The limited therapeutic options and the recent emergence of multidrug-resistant Candida species present a significant challenge to human medicine and underscore the need for novel therapeutic approaches. Drug repurposing appears as a promising tool to augment the activity of current azole antifungals, especially against multidrug-resistant Candida auris. In this study, we evaluated the fluconazole chemosensitization activities of 1,547 FDA-approved drugs and clinical molecules against azole-resistant C. auris.
KEYWORDS: HIV protease inhibitors, Candida auris, glucose transporters, glucose-induced acidification assay, ATP bioluminescence assay, Nile red efflux assay, azole resistance
ABSTRACT
The limited therapeutic options and the recent emergence of multidrug-resistant Candida species present a significant challenge to human medicine and underscore the need for novel therapeutic approaches. Drug repurposing appears as a promising tool to augment the activity of current azole antifungals, especially against multidrug-resistant Candida auris. In this study, we evaluated the fluconazole chemosensitization activities of 1,547 FDA-approved drugs and clinical molecules against azole-resistant C. auris. This led to the discovery that lopinavir, an HIV protease inhibitor, is a potent agent capable of sensitizing C. auris to the effect of azole antifungals. At a therapeutically achievable concentration, lopinavir exhibited potent synergistic interactions with azole drugs, particularly with itraconazole against C. auris (fractional inhibitory concentration index [ΣFICI] ranged from 0.04 to 0.09). Additionally, the lopinavir/itraconazole combination enhanced the survival rate of C. auris-infected Caenorhabditis elegans by 90% and reduced the fungal burden in infected nematodes by 88.5% (P < 0.05) relative to that of the untreated control. Furthermore, lopinavir enhanced the antifungal activity of itraconazole against other medically important Candida species, including C. albicans, C. tropicalis, C. krusei, and C. parapsilosis. Comparative transcriptomic profiling and mechanistic studies revealed that lopinavir was able to significantly interfere with the glucose permeation and ATP synthesis. This compromised the efflux ability of C. auris and consequently enhanced the susceptibility to azole drugs, as demonstrated by Nile red efflux assays. Altogether, these findings present lopinavir as a novel, potent, and broad-spectrum azole-chemosensitizing agent that warrants further investigation against recalcitrant Candida infections.
INTRODUCTION
Since its emergence in 2009, Candida auris has been implicated in several global outbreaks of serious invasive infections that are usually associated with high mortality rates (40 to 60%) (1–5). Phylogenetically, C. auris isolates have been grouped into four distinct clades, South Asian (clade I), East Asian (clade II), African (clade III), and South American (clade IV) (6, 7). These clades differ significantly in several attributes, including geographic prevalence, susceptibility to antifungal drugs, mechanisms of drug resistance, and pathogenesis. In contrast to those in clade II, isolates belonging to clades I, III, and IV demonstrate a higher propensity for invasive infections and are more resistant to antifungal drugs (8). Additionally, isolates belonging to clades I and IV were found to be more virulent in animal models of invasive candidemia than other isolates (6, 9). The remarkable resistance to standard antifungal agents, the ability to withstand commonly used disinfectants, the ability to persist on abiotic surfaces for extended periods, and the efficient transmissibility among patients are key elements underlying the global threat posed by C. auris (1, 10–12). In recognition of this threat, the U.S. Centers for Disease Control and Prevention (CDC) has recently classified C. auris as an urgent threat that requires immediate action (13).
Unfortunately, only three classes of antifungal drugs—azoles, polyenes, and echinocandins—are currently in use for the treatment of invasive Candida infections (14) According to the CDC, approximately 90% of C. auris isolates in the United States were reported to be fluconazole resistant, ∼30% demonstrated resistance to amphotericin B, and ∼5% were resistant to echinocandins (15). Due to their safety profile, oral bioavailability, low cost, and broad-spectrum antifungal activity, azole drugs have gained preference as a vital antifungal therapy (16, 17). Thus, there is a pressing need to preserve the clinical utility of azole drugs by enhancing their antifungal activity against azole-resistant species. Candida species utilize various mechanisms to resist the antifungal activity of azole drugs. These mechanisms are primarily attributed to overproduction or mutation of the azole target (ERG11), hyperactivity of the membrane efflux transporters, aneuploidy, altered sterol composition, and increased uptake of exogenous sterols (18, 19). In C. auris, hyperactivity of the membrane efflux transporters and mutations in the azole target site (ERG11) were reported to be predominant in isolates belonging to clades I, III, and IV, while increased copy numbers of the ERG11 were reported to be widespread among clade III isolates (6, 20–23).
Using an adjuvant to resensitize/enhance the susceptibility of drug-resistant Candida species to the antifungal activity of current azoles is an approach that warrants further investigation. This approach has been successfully implemented to control various bacterial and viral infections and has been also used in the treatment of cancer (24–27). However, to a large extent, drug combinations are still inadequately exploited as a therapeutic strategy to treat systemic fungal infections. We previously reported that several FDA-approved drugs such as sulfa drugs, pitavastatin, and ospemifene interacted synergistically and enhanced the activity of azoles against Candida species (16, 28–30). However, sulfa drugs displayed limited azole-chemosensitizing activity against C. auris. Pitavastatin and ospemifene were able to sensitize C. auris to the effect of azoles but at concentrations that are difficult to achieve in human serum. In this study, we conducted a whole-cell screening assay designed specifically to identify potent chemosensitizing agents capable of restoring the antifungal activity of fluconazole in C. auris. The most potent hit identified, lopinavir (LPV), was further assessed with different azole drugs against multiple Candida species. Transcriptome sequencing (RNA-Seq) analysis was utilized to investigate the potential mechanism underlying the synergistic interactions between lopinavir and azole drugs.
RESULTS
Screening of the JHCCL and identification of hits.
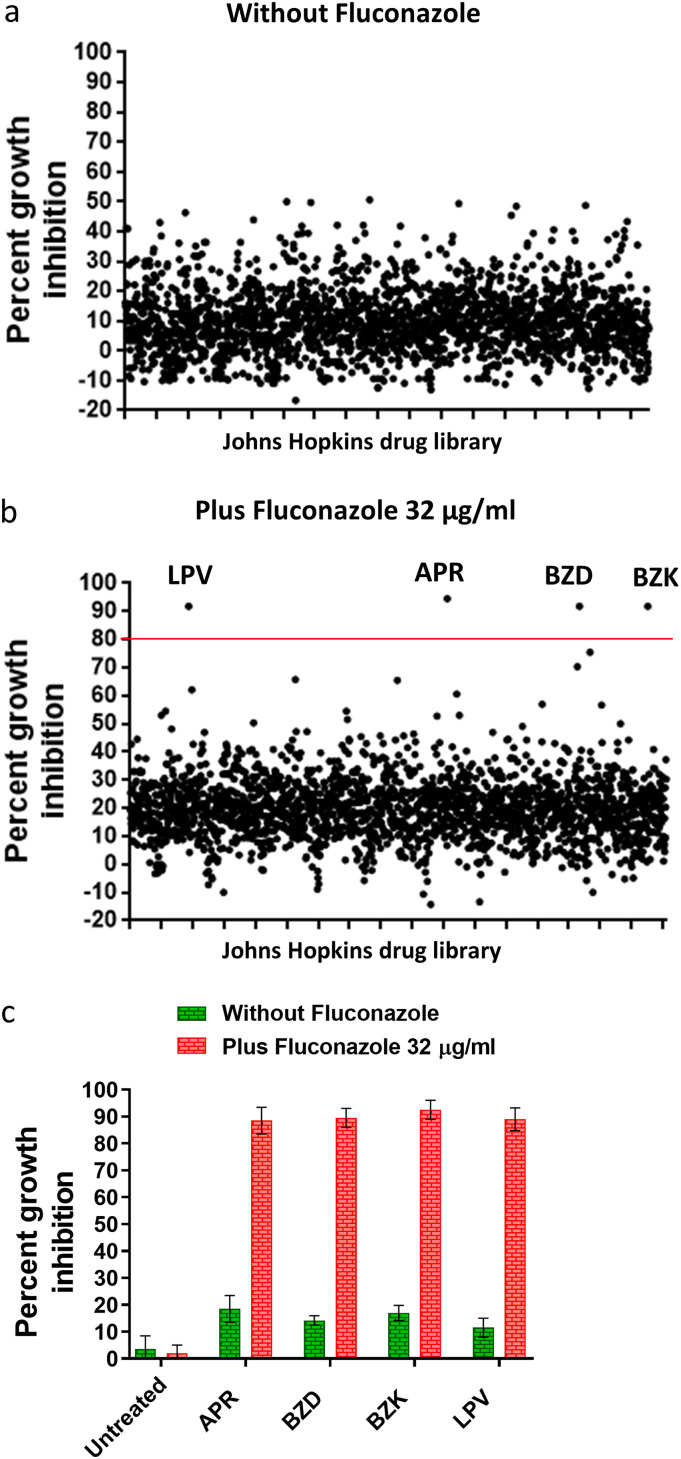
To identify drugs hit capable of restoring the antifungal activity of fluconazole against azole-resistant C. auris, the Johns Hopkins clinical compound library (JHCCL) was screened at a fixed concentration (16 μM) against C. auris AR0390 in the presence or absence of fluconazole (32 μg/ml). Positive hits were assigned for those compounds which inhibited fungal growth by ≥80% only in the presence of fluconazole. Only four hits, lopinavir (LPV), aprepitant (APR), benzalkonium chloride (BZK), and benzododecinium chloride (BZD), were effective in restoring susceptibility of the test isolate to fluconazole (Fig. 1a and b). Positive hits were initially determined by visual inspection and then further confirmed spectrophotometrically by measuring the optical density of C. auris cultures at 490 nm (OD490). As expected, all hit compounds were able to reduce fungal growth by 90 to 95% relative to that of the untreated control (Fig. 1c). Since the focus of this work was to identify fluconazole adjuvants suitable for treating systemic C. auris infections, the two antiseptic compounds (BZK and BZD) were excluded from further analysis. Both lopinavir (an HIV antiviral) and aprepitant (antiemetic) exhibited potent fluconazole-chemosensitizing activities. In this study, we focused our attention on lopinavir and characterized its activity in combination with different azole drugs.
FIG 1.
Identification of the primary screening hits that sensitized C. auris AR0390 to fluconazole. The Johns Hopkins clinical compound library was screened at 16 μM against the azole-resistant C. auris AR0390 in the absence of fluconazole (a). Hit compounds identified included lopinavir (LPV), aprepitant (APR), benzododecinium chloride (BZD), and benzalkonium chloride (BZK), which inhibited fungal growth by more than 80% in the presence of 32 μg/ml of fluconazole (b). Cultures treated with hit compounds were validated spectrophotometrically by measuring the absorbance at OD490 nm (c).
Lopinavir restores the fungistatic activity of fluconazole against C. auris AR0390.
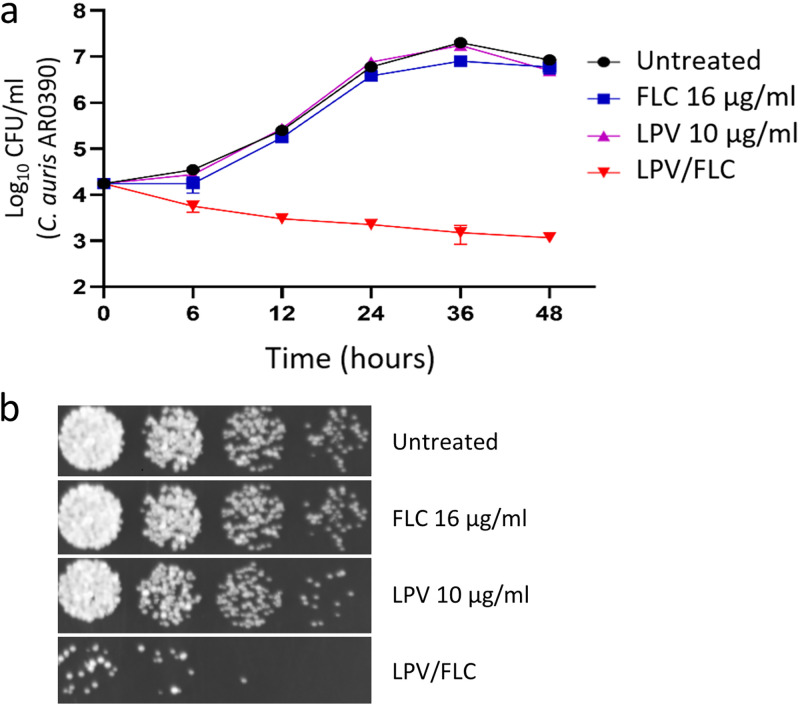
To study the killing kinetics of the lopinavir/fluconazole combination against C. auris, a time-kill assay was conducted. As shown in Fig. 2a, neither lopinavir (10 μg/ml) nor fluconazole (16 μg/ml) was solely able to exert any noticeable antifungal activity against the test isolate (AR0390). However, the combination of the two drugs at the same concentration displayed a fungistatic effect against the test isolate. Additionally, a spotting assay was used to further illustrate the fluconazole-chemosensitizing activity of lopinavir. As shown in Fig. 2b and c, C. auris cultures treated with either lopinavir (10 μg/ml) or fluconazole (16 μg/ml) grew normally on yeast-peptone-dextrose (YPD) agar plates. In contrast, prominent growth inhibition was observed only in cultures treated with the lopinavir/fluconazole combination (Fig. 2b).
FIG 2.
Time-kill analysis of lopinavir at 10 μg/ml, fluconazole (FLC) at 16 μg/ml, or a combination of the two drugs. Test agents were evaluated against C. auris AR0390 over a 48-h incubation period at 35°C. DMSO served as a negative untreated control (a). Cultures of C. auris AR0390 treated with lopinavir (at 10 μg/ml), either alone or in combination with fluconazole (at 16 μg/ml), were spotted onto YPD agar plates and incubated for 24 h before being scanned (b).
Interactions between lopinavir and azole drugs against C. auris isolates.
The ability of lopinavir to restore the antifungal activity of fluconazole against the C. auris AR0390 isolate encouraged us to explore the potential interactions between lopinavir and different azole drugs (fluconazole, voriconazole, and itraconazole) against a panel of 10 C. auris clinical isolates (Table 1). These isolates represent the four major clades of C. auris and were reported to exhibit differences in susceptibility to azole drugs and the utilized mechanisms of azole resistance (21–23). Consistent with these reports, all isolates belonging to clades I, III, and IV (except for strains AR0382 and AR0387) were highly resistant to fluconazole (MIC ≥ 128 μg/ml) and in general displayed low susceptibility to voriconazole and itraconazole (MIC ≥ 0.5 μg/ml), as shown in Table 1. Against all tested isolates, the individual treatment with lopinavir did not exhibit any observable antifungal activity (MIC > 128 μg/ml) (Table 1). Next, standard microdilution checkerboard assays were performed and the fractional inhibitory concentration indices (ƩFICI) were calculated to assess the potential interactions between lopinavir and azole drugs. The results presented in Table 2 indicated that lopinavir was able to display a synergistic relationship with fluconazole against three isolates (ƩFICI ranged from 0.13 to 0.31), while an indifference effect was observed against the remaining seven isolates (ƩFICI ranged from 0.53 to 1.06). We noticed that all the three isolates that responded synergistically to the lopinavir/fluconazole combination belong to clade I and that two of them were highly resistant to fluconazole (MIC = 256 μg/ml). When tested with voriconazole, lopinavir was able to display synergistic interactions against six isolates (ƩFICI ranged from 0.19 to 0.31), while an indifference effect was observed against the remaining four isolates (ƩFICI = 0.53). Notably, the lopinavir/voriconazole combination was able to display synergistic interactions only against isolates of clades I and IV which displayed reduced susceptibility to voriconazole (MIC ≥ 0.5 μg/ml). Interestingly, when lopinavir was combined with itraconazole, potent synergistic interactions against all tested isolates were observed (ƩFICI ranged from 0.04 to 0.09), including those exhibiting reduced susceptibility to itraconazole (MIC ≥ 0.5 μg/ml). These potent synergistic interactions were responsible for significant reductions, ranging from 32- to 256-fold, in the MICs of itraconazole. Since lopinavir interacted more favorably with itraconazole, the combination of lopinavir and itraconazole was selected for further investigation.
TABLE 1.
Clade classification, mechanism of azole resistance, and susceptibility of C. auris to lopinavir, fluconazole, voriconazole, and itraconazole
|
C. auris isolate |
Clade | Mechanism(s) of azole resistance | MIC (μg/ml)a |
|||
|---|---|---|---|---|---|---|
| LPV | FLC | VRC | ITC | |||
| AR0381 | II | None | >128 | 1 | 0.0078 | 0.125 |
| AR0382 | I | None | >128 | 1 | 0.0625 | 0.25 |
| AR0383 | III | ERG11 mutation (F126L, V125A), overexpression of CDR1 | >128 | 256 | 0.5 | 0.5 |
| AR0384 | III | ERG11 mutation (F126L, V125A), overexpression of CDR1 | >128 | 128 | 0.5 | 0.25 |
| AR0385 | IV | ERG11 mutation (Y132F, K177R, N335S, E343D), overexpression of CDR1 | >128 | 256 | 4 | 0.5 |
| AR0386 | IV | ERG11 mutation (Y132F, K177R, N335S, E343D), overexpression of CDR1 | >128 | 256 | 2 | 0.5 |
| AR0387 | I | None | >128 | 1 | 0.0312 | 0.125 |
| AR0388 | I | ERG11 mutation (K143R), overexpression of CDR1 and MDR1 | >128 | 256 | 0.5 | 1 |
| AR0389 | I | ERG11 mutation (Y132F), overexpression of CDR1 | >128 | 256 | 2 | 1 |
| AR0390 | I | ERG11 mutation (K143R), overexpression CDR1 and MDR1 | >128 | 256 | 0.5 | 1 |
LPV, lopinavir (LPV); FLC, fluconazole; VRC, voriconazole; ITC, itraconazole.
TABLE 2.
Effects of lopinavir on the antifungal activity of fluconazole, voriconazole, and itraconazole against C. auris
|
C. auris isolate |
LPV/FLC combination |
LPV/VRC combination |
LPV/ITC combination |
||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | ƩFICa | Mode | MIC (μg/ml) | ƩFIC | Mode | MIC (μg/ml) | ƩFIC | Mode | |
| AR0381 | 0.5/0.5 | 0.53 | IND | 8/0.0039 | 0.53 | IND | 2/0.00098 | 0.04 | SYN |
| AR0382 | 1/0.25 | 0.31 | SYN | 8/0.0156 | 0.31 | SYN | 8/0.0019 | 0.08 | SYN |
| AR0383 | 2/128 | 0.56 | IND | 8/0.25 | 0.53 | IND | 8/0.0019 | 0.07 | SYN |
| AR0384 | 2/128 | 1.06 | IND | 8/0.25 | 0.53 | IND | 8/0.0078 | 0.09 | SYN |
| AR0385 | 2/128 | 0.56 | IND | 4/0.5 | 0.19 | SYN | 8/0.0078 | 0.05 | SYN |
| AR0386 | 16/128 | 0.53 | IND | 4/0.25 | 0.19 | SYN | 8/0.0078 | 0.05 | SYN |
| AR0387 | 0.5/1 | 1.03 | IND | 4/0.0156 | 0.53 | IND | 8/0.00098 | 0.04 | SYN |
| AR0388 | 8//32 | 0.19 | SYN | 8/0.0625 | 0.19 | SYN | 2/0.0039 | 0.07 | SYN |
| AR0389 | 8/128 | 0.56 | IND | 8/0.5 | 0.31 | SYN | 2/0.0078 | 0.07 | SYN |
| AR0390 | 8//16 | 0.13 | SYN | 8/0.0625 | 0.19 | SYN | 4/0.0039 | 0.07 | SYN |
The fractional inhibitory concentration index (ΣFICI) was used to quantify the interactions between the tested combinations based on the following definitions: synergy (SYN) at ΣFICI values of ≤0.5, indifference (IND) at ΣFICI values ranging from >0.5 to ≤4, and antagonism at ΣFICI values of >4.
Effect of lopinavir/itraconazole against other Candida species.
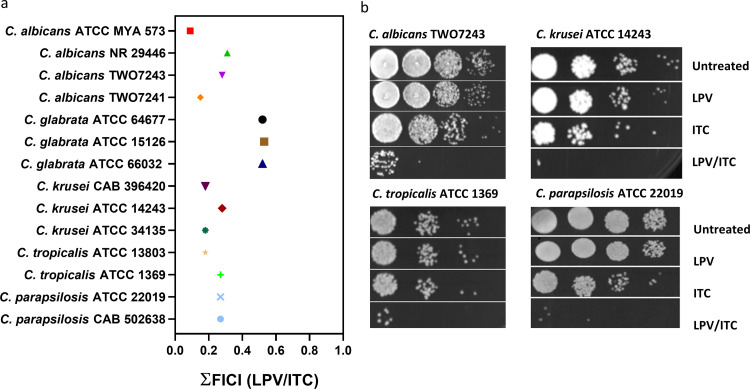
As lopinavir possessed a potent synergistic relationship with itraconazole against all tested C. auris isolates, we next moved to examine whether lopinavir would have a similar effect against other clinically important Candida species. Using standard microdilution checkerboard assays, we examined the interaction between lopinavir and itraconazole against a panel of 14 isolates, including C. albicans (n = 4), C. glabrata (n = 3), C. krusei (n = 3), C. tropicalis (n = 2), and C. parapsilosis (n = 2). As shown in Fig. 3a, lopinavir exhibited broad-spectrum synergistic interactions with itraconazole against all tested isolates, except for C. glabrata, as demonstrated by the low ƩFICI values (ranged from 0.09 to 0.31). The observed synergistic interactions were further demonstrated by spotting the treated cultures onto YPD agar plates and visually observing the growth of CFU. As shown in Fig. 3b, individual treatments with either lopinavir (10 μg/ml) or itraconazole (0.25× MIC) failed to significantly interfere with the visual growth of all tested isolates. However, the growth of C. albicans (TWO7243), C. krusei (ATCC 14243), C. tropicalis (ATCC 1369), and C. parapsilosis (ATCC 22019) was considerably inhibited when fungal cultures were treated with the lopinavir/itraconazole combination.
FIG 3.
Broad-spectrum synergistic interactions of lopinavir and itraconazole (ITC) against different Candida species. Shown are fractional inhibitory concentration index (ƩFICI) values as calculated from checkerboard assays (a). Cultures of C. albicans TWO743, C. krusei ATCC 14243, C. tropicalis ATCC 1369, and C. parapsilosis ATCC 22019 were treated with LPV (10 μg/ml) and ITC (0.25× MIC), either alone or in combination. Cultures were incubated at 35°C for 24 h and then were spotted onto YPD agar plates and incubated for 24 h before being scanned (b).
Effect of the lopinavir/itraconazole combination on the transcriptome of C. auris.
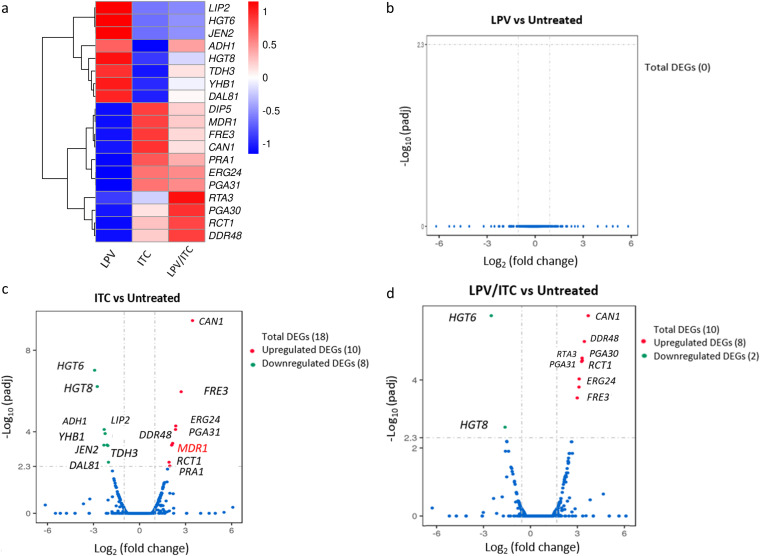
To gain insight into the molecular mechanisms underlying the synergistic relationship between lopinavir and azole drugs, we performed comparative transcriptomic analysis of C. auris isolate AR0390 treated with either dimethyl sulfoxide (DMSO; 1%), lopinavir (10 μg/ml), itraconazole (1 μg/ml), or a combination of the last two drugs at the same concentration. A total of 84.2 million reads were obtained from the four samples. Differentially expressed genes (DEGs) were identified between the tested groups using edgeR (31). Genes that showed a >2-fold (up or down) difference in their expression were considered differentially expressed, and the cutoff for statistical significance was adjusted to a P value of 0.05. As presented in Fig. 4a, a total of 19 DEGs were identified in samples treated with either itraconazole or the lopinavir/itraconazole combination. While no DEGs were identified in the sample receiving lopinavir alone (Fig. 4b), a total of 18 DEGs (10 upregulated plus 8 downregulated) were detected in the itraconazole-treated sample (Fig. 4c and Table 3). Among the upregulated genes in the itraconazole group were the drug transporter MDR1, amino acid permeases (DIP5 and CAN1), stress-associated genes (DDR48 and PGA31), and genes involved in ergosterol biosynthesis (ERG24) and ferric reductase (FRE3). On the other hand, itraconazole treatment resulted in significant downregulation of genes for high-affinity glucose transporters (HGT6 and HGT8), alcohol dehydrogenase (ADH1), and a secreted lipase (LIP2).
FIG 4.
Effects of lopinavir, itraconazole, and a combination of the two drugs on the C. auris transcriptome. Shown is a transcriptional comparison of C. auris AR0390 treated with LPV, ITC, or a combination of both drugs versus the untreated control. (a) Heat map of FPKM values of DEGs of each treatment versus the untreated control, scaled by row. Genes were clustered using hierarchical clustering based on Euclidean distance. (b) Volcano plot of DEGs from C. auris AR0390 treated with LPV at 10 μg/ml. (c) Volcano plot of DEGs from C. auris AR0390 treated with ITC at 1 μg/ml. (d) Volcano plot of DEGs from C. auris AR0390 treated with the lopinavir/itraconazole (LPV/ITC) combination.
TABLE 3.
List of differentially expressed genes for C. auris AR0390 treated with itraconazole (1 μg/ml)
| ID | Name | Log2 fold change | P value | CGDa description |
|---|---|---|---|---|
| 40028953 | CAN1 | 3.4505 | 3.5 × 10−10 | Basic amino acid permease |
| 40025718 | FRE3 | 2.7106 | 1.09 × 10−6 | Protein with similarity to ferric reductase Fre10p |
| 40028243 | ERG24 | 2.3585 | 5.2 × 10−5 | C-14 sterol reductase with a role in ergosterol biosynthesis |
| 40030120 | PGA31 | 2.3457 | 7.7 × 10−5 | Protein associated with cellular response to chemical stimulus |
| 40030199 | DDR48 | 2.1539 | 3.7 × 10−4 | Stress-associated protein, induced by benomyl, caspofungin, and ketoconazole |
| 40029187 | MDR1 | 2.1045 | 4.4 × 10−4 | Plasma membrane MDR/MFS multidrug efflux pump |
| 40027842 | RCT1 | 2.1026 | 4.5 × 10−4 | Fluconazole-induced protein, required for caspofungin tolerance |
| 40028209 | PRA1 | 2.0941 | 4.5 × 10−4 | Cell surface protein that sequesters zinc from host tissue |
| 40026925 | DIP5 | 1.9692 | 4.7 × 10−4 | Dicarboxylic amino acid permease |
| 40025268 | PGA30 | 1.9233 | 3.1 × 10−4 | Predicted GPI-anchored protein |
| 40030546 | DAL81 | −2.0262 | 3.7 × 10−4 | RNA polymerase II repressing transcription factor binding activity |
| 40028527 | TDH3 | −2.0678 | 4.8 × 10−4 | NAD-linked glyceraldehyde-3-phosphate dehydrogenase |
| 40029413 | JEN2 | −2.1117 | 4.4 × 10−4 | Dicarboxylic acid transporter |
| 40027889 | YHB1 | −2.245 | 1.2 × 10−4 | Nitric oxide dioxygenase |
| 40029539 | ADH1 | −2.3056 | 7.7 × 10−5 | Alcohol dehydrogenase |
| 40029385 | LIP2 | −2.3233 | 4.5 × 10−4 | Secreted lipase |
| 40027170 | HGT8 | −2.7557 | 6.1 × 10−7 | High-affinity glucose transporter of the major facilitator superfamily |
| 40027169 | HGT6 | −2.9273 | 9.6 × 10−8 | Putative MFS glucose transporter |
Descriptions of the identified differentially expressed genes (DEGs) were obtained from the Candida Genome Database (CGD) supplemented by a tBLASTx search. ID, identifier; GPI, glycosylphosphatidylinositol.
Of interest, a total of 10 DEGs (8 upregulated plus 2 downregulated) were identified in the lopinavir/itraconazole treatment group (Fig. 4d and Table 4). Similar to the case with the sample treated with itraconazole alone, we noticed that genes involved in ergosterol function (ERG24), stress response (DDR48), amino acid permeases (DIP5 and CAN1), and ferric reductase (FRE3) were upregulated in the lopinavir/itraconazole treatment group. However, in contrast to the case with itraconazole treatment, no significant upregulation of the drug transporter MDR1 was observed in the lopinavir/itraconazole-treated group. Additionally, an upregulation of the lipid translocase encoded by RTA3, whose expression is indicative of stress response, was detected only in the lopinavir/itraconazole treatment. On the other hand, similar to the case with the sample treated with itraconazole alone, two high-affinity glucose transporters (HGT6 and HGT8) were found to be downregulated in the lopinavir/itraconazole sample.
TABLE 4.
List of differentially expressed genes for C. auris AR0390 treated with a combination of lopinavir (10 μg/ml) and itraconazole (1 μg/ml)
| ID | Name | Log2 fold change | P value | CGDa description |
|---|---|---|---|---|
| 40025268 | PGA30 | 2.4588 | 2.6 × 10−5 | Protein with unknown function |
| 40025718 | FRE3 | 2.1689 | 3.4 × 10−4 | Protein with similarity to ferric reductase Fre10p |
| 40028953 | CAN1 | 2.7975 | 1.3 × 10−6 | Basic amino acid permease |
| 40030199 | DDR48 | 2.5766 | 7.4 × 10−6 | Stress-associated protein, induced by benomyl, caspofungin, and ketoconazole |
| 40027842 | RCT1 | 2.4473 | 2.3 × 10−5 | Fluconazole-induced protein, required for caspofungin tolerance |
| 40029690 | RTA3 | 2.4239 | 2.9 × 10−5 | Putative lipid translocase that influences the susceptibility of C. albicans to fluconazole |
| 40028243 | ERG24 | 2.2803 | 9.3 × 10−5 | C-14 sterol reductase with a role in ergosterol biosynthesis |
| 40030120 | PGA31 | 2.2697 | 1.6 × 10−4 | Cellular response to chemical stimulus |
| 40027170 | HGT8 | −1.9543 | 2.4 × 10−4 | High-affinity glucose transporter of the major facilitator superfamily |
| 40027169 | HGT6 | −2.7443 | 1.3 × 10−6 | Putative MFS glucose transporter |
Descriptions of the identified DEGs were obtained from the Candida Genome Database supplemented by a tBLASTx search.
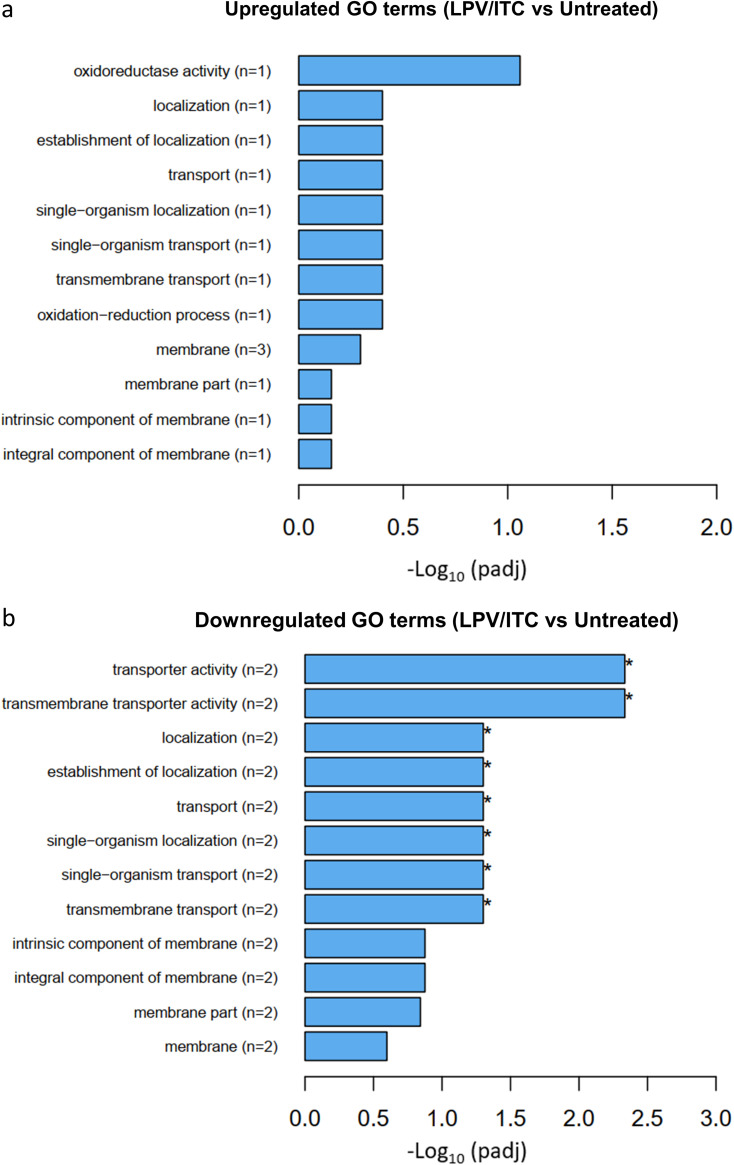
In order to gain an overall understanding of the impact of the lopinavir/itraconazole treatment on the transcriptome of C. auris, we performed Gene Ontology (GO) analysis of the identified DEGs. Interestingly, eight GO terms were found to be significantly overrepresented (enriched) only in the sample treated with lopinavir/itraconazole, while no significant GO terms were identified in the sample treated with itraconazole alone. Of note, all identified GO terms in the lopinavir/itraconazole-treated group were found to be downregulated, of which five are associated with membrane transport activities (Fig. 5). All significant downregulated GO terms are provided in Table S4 in the supplemental material.
FIG 5.
Gene Ontology (GO) enrichment analysis of differentially expressed genes in the lopinavir/itraconazole-treated sample. GO analysis was implemented by the Cluster Profiler R package, and a P value of ≤0.05 was used as the cutoff parameter. Upregulated GO terms (a) and downregulated GO terms (b) are shown.
Validation of RNA-Seq data.
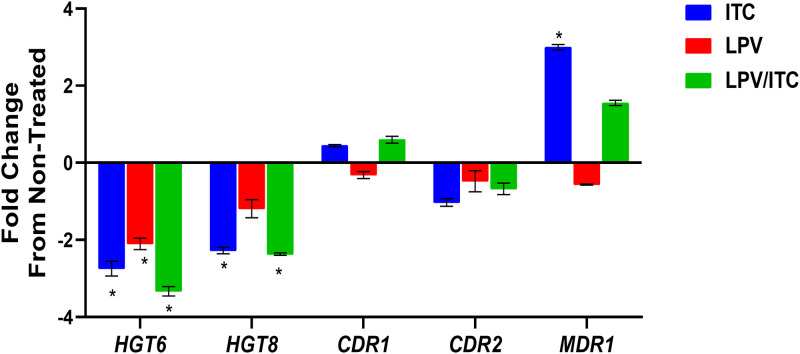
To validate the RNA-Seq data, we selected five DEGs exhibiting a broad range of differential expression for analysis by quantitative real-time reverse transcription-PCR (RT-qPCR). The five DEGs selected were the two glucose transporters (HGT6 and HGT8) and azole resistance-related efflux genes (CDR1, CDR2, and MDR1). As shown in Fig. 6 and consistent with the RNA-Seq data, we did not detect a significant difference in the expression of the drug transporters CDR1 and CDR2 for all treatment groups. However, a significant increase in the expression level of MDR1 was observed in the sample treated with itraconazole alone. Additionally, both itraconazole alone and the lopinavir/itraconazole combination resulted in significant downregulation of the glucose transporters HGT6 and HGT8. Interestingly, lopinavir treatment alone was able to significantly reduce the expression of the glucose transporter HGT6 relative to that of the untreated control. Moreover, the impact of the lopinavir/itraconazole combination on the expression of HGT6 was more pronounced than the effect observed for itraconazole treatment alone.
FIG 6.
Effect of itraconazole, lopinavir (LPV), or a combination of the two drugs on the mRNA expression levels of five selected genes involved in glucose transport and efflux in C. auris. Exponentially grown C. auris AR0390 cells were treated with either LPV (10 μg/ml), ITC (1 μg/ml), or a combination of the two drugs for 3 h. Following treatment, cells were harvested and lysed, and RNA was extracted. The expression of genes encoding glucose transport (HGT6 and HGT8) and the azole resistance-related efflux genes (CDR1, CDR2, and MDR1) was internally normalized to ACT1 and compared to that of the untreated control. Asterisks indicate significant changes in expression of the examined genes (at least 2-fold differences, up or down) relative to that of the untreated control. The results are presented as means ± SD.
Lopinavir interferes with glucose transport, ATP content, and efflux activity in C. auris.
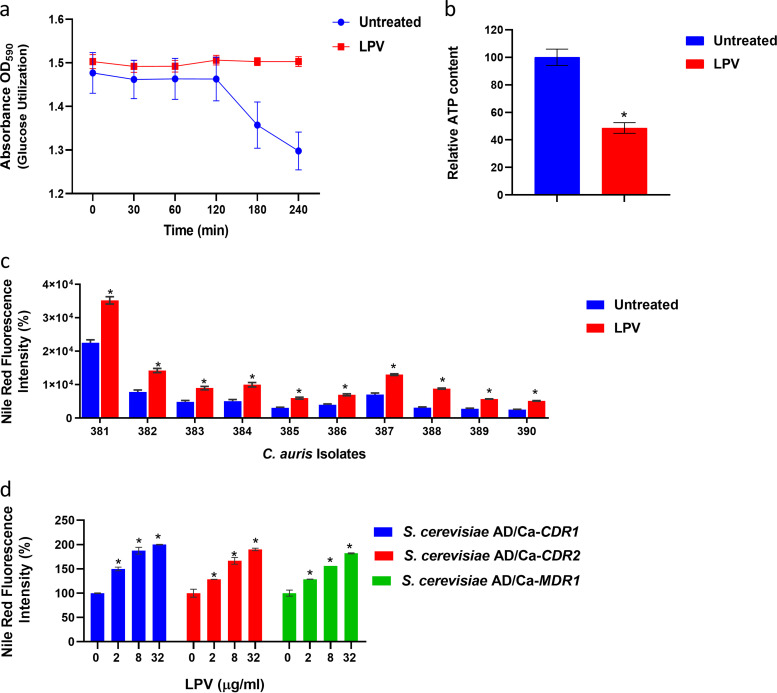
Since lopinavir was able to interfere significantly with the mRNA levels of the glucose transporter HGT6, we were curious to study its effect on glucose utilization, ATP synthesis, and efflux activities in C. auris. To examine the effect on glucose utilization, we used a glucose-induced acidification assay for which bromophenol blue was used as a pH indicator. As shown in Fig. 7a, as expected, untreated C. auris AR0390 cells were able to utilize glucose present in the assay medium, resulting in a significant drop in pH (resulting in lower OD595 values). However, lopinavir (at 10 μg/ml) significantly interfered with the glucose utilization ability of C. auris, and no observable reduction in OD595 values was detected. Next, we assessed the effect of lopinavir treatment on the cellular ATP levels. At 10 μg/ml, lopinavir significantly reduced cellular ATP levels in C. auris AR0390, by ∼52% ± 3.9%, compared to those in the untreated control (Fig. 7b).
FIG 7.
Effect of lopinavir on the glucose utilization, ATP content, and efflux activity of C. auris. (a) Effect of LPV on the glucose utilization ability of C. auris. Cultures of C. auris AR0390 were treated with DMSO (1%) or lopinavir (LPV) at 10 μg/ml, and then the ability of C. auris to utilize externally supplemented glucose and acidify the assay medium was detected by the decreased absorbance of bromophenol blue at 590 nm. (b) Effect of lopinavir on the ATP content of C. auris. Exponentially grown C. auris AR0390 cells were treated with DMSO (1%) or LPV (10 μg/ml) for 3 h at 35°C before being lysed, and ATP content was determined. Asterisks represent a statistical difference (P < 0.05) in ATP content between treated cells and the untreated control, as determined by unpaired t test. (c) Effect of lopinavir (10 μg/ml) on Nile red efflux from 10 different C. auris isolates. (d) Effect of lopinavir (at 2, 8, or 32 μg/ml) on Nile red efflux from recombinant S. cerevisiae strains expressing individual efflux genes CDR1, CDR2, or MDR1 from C. albicans. Data represent means + SD from triplicate measurements. Asterisks represent a statistical difference (P < 0.05) in the efflux of Nile red for LPV-treated cells compared to that of the untreated control, as determined by multiple t tests using the Holm-Sidak method for multiple comparisons.
Next, we utilized glucose-induced Nile red efflux assays to assess the effect of lopinavir on the efflux activity of C. auris. As presented in Fig. 7c, lopinavir treatment, at 10 μg/ml, significantly interfered with the efflux of Nile red from all 10 isolates of C. auris tested. Significant increases in Nile red fluorescence intensity, by 56 to 184% (P < 0.05) depending on the tested isolates, were observed compared to that in the untreated cultures. We also assessed the effect of lopinavir treatment at different concentrations (2, 8, and 32 μg/ml) on Nile red efflux from three recombinant Saccharomyces cerevisiae strains exclusively expressing individual efflux genes of C. albicans (CDR1, CDR2, or MDR1); our data indicate that lopinavir, in a dose-dependent manner, interfered significantly (P < 0.05) with Nile red efflux from all tested recombinant strains (Fig. 7d).
Caenorhabditis elegans.
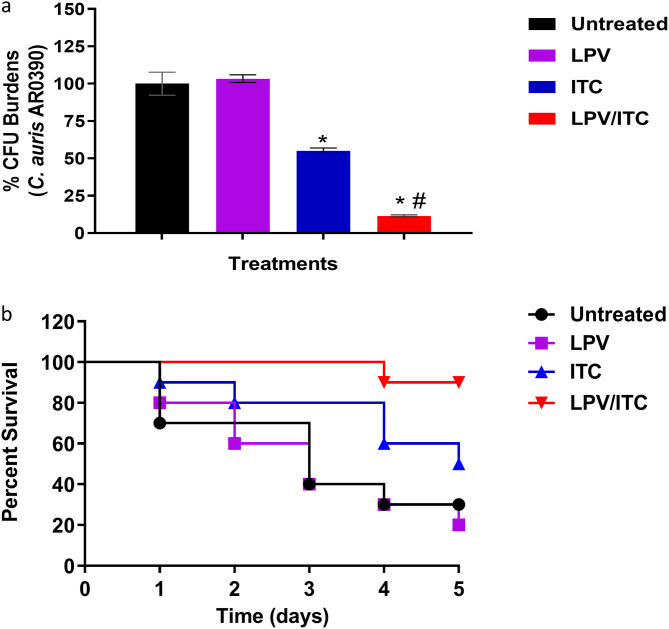
To further corroborate the in vitro activity of the lopinavir/itraconazole combination, we evaluated the combination’s in vivo efficacy using a C. elegans infection model. As shown in Fig. 8a, treatment with lopinavir (10 μg/ml) alone failed to reduce the CFU burden of C. auris in infected nematodes. Itraconazole (at 1 μg/ml, 1× MIC) was able to reduce the burden of C. auris by ∼45% ± 1.8% in infected nematodes compared to that in the untreated control. The lopinavir/itraconazole combination, at the same tested concentrations, significantly reduced C. auris CFU, by ∼88.5% ± 0.68%, compared to that in the untreated control.
FIG 8.
In vivo efficacy of the lopinavir/itraconazole combination using a Caenorhabditis elegans infection model. Nematodes infected with C. auris AR0390 were treated with LPV at 10 μg/ml and ITC at 1 μg/ml, either alone or in combination. Untreated worms served as a negative control. Effects of LPV, ITC, and the lopinavir/itraconazole combination on reducing fungal burden (CFU) after 24 h of treatment are shown (a). Asterisks indicate a statistical significance (P < 0.05) compared to the untreated control, while a pound sign indicates a statistical significance for the combination treatment compared to treatment with ITC alone (P value < 0.05, as determined by one-way analysis of variance [ANOVA] using Dunnett’s test for multiple comparisons). A Kaplan-Meier survival curve, assessed by log-rank test for significance, to evaluate the ability of the lopinavir/itraconazole combination to enhance survival of C. elegans infected with C. auris AR0390 was performed (b).
Next, we assessed the effect of the lopinavir/itraconazole combination on enhancing the survival of C. elegans infected with C. auris AR0390. As shown in Fig. 8b, 5 days postinfection, only 20% of untreated C. elegans organisms survived infection with C. auris. Lopinavir alone, at 10 μg/ml, did not improve the survival of C. elegans, while itraconazole at 1 μg/ml (1× MIC) enhanced the survival rate of infected worms by ∼50% (P < 0.05). However, the combination of lopinavir/itraconazole significantly improved the survival of C. elegans, by ∼90% (P < 0.05).
DISCUSSION
The recent emergence of multidrug-resistant C. auris poses a significant threat to public health and imposes the need for immediate efforts to explore novel antifungal agents and to uncover alternative therapeutic approaches. In this study, we utilized drug repurposing as a promising approach to identify novel adjuvants capable of enhancing the antifungal activity of azole drugs against the multidrug-resistant C. auris. The Johns Hopkins clinical compound library (JHCCL), which contains 1,547 FDA-approved drugs and clinical molecules, was screened against the multidrug-resistant isolate C. auris AR0390 in the presence or absence of a subinhibitory concentration of fluconazole (32 μg/ml). AR0390 is a multidrug-resistant isolate and thus was used to screen the JHCCL to identify new adjuvants with potent azole-chemosensitizing activity. The primary screen revealed four compounds were able to inhibit the growth of C. auris AR0390 only in the presence of fluconazole. In this study, we decided to focus our attention on lopinavir, an HIV protease inhibitor, which restored the fungistatic activity of fluconazole against the test isolate at a clinically achievable concentration, as shown in the time-kill study. Indeed, previous studies indicated that lopinavir can reach up to ∼12 μg/ml in human serum with standard doses as an antiviral agent (32–34). Next, checkerboard assays were utilized to assess the interactions between lopinavir and different azole drugs against a panel of 10 C. auris isolates. Our data indicate that different C. auris clades responded distinctly to the combination of lopinavir with either fluconazole or voriconazole. All isolates that responded synergistically to the lopinavir/fluconazole combination were found to belong to clade I. Interestingly, isolates AR0388 and AR0390, which exhibited extensive resistance to fluconazole, were highly sensitive to the lopinavir/fluconazole combination. We noticed that the two isolates shared the same azole resistance mechanism, a single point mutation in ERG11 (K143R), and overexpression of the efflux transporters CDR1 and MDR1 (21, 23). However, the clade I isolate AR0389, which also has a single mutation in ERG11 (Y132F), did not respond to the lopinavir/fluconazole combination, probably due to its extensive CDR1 overexpression compared to those of AR0388 and AR0390, as previously reported (22, 23). Moreover, all isolates with multiple ERG11 mutations did not respond to the lopinavir/fluconazole combination. We also noticed that the lopinavir/voriconazole combination was able to display synergistic interactions only against isolates of clades I and IV, which displayed reduced susceptibility to voriconazole (MIC ≥ 0.5 μg/ml). In contrast, clade III isolates did not respond to the lopinavir/voriconazole combination. These findings suggest that the lopinavir/voriconazole combination is able to overcome azole resistance mechanisms that involve reduced affinity to mutated ERG11 or increased efflux activities due to overexpression of CDR1 and/or MDR1. In addition, the lack of activity against clade III isolates may be attributable to the existence of additional resistance mechanisms such as increased copy number of ERG11, which was reported to be predominant among clade III isolates (6). Collectively, these data indicate that the genetic variability among C. auris clades and the underlying mechanisms of azole resistance play critical roles in dictating the efficacies of these drug combinations.
Interestingly, the combination of lopinavir and itraconazole displayed potent synergistic interactions against all tested isolates, regardless of clades’ differences and the various azole resistance mechanisms utilized. These potent synergistic interactions remarkably reduced the MICs of itraconazole, by 32- to 256-fold. In this regard, lopinavir surpassed several known azole-chemosensitizing agents, including sulfamethoxazole, clorgyline, and cyclosporine (16, 35, 36). Sulfamethoxazole displayed synergistic interactions with itraconazole against only three C. auris isolates, as previously reported (16). However, clorgyline interacted synergistically with itraconazole against only one isolate, while cyclosporine displayed synergistic interactions with itraconazole against all 10 isolates but with higher ƩFICI values (Table S5). The synergistic interaction between lopinavir and itraconazole was further validated in vivo using C. elegans as an infection model. Lopinavir at 10 μg/ml significantly reduced the burden of C. auris AR0390 in the infected nematodes and improved their survival compared to the results with the single treatment with itraconazole. These results suggest that lopinavir has the potential to be used clinically as an antifungal adjuvant for overcoming azole resistance in C. auris, though further pharmacokinetic studies and in vivo assessment in rodents and humans are needed.
To explore the potential downstream mechanism by which lopinavir enhances the activity of azole drugs, we performed a global transcriptomic analysis of C. auris AR0390 treated with lopinavir or itraconazole (either alone or in combination). Interestingly, a significant upregulation of the azole exporter gene MDR1 was observed only when C. auris was exposed to itraconazole; however, cells treated with the lopinavir/itraconazole combination did not exhibit a significant increase in the expression of MDR1. Additionally, GO enrichment analysis indicated that the lopinavir/itraconazole combination exerted a broad-ranging inhibitory effect against several MFS membrane transporters, including MDR1 and the glucose transporters HGT6 and HGT8. To validate the RNA-Seq data, RT-qPCR was used to measure the mRNA levels of MDR1, CDR1, and CDR2, whose overexpression is known to be a major cause of azole resistance in Candida species (18, 37, 38). Also, we evaluated the mRNA expression levels of the glucose transporters HGT6 and HGT8. Consistent with our RNA-Seq data, MDR1 was significantly upregulated only in the itraconazole-treated group. Additionally, we did not detect any significant difference in the expression of CDR1 and CDR2 in all treatment groups, in agreement with RNA-Seq data. However, RT-qPCR data regarding the expression of glucose transporters were not exactly the same as the data obtained from RNA-Seq. Though both itraconazole and the lopinavir/itraconazole combination resulted in a significant downregulation of HGT6 and HGT8, we noticed that lopinavir was also able to significantly interfere with the expression of HGT6. In addition, the negative impact on HGT6 expression was found to be more significant in cells treated with the lopinavir/itraconazole combination than with the single treatment with either itraconazole or lopinavir. Together, these observations indicate that lopinavir, by itself, was able to interfere with glucose transport, and that its combination with itraconazole interfered more significantly with the expression of several MFS transporters, including those responsible for glucose permeation and the azole-related efflux transporter MDR1, via a mechanism that still needs to be investigated. Notably, several HIV protease inhibitors, including lopinavir, were previously shown to interfere with glucose transport in human cells and also in the malarial pathogen Plasmodium falciparum (39–42). Given the highly conserved nature of glucose permeases, it is conceivable that lopinavir could have a similar effect against glucose transport in Candida species, though further molecular studies are needed to confirm this point.
Next, we assessed the effect of lopinavir on the ability of C. auris to utilize externally added glucose and the subsequent effect on cellular ATP levels. Consistent with the transcriptomic data, lopinavir significantly interfered with the ability of C. auris to utilize glucose and consequently resulted in a significant reduction in the ATP content. We hypothesized that this effect could defuse energy-dependent drug resistance mechanisms such as efflux hyperactivity, particularly ABC-mediated efflux activity, which is a vital azole resistance mechanism in C. auris. Indeed, previously reported whole-genome sequencing data revealed that C. auris contains large proportions of efflux transporters (43). In the same vein, abrogation of azole-related efflux genes, particularly CDR1, was shown to restore the antifungal activity of azole drugs against C. auris (21). To test our hypothesis, we examined the effect of lopinavir on the efflux activity of C. auris using dye efflux assays. Consistent with our hypothesis, lopinavir was able to interfere significantly with the Nile red efflux from all tested isolates. Moreover, we noticed that lopinavir was also capable of interfering with the Nile red efflux from recombinant S. cerevisiae mutants, exclusively expressing the individual efflux genes of C. albicans, CDR1, CDR2, and MDR1. Since a high degree of homology exists between the efflux genes of C. auris and C. albicans (21), we assume that lopinavir would have similar inhibitory effects on C. auris transporters. Collectively, these results suggest that the mechanism by which lopinavir interacts synergistically with azole drugs is mediated by significant interference with Candida’s efflux activities.
In summary, this study utilized drug repurposing as a powerful tool to identify potent compounds capable of restoring/enhancing the antifungal activities of azole drugs, especially against drug-resistant Candida. This led to the discovery that lopinavir displayed variable azole-chemosensitizing activities against C. auris depending on the azole agent and nature of the tested isolate. The lopinavir/itraconazole combination displayed the most potent synergistic relationship and was effective against major clinically important Candida species, including the emergent multidrug-resistant C. auris. A potential drawback of this novel combination is that the commercial availability of itraconazole can be a concern in some geographic locations, where fluconazole is the only available azole agent. However, according to the current distribution maps of antifungals, relatively few countries, mainly clustered in central and west Africa, do not have itraconazole in their drug markets (44). Finally, it should be emphasized that previous studies indicated that HIV antiviral agents were able to interfere with the fungal aspartyl proteases and were presented as potent inhibitors of several key virulence attributes in Candida species, such as hypha formation, adherence, biofilm formation, and phenotypic switching (45–49). These studies and the data derived from this work further support the clinical potential of lopinavir as a promising antifungal adjuvant and open the door for a more comprehensive study to investigate the chemosensitizing activities of various HIV protease inhibitors.
MATERIALS AND METHODS
Fungal strains, reagents, and chemicals.
Fungal strains used in this study are listed in Table S1. RPMI 1640 powder with glutamine, but without NaHCO3, was purchased from Thermo Fisher Scientific (Waltham, MA). 3-(N-Morpholino)propanesulfonic acid (MOPS) was obtained from Sigma-Aldrich (St. Louis, MO). Yeast-peptone-dextrose (YPD) broth and YPD agar were obtained from Becton, Dickinson and Company (Franklin Lakes, NJ). The Johns Hopkins clinical compound library (JHCCL) was purchased from The Johns Hopkins University School of Medicine, delivered in microplates (10 mM, dissolved in DMSO), and stored at −80°C until use. Nile red, voriconazole, and itraconazole were obtained from TCI America (Portland, OR). Lopinavir was obtained from Sigma-Aldrich. Fluconazole was obtained from Fisher Scientific (Pittsburgh, PA). Gentamicin sulfate was purchased from Chem-Impex International Inc. (Wood Dale, IL).
Screening of the JHCCL library.
The drug library was screened at a fixed concentration (16 μM) against a multidrug-resistant C. auris strain, AR0390, in the presence or absence of fluconazole (32 μg/ml), following the guidelines of CLSI M27A3 (69), with modifications. Briefly, a fresh culture of C. auris AR0390 was diluted to approximately 2.5 × 103 CFU/ml in RPMI 1640 medium buffered with 0.165 M MOPS reagent. Aliquots (at 100-μl volumes) of the fungal suspension were transferred to the wells of a round-bottomed 96-well microtiter plate containing a 16 μM concentration of each drug. The plates were then incubated for 24 h at 35°C. Positive hits were assigned for compounds that prominently reduced the visual growth of the test isolate only in the presence of fluconazole and were further validated spectrophotometrically by reducing the OD490 value of the treated culture by at least 80% relative to that of the untreated control.
Time-kill and spotting assays.
To study the killing kinetics of the lopinavir/fluconazole combination against C. auris, a time-kill assay was performed as previously described (50–52). Briefly, exponential-phase C. auris AR0390 cells were diluted to ∼1 × 105 CFU/ml in RPMI 1640 medium containing lopinavir (10 μg/ml), fluconazole (16 μg/ml), or a combination of the two. Cells treated with DMSO (1%), the solvent of the drugs, served as an untreated control. Test agents were added at the respective concentrations and incubated at 35°C for 24 h. The number of viable cells was monitored by counting CFU at specific time points (0, 6, 12, and 24 h). Killing curves were constructed by plotting CFU from different treatments versus the specified time points. The results are presented as the mean values of triplicate measurements obtained from three independent experiments. The fluconazole-chemosensitizing activity of lopinavir was further validated by spotting 5-μl aliquots of treated and nontreated cultures on YPD agar plates. The plates were incubated at 35°C for another 24 h before they were scanned.
Microdilution checkerboard assays.
The interaction between lopinavir and different azole antifungal drugs (fluconazole, voriconazole, and itraconazole) was assessed using microdilution checkerboard assays, as previously reported (53–55). The fractional inhibitory concentration index (ΣFICI) was used to assess the interactions between the tested drug combinations. ΣFICI interpretations corresponded to the following definitions: synergy at ΣFICI values of ≤0.5, indifference at ΣFICI values ranging from >0.5 to ≤4, and antagonism at ΣFICI values of >4 (56–58).
Effect of the lopinavir/itraconazole combination against other Candida species.
To evaluate the potential interaction between lopinavir and itraconazole against other Candida species, we utilized checkerboard assays and calculated the ΣFICI, as described above. The lopinavir/itraconazole combination was assessed against 14 isolates of clinically important Candida species, including C. albicans (n = 4), C. glabrata (n = 3), C. krusei (n = 3), C. tropicalis (n = 2), and C. parapsilosis (n = 2). Additionally, 5-μl aliquots from treated and untreated cultures of representative Candida strains that were sensitive to the lopinavir/itraconazole combination were spotted onto YPD agar plates and incubated for 24 h at 35°C before the plates were scanned.
RNA extraction.
Exponential-stage cultures of C. auris AR0390 were treated with either DMSO (1%), lopinavir (10 μg/ml), itraconazole (1 μg/ml), or a combination of the two drugs. For each treatment condition, duplicate samples were prepared. All treated cultures were incubated for 3 h at 35°C. Cells were subsequently pelleted and washed twice with phosphate-buffered saline (PBS), and RNA was extracted using an Ambion RiboPure yeast kit, following the manufacturer’s guidelines. The RNA quality was checked on a bioanalyzer nanochip (Agilent), and all RNAs used for downstream experiments were determined to have RNA integrity numbers (RIN) of 9.5 and above. The SuperScript III first-strand kit (Invitrogen) was used to prepare cDNA, following the manufacturer’s guidelines.
RNA sequencing and enrichment analysis of differentially expressed genes (DEGs).
An Illumina NovaSeq6000 was utilized to sequence 150-bp paired-end reads. Next, adapters were removed and were then trimmed based on quality using the program Fastp. Trimmed reads were aligned to the C. auris NCBI reference genome version B11221 (https://www.ncbi.nlm.nih.gov/assembly/GCF_002775015.1) using the aligner HISAT2 (v2.1) (59). FeatureCounts (v2.1) used the GTF file associated with the reference genome to generate a count matrix of reads mapping to each gene (60). The fragments per kilobase per million (FPKM) of each gene was calculated based on the length of the gene and read count mapped to this gene. Differential expression analysis was performed using the package edgeR (v3.16.5) (31). The P values were adjusted for multiple testing using the Benjamini and Hochberg method (61). A corrected P value of 0.005 and an absolute fold change of two were set as the thresholds for significant differential expression. Gene names and descriptions were assigned to the identified DEGs by BLAST searching gene sequences using the Candida Genome Database (CGD) and by using tBLASTx against C. albicans SC5314 (taxid 237561) with an E value cutoff of 0.01 (62). All gene sequences are listed in Table S2.
Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the ClusterProfiler Bioconductor package (v2.4.3), and GO terms with a corrected P value of less than 0.05 were considered significantly enriched (63).
RT-qPCR.
To validate the RNA-Seq results, we selected five genes (HGT6, HGT8, CDR1, CDR2, and MDR1) for quantitative real-time reverse transcription-PCR (RT-qPCR) analysis. Gene expression levels were calculated using the threshold cycle (2−ΔΔCT) method, as previously described (64). The primers used in this study are listed in Table S3. The gene expression of C. auris AR0390 treated with lopinavir, itraconazole, or a combination of the two was internally normalized to ACT1 and compared to the untreated control (1% DMSO).
Glucose acidification assay.
The effect of lopinavir on the glucose utilization ability of C. auris AR0390 was assessed, as described previously (65). Briefly, exponentially grown C. auris AR0390 cells were collected, suspended in 0.1 M KCl, and incubated for 1 h at 35°C, and then the yeast suspension was adjusted to an OD600 of 2 to 3 and incubated overnight at 4°C. Aliquots (20 μl) of yeast suspension were added to 155 μl of buffer (0.1 M KCl, 50 μg/ml of bromophenol blue [pH 5]) with either lopinavir (10 μg/ml) or DMSO (1%). Medium acidification was started by adding 25 μl of 20% glucose to the yeast cultures. The reaction was monitored spectrophotometrically by measuring the OD590 every 30 min for 4 h. The reduction in OD590 values for the untreated control (DMSO) is indicative of the glucose-utilizing ability of C. auris.
Cellular ATP content assay.
The effect of lopinavir on fungal cellular ATP content was determined as described previously, with the following modifications (66). Briefly, an exponential-stage culture of C. auris AR0390 was adjusted to ∼1 × 107 cells/ml in RPMI 1640 medium. The yeast suspension was then treated with either lopinavir (10 μg/ml) or DMSO (1%) and incubated at 35°C for 3 h. Following incubation, cells were harvested, washed twice with PBS, and then lysed by bead beating to release the cellular ATP molecules. Aliquots of cell lysates were mixed with equal volumes of BacTiter-Glo reagent (Promega Corporation, Madison, WI) and incubated for 10 min in the dark at room temperature before recording of the luminescence intensity. Control cell-free wells were used to determine the background luminescence. Signals represented the means of three separate experiments, and data represent the percentage ATP content in the lopinavir-treated wells relative to that of the untreated control.
Nile red efflux assay.
To evaluate the effect of lopinavir on Nile red efflux from 10 C. auris isolates and three S. cerevisiae strains exclusively expressing C. albicans efflux genes CDR1, CDR2, and MDR1, we used Nile red efflux assays as previously described (16, 67, 68). Briefly, exponential state cells were harvested, washed twice with PBS, and then incubated for an additional 2 h at 35°C with shaking (200 rpm) to starve cells. Cells were incubated overnight on ice and were then adjusted to ∼1 × 107/ml in 50 mM HEPES-NaOH buffer (pH 7.0) containing 7.5 mM Nile red for 30 min at 35°C. Nile red-loaded cells were washed three times with cold HEPES buffer and then transferred onto opaque 96-well plates containing lopinavir (final concentration of 10 μg/ml) or DMSO (1%). Efflux was initiated by adding glucose at a final concentration of 10 mM to all wells, and plates were incubated at room temperature for 10 min before recording of the fluorescence signal. Nile Red fluorescence intensity was measured at 485/528 nm using a SpectraMax i3x microplate reader (Molecular Devices, San Jose, CA).
Caenorhabditis elegans infection model.
To assess the in vivo efficacy of the lopinavir/itraconazole combination, a C. elegans infection model was utilized, as previously described (14, 28). Briefly, synchronized worms [strain AU37 genotype glp-4(bn2) I; sek-1(km4) X] at larval stage 4 (L4) were incubated with 5 ml of YPD suspension containing log-phase C. auris AR0390 (∼1 × 106 CFU/ml) for 3 h at room temperature. Following infection, nematodes were washed five times with PBS to remove noningested cells. The nematodes were resuspended in M9 buffer (containing 20% RPMI 1640 medium) and distributed into groups (n = 20 worms), and then DMSO (1%), lopinavir (10 μg/ml), itraconazole (1 μg/ml), or a combination of the two drugs was added. All treatments (in triplicates) were maintained for 24 h at 25°C before being removed with repetitive washing with PBS. To release the ingested C. auris cells, worms were subjected to vigorous vortexing with silicon carbide beads for at least 2 min. The C. elegans homogenates were then serially diluted and plated onto YPD agar plates containing gentamicin (100 μg/ml). Plates were incubated for 24 h at 35°C before CFU per worm were determined.
In a subsequent experiment, worms were infected with a large inoculum of C. auris AR0390 (∼1 × 107 CFU/ml) as described above. Infected worms were treated with DMSO (1%), lopinavir (10 μg/ml), itraconazole (1 μg/ml), or a combination of the two drugs, and survival of nematodes was monitored and recorded for 5 days. Data are presented as percent survival of infected C. elegans using a Kaplan-Meier survival curve generated using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA).
Data availability.
The sequence reads of all samples were deposited in the NCBI database (Gene Expression Omnibus [GEO]) under the accession number GSE148341.
Supplementary Material
ACKNOWLEDGMENTS
We thank BEI Resources and the U.S. Centers for Disease Control and Prevention (CDC) for providing the clinical isolates used in this study. We thank Richard D. Cannon (University of Otago, New Zealand) for kindly providing the recombinant Saccharomyces cerevisiae strains. We thank Theodore White (University of Missouri—Kansas City) for providing us with isolates TWO7241 and TWO7241. Finally, we thank Haroon Mohammad (Purdue University) for proofreading the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nett JE. 2019. Candida auris: an emerging pathogen “incognito”? PLoS Pathog 15:e1007638. doi: 10.1371/journal.ppat.1007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Cassia Orlandi Sardi J, Silva DR, Soares Mendes-Giannini MJ, Rosalen PL. 2018. Candida auris: epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb Pathog 125:116–121. doi: 10.1016/j.micpath.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, Vallabhaneni S. 2019. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol 57:1–12. doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 5.Lone SA, Ahmad A. 2019. Candida auris—the growing menace to global health. Mycoses 62:620–637. doi: 10.1111/myc.12904. [DOI] [PubMed] [Google Scholar]
- 6.Chow NA, Munoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Arauz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandon P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. doi: 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szekely A, Borman AM, Johnson EM. 2019. Candida auris isolates of the Southern Asian and South African lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J Clin Microbiol 57:e2055-18. doi: 10.1128/JCM.02055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. 2019. Insights into the unique nature of the East Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol 57:e00007-19. doi: 10.1128/JCM.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forgacs L, Borman AM, Prepost E, Toth Z, Kardos G, Kovacs R, Szekely A, Nagy F, Kovacs I, Majoros L. 2020. Comparison of in vivo pathogenicity of four Candida auris clades in a neutropenic bloodstream infection murine model. Emerg Microbes Infect 9:1160–1169. doi: 10.1080/22221751.2020.1771218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caceres DH, Forsberg K, Welsh RM, Sexton DJ, Lockhart SR, Jackson BR, Chiller T. 2019. Candida auris: a review of recommendations for detection and control in healthcare settings. J Fungi (Basel) 5:111. doi: 10.3390/jof5040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortegiani A, Misseri G, Fasciana T, Giammanco A, Giarratano A, Chowdhary A. 2018. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J Intensive Care 6:69. doi: 10.1186/s40560-018-0342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CDC. 2019. Antibiotic resistance threats in the United States. CDC, Atlanta, GA. [Google Scholar]
- 14.Mohammad H, Elghazawy NH, Eldesouky HE, Hegazy YA, Younis W, Avrimova L, Hazbun T, Arafa RK, Seleem MN. 2018. Discovery of a novel dibromoquinoline compound exhibiting potent antifungal and antivirulence activity that targets metal ion homeostasis. ACS Infect Dis 4:403–414. doi: 10.1021/acsinfecdis.7b00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC. Antifungal susceptibility testing and interpretation in Candida auris, https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html. Accessed on 25 September 2020.
- 16.Eldesouky HE, Li X, Abutaleb NS, Mohammad H, Seleem MN. 2018. Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int J Antimicrob Agents 52:754–761. doi: 10.1016/j.ijantimicag.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Allen D, Wilson D, Drew R, Perfect J. 2015. Azole antifungals: 35 years of invasive fungal infection management. Expert Rev Anti Infect Ther 13:787–798. doi: 10.1586/14787210.2015.1032939. [DOI] [PubMed] [Google Scholar]
- 18.Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. 2016. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida Species. Front Microbiol 7:2173. doi: 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teo JQ, Lee SJ, Tan AL, Lim RS, Cai Y, Lim TP, Kwa AL. 2019. Molecular mechanisms of azole resistance in Candida bloodstream isolates. BMC Infect Dis 19:63. doi: 10.1186/s12879-019-3672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkow EL, Lockhart SR. 2017. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 10:237–245. doi: 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. 2019. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother 63:e00057-19. doi: 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SH, Iyer KR, Pardeshi L, Munoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. 2019. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 10:e00346-19. doi: 10.1128/mBio.00346-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, Lee SY, Chun S, Lee JH, Choi HJ, Kee SJ, Kim SH, Shin MG. 2019. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol 57:e01624-18. doi: 10.1128/JCM.01624-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worthington RJ, Melander C. 2013. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang CY, Uzoma I, Moore RT, Gilbert M, Duplantier AJ, Panchal RG. 2018. Mitigating the impact of antibacterial drug resistance through host-directed therapies: current progress, outlook, and challenges. mBio 9:e01932-17. doi: 10.1128/mBio.01932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizzorno A, Padey B, Terrier O, Rosa-Calatrava M. 2019. Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy. Front Immunol 10:531. doi: 10.3389/fimmu.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurunathan S, Kang MH, Qasim M, Kim JH. 2018. Nanoparticle-mediated combination therapy: two-in-one approach for cancer. Int J Mol Sci 19:3264. doi: 10.3390/ijms19103264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eldesouky HE, Mayhoub A, Hazbun TR, Seleem MN. 2018. Reversal of azole resistance in Candida albicans by sulfa antibacterial drugs. Antimicrob Agents Chemother 62:e00701-17. doi: 10.1128/AAC.00701-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eldesouky HE, Salama EA, Hazbun TR, Mayhoub AS, Seleem MN. 2020. Ospemifene displays broad-spectrum synergistic interactions with itraconazole through potent interference with fungal efflux activities. Sci Rep 10:6089. doi: 10.1038/s41598-020-62976-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eldesouky HE, Salama EA, Li X, Hazbun TR, Mayhoub AS, Seleem MN. 2020. Repurposing approach identifies pitavastatin as a potent azole chemosensitizing agent effective against azole-resistant Candida species. Sci Rep 10:7525. doi: 10.1038/s41598-020-64571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puthanakit T, van der Lugt J, Bunupuradah T, Ananworanich J, Gorowara M, Phasomsap C, Jupimai T, Boonrak P, Pancharoen C, Burger D, Ruxrungtham K. 2009. Pharmacokinetics and 48 week efficacy of low-dose lopinavir/ritonavir in HIV-infected children. J Antimicrob Chemother 64:1080–1086. doi: 10.1093/jac/dkp322. [DOI] [PubMed] [Google Scholar]
- 33.Kityo C, Walker AS, Dickinson L, Lutwama F, Kayiwa J, Ssali F, Nalumenya R, Tumukunde D, Munderi P, Reid A, Gilks CF, Gibb DM, Khoo S, Dart Trial Team. 2010. Pharmacokinetics of lopinavir-ritonavir with and without nonnucleoside reverse transcriptase inhibitors in Ugandan HIV-infected adults. Antimicrob Agents Chemother 54:2965–2973. doi: 10.1128/AAC.01198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalik DE, Jackson-Alvarez JT, Flores R, Tolentino-Baldridge C, Batra JS. 2015. Low third-trimester serum levels of lamivudine/zidovudine and lopinavir/ritonavir in an HIV-infected pregnant woman with gastric bypass. J Int Assoc Provid AIDS Care 14:116–119. doi: 10.1177/2325957414555231. [DOI] [PubMed] [Google Scholar]
- 35.Marchetti O, Moreillon P, Glauser MP, Bille J, Sanglard D. 2000. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob Agents Chemother 44:2373–2381. doi: 10.1128/AAC.44.9.2373-2381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holmes AR, Keniya MV, Ivnitski-Steele I, Monk BC, Lamping E, Sklar LA, Cannon RD. 2012. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrob Agents Chemother 56:1508–1515. doi: 10.1128/AAC.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41:1482–1487. doi: 10.1128/AAC.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. 2014. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med 5:a019752. doi: 10.1101/cshperspect.a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murata H, Hruz PW, Mueckler M. 2002. Indinavir inhibits the glucose transporter isoform Glut4 at physiologic concentrations. AIDS 16:859–863. doi: 10.1097/00002030-200204120-00005. [DOI] [PubMed] [Google Scholar]
- 40.Hresko RC, Hruz PW. 2011. HIV protease inhibitors act as competitive inhibitors of the cytoplasmic glucose binding site of GLUTs with differing affinities for GLUT1 and GLUT4. PLoS One 6:e25237. doi: 10.1371/journal.pone.0025237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraft TE, Armstrong C, Heitmeier MR, Odom AR, Hruz PW. 2015. The glucose transporter PfHT1 is an antimalarial target of the HIV protease inhibitor lopinavir. Antimicrob Agents Chemother 59:6203–6209. doi: 10.1128/AAC.00899-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heitmeier MR, Hresko RC, Edwards RL, Prinsen MJ, Ilagan MXG, Odom John AR, Hruz PW. 2019. Identification of druggable small molecule antagonists of the Plasmodium falciparum hexose transporter PfHT and assessment of ligand access to the glucose permeation pathway via FLAG-mediated protein engineering. PLoS One 14:e0216457. doi: 10.1371/journal.pone.0216457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. 2016. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Global Action Fund For Fungal Infections (Gaffi). Antifungal drug maps. https://antifungalsavailability.org/maps/map/itraconazole. Accessed 25 September 2020.
- 45.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/mmbr.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Bernardis F, Tacconelli E, Mondello F, Cataldo A, Arancia S, Cauda R, Cassone A. 2004. Anti-retroviral therapy with protease inhibitors decreases virulence enzyme expression in vivo by Candida albicans without selection of avirulent fungus strains or decreasing their anti-mycotic susceptibility. FEMS Immunol Med Microbiol 41:27–34. doi: 10.1016/j.femsim.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Migliorati CA, Birman EG, Cury AE. 2004. Oropharyngeal candidiasis in HIV-infected patients under treatment with protease inhibitors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 98:301–310. doi: 10.1016/j.tripleo.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 48.Asencio MA, Garduno E, Perez-Giraldo C, Blanco MT, Hurtado C, Gomez-Garcia AC. 2005. Exposure to therapeutic concentrations of ritonavir, but not saquinavir, reduces secreted aspartyl proteinase of Candida parapsilosis. Chemotherapy 51:252–255. doi: 10.1159/000087252. [DOI] [PubMed] [Google Scholar]
- 49.Lohse MB, Gulati M, Craik CS, Johnson AD, Nobile CJ. 2020. Combination of antifungal drugs and protease inhibitors prevent Candida albicans biofilm formation and disrupt mature biofilms. Front Microbiol 11:1027. doi: 10.3389/fmicb.2020.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohammad H, Kyei-Baffour K, Younis W, Davis DC, Eldesouky H, Seleem MN, Dai M. 2017. Investigation of aryl isonitrile compounds with potent, broad-spectrum antifungal activity. Bioorg Med Chem 25:2926–2931. doi: 10.1016/j.bmc.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohammad H, Eldesouky HE, Hazbun T, Mayhoub AS, Seleem MN. 2019. Identification of a phenylthiazole small molecule with dual antifungal and antibiofilm activity against Candida albicans and Candida auris. Sci Rep 9:18941. doi: 10.1038/s41598-019-55379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thangamani S, Eldesouky HE, Mohammad H, Pascuzzi PE, Avramova L, Hazbun TR, Seleem MN. 2017. Ebselen exerts antifungal activity by regulating glutathione (GSH) and reactive oxygen species (ROS) production in fungal cells. Biochim Biophys Acta 1861:3002–3010. doi: 10.1016/j.bbagen.2016.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu WR, Guo DM, Zhang LP, Xu DM, Sun SJ. 2016. The synergistic effect of azoles and fluoxetine against resistant Candida albicans strains is attributed to attenuating fungal virulence. Antimicrob Agents Chemother 60:6179–6188. doi: 10.1128/AAC.03046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun LM, Liao K, Liang S, Yu PH, Wang DY. 2015. Synergistic activity of magnolol with azoles and its possible antifungal mechanism against Candida albicans. J Appl Microbiol 118:826–838. doi: 10.1111/jam.12737. [DOI] [PubMed] [Google Scholar]
- 55.Chen YL, Lehman VN, Averette AF, Perfect JR, Heitman J. 2013. Posaconazole exhibits in vitro and in vivo synergistic antifungal activity with caspofungin or FK506 against Candida albicans. PLoS One 8:e57672. doi: 10.1371/journal.pone.0057672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmadi A, Mahmoudi S, Rezaie S, Hashemi SJ, Dannaoui E, Badali H, Ghaffari M, Aala F, Izadi A, Maleki A, Meis JF, Khodavaisy S. 2020. In vitro synergy of echinocandins with triazoles against fluconazole-resistant Candida parapsilosis complex isolates. J Glob Antimicrob Resist 21:331–334. doi: 10.1016/j.jgar.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Hooper RW, Ashcraft DS, Pankey GA. 2019. In vitro synergy with fluconazole plus doxycycline or tigecycline against clinical Candida glabrata isolates. Med Mycol 57:122–126. doi: 10.1093/mmy/myy008. [DOI] [PubMed] [Google Scholar]
- 58.Bidaud AL, Botterel F, Chowdhary A, Dannaoui E. 2019. In vitro antifungal combination of flucytosine with amphotericin B, voriconazole, or micafungin against Candida auris shows no antagonism. Antimicrob Agents Chemother 63:e01393-19. doi: 10.1128/AAC.01393-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 61.Benjamini YH, Yosef H. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 62.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. 2004. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother 48:2124–2131. doi: 10.1128/AAC.48.6.2124-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koselny K, Green J, DiDone L, Halterman JP, Fothergill AW, Wiederhold NP, Patterson TF, Cushion MT, Rappelye C, Wellington M, Krysan DJ. 2016. The celecoxib derivative AR-12 has broad-spectrum antifungal activity in vitro and improves the activity of fluconazole in a murine model of cryptococcosis. Antimicrob Agents Chemother 60:7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li SX, Wu HT, Liu YT, Jiang YY, Zhang YS, Liu WD, Zhu KJ, Li DM, Zhang H. 2018. The F1Fo-ATP synthase beta subunit is required for Candida albicans pathogenicity due to its role in carbon flexibility. Front Microbiol 9:1025. doi: 10.3389/fmicb.2018.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keniya MV, Fleischer E, Klinger A, Cannon RD, Monk BC. 2015. Inhibitors of the Candida albicans major facilitator superfamily transporter Mdr1p responsible for fluconazole resistance. PLoS One 10:e0126350. doi: 10.1371/journal.pone.0126350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ivnitski-Steele I, Holmes AR, Lamping E, Monk BC, Cannon RD, Sklar LA. 2009. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394:87–91. doi: 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.CLSI . 2017. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th ed. CLSI standard M27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence reads of all samples were deposited in the NCBI database (Gene Expression Omnibus [GEO]) under the accession number GSE148341.