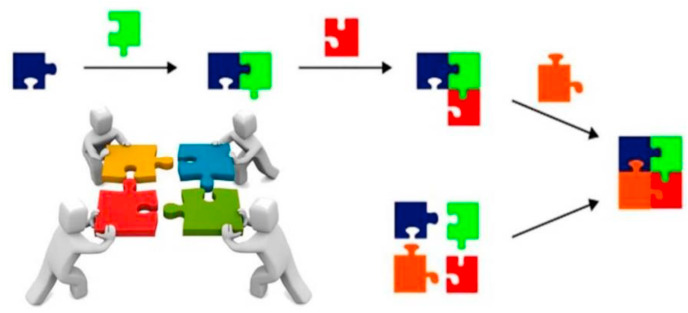
In recent decades, multicomponent reactions (MCRs) have gained much attention in the synthesis of drugs, heterocyclic compounds, ligands and biologically active molecules. In addition, this approach can effectively help the green chemistry and sustainable rules (Graebin et al., 2019; Shaabani and Hooshmand, 2018). MCRs have recently been applied to prepare functionalized nanomaterials, which are used as biosensors, nanocarriers for drug delivery, bioimaging reporters, antibacterial materials and so on (Afshari and Shaabani, 2018). MCRs are single-step and one-pot reactions (most of them recently focused on green synthesized approaches by using water as the solvent) that include three or more commercially available or easily accessible starting materials, and which allow the construction of complex scaffolds. Nowadays, MCRs are becoming common within the synthetic organic chemistry community to increase the overall efficiency of synthetic reactions. One common type of MCRs, is isocyanide-based multicomponent reactions (IMCRs) such as Ugi, Passerini and also the Groebke–Blackburn–Bienayme reactions (Alvim et al., 2014). The Ugi four-component reaction involves an amine, a carbonyl compound, a carboxylic acid, and an isocyanide all reacting together in a one-pot manner to form a pseudeopeptide-like framework is one of the most effective and practical examples (Yang et al., 2015). The use of MCRs enables chemists and biologists to design a biosensor that simultaneously contains different several bioreceptors, such as enzymes, aptamers and antibodies. MCRs have the advantages of being single-pot with short reaction times, and allowing diversity in the selection of substrates, for the development of cutting-edge biosensors (Fig. 9 ).
Fig. 9.
Schematic comparison between of convention synthetic pathway and MCR strategy. Reproduced with permission from ref. (Alvim et al., 2014). Copyright (2014) Royal Society of Chemistry.
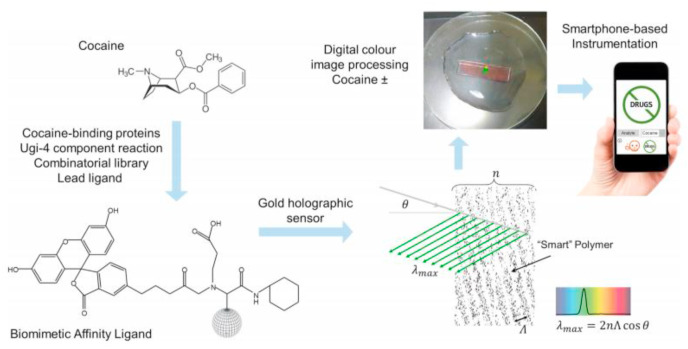
Camacho and co-workers devised an innovative synthetic strategy for the construction of an alginate modified-gold electrode for immobilizing horseradish peroxidase (HRP) enzyme using an Ugi four-component reaction (Camacho et al., 2007). In this procedure, the alginate-coated Au electrode was modified using the Ugi MCR method to produce a support for immobilizing the enzyme. Ultimately, the enzyme-modified Au electrode was a highly sensitive and rapid electroanalytical biosensor for detection of H2O2. Another approach for the construction of biosensors based on the MCR strategy was developed by Vrbova and Marek, who used collagen as a natural material to fabricate an enzyme-catalyzed electrode for d-galactose determination. They immobilized galactose oxidase or co-immobilized galactose oxidase plus catalase to a Clark-type oxygen sensor using the Ugi-four component MCR (Vrbová and Marek, 1990). In this approach, the prepared biosensor was investigated for the detection of d-galactose in real samples of plasma and serum from patients with suspected galactosemia. In 2018, Lowe et al. described an efficient route for the development of a holographic sensor modified with Ugi-4MCR ligands for cocaine determination using a smart-phone based device (Oliveira et al., 2018). Several ligands were prepared to perform protein−cocaine binding, especially human carboxylesterase-1 and the catalytic monoclonal antibody GNL7A1. The Ugi-4CR reaction involved β-alanine, γ-aminobutyric acid, and cyclohexyl isocyanide below the surface of a hybrid hydrogel of poly(2-hydroxyethyl-methacrylate−co-ethylene dimethacrylate-co-methacrolein) as the carbonyl source. The efficiency of the MCR platform for the rapid detection of cocaine in the same range as clinical drug samples was demonstrated (Fig. 10 ).
Fig. 10.
Development of Ugi-holographic biosensor for cocaine detection applying a smartphone-based device. Reproduced with permission from ref. (Oliveira et al., 2018). Copyright (2018) Elsevier.
Based on the above-mentioned data, MCRs could be used as versatile and flexible synthetic tools to design an effective biosensor for the rapid detection of COVID-19. On account of the fact that MCRs are able to incorporate several different bioreceptors within a single platform, this approach could produce a highly sensitive and rapid optical biosensor.
5.2. Computational methods
In recent years, various computational approaches have been employed in drug discovery and biosensor development. These encompass QSAR, virtual screening, molecular docking, homology modelling, and machine learning approaches (Gonzalez-Navarro et al., 2016; Green et al., 2019). Some of these methods have their own advantages and drawbacks. Docking simulations can pinpoint and optimize the 3D structure of small molecules as well as identify potential binding sites. However, the docking simulation process is somewhat tedious, particularly for macromolecular compounds (Bagherian et al., 2020). Methods which employ data from more computationally rigorous, and physically accurate calculations, such as molecular dynamics simulations (MD), are limited in applicability due to their computational cost and poor accuracy (Ellingson et al., 2020). Machine learning techniques have revolutionized the use of computational methods in systems biology. Machine learning can accurately analyze the correlation between chemical structure and interactions with known proteins to form ligand pairs, to provide statistical models to predict the activity of other unknown compounds. The prediction of binding energy requires a regression algorithm, and predicting the likelihood of an interaction could be achieved (Kundu et al., 2018). In addition, a statistical learning approach has very recently been applied for the classification of G-protein coupled receptors and DNA-binding proteins. This approach has been used with several protein structures to predict interactions, including protein–protein interactions (Bock and Gough, 2001), fold recognition (Dubchak et al., 1995), and also structural predictions (Cai et al., 2002).
In order to discover new bioreceptors which might bind to specific targets in the COVID-19 virus, a machine learning approach could provide a more reliable and rapid solution.
6. Conclusions and future directions
The rapid and sensitive detection of pandemic COVID-19 virus, could be the key to limit the transmission throughout the world. Various diagnostic techniques have already been developed for detecting COVID-19, and some have even been commercialized. However, the most important point to consider for diagnosis in different countries, is to develop techniques that are inexpensive, simple and user-friendly. It is important to reduce the cost of diagnosis to produce the best result in the shortest possible time, as well as to reduce the need for travel to hospitals or clinics by developing POC systems. One important example that has not so far received much attention, concerns the plasmonic and optical properties of nanomaterials, such as AuNPs which could be incorporated into face-masks or PPE in the form of nanotechnology-enabled functional clothing. In the presence of SARS-CoV-2, the color of the NPs could significantly change, to allow quick and easy virus detection and prevent the further spread of the virus.
Given the widespread prevalence of COVID-19 worldwide and the increasing mortality, early and highly accurate detection of suspected or infected individuals can play an important role in reducing mortality and preventing other people from becoming infected. Various studies by researchers around the world have led to the development of diagnostic systems based on optical nanosensors, with an emphasis on the development of POC devices. New creative ideas can lead to more innovative approaches, which could significantly reduce the prevalence and mortality of this pandemic disease.
Authors contribution
Sepideh Ahmadi: Writing the sections 1, 2; review and edit whole of the manuscript.
Navid Rabiee: Writing the sections 1, 2; review and edit whole of the manuscript.
Yousef Fatahi: Writing the sections 3.1, 3.2, 3.3.; designing the graphics.
Seyyed Emad Hooshmand: Writing the section 5; review and edit whole of the manuscript.
Mojtaba Bagherzadeh: Writing the sections 3, 3.3.4.; review and edit whole of the manuscript.
Mohammad Rabiee: Writing the sections 2, 3.5.; review and edit whole of the manuscript.
Vahid Jajarmi: Writing the section 4, 4.1.2.; review and edit whole of the manuscript.
Rassoul Dinarvand: Writing the section 4, 4.1.2.; review and edit whole of the manuscript.
Sajjad Habibzadeh: Writing the section 4, 4.3.4.; review and edit whole of the manuscript.
Mohammad Reza Saeb: Writing the section 4, 4.3.4.; review and edit whole of the manuscript.
Rajender S. Varma: Supervision, providing special advice, review and edit whole of the manuscript.
Mohammadreza Shokouhimehr: Writing the section 6; Supervision, providing special advice, review and edit whole of the manuscript.
Michael R Hamblin: Supervision, providing special advice, review and edit whole of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The financial support of the Future Material Discovery Program (2016M3D1A1027666) and the Basic Science Research Program (2017R1A2B3009135) through the National Research Foundation of Korea are appreciated. MRH was supported by US NIH Grants R01AI050875 and R21AI121700.
References
- Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., Pande T., Endy D., La Russa M.F., Lewis D.B., Qi L.S. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876. doi: 10.1016/j.cell.2020.04.020. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adegoke O., Morita M., Kato T., Ito M., Suzuki T., Park E.Y. Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays. Biosens. Bioelectron. 2017;94:513–522. doi: 10.1016/j.bios.2017.03.046. [DOI] [PubMed] [Google Scholar]
- Adegoke O., Pereira-Barros M.A., Zolotovskaya S., Abdolvand A., Daeid N.N. Aptamer-based cocaine assay using a nanohybrid composed of ZnS/Ag2Se quantum dots, graphene oxide and gold nanoparticles as a fluorescent probe. Microchim. Acta. 2020;187:104. doi: 10.1007/s00604-019-4101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshari R., Shaabani A. Materials functionalization with multicomponent reactions: state of the art. ACS Comb. Sci. 2018;20:499–528. doi: 10.1021/acscombsci.8b00072. [DOI] [PubMed] [Google Scholar]
- Ahmadi S., Kamaladini H., Haddadi F., Sharifmoghadam M.R. Thiol-capped gold nanoparticle biosensors for rapid and sensitive visual colorimetric detection of Klebsiella pneumoniae. J. Fluoresc. 2018;28:987–998. doi: 10.1007/s10895-018-2262-z. [DOI] [PubMed] [Google Scholar]
- Ahmadi S., Rabiee N., Rabiee M. Application of aptamer-based hybrid molecules in early diagnosis and treatment of diabetes mellitus: from the concepts towards the future. Curr. Diabetes Rev. 2019;15:309–313. doi: 10.2174/1573399814666180607075550. [DOI] [PubMed] [Google Scholar]
- Ahmed S.R., Kang S.W., Oh S., Lee J., Neethirajan S. Chiral zirconium quantum dots: a new class of nanocrystals for optical detection of coronavirus. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrberg C.D., Manz A., Chung B.G. Polymerase chain reaction in microfluidic devices. Lab Chip. 2016;16:3866–3884. doi: 10.1039/c6lc00984k. [DOI] [PubMed] [Google Scholar]
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa-Ogawa Y., Nagai H., Hagihara Y. Heat denaturation of the antibody, a multi-domain protein. Biophys. Rev. 2018;10:255–258. doi: 10.1007/s12551-017-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvim H.G.O., Da Silva Júnior E.N., Neto B.a.D. What do we know about multicomponent reactions? Mechanisms and trends for the Biginelli, Hantzsch, Mannich, Passerini and Ugi MCRs. RSC Adv. 2014;4:54282–54299. [Google Scholar]
- Amanat F., Nguyen T., Chromikova V., Strohmeier S., Stadlbauer D., Javier A., Jiang K., Asthagiri-Arunkumar G., Polanco J., Bermudez-Gonzalez M., Caplivski D., Cheng A., Kedzierska K., Vapalahti O., Hepojoki J., Simon V., Krammer F. medRxiv; 2020. A Serological Assay to Detect SARS-CoV-2 Seroconversion in Humans. 2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiba H., Sugiyama Y., Wang X., Shirato H., Higo-Moriguchi K., Taniguchi K., Ohki Y., Fujimaki M. Detection of norovirus virus-like particles using a surface plasmon resonance-assisted fluoroimmunosensor optimized for quantum dot fluorescent labels. Biosens. Bioelectron. 2017;93:260–266. doi: 10.1016/j.bios.2016.08.099. [DOI] [PubMed] [Google Scholar]
- Bagherian M., Sabeti E., Wang K., Sartor M.A., Nikolovska-Coleska Z., Najarian K. 2020. Machine Learning Approaches and Databases for Prediction of Drug-Target Interaction: a Survey Paper. Brief Bioinform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H., Wang R., Hargis B., Lu H., Li Y. A SPR Aptasensor for detection of avian influenza virus H5N1. Sensors. 2012;12:12506–12518. doi: 10.3390/s120912506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran K., Rentsch C.T., Mackenna B., Schultze A., Mehrkar A., Bates C.J., Eggo R.M., Morton C.E., Bacon S.C.J., Inglesby P., Douglas I.J., Walker A.J., Mcdonald H.I., Cockburn J., Williamson E.J., Evans D., Forbes H.J., Curtis H.J., Hulme W.J., Parry J., Hester F., Harper S., Evans S.J.W., Smeeth L., Goldacre B. HIV infection and COVID-19 death: a population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. Lancet HIV. 2021;8:e24–e32. doi: 10.1016/S2352-3018(20)30305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock J.R., Gough D.A. Predicting protein–protein interactions from primary structure. Bioinformatics. 2001;17:455–460. doi: 10.1093/bioinformatics/17.5.455. [DOI] [PubMed] [Google Scholar]
- Bolles M., Donaldson E., Baric R. SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr. Opin. Virol. 2011;1:624–634. doi: 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y.-D., Liu X.-J., Xu X.-B., Chou K.-C. Prediction of protein structural classes by support vector machines. Comput. Chem. 2002;26:293–296. doi: 10.1016/s0097-8485(01)00113-9. [DOI] [PubMed] [Google Scholar]
- Camacho C., Matías J.C., García D., Simpson B.K., Villalonga R. Amperometric enzyme biosensor for hydrogen peroxide via Ugi multicomponent reaction. Electrochem. Commun. 2007;9:1655–1660. [Google Scholar]
- Camacho S.A., Sobral-Filho R.G., Aoki P.H.B., Constantino C.J.L., Brolo A.G. Zika immunoassay based on surface-enhanced Raman scattering nanoprobes. ACS Sens. 2018;3:587–594. doi: 10.1021/acssensors.7b00639. [DOI] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-Y., Ma R.-T., Ha W., Shi Y.-P. Direct colorimetric detection of aspartic acid in rat brain based on oriented aggregation of Janus gold nanoparticle. Sensor. Actuator. B Chem. 2018;274:668–675. [Google Scholar]
- Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic testing for severe acute respiratory syndrome–related coronavirus-2. Ann. Intern. Med. 2020;172(11):726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow D.S. Next-generation diagnostics with CRISPR. Science. 2018;360:381. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- Christodouleas D., Fotakis C., Economou A., Papadopoulos K., Timotheou-Potamia M., Calokerinos A. Flow-based methods with chemiluminescence detection for Food and environmental analysis: a review. Anal. Lett. 2011;44:176–215. [Google Scholar]
- Çinkooğlu A., Bayraktaroğlu S., Savaş R. Lung changes on chest CT during 2019 novel coronavirus (COVID-19) pneumonia. Eur. J. Breast Health. 2020:16. doi: 10.5152/ejbh.2020.010420. [Online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compostella F., Pitirollo O., Silvestri A., Polito L. Glyco-gold nanoparticles: synthesis and applications. Beilstein J. Org. Chem. 2017;13:1008–1021. doi: 10.3762/bjoc.13.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.L., Devine P.W.A., Phillips J.J., Higazi D.R., Lloyd C., Popovic B., Arnold J., Buchanan A., Lewis A., Goodman J., Van Der Walle C.F., Thornton P., Vinall L., Lowne D., Aagaard A., Olsson L.-L., Ridderstad Wollberg A., Welsh F., Karamanos T.K., Pashley C.L., Iadanza M.G., Ranson N.A., Ashcroft A.E., Kippen A.D., Vaughan T.J., Radford S.E., Lowe D.C. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci. Rep. 2016;6:38644. doi: 10.1038/srep38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M., Boesecke C., Schulte B., Diegmann C., Sib E., Richter E., Eschbach-Bludau M., Aldabbagh S., Marx B., Eis-Hübinger A.M., Schmithausen R.M., Streeck H. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Publ. Health. 2020;182:170–172. doi: 10.1016/j.puhe.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I., Muchnik I., Holbrook S.R., Kim S.H. Prediction of protein folding class using global description of amino acid sequence. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8700–8704. doi: 10.1073/pnas.92.19.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson S.R., Davis B., Allen J. Machine learning and ligand binding predictions: a review of data, methods, and obstacles. Biochim. Biophys. Acta Gen. Subj. 2020;1864:129545. doi: 10.1016/j.bbagen.2020.129545. [DOI] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadkar Vijay J., Goldfarb David M., Gantt Soren, Tilley P.a.G. Real-time detection and monitoring of loop mediated amplifcation (LAMP) reaction using self-quenching and dequenching fluorogenic probes. Sci. Rep. 2018;8:5548. doi: 10.1038/s41598-018-23930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Ghoshal U., Patel S.S., Singh D.V., Arya A.K., Vasanth S., Pandey A., Srivastava N. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J. Med. Virol. 2020;93(4):2281–2286. doi: 10.1002/jmv.26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi A., Rabiee N., Ahmadi S., Hashemzadeh S., Lolasi F., Bozorgomid M., Kalbasi A., Nasseri B., Shiralizadeh Dezfuli A., Aref A.R., Karimi M., Hamblin M.R. Optical assays based on colloidal inorganic nanoparticles. Analyst. 2018;143:3249–3283. doi: 10.1039/c8an00731d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Navarro F.F., Stilianova-Stoytcheva M., Renteria-Gutierrez L., Belanche-Muñoz L.A., Flores-Rios B.L., Ibarra-Esquer J.E. Glucose oxidase biosensor modeling and predictors optimization by machine learning methods. Sensors. 2016;16 doi: 10.3390/s16111483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebin C.S., Ribeiro F.V., Rogério K.R., Kümmerle A.E. Multicomponent reactions for the synthesis of bioactive compounds: a review. Curr. Org. Synth. 2019;16:855–899. doi: 10.2174/1570179416666190718153703. [DOI] [PubMed] [Google Scholar]
- Granger J.H., Schlotter N.E., Crawford A.C., Porter M.D. Prospects for point-of-care pathogen diagnostics using surface-enhanced Raman scattering (SERS) Chem. Soc. Rev. 2016;45:3865–3882. doi: 10.1039/c5cs00828j. [DOI] [PubMed] [Google Scholar]
- Green E.M., Van Mourik R., Wolfus C., Heitner S.B., Dur O., Semigran M.J. Machine learning detection of obstructive hypertrophic cardiomyopathy using a wearable biosensor. NPJ Digit. Med. 2019;2:57. doi: 10.1038/s41746-019-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy M.E., Del Carlo M., Hussein H.A., Salah T.A., El-Deeb A.H., Emara M.M., Pezzoni G., Compagnone D. Development of gold nanoparticles biosensor for ultrasensitive diagnosis of foot and mouth disease virus. J. Nanobiotechnol. 2018;16:48. doi: 10.1186/s12951-018-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Gao X., Su J.Z., Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- Higashi T., Morita K., Song X., Zhu J., Tamura A., Yui N., Motoyama K., Arima H., Li J. One-pot synthesis of cyclodextrin-based radial poly [n] catenanes. Commun. Chem. 2019;2:1–8. [Google Scholar]
- Huang X., Jain P.K., El-Sayed I.H., El-Sayed M.A. Plasmonic photothermal therapy (PPTT) using gold nanoparticles. Laser Med. Sci. 2007;23:217. doi: 10.1007/s10103-007-0470-x. [DOI] [PubMed] [Google Scholar]
- Jeong Y., Kook Y.-M., Lee K., Koh W.-G. Metal enhanced fluorescence (MEF) for biosensors: general approaches and a review of recent developments. Biosens. Bioelectron. 2018;111:102–116. doi: 10.1016/j.bios.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M.-L.W., Kim N.-G., Yu X., Li J., Walker B.D., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. medRxiv; 2020. Point-of-care Testing for COVID-19 Using SHERLOCK Diagnostics. the preprint server for health sciences, 2020.05.04.20091231. [Google Scholar]
- Jung W., Han J., Choi J.-W., Ahn C.H. Point-of-care testing (POCT) diagnostic systems using microfluidic lab-on-a-chip technologies. Microelectron. Eng. 2015;132:46–57. [Google Scholar]
- Kaneda S., Ono K., Fukuba T., Nojima T., Yamamoto T., Fujii T. Modification of the glass surface property in PDMS-glass hybrid microfluidic devices. Anal. Sci. 2012;28 doi: 10.2116/analsci.28.39. 39-39. [DOI] [PubMed] [Google Scholar]
- Kaya S.I., Karadurmus L., Ozcelikay G., Bakirhan N.K., Ozkan S.A. Nanosensors for Smart Cities; 2020. Electrochemical Virus Detections with Nanobiosensors; pp. 303–326. [Google Scholar]
- Khan M., Wang R., Li B., Liu P., Weng Q., Chen Q. Comparative evaluation of the LAMP assay and PCR-based assays for the rapid detection of Alternaria solani. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Park M., Hwang J., Kim J.H., Chung D.-R., Lee K.-S., Kang M. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019;4:1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- Kozel T.R., Burnham-Marusich A.R. Point-of-Care testing for infectious diseases: past, present, and future. J. Clin. Microbiol. 2017;55:2313. doi: 10.1128/JCM.00476-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., Derisi J., Yang J.Y., Cox N., Hughes J.M., Leduc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kundu I., Paul G., Banerjee R. A machine learning approach towards the prediction of protein–ligand binding affinity based on fundamental molecular properties. RSC Adv. 2018;8:12127–12137. doi: 10.1039/c8ra00003d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H., Li G., Xu F., Zhang Z. Metal–organic frameworks: opportunities and challenges for surface-enhanced Raman scattering – a review. J. Mater. Chem. 2020;8:2952–2963. [Google Scholar]
- Laing S., Gracie K., Faulds K. Multiplex in vitro detection using SERS. Chem. Soc. Rev. 2016;45:1901–1918. doi: 10.1039/c5cs00644a. [DOI] [PubMed] [Google Scholar]
- Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C.Y., Fomsgaard A., Krogfelt K.A., Jørgensen C.S. medRxiv; 2020. Evaluation of Nine Commercial SARS-CoV-2 Immunoassays. 2020.04.09.20056325. [Google Scholar]
- Lee J., Takemura K., Park Y.E. Plasmonic nanomaterial-based optical biosensing platforms for virus detection. Sensors. 2017;17 doi: 10.3390/s17102332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N.H.L., Chu D.K.W., Shiu E.Y.C., Chan K.-H., Mcdevitt J.J., Hau B.J.P., Yen H.-L., Li Y., Ip D.K.M., Peiris J.S.M., Seto W.-H., Leung G.M., Milton D.K., Cowling B.J. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020;26:676–680. doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li Y., Hong M., Qiu B., Lin Z., Cai Z., Chen Y., Chen G. A highly sensitive chemiluminescent metalloimmunoassay for H1N1 influenza virus detection based on a silver nanoparticle label. Chem. Comm. 2013;49:10563–10565. doi: 10.1039/c3cc45329d. [DOI] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm Anal. 2020;10(2) doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of A rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;27:25727. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020;526:165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghsoudi S., Shahraki B.T., Rabiee N., Afshari R., Fatahi Y., Dinarvand R., Ahmadi S., Bagherzadeh M., Rabiee M., Tayebi L., Tahriri M. Recent Advancements in aptamer-bioconjugates: sharpening Stones for breast and prostate cancers targeting. J. Drug Deliv. Sci. Technol. 2019;53:101146. [Google Scholar]
- Malekzad H., Zangabad P.S., Mohammadi H., Sadroddini M., Jafari Z., Mahlooji N., Abbaspour S., Gholami S., Ghanbarpoor M., Pashazadeh R., Beyzavi A., Karimi M., Hamblin M.R. Noble metal nanostructures in optical biosensors: basics, and their introduction to anti-doping detection. Trac. Trends Anal. Chem. 2018;100:116–135. doi: 10.1016/j.trac.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M.J., Rashid A., Rejzek M., Fairhurst S.A., Wharton S.A., Martin S.R., Mccauley J.W., Wileman T., Field R.A., Russell D.A. Glyconanoparticles for the plasmonic detection and discrimination between human and avian influenza virus. Org. Biomol. Chem. 2013;11:7101–7107. doi: 10.1039/c3ob41703d. [DOI] [PubMed] [Google Scholar]
- Masson J.-F. Surface plasmon resonance clinical biosensors for medical diagnostics. ACS Sens. 2017;2:16–30. doi: 10.1021/acssensors.6b00763. [DOI] [PubMed] [Google Scholar]
- Maurin G., Serre C., Cooper A., Férey G. The new age of MOFs and of their porous-related solids. Chem. Soc. Rev. 2017;46:3104–3107. doi: 10.1039/c7cs90049j. [DOI] [PubMed] [Google Scholar]
- Miller S.E., Teplensky M.H., Moghadam P.Z., Fairen-Jimenez D. Metal-organic frameworks as biosensors for luminescence-based detection and imaging. Interface Focus. 2016;6:20160027. doi: 10.1098/rsfs.2016.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed E.-T., Haim H.B., Jinzhao S. 2020. A Single and Two-Stage, Closed-Tube, Molecular Test for the 2019 Novel Coronavirus (COVID-19) at Home, Clinic, and Points of Entry. [Google Scholar]
- Mosier-Boss A.P. Review of SERS substrates for chemical sensing. Nanomaterials. 2017;7 doi: 10.3390/nano7060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Nasseri B., Soleimani N., Rabiee N., Kalbasi A., Karimi M., Hamblin M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018;117:112–128. doi: 10.1016/j.bios.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nge P.N., Rogers C.I., Woolley A.T. Advances in microfluidic materials, functions, integration, and applications. Chem. Rev. 2013;113:2550–2583. doi: 10.1021/cr300337x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Duong Bang D., Wolff A. 2019 novel coronavirus disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines. 2020;11 doi: 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaeen G., Abbaszadeh S., Yousefinejad S. Application of nanomaterials in treatment, anti-infection and detection of coronaviruses. Nanomedicine. 2020;15(15) doi: 10.2217/nnm-2020-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira N.C.L., Khoury G.E., Versnel J.M., Moghaddam G.K., Leite L.S., Lima-Filho J.L., Lowe C.R. A holographic sensor based on a biomimetic affinity ligand for the detection of cocaine. Sensor. Actuator. B Chem. 2018;270:216–222. [Google Scholar]
- Otten L., Vlachou D., Richards S.-J., Gibson M.I. Glycan heterogeneity on gold nanoparticles increases lectin discrimination capacity in label-free multiplexed bioassays. Analyst. 2016;141:4305–4312. doi: 10.1039/c6an00549g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Zhou Z., Chen Z., Tan H. Optical diagnostic based on functionalized gold nanoparticles. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20184346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Rong Z., Wang J., Xiao R., Wang S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF) Biosens. Bioelectron. 2014;66C:527–532. doi: 10.1016/j.bios.2014.10.052. [DOI] [PubMed] [Google Scholar]
- Parrish C.R., Holmes E.C., Morens D.M., Park E.C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzada M., Altintas Z. Recent progress in optical sensors for biomedical diagnostics. Micromachines. 2020;11 doi: 10.3390/mi11040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabowo B.A., Wang R.Y.L., Secario M.K., Ou P.-T., Alom A., Liu J.-J., Liu K.-C. Rapid detection and quantification of Enterovirus 71 by a portable surface plasmon resonance biosensor. Biosens. Bioelectron. 2017;92:186–191. doi: 10.1016/j.bios.2017.01.043. [DOI] [PubMed] [Google Scholar]
- Qiao X., Su B., Liu C., Song Q., Luo D., Mo G., Wang T. Selective surface enhanced Raman scattering for quantitative detection of lung cancer biomarkers in superparticle@MOF structure. Adv. Mater. 2018;30:1702275. doi: 10.1002/adma.201702275. [DOI] [PubMed] [Google Scholar]
- Qin J., You C., Lin Q., Hu T., Yu S., Zhou X.-H. medRxiv; 2020. Estimation of Incubation Period Distribution of COVID-19 Using Disease Onset Forward Time: a Novel Cross-Sectional and Forward Follow-Up Study. the preprint server for health sciences, 2020.03.06.20032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5) doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Quesada-González D., Merkoçi A. Nanomaterial-based devices for point-of-care diagnostic applications. Chem. Soc. Rev. 2018;47:4697–4709. doi: 10.1039/c7cs00837f. [DOI] [PubMed] [Google Scholar]
- Rao S.-J., Zhang Q., Mei J., Ye X.-H., Gao C., Wang Q.-C., Qu D.-H., Tian H. One-pot synthesis of hetero [6] rotaxane bearing three different kinds of macrocycle through a self-sorting process. Chem. Sci. 2017;8:6777–6783. doi: 10.1039/c7sc03232c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccò R., Liang W., Li S., Gassensmith J.J., Caruso F., Doonan C., Falcaro P. Metal-organic frameworks for cell and virus biology: a perspective. ACS Nano. 2018;12:13–23. doi: 10.1021/acsnano.7b08056. [DOI] [PubMed] [Google Scholar]
- Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomed.. 2012;7:2173–2179. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C., Jo S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. 2011;86:1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S., Putalun W., Vimolmangkang S., Phoolcharoen W., Shoyama Y., Tanaka H., Morimoto S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018;72:32–42. doi: 10.1007/s11418-017-1144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F., Vossmeyer T., Bastús N.G., Weller H. Effect of the spacer structure on the stability of gold nanoparticles functionalized with monodentate thiolated poly(ethylene glycol) ligands. Langmuir. 2013;29:9897–9908. doi: 10.1021/la401956c. [DOI] [PubMed] [Google Scholar]
- Sebba D., Lastovich A.G., Kuroda M., Fallows E., Johnson J., Ahouidi A., Honko A.N., Fu H., Nielson R., Carruthers E., Diédhiou C., Ahmadou D., Soropogui B., Ruedas J., Peters K., Bartkowiak M., Magassouba N., Mboup S., Ben Amor Y., Connor J.H., Weidemaier K. A point-of-care diagnostic for differentiating Ebola from endemic febrile diseases. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat0944. [DOI] [PubMed] [Google Scholar]
- Shaabani A., Hooshmand S.E. Diversity-oriented catalyst-free synthesis of pseudopeptides containing rhodanine scaffolds via a one-pot sequential isocyanide-based six-component reactions in water using ultrasound irradiation. Ultrason. Sonochem. 2018;40:84–90. doi: 10.1016/j.ultsonch.2017.06.030. [DOI] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. 2019. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon W.W., Hariharan M., Snyder M.P. High-throughput sequencing for biology and medicine. Mol. Syst. Biol. 2013;9:61. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soum V., Park S., Brilian I.A., Kwon O.-S., Shin K. Programmable paper-based microfluidic devices for biomarker detections. Micromachines. 2019;10 doi: 10.3390/mi10080516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.L., Leo Y.S., Zhou X., Ng W., Wong T.I., Deng J. Localized surface plasmon resonance based point-of-care system for sepsis diagnosis. Mater. Sci. Eng. Technol. 2020;3:274–281. [Google Scholar]
- Takemura K., Adegoke O., Suzuki T., Park E.Y. A localized surface plasmon resonance-amplified immunofluorescence biosensor for ultrasensitive and rapid detection of nonstructural protein 1 of Zika virus. PloS One. 2019;14 doi: 10.1371/journal.pone.0211517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura K., Lee J., Suzuki T., Hara T., Abe F., Park E.Y. Ultrasensitive detection of norovirus using a magnetofluoroimmunoassay based on synergic properties of gold/magnetic nanoparticle hybrid nanocomposites and quantum dots. Sensor. Actuator. B Chem. 2019;296:126672. [Google Scholar]
- Tam Y., Zeenathul N., Akhavan Rezaei M., Mustafa N., Mohd Lila M., Bahaman A., Sewn Cen L., Joo Shun T., Hani H., Abdullah R. Wide dynamic range of surface-plasmon-resonance-based assay for Hepatitis-B-surface-antigen-antibody optimal detection in comparison with ELISA. Biotechnol. Appl. Biochem. 2016;64 doi: 10.1002/bab.1528. [DOI] [PubMed] [Google Scholar]
- Tokel O., Inci F., Demirci U. Advances in plasmonic technologies for point of care applications. Chem. Rev. 2014;114:5728–5752. doi: 10.1021/cr4000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás A.L., De Almeida M.P., Cardoso F., Pinto M., Pereira E., Franco R., Matos O. Development of a gold nanoparticle-based lateral-flow immunoassay for pneumocystis pneumonia serological diagnosis at point-of-care. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Ujike M., Taguchi F. 2015. Incorporation of Spike and Membrane Glycoproteins into Coronavirus Virions. Viruses 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kasteren P.B., Van Der Veer B., Van Den Brink S., Wijsman L., De Jonge J., Van Den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist K.S. Vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics. 2020;10 doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrbová E., Marek M. Application of the Ugi reaction for the preparation of enzyme electrodes. Anal. Chim. Acta. 1990;239:263–268. [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., Mcguire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F., Anderson D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019;34:79–89. doi: 10.1016/j.coviro.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.E., Zhang Y., Cai J., Cai W., Gao T. Aptamer-based fluorescent biosensors. Curr. Med. Chem. 2011;18:4175–4184. doi: 10.2174/092986711797189637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J., Xiao Y., Wang H. Limits of detection of 6 approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2) Clin. Chem. 2020;66:977–979. doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramaratna P., Paton R.S., Ghafari M., Lourenco J. medRxiv; 2020. Estimating False-Negative Detection Rate of SARS-CoV-2 by RT-PCR. 2020.04.05.20053355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26(6) doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Wang X. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 2020;17:555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zhao Y., Wei Y., Fu C., Tao L. The Ugi reaction in polymer chemistry: syntheses, applications and perspectives. Polym. Chem. 2015;6:8233–8239. [Google Scholar]
- Zengin A., Tamer U., Caykara T. SERS detection of hepatitis B virus DNA in a temperature-responsive sandwich-hybridization assay. J. Raman Spectrosc. 2017;48:668–672. [Google Scholar]
- Zhang F., Oo Abudayyeh, Js G. vol. 8. 2020. (A Protocol for Detection of COVID-19 Using CRISPR Diagnostics). [Google Scholar]
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. medRxiv; 2020. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. 2020.02.26.20028373. [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71(16) doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Cui H., Song W., Ru X., Zhou W., Yu X. bioRxiv; 2020. A Simple Magnetic Nanoparticles-Based Viral RNA Extraction Method for Efficient Detection of SARS-CoV-2. 2020.02.22.961268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Long Z., Tian Y., Ding X., Wu L., Hou X. A chemiluminescence metalloimmunoassay for sensitive detection of alpha-fetoprotein in human serum using Fe-MIL-88B-NH2 as a label. Appl. Spectrosc. Rev. 2016;51:517–526. [Google Scholar]
- Zhu Y., Hong H., Xu Z.P., Li Z., Cai W. Quantum dot-based nanoprobes for in vivo targeted imaging. Curr. Mol. Med. 2013;13:1549–1567. doi: 10.2174/1566524013666131111121733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]