Abstract
The coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread into a global pandemic. Early and accurate diagnosis and quarantine remain the most effective mitigation strategy. Although reverse transcriptase polymerase chain reaction (RT-qPCR) is the gold standard for COVID-19 diagnosis, recent studies suggest that nucleic acids were undetectable in a significant number of cases with clinical features of COVID-19. Serologic assays that detect human antibodies to SARS-CoV-2 serve as a complementary method to diagnose these cases, as well as to identify asymptomatic cases and qualified convalescent serum donors. However, commercially available enzyme-linked immunosorbent assays (ELISA) are laborious and non-quantitative, while point-of-care assays suffer from low detection accuracy. To provide a serologic assay with high performance and portability for potential point-of-care applications, we developed DNA-assisted nanopore sensing for quantification of SARS-CoV-2 related antibodies in human serum. Different DNA structures were used as detection reporters for multiplex quantification of immunoglobulin M (IgM) and immunoglobulin G (IgG) antibodies against the nucleocapsid protein of SARS-CoV-2 in serum specimens from patients with conformed or suspected infection. Comparing to a clinically used point-of-care assay and an ELISA assay, our technology can reliably quantify SARS-CoV-2 antibodies with higher accuracy, large dynamic range, and potential for assay automation.
Keywords: COVID-19, SARS-CoV-2, Antibody, In vitro diagnostics, Nanopore
1. Introduction
In December 2019, China reported a new coronavirus that causes an acute respiratory disease named as coronavirus disease 19 (COVID-19) (Zhou et al., 2020). The virus was named SARS-CoV-2 as it was identified to be a betacoronavirus related to severe acute respiratory syndrome coronavirus (SARS-CoV) (Gorbalenya et al., 2020). As of January 2021, the COVID-19 pandemic has caused nearly two million death among 91 million confirmed cases worldwide. Before the large-scale deployment of vaccination, early detection and quarantine of asymptomatic or mild-symptomatic cases, as well as critical care for severely ill patients are key steps for mitigating the pandemic.
To date, amplification of the viral RNA from clinical specimens (i.e. nasal swabs, pharyngeal swabs, etc.) by RT-qPCR is still the gold-standard for COVID-19 diagnosis (Corman et al., 2020; Huang et al., 2020; Udugama et al., 2020). However, due to complexities in sample collection and processing, false results are commonly seen in clinical practice (Ai et al., 2020; Alvin et al., 2020; Fang et al., 2020; Xiao et al., 2020). Comparing to RT-qPCR and imaging tests, immunoassay based COVID-19 antigen/antibody tests are often faster, inexpensive, and user-friendly to medical staffs with minimal to no laboratory training (Xiang et al., 2020). In addition, serology analysis also supports a number of highly relevant applications: (1) detection of asymptomatic cases to reduce transmission (Bai et al., 2020); (2) identification of individuals with strong antibody responses who could serve as donors for convalescent serum therapies (Casadevall and Pirofski 2020; Shen et al., 2020; Tan et al., 2020); (3) quantitative evaluation of patients’ immune responses to SARS-CoV-2, which will inform prognosis, treatment and quarantine plans, etc. (Amanat et al., 2020; Zhao et al., 2020); (4) estimation of infection rate in affected areas to inform public health decisions (Okba et al., 2020). Current commercially available COVID-19 antibody assays mostly employ ELISA or lateral flow assay (LFA) technologies for lab-based and point-of-care testing (POCT) applications, respectively. Each of these technologies has its pros and cons: ELISA is more sensitive and widely used by clinical labs worldwide but requires labor-intensive hands-on procedures. Deployment of ELISA assays to the field and operations by inexperienced personnel may result in significantly reduced sensitivity (Dysinger et al., 2017; Elshal and McCoy 2006; Lewis et al., 2015). LFA does not require instruments, and is easy to use, store, and transport in resource-limited settings. However, its sensitivity suffer from the insufficient reaction time between the biorecognition element and the target, as well as from the simple readout technology (Sajid et al., 2015). To fill these gaps, there is an urgent need for a POCT technology with analytical performance comparable to or exceeding that of lab testing technologies.
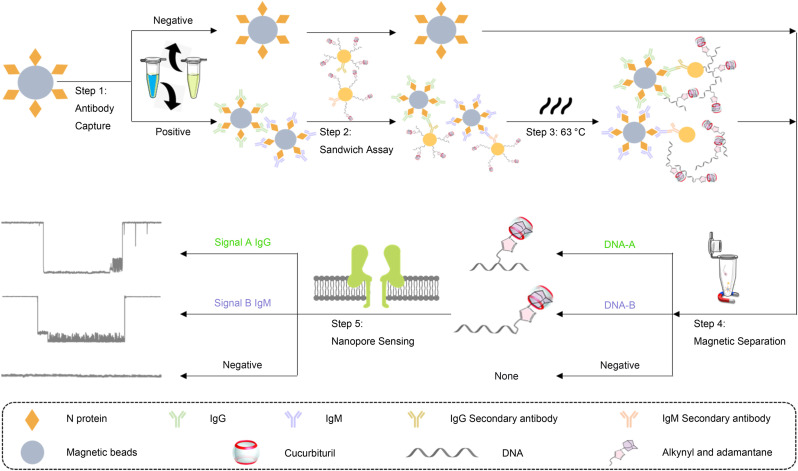
Since the introduction of electrical resistive pulse nanopore sensing, its single-molecule sensitivity and robustness suggest many potential applications in biosensing (Lenhart et al., 2020; Stoloff and Wanunu 2013). Consequently, nanopore based nucleic acid sequencing has been successfully commercialized (Callaway 2018; Deamer et al., 2016; Garalde et al., 2018; Lu et al., 2016; Samson et al., 2019). A nanopore sensor operates as a single-molecule level Coulter counter: under a voltage applied across a single nano-sized pore on a membrane that separates an ionic solution, an analyte driven through the nanopore (i.e. translocation) can cause a ionic current blockade which provides information of the molecule (Asandei et al., 2015; Howorka and Siwy 2009; Kasianowicz et al., 2008; Liu et al., 2018; Wanunu 2012; Wei et al. 2020a, 2020b; Wilson et al., 2016). Comparing to other delicate analytical instruments, its portability, robustness, and cost efficiency renders the nanopore technology great potentials for POCT (Eisenstein 2017). However, due to the nature of the electrophoresis-based stochastic sensing mechanism, native nanopores do not have specificity towards any analytes and is only suitable for detecting charged macromolecules (i.e. DNA) with a comparable size to the sensing region (i.e. constriction) in the pore. Based on DNA-assisted nanopore sensing (Guo et al. 2018, 2020a; Li et al., 2015; Liu et al., 2018; Wei et al., 2020c; You et al., 2019; Zhang et al., 2019), here we report a biosensing platform for accurate, multiplex, quantitative detection of SARS-CoV-2 specific antibodies with large dynamic ranges, and demonstrate its clinical utility by quantifying IgG and IgM against SARS-CoV-2 nucleocapsid (N) proteins in serum samples from confirmed and unconfirmed COVID-19 patients. The sensing principle of the assay involves: (1) capture of target antibodies with N protein functionalized magnetic beads (MBs); (2) labeling IgG or IgM antibodies with probe DNA modified gold nanoparticles (AuNPs); (3) thermal dehybridization of the probe DNAs from the AuNPs; (4) magnetic separation of probe DNAs from the sandwich structures; and (5) quantification of probe DNAs using the nanopore sensor (Fig. 1 ). Instead of directly detecting IgG or IgM antibodies, this approach uses two modified probe DNAs with different “host-guest” structures as detection surrogates. Translocation of these probes in the nanopore induces two unique oscillating patterns in the signal that are clearly different from signals of any other known biomolecules, which will afford highly specific detection. Comparing to clinically used LFA and ELISA assays, the nanopore biosensor can simultaneously quantify SARS-CoV-2 antibodies with higher accuracy and large dynamic range, as well as potential for assay automation.
Fig. 1.
Schematic representation of the DNA-assisted Nanopore Assay for multiplex quantification of SARS-CoV-2 antibodies. Step 1: IgG and IgM captured by the N-protein modified MBs. Step 2: Formation of the sandwich structure between MBs, IgG or IgM antibody, and probe DNA modified AuNPs. Step 3: Dehybridization of the probe DNAs from the AuNPs. Step 4: Magnetic separation of probe DNAs from the remaining sandwich complex. Step 5: Quantification of probe DNAs to derive the concentration of IgG and IgM, respectively.
2. Materials and methods
2.1. Reagents and buffers
The human COVID-19 IgG/IgM antibody ELISA kits, anti-COVID-19 Nucleocapsid humanized coronavirus monoclonal antibodies (IgG: MBS355908, IgM: MBS355899), and anti-human IgG Secondary antibody (MBS355657) were purchased from MyBiosource (San Diego, CA, USA). Anti-human IgM Secondary antibody (ab97201) was purchased from Abcam (Cambridge, UK). SARS-CoV-2 Nucleocapsid Protein (230–30164) and the Novel Coronavirus IgG/IgM antibody detection LFA kits were obtained from RayBiotech (Peachtree Corners, GA, USA). DNA samples with alkynyl modifications at different locations were purchased from Sangon Biotechnology (Shanghai, China). Bovine serum albumin (BSA) and Tween 20 (for molecular biology) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Lipid (1,2 diphytanoyl-sn-glycero-3-phosphocholine) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). Magnetic beads M-270 carboxylic acid were obtained from Invitrogen (Carlsbad, CA, USA). Gold colloids solution (PELCO® NanoXact™ Gold Colloids, 30 nm) was purchased from TED PELLA (Redding, CA, USA). Micro Bio-Spin P6 gel columns (Tris buffer) were purchased from Bio-Rad (Hercules, CA, USA). All reagents were used directly unless otherwise stated.
The assay buffer consists of 0.1 M NaCl, 0.025% Tween 20, 0.1% BSA and 10 mM PB at pH 7.2. The washing buffer consists of 0.15 M NaCl and 10 mM PB at pH 8.0. The detection buffer consists of 3 M KCl and 10 mM Tris-HCl at pH 8.0.
2.2. Preparation of functionalized AuNPs
Alkynyl modified DNA (100 μM, 3.3 μL) was first added into HEPES buffer (2 μL). After mixing, 1.2 μL deionized (DI) water, 2 μL Azidoadamantane (200 mM, dissolved in dimethyl sulfoxide), and 1 μL sodium ascorbate (20 mM, dissolved in water) were added into the HEPES buffer. Finally, 0.5 μL copper nitrate solution (20 mM, dissolved in water) was added and thoroughly mixed. The “click” reaction was allowed to take place for 8 h at 50 °C until stopped by the addition of 2 μL EDTA (100 mM). Micro Bio-Spin P6 gel columns were used to desalt the DNA products. The final probe DNAs were obtained by incubating with 10 μL cucurbituril [6] (5 mM) and 1 μL 5' thiol-modified DNA (100 mM in PBS) for 2 h (Liu et al., 2018; Wei et al., 2020c). To functionalize AuNPs, 1 mL Au colloid solution (pH 9.2) was mixed with 4 μg anti-human IgG or anti-human IgM antibodies and incubated at 10 °C for 30 min. Following this, respective probe DNAs were added to each type of antibody modified AuNPs and reacted at 4 °C for 2 h. After that, to facilitate the separation of AuNPs, the reaction solution was titrated with 100 μL NaCl solution (1.5 M) by adding 20 μL every 30 min. Finally, functionalized AuNPs were centrifuged at 14,000 g for 10 min, removed from supernatant, resuspended in 200 μL assay buffer, and kept at 4 °C (Li et al., 2015).
2.3. Preparation of functionalized MBs
The N protein functionalization of MBs is based on the reaction between carboxylic acid and carbodiimide. MBs were first activated following the protocol of the kit. After activation, 60 μg N protein was added to 100 μL MES buffer (25 mM, pH 5.0) and thoroughly mixed, followed by a 30 min incubation at room temperature. After incubation, the tube was placed on a magnet and the supernatant was removed. Functionalized MBs were then washed with 100 μL PBS for 4 times. The final product was blocked using BSA protein (0.1%), resuspended in 100 μL PBS, and stored at 4 °C for further use.
2.4. General procedures for antibody detection in human serum
The detection of human IgG and IgM follows similar protocols. N protein functionalized MBs (50 μL) were first washed with 500 μL assay buffer for five times, and then dissolved in 500 μL assay buffer. Each human serum sample was mixed with functionalized MBs (50 μL) and incubated for 1 h with vortex at room temperature. After the reaction, MBs were washed for seven times and resuspended in 200 μL assay buffer. Following this, functionalized AuNPs (with anti-human IgG or anti-human IgM antibodies) were added to the solution, and incubated with MBs for 2 h at 4 °C to form the sandwich structure between MBs and AuNPs. MBs were then separated from the solution and washed with assay buffer for five times. To avoid the influence of Tween-20 on the stability of the lipid bilayer that supports the nanopore, MBs were washed again using washing buffer. To dehybridize probe DNAs from the sandwich structures, MBs were added to 300 μL DI water and incubated at 63 °C for 15 min in water bath, followed by the separation and collection of the supernatant. Dehybridization was repeated for 3 times, and the supernatants were pooled and concentrated by ultra-centrifugation using Amicon Ultra-0.5 centrifugal Filter (3 KD) at 14,000 g for 15 min. The obtained single stranded probe DNA solution was kept for electrical resistive pulse nanopore sensing experiments. Standard curves were established using serum samples (200 μL) from healthy donors spiked with IgG antibody of different concentrations (1000 μg/mL, 100 μg/mL, 10 μg/mL, 1 μg/mL, 100 ng/mL,10 ng/mL) and IgM antibody of different concentrations (10 μg/mL, 5 μg/mL, 1 μg/mL, 100 ng/mL, 50 ng/mL).
2.5. Electrical resistive pulse nanopore sensing and data analysis
The formation of nanopore sensor, data recording and analysis were previously described (Wei et al. 2020a, 2020b, 2020c). For each experiment, a lipid bilayer was formed by 1,2-Diphytanoyl-sn-glycero3-phosphocholine in an 200 μm orifice punctured on a 25 μm thick Delrin wall that separates the cis (grounded) and the trans chambers of a flow cell. Both cis and trans contained 1 mL buffer (3 M KCl, 10 mM Tris, pH 8.0). A nanopore sensor was formed by inserting a single α-Hemolysin protein into the lipid bilayer from the cis side under 100 mV trans voltage. After adding a probe DNA sample to cis, electrical resistive pulse data was recorded using a patch clamp amplifier (Warner Instruments) at a holding potential of 200 mV. Each sample was measured in three replicates with fresh nanopores for at least 40 min total duration. The ionic current was sampled at 100 kHz using a Digidata 1440A analog-to-digital converter (Molecular Devices) and processed with pClamp11 software (Molecular Devices). The current blockade represents the capture and the translocation of a probe DNA in the nanopore and is defined as I/I 0 (I = I 0 -I b, I b: the average current measured with the DNA inside the pore; I 0: the average baseline current in absence of analytes). Dwell time (duration) represents the effective interaction time between nanopore and the probe DNA. Frequencies of characteristic oscillating events were calculated by visual inspections of raw data recordings.
2.6. Antibody detection using LFA and ELISA
The detection of IgG and IgM antibodies using LFA and ELISA was performed following the protocols provided with the commercialized kits. For LFA, each sample (25 μL) was mixed thoroughly with the sample diluent buffer, followed by the addition of 2–3 drops of the mixture to the release pad section (S) of the detection cassette. Results were obtained by visual inspection of the test and the control lines in 7 min. For ELISA, each sample (40 × dilution for IgG; 100 × dilution for IgM) was added to a pre-coated microtiter plate in three replicates (100 μL in each well). After incubation and washing steps, 100 μL of the conjugate solution was added to each well and incubated. Next, 100 μL substrate solution was added and incubated without light. Finally, the plate was treated using stop solution, and the optical density at 450 nm was determined by a microplate reader for each well.
2.7. Clinical samples and data sources
Serum samples were obtained from subjects enrolled at Prisma Health Richland Hospital from May to June 2020 (Approval # Pro00098873). Positive and negative subjects were confirmed by SARS-CoV-2 specific RT-qPCR. Possible (unconfirmed) subjects were characterized by symptoms and contact history, and were treated as COVID-19 cases.
3. Results
3.1. DNA-assisted nanopore enabled COVID-19 antibody assay
Human immune response against SARS-CoV-2 is similar to it against other viral pathogens. In 5–10 days after infection, IgM appears in blood first, followed by the generation of IgG for sustained immune suppression (Guo et al., 2020b; Ravi et al., 2020). Accurate longitudinal quantification of SARS-CoV-2 specific IgG and IgM may greatly improve COVID-19 diagnosis and prognosis. While antibodies induced by S proteins are known for neutralizing effects in convalescent plasma that is suitable for treatment purposes (Amanat et al., 2020), other studies indicated that antibodies to N proteins is more abundant during active infection (Dutta et al., 2020), and thus are more suitable for detection (Burbelo et al., 2020a, b).
In this study, we selected SARS-CoV-2 N protein specific IgG and IgM antibodies as biomarkers to demonstrate a multiplex quantitative nanopore assay, aiming to achieve high sensitivity and high specificity comparable to lab-based technologies. First, IgG and IgM antibodies were captured and enriched from a human serum sample using N protein modified MBs; two different sandwich structures were then formed for IgG and IgM MBs using two types of AuNPs co-functionalized by anti-IgG with DNA-A or anti-IgM with DNA-B, respectively; finally, after dehybridization and separation from the sandwich structures, DNA-A and -B were quantified by a α-HL nanopore biosensor to indicate IgG and IgM concentrations, respectively (Fig. 1).
3.2. Characterization of probe DNAs for multiplex quantification
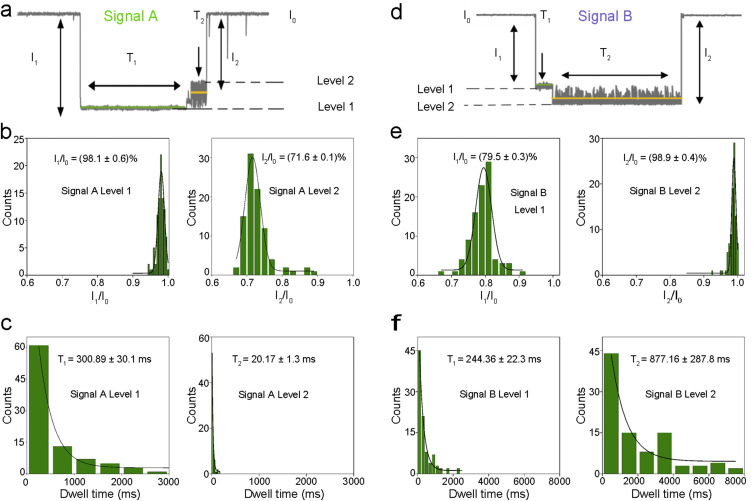
The highly specific multiplex quantification of SARS-CoV-2 antibodies is based on the two different characteristic signals by translocations of adamantane-cucurbituril[6]-modified probe DNAs through α-hemolysin (α-HL). First, two DNAs (DNA-A and DNA-B) with alkynyl modifications at different locations (center and 5’ end) were “clicked” with azidoadamantane and characterized by mass spectrometry (Fig. S1). Next, probe DNAs were reacted with cucurbituril [6] and characterized by nanopore measurement. For the generation of signal A, the translocation of DNA-A can be described as a series of processes (Fig. S2a). When a probe DNA-A enters the nanopore from the cis side, the front end of the single-stranded DNA (ssDNA) passes through, while the host-guest modification sites was blocked by the narrow entrance of the β-barrel region and stayed in the lumen region. Under the applied voltage, the ssDNA molecule continues to be driven through the β-barrel completely, leading to the separation of the host molecule and the guest molecule. This process is characterized by the level 1 of signal A (Fig. 2 a). Next, after separation, the modified ssDNA exit the nanopore to the trans side, while cucurbituril[6] oscillates inside the lumen for a period of time before exiting back to the cis side of the nanopore (Fig. S2a). This process corresponds to the level 2 of signal A (Fig. 2a). Upon analyzing ~100 events of signal A in histograms, the average current blockade of level 1 (I1/I0) and level 2 (I2/I0) were found to be (98.1 ± 0.6)% and (71.6 ± 0.1)% with Gaussian distribution, respectively (Fig. 2b). The average dwell time for level 1 (T1) and level 2 (T2) were (300.89 ± 30.1) ms and (20.17 ± 1.3) ms, respectively, corresponding to exponential fitting (Fig. 2c).
Fig. 2.
Statistical characterization of signals by two different probe DNAs. a: Signal A is generated by DNA-A. b: Histograms of current blockades of level 1 and level 2 in Signal A. The solid lines are Gaussian fit to the histograms. c: Histograms of dwell times of level 1 and level 2 in Signal A. The solid lines are single exponential fit to the histograms. d: Signal B is generated by DNA-B. e: Histograms of current blockades of level 1 and level 2 in Signal B. The solid lines are Gaussian fit to the histograms. f: Histograms of dwell times of level 1 and level 2 in Signal B. The solid lines are single exponential fit to the histograms. All electrical resistive pulse nanopore sensing data was acquired in 3 M KCl, 10 mM tris buffer, pH 8.0, n = 3.
The difference between signal A and signal B is caused by the different modification sites on the two types of ssDNA molecules. For DNA-A, the modification site is at the center of the ssDNA, while the modification site of DNA-B is at the 5′ end of the ssDNA. Similar to DNA-A, the translocation of DNA-B in the nanopore also involves the separation of the host-guest complex and the oscillation of cucurbituril[6], which subsequently generate levels 1 and 2 in signal B (Fig. S2b). However, due to the different structure of DNA-B, the pattern of signal B is significantly different from that of signal A (Fig. 2d). In signal B events, the average current blockade of level 1 (I1/I0) and level 2 (I2/I0) were found to be (79.5 ± 0.3) % and (98.9 ± 0.4) % with Gaussian distribution, respectively (Fig. 2e). The average dwell time for level 1 (T1) and level 2 (T2) were (244.36 ± 22.3) ms and (877.16 ± 287.8) ms, respectively, with exponential fitting (Fig. 2f). We anticipate that such difference in signal patterns was caused by the early separation of the host-guest complex when 5’ modified probe DNAs (DNA-B) traverse the nanopore.
3.3. Quantification of SARS-CoV-2 specific IgG and IgM in human sera
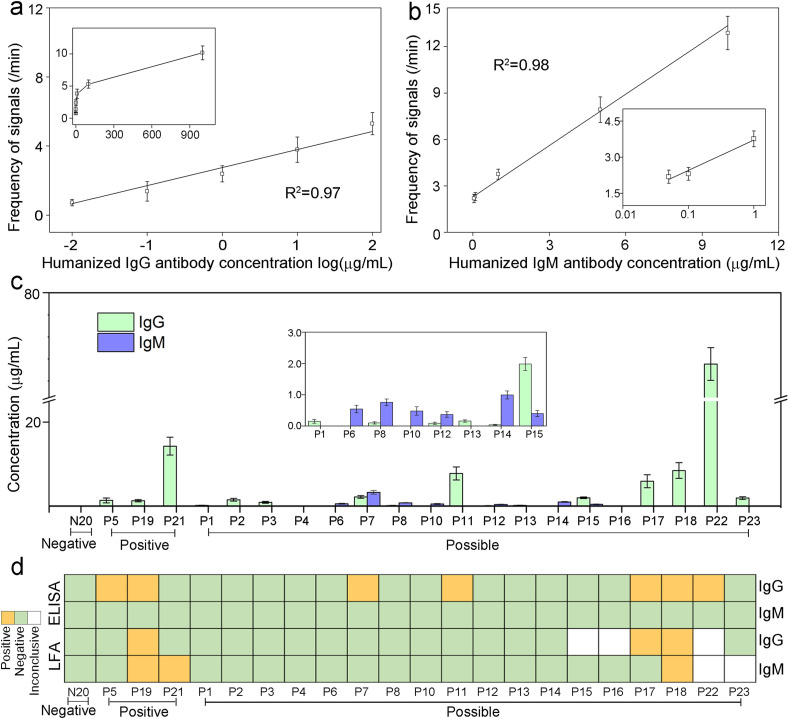
To quantitatively measure SARS-CoV-2 IgG and IgM in patient samples, standard curves were first established using healthy donors’ sera spiked with various amount of humanized COVID-19 monoclonal antibodies. Concentration ranges of IgG and IgM on the curves were determined based on previous clinical observations (Roy et al., 2020; Li et al., 2020; Ma et al., 2020; Iyer et al., 2020). Both signal A and B frequencies displayed excellent linear relationships with IgG and IgM concentrations, respectively (Fig. 3 a&b, Fig. S3&S4). Based on calculation from limit of blanks (Armbruster and Pry 2008), the limits of detection (LOD) for IgG and IgM are 10 ng/mL and 50 ng/mL, respectively, with large dynamic range up to the μg/mL level, indicating suitability to antibody quantification at all stages of infection.
Fig. 3.
Quantification of SARS-CoV-2 IgG and IgM in human serum samples. a: Standard curve of correlation between signal frequencies and concentrations of humanized IgG spiked in blank human serum (0.01–100 μg/mL). Inset shows the curve at 0–1000 μg/mL. b: Standard curve of correlation between signal frequencies and concentrations of humanized IgM spiked in blank human serum (0.05–10 μg/mL). Inset shows the curve at lower concentration ranges. c: IgG and IgM concentrations measured by the DNA-assisted Nanopore Assay for 1 negative sample, 3 positive samples, and 18 possible samples. If the calculated concentration was below the LOD, the concentration is marked as 0 μg/mL. Data recording conditions are the same as used in Fig. 2d: Qualitative IgG and IgM results for matching samples measured by the ELISA kit and the LFA kit.
For assay validation and feasibility assessment, we further applied the DNA-assisted nanopore sensing assay to quantify IgG and IgM antibodies in serum samples from confirmed and unconfirmed patients enrolled at Prisma Health Richland Hospital (Fig. 3c), and compared to the results from a LFA kit and an ELISA kit used at the hospital (Fig. 3d, Fig. S5&S6 and Table S1). In three COVID-19 positive patients confirmed by RT-qPCR, the LFA kit detected one with both IgG and IgM and one with IgM only, the ELISA kit detected two with IgG only, whereas the DNA-assisted nanopore assay detected all three of them with IgG only. In eighteen possible COVID-19 patients, the LFA kit detected two with IgG and/or IgM, the ELISA kit detected five with IgG only. However, using the DNA-assisted nanopore assay, we were able to detect IgG and/or IgM in sixteen of them. It is worthy to notice that among all possible patients, the five patients that are detectable by IgG ELISA also exhibited highest IgG levels according to our nanopore assay results, indicating nanopore assay's excellent correlation with traditional lab-based methods and its superior detection sensitivity. None of the sensing methods used in this study showed false-positive results for the confirmed negative control individual. To further evaluate its specificity in extreme conditions, the nanopore assay was used to test serum samples from healthy donors and a patient spiked with extra amount of common assay interferents (Hemolysate, Triglyceride, Bilirubin, and Human Albumin). Although a matrix effect was observed for both positive and negative samples, we do not anticipate significant limitation to the quantitative capacity of the nanopore assay, as such high amount of interferents are not commonly seen in clinical serum samples (Fig. S7).
4. Discussion
The COVID-19 outbreak reinforces the pressing needs for more effective diagnostics for infectious diseases (Zhou et al., 2020). Due to the fast-spreading nature of most infectious diseases, technologies that can enable rapid and easy detections at the point of care is highly desired. However, these are usually achieved at the cost of significant loss of detection accuracy and quantification features (Dysinger et al., 2017; Elshal and McCoy 2006; Lewis et al., 2015; Sajid et al., 2015). To mitigate the current pandemic and to ready our medical forces for the next public health emergency, there is an urgent unmet need for a POCT technology with analytical performance comparable to or exceeds lab testing technologies.
In order to reliably detect biomarkers from complex biological samples using nanopore biosensors, previous studies have attempted to improve the sensitivity and/or the specificity via modifications to the biomarkers (Lin et al., 2017; Tang et al., 2020; Zhang et al., 2017), engineering the nanopores (Wang et al., 2013), and combination with signal amplification mechanisms (Nouri et al., 2020). In this study, we developed a DNA-assisted nanopore biosensing assay to enable multiplex quantification of biomarkers from human sera. The single-molecule level detection capacity and the unique oscillation signals of probe DNAs afford both high sensitivity and high specificity of the assay. Although the current platform still requires manual operations, the robustness of nanopore sensing mechanism and the magnetic beads-based immunoprecipitation can ensure easy adoption of assay automation. Upon incorporation with microfluidics and scaled-up production, the cost of this nanopore assay is expected to be slightly lower than that of ELISA (~$8 per test). Notably, this assay can also be readily modified with minimal optimization to quantify SARS-CoV-2 antigens in nasopharyngeal swab samples.
In the performance evaluation study using patients’ samples, the DNA-assisted nanopore assay was applied to detect IgG and IgM antibodies against the N protein. For comparison, a LFA and an ELISA made using the same biorecognition element that targets the same antibodies were also used to test the same human samples. Among the three technologies, only the DNA-assisted nanopore assay shows quantitative capacity with low LOD and large dynamic range. The ELISA assay did not detect IgM in any samples, but detected IgG in seven positive and possible samples (P5, P19, P7, P11, P17, P18, P22). The DNA-assisted nanopore assay confirmed that IgG levels in these samples are among the highest in all tested samples. However, LFA was only able to detect IgG in three of these seven samples (P19, P17, P18). In addition, the nanopore assay also detected lower levels of IgG in nine positive and possible samples that are undetectable by ELISA and LFA. Among the three samples with detectable IgM by LFA (P19, P21, P18), no detectable IgM result was observed by the nanopore assay. Seven possible samples (P6, P7, P8, P10, P12, P14, P15) exhibited detectable IgM by the nanopore assay. The overall low IgM positivity could be attributed to the decrease of IgM after the acute phase of the infection. However, due to the difficulty of contact tracing in the US during the early phases of the pandemic, precise infection time is not available for these samples.
In a review of seven other newly reported COVID-19 antibody testing technologies, several were found to be semi-quantitative (Mairesse et al., 2020; Soleimani et al., 2021), while others have narrow dynamic range or lack clinically validation (Table S2) (Tan et al., 2020; Shaw et al., 2020; Roy et al., 2020; Liu et al., 2020; Dzimianski et al., 2020). In general, we believe that the DNA-assisted nanopore assay can simultaneously quantify SARS-CoV-2 antibodies with higher accuracy and larger dynamic range, while correlating with results from the “gold standard diagnostic technology” ELISA.
5. Conclusions
The DNA-assisted nanopore sensing assay provides a sensitive and robust strategy for multiplex quantification of SARS-CoV-2 specific IgG and IgM antibodies in human serum. Although the current assay protocol still involves multiple manual steps and long incubations, we envision that, with the incorporation of microfluidics-based assay automation, this method may be applied in the future as a highly accurate POCT in large scale testing efforts for COVID-19 diagnosis and monitoring in resource-limited settings. While the proof-of-concept results appear to be promising, this assay warrants further investigation using well-organized patient cohorts with longitudinal sampling. We are currently recruiting a larger validation cohort with more rigorous enrollment criteria on contact tracing, and is developing a palm-sized, battery-powered, automatic nanopore assay device.
CRediT authorship contribution statement
Zehui Zhang: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft. Xiaoqin Wang: Methodology, Investigation. Xiaojun Wei: Formal analysis, Visualization. Sophia W. Zheng: Formal analysis, Writing – review & editing. Brian J. Lenhart: Formal analysis. Peisheng Xu: Resources, Formal analysis. Jie Li: Resources, Formal analysis. Jing Pan: Formal analysis, Writing – review & editing. Helmut Albrecht: Resources, Formal analysis, Writing – review & editing. Chang Liu: Conceptualization, Funding acquisition, Resources, Supervision, Methodology, Formal analysis, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work is supported by the COVID-19 Research Initiative of the University of South Carolina Office of the Vice President for Research. C. Liu acknowledges financial supports from: the Biomedical Engineering Program and the Department of Chemical Engineering of the University of South Carolina, the National Institute of Allergy and Infectious Diseases (NIAID) award K22AI136686, the South Carolina IDeA Networks of Biomedical Research Excellence Developmental Research Project by the National Institute of General Medical Sciences (NIGMS) award P20GM103499, and the National Science Foundation (NSF) CAREER Award 2047503. The authors are grateful to the study participants and the medical professionals at Prisma Health and Palmetto Health USC Medical Group.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113134.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvin M.D., George E., Deng F., Warhadpande S., Lee S.I. The impact of COVID-19 on radiology trainees. Radiology. 2020;296(2):246–248. doi: 10.1148/radiol.2020201222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster D.A., Pry T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008;29(Suppl. 1):S49–S52. [PMC free article] [PubMed] [Google Scholar]
- Asandei A., Chinappi M., Lee J.K., Seo C.H., Mereuta L., Park Y., Luchian T. Placement of oppositely charged aminoacids at a polypeptide termini determines the voltage-controlled braking of polymer transport through nanometer-scale pores. Sci. Rep. 2015;5:13. doi: 10.1038/srep10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Cohen J.I. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. medRxiv. 2020 2020.2004.2020.20071423. [Google Scholar]
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Jr., Cohen J.I. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222(2):206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Flu virus finally sequenced in its native form Technique could help to unpick role of enigmatic chemical modifications in genetic material. Nature. 2018;556(7702) doi: 10.1038/d41586-018-04908-5. 420-420. [DOI] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.-a. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130(4):1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D., Akeson M., Branton D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016;34(5):518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J. Virol. 2020;94(13) doi: 10.1128/JVI.00647-20. e00647-00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dysinger M., Marusov G., Fraser S. Quantitative analysis of four protein biomarkers: an automated microfluidic cartridge-based method and its comparison to colorimetric ELISA. J. Immunol. Methods. 2017;451:1–10. doi: 10.1016/j.jim.2017.08.009. [DOI] [PubMed] [Google Scholar]
- Dzimianski J.V., Lorig-Roach N., O'Rourke S.M., Alexander D.L., Kimmey J.M., DuBois R.M. Rapid and sensitive detection of SARS-CoV-2 antibodies by biolayer interferometry. Sci. Rep. 2020;10(1):21738. doi: 10.1038/s41598-020-78895-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M. An ace in the hole for DNA sequencing. Nature. 2017;550(7675):285–288. doi: 10.1038/550285a. [DOI] [PubMed] [Google Scholar]
- Elshal M.F., McCoy J.P. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods. 2006;38(4):317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A., Jordan M., Ciccone J., Serra S., Keenan J., Martin S., McNeill L., Wallace E.J., Jayasinghe L., Wright C., Blasco J., Young S., Brocklebank D., Juul S., Clarke J., Heron A.J., Turner D.J. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018;15(3):201. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of, V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Sheng Y., Zhou K., Liu Q., Liu L., Wu H.-C. Analyte-triggered DNA-probe release from a triplex molecular beacon for nanopore sensing. Angew. Chem. Int. Ed. 2018;57(14):3602–3606. doi: 10.1002/anie.201711690. [DOI] [PubMed] [Google Scholar]
- Guo B., Song P., Zhou K., Liu L., Wu H.-C. Simultaneous sensing of multiple cancer biomarkers by a single DNA nanoprobe in a nanopore. Anal. Chem. 2020;92(13):9405–9411. doi: 10.1021/acs.analchem.0c01909. [DOI] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.-W., Xu S.-Y., Zhu H.-D., Xu Y.-C., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S., Siwy Z. Nanopore analytics: sensing of single molecules. Chem. Soc. Rev. 2009;38(8):2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A. vol. 5. 2020. (Persistence and Decay of Human Antibody Responses to the Receptor Binding Domain of SARS-CoV-2 Spike Protein in COVID-19 Patients). 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J.J., Robertson J.W.F., Chan E.R., Reiner J.E., Stanford V.M. Nanoscopic porous sensors. Annu. Rev. Anal. Chem. 2008;1:737–766. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- Lenhart B., Wei X., Zhang Z., Wang X., Wang Q., Liu C. Nanopore fabrication and application as biosensors in neurodegenerative diseases. Crit. Rev. Biomed. Eng. 2020;48(1):29–62. doi: 10.1615/CritRevBiomedEng.2020033151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.M., Macpherson P., Adams E.R., Ochodo E., Sands A., Taegtmeyer M. Field accuracy of fourth-generation rapid diagnostic tests for acute HIV-1: a systematic review. AIDS. 2015;29(18):2465–2471. doi: 10.1097/QAD.0000000000000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu L., Li Y., Xie J., Wu H.-C. A universal strategy for aptamer-based nanopore sensing through host–guest interactions inside α-hemolysin. Angew. Chem. Int. Ed. 2015;54(26):7568–7571. doi: 10.1002/anie.201502047. [DOI] [PubMed] [Google Scholar]
- Li K., Huang B., Wu M., Zhong A., Li L., Cai Y., Wang Z., Wu L., Zhu M., Li J., Wang Z., Wu W., Li W., Bosco B., Gan Z., Qiao Q., Wu J., Wang Q., Wang S., Xia X. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat. Commun. 2020;11(1):6044. doi: 10.1038/s41467-020-19943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Ying Y.-L., Shi X., Liu S.-C., Long Y.-T. Direct sensing of cancer biomarkers in clinical samples with a designed nanopore. Chem. Commun. 2017;53(84):11564–11567. doi: 10.1039/c7cc06775e. [DOI] [PubMed] [Google Scholar]
- Liu L., Li T., Zhang S., Song P., Guo B., Zhao Y., Wu H.C. Simultaneous quantification of multiple cancer biomarkers in blood samples through DNA-assisted nanopore sensing. Angew. Chem. 2018;57(37):11882–11887. doi: 10.1002/anie.201803324. [DOI] [PubMed] [Google Scholar]
- Liu T., Hsiung J., Zhao S., Kost J., Sreedhar D., Hanson C.V., Olson K., Keare D., Chang S.T., Bliden K.P., Gurbel P.A., Tantry U.S., Roche J., Press C., Boggs J., Rodriguez-Soto J.P., Montoya J.G., Tang M., Dai H. Quantification of antibody avidities and accurate detection of SARS-CoV-2 antibodies in serum and saliva on plasmonic substrates. Nature Biomedical Engineering. 2020;4(12):1188–1196. doi: 10.1038/s41551-020-00642-4. [DOI] [PubMed] [Google Scholar]
- Lu H.Y., Giordano F., Ning Z.M. a3 oxford nanopore MinION sequencing and genome assembly. Dev. Reprod. Biol. 2016;14(5):265–279. doi: 10.1016/j.gpb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17(7):773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairesse A., Favresse J., Eucher C., Elsen M., Tré-Hardy M., Haventith C., Gruson D., Dogné J.-M., Douxfils J., Göbbels P. High clinical performance and quantitative assessment of antibody kinetics using a dual recognition assay for the detection of SARS-CoV-2 IgM and IgG antibodies. Clin. Biochem. 2020;86:23–27. doi: 10.1016/j.clinbiochem.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri R., Jiang Y., Lian X.L., Guan W. Sequence-specific recognition of HIV-1 DNA with solid-state CRISPR-cas12a-assisted nanopores (SCAN) ACS Sens. 2020;5(5):1273–1280. doi: 10.1021/acssensors.0c00497. [DOI] [PubMed] [Google Scholar]
- Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv. 2020 doi: 10.3201/eid2607.200841. 2020.2003.2018.20038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy V., Fischinger S., Atyeo C., Slein M., Loos C., Balazs A., Luedemann C., Astudillo M.G., Yang D., Wesemann D.R., Charles R., Lafrate A.J., Feldman J., Hauser B., Caradonna T., Miller T.E., Murali M.R., Baden L., Nilles E., Ryan E., Lauffenburger D., Beltran W.G., Alter G. SARS-CoV-2-specific ELISA development. J. Immunol. Methods. 2020;484–485:112832. doi: 10.1016/j.jim.2020.112832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid M., Kawde A.-N., Daud M. Designs, formats and applications of lateral flow assay: a literature review. Journal of Saudi Chemical Society. 2015;19(6):689–705. [Google Scholar]
- Samson R., Shah M., Yadav R., Sarode P., Rajput V., Dastager S.G., Dharne M.S., Khairnar K. Metagenomic insights to understand transient influence of Yamuna River on taxonomic and functional aspects of bacterial and archaeal communities of River Ganges. Sci. Total Environ. 2019;674:288–299. doi: 10.1016/j.scitotenv.2019.04.166. [DOI] [PubMed] [Google Scholar]
- Shaw A.M., Hyde C., Merrick B., James-Pemberton P., Squires B.K., Olkhov R.V., Batra R., Patel A., Bisnauthsing K., Nebbia G., MacMahon E., Douthwaite S., Malim M., Neil S., Martinez Nunez R., Doores K., Mark T.K.I., Signell A.W., Betancor G., Wilson H.D., Galão R.P., Pickering S., Edgeworth J.D. Real-world evaluation of a novel technology for quantitative simultaneous antibody detection against multiple SARS-CoV-2 antigens in a cohort of patients presenting with COVID-19 syndrome. Analyst. 2020;145(16):5638–5646. doi: 10.1039/d0an01066a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani R., Khourssaji M., Gruson D., Rodriguez-Villalobos H., Berghmans M., Belkhir L., Yombi J.-C., Kabamba-Mukadi B. Clinical usefulness of fully automated chemiluminescent immunoassay for quantitative antibody measurements in COVID-19 patients. J. Med. Virol. 2021;93(3):1465–1477. doi: 10.1002/jmv.26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoloff D.H., Wanunu M. Recent trends in nanopores for biotechnology. Curr. Opin. Biotechnol. 2013;24(4):699–704. doi: 10.1016/j.copbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Krel M., Dolgov E., Park S., Li X., Wu W., Sun Y.-L., Zhang J., Khaing Oo M.K., Perlin D.S., Fan X. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens. Bioelectron. 2020;169:112572. doi: 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wang H., Yang C., Zhao D., Qian Y., Li Y. Nanopore-based strategy for selective detection of single carcinoembryonic antigen (CEA) molecules. Anal. Chem. 2020;92(4):3042–3049. doi: 10.1021/acs.analchem.9b04185. [DOI] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Wang S., Haque F., Rychahou P.G., Evers B.M., Guo P. Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano. 2013;7(11):9814–9822. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M. Nanopores: a journey towards DNA sequencing. Phys. Life Rev. 2012;9(2):125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Ma D., Jing L., Wang L.Y., Wang X., Zhang Z., Lenhart B.J., Yin Y., Wang Q., Liu C. Enabling nanopore for sensing individual amino acids by a derivatization strategy. J. Mater. Chem. B. 2020 doi: 10.1039/d0tb00895h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Ma D., Zhang Z., Wang L.Y., Gray J.L., Zhang L., Zhu T., Wang X., Lenhart B.J., Yin Y., Wang Q., Liu C. N-terminal derivatization-assisted identification of individual amino acids using a biological nanopore sensor. ACS Sens. 2020 doi: 10.1021/acssensors.0c00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Zhang Z., Wang X., Lenhart B., Gambarini R., Gray J., Liu C. Insight into the effects of electrochemical factors on host-guest interaction induced signature events in a biological nanopore. Nanotechnology and Precision Engineering. 2020;3(1):2–8. doi: 10.1016/j.npe.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J., Sloman L., He Z.R., Aksimentiev A. Graphene nanopores for protein sequencing. Adv. Funct. Mater. 2016;26(27):4830–4838. doi: 10.1002/adfm.201601272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H., Wei X., Cai P., Ma W.-L. Clinical Infectious Diseases; 2020. Antibody Detection and Dynamic Characteristics in Patients with Coronavirus Disease 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A.T., Tong Y.X., Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J. Med. Virol. 2020;92(10):1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Zhou K., Guo B., Liu Q., Cao Z., Liu L., Wu H.-C. Measuring binding constants of cucurbituril-based host–guest interactions at the single-molecule level with nanopores. ACS Sens. 2019;4(3):774–779. doi: 10.1021/acssensors.9b00408. [DOI] [PubMed] [Google Scholar]
- Zhang J.-H., Liu X.-L., Hu Z.-L., Ying Y.-L., Long Y.-T. Intelligent identification of multi-level nanopore signatures for accurate detection of cancer biomarkers. Chem. Commun. 2017;53(73):10176–10179. doi: 10.1039/c7cc04745b. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Li T., Sheng Y., Liu L., Wu H.-C. Enhanced sensitivity in nanopore sensing of cancer biomarkers in human blood via click chemistry. Small. 2019;15(2):1804078. doi: 10.1002/smll.201804078. [DOI] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Clinical Infectious Diseases; 2020. Antibody Responses to SARS-CoV-2 in Patients with Novel Coronavirus Disease 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.