Candida auris is a newly emerging fungal pathogen of humans and has attracted considerable attention from both the clinical and basic research communities. Clinical isolates of C. auris are often resistant to one or more antifungal agents. To explore how antifungal resistance develops, we performed experimental evolution assays using a fluconazole-susceptible isolate of C. auris (BJCA001). After a series of passages through medium containing increasing concentrations of fluconazole, fungal cells acquired resistance.
KEYWORDS: Candida auris, aneuploidy, antifungal resistance, experimental evolution, fluconazole
ABSTRACT
Candida auris is a newly emerging fungal pathogen of humans and has attracted considerable attention from both the clinical and basic research communities. Clinical isolates of C. auris are often resistant to one or more antifungal agents. To explore how antifungal resistance develops, we performed experimental evolution assays using a fluconazole-susceptible isolate of C. auris (BJCA001). After a series of passages through medium containing increasing concentrations of fluconazole, fungal cells acquired resistance. By sequencing and comparing the genomes of the parental fluconazole-susceptible strain and 26 experimentally evolved strains of C. auris, we found that a portion of fluconazole-resistant strains carried one extra copy of chromosome V. In the absence of fluconazole, C. auris cells rapidly became susceptible and lost the extra copy of chromosome V. Genomic and transcriptome sequencing (RNA-Seq) analyses indicate that this chromosome carries a number of drug resistance-related genes, which were transcriptionally upregulated in the resistant, aneuploid strains. Moreover, missense mutations were identified in the genes TAC1B, RRP6, and SFT2 in all experimentally evolved strains. Our findings suggest that the gain of an extra copy of chromosome V is associated with the rapid acquisition of fluconazole resistance and may represent an important evolutionary mechanism of antifungal resistance in C. auris.
INTRODUCTION
The emerging fungal pathogen Candida auris was first described in Japan in 2009 and is becoming a serious global threat to human health (1–4). C. auris emerged simultaneously on different continents and spread around the world rapidly. Infections with C. auris have been reported in at least 30 countries across six continents (5, 6). There were 1,092 confirmed cases in the United States alone, as of March 2020 (CDC data [https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html]). Recently, several large-scale nosocomial outbreaks were reported (7–9).
C. auris is often multidrug resistant, and some clinical isolates are even resistant to all three major classes of antifungals: azoles, polyenes, and echinocandins (2). Most clinical isolates are resistant to fluconazole, one of the first-line drugs; more than 10% of clinical isolates are resistant to two or three classes of antifungals (2, 3, 10). Comparative genomic analysis indicates that there are a number of conserved orthologs associated with antifungal resistance in C. auris and the most common fungal pathogen, Candida albicans (11). For example, genes of the ergosterol biosynthesis pathway and drug transporters are conserved in the genomes of fungi.
The lanosterol 14-alpha-demethylase Erg11 is a key enzyme of the ergosterol biosynthesis pathway and the primary target of azole drugs (12). Three major mutations, VF125AL (commonly referred to as F126L), Y132F, and K143R, have been found in Erg11 and are associated with antifungal resistance in clinical isolates of C. auris (3, 11, 13, 14). However, it has been reported that the expression of ERG11 in clinical fluconazole-resistant strains was not significantly increased (15), although its expression level was upregulated 8-fold in older cells (16). Heterologous expression of C. auris ERG11 alleles with Y132F or K143R substitutions increased azole MICs in Saccharomyces cerevisiae (14). Although most clinical isolates of C. auris are resistant to fluconazole, many strains that show a relatively high MIC for fluconazole do not carry any mutations of the three hot spots on Erg11 (3, 15), and it has not been shown that the three major ERG11 mutations fully explain the high resistance levels of isolates carrying these changes. Therefore, both Erg11-dependent and Erg11-independent mechanisms could be involved in the antifungal resistance of C. auris.
Aneuploidy is a common phenomenon in fungi, in which the number of one or more chromosomes is abnormal (17, 18). In C. albicans and Cryptococcus neoformans, aneuploidy is often associated with drug resistance (17, 19). At least 50% of fluconazole-resistant isolates of C. albicans are aneuploid (19). In vitro experimental assays demonstrated that antifungal and environmental stresses drive the formation of aneuploidy in fungi (20). In the present study, we set out to explore the evolutionary mechanism of antifungal resistance in C. auris. Taking advantage of a fluconazole-susceptible strain (BJCA001) isolated in Beijing, China (21), we performed experimental evolution assays. Through a series of passages in medium containing increasing concentrations of fluconazole, we obtained a set of fluconazole-resistant strains of C. auris. Whole-genome sequencing analysis indicates that a portion of experimentally evolved fluconazole-resistant strains carry one extra chromosome V (or scaffold 6 of the B8441 reference genome). In addition, transcriptome sequencing (RNA-Seq) analysis indicates that potential fluconazole resistance or ergosterol biosynthesis-related genes, such as ERG9 and OPT1/2, on this chromosome were upregulated, which suggests that the gain of this extra chromosome is associated with the development of fluconazole resistance in these C. auris strains.
RESULTS
Induction of resistance in a fluconazole-susceptible isolate of C. auris.
In a recent study, we reported the first isolate of C. auris in China (BJCA001, belonging to south Asia clade I). The MIC of fluconazole for BJCA001 is 2.0 mg/liter when this strain is incubated for 24 h (21). Interestingly, cells of C. auris were able to grow in the presence of 32 mg/liter of fluconazole when the incubation time was extended to 48 to 72 h. However, cells of the C. albicans control (SC5314) were unable to grow under the same culture conditions (Fig. S1). We predicted that the growth of C. auris in the presence of fluconazole could be due to (i) the effect of trailing growth, a commonly observed phenomenon in Candida spp. (22), or (ii) the gain of fluconazole resistance in fungal cells.
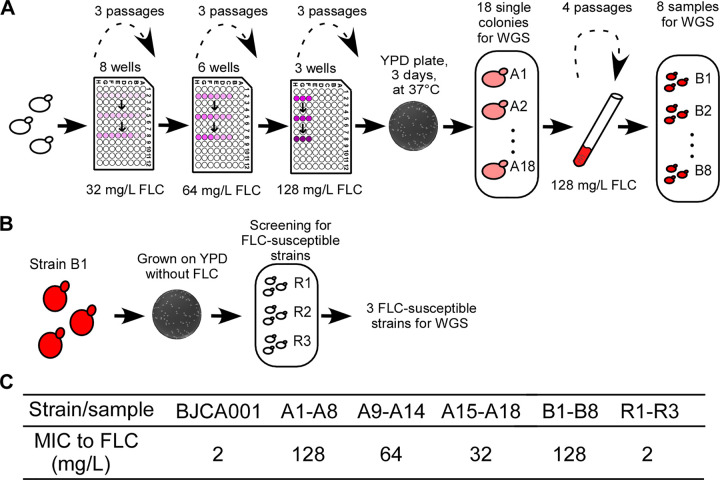
To investigate whether the treatment of fluconazole could induce the development of resistance in C. auris, we designed a strategy of experimental evolution (Fig. 1A). Cells of C. auris were passaged in RPMI 1640 medium containing a series of increased concentrations of fluconazole (32 mg/liter, 64 mg/liter, or 128 mg/liter). After three passages in each culture, cells were plated on yeast extract-peptone-dextrose (YPD) medium and cultured at 37°C for 3 days. Eighteen single colonies (A1 to A18) from the YPD plates were collected and tested for MIC of fluconazole (Fig. 1). Of note, as we describe in Materials and Methods, some of these samples (A1 to A18) could be clonal, since they were derived from three different lineages. The MICs for samples A1 to A8, A9 to A14, and A15 to A18 were 128 mg/liter, 64 mg/liter, and 32 mg/liter, respectively. To further stabilize the antifungal resistance, samples A1 to A8 were subjected to four additional passages in RPMI 1640 medium containing 128 mg/liter of fluconazole (Fig. 1B and C), generating strains B1 to B8. The MICs for B1 to B8 were 128 mg/liter.
FIG 1.
Schematics of the experimental evolution strategies of fluconazole resistance in C. auris. (A) Eight independent samples (containing 4 × 105 cells) of C. auris BJCA001 were initially cultured in 0.2 ml RPMI 1640 medium (OD600 ≈ 0.1) containing 32 mg/liter fluconazole (FLC) and grown at 35°C for 48 h. After two more passages under the same conditions, cells were inoculated and grown in YPD medium containing 64 mg/liter and then 128 mg/liter fluconazole for three passages each. Two and three samples failed to grow after treated with 32 mg/liter and 64 mg/liter fluconazole, respectively. Three isolates that grew in the medium containing 128 mg/liter fluconazole were then plated on YPD medium for 3 days of incubation at 37°C. Eighteen independent colonies (A1 to A18) with MICs of ≥32 mg/liter were picked and subjected to whole-genome sequencing (WGS). To strengthen the fluconazole-resistant feature, samples A1 to A8 were inoculated into RPMI 1640 medium containing 128 mg/liter fluconazole for four additional passages and then subjected to WGS (samples B1 to B8). (B) Induction of the loss of fluconazole resistance strain B1. Cells of the fluconazole-resistant strain B1 were inoculated and grown on YPD plates without fluconazole at 25°C for 1 month. Colonies were then subjected to MIC testing. (C) MICs of fluconazole for BJCA001 and strains A1 to A18, B1 to B8, and R1 to R3.
Aneuploidy for chromosome V (scaffold 6) is associated with fluconazole resistance of experimentally evolved strains.
To explore the rapid development of fluconazole resistance in C. auris, we resequenced the genomes of the parental fluconazole-sensitive strain BJCA001 and 26 experimentally evolved strains (A1 to A18 and B1 to B8) (Data Set S1). We found that the evolved isolates do not carry known drug resistance mutations, such as F126L, Y132F, or K143R in ERG11, which could not explain the resistant phenotype in these isolates. Single nucleotide polymorphism (SNP) analysis identified 40 missense and 3 stop mutations in the 26 experimentally evolved strains (Data Set S1). Two stop mutations were found in samples A2 and A17 (GenBank accession no. PIS51396.1 and PIS52414.1) and one in sample A18 (accession no. PIS55644.1). PIS51396.1 encodes a homolog of fungal Ime2, which is a serine/threonine protein kinase involved in the regulation of meiosis. In Candida albicans, mutation of Ime2 leads to hypersensitivity to amphotericin B (23). PIS52414.1 encodes a homolog of C. albicans cyclic AMP (cAMP)-dependent protein kinase catalytic subunit (Tpk1). Tpk1 plays a global role in the regulation of morphological transitions, stress response, and antifungal resistance in C. albicans (24). PIS55644.1 encodes a homolog of the Tcc1 transcription factor that regulates filamentation and virulence in C. albicans (25). Tcc1 is repressed by the antifungal flucytosine in C. albicans (26), suggesting that it could be involved in antifungal resistance.
Three missense site mutations (PIS52262.1_L564R, PIS58786.1_I162L, PIS49793.1_S195R; the designations correspond to the GenPept accession number plus the mutation) were found in all 26 experimentally evolved strains. PIS49793.1 is a homolog of C. albicans TAC1 encoding a transcriptional activator of drug transporter-encoding genes CDR1 and CDR2 (27). TAC1B is located in chromosome V in C. auris. A recent study demonstrated that missense mutations in TAC1B (PIS49793.1 mutations R495G and F214S) are associated with fluconazole resistance, while the A640V mutation could directly confer resistance in C. auris (28). Another study indicates that deletion of TAC1B results in the increased susceptibility to fluconazole and voriconazole in C. auris (29). PIS52262.1 is a homolog of C. albicans RRP6, encoding a nuclear exosome exonuclease component, whereas PIS58786.1 is a homolog of C. albicans SFT2, encoding a putative membrane protein. However, no evidence implies that RRP6 and SFT2 are related to drug resistance based on the reported functions of their orthologs in C. albicans and other fungal species.
Moreover, some other missense site mutations of genes involved in drug resistance, cell wall maintenance, and oxidative metabolisms were found in a subset of evolved strains (Data Set S1). For example, a mutation of PIS58450.1 was found in two samples (A11 and A15). PIS58450.1 encodes a homolog of C. albicans Age3, which is an ADP-ribosylation factor GTPase-activating protein. Mutation of AGE3 in C. albicans affects chemical and drug resistance (30). Some strains carried mutations of homologs of C. albicans genes for Ald5, Amo2, and Taf4. It has been demonstrated that the expression of Ald5 is regulated by fluconazole (31), whereas mutation of Amo2 and Taf4 in C. albicans results in hypersensitivity to toxic ergosterol analog (23). These findings indicate that these SNPs of the experimentally evolved strains of C. auris could also affect fluconazole resistance.
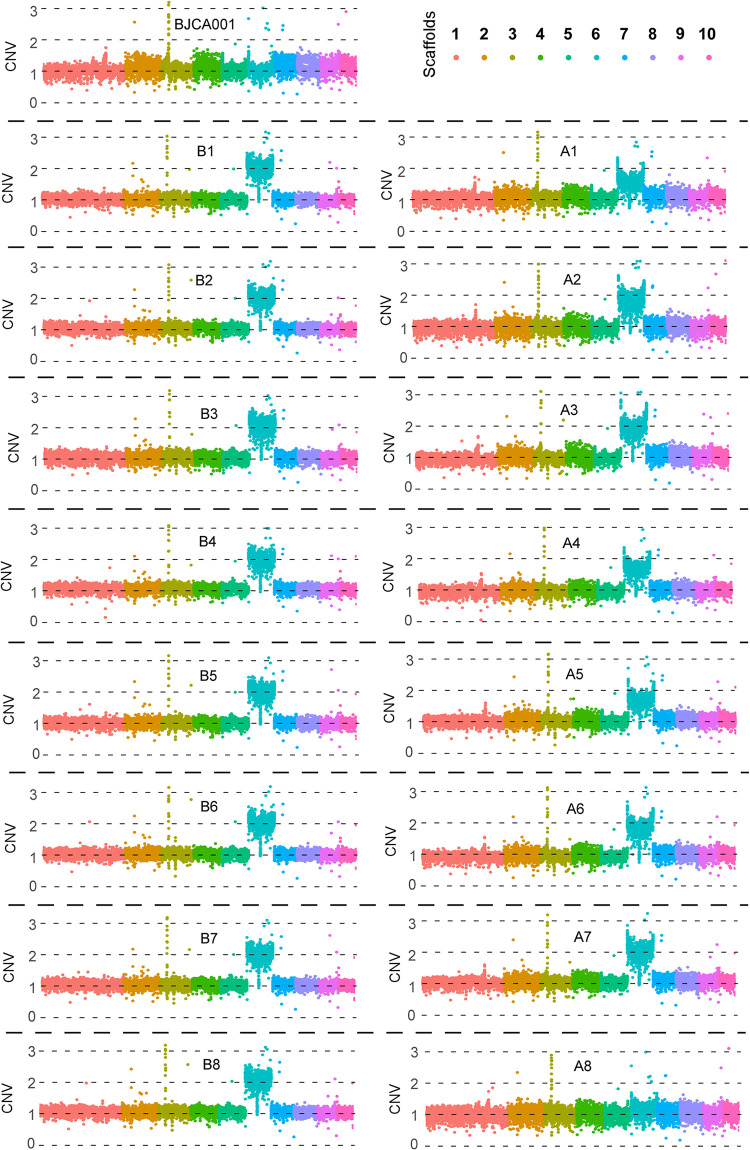
C. auris typically exists as a haploid organism (11, 32). Muñoz et al. assembled the genomic DNA of two C. auris strains (B8441 and B11221) into seven chromosomes (11). Using the reported genome assembly of strain B8441 as a reference, we analyzed the copy number variation (CNV) of each scaffold based on our resequencing data for strains BJCA001, A1 to A18, and B1 to B8. As shown in Fig. S2, C. auris strain BJCA001 is a typical haploid isolate and contains only one copy of each scaffold. The copy numbers of scaffold 6 (chromosome V) in A1 to A18 and B1 to B8 varied from 1.00 to 2.03. For B1 to B8 and A7, the copy number of chromosome V was 2 or close to 2, implying that these strains carried an extra copy of this chromosome. The copy numbers of chromosome V of several strains were nonintegers, including those of strains A1 to A7, which exhibited a high MIC of fluconazole (128 mg/liter), and A9, A13, A14, and A18, which exhibited moderate MICs (64 mg/liter and 32 mg/liter). We hypothesized that there were two potential factors leading to these noninteger copy numbers. First, only a portion of cells in the population of these samples (A1 to A7, A9, A13, A14, and A18) carried an extra copy of chromosome V. Samples A1 to A18 were grown on YPD medium without fluconazole before collection for genomic sequencing. Although single colonies were used, a portion of cells in the colonies could have lost the extra chromosome during the growth in the absence of fluconazole stress. Second, these strains (A1 to A7, A9, A13, A14, and A18) might carry an isochromosome rather than a full-length chromosome. To uncover potential reasons for the noninteger copy numbers of chromosome V, we examined the average read depth for nonoverlapping 500-bp windows throughout chromosome V for strains BJCA001, A1 to A18, and B1 to B8. Of note, B1 to B8 were derived from cultures A1 to A8 and subjected to an enrichment for drug-resistant cells through four additional passages in the presence of 128 mg/liter fluconazole (Fig. 1). As shown in Fig. 2, Fig. S3, and Data Set S2, enriched samples B1 to B8 all carried a whole extra copy of chromosome V, suggesting that additional passages favored the growth of aneuploid cells. For samples A1 to A7, A9, A13, A14, and A18, the CNV of each window showed increased copy numbers across chromosome V, with an average centered between haploid and diploid levels, suggesting that they were heterozygous populations and only a portion of cells carried an extra copy of chromosome V.
FIG 2.
Copy-number variations (CNV) of C. auris after a series of passages in the fluconazole-containing medium. Fluconazole-susceptible strain BJCA001 served as a reference. Experimentally evolved strains A1 to A8 and B1 to B8 (Fig. 1) were tested. A comparative genomic analysis shows a copy number variation at scaffold 6 (chromosome V) using the B8441 genome assembly as a reference. The data for strains A9 to A18 are presented in Fig. S1. The x axis indicates scaffolds 1 to 10. The y axis shows the copy number of each scaffold. Each spot represents the sequence depth for a genomic segment of 500 bp across the chromosomes.
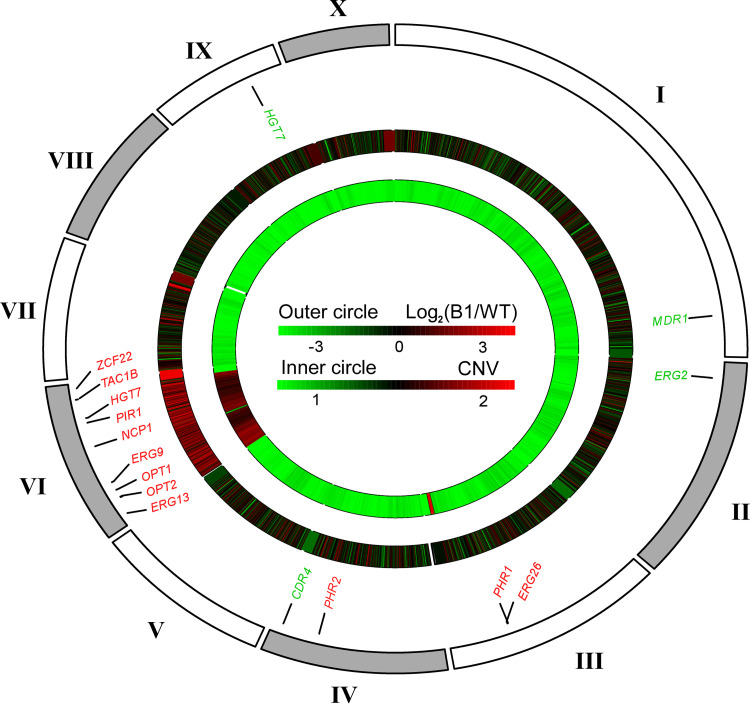
Interestingly, genomic analysis indicated that at least 23 genes associated with drug resistance or ergosterol biosynthesis, including TAC1B, NCP1, ERG9, and ERG13, are located on chromosome V of C. auris (Fig. 3 and Data Set S3), suggesting that this chromosome is associated with antifungal resistance.
FIG 3.
Genome-wide profile of differential gene expression between fluconazole-resistant strain B1 and fluconazole-susceptible strain BJCA001 and scaffold copy number variation for B1. Different scaffolds are shown outside the circle. The differential gene expression and copy number changes are shown in the middle and inner rings. Several genes that may be involved in drug resistance are shown between the outside and middle circles; genes in red and green are upregulated and downregulated, respectively, in B1 compared with BJCA001.
C. auris cells become fluconazole susceptible and return to the euploid state.
To confirm that the extra copy of chromosome V of resistant strains of C. auris was associated with fluconazole resistance, we next grew the fluconazole-resistant strain B1 on YPD medium plates containing no antifungal drug. We obtained a set of fluconazole-susceptible strains, indicating that the resistant strain could return to the susceptible state in the absence of antifungal drug stress. Whole-genome sequencing analysis indicated that the recovered fluconazole-susceptible strains R1 to R3 (MIC = 2 mg/liter) lost the extra copy of chromosome V (Fig. S2 and S3 and Data Set S1). Moreover, strains R1 to R3 still possessed the SNPs in the genes TAC1B, RRP6, and SFT2, suggesting that these mutations do not play a major role in resistance of our evolved strains. Three new SNPs (PIS51247.1_K357E, PIS48789.1_S115L, and PIS51736.1_N251I) were observed in strains R1 to R3. PIS51247.1 is an ortholog of C. albicans orf19.2008 (encoding an S-methyl-5-thioribose-1-phosphate isomerase). No orthologs of PIS48789.1 and PIS51736.1 were found in C. albicans and S. cerevisiae. Taken together, our results suggest that aneuploidy of this chromosome is associated with the acquisition of fluconazole resistance in C. auris.
Fluconazole-resistant strains exhibit a low growth rate.
Both aneuploidy and drug resistance affect cellular physiology in the absence of antifungal stress (17, 33, 34). For example, aneuploid strains proliferate more slowly than do wild-type cells of S. cerevisiae (35). We next compared the growth rates of the original fluconazole-sensitive strain BJCA001, experimentally evolved strains B1 to B8, and recovery strains R1 to R3 (fluconazole sensitive). As shown in Fig. 4, all experimentally evolved fluconazole-resistant strains exhibit a lower growth rate than BJCA001 and recovery susceptible strains, implying that there is a trade-off between growth rate and resistance to fluconazole in C. auris conferred by aneuploidy.
FIG 4.
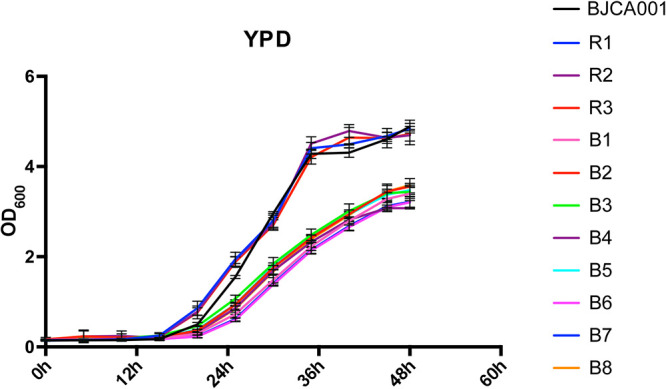
Growth curves of strain BJCA001, experimentally evolved fluconazole-resistant strains B1 to B8, and recovery strains R1 to R3 in YPD medium. Cells were cultured with an initial inoculum with an OD600 of 0.2 at 30°C with shaking for 48 h. Three biological repeats were performed.
Fluconazole-resistant and fluconazole-susceptible strains exhibit globally differential transcriptional profiles.
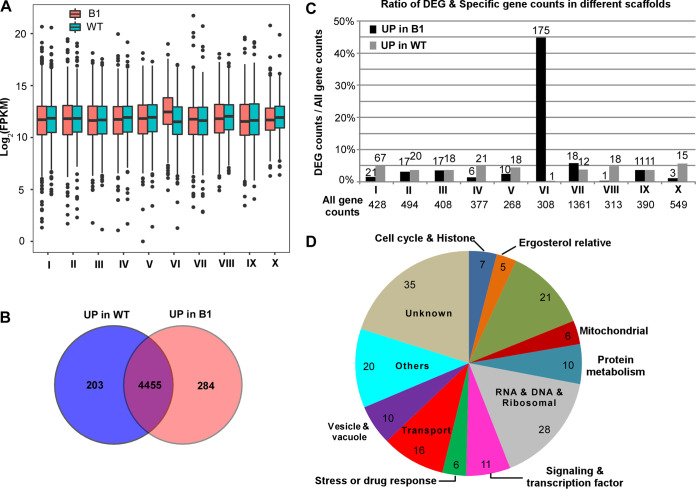
As mentioned above, at least some drug resistance-related genes are located on chromosome V of C. auris. To investigate whether the dosage effect of this chromosome affected the transcriptional profile of genes on it, we performed RNA-Seq analysis with the parental fluconazole-susceptible strain BJCA001 and experimentally evolved fluconazole-resistant strain B1. As mentioned above, strain B1 carried an extra copy of chromosome V (Fig. 2). Compared to BJCA001, we found that 285 genes were upregulated and 203 genes downregulated in cells of B1 (2-fold change cutoff; adjusted P value, <0.05; three replicates tested) (Fig. 5 and Data Set S3). Of the 285 genes upregulated in B1, 175 genes are located on chromosome V (representing 44.9% of 390 genes on this chromosome) (Fig. 5 and Data Set S3).
FIG 5.
Differentially expressed global gene profiles between the fluconazole-resistant strain B1 and original strain BJCA001. (A) Box plot of distribution of gene expression values for each scaffold. (B) Differentially expressed genes (DEG) in strains B1 and BJCA001. (C) Upregulated and downregulated genes in strains B1 and BJCA001 for each scaffold. (D) Functional classification of DEG at scaffold 6. Genes were classified based on C. albicans gene annotation and ontology (http://www.candidagenome.org).
Interestingly, a number of genes related to antifungal resistance or ergosterol biosynthesis on chromosome V of C. auris were upregulated in the evolved resistant strain (Table 1). There was only one gene (PIS49750.1) on chromosome V that was downregulated in B1 cells. Four genes on chromosome V (NCP1, ERG9, ERG13, and ZCF22) and two genes related to ergosterol or sterol biosynthesis on chromosomes II and III (ERG11 and ERG1, respectively) were transcriptionally upregulated in strain B1 based on RNA-Seq (Fig. 5 and Data Set S3) or qRT-PCR (Fig. S4). ERG13 encodes a hydroxymethylglutaryl coenzyme A (hydroxymethylglutaryl-CoA) synthase to yield 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) (36). ERG9 and ERG1 encode squalene synthase and squalene epoxidase, respectively; the first enzyme is dedicated to production of squalene, which is the precursor of all steroids (37). ERG11 encodes a microsomal and membrane-bound protein, which functions as a lanosterol 14,α-demethylase of the cytochrome P450 family and the primary target of azole drugs (12). NCP1 encodes a coenzyme of Erg11, and deletion of NCP1 in C. albicans leads to a 200-fold increase of susceptibility to ketoconazole (38, 39). ZCF22 is a predicted Zn(II)2Cys6 transcription factor-encoding gene (40). Zcf22 protein has domains similar to those of Upc2, which is a regulator of sterol biosynthesis (41). Sequence analysis indicates that both Upc2 and Zcf22 of C. auris belong to the Upc2 family. However, it remains to be investigated whether Zcf22 plays a role similar to that of Upc2 and is involved in the regulation of sterol biosynthesis.
TABLE 1.
Differentially expressed genes of interest on chromosome V of C. aurisa
| Functional description | C. auris gene accession no. | C. albicans homolog | Log2(WT/B1) | FDR | Reference(s) |
|---|---|---|---|---|---|
| Ergosterol biosynthesis | PIS49477.1 | ERG13 | −0.61 | 1.27E−06 | 26, 55 |
| PIS49563.1 | ERG9 | −1.18 | 5.61E−26 | 26, 55 | |
| PIS49684.1 | NCP1 | −2.06 | 3.72E−54 | 39 | |
| PIS49856.1 | ZCF22 | −1.12 | 3.64E−15 | 40, 41 | |
| CDR or MDR family | PIS49792.1 | TAC1A | −1.61 | 3.52E−33 | 27 |
| PIS49793.1 | TAC1B | −0.59 | 8.32E−05 | 27 | |
| PIS49714.1 | orf19.1449 | −0.66 | 5.79E−04 | 56 | |
| PIS49547.1 | orf19.2473 | −0.62 | 4.14E−10 | 55 | |
| Oligopeptide transporters | PIS49539.1 | OPT1 | −1.48 | 3.22E−04 | 11 |
| PIS49578.1 | OPT1 | 0.47 | 2.59E−05 | 11 | |
| PIS49456.1 | OPT2 | −0.32 | 3.30E−03 | 11 | |
| PIS49514.1 | OPT2 | −0.61 | 3.71E−03 | 11 | |
| PIS49515.1 | OPT2 | −0.16 | 5.62E−01 | 11 | |
| PIS49521.1 | OPT2 | −1.05 | 3.86E−12 | 11 | |
| PIS49522.1 | OPT2 | −1.62 | 1.66E−19 | 11 | |
| PIS49537.1 | OPT2 | −0.55 | 1.74E−03 | 11 | |
| Other major facilitator superfamily | PIS49770.1 | HGT7 | −1.97 | 3.84E−53 | 57 |
| PIS49560.1 | orf19.4090 | −0.82 | 1.11E−05 | 57 | |
| PIS49745.1 | orf19.4905 | −1.17 | 1.77E−33 | 57 | |
| PIS49665.1 | orf19.7666 | −0.62 | 1.18E−04 | 57 | |
| Fluconazole resistance related | PIS49757.1 | orf19.2893 | −1.25 | 4.24E−24 | 58 |
| PIS49801.1 | GRP2 | −1.99 | 1.94E−10 | 58 | |
| PIS49656.1 | orf19.6502 | −0.61 | 6.22E−07 | 58 |
This table is associated with Data Set S3. Log2(WT/B1) is the log2 ratio of the FPKM value of the WT to that of B1. FDR (false discovery rate) reflects adjusted P values. CDR, Candida drug resistance genes; MDR, multidrug resistance genes. The WT strain was BJCA001; B1 is a fluconazole-resistant strain derived from BJCA001.
Moreover, 16 transporter-encoding genes, including OPT2, OPT1, EMP46, SEC24, and MEP2 were transcriptionally upregulated in B1 (Data Set S3). These transporters could play a role in efflux pumps of drugs. For example, OPT2 encodes an oligopeptide transporter which is induced in the presence of fluconazole in C. albicans (42). In C. auris and other drug-resistant species of the CTG clade, there is an expansion of OPT transporters, including over 10 copies of OPT2 in the genome (11). There are 8 OPT genes on chromosome V of C. auris. Of them, 7 exhibited increased expression in strain B1. We noted that the transcriptional expression of CDR1 was not significantly changed (1.03-fold) and that of CDR4 was downregulated in B1 (2.08-fold), suggesting that the mutation site of PIS49793.1_S195R (the homolog of C. albicans TAC1) does not obviously affect its regulatory function on the two genes. Taken together, these results imply that the increased expression of genes on chromosome V due to copy number variation is associated with the fluconazole resistance of the experimentally evolved strains of C. auris.
In addition, TAC1A and MDR1 regulate antifungal resistance in other fungi and were differentially expressed in the wild-type (WT) and B1 strains. ERG3, UPC2, TAC1B, and MRR1 also exhibited 1.3- to 2.0-fold changes (Data Set S3). Both TAC1A and TAC1B are the homologs of C. albicans TAC1 and are located on chromosome V of C. auris. Recently, it was demonstrated that mutation of TAC1B could confer antifungal resistance in C. auris (28). ERG3 encodes a sterol desaturase that is involved in one of the final reactions in the ergosterol biosynthetic pathway in fungi. Some clinical isolates of C. albicans carrying Erg3 mutations exhibit increased azole resistance (43). As mentioned earlier, the Upc2 transcription factor functions as a regulator of ergosterol biosynthetic genes and sterol uptake (41), whereas the Mrr1 transcription factor plays a role in the regulation of the expression of the multidrug resistance-related gene MDR1 in C. albicans (44). Taken together, the results indicate that the differential expression of these genes in the WT and experimentally evolved B1 strains could contribute to the difference in fluconazole resistance.
DISCUSSION
The development of drug resistance is an important topic of both the public health and research communities. The newly emerging fungal pathogen C. auris has attracted much attention from the public and research community due to its multidrug resistance feature. It has been proposed that the widespread use of antifungal drugs may play a critical role in the emergence of this fungus (4). Most clinical isolates of C. auris are resistant to the first-line drug fluconazole (3, 4). How does C. auris develop antifungal resistance? To explore the mechanism, in this study, we took advantage of a fluconazole-sensitive strain of C. auris BJCA001 and performed in vitro experimental evolution assays. After a series of passages in the presence of fluconazole, cells of C. auris BJCA001 acquired resistance to the drug (Fig. 1). Genomic sequencing analysis indicates that the gain of an extra chromosome V is associated with the development of resistance to fluconazole in the experimentally evolved strains. As described in Results and in Materials and Methods, the design of our experiments had some limitations. First, the A samples (A1 to A18) were derived from three different lineages, and some of them could be clonal or duplications, although they carried various copy numbers of chromosome V and different SNPs. Second, we performed the in vitro experimental evolution assays using only a single strain of genetic clade I. It would be worthwhile to examine whether the gain of the extra chromosome is a general feature in the other genetic clades. Third, we did not test whether this aneuploidy is a feature of naturally fluconazole-resistant isolates from clinical settings.
The formation of aneuploidy is a general adaptive mechanism of fungi in the presence of antifungal stress (17, 19). For example, the gain of a partial or full extra chromosome V in C. albicans is related to fluconazole resistance (19). Chromosome V of C. albicans houses genes encoding the enzyme (Erg11) of the ergosterol pathway targeted by azole drugs and efflux proteins. In C. auris, increased copy number of the chromosomal region encompassing ERG11 had been reported as another mechanism of azole resistance (11). In the present study, we found that a portion of experimental evolved fluconazole-resistant strains contained an aneuploidy for chromosome V that also bears a range of antifungal resistance-associated genes (Fig. 3; Table 1).
Aneuploidy often results in the changes of cellular physiology and gene expression profile. It has been observed in S. cerevisiae that most genes on aneuploid chromosomes were expressed at higher levels consistent with a dosage effect (35). To explore the effect of aneuploidy on the transcriptional profile in C. auris, we performed RNA-Seq analysis (Fig. 5). As expected, 50% genes located on chromosome V were significantly upregulated in the fluconazole-resistant strain harboring an extra copy of chromosome V (Fig. 5, Data Set S3, and Table 1). Several genes (for example, OPT [oligopeptide transporter] family transporter genes, ERG9, and NCP1) could contribute to the development of resistance in C. auris. The increased expression of ERG13, ERG9, and NCP1 could result in an increase of the cellular level of ergosterol (Fig. S4). Moreover, some genes associated with ergosterol biosynthesis (e.g., ERG1, ERG7, ERG8, ERG11, and ERG26) were also upregulated in fluconazole-resistant strain B1 (Data Set S3).
Although aneuploidy could occur spontaneously in fungi, our study demonstrates that the spontaneous rate of aneuploidy formation in C. auris could be significantly increased in the presence of fluconazole stress. Under the antifungal stress, aneuploid cells of C. auris were still able to proliferate. However, under the same culture condition, cells of C. albicans strain SC5314 failed to grow in the presence of fluconazole (Fig. S1). A recent study suggested that aneuploidy is uncommon in clinical isolates of C. auris (32), and the authors suggest that ploidy changes do not appear to be a mechanism of environmental adaption in C. auris. However, our study implies that aneuploidy may be unstable and would be quickly lost in the absence of drug pressure, and it may therefore be missed when strains are cultured in the absence of drug but could still be important for response to antifungal agent in C. auris. The loss of the extra chromosome V was repeatable in different experimentally evolved strains after growth on medium without fluconazole. This genomic plasticity could be a common feature of pathogenic fungi (45). Together with the fact that most clinical isolates of C. auris are fluconazole resistant, these findings indicate that the genome of C. auris could be more plastic than that of C. albicans under stressful conditions.
Although the feature of antifungal resistance provides an advantage to C. auris cells in the presence of drug stress, we found that all fluconazole-resistant strains grew more slowly than the susceptible counterpart (Fig. 4). Similar to the case in C. albicans, there could be a fitness trade-off between cell growth and antifungal resistance in C. auris. Aneuploidy may result in unbalanced gene expression and protein stoichiometry (35). The perturbation of cellular homeostasis could lead to a delay of the G1 phase. Consistent with our results, a recent study reported that overexpression of nine genes of the ergosterol biosynthesis pathway (HMG1, ERG9, ERG1, NCP1, ERG25, ERG27, ERG28, ERG6, and ERG2) led to a lowered growth rate in Saccharomyces cerevisiae (46).
Taken together, our results reveal a new mechanism of the development of fluconazole resistance in C. auris using an integrated strategy that combined the experimental evolution and systems biology approaches. Formation of aneuploidy could be one of the antifungal resistance mechanisms in this fungus. Our findings provide an example of drug resistance development in fungi and shed new light on the global emergence of C. auris.
MATERIALS AND METHODS
Strains and culture conditions.
YPD medium (10 g/liter yeast extract, 20 g/liter peptone, and 20 g/liter glucose) was used for routine growth of C. auris and C. albicans. C. auris strain BJCA001 and C. albicans strain SC5314 were used in this study. To determine the growth curves of C. auris and C. albicans strains, we cultured cells in YPD medium for 48 h using 96-well plates. The growth intensity (measured as optical density at 600 nm [OD600]) was read with a Synergy 4 Gen5 plate reader (Biotek, Potton, United Kingdom).
MIC assay.
MIC assays for all strains (BJCA001, A1 to A18, B1 to B8, and R1 to R3) used in this study were performed according to the CLSI standard M27 (47) and a previous report (48). Three repeats were performed for each strain. Briefly, C. auris cells of each strain were initially patched on synthetic complete defined (SD) medium plates at 35°C for 24 h. Approximately 500 fungal cells in 0.1 ml RPMI 1640 medium (1.04% [wt/vol] RPMI 1640, 3.45% [wt/vol] MOPS [morpholinepropanesulfonic acid]S, NaOH used for pH adjustment to 7.0) were mixed with 0.1 ml RPMI 1640 medium with different concentrations of antifungal drugs in a 96-well plate for MIC assays. A series of fluconazole concentrations (from 0.5 to 128 μg/ml) was used. Cells were incubated at 35°C in air for 24 h. Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 served as quality controls.
Induction of fluconazole resistance in C. auris.
The fluconazole-susceptible strain BJCA001 (MIC of fluconazole = 2) was used as the starting strain. All cultures were performed in 96-well plates, and the growth intensity (OD600) was read with a Synergy 4 Gen5 plate reader (Biotek, Potton, United Kingdom). To induce fluconazole resistance, eight independent samples of C. auris strain BJCA001 were treated with fluconazole as demonstrated in Fig. 1. Eight independent cell samples (colonies) of C. auris BJCA001 were initially treated with 32 mg/liter fluconazole for three passages. Then, 4 × 105 cells of each sample in 0.2 ml RPMI 1640 medium were incubated in each well of 96-well plates at 35°C for 48 h. Two samples failed to grow in the presence of 32 mg/liter fluconazole. The other six samples (4 × 105 cells for each treatment) were subsequently treated with 64 mg/liter fluconazole for three passages. Three samples failed to grow in the presence of 64 mg/liter fluconazole. The other three samples were further treated with 128 mg/liter fluconazole for three passages and then plated on YPD medium (without fluconazole). Eighteen representative single colonies (A1 to A18) from the three independent samples were tested for the MIC of fluconazole and subjected to genomic sequencing. Of note, these samples (A1 to A18) represent three different lineages, and many of them were clonal. To obtain samples B1 to B8, strains A1 to A8 were subjected to four additional passages in RPMI 1640 medium containing 128 mg/liter fluconazole. To obtain fluconazole-susceptible recovery strains, the fluconazole-resistant strain B1 was grown on YPD plates (without fluconazole) at 25°C for 1 month. To screen fluconazole-susceptible strains, the cells of different colonies were reinoculated on YPD plates with or without 128 mg/liter fluconazole. Colonies that grew on YPD plates without fluconazole but not on YPD plates with 128 mg/liter fluconazole were subjected to MIC tests.
Whole-genome resequencing analysis.
Single colonies of C. auris A1 to A18, R1 to R3, and BJCA001 were grown in liquid YPD medium at 37°C for 24 h. Cells were subjected to MIC testing to ensure that they maintained the original antifungal-resistant features and then were collected for genomic DNA extraction. To generate strains B1 to B8, cells of single colonies of A1 to A8 from YPD plates were inoculated into liquid YPD medium containing 128 mg/liter fluconazole and passaged four times. Genomic DNA of B1 to B8 was then extracted for sequencing. Genomic DNA was extracted using the TIANamp yeast DNA kit (TianGen Biotech, Beijing, China) according to the company’s recommended protocols. The construction of libraries and genomic sequencing was performed by Berry Genomics Co., Beijing, China, according to the company’s protocols. Sequencing libraries were generated using a NEBNext Ultra DNA library prep kit for Illumina (NEB, USA). The DNA samples were fragmented by sonication to a size of ∼300 bp. DNA fragments were then end polished, A tailed, and ligated with the full-length adaptor for Illumina sequencing with further PCR amplification. Purified PCR products were subjected to size distribution analysis with an Agilent 2100 Bioanalyzer. Each sample was then sequenced using the Illumina NovaSeq platform with 2 × 150-bp reads and 450× minimum coverage.
Reference-based alignment and variant calling assays.
To analyze genomic sequence variations of C. auris strains, we compared the sequences of 30 samples (BJCA001, A1 to A18, B1 to B8, and R1 to R3). The FASTX toolkit v0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/index.html) was used to remove low-quality (Phred score ≤ 10), ambiguous, and adaptor bases from raw reads. The clean reads were then mapped to the previously published genomic assembly of C. auris strain B8441 (NCBI accession number GCA_002759435.2 (11)) using BWA mem 0.7.17 software with default settings (49). SAMtools v1.361 (22) was employed to convert the alignment results into the BAM format, and Picard Tools v1.56 (https://github.com/broadinstitute/picard) was used to sort and mark duplicated reads with the commands “SortSam” and “MarkDuplicates,” respectively. The variable sequence sites were detected with the SAMtools and Genome Analysis Toolkit (GATK v2.7.2) (50). For GATK, the program HaplotypeCaller was used for SNP calling. The ploidy of the genome of BJCA001 was set as 1. The strand_call_conf (thresholds for low- and high-quality variation loci) and strand_emit_conf (minimum Phred-scaled confidence threshold) parameters were set to 50.0 and 20.0, respectively. High-quality SNPs from SAMtools and GATK were merged with the GATK program SelectVariants. Sites with total quality by depth of <2.00, mapping quality of <40, genotype quality of <30, and genotype depth of <5 were filtered out using GATK Variant Filtration. The variant sites with a coverage depth of ≥20 were used for subsequent analyses and final SNP extraction.
Copy number variation analysis.
The BAM data sets with duplications removed were used to estimate copy number variation (CNV) for each isolate. Genomic regions with CNV were identified with the Splint script to avoid the “smiley pattern” bias (51). The read depth obtained with 500-bp nonoverlapping windows across the genome was generated with default internal parameters. The relative copy number values of each window were normalized by the median value of nonoverlapping 500-bp windows and visualized using R script. The predicted copy number of each chromosome was determined based on the mean read depth coverage in a given chromosome and normalized by the whole-genome coverage.
Q-RT-PCR and RNA-Seq assays.
Quantitative real-time PCR (Q-RT-PCR) assays were performed according to our previous publication, with modifications (52). Briefly, fungal cells were collected from cultures grown on solid plates. Cells were subjected to MIC testing to ensure that they maintained the original antifungal-resistant features. One microgram of total RNA per sample was used to synthesize cDNA with RevertAid H Minus reverse transcriptase (Thermo Scientific, Inc., Beijing, China). Quantification of transcripts was performed in a Bio-Rad CFX96 real-time PCR detection system using SYBR green. The signal from each experimental sample was normalized to the expression level of the C. auris ACT1 gene.
For RNA-Seq assays, C. auris cells grown on YPD medium at 37°C for 24 h were used for total RNA extraction and RNA-Seq analysis. The libraries were sequenced using the Illumina NovaSeq platform (by Berry Genomics Co., Beijing, China). Approximately 6 billion reads were obtained by sequencing each library. Low-quality (Phred score ≤ 10), ambiguous, and adaptor bases were removed from the raw reads using the FASTX-Toolkit v0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/index.html). The clean reads were aligned to the transcriptome of C. auris (from the NCBI database; accession number GCA_002759435.2) (11) using the software HiSat2 v2.0.5 with default parameters. Transcriptional expression of different samples was estimated with StringTie v1.3.3b with default parameters (53). Differentially expressed genes were analyzed with the DESeq2 R package (54). Differentially expressed genes must satisfy three criteria: (i) an FPKM (fragments per kilobase per million) value for at least one sample of >20; (ii) a fold change of ≥2; (iii) false discovery rates (FDRs) of ≤0.05.
Data availability.
All data were deposited in the Sequence Read Archive (SRA) database (accession no. SRR9325914 to SRR9325939 and SRR9316737). The RNA-Seq data set has been deposited in the NCBI Gene Expression Omnibus (GEO) portal (accession no. GSE136768).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China awards 31930005 and 31625002 (to G.H.) and National Science and Technology Major Project (http://program.most.gov.cn/; grant number 2018ZX10101004-003-002).
We thank all members of the Huang lab for insightful discussions on the study.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Spivak ES, Hanson KE. 2017. Candida auris: an emerging fungal pathogen. J Clin Microbiol 56:e01588-17. doi: 10.1128/JCM.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saris K, Meis JF, Voss A. 2018. Candida auris. Curr Opin Infect Dis 31:334–340. doi: 10.1097/QCO.0000000000000469. [DOI] [PubMed] [Google Scholar]
- 3.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lone SA, Ahmad A. 2019. Candida auris—the growing menace to global health. Mycoses 62:620–637. doi: 10.1111/myc.12904. [DOI] [PubMed] [Google Scholar]
- 6.Vallabhaneni S, Jackson BR, Chiller TM. 2019. Candida auris: an emerging antimicrobial resistance threat. Ann Intern Med 171:432. doi: 10.7326/M19-2205. [DOI] [PubMed] [Google Scholar]
- 7.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borman AM, Szekely A, Johnson EM. 2017. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 55:563–567. doi: 10.1093/mmy/myw147. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Gaitan A, Moret AM, Tasias-Pitarch M, Aleixandre-Lopez AI, Martinez-Morel H, Calabuig E, Salavert-Lleti M, Ramirez P, Lopez-Hontangas JL, Hagen F, Meis JF, Mollar-Maseres J, Peman J. 2018. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses 61:498–505. doi: 10.1111/myc.12781. [DOI] [PubMed] [Google Scholar]
- 10.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Manuel R, Brown CS, Candida auris Incident Management Team. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029-17. doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muñoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, Farrer RA, Litvintseva AP, Cuomo CA. 2018. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 9:5346. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitchcock CA, Dickinson K, Brown SB, Evans EG, Adams DJ. 1990. Interaction of azole antifungal antibiotics with cytochrome P-450-dependent 14 alpha-sterol demethylase purified from Candida albicans. Biochem J 266:475–480. doi: 10.1042/bj2660475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, Fisher MC, Schelenz S. 2018. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya S, Holowka T, Orner EP, Fries BC. 2019. Gene duplication associated with increased fluconazole tolerance in Candida auris cells of advanced generational age. Sci Rep 9:5052. doi: 10.1038/s41598-019-41513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon-Chung KJ, Chang YC. 2012. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog 8:e1003022. doi: 10.1371/journal.ppat.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todd RT, Forche A, Selmecki A. 2017. Ploidy variation in fungi: polyploidy, aneuploidy, and genome evolution. Microbiol Spectr 5:FUNK-0041-2016. doi: 10.1128/microbiolspec.FUNK-0051-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerstein AC, Lim H, Berman J, Hickman MA. 2017. Ploidy tug-of-war: evolutionary and genetic environments influence the rate of ploidy drive in a human fungal pathogen. Evolution 71:1025–1038. doi: 10.1111/evo.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Bing J, Zheng Q, Zhang F, Liu J, Yue H, Tao L, Du H, Wang Y, Wang H, Huang G. 2018. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect 7:93. doi: 10.1038/s41426-018-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal D, Patterson TF, Rinaldi MG, Revankar SG. 2007. Trailing end-point phenotype of Candida spp. in antifungal susceptibility testing to fluconazole is eliminated by altering incubation temperature. J Med Microbiol 56:1003–1004. doi: 10.1099/jmm.0.47168-0. [DOI] [PubMed] [Google Scholar]
- 23.Xu D, Jiang B, Ketela T, Lemieux S, Veillette K, Martel N, Davison J, Sillaots S, Trosok S, Bachewich C, Bussey H, Youngman P, Roemer T. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog 3:e92. doi: 10.1371/journal.ppat.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang G, Huang Q, Wei Y, Wang Y, Du H. 2019. Multiple roles and diverse regulation of the Ras/cAMP/protein kinase A pathway in Candida albicans. Mol Microbiol 111:6–16. doi: 10.1111/mmi.14148. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko A, Umeyama T, Utena-Abe Y, Yamagoe S, Niimi M, Uehara Y. 2006. Tcc1p, a novel protein containing the tetratricopeptide repeat motif, interacts with Tup1p to regulate morphological transition and virulence in Candida albicans. Eukaryot Cell 5:1894–1905. doi: 10.1128/EC.00151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu TT, Lee RE, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother 49:2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell 3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rybak JM, Munoz JF, Barker KS, Parker JE, Esquivel BD, Berkow EL, Lockhart SR, Gade L, Palmer GE, White TC, Kelly SL, Cuomo CA, Rogers PD. 2020. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio 11:e00365-20. doi: 10.1128/mBio.00365-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayr EM, Ramirez-Zavala B, Kruger I, Morschhauser J. 2020. A zinc cluster transcription factor contributes to the intrinsic fluconazole resistance of Candida auris. mSphere 5:e00279-20. doi: 10.1128/mSphere.00279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epp E, Vanier G, Harcus D, Lee AY, Jansen G, Hallett M, Sheppard DC, Thomas DY, Munro CA, Mullick A, Whiteway M. 2010. Reverse genetics in Candida albicans predicts ARF cycling is essential for drug resistance and virulence. PLoS Pathog 6:e1000753. doi: 10.1371/journal.ppat.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers PD, Barker KS. 2002. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob Agents Chemother 46:3412–3417. doi: 10.1128/aac.46.11.3412-3417.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bravo Ruiz G, Ross ZK, Holmes E, Schelenz S, Gow NAR, Lorenz A. 2019. Rapid and extensive karyotype diversification in haploid clinical Candida auris isolates. Curr Genet 65:1217–1228. doi: 10.1007/s00294-019-00976-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill JA, O'Meara TR, Cowen LE. 2015. Fitness trade-offs associated with the evolution of resistance to antifungal drug combinations. Cell Rep 10:809–819. doi: 10.1016/j.celrep.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. 2013. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol 11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 36.Miziorko HM. 2011. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys 505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karst F, Lacroute F. 1977. Ertosterol biosynthesis in Saccharomyces cerevisiae: mutants deficient in the early steps of the pathway. Mol Gen Genet 154:269–277. doi: 10.1007/BF00571282. [DOI] [PubMed] [Google Scholar]
- 38.Park HG, Lim YR, Eun CY, Han S, Han JS, Cho KS, Chun YJ, Kim D. 2010. Candida albicans NADPH-P450 reductase: expression, purification, and characterization of recombinant protein. Biochem Biophys Res Commun 396:534–538. doi: 10.1016/j.bbrc.2010.04.138. [DOI] [PubMed] [Google Scholar]
- 39.Venkateswarlu K, Kelly DE, Manning NJ, Kelly SL. 1998. NADPH cytochrome P-450 oxidoreductase and susceptibility to ketoconazole. Antimicrob Agents Chemother 42:1756–1761. doi: 10.1128/AAC.42.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maicas S, Moreno I, Nieto A, Gomez M, Sentandreu R, Valentin E. 2005. In silico analysis for transcription factors with Zn(II)(2)C(6) binuclear cluster DNA-binding domains in Candida albicans. Comp Funct Genomics 6:345–356. doi: 10.1002/cfg.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vik A, Rine J. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol 21:6395–6405. doi: 10.1128/mcb.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh V, Sinha I, Sadhale PP. 2005. Global analysis of altered gene expression during morphogenesis of Candida albicans in vitro. Biochem Biophys Res Commun 334:1149–1158. doi: 10.1016/j.bbrc.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Rolley N, Kelly DE, Kelly SL. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob Agents Chemother 54:4527–4533. doi: 10.1128/AAC.00348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morschhauser J, Barker KS, Liu TT, Bla BWJ, Homayouni R, Rogers PD. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog 3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrow CA, Fraser JA. 2013. Ploidy variation as an adaptive mechanism in human pathogenic fungi. Semin Cell Dev Biol 24:339–346. doi: 10.1016/j.semcdb.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Bhattacharya S, Esquivel BD, White TC. 2018. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio 9:e01291-18. doi: 10.1128/mBio.01291-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard M27-A3, 3rd ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 48.Espinel-Ingroff A, Barchiesi F, Cuenca-Estrella M, Pfaller MA, Rinaldi M, Rodriguez-Tudela JL, Verweij PE. 2005. International and multicenter comparison of EUCAST and CLSI M27-A2 broth microdilution methods for testing susceptibilities of Candida spp. to fluconazole, itraconazole, posaconazole, and voriconazole. J Clin Microbiol 43:3884–3889. doi: 10.1128/JCM.43.8.3884-3889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallone B, Steensels J, Prahl T, Soriaga L, Saels V, Herrera-Malaver B, Merlevede A, Roncoroni M, Voordeckers K, Miraglia L, Teiling C, Steffy B, Taylor M, Schwartz A, Richardson T, White C, Baele G, Maere S, Verstrepen KJ. 2016. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166:1397–1410 E16. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao C, Wu M, Bing J, Tao L, Ding X, Liu X, Huang G. 2017. Global regulatory roles of the cAMP/PKA pathway revealed by phenotypic, transcriptomic and phosphoproteomic analyses in a null mutant of the PKA catalytic subunit in Candida albicans. Mol Microbiol 105:46–64. doi: 10.1111/mmi.13681. [DOI] [PubMed] [Google Scholar]
- 53.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henry KW, Nickels JT, Edlind TD. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob Agents Chemother 44:2693–2700. doi: 10.1128/aac.44.10.2693-2700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaur M, Puri N, Manoharlal R, Rai V, Mukhopadhayay G, Choudhury D, Prasad R. 2008. MFS transportome of the human pathogenic yeast Candida albicans. BMC Genomics 9:579. doi: 10.1186/1471-2164-9-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers PD, Barker KS. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother 47:1220–1227. doi: 10.1128/aac.47.4.1220-1227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data were deposited in the Sequence Read Archive (SRA) database (accession no. SRR9325914 to SRR9325939 and SRR9316737). The RNA-Seq data set has been deposited in the NCBI Gene Expression Omnibus (GEO) portal (accession no. GSE136768).