Fluoroquinolones (FQ) are crucial components of multidrug-resistant tuberculosis (MDR TB) treatment. Differing levels of resistance are associated with specific mutations within the quinolone-resistance-determining region (QRDR) of gyrA. We sequenced the QRDR from serial isolates of MDR TB patients in the Preserving Effective TB Treatment Study (PETTS) with baseline FQ resistance (FQR) or acquired FQ resistance (FQACQR) using an Ion Torrent Personal Genome Machine (PGM) to a depth of 10,000× and reported single nucleotide polymorphisms in ≥1% of reads.
KEYWORDS: multidrug-resistant tuberculosis, fluoroquinolone, levofloxacin, acquired drug resistance, QRDR, moxifloxacin
ABSTRACT
Fluoroquinolones (FQ) are crucial components of multidrug-resistant tuberculosis (MDR TB) treatment. Differing levels of resistance are associated with specific mutations within the quinolone-resistance-determining region (QRDR) of gyrA. We sequenced the QRDR from serial isolates of MDR TB patients in the Preserving Effective TB Treatment Study (PETTS) with baseline FQ resistance (FQR) or acquired FQ resistance (FQACQR) using an Ion Torrent Personal Genome Machine (PGM) to a depth of 10,000× and reported single nucleotide polymorphisms in ≥1% of reads. FQR isolates harbored 15 distinct alleles with 1.3 (maximum = 6) on average per isolate. Eighteen alleles were identified in FQACQR isolates with an average of 1.6 (maximum = 9) per isolate. Isolates from 78% of FQACQR individuals had mutant alleles identified within 6 months of treatment initiation. Asp94Gly was the predominant allele in the initial FQ-resistant isolates followed by Ala90Val. Seventy-seven percent (36/47) of FQACQR group patients had isolates with FQ resistance alleles prior to changes to the FQ component of their treatment. Unlike the individuals treated initially with other FQs, none of the 21 individuals treated initially with levofloxacin developed genotypic or phenotypic FQ resistance, although country of residence was likely a contributing factor since 69% of these individuals were from a single country. Initial detection of phenotypic resistance and genotypic resistance occurred simultaneously for most; however, phenotypic resistance occurred earlier in isolates harboring mixtures of alleles of very low abundance (<1% of reads), whereas genotypic resistance often occurred earlier for alleles associated with low-level resistance. Understanding factors influencing acquisition and evolution of FQ resistance could reveal strategies for improved treatment success.
INTRODUCTION
Mycobacterium tuberculosis resistance to fluoroquinolones (FQs) is largely attributed to mutations in gyrA, which encodes one subunit of DNA gyrase, an enzyme required for DNA replication. These mutations typically occur within a defined region of gyrA referred to as the quinolone-resistance-determining region (QRDR; nucleotides 262 to 285 or codons 88 to 95), most commonly at codons 90 and 94 (1, 2). Specific mutations result in differing levels of FQ resistance (FQR), and some substitutions can be associated with either low- or high-level resistance, although the specific MIC levels are dependent on the FQ in question (3). To date, most research into the genetics of FQ resistance in M. tuberculosis has relied on results from Sanger sequencing. However, FQ resistance prediction based on Sanger sequencing is problematic due to the phenomenon of heteroresistance, or the presence of multiple variants of a resistance-associated locus within a single isolate (4). FQ-heteroresistant isolates may contain multiple mutant QRDR alleles alone or in combination with wild-type (WT) alleles in various proportions (4–6). Sanger sequencing results in high-level consensus data reflecting the genetics of a pool of sequenced DNA. Alleles must constitute at least 15% to 20% of the total reads generated to be detected by Sanger sequencing; thus, identification of minor alleles is challenging (7). However, next-generation-sequencing (NGS) techniques in combination with bioinformatics tools enable genetic visualization in exquisite detail (8). Using these tools, we can now determine the exact sequence of individual strands of DNA, enabling discrimination of mutation phasing (i.e., localization of multiple mutations on the same piece of DNA). NGS is the ideal research tool to study the acquisition and evolution of FQ resistance in individual patients. A more comprehensive understanding of the development of FQ resistance will facilitate more-rapid and more-accurate diagnosis and enable clinicians to design treatment regimens that are more effective at killing M. tuberculosis and less likely to contribute to FQ resistance.
We performed ultradeep NGS sequencing to evaluate genotypic FQ resistance in serial M. tuberculosis isolates from patients residing in eight countries with locally confirmed multidrug-resistant tuberculosis (MDR TB) who were enrolled in the Preserving Effective TB Treatment Study (PETTS) from 2005 to 2010 (9). Baseline and monthly follow-up sputum specimens were collected from patients along with clinical and demographic information throughout the course of treatment for MDR TB. Drug susceptibility testing (DST) using agar proportion was performed at the U.S. Centers for Disease Control and Prevention for 11 first- and second-line drugs on baseline and final isolates (9). DST was also performed on intermediate isolates for individuals whose final isolate demonstrated acquired resistance to a second-line drug (ciprofloxacin [CIP], ofloxacin [OFX], kanamycin [KAN], amikacin [AMK], or capreomycin [CAP]). In this study, we used NGS to evaluate the acquisition and evolution of mutations associated with FQ resistance in PETTS patients and compared results with phenotypic DST.
RESULTS
Sample set.
For this study, individuals were classified into one of three groups based on agar proportion DST results for OFX (2 mg/ml) as follows. In group 1, the FQ-susceptible (FQS) group, baseline and final isolates were susceptible to OFX. In group 2, the FQ-resistant (FQR) group, the baseline isolate was resistant to OFX. In group 3, the FQACQR (acquired FQ resistance) group, the baseline isolate was susceptible to OFX, but the final isolate was resistant (9). A variable number of intermediate isolates were available for each patient. To assess development and evolution of resistance, for the FQR and FQACQR groups, we included individuals with a baseline isolate, a final isolate, and at least two intermediate isolates available (FQR n = 55; FQACQR n = 56). We also included 168 randomly selected individuals who were classified as FQS (only baseline and final isolates were tested for the FQS group). A total of 447 isolates were available from FQACQR individuals (n = 56), with a mean of 8.0 isolates (range, 4 to 18) per patient, whereas 391 isolates were available from FQR individuals (n = 55), with a mean of 7.1 isolates per patient (range, 4 to 17). Individuals were from Estonia, South Korea, Latvia, Peru, Philippines, Russia, South Africa, and Thailand (Table 1). Baseline and final isolates had been genotyped previously by 24-locus mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) analysis when an enrollee’s baseline and final cultures had differing susceptibility profiles with respect to FQs, injectable drugs (amikacin, KAN, and CAP), isoniazid, or rifampin (9). All isolates from individuals whose baseline and final isolates had discordant MIRU-VNTR genotypes were excluded from the analysis.
TABLE 1.
Number of Mycobacterium tuberculosis isolates by country of enrollment for individuals in each fluoroquinolone groupa
| Patient group |
No. (%) of isolates |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Estonia (n = 22) |
South Korea (n = 36) |
Latvia (n = 23) |
Peru (n = 17) |
Philippines (n = 26) |
Russia (n = 27) |
South Africa (n = 122) |
Thailand (n = 6) |
Total (n = 279) |
|
| FQACQR | 6 (10.7) | 1 (1.8) | 5 (8.9) | 4 (7.1) | 3 (5.4) | 2 (3.6) | 34 (60.7) | 1 (1.8) | 56 |
| FQR | 5 (9.1) | 8 (14.5) | 4 (7.3) | 2 (3.6) | 8 (14.5) | 10 (18.2) | 17 (30.9) | 1 (1.8) | 55 |
| FQS | 11 (6.5) | 27 (16.1) | 14 (8.3) | 11 (6.5) | 15 (8.9) | 15 (8.9) | 71 (42.3) | 4 (2.4) | 168 |
FQACQR, the baseline isolate was susceptible to ofloxacin (OFX), but the final isolate was resistant; FQR, the baseline isolate was resistant to OFX; FQS, baseline and final isolates were susceptible to OFX. The percentage of individuals from each country making up each susceptibility group is indicated in parentheses. FQ, fluoroquinolone.
SNP detection.
We interrogated the gyrA QRDR of baseline (FQR, FQACQR, FQS), final (FQR, FQACQR, FQS), and intermediate (FQR, FQACQR) isolates at a minimum depth of 10,000×. A single nucleotide polymorphism (SNP) was reported when present in greater than 1% of reads. FQ resistance is commonly complicated by heteroresistance, in which individual isolates have multiple QRDR alleles (4). SNP detection was performed exclusively on reads spanning the entire QRDR to distinguish between phased mutations (multiple SNPs on a single strand of DNA) and unphased mutations (multiple SNPs located independently on different strands of DNA). We refer to each unique gyrA QRDR sequence as an allele; therefore, some alleles contain a single mutation and others contain multiple phased mutations. Although mutations affected only 6 codons (80, 88, 90, 91, 94, and 95) within the QRDR, multiple different nonsynonymous substitutions were detected at many of these codons. In total, 27 unique mutant alleles were identified among all baseline, final, and intermediate isolates containing either a single amino acid change (16 alleles) or double amino acid changes (11 alleles). Isolates often had a combination of alleles (heteroresistance) at any time point, including different amino acid substitutions at the same codon position. The SNPs identified have all been previously associated with FQ resistance, with the exception of Gly88Val and Asp94Cys, which, to our knowledge, have not been reported. It is likely that these SNPs result in FQ resistance since multiple other substitutions at these positions are known to be associated with resistance. Both mutations were identified in isolates from the FQACQR group, and each isolate was heteroresistant, harboring additional gyrA alleles with mutations that have been previously associated with FQ resistance. Eleven different phased, double mutations (two SNPs present on the same piece of DNA) were identified; however, alleles with phased, double mutations were infrequent overall, especially in the FQR group.
In the FQS group, the majority of baseline and final isolates exclusively contained WT QRDR alleles (Table 2). However, isolates from 11 FQS individuals (11/168, 6.5%) had mutant alleles present in either their baseline isolates (n = 1) or final isolates (n = 5) or both (n = 5). For individuals with mutant alleles in both baseline and final isolates, the same allele was identified in both isolates. Three (3/5) had SNPs with established associations with low-level FQ resistance (Asp94Ala, n = 2; Ala90Val, n = 1). The remaining two had Gly88Ala mutations. Fewer data are available concerning the effects of Gly88Ala mutations on FQ susceptibility, but it has been identified in a small number of clinical isolates with low-level FQ resistance (10–13). In one individual in the FQS group, Asp94Gly was identified in a minority of QRDR-spanning reads from the baseline isolate but not in those from the corresponding final isolate (Table 2). The five non-WT final isolates from the FQS group harbored low-level-resistance-associated mutations (Ala90Val, Asp94Ala), a Gly88Ala substitution, or Asp94Tyr (Table 2). Asp94Tyr has been associated with both low-level and high-level FQ resistance, but it is more commonly considered a mutation conferring high-level resistance (14–19). One final isolate from an individual in the FQS group had a combination of a mutation associated with low-level resistance (Asp94Ala, 93.5% of reads) and a mutation associated with high-level resistance (Asp94Gly, 4.7% of reads). A complementary analysis of baseline isolates from individuals in the FQR group identified fully WT QRDR alleles in only two individuals (2/55, 3.6%; Table 2). Furthermore, no mutation within the gyrA QRDR was detected in any of the serial isolates from either of these individuals. Evaluation of baseline isolates from 56 individuals in the FQACQR group revealed a fully WT QRDR profile for 50 individuals, as expected, while mutant QRDR alleles were identified in baseline isolates from 6 individuals (6/56, 10.7%; Table 2). Five isolates had low-level-resistance-associated mutations Ala90Val and Asp94Ala (n = 3 and n = 2, respectively), and one isolate had a Gly88Ala mutation. Despite phenotypic FQ resistance, none of the isolates from one individual in the FQACQR group (1/56, 1.8%) had a detectable mutant gyrA QRDR allele, raising the possibility of a mutation outside the gyrA QRDR, possibly in gyrB. The final isolate from another individual did not contain a detectable mutant allele, although QRDR mutations were identified in intermediate isolates from this individual.
TABLE 2.
GyrA QRDR alleles identified in baseline and final isolates from individuals in each groupa
| Patient group | Concordant | Discordant (% of total reads in discordant isolates) |
|---|---|---|
| FQS (n = 168) | WT baseline and final (n = 157) | Mutant baseline (n = 1), Asp94Glyb (1%); mutant final (n = 5), Gly88Ala (98.5%), Ala90Val (2.5%), Asp94Ala (98.7%), Asp94Tyrb (9.4%), Asp94Ala (93.5%) and Asp94Glyb (4.7%); mutant baseline and final (n = 5), Gly88Ala (>98%), Gly88Ala (>97%), Ala90Val (>97%), Asp94Ala (>95%), Asp94Ala (>94%) |
| FQACQR (n = 56) | WT baseline and mutant final (n = 49) | Mutant baseline (n = 6), Gly88Ala (13.3%), Ala90Val (95.9%), Ala90Val (46.1%), Ala90Val (97.6%), Asp94Ala (95.7%), Asp94Ala (98.5%); WT final (n = 1) |
| FQR (n = 55) | Mutant baseline and mutant final (n = 53) | WT baseline and final (n = 2) |
FQACQR, the baseline isolate was susceptible to ofloxacin (OFX), but the final isolate was resistant; FQR, the baseline isolate was resistant to OFX; FQS, baseline and final isolates were susceptible to OFX; WT, wild-type gyrA QRDR (quinolone resistance-determining region); Concordant, expected QRDR alleles present in baseline and/or final isolate; Discordant, unexpected QRDR alleles present in baseline and/or final isolate.
Mutation associated with high-level resistance.
Baseline isolates from the FQR group and the first isolate from the FQACQR group in which a mutant allele was detected (referred to here as the FMA isolate) frequently contained a WT allele in combination with a mutant allele(s), with heterogeneity particularly pronounced in the FQACQR group (Table 3). In total, 15 unique mutant alleles were present in isolates from the FQR group. Isolates from individuals in this group had an average of 1.3 alleles per time point, with a maximum of 6 alleles in a single isolate at a single time point (Table 4). The FQACQR group had an even larger variety of mutant alleles (n = 18), with an average of 1.6 mutant alleles per time point and a maximum of 9 mutant alleles in one isolate (Table 4). Asp94Gly was the predominant substitution in FQR baseline isolates, FQACQR FMA isolates, and final isolates from both groups (for FQR baseline isolates, 36.4% [20/55]; for FQR final isolates, 34.5% [19/55]; for FQACQR FMA isolates, 62.5% [35/56]; for FQACQR final isolates, 69.6% [39/56]; Fig. 1); Ala90Val was the second most abundant allele in both groups (for FQR baseline isolates, 29.1% [16/55]; for FQR final isolates, 30.9% [17/55]; for FQACQR FMA isolates, 44.6% [25/56]; for FQACQR final isolates, 48.2% [27/56]; Fig. 1). The total number of high-level FQ resistance mutations (Asp94Gly, Asp94Asn, Asp94Tyr) was lower than the combined total number of all other mutations in the month in which the first mutation was detected as well as in final isolates in both the FQACQR and FQR groups (Fig. 2).
TABLE 3.
Percentages of individuals with an isolate harboring a mutant QRDR allele(s) either alone or in combination with a WT QRDR allele at baseline or in the FMA isolatea
| Patient group | Mutant allele(s) only |
Mutant allele(s) + WT allele |
|---|---|---|
| FQACQRb | 36.4% (n = 20) | 63.6% (n = 35) |
| FQRb | 81.0% (n = 43) | 19.0% (n = 10) |
FQACQR, the baseline isolate was susceptible to ofloxacin (OFX), but the final isolate was resistant; FQR, the baseline isolate was resistant to OFX; FMA isolate, the first serial isolate from an individual in the FQACQR group with a mutant gyrA allele; WT, wild type; QRDR, quinolone resistance-determining region.
One individual in the FQACQR group (1/56) and two individuals in FQR group (2/55) had WT QRDR alleles.
TABLE 4.
Number of mutant QRDR alleles in all isolates or in individual isolates by groupa
| Patient group |
All isolates |
Individual isolates |
||
|---|---|---|---|---|
| No. of isolates |
Total no. of unique alleles |
Mean no. of mutant alleles |
Range of mutant alleles |
|
| FQR | 391 | 15 | 1.3 | 0–6 |
| FQACQR | 447 | 18 | 1.6 | 0–9 |
FQACQR, the baseline isolate was susceptible to ofloxacin (OFX), but the final isolate was resistant; FQR, the baseline isolate was resistant to OFX; QRDR, quinolone resistance-determining region.
FIG 1.
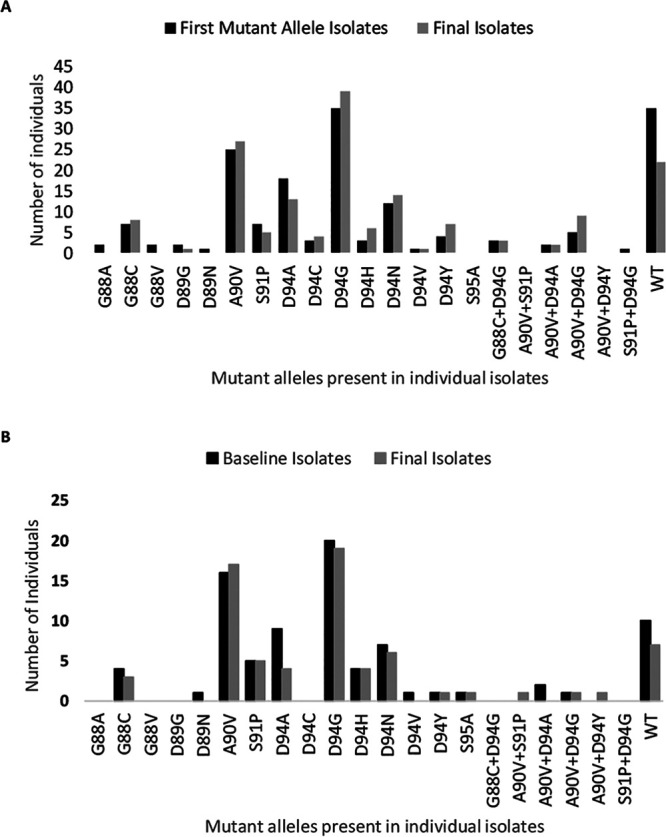
Number of FMA isolates (FQACQR; A), baseline isolates (FQR; B), and final isolates (FQACQR, A; FQR, B) harboring each quinolone-resistance-determining region. FQACQR, baseline isolate was susceptible to ofloxacin (OFX), but final isolate was resistant; FQR, baseline isolate was resistant to OFX; FMA isolate, the first serial isolate from an individual in the FQACQR group with a mutant gyrA allele.
FIG 2.
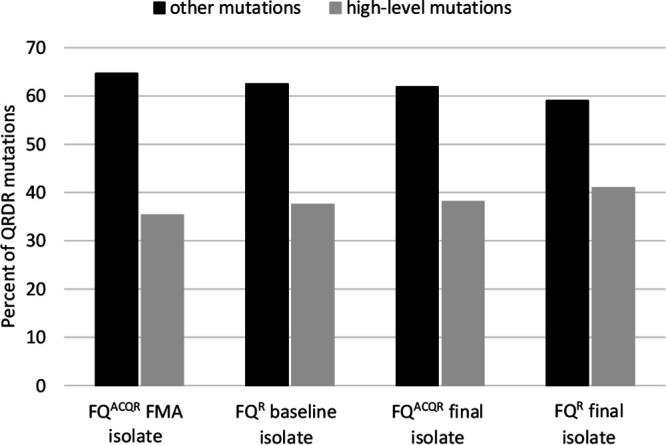
Percentage of QRDR (quinolone-resistance-determining region) mutations that are associated with high-level fluoroquinolone (FQ) resistance (Asp94Gly, Asp94Asn, Asp94Tyr) compared to all other QRDR mutations. FQACQR, baseline isolate was susceptible to ofloxacin (OFX), but final isolate was resistant; FQR, baseline isolate was resistant to OFX; FMA isolate, the first serial isolate from an individual in the FQACQR group with a mutant gyrA allele.
Timing of detection of FQ resistance by molecular versus phenotypic methods.
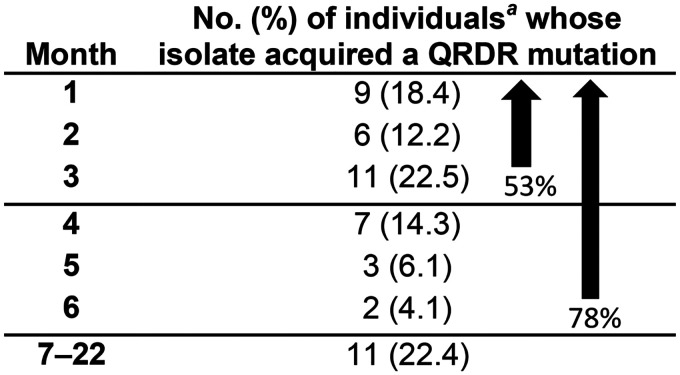
Isolates from individuals in the FQACQR group developed QRDR mutations early in treatment. Isolates from 53% of individuals in this group with previously WT gyrA acquired at least one mutant QRDR allele within 3 months and 78% by 6 months following treatment initiation (Table 5). For individuals whose serial isolates acquired FQ resistance, we compared results from the month in which the FMA was detected with results from the month in which phenotypic FQ resistance was initially reported. In 40 (40/55; 72.3%) individuals, the initial occurrence of FQ resistance was detected in the same isolate using either phenotypic or molecular detection methods. However, FQ resistance was detected earlier using molecular methods in isolates from 11 individuals (11/55; 20%, Table 6), including six baseline isolates that, despite demonstrating phenotypic FQ susceptibility, contained mutant QRDR alleles. Eight isolates with earlier resistance detection by molecular methods harbored alleles associated with low-level resistance (Ala90Val, Asp94Ala), an isolate from one individual had a Gly88Ala mutation (13.3% of reads), an isolate from one individual had a mixture of Ala90Val and Ser91Pro (96.3% and 1.1% of reads, respectively), and an isolate from one individual had a mixture of WT gyrA and Asp94Gly (95.4% and 1% of reads, respectively) (Table 6). In this group, detection by molecular methods preceded detection by phenotypic methods by up to 15 months (mean = 7.2, median = 9), although resistance detection by either method could have potentially occurred earlier in a few cases where consecutive monthly isolates were not available. FQ resistance was detected earlier via phenotypic testing in four individuals (4/55; 7.3%, Table 6). For one individual, phenotypic resistance was detected at month 2, while the FMA isolate (a mixture of gyrA alleles) was not detected until month 8; however, isolates from months 3 to 7 gave phenotypically susceptible test results for this patient. In all other individuals for whom resistance was detected earlier using phenotypic methods, an FMA result was identified in the next consecutive isolate tested and consisted of a mixture of mutant gyrA alleles, with Gly88Cys predominating.
TABLE 5.
Month after treatment initiation when the first mutant QRDR allele was detected in isolates from FQACQR individualsb
Despite phenotypic susceptibility, baseline isolates from six individuals (6/56) had mutant QRDR alleles and are not included.
FQACQR, the baseline isolate was susceptible to ofloxacin (OFX), but the final isolate was resistant; FMA isolate, the first serial isolate from an individual in the FQACQR group with a mutant gyrA allele; QRDR, quinolone resistance-determining region.
TABLE 6.
Time of detection of phenotypic and genotypic fluoroquinolone resistance when it did not occur in the same isolatea
| Method of initial detection of FQ resistance |
Sample ID |
Mo of phenotypic resistance detection |
Mo of genotypic resistance detection |
Allele(s) present in FMA isolates (% of reads) |
|---|---|---|---|---|
| Molecular | 3041 | 5 | 1 | Ala90Val |
| 4035 | 1 | 0 | Ala90Val | |
| 12239 | 4 | 0 | Ala90Val | |
| 13059 | 12 | 0 | Ala90Val | |
| 8001 | 12 | 3 | Ala90Val (96.3), S91Pro (1.1) | |
| 2028 | 9 | 0 | Asp94Ala | |
| 2042 | 11 | 0 | Asp94Ala | |
| 2045 | 22 | 13 | Asp94Ala | |
| 11824 | 12 | 11 | Asp94Ala | |
| 11506 | 21 | 6 | Asp94Gly (1), WT (95.4) | |
| 3002 | 4 | 0 | Gly88Ala | |
| Phenotypic | 2008 | 2 | 8 | Asp94Asn |
| 11963 | 2 | 3 | Gly88Cys, Gly88Cys+Asp94Gly, Asp94Gly | |
| 11876 | 2 | 3 | Gly88Cys, Gly88Cys+Asp94Gly, Ala90Val, Asp94Gly, Asp94His | |
| 11225 | 3 | 5 | Gly88Cys, Gly88Val, Asp89Gly, Ala90Val, Asp94Gly | |
FQACQR, the baseline isolate was susceptible to ofloxacin (OFX), but the final isolate was resistant; FMA isolate, the first serial isolate from an individual in the FQACQR group with a mutant gyrA allele; FQ, fluoroquinolone; ID, identifier; Mo, month.
Treatment with fluoroquinolones and FQ resistance.
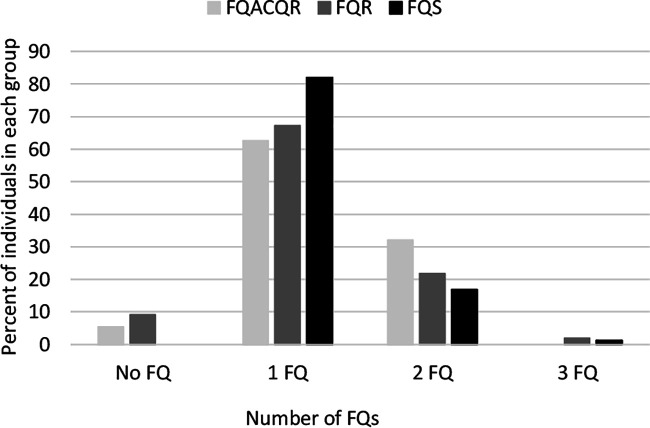
Treatment regimens for individuals enrolled in PETTS were dependent on a number of factors, including country of residence, treatment history, DST results, and drug availability (9). Three of 56 individuals (5.4%) in the FQACQR group and 5 of 55 individuals (9.1%) in the FQR group were not treated with a FQ during this study. Ciprofloxacin (CIP), ofloxacin (OFX), levofloxacin (LEV), gatifloxacin (GAT), and/or moxifloxacin (MOX) was used to treat all other individuals. Some individuals (FQACQR group = 32.1%, FQS group = 18.0%, FQR group = 23.6%) were treated with multiple FQs over the course of the study (Fig. 3). Eight individuals (14.2%) in the FQACQR group had isolates demonstrating genotypic resistance prior to initiation of a FQ-containing treatment regimen. It was common for individuals to have changes to or interruptions in treatment. We specifically looked at times when individuals switched from one FQ to a different FQ or when there was an interruption in treatment with a FQ. We defined each time that such a modification or interruption occurred as a new FQ treatment episode. Individuals in all groups experienced between zero and eight FQ treatment episodes, with a median of two episodes. Interestingly, 76.6% (36/47) of patients in the FQACQR group who acquired genotypic resistance following initiation of a FQ-containing treatment regimen did so during their first FQ treatment episode.
FIG 3.
Number of fluoroquinolones (FQs) to which patients were exposed over the duration of treatment. FQACQR, baseline isolate was susceptible to ofloxacin (OFX), but final isolate was resistant; FQR, baseline isolate was resistant to OFX; FQS, baseline and final isolates were susceptible to OFX; No FQ, patient was not exposed to any FQ.
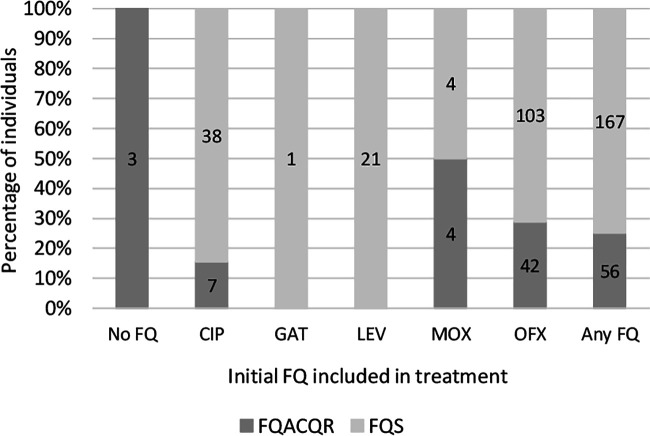
We compared treatment regimens for FQS and FQACQR individuals to determine whether development of FQ resistance might be influenced by the initial FQ used for treatment. Of 45 individuals for whom CIP was the first or only FQ used, serial isolates from 17.8% (8/45) acquired gyrA QRDR mutations and 11.1% (5/45) became phenotypically FQ resistant (Fig. 4). Of 145 individuals who were initially treated with OFX, isolates from 30.3% (44/145) acquired mutations associated with FQ resistance at some point during PETTS whereas 29% (42/145) acquired phenotypic resistance to FQs. A single individual was initially treated with GAT, and isolates from this individual maintained a WT QRDR and phenotypic susceptibility. Isolates collected prior to treatment initiation from five of eight (FQS = 2, FQACQR = 3) individuals for whom MOX was the initial FQ used harbored QRDR mutations, and serial isolates from one individual (1/8) acquired QRDR mutations following initiation of MOX treatment. Serial isolates from five individuals initially treated with MOX developed phenotypic resistance. No serial isolates from the 21 individuals for whom LEV was the first FQ used developed genotypic or phenotypic resistance.
FIG 4.
Percentage of individuals whose isolates developed phenotypic fluoroquinolone (FQ) resistance by the initial FQ included in treatment. CIP, ciprofloxacin; GAT, gatifloxacin; LEV, levofloxacin; MOX, moxifloxacin; OFX, ofloxacin, FQACQR, baseline isolate was susceptible to OFX, but final isolate was resistant; FQS, baseline and final isolates were susceptible to OFX; No Fq, patient was not exposed to any FQ; Any FQ, patient was treated with any FQ. Numbers within bars indicate the actual number of individuals in each category.
Other factors may have influenced the lack of acquired FQ resistance in individuals whose first FQ was LEV. Sixty-nine percent (18/21) of these individuals were from one country. Previous analysis of the PETTS cohort demonstrated that the strongest risk factor for acquisition of resistance to a FQ was the absence of an effective injectable drug (AMK, CAP, or KAN) in a treatment regimen (20). Inclusion of an effective injectable drug did not explain the propensity for development of FQ resistance among individuals treated with any other FQ versus those treated with LEV since 86% of the former were also treated with an effective injectable drug compared to only 52.4% individuals for whom LEV was the first FQ.
DISCUSSION
Treatment with an effective FQ is critical for successful outcomes for individuals with MDR TB (21). LEV and MOX are listed as group A agents in WHO MDR TB treatment guidelines and therefore should be included in longer MDR-TB treatment regimens whenever possible (22). Thus, protection of FQ susceptibility and rapid detection of FQ resistance are crucial. We undertook this investigation to visualize the development of resistance to FQs during MDR TB treatment from a molecular perspective. Our study set for this retrospective analysis was chosen to include individuals whose isolates maintained phenotypic FQ susceptibility throughout treatment (FQS group), those whose isolates were FQ resistant upon treatment initiation (FQR group), and those whose isolates acquired FQ resistance during PETTS (FQACQR group). We selected 224 individuals (FQS and FQACQR individuals) whose isolates were phenotypically FQ susceptible and 55 individuals (FQR individuals) whose isolates were phenotypically FQ resistant at the time of enrollment. Isolates from 56 individuals (FQACQR individuals) developed phenotypic FQ resistance during treatment, while isolates from 168 individuals (FQS individuals) remained FQ susceptible. We performed ultradeep sequencing on the gyrA QRDR of baseline and final isolates for all individuals and additionally sequenced the gyrA QRDR for intermediate isolates from those individuals whose serial isolates either acquired FQ resistance during treatment (FQACQR group) or had FQ resistance at the time of enrollment (FQR group). Thus, we were able to pinpoint the time period during which individual resistance allele appeared and evaluate this in the context of treatment.
During this study, isolates acquired mutant gyrA alleles soon after initiation of treatment, with over half developing at least one mutant allele within the first 3 months and three-quarters acquiring genotypic FQ resistance within 6 months of treatment initiation. A similar timeline was seen for acquisition of phenotypic resistance. Surprisingly, resistance development did not appear to be associated with FQ nonadherence or changes in the FQ component of treatment regimens in this cohort given that isolates from 77% of individuals acquired FQ resistance during their first FQ treatment episode. Since resistance primarily emerged during initial treatment episodes, we evaluated whether the initial FQ that an individual was treated with had any effect on resistance development. Remarkably, serial isolates from all 21 individuals for whom LEV was the initial FQ administered retained a WT gyrA QRDR throughout the study, versus 82.2%, 69.1%, and 66.6% of isolates from individuals for whom CIP (n = 45), OFX (n = 139), or MOX (n = 3) was the earliest FQ administered. Further, as previous studies have shown that treatment with an effective injectable drug is protective for FQ susceptibility (20), we evaluated the frequency with which individuals initially treated with LEV were also treated with an effective injectable and found that only 52% had been treated with an effective injectable drug compared with 86% of all other individuals whose isolates acquired FQ resistance. Of note, the majority of individuals initially treated with LEV were from a single country, and while LEV was the initial FQ prescribed to 66.7% of individuals from that country enrolled in PETTS, 96.3% of all isolates from all individuals from this country remained FQ susceptible. Therefore, it is possible that another factor(s), including country of residence, influenced the lack of acquired FQ resistance. It is also possible that selection bias played a role in this finding as availability of at least four isolates, including baseline and final isolates, was a criterion for patient inclusion in our analysis and individuals who achieved cure generally had fewer isolates.
Some mutations within the gyrA QRDR are associated with either low-level or high-level FQ resistance. To evaluate whether initial development of low-level FQ resistance precedes acquisition of high-level resistance in MTBC, we compared the specific alleles identified in isolates from individuals in the FQACQR group with those found in isolates from individuals in the FQR group, who presumably had more-established infections. In this study set, resistance-associated mutations were identified in 6 different amino acid positions within the gyrA QRDR; however, a total of 27 different gyrA alleles were identified, including two mutations (Gly88Val, Asp94Cys) that to our knowledge were previously unpublished. Overall, a greater variety of QRDR alleles was identified within FQACQR group. All mutations except for Ser95Ala (present in one isolate) identified in individuals in the FQR group were also identified in the FQACQR group. As reported from other studies, Asp94Gly (high-level resistance) and Ala90Val (low-level resistance) were the first and second most common mutations identified in both groups, indicating that there was no bias for mutations associated with high-level FQ resistance in individuals with established versus developing FQ resistance. We also compared prevalences of specific alleles in baseline/FMA isolates versus final isolates within the FQR and FQACQR groups (Fig. 1) and found very little variation in the specific alleles present in each group of isolates. Moreover, the combined number of alleles known to be associated with high-level resistance (Asp94Asn, Asp94Gly, Asp94Tyr) was less than the total number of other alleles in baseline/FMA and final isolates from individuals in both groups (Fig. 2). Thus, there does not appear to be a shift from alleles associated with low-level resistance to those associated with high-level resistance over time.
While use of molecular techniques for identification of drug resistance is becoming more commonplace, phenotypic drug susceptibility testing is still considered the gold standard in most of the world. Phenotypic testing is based on identification of resistance that is present in greater than 1% of the bacterial population. In the current study, we used ultradeep sequencing to identify gyrA QRDR mutations present in at least 1% of reads from an individual isolate; thus, similar sensitivities are expected for the two methods. We compared time to initial detection of phenotypic resistance versus time to initial detection of genotypic resistance in the FQACQR group. Overall, the results obtained with the two methods correlated well and initial resistance was detected in the same month using either method in isolates from 72.3% of individuals (n = 40); otherwise, resistance was detected first via genotypic methods in isolates from 20% of individuals (n = 11) and via phenotypic methods for isolates from 7.3% of individuals (n = 4).
Examination of serial isolates from individuals in which initial resistance detection occurred in different months via the two methods provides insight into the strengths and weaknesses of each method. When phenotypic resistance was detected prior to genotypic resistance, FMA isolates always harbored a mixture of very-low-abundance gyrA alleles, highlighting the fact that molecular detection of resistance does not occur unless a single allele is present in a number of reads that equals or exceeds the sensitivity limit of the molecular method in use (1% of reads in this study). However, since all alleles in an isolate contribute to phenotypic resistance, any single allele or any combination of alleles present in at least 1% of the bacterial population will likely signal phenotypic resistance. While FQ heteroresistance is not uncommon, the frequency of the occurrence of an isolate harboring very-low-abundance alleles during an actual infection is unknown. In this study, for example, we identified genetic markers for resistance concurrent with/prior to detection of phenotypic resistance 91% (51/56) of the time and within 1 month of detection of phenotypic resistance 96% (54/56) of the time. However, we used an exceptionally sensitive technique capable of detecting mutant alleles present in as few as 1% of reads. Others may use less-sensitive molecular techniques; thus, to simulate a less sensitive molecular method, we adjusted our sensitivity cutoff to detect alleles present in at least 10% of reads. Under these conditions, molecular detection of resistance in 6 individuals in this study set would be delayed compared to detection with a 1% sensitivity rate. Mutant QRDR alleles would, however, reach the limit of detection in isolates from 5 of these individuals in their next available isolate. Therefore, in isolates harboring a very-low-abundance (<1%) resistance-associated allele(s), phenotypic methods would have sensitivity superior to molecular methods, possibly leading to resistance being detected slightly earlier than with the molecular methods.
Comparing the timing of initial detection of FQ resistance by phenotypic methods to that by molecular methods, detection occurred earlier by ultradeep sequencing more frequently than by phenotypic methods. In this case, most (10 of 11) isolates harbored low-level-resistance-associated mutations either alone or as the predominant component of a mixture of alleles, suggesting that detection of alleles associated with low-level FQ resistance may be a strength of genotypic versus phenotypic DST as has been reported previously (6). It is also noteworthy that GyrA mutations Gly88Ala, Ala90Val, and Asp94Ala associated with low-level FQ resistance were identified alone or as the majority mutant alleles in 15 isolates (4, 5, and 6 isolates, respectively) categorized as susceptible by phenotypic testing, providing further evidence for this supposition. Possibly, the nearness of the MIC associated with low-level-resistance alleles to the critical concentration used for susceptibility testing can result in false susceptibility results during phenotypic testing. Therefore, individuals with low-level-resistance mutations may be at risk for false predictions of susceptibility when phenotypic methods are the sole techniques employed for FQ resistance detection.
In this study, we analyzed a large number of diverse isolates from different geographic regions, but the study did have some limitations. Individuals with <4 isolates were excluded from our study set, possibly resulting in a bias against those with rapid cure or with poor adherence to monthly follow-up. Moreover, monthly follow-up isolates were not available for all individuals for every month, resulting in gaps in the timeline. Conceivably, the absence of those isolates may have delayed resistance detection and, furthermore, the missing isolates may have harbored additional gyrA alleles. Initial susceptibility group assignments were based on agar proportion DST using OFX at 2 μg/ml. Although these DST methods are well validated and generally robust, we cannot rule out the possibility that had DST been performed using alternative methods or a different FQ, phenotypic results or times of detection or both might have differed. Also, although some mutations in gyrB are associated with resistance to FQs, these mutations occur at a much lower rate than gyrA mutations and were not considered here. Finally, most individuals within our study set were initially treated with either OFX or CIP; thus, conclusions regarding individuals initially treated with MOX and LEV are underpowered and warrant further study.
Based on the results of this study, it appears that FQ-resistant isolates develop rapidly following the initiation of treatment in individuals with MDR TB and, although further studies are warranted, that persons treated with LEV initially might be less likely to develop isolates with FQ resistance compared to those treated with other FQs. Although many isolates from many individuals harbored multiple alleles over the course of the study, we did not identify a trend for progression from low-level to high-level FQ resistance. Detection of FQ-resistant isolates occurred in the same month by the ultradeep sequencing and phenotypic methods for most individuals, although each technique has advantages under specific circumstances. Phenotypic DST can detect mixtures of alleles present in very low abundance, possibly below the limit of detection for some molecular methods; however, low-level-resistance-associated mutations were more likely to be detected earlier by molecular DST. Although the reduced sensitivity for detection of very-low-abundance alleles by molecular DST compared to phenotypic DST is concerning, the delay in identification of low-level-resistance-associated alleles may be a more significant issue since the delay was longer and the frequency of low-level mutations was greater than the frequency of low-abundance mutations. It is possible that this shortcoming could be overcome by testing multiple concentrations of FQ when using phenotypic methods. In order to increase the opportunity for a good outcome, individuals with MDR TB should be monitored closely using both highly sensitive molecular methods and phenotypic methods, if possible, for development of FQ resistance during the first months of treatment so that treatment regimens can be adjusted in a timely manner.
MATERIALS AND METHODS
Informed consent for participation in the study was obtained from all patients.
Sequencing.
A 30-μl aliquot from a broth culture of each M. tuberculosis isolate was heat inactivated at 97°C for 30 min and stored at 4°C until needed. The QRDR region of gyrA (nucleotides 172 to 395, amino acids 58 to 132) was amplified using a forward primer composed of an Ion Torrent A adaptor, an IonXpress barcode sequence, and a gene-specific sequence (5′-GCAATGTTCGATTCCGGCTTC) and a reverse primer composed of an Ion Torrent P1 adaptor followed by the gene-specific sequence (5′-GCTTCGGTGTACCTCATCGC). PCR mixtures (25 μl) included 1 μl of heat-inactivated isolate (diluted 1:2 with water), 400 nM (final concentration) forward and reverse primers, and 12.5 μl GoTaq 2× master mix (Promega, Madison, WI). Cycling conditions were as follows: 5 min denaturation at 97°C, followed by 40 cycles of 30 s at 97°C, 30 s at 55°C, and 30 s at 72°C, with a final 5-min extension at 72°C. Equal volumes of up to 91 PCR mixtures from different M. tuberculosis isolates were pooled, and 65 μl of the pool was purified using 1.8× AMPure XP magnetic beads (Beckman Coulter, Indianapolis, IN) and eluted with 40 μl Tris-low EDTA (10 mM Tris [pH 8.0], 0.1 mM EDTA). Purified pools were quantified, diluted to 50 pM, and loaded onto an Ion Chef instrument (Thermo Fisher Scientific, Asheville, NC) to generate libraries. Libraries were loaded onto an Ion Torrent PGM sequencer (Thermo Fisher Scientific, Asheville, NC) and sequenced using an Ion PGM Hi-Q sequencing kit and Ion 316 v2 chips. Sequence reads were generated with Torrent Suite software.
Sequence analysis.
Following sequencing, BAM files were exported using the File Exporter plugin of Torrent Suite software. The BAM files were analyzed using Amplicon Sequencing Analysis Pipeline (ASAP; TGen) (23, 24). A “region of interest” (ROI) assay was created for the QRDR at positions 7563 to 7586 of the H37Rv reference (GenBank accession number NC_000962.3) used in the BAM files. ASAP inspects each read aligning to the ROI, extracts the nucleotide sequence, and translates it into the amino acid sequence. Reads that do not have complete coverage of the ROI are discarded. Remaining amino acid sequences are added to a counter variable which tracks the number of occurrences for each unique sequence. Sequences occurring in at least 1% of the total read depth at the QRDR are output in the final report for each isolate. For easier interpretation, each amino acid sequence was added to a lookup table to convert the sequence into a haplotype name listing the amino acid substitution(s) present in the sequence, such as A90V/D94Y. ASAP additionally analyzes the full pileup and reports each SNP occurring at a rate of greater than 1% throughout the amplicon sequence. Sequencing and analysis were repeated for isolates with less than 10,000× coverage. Isolates were excluded from the analysis if repeat sequencing failed to yield coverage greater than 10,000×.
To determine the expected error rate associated with single nucleotide polymorphism (SNP) calling, we prepared libraries from 96 M. tuberculosis H37Rv aliquots in duplicate and performed sequencing as described above. Sequences were aligned to an H37Rv reference sequence, and the error rate was determined to be 0.2%. For the study isolates, the threshold for SNP calling was set at 1%, 5 times greater than the previously determined error rate (0.2%). Therefore, only SNPs present in ≥1% of reads were considered.
ACKNOWLEDGMENTS
Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the U.S. Public Health Service, or the CDC. The findings and conclusions in this report are ours and do not necessarily represent the official views of the Centers for Disease Control and Prevention.
The study was approved by the U.S. Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA) Institutional Review Board and by institutional review boards at all participating sites.
REFERENCES
- 1.Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM, Kreiswirth B, Cole ST, Jacobs WR, Telenti A. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother 38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. 2012. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 67:819–831. doi: 10.1093/jac/dkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willby M, Sikes RD, Malik S, Metchock B, Posey JE. 2015. Correlation between GyrA substitutions and ofloxacin, levofloxacin, and moxifloxacin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:5427–5434. doi: 10.1128/AAC.00662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eilertson B, Maruri F, Blackman A, Herrera M, Samuels DC, Sterling TR. 2014. High proportion of heteroresistance in gyrA and gyrB in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 58:3270–3275. doi: 10.1128/AAC.02066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Operario DJ, Koeppel AF, Turner SD, Bao Y, Pholwat S, Banu S, Foongladda S, Mpagama S, Gratz J, Ogarkov O, Zhadova S, Heysell SK, Houpt ER. 2017. Prevalence and extent of heteroresistance by next generation sequencing of multidrug-resistant tuberculosis. PLoS One 12:e0176522. doi: 10.1371/journal.pone.0176522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelthaler DM, Streicher EM, Kelley EJ, Allender CJ, Wiggins K, Jimenez D, Lemmer D, Vittinghoff E, Theron G, Sirgel FA, Warren RM, Metcalfe JZ. 2019. Minority Mycobacterium tuberculosis genotypic populations as an indicator of subsequent phenotypic resistance. Am J Respir Cell Mol Biol 61:789–791. doi: 10.1165/rcmb.2019-0178LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. 2010. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn 12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colman RE, Schupp JM, Hicks ND, Smith DE, Buchhagen JL, Valafar F, Crudu V, Romancenco E, Noroc E, Jackson L, Catanzaro DG, Rodwell TC, Catanzaro A, Keim P, Engelthaler DM. 2015. Detection of low-level mixed-population drug resistance in Mycobacterium tuberculosis using high fidelity amplicon sequencing. PLoS One 10:e0126626. doi: 10.1371/journal.pone.0126626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton T, Cegielski P, Akksilp S, Asencios L, Campos Caoili J, Cho SN, Erokhin VV, Ershova J, Gler MT, Kazennyy BY, Kim HJ, Kliiman K, Kurbatova E, Kvasnovsky C, Leimane V, van der Walt M, Via LE, Volchenkov GV, Yagui MA, Kang H, Global PETTS Investigators, Akksilp R, Sitti W, Wattanaamornkiet W, Andreevskaya SN, Chernousova LN, Demikhova OV, Larionova EE, Smirnova TG, Vasilieva IA, Vorobyeva AV, Barry CE, III, Cai Y, Shamputa IC, Bayona J, Contreras C, Bonilla C, Jave O, Brand J, Lancaster J, Odendaal R, Chen MP, Diem L, Metchock B, Tan K, Taylor A, Wolfgang M, Cho E, et al. 2012. Prevalence of and risk factors for resistance to second-line drugs in people with multidrug-resistant tuberculosis in eight countries: a prospective cohort study. Lancet 380:1406–1417. doi: 10.1016/S0140-6736(12)60734-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kateete DP, Kamulegeya R, Kigozi E, Katabazi FA, Lukoye D, Sebit SI, Abdi H, Arube P, Kasule GW, Musisi K, Dlamini MG, Khumalo D, Joloba ML. 2019. Frequency and patterns of second-line resistance conferring mutations among MDR-TB isolates resistant to a second-line drug from Eswatini, Somalia and Uganda (2014–2016). BMC Pulm Med 19:124. doi: 10.1186/s12890-019-0891-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nosova EY, Bukatina AA, Isaeva YD, Makarova MV, Galkina KY, Moroz AM. 2013. Analysis of mutations in the gyrA and gyrB genes and their association with the resistance of Mycobacterium tuberculosis to levofloxacin, moxifloxacin and gatifloxacin. J Med Microbiol 62:108–113. doi: 10.1099/jmm.0.046821-0. [DOI] [PubMed] [Google Scholar]
- 12.Huo F, Zhang F, Xue Y, Shang Y, Liang Q, Ma Y, Li Y, Zhao L, Pang Y. 2020. Increased prevalence of levofloxacin-resistant Mycobacterium tuberculosis in China is associated with specific mutations within the gyrA gene. Int J Infect Dis 92:241–246. doi: 10.1016/j.ijid.2020.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Matrat S, Veziris N, Mayer C, Jarlier V, Truffot-Pernot C, Camuset J, Bouvet E, Cambau E, Aubry A. 2006. Functional analysis of DNA gyrase mutant enzymes carrying mutations at position 88 in the A subunit found in clinical strains of Mycobacterium tuberculosis resistant to fluoroquinolones. Antimicrob Agents Chemother 50:4170–4173. doi: 10.1128/AAC.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niward K, Angeby K, Chryssanthou E, Paues J, Bruchfeld J, Jureen P, Giske CG, Kahlmeter G, Schon T. 2016. Susceptibility testing breakpoints for Mycobacterium tuberculosis categorize isolates with resistance mutations in gyrA as susceptible to fluoroquinolones: implications for MDR-TB treatment and the definition of XDR-TB. J Antimicrob Chemother 71:333–338. doi: 10.1093/jac/dkv353. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Lu J, Wang Y, Pang Y, Zhao Y. 2014. Prevalence and molecular characterization of fluoroquinolone-resistant Mycobacterium tuberculosis isolates in China. Antimicrob Agents Chemother 58:364–369. doi: 10.1128/AAC.01228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kam KM, Yip CW, Cheung TL, Tang HS, Leung OC, Chan MY. 2006. Stepwise decrease in moxifloxacin susceptibility amongst clinical isolates of multidrug-resistant Mycobacterium tuberculosis: correlation with ofloxacin susceptibility. Microb Drug Resist 12:7–11. doi: 10.1089/mdr.2006.12.7. [DOI] [PubMed] [Google Scholar]
- 17.Von Groll A, Martin A, Jureen P, Hoffner S, Vandamme P, Portaels F, Palomino JC, da Silva PA. 2009. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob Agents Chemother 53:4498–4500. doi: 10.1128/AAC.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi R, Zhang J, Li C, Kazumi Y, Sugawara I. 2006. Emergence of ofloxacin resistance in Mycobacterium tuberculosis clinical isolates from China as determined by gyrA mutation analysis using denaturing high-pressure liquid chromatography and DNA sequencing. J Clin Microbiol 44:4566–4568. doi: 10.1128/JCM.01916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan RC, Hui M, Chan EW, Au TK, Chin ML, Yip CK, AuYeang CK, Yeung CY, Kam KM, Yip PC, Cheng AF. 2007. Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J Antimicrob Chemother 59:866–873. doi: 10.1093/jac/dkm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cegielski JP, Dalton T, Yagui M, Wattanaamornkiet W, Volchenkov GV, Via LE, Van Der Walt M, Tupasi T, Smith SE, Odendaal R, Leimane V, Kvasnovsky C, Kuznetsova T, Kurbatova E, Kummik T, Kuksa L, Kliiman K, Kiryanova EV, Kim H, Kim C-k, Kazennyy BY, Jou R, Huang W-L, Ershova J, Erokhin VV, Diem L, Contreras C, Cho SN, Chernousova LN, Chen MP, Caoili JC, Bayona J, Akksilp S, Calahuanca GY, Wolfgang M, Viiklepp P, Vasilieva IA, Taylor A, Tan K, Suarez C, Sture I, Somova T, Smirnova TG, Sigman E, Skenders G, Sitti W, Shamputa IC, Riekstina V, Pua KR, et al. 2014. Extensive drug resistance acquired during treatment of multidrug-resistant tuberculosis. Clin Infect Dis 59:1049–1063. doi: 10.1093/cid/ciu572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad N, Ahuja SD, Akkerman OW, Alffenaar JC, Anderson LF, Baghaei P, Bang D, Barry PM, Bastos ML, Behera D, Benedetti A, Bisson GP, Boeree MJ, Bonnet M, Brode SK, Brust JCM, Cai Y, Caumes E, Cegielski JP, Centis R, Chan PC, Chan ED, Chang KC, Charles M, Cirule A, Dalcolmo MP, D'Ambrosio L, de Vries G, Dheda K, Esmail A, Flood J, Fox GJ, Frechet-Jachym M, Fregona G, Gayoso R, Gegia M, Gler MT, Gu S, Guglielmetti L, Holtz TH, Hughes J, Isaakidis P, Jarlsberg L, Kempker RR, Keshavjee S, Khan FA, Kipiani M, Koenig SP, Koh WJ, Kritski A, et al. 2018. Treatment correlates of successful outcomes in pulmonary multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet 392:821–834. doi: 10.1016/S0140-6736(18)31644-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization. 2019. WHO consolidated guidelines on drug-resistant tuberculosis treatment. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 23.Colman RE, Anderson J, Lemmer D, Lehmkuhl E, Georghiou SB, Heaton H, Wiggins K, Gillece JD, Schupp JM, Catanzaro DG, Crudu V, Cohen T, Rodwell TC, Engelthaler DM. 2016. Rapid drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates directly from clinical samples by use of amplicon sequencing: a proof-of-concept study. J Clin Microbiol 54:2058–2067. doi: 10.1128/JCM.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowers JR, Lemmer D, Sahl JW, Pearson T, Driebe EM, Wojack B, Saubolle MA, Engelthaler DM, Keim P. 2016. KlebSeq, a diagnostic tool for surveillance, detection, and monitoring of Klebsiella pneumoniae. J Clin Microbiol 54:2582–2596. doi: 10.1128/JCM.00927-16. [DOI] [PMC free article] [PubMed] [Google Scholar]