Abstract
The unprecedented demand for rapid diagnostics in response to the COVID‐19 pandemic has brought the spotlight onto clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated systems (Cas)‐assisted nucleic acid detection assays. Already benefitting from an elegant detection mechanism, fast assay time, and low reaction temperature, these assays can be further advanced via integration with powerful, digital‐based detection. Thus motivated, the first digital CRISPR/Cas‐assisted assay—coined digitization‐enhanced CRISPR/Cas‐assisted one‐pot virus detection (deCOViD)—is developed and applied toward SARS‐CoV‐2 detection. deCOViD is realized through tuning and discretizing a one‐step, fluorescence‐based, CRISPR/Cas12a‐assisted reverse transcription recombinase polymerase amplification assay into sub‐nanoliter reaction wells within commercially available microfluidic digital chips. The uniformly elevated digital concentrations enable deCOViD to achieve qualitative detection in <15 min and quantitative detection in 30 min with high signal‐to‐background ratio, broad dynamic range, and high sensitivity—down to 1 genome equivalent (GE) µL−1 of SARS‐CoV‐2 RNA and 20 GE µL−1 of heat‐inactivated SARS‐CoV‐2, which outstrips its benchtop‐based counterpart and represents one of the fastest and most sensitive CRISPR/Cas‐assisted SARS‐CoV‐2 detection to date. Moreover, deCOViD can detect RNA extracts from clinical samples. Taken together, deCOViD opens a new avenue for advancing CRISPR/Cas‐assisted assays and combating the COVID‐19 pandemic and beyond.
Keywords: CRISPR/Cas, diagnostics, digital, nucleic acid, SARS‐CoV‐2
A digital clustered regularly interspaced short palindromic repeats (CRISPR)/Cas‐assisted nucleic acid detection assay is created by discretizing a one‐step, fluorescence‐based, CRISPR/Cas12a‐assisted reverse transcription recombinase polymerase amplification assay within sub‐nanoliter reaction wells of a commercial microfluidic digital chip, which enables quantitative detection of 1 genome equivalent µL−1 of SARS‐CoV‐2 RNA in <30 min—one of the fastest and most sensitive CRISPR/Cas‐assisted detection to date.
The crucial but unmet need for rapid and sensitive nucleic acid detection assays for diagnostic testing of highly infectious diseases has become front and center as the COVID‐19 pandemic[ 1 , 2 , 3 ] continues to devastate. The global scientific community has responded with unprecedented urgency to develop assays that can rapidly and sensitively detect the causative SARS‐CoV‐2 virus and curb the spread of COVID‐19.[ 4 , 5 , 6 ] As a result, a plethora of assays,[ 7 , 8 , 9 , 10 , 11 ] including standardized assays based on reverse transcription PCR (RT‐PCR)[ 12 , 13 ] and emerging isothermal assays based on reverse transcription loop mediated isothermal amplification (RT‐LAMP),[ 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 ] have been reported within a remarkably short period of time. Among the emerging assays, those incorporating clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated systems (Cas)[ 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 ] (e.g., DETECTR[ 22 ] and SHERLOCK[ 31 , 32 ]) have attracted particular attention due to their elegant detection mechanism, fast turnaround time, and potential circumvention of instrument‐intensive thermocycling. However, despite these advantages and rapid advances, with the exception of two recently reported one‐step CRISPR/Cas‐assisted assays,[ 23 , 31 ] current CRISPR/Cas‐assisted assays remain hampered by a requisite but separate preamplification step. The resulting multipot and multistep assay format has precluded them from digital detection—a powerful detection approach that has enhanced the diagnostic capabilities of PCR[ 33 , 34 , 35 , 36 ] and LAMP.[ 37 ] Indeed, to date, no CRISPR/Cas‐assisted assay has been implemented in digital detection format, nor have the potential enhancements from digitization been realized or explored.
In response, we have developed digitization‐enhanced CRISPR/Cas‐assisted one‐pot virus detection (deCOViD)—the first digital CRISPR/Cas‐assisted assay that can detect SARS‐CoV‐2 RNA and heat‐inactivated SARS‐CoV‐2 (Figure 1 ). In deCOViD, SARS‐CoV‐2 RNA and inactivated SARS‐CoV‐2 can be fluorescently detected by a single‐step assay that integrates reverse transcription, recombinase polymerase amplification (RPA),[ 38 ] and CRSIPR/Cas12a‐based detection.[ 39 ] Within the assay, RNA targets are reverse transcribed and amplified via RT‐RPA into DNA amplicons, which activate Cas12a‐guide RNA complexes to cleave single‐stranded DNA fluorogenic reporters and yield fluorescence.[ 39 ] This single‐step assay is then fine‐tuned to ensure that it can be easily loaded into commercially available microfluidic digital chips and reliably discretized within digital reaction wells. Upon assay digitization within digital reaction wells, every copy of the target is isolated at a locally elevated concentration, facilitating rapid amplification that is independent of the sample concentration. This advantageous mechanism therefore empowers deCOViD to outperform its bulk counterpart in detecting SARS‐CoV‐2 and potentially other RNA and even DNA targets with accelerated detection time, enhanced signal‐to‐background ratio, widened dynamic range, and improved sensitivity.
Figure 1.
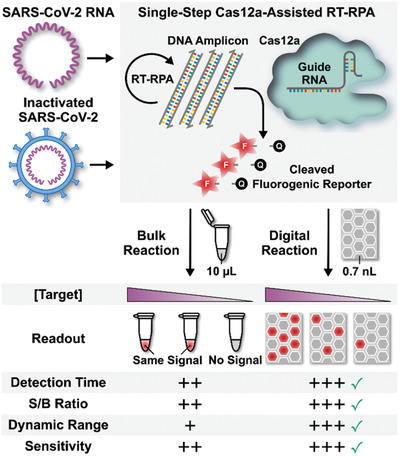
Overview of digitization‐enhanced CRISPR/Cas‐assisted one‐pot virus detection (deCOViD). SARS‐CoV‐2 RNA and inactivated SARS‐CoV‐2 virus can be detected by a single‐step CRISPR/Cas12a‐assisted reverse transcription recombinase polymerase amplification (RT‐RPA) assay, which produces DNA amplicons from RNA targets to activate Cas12a‐based cleavage of fluorogenic reporters in a single step. Importantly, compared to microliter‐scale bulk assay, deCOViD takes advantage of assay digitization to elevate the RNA concentration in sub‐nanoliter digital reaction wells and facilitate rapid amplification, leading to faster detection time for low target concentrations, higher signal‐to‐background (S/B) ratio, wider dynamic range, and better sensitivity.
We first established a prerequisite bulk‐based single‐step CRISPR/Cas12a‐assisted RT‐RPA assay that can detect SARS‐CoV‐2. We adopted the RPA primers and the Cas12a guide RNAs (Table S1, Supporting Information) from a recently reported one‐pot CRISPR/Cas12a‐assisted RT‐RPA assay that uses two Cas12a guide RNAs to detect a fragment of the SARS‐CoV‐2 N gene[ 23 ] and employed Alexa647‐labeled fluorogenic reporter from our previous work.[ 40 ] We elected to test this assay against standardized synthetic SARS‐CoV‐2 RNA from NIAID BEI Resources (NR‐52358) rather than an RNA sequence in vitro transcribed in house. Performed in a benchtop real‐time qPCR instrument under an isothermal condition, this bulk assay successfully detected 200, 100, and 50 genome equivalents (GE) µL−1 RNA, as indicated by strong fluorescence signals above the no‐RNA control (i.e., 0 GE µL−1) at 60 min (Figure 2a). In addition to detecting the standardized SARS‐CoV‐2 RNA for the first time, we made two notable improvements for this bulk assay. First, we elevated the reaction temperature to 42 °C (Figure S1a, Supporting Information) to improve reverse transcription efficiency in the assay. We also added 0.01 mg mL−1 bovine serum albumin to our assay (Figure S1b, Supporting Information) to reduce adsorption of enzymes to reaction tube walls.
Figure 2.
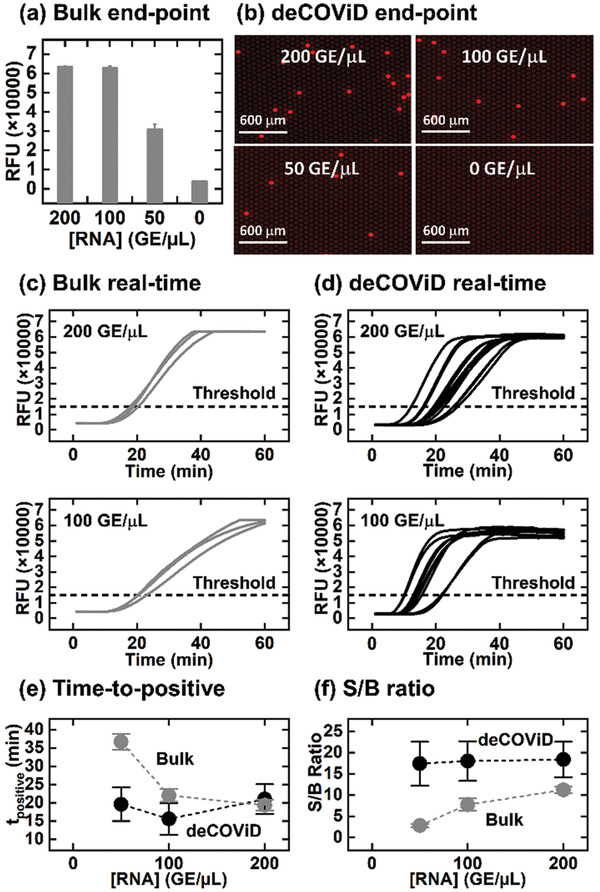
Comparison of bulk CRISPR/Cas12a‐assisted RT‐RPA and deCOViD. Detection of various concentrations of SARS‐CoV‐2 RNA (in genome equivalent (GE) µL−1) is successful at 60 min via both a) bulk CRISPR/Cas12a‐assisted RT‐RPA, which yields higher fluorescence signals than the no‐RNA control, and b) deCOViD (i.e., digital CRISPR/Cas12a‐assisted RT‐RPA), which outputs serially reduced number of positive reaction wells with strong fluorescence and no positive reaction wells for the no‐RNA control. Real‐time amplifications curves from samples with 200 and 100 GE µL−1 RNA reveal that c) the bulk assay speed decreases from the reduced sample concentration, but d) deCOViD maintains consistent assay speed from both sample RNA concentrations, while also allowing qualitative detection in <15 min by detecting the early positive reaction wells. Compared to the bulk assay, assay digitization in deCOViD facilitates e) faster and RNA concentration‐independent time‐to‐positive (i.e., the time at which the sample fluorescence signal surpasses the predefined threshold) and f) higher signal‐to‐background (S/B) ratios at 30 min reaction time across various sample concentrations. Data in (a), (e), and (f) presented as mean ± SD, n = 3 (bulk) or n > 3 (deCOViD).
For actualizing deCOViD, we digitized the bulk CRISPR/Cas12a‐assisted RT‐RPA assay in QuantStudio 3D Digital PCR 20K Chip with 0.7 nL digital reaction wells. QuantStudio digital chip offers an attractive option for assay digitization thanks to its commercial availability, simple and fast digitization workflow, and compatibility with fluorescence microscopy—our preferred detection method for this proof‐of‐concept study. Robust assay loading and discretization into QuantStudio chips could be completed with ≈5 min hands‐on time per chip, and was further facilitated by adding 0.1% Tween‐20 in the reaction mix, which reduced its viscosity without compromising its detection performance (Figure S2, Supporting Information). We also verified that no amplification would occur at room temperature during assay digitization (Figure S3, Supporting Information). After digitizing 200, 100, and 50 GE µL−1 RNA into QuantStudio chips and heating at 42 °C for 60 min, we detected digital reaction wells with strong fluorescence (i.e., positive) via fluorescence microscopy. Importantly, we found fewer positive reaction wells as the RNA concentration decreased, indicating that single copies of RNA were indeed digitized and amplified (Figure 2b and Figure S4, Supporting Information). Finally, we detected 0 positive reaction well from the no‐RNA control. These results offer strong initial validation for deCOViD.
We subsequently acquired and compared real‐time amplification curves from both the bulk assay and deCOViD at different RNA concentrations to illustrate the benefit of assay digitization. For the bulk assay, we used the benchtop real‐time qPCR instrument to acquire the amplification curves. Samples with 200 RNA GE µL−1 yielded amplification curves that plateaued at ≈40 min, whereas samples with 100 RNA GE µL−1 yielded noticeably flatter amplification curves that plateaued at ≈60 min (Figure 2c). These results suggest that the bulk reaction rate slows with reduced RNA concentration. For deCOViD, we employed a custom‐assembled miniature heater that is compatible with fluorescence microscopy (Figure S5, Supporting Information) to simultaneously measure the fluorescence intensities from ≈800 digital reaction wells as a function of time (Figure S6, Supporting Information). All resulting deCOViD amplification curves from both 200 and 100 RNA GE µL−1 rose sharply and plateaued in ≈40 min, suggesting comparable amplification speeds between the two target concentrations (Figure 2d). The contrasting bulk and deCOViD results illustrate that, by digitizing each RNA molecule within a digital reaction well at a uniform concentration, the amplification speed of deCOViD becomes independent of the sample RNA concentration.
Quantitative analysis of real‐time detection results further confirms the advantages of deCOViD in amplification speed and signal‐to‐background ratio. As the RNA concentration decreased from 200, 100 to 50 GE µL−1, the average time‐to‐positive (i.e., the time at which the sample fluorescence signal surpasses the predefined threshold) for the bulk assay increased from 19.3 ± 1.5, 22.0 ± 1.7 to 36.7 ± 2.1 min (Figure 2e and Figure S7a, Supporting Information). In contrast, the average time‐to‐positive for deCOViD remained consistent across the three RNA concentrations at 21.0 ± 4.1, 15.6 ± 4.3, and 19.6 ± 4.6 min (Figure 2e and Figure S7b, Supporting Information). These results show that deCOViD retains consistently rapid detection even as the RNA concentration decreases. This advantage is especially useful for qualitative detection, which could be achieved in <15 min—as soon as the first few reaction wells became positive—even for 50 RNA GE µL−1 (Figure S7b, Supporting Information). We further note that, as positive reaction wells across different RNA concentrations were all detectable within 30 min (Figure S8, Supporting Information), we could shorten the assay time to 30 min for quantitative detection and deCOViD could still retain higher signal‐to‐background ratio than the bulk assay and achieve robust detection (Figure 2f).
With the shortened 30 min assay time, we proceeded to demonstrate improved detection capability of deCOViD by challenging both the bulk assay and deCOViD with titrations of synthetic SARS‐CoV‐2 RNA. Relying on fluorescence intensity measurements, the bulk assay could detect down to 10 GE µL−1 (Figure 3a, Bulk). On the other hand, deCOViD quantifies the fraction of positive reaction wells (i.e., percent positive) and was able to detect 1 GE µL−1 (equivalent to ≈15 GE), or a tenfold improvement in the limit of detection (Figure 3a, deCOViD). The speed and sensitivity of deCOViD outstrip most CRISPR/Cas‐assisted SARS‐CoV‐2 assays, including DETECTR[ 22 ] and SHERLOCK[ 31 , 32 ] (Table S2, Supporting Information). Of note, we found that deCOViD was ≈4–5‐fold less sensitive than digital RT‐PCR (Figure S9, Supporting Information), but its 30 min assay time and isothermal reaction condition significantly offer significant advantages. Moreover, deCOViD widened the dynamic range for quantitative detection. The signal from the bulk assay saturated at ≈280 GE µL−1 (Figure S10a, Supporting Information), rendering quantitative detection above this concentration impossible. On the other hand, deCOViD displayed linearly correlated detection beyond 500 GE µL−1 (Figure S10b, Supporting Information). These results show that deCOViD enhances the assay sensitivity, quantification, and dynamic range—a clear illustration for the benefits of digitization.
Figure 3.
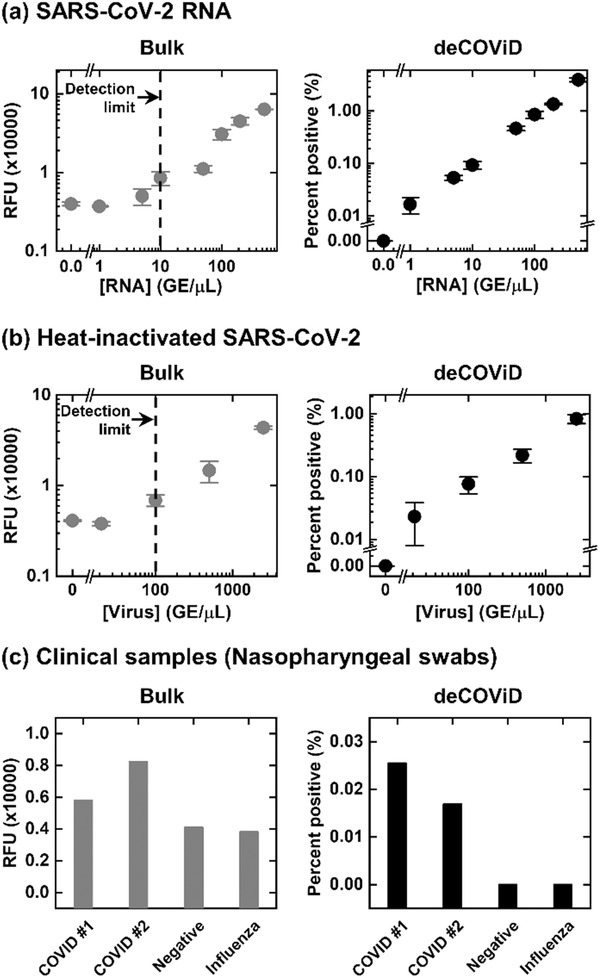
Detection of SARS‐CoV‐2 RNA, heat‐inactivated SARS‐CoV‐2, and clinical samples in 30 min. a) In detecting SARS‐CoV‐2 RNA, the bulk assay can detect ten genome equivalent (GE) µL−1, while deCOViD can detect 1 GE µL−1, or a tenfold improvement. This RNA detection sensitivity also outstrips existing CRISPR/Cas‐assisted assays. b) In detecting heat‐inactivated SARS‐CoV‐2, the bulk assay can detect 100 GE µL−1, while deCOViD can detect 20 GE µL−1, or a fivefold improvement. Notably, both assays can readily detect heat‐inactivated SARS‐CoV‐2 without RNA extraction. c) When testing RNA extracts from clinical nasopharyngeal swabs, both the bulk assay and deCOViD can differentiate positive samples from the negative sample and the influenza sample. Data in (a) and (b) presented as mean ± SD, n = 3.
We also showed that deCOViD outperformed the bulk assay in directly detecting heat‐inactivated SARS‐CoV‐2 without RNA extraction. For this demonstration, we spiked 2500, 500, 100, and 20 GE µL−1 of heat‐inactivated SARS‐CoV‐2 from NIAID BEI Resources (NR‐52350) directly into both the bulk assay and deCOViD. The bulk assay detected 100 GE µL−1 (Figure 3b, bulk), while deCOViD detected 20 GE µL−1 (Figure 3b, deCOViD), or a fivefold improvement. We note that heat‐inactivated SARS‐CoV‐2 was directly detected by both the bulk assay and deCOViD without any preparatory RNA extraction steps. Our results reveal the feasibility of directly detecting heat‐inactivated SARS‐CoV‐2 with a CRISPR/Cas‐assisted assay, a capability similar to previously reported RNA extraction‐free detection of heat‐inactivated SARS‐CoV‐2 via RT‐PCR.[ 41 ]
Finally, we demonstrated the feasibility of clinical sample testing. For this initial demonstration, we tested four RNA extracts from clinical nasopharyngeal swabs via both the bulk assay and deCOViD. The four samples, including two SARS‐CoV‐2 positive samples, one SARS‐CoV‐2 negative sample, and one influenza sample, were first confirmed via an in‐house RT‐qPCR assay that uses the US CDC‐approved SARS‐CoV‐2 primers and probes[ 42 ] (Figure S11, Supporting Information). Both the bulk assay and deCOViD yielded higher signals from the two positive samples than those from the negative sample—indicating successful detection results that agree with RT‐qPCR (Figure 3c). In addition, for the influenza sample, both the bulk assay and deCOViD yielded signals that are indistinguishable from those from the negative sample, which matches RT‐qPCR while also illustrating the specificity of both methods (Figure 3c and Figure S11, Supporting Information). Finally, as a demonstration, we diluted one of the positive samples by twofold and challenged both the bulk assay and deCOViD. This diluted sample became undetectable to the bulk assay but was detected by deCOViD (Figure S12, Supporting Information). These results thus provide additional support for the improved sensitivity of deCOViD over the bulk assay.
In conclusion, we have developed the first digital CRISPR/Cas‐assisted assay, deCOViD, that can rapidly and sensitively detect both SARS‐CoV‐2 RNA and heat‐inactivated SARS‐CoV‐2. Successful digitization of a fine‐tuned single‐step CRISPR/Cas12a‐assisted RT‐RPA assay in commercial QuantStudio digital chip, coupled to successful development of digital real‐time detection capability by incorporating a miniature heater, allows us to realize deCOViD and demonstrate enhancements in assay time, signal‐to‐background ratio, dynamic range, and sensitivity over the bulk assay. As a result, deCOViD can accelerate qualitative detection to <15 min and achieve quantitative detection down to 1 GE µL−1 RNA and 20 GE µL−1 heat‐inactivated SARS‐CoV‐2 without RNA extraction in 30 min—among the fastest and the most sensitive CRISPR/Cas‐assisted SARS‐CoV‐2 assays to date. In addition to building upon our initial demonstration of clinical sample testing and conducting a more thorough clinical validation, we see several routes for advancing deCOViD. First, we can further improve the speed and sensitivity of deCOViD by optimizing the concentrations of RPA primers, Cas12a effector, Cas12‐guide RNAs, and fluorogenic reporter in custom digital chips with greater total analysis volume and rapid digitization workflow.[ 43 , 44 , 45 ] We also envision enhancing the user‐friendliness of deCOViD by predigitizing and drying magnesium acetate (the chemical that initiates RPA) in digital chips.[ 46 ] Second, we can replace fluorescence microscopy with mobile phone‐based fluorescence detection[ 47 , 48 ] and couple to our miniature heater to make a portable device with the potential for point‐of‐care use. Third, as the relationship between SARS‐CoV‐2 viral load and COVID‐19 state remains incompletely understood, we suspect that our highly quantitative deCOViD may provide a new tool in such studies. Finally, as deCOViD can be readily designed for other DNA or RNA targets, we foresee applying deCOViD toward other diseases. Based on the encouraging results and potential for improvement, we believe deCOViD can open a new avenue for advancing CRISPR/Cas‐assisted diagnostic assays and provide a new tool for combating the COVID‐19 pandemic and beyond.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Acknowledgements
J.S.P. and K.H. contributed equally to this work. The authors are grateful for the financial support from the National Institutes of Health (R01AI138978 and R01AI137272) and the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases at Johns Hopkins University (FCDP‐010ZHA2020).
Park J. S., Hsieh K., Chen L., Kaushik A., Trick A. Y., Wang T.‐H., Digital CRISPR/Cas‐Assisted Assay for Rapid and Sensitive Detection of SARS‐CoV‐2. Adv. Sci. 2021, 8, 2003564. 10.1002/advs.202003564
References
- 1. Fauci A. S., Lane H. C., Redfield R. R., N. Engl. J. Med. 2020, 382, 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang C., Horby P. W., Hayden F. G., Gao G. F., Lancet 2020, 395, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., N. Engl. J. Med. 2020, 382, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bedford J., Enria D., Giesecke J., Heymann D. L., Ihekweazu C., Kobinger G., Lane H. C., Memish Z., Oh M.‐D., Sall A. A., Schuchat A., Ungchusak K., Wieler L. H., Lancet 2020, 395, 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esbin M. N., Whitney O. N., Chong S., Maurer A., Darzacq X., Tjian R., RNA 2020, 26, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weissleder R., Lee H., Ko J., Pittet M. J., Sci. Transl. Med. 2020, 12, eabc1931. [DOI] [PubMed] [Google Scholar]
- 7. Carter L. J., Garner L. V., Smoot J. W., Li Y., Zhou Q., Saveson C. J., Sasso J. M., Gregg A. C., Soares D. J., Beskid T. R., Jervey S. R., Liu C., ACS Cent. Sci. 2020, 6, 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feng W., Newbigging A. M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.‐B., Cui M., Tao J., Tyrrell D. L., Zhang X.‐E., Zhang H., Le X. C., Anal. Chem. 2020, 92, 10196. [DOI] [PubMed] [Google Scholar]
- 9. Ji T., Liu Z., Wang G., Guo X., khan S. A., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q., Biosens. Bioelectron. 2020, 166, 112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang Y.‐W., Schmitz J. E., Persing D. H., Stratton C. W., J. Clin. Microbiol. 2020, 58, e00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Udugama B., Kadhiresan P., Kozlowski H. N., Malekjahani A., Osborne M., Li V. Y. C., Chen H., Mubareka S., Gubbay J. B., Chan W. C. W., ACS Nano 2020, 14, 3822. [DOI] [PubMed] [Google Scholar]
- 12. Chu D. K. W., Pan Y., Cheng S. M. S., Hui K. P. Y., Krishnan P., Liu Y., Ng D. Y. M., Wan C. K. C., Yang P., Wang Q., Peiris M., Poon L. L. M., Clin. Chem. 2020, 66, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K., Bleicker T., Brünink S., Schneider J., Schmidt M. L., Mulders D. G., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.‐L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P., Drosten C., Eurosurveillance 2020, 25, 2000045. [Google Scholar]
- 14. Baek Y. H., Um J., Antigua K. J. C., Park J.‐H., Kim Y., Oh S., Kim Y.‐I., Choi W.‐S., Kim S. G., Jeong J. H., Chin B. S., Nicolas H. D. G., Ahn J.‐Y., Shin K. S., Choi Y. K., Park J.‐S., Song M.‐S., Emerging Microbes Infect. 2020, 9, 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thi V. L. D., Herbst K., Boerner K., Meurer M., Kremer L. P. M., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M. L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I. B., Schnitzler P., Kräusslich H.‐G., Knop M., Anders S., Sci. Transl. Med. 2020, 12, eabc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang W. E., Lim B., Hsu C.‐C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z., Microb. Biotechnol. 2020, 13, 950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee J. Y. H., Best N., McAuley J., Porter J. L., Seemann T., Schultz M. B., Sait M., Orlando N., Mercoulia K., Ballard S. A., Druce J., Tran T., Catton M. G., Pryor M. J., Cui H. L., Luttick A., McDonald S., Greenhalgh A., Kwong J. C., Sherry N. L., Graham M., Hoang T., Herisse M., Pidot S. J., Williamson D. A., Howden B. P., Monk I. R., Stinear T. P., bioRxiv 2020, 2020.04.28.067363. [Google Scholar]
- 18. Park G.‐S., Ku K., Baek S.‐H., Kim S.‐J., Kim S. I., Kim B.‐T., Maeng J.‐S., J. Mol. Diagn. 2020, 22, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J., Clin. Microbiol. Infect. 2020, 26, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.‐H., Yin X., Clin. Chem. 2020, 66, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ganguli A., Mostafa A., Berger J., Aydin M. Y., Sun F., Ramirez S. A. S. d., Valera E., Cunningham B. T., King W. P., Bashir R., Proc. Natl. Acad. Sci. USA 2020, 202014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V., Singh J., Miao X., Streithorst J. A., Granados A., Sotomayor‐Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.‐Y., Guevara H., Wadford D. A., Chen J. S., Chiu C. Y., Nat. Biotechnol. 2020, 38, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding X., Yin K., Li Z., Lalla R. V., Ballesteros E., Sfeir M. M., Liu C., Nat. Commun. 2020, 11, 4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R. R., Zhou Q., Li W., Cell Discovery 2020, 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Z., Tian D., Liu Y., Lin Z., Lyon C. J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B., Biosens. Bioelectron. 2020, 164, 112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lucia C., Federico P.‐B., Alejandra G. C., bioRxiv 2020, 2020.02.29.971127. [Google Scholar]
- 27. Ooi K. H., Tay J. W. D., Teo S. Y., Liu M. M., Kaewsapsak P., Jin S., Gao Y.‐G., Tan M. H., bioRxiv 2020, 2020.07.03.185850. [Google Scholar]
- 28. Wang X., Zhong M., Liu Y., Ma P., Dang L., Meng Q., Wan W., Ma X., Liu J., Yang G., Yang Z., Huang X., Liu M., Sci. Bull. 2020, 65, 1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoshimi K., Takeshita K., Yamayoshi S., Shibumura S., Yamauchi Y., Yamamoto M., Yotsuyanagi H., Kawaoka Y., Mashimo T., medRxiv 2020, 2020.06.02.20119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Gu B., Wang J., Xu T., PLoS Pathog. 2020, 16, e1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joung J., Ladha A., Saito M., Kim N.‐G., Woolley A. E., Segel M., Barretto R. P. J., Ranu A., Macrae R. K., Faure G., Ioannidi E. I., Krajeski R. N., Bruneau R., Huang M.‐L. W., Yu X. G., Li J. Z., Walker B. D., Hung D. T., Greninger A. L., Jerome K. R., Gootenberg J. S., Abudayyeh O. O., Zhang F., N. Engl. J. Med. 2020, 383, 1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J. S., Abudayyeh O. O., Zhang F., Horthongkham N., Uttamapinant C., Nat. Biomed. Eng. 2020. [DOI] [PubMed] [Google Scholar]
- 33. Sedlak R. H., Jerome K. R., Expert Rev. Mol. Diagn. 2014, 14, 501. [DOI] [PubMed] [Google Scholar]
- 34. Kuypers J., Jerome K. R., J. Clin. Microbiol. 2017, 55, 1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quan P. L., Sauzade M., Brouzes E., Sensors 2018, 18, 1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salipante S. J., Jerome K. R., Clin. Chem. 2020, 66, 117. [DOI] [PubMed] [Google Scholar]
- 37. Yuan H., Chao Y., Shum H. C., Small 2020, 16, 1904469. [DOI] [PubMed] [Google Scholar]
- 38. Piepenburg O., Williams C. H., Stemple D. L., Armes N. A., PLoS Biol. 2006, 4, e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen J. S., Ma E. B., Harrington L. B., Da Costa M., Tian X. R., Palefsky J. M., Doudna J. A., Science 2018, 360, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsieh K., Zhao G., Wang T.‐H., Analyst 2020, 145, 4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lubke N., Senff T., Scherger S., Hauka S., Andree M., Adams O., Timm J., Walker A., J. Clin. Virol. 2020, 130, 104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu X., Wang L., Sakthivel S. K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S. A., Harcourt J., Tamin A., Thornburg N. J., Villanueva J. M., Lindstrom S., Emerging Infect. Dis. 2020, 26, 1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsieh K., Zec H. C., Chen L., Kaushik A. M., Mach K. E., Liao J. C., Wang T.‐H., Anal. Chem. 2018, 90, 9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Keefe C. M., Pisanic T. R., Zec H., Overman M. J., Herman J. G., Wang T.‐H., Sci. Adv. 2018, 4, eaat6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Athamanolap P., Hsieh K., O'Keefe C. M., Zhang Y., Yang S., Wang T.‐H., Anal. Chem. 2019, 91, 12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yeh E.‐C., Fu C.‐C., Hu L., Thakur R., Feng J., Lee L. P., Sci. Adv. 2017, 3, e1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu H., Yaglidere O., Su T.‐W., Tseng D., Ozcan A., Lab Chip 2011, 11, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gou T., Hu J. M., Wu W. S., Ding X., Zhou S. F., Fang W. B., Mu Y., Biosens. Bioelectron. 2018, 120, 144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


